 Open Access Article
Open Access ArticleFunctionally active modulators targeting the LRRK2 WD40 repeat domain identified by FRASE-bot in CACHE Challenge #1†
Akhila
Mettu
a,
Marta
Glavatskikh
b,
Xiaowen
Wang
a,
Antonio Jesús
Lara Ordóñez
 c,
Fengling
Li
d,
Irene
Chau
d,
Suzanne
Ackloo
c,
Fengling
Li
d,
Irene
Chau
d,
Suzanne
Ackloo
 d,
Cheryl
Arrowsmith
dfg,
Albina
Bolotokova
d,
Pegah
Ghiabi
d,
Elisa
Gibson
d,
Cheryl
Arrowsmith
dfg,
Albina
Bolotokova
d,
Pegah
Ghiabi
d,
Elisa
Gibson
 d,
Levon
Halabelian
de,
Scott
Houliston
g,
Rachel J.
Harding
deh,
Ashley
Hutchinson
d,
Peter
Loppnau
d,
Sumera
Perveen
d,
Almagul
Seitova
d,
Hong
Zeng
d,
Matthieu
Schapira
de,
Jean-Marc
Taymans
d,
Levon
Halabelian
de,
Scott
Houliston
g,
Rachel J.
Harding
deh,
Ashley
Hutchinson
d,
Peter
Loppnau
d,
Sumera
Perveen
d,
Almagul
Seitova
d,
Hong
Zeng
d,
Matthieu
Schapira
de,
Jean-Marc
Taymans
 c and
Dmitri
Kireev‡
c and
Dmitri
Kireev‡
 *ab
*ab
aUniversity of Missouri – Columbia, Department of Chemistry, USA. E-mail: dmitri.kireev@missouri.edu
bUniversity of North Carolina at Chapel Hill, Center for Integrative Chemical Biology and Drug Discovery, USA
cUniv. Lille, Inserm, CHU Lille, UMR-S 1172 – LilNCog – Lille Neuroscience & Cognition, F-59000, France
dStructural Genomics Consortium, University of Toronto, Toronto, ON M5G 1L7, Canada
eDepartment of Pharmacology and Toxicology, University of Toronto, Toronto, Ontario M5S 1A8, Canada
fMedical Biophysics, University of Toronto, Toronto, Ontario M5G 1L7, Canada
gPrincess Margaret Cancer Centre, University Health Network, Toronto, Ontario M5G 2C4, Canada
hLeslie Dan Faculty of Pharmacy, University of Toronto, Toronto, Ontario M5S 3M2, Canada
First published on 9th January 2025
Abstract
Critical Assessment of Computational Hit-Finding Experiments (CACHE) Challenges emerged as real-life stress tests for computational hit-finding strategies. In CACHE Challenge #1, 23 participants contributed their original workflows to identify small-molecule ligands for the WD40 repeat (WDR) of LRRK2, a promising Parkinson's target. We applied the FRASE-based hit-finding robot (FRASE-bot), a platform for interaction-based screening allowing a drastic reduction of the explorable chemical space and a concurrent detection of putative ligand-binding sites. In two screening rounds, 84 compounds were procured for experimental testing and 8 were confirmed to bind LRRK2-WDR with dissociation constants (Kd) ranging from 3 to 41 μM. To investigate the functional effect of WDR ligands, they were tested for their ability to modify the LRRK2 activity markers in HEK293T cells. Two compounds showed statistically significant increases in the kinase activity of WT LRRK2, and two compounds affected the conformation and kinase activity of major LRRK2 mutants.
Introduction
Parkinson's disease (PD) stands as the second-most prevalent neurodegenerative disorder, afflicting nearly one million individuals in the United States alone, and the worldwide genetic prevalence was estimated to be 18 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 individuals with its incidence persistently escalating despite available therapeutic interventions.1 Among the novel targets emerging in PD research, leucine-rich repeat kinase 2 (LRRK2) gained particular attention.2 LRRK2 is a seven-domain protein primarily composed of Ras of complex (ROC) and kinase domains and a C-terminal WD40 domain.3,4 Mutations within various LRRK2 domains have been implicated as potential drivers of PD pathology, leading to hyperphosphorylation of its substrate Rab proteins, a pivotal event in disease progression.5–7 The formation of LRRK2 filaments is catalyzed by the interaction of LRRK2 with microtubules and is likely caused by the kinase hyperactivity exacerbated by the PD-associated LRRK2 mutations.8 Notably, most chemical compounds under preclinical and clinical investigation for PD therapy target the kinase domain, exemplified by DNL201, and DNL151.9 While the clinical efficacy of kinase inhibitors is yet to be demonstrated, it would be of interest to consider alternative options for pharmaceutical intervention. In particular, the WD40 repeat domain (WDR) of LRRK2 has also been shown to undergo PD-associated mutations and putatively play a pivotal role in tubulin binding or recruitment of other signaling partners.3,4 Small-molecule chemical probes binding to the WDR might be a useful tool to further explore its potential as a target for PD therapeutics. However, no small-molecule ligands have been reported yet for LRRK2-WDR. Moreover, the location of the potential binding site for such ligands is unknown.
000 individuals with its incidence persistently escalating despite available therapeutic interventions.1 Among the novel targets emerging in PD research, leucine-rich repeat kinase 2 (LRRK2) gained particular attention.2 LRRK2 is a seven-domain protein primarily composed of Ras of complex (ROC) and kinase domains and a C-terminal WD40 domain.3,4 Mutations within various LRRK2 domains have been implicated as potential drivers of PD pathology, leading to hyperphosphorylation of its substrate Rab proteins, a pivotal event in disease progression.5–7 The formation of LRRK2 filaments is catalyzed by the interaction of LRRK2 with microtubules and is likely caused by the kinase hyperactivity exacerbated by the PD-associated LRRK2 mutations.8 Notably, most chemical compounds under preclinical and clinical investigation for PD therapy target the kinase domain, exemplified by DNL201, and DNL151.9 While the clinical efficacy of kinase inhibitors is yet to be demonstrated, it would be of interest to consider alternative options for pharmaceutical intervention. In particular, the WD40 repeat domain (WDR) of LRRK2 has also been shown to undergo PD-associated mutations and putatively play a pivotal role in tubulin binding or recruitment of other signaling partners.3,4 Small-molecule chemical probes binding to the WDR might be a useful tool to further explore its potential as a target for PD therapeutics. However, no small-molecule ligands have been reported yet for LRRK2-WDR. Moreover, the location of the potential binding site for such ligands is unknown.
To address the need for WDR-targeting chemical probes, the Michael J. Fox Foundation for Parkinson's Research and the Critical Assessment of Computational Hit-Finding Experiments (CACHE) initiative announced CACHE Challenge #1 (ref. 10 and 11) (35 teams responded to this inaugural challenge, 23 of which were selected to participate). According to the CACHE Challenge terms, participants were allowed to submit a list of up to 100 virtual hits in Round-1. The teams that identified any experimentally confirmed Round-1 hits were invited to submit a Round-2, hit-expansion list to explore preliminary structure–activity relationships (SARs).
Finding ligands for the WDR represents two distinct problems: (i) locating the binding site and (ii) docking/scoring the ligands. There are a variety of options for both tasks. While potent ligands typically exploit the central cavity of other donut shaped WDR domains,12,13 an unbiased approach would be to identify ligand-binding pockets through cavity sensing algorithms such as SiteMap (Schrodinger suite14), GRID,15 POCKET,16 or SurfNet.17 Next, ligands can be placed into the detected pockets and assessed for their binding affinity using one of many docking techniques available either in commercial molecular modeling suites or using open-source software, such as Autodock18 and its popular forks, e.g., Autodock Vina19 (it is not our intention here to provide a comprehensive review of the vast universe of docking algorithms). In recent years, machine learning (ML) scoring functions (for instance, GNINA,20 OnionNet,21 Pafnucy,22 or TopologyNet23) allowed, to some extent, the increase of the accuracy of binding affinity predictions. Moreover, deep active learning strategies, e.g., Deep Docking,24 extended the scope of docking-based screens to billions of compounds. Finally, blind docking techniques, such as DiffDock,25 EquiBind,26 or TANKBind,27 do not need a prior assumption about the binding pocket location and hence solve both problems (pocket identification and docking) at once. However, despite the recent progress, hit finding for novel ligand-orphan targets still suffers from the same old problem, an excessive false-positive rate (which may be further exacerbated when the binding site is not known), that often prevents finding any hits. Hence, new strategies are welcome. We recently developed the FRASE-based hit-finding robot (FRASE-bot)28 that takes advantage of the FRAgments in Structural Environments (FRASE)29 concept. Similar to blind docking methods, FRASE-bot does not require to specify the binding site. Instead, the structure of the protein of interest is seeded with ligand fragments transferred from experimental protein–ligand complexes (as described in ref. 29). The seeded fragments can then be exploited in virtual screening, e.g., to build a pharmacophore model or to run a substructure search (the option used in this work). Such fragment-based primary screens can be efficiently run on billion-scale databases and result in focused compound sets enriched in actives, thus improving the odds of success for the downstream docking-based screens. Binding sites for docking algorithms can be defined as the protein regions with the highest densities of the seeded fragments. Alternatively, the target structures and poses could be plugged into a molecular generator as a source of structural constraints. In CACHE Challenge #1, our hit-finding platform enabled identification of 8 WDR ligands in a low micromolar range, thus placing us among the winners of the competition.
Results
FRASE screening identifies ligand fragments seeded in the WDR structure
Fast screening of the FRASE database (as described in ref. 28) was performed against an X-ray structure of the LRRK2 WDR domain (PDB: 6DLO). The screening resulted in 2544 fragments seeded in multiple regions of the protein structure (Fig. 1A). Two filters were applied to eliminate flawed fragment poses, i.e., poses displaying “collisions” (distances of 1 Å or closer) between the ligand and the protein atoms and poses that are not sufficiently “buried” within the protein (that is, each ligand atom has an average of less than 5 protein atoms within 5 Å). The application of both filters reduced the number of fragments to 348. Finally, a previously developed neural network model28 was applied to calculate fitness scores for the selected poses (the model was trained to distinguish between true FRASEs and decoys). The threshold of 0.4 was applied to further reduce the number of fragments to 85 (Fig. 1B).Ligand fragments form dense clusters in putative ligand-binding pockets
To assess the collective value of the 85 fragments resulting from the FRASE screen, we analyzed high-density regions of the fragment population. To this end, the fragments were represented by the Cartesian coordinates of their geometric centers and clustered using the K-means technique. The clustering yielded five spatially distinct groups (designated in Fig. 2B as clusters 1–5), potentially representing two binding pockets for docking-based screens. Cluster centers of 1–3 form an approximately equilateral triangle with sides of ∼10 Å (Fig. 1B), suggesting that a small-molecule ligand may potentially bind any two of the sub-pockets occupied by clusters 1–3 but not all three (this would require a ligand of considerable size). Clusters 4 and 5 are close to each other (5 Å between the centers) and far away (∼25 Å) from clusters 1–3 (Fig. 1B). It is hence unlikely for a single ligand to simultaneously bind the pocket occupied by 1–3 and the pocket occupied by 4–5. The fragments populating each of the respective sub-pockets also reflect the pockets' nature. In particular, cluster 1, embedded in the body of the WDR “donut”, is predominantly composed of non-polar, aromatic, halogen-substituted rings (Fig. 1C); cluster 2, located on the outer surface of the “donut”, mainly includes heterocycles, with many of them featuring hydrogen-bond donor (HBD) substituents (Fig. 1C); cluster 3, occupying the rim of the donut's central cavity, represents a mixture of polar heterocycles, such as diazoles and triazoles, and non-polar aromatic fragments (Fig. 1C); cluster 4, entangled within the β-strands on the opposite side of the donut (Fig. 1C), mixes oxygen- and nitrogen-containing heterocycles, and halogen-substituted non-polar aromatics; and cluster 5, adjacent to cluster 4, is largely non-polar aromatic. | ||
| Fig. 2 The experimentally confirmed Round-1 hits. Chemical structures, LRRK2 Kd values, solubility, and SPR dose–response data for the Round-1 hits 1 (A) and 2 (B); docking poses for 1 (C) and 2 (D). | ||
Seeded fragments enable selection of a focused compound set
Both structural and spatial information provided by the 85 seeded fragments was exploited to pre-filter a collection of commercially available compounds – the Advanced Screening Collection (Enamine) of ∼900![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 molecules. In particular, we sought to retrieve the compounds potentially binding to at least two identified sub-pockets. To this end, combinations of fragment structures were used to compose substructure queries under the following conditions: (i) fragments for the same query must be pulled from two different clusters and (ii) the two clusters must be within ∼10 Å from each other. That is, the queries may combine fragments from clusters 1 and 2, 1 and 3, 2 and 3, and 4 and 5. These multiple substructure searches resulted in a combined set of ∼100
000 molecules. In particular, we sought to retrieve the compounds potentially binding to at least two identified sub-pockets. To this end, combinations of fragment structures were used to compose substructure queries under the following conditions: (i) fragments for the same query must be pulled from two different clusters and (ii) the two clusters must be within ∼10 Å from each other. That is, the queries may combine fragments from clusters 1 and 2, 1 and 3, 2 and 3, and 4 and 5. These multiple substructure searches resulted in a combined set of ∼100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds enriched in potential LRRK2 WDR binders.
000 compounds enriched in potential LRRK2 WDR binders.
Docking-based screening designates putative LRRK2 binders
The ∼100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds resulting from the substructure search were docked using Autodock Vina19 to the two binding pockets corresponding to clusters 1/2/3 and 4/5, respectively. The top 10% hits from Autodock Vina were then subjected to docking using the Glide algorithm.30 The top 1000 Glide hits were processed through a min–max diversity filter to remove structurally redundant compounds (see the Methods for more details). Next, binding free energies (BFE) were calculated for the remaining ∼600 compounds using the MM-GBSA31 method (as implemented in the Schrodinger software suite14). Finally, the top 200 docked ligands were visually inspected. The decision to keep a hit was made based on three scores (Vina, Glide, and MM-PBSA BFE) and visually perceived factors such as how well-buried the ligand is, how flexible it is, and how “natural” its conformation looks like. A total of 54 compounds were retained at this step, of which 37 were available for purchase on the day of the order and submitted to the CACHE challenge organizers as Round-1 hits.
000 compounds resulting from the substructure search were docked using Autodock Vina19 to the two binding pockets corresponding to clusters 1/2/3 and 4/5, respectively. The top 10% hits from Autodock Vina were then subjected to docking using the Glide algorithm.30 The top 1000 Glide hits were processed through a min–max diversity filter to remove structurally redundant compounds (see the Methods for more details). Next, binding free energies (BFE) were calculated for the remaining ∼600 compounds using the MM-GBSA31 method (as implemented in the Schrodinger software suite14). Finally, the top 200 docked ligands were visually inspected. The decision to keep a hit was made based on three scores (Vina, Glide, and MM-PBSA BFE) and visually perceived factors such as how well-buried the ligand is, how flexible it is, and how “natural” its conformation looks like. A total of 54 compounds were retained at this step, of which 37 were available for purchase on the day of the order and submitted to the CACHE challenge organizers as Round-1 hits.
Binding confirmation for the Round-1 hits
Binding for the 37 sourced compounds was evaluated using a Surface Plasmon Resonance (SPR) assay with biotinylated LRRK2-WDR immobilized on a streptavidin chip, thus facilitating the capture of target molecules (see Fig. 2A and B and ESI file 1†). Moreover, binding to the NSD2 PWWP domain, an unrelated target, was assessed to monitor unspecific protein binding. Dynamic Light Scattering (DLS) was used to evaluate solubility and aggregation at 200 μM. Initially, all compounds underwent rapid SPR screening at concentrations of 50 μM and 100 μM. Two compounds, 1 and 2, exhibited a binding signal ranging between 50% and 200% of the expected value and were selected for dose–response experiments. Both 1 and 2 demonstrated dose-dependent binding with Kd of 16 μM and 32 μM, respectively. According to DLS data, both compounds were 30% soluble at a high concentration of 200 μM and 1 showed signs of aggregation. While 1 appeared selective for LRRK2, 2 displayed binding to the unrelated NSD2 PWWP domain with a Kd of 42 μM. Although, the 3D structures of the LRRK2 WDR in complex with 1 and 2 are yet to be solved, the respective docking poses (Fig. 2C and D) can be used as a template for further hit expansion, as well as initial exploration of structure–activity relationships (SARs). Both 1 and 2 occupy the pocket corresponding to cluster 3 (Fig. 1C, 2C and D) and can potentially be extended toward pockets 1 or 2.Round 2: hit expansion enables preliminary structure–activity relationships
In Round-2, pharmacophore screening and similarity search were employed to identify structural analogs of 1 and 2 in Enamine REAL, an ultra-large screening collection of 6.5 billion compounds.32 Since the full database is out of reach for a 3D pharmacophore screening, it was prefiltered using 2D pharmacophore fingerprints (as described in the Methods). The top 70 million 2D-pharmacophore hits were then subjected to 3D pharmacophore screening using Phase,33 with pharmacophore models derived from the docking poses of 1 and 2, resulting in a combined pool of 1122 compounds. Concurrently, a similarity search on the full Enamine REAL database with 1 and 2 as queries and a Tanimoto threshold of 0.8 (as described in the Methods) yielded 190 molecules. Next, the 1312 (1122 + 190) compounds were docked using Glide30 and clustered using the Butina method.34 We then selected two overlapping lists for the final triage: (i) 150 top-scored cluster representatives and (ii) 50 top-scored ligands overall (based on Glide scores). The poses of these selected ligands were visually inspected to assess their structural alignment with the parent compounds 1 and 2. A curated set of 47 analogs (43 for parent 1 and 4 for parent 2) was submitted for experimental testing (see ESI file 2†).Six analogs (3–8) of the parent hit 1 exhibited dose-dependent binding to the LRRK2 WDR domain in the range of 3 μM to 44 μM (Fig. 3). Apart from 7, which showed signs of aggregation at 50 μM and was only 75% soluble at 100 μM, compounds behaved well up to 100 μM as measured by DLS and all of them displayed no binding to the unrelated target NSD2 PWWP domain. The analogs' activity helped to further validate the 1-arylmethyl-6-phenyl-1H-pyrazolo[3,4-b]pyridine chemical series and inform early SARs.
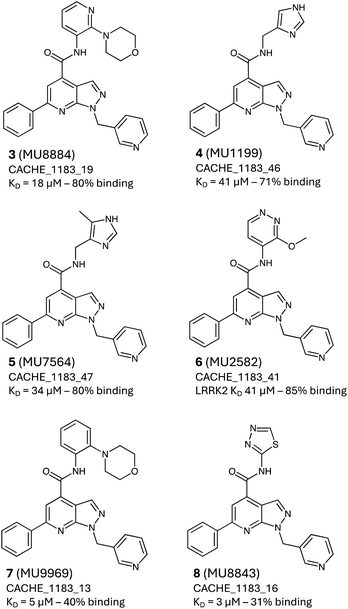 | ||
| Fig. 3 The experimentally confirmed Round-2 hits. Chemical structures and LRRK2 Kd values, and solubility for the Round-3 hits 3–8. | ||
Effect in cell-based assays
Round-1 and Round-2 hits were tested in cells to identify possible changes in LRRK2 kinase activity or its phosphorylation status. As an initial approach and based on the solubility data mentioned above, we treated HEK293T cells transfected with a WT or Y1699C pathogenic LRRK2 mutant with the different hits at a concentration of 30 μM. After 2 hours of treatment, we could observe subtle changes in the phosphorylation status of LRRK2 and its kinase activity (Fig. 4). Two analogs (5 and 6) showed a significant increase in the phosphorylation level of residue S935 in WT LRRK2 but not in YC pathogenic mutant transfected cells (Fig. 4B). When we analyzed the kinase activity of LRRK2 in the presence of the hits, we observed compounds 1 and 6 to induce a slight increase in the LRRK2 kinase activity of the Y1699C pathogenic mutant compared to DMSO treated cells (Fig. 4C and D).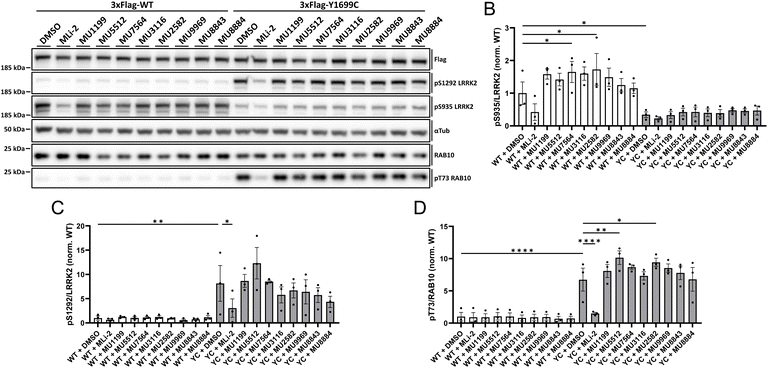 | ||
| Fig. 4 Cellular effects of compounds on LRRK2 kinase activity. (A) Example of HEK293T cells transfected with a 3×Flag-tagged LRRK2 WT or Y1699C pathogenic mutant and treated with DMSO, 30 nM MLi-2, a potent kinase domain inhibitor (ref. 35), or 30 μM of the different hits for 2 h, cell lysates analyzed by western blotting for Flag, pS1292 LRRK2, pS935 LRRK2, RAB10, pT73 RAB10 and α-tubulin as the loading control. (B) Quantification of the pS935 LRRK2/LRRK2 ratio of blots depicted in (A). *p < 0.05. (C) Quantification of the pS1292 LRRK2/LRRK2 ratio of blots depicted in (A). *p < 0.05, **p < 0.01. (D) Quantification of the pT73 RAB10/RAB10 ratio of blots depicted in (A). *p < 0.05, **p < 0.01, and ****p < 0.001. | ||
Discussion
CACHE Challenge10,11 was a long-needed benchmark for computational hit-finding methods. Certainly, the challenges come at a high cost due to the substantial amount of experimental work required for confirmation of virtual hits submitted by all selected participants. But this is a fair price to pay for excluding any conscious and subconscious bias that may influence hit-finding studies on a protein with prior knowledge of ligands. Challenge #1 will provide a triple benefit to the drug/probe discovery community: (i) inform the computational hit-finding community on which methods are most efficient; (ii) facilitate access to these most efficient methods by encouraging the use of original open-source software; and (iii) catalyze the development of publicly accessible chemical probes for promising but difficult therapeutic targets, such as LRRK2 WDR.The LRRK2 WDR is the first genuine ligand-orphan target to which FRASE-bot was applied. Previously, the concept of fragments in structural environments was introduced to optimize selectivity profiles in a “model system” of three enzymes (TYRO3, AXL, and MERTK) belonging to a well-established superfamily of protein kinases.29 Recently, a more challenging Calcium and Integrin Binding protein 1 (CIB1) belonging to a small family of four proteins was successfully targeted by FRASE-bot.36 Although no small-molecule ligands were known for CIB1, a putative ligand-binding site was previously identified through the discovery of peptide binders by phage display screens.37–40 For the LRRK2 WDR, neither the ligands nor their potential binding sites were known, and hence, both tasks – site and ligand identification – needed to be addressed by FRASE-bot.
In CACHE Challenge #1,10,41 FRASE-bot was applied to identify LRRK2 WDR ligands among many other original workflows. Eventually, only 7 participants out of 23 were able to identify any hits. In two rounds of the challenge, we identified 8 experimentally confirmed LRRK2 WDR binders (out of 85 submitted compounds, thus showing a 9% success rate) with Kd ranging from 3 μM to 44 μM as determined by surface plasmon resonance.42 This study demonstrates the high potential of the FRASE-based strategy in the intended setting, that is, hit-finding for a difficult non-conventional target. Conceptually, FRASE-based screening and design can be described as an interaction-based approach, in which the activity of new ligands is established based on the similarity of structural patterns of ligand–protein interaction to those of known active ligands. Such similarity can be defined either for a whole ligand, e.g., through Structural Protein–Ligand Interaction Fingerprints (SPLIF),43,44 or fragment-wise, by means of FRASE screening.36
The LRRK2 WDR binders resulting from the challenge will benefit the Parkinson's research. After optimization for an increased potency, cell membrane permeability, and pharmacokinetics, WDR probes will help to further investigate the biology of LRRK2 and its role in Parkinson's disease. This is of particular interest in light of our fragmentary knowledge on how the LRRK2 WD40 domain affects LRRK2 functions. For instance, it has previously been shown that the C-terminal domain of LRRK2, where the WDR is located, is essential for its kinase activity.45 Recent studies showed that a helix extending from the WDR domain acts as a dynamic regulator of LRRK2 kinase activity, alternatively binding LRRK2 kinase or LRRK2 N-terminal domains.46 Therefore, WDR binding molecules may affect this regulation, impacting both LRRK2 kinase activity and overall LRRK2 conformation. In our preliminary cellular studies, we were able to probe for both of these effects using the LRRK2 kinase activity markers pT73-Rab10 and pS1292-LRRK2 as well as the LRRK2 conformational marker pS935-LRRK2. Our results show that the three compounds affect the LRRK2 function in different ways. Two of them (5 and 6) induce weak but observable increases in pS935-LRRK2 levels for WT LRRK2, suggesting that some compounds may affect LRRK2 conformation, thereby affecting the phosphorylation/dephosphorylation equilibrium. Interestingly, two compounds also led to slight increases in the kinase activity marker pT73-Rab10 in the Y1699C-LRRK2 mutant but not WT (1 and 6), suggesting that the WDR domain ligands may regulate LRRK2 kinase activity. It remains to be determined if this effect is limited to certain disease mutants and whether these can affect not only kinase activation but also kinase inactivation. Further testing will be needed to progress the current compounds toward potential chemical probes of WDR function. In addition, further optimization would be needed to determine whether some WDR binding candidates may show potential as Parkinson's drugs.
Methods
Computational methods
Surface plasmon resonance (SPR)
The binding affinity of compounds was assessed by surface plasmon resonance (SPR, Biacore™ 8K, Cytiva Inc.) at 25 °C. Biotinylated LRRK2 (2141-2527aa – https://www.addgene.org/210899/) was captured onto flow cells of a streptavidin-conjugated SA chip at approximately 5000 response units (RUs) (according to the manufacturer's protocol). Compounds were dissolved in 100% DMSO (30 mM stock) and diluted to 10 mM before serial dilutions were prepared in 100% DMSO (a dilution factor of 0.33 was used to yield 5 concentrations). For SPR analysis, the serially titrated compound was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 in HBS buffer (10 mM HEPES pH 7.4, 150 mM NaCl, and 0.01% Tween-20) to a final concentration of 2% DMSO. Experiments were performed using the same buffer containing 2% DMSO and multi-cycle kinetics with a 60 s contact time and a dissociation time of 120 s at a flow rate of 40 μL min−1. Kinetic curve fittings and KD value calculations were done with a 1
50 in HBS buffer (10 mM HEPES pH 7.4, 150 mM NaCl, and 0.01% Tween-20) to a final concentration of 2% DMSO. Experiments were performed using the same buffer containing 2% DMSO and multi-cycle kinetics with a 60 s contact time and a dissociation time of 120 s at a flow rate of 40 μL min−1. Kinetic curve fittings and KD value calculations were done with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model using Biacore Insight Evaluation Software (Cytiva Inc.).
1 binding model using Biacore Insight Evaluation Software (Cytiva Inc.).
Dynamic light scattering (DLS)
The solubility of compounds was estimated by DLS that directly measures compound aggregates and laser power in solution. Compounds were serially diluted directly from DMSO stocks and then diluted 50× into filtered 10 mM HEPES pH 7.4 and 150 mM NaCl (2% DMSO final). The resulting samples were then distributed into 384-well plates (black with a clear bottom, Corning 3540), with 20 μL in each well. The sample plate was centrifuged at 3500 rpm for 5 minutes before loading into the DynaPro DLS Plate Reader III (Wyatt Technology) and analyzed as previously described.50,51Cell assays
The activity of compounds was determined in HEK293T cells (ATCC, CRL-3216) cultured as previously described (Marchand et al., 2022). The cells were subcultured in 6-well plates and transfected at 70–80% confluence with 2 μg of LRRK2 WT or LRRK2 Y1699C pathogenic mutant constructs using branched polyethyleneimine (Clinisciences, 19850). The next day, transfected cells were subcultured into 12-well plates and subjected to western blot 48 h post-transfection. The cells were treated with DMSO (Sigma-Aldrich, D2438), 30 nM of MLi-2, a kinase domain inhibitor35 (Tocris, 5756) or 30 μM of the different compounds for 2 hours prior to collection.The cells were collected and washed in PBS, followed by resuspension in lysis buffer ((20 mM Tris pH 7.5, 150 mM NaCl, 0.2% Triton-X100, 5 mM MgCl2, and 10% glycerol) + cOmplete EDTA-free protease inhibitor cocktail + PhosSTOP phosphatase inhibitor cocktail) and incubation on a rotary wheel for 30 minutes at 4 °C. Lysates were then centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 rpm for 10 minutes at 4 °C, and the protein concentration of supernatants was estimated by BCA assay (Takara, T9300A). Around 30 μg of protein were mixed with an appropriate amount of 4× NuPAGE™ LDS Sample Buffer (Invitrogen, NP0008) and 10× Bolt™ Sample Reducing Agent (Invitrogen, B0009), boiled at 70 °C for 10 minutes, and resolved in NuPAGE™ 4–12% Bis–Tris Midi Gels (Invitrogen, WG1403) at 100 V for 2 hours with NuPAGE™ MOPS SDS running buffer (Invitrogen, NP0001). Proteins were electrophoretically transferred onto 0.45 μm nitrocellulose membranes (Cytiva, 10600002) at 40 mA overnight at 4 °C in transfer buffer (20 mM Tris pH 8.6, 122 mM glycine, and 5% (v/v) MeOH). The membranes were blocked with 5% (w/v) skim milk powder in 0.1% Tween-20/PBS for 1 hour at room temperature and incubated with primary antibodies diluted in blocking solution overnight at 4 °C. Primary antibodies included rabbit anti-pS1292 LRRK2 [MJFR-19-7-8] (1
200 rpm for 10 minutes at 4 °C, and the protein concentration of supernatants was estimated by BCA assay (Takara, T9300A). Around 30 μg of protein were mixed with an appropriate amount of 4× NuPAGE™ LDS Sample Buffer (Invitrogen, NP0008) and 10× Bolt™ Sample Reducing Agent (Invitrogen, B0009), boiled at 70 °C for 10 minutes, and resolved in NuPAGE™ 4–12% Bis–Tris Midi Gels (Invitrogen, WG1403) at 100 V for 2 hours with NuPAGE™ MOPS SDS running buffer (Invitrogen, NP0001). Proteins were electrophoretically transferred onto 0.45 μm nitrocellulose membranes (Cytiva, 10600002) at 40 mA overnight at 4 °C in transfer buffer (20 mM Tris pH 8.6, 122 mM glycine, and 5% (v/v) MeOH). The membranes were blocked with 5% (w/v) skim milk powder in 0.1% Tween-20/PBS for 1 hour at room temperature and incubated with primary antibodies diluted in blocking solution overnight at 4 °C. Primary antibodies included rabbit anti-pS1292 LRRK2 [MJFR-19-7-8] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, ab203181), mouse anti-Flag® M2 (1
1000, Abcam, ab203181), mouse anti-Flag® M2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, Sigma-Aldrich, F1804), mouse anti-α-tubulin (1
500, Sigma-Aldrich, F1804), mouse anti-α-tubulin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Bio-Techne, NB100-690), rabbit anti-pT73 RAB10 [MJF-R21] (1
000, Bio-Techne, NB100-690), rabbit anti-pT73 RAB10 [MJF-R21] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Abcam, ab230261), and mouse anti-RAB10 [4E2] (1
1000, Abcam, ab230261), and mouse anti-RAB10 [4E2] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Thermo Scientific, MA515670). The following day, the membranes were washed twice in 0.1% Tween-20/PBS for 10 minutes at room temperature and incubated with secondary antibodies diluted in 0.1% Tween-20/PBS for 1 hour at room temperature. Secondary antibodies included HRP-conjugated goat anti-mouse (1
1000, Thermo Scientific, MA515670). The following day, the membranes were washed twice in 0.1% Tween-20/PBS for 10 minutes at room temperature and incubated with secondary antibodies diluted in 0.1% Tween-20/PBS for 1 hour at room temperature. Secondary antibodies included HRP-conjugated goat anti-mouse (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Cell Signaling, 7076S) and goat anti-rabbit (1
000, Cell Signaling, 7076S) and goat anti-rabbit (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, Cell Signaling, 7074S). After two additional washes, the membranes were revealed using ECL Standard Western Blotting Detection Reagent (Cytiva, RPN2209) for 1 minute, and the chemiluminescent reaction was detected with Amersham ImageQuant™ 800 Western blot imaging systems (Cytiva, USA).
5000, Cell Signaling, 7074S). After two additional washes, the membranes were revealed using ECL Standard Western Blotting Detection Reagent (Cytiva, RPN2209) for 1 minute, and the chemiluminescent reaction was detected with Amersham ImageQuant™ 800 Western blot imaging systems (Cytiva, USA).
Statistical analysis
Data were checked for normal distribution using the Shapiro–Wilk test. Normally distributed data were analyzed by one-way ANOVA and Fisher's LSD multiple comparison. Significance values for all data are indicated in figure legends. Statistical analysis and graphs were generated using GraphPad Prism software version 10.2 (San Diego, CA, USA).Code availability
The source code used to perform the current study is shared as a supplementary archive file and through the GitHub repository https://www.github.com/kireevlab/FRASE-bot-Pipeline-Pilot.47 The latest version of FRASE-bot implemented as a Python code is available at https://www.github.com/kireevlab/FRASE-bot-RDKit.52Data availability
A spreadsheet with SPR data for experimentally tested compounds selected by virtual screening is shared as ESI files 1 and 2† (respectively, “t_1183_CACHE1-Round1-confirmation.xlsx” and “t_1183_CACHE1-Round2-confirmation”). Source data for the figures are provided with this paper.Author contributions
Methodology, investigation, analyses, writing, and editing: A. M., M. G., X. W., A. J. L. O., F. L., I. C., S. A., C. A., A. B., P. G., E. G., L. H., S. H., R. J. H., A. H., P. L., S. P., A. S., H. Z., M. S., J.-M. T., and D. K.; conceptualization, writing, review, supervision, resources, and project administration: M. S., J.-M. T. and D. K.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by a startup fund provided by the University of Missouri and by grant 5R01GM132299 from NIH, National Institute of General Medical Sciences (NIGMS), to D. K. A. J. L. O. and J.-M. T. are profoundly grateful for the funding from the Agence Nationale de Recherche (ANR-21-CE16-0003-01, project PARK-PEP). Experimental testing was supported by an Open Science Drug Discovery grant from Canada's Strategic Innovation Fund (SIF Stream 5) administered by Conscience and the Michael J. Fox Foundation and conducted at the Structural Genomics Consortium, a registered charity (no: 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through the Ontario Genomics Institute [OGI-196], Janssen, Merck KGaA (aka EMD in Canada and USA), Pfizer and Takeda.References
- S. Rong, G. Xu, B. Liu, Y. Sun, L. G. Snetselaar, R. B. Wallace, B. Li, J. Liao and W. Bao, Trends in Mortality From Parkinson Disease in the United States, 1999–2019, Neurology, 2021, 97, e1986–e1993 CrossRef PubMed.
- J.-M. Taymans, M. Fell, T. Greenamyre, W. D. Hirst, A. Mamais, S. Padmanabhan, I. Peter, H. Rideout and A. Thaler, Perspective on the current state of the LRRK2 field, npj Parkinson's Dis., 2023, 9, 104 CrossRef CAS PubMed.
- P. Zhang, Y. Fan, H. Ru, L. Wang, V. G. Magupalli, S. S. Taylor, D. R. Alessi and H. Wu, Crystal structure of the WD40 domain dimer of LRRK2, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 1579–1584 CrossRef CAS PubMed.
- C. K. Deniston, J. Salogiannis, S. Mathea, D. M. Snead, I. Lahiri, M. Matyszewski, O. Donosa, R. Watanabe, J. Böhning, A. K. Shiau, S. Knapp, E. Villa, S. L. Reck-Peterson and A. E. Leschziner, Structure of LRRK2 in Parkinson's disease and model for microtubule interaction, Nature, 2020, 588, 344–349 CrossRef CAS PubMed.
- D. R. Alessi and E. Sammler, LRRK2 kinase in Parkinson's disease, Science, 2018, 360, 36–37 CrossRef CAS.
- G. Ito and N. Utsunomiya-Tate, Overview of the Impact of Pathogenic LRRK2 Mutations in Parkinson's Disease, Biomolecules, 2023, 13, 845 CrossRef CAS PubMed.
- S. Wang, Z. Liu, T. Ye, O. S. Mabrouk, T. Maltbie, J. Aasly and A. B. West, Elevated LRRK2 autophosphorylation in brain-derived and peripheral exosomes in LRRK2 mutation carriers, Acta Neuropathol. Commun., 2017, 5, 86 CrossRef PubMed.
- C. K. Deniston, J. Salogiannis, S. Mathea, D. M. Snead, I. Lahiri, M. Matyszewski, O. Donosa, R. Watanabe, J. Böhning, A. K. Shiau, S. Knapp, E. Villa, S. L. Reck-Peterson and A. E. Leschziner, Structure of LRRK2 in Parkinson's disease and model for microtubule interaction, Nature, 2020, 588, 344–349 CrossRef CAS PubMed.
- T. Müller, DNL151, DNL201, and BIIB094: experimental agents for the treatment of Parkinson's disease, Expert Opin. Invest. Drugs, 2023, 32, 787–792 CrossRef.
- S. Ackloo, R. Al-awar, R. E. Amaro, C. H. Arrowsmith, H. Azevedo, R. A. Batey, Y. Bengio, U. A. K. Betz, C. G. Bologa, J. D. Chodera, W. D. Cornell, I. Dunham, G. F. Ecker, K. Edfeldt, A. M. Edwards, M. K. Gilson, C. R. Gordijo, G. Hessler, A. Hillisch, A. Hogner, J. J. Irwin, J. M. Jansen, D. Kuhn, A. R. Leach, A. A. Lee, U. Lessel, M. R. Morgan, J. Moult, I. Muegge and T. I. Oprea, et al., CACHE (Critical Assessment of Computational Hit-finding Experiments): A public–private partnership benchmarking initiative to enable the development of computational methods for hit-finding, Nat. Rev. Chem, 2022, 6, 287–295 CrossRef PubMed.
- F. Li, S. Ackloo, C. H. Arrowsmith, F. Ban, C. J. Barden, H. Beck, J. Beránek, F. Berenger, A. Bolotokova, G. Bret, M. Breznik, E. Carosati, I. Chau, Y. Chen, A. Cherkasov, D. D. Corte, K. Denzinger, A. Dong, S. Draga, I. Dunn, K. Edfeldt, A. Edwards, M. Eguida, P. Eisenhuth, L. Friedrich, A. Fuerll, S. S. Gardiner, F. Gentile, P. Ghiabi, E. Gibson, et al., CACHE Challenge #1: targeting the WDR domain of LRRK2, a Parkinson's Disease associated protein, bioRxiv, 2024, preprint, DOI:10.1101/2024.07.18.603797.
- R. Song, Z.-D. Wang and M. Schapira, Disease Association and Druggability of WD40 Repeat Proteins, J. Proteome Res., 2017, 16, 3766–3773 CrossRef CAS.
- M. Schapira, M. Tyers, M. Torrent and C. H. Arrowsmith, WD40 repeat domain proteins: a novel target class?, Nat. Rev. Drug Discovery, 2017, 16, 773–786 CrossRef CAS PubMed.
- F. Li, S. Ackloo, C. H. Arrowsmith, F. Ban, C. J. Barden, H. Beck, J. Beránek, F. Berenger, A. Bolotokova, G. Bret, M. Breznik, E. Carosati, I. Chau, Y. Chen, A. Cherkasov, D. Della Corte, K. Denzinger, A. Dong, S. Draga, I. Dunn, K. Edfeldt, A. Edwards, M. Eguida, P. Eisenhuth, L. Friedrich, A. Fuerll, S. S. Gardiner, F. Gentile, P. Ghiabi, E. Gibson, M. Glavatskikh, C. Gorgulla, J. Guenther, A. Gunnarsson, F. Gusev, E. Gutkin, L. Halabelian, R. J. Harding, A. Hillisch, L. Hoffer, A. Hogner, S. Houliston, J. J Irwin, O. Isayev, A. Ivanova, C. Jacquemard, A. J Jarrett, J. H. Jensen, D. Kireev, J. Kleber, S. B. Koby, D. Koes, A. Kumar, M. G. Kurnikova, A. Kutlushina, U. Lessel, F. Liessmann, S. Liu, W. Lu, J. Meiler, A. Mettu, G. Minibaeva, R. Moretti, C. J Morris, C. Narangoda, T. Noonan, L. Obendorf, S. Pach, A. Pandit, S. Perveen, G. Poda, P. Polishchuk, K. Puls, V. Pütter, D. Rognan, D. Roskams-Edris, C. Schindler, F. Sindt, V. Spiwok, C. Steinmann, R. L. Stevens, V. Talagayev, D. Tingey, O. Vu, W. P. Walters, X. Wang, Z. Wang, G. Wolber, C. A. Wolf, L. Wortmann, H. Zeng, C. A. Zepeda, K. Y. J. Zhang, J. Zhang, S. Zheng, M. Schapira, Life Science Schrödinger Suite Release 2022, Schrodinger LLC, 2022 Search PubMed.
- P. J. Goodford, A computational procedure for determining energetically favorable binding sites on biologically important macromolecules, J. Med. Chem., 1985, 28, 849–857 CrossRef CAS PubMed.
- D. G. Levitt and L. J. Banaszak, POCKET: A computer graphics method for identifying and displaying protein cavities and their surrounding amino acids, J. Mol. Graphics, 1992, 10, 229–234 CrossRef CAS PubMed.
- R. A. Laskowski, SURFNET: A program for visualizing molecular surfaces, cavities, and intermolecular interactions, J. Mol. Graphics, 1995, 13, 323–330 CrossRef CAS.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- A. T. McNutt, P. Francoeur, R. Aggarwal, T. Masuda, R. Meli, M. Ragoza, J. Sunseri and D. R. Koes, GNINA 1.0: molecular docking with deep learning, J. Cheminf., 2021, 13, 43 Search PubMed.
- L. Zheng, J. Fan and Y. Mu, OnionNet: a Multiple-Layer Intermolecular-Contact-Based Convolutional Neural Network for Protein–Ligand Binding Affinity Prediction, ACS Omega, 2019, 4, 15956–15965 CrossRef CAS.
- M. M. Stepniewska-Dziubinska, P. Zielenkiewicz and P. Siedlecki, Development and evaluation of a deep learning model for protein–ligand binding affinity prediction, Bioinformatics, 2018, 34, 3666–3674 CrossRef CAS PubMed.
- Z. Cang and G.-W. Wei, TopologyNet: Topology based deep convolutional and multi-task neural networks for biomolecular property predictions, PLoS Comput. Biol., 2017, 13, e1005690 CrossRef PubMed.
- A. Ton, F. Gentile, M. Hsing, F. Ban and A. Cherkasov, Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds, Mol. Inf., 2020, 39, 2000028 CrossRef CAS.
- G. Corso, H. Stärk, B. Jing, R. Barzilay and T. Jaakkola, DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking, arXiv, 2022, preprint, arXiv:2210.01776, DOI:10.48550/arXiv.2210.01776.
- H. Stärk, O. Ganea, L. Pattanaik, R. Barzilay and T. Jaakkola, Equibind: geometric deep learning for drug binding structure prediction, in International Conference on Machine Learning 20503–20521, PMLR, 2022 Search PubMed.
- W. Lu, Q. Wu, J. Zhang, J. Rao, C. Li and S. Zheng, Tankbind: Trigonometry-aware neural networks for drug-protein binding structure prediction, Adv. Neural Inf. Process. Syst., 2022, 35, 7236–7249 Search PubMed.
- Y. An, J. Lim, M. Glavatskikh, X. Wang, J. Norris, P. B. Hardi, T. Leisner, K. Pearce and D. Kireev, Machine Learning-driven Fragment-Based Discovery of CIB1-directed Anti-Tumor Agents by FRASE-bot, Nat. Commun., 2024, 15(1), 5564 CrossRef CAS PubMed.
- C. Da, D. Zhang, M. Stashko, E. Vasileiadi, R. E. Parker, K. A. Minson, M. G. Huey, J. M. Huelse, D. Hunter, T. S. K. Gilbert, J. Norris-Drouin, M. Miley, L. E. Herring, L. M. Graves, D. Deryckere, H. S. Earp, D. K. Graham, S. V. Frye, X. Wang and D. Kireev, Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology, J. Am. Chem. Soc., 2019, 141, 15700–15709 CrossRef CAS.
- R. A. Friesner, J. L. Banks, R. B. Murphy, T. A. Halgren, J. J. Klicic, D. T. Mainz, M. P. Repasky, E. H. Knoll, M. Shelley, J. K. Perry, D. E. Shaw, P. Francis and P. S. Shenkin, Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy, J. Med. Chem., 2004, 47, 1739–1749 CrossRef CAS.
- G. Rastelli, A. Del Rio, G. Degliesposti and M. Sgobba, Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA, J. Comput. Chem., 2010, 31, 797–810 CrossRef CAS PubMed.
- Enamine REAL library.
- S. L. Dixon, A. M. Smondyrev, E. H. Knoll, S. N. Rao, D. E. Shaw and R. A. Friesner, PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results, J. Comput.-Aided Mol. Des., 2006, 20, 647–671 CrossRef CAS PubMed.
- D. Butina, Unsupervised Data Base Clustering Based on Daylight's Fingerprint and Tanimoto Similarity: A Fast and Automated Way To Cluster Small and Large Data Sets, J. Chem. Inf. Comput. Sci., 1999, 39, 747–750 CrossRef CAS.
- M. J. Fell, C. Mirescu, K. Basu, B. Cheewatrakoolpong, D. E. DeMong, J. M. Ellis, L. A. Hyde, Y. Lin, C. G. Markgraf, H. Mei, M. Miller, F. M. Poulet, J. D. Scott, M. D. Smith, Z. Yin, X. Zhou, E. M. Parker, M. E. Kennedy and J. A. Morrow, MLi-2, a Potent, Selective, and Centrally Active Compound for Exploring the Therapeutic Potential and Safety of LRRK2 Kinase Inhibition, J. Pharmacol. Exp. Ther., 2015, 355, 397–409 CrossRef CAS.
- Y. An, J. Lim, M. Glavatskikh, X. Wang, J. Norris-Drouin, P. B. Hardy, T. M. Leisner, K. H. Pearce and D. Kireev, In silico fragment-based discovery of CIB1-directed anti-tumor agents by FRASE-bot, Nat. Commun., 2024, 15, 5564 CrossRef CAS PubMed.
- A. C. Puhl, J. W. Bogart, V. A. Haberman, J. E. Larson, A. S. Godoy, J. L. Norris-Drouin, S. H. Cholensky, T. M. Leisner, S. V. Frye, L. V. Parise, A. A. Bowers and K. H. Pearce, Discovery and Characterization of Peptide Inhibitors for Calcium and Integrin Binding Protein 1, ACS Chem. Biol., 2020, 15, 1505–1516 CrossRef CAS PubMed.
- V. Haberman, S. Fleming, T. Leisner, A. Puhl, E. Feng, L. Xie, X. Chen, Y. Goto, H. Suga, L. Parise, D. Kireev, K. Pearce and A. Bowers, Discovery and Development of Cyclic Peptide Inhibitors of CIB1, ACS Med. Chem. Lett., 2021, 12(11), 1832–1839 CrossRef CAS PubMed.
- T. C. Freeman, J. L. Black, H. G. Bray, O. Dagliyan, Y. I. Wu, A. Tripathy, N. V. Dokholyan, T. M. Leisner and L. V. Parise, Identification of Novel Integrin Binding Partners for Calcium and Integrin Binding Protein 1 (CIB1): Structural and Thermodynamic Basis of CIB1 Promiscuity, Biochemistry, 2013, 52, 7082–7090 CrossRef CAS PubMed.
- V. A. Haberman, S. R. Fleming, T. M. Leisner, A. C. Puhl, E. Feng, L. Xie, X. Chen, Y. Goto, H. Suga, L. V. Parise, D. Kireev, K. H. Pearce and A. A. Bowers, Discovery and Development of Cyclic Peptide Inhibitors of CIB1, ACS Med. Chem. Lett., 2021, 12, 1832–1839 CrossRef CAS PubMed.
- Critical Assessment of Computational Hit-Finding Experiments. Results of CACHE Challenge #1, 2023, https://www.cache-challenge.org/results-cache-challenge-1, accessed 18th May 2024.
- R. B. M. Schasfoort, Introduction to surface plasmon resonance, in Handbook of Surface Plasmon Resonance, The Royal Society of Chemistry, 2017, pp. 1–26, 10.1039/9781788010283-00001.
- Z. Deng, C. Chuaqui and J. Singh, Structural Interaction Fingerprint (SIFt): A Novel Method for Analyzing Three-Dimensional Protein-Ligand Binding Interactions, J. Med. Chem., 2003, 47, 337–344 CrossRef PubMed.
- C. Da and D. Kireev, Structural Protein–Ligand Interaction Fingerprints (SPLIF) for Structure-Based Virtual Screening: Method and Benchmark Study, J. Chem. Inf. Model., 2014, 54, 2555–2561 CrossRef CAS PubMed.
- N. D. Jorgensen, Y. Peng, C. C.-Y. Ho, H. J. Rideout, D. Petrey, P. Liu and W. T. Dauer, The WD40 Domain Is Required for LRRK2 Neurotoxicity, PLoS One, 2009, 4, e8463 CrossRef PubMed.
- P. K. Sharma, J.-H. Weng, J. T. Manschwetus, J. Wu, W. Ma, F. W. Herberg and S. S. Taylor, Role of the leucine-rich repeat protein kinase 2 C-terminal tail in domain cross-talk, Biochem. J., 2024, 481, 313–327 CrossRef CAS.
- D. Kireev & M. Glavatskikhkireevlab/FRASE-bot-Pipeline-Pilot: In silico Fragment-based Discovery of CIB1-directed Anti-Tumor Agents by FRASE-bot. ( 2021). DOI:10.5281/zenodo.11372285.
- R. Zhou, F. RA, A. Ghosh, R. RC, J. WL and L. RM, New linear interaction method for binding affinity calculations using a continuum solvent model, J. Phys. Chem. B, 2001, 105, 10388 CrossRef CAS.
- J. Li, R. Abel, K. Zhu, Y. Cao, S. Zhao and R. A. Friesner, The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling, Proteins: Struct., Funct., Bioinf., 2011, 79, 2794–2812 CrossRef CAS PubMed.
- S. Aleandri, A. Vaccaro, R. Armenta, A. C. Völker and M. Kuentz, Dynamic Light Scattering of Biopharmaceutics—Can Analytical Performance Be Enhanced by Laser Power?, Pharmaceutics, 2018, 10, 94 CrossRef CAS PubMed.
- S. J. Allen, C. M. Dower, A. X. Liu and K. J. Lumb, Detection of Small-Molecule Aggregation with High-Throughput Microplate Biophysical Methods, Curr. Protoc. Chem. Biol., 2020, 12(1), e78 CrossRef CAS PubMed.
- X. Wang and D. Kireev, kireevlab/FRASE-bot-RDKit: In silico Fragment-based Discovery of CIB1-directed Anti-Tumor Agents by FRASE-bot, 2023, DOI:10.5281/zenodo.11372361.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc07532c |
| ‡ Lead contact. |
| This journal is © The Royal Society of Chemistry 2025 |

