Open Access Article
Open Access ArticleFormation of rippled β-sheets from mixed chirality linear and cyclic peptides—new structural motifs based on the pauling-corey rippled β-sheet†
Amaruka
Hazari‡
*a,
Michael R.
Sawaya‡
b,
Hyeonju
Lee
cd,
Maria
Sajimon
a,
Hyungjun
Kim
d,
William A.
Goddard III
 c,
David
Eisenberg
b and
Jevgenij A.
Raskatov
c,
David
Eisenberg
b and
Jevgenij A.
Raskatov
 *a
*a
aDept. of Chemistry and Biochemistry, UCSC, 1156 High Street, Santa Cruz, CA 95064, USA. E-mail: jraskato@ucsc.edu; amhazari@ucsc.edu
bDept. of Chemistry and Biochemistry, UCLA, 607 Charles E. Young Drive East Box 951569, Los Angeles, CA 90095-1569, USA
cMaterials and Process Simulation Center, California Institute of Technology, Pasadena, CA 91125, USA
dDepartment of Chemistry, Korea Advanced Institute of Science and Technology (KAIST), 291 Daehak-Ro, Yuseong-Gu, Daejeon, 34141, Republic of Korea
First published on 6th March 2025
Abstract
The rippled β-sheet is a structural motif formed by certain racemic peptides that is distinct from the commonly known pleated β-sheet. Although the structure was predicted in 1953, unambiguous crystallographic observation of a rippled β-sheet was not reported until 2022. The structural foundation of the rippled β-sheet field continues to expand, stimulating new research questions, both fundamental and applied. Recent studies found that racemic peptides of varied length and amino acid composition assemble into rippled β-sheets. Intriguingly, certain rippled sheets were found to encapsulate small molecules in ways that could become useful in drug delivery, or to trap harmful substances. These and many other potential applications hinge on the development of a comprehensive structural foundation based on both experiment and theory. In this paper we introduce the concept of the single-component rippled-sheet, composed of joined segments of L and D chirality. The scope of rippled sheet-forming motifs is expanded to include two unexplored classes of rippled sheets: single-component cyclic and linear peptide chimeras. We report on the design, synthesis, and crystal structural characterization of eight self-assembling peptide systems. All five linear systems, in which amino acid sequence, charge and chirality were varied, formed rippled β-sheets with distinct two- and three-dimensional lattices. Of the three cyclic peptides, however, only one system formed a rippled β-sheet, while the other two formed pleated β-sheets. Molecular modeling is used to better understand chiral selection in cyclic systems.
Introduction
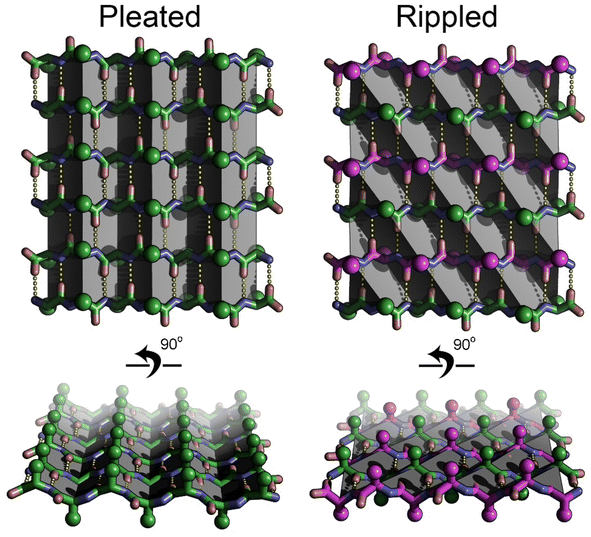
The rippled β-sheet is a unique structural motif and an emerging area within the field of peptide self-assembly. Unlike the more commonly known pleated β-sheet composed of peptide strands of the same chirality, the rippled β-sheet is composed of mirror-image peptide strands assembled in strictly alternating fashion (Fig. 1). Despite being predicted over 70 years ago, by Pauling and Corey, the motif remained largely neglected until recently.1,2 Studies, conducted independently in the laboratories of Schneider, Nilsson, and Raskatov form the experimental foundation of the field. Rippled β-sheet formation leads to the formation of hydrogels with enhanced properties,3–6 and promotes amyloid beta chiral inactivation where mirror-image amyloid drives oligomer-to-fibril conversion attenuating oligomer-associated Aβ neurotoxicity.7–10 A detailed review of the history of the field, including its unusually long induction phase, was published jointly by the three investigators in 2021.11Structural characterization of the rippled β-sheet was limited to biophysical methods until 2022 when we obtained the first atomic resolution structures. We demonstrated that racemic mixtures of tripeptides composed of aromatic amino acids (Phe, Tyr and Trp) formed rippled β-sheets.12,13 In subsequent work, we expanded the crystallographic foundation to include sequences of greater length (up to 7-mer) and complexity.14 We showed that the rippled β-sheet framework could support a wide range of amino acids including charged (Lys and Glu), aliphatic (Leu, Ile, Val, Ala, Gly and Met) and small polar residues (Ser). In total, the number of ripple-genic residues now stands at twelve, more than half the canonical amino acid alphabet.
In this work, we describe the results of a combined experimental/theoretical approach designed to further our fundamental knowledge of the rippled β-sheet. In contrast to our previous work where we focused on incorporating a diverse range of amino acids into linear peptides, here we examine the effects of charge, chirality and peptide conformation (cyclic vs. linear) on rippled sheet formation. Our first objective was to assess how small modifications such as single amino-acid modifications and charge (cationic vs. zwitterionic) influence the structure of the rippled β-sheet. A second objective was to determine whether peptides composed of joined segments of L and D chirality, were capable of forming rippled β-sheets. The development of single-component systems would simplify self-assembly and expand the rippled β-sheet structure–function space. A third objective was to establish whether cyclic peptides could form rippled β-sheets. Rippled β-sheet cyclic peptides represent a new, unexplored structural class with the potential for unique properties and applications. Addressing these questions with a structural approach that combines experiment with theory will expand our ability to rationally design rippled sheets, for which a plethora of applications have been proposed by us and others.11,15
Results
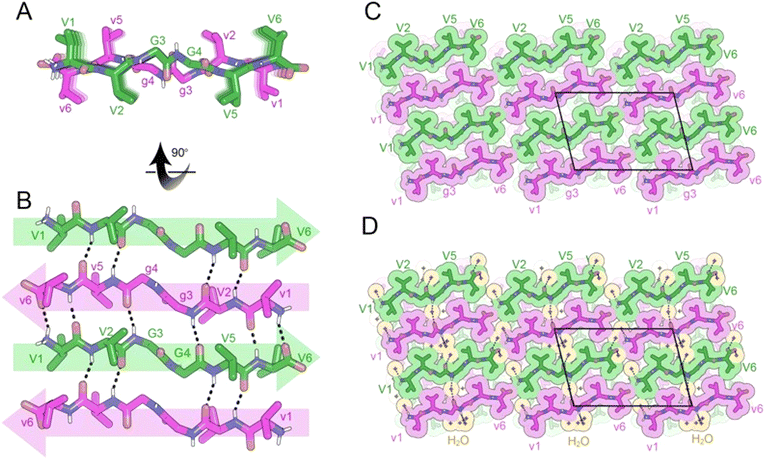
In previous work, we demonstrated that the rippled β-sheets formed from MVGGVV and its enantiomer mvggvv included solvent in the interface between sheets.14 The inclusion of solvent in the interface led to fibril architectures distinct from the dry-interfaces typically observed in enantiopure amyloid-like crystals. A comparison of the crystal structures of enantiopure MVGGVV16 and racemic MVGGVV:mvggvv revealed that the central diglycine bridge is highly flexible. Accordingly, we incorporated this feature into our designs of linear, cyclic and single-component mixed chirality peptides.To examine the effect of a single amino acid modification on rippled β-sheet structure, we replaced the methionine residue of MVGGVV with a valine (VVGGVV). A racemic mixture of VVGGVV and vvggvv was crystallized from a solution of trifluoroethanol (TFE) in water, yielding needles of antiparallel rippled β-sheet layers as determined by X-ray crystallography (Fig. 2A and B). As observed in the previously reported structures of the MVGGVV:mvggvv polymorphs,14 the individual hexapeptides stack in the H-bonding dimension, forming extended antiparallel rippled β-sheet layers, in which mirror image peptide strands are arranged in strictly alternating fashion. Each L-hexapeptide (Fig. 2B, green) is sandwiched between two D-hexapeptides (Fig. 2B, purple), and each D-hexapeptide is sandwiched between two L-hexapeptides in periodic fashion, with H-bond distances ranging from 1.99 to 2.97 Å. Each hexapeptide has six H-bonds to one of its two direct neighbors in the layer, forming a tight dimer, and four H-bonds to the other, forming a loose dimer (Fig. 2B). The rippled interface is partially interrupted at the G3–G4 junction. As a consequence, the amide backbone at G3–G4 does not participate in the stabilization of the rippled β-sheet.
The torsional angles for the glycine residues G3 and G4 are measured as φ = −75.4 and ψ = 156.6 and, φ = −81.4 and ψ = 169.9, respectively for the L-hexapeptide, and φ = 75.4 and ψ = −156.6 and, φ = 81.4 and ψ = −169.9, respectively, for the inversion-related D-hexapeptide. The G3 and G4 φ and ψ angles are comparable to those found in the structures of both MVGGVV:mvggvv polymorphs.14 As observed in the MVGGVV:mvggvv structures, the G3–G4 peptide plane is rotated by ∼90° from its standard position in a β-sheet. This rotation positions the V2 and V5 sidechains on the same face of the β-sheet (Fig. 2A).
In the three-dimensional lattice, the rippled β-sheets are packed closely together (5.5 Å) forming a small cavity, which is occupied by a cluster of water molecules (Fig. 2C and D). The water molecules hydrogen bond to the amide backbone of G3 and G4. The valine residues are more tightly packed; L-valine residues mate with D-valine residues, forming a dry interface between pairs of sheets.
To further investigate the effect of a single amino acid modification on rippled β-sheet structure, we substituted the first valine residue of VVGGVV with the polar residue glutamine (QVGGVV), which had not previously been observed as part of a rippled β-sheet. Crystals of racemic QVGGVV:qvggvv, suitable for X-ray diffraction, were grown from a solution of isopropanol and water. Structural analysis revealed the formation of an antiparallel rippled β-sheet (Fig. 3A and B), closely resembling the two-dimensional layer structure of VVGGVV:vvggvv.
The three-dimensional packing of the individual rippled sheet layers of QVGGVV:qvggvv also resembles that of VVGGVV:vvggvv in terms of the close packing of the rippled β-sheets (Fig. 3C and D). A key difference is the organization of pairs of sheets. Unlike VVGGVV:vvggvv where L-residues mate with D-residues across the dry interface, in QVGGVV:qvggvv, L-residues mate with L-residues and D-residues mate with D-residues, to form the interface between pairs of sheets.
In contrast to the structures of both MVGGVV:mvggvv polymorphs14 in which the three-dimensional lattices are loosely packed, the rippled β-sheets in the three-dimensional lattices of VVGGVV:vvggvv and QVGGVV:qvggvv are packed closely together. This observation led us to explore the role of the methionine residue in generating more openly spaced three-dimensional lattices. To accomplish this objective, we generated quasi-racemic systems by combing L-XVGGVV with D-mvggvv. The quasi-racemic system VVGGVV:mvggvv failed to yield crystals suitable for X-ray diffraction. Combining FVGGVV with mvggvv, however, led to the formation of the anti-parallel rippled β-sheet, FVGGVV:mvggvv (Fig. 4). In the two-dimensional lattice, the hydrogen bonding pattern and torsional angles around G3 and G4 are similar to those observed in other XVGGVV:xvggvv type systems (Fig. 4A and B). The three-dimensional lattice, however, features loosely packed sheets which encapsulate HFIP molecules (Fig. 4C and D), as observed in the MVGGVV:mvggvv HFIP polymorph.
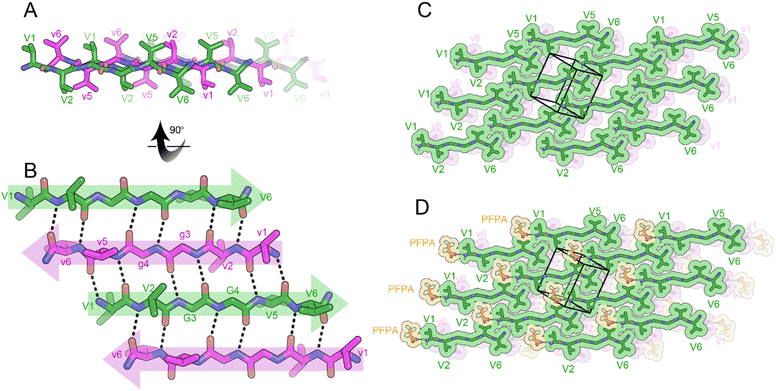
To determine the effect of charge on rippled β-sheet structure, we amidated the C-termini of VVGGVV and vvggvv to form cationic peptides. A racemic mixture of VVGGVV-NH2 and vvggvv-NH2 was crystallized from a solution of pentafluoropropionic acid (PFPA) in water, yielding needles of antiparallel rippled β-sheets. The individual hexapeptides stack in the H-bonding dimension, forming extended antiparallel rippled β-sheet layers (Fig. 5A). The hexapeptide strands form an out-of-register sheet resulting in only tight dimers between strands (Fig. 5B). Each hexapeptide strand forms six hydrogen bonds with each of its neighbors with H-bond distances ranging from 1.91 to 2.85 Å.
The torsional angles for the glycine residues G3 and G4 are measured as φ = −150.2 and ψ = 160.3, φ = −151.5 and ψ = 146.0, respectively for the L-peptide and, φ = 150.2 and ψ = −160.3 and, φ = 151.5 and ψ = −146.0, respectively for the D-peptide. The flattened configuration has the effect of positioning V2 and V5 sidechains on opposite sides of the β-sheet (Fig. 5A).
The rippled sheets pack face-to-back with a staggered pattern (Fig. 5C) such that V5 and V6 form a tight dry interface with V1 and V2 of a neighboring rippled dimer in the vertical direction. PFPA molecules are bound to the N-termini of the hexapeptides, only partially occupying gaps in the lattice (Fig. 5D).
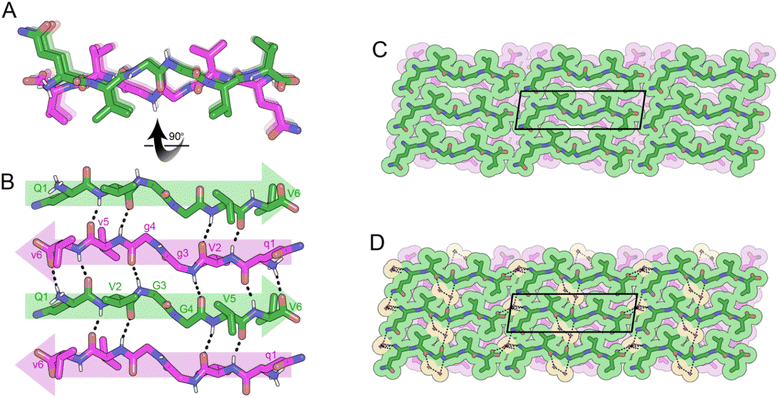
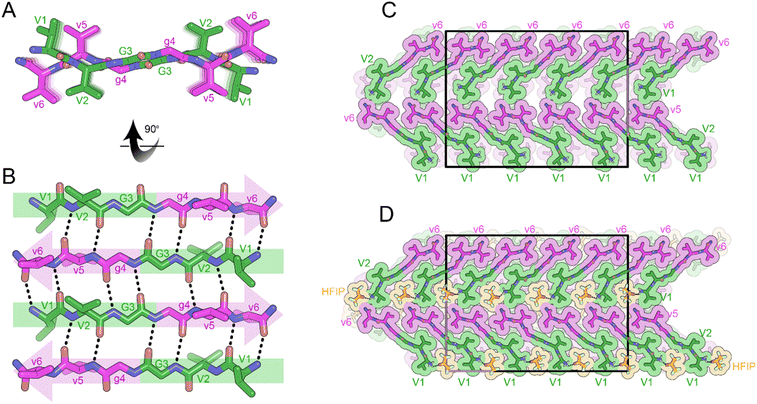
The rippled sheet crystal structures reported here as well as all previously reported structures12–14 were constructed from equimolar racemic mixtures of L and D-peptides. Given our success at obtaining atomic resolution structures from XVGGVV:xvggvv type systems, we used this framework as the basis for the design of a single-component system. The (L,L)-divaline unit was formally connected via diglycine to (D,D)-divaline unit, yielding the peptide with the sequence VVGgvv-NH2. While the L- and D-segments are not strictly racemic due to their directionality, the system still formed anti-parallel rippled β-sheets upon crystallization from a solution of HFIP in water. Each hexapeptide strand forms 6-H bonds to each of its neighbors with H-bond distances between the rippled dimers ranging from 1.81 to 2.32 Å (Fig. 6A and B). The torsional angles for the glycine residues G3 and G4 are measured as φ = −159.3 and ψ = 153.4 and φ = 178.9 and ψ = −175.4, respectively. The backbone is highly extended with both G4 φ and ψ angles approaching 180°. When viewed along the hydrogen bonding direction rippled sheets mate at alternating angles of +45° and −45° such that a tight dry interface forms between V1 and V5 and V2 and V6, respectively (Fig. 6C). The gaps in the lattice are occupied by HFIP molecules which bind to the N-terminus of one dimer and the C-terminus of a neighboring dimer, bridging the sheets together (Fig. 6D).
Inspired by this discovery, we sought to determine whether a cyclic rippled β-sheet could be formed from a single-component system. We designed a closely related cyclic peptide, the N-to-C cyclized octapeptide, VVGGvvgg, which is truly internally racemic (Fig. 7A). Crystals of cyclic VVGGvvgg were grown from a solution of dimethylsulfoxide (DMSO) in water but did not yield rippled β-sheets. Instead, the chiral domains engaged in homochiral self-sorting with L-V1 hydrogen bonding with L-V1, and D-v5 hydrogen bonding with D-v5 (Fig. 7B). In a rippled β-sheet L-Val would be expected to hydrogen bond to D-val. The self-sorted system features a unique hydrogen bonding pattern. The valine residues stack parallel, in-register, whereas the glycine residues form a discontinuous β-sheet which runs at a different angle to the stacked valine residues. Within the β-sheet, 4 H bonds are observed between neighboring cyclic monomers with H-bond distances ranging from 1.90 to 2.50 Å.
The three-dimensional lattice is stabilized by hydrogen bonding between sheets in the vertical direction; G3 bonds to G8 and G4 bonds to G7, with H bond distances ranging from 2.11 to 2.20 Å. In the lateral direction, a tight interface between V1 and V6 and V2 and V5 is observed (Fig. 7C). Cavities within the sheets are occupied by water molecules (Fig. 7D).
Attempts to form a cyclic rippled sheet from a racemic mixture of cyclic VVGGVVGG and vvggvvgg led to a similar outcome, with the mixture self-sorting into enantiopure pleated sheets, rather than the desired rippled sheets (Fig. S1†).
We hypothesized that a larger decapeptide may be less strained and thus more likely to form cyclic rippled β-sheets. We devised a cyclic decapeptide, in which the diglycine linking units were used to formally connect the L-tripeptide FKF and the D-tripeptide fef, producing the zwitterionic cyclic framework FKFGGfefgg. The tripeptide segments of the cycle were inspired by our studies with aromatic tripeptides but employed charged residues to enhance solubility.
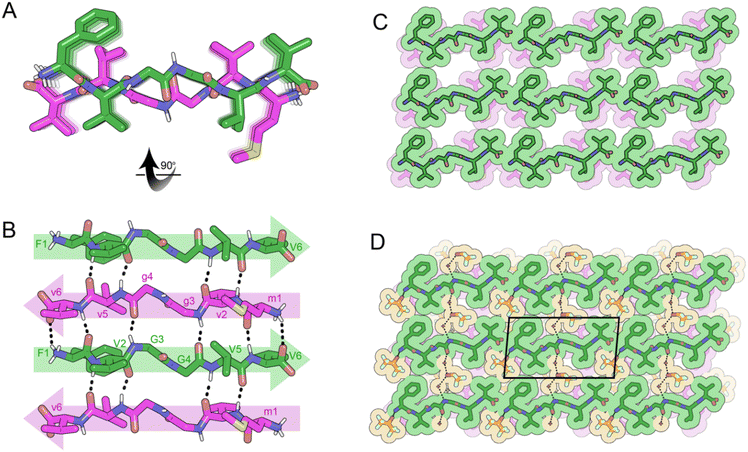
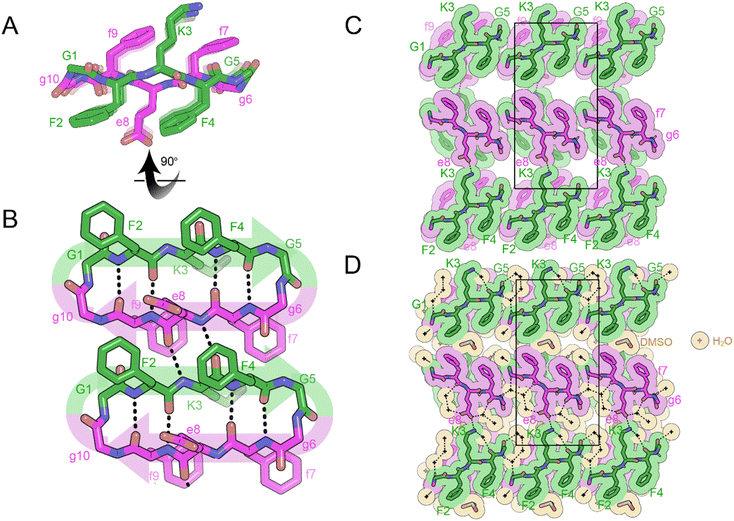
Crystals of the cyclic single-component system FKFGGfefgg were grown from a mixture of DMSO and water and yielded cyclic, self-rippling β-sheets (Fig. 8A). The cyclic peptide forms a rippled monomer which is stabilized by intramolecular hydrogen bonding; 4 hydrogen bonds are observed between the amide backbones of L-F2 and D-f9 and L-F4 and D-f7, ranging from 1.96 to 2.24 Å. Each rippled monomer forms two hydrogen bonds to a neighboring monomer with H-bonding distances of 2.12 and 2.39 Å (Fig. 8B). The periodic alternation of intra and intermolecular hydrogen bonds results in a tight/loose hydrogen bonding arrangement mirroring that observed in the linear rippled sheets VVGGVV:vvggvv (Fig. 2B) and QVGGVV:qvggvv (Fig. 3B).
In the three-dimensional lattice the sheets are aligned face-to-face at an angle that enables the oppositely charged termini of residues D-e8 and L-K3 to hydrogen bond and allows a tight interface to form between the phenyl rings of F2 and f9 (Fig. 8C). Clusters of water molecules occupy gaps in the lattice and form hydrogen bonding networks with the termini of D-e8 and L-K3 as well as with oxygen atoms from the amide backbone. Additional gaps in the lattice are filled with DMSO molecules (Fig. 8D).
Discussion
Structural variation in racemic and single-component linear rippled β-sheets
Our results show that a single amino-acid modification to the peptide sequence can have a profound impact on the structure of the lattice. While the structures of the rippled dimers of VVGGVV:vvggvv, QVGGVV:qvggvv, FVGGVV:mvggvv and the previously reported MVGGVV:mvggvv polymorphs14 are alike, the three-dimensional lattices differ. The methionine containing structures, MVGGVV:mvggvv and FVGGVV:mvggvv form three-dimensional lattices capable of encapsulating fluorinated solvents in the cavities between sheets. In contrast, the rippled β-sheets in VVGGVV:vvggvv and QVGGVV:qvggvv are packed too closely together (5.5 Å in VVGGVV:vvggvv vs. 10.0 Å in MVGGVV:mvggvv HFIP) for the cavity between the sheets to accommodate fluorinated solvent (Fig. S2†).Single amino-acid modifications may also influence the organization of rippled sheet layers into periodic three-dimensional architectures. The most common arrangement involves L-residues mating with L-residues across the rippled β-sheet interface and in the lateral direction, such that the β-sheets are aligned with the vertical and horizontal axes. An exception is the symmetric system, VVGGVV:vvggvv, where L-residues mate with D-residues across both the hydrophobic and the lateral (coulombic) interfaces, such that the β-sheets are offset from the horizontal axis.
Furthermore, we demonstrate that charge can influence the structure of both the rippled dimer and the three-dimensional lattice. A key difference between the cationic system, VVGGVV-NH2:vvggvv-NH2, and the zwitterionic XVGGVV:xvggvv systems, is the hydrogen bonding between the individual strands of the rippled dimer; in VVGGVV-NH2:vvggvv-NH2 the hexapeptide strands form an out-of-register sheet resulting in only tight dimers between strands (Fig. 5B) as opposed to the tight/loose dimer arrangement observed in the in-register sheets formed by VVGGVV:vvggvv (Fig. 1B), QVGGVV:qvggvv (Fig. 2B) and MVGGVV:mvggvv (Fig. S3†). An out-of-register rippled β-sheet was also observed in the crystal structure of the previously reported cationic peptide KLVFFA:klvffa.14
An additional distinguishing feature of VVGGVV-NH2:vvggvv-NH2 rippled dimer is the conformation of the backbone. The φ angles in the cationic system vary substantially from those observed in neutral XVGGVV:xvggvv systems, resulting in a more extended backbone conformation at the G3 and G4 residues. Consequently, the V2 and V5 sidechains lie on opposite sides of the β-sheet as opposed to the same side as in the XVGGVV:xvggvv systems.
The three-dimensional lattice in VVGGVV-NH2:vvggvv-NH2 also differs from that observed in other XVGGVV:xvggvv systems. The sheets pack in a staggered pattern as opposed to the face-to-face arrangements seen in VVGGVV:vvggvv, QVGGVV:qvgggvv and MVGGVV:mvggvv-HFIP, respectively and the face-to-edge arrangements observed in MVGGVV:mvggvv-PFPA (Fig. S4†). The staggered pattern and binding of PFPA anions to the N-termini of individual peptide strands results in a more open lattice than observed in other XVGGVV:xvggvv type structures.
Further structural variation was observed upon analysis of the crystal structure of the single-component system VVGGvv-NH2. In comparison to the racemic XVGGVV:xvggvv type structures, the backbone of VVGGvv-NH2 is more extended with both G4 φ and ψ angles approaching 180° (Fig. S5†). The three-dimensional lattice is also distinctive, with rippled sheets mating at alternating angles of +45° and −45°.
It is important to note that in three of the linear rippled β-sheet systems the two and three-dimensional lattices formed by each system are distinct, despite being composed of the same amino acid sequence. In addition to charge and chirality, the diglycine bridge is a contributing element, adopting a range of angles which influence the structure of both the rippled dimer and the three-dimensional lattice.
Computational analysis of the VVGGvvgg cyclic octapeptide
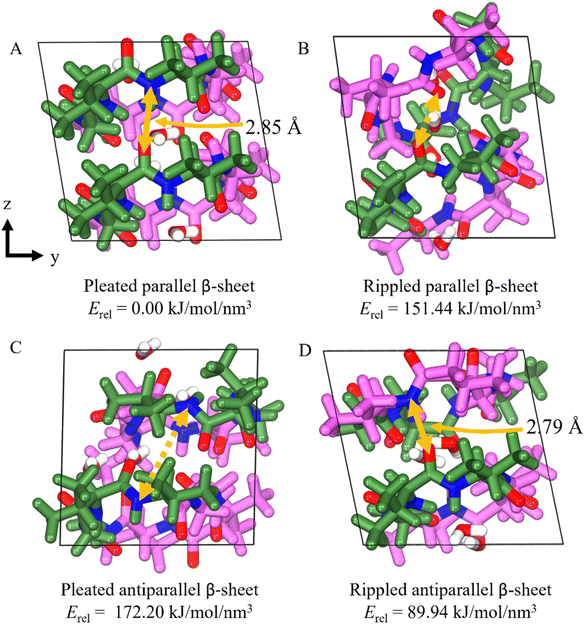
We were surprised to discover that the cyclic octapeptide VVGGvvgg formed pleated β-sheets rather than rippled β-sheets despite the sequence similarity with the “self-rippling” linear system VVGGvv. These observations are consistent with a recent solution state NMR study that shows that certain cyclic peptides favor pleated β-sheets over rippled β-sheets.17 The self-sorting behavior of cyclic VVGGvvgg offered us an opportunity to expand our theoretical understanding of the rippled β-sheet motif.18 We used molecular modelling to compare the cohesive energy differences of the experimentally observed pleated arrangement with the hypothetical alternatives: anti-parallel pleated, anti-parallel rippled and parallel rippled.Our results show that the experimentally observed pleated parallel system is the most thermodynamically stable system (Fig. 9). This stability can be attributed to the centrosymmetric arrangement of the cycle, which facilitates H-bond interactions between the sides of the backbone, as indicated by the yellow arrows in Fig. 9. The H-bond interactions between the backbones of the cyclic peptide play a crucial role in stabilizing and maintaining β-sheet structure. Interestingly, the rippled antiparallel system is more thermodynamically stable than the rippled parallel and pleated antiparallel systems. The rippled antiparallel system also features a centrosymmetric arrangement which facilitates hydrogen bonding and allows it to retain its initial structure. The centrosymmetric arrangement is lacking in both the rippled parallel and pleated antiparallel systems, contributing to their lower thermodynamic stability.
Comparing the lattice parameters from X-ray crystallography with the DFT-optimized cells, the pleated parallel and rippled antiparallel systems show a maximum error of 1.27% and 2.71%, respectively, indicating that the structures are well described (Table S1†). The rippled parallel and pleated antiparallel systems not observed by crystallography, however, exhibit volume increases of 13.9% and 14.4%, respectively, as the structures shift to increase the number of H-bonds from 0 to 4 between backbones, leading to the disruption of the unit cell (Table 1).
| X-ray crystallography | Pleated parallel | Rippled parallel | Pleated antiparallel | Rippled antiparallel | |||||
|---|---|---|---|---|---|---|---|---|---|
| DFT | Error (%) | DFT | Error (%) | DFT | Error (%) | DFT | Error (%) | ||
| a (Å) | 17.59 | 17.62 | 0.14 | 17.08 | 2.89 | 17.94 | 1.98 | 17.88 | 1.63 |
| b (Å) | 10.36 | 10.23 | 1.27 | 10.10 | 2.53 | 10.01 | 3.35 | 10.08 | 2.71 |
| c (Å) | 9.66 | 9.70 | 0.37 | 10.45 | 8.17 | 10.54 | 9.10 | 9.85 | 1.98 |
| α (°) | 98.28 | 98.43 | 0.15 | 99.15 | 0.88 | 85.90 | 12.60 | 99.73 | 1.48 |
| β (°) | 95.07 | 94.45 | 0.65 | 81.89 | 13.87 | 108.77 | 14.41 | 97.06 | 2.10 |
| γ (°) | 100.37 | 99.83 | 0.54 | 101.39 | 1.01 | 98.59 | 1.77 | 101.52 | 1.15 |
Ring size vs. charge as determinants in cyclic systems
Crystallization of the cyclic octapeptides VVGGvvgg and racemic VVGGVVGG/vvggvvgg unexpectedly resulted in the formation of pleated β-sheets whereas the cyclic decapeptide FKFGGfefgg formed a rippled β-sheets. Based on these observations, our hypothesis is that ring size is a determinant in whether cyclic peptides prefer pleated or rippled β-sheets; cyclic octapeptides favor pleated sheets whereas decapeptides favor rippled sheets. The role of charge, however, cannot be discounted as cyclic FKFGGfefgg is intrinsically locked into a rippled β-sheet.Future studies focused on systematically varying the residues in both ring systems will further our understanding of the respective roles of ring size and charge in determining whether a cyclic peptide forms a pleated or rippled β-sheet.
Conclusions
Herein, we presented the atomic resolution structures of eight distinct peptide systems. Six of the systems assemble into rippled β-sheets while two of the systems self-sort into pleated β-sheets. We demonstrate that both cyclic and linear rippled β-sheets can be formed from single-component systems composed of joined segments of L and D chirality. In the linear systems, small modifications such as single-amino acid substitutions or variations in charge or chirality lead to significant structural changes in both the rippled dimer and the three-dimensional lattice. In two of the cyclic systems, preferential self-sorting into pleated β-sheets is observed. To help understand these preferences, we applied molecular dynamics (MD) to predict chiral selection. Our theoretical findings are in good agreement with the experimental results. Our findings expand both the types of fibril architectures formed by rippled β-sheets and the classes of peptides capable of forming rippled β-sheets, thereby reinforcing the generality of the rippled β-sheet. Moreover, combining experiment with theory provides a foundation for understanding how to design self-rippling systems to control chiral selection at the molecular level.Data availability
Crystallographic data for [compound number] has been deposited at the PBD under 9DYW, 9DYZ, 9DYY, 9DYZ, 9Z0, 9DZ1, 9N31, and 9N35.Author contributions
A. H. and J. A. R. designed research; A. H., M. R. S., H. L., M. S. and J. A. R. performed research; A. H., M. R. S., H. L., W. A. G. and J. A. R. analyzed data; A. H., M. R. S.., H. L., M. S., H. K., W. A. G., D. E. and J. A. R. wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. H. and J. A. R. thank the Seaver Institute for a generous gift. We acknowledge the NIH R01AG070895, R01AG048120, RF1AG065407, R01AG074954. We thank the NSF (MCB1616265), and the California NanoSystems Institute at the University of California, Los Angeles. The authors also acknowledge the Department of Energy Grant DE-FC02-02ER63421 for support. This research used beamline 17-ID-2 at the National Synchrotron Light Source II, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893) and by the DOE Office of Biological and Environmental Research (KP1607011). This work is also based upon research conducted at the Northeastern Collaborative Access Team beamline 24-ID-E, which is funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30GM124165). The Eiger 16M detector is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank Duilio Cascio for support during data collection and refinement. WAG thanks NSF (CBET 2311117) and NIH (R01HL155532) for support.References
- L. Pauling and R. B. Corey, Two rippled-sheet configurations of polypeptide chains, and a note about the pleated sheets, Proc. Natl. Acad. Sci. U. S. A., 1953, 39(4), 253–256, DOI:10.1073/pnas.39.4.253.
- J. A. Raskatov, J. P. Schneider and B. L. Nilsson, Defining the landscape of the pauling-corey rippled sheet: an orphaned motif finding new homes, Acc. Chem. Res., 2021, 54(10), 2488–2501, DOI:10.1021/acs.accounts.1c00084.
- K. Nagy-Smith, P. J. Beltramo, E. Moore, R. Tycko, E. M. Furst and J. P. Schneider, Molecular, local, and network-level basis for the enhanced stiffness of hydrogel networks formed from coassembled racemic peptides: predictions from pauling and corey, ACS Cent. Sci., 2017, 3(6), 586–597, DOI:10.1021/acscentsci.7b00115.
- K. J. Nagy, M. C. Giano, A. Jin, D. J. Pochan and J. P. Schneider, Enhanced mechanical rigidity of hydrogels formed from enantiomeric peptide assemblies, J. Am. Chem. Soc., 2011, 133(38), 14975–14977, DOI:10.1021/ja206742m.
- J. M. Urban, J. Ho, G. Piester, R. Fu and B. L. Nilsson, Rippled β-sheet formation by an amyloid-β fragment indicates expanded scope of sequence space for enantiomeric β-sheet peptide coassembly, Molecules, 2019, 24(10), 1983 CrossRef CAS.
- R. J. Swanekamp, J. T. M. DiMaio, C. J. Bowerman and B. L. Nilsson, Coassembly of enantiomeric amphipathic peptides into amyloid-inspired rippled beta-sheet fibrils, J. Am. Chem. Soc., 2012, 134(12), 5556–5559, DOI:10.1021/ja301642c.
- S. Dutta, A. R. Foley, C. J. A. Warner, X. Zhang, M. Rolandi, B. Abrams and J. A. Raskatov, Suppression of oligomer formation and formation of non-toxic fibrils upon addition of mirror-image aβ42 to the natural L-enantiomer, Angew. Chem., Int. Ed., 2017, 56, 11506–11510 CrossRef CAS.
- J. A. Raskatov, Chiral inactivation: an old phenomenon with a new twist, Chem.–Eur. J., 2017, 23, 16920–16923 CrossRef CAS.
- S. Dutta, A. Rodriguez Foley, A. Kuhn, B. Abrams, H.-W. Lee and J. A. Raskatov, New insights into differential aggregation of enantiomerically pure and racemic Aβ40 systems, Peptide Sci., 2019, 111, e24139 CrossRef CAS.
- J. A. Raskatov, A. R. Foley, J. M. Louis, W.-M. Yau and R. Tycko, Constraints on the structure of fibrils formed by a racemic mixture of amyloid-β peptides from solid-state NMR, electron microscopy, and theory, J. Am. Chem. Soc., 2021, 143(33), 13299–13313, DOI:10.1021/jacs.1c06339.
- J. A. Raskatov, J. P. Schneider and B. L. Nilsson, Defining the landscape of the Pauling–Corey rippled sheet: an orphaned motif finding new homes, Acc. Chem. Res., 2021, 54(10), 2488–2501, DOI:10.1021/acs.accounts.1c00084.
- A. J. Kuhn, B. Ehlke, T. C. Johnstone, S. R. J. Oliver and J. A. Raskatov, A crystal-structural study of Pauling–Corey rippled sheets, Chem. Sci., 2022, 13(3), 671–680, 10.1039/D1SC05731F.
- A. Hazari, M. R. Sawaya, N. Vlahakis, T. C. Johnstone, D. Boyer, J. Rodriguez, D. Eisenberg and J. A. Raskatov, The rippled β-sheet layer configuration—a novel supramolecular architecture based on predictions by Pauling and Corey, Chem. Sci., 2022, 13(31), 8947–8952, 10.1039/D2SC02531K.
- A. Hazari, M. R. Sawaya, M. Sajimon, N. Vlahakis, J. Rodriguez, D. Eisenberg and J. A. Raskatov, Racemic peptides from amyloid β and amylin form rippled β-sheets rather than pleated β-sheets, J. Am. Chem. Soc., 2023, 145(47), 25917–25926, DOI:10.1021/jacs.3c11712.
- R. Saklani and A. J. Domb, Peptide and protein stereocomplexes, ACS Omega, 2024, 9(16), 17726–17740, DOI:10.1021/acsomega.4c00178.
- M. R. Sawaya, S. Sambashivan, R. Nelson, M. I. Ivanova, S. A. Sievers, M. I. Apostol, M. J. Thompson, M. Balbirnie, J. J. W. Wiltzius and H. T. McFarlane, et al., Atomic structures of amyloid cross-β spines reveal varied steric zippers, Nature, 2007, 447(7143), 453–457, DOI:10.1038/nature05695.
- X. Li, S. E. Rios and J. S. Nowick, Enantiomeric β-sheet peptides from Aβ form homochiral pleated β-sheets rather than heterochiral rippled β-sheets, Chem. Sci., 2022, 13(26), 7739–7746, 10.1039/D2SC02080G.
- H. Lee, M. Y. Yang, J. A. Raskatov, H. Kim and W. A. Goddard III, Molecular dynamics studies of atomistically determined fibrillar assemblies: comparison of the rippled β-sheet, pleated β-sheet, and herringbone structures, J. Phys. Chem. Lett., 2024, 15(17), 4568–4574, DOI:10.1021/acs.jpclett.4c00101.
Footnotes |
| † Electronic supplementary information (ESI) available: Crystallization protocols, structural refinement parameters and additional tables are provided free of charge; the rippled sheet structural files have been deposited in the PDB with accession codes 9DYW, 9DYZ, 9DYY, 9DYZ, 9Z0, 9N31, 9N35 and 9DZ1; X-ray diffraction data quality, atomic refinement methods and statistics, as well as computational details are provided. See DOI: https://doi.org/10.1039/d4sc08079c |
| ‡ Denotes equal contribution. |
| This journal is © The Royal Society of Chemistry 2025 |