Open Access Article
Open Access ArticleNew coumarin–chalcone–triazole hybrids as promising anti-diabetic agents: from molecular design to in vivo validation
Fariba Peytamab,
Maryam Norouzbaharic,
Mahsa Akbarib,
Hayrettin Ozan Gülcand,
Mahfam Moradia,
Somayeh Mojtabavi e,
Mohammad Ali Faramarzi
e,
Mohammad Ali Faramarzi e,
Fahimeh Ghasemif,
Mohammadreza Torabif,
Maliheh Barazandeh Tehrania,
Vahid Sheibanig,
Loghman Firoozpour*ab and
Alireza Foroumadi
e,
Fahimeh Ghasemif,
Mohammadreza Torabif,
Maliheh Barazandeh Tehrania,
Vahid Sheibanig,
Loghman Firoozpour*ab and
Alireza Foroumadi *ab
*ab
aDepartment of Medicinal Chemistry, Faculty of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran
bDrug Design and Development Research Center, The Institute of Pharmaceutical Sciences (TIPS), Tehran University of Medical Sciences, Tehran, Iran
cFaculty of Pharmacy, Final International University, Kyrenia via Mersin 10 Turkey, TRNC, Catalkoy, Turkey
dEastern Mediterranean University, Faculty of Pharmacy, TRNC, via Mersin 10 Turkey, Famagusta, Cyprus
eDepartment of Pharmaceutical Biotechnology, Faculty of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran
fDepartment of Bioinformatics and Systems Biology, School of Advanced Technologies in Medicine, Isfahan University of Medical Sciences, Isfahan, Iran
gNeuroscience Research Center, Institute of Neuropharmacology, Kerman University of Medical Sciences, Kerman, Iran
First published on 19th December 2025
Abstract
A series of coumarin–chalcone-1,2,3-triazoles were designed and synthesized as potential antidiabetic agents targeting α-glucosidase. Among them, compound 20q exhibited exceptional inhibitory potency (IC50 = 0.50 ± 0.04 µM), significantly outperforming acarbose (IC50 = 750.08 ± 1.52 µM). Kinetic analyses confirmed a competitive inhibition mechanism, and computational studies—including deep-learning prediction, molecular docking, and molecular dynamics simulations—revealed strong and stable interactions of 20q with the enzyme active site, supporting its efficacy. This compound showed no cytotoxicity and α-amylase inhibition even at high concentrations, indicating its favorable safety profile with high selectivity. CD and fluorescence studies demonstrated that its binding induced a more ordered enzyme conformation (increased α-helix, reduced β-sheet/coil) through static, electrostatic interactions. In vivo assessments with compound 20q showed no acute toxicity at doses up to 1000 mg kg−1 and a dose-dependent antihyperglycemic effect, restoring fasting blood glucose and HbA1c levels to near-normal values, and improving liver and pancreas histopathology at 8 mg kg−1 BW, outperforming acarbose at a comparable dose. These comprehensive findings identify compound 20q as a highly potent, selective, and safe α-glucosidase inhibitor with significant potential for further development as an antidiabetic agent.
1 Introduction
Diabetes mellitus represents a significant global health challenge, marked by the body's inability to regulate blood glucose levels effectively.1 As of 2021, it affected over half a billion individuals globally, with projections suggesting a rise to 783 million by 2045 which equates to 1 in 8 adults living with the condition.2 The abnormal elevation of the blood glucose levels, contributes to a range of serious complications, including cardiovascular diseases, hypertension, nephropathy, and neuropathy, imposing a considerable economic burden on healthcare systems worldwide.3Type 2 diabetes mellitus (T2DM) is the most prevalent form of diabetes, accounting for over 90% of all cases.4 It is characterized by insulin resistance and relative insulin deficiency. In contrast to Type 1 diabetes mellitus, which primarily results from autoimmune destruction of insulin-producing cells and typically manifests in childhood,5 T2DM commonly develops in individuals above 65 years of age and is strongly influenced by lifestyle factors such as a decline in physical activity, coupled with a rising prevalence of obesity.4 The highest rates of T2DM are observed in the Middle East and North Africa, reflecting the influence of regional lifestyle and genetic predispositions on the disease's distribution.6
Current therapeutic approaches for T2DM focus on controlling hyperglycemia, with α-glucosidase inhibitors playing a significant role.7 This enzyme, located on the intestinal brush-border membrane, catalyzes the hydrolysis of dietary oligosaccharides; thus, its inhibition delays carbohydrate digestion and absorption, effectively reducing postprandial glucose elevations. Acarbose, voglibose, and miglitol are the primary α-glucosidase inhibitors used in clinical practice, with acarbose being the most extensively researched and utilized. However, their complex syntheses and gastrointestinal side effects limit patient adherence and overall therapeutic efficacy.8 Consequently, the identification and development of novel α-glucosidase inhibitors with enhanced potency and selectivity have emerged as a critical focus in medicinal chemistry and drug discovery efforts.9–22
Hybrid compounds have emerged as valuable platforms for drug discovery due to their numerous advantages, including high efficacy, low side effects, targeted receptor interactions, and favorable pharmacokinetics.23,24 Therefore, hybridization strategies that combine two or more pharmacophores are widely employed. To this aim, numerous coumarins and chalcones have been hybridized to enhance their biological potencies. These pharmacophores are connected either directly or via a linker, such as 1,2,3-triazole. Notably, this triazole ring possesses unique structural abilities to form pivotal protein–ligand interactions, including hydrogen bonding and pi-based interactions, making it a crucial structural motif in the design of various therapeutic agents.25
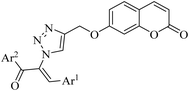
Numerous coumarin–chalcone hybrids have already been developed, demonstrating diverse biological activities, such as antioxidant,26,27 antimicrobial,28 anticancer,29 , antimalarial,30,31 neurodegenerative32 and anti-inflammatory33 effects. While individual coumarins17,34–41 and chalcones42–47 have been extensively explored for their α-glucosidase inhibitory potencies, making them attractive candidates for drug development in T2DM management (some of them are shown in Fig. 1A–F), studies investigating the α-glucosidase inhibitory potential of coumarin–chalcone hybrids remain limited (Fig. 1G).48 To address this gap, and as part of our ongoing efforts to identify potent α-glucosidase inhibitors,49–54 we introduce a novel hybrid scaffold integrating coumarin, chalcone, and 1,2,3-triazole. This innovative hybridizing approach was designed to utilize the synergistic effects of these pharmacophores, aiming to synthesize compounds with enhanced inhibitory efficacy against α-glucosidase.
 | ||
| Fig. 1 Design strategy for the synthesis of novel coumarin–chalcone hybrids as potential α-glucosidase inhibitors. | ||
2 Results and discussion
2.1 Chemistry
As outlined in Scheme 1, an efficient and straightforward synthetic strategy was employed to synthesize novel conjugates of coumarin–chalcone-1,2,3-triazole. This protocol was initiated by the preparation of α-azidochalcones 7, which have been utilized as valuable starting materials for synthesizing various nitrogen-containing skeletons over the past decade.55–65 The synthesis of this adduct involved three steps: first, a condensation reaction between arylaldehyde 1 and 1-(aryl)ethan-1-one 2 in basic conditions produced the corresponding chalcone 3. This intermediate then went through the bromination of olefin (compound 5), followed by the nucleophilic substitution using azide, resulting in the desired substituted α-azidochalcones 7. In parallel, several hydroxy-substituted coumarins (compounds 8–11) were activated with a base for the nucleophilic substitution with propargyl bromide 12 to produce compounds 13–16.Finally, a copper-catalyzed azide–alkyne cycloaddition (CuAAC) “click” reaction between the azide-functionalized chalcone 7 and the alkyne-bearing coumarins 13–16 was performed, affording the coumarin–chalcone-1,2,3-triazole hybrids 17–20. Furthermore, to investigate the generality of this strategy, various derivatives of α-azidochalcones 7 (unsubstituted phenyl, those with electron-donating groups, chloro- or bromo-substituted phenyl rings, and heteroaryl) were employed under the optimized conditions. This approach successfully generated a large library of targeted compounds 20a–20x in 65–92% yields.
The structures of all isolated products were completely deduced on the basis of their IR, 1H and 13C NMR spectroscopy, high-resolution mass spectrometry (HRMS), and elemental analysis. Partial assignments of these resonances are given in the Experimental Part.
For instance, the 1H NMR spectrum of the representative compound 20e (see SI, page S34) displayed key resonances, such as a sharp singlet at δ 5.40 ppm, corresponding to the methylene bridge (H-10). The coumarin core protons H-2 and H-3 appeared as distinct doublets at δ 6.30 ppm and 8.00 ppm. The other aromatic protons of the coumarin ring, H-5, H-6, and H-8, were observed as a doublet at δ 7.65 ppm, a doublet of doublets at δ 7.04 ppm, and a doublet at δ 7.15 ppm, respectively. The triazole ring proton (H-12) resonated as a sharp singlet at δ 7.83 ppm. In the chalcone moiety, the olefinic proton (H-17) gave a singlet at δ 8.52 ppm. The phenyl ring protons (H-13, H-14, and H-15) were observed as a triplet (at δ 7.36 ppm), triplet (at δ 7.21 ppm), and doublet (at δ 6.85 ppm), respectively, while the 4-bromophenyl ring protons (H-21 and H-22) appeared as a multiplet in the range of δ 7.72 to 7.79 ppm. The 13C NMR spectrum of 20e showed two significant signals at 61.86 ppm for carbon 10, and 190.32 ppm for carbon 19, along with other 21 distinct peaks, all in full agreement with the proposed structure.
The geometry of the olefinic double bond in the chalcone moiety was unequivocally assigned for a representative compound 20e using 2D NMR spectroscopy. As illustrated in the SI (pages S17 and S18), the NOESY spectrum revealed critical through-space correlations in addition to the through-bond connectivity. The key NOE interactions for compound 20e are summarized as follows:
A strong NOE cross-peak was observed between the triazole proton (H-12, δ 7.85 ppm) and the ortho-protons of the phenyl ring (H-15, δ 6.85 ppm). This through-space interaction is only feasible if these protons are in close spatial proximity, which definitively confirms the Z configuration. This assignment is further supported by the absence of any NOE correlation between the olefinic proton (H-17, δ 8.54 ppm) and the triazole proton (H-12, δ 7.85 ppm). The same Z-configuration is expected for the entire series, given the analogous synthetic pathway used. Additional NOE correlations were the correlations between the phenyl ring protons H-15 (δ 6.86 ppm) and the olefinic proton H-17 (δ 8.54 ppm), as well as the correlations between the methylene bridge (H-10) and the coumarin protons H-6 (δ 7.05 ppm) and H-8 (δ 7.17 ppm), indicating spatial proximity suggesting conformational flexibility around the –O–CH2– (C-10) linker.
2.2 α-Glucosidase inhibitory activity
In the present study, novel coumarin–chalcone–triazole hybrids 17, 18, 19, and 20a–20x were synthesized to assess their in vitro inhibitory activities against Saccharomyces cerevisiae α-glucosidase and compare their results with acarbose as the standard drug. To present a comprehensive structure–activity relationship (SAR) analysis, the binding position of the chalcone–triazole conjugated to coumarin backbone (as summarized in Table 1) as well as the presence of various substituents on the aromatic rings of chalcone moiety (as summarized in Table 2) were investigated.| Compound | Ar1 | Ar2 | IC50a (µM) |
|---|---|---|---|
| a Values are expressed as mean ± SD. All experiments were performed at least three times. | |||
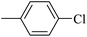
| 20a | 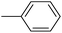 |
 |
3.60 ± 0.08 |
| 20b | 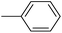 |
 |
11.48 ± 0.54 |
| 20c | 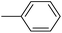 |
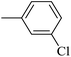 |
26.73 ± 0.23 |
| 20d | 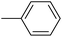 |
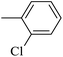 |
44.74 ± 1.12 |
| 20e | 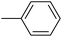 |
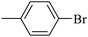 |
38.63 ± 0.55 |
| 20f | 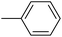 |
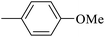 |
1.34 ± 0.08 |
| 20g | 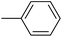 |
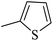 |
90.82 ± 0.44 |
| 20h | 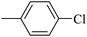 |
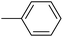 |
84.33 ± 0.44 |
| 20i | 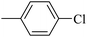 |
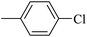 |
136.60 ± 1.46 |
| 20j | 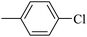 |
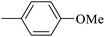 |
110.89 ± 2.28 |
| 20k | 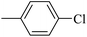 |
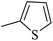 |
285.52 ± 0.73 |
| 20l | 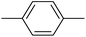 |
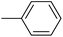 |
1.07 ± 0.29 |
| 20m | 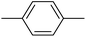 |
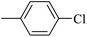 |
12.23 ± 0.67 |
| 20n | 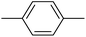 |
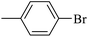 |
42.48 ± 0.32 |
| 20o | 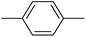 |
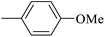 |
4.84 ± 0.16 |
| 20p | 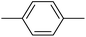 |
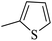 |
46.41 ± 1.23 |
| 20q | 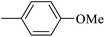 |
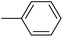 |
0.50 ± 0.04 |
| 20r | 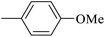 |
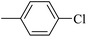 |
3.35 ± 0.14 |
| 20s | 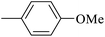 |
 |
1.50 ± 0.08 |
| 20t | 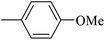 |
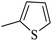 |
21.70 ± 0.19 |
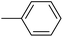
| 20u | 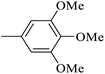 |
 |
14.63 ± 0.21 |
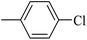
| 20v | 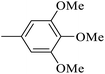 |
 |
39.84 ± 0.16 |
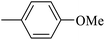
| 20w | 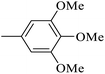 |
 |
26.92 ± 0.12 |
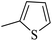
| 20x | 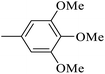 |
 |
183.69 ± 0.08 |
| Acarbose | — | — | 750.08 ± 1.52 |
Initially, various hydroxycoumarin derivatives 8–11 underwent the SN2 reaction with propargyl bromide 13, followed by click reaction with 2-azido-1,3-diphenylprop-2-en-1-one 7a, to obtain compounds 17, 18, 19, and 20a. Our aim was to maintain unsubstituted phenyl rings on the chalcone moiety and thereby identify the optimal position on the coumarin backbone. As presented in Table 1, the most favorable position was C-7, as compound 20a exhibited the best α-glucosidase inhibitory activity with IC50 value of 3.60 ± 0.08 µM. Molecular docking validated the relevance of this position, and we next examined how substituents on the chalcone phenyl rings affected activity.
As presented in Table 2, all compounds 20a–20x demonstrated significant inhibitory activities with IC50 values, ranging from 0.50 ± 0.04 µM to 285.52 ± 0.73 µM. All compounds were substantially more potent than the reference drug acarbose (IC50 = 750.08 ± 1.52 µM). To describe the SAR analysis, compounds were subdivided into five groups based on the nature and position of substituents on the Ar1 of chalcone moiety: (1) unsubstituted derivatives (20a–20g); (2) 4-chlorophenyl derivatives (20h–20k); (3) 4-methyl derivatives (20l–20p); (4) 4-methoxyphenyl derivatives (20q–20t); (5) 3,4,5-trimethoxyphenyl derivatives (20u–20x).
Across the first category, compound 20a without any substituent on both Ar1 and Ar2 rings exhibited significant inhibitory potency (IC50 = 3.60 ± 0.08 µM). Introducing an electron-withdrawing group like chlorine and bromine on the different position of Ar2 (compounds 20b–20e) led to detrimental effects on the activities. While the presence of methoxy at C-4 position of this phenyl ring (compound 20f) improved the inhibitory potency against α-glucosidase (IC50 = 1.34 ± 0.08 µM). Furthermore, the replacement of Ar2 phenyl ring with thiophene (compound 20g) caused a significant decrease in activity (IC50 = 90.82 ± 0.44 µM).
In the second group, a chlorine substituent at C-4 position of Ar1 generally caused detrimental effects on inhibitory activity. For example, compound 20h (IC50 = 84.33 ± 0.44 µM) exhibits significantly lower activity, being several times less potent than compound 20a. The electron-withdrawing nature of chlorine might disrupt optimal binding interactions, reducing the compounds' effectiveness. Compound 20i (IC50 = 136.60 ± 1.46 µM) with a 4-Cl substituent on Ar2 showed even more reduced activity, indicating that multiple electron-withdrawing groups can synergistically diminish inhibitory potency. The presence of a chlorine group at C-4 position of Ar1 was so detrimental that even adding a methoxy substituent failed to restore activity (as seen in compound 20j).
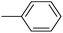
In the third group, the introduction of a methyl group on Ar1 reduced the inhibitory activity, though this trend was not absolute. For instance, compound 20l (IC50 = 1.07 ± 0.29 µM) was more potent than its unsubstituted analogue 20a (IC50 = 3.60 ± 0.08 µM). However, the overall inhibitory potency also depended significantly on the nature and position of substituents on Ar2 ring. Compound 20o (IC50 = 4.84 ± 0.16 µM) possessed notable inhibitory activity, which can be attributed to the combination effects of hydrophobic methyl and electron-donating methoxy groups. Similar to the observed trend across the first category, compounds 20m, 20n, and 20p demonstrated lower activity, confirming the adverse role of chlorine, bromine, and thiophene ring on Ar2 ring.
The fourth group included the most potent inhibitors in the present study, highlighting the significant impact of methoxy substituents on the overall activity. For instance, compound 20q (with an IC50 value of 0.50 ± 0.04 µM) emerged as the most potent α-glucosidase inhibitor, with a potency nearly 1500-fold greater than that of acarbose (IC50 = 750.08 ± 1.52 µM). The electron-donating character and hydrogen-bond-accepting capacity of the methoxy groups are proposed to facilitate stronger, more favorable interactions within the enzyme's active site. Compound 20s (IC50 = 1.50 ± 0.08 µM) further demonstrated that methoxy groups on both Ar1 and Ar2 positions could enhance inhibitory activity.
Given the remarkable role of methoxy in inhibiting α-glucosidase activity, compounds 20u–20x bearing additional groups were synthesized and evaluated. Although they generally showed great inhibitory potencies, their activities were weaker than those of their analogues in the fourth series (compounds 20q and 20s).
In summary, our SAR analysis revealed that incorporating chalcone–triazole moiety at C-7 position of coumarin as well as methoxy group on at C-4 position of phenyl ring of Ar1 significantly enhanced the α-glucosidase inhibitory activity. Substituent variations on the Ar2 ring also played a critical role, with unsubstituted phenyl and methoxy-containing derivatives exhibiting the highest potency. However, introducing EWGs (like chlorine and bromine) at different positions of this phenyl ring as well using thiophene as a heteroaryl group on Ar2 caused a detrimental effect on α-glucosidase inhibitory potency. Fig. 2 presents an overall SAR trend overview.
Based on the enzymatic results, compound 20q emerged as the most potent inhibitor, possessing the IC50 value of 0.50 ± 0.04 µM, which was almost 1500 times greater than acarbose. Accordingly, this compound was selected for a promising candidate for further evaluation.
2.3 α-Glucosidase kinetic studies
To identify the inhibition mode of coumarin–chalcone–triazole hybrids against α-glucosidase, compound 20q was investigated in a kinetic study. Initial reaction rates were measured at varying concentrations of the substrate, p-nitrophenyl α-D-glucopyranoside (1–16 mM), and the inhibitor (0, 0.125, 0.25, and 0.5 µM). Lineweaver–Burk analysis revealed an increasing apparent Km with no significant change in Vmax upon addition of 20q, characteristic of a competitive inhibition mechanism (Fig. 3A). This indicates this compound competes with the substrate for the enzyme's active site. The binding constant (Ki) was calculated from a secondary plot of Km versus inhibitor concentration (Fig. 3B), yielding a value of 0.5 µM. This low Ki value confirms the potent binding affinity of hybrid 20q for α-glucosidase.2.4 α-Amylase inhibitory activity
α-Amylase is another key enzyme involved in carbohydrate digestion, which, in contrast to α-glucosidase, is responsible for the breakdown of starches into smaller polysaccharides in the saliva and pancreas. Selective inhibition of α-glucosidase without affecting α-amylase can be advantageous in minimizing adverse effects. Inhibition of α-amylase is associated with gastrointestinal discomforts, such as bloating, flatulence, and diarrhea, due to excessive fermentation of undigested starches by gut microbiota. Therefore, compounds that selectively inhibit α-glucosidase while sparing α-amylase are preferred for achieving glycemic control with reduced gastrointestinal side effects.66,67 In this context, the α-amylase inhibitory activity of compound 20q was evaluated and found to be negligible, with no activity observed even at concentration up to 100 µM. This highlights its potential with high selectivity and reduced risk of gastrointestinal side effects—a significant limitation of current α-glucosidase inhibitors.2.5 Circular dichroism (CD) spectroscopy assessment
Circular dichroism (CD) spectroscopy was employed to probe conformational changes in α-glucosidase upon binding to inhibitor 20q. Far-UV CD spectra (190–240 nm) revealed a significant alteration in the enzyme's secondary structure (Table 3). Under native conditions, the enzyme was predominantly disordered, composed of 60% random coil and 32.4% β-sheet, with no detectable α-helix.| Entry | α-Helix | β-Sheet | β-Turn | Random coils |
|---|---|---|---|---|
| a All numbers are expressed as percentages; conditions 1 was recorded with native α-glucosidase, and conditions 2 was measured using α-glucosidase in the presence of compound 20q. | ||||
| 1 | 0 | 32.4 | 7.6 | 60 |
| 2 | 38.5 | 0 | 49.4 | 12 |
Binding with compound 20q induced a major structural reorganization:
α-Helix content increased from 0% to 38.5%, indicating a transition toward a more ordered and stabilized secondary structure. β-Sheet structures were completely removed, suggesting disruption or replacement of these regions by alternative folding motifs. β-Turns increased significantly from 7.6% to 49.4%, indicating enhanced local folding and potential stabilization of loop regions. Random coil content decreased sharply from 60% to 12%, further supporting the notion of structural ordering induced by compound 20q.
These results proved that compound 20q formed strong interactions with α-glucosidase, inducing a conformational shift toward a more compact and ordered structure. The emergence of α-helical content and the reduction in disordered regions exhibited a stabilizing effect, which may correlate with the compound's inhibitory potency. Moreover, the loss of β-sheet structures, often associated with enzymatic activity, further supports the hypothesis that compound 20q effectively disrupted the native conformation required for catalytic function.
2.6 Fluorescence spectroscopy measurements
Fluorescence spectroscopy measurement is an analytical technique used to monitor the alteration of fluorescence characteristics and tertiary structure as a result of a formation of enzyme-inhibitor complexes. As the inhibitors bind to the enzyme, the intensity of fluorescence from fluorophores present in the active site of the enzyme decreases, causing fluorescence quenching. As inhibitors bind near fluorophores in the enzyme's active site, a decrease in fluorescence intensity—known as fluorescence quenching—occurs. Analysis of quenching mechanisms (static, dynamic, or resonance energy transfer) provides insight into the nature of the binding interaction.The interaction between coumarin–chalcone–triazole hybrid 20q and α-glucosidase was investigated by monitoring the intrinsic fluorescence quenching of the enzyme. Fluorescence emission spectra were recorded using a Synergy HTX multi-mode reader (Biotek Instruments, Winooski, VT, USA) with a 10 mm quartz cuvette. As depicted in Fig. 4, a maximum fluorescence intensity was observed at 340 nm, corresponding to the known emission wavelength of tryptophan. This result indicated that the inhibitor 20q is likely positioned near to this residue, facilitating significant interactions, which may lead to alterations in the enzyme's tertiary structure.
 | ||
| Fig. 4 Fluorescence spectra of α-glucosidase at 20–50 °C: (A) in the absence of any inhibitor (control); (B) enzyme exposed to compound 20q at the concentration of 0.5 µM. | ||
Another valuable application of fluorescence spectroscopy assessment is the calculation of binding constant, number of binding sites, and thermodynamic parameters of the studied interactions using the following equations. Based on the findings, the coumarin–chalcone–triazole hybrid 20q exhibited static fluorescence quenching.
The reaction is outlined as P + D → DnP; where P, D, and DnP represent the protein, drug (inhibitor), and resulting complex molecule, respectively. Using eqn (1), the binding constant of this complex, denoted as KA, is calculated.
 | (1) |
The number of binding sites is denoted as “n” and remains unchanged in the static quenching mechanism. The number of the binding site of protein and drug is n and 1, respectively. Therefore, the equivalent concentration of the complex DnP is n[DnP]. The equivalent concentration of the protein is n[P], and the equivalent concentration of the drug is [D].
The total concentration of protein is [Pt], and [Pt] is [Pf] + [DnP]. The total concentration of the drug is [Dt], and [Df] is [Dt] − n[DnP]. Since protein (P) is the only fluorescence species in the present study; therefore,
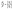 | (2) |
The fluorescence measurements were performed under the chosen experimental conditions, maintaining a constant total concentration of α-glucosidase at 46 nM while varying the concentrations of the compounds. The experimental data were then recorded. The fluorescence intensity of protein in the presence and absence of drug is F and F0. The correlation between these intensities and [Dt] is calculated in eqn (4), by which the plot of F0/F vs. [Dt] F0/(F0 − F) is outlined at 20 °C for compound 20q (Fig. 5). Moreover, using this equation, important parameters, including n and r at 20 °C, as well as KA at 20 °C and 50 °C, are calculated, as listed in Table 4.
 | ||
| Fig. 5 The plots F0/F vs. function of [Dt] F0/(F0 − F) at 20 °C for coumarin–chalcone–triazole hybrid 20q. | ||
The data in this Table 3 is graphed against temperature and binding constants, and important thermodynamic profile, including ΔG (free energy change), ΔH (enthalpy change), and ΔS (entropy change), could be computed through the equations as follow:
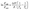 | (3) |
ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KA = ΔH − TΔS KA = ΔH − TΔS
| (4) |
The obtained results are presented in Table 5:
These figures are of great significance to determine the type of non-covalent forces between drug and enzyme's binding site, which are categorized into four groups: hydrophobic interaction, hydrogen bond, van der Waals forces, and electrostatic attraction. To identify the type of this interaction, the ΔH and ΔS values play a determining role, as follows: (1) ΔH > 0, ΔS > 0 indicating hydrophobic interactions; (2) ΔH < 0, ΔS < 0 indicating hydrogen bond and van der Waals interactions; (3) ΔH < 0, ΔS > 0 indicating electrostatic interactions. Considering the signs of ΔH and ΔS as presented in Table 5, electrostatic forces primarily formed between compound 20q with the active site of α-glucosidase.
2.7 Computational studies
To ensure a comprehensive predictive framework, over 1500 α-glucosidase inhibitors were collected and systematically compared to acarbose (IC50 = 750 µM), as a positive control. Additionally, pre-trained models from the Hugging Face repository, including those by Seyone Chithrananda and DeepChem, were fine-tuned to improve training efficiency and predictive accuracy. The “AutoModelForSequenceClassification” method was used to refine these models, with PC10M-396_250 and PC10M-450k emerging as the top performers based on key evaluation metrics.
The model's performance was assessed using Accuracy (ACC) and Matthews Correlation Coefficient (MCC) metrics, calculated as follows:
| Accuracy = (TP + TN)/(TP + TN + FP + FN) | (5) |
 | (6) |
These metrics provided a reliable assessment of the classification model, addressing challenges like imbalanced datasets.
The results demonstrated that the 10 M-MTR models achieved the highest MCC values (Fig. 6), indicating their superior predictive capabilities. Compounds with higher prediction scores consistently exhibited lower IC50 values, reinforcing the accuracy of the model (Table 6). For instance, compound 20q, with the highest prediction score (3.950), demonstrated the most potent inhibitory activity (IC50 = 0.50 ± 0.04 µM). The consistency of the model's predictions was confirmed across multiple iterations, highlighting its reliability.
| Compound | Predictions score | Count models | IC50 | Compound | Predictions score | Count models | IC50 |
|---|---|---|---|---|---|---|---|
| 20a | −0.204 | 6 | 3.60 ± 0.08 | 20m | 1.489 | 9 | 12.23 ± 0.67 |
| 20b | 0.791 | 9 | 11.48 ± 0.54 | 20n | 0.154 | 6 | 42.48 ± 0.32 |
| 20c | 0.93 | 9 | 26.73 ± 0.23 | 20o | 1.981 | 10 | 4.84 ± 0.16 |
| 20d | 0.241 | 9 | 44.74 ± 1.12 | 20p | 2.639 | 10 | 46.41 ± 1.23 |
| 20e | −0.361 | 5 | 38.63 ± 0.55 | 20q | 3.950 | 10 | 0.50 ± 0.04 |
| 20f | 0.179 | 5 | 1.34 ± 0.08 | 20r | 0.511 | 6 | 3.35 ± 0.14 |
| 20g | 0.701 | 6 | 90.82 ± 0.44 | 20s | 0.281 | 5 | 1.50 ± 0.08 |
| 20h | 1.031 | 9 | 84.33 ± 0.44 | 20t | 1.246 | 9 | 21.70 ± 0.19 |
| 20i | 1.252 | 9 | 136.60 ± 1.46 | 20u | 0.017 | 4 | 14.63 ± 0.21 |
| 20j | 0.519 | 8 | 110.89 ± 2.28 | 20v | −0.605 | 3 | 39.84 ± 0.16 |
| 20k | 1.693 | 10 | 285.52 ± 0.73 | 20w | 1.431 | 9 | 26.92 ± 0.12 |
| 20l | 0.689 | 8 | 1.07 ± 0.29 | 20x | 2.519 | 10 | 183.69 ± 0.08 |
| Acarbose | — | — | 750.08 ± 1.52 | Acarbose | — | — | 750.08 ± 1.52 |
The calculated binding energies for 17, 18, 19, 20a, and acarbose were −9.81, −9.37, −9.64, −11.94, −9.04, respectively, demonstrating the superior binding affinity of 20a among these compounds. Notably, compound 20q achieved the notable binding energy of −9.91, highlighting its potential as an α-glucosidase inhibitor.
The ligand RMSD, calculated after fitting on the protein backbone, began below 1.0 Å, indicating a well-positioned initial binding pose. It increased moderately to a stable range of ∼1.5–2.0 Å for the remainder of the simulation, demonstrating that 20q maintains a consistent binding orientation within the active site without major dissociation events. A transient fluctuation observed in both the protein and ligand trajectories around 100–120 ns signifies a short-lived conformational adjustment before the system re-established a stable equilibrium.
A comparison of the ligand RMSD profiles for 20q and acarbose is presented in Fig. 8B. The acarbose-bound complex exhibited significantly higher ligand RMSD values, ranging from 5 to 8 Å with peaks exceeding 10 Å, which indicates substantial internal flexibility and repositioning within the binding pocket. In contrast, ligand 20q displayed lower and more stable RMSD values (approximately 4–7 Å), with a reduced fluctuation amplitude throughout the 200 ns trajectory. This comparatively restrained mobility suggests a more stable interaction network and a tighter fit within the active site than acarbose.
Overall, the RMSD analyses demonstrate that ligand 20q forms a more dynamically stable complex with α-glucosidase, exhibiting less conformational drift and more consistent binding behavior compared to acarbose.
The root-mean-square fluctuation (RMSF) of the Cα atoms of α-glucosidase in complex with ligand 20q was analyzed to assess local flexibility (Fig. 9). The enzyme backbone remained predominantly stable during the 200 ns simulation, with most residues exhibiting RMSF values below ∼1.5 Å, indicating a rigid structural core. Several localized peaks were observed—notably around residue ranges ∼150, ∼210–230, ∼300, and ∼400—where fluctuations increased to 2–4 Å. These regions likely correspond to flexible loops or surface-exposed segments undergoing natural dynamics. Crucially, the residues directly participating in ligand binding showed significantly reduced fluctuations (<1.0–1.5 Å), highlighting their stabilization upon interaction with 20q. This pattern indicates that ligand binding effectively restricts mobility in the active-site region while permitting flexibility in distal loops.
2.8 In vivo anti-diabetic studies of compound 20q
As depicted in Fig. 10, compared to the diabetic control, which kept high blood glucose throughout the study, treatment with compound 20q provided a remarkable and time-dependent reduction in FBG levels. This antihyperglycemic effect became noticeable after the second week, when FBG levels in the treated groups started to show a clear difference from those in the diabetic control group. Furthermore, the administration of compound 20q at different doses exhibited a dose-dependent hypoglycemic effect over the 28 days treatment period in diabetic rats. Most notably, 20q at a dose of 8 mg kg−1 led to a substantial 71% reduction in FBG, restoring levels to near-normoglycemia (115.17 ± 4.28 mg dL−1) by the end of the study. This final value was statistically comparable to that of the healthy control group (105.17 ± 3.11 mg dL−1; p < 0.05). In contrast, the standard drug acarbose (10 mg kg−1) achieved a significantly lower reduction of only 22.1%, which was less effective than even the lowest dose of 20q (48.0%).
Overall, this lead compound showed superior potency, with the 8 mg kg−1 dose lowering FBG by about 195 mg dL−1 more than acarbose, highlighting its outstanding antihyperglycemic potential.
 | ||
| Fig. 11 Blood glucose levels (mg dL−1) in OGTT; ns: P < 0.05 for comparisons between every treatment group and control group at each time point. | ||
The area under the curve (AUC) of the OGTT was calculated using the trapezoidal rule to quantify the total glucose exposure over the 120 minutes test. As expected, the diabetic control group exhibited the highest AUC value (58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 274 mg dL−1 min−2), showing severe glucose intolerance. Treatment with compound 20q resulted in a remarkable, dose-dependent reduction in the AUC. The values were 32
274 mg dL−1 min−2), showing severe glucose intolerance. Treatment with compound 20q resulted in a remarkable, dose-dependent reduction in the AUC. The values were 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081, 25
081, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 935, and 18
935, and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 429 mg dL−1 min−2 for the 2, 4, and 8 mg kg−1 doses, respectively. Moreover, the AUC in the group 3 was not significantly different from that of the group 1 (15
429 mg dL−1 min−2 for the 2, 4, and 8 mg kg−1 doses, respectively. Moreover, the AUC in the group 3 was not significantly different from that of the group 1 (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 869 mg dL−1 min−2), indicating a near-complete normalization of glucose tolerance. In contrast, the standard drug acarbose (10 mg kg−1) was substantially less effective, yielding an AUC of 41
869 mg dL−1 min−2), indicating a near-complete normalization of glucose tolerance. In contrast, the standard drug acarbose (10 mg kg−1) was substantially less effective, yielding an AUC of 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 788 mg dL−1 min−2, which was even higher than that of group 5.
788 mg dL−1 min−2, which was even higher than that of group 5.
These results highlighted the potent and dose-dependent antihyperglycemic activity of compound 20q in diabetic rats, confirming its promising potential in restoring impaired glucose tolerance more effectively than acarbose, the reference anti-diabetic drug in this study.
| Group | HbA1c (%) | Group | HbA1c (%) |
|---|---|---|---|
| Normal control | 4.0 ± 0.15 | Treated with 20q (4 mg kg−1) | 5.0 ± 0.18 |
| Diabetic control | 8.1 ± 0.11 | Treated with 20q (2 mg kg−1) | 6.4 ± 0.22 |
| Treated with 20q (8 mg kg−1) | 4.2 ± 0.28 | Treated with acarbose (10 mg kg−1) | 7.8 ± 0.29 |
The normal HbA1c range in rats is between 3.5% and 4.5%. Animals in a prediabetic state typically possess HbA1c levels below 6.5%, whereas diabetic rats generally exhibit values ranging from 6.5% to 8.5%. As presented in Table 7, the HbA1c level in the diabetic control group was 8.1%, confirming the successful induction of diabetes. The normal control group showed an HbA1c level of 4.0%, while treatment with compound 20q at a dose of 8 mg kg−1 BW resulted in a value of 4.2%, close to that of the normal group. Using the comparable dose of acarbose (10 mg kg−1) had only a partial blood glucose-lowering effect, and the animals remained diabetic (HbA1c level of 7.8%). The group receiving 4 mg kg−1 BW of the compound 20q exhibited an HbA1c level of 5.0%, categorized into the prediabetic range. Furthermore, the group treated with 2 mg kg−1 BW had a value of 6.4%, approaching the diabetic threshold.
Overall, HbA1c levels significantly decreased in diabetic rats treated with compound 20q in a dose-dependent manner. Moreover, administration of compound 20q at a dose of 8 mg kg−1 BW restored HbA1c to a near-normal level (4.2 ± 0.28%), which was remarkably comparable to that of the healthy control group (4.0 ± 0.15%) and substantially lower than both the diabetic control (8.1 ± 0.11%) and acarbose-treated (7.8 ± 0.29%) groups. These results highlight the remarkable long-term antihyperglycemic potency of compound 20q, surpassing the efficacy of the standard drug acarbose.
 | ||
| Fig. 12 Liver sections of rats (stained with H&E ×40) from (A) group 1; (B) group 2; (C) group 3; (D) group 4; (E) group 5; (F) group 6. | ||
 | ||
| Fig. 13 Pancreas sections of rats (stained with H&E ×40) from (A) group 1; (B) group 2; (C) group 3; (D) group 4; (E) group 5; (F) group 6. | ||
As presented in Fig. 12, the liver tissue from the normal control group (A) exhibited a typical hepatic architecture, characterized by well-preserved hepatocytes arranged in cords, normal sinusoidal spaces, and distinct central veins, all indicative of a physiologically healthy liver. In contrast, the diabetic control group (B) showed severe histological damages, including hepatocellular swelling, prominent lipid droplet accumulation (steatosis), and inflammatory cell infiltration, particularly around the central vein. Disruption of the sinusoidal architecture was also evident. These findings correlate with the elevated FBG and HbA1c levels, confirming diabetes-induced hepatic damage.
Treatment with compound 20q at 8 mg kg−1 (C) restored the liver histology close to normal, with minimal cytoplasmic changes and a reappearance of organized sinusoidal patterns. This strong hepatoprotective effect correlated with significant reductions in FBG and HbA1c. Treatment with compound 20q at 4 mg kg−1 (D) led to a moderate improvement with reduced inflammation and partially restored sinusoids, though mild swelling and lipid accumulation persisted. 2 mg kg−1 group (E) showed only mild improvement, with residual steatosis, occasional inflammation, and persistent sinusoidal disruption, reflecting limited efficacy, in line with the borderline HbA1c values. Acarbose (F) Provided only partial recovery, with moderate reduction in steatosis and inflammation, but significant architectural and sinusoidal irregularities remained, indicating that acarbose was less effective than compound 20q, even at its lowest dose.
As illustrated in Fig. 13, pancreatic sections from the normal control group (A) exhibited characteristic healthy islet architecture. In contrast, the diabetic control (B) showed severe atrophy, ill-defined borders, decreased cellular density, and β-cell necrosis. Treatment with compound 20q improved pancreas morphology in a dose-dependent manner: the high dose (8 mg kg−1, C) almost completely preserved islet size, density, and organization; the medium dose (4 mg kg−1, D) showed significant preservation of islet size and integrity; and the low dose (2 mg kg−1, E) resulted only in slight improvement with residual atrophy. Conversely, the acarbose group (F) showed no significant improvement, closely resembling the diabetic control, which aligns with its limited efficacy in controlling hyperglycemia.
The remarkable potential of compound 20q is shown by its ability to normalize key glycemic parameters (FBG, OGTT, HbA1c) and also to repair diabetes-related damage in the liver and pancreas. The dose-dependent restoration of liver structure and pancreatic islets supports the biochemical improvements in this study. This clear link between functional recovery and tissue protection shows that compound 20q is not just controlling symptoms, like acarbose, but actually modifying the disease itself.
3 Conclusion
In summary, this study reports the design, synthesis, and comprehensive biological evaluation of a novel series of coumarin–chalcone-1,2,3-triazole hybrids as potential anti-diabetic agents. Systematic SAR investigations identified the critical structural features for enhancing inhibitory activity, the necessary presence of substituents on the C-7 position of the coumarin backbone and electron-donating methoxy groups on the chalcone.Among the synthesized library, compound 20q exhibited exceptional potency against α-glucosidase (IC50 = 0.50 ± 0.04 µM, compared to acarbose), high selectivity over α-amylase, and an excellent safety profile with no cytotoxicity. Further mechanistic studies revealed that compound 20q competitively bonded to α-glucosidase and induced substantial conformational rearrangements, as proved by circular dichroism and fluorescence spectroscopy, which demonstrated the emergence of α-helical content, disruption of β-sheets, and strong electrostatic interactions with active site residues. These findings correlated with our computational analyses, including a deep-learning predictive model, molecular docking (which showed key interactions with the active site residues), and molecular dynamics simulations (which confirmed the stability of the enzyme-inhibitor complex). Furthermore, in vivo studies in diabetic rats showed dose-dependent reductions in FBG and HbA1c, restoration of glucose tolerance, and protecting the histopathological damage in the liver and pancreas. These results were significantly superior to those of acarbose. This compound also showed an acceptable acute oral safety profile.
The comprehensive in vitro, in silico, and in vivo data collectively identified compound 20q as a promising candidate for further investigation into its antidiabetic potential and also structural modifications to develop more potent agents.
4 Experimental
All chemicals were purchased from Merck (Germany) and were used without further purification. The reaction progress and the purity of synthesized compounds were monitored by thin-layer chromatography (TLC) on silica gel 250-micron F254 plastic sheets; zones were detected visually under UV light (254 nm). Melting points were measured on an Electrothermal 9100 apparatus. IR spectra were recorded on a Shimadzu IR-460 spectrometer. 1H and 13C NMR spectra were measured (DMSO-d6 solution) with Bruker DRX-400 AVANCE (at 400.1 and 100.1 MHz) and Bruker DRX-500 AVANCE (at 500.1 and 125.1 MHz) instruments. 2D NMR spectrum was recorded on the Bruker DRX-500 AVANCE. Chemical shifts were reported in parts per million (ppm), downfield from tetramethylsilane (TMS). Proton coupling patterns were described as singlet (s), doublet (d), triplet (t), and multiplet (m). HRMS analysis was performed using a Waters Synapt G1 HDMS High Definition mass spectrometer equipped with an electrospray ionization (ESI) source. The samples were prepared by diluting the isolated compounds in methanol to a final concentration of 10 µg mL−1. The analysis was conducted in negative ion mode with a mass range of m/z 50–1000. Elemental analyses for C, H and N were performed using a Heraeus CHN-O-Rapid analyzer.4.1 General synthetic procedures
4.1.5.1 4-((1-(3-Oxo-1,3-diphenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (17). Milky solid, mp 189–188 °C, yield: 81%. IR (KBr) (νmax/cm−1): 3210 and 3102, 1723 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1674 (C
O, coumarin), 1674 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1587, 1502, 1467, 1423, 1356, 1277, 1212, 1108, 1023, 978, 897, 855, 798, 671, 635. 1H NMR (500.1 MHz, DMSO-d6): δ 8.68 (s, 1H, CH), 7.82–7.65 (m, 7H, 7CH), 7.60 (t, J = 7.5 Hz, 1H, CH), 7.49 (d, J = 8.0 Hz, 1H, CH), 7.44–7.34 (m, 2H, 2CH), 7.27 (t, J = 7.2 Hz, 2H, 2CH), 6.92 (d, J = 7.5 Hz, 2H, 2CH), 6.23 (s, 1H, CH), 5.55 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.56, 162.80, 160.71, 153.68, 143.58, 142.57, 137.49, 132.6, 132.34, 131.41, 130.65, 130.11, 129.68, 129.15, 128.57, 128.20, 128.08, 127.83, 126.88, 117.59, 116.39, 91.89, 61.84. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1306. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 72.28; H, 4.44; N, 9.48%.
O, chalcone), 1587, 1502, 1467, 1423, 1356, 1277, 1212, 1108, 1023, 978, 897, 855, 798, 671, 635. 1H NMR (500.1 MHz, DMSO-d6): δ 8.68 (s, 1H, CH), 7.82–7.65 (m, 7H, 7CH), 7.60 (t, J = 7.5 Hz, 1H, CH), 7.49 (d, J = 8.0 Hz, 1H, CH), 7.44–7.34 (m, 2H, 2CH), 7.27 (t, J = 7.2 Hz, 2H, 2CH), 6.92 (d, J = 7.5 Hz, 2H, 2CH), 6.23 (s, 1H, CH), 5.55 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.56, 162.80, 160.71, 153.68, 143.58, 142.57, 137.49, 132.6, 132.34, 131.41, 130.65, 130.11, 129.68, 129.15, 128.57, 128.20, 128.08, 127.83, 126.88, 117.59, 116.39, 91.89, 61.84. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1306. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 72.28; H, 4.44; N, 9.48%.
4.1.5.2 6-((1-(3-Oxo-1,3-diphenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (18). Milky solid, mp 222–225 °C, yield: 84%. IR (KBr) (νmax/cm−1): 3187 and 3118, 1739 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1668 (C
O, coumarin), 1668 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1557, 1498, 1458, 1399, 1342, 1286, 1198, 1105, 1025, 999, 875, 843, 776, 652, 623. 1H NMR (500.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.05 (d, J = 9.2 Hz, 1H, CH), 7.94–7.88 (m, 3H, 3CH), 7.62 (t, J = 7.7 Hz, 1H, CH), 7.57 (t, J = 7.6 Hz, 2H, 2CH), 7.36 (t, J = 7.4 Hz, 1H, CH), 7.30–7.25 (m, 3H, 3CH), 7.16 (d, J = 7.5 Hz, 1H, CH), 7.00 (d, J = 7.5 Hz, 1H, CH), 6.92 (d, J = 7.7 Hz, 2H, 2CH), 6.29 (d, J = 9.2 Hz, 1H, CH), 5.38 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 191.61, 161.16, 157.89, 152.96, 143.83, 143.60, 143.05, 136.48, 133.54, 133.14, 133.04, 131.82, 130.78, 130.00, 129.91, 129.31, 128.77, 117.76, 115.81, 115.18, 113.18, 108.39, 61.88. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1303. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 71.98; H, 4.09; N, 9.19%.
O, chalcone), 1557, 1498, 1458, 1399, 1342, 1286, 1198, 1105, 1025, 999, 875, 843, 776, 652, 623. 1H NMR (500.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.05 (d, J = 9.2 Hz, 1H, CH), 7.94–7.88 (m, 3H, 3CH), 7.62 (t, J = 7.7 Hz, 1H, CH), 7.57 (t, J = 7.6 Hz, 2H, 2CH), 7.36 (t, J = 7.4 Hz, 1H, CH), 7.30–7.25 (m, 3H, 3CH), 7.16 (d, J = 7.5 Hz, 1H, CH), 7.00 (d, J = 7.5 Hz, 1H, CH), 6.92 (d, J = 7.7 Hz, 2H, 2CH), 6.29 (d, J = 9.2 Hz, 1H, CH), 5.38 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 191.61, 161.16, 157.89, 152.96, 143.83, 143.60, 143.05, 136.48, 133.54, 133.14, 133.04, 131.82, 130.78, 130.00, 129.91, 129.31, 128.77, 117.76, 115.81, 115.18, 113.18, 108.39, 61.88. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1303. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 71.98; H, 4.09; N, 9.19%.
4.1.5.3 8-((1-(3-Oxo-1,3-diphenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (19). Milky solid, mp 193–195 °C, yield: 69%. IR (KBr) (νmax/cm−1): 3162 and 3099, 1725 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1649 (C
O, coumarin), 1649 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1556, 1487, 1429, 1379, 1320, 1279, 1185, 1132, 1076, 957, 872, 830, 752, 649. 1H NMR (500.1 MHz, DMSO-d6): δ 8.64 (s, 1H, CH), 8.12 (d, J = 9.4 Hz, 1H, CH), 7.94–7.83 (m, 3H, 3CH), 7.70 (t, J = 7.7 Hz, 1H, CH), 7.57 (t, J = 7.6 Hz, 2H, 2CH), 7.50 (d, J = 8.0 Hz, 1H, CH), 7.36 (t, J = 7.5 Hz, 1H, CH), 7.24 (t, J = 7.4 Hz, 2H, 2CH), 7.10 (t, J = 7.8 Hz, 1H, CH), 7.00 (d, J = 7.6 Hz, 1H, CH), 6.86 (d, J = 8.0 Hz, 2H, 2CH), 6.36 (d, J = 9.4 Hz, 1H, CH), 5.30 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 189.75, 161.61, 155.77, 154.53, 144.77, 143.55, 142.18, 136.35, 132.89, 131.97, 131.71, 130.98, 129.98, 129.76, 129.26, 129.21, 128.10, 119.66, 113.38, 113.13, 113.03, 101.99, 62.05. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1302. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 72.29; H, 4.38; N, 9.55%.
O, chalcone), 1556, 1487, 1429, 1379, 1320, 1279, 1185, 1132, 1076, 957, 872, 830, 752, 649. 1H NMR (500.1 MHz, DMSO-d6): δ 8.64 (s, 1H, CH), 8.12 (d, J = 9.4 Hz, 1H, CH), 7.94–7.83 (m, 3H, 3CH), 7.70 (t, J = 7.7 Hz, 1H, CH), 7.57 (t, J = 7.6 Hz, 2H, 2CH), 7.50 (d, J = 8.0 Hz, 1H, CH), 7.36 (t, J = 7.5 Hz, 1H, CH), 7.24 (t, J = 7.4 Hz, 2H, 2CH), 7.10 (t, J = 7.8 Hz, 1H, CH), 7.00 (d, J = 7.6 Hz, 1H, CH), 6.86 (d, J = 8.0 Hz, 2H, 2CH), 6.36 (d, J = 9.4 Hz, 1H, CH), 5.30 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 189.75, 161.61, 155.77, 154.53, 144.77, 143.55, 142.18, 136.35, 132.89, 131.97, 131.71, 130.98, 129.98, 129.76, 129.26, 129.21, 128.10, 119.66, 113.38, 113.13, 113.03, 101.99, 62.05. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1302. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 72.29; H, 4.38; N, 9.55%.
4.1.5.4 7-((1-(3-Oxo-1,3-diphenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20a). Milky solid, mp 216–218 °C, yield: 78%. IR (KBr) (νmax/cm−1): 3156 and 3089, 1748 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1668 (C
O, coumarin), 1668 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1525, 1488, 1444, 1369, 1299, 1235, 1110, 1045, 966, 914, 872, 823, 752, 699, 648, 623. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.89–7.79 (m, 3H, 3CH), 7.71–7.63 (m, 2H, 2CH), 7.57 (t, J = 7.5 Hz, 2H, 2CH), 7.36 (t, J = 7.4 Hz, 1H, CH), 7.22 (t, J = 7.6 Hz, 2H, 2CH), 7.16 (s, 1H, CH), 7.05 (d, J = 8.5 Hz, 1H, CH), 6.85 (d, J = 7.8 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.41 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 191.08, 161.33, 160.71, 155.72, 144.77, 143.30, 142.78, 136.73, 133.43, 132.31, 131.79, 131.58, 130.60, 129.98, 129.75, 129.27, 129.21, 127.51, 113.62, 113.19, 113.12, 102.17, 61.86. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1299. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 72.38; H, 4.12; N, 9.52%.
O, chalcone), 1599, 1525, 1488, 1444, 1369, 1299, 1235, 1110, 1045, 966, 914, 872, 823, 752, 699, 648, 623. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.89–7.79 (m, 3H, 3CH), 7.71–7.63 (m, 2H, 2CH), 7.57 (t, J = 7.5 Hz, 2H, 2CH), 7.36 (t, J = 7.4 Hz, 1H, CH), 7.22 (t, J = 7.6 Hz, 2H, 2CH), 7.16 (s, 1H, CH), 7.05 (d, J = 8.5 Hz, 1H, CH), 6.85 (d, J = 7.8 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.41 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 191.08, 161.33, 160.71, 155.72, 144.77, 143.30, 142.78, 136.73, 133.43, 132.31, 131.79, 131.58, 130.60, 129.98, 129.75, 129.27, 129.21, 127.51, 113.62, 113.19, 113.12, 102.17, 61.86. HRMS (ESI) m/z for C27H18N3O4− [M–H]−, calculated: 448.1303, found: 448.1299. Anal. calcd. for C27H19N3O4: C, 72.15; H, 4.26; N, 9.35; found: C, 72.38; H, 4.12; N, 9.52%.
4.1.5.5 7-((1-(3-(4-Chlorophenyl)-3-oxo-1-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20b). Milky solid, mp 272–275 °C, yield: 82%. IR (KBr) (νmax/cm−1): 3197 and 3054, 1748 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1656 (C
O, coumarin), 1656 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1603, 1525, 1488, 1424, 1389, 1333, 1298, 1245, 1136, 1073, 1025, 985, 912, 872, 854, 774, 696, 638. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.01 (d, J = 8.9 Hz, 1H, CH), 7.91–7.81 (m, 3H, 3CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.63 (d, J = 8.2 Hz, 2H, 2CH), 7.37 (t, J = 7.3 Hz, 1H, CH), 7.23 (t, J = 7.6 Hz, 2H, 2CH), 7.17 (s, 1H, CH), 7.05 (d, J = 8.6 Hz, 1H, CH), 6.86 (d, J = 7.6 Hz, 2H, 2CH), 6.32 (d, J = 8.9 Hz, 1H, CH), 5.42 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.14, 161.32, 160.71, 155.72, 144.77, 143.30, 143.16, 138.25, 135.50, 132.09, 131.91, 131.65, 131.58, 130.69, 129.99, 129.32, 129.28, 127.46, 113.60, 113.21, 113.13, 102.18, 61.85. HRMS (ESI) m/z for C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0914. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 67.29; H, 3.52; N, 8.90%.
O, chalcone), 1603, 1525, 1488, 1424, 1389, 1333, 1298, 1245, 1136, 1073, 1025, 985, 912, 872, 854, 774, 696, 638. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.01 (d, J = 8.9 Hz, 1H, CH), 7.91–7.81 (m, 3H, 3CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.63 (d, J = 8.2 Hz, 2H, 2CH), 7.37 (t, J = 7.3 Hz, 1H, CH), 7.23 (t, J = 7.6 Hz, 2H, 2CH), 7.17 (s, 1H, CH), 7.05 (d, J = 8.6 Hz, 1H, CH), 6.86 (d, J = 7.6 Hz, 2H, 2CH), 6.32 (d, J = 8.9 Hz, 1H, CH), 5.42 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.14, 161.32, 160.71, 155.72, 144.77, 143.30, 143.16, 138.25, 135.50, 132.09, 131.91, 131.65, 131.58, 130.69, 129.99, 129.32, 129.28, 127.46, 113.60, 113.21, 113.13, 102.18, 61.85. HRMS (ESI) m/z for C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0914. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 67.29; H, 3.52; N, 8.90%.
4.1.5.6 7-((1-(3-(3-Chlorophenyl)-3-oxo-1-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20c). Milky solid, mp 238–240 °C, yield: 69%. IR (KBr) (νmax/cm−1): 3136 and 3088, 1757 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1664 (C
O, coumarin), 1664 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1598, 1556, 1492, 1422, 1356, 1307, 1252, 1232, 1108, 1077, 948, 906, 878, 830, 765, 740, 696, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.56 (s, 1H, CH), 8.00 (d, J = 9.5 Hz, 1H, CH), 7.86 (d, J = 7.5 Hz, 2H, 2CH), 7.83 (s, 1H, CH), 7.70 (t, J = 7.6 Hz, 1H, CH), 7.64 (d, J = 8.5 Hz, 1H, 1CH), 7.57 (t, J = 7.5 Hz, 2H, 2CH), 7.42 (d, J = 7.7 Hz, 1H, CH), 7.24 (t, J = 7.9 Hz, 1H, CH), 7.16 (s, 1H, CH), 7.10–6.98 (m, 1H, CH), 6.97 (s, 1H, CH), 6.80 (d, J = 7.9 Hz, 1H, CH), 6.31 (d, J = 9.5 Hz, 1H, CH), 5.39 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.84, 161.34, 160.71, 155.75, 144.78, 140.92, 139.67, 136.45, 133.90, 133.79, 133.63, 133.18, 131.16, 130.96, 130.07, 129.99, 129.90, 129.25, 128.75, 127.57, 113.56, 113.17, 113.12, 102.08, 61.72. HRMS (ESI) m/z for C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0913. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 67.34; H, 3.88; N, 8.84%.
O, chalcone), 1598, 1556, 1492, 1422, 1356, 1307, 1252, 1232, 1108, 1077, 948, 906, 878, 830, 765, 740, 696, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.56 (s, 1H, CH), 8.00 (d, J = 9.5 Hz, 1H, CH), 7.86 (d, J = 7.5 Hz, 2H, 2CH), 7.83 (s, 1H, CH), 7.70 (t, J = 7.6 Hz, 1H, CH), 7.64 (d, J = 8.5 Hz, 1H, 1CH), 7.57 (t, J = 7.5 Hz, 2H, 2CH), 7.42 (d, J = 7.7 Hz, 1H, CH), 7.24 (t, J = 7.9 Hz, 1H, CH), 7.16 (s, 1H, CH), 7.10–6.98 (m, 1H, CH), 6.97 (s, 1H, CH), 6.80 (d, J = 7.9 Hz, 1H, CH), 6.31 (d, J = 9.5 Hz, 1H, CH), 5.39 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.84, 161.34, 160.71, 155.75, 144.78, 140.92, 139.67, 136.45, 133.90, 133.79, 133.63, 133.18, 131.16, 130.96, 130.07, 129.99, 129.90, 129.25, 128.75, 127.57, 113.56, 113.17, 113.12, 102.08, 61.72. HRMS (ESI) m/z for C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0913. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 67.34; H, 3.88; N, 8.84%.
4.1.5.7 7-((1-(3-(2-Chlorophenyl)-3-oxo-1-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20d). Milky solid, mp 220–223 °C, yield: 65%. IR (KBr) (νmax/cm−1): 3155 and 3049, 1753 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1678 (C
O, coumarin), 1678 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1606, 1548, 1493, 1458, 1429, 1382, 1313, 1298, 1236, 1203, 1158, 1115, 1078, 975, 904, 874, 829, 755, 703, 644. 1H NMR (400.1 MHz, DMSO-d6): δ 8.54 (s, 1H, CH), 8.01 (d, J = 9.4 Hz, 1H, CH), 7.89 (d, J = 7.4 Hz, 2H, 2CH), 7.82 (s, 1H, CH), 7.74–7.49 (m, 5H, 5CH), 7.36 (t, J = 7.5 Hz, 1H, CH), 7.13 (s, 1H, CH), 7.07 (t, J = 7.2 Hz, 1H, CH), 7.02 (d, J = 8.2 Hz, 1H, CH), 6.62 (d, J = 7.6 Hz, 1H, CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.35 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.54, 161.30, 160.72, 155.70, 144.77, 142.89, 139.60, 137.59, 136.27, 134.37, 133.84, 132.46, 130.43, 130.23, 129.96, 129.88, 129.73, 129.28, 128.37, 127.72, 113.56, 113.19, 113.11, 102.10, 61.76. HRMS (ESI) m/z for C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0915. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 66.86; H, 3.98; N, 8.92%.
O, chalcone), 1606, 1548, 1493, 1458, 1429, 1382, 1313, 1298, 1236, 1203, 1158, 1115, 1078, 975, 904, 874, 829, 755, 703, 644. 1H NMR (400.1 MHz, DMSO-d6): δ 8.54 (s, 1H, CH), 8.01 (d, J = 9.4 Hz, 1H, CH), 7.89 (d, J = 7.4 Hz, 2H, 2CH), 7.82 (s, 1H, CH), 7.74–7.49 (m, 5H, 5CH), 7.36 (t, J = 7.5 Hz, 1H, CH), 7.13 (s, 1H, CH), 7.07 (t, J = 7.2 Hz, 1H, CH), 7.02 (d, J = 8.2 Hz, 1H, CH), 6.62 (d, J = 7.6 Hz, 1H, CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.35 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.54, 161.30, 160.72, 155.70, 144.77, 142.89, 139.60, 137.59, 136.27, 134.37, 133.84, 132.46, 130.43, 130.23, 129.96, 129.88, 129.73, 129.28, 128.37, 127.72, 113.56, 113.19, 113.11, 102.10, 61.76. HRMS (ESI) m/z for C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0915. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 66.86; H, 3.98; N, 8.92%.
4.1.5.8 7-((1-(3-(4-Bromophenyl)-3-oxo-1-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20e). Milky solid, mp 296–297 °C, yield: 92%. IR (KBr) (νmax/cm−1): 3189 and 3023, 1729 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1658 (C
O, coumarin), 1658 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1590, 1538, 1509, 1482, 1451, 1386, 1323, 1238, 1188, 1142, 1105, 1078, 1042, 977, 923, 829, 738, 699, 626. 1H NMR (500.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.00 (d, J = 9.4 Hz, 1H, CH), 7.83 (s, 1H, CH), 7.79–7.72 (m, 4H, 4CH), 7.65 (d, J = 8.6 Hz, 1H, CH), 7.36 (t, J = 7.4 Hz, 1H, CH), 7.21 (t, J = 7.9 Hz, 2H, 2CH), 7.15 (d, J = 2.5 Hz, 1H, CH), 7.06 (dd, J = 8.7, 2.5 Hz, 1H, CH), 6.85 (d, J = 7.1 Hz, 2H, 2CH), 6.30 (d, J = 9.4 Hz, 1H, CH), 5.40 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.32, 161.33, 160.71, 155.72, 144.77, 143.31, 143.22, 135.85, 132.25, 132.07, 131.91, 131.75, 131.58, 130.70, 129.99, 129.28, 127.47, 127.37, 113.59, 113.21, 113.13, 102.18, 61.86. HRMS (ESI) m/z for C27H17BrN3O4− [M–H]−, calculated: 526.0408, found: 526.0411. Anal. calcd. for C27H18BrN3O4: C, 61.38; H, 3.43; N, 7.95; found: C, 61.62; H, 3.27; N, 8.12%.
O, chalcone), 1590, 1538, 1509, 1482, 1451, 1386, 1323, 1238, 1188, 1142, 1105, 1078, 1042, 977, 923, 829, 738, 699, 626. 1H NMR (500.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.00 (d, J = 9.4 Hz, 1H, CH), 7.83 (s, 1H, CH), 7.79–7.72 (m, 4H, 4CH), 7.65 (d, J = 8.6 Hz, 1H, CH), 7.36 (t, J = 7.4 Hz, 1H, CH), 7.21 (t, J = 7.9 Hz, 2H, 2CH), 7.15 (d, J = 2.5 Hz, 1H, CH), 7.06 (dd, J = 8.7, 2.5 Hz, 1H, CH), 6.85 (d, J = 7.1 Hz, 2H, 2CH), 6.30 (d, J = 9.4 Hz, 1H, CH), 5.40 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.32, 161.33, 160.71, 155.72, 144.77, 143.31, 143.22, 135.85, 132.25, 132.07, 131.91, 131.75, 131.58, 130.70, 129.99, 129.28, 127.47, 127.37, 113.59, 113.21, 113.13, 102.18, 61.86. HRMS (ESI) m/z for C27H17BrN3O4− [M–H]−, calculated: 526.0408, found: 526.0411. Anal. calcd. for C27H18BrN3O4: C, 61.38; H, 3.43; N, 7.95; found: C, 61.62; H, 3.27; N, 8.12%.
4.1.5.9 7-((1-(3-(4-Methoxyphenyl)-3-oxo-1-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20f). Milky solid, mp 279–282 °C, yield: 78%. IR (KBr) (νmax/cm−1): 3173 and 2994, 1736 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1639 (C
O, coumarin), 1639 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1532, 1494, 1436, 1386, 1326, 1294, 1258, 1140, 1110, 1056, 1025, 970, 935, 886, 848, 762, 679, 633. 1H NMR (400.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.01 (d, J = 9.4 Hz, 1H, CH), 7.85 (d, J = 8.7 Hz, 2H, 2CH), 7.77 (s, 1H, CH), 7.65 (d, J = 8.6 Hz, 1H, CH), 7.35 (t, J = 7.4 Hz, 1H, CH), 7.21 (t, J = 7.6 Hz, 2H, 2CH), 7.16 (d, J = 1.6 Hz, 1H, CH), 7.09 (d, J = 8.7 Hz, 2H, 2CH), 7.05 (dd, J = 8.6, 2.0 Hz, 1H, CH), 6.86 (d, J = 7.7 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.42 (s, 2H, CH2), 3.86 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 189.39, 163.72, 161.32, 160.72, 155.71, 144.76, 143.24, 141.29, 132.35, 132.24, 131.72, 131.50, 130.43, 129.97, 129.23, 128.98, 127.50, 114.58, 113.59, 113.18, 113.11, 102.16, 61.82, 56.09. HRMS (ESI) m/z for C28H20N3O5− [M–H]−, calculated: 478.1408, found: 478.1403. Anal. calcd. for C28H21N3O5: C, 70.14; H, 4.41; N, 8.76; found: C, 70.29; H, 4.65; N, 8.98%.
O, chalcone), 1599, 1532, 1494, 1436, 1386, 1326, 1294, 1258, 1140, 1110, 1056, 1025, 970, 935, 886, 848, 762, 679, 633. 1H NMR (400.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.01 (d, J = 9.4 Hz, 1H, CH), 7.85 (d, J = 8.7 Hz, 2H, 2CH), 7.77 (s, 1H, CH), 7.65 (d, J = 8.6 Hz, 1H, CH), 7.35 (t, J = 7.4 Hz, 1H, CH), 7.21 (t, J = 7.6 Hz, 2H, 2CH), 7.16 (d, J = 1.6 Hz, 1H, CH), 7.09 (d, J = 8.7 Hz, 2H, 2CH), 7.05 (dd, J = 8.6, 2.0 Hz, 1H, CH), 6.86 (d, J = 7.7 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.42 (s, 2H, CH2), 3.86 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 189.39, 163.72, 161.32, 160.72, 155.71, 144.76, 143.24, 141.29, 132.35, 132.24, 131.72, 131.50, 130.43, 129.97, 129.23, 128.98, 127.50, 114.58, 113.59, 113.18, 113.11, 102.16, 61.82, 56.09. HRMS (ESI) m/z for C28H20N3O5− [M–H]−, calculated: 478.1408, found: 478.1403. Anal. calcd. for C28H21N3O5: C, 70.14; H, 4.41; N, 8.76; found: C, 70.29; H, 4.65; N, 8.98%.
4.1.5.10 7-((1-(3-Oxo-1-phenyl-3-(thiophen-2-yl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20g). Milky solid, mp 216–219 °C, yield: 68%. IR (KBr) (νmax/cm−1): 3209 and 3044, 1753 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1646 (C
O, coumarin), 1646 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1536, 1490, 1452, 1390, 1289, 1224, 1163, 1116, 1088, 1037, 977, 924, 885, 832, 741, 723, 638. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.13 (d, J = 4.7 Hz, 1H, CH), 8.10 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 2H, 2CH), 7.50 (d, J = 3.3 Hz, 1H, CH), 7.37 (t, J = 7.4 Hz, 1H, CH), 7.27–7.19 (m, 3H, 3CH), 7.17 (d, J = 1.9 Hz, 1H, CH), 7.05 (dd, J = 8.6, 2.1 Hz, 1H, CH), 6.89 (d, J = 7.7 Hz, 1H, CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.42 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 181.22, 161.26, 160.69, 155.71, 144.77, 143.59, 141.43, 140.99, 136.96, 135.11, 131.72, 131.67, 131.61, 130.63, 129.99, 129.46, 129.28, 127.83, 113.66, 113.22, 113.15, 102.22, 61.78. HRMS (ESI) m/z for C25H16N3O4S− [M–H]−, calculated: 454.0867, found: 454.0867. Anal. calcd. for C25H17N3O4S: C, 65.92; H, 3.76; N, 9.23; found: C, 66.12; H, 3.63; N, 9.42%.
O, chalcone), 1599, 1536, 1490, 1452, 1390, 1289, 1224, 1163, 1116, 1088, 1037, 977, 924, 885, 832, 741, 723, 638. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.13 (d, J = 4.7 Hz, 1H, CH), 8.10 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 2H, 2CH), 7.50 (d, J = 3.3 Hz, 1H, CH), 7.37 (t, J = 7.4 Hz, 1H, CH), 7.27–7.19 (m, 3H, 3CH), 7.17 (d, J = 1.9 Hz, 1H, CH), 7.05 (dd, J = 8.6, 2.1 Hz, 1H, CH), 6.89 (d, J = 7.7 Hz, 1H, CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.42 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 181.22, 161.26, 160.69, 155.71, 144.77, 143.59, 141.43, 140.99, 136.96, 135.11, 131.72, 131.67, 131.61, 130.63, 129.99, 129.46, 129.28, 127.83, 113.66, 113.22, 113.15, 102.22, 61.78. HRMS (ESI) m/z for C25H16N3O4S− [M–H]−, calculated: 454.0867, found: 454.0867. Anal. calcd. for C25H17N3O4S: C, 65.92; H, 3.76; N, 9.23; found: C, 66.12; H, 3.63; N, 9.42%.
4.1.5.11 7-((1-(1-(4-Chlorophenyl)-3-oxo-3-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20h). Milky solid, mp 236–239 °C, yield: 79%. IR (KBr) (νmax/cm−1): 3152 and 3023, 1752 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1655 (C
O, coumarin), 1655 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1605, 1595, 1547, 1478, 1436, 1382, 1293, 1229, 1195, 1154, 1107, 1048, 962, 904, 878, 828, 752, 701, 639. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.87–7.81 (m, 3H, 3CH), 7.72–7.65 (m, 2H, 2CH), 7.57 (t, J = 7.6 Hz, 2H, 2CH), 7.27 (d, J = 8.5 Hz, 2H, 2CH), 7.16 (d, J = 1.9 Hz, 1H, CH), 7.06 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.86 (d, J = 8.5 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.41 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.87, 161.28, 160.69, 155.71, 144.74, 143.37, 141.26, 136.58, 136.34, 133.53, 132.70, 132.12, 130.59, 129.98, 129.81, 129.31, 129.22, 127.48, 113.57, 113.25, 113.14, 102.22, 61.81. HRMS (ESI) m/z C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0911. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 67.15; H, 3.98; N, 8.51%.
O, chalcone), 1605, 1595, 1547, 1478, 1436, 1382, 1293, 1229, 1195, 1154, 1107, 1048, 962, 904, 878, 828, 752, 701, 639. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.87–7.81 (m, 3H, 3CH), 7.72–7.65 (m, 2H, 2CH), 7.57 (t, J = 7.6 Hz, 2H, 2CH), 7.27 (d, J = 8.5 Hz, 2H, 2CH), 7.16 (d, J = 1.9 Hz, 1H, CH), 7.06 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.86 (d, J = 8.5 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.41 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 190.87, 161.28, 160.69, 155.71, 144.74, 143.37, 141.26, 136.58, 136.34, 133.53, 132.70, 132.12, 130.59, 129.98, 129.81, 129.31, 129.22, 127.48, 113.57, 113.25, 113.14, 102.22, 61.81. HRMS (ESI) m/z C27H17ClN3O4− [M–H]−, calculated: 482.0913, found: 482.0911. Anal. calcd. for C27H18ClN3O4: C, 67.02; H, 3.75; N, 8.68; found: C, 67.15; H, 3.98; N, 8.51%.
4.1.5.12 7-((1-(1,3-Bis(4-chlorophenyl)-3-oxoprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20i). Milky solid, mp 246–249 °C, yield: 86%. IR (KBr) (νmax/cm−1): 3163 and 2934, 1736 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1668 (C
O, coumarin), 1668 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1607, 1545, 1492, 1446, 1384, 1297, 1248, 1205, 1138, 1107, 998, 836, 771, 738, 691, 628. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.87–7.82 (m, 3H, 3CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.63 (d, J = 8.4 Hz, 2H, 2CH), 7.28 (d, J = 8.5 Hz, 2H, 2CH), 7.17 (d, J = 2.0 Hz, 1H, CH), 7.06 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.86 (d, J = 8.5 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.41 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 189.96, 161.27, 160.69, 155.71, 144.75, 143.42, 141.67, 138.36, 136.47, 135.35, 132.48, 132.20, 131.71, 130.58, 129.99, 129.34, 129.33, 127.47, 113.56, 113.26, 113.14, 102.23, 61.79. HRMS (ESI) m/z C27H16Cl2N3O4− [M–H]−, calculated: 516.0523, found: 516.0529. Anal. calcd. for C27H17Cl2N3O4: C, 62.56; H, 3.31; N, 8.11; found: C, 62.79; H, 3.52; N, 8.37%.
O, chalcone), 1607, 1545, 1492, 1446, 1384, 1297, 1248, 1205, 1138, 1107, 998, 836, 771, 738, 691, 628. 1H NMR (400.1 MHz, DMSO-d6): δ 8.55 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.87–7.82 (m, 3H, 3CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.63 (d, J = 8.4 Hz, 2H, 2CH), 7.28 (d, J = 8.5 Hz, 2H, 2CH), 7.17 (d, J = 2.0 Hz, 1H, CH), 7.06 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.86 (d, J = 8.5 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.41 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 189.96, 161.27, 160.69, 155.71, 144.75, 143.42, 141.67, 138.36, 136.47, 135.35, 132.48, 132.20, 131.71, 130.58, 129.99, 129.34, 129.33, 127.47, 113.56, 113.26, 113.14, 102.23, 61.79. HRMS (ESI) m/z C27H16Cl2N3O4− [M–H]−, calculated: 516.0523, found: 516.0529. Anal. calcd. for C27H17Cl2N3O4: C, 62.56; H, 3.31; N, 8.11; found: C, 62.79; H, 3.52; N, 8.37%.
4.1.5.13 7-((1-(1-(4-Chlorophenyl)-3-(4-methoxyphenyl)-3-oxoprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20j). Milky solid, mp 239–240 °C, yield: 72%. IR (KBr) (νmax/cm−1): 3146 and 3057, 1749 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1654 (C
O, coumarin), 1654 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1602, 1531, 1475, 1402, 1386, 1309, 1246, 1187, 1109, 1022, 986, 918, 836, 766, 739, 693, 638. 1H NMR (400.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.85 (d, J = 8.7 Hz, 2H, 2CH), 7.78 (s, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.26 (d, J = 8.5 Hz, 2H, 2CH), 7.19 (d, J = 1.9 Hz, 1H, CH), 7.12–7.03 (m, 3H, 3CH), 6.86 (d, J = 8.5 Hz, 2H, 2CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.39 (s, 2H, CH2), 3.86 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 189.17, 163.81, 161.26, 160.70, 155.70, 144.73, 143.32, 139.77, 136.04, 132.65, 132.41, 131.96, 130.73, 129.97, 129.26, 128.85, 127.46, 114.59, 113.54, 113.24, 113.13, 102.21, 61.77, 56.10. HRMS (ESI) m/z C28H19ClN3O5− [M–H]−, calculated: 512.1019, found: 512.1019. Anal. calcd. for C28H20ClN3O5: C, 65.44; H, 3.92; N, 8.18; found: C, 65.57; H, 4.18; N, 8.33%.
O, chalcone), 1602, 1531, 1475, 1402, 1386, 1309, 1246, 1187, 1109, 1022, 986, 918, 836, 766, 739, 693, 638. 1H NMR (400.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.85 (d, J = 8.7 Hz, 2H, 2CH), 7.78 (s, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.26 (d, J = 8.5 Hz, 2H, 2CH), 7.19 (d, J = 1.9 Hz, 1H, CH), 7.12–7.03 (m, 3H, 3CH), 6.86 (d, J = 8.5 Hz, 2H, 2CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.39 (s, 2H, CH2), 3.86 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 189.17, 163.81, 161.26, 160.70, 155.70, 144.73, 143.32, 139.77, 136.04, 132.65, 132.41, 131.96, 130.73, 129.97, 129.26, 128.85, 127.46, 114.59, 113.54, 113.24, 113.13, 102.21, 61.77, 56.10. HRMS (ESI) m/z C28H19ClN3O5− [M–H]−, calculated: 512.1019, found: 512.1019. Anal. calcd. for C28H20ClN3O5: C, 65.44; H, 3.92; N, 8.18; found: C, 65.57; H, 4.18; N, 8.33%.
4.1.5.14 7-((1-(1-(4-Chlorophenyl)-3-oxo-3-(thiophen-2-yl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20k). Milky solid, mp 206–209 °C, yield: 68%. IR (KBr) (νmax/cm−1): 3146 and 3028 (2NH), 1758 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1644 (C
O, coumarin), 1644 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1537, 1495, 1423, 1385, 1309, 1221, 1169, 1138, 1109, 1078, 1034, 933, 889, 764, 733, 692, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.13 (d, J = 4.7 Hz, 1H, CH), 8.09 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.53 (d, J = 3.4 Hz, 1H, CH), 7.27 (d, J = 8.5 Hz, 2H, 2CH), 7.22 (t, J = 4.3 Hz, 1H, CH), 7.16 (d, J = 1.8 Hz, 1H, CH), 7.06 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.87 (d, J = 8.5 Hz, 2H, 2CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.42 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 181.09, 161.20, 160.69, 155.69, 144.74, 143.70, 141.33, 139.56, 137.12, 136.29, 135.33, 132.14, 132.02, 130.65, 129.99, 129.50, 129.32, 127.79, 113.59, 113.27, 113.16, 102.27, 61.72. HRMS (ESI) m/z C25H15ClN3O4S− [M–H]−, calculated: 488.0477, found: 488.0477. Anal. calcd. for C25H16ClN3O4S: C, 61.29; H, 3.29; N, 8.58; found: C, 61.47; H, 3.50; N, 8.82%.
O, chalcone), 1599, 1537, 1495, 1423, 1385, 1309, 1221, 1169, 1138, 1109, 1078, 1034, 933, 889, 764, 733, 692, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.13 (d, J = 4.7 Hz, 1H, CH), 8.09 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.53 (d, J = 3.4 Hz, 1H, CH), 7.27 (d, J = 8.5 Hz, 2H, 2CH), 7.22 (t, J = 4.3 Hz, 1H, CH), 7.16 (d, J = 1.8 Hz, 1H, CH), 7.06 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.87 (d, J = 8.5 Hz, 2H, 2CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.42 (s, 2H, CH2). 13C NMR (100.1 MHz, DMSO-d6): δ 181.09, 161.20, 160.69, 155.69, 144.74, 143.70, 141.33, 139.56, 137.12, 136.29, 135.33, 132.14, 132.02, 130.65, 129.99, 129.50, 129.32, 127.79, 113.59, 113.27, 113.16, 102.27, 61.72. HRMS (ESI) m/z C25H15ClN3O4S− [M–H]−, calculated: 488.0477, found: 488.0477. Anal. calcd. for C25H16ClN3O4S: C, 61.29; H, 3.29; N, 8.58; found: C, 61.47; H, 3.50; N, 8.82%.
4.1.5.15 7-((1-(3-Oxo-3-phenyl-1-(p-tolyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20l). Milky solid, mp 246–248 °C, yield: 75%. IR (KBr) (νmax/cm−1): 3138 and 2922, 1747 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1657 (C
O, coumarin), 1657 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1603, 1588, 1527, 1494, 1426, 1388, 1308, 1256, 1205, 1111, 1034, 989, 923, 829, 742, 699, 646. 1H NMR (400.1 MHz, DMSO-d6): δ 8.53 (s, 1H, CH), 8.04 (d, J = 9.4 Hz, 1H, CH), 7.83 (d, J = 8.8 Hz, 2H, 2CH), 7.77 (s, 1H, CH), 7.70–7.60 (m, 2H, 2CH), 7.55 (t, J = 7.7 Hz, 2H, 2CH), 7.16 (s, 1H, CH), 7.05 (d, J = 8.6 Hz, 1H, CH), 6.97 (d, J = 8.8 Hz, 2H, 2CH), 6.71 (d, J = 8.8 Hz, 2H, 2CH), 6.34 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 2.19 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 191.66, 161.15, 160.48, 155.82, 143.99, 143.02, 141.78, 136.57, 132.57, 132.50, 132.46, 131.10, 130.77, 129.88, 129.43, 129.32, 128.64, 128.48, 113.84, 113.44, 113.00, 102.54, 61.75, 21.90. HRMS (ESI) m/z for C28H20N3O4− [M–H]−, calculated: 462.1459, found: 462.1463. Anal. calcd. for C28H21N3O4: C, 72.56; H, 4.57; N, 9.07; found: C, 72.68; H, 4.74; N, 8.89%.
O, chalcone), 1603, 1588, 1527, 1494, 1426, 1388, 1308, 1256, 1205, 1111, 1034, 989, 923, 829, 742, 699, 646. 1H NMR (400.1 MHz, DMSO-d6): δ 8.53 (s, 1H, CH), 8.04 (d, J = 9.4 Hz, 1H, CH), 7.83 (d, J = 8.8 Hz, 2H, 2CH), 7.77 (s, 1H, CH), 7.70–7.60 (m, 2H, 2CH), 7.55 (t, J = 7.7 Hz, 2H, 2CH), 7.16 (s, 1H, CH), 7.05 (d, J = 8.6 Hz, 1H, CH), 6.97 (d, J = 8.8 Hz, 2H, 2CH), 6.71 (d, J = 8.8 Hz, 2H, 2CH), 6.34 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 2.19 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 191.66, 161.15, 160.48, 155.82, 143.99, 143.02, 141.78, 136.57, 132.57, 132.50, 132.46, 131.10, 130.77, 129.88, 129.43, 129.32, 128.64, 128.48, 113.84, 113.44, 113.00, 102.54, 61.75, 21.90. HRMS (ESI) m/z for C28H20N3O4− [M–H]−, calculated: 462.1459, found: 462.1463. Anal. calcd. for C28H21N3O4: C, 72.56; H, 4.57; N, 9.07; found: C, 72.68; H, 4.74; N, 8.89%.
4.1.5.16 7-((1-(3-(4-Chlorophenyl)-3-oxo-1-(p-tolyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20m). Milky solid, mp 225–227 °C, yield: 83%. IR (KBr) (νmax/cm−1): 3187 and 3024, 1733 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1669 (C
O, coumarin), 1669 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1524, 1488, 1424, 1367, 1323, 1299, 1225, 1110, 1066, 968, 914, 872, 833, 752, 699, 623. 1H NMR (400.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.82 (s, 1H, CH), 7.79 (d, J = 8.2 Hz, 2H, 2CH), 7.68 (d, J = 8.6 Hz, 1H, CH), 7.62 (d, J = 8.2 Hz, 2H, 2CH), 7.16 (s, 1H, CH), 7.06 (d, J = 8.6 Hz, 1H, CH), 7.00 (d, J = 7.1 Hz, 2H, 2CH), 6.69 (d, J = 7.1 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 2.23 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 188.44, 161.30, 160.68, 155.72, 144.78, 142.52, 141.21, 136.82, 132.22, 131.62, 131.51, 131.39, 130.84, 129.98, 129.91, 129.29, 128.73, 127.41, 113.68, 113.21, 113.14, 102.25, 61.85, 21.49. HRMS (ESI) m/z for C28H19ClN3O4− [M–H]−, calculated: 496.1070, found: 496.1072. Anal. calcd. for C28H20ClN3O4: C, 67.54; H, 4.05; N, 8.44; found: C, 67.69; H, 4.20; N, 8.62%.
O, chalcone), 1599, 1524, 1488, 1424, 1367, 1323, 1299, 1225, 1110, 1066, 968, 914, 872, 833, 752, 699, 623. 1H NMR (400.1 MHz, DMSO-d6): δ 8.52 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.82 (s, 1H, CH), 7.79 (d, J = 8.2 Hz, 2H, 2CH), 7.68 (d, J = 8.6 Hz, 1H, CH), 7.62 (d, J = 8.2 Hz, 2H, 2CH), 7.16 (s, 1H, CH), 7.06 (d, J = 8.6 Hz, 1H, CH), 7.00 (d, J = 7.1 Hz, 2H, 2CH), 6.69 (d, J = 7.1 Hz, 2H, 2CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 2.23 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 188.44, 161.30, 160.68, 155.72, 144.78, 142.52, 141.21, 136.82, 132.22, 131.62, 131.51, 131.39, 130.84, 129.98, 129.91, 129.29, 128.73, 127.41, 113.68, 113.21, 113.14, 102.25, 61.85, 21.49. HRMS (ESI) m/z for C28H19ClN3O4− [M–H]−, calculated: 496.1070, found: 496.1072. Anal. calcd. for C28H20ClN3O4: C, 67.54; H, 4.05; N, 8.44; found: C, 67.69; H, 4.20; N, 8.62%.
4.1.5.17 7-((1-(3-(4-Bromophenyl)-3-oxo-1-(p-tolyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20n). Milky solid, mp 287–291 °C, yield: 89%. IR (KBr) (νmax/cm−1): 3148 and 2998, 1748 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1658 (C
O, coumarin), 1658 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1602, 1573, 1518, 1495, 1443, 1385, 1302, 1286, 1211, 1122, 1087, 978, 915, 873, 823, 741, 698, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.53 (s, 1H, CH), 8.03 (d, J = 9.5 Hz, 1H, CH), 7.81 (s, 1H, CH), 7.77 (d, J = 8.8 Hz, 2H, 2CH), 7.74 (d, J = 8.8 Hz, 2H, 2CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.17 (d, J = 2.0 Hz, 1H, CH), 7.07 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.99 (d, J = 8.0 Hz, 2H, 2CH), 6.69 (d, J = 8.0 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.44 (s, 2H, CH2), 2.23 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 190.32, 161.30, 160.70, 155.72, 144.79, 143.50, 143.35, 142.54, 136.01, 132.23, 131.64, 131.39, 130.85, 130.00, 129.91, 128.73, 127.41, 127.18, 113.69, 113.21, 113.13, 102.25, 61.85, 21.50. HRMS (ESI) m/z C28H19BrN3O4− [M–H]−, calculated: 540.0564, found: 540.0563. Anal. calcd. for C28H20BrN3O4: C, 62.00; H, 3.72; N, 7.75; found: C, 62.15; H, 3.98; N, 7.92%.
O, chalcone), 1602, 1573, 1518, 1495, 1443, 1385, 1302, 1286, 1211, 1122, 1087, 978, 915, 873, 823, 741, 698, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.53 (s, 1H, CH), 8.03 (d, J = 9.5 Hz, 1H, CH), 7.81 (s, 1H, CH), 7.77 (d, J = 8.8 Hz, 2H, 2CH), 7.74 (d, J = 8.8 Hz, 2H, 2CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.17 (d, J = 2.0 Hz, 1H, CH), 7.07 (dd, J = 8.6, 2.2 Hz, 1H, CH), 6.99 (d, J = 8.0 Hz, 2H, 2CH), 6.69 (d, J = 8.0 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.44 (s, 2H, CH2), 2.23 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 190.32, 161.30, 160.70, 155.72, 144.79, 143.50, 143.35, 142.54, 136.01, 132.23, 131.64, 131.39, 130.85, 130.00, 129.91, 128.73, 127.41, 127.18, 113.69, 113.21, 113.13, 102.25, 61.85, 21.50. HRMS (ESI) m/z C28H19BrN3O4− [M–H]−, calculated: 540.0564, found: 540.0563. Anal. calcd. for C28H20BrN3O4: C, 62.00; H, 3.72; N, 7.75; found: C, 62.15; H, 3.98; N, 7.92%.
4.1.5.18 7-((1-(3-(4-Methoxyphenyl)-3-oxo-1-(p-tolyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20o). Milky solid, mp 249–252 °C, yield: 72%. IR (KBr) (νmax/cm−1): 3124 and 3028, 1752 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1669 (C
O, coumarin), 1669 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1552, 1494, 1448, 1384, 1308, 1249, 1216, 1176, 1108, 1072, 1033, 968, 922, 847, 788, 739, 698, 637. 1H NMR (400.1 MHz, DMSO-d6): δ 8.51 (s, 1H, CH), 8.03 (d, J = 9.5 Hz, 1H, CH), 7.80 (d, J = 8.7 Hz, 2H, 2CH), 7.76 (s, 1H, CH), 7.68 (d, J = 8.6 Hz, 1H, CH), 7.16 (s, 1H, CH), 7.09 (d, J = 8.7 Hz, 2H, 2CH), 7.01 (d, J = 8.6 Hz, 1H, CH), 6.98 (d, J = 7.5 Hz, 2H, 2CH), 6.72 (d, J = 7.5 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.48 (s, 2H, CH2), 3.68 (s, 3H, CH3), 2.32 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 187.87, 161.79, 160.68, 155.87, 153.66, 144.89, 142.18, 141.15, 139.50, 132.45, 132.13, 130.42, 130.27, 130.00, 129.15, 129.09, 128.47, 127.95, 114.73, 114.09, 113.90, 102.64, 61.44, 55.56, 21.03. HRMS (ESI) m/z C29H22N3O5− [M–H]−, calculated: 492.1565, found: 492.1565. Anal. calcd. for C29H23N3O5: C, 70.58; H, 4.70; N, 8.51; found: C, 70.71; H, 4.98; N, 8.36%.
O, chalcone), 1599, 1552, 1494, 1448, 1384, 1308, 1249, 1216, 1176, 1108, 1072, 1033, 968, 922, 847, 788, 739, 698, 637. 1H NMR (400.1 MHz, DMSO-d6): δ 8.51 (s, 1H, CH), 8.03 (d, J = 9.5 Hz, 1H, CH), 7.80 (d, J = 8.7 Hz, 2H, 2CH), 7.76 (s, 1H, CH), 7.68 (d, J = 8.6 Hz, 1H, CH), 7.16 (s, 1H, CH), 7.09 (d, J = 8.7 Hz, 2H, 2CH), 7.01 (d, J = 8.6 Hz, 1H, CH), 6.98 (d, J = 7.5 Hz, 2H, 2CH), 6.72 (d, J = 7.5 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.48 (s, 2H, CH2), 3.68 (s, 3H, CH3), 2.32 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 187.87, 161.79, 160.68, 155.87, 153.66, 144.89, 142.18, 141.15, 139.50, 132.45, 132.13, 130.42, 130.27, 130.00, 129.15, 129.09, 128.47, 127.95, 114.73, 114.09, 113.90, 102.64, 61.44, 55.56, 21.03. HRMS (ESI) m/z C29H22N3O5− [M–H]−, calculated: 492.1565, found: 492.1565. Anal. calcd. for C29H23N3O5: C, 70.58; H, 4.70; N, 8.51; found: C, 70.71; H, 4.98; N, 8.36%.
4.1.5.19 7-((1-(3-Oxo-3-(thiophen-2-yl)-1-(p-tolyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20p). Milky solid, mp 223–227 °C, yield: 68%. IR (KBr) (νmax/cm−1): 3138 and 2994, 1728 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1676 (C
O, coumarin), 1676 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1606, 1528, 1488, 1414, 1334, 1277, 1239, 1175, 1109, 1031, 989, 903, 833, 743, 711, 662, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.10 (d, J = 4.8 Hz, 1H, CH), 8.05 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.42 (d, J = 3.4 Hz, 1H, CH), 7.20 (t, J = 4.4 Hz, 1H, CH), 7.16 (d, J = 1.5 Hz, 1H, CH), 7.06 (dd, J = 8.6, 1.8 Hz, 1H, CH), 6.99 (d, J = 8.0 Hz, 2H, 2CH), 6.71 (d, J = 8.0 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.44 (s, 2H, CH2), 2.23 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 181.10, 161.22, 160.70, 155.70, 144.78, 143.70, 142.31, 141.49, 141.25, 136.72, 134.79, 130.86, 130.78, 129.99, 129.91, 129.41, 128.82, 127.81, 113.72, 113.21, 113.14, 102.28, 61.76, 21.48. HRMS (ESI) m/z C26H18N3O4S− [M–H]−, calculated: 468.1024, found: 468.1024. Anal. calcd. for C26H19N3O4S: C, 66.51; H, 4.08; N, 8.95; found: C, 66.68; H, 4.24; N, 9.08%.
O, chalcone), 1606, 1528, 1488, 1414, 1334, 1277, 1239, 1175, 1109, 1031, 989, 903, 833, 743, 711, 662, 636. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.10 (d, J = 4.8 Hz, 1H, CH), 8.05 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.42 (d, J = 3.4 Hz, 1H, CH), 7.20 (t, J = 4.4 Hz, 1H, CH), 7.16 (d, J = 1.5 Hz, 1H, CH), 7.06 (dd, J = 8.6, 1.8 Hz, 1H, CH), 6.99 (d, J = 8.0 Hz, 2H, 2CH), 6.71 (d, J = 8.0 Hz, 2H, 2CH), 6.33 (d, J = 9.5 Hz, 1H, CH), 5.44 (s, 2H, CH2), 2.23 (s, 3H, CH3). 13C NMR (100.1 MHz, DMSO-d6): δ 181.10, 161.22, 160.70, 155.70, 144.78, 143.70, 142.31, 141.49, 141.25, 136.72, 134.79, 130.86, 130.78, 129.99, 129.91, 129.41, 128.82, 127.81, 113.72, 113.21, 113.14, 102.28, 61.76, 21.48. HRMS (ESI) m/z C26H18N3O4S− [M–H]−, calculated: 468.1024, found: 468.1024. Anal. calcd. for C26H19N3O4S: C, 66.51; H, 4.08; N, 8.95; found: C, 66.68; H, 4.24; N, 9.08%.
4.1.5.20 7-((1-(1-(4-Methoxyphenyl)-3-oxo-3-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20q). Milky solid, mp 242–247 °C, yield: 80%. IR (KBr) (νmax/cm−1): 3181 and 3028, 1764 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1666 (C
O, coumarin), 1666 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1598, 1538, 1509, 1478, 1417, 1387, 1325, 1242, 1214, 1138, 1138, 1068, 1012, 982, 838, 767, 740, 699, 625. 1H NMR (400.1 MHz, DMSO-d6): δ 8.54 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.79 (d, J = 7.6 Hz, 2H, 2CH), 7.79 (s, 1H, CH), 7.70–7.62 (m, 2H, 2CH), 7.55 (t, J = 7.5 Hz, 1H, CH), 7.18 (d, J = 1.5 Hz, 1H, CH), 7.07 (dd, J = 8.6, 1.6 Hz, 2H, 2CH), 6.80–6.68 (m, 4H, 4CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 3.73 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 191.11, 162.40, 161.33, 160.70, 155.73, 144.75, 143.22, 137.15, 133.08, 133.06, 131.17, 130.31, 129.98, 129.54, 129.15, 127.51, 123.79, 114.91, 113.62, 113.20, 113.12, 102.21, 61.87, 55.93. HRMS (ESI) m/z C28H20N3O5− [M–H]−, calculated: 478.1408, found: 478.1405. Anal. calcd. for C28H21N3O5: C, 70.14; H, 4.41; N, 8.76; found: C, 70.26; H, 4.63; N, 8.93%.
O, chalcone), 1598, 1538, 1509, 1478, 1417, 1387, 1325, 1242, 1214, 1138, 1138, 1068, 1012, 982, 838, 767, 740, 699, 625. 1H NMR (400.1 MHz, DMSO-d6): δ 8.54 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.79 (d, J = 7.6 Hz, 2H, 2CH), 7.79 (s, 1H, CH), 7.70–7.62 (m, 2H, 2CH), 7.55 (t, J = 7.5 Hz, 1H, CH), 7.18 (d, J = 1.5 Hz, 1H, CH), 7.07 (dd, J = 8.6, 1.6 Hz, 2H, 2CH), 6.80–6.68 (m, 4H, 4CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 3.73 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 191.11, 162.40, 161.33, 160.70, 155.73, 144.75, 143.22, 137.15, 133.08, 133.06, 131.17, 130.31, 129.98, 129.54, 129.15, 127.51, 123.79, 114.91, 113.62, 113.20, 113.12, 102.21, 61.87, 55.93. HRMS (ESI) m/z C28H20N3O5− [M–H]−, calculated: 478.1408, found: 478.1405. Anal. calcd. for C28H21N3O5: C, 70.14; H, 4.41; N, 8.76; found: C, 70.26; H, 4.63; N, 8.93%.
4.1.5.21 7-((1-(3-(4-Chlorophenyl)-1-(4-methoxyphenyl)-3-oxoprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20r). Milky solid, mp 256–258 °C, yield: 84%. IR (KBr) (νmax/cm−1): 3137 and 3024, 1743 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1653 (C
O, coumarin), 1653 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1538, 1492, 1385, 1308, 1288, 1215, 1137, 1107, 1034, 987, 944, 877, 838, 769, 656, 620. 1H NMR (400.1 MHz, DMSO-d6): δ 8.54 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.83–7.76 (m, 3H, 3CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.61 (d, J = 8.3 Hz, 2H, 2CH), 7.18 (d, J = 1.4 Hz, 1H, CH), 7.07 (dd, J = 8.6, 1.5 Hz, 1H, CH), 6.82–6.67 (m, 4H, 4CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 3.74 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 190.14, 162.51, 161.32, 160.70, 155.73, 144.75, 143.57, 143.41, 137.85, 135.92, 133.21, 131.44, 130.08, 129.98, 129.26, 127.47, 123.81, 114.93, 113.60, 113.21, 113.13, 102.22, 61.86, 55.94. HRMS (ESI) m/z C28H19ClN3O5− [M–H]−, calculated: 512.1019, found: 512.1018. Anal. calcd. for C28H20ClN3O5: C, 65.44; H, 3.92; N, 8.18; found: C, 65.28; H, 4.09; N, 7.97%.
O, chalcone), 1599, 1538, 1492, 1385, 1308, 1288, 1215, 1137, 1107, 1034, 987, 944, 877, 838, 769, 656, 620. 1H NMR (400.1 MHz, DMSO-d6): δ 8.54 (s, 1H, CH), 8.02 (d, J = 9.4 Hz, 1H, CH), 7.83–7.76 (m, 3H, 3CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.61 (d, J = 8.3 Hz, 2H, 2CH), 7.18 (d, J = 1.4 Hz, 1H, CH), 7.07 (dd, J = 8.6, 1.5 Hz, 1H, CH), 6.82–6.67 (m, 4H, 4CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 5.43 (s, 2H, CH2), 3.74 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 190.14, 162.51, 161.32, 160.70, 155.73, 144.75, 143.57, 143.41, 137.85, 135.92, 133.21, 131.44, 130.08, 129.98, 129.26, 127.47, 123.81, 114.93, 113.60, 113.21, 113.13, 102.22, 61.86, 55.94. HRMS (ESI) m/z C28H19ClN3O5− [M–H]−, calculated: 512.1019, found: 512.1018. Anal. calcd. for C28H20ClN3O5: C, 65.44; H, 3.92; N, 8.18; found: C, 65.28; H, 4.09; N, 7.97%.
4.1.5.22 7-((1-(1,3-Bis(4-methoxyphenyl)-3-oxoprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20s). Milky solid, mp 248–252 °C, yield: 76%. IR (KBr) (νmax/cm−1): 3108 and 2984, 1738 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1663 (C
O, coumarin), 1663 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1539, 1494, 1428, 1388, 1304, 1262, 1206, 1111, 1043, 953, 902, 841, 757, 697, 626. 1H NMR (400.1 MHz, DMSO-d6): δ 8.51 (s, 1H, CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.79 (d, J = 8.7 Hz, 2H, 2CH), 7.73 (s, 1H, CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.18 (d, J = 1.8 Hz, 1H, CH), 7.11–7.03 (m, 3H, 3CH), 6.77–6.70 (m, 4H, 4CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.41 (s, 2H, CH2), 3.86 and 3.73 (2s, 6H, 2OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 189.49, 163.43, 162.14, 161.32, 160.71, 155.72, 144.74, 143.32, 141.85, 132.83, 132.12, 130.26, 129.97, 129.34, 127.52, 123.94, 114.85, 114.51, 113.60, 113.20, 113.11, 102.20, 61.84, 56.04, 55.89. HRMS (ESI) m/z C29H22N3O6− [M–H]−, calculated: 508.1514, found: 508.1514. Anal. calcd. for C29H23N3O6: C, 68.36; H, 4.55; N, 8.25; found: C, 68.61; H, 4.35; N, 8.11%.
O, chalcone), 1599, 1539, 1494, 1428, 1388, 1304, 1262, 1206, 1111, 1043, 953, 902, 841, 757, 697, 626. 1H NMR (400.1 MHz, DMSO-d6): δ 8.51 (s, 1H, CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.79 (d, J = 8.7 Hz, 2H, 2CH), 7.73 (s, 1H, CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.18 (d, J = 1.8 Hz, 1H, CH), 7.11–7.03 (m, 3H, 3CH), 6.77–6.70 (m, 4H, 4CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.41 (s, 2H, CH2), 3.86 and 3.73 (2s, 6H, 2OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 189.49, 163.43, 162.14, 161.32, 160.71, 155.72, 144.74, 143.32, 141.85, 132.83, 132.12, 130.26, 129.97, 129.34, 127.52, 123.94, 114.85, 114.51, 113.60, 113.20, 113.11, 102.20, 61.84, 56.04, 55.89. HRMS (ESI) m/z C29H22N3O6− [M–H]−, calculated: 508.1514, found: 508.1514. Anal. calcd. for C29H23N3O6: C, 68.36; H, 4.55; N, 8.25; found: C, 68.61; H, 4.35; N, 8.11%.
4.1.5.23 7-((1-(1-(4-Methoxyphenyl)-3-oxo-3-(thiophen-2-yl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20t). Milky solid, mp 196–199 °C, yield: 65%. IR (KBr) (νmax/cm−1): 3157 and 3022, 1729 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1655 (C
O, coumarin), 1655 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1598, 1538, 1494, 1425, 1394, 1279, 1235, 1198, 1148, 1074, 999, 960, 878, 830, 751, 689, 629. 1H NMR (400.1 MHz, DMSO-d6): δ 8.59 (s, 1H, CH), 8.11–8.03 (m, 2H, 2CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.35 (d, J = 3.5 Hz, 1H, CH), 7.21–7.14 (m, 2H, 2CH), 7.07 (dd, J = 8.5, 1.9 Hz, 1H, CH), 6.80–6.70 (m, 4H, 4CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.44 (s, 2H, CH2), 3.74 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 180.99, 162.35, 161.23, 160.71, 155.70, 144.74, 143.78, 141.63, 141.40, 136.40, 134.42, 133.12, 129.99, 129.47, 129.34, 127.92, 123.93, 114.92, 113.66, 113.22, 113.14, 102.26, 61.76, 55.92. HRMS (ESI) m/z C26H18N3O5S− [M–H]−, calculated: 484.0973, found: 484.0975. Anal. calcd. for C26H19N3O5S: C, 62.27; H, 3.82; N, 8.38; found: C, 62.55; H, 4.07; N, 8.23%.
O, chalcone), 1598, 1538, 1494, 1425, 1394, 1279, 1235, 1198, 1148, 1074, 999, 960, 878, 830, 751, 689, 629. 1H NMR (400.1 MHz, DMSO-d6): δ 8.59 (s, 1H, CH), 8.11–8.03 (m, 2H, 2CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.67 (d, J = 8.6 Hz, 1H, CH), 7.35 (d, J = 3.5 Hz, 1H, CH), 7.21–7.14 (m, 2H, 2CH), 7.07 (dd, J = 8.5, 1.9 Hz, 1H, CH), 6.80–6.70 (m, 4H, 4CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 5.44 (s, 2H, CH2), 3.74 (s, 3H, OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 180.99, 162.35, 161.23, 160.71, 155.70, 144.74, 143.78, 141.63, 141.40, 136.40, 134.42, 133.12, 129.99, 129.47, 129.34, 127.92, 123.93, 114.92, 113.66, 113.22, 113.14, 102.26, 61.76, 55.92. HRMS (ESI) m/z C26H18N3O5S− [M–H]−, calculated: 484.0973, found: 484.0975. Anal. calcd. for C26H19N3O5S: C, 62.27; H, 3.82; N, 8.38; found: C, 62.55; H, 4.07; N, 8.23%.
4.1.5.24 7-((1-(3-Oxo-3-phenyl-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20u). Milky solid, mp 266–269 °C, yield: 79%. IR (KBr) (νmax/cm−1): 3358 and 2921, 1738 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1672 (C
O, coumarin), 1672 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1528, 1488, 1404, 1394, 1313, 1271, 1233, 1118, 1026, 988, 934, 824, 789, 699, 651. 1H NMR (400.1 MHz, DMSO-d6): δ 8.59 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.85 (d, J = 8.6 Hz, 2H, 2CH), 7.77 (s, 1H, CH), 7.73–7.62 (m, 2H, 2CH), 7.60–7.50 (m, 2H, 2CH), 7.21 (d, J = 1.8 Hz, 1H, CH), 7.05 (dd, J = 8.6, 2.0 Hz, 1H, CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 6.32 (s, 2H, 2CH), 5.37 (s, 2H, CH2), 3.66 and 3.54 (2s, 9H, 3OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 187.77, 161.48, 160.72, 153.55, 153.12, 144.12, 142.13, 140.47, 140.00, 132.27, 131.40, 130.98, 130.83, 130.06, 129.19, 127.88, 126.77, 113.34, 113.20, 113.12, 108.22, 101.86, 61.71, 60.60, 56.58. HRMS (ESI) m/z C30H24N3O7− [M–H]−, calculated: 538.1620, found: 538.1624. Anal. calcd. for C30H25N3O7: C, 66.78; H, 4.67; N, 7.79; found: C, 66.91; H, 4.84; N, 7.86%.
O, chalcone), 1599, 1528, 1488, 1404, 1394, 1313, 1271, 1233, 1118, 1026, 988, 934, 824, 789, 699, 651. 1H NMR (400.1 MHz, DMSO-d6): δ 8.59 (s, 1H, CH), 8.02 (d, J = 9.5 Hz, 1H, CH), 7.85 (d, J = 8.6 Hz, 2H, 2CH), 7.77 (s, 1H, CH), 7.73–7.62 (m, 2H, 2CH), 7.60–7.50 (m, 2H, 2CH), 7.21 (d, J = 1.8 Hz, 1H, CH), 7.05 (dd, J = 8.6, 2.0 Hz, 1H, CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 6.32 (s, 2H, 2CH), 5.37 (s, 2H, CH2), 3.66 and 3.54 (2s, 9H, 3OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 187.77, 161.48, 160.72, 153.55, 153.12, 144.12, 142.13, 140.47, 140.00, 132.27, 131.40, 130.98, 130.83, 130.06, 129.19, 127.88, 126.77, 113.34, 113.20, 113.12, 108.22, 101.86, 61.71, 60.60, 56.58. HRMS (ESI) m/z C30H24N3O7− [M–H]−, calculated: 538.1620, found: 538.1624. Anal. calcd. for C30H25N3O7: C, 66.78; H, 4.67; N, 7.79; found: C, 66.91; H, 4.84; N, 7.86%.
4.1.5.25 7-((1-(3-(4-Chlorophenyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20v). Milky solid, mp 302–305 °C, yield: 84%. IR (KBr) (νmax/cm−1): 3188 and 3023, 1729 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1658 (C
O, coumarin), 1658 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1600, 1578, 1509, 1473, 1427, 1358, 1316, 1247, 1204, 1123, 1080, 1015, 830, 799, 743, 697, 626. 1H NMR (400.1 MHz, DMSO-d6): δ 8.61 (s, 1H, CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.89–7.75 (m, 3H, 3CH), 7.66 (d, J = 8.5 Hz, 1H, CH), 7.60 (d, J = 7.8 Hz, 2H, 2CH), 7.20 (s, 1H, CH), 7.03 (d, J = 8.6 Hz, 1H, CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 6.23 (s, 2H, 2CH), 5.38 (s, 2H, CH2), 3.67 and 3.54 (2s, 9H, 3OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 191.11, 161.49, 160.67, 155.81, 153.13, 144.75, 143.44, 143.32, 140.76, 136.95, 133.26, 131.52, 130.06, 129.66, 129.24, 127.86, 126.59, 113.33, 113.20, 113.13, 108.43, 101.88, 61.73, 60.61, 56.11. HRMS (ESI) m/z C30H23ClN3O7− [M–H]−, calculated: 572.1230, found: 572.1234. Anal. calcd. for C30H24ClN3O7: C, 62.78; H, 4.21; N, 7.32; found: C, 62.93; H, 4.04; N, 7.55%.
O, chalcone), 1600, 1578, 1509, 1473, 1427, 1358, 1316, 1247, 1204, 1123, 1080, 1015, 830, 799, 743, 697, 626. 1H NMR (400.1 MHz, DMSO-d6): δ 8.61 (s, 1H, CH), 8.01 (d, J = 9.5 Hz, 1H, CH), 7.89–7.75 (m, 3H, 3CH), 7.66 (d, J = 8.5 Hz, 1H, CH), 7.60 (d, J = 7.8 Hz, 2H, 2CH), 7.20 (s, 1H, CH), 7.03 (d, J = 8.6 Hz, 1H, CH), 6.32 (d, J = 9.5 Hz, 1H, CH), 6.23 (s, 2H, 2CH), 5.38 (s, 2H, CH2), 3.67 and 3.54 (2s, 9H, 3OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 191.11, 161.49, 160.67, 155.81, 153.13, 144.75, 143.44, 143.32, 140.76, 136.95, 133.26, 131.52, 130.06, 129.66, 129.24, 127.86, 126.59, 113.33, 113.20, 113.13, 108.43, 101.88, 61.73, 60.61, 56.11. HRMS (ESI) m/z C30H23ClN3O7− [M–H]−, calculated: 572.1230, found: 572.1234. Anal. calcd. for C30H24ClN3O7: C, 62.78; H, 4.21; N, 7.32; found: C, 62.93; H, 4.04; N, 7.55%.
4.1.5.26 7-((1-(3-(4-Methoxyphenyl)-3-oxo-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20w). Milky solid, mp 286–288 °C, yield: 79%. IR (KBr) (νmax/cm−1): 3168 and 3049, 1745 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1664 (C
O, coumarin), 1664 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1563, 1522, 1486, 1432, 1368, 1293, 1243, 1217, 1116, 1030, 989, 922, 866, 826, 776, 744, 693, 622. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.03 (d, J = 9.3 Hz, 1H, CH), 7.88 (s, 1H, CH), 7.83 (d, J = 8.2 Hz, 2H, 2CH), 7.68 (d, J = 8.6 Hz, 1H, CH), 7.14 (s, 1H, CH), 7.10 (d, J = 8.2 Hz, 2H, 2CH), 7.03 (d, J = 8.6 Hz, 1H, CH), 6.31 (d, J = 9.3 Hz, 1H, CH), 6.18 (s, 2H, 2CH), 5.31 (s, 2H, CH2), 3.71 and 3.61 (2s, 12H, 4OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 187.58, 161.74, 160.52, 155.31, 153.98, 153.14, 144.56, 143.61, 143.16, 140.32, 132.09, 131.04, 130.15, 129.42, 129.11, 127.73, 114.53, 113.85, 113.57, 113.29, 108.13, 101.72, 61.64, 60.28, 56.54, 55.29. HRMS (ESI) m/z C31H26N3O8− [M–H]−, calculated: 568.1725, found: 568.1725. Anal. calcd. for C31H27N3O8: C, 65.37; H, 4.78; N, 7.38; found: C, 65.49; H, 4.94; N, 7.26%.
O, chalcone), 1599, 1563, 1522, 1486, 1432, 1368, 1293, 1243, 1217, 1116, 1030, 989, 922, 866, 826, 776, 744, 693, 622. 1H NMR (400.1 MHz, DMSO-d6): δ 8.58 (s, 1H, CH), 8.03 (d, J = 9.3 Hz, 1H, CH), 7.88 (s, 1H, CH), 7.83 (d, J = 8.2 Hz, 2H, 2CH), 7.68 (d, J = 8.6 Hz, 1H, CH), 7.14 (s, 1H, CH), 7.10 (d, J = 8.2 Hz, 2H, 2CH), 7.03 (d, J = 8.6 Hz, 1H, CH), 6.31 (d, J = 9.3 Hz, 1H, CH), 6.18 (s, 2H, 2CH), 5.31 (s, 2H, CH2), 3.71 and 3.61 (2s, 12H, 4OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 187.58, 161.74, 160.52, 155.31, 153.98, 153.14, 144.56, 143.61, 143.16, 140.32, 132.09, 131.04, 130.15, 129.42, 129.11, 127.73, 114.53, 113.85, 113.57, 113.29, 108.13, 101.72, 61.64, 60.28, 56.54, 55.29. HRMS (ESI) m/z C31H26N3O8− [M–H]−, calculated: 568.1725, found: 568.1725. Anal. calcd. for C31H27N3O8: C, 65.37; H, 4.78; N, 7.38; found: C, 65.49; H, 4.94; N, 7.26%.
4.1.5.27 7-((1-(3-Oxo-3-(thiophen-2-yl)-1-(3,4,5-trimethoxyphenyl)prop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20x). Milky solid, mp 249–251 °C, yield: 69%. IR (KBr) (νmax/cm−1): 3146 and 2998, 1749 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, coumarin), 1673 (C
O, coumarin), 1673 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, chalcone), 1599, 1556, 1494, 1437, 1384, 1312, 1258, 1204, 1118, 1065, 999, 954, 878, 833, 796, 735, 686, 624. 1H NMR (400.1 MHz, DMSO-d6): δ 8.66 (s, 1H, CH), 8.12 (d, J = 4.8 Hz, 1H, CH), 8.09 (s, 1H, CH), 8.01 (d, J = 9.4 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.47 (d, J = 3.4 Hz, 1H, CH), 7.24 (t, J = 3.1 Hz, 1H, CH), 7.17 (d, J = 1.7 Hz, 1H, CH), 7.03 (dd, J = 8.6, 2.0 Hz, 1H, CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 6.26 (s, 2H, 2CH), 5.39 (s, 2H, CH2), 3.67 and 3.56 (2s, 9H, 3OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 181.07, 161.41, 160.73, 155.77, 153.14, 144.76, 141.64, 141.43, 140.72, 136.68, 134.84, 130.73, 130.07, 129.96, 129.45, 128.26, 126.70, 113.37, 113.21, 113.14, 108.43, 101.91, 61.65, 60.63, 56.10. HRMS (ESI) m/z C28H22N3O7S− [M–H]−, calculated: 544.1184, found: 544.1184. Anal. calcd. for C28H23N3O7S: C, 61.64; H, 4.25; N, 7.70; found: C, 61.75; H, 4.42; N, 7.89%.
O, chalcone), 1599, 1556, 1494, 1437, 1384, 1312, 1258, 1204, 1118, 1065, 999, 954, 878, 833, 796, 735, 686, 624. 1H NMR (400.1 MHz, DMSO-d6): δ 8.66 (s, 1H, CH), 8.12 (d, J = 4.8 Hz, 1H, CH), 8.09 (s, 1H, CH), 8.01 (d, J = 9.4 Hz, 1H, CH), 7.66 (d, J = 8.6 Hz, 1H, CH), 7.47 (d, J = 3.4 Hz, 1H, CH), 7.24 (t, J = 3.1 Hz, 1H, CH), 7.17 (d, J = 1.7 Hz, 1H, CH), 7.03 (dd, J = 8.6, 2.0 Hz, 1H, CH), 6.32 (d, J = 9.4 Hz, 1H, CH), 6.26 (s, 2H, 2CH), 5.39 (s, 2H, CH2), 3.67 and 3.56 (2s, 9H, 3OCH3). 13C NMR (100.1 MHz, DMSO-d6): δ 181.07, 161.41, 160.73, 155.77, 153.14, 144.76, 141.64, 141.43, 140.72, 136.68, 134.84, 130.73, 130.07, 129.96, 129.45, 128.26, 126.70, 113.37, 113.21, 113.14, 108.43, 101.91, 61.65, 60.63, 56.10. HRMS (ESI) m/z C28H22N3O7S− [M–H]−, calculated: 544.1184, found: 544.1184. Anal. calcd. for C28H23N3O7S: C, 61.64; H, 4.25; N, 7.70; found: C, 61.75; H, 4.42; N, 7.89%.
4.2 α-Glucosidase inhibition assay
α-Glucosidase enzyme (EC3.2.1.20, Saccharomyces cerevisiae, 20 U mg−1) and substrate (p-nitrophenyl glucopyranoside) were purchased from Sigma-Aldrich. Enzyme was prepared in potassium phosphate buffer (pH 6.8, 50 mM), as well as coumarin–chalcone–triazole hybrids 17, 18, 19, and 20a–20x were dissolved in DMSO (10% final concentration). The various concentrations of these compounds (20 mL), enzyme solution (20 mL) and potassium phosphate buffer (135 mL) were added in the 96-well plate and incubated at 37 °C for 10 min. Afterwards, the substrate (25 mL, 4 mM) was added to the mentioned mixture and allowed to incubate at 37 °C for 20 min. Finally, the change in absorbance was measured at 405 nm by using spectrophotometer (Gen5, Power wave xs2, BioTek, America). The percentage of enzyme inhibition was calculated using eqn (1) and IC50 values were obtained from non-linear regression curve using the Logit method.| % Inhibition = [(Abscontrol − Abssample)/Abscontrol] × 1006 | (7) |
4.3 Kinetic studies
The kinetic analysis was performed for the most potent derivative 20q to reveal the inhibition mode against α-glucosidase. The 20 mL of enzyme solution (1 U mL−1) was incubated with different concentrations (0, 3.1, 6.2, and 12.4 µM) of this compound for 15 min at 30 °C. Afterwards, various concentrations of substrate (p-nitrophenyl glucopyranoside, 1 to 10 mM) was added to measure the change of absorbance for 20 min at 405 nm by using spectrophotometer (Gen5, Power wave xs2, BioTek, America).In the presence of a competitive inhibitor, Km increases while Vmax does not change. Michaelis–Menten saturation curve for an enzyme reaction shows the relation between the substrate concentration and reaction rate as bellow:
 | (8) |
According to Michaelis–Menten graph, Kmapp is also defined as:
 | (9) |
[I] is the concentration of inhibitor.
Lineweaver Burk plot that provides a useful graphical method for analysis of the Michaelis–Menten is represented as:
 | (10) |
Therefore, the slope of Lineweaver Burk plot is equal to:
 | (11) |
The Kmapp value is calculated by eqn (5):
 | (12) |
Therefore, from replot of Kmapp vs. [I], eqn (7) can be used for the calculation of KI:68,69
 | (13) |
4.4 Fluorescence spectroscopy measurements
This assay was carried out for the most potent derivative 20q to measure the fluorescence intensity. To this aim, different solutions containing different concentrations (0 to 1.0 µM) of the inhibitor and α-glucosidase (3 mL, 0.1 U mL−1) were held for 10 min to equilibrate before measurements. Moreover, the fluorescence of the buffer containing compound 20q in the absence of the enzyme were subtracted as the background fluorescence. Afterwards, at the excitation wavelength of 280 nm, the fluorescence emission spectra were measured from 300 to 450 nm using a Synergy HTX multi-mode reader (Biotek Instruments, Winooski, VT, USA) equipped with a 1.0 cm quartz cell holder.704.5 Deep learning model
Deep learning has markedly advanced computational drug discovery by cutting both time and cost while providing highly expressive models for predicting molecular activity. In this study, we developed a transfer-learning-based predictive framework to deepen understanding of α-glucosidase inhibitors. Starting from a BERT architecture originally created for natural-language tasks, we reinterpreted its tokenization and attention mechanisms to operate on SMILES strings so the model could learn contextualized, chemically meaningful substructures instead of isolated descriptors. The model was first pretrained (self-supervised) on a large corpus of unlabeled SMILES to learn general chemical grammar, then fine-tuned on a labeled dataset of molecules with measured α-glucosidase inhibition to optimize predictions for this specific bioactivity. During fine-tuning we applied standard practices—data augmentation of SMILES, stratified splitting, and early stopping—to improve robustness and avoid overfitting. Performance was evaluated using hold-out testing and metrics appropriate for imbalanced bioactivity data (ROC-AUC, precision-recall, and calibration), and model attention maps were inspected to provide interpretable clues about substructures driving predictions. By leveraging transfer learning, our approach reduces dependence on large labeled datasets and yields a flexible, interpretable tool for prioritizing candidate inhibitors and guiding medicinal-chemistry hypotheses.714.6 Molecular docking
Molecular docking study was performed with Schrodinger Suite 14.0, utilizing the Glide tool to assess interactions between potential drug candidates and the target protein (PDB ID: 3 A4A). Initially, the protein structure was downloaded from the Protein Data Bank (PDB) and prepared via the Protein Preparation Wizard This preparation process included adding hydrogen atoms, defining bond orders, and optimizing the hydrogen bonding network to achieve a realistic protein conformation. Ligand structures were processed through the LigPrep module to generate ionization states at a physiological pH of 7.0 ± 2.0. Docking grids were established around the active site of 3A4A, using a 36 Å × 36 Å × 36 Å grid box centered at (x = 14.5, y = −11.1, z = 18.2). Docking was performed using both standard precision (SP) and extra precision (XP) modes, with poses ranked according to GlideScore, which estimates ligand binding affinity. Post-docking minimization steps were conducted to refine the poses, and the interactions were examined through the XP Visualizer.4.7 Molecular dynamics
The molecular dynamics (MD) simulation was performed using Desmond v2021.1 from the Schrödinger 2021-1 suite, utilizing GPU acceleration with an NVIDIA RTX 3060. Maestro (for academic use only) facilitated system setup and data analysis. The protein–ligand complex was embedded in an orthorhombic box with TIP3P water molecules under periodic boundary conditions. To stabilize the environment and simulate physiological conditions, counter-ions and a 0.15 M NaCl solution were introduced to neutralize the system. The MD procedure encompassed minimization, equilibration, and production stages. The minimization phase relaxed the system for 2500 steps using the steepest descent algorithm. During equilibration, the temperature was progressively increased from 0 K to 300 K with a small restraining force on the enzyme. Production simulations followed, applying the NPT ensemble to sustain a temperature of 300 K and a pressure of 1.01325 bar. A Nose–Hoover chain thermostat with a 1.0 ps relaxation time and a Martyna–Tobias–Klein barostat with a 2.0 ps relaxation time were used, while long-range electrostatics were calculated with the particle-mesh Ewald method using a 9.0 Å cutoff. The production run spanned 200 ns, with frames recorded every 1 ps to track dynamic behavior and structural variations in the system.4.8 Cytotoxicity assay
The MTT method was used to evaluate the cytotoxicity of 7-((1-(1-(4-methoxyphenyl)-3-oxo-3-phenylprop-1-en-2-yl)-1H-1,2,3-triazol-4-yl)methoxy)-2H-chromen-2-one (20q) on the MCF-7 (breast cancer cell line) and HT-29 (human colon adenocarcinoma cell line) according with our previous studies.72 Cells were seeded at a density of 5 × 103 per well in 96-well plates and incubated in a humidified atmosphere with 5% CO2 at 37 °C for 24 h. Various concentrations of compound 20q (up to 100 µM) were added to the cells in a medium containing 0.1% DMSO. The solvent served as the negative control. Following 72 h of treatment, a solution of 5 mg mL−1 of MTT in PBS was added to each well. The cells were incubated with MTT for 3 h at 37 °C before the excess MTT was removed. Formazan crystals in each well were dissolved in 150 µL of DMSO. The absorbance of the dissolved formazan was then measured at 570 and 630 nm using a Biotek Epoch™ microplate reader.4.9 In vivo antidiabetic study
Adult male albino Wistar rats (240–260 g) were procured from the Experimental Animal Laboratory, School of Pharmacy, Tehran University of Medical Sciences, Tehran, Iran. The animals were maintained under standard environmental conditions (25 ± 2 °C, relative humidity 65 ± 5%, and a 12 h light/dark cycle), with free access to standard pellet diet and water. All animal experiments were performed in strict compliance with the national and institutional guidelines for the care and use of laboratory animals. The study protocols were reviewed and approved by the Animal Ethics Committee of Tehran University of Medical Sciences, under approval number IR.TUMS.BLC.1404.079.Throughout the study, all efforts were made to minimize animal suffering and to use the minimum number of animals necessary to achieve statistical significance. The housing, feeding, and environmental enrichment conditions were in accordance with standard laboratory animal welfare protocols.
4.10 Acute oral toxicity study
The acute oral toxicity of compound 20q was determined using Lorke's method. Compound 20q was administrated orally through gastric gavage at different doses ranging from 10 to 1000 mg kg−1 body weight (BW). Subsequently, animals were carefully monitored over a period of two weeks for any signs of toxicity and mortality, leading to estimate the lethal dose (LD50).734.11 Induction of diabetes
Diabetes was induced in rats using a high-fat diet and streptozotocin (STZ).74 To this aim, animals in diabetic groups (2 to 6) were fed with high-fat diet for four consecutive weeks. After an overnight fast, a single dose of STZ (40 mg kg−1 BW) dissolved in freshly-prepared cold citrate buffer (0.1 M, pH 4.5) was injected intraperitoneally. To prevent hypoglycemia, animals received a 10% glucose solution immediately after STZ injection. It must be noted that non-diabetic control rats (healthy animals, group 1) received an intraperitoneal injection of buffer. After 72 hours, fasting blood glucose levels were measured using a glucometer (Accu–Chek, Roche), and animals with glucose levels upper than 250 mg dL−1 were considered diabetic and selected for further experimentation. During the study period, all animals had free access to food and water, except before measuring the blood glucose.4.12 Experimental design
Rats were randomly divided into six groups, each consisting of six animals (n = 6):• Group 1: normal healthy rats, serving as the non-diabetic control group.
• Group 2: diabetic rats that received no treatment, serving as the diabetic control group.
Groups 3–5: diabetic rats treated orally with scalar doses of compound 20q at 8, 4, and 2 mg kg−1 BW, respectively, once daily for 28 consecutive days.
• Group 3: compound 20q (8 mg kg−1 BW)
• Group 4: compound 20q (4 mg kg−1 BW)
• Group 5: compound 20q (2 mg kg−1 BW)
• Group 6: diabetic rats treated orally with acarbose (10 mg kg−1 BW) once daily for 28 days, serving as the positive control group.
All treatments were administered orally via gavage on a daily basis. Fasting blood glucose levels were measured weekly by collecting fresh blood from the tail veins of the rats to evaluate the hyperglycemic effect of the compound 20q in comparison with acarbose.
4.13 Oral glucose tolerance test (OGTT)
To evaluate the effect of compound 20q on postprandial glucose levels, an oral glucose tolerance test (OGTT) was performed on day 28 of the study. Following an overnight fast, FBG levels were measured, and rats from each group received their respective treatments orally via gavage. After 10 minutes, a sucrose solution (2 g kg−1 BW) was administered orally. Blood samples were collected from the tail vein at 0, 15, 30, 60, 90, and 120 minutes following sucrose administration. Subsequently, the blood glucose levels were measured using a glucometer to assess glucose tolerance and to compare the anti-hyperglycemic potential of compound 20q with that of the standard drug (acarbose).4.14 Histological evaluation
On the final day of the study, the rats were anesthetized via intraperitoneal injection of ketamine (5 mg per 100 g body weight) and subsequently euthanized. The liver was carefully excised and fixed in 10% neutral buffered formalin for histological processing. The preserved tissues were embedded in paraffin, sliced into sections approximately 5 µm thick, stained with hematoxylin and eosin (H&E), and examined microscopically for histopathological assessment.Author contributions
Loghman Firoozpour and Alireza Foroumadi: conceptualization, project administration, resources, and supervision; Fariba Peytam, Maryam Norouzbahari, Mahsa Akbari, Hayrettin Ozan Gülcan, and Mahfam Moradi: data curation, formal analysis, investigation, and writing – original draft; Somayeh Mojtabavi, Mohammad Ali Faramarzi, and Vahid Sheibani: data curation and formal analysis; Fahimeh Ghasemi and Mohammadreza Torabi: software and visualization; Maliheh Barazandeh Tehrani: validation and writing – review & editing.Conflicts of interest
The authors declare that they have no conflicts of interest.Data availability
The authors confirm that the data supporting the finding of this study are available within the manuscript and Supplementary information (SI). Supplementary information: the spectra images of the final compounds are provided in the SI. See DOI: https://doi.org/10.1039/d5ra07254a.Acknowledgements
This work was supported and funded by grants No. 1402-1-104-64961 and 1404-1-263-91722 from the research council of Tehran University of Medical Sciences, Tehran, Iran.References
- S. A. Antar, N. A. Ashour, M. Sharaky, M. Khattab, N. A. Ashour and R. T. Zaid, et al., Diabetes mellitus: Classification, mediators, and complications; A gate to identify potential targets for the development of new effective treatments, Biomed. Pharmacother., 2023, 168, 115734 CrossRef CAS PubMed
.
- H. Sun, P. Saeedi, S. Karuranga, M. Pinkepank, K. Ogurtsova and B. B. Duncan, et al., IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045, Diabetes Res. Clin. Pract., 2022, 183, 109119 CrossRef PubMed
.
- U. Hossain, A. K. Das, S. Ghosh and P. C. Sil, An overview on the role of bioactive α-glucosidase inhibitors in ameliorating diabetic complications, Food Chem. Toxicol., 2020, 145, 111738 CrossRef CAS PubMed
.
- M. Roden and G. I. Shulman, The integrative biology of type 2 diabetes, Nature, 2019, 576(7785), 51–60 CrossRef CAS PubMed
.
- R. I. Holt, J. H. DeVries, A. Hess-Fischl, I. B. Hirsch, M. S. Kirkman and T. Klupa, et al., The management of type 1 diabetes in adults. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD), Diabetes Care, 2021, 44(11), 2589–2625 CrossRef CAS PubMed
.
- I. M. El-Kebbi, N. H. Bidikian, L. Hneiny and M. P. Nasrallah, Epidemiology of type 2 diabetes in the Middle East and North Africa: Challenges and call for action, World J. Diabetes, 2021, 12(9), 1401 CrossRef PubMed
.
- D. M. Nathan, J. B. Buse, M. B. Davidson, E. Ferrannini, R. R. Holman and R. Sherwin, et al., Medical management of hyperglycemia in type 2 diabetes: a consensus algorithm for the initiation and adjustment of therapy: a consensus statement of the American Diabetes Association and the European Association for the Study of Diabetes, Diabetes Care, 2009, 32(1), 193–203 CrossRef CAS PubMed
.
- J. J. DiNicolantonio, J. Bhutani and J. H. O'Keefe, Acarbose: safe and effective for lowering postprandial hyperglycaemia and improving cardiovascular outcomes, Open heart, 2015, 2(1), e000327 CrossRef PubMed
.
- M. Adib, F. Peytam, R. Shourgeshty, M. Mohammadi-Khanaposhtani, M. Jahani and S. Imanparast, et al., Design and synthesis of new fused carbazole-imidazole derivatives as anti-diabetic agents: In vitro α-glucosidase inhibition, kinetic, and in silico studies, Bioorg. Med. Chem. Lett., 2019, 29(5), 713–718 CrossRef CAS PubMed
.
- F. Peytam, M. Adib, R. Shourgeshty, M. Mohammadi-Khanaposhtani, M. Jahani and S. Imanparast, et al., Design and synthesis of new imidazo [1, 2-b] pyrazole derivatives, in vitro α-glucosidase inhibition, kinetic and docking studies, Mol. Diversity, 2020, 24, 69–80 CrossRef CAS PubMed
.
- A. Mushtaq, U. Azam, S. Mehreen and M. M. Naseer, Synthetic α-glucosidase inhibitors as promising anti-diabetic agents: Recent developments and future challenges, Eur. J. Med. Chem., 2023, 249, 115119 CrossRef CAS PubMed
.
- S. N. M. Radzuan, L. Phongphane, M. H. A. Bakar, M. T. C. Omar, N. S. N. Shahril and U. Supratman, et al., Synthesis, biological activities, and evaluation molecular docking-dynamics studies of new phenylisoxazole quinoxalin-2-amine hybrids as potential α-amylase and α-glucosidase inhibitors, RSC Adv., 2024, 14(11), 7684–7698 RSC
.
- M. I. Abdjan, N. S. Aminah, A. N. Kristanti, I. Siswanto, B. Ilham and A. P. Wardana, et al., Structure-based approach: molecular insight of pyranocumarins against α-glucosidase through computational studies, RSC Adv., 2023, 13(6), 3438–3447 RSC
.
- R. Zhang, Y. Zhang, G. Huang, X. Xin, L. Tang and H. Li, et al., Chemical synthesis, inhibitory activity and molecular mechanism of 1-deoxynojirimycin–chrysin as a potent α-glucosidase inhibitor, RSC Adv., 2021, 11(61), 38703–38711 RSC
.
- F. Peytam, M. Adib, R. Shourgeshty, M. Mohammadi-Khanaposhtani, M. Jahani and S. Imanparast, et al., Synthesis and biological evaluation of new dihydroindolizino
[8, 7-b] indole derivatives as novel α-glucosidase inhibitors, J. Mol. Struct., 2021, 1224, 129290 CrossRef CAS
.
- N. Nivetha, R. M. Martiz, S. M. Patil, R. Ramu, S. Sreenivasa and S. Velmathi, Benzodioxole grafted spirooxindole pyrrolidinyl derivatives: Synthesis, characterization, molecular docking and anti-diabetic activity, RSC Adv., 2022, 12(37), 24192–24207 RSC
.
- H. Z. Tariq, A. Saeed, S. Ullah, N. Fatima, S. A. Halim and A. Khan, et al., Synthesis of novel coumarin–hydrazone hybrids as α-glucosidase inhibitors and their molecular docking studies, RSC Adv., 2023, 13(37), 26229–26238 RSC
.
- M. S. Ayoup, N. Khaled, H. Abdel-Hamid, D. A. Ghareeb, S. A. Nasr and A. Omer, et al., Novel sulfonamide derivatives as multitarget antidiabetic agents: design, synthesis, and biological evaluation, RSC Adv., 2024, 14(11), 7664–7675 RSC
.
- A. R. Pasha, S. Ullah, A. Khan, S. A. Halim, J. Hussain and T. Rehman, et al., Synthesis, in vitro and in silico study of novel 1, 3-diphenylurea derived Schiff bases as competitive α-glucosidase inhibitors, RSC Adv., 2024, 14(40), 29288–29300 RSC
.
- A. Saeed, A. Ahmed, M. B. Haider, H. Ismail, K. Hayat and G. Shabir, et al., Novel pyrazoline linked acyl thiourea pharmacophores as antimicrobial, urease, amylase and α-glucosidase inhibitors: design, synthesis, SAR and molecular docking studies, RSC Adv., 2024, 14(2), 1018–1033 RSC
.
- R. R. Mudireddy, R. Gundla, B. B. Shaik, A. Bodapati, P. Mahesh and S. S. Naidu, et al., Synthesis, characterization, and in vitro and in silico α-glucosidase inhibitory evolution of novel N′-(2-cyclopentyl-2-phenylacetyl) cinnamohydrazide derivatives, RSC Adv., 2025, 15(22), 17118–17129 RSC
.
- S. Mehreen, M. I. Ali, S. tul Muntha, M. Zia, A. Ullah and S. Ullah, et al., Synthesis, structural insights and bio-evaluation of N-phenoxyethylisatin hydrazones as potent α-glucosidase inhibitors, RSC Adv., 2025, 15(19), 14717–14729 RSC
.
- C. Viegas-Junior, A. Danuello, V. da Silva Bolzani, E. J. Barreiro and C. A. M. Fraga, Molecular hybridization: a useful tool in the design of new drug prototypes, Curr. Med. Chem., 2007, 14(17), 1829–1852 CrossRef CAS PubMed
.
- A. H. Alkhzem, T. J. Woodman and I. S. Blagbrough, Design and synthesis of hybrid compounds as novel drugs and medicines, RSC Adv., 2022, 12(30), 19470–19484 RSC
.
- R. Carlucci, M.-N. Lisa and G. R. Labadie, 1, 2, 3-Triazoles in Biomolecular Crystallography: A Geometrical Data-Mining Approach, J. Med. Chem., 2023, 66(21), 14377–14390 CrossRef CAS PubMed
.
- G. Moya-Alvarado, O. Yañez, N. Morales, A. González-González, C. Areche and M. T. Núñez, et al., Coumarin-chalcone hybrids as inhibitors of MAO-B: Biological activity and in silico studies, Molecules, 2021, 26(9), 2430 CrossRef CAS PubMed
.
- M. Mohammadnia, Z. Emamgholipour, F. Peytam, M. Nikbakhtzadeh, S. Hosseindoost and S. B. Alsaeed, et al., Coumarin-Chalcone derivatives as promising antioxidant agents targeting ischemia/reperfusion injury through Nrf2 pathway activation, Bioorg. Chem., 2025, 108790 CrossRef CAS PubMed
.
- S. V. Rodriguez, R. Guíñez, M. Matos, C. Azar, J. Maya and L. Santana, et al., Synthesis and trypanocidal properties of new coumarin-chalcone derivatives, Med. Chem., 2015, 5, 173–177 Search PubMed
.
- N. A. Alhakamy, M. Saquib, K. M. F. Sanobar, W. A. Ansari and D. O. Arif, et al., Natural product-inspired synthesis of coumarin–chalcone hybrids as potential anti-breast cancer agents, Front. Pharmacol, 2023, 14, 1231450 CrossRef CAS PubMed
.
- R. Pingaew, A. Saekee, P. Mandi, C. Nantasenamat, S. Prachayasittikul and S. Ruchirawat, et al., Synthesis, biological evaluation and molecular docking of novel chalcone–coumarin hybrids as anticancer and antimalarial agents, Eur. J. Med. Chem., 2014, 85, 65–76 CrossRef CAS PubMed
.
- K. Patel, C. Karthikeyan, N. Hari Narayana Moorthy, G. S. Deora, V. R. Solomon and H. Lee, et al., Design, synthesis and biological evaluation of some novel 3-cinnamoyl-4-hydroxy-2H-chromen-2-ones as antimalarial agents, Med. Chem. Res., 2012, 21, 1780–1784 CrossRef CAS
.
- A. Kumar and S. Kumar, Coumarin-chalcone hybrids for biological potentials: a strategy of molecular hybridization for drug design, Int. J. Pharm. Sci. Rev. Res., 2020, 64(2), 146–151 CrossRef CAS
.
- K. V. Sashidhara, M. Kumar, R. K. Modukuri, R. Sonkar, G. Bhatia and A. Khanna, et al., Synthesis and anti-inflammatory activity of novel biscoumarin–chalcone hybrids, Bioorg. Med. Chem. Lett., 2011, 21(15), 4480–4484 CrossRef CAS PubMed
.
- M. Taha, S. A. A. Shah, M. Afifi, S. Imran, S. Sultan and F. Rahim, et al., Synthesis, α-glucosidase inhibition and molecular docking study of coumarin based derivatives, Bioorg. Chem., 2018, 77, 586–592 CrossRef CAS PubMed
.
- P. Singh, N. Ngcoya, R. Mopuri, N. Kerru, N. Manhas and O. Ebenezer, et al., α-Glucosidase inhibition, antioxidant and docking studies of hydroxycoumarins and their mono and bis O-alkylated/acetylated analogs, Lett. Drug Des. Discovery, 2018, 15(2), 127–135 CrossRef CAS
.
- Y. Hu, B. Wang, J. Yang, T. Liu, J. Sun and X. Wang, Synthesis and biological evaluation of 3-arylcoumarin derivatives as potential anti-diabetic agents, J. Enzyme Inhib. Med. Chem., 2019, 34(1), 15–30 CrossRef CAS PubMed
.
- E. Menteşe, N. Baltaş and O. Bekircan, Synthesis and kinetics studies of N′-(2-(3, 5-disubstituted-4H-1, 2, 4-triazol-4-yl) acetyl)-6/7/8-substituted-2-oxo-2H-chromen-3-carbohydrazide derivatives as potent antidiabetic agents, Arch. Pharm., 2019, 352(12), 1900227 CrossRef PubMed
.
- X.-T. Xu, X.-Y. Deng, J. Chen, Q.-M. Liang, K. Zhang and D.-L. Li, et al., Synthesis and biological evaluation of coumarin derivatives as α-glucosidase inhibitors, Eur. J. Med. Chem., 2020, 189, 112013 CrossRef PubMed
.
- X. Zhang, Y.-Y. Zheng, C.-M. Hu, X.-Z. Wu, J. Lin and Z. Xiong, et al., Synthesis and biological evaluation of coumarin derivatives containing oxime ester as α-glucosidase inhibitors, Arabian J. Chem., 2022, 15(9), 104072 CrossRef CAS
.
- A. Singh, K. Singh, K. Kaur, A. Sharma, P. Mohana and J. Prajapati, et al., Discovery of triazole tethered thymol/carvacrol-coumarin hybrids as new class of α-glucosidase inhibitors with potent in vivo antihyperglycemic activities, Eur. J. Med. Chem., 2024, 263, 115948 CrossRef CAS PubMed
.
- W. Yang, J. Chen, Z. Peng and G. Wang, Design, synthesis and enzymatic inhibition evaluation of novel 4-Hydroxy Pd-C-III derivatives as α-glucosidase and PTP1B dual-target inhibitors, Eur. J. Med. Chem., 2024, 116938 CrossRef CAS PubMed
.
- W. D. Seo, J. H. Kim, J. E. Kang, H. W. Ryu, M. J. Curtis-Long and H. S. Lee, et al., Sulfonamide chalcone as a new class of α-glucosidase inhibitors, Bioorg. Med. Chem. Lett., 2005, 15(24), 5514–5516 CrossRef CAS PubMed
.
- R. Ranga Rao, A. K. Tiwari, P. Prabhakar Reddy, K. Suresh Babu, G. Suresh and A. Z. Ali, et al., Synthesis of antihyperglycemic, α-glucosidase inhibitory, and DPPH free radical scavenging furanochalcones, Med. Chem. Res., 2012, 21, 760–774 CrossRef CAS
.
- A. Rammohan, B. V. Bhaskar, N. Venkateswarlu, W. Gu and G. V. Zyryanov, Design, synthesis, docking and biological evaluation of chalcones as promising antidiabetic agents, Bioorg. Chem., 2020, 95, 103527 CrossRef CAS PubMed
.
- F. Saleem, K. M. Khan, S. Chigurupati, M. Solangi, A. R. Nemala and M. Mushtaq, et al., Synthesis of azachalcones, their α-amylase, α-glucosidase inhibitory activities, kinetics, and molecular docking studies, Bioorg. Chem., 2021, 106, 104489 CrossRef CAS PubMed
.
- S. Y. Lv and L. P. Cheng, Design, synthesis and inhibition evaluation of novel chalcone amide α-glucosidase inhibitors, Future Med. Chem., 2024, 1–13 CAS
.
- N. Hosseini Nasab, H. Raza, Y. S. Eom, F. H. Shah, J.-H. Kwak and S. J. Kim, Exploring chalcone-sulfonyl piperazine hybrids as anti-diabetes candidates: design, synthesis, biological evaluation, and molecular docking study, Mol. Diversity, 2024, 1–17 Search PubMed
.
- C.-M. Hu, Y.-X. Luo, W.-J. Wang, J.-P. Li, M.-Y. Li and Y.-F. Zhang, et al., Synthesis and evaluation of coumarin-chalcone derivatives as α-glucosidase inhibitors, Front. Chem., 2022, 10, 926543 CrossRef CAS PubMed
.
- F. Peytam, M. Adib, R. Shourgeshty, L. Firoozpour, M. Rahmanian-Jazi and M. Jahani, et al., An efficient and targeted synthetic approach towards new highly substituted 6-amino-pyrazolo [1, 5-a] pyrimidines with α-glucosidase inhibitory activity, Sci. Rep., 2020, 10(1), 2595 CrossRef CAS PubMed
.
- F. Peytam, G. Takalloobanafshi, T. Saadattalab, M. Norouzbahari, Z. Emamgholipour and S. Moghimi, et al., Design, synthesis, molecular docking, and in vitro α-glucosidase inhibitory activities of novel 3-amino-2, 4-diarylbenzo [4, 5] imidazo [1, 2-a] pyrimidines against yeast and rat α-glucosidase, Sci. Rep., 2021, 11(1), 11911 CrossRef CAS PubMed
.
- F. Peytam, F. S. Hosseini, M. Hekmati, B. Bayati, M. S. Moghadam and Z. Emamgholipour, et al., Imidazo [1, 2-c] quinazolines as a novel and potent scaffold of α-glucosidase inhibitors: design, synthesis, biological evaluations, and in silico studies, Sci. Rep., 2023, 13(1), 15672 CrossRef CAS PubMed
.
- F. Peytam, F. S. Hosseini, R. Fathimolladehi, M. J. D. Nayeri, M. S. Moghadam and B. Bayati, et al., Design, synthesis, and evaluation of novel substituted imidazo [1, 2-c] quinazoline derivatives as potential α-glucosidase inhibitors with bioactivity and molecular docking insights, Sci. Rep., 2024, 14(1), 27507 CrossRef CAS PubMed
.
- F. Peytam, P. Foroumadi, H. O. Gulcan, M. Norouzbahari, S. Mojtabavi and M. A. Faramarzi, et al., Design, synthesis, and evaluation of triazolo [1, 5-a] pyridines as novel and potent α-glucosidase inhibitors, Sci. Rep., 2025, 15(1), 17813 CrossRef CAS PubMed
.
- F. Peytam, M. Norouzbahari, H. O. Gulcan, F. S. Hosseini, M. S. Moghadam and S. Mojtabavi, et al., Identification of novel triazolopyrimidines as potent α-glucosidase inhibitor through design, synthesis, biological evaluations, and computational analysis, Sci. Rep., 2025, 15(1), 39667 CrossRef PubMed
.
- J. Shao, X. Liu, K. Shu, P. Tang, J. Luo and W. Chen, et al., Tuning the annulation reactivity of vinyl azides and carbazates: A divergent synthesis of aza-pyrimidinones and imidazoles, Org. Lett., 2015, 17(18), 4502–4505 CrossRef CAS PubMed
.
- K. Shu, J. Shao, H. Li, B. Chen, P. Tang and X. Liu, et al., Base-mediated synthesis of highly functionalized 2-aminonicotinonitriles from α-keto vinyl azides and α, α-dicyanoalkenes, RSC Adv., 2016, 6(54), 49123–49126 RSC
.
- M. Adib, F. Peytam, M. Rahmanian-Jazi, H. R. Bijanzadeh and M. Amanlou, A new synthetic strategy towards 2, 4, 5-trisubstituted 1H-imidazoles and highly substituted pyrrolo [1, 2-c] imidazoles by use of α-azidochalcones via Michael addition-cyclization followed by Wittig reaction, Tetrahedron, 2017, 73(48), 6696–6705 CrossRef CAS
.
- K. Rajaguru, A. Mariappan, S. Muthusubramanian and N. Bhuvanesh, Divergent reactivity of α-azidochalcones with metal β-diketonates: Tunable synthesis of substituted pyrroles and indoles, Org. Chem. Front., 2017, 4(1), 124–129 RSC
.
- M. Adib, F. Peytam, M. Rahmanian-Jazi, M. Mohammadi-Khanaposhtani, S. Mahernia and H. R. Bijanzadeh, et al., Design, synthesis and in vitro α-glucosidase inhibition of novel coumarin-pyridines as potent antidiabetic agents, New J. Chem., 2018, 42(21), 17268–17278 RSC
.
- M. Adib, F. Peytam, M. Rahmanian-Jazi, S. Mahernia, H. R. Bijanzadeh and M. Jahani, et al., New 6-amino-pyrido [2, 3-d] pyrimidine-2, 4-diones as novel agents to treat type 2 diabetes: A simple and efficient synthesis, α-glucosidase inhibition, molecular modeling and kinetic study, Eur. J. Med. Chem., 2018, 155, 353–363 CrossRef CAS PubMed
.
- M. Adib and F. Peytam, An efficient synthesis of fully substituted pyrazolo [3, 4-b] pyridin-5-amines from α-azidochalcones, Tetrahedron, 2018, 74(20), 2414–2420 CrossRef CAS
.
- S. Borra, D. Chandrasekhar, U. D. Newar and R. A. Maurya, Access to 2, 3-fused pyrroles via visible light driven coupling of α-azidochalcones with 1/2-naphthols, or 2-hydroxy-1, 4-naphthoquinone, J. Org. Chem., 2018, 84(2), 1042–1052 CrossRef PubMed
.
- F. Peytam, M. Adib, R. Shourgeshty, M. Rahmanian-Jazi, M. Jahani and B. Larijani, et al., A one-pot and three-component synthetic approach for the preparation of asymmetric and multi-substituted 1, 4-dihydropyrazines, Tetrahedron Lett., 2019, 60(47), 151257 CrossRef
.
- U. D. Newar, S. Borra and R. A. Maurya, Visible-light 2, 4-dinitrophenol-mediated photoannulation of α-azidochalcones into 2, 5-diaryloxazoles, Org. Lett., 2022, 24(24), 4454–4458 CrossRef CAS PubMed
.
- U. Devi Newar, D. Jyoti Boruah and R. Awatar Maurya, Recent Advances in the Use of α-Azidochalcones in Heterocycle Synthesis, Adv. Synth. Catal., 2024, 366(13), 2859–2897 CrossRef CAS
.
- N. Mahmood, A review of α-amylase inhibitors on weight loss and glycemic control in pathological state such as obesity and diabetes, Comp. Clin. Pathol., 2016, 25(6), 1253–1264 CrossRef CAS
.
- T.-P. Lam, N.-V. N. Tran, L.-H. D. Pham, N. V.-T. Lai, B.-T. N. Dang and N.-L. N. Truong, et al., Flavonoids as dual-target inhibitors against α-glucosidase and α-amylase: a systematic review of in vitro studies, Nat. Prod. Bioprospect., 2024, 14(1), 4 CrossRef CAS PubMed
.
- M. Dixon, The determination of enzyme inhibitor constants, Biochem. J., 1953, 55(1), 170 CrossRef CAS PubMed
.
- M. J. Todd and R. Hausinger, Competitive inhibitors of Klebsiella aerogenes urease: mechanisms of interaction with the nickel active site, J. Biol. Chem., 1989, 264(27), 15835–15842 CrossRef CAS PubMed
.
- M. K. Barker and D. R. Rose, Specificity of processing α-glucosidase I is guided by the substrate conformation: crystallographic and in silico studies, J. Biol. Chem., 2013, 288(19), 13563–13574 CrossRef CAS PubMed
.
- M. Torabi, I. Haririan, A. Foroumadi, H. Ghanbari and F. Ghasemi, A deep learning model based on the BERT pre-trained model to predict the antiproliferative activity of anti-cancer chemical compounds, SAR QSAR Environ. Res., 2024, 35(11), 971–992 CrossRef CAS PubMed
.
- Z. Hasanvand, T. O. Bakhshaiesh, F. Peytam, L. Firoozpour, E. Hosseinzadeh and R. Motahari, et al., Imidazo [1, 2-a] quinazolines as novel, potent EGFR-TK inhibitors: Design, synthesis, bioactivity evaluation, and in silico studies, Bioorg. Chem., 2023, 133, 106383 CrossRef CAS PubMed
.
- D. Lorke, A new approach to practical acute toxicity testing, Arch. Toxicol., 1983, 54(4), 275–287 CrossRef CAS PubMed
.
- D. A. D. Magalhães, W. T. Kume, F. S. Correia, T. S. Queiroz, E. W. Allebrandt and M. P. D. Santos, et al., High-fat diet and streptozotocin in the induction of type 2 diabetes mellitus: a new proposal, An. Acad. Bras. Cienc., 2019, 91(01), e20180314 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2025 |