Open Access Article
Open Access ArticleInhibition of the cGAS–STING pathway via an endogenous copper ion-responsive covalent organic framework nanozyme for Alzheimer's disease treatment†
Haochen
Zhang
ab,
Junlin
Ya
ab,
Mengyu
Sun
ab,
Xiubo
Du
c,
Jinsong
Ren
 ab and
Xiaogang
Qu
ab and
Xiaogang
Qu
 *ab
*ab
aLaboratory of Chemical Biology and State Key Laboratory of Rare Earth Resource Utilization, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, Jilin 130022, China. E-mail: xqu@ciac.ac.cn
bSchool of Applied Chemistry and Engineering, University of Science and Technology of China, Hefei, Anhui 230029, China
cCollege of Life Sciences and Oceanography, Shenzhen Key Laboratory of Microbial Genetic Engineering, Shenzhen University, Shenzhen 518060, China
First published on 11th March 2025
Abstract
Inhibition of cGAS–STING overactivation has recently emerged as a promising strategy to counteract Alzheimer's disease (AD). However, current cGAS–STING inhibitors as immunosuppressants suffer from instability, non-specific targeting, and innate immune disruption. Here, an endogenous AD brain copper ion-responsive covalent organic framework (COF)-based nanozyme (denoted as TP@PB-COF@NADH) has been designed for targeted inhibition of the cGAS–STING pathway for AD treatment. The effective trapping of excess brain endogenous copper ions by TP@PB-COF@NADH not only inhibits the Cu2+-induced harmful reactive oxygen species (ROS) production which is one of the mediators of cGAS–STING activation, but also activates the nanozyme activity of TP@PB-COF@NADH. Furthermore, the well-prepared nanozyme catalytically generates NAD+ and consumes hydrogen peroxide (H2O2) through second near-infrared (NIR-II) enhanced nicotinamide adenine dinucleotide (NADH) peroxidase (NPX)-like activity, realizing the efficient inhibition of the cGAS–STING pathway and associated neuroinflammation. Moreover, replenishing NAD+ levels efficiently restores mitochondrial function and ATP supply. In vivo studies demonstrate that TP@PB-COF@NADH with NIR-II irradiation significantly improves cognitive function in 3× Tg-AD mice, with a reduction in amyloid-β (Aβ) plaque, neuroinflammation and neuronal damage. Collectively, this work presents a promising approach for AD treatment by using an AD brain harmful excess endogenous copper ion-responsive and efficient nanozyme.
Introduction
Alzheimer's disease (AD) is one of the most prevalent fatal disorders, pathologically featured by the presence of dense amyloid plaques, chronic neuroinflammation, oxidative stress and disturbances in metal ion homeostasis.1–3 Notably, elevated levels of copper have been observed in the serum of AD patients,4 with copper in amyloid plaques reaching nearly 5.7-fold elevated concentrations compared to healthy brain tissue.5 The redox-active nature of copper ions promotes the generation of reactive oxygen species (ROS), contributing to the oxidative stress mechanisms characteristic of AD pathogenesis.4,5 Moreover, elevated copper levels have been shown to stimulate neuroinflammatory cascades, potentially exacerbating the aggregation and deposition of Aβ plaques.6,7 Collectively, targeting copper ion dyshomeostasis represents a highly promising therapeutic strategy for AD.8,9Emerging evidence has suggested that overstimulation of the cyclic GMP–AMP synthase (cGAS)-stimulator of the interferon genes (STING) pathway and its associated neuroinflammation are implicated in the progression of AD.10–15 Thus, inhibition of the cGAS–STING pathway has been considered as a new therapeutic strategy.10,16 Nevertheless, research on cGAS–STING inhibitors is currently still in its nascent stages and faces numerous challenges. In particular, cGAS–STING inhibitors acting as immunosuppressants may pose risks by disrupting the innate immune balance in normal organs, potentially leading to infections.17,18 Moreover, the therapeutic efficacy of current cGAS–STING inhibitors is often restricted by off-target effects, instability, rapid clearance from circulation, blood–brain barrier (BBB) permeability and poor bioavailability.19–21 It is also crucial to avoid unpredictable side effects from over-inhibition and frequent administration.17 Therefore, developing a novel and effective therapeutic strategy to down-regulate the over-activated cGAS–STING pathway is highly desirable for AD treatment and remains a great challenge.
Nicotinamide adenine dinucleotide (NAD+), a critical regulator of metabolic networks, is crucial for mitochondrial health, genome integrity and neuronal stress resistance.22–24 Emerging evidence has demonstrated that elevating NAD+ can inhibit the cGAS–STING pathway.25–27 However, lower levels of NAD+ in AD brains have been identified.16,27–29 Notably, as the bioavailability of NAD+ declines, enzymes that depend on NAD+ compete for a limited supply of NAD+, which may restrict their cellular functionality.30 Besides, due to the relatively high energy requirements of neurons, they are highly vulnerable to deficiencies in NAD+ levels and ATP production impairment.31,32 Despite substantial efforts, current approaches for NAD+ supplementation with precursors exhibit limited effectiveness owing to inefficiency, lack of targeting, tissue-specific cytotoxicity and suboptimal oral bioavailability.27,33–36 Additionally, the catalytic methods encounter obstacles such as heavy metal toxicity, low stability in harsh biological fluids, and high cost to scale up.34,36–39 Hence, there is an urgent need to explore an efficient strategy to elevate NAD+ levels for cGAS–STING inhibition.
Nanozymes, an emerging class of artificial enzymes, possess distinctive physicochemical attributes of nanomaterials coupled with enzyme-mimicking catalytic capabilities.40–42 Numerous nanomaterials have been engineered to mimic the biocatalytic functions of natural enzymes.43–48 Compared to natural enzymes, which suffer from limited yields and struggle to cross the BBB, nanozymes possess many advantages including remarkable stability, tunable catalysis, and ease of preparation and functional modifications.49,50 However, the metal in nanozymes has a potential risk of dissolving in lysosomes, resulting in metal ion accumulation and exacerbation of metal dyshomeostasis in the context of neurodegenerative diseases, ultimately causing inflammation and cell death.5,51–53 Furthermore, the high reactivity of nanozymes may lead to undesirable off-target effects during systemic circulation and incorrect site-specific activation in normal tissues.54–56 Considering the constraints above, covalent organic frameworks (COFs), a significant metal-free platform with tunable compositions, are ideal candidates for biocompatible nanozymes with specific activation capabilities and high catalytic performance. With the advantages of favorable biocompatibility, high stability, and flexible structure, COFs have been extensively used in various catalytic and biomedical applications.57–63 Multiple COF-derived materials exhibit exceptional catalytic properties, while also enhancing the capture of substrate species.64,65 As enzyme-mimetic catalysts, the catalytic activity of COFs can be regulated through adjusting functional groups.66,67 By leveraging the distinctive characteristics, we hypothesize that COFs can be harnessed as metal-free nanozyme systems capable of inhibiting cGAS–STING, thereby offering a potential AD therapeutic approach.
Herein, prompted by the biofunctionality of natural NADH peroxidase (NPX),68 we report a rationally designed COF-based endogenous copper ion-responsive nanozyme (TP@PB-COF@NADH) to achieve targeted inhibition of the overactivated cGAS–STING pathway for AD treatment (Scheme 1). Benefiting from the merits of COFs with tunable functional groups and high surface area, the predesigned TP@PB-COF@NADH with 2,2′-bipyridine (2,2′-Bpy) groups could trap the excess accumulated Cu2+ ions in AD. Capturing and redox silencing Cu2+ ions with TP@PB-COF@NADH can decrease the ROS production and toxic effects induced by Cu2+ ions, reducing the abnormal activation of the cGAS–STING pathway. Additionally, given that COFs possess extended π-conjugated frameworks and offer adequate contact with reactant substrate molecules, COF-based porous materials exhibit shortened electron transportation pathways and promoted electron transport, thereby improving the catalytic efficiency.69,70 Particularly, the NPX-like activity of TP@PB-COF@NADH, activated by in situ Cu2+ ions of AD, is significantly enhanced by external NIR-II stimulation. Therefore, our designed nanozyme can effectively improve NAD+ levels and consume H2O2, inhibiting the over-activation of the cGAS–STING pathway and alleviating the associated inflammation. Moreover, the supplement of NAD+ effectively reduced oxidative stress, improved mitochondrial function, and restored ATP production. Owing to the covalently grafted target peptides of KLVFFAED, TP@PB-COF@NADH can pass through the BBB, target Aβ and achieve high accumulation in the brain.71–74In vivo studies have shown that TP@PB-COF@NADH with NIR-II irradiation can reverse cognitive deficits, reduce neuronal injury, decrease Aβ plaque deposition and alleviate neuroinflammation in APPSwe/PS1M146 V/tauP301L triple-transgenic AD (3× Tg-AD) model mice. Beyond the dual capacity for Cu2+ chelation and oxidative stress mitigation, our system synergistically integrates NAD+-mediated energy homeostasis restoration and cGAS–STING-driven neuroinflammation attenuation, establishing a combinatorial strategy addressing oxidative stress, energy homeostasis, and inflammation that is previously unexplored in copper-targeting AD therapies.8,9 Our study provides a new perspective on inhibiting the cGAS–STING pathway by nanozyme catalysis as a potential therapeutic approach for AD.
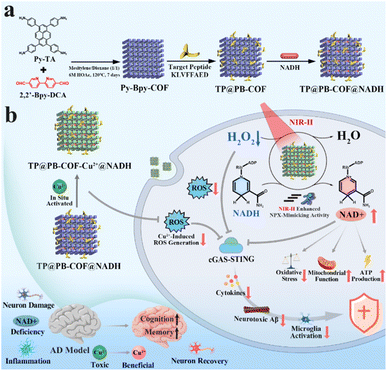 | ||
| Scheme 1 (a) Schematic chart of TP@PB-COF@NADH synthesis and (b) therapeutic mechanism of TP@PB-COF@NADH in AD. | ||
Results and discussion
The Py-Bpy-COF was synthesized via a solvothermal approach utilizing the Schiff-base condensation method, employing 1,3,6,8-tetrakis(4-aminophenyl)pyrene (Py-TA) and 2,2′-bipyridyl-5,5′-dialdehyde (BPy-DCA) as the precursors (Scheme 1a). The synthetic process was carried out in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio mixture of mesitylene and dioxane at 120 °C for seven days to obtain the Py-Bpy-COF, with 6 M aqueous acetic acid (HOAc) serving as the catalyst.75 Furthermore, the utilization of the HOAc solution facilitated the transformation of BPy-DCA into a more stable cis conformer. The predicted pKa of BPy-DCA (3.83) exceeded the pH of the reaction mixture (3.15), suggesting that BPy-DCA existed predominantly in the cis configuration during the reaction, attributed to the creation of the intramolecular H-bonding and π-conjugation.76 To further verify the preferred configuration of BPy-DCA under reaction conditions, UV-vis absorption spectra were recorded. Upon the addition of 6 M HOAc to the BPy-DCA solution, a distinct band emerged at 312 nm, exhibiting a red shift compared to the original solution (Fig. S1†). The peaks at 312 nm and 256 nm corresponded to the characteristic absorption of the monocationic BPy-DCA species, indicating the substantial incorporation of cis-configured 2,2′-Bpy into the Py-Bpy-COF during the synthesis. Moreover, the extended π-interaction between stacked layers stabilized the cis conformation of 2,2′-Bpy in the Py-Bpy-COF, which is crucial for subsequent post-synthetic modifications. The successful synthesis of the Py-Bpy-COF was characterized using Fourier transform infrared (FT-IR) spectroscopy. As illustrated in Fig. 1a, the characteristic peak at 1697 cm−1 (C
1 volume ratio mixture of mesitylene and dioxane at 120 °C for seven days to obtain the Py-Bpy-COF, with 6 M aqueous acetic acid (HOAc) serving as the catalyst.75 Furthermore, the utilization of the HOAc solution facilitated the transformation of BPy-DCA into a more stable cis conformer. The predicted pKa of BPy-DCA (3.83) exceeded the pH of the reaction mixture (3.15), suggesting that BPy-DCA existed predominantly in the cis configuration during the reaction, attributed to the creation of the intramolecular H-bonding and π-conjugation.76 To further verify the preferred configuration of BPy-DCA under reaction conditions, UV-vis absorption spectra were recorded. Upon the addition of 6 M HOAc to the BPy-DCA solution, a distinct band emerged at 312 nm, exhibiting a red shift compared to the original solution (Fig. S1†). The peaks at 312 nm and 256 nm corresponded to the characteristic absorption of the monocationic BPy-DCA species, indicating the substantial incorporation of cis-configured 2,2′-Bpy into the Py-Bpy-COF during the synthesis. Moreover, the extended π-interaction between stacked layers stabilized the cis conformation of 2,2′-Bpy in the Py-Bpy-COF, which is crucial for subsequent post-synthetic modifications. The successful synthesis of the Py-Bpy-COF was characterized using Fourier transform infrared (FT-IR) spectroscopy. As illustrated in Fig. 1a, the characteristic peak at 1697 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) for BPy-DCA significantly diminished, and a new peak for the characteristic stretching vibrations of imine (C
O) for BPy-DCA significantly diminished, and a new peak for the characteristic stretching vibrations of imine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) groups appeared at 1609 cm−1, indicating the formation of imine linkages in the Py-Bpy-COF.77 The powder X-ray diffraction pattern (PXRD) analysis further demonstrated the successful synthesis of the Py-Bpy-COF (Fig. S2†). The characteristic peaks at 3.15°, 6.4°, and 9.55° are assigned to the (100), (220), and (330) lattice facets, respectively.75 Subsequently, scanning electron microscopy (SEM) images and dynamic light scattering (DLS) analysis revealed that Py-Bpy-COF nanoparticles were approximately 100 nm in diameter (Fig. 1b). Furthermore, the porosity of the Py-Bpy-COF was demonstrated through CO2 adsorption isotherm analysis (Fig. S3†).78 The isotherm exhibits a Type I adsorption behavior, reaching a CO2 adsorption capacity of 26.76 cm3 g−1 at P/P0 = 0.03. The Brunauer–Emmett–Teller (BET) surface area was calculated as 225.4 m2 g−1. Notably, the absence of a hysteresis between adsorption and desorption branches indicates reversible physisorption behaviour and structural stability under CO2 loading.
N) groups appeared at 1609 cm−1, indicating the formation of imine linkages in the Py-Bpy-COF.77 The powder X-ray diffraction pattern (PXRD) analysis further demonstrated the successful synthesis of the Py-Bpy-COF (Fig. S2†). The characteristic peaks at 3.15°, 6.4°, and 9.55° are assigned to the (100), (220), and (330) lattice facets, respectively.75 Subsequently, scanning electron microscopy (SEM) images and dynamic light scattering (DLS) analysis revealed that Py-Bpy-COF nanoparticles were approximately 100 nm in diameter (Fig. 1b). Furthermore, the porosity of the Py-Bpy-COF was demonstrated through CO2 adsorption isotherm analysis (Fig. S3†).78 The isotherm exhibits a Type I adsorption behavior, reaching a CO2 adsorption capacity of 26.76 cm3 g−1 at P/P0 = 0.03. The Brunauer–Emmett–Teller (BET) surface area was calculated as 225.4 m2 g−1. Notably, the absence of a hysteresis between adsorption and desorption branches indicates reversible physisorption behaviour and structural stability under CO2 loading.
As a versatile building block, 2,2′-Bpy is widely utilized in complexation due to customizable functionality and good redox stability.79 The complexation of transition metals with 2,2′-Bpy-based COFs significantly broadens their applications in catalytic fields.80 When the metal-free Py-Bpy-COF reacts with CuCl2 in an aqueous solution, PB-COF-Cu2+ can be formed. Inductively coupled plasma atomic mass spectrometry (ICP-MS) analysis showed that the Cu species loading in PB-COF-Cu2+ was about 6.0 wt%. The FT-IR spectra of PB-COF-Cu2+ revealed that the chemical functionalities of the Py-Bpy-COF were largely preserved, with the exception of a noticeable blue shift in the C–N stretching vibration peak (Fig. S4†), suggesting that Cu2+ coordinates to the bipyridinic N atoms in the Py-Bpy-COF.81 Furthermore, the zeta potential of the Py-Bpy-COF was increased after the addition of Cu2+ (Fig. S5†). X-ray photoelectron spectroscopy (XPS) provided further structural details about the coordination of Cu2+ ions in PB-COF-Cu2+. The presence of Cu2+ in PB-COF-Cu2+ was verified by the full scan XPS spectrum, which demonstrated the effective incorporation of Cu2+ into the frameworks (Fig. S6 and S7†). The N 1s XPS spectrum of PB-COF-Cu2+ showed a slight shift in the N 1s peaks compared to the Py-Bpy-COF, which was attributed to the coordination with CuII (Fig. 1c).82 High-resolution Cu 2p XPS spectra indicated the presence of divalent CuII in PB-COF-Cu2+ (Fig. 1d). These results suggested robust interactions between the Cu species and bipyridine groups in PB-COF-Cu2+. Moreover, the SEM image and DLS results revealed that the grain sizes of PB-COF-Cu2+ were comparable to those of the Py-Bpy-COF, indicating that the framework structures remained intact after the incorporation of Cu2+ (Fig. S8 and S9†). The PXRD profile of PB-COF-Cu2+ was comparable to that of the Py-Bpy-COF, confirming that the Py-Bpy-COF skeletons were preserved following the addition of Cu2+ (Fig. S2†).83
Given that Cu2+ accumulates in Aβ plaques (up to 0.4 mM) and induces ROS production and enhanced Aβ toxicity,5 sequestering Cu2+ from Aβ has been considered as a crucial approach for AD treatment.84 Due to the high binding affinity of 2,2′-Bpy to Cu2+,85 we evaluated the Cu2+ sequestration ability of the Py-Bpy-COF utilizing the intrinsic fluorescence of Tyr10 in Aβ42 by fluorescence spectroscopy. As depicted in Fig. 2a, the Tyr10 fluorescence was quenched, suggesting the binding of Cu2+ to Aβ42 in close proximity.86 Notably, the recovery of Tyr10 fluorescence (308 nm) demonstrated that the Py-Bpy-COF could effectively remove Cu2+ from the Aβ42-Cu2+ complex. Next, the selectivity of the Py-Bpy-COF for binding and sequestering Cu2+ in the presence of other metal ions was examined. As shown in Fig. S10,† the Tyr10 fluorescence of Aβ42-Cu2+ was restored in the presence of Zn2+, Fe3+, and Ca2+ after incubation with the Py-Bpy-COF, indicating that the Py-Bpy-COF selectively bound and sequestered Cu2+ from the Aβ42-Cu2+ complex.
As the Aβ-Cu2+ complex is well known to induce the generation of ROS that is harmful to AD,87 we performed the ascorbate consumption assay to determine the impact of the Py-Bpy-COF on the redox cycling of Cu2+-ascorbate and the generation of ROS. As shown in Fig. 2b, ascorbate underwent rapid consumption in the presence of Cu2+. Notably, the consumption of ascorbate was significantly reduced when Cu2+ was pre-mixed with the Py-Bpy-COF, suggesting that the Py-Bpy-COF possessed the ability to bind to Cu2+ and effectively impede redox cycling. In order to mimic conditions more comparable to AD, we further carried out the ascorbate consumption assay with the addition of Aβ. The Py-Bpy-COF significantly reduced ascorbate consumption in the presence of Aβ (Fig. 2c), suggesting that the binding of the Py-Bpy-COF to Cu2+ can slow down the Cu2+-ascorbate redox process even with Aβ present. Furthermore, the ability of the Py-Bpy-COF to hinder ROS generation induced by Cu2+ was verified by monitoring the production of hydroxyl radicals (˙OH), which was measured through the fluorescence intensity of 7-hydroxycoumarin-3-carboxylic acid (7-OH-CCA).88 As shown in Fig. 2b–e, when the Py-Bpy-COF was introduced, the fluorescence intensity significantly increased in the presence of both ascorbate and Cu2+, confirming the effective mitigation of ˙OH formation by the Py-Bpy-COF. Above all, these findings revealed that the Py-Bpy-COF could sequester Cu2+ and consequently limit the production of ROS caused by Cu2+.
It has been reported that extensive π-electronic conjugation and π–π interactions between interlayers impart keto-enamine-linked COFs enhanced photothermal conversion capability.75,89 Next, the photothermal performance of the Py-Bpy-COF was investigated. To assess the photothermal conversion efficiency of the Py-Bpy-COF, temperature changes were recorded during irradiation with the NIR-I laser and NIR-II laser. The longer-wavelength NIR-II laser offers superior tissue penetration and reduced background autofluorescence emission,90 making it ideal for deep brain therapy through the scalp and skull.74 As illustrated in Fig. 2f and h, the temperature of the Py-Bpy-COF rapidly increased after 5 minutes irradiation with 808 nm and 1064 nm lasers, with temperature changes of approximately 11 °C and 9 °C, respectively. These findings indicated that the Py-Bpy-COF could effectively convert NIR-I and NIR-II energy into thermal energy. Additionally, the results showed that PB-COF-Cu2+ retained similar photothermal conversion properties to the Py-Bpy-COF. Moreover, the photothermal cycling stability of the Py-Bpy-COF and PB-COF-Cu2+ under NIR laser irradiation was also evaluated. As illustrated in Fig. 2g and i, the heating process remained stable for both the Py-Bpy-COF and PB-COF-Cu2+ when NIR-I and NIR-II lasers were alternately turned on and off in each cycle, demonstrating the outstanding photothermal stability of both the Py-Bpy-COF and PB-COF-Cu2+.
Since Cu often serves as the catalytic center of multiple enzymes, we subsequently investigated the enzyme-like properties of PB-COF-Cu2+ towards NADH oxidation and its catalytic performance under NIR-II irradiation (Fig. 3a). The NPX-mimicking catalytic activity was assessed through monitoring the UV-vis absorption spectrum of NADH. The characteristic band of NADH (340 nm) gradually reduced as the reaction time increased in the presence of PB-COF-Cu2+ (Fig. 3b). This notable change corresponded to the conversion of NADH to NAD+, demonstrating that PB-COF-Cu2+ could catalyze NADH oxidation with NPX-mimicking activity. Moreover, to gain further insights into the NPX-mimicking activity of PB-COF-Cu2+, we analyzed the enzyme kinetics for the catalytic oxidation of NADH. Fig. 3c exhibits the time-dependent absorbance changes at a wavelength of 340 nm with differing levels of NADH (0, 0.05, 0.1, 0.2, 0.3, and 0.4 mM). The initial reaction rates (v0) of NADH oxidation were calculated from the absorbance changes. Michaelis–Menten curves were then generated by plotting v0 against the NADH concentrations (Fig. 3d). The Vmax and Km values for the catalytic reaction of PB-COF-Cu2+ nanozymes were 19.28 μM min−1 and 1.7 μM, respectively. Collectively, these outcomes indicated PB-COF-Cu2+ possessed NPX-like activity. Furthermore, as a keto-enamine-linked COF with good thermal properties, PB-COF-Cu2+ exhibited the potential for enhanced catalytic performance under NIR-II irradiation. We assessed the NPX catalytic activity of PB-COF-Cu2+ under different durations of NIR-II irradiation. As shown in Fig. 3e, the catalytic activity of PB-COF-Cu2+ significantly increased with prolonged irradiation with a 1064 nm laser compared with the group without the laser, indicating that the NPX-like activity of PB-COF-Cu2+ could be enhanced by NIR-II irradiation.
Next, a standard methyl thiazolyl tetrazolium (MTT) assay was conducted to assess the biosafety of the Py-Bpy-COF. The viability of hippocampal neuronal cell line (HT22) cells remained above 90% even at Py-Bpy-COF concentrations up to 180 μg·mL−1, demonstrating the outstanding biocompatibility and biosafety of the Py-Bpy-COF in vitro (Fig. S11†). Apoptosis analysis via annexin V-FITC/PI flow cytometry revealed a low level of apoptosis in the Py-Bpy-COF group, further confirming its biocompatibility (Fig. S12†).
Since NAD+ is essential for cellular metabolism, mitochondrial homeostasis, and genome integrity,24 emerging evidence indicates that lower levels of NAD+ are associated with AD pathology.16 Elevating NAD+ levels has been shown to inhibit cGAS–STING signaling, restore mitochondrial homeostasis, and reduce neuroinflammation.16 However, previous attempts to increase NAD+ levels, such as orally administering precursors and directly supplementing cells with NAD+ synthesis precursors, have demonstrated poor efficacy.36 Therefore, for effective catalytic oxidation of NADH to NAD+, NADH was introduced in the Py-Bpy-COF (referred to as PB-COF@NADH). The incorporation amount of PB-COF@NADH was estimated to be 6.3%. Subsequently, we investigated whether PB-COF@NADH could catalyze the conversion of NADH to NAD+ through its NPX-like catalytic activity by binding to Cu2+ and increase cellular NAD+ levels. As shown in Fig. 4a, the intracellular relative ratio of NAD+/NADH in the Aβ-Cu2+ + PB-COF@NADH treated group displayed a marked increase compared to the group incubated with Aβ-Cu2+ solely. Moreover, an even stronger restoration was observed under 1064 nm laser irradiation, indicating that NAD+ homeostasis was significantly restored through the NIR-II enhanced NPX-mimicking activity of PB-COF@NADH.
Then, we investigated the potential usage of PB-COF@NADH for the mitigation of oxidative stress. It has been reported that up-regulating NAD+ can reduce oxidative damage, which is a major pathogenic factor in AD.27 Thus, the 2′,7′-dichlorofluorescin diacetate (DCFH-DA) test was conducted to measure cellular ROS accumulation under various conditions. As illustrated in Fig. S13,† compared to the control group, Aβ-Cu2+-treated cells displayed high fluorescent signals, indicating Aβ-Cu2+ induced oxidative stress. Notably, cells treated with Aβ-Cu2+ + PB-COF@NADH + NIR-II showed a significant reduction in ROS generation. Besides, similar results were obtained via the flow cytometry analysis (Fig. 4b), as ROS levels in the PB-COF@NADH + NIR-II – treated group were reduced to one-third of those in the Aβ-Cu2+-treated group according to quantitative analysis (Fig. 4c). Collectively, these outcomes suggested that oxidative stress could be reduced by the NIR-II-enhanced NPX-like catalytic activities of PB-COF@NADH. Additionally, the redox silencing of Cu2+ by PB-COF@NADH further contributed to the mitigation of oxidative stress.
Previous studies have reported that oxidative stress and NAD+ depletion cause increased activation of the cGAS–STING pathway, which accelerates the progression of AD.10 Next, we evaluated the effect of PB-COF@NADH on the inhibition of cGAS–STING overactivation by western blot. The expression levels of cGAS and STING proteins in HMC3 human microglial cells were considered as the quantitative indicators. As shown in Fig. 4d, the cells treated with Aβ-Cu2+ exhibited significantly higher levels of cGAS and STING expression compared to PBS-treated cells. As expected, the expressions of cGAS and STING proteins in Aβ-Cu2+ + PB-COF@NADH + NIR-II treated cells markedly decreased, respectively. These results demonstrated that the overactivated cGAS–STING pathway was inhibited by PB-COF@NADH through replenishing NAD+ levels and decreasing ROS, mediated by the NIR-II-enhanced NPX-mimicking activity.
Given the crucial role of the cGAS–STING pathway in various autoinflammatory, autoimmune and degenerative diseases,91 we next investigated the ability of PB-COF@NADH to decrease STING-dependent neuroinflammation. The expressions of neuroinflammation-related markers in HMC3 cells were subsequently studied. The secretion of key proinflammatory markers, IL-1β and TNF-α, was evaluated using enzyme-linked immunosorbent assay (ELISA). The data in Fig. 4e and f demonstrated that the secretion of IL-1β and TNF-α significantly decreased following PB-COF@NADH + NIR-II treatment, while the production of IL-1β and TNF-α increased in the Aβ-Cu2+ treated group. Subsequently, the level of integrin CD11b was detected by flow cytometry, as CD11b is expressed on the microglial surface during activation (Fig. 4g). Quantitative analysis revealed an obvious decline in the expression of CD11b in the Aβ-Cu2+ + PB-COF@NADH + NIR-II group compared to the Aβ-Cu2+ treated group (Fig. 4h). Taken together, the above results indicated that PB-COF@NADH + NIR-II treatment mitigated neuroinflammation. PB-COF@NADH possessed a multifaceted therapeutic potential that involved the in situ activation of NIR-II-enhanced NPX-mimicking activity and redox silencing of Cu2+. This resulted in the elevation of NAD+ levels, mitigation of oxidative stress, inhibition of the overactivated cGAS–STING pathway, and ultimately attenuation of neuroinflammation.
NAD+ depletion and oxidative stress have been reported to be associated with mitochondrial dysregulation, which is a hallmark of AD.27 To further explore the restorative effects of PB-COF@NADH, we used the JC-1 fluorescent probe (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl-carbocyanine iodide) to assess its protective effect on mitochondrial function.92 As presented in Fig. 4i, the Aβ-Cu2+ treated group exhibited intense green fluorescence, indicating significant depolarization of the mitochondrial membrane potential (MMP), which led to the loss of mitochondrial function. Importantly, treatment with PB-COF@NADH + NIR-II prevented the depolarization of MMP, as evidenced by decreased green fluorescence and increased red fluorescence (Fig. 4j). These results suggested that PB-COF@NADH could restore mitochondrial homeostasis by elevating NAD+ levels and reducing oxidative stress through NPX-mimicking activity.
Given that mitochondria are regarded as the cellular energy factories and responsible for synthesizing ATP, the ability of PB-COF@NADH to restore ATP production was then investigated. Following the incubation with PB-COF@NADH nanozymes, the cellular ATP production was considerably restored, showing an increase of approximately 73.3% compared to the Aβ-Cu2+-treated group (Fig. 4k). ATP levels were elevated by about 93.2% under 1064 nm laser irradiation. These results suggested that the PB-COF@NADH nanozyme, with NIR-II-promoted NPX-mimicking activity, could effectively restore the NAD+/NADH ratio and mitochondrial function, thereby enhancing ATP synthesis.
Subsequently, the impact of PB-COF@NADH on neuronal survival under Aβ-Cu2+ toxicity was evaluated. The results indicated a notable decrease in cell viability following Aβ-Cu2+ treatment, indicating that Aβ-Cu2+ caused cell death (Fig. 4l). In contrast, co-treatment with Aβ-Cu2+ and PB-COF@NADH significantly increased cell viability, which was further enhanced with 1064 nm laser irradiation. These results demonstrated that PB-COF@NADH effectively mitigated the Aβ-Cu2+-induced cytotoxicity, attributed to the therapeutic effects of redox silencing of Cu2+ and NIR-II enhanced NPX-mimicking activity of PB-COF@NADH.
The BBB is well recognized as a major obstacle hindering the majority of drugs from entering the brain.71 Recently, nanoparticles modified with the 8 amino-acid peptide KLVFFAED have been designed to improve BBB penetration for the treatment of AD.72 The interaction between KLVFFAED and the receptor for advanced glycation end-products (RAGE) increases the efficiency of nanoparticle traversal through the BBB.74,93 Accordingly, we propose that PB-COF@NADH could cross the BBB after modification of KLVFFAED. Besides, KLVFFAED can enable PB-COF@NADH to specifically target Aβ.73 Furthermore, given the photothermal properties of PB-COF@NADH, its ability to permeate the BBB could be enhanced under NIR irradiation.94 The KLVFFAED peptide was covalently modified on the Py-Bpy-COF (referred to as TP@PB-COF). The FT-IR spectra of the TP@PB-COF exhibited characteristic stretching vibrations of N–H and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O from KLVFFAED, as well as C
O from KLVFFAED, as well as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N from the Py-Bpy-COF, confirming the successful synthesis of the TP@PB-COF (Fig. S14†).8,74 The DLS analysis of TP@PB-COF@NADH displayed a hydrodynamic diameter of about 200 nm (Fig. S15†). Next, the MTT assay demonstrated the outstanding biocompatibility of TP@PB-COF@NADH in vitro, with cell viability exceeding 90% at concentrations up to 160 μg·mL−1 (Fig. S16†). In subsequent in vivo experiments, the traversal of TP@PB-COF@NADH across the BBB was assessed. Cyanine3 was incorporated into the TP@PB-COF@NADH (denoted as TP@PB-COF@NADH@Cy3) to track BBB transport. Following intraperitoneal administration of TP@PB-COF@NADH@Cy3 for 24 hours, the AD model mice were euthanized. Their brains along with major organs were harvested for fluorescence imaging. A significant fluorescence signal was detected in the brain of the 3× Tg-AD mouse model treated with TP@PB-COF@NADH@Cy3 (Fig. S17a and b†). Conversely, the fluorescence signal from the brain of the mice treated with PB-COF@NADH@Cy3 without KLVFFAED significantly decreased (Fig. S17c and d†). These findings demonstrated that KLVFFAED facilitated active targeting to the brain, thereby enhancing the penetration of TP@PB-COF@NADH across the BBB.74 Next, we proceeded to assess the in vivo therapeutic efficacy of TP@PB-COF@NADH in 3× Tg-AD mice, which involved intraperitoneal injection once every three days for six times and NIR-II (1064 nm, 0.7 W·cm−2) irradiation for 10 minutes daily (Fig. 5a). After 2 weeks of continuous irradiation following the injections, the Morris water maze (MWM) experiment was conducted for assessment of the cognitive abilities of the mice. As illustrated in Fig. 5b–f, after a 4 days training process, the mice with TP@PB-COF@NADH treatment in combination with NIR-II irradiation exhibited significant improvements in memory performance, as evidenced by the greater proportion of time spent within the target area, increased swimming velocity, higher frequency of platform crossings, and reduced latency to reach the platform compared to the other groups. These results suggested that TP@PB-COF@NADH with NIR-II light radiation could improve cognitive functions in 3× Tg-AD mice.
N from the Py-Bpy-COF, confirming the successful synthesis of the TP@PB-COF (Fig. S14†).8,74 The DLS analysis of TP@PB-COF@NADH displayed a hydrodynamic diameter of about 200 nm (Fig. S15†). Next, the MTT assay demonstrated the outstanding biocompatibility of TP@PB-COF@NADH in vitro, with cell viability exceeding 90% at concentrations up to 160 μg·mL−1 (Fig. S16†). In subsequent in vivo experiments, the traversal of TP@PB-COF@NADH across the BBB was assessed. Cyanine3 was incorporated into the TP@PB-COF@NADH (denoted as TP@PB-COF@NADH@Cy3) to track BBB transport. Following intraperitoneal administration of TP@PB-COF@NADH@Cy3 for 24 hours, the AD model mice were euthanized. Their brains along with major organs were harvested for fluorescence imaging. A significant fluorescence signal was detected in the brain of the 3× Tg-AD mouse model treated with TP@PB-COF@NADH@Cy3 (Fig. S17a and b†). Conversely, the fluorescence signal from the brain of the mice treated with PB-COF@NADH@Cy3 without KLVFFAED significantly decreased (Fig. S17c and d†). These findings demonstrated that KLVFFAED facilitated active targeting to the brain, thereby enhancing the penetration of TP@PB-COF@NADH across the BBB.74 Next, we proceeded to assess the in vivo therapeutic efficacy of TP@PB-COF@NADH in 3× Tg-AD mice, which involved intraperitoneal injection once every three days for six times and NIR-II (1064 nm, 0.7 W·cm−2) irradiation for 10 minutes daily (Fig. 5a). After 2 weeks of continuous irradiation following the injections, the Morris water maze (MWM) experiment was conducted for assessment of the cognitive abilities of the mice. As illustrated in Fig. 5b–f, after a 4 days training process, the mice with TP@PB-COF@NADH treatment in combination with NIR-II irradiation exhibited significant improvements in memory performance, as evidenced by the greater proportion of time spent within the target area, increased swimming velocity, higher frequency of platform crossings, and reduced latency to reach the platform compared to the other groups. These results suggested that TP@PB-COF@NADH with NIR-II light radiation could improve cognitive functions in 3× Tg-AD mice.
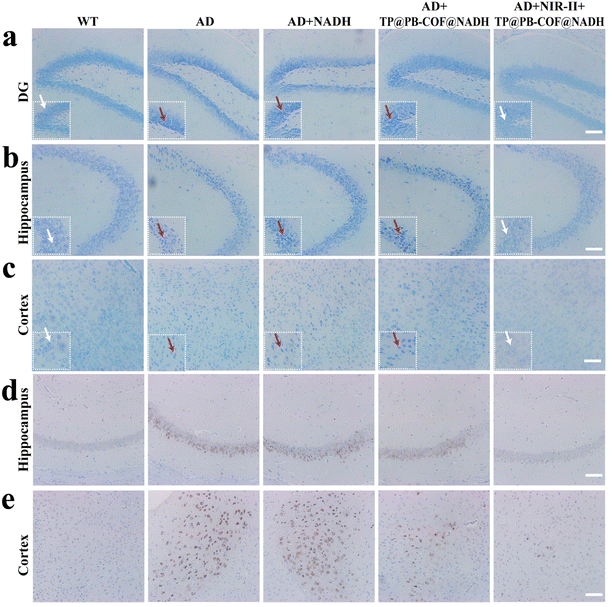
Finally, following the MWM test, all mice underwent euthanasia, and their major organs including brains were harvested for subsequent evaluation. Nissl staining was performed to evaluate the neuroprotective effects of our approach. As depicted in Fig. 6a–c, compared to the healthy wild-type mice, the untreated 3× Tg-AD mice exhibited neuronal loss and noticeable neuron nuclear shrinkage with deep staining (indicated by red arrows) in the hippocampus, dentate gyrus (DG) and cortex.95,96 Conversely, AD mice treated with TP@PB-COF@NADH under NIR-II irradiation showed intact, full-shaped neuronal cell bodies with faint blue staining (indicated by white arrows), indicating the remarkable neuroprotective effect of the COF-based nanozyme as a cGAS–STING inhibitor. These results were further validated by hematoxylin and eosin (H&E) staining (Fig. S18†). Likewise, immunohistochemical analysis for Aβ in the hippocampal and cortical regions revealed that the “TP@PB-COF@NADH + NIR-II” group exhibited the greatest decrease in Aβ plaques compared to the other groups (Fig. 6d and e). This indicated that TP@PB-COF@NADH could reduce Aβ plaques in the lesion areas through the cGAS–STING pathway.10 Moreover, the levels of proinflammatory cytokines TNF-α and IL-1β significantly decreased in the brain of AD mice treated with TP@PB-COF@NADH + NIR-II (Fig. S19 and S20†), indicating that TP@PB-COF@NADH synergistically alleviated the cerebral inflammation in 3× Tg-AD model mice. Furthermore, H&E staining images showed no significant injury to the major organs in AD mice treated with TP@PB-COF@NADH and NIR-II irradiation (Fig. S21†), demonstrating the excellent biocompatibility of TP@PB-COF@NADH. The hemocompatibility of TP@PB-COF@NADH was evaluated through in vitro hemolytic testing according to ISO 10993-4.97 The average hemolysis rate was below the 5% safety threshold, indicating negligible erythrocyte disruption and superior hemocompatibility of TP@PB-COF@NADH (Fig. S22†).
Conclusion
In summary, we have developed a promising therapeutic approach to targeting the cGAS–STING pathway for the treatment of AD. This approach utilizes an endogenous copper ion-responsive COF nanozyme (TP@PB-COF@NADH) with NIR-II-enhanced activity. Excessive Cu2+ in AD could be sequestered and redox silenced by TP@PB-COF@NADH, resulting in decreased generation of ROS, one of the trigger factors of cGAS–STING activation. Besides, during the treatment process, TP@PB-COF@NADH exhibited a significant ability to generate NAD+ and consume H2O2 through NIR-II augmented NPX-like catalytic activities in situ activated by Cu2+. This mechanism effectively prevented the overactivation of the cGAS–STING pathway, reduced neuroinflammation, and restored mitochondrial function. Furthermore, in vivo studies demonstrated that the designed nanozyme inhibitor significantly alleviated cognitive impairment in 3 × Tg AD mice, suggesting the efficacy of this therapeutic approach. Overall, our study provides new insights into the inhibition of cGAS–STING overactivation for treatment of AD by using an AD brain harmful excess endogenous copper ion-responsive and efficient nanozyme.Ethical statement
Animal Model: the APPswe/PS1M146V/tauP301L transgenic model mice (3× Tg-AD mice) were generously provided by the Xiubo Du group at Shenzhen University (Shenzhen, China). Control mice (C57BL/6 mice) were obtained from Jilin University. The Changchun Institute of Applied Chemistry Animal Care and Use Committee authorized all animal studies, which were conducted in compliance with the NIH Publication No. 85–23 Rev. 1985 standards for the care and use of laboratory animals. Additionally, all animal care and handling practices followed the rules that the Changchun Institute of Applied Chemistry's ethical committee had authorized.Data availability
The data are available upon request from the authors.Author contributions
X. Q. and J. R. conceived and designed the experiments, supervised the study and revised the manuscript. H. Z. performed the experiments, analyzed the data, wrote the original manuscript; J. Y., M. S. and X. D. performed the experiments and discussion.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key R&D Program of China (2019YFA0709202 and 2021YFF1200700) and National Natural Science Foundation of China (22437006 and 22237006).References
- D. S. Knopman, H. Amieva, R. C. Petersen, G. Chetelat, D. M. Holtzman, B. T. Hyman, R. A. Nixon and D. T. Jones, Nat. Rev. Dis. Primers, 2021, 7, 33 Search PubMed.
- P. Scheltens, K. Blennow, M. M. B. Breteler, B. de Strooper, G. B. Frisoni, S. Salloway and W. M. Van der Flier, Lancet, 2016, 388, 505–517 Search PubMed.
- Y. Huang and L. Mucke, Cell, 2012, 148, 1204–1222 Search PubMed.
- T. Ho, S. Ahmadi and K. Kerman, Free Radicals Biol. Med., 2022, 181, 180–196 Search PubMed.
- Y. Liu, M. Nguyen, A. Robert and B. Meunier, Acc. Chem. Res., 2019, 52, 2026–2035 Search PubMed.
- Z. Zhu, M. Song, J. Ren, L. Liang, G. Mao and M. Chen, Cell Death Dis., 2024, 15, 850 Search PubMed.
- M. Zubillaga, X. A. Calaf, A. Colell, M. J. Bellini and N. Arnal, Alzheimer's Dementia, 2023, 19, e076143 Search PubMed.
- Q. Ren, H. Chen, Y. Chen, Z. Song, S. Ouyang, S. Lian, J. Tao, Y. Song and P. Zhao, ACS Appl. Mater. Interfaces, 2023, 15, 4947–4958 Search PubMed.
- C. Du, W. Feng, X. Dai, J. Wang, D. Geng, X. Li, Y. Chen and J. Zhang, Small, 2022, 18, e2203031 Search PubMed.
- X. Xie, G. Ma, X. Li, J. Zhao, Z. Zhao and J. Zeng, Nat. Aging, 2023, 3, 202–212 Search PubMed.
- M. F. Gulen, N. Samson, A. Keller, M. Schwabenland, C. Liu, S. Gluck, V. V. Thacker, L. Favre, B. Mangeat, L. J. Kroese, P. Krimpenfort, M. Prinz and A. Ablasser, Nature, 2023, 620, 374–380 Search PubMed.
- B. J. L. Eggen, Nature, 2023, 620, 280–282 Search PubMed.
- X. Xia, X. He, T. Zhao, J. Yang, Z. Bi, Q. Fu, J. Liu, D. Ao, Y. Wei and X. Wei, Cell Proliferation, 2024, 57, e13529 Search PubMed.
- Z. P. Van Acker, A. Perdok, R. Hellemans, K. North, I. Vorsters, C. Cappel, J. Dehairs, J. V. Swinnen, R. Sannerud, M. Bretou, M. Damme and W. Annaert, Nat. Commun., 2023, 14, 2847 Search PubMed.
- T. J. Nelson and Y. Xu, Sci. Rep., 2023, 13, 8304 Search PubMed.
- Y. Hou, Y. Wei, S. Lautrup, B. Yang, Y. Wang, S. Cordonnier, M. P. Mattson, D. L. Croteau and V. A. Bohr, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2011226118 Search PubMed.
- A. Decout, J. D. Katz, S. Venkatraman and A. Ablasser, Nat. Rev. Immunol., 2021, 21, 548–569 Search PubMed.
- M. C. C. Canesso, L. Lemos, T. C. Neves, F. M. Marim, T. B. R. Castro, E. S. Veloso, C. P. Queiroz, J. Ahn, H. C. Santiago, F. S. Martins, J. Alves-Silva, E. Ferreira, D. C. Cara, A. T. Vieira, G. N. Barber, S. C. Oliveira and A. M. C. Faria, Mucosal Immunol., 2018, 11, 820–834 Search PubMed.
- Y. Huang, B. Liu, S. C. Sinha, S. Amin and L. Gan, Mol. Neurodegener., 2023, 18, 79 Search PubMed.
- S. M. Haag, M. F. Gulen, L. Reymond, A. Gibelin, L. Abrami, A. Decout, M. Heymann, F. G. van der Goot, G. Turcatti, R. Behrendt and A. Ablasser, Nature, 2018, 559, 269–273 Search PubMed.
- M. Govindarajulu, S. Ramesh, M. Beasley, G. Lynn, C. Wallace, S. Labeau, S. Pathak, R. Nadar, T. Moore and M. Dhanasekaran, Int. J. Mol. Sci., 2023, 24, 8151 Search PubMed.
- E. Verdin, Science, 2015, 350, 1208–1213 Search PubMed.
- M. S. Bonkowski and D. A. Sinclair, Nat. Rev. Mol. Cell Biol., 2016, 17, 679–690 Search PubMed.
- E. F. Fang, S. Lautrup, Y. Hou, T. G. Demarest, D. L. Croteau, M. P. Mattson and V. A. Bohr, Trends Mol. Med., 2017, 23, 899–916 Search PubMed.
- W. Luo, X. Zou, Y. Wang, Z. Dong, X. Weng, Z. Pei, S. Song, Y. Zhao, Z. Wei, R. Gao, B. Zhang, L. Liu, P. Bai, J. Liu, X. Wang, T. Gao, Y. Zhang, X. Sun, H. Chen, K. Hu, S. Du, A. Sun and J. Ge, Circ. Res., 2023, 132, e223–e242 Search PubMed.
- K. Myakala, X. X. Wang, N. V. Shults, E. Krawczyk, B. A. Jones, X. Yang, A. Z. Rosenberg, B. Ginley, P. Sarder, L. Brodsky, Y. Jang, C. H. Na, Y. Qi, X. Zhang, U. Guha, C. Wu, S. Bansal, J. Ma, A. Cheema, C. Albanese, M. D. Hirschey, T. Yoshida, J. B. Kopp, J. Panov and M. Levi, J. Biol. Chem., 2023, 299, 104975 Search PubMed.
- Y. Hou, S. Lautrup, S. Cordonnier, Y. Wang, D. L. Croteau, E. Zavala, Y. Zhang, K. Moritoh, J. F. O'Connell, B. A. Baptiste, T. V. Stevnsner, M. P. Mattson and V. A. Bohr, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E1876–E1885 Search PubMed.
- A. J. Covarrubias, R. Perrone, A. Grozio and E. Verdin, Nat. Rev. Mol. Cell Biol., 2021, 22, 119–141 Search PubMed.
- Y. Dong and G. J. Brewer, J. Alzheim. Dis., 2019, 71, 119–140 Search PubMed.
- C. Canto, A. A. Sauve and P. Bai, Mol. Aspects Med., 2013, 34, 1168–1201 Search PubMed.
- J. S. Kerr, B. A. Adriaanse, N. H. Greig, M. P. Mattson, M. Z. Cader, V. A. Bohr and E. F. Fang, Trends Neurosci., 2017, 40, 151–166 Search PubMed.
- E. F. Fang, M. Scheibye-Knudsen, L. E. Brace, H. Kassahun, T. SenGupta, H. Nilsen, J. R. Mitchell, D. L. Croteau and V. A. Bohr, Cell, 2014, 157, 882–896 Search PubMed.
- T. Nacarelli, L. Lau, T. Fukumoto, J. Zundell, N. Fatkhutdinov, S. Wu, K. M. Aird, O. Iwasaki, A. V. Kossenkov, D. Schultz, K. I. Noma, J. A. Baur, Z. Schug, H. Y. Tang, D. W. Speicher, G. David and R. Zhang, Nat. Cell Biol., 2019, 21, 397–407 Search PubMed.
- J. Yoshino, J. A. Baur and S. I. Imai, Cell Metab., 2018, 27, 513–528 Search PubMed.
- A. Poyan Mehr, M. T. Tran, K. M. Ralto, D. E. Leaf, V. Washco, J. Messmer, A. Lerner, A. Kher, S. H. Kim, C. C. Khoury, S. J. Herzig, M. E. Trovato, N. Simon-Tillaux, M. R. Lynch, R. I. Thadhani, C. B. Clish, K. R. Khabbaz, E. P. Rhee, S. S. Waikar, A. H. Berg and S. M. Parikh, Nat. Med., 2018, 24, 1351–1359 Search PubMed.
- O. K. Reiten, M. A. Wilvang, S. J. Mitchell, Z. Hu and E. F. Fang, Mech. Ageing Dev., 2021, 199, 111567 Search PubMed.
- X. Liu, J. Li, A. Zitolo, M. Gao, J. Jiang, X. Geng, Q. Xie, D. Wu, H. Zheng, X. Cai, J. Lu, F. Jaouen and R. Li, J. Am. Chem. Soc., 2023, 145, 3108–3120 Search PubMed.
- A. Patgiri, O. S. Skinner, Y. Miyazaki, G. Schleifer, E. Marutani, H. Shah, R. Sharma, R. P. Goodman, T. L. To, X. Robert Bao, F. Ichinose, W. M. Zapol and V. K. Mootha, Nat. Biotechnol., 2020, 38, 309–313 Search PubMed.
- R. P. Goodman, A. L. Markhard, H. Shah, R. Sharma, O. S. Skinner, C. B. Clish, A. Deik, A. Patgiri, Y. H. Hsu, R. Masia, H. L. Noh, S. Suk, O. Goldberger, J. N. Hirschhorn, G. Yellen, J. K. Kim and V. K. Mootha, Nature, 2020, 583, 122–126 Search PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 Search PubMed.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48, 1004–1076 Search PubMed.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 Search PubMed.
- M. Sun, L. Xu, A. Qu, P. Zhao, T. Hao, W. Ma, C. Hao, X. Wen, F. M. Colombari, A. F. de Moura, N. A. Kotov, C. Xu and H. Kuang, Nat. Chem., 2018, 10, 821–830 Search PubMed.
- L. Ma, J. J. Zheng, N. Zhou, R. Zhang, L. Fang, Y. Yang, X. Gao, C. Chen, X. Yan and K. Fan, Nat. Commun., 2024, 15, 233 Search PubMed.
- S. Zhao, H. Li, R. Liu, N. Tao, L. Deng, Q. Xu, J. Hou, J. Sheng, J. Zheng, L. Wang, W. Chen, S. Guo and Y. N. Liu, J. Am. Chem. Soc., 2023, 145, 10322–10332 Search PubMed.
- M. Li, J. Chen, W. Wu, Y. Fang and S. Dong, J. Am. Chem. Soc., 2020, 142, 15569–15574 Search PubMed.
- Y. Sang, F. Cao, W. Li, L. Zhang, Y. You, Q. Deng, K. Dong, J. Ren and X. Qu, J. Am. Chem. Soc., 2020, 142, 5177–5183 Search PubMed.
- W. H. Chen, M. Vázquez-González, A. Kozell, A. Cecconello and I. Willner, Small, 2017, 14, 1703149 Search PubMed.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 Search PubMed.
- L. Zhang, H. Wang and X. G. Qu, Adv. Mater., 2023, 2211147, DOI:10.1002/adma.202211147.
- L. Yan, F. Zhao, J. Wang, Y. Zu, Z. Gu and Y. Zhao, Adv. Mater., 2019, 31, e1805391 Search PubMed.
- A. E. Nel, L. Madler, D. Velegol, T. Xia, E. M. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 Search PubMed.
- C. Kaweeteerawat, C. H. Chang, K. R. Roy, R. Liu, R. Li, D. Toso, H. Fischer, A. Ivask, Z. Ji, J. I. Zink, Z. H. Zhou, G. F. Chanfreau, D. Telesca, Y. Cohen, P. A. Holden, A. E. Nel and H. A. Godwin, ACS Nano, 2015, 9, 7215–7225 Search PubMed.
- W. Zhang, M. Wang, B. Liu, H. Chen, J. Tan, Q. Meng, J. Li, B. Ding, P. Ma and J. Lin, Angew. Chem., Int. Ed., 2024, 63, e202402397 Search PubMed.
- C. Chen, Y. Chen, X. Wang, L. Zhang, Y. Luo, Q. Tang, Y. Wang, X. Liang and C. Ma, iScience, 2023, 26, 106066 Search PubMed.
- L. Zhang, L. Zhang, H. Deng, H. Li, W. Tang, L. Guan, Y. Qiu, M. J. Donovan, Z. Chen and W. Tan, Nat. Commun., 2021, 12, 2002 Search PubMed.
- W. Zhen, D. W. Kang, Y. Fan, Z. Wang, T. Germanas, G. T. Nash, Q. Shen, R. Leech, J. Li, G. S. Engel, R. R. Weichselbaum and W. Lin, J. Am. Chem. Soc., 2024, 146, 16609–16618 Search PubMed.
- Y. H. Wijesundara, T. S. Howlett, S. Kumari and J. J. Gassensmith, Chem. Rev., 2024, 124, 3013–3036 Search PubMed.
- J. Han, J. Feng, J. Kang, J.-M. Chen, X.-Y. Du, S.-Y. Ding, L. Liang and W. Wang, Science, 2024, 383, 1014–1019 Search PubMed.
- L. L. Zhou, Q. Guan and Y. B. Dong, Angew. Chem., Int. Ed., 2023, 63, e202314763 Search PubMed.
- L. Zhang, A. Song, Q.-C. Yang, S.-J. Li, S. Wang, S.-C. Wan, J. Sun, R. T. K. Kwok, J. W. Y. Lam, H. Deng, B. Z. Tang and Z.-J. Sun, Nat. Commun., 2023, 14, 5355 Search PubMed.
- Y. Duan, Y. Yu, P. Liu, Y. Gao, X. Dai, L. Zhang, L. Chen and Y. Chen, Angew. Chem., Int. Ed., 2023, 62, e202302146 Search PubMed.
- L. Liang, R. Yang, J. Wu, Y. Qin, Y. Jiang, S. Zhao and F. Ye, Anal. Chem., 2024, 96, 18545–18554 Search PubMed.
- W. Sun, X. Tang, Q. Yang, Y. Xu, F. Wu, S. Guo, Y. Zhang, M. Wu and Y. Wang, Adv. Mater., 2019, 31, 1903176 Search PubMed.
- F. M. Zhang, J. L. Sheng, Z. D. Yang, X. J. Sun, H. L. Tang, M. Lu, H. Dong, F. C. Shen, J. Liu and Y. Q. Lan, Angew. Chem., Int. Ed., 2018, 57, 12106–12110 Search PubMed.
- L. Zhang, Z. Liu, Q. Deng, Y. Sang, K. Dong, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2020, 60, 3469–3474 Search PubMed.
- Q. Sun, C. W. Fu, B. Aguila, J. Perman, S. Wang, H. Y. Huang, F. S. Xiao and S. Ma, J. Am. Chem. Soc., 2018, 140, 984–992 Search PubMed.
- D. Parsonage, H. Miller, R. P. Ross and A. Claiborne, J. Biol. Chem., 1993, 268, 3161–3167 Search PubMed.
- C. S. Diercks, S. Lin, N. Kornienko, E. A. Kapustin, E. M. Nichols, C. Zhu, Y. Zhao, C. J. Chang and O. M. Yaghi, J. Am. Chem. Soc., 2018, 140, 1116–1122 Search PubMed.
- Q. Guan, L. L. Zhou, F. H. Lv, W. Y. Li, Y. A. Li and Y. B. Dong, Angew. Chem., Int. Ed., 2020, 59, 18042–18047 Search PubMed.
- I. U. Ali and X. Chen, ACS Nano, 2015, 9, 9470–9474 Search PubMed.
- Z. Liu, Q. Deng, G. Qin, J. Yang, H. Zhang, J. Ren and X. Qu, Chem, 2023, 9, 2016–2038 Search PubMed.
- H. Zhang, D. Yu, S. Liu, C. Liu, Z. Liu, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2022, 61, e202109068 Search PubMed.
- M. Ma, N. Gao, X. Li, Z. Liu, Z. Pi, X. Du, J. Ren and X. Qu, ACS Nano, 2020, 14, 9894–9903 Search PubMed.
- Z. Mi, P. Yang, R. Wang, J. Unruangsri, W. Yang, C. Wang and J. Guo, J. Am. Chem. Soc., 2019, 141, 14433–14442 Search PubMed.
- K. Nakamoto, J. Phys. Chem., 1960, 64, 1420–1425 Search PubMed.
- X. Yang, Q. An, X. Li, Y. Fu, S. Yang, M. Liu, Q. Xu and G. Zeng, Nat. Commun., 2024, 15, 1889 Search PubMed.
- L. Zou, Z. A. Chen, D. H. Si, S. L. Yang, W. Q. Gao, K. Wang, Y. B. Huang and R. Cao, Angew. Chem., Int. Ed., 2023, 62, e202309820 Search PubMed.
- C. Kaes, A. Katz and M. W. Hosseini, Chem. Rev., 2000, 100, 3553–3590 Search PubMed.
- Q. Guan, L.-L. Zhou and Y.-B. Dong, Chem. Soc. Rev., 2022, 51, 6307–6416 Search PubMed.
- G. Wen, Q. Peng, C. Yuan, J. He and X. Hou, Nanoscale, 2024, 17, 322–332 Search PubMed.
- Q. Sun, B. Aguila, J. Perman, N. Nguyen and S. Ma, J. Am. Chem. Soc., 2016, 138, 15790–15796 Search PubMed.
- J. Li, D. Ma, Q. Huang, Y. Du, Q. He, H. Ji, W. Ma and J. Zhao, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2302761120 Search PubMed.
- L. E. Cassagnes, V. Hervé, F. Nepveu, C. Hureau, P. Faller and F. Collin, Angew. Chem., Int. Ed., 2013, 52, 11110–11113 Search PubMed.
- Y. Ji, H. J. Lee, M. Kim, G. Nam, S. J. C. Lee, J. Cho, C. M. Park and M. H. Lim, Inorg. Chem., 2017, 56, 6695–6705 Search PubMed.
- B. Alies, E. Renaglia, M. Rózga, W. Bal, P. Faller and C. Hureau, Anal. Chem., 2013, 85, 1501–1508 Search PubMed.
- A. Conte-Daban, M. Beyler, R. Tripier and C. Hureau, Chem.–Eur. J., 2018, 24, 8447–8452 Search PubMed.
- A. E. Behar, L. Sabater, M. Baskin, C. Hureau and G. Maayan, Angew. Chem., Int. Ed., 2021, 60, 24588–24597 Search PubMed.
- J. Tan, S. Namuangruk, W. Kong, N. Kungwan, J. Guo and C. Wang, Angew. Chem., Int. Ed., 2016, 55, 13979–13984 Search PubMed.
- G. Hong, A. L. Antaris and H. Dai, Nat. Biomed. Eng., 2017, 1, 0010 Search PubMed.
- Z. Zhang, D. Zhou, Z. Li, X. Luan, J. Yang, S. Tang and Y. Song, Angew. Chem., Int. Ed., 2024, 63, e202316007 Search PubMed.
- P. Yang, Q. Huang, J. Zhang, Y. Li, H. Gao and Z. Gu, Adv. Mater., 2024, 36, e2308393 Search PubMed.
- E. Cuevas, H. Rosas-Hernandez, S. M. Burks, M. A. Ramirez-Lee, A. Guzman, S. Z. Imam, S. F. Ali and S. Sarkar, Metab. Brain Dis., 2019, 34, 1365–1374 Search PubMed.
- W. Chen, J. Ouyang, X. Yi, Y. Xu, C. Niu, W. Zhang, L. Wang, J. Sheng, L. Deng, Y. N. Liu and S. Guo, Adv. Mater., 2017, 30, 1703458 Search PubMed.
- J. Shen, Z. Lu, J. Wang, Q. Hao, W. Ji, Y. Wu, H. Peng, R. Zhao, J. Yang, Y. Li, Z. Shi and X. Zhang, Adv. Mater., 2021, 33, e2101993 Search PubMed.
- M. Huang, M. Zheng, Q. Song, X. Ma, Q. Zhang, H. Chen, G. Jiang, S. Zhou, H. Chen, G. Wang, C. Dai, S. Li, P. Li, H. Wang, A. Zhang, Y. Huang, J. Chen and X. Gao, Adv. Mater., 2024, 36, e2311420 Search PubMed.
- L. Hou, Z. Li, H. Zhao, Y. Pan, S. Pavlinich, X. Liu, X. Li, Y. Zheng and L. Li, J. Mater. Sci. Technol., 2016, 32, 874–882 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc07963a |
| This journal is © The Royal Society of Chemistry 2025 |