 Open Access Article
Open Access ArticleRemoving redundancy of the NCN codons in vitro for maximal sense codon reassignment†
Clark A.
Jones
ab,
Chelsea A.
Makovsky
ab,
Aidan K.
Haney
a,
Alba C.
Dutra
ab,
Clinton A. L.
McFeely
ab,
T. Ashton
Cropp
 a and
Matthew C. T.
Hartman
a and
Matthew C. T.
Hartman
 *ab
*ab
aDepartment of Chemistry, Virginia Commonwealth University, Box 842006, 1001 W. Main St, Richmond, 23284-2006, VA, USA. E-mail: mchartman@vcu.edu
bMassey Comprehensive Cancer Center, Virginia Commonwealth University, Box 980037, 401 College St, Richmond, 23298-0037, VA, USA
First published on 22nd April 2025
Abstract
Expanding the genetic code affords exciting opportunities for synthetic biology, studies of protein function, and creation of diverse peptide libraries by mRNA display. Maximal expansion with the standard 64 codon code requires breaking the degeneracy of the 61 sense codons which encode for only 20 amino acids. In E. coli these 61 codons are decoded by 46 different tRNAs. Moreover, many codons are decoded by multiple tRNAs, further complicating efforts to break this redundancy. The overlapping decoding patterns of the 11 tRNAs in E. coli which read the 16 codons that encode serine, proline, threonine, and alanine codons exemplify this difficulty. Here we tackle this challenge by first outlining a general process to evaluate codons for their potential for reassignment. We then use this knowledge to assign these 16 codons to 10 different amino acids, more than doubling their encoding potential. Our work highlights the expanded potential of sense codon reassignment and points the way to a dramatically expanded code containing more than 30 monomers.
Introduction
Expansion of the genetic code is accomplished by increasing the number of amino acids (AAs) that can be encoded beyond the canonical 20.1,2 A popular technique to encode for additional amino acids is stop codon suppression, which adds additional amino acids to the genetic code via the transformation of stop codons into sense codons capable of encoding non-canonical amino acids (ncAAs).3 This technique has enabled the introduction of post-translational modifications and site-specific labels, leading to advances in our understanding of protein function.4,5 However, the potential of stop codon suppression is limited by the small number of stop codons.6 Sense codon reassignment (SCR), which instead reassigns codons from canonical to non-canonical amino acids, has a much higher theoretical potential for genetic code expansion.2,7–9 The genetic code contains innate redundancy, with three of the 20 canonical amino acids encoding for up to six codons each (leucine, serine, and arginine), and five amino acids account for four codons each (valine, proline, threonine, alanine, and glycine). If the degeneracy of these codon boxes can be reduced, a dramatically expanded genetic code could be created. Such a code would enable the development of designer organisms with novel functions, expression of proteins with elaborate modifications, and the discovery of shorter drug-like macrocyclic peptides from diverse libraries via mRNA display.10–13The redundancy of the genetic code is controlled by the aminoacyl-tRNA synthetases (AARS) and their specificity for tRNAs as well as the ribosome's ability to select the tRNAs that correspond to their cognate codons. To break this degeneracy first requires loading the tRNAs that read a given codon box with additional ncAAs. Mature technologies for charging ncAAs onto unique tRNAs have been developed in vitro and in vivo,14–23 leading to modest genetic code expansion.2,12,24,25 However, a remaining key roadblock for sense codon reassignment is the overlapping codon readings of tRNA isoacceptors.26–31 This overlap limits discrete reassignment because tRNAs bearing different AAs will compete for the same codon. Attempts to break the degeneracy of the 4-fold degenerate codon boxes epitomize the challenges of overlapping reading. To date, no one has been able to break the degeneracy of these codon boxes more than 2-fold.2
Most of the work in this field has been carried out under the assumption that codons read by more than one tRNA cannot be discretely reassigned. However, overlapping reading does not mean that each tRNA reads the codon with equal efficiency. To understand the competitive nature of this codon reading, we recently developed an isotopic competition assay where each tRNA isoacceptor decoding codons in a given codon box is charged with an isotopically labeled canonical amino acid of distinct mass. The decoding percentage of each tRNA at each codon is assessed by measuring at the isotopic distribution of the resulting peptides via mass spectrometry.32,33
Guided by this assay, we showed that by using in vitro transcribed tRNAs and hyperaccurate ribosomes we were able to split the six leucine codons to encode for five unique amino acids.32–34 It is not, however, clear that such a dramatic genetic code expansion will extend beyond the leucine codons. Here we focus on investigating the fourfold degenerate NCN codon boxes that encode for serine (UCN), proline (CCN), threonine (ACN), and alanine (GCN). Previous attempts at sense codon reassignment have led to the splitting of both the UCN and ACN codon boxes to encode two amino acids each.12 To our knowledge, no one has been able to break the degeneracy of the proline (CCN) or alanine (GCN) codons. By comparison, here we show that these 16 codons can encode for 10 unique amino acids, demonstrating that the genetic code as a whole is much more pliable than previously thought. Our work paves the way for the future creation of peptide libraries and organisms with a dramatically expanded genetic code.
Results and discussion
Re-coding the UCN serine codons
The rules governing the capabilities of a given tRNA to read a codon are well-understood.26,35 However, when considering sense codon reassignment, a more important metric is a competitive ranking of their abilities to read a given codon. For this purpose, we used an isotopic competition assay which gives the relative decoding efficiency of a given codon for all competing tRNAs.32,33 With an eye towards more expansive genetic code reassignment, we utilized hyperaccurate ribosomes in these experiments.34,36,37 These ribosomes have shown enhanced ability to distinguish near-cognate tRNAs and tRNAs within a codon box.4,33We aminoacylated each of the three serine tRNAs purported to read the UCN codons (Ser2CGA, Ser1UGA, and Ser5GGA) with different isotopically labeled serine analogs of unique mass (serine, serine-d3, and serine-d3-13C3–15N1, respectively). We mixed these charged tRNAs together, and used this mixture in separate in vitro translation assays with four different mRNAs, each containing one of the UCN codons (Fig. 1A and S1†). These translations lacked SerRS, but contained the other enzymes necessary to create the final peptides. The percentages of incorporation of each isotope were determined by comparison of the intensity of the peptides of varying isotopic mass (Fig. 1B). Since the AAs are isotopologues, they should ionize identically in MS and also be recognized equally by the translation machinery. The UCA, UCC, and UCG codons all showed >80% selectivity for a single tRNA. The UCU mRNA was read by two different tRNAs, Ser5GGA and Ser1UGA. These results were converted to the heat map shown in Fig. 1C.
 | ||
| Fig. 1 Breaking the degeneracy of the UCN serine codon box. (A) mRNA templates used for the in vitro translation testing the codon readthrough of each tRNA. Targeted codons highlighted in blue. mRNA 2 included a different histidine codon (CUC) which improved the yield compared to the CAU containing mRNA as predicted by Kim and Jung.38 (B) Representative mass spectra showing different isotopic incorporations on serine codons. Translations incubated at 37 °C for 30 min with 5 μM serine-Ser2CGA(L-serine), serine-d3-Ser1UGA and serine-d3-13C3–15N1-Ser5GGA. All MS data used for these experiments are shown in Fig. S1.† Calculated masses for the peptides are: Ser (Ser2CGA, 1728.76); d3-ser (Ser1UGA, 1731.78), and d3-13C3–15N1-Ser (Ser5GGA, 1735.80). (C) Heat map demonstrating the observed incorporation percentages for each tRNA with their respective mRNA codons. The isotopically labeled serines are shown above their respective tRNAs. * indicates isotopically labeled 13C and 15N. Error bars represent the mean of 3 experiments. (D) Representative MALDI spectra showing incorporation of ncAAs/AAs on their respective mRNA codons. Each tRNA was added at 15 μM concentration and the translations were carried out for 30 min. Additional replicates are shown in Fig. S5.† | ||
Using this heatmap as a guideline, we charged each tRNA with a unique amino acid using enzymatic charging or flexizyme (Fig. 1D, S2 and S3†). Ser1UGA was charged with O-methyl serine (Ser(Me)),11 Ser2CGA with serine, and Ser5CGA with allylglycine (AllylG), and these tRNAs were used in in vitro translations with each of the mRNAs. When each was used at 15 μM concentration, the UCA, UCC, and UCG codons were predominantly read by the expected tRNA, in line with our isotopic competition experiment. Also, as expected, the UCU mRNA gave a mixture of peaks, due to both Ser1UGA and Ser5GGA decoding with Ser5GGA as the major peak (Fig. S4†). Increasing the concentration of Ser5GGA from 15 μM to 30 μM significantly decreased the incorporation of the Ser1UGA tRNA on the UCU codon and enabled predominant reading of all four serine codons by a single tRNA species (Fig. 1D and S5†).
Re-coding the CCN proline codons
We then used the same protocol to investigate the proline codon box (Fig. 2). The three proline tRNAs purported to read the CCN codons (Pro1CGG, Pro2GGG, and Pro3UGG) were aminoacylated with proline, proline-d3, and proline-d7, respectively. | ||
| Fig. 2 Breaking the degeneracy of the CCN proline codon box. (A) mRNA templates used for the in vitro translation testing the codon readthrough of each tRNA. Targeted codons highlighted in red. mRNA 4 included a different histidine codon (CAC) and phenylalanine codon (UUC) which improved the yield compared to the CAC/UUC containing mRNA as predicted by Kim and Jung.38 (B) Heat map demonstrating the observed incorporation percentages for each tRNA with their respective mRNA codons. Error bars represent the mean of 3 experiments. All MS data used for this experiment are shown in Fig. S6.† (C) Representative MALDI spectra showing incorporation of ncAAs/AAs on their respective mRNA codons. Each tRNA was added at 15 μM concentration and the translations were carried out for 30 min. Additional replicates are shown in Fig. S7.† *An unknown +3 Da peak that results from a contaminant in the O-Me glutamic acid dinitrobenzyl ester amino acid flexizyme substrate. | ||
The CCA, CCC, and CCG codons all showed >78% selectivity for a single tRNA (Fig. 2B and S6†). The CCU mRNA was read by both Pro2GGG and Pro3UGG. The results were converted into a heatmap (Fig. 2B). We proceeded with sense codon reassignment using the same protocol previously mentioned. For this experiment we aminoacylated Pro1CGG with 1-aminocyclopropane-1-carboxylic acid (Acp),39 Pro2GGG with pipecolic acid (Pip),40 and Pro3UGG with glutamic acid γ-methyl ester (Glu(Me))25 (Fig. S3†).
Each of the 4 codons was predominantly read by a single tRNA (Fig. 2C and S7†), in line with our isotopic competition experiment. In contrast to our isotopic competition experiment, however, no co-reading of the CCU codon by Pro3UGG was observed, with the CCU codon being primarily read by Pro2GGG alone. In order to test the impact of the ncAA structure on codon reading we repeated this experiment with a different set of ncAAs (Fig. S6†). These experiments also showed excellent orthogonality, showing that the codon reading pattern is not dependent on the ncAA attached.
Re-coding the ACN threonine codons
We continued to use the same protocol to investigate the threonine codon box using mRNAs 9–12 (Fig. 3A). The three threonine tRNAs purported to read the ACN codons (Thr1GGU, Thr2CGU, and Thr4UGU) were aminoacylated with threonine, threonine-13C4–15N1, and threonine-d5-13C4–15N1, respectively. | ||
| Fig. 3 Breaking the degeneracy of the ACN threonine codon box. (A) mRNA templates used for the in vitro translation testing the codon readthrough of each tRNA. Targeted codons highlighted in green. (B) Heat map demonstrating the observed incorporation percentages for each tRNA with their respective mRNA codons. * indicates isotopically labeled 13C and 15N. Error bars represent the mean of 3 experiments. All MS data used for this experiment are shown in Fig. S9.† (C) Representative MALDI spectra showing incorporation of ncAAs/AAs on their respective mRNA codons. Each tRNA was added at 15 μM concentration and the translations were carried out for 30 min. Additional replicates are shown in Fig. S10.† *Observed misincorporation of methionine (expected: 1773.68, observed: 1774.08). | ||
The competitive codon-reading data (Fig. S9†) were converted into a heatmap (Fig. 3B). The ACA and ACG codons both showed >90% selectivity for a single tRNA. The ACC codon was read predominantly by Thr1GGU (82%) but was also read at lower percentages by Thr2CGU and Thr4UGU. The ACU codon was decoded by Thr1GGU (71%), as well as Thr2CGU (10%) and Thr4UGU (19%) (Fig. S9†).
We proceeded with sense codon reassignment. For this experiment we aminoacylated Thr1GGU with L-2-aminobutyric acid (Abu), Thr2CGU with cyclopentyl glycine (CPG) and Thr4UGU with propargyl glycine (PropG) (Fig. S3†). Each of the four threonine codons were predominantly read by a single tRNA. This selectivity of codons for their respective tRNAs was observed with non-canonical amino acids as well (Fig. 3D and S10†).
Re-coding the GCN alanine codons
The two alanine tRNAs (Ala1UGC and Ala2GGC) were aminoacylated with alanine and +8 alanine, respectively. The GCA and GCG codons showed >90% selectivity for Ala1UGC, and the GCC and GCU codons were read by both Ala1UGC and Ala2GGC (Fig. 4). These results were converted into a heatmap (Fig. 4). The dominant nature of the Ala1UGC tRNA in these experiments suggests that sense-codon reassignment of these codons will require some optimization and we decided not to pursue addressing this codon box with ncAAs. | ||
| Fig. 4 Attempted breaking of the degeneracy of the alanine codon box. Heat map demonstrating the observed incorporation percentages for both alanine tRNAs with their respective mRNA codons. * indicates isotopically labeled 13C and 15N. Error bars represent the mean of 3 experiments. All MS data used for this experiment are shown in Fig. S11.† | ||
Orthogonality with multiple incorporations
Our experiments above investigated single ncAA incorporation events. To be useful downstream for library generation via mRNA display it is important that multiple ncAAs can be incorporated into a single template with high fidelity. To test this, we designed 3 mRNAs, each containing three NC(A/G/C) codons, Fig. 5A–C and S12.† To each translation experiment, we added a mixture of the three tRNAs bearing ncAAs expected to read those codons using the same coding scheme as described in Fig. 1–3. In each case, the peak of the desired mass was produced with high fidelity, Fig. 5 and S12.† The triple-proline template (Fig. 5B) gave poorer signal-to-noise compared to the other two templates which is consistent with the lowered efficiency of translation with α,α-disubstituted amino acids17,34,41–43 and Acp in particular.44 | ||
| Fig. 5 Assessing translational fidelity with multiple incorporations of ncAAs in a single mRNA. (A) Serine codons: UCA, UCC, and UCG. (B) Proline codons: CCC, CCG and CCA. (C) Threonine codons: ACA, ACC, and ACG. Representative MALDI spectra are shown for each experiment. Additional replicates are shown in Fig. S12.† tRNA-ncAA pairings are identical to those in Fig. 1–3. The Ser and Thr AA-tRNAs were added at 15 μM concentration, and the Pro AA-tRNAs were added at 30 μM. The translations were carried out for 60 min at 37 °C. | ||
In this work, through the use of flexizyme-charged in vitro transcribed tRNAs in conjunction with hyperaccurate ribosomes, the UCN, CCN, and ACN (serine, proline, and threonine) codon boxes were reassigned to encode for three different ncAAs/AAs each, thus tripling the encoding potential for all three codon boxes (Fig. 6A). The tRNA readings for these three boxes share a common pattern (Fig. 6B). For the third codon position Watson Crick base pairs are preferred for G:C (codon:tRNA), A:U, and C:G. The codon ending in U is also paired with a G, forming a wobble pair.45,46 The clean orthogonality is in contrast to expectations with these codon boxes, which show a web of overlapping tRNA reading2,26,27 in particular from the tRNAs that have a U in the anticodon (Fig. 6 B). The improved orthogonality likely results from our use of hyperaccurate ribosomes.34
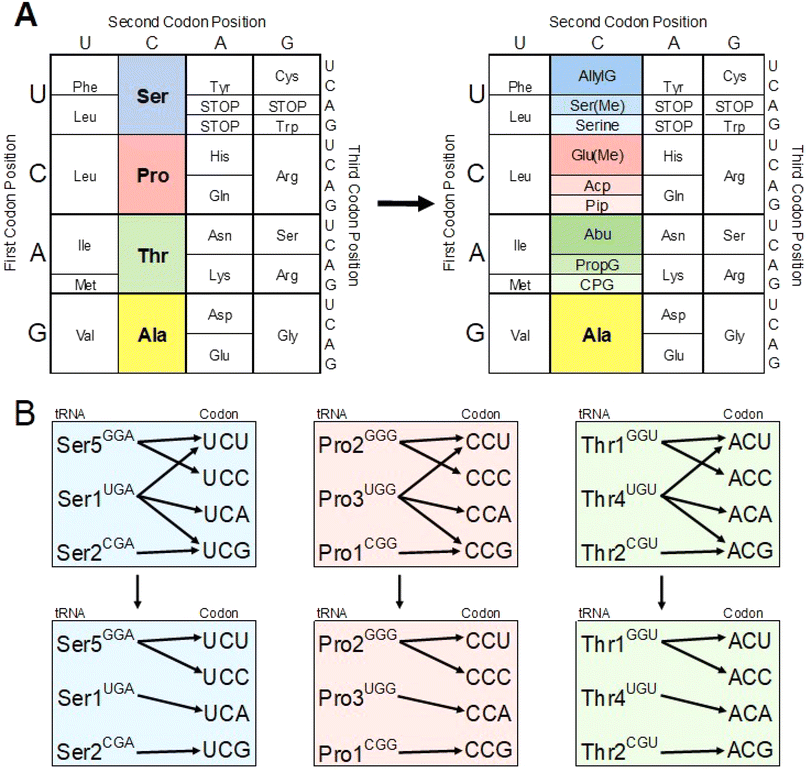 | ||
| Fig. 6 Re-encoding the NCN codons. (A) Codon chart demonstrating the reassignment of the UCN, CCN, and ACN codon boxes. On the left is the natural codon chart and on the right is our maximized codon chart. (B) Change in tRNA isoacceptor reading. Top is the expected codon reading pattern26 and bottom is our maximized codon readthrough pattern. | ||
In the case of the alanine and proline codons, the presence of two G:C pairs at the first and second codon positions makes these “strong” codons as defined by Grosjean and Westhof.47 It is interesting that it is easier to divide the proline (CCN) codon box (Fig. 2B) than the alanine (GCN) codon box (Fig. 4). The alanine GCN codon box is unique among the NCN codon boxes in E. coli because it is decoded by 2, rather than 3 tRNAs, although other bacteria do have 3 tRNAs that read these codons.48E. coli also has only two tRNAs that read the valine GUN codon box, and it was also difficult to reassign this codon box to encode for three amino acids.33 Conversely, E. coli uses 5 tRNAs to read the 6 leucine codons, and it was possible to extensively break the degeneracy of these codons.33 Taken together, these data suggest that the number of tRNAs used by an organism to decode a specific codon box may be a useful feature to predict the ability to break the degeneracy of those codons.
Presumably, the rules for breaking of the NCN sense codons should also be applicable in living E. coli that are engineered to have hyperaccurate ribosomes. To do so will first require recoding the genome of E. coli to eliminate the extra codons and the native tRNAs that read them. Recent progress in genome synthesis49 should enable such a strategy. Indeed, the first examples of sense codon liberation have been described.50,51 These codons can then be re-encoded using an orthogonal synthetase/tRNA pair.51 There are a growing number of such pairs under development.52,53
Although there are many postulates for how the genetic code evolved into its current form, there are strong reasons to believe that alanine and proline were among the original entrants into the code (along with glycine and a positively charged amino acid).54,55 Our isotopic competition assay and in vitro translation system could be useful to researchers testing the potential basis for such a minimal code.
In addition to establishing codon orthogonality, this translation system successfully encoded for multiple ncAA monomers with unique functional groups not present in the canonical amino acids (Fig. S2†). Acp44 is an α,α-disubstituted amino acid that exerts unique conformational constraints when incorporated into a peptide.56 PropG, AllylG, and Ser(Me) introduce alkynes, alkenes, and ethers into the genetic code. PropG's alkyne in particular can permit the introduction of click-chemistry reactions into peptides. Similarly, Glu(Me) provides a side chain ester that could be amenable to a variety of chemical transformations.57 Additionally, to our knowledge, both AllylG and CPG have never been used in translation previously.
Conclusion
Taken together, our work demonstrates the capability of re-encoding the 16 NCN codons to encode for 10 different amino acids, a dramatic expansion from the four canonical amino acids. When combined with other codons, it is likely that a genetic code responding to upwards of 35 monomers is increasingly likely.Data availability
All data for the manuscript are found in the ESI.†Author contributions
Conceptualization: M. C. T. H., C. A. L. M.; methodology: M. C. T. H., C. A. J.; investigation: C. A. J., C. A. M., A. K. H., A. G. D., M. C. T. H., T. A. C.; writing – original draft: C. A. J., M. C. T. H.; writing – review and editing: M. C. T. H., C. A. J., T. A. C.; visualization: C. A. J.; supervision: M. C. T. H.; T. A. C.; project administration: M. C. T. H.; funding acquisition: M. C. T. H., T. A. C.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors would like to acknowledge the support of the NIGMS (R01GM143396) to M. C. T. H. and T. A. C. The graphical abstract was created in BioRender. Hartman, M., 2024, http://BioRender.com/x55m334.References
- M. A. Shandell, Z. Tan and V. W. Cornish, Biochemistry, 2021, 3455–3469 CrossRef CAS PubMed.
- C. A. Jones and M. C. T. Hartman, Isr. J. Chem., 2024, e202400026 CrossRef.
- J. W. Chin, T. A. Cropp, J. C. Anderson, M. Mukherji, Z. Zhang and P. G. Schultz, Science, 2003, 301, 964–967 CrossRef CAS PubMed.
- D. P. Nguyen, M. Mahesh, S. J. Elsässer, S. M. Hancock, C. Uttamapinant and J. W. Chin, J. Am. Chem. Soc., 2014, 136, 2240–2243 CrossRef CAS PubMed.
- E. A. Lemke, D. Summerer, B. H. Geierstanger, S. M. Brittain and P. Schultz, Nat. Chem. Biol., 2007, 3, 769–772 CrossRef CAS PubMed.
- A. O. Osgood, Y. Zheng, S. J. S. Roy, N. Biris, M. Hussain, C. Loynd, D. Jewel, J. S. Italia and A. Chatterjee, Angew. Chem., Int. Ed., 2023, 62, e202219269 CrossRef CAS PubMed.
- A. J. Link and D. A. Tirrell, Methods, 2005, 36, 291–298 CrossRef CAS PubMed.
- W. Biddle, D. G. Schwark, M. A. Schmitt and J. D. Fisk, Front. Chem., 2022, 10, 815788 CrossRef CAS PubMed.
- S. L. Richardson, K. K. Dods, N. A. Abrigo, E. S. Iqbal and M. C. Hartman, Curr. Opin. Chem. Biol., 2018, 46, 172–179 CrossRef CAS PubMed.
- S. E. Iskandar, J. M. Pelton, E. T. Wick, D. L. Bolhuis, A. S. Baldwin, M. J. Emanuele, N. G. Brown and A. A. Bowers, J. Am. Chem. Soc., 2023, 145, 1512–1517 CrossRef CAS PubMed.
- T. Passioura, W. Liu, D. Dunkelmann, T. Higuchi and H. Suga, J. Am. Chem. Soc., 2018, 140, 11551–11555 CrossRef CAS PubMed.
- C. Lin, M. J. Harner, A. E. Douglas, V. Lafont, F. Yu, V. G. Lee, M. A. Poss, J. F. Swain, M. Wright and D. Lipovšek, Angew. Chem., Int. Ed., 2021, 133, 22640–22645 CrossRef PubMed.
- L. Davis and J. W. Chin, Nat. Rev. Mol. Cell Biol., 2012, 13, 168–182 CrossRef CAS PubMed.
- W. Biddle, M. A. Schmitt and J. D. Fisk, Biochemistry, 2015, 54, 7355–7364 CrossRef CAS PubMed.
- X. Gong, H. Zhang, Y. Shen and X. Fu, J. Bacteriol., 2023, 205, e003855–22 CrossRef PubMed.
- Y. Goto, T. Katoh and H. Suga, Nat. Protoc., 2011, 6, 779–790 CrossRef CAS PubMed.
- M. C. T. Hartman, ChemBioChem, 2022, 23, e202100299 CrossRef CAS PubMed.
- M. C. T. Hartman, K. Josephson and J. W. Szostak, Proc. Natl. Acad. Sci. U.S.A., 2006, 4356–4361 CrossRef CAS PubMed.
- C. Merryman and R. Green, Chem. Biol., 2004, 11, 575–582 CrossRef CAS PubMed.
- T. G. Heckler, L. H. Chang, Y. Zama, T. Naka, M. S. Chorghade and S. M. Hecht, Biochemistry, 1984, 23, 1468–1473 CrossRef CAS PubMed.
- S. A. Robertson, J. A. Ellman and P. G. Schultz, J. Am. Chem. Soc., 1991, 113, 2722–2729 CrossRef CAS.
- N. Budisa, Angew. Chem., Int. Ed., 2004, 43, 6426–6463 CrossRef CAS PubMed.
- J. Andrews, Q. Gan and C. Fan, Protein Sci., 2023, 32, e4559 CrossRef CAS PubMed.
- Y. Iwane, A. Hitomi, H. Murakami, T. Katoh, Y. Goto and H. Suga, Nat. Chem., 2016, 8, 317–325 CrossRef CAS PubMed.
- T. Passioura, W. Liu, D. Dunkelmann, T. Higuchi and H. Suga, J. Am. Chem. Soc., 2018, 140, 11551–11555 CrossRef CAS PubMed.
- H. Dong, L. Nilsson and C. G. Kurland, J. Mol. Biol., 1996, 260, 649–663 CrossRef CAS PubMed.
- Z. Cui, V. Stein, Z. Tnimov, S. Mureev and K. Alexandrov, J. Am. Chem. Soc., 2015, 137, 4404–4413 CrossRef CAS PubMed.
- K. Hibi, K. Amikura, N. Sugiura, K. Masuda, S. Ohno, T. Yokogawa, T. Ueda and Y. Shimizu, Commun. Biol., 2020, 3, 350 CrossRef CAS PubMed.
- K. Takai, H. Takaku and S. Yokoyama, Nucleic Acids Res., 1996, 24, 2894–2899 CrossRef CAS PubMed.
- D. G. Schwark, M. A. Schmitt, W. Biddle and J. D. Fisk, ChemBioChem, 2020, 21, 2274–2286 CrossRef CAS PubMed.
- I. Kwon, K. Kirshenbaum and D. A. Tirrell, J. Am. Chem. Soc., 2003, 125, 7512–7513 CrossRef CAS PubMed.
- C. A. L. Mcfeely, K. K. Dods, S. S. Patel and M. C. T. Hartman, Nucleic Acids Res., 2022, 50, 11374–11386 CrossRef CAS PubMed.
- C. A. L. Mcfeely, B. Shakya, C. A. Makovsky, A. K. Haney, T. A. Cropp and M. C. Hartman, Nat. Commun., 2023, 14, 5008 CrossRef CAS PubMed.
- B. Shakya, O. G. Joyner and M. C. T. Hartman, ACS Synth. Biol., 2022, 11, 2193–2201 CrossRef CAS PubMed.
- K. Takai, H. Takaku and S. Yokoyama, Biochem. Biophys. Res. Commun., 1999, 257, 662–667 CrossRef CAS PubMed.
- H. S. Zaher and R. Green, Mol. Cell, 2010, 39, 110–120 CrossRef CAS PubMed.
- E. B. Kramer and P. J. Farabaugh, RNA, 2007, 13, 87–96 CrossRef CAS PubMed.
- S. W. Seo, J. S. Yang, I. Kim, J. Yang, B. E. Min, S. Kim and G. Y. Jung, Metab. Eng., 2013, 15, 67–74 CrossRef CAS PubMed.
- E. Adaligil, A. Song, K. K. Hallenbeck, C. N. Cunningham and W. J. Fairbrother, ACS Chem. Biol., 2021, 16, 1011–1018 CrossRef CAS PubMed.
- X. D. Zhang, Y. S. Wang, H. Xiang, L. W. Bai, P. Cheng, K. Li, R. Huang, X. Wang and X. Lei, Chem. Commun., 2024, 60, 1607–1610 RSC.
- E. S. Iqbal, K. K. Dods and M. C. Hartman, Org. Biomol. Chem., 2018, 16, 1073–1078 RSC.
- M. C. Hartman, K. Josephson, C.-W. Lin and J. W. Szostak, PLoS One, 2007, 2, e972 CrossRef PubMed.
- T. Katoh, Y. Iwane and H. Suga, Nucleic Acids Res., 2017, 45, 12601–12610 CrossRef CAS PubMed.
- A. I. Chan, M. S. Sawant, D. J. Burdick, J. Tom, A. Song and C. N. Cunningham, ACS Chem. Biol., 2023, 18, 81–90 CrossRef CAS PubMed.
- S. J. Näsvall, P. Chen and G. R. Björk, RNA, 2007, 13, 2151–2164 CrossRef PubMed.
- P. F. Agris, Nucleic Acids Res., 2004, 32, 223–238 CrossRef CAS PubMed.
- H. Grosjean and E. Westhof, Nucleic Acids Res., 2016, 44, 8020–8040 CrossRef CAS PubMed.
- M. E. Saks and J. S. Conery, RNA, 2007, 13, 651–660 CrossRef CAS PubMed.
- J. F. Zürcher, A. A. Kleefeldt, L. F. Funke, J. Birnbaum, J. Fredens, S. Grazioli, K. C. Liu, M. Spinck, G. Petris and P. Murat, Nature, 2023, 619, 555–562 CrossRef PubMed.
- N. Bohlke and N. Budisa, FEMS Microbiol. Lett., 2014, 351, 133–144 CrossRef CAS PubMed.
- W. E. Robertson, L. F. Funke, D. de la Torre, J. Fredens, T. S. Elliott, M. Spinck, Y. Christova, D. Cervettini, F. L. Böge and K. C. Liu, Science, 2021, 372, 1057–1062 CrossRef CAS PubMed.
- A. Costello, A. A. Peterson, D. L. Lanster, Z. Li, G. D. Carver and A. H. Badran, Nat. Biotechnol., 2024, 1–12 Search PubMed.
- D. Cervettini, S. Tang, S. D. Fried, J. C. Willis, L. F. Funke, L. J. Colwell and J. W. Chin, Nat. Biotechnol., 2020, 38, 989–999 CrossRef CAS PubMed.
- V. Kubyshkin and N. Budisa, Int. J. Mol. Sci., 2019, 20, 5507 CrossRef CAS PubMed.
- H. Hartman and T. F. Smith, Life, 2014, 4, 227–249 CrossRef PubMed.
- C. Cativiela and M. D. Díaz-De-Villegas, Tetrahedron:Asymmetry, 2000, 11, 645–732 CrossRef CAS.
- C. G. Pappas, N. Wijerathne, J. K. Sahoo, A. Jain, D. Kroiss, I. R. Sasselli, A. S. Pina, A. Lampel and R. V. Ulijn, ChemSystemsChem, 2020, 2, e2000013 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary figures (S1–S12) and supplementary tables (S1 and S2) as well as synthetic details and characterization. See DOI: https://doi.org/10.1039/d4sc06740a |
| This journal is © The Royal Society of Chemistry 2025 |
