Open Access Article
Open Access ArticleLC-MS-based metabolomic characterization of Tinospora crispa extracts: impact of solvent selection on phytochemical composition and antioxidant properties
Nattha Muangritdech a,
Piya Prajumwongsa,
Nisana Namwat
a,
Piya Prajumwongsa,
Nisana Namwat ab,
Poramate Klanritab,
Arporn Wangwiwatsinab,
Hasaya Dokduang
ab,
Poramate Klanritab,
Arporn Wangwiwatsinab,
Hasaya Dokduang d,
Sirinya Sitthirake,
Attapol Titapunac and
Watcharin Loilome*ab
d,
Sirinya Sitthirake,
Attapol Titapunac and
Watcharin Loilome*ab
aCholangiocarcinoma Research Institute, Khon Kaen University, Khon Kaen 40002, Thailand. E-mail: nattha.m@kkumail.com; watclo@kku.ac.th
bDepartment of Systems Biosciences and Computational Medicine, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand
cDepartment of Surgery, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand
dFaculty of Medicine, Mahasarakham University, Mahasarakham 44000, Thailand
eDepartment of Medical Technology, School of Allied Health Sciences, Walailak University, Nakhon Si Thammarat 80161, Thailand
First published on 18th December 2025
Abstract
The medicinal plant Tinospora crispa (T. crispa) contains a wide array of bioactive compounds known for their antidiabetic, antioxidant, and anticancer properties. This study aimed to evaluate the effect of various solvents on the efficiency of metabolite extraction, and to assess the antioxidant activity of the respective extracts. T. crispa was extracted using ethanol, ethyl acetate, and water via maceration. Antioxidant capacity was measured using total phenolic content (TPC), 2,2-diphenyl-1-picrylhydrazyl assay (DPPH), and ferric reducing antioxidant power assay (FRAP). Metabolite profiling was analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS), and correlation analysis was performed between metabolite profiles and antioxidant parameters to identify potential antioxidant-related compounds. Metabolomic analysis revealed distinct chemical compositions across the three solvent extracts. The ethanol extract demonstrated the highest antioxidant activity in DPPH and FRAP assays, while the ethyl acetate extract exhibited the greatest TPC value. A total of 20 metabolites showed moderate to strong positive correlations (r ≥ 0.4) with three antioxidant capacity assays, indicating their potential contribution to antioxidant properties. These metabolites were predominantly flavonoids, alkaloids, and other plant-derived secondary metabolites. Overall, ethanol was the most efficient solvent for extracting bioactive compounds related to antioxidant activity. This study highlights the critical role of solvent selection in optimizing extraction protocols for medicinal plants and underscores the potential of T. crispa as a natural antioxidant source. Furthermore, the combined use of LC-MS/MS-based metabolomics and correlation analysis provides a powerful approach for identifying bioactive metabolites, suggesting promising directions for future pharmacological research and therapeutic applications.
1 Introduction
Tinospora crispa (T. crispa), a medicinal plant widely distributed across Southeast Asia, has been extensively utilized in traditional medicine for its therapeutic properties.1 Traditionally, the stems of T. crispa have been prepared as decoctions or infusions and used in Southeast Asian folk medicine to reduce fever, alleviate inflammation, regulate metabolism, and support liver health. These ethnopharmacological applications underscore its longstanding role in traditional healing practices and provide a foundation for its modern pharmacological investigations.2,3 Renowned for its efficacy in treating a variety of health conditions, including hypertension, type 2 diabetes mellitus, inflammation, and microbial infections,3–5 this plant derives its pharmacological potential from its rich profile of bioactive compounds. These compounds, which include flavonoids, phenolics, alkaloids, glycosides, diterpenoids, triterpenoids compounds,2,6,7 collectively contribute to its diverse biological activities. Among these, specific metabolites belonging to groups such as glycosides and alkaloids have been reported to play pivotal roles in its antidiabetic, anti-inflammatory, and antioxidant effects. For instance, certain glycosides have been shown to enhance glucose uptake in insulin-resistant cells,8 whereas alkaloids have exhibited beneficial antioxidant properties by diminishing pro-inflammatory cytokines in animal models. T. crispa extracts have demonstrated free radical scavenging activity, thereby reducing oxidative stress in cellular models.2 Animal studies show that T. crispa extracts protect against oxidative stress and mitigate liver and kidney damage in diabetic rats.5,9,10 Beyond its antidiabetic and antioxidant effects, T. crispa exhibits anticancer potential, with bioactive compounds modulating oxidative stress pathways such as Nrf2/ARE and alkaloids enhancing chemosensitivity in drug-resistant cancer cells by inhibiting multidrug resistance-related transport proteins.11,12 These results suggest that T. crispa may act as an adjuvant in cancer therapy, which may improve the effectiveness of chemotherapy against resistant cancer cells.The extraction of bioactive compounds from T. crispa is strongly influenced by solvent polarity, as solvents selectively dissolve specific classes of phytochemicals. Along the polarity gradient, non-polar solvents (e.g., hexane) primarily extract lipophilic compounds such as fats, waxes, and some terpenes; semi-polar solvents (e.g., ethanol and ethyl acetate) target moderately polar compounds including certain terpenoids, flavonoid aglycones, and low-molecular-weight phenolics; and polar solvents (and water) solubilize a broad range of bioactive compounds.13–16 Ethanol is particularly effective for polyphenols, tannins, and alkaloids, whereas water is optimal for hydrophilic compounds such as glycosides, polysaccharides, and organic acids.17 Thus, solvent selection is critical for isolating pharmacologically active constituents.
Advanced techniques such as LC-MS/MS enable precise identification and quantification of T. crispa bioactive compounds, offering high sensitivity, specificity, and suitability for complex mixtures. This approach not only compares solvent extraction efficiency but also links chemical profiles to pharmacological activities, guiding standardized extraction methods and ensuring consistent therapeutic efficacy. Therefore, our study aims to investigate the influence of ethyl acetate, ethanol, and water on the extraction efficiency of T. crispa bioactive compounds. Antioxidant activity was evaluated for all extracts and LC-MS/MS was used to identify and quantify their constituents. Extracts with the highest antioxidant potential were further analyzed to correlate metabolic profiles with antioxidant activity, aiming to identify metabolites responsible for the observed biological effects.
2 Materials and methods
2.1 Sample collection, chemical and reagents
A powdered sample of T. crispa was obtained from local herbal stores located in Pathum Thani Province, Thailand (approximate coordinates: lat. 14.02° N, long. 100.53° E) during August 2024. The collection sites were documented to ensure reproducibility and traceability. The extraction solvents including ethanol (purity 99.7%) and ethyl acetate (purity 99%) were purchased from Carlo Erba Reagents Co., Ltd, Barcelona, Spain. Dimethylsulfoxide (DMSO) was purchased from Sigma-Aldrich (St. Louis, MO, USA).2.2 Preparation of crude extract from T. crispa powder
The extraction of bioactive compounds from dried T. crispa powder was carried out using the maceration method. The dried T. crispa powder was extracted with a solvent at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (w/v). Specifically, a 50 g amount of T. crispa powder was mixed with 200 mL of solvent (ethyl acetate, ethanol, and water). The mixtures were agitated using an incubator shaker at room temperature, operating at 100 rpm for 6 h per day over 7 consecutive days, protected from light. After that the extracts were filtered to remove the particulate matter using Whatman® No. 4 filter paper. For ethanol and ethyl acetate extract fraction, the filtrates were then evaporated to remove the solvent using a rotary evaporator at 40 °C. and further dried using a rotary evaporator vacuum pump (Rotavapor® R-100 Rotary Evaporator, Buchi, Switzerland) until fully dry. For the water extract fraction, the filtrate was frozen at −80 °C and lyophilized by a freeze dryer (Labconco/FreeZone 4.5, Labconco Corporation, USA). The crude extracts obtained were stored at −20 °C until used.
4 (w/v). Specifically, a 50 g amount of T. crispa powder was mixed with 200 mL of solvent (ethyl acetate, ethanol, and water). The mixtures were agitated using an incubator shaker at room temperature, operating at 100 rpm for 6 h per day over 7 consecutive days, protected from light. After that the extracts were filtered to remove the particulate matter using Whatman® No. 4 filter paper. For ethanol and ethyl acetate extract fraction, the filtrates were then evaporated to remove the solvent using a rotary evaporator at 40 °C. and further dried using a rotary evaporator vacuum pump (Rotavapor® R-100 Rotary Evaporator, Buchi, Switzerland) until fully dry. For the water extract fraction, the filtrate was frozen at −80 °C and lyophilized by a freeze dryer (Labconco/FreeZone 4.5, Labconco Corporation, USA). The crude extracts obtained were stored at −20 °C until used.
2.3 Metabolite analysis by LC-MS/MS
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, and the supernatant was transferred to a polypropylene vial for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.
000 rpm for 10 min, and the supernatant was transferred to a polypropylene vial for liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis.Mass spectrometric (MS) detection was conducted using electrospray ionization (ESI) in both positive (ESI+) and negative (ESI−) modes with a mass range of 100–1700 m/z. The QTOF detector was calibrated in a high-resolution mode against 6 reference mass of ESI-L tunning mix (Agilent Technologies, part no. G1969-85000) right before sample acquisition in both positive and negative modes. The MS parameters were set as follows: gas temperature, 325 °C; nebulizer, 45 psi; drying gas flow rate, 13 L min−1; sheath gas temperature, 275 °C; sheath gas flow rate, 12 L min−1; nozzle voltage, 500 V; fragmentor voltage, 175 V; skimmer voltage, 65 V; capillary voltage, 4000 and 3000 V for positive and negative mode, respectively. Data acquisition was performed at a rate of 3.35 spectra s−1. A maximum of 10 precursor ions per cycle were selected for MS/MS fragmentation, with collision energies of 20 eV (positive mode) and 10 eV (negative mode). Trifluoroacetic acid anion, purine, HP-0921 (Agilent Technologies, part no. G1969-85001) were used as reference compounds. The reference masses were at m/z 121.0509 and 922.0098 in the positive mode and at m/z 112.9596 and 1033.9881 in the negative mode, respectively. Data were acquired in centroid mode. Each sample was injected in triplicate.
Chromatographic deconvolution was performed using the ADAP wavelets algorithm with a signal-to-noise ratio threshold of 10. Isotopic features were grouped using a 5 ppm m/z and 0.05 min RT tolerance. Features were aligned using the join aligner with 5 ppm m/z and 0.10 min RT tolerances, weighted 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 for m/z and RT. Gap filling was performed using the peak finder algorithm. Features found in blank samples were excluded, and only those present in at least one real sample or ≥10% of the sample set were retained.
30 for m/z and RT. Gap filling was performed using the peak finder algorithm. Features found in blank samples were excluded, and only those present in at least one real sample or ≥10% of the sample set were retained.
Metabolite identification was based on library matching of experimental MS/MS spectra against multiple public databases (GNPS, MassBank, HMDB, and MoNA, as of March 26, 2025). A cosine similarity score ≥0.5, precursor m/z tolerance of ±0.015 (20 ppm), and at least four matched fragment ions were required. Only high-confidence annotations based on MS/MS spectral similarity were retained.
Duplicate metabolite annotations were resolved by retaining the metabolite with the highest peak intensity.21–23 All metabolite assignments were manually checked for biological relevance; structurally complex or ambiguous compounds that could not be confidently identified were excluded from downstream analyses.21,23 After filtering, a total of 106 and 53 annotated metabolites in positive and negative ionization modes, respectively, were used for statistical analyses.
2.4 Total phenolic content and antioxidant property
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. A 10 µL aliquot of each extract was added to a 96-well plate, followed by 190 µL of FRAP reagent. The reaction mixture was incubated at room temperature for 30 min, while being protected from light. Absorbance was measured at 593 nm using an EZ Read 2000 microplate reader (Biochrom, US). Trolox was used as the standard, and antioxidant activity was expressed as micrograms of Trolox equivalent per milligram of sample (µg TEAC mg−1 extract).
1 ratio. A 10 µL aliquot of each extract was added to a 96-well plate, followed by 190 µL of FRAP reagent. The reaction mixture was incubated at room temperature for 30 min, while being protected from light. Absorbance was measured at 593 nm using an EZ Read 2000 microplate reader (Biochrom, US). Trolox was used as the standard, and antioxidant activity was expressed as micrograms of Trolox equivalent per milligram of sample (µg TEAC mg−1 extract).2.5 Statistical analyses
All results from antioxidant assays (TPC, DPPH, FRAP) were presented as mean ± standard deviation (SD) from three independent replicates. Statistical analyses were conducted using SPSS software (version 29.0; IBM Corp., Chicago, Illinois, USA). Group comparisons were performed using one-way analysis of variance (ANOVA) follow by Bonferroni post hoc test for parametric variables. For non-parametric variables, the Kruskal–Wallis H test was applied, followed by pairwise comparisons using Mann–Whitney U tests with Bonferroni-adjusted p-values. Graphs were generated using GraphPad Prism (version 8.0; GraphPad Software, Inc., San Diego, CA, USA). A p < 0.05 was considered statistically significant.3 Results
3.1 LC-MS/MS-based metabolomics analysis of Tinospora crispa (T. crispa) extracts
Tinosporia crispa (T. crispa), is wide spectrum of pharmacological activities including antioxidant, anti-inflammatory, antimicrobial, and anticancer effects. These therapeutic benefits are primarily attributed to its rich phytochemical composition, consisting of alka-loids, terpenoids, flavonoids, phenolic acids, and glycosides.2,3,31,32 This study aimed to evaluate the effect of different extraction solvents on the chemical composition of T. crispa extracts. The samples were analyzed using LC-MS/MS to assess the metabolomics profile in three different solvents. The preliminary metabolite identification results were subjected to a filtering process to ensure biological relevance and data quality. We applied the following exclusion criteria to select candidate metabolites: (1) in cases of duplicate identifications, the metabolite with the highest match score was retained; (2) only metabolites known to be present in plants or herbs were included; and (3) structurally complex or ambiguous metabolites that could not be clearly identified were excluded. Based on these criteria, a total of 106 and 53 putative metabolites were retained in the positive and negative ionization modes (Table S1), respectively. Subsequently, the curated metabolite datasets were subjected to chemometric analysis using MetaboAnalyst to compare the metabolic profiles of T. crispa extracts prepared with ethyl acetate, ethanol, and water as solvents.Heatmap analysis was conducted to visualize the distribution and abundance of metabolites across different extraction solvents. The hierarchical clustering heatmaps revealed distinct metabolite expression patterns among the three extract groups (Fig. 1A and B). Both ethyl acetate and ethanol extracts exhibited higher numbers of strongly expressed metabolites compared to the water extract in both positive and negative ionization modes. Notably, ethyl acetate and ethanol extracts clustered more closely together, indicating a similar metabolic profile. This finding suggested that semi-polarity (ethyl acetate and ethanol) ability of solvent contributed a more efficient to extract a broader range of bioactive compounds from T. crispa.13–16
Principal component analysis (PCA) was conducted to further evaluate the chemical separation between groups. The PCA scores plots demonstrated a clear separation among the ethyl acetate, ethanol, and water extract groups in both ion modes. In positive mode (Fig. 1B), the first two principal components accounted for 47.1% (PC1) and 22.7% (PC2) of the total variance. Similarly, in negative mode (Fig. 1H), PC1 and PC2 explained 52.0% and 14.0% of the variance, respectively, highlighting substantial chemical diversity among the solvent extracts. The corresponding PCA loading plots illustrate the contribution of individual metabolites to the observed group separation. In positive ion mode, metabolites such as D-Tetrahydropalmatine, Apigenin, Piperine, 13-OxoODE, and N-trans-P-coumaroyltyramine showed high absolute loadings, strongly influencing the separation among ethyl acetate, ethanol, and water extracts, while other metabolites also contributed to the differentiation but are not highlighted here (Fig. S1A). Similarly, in negative ion mode, norstictic acid, FA 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 + 1°, suberic acid (Z)-5,8,11-trihydroxyoctadec-9-enoic acid, and α-linolenic acid (18
2 + 1°, suberic acid (Z)-5,8,11-trihydroxyoctadec-9-enoic acid, and α-linolenic acid (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 + 1°) were major contributors to the group separation, with additional influential metabolites not highlighted for brevity (Fig. S1B).
3 + 1°) were major contributors to the group separation, with additional influential metabolites not highlighted for brevity (Fig. S1B).
To evaluate the differences among T. crispa extracts prepared using three different solvents and to identify the metabolites contributing most to group separation, supervised partial least squares discriminant analysis (PLS-DA) was performed. In positive mode, latent variable 1 (LV1) accounted for 47.0% and latent variable 2 (LV2) for 22.1%, illustrating clear separation among the three extract groups (Fig. 1C and S2A). In negative mode, LV1 accounted for 51.9% and LV2 for 11.7%, also showing distinct group separation (Fig. 1I and S2B). The robustness and stability of the PLS-DA models were assessed using 10-fold cross-validation and permutation tests. In positive ion mode, the eight-component model achieved R2 = 0.99764, Q2 = 0.98535, and accuracy = 1.00 (Fig. 1D), while the seven-component model in negative ion mode reached R2 = 0.99419, Q2 = 0.94732, and accuracy = 0.96333 (Fig. 1J). Notably, the first two components alone captured the model structure with R2 > 0.90, Q2 > 0.85, and accuracy > 0.90 in both ion modes, demonstrating clear group separation and strong predictive performance. Permutation tests (n = 100) further confirmed that group discrimination was statistically significant (empiric p < 0.001; Fig. 1E and K), indicating the models were robust and not overfitted.
Overall, these results indicate that the metabolomic profiles of T. crispa extracts were strongly influenced by the type of extraction solvent, and the PLS-DA models successfully captured these differences while identifying key variables contributing to group discrimination.
The VIP scores showed top 15 impact metabolites in the PLS-DA model both positive and negative modes (Fig. 1F and L). Subsequently, metabolites with a PLS-DA VIP score ≥1 and an FDR-adjusted p-value < 0.05 between groups were selected, comprising 21 metabolites in the positive ionization mode and 11 in the negative ionization mode, for further analysis. We also examined the corresponding PLS-DA loading plots for both ion modes, which confirmed that the metabolites most influential in driving group separation were largely consistent with those identified by VIP scores. For example, in positive mode, D-tetrahydropalmatine, apigenin, and piperine were among the key metabolites influencing the model (Fig. S2A), whereas in negative mode, norstictic acid, suberic acid, and α-linolenic acid (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 + 1°) were important contributors to group differentiation (Fig. S2B). These results further validate the selection of metabolites that significantly influence the chemometric separation across different solvent extracts.
3 + 1°) were important contributors to group differentiation (Fig. S2B). These results further validate the selection of metabolites that significantly influence the chemometric separation across different solvent extracts.
Notably, the key metabolites contributing to group separation were largely consistent between the PCA and PLS-DA models, indicating that the same metabolites predominantly drive the differentiation among ethyl acetate, ethanol, and water extracts. This concordance further highlights the strong effect of solvent polarity on the selective extraction of specific metabolite classes in T. crispa.
Consistent with these observations, solvent polarity played a critical role in deter-mining extraction efficiency. Both ethanol and ethyl acetate, with intermediate polarity indices (polarity indices ≈ 5.2 and 4.4), were more effective than water (polarity index ≈ 10 in solubilizing a broad spectrum of phytochemicals, including flavonoids, phenolic acids, alkaloids, and terpenoids.3,6 Distinct patterns of metabolite enrichment were observed in each solvent based on the observed MS features and clustering patterns: the semi-polar ethyl acetate extract was enriched in a broad range of metabolites with moderate to low polarity, mainly lipids, less polar flavonoids, terpenoids, and hydrophobic phenolic compounds. Furthermore, highly polar compounds such as amino acids and sugars were markedly depleted in this extract. The moderately polar ethanol extract was enriched in a wide array of flavonoids and phenolic compounds, with several alkaloids and glycosides also effectively extracted.
Finally, the highly polar water extract was distinctively enriched in highly polar metabolites, including amino acids, sugars, organic acids, and highly polar phenolic compounds, while showing a significant depletion of hydrophobic compounds like lipids and less polar terpenoids, strongly supporting the link between solvent polarity and metabolite class enrichment. This finding agrees with previous studies, such as those reporting ethanol as a superior solvent for polyphenolic extraction from Mentha longifolia and Portulaca oleracea, which also resulted in enhanced antioxidant activity.33,34 Tourabi et al. Investigated the influence of solvent polarity (ethanol 70%, ethyl acetate, and water) and extraction techniques (Soxhlet, ultrasonic-assisted extraction, and cold maceration) on Mentha longifolia extracts using LC-MS-based analysis. They found that ethanol consistently yielded the highest total phenolic and flavonoid contents, while ethyl acetate selectively extracted specific phenolic acids such as gallic acid and kaempferol. Water extracts contained predominantly hydrophilic compounds with lower antioxidant potential.33 Similarly, Chen et al. applied LC-MS-based technology to study Portulaca oleracea, demonstrating that solvent polarity strongly influences the recovery of bioactive metabolites. Semi-polar solvents, particularly ethanol and acetone, yielded the highest levels of total phenolics and flavonoids, as well as the strongest antioxidant activities, compared with nonpolar or more polar solvents. These findings indicate that semi-polar solvents are especially effective at extracting diverse antioxidant-related metabolites.35 In addition, LC-MS/MS analysis of Cassia auriculata showed that solvent polarity strongly affects extraction efficiency and metabolite profile. Semi-polar solvents, particularly methanol and ethanol, extracted the widest range of phenolic and flavonoid compounds with the highest peak intensities, while water showed the lowest efficiency. Ethyl acetate performed moderately, better than water but less than ethanol, highlighting the advantage of semi-polar solvents for recovering diverse bioactive metabolites.36
These results demonstrate that broad metabolite classes are preferentially extracted according to solvent polarity and emphasize the effectiveness of semi-polar solvents in recovering diverse bioactive metabolites from medicinal plants, especially T. crispa.
3.2 Antioxidant activity of T. crispa extracts
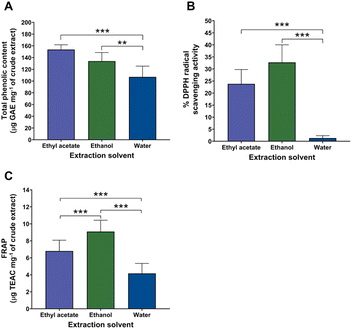
The antioxidant properties of T. crispa extracts obtained using ethyl acetate, ethanol, and water were compared based on their total phenolic content (TPC), DPPH radical scavenging activity, and ferric reducing antioxidant power (FRAP), as summarized in Table S2 and illustrated in Fig. 2A–C. The ethyl acetate extract exhibited the highest TPC (153.72 ± 8.01 µg GAE mg−1 extract), followed by ethanol (133.97 ± 14.61 µg GAE mg−1 extract) and water (107.04 ± 18.37 µg GAE mg−1 extract), with all differences being statistically significant (p < 0.05) (Fig. 2A). Conversely, the antioxidant activity measured by DPPH assay revealed a different trend. The ethanol extract exhibited the highest free radical scavenging activity (32.68 ± 7.36%), followed by ethyl acetate (23.81 ± 5.87%) and water (1.41 ± 0.93%), significant differences were observed among all groups (p < 0.05) (Fig. 2B). Similarly, the FRAP assay indicated that the ethanol extract possessed the highest reducing power (9.09 ± 1.34 µg TEAC mg−1 extract) compared to the ethyl acetate and water extracts which significantly greater than those of ethyl acetate (6.80 ± 1.27 µg TEAC mg−1 extract) and water (4.17 ± 1.18 µg TEAC mg−1 extract) (Fig. 2C). These results suggested that although the ethyl acetate extract contained the highest phenolic content, the ethanol extract exhibited superior antioxidant performance in both DPPH and FRAP assays, highlighting the influence of solvent polarity on the extraction efficiency of bioactive antioxidant compounds from T. crispa. These results indicated that while phenolic compounds play a major role in antioxidant capacity, other co-extracted metabolites in ethanol and ethyl acetate extracts may synergistically enhance its activity. The superior performance of ethanol and ethyl acetate was likely attributed to its ability to extract a wide range of flavonoids and phenolic acids with strong redox potential, such as apigenin, luteolin, and ferulic acid. This aligns with previous reports demonstrating moderately polar sovents superiority in recovering antioxidant-rich metabolites from medicinal plants.33,34,37 Conversely, the weak activity of the water extract reflects its limited phenolic composition, consistent with its hydrophilic metabolite dominance. Together, these findings highlight a strong interplay between solvent polarity, metabolite diversity, and antioxidant efficiency in T. crispa.3.3 Correlation between metabolite composition and antioxidant activity and identification of potentially candidate metabolites in T. crispa extracts
To gain insight into the potential contribution of individual metabolites to the antioxidant properties of the T. crispa extracts, a correlation analysis was performed between LC-MS/MS-based metabolic profiles and experimental antioxidant parameters, including total phenolic content (TPC), and antioxidant activities measured by DPPH and FRAP assays. The analysis was conducted using the MetaboAnalyst platform, applying Spearman's correlation coefficient |r| to evaluate the linear associations. A correlation heatmap was generated to visualize the strength and direction of these association.Correlation strength was categorized according to established criteria: moderate (r = 0.40–0.69), strong (r = 0.70–0.89), and very strong (r = 0.90–1.00), following a previously reported guideline.38 To ensure biological relevance, only metabolites exhibiting at least moderate correlation (r ≥ 0.4) with all three antioxidant assays (TPC, DPPH, and FRAP) were considered potential bioactive constituents.
The results were presented as heatmaps for both positive and negative ionization modes (Fig. 3). In the heatmap, red indicated positive correlations that met the cut-off threshold, with deeper red reflecting stronger correlations. Green indicated negative correlations, and black denoted weak or non-significant correlations (r < 0.4).
In the positive ionization mode, 12 out of 21 significant metabolites exhibited positive correlations (Spearman's r ≥ 0.4) with all three antioxidant assays (Fig. 3). Similarly, in the negative ionization mode, 8 out of 12 metabolites were positively correlated above the threshold level, indicating a moderate to strong positive association with antioxidant parameters in all three antioxidant assays (Fig. 3). These findings suggested that several metabolites detected under both ionizations conditions may play a substantial role in the antioxidant capacity of T. crispa extracts.
Correlation analysis revealed 20 metabolites with moderate-to-strong positive correlations (Spearman's r ≥ 0.4) across all three antioxidant assays—TPC, DPPH, and FRAP. These metabolites were considered candidate bioactive compounds due to their consistent associations across all three assays. Our metabolomic profiling revealed a diverse array of metabolites classified into several major super classes according to the PubChem compound database,37 including flavonoids, phenylpropanoids and polyketides, fatty acids derivatives, alkaloids and derivatives and terpenoids (Table 1).
| No | Metabolites | Molecular formula | Adduct ion | VIP Scores | RT (min) | Observative mass (m/z) | Theoretical mass (m/z) | Compound class | PubChem Id | Correlation coefficient (r ≥ 0.4) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DPPH | FRAP | TPC | ||||||||||
| Positive mode | ||||||||||||
| 1 | Apigenin | C15H10O5 | [M + H]+ | 4.69 | 7.600 | 271.0615 | 270.24 | Flavonoids | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 443 443 |
0.716 | 0.546 | 0.792 |
| 2 | Naringenin chalcone | C15H12O5 | [M + H]+ | 3.10 | 7.512 | 273.0758 | 272.25 | Flavonoids | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 960 960 |
0.758 | 0.521 | 0.620 |
| 3 | N-trans-P-coumaroyltyramine | C17H17NO3 | [M + H]+ | 2.50 | 6.415 | 284.1287 | 283.32 | Phenylpropanoids | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 372 372![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 945 945 |
0.532 | 0.765 | 0.482 |
| 4 | Feruloyl dehydrotyramine (isomer of 1654) | C18H17NO4 | [M + H]+ | 2.13 | 5.464 | 312.1232 | 311.30 | Phenylpropanoids | 139![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 292 292![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 013 013 |
0.741 | 0.465 | 0.821 |
| 5 | trans-ferulic acid | C10H10O4 | [M + H–H2O]+ | 2.06 | 4.077 | 177.0546 | 194.18 | Phenylpropanoids | 445![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 858 858 |
0.634 | 0.528 | 0.656 |
| 6 | Vanillin | C8H8O3 | [M + H]+ | 2.08 | 4.724 | 153.0546 | 152.15 | Phenylpropanoids | 1183 | 0.813 | 0.417 | 0.786 |
| 7 | N-methylcorydaldine | C12H15NO3 | [M + H]+ | 2.27 | 5.899 | 222.1125 | 221.25 | Alkaloids and derivatives | 303![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 906 906 |
0.631 | 0.488 | 0.775 |
| 8 | Piperine | C17H19NO3 | [M + H]+ | 1.53 | 10.325 | 286.1450 | 285.34 | Alkaloids and derivatives | 638![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 024 024 |
0.775 | 0.654 | 0.569 |
| 9 | Reticuline | C19H24NO4 | [M + H]+ | 1.20 | 3.937 | 330.1689 | 329.40 | Alkaloids and derivatives | 439![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 653 653 |
0.828 | 0.458 | 0.818 |
| 10 | D-tetra-hydropalmatine | C21H25NO4 | [M + H]+ | 1.04 | 6.039 | 356.1857 | 355.40 | Alkaloids and derivatives | 969![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 488 488 |
0.473 | 0.940 | 0.542 |
| 11 | 13-OxoODE | C18H30O3 | [M + Na]+ | 2.19 | 12.844 | 295.2267 | 294.40 | Fatty acid derivative | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 283 283![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 012 |
0.738 | 0.570 | 0.643 |
| 12 | Loliolide | C11H16O3 | [M + H]+ | 1.43 | 5.591 | 197.1171 | 196.24 | Terpenoids | 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 332 332 |
0.762 | 0.436 | 0.807 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||||||
| Negative mode | ||||||||||||
| 13 | Eriodictyol | C15H12O6 | [M–H]− | 1.93 | 6.700 | 287.05202 | 288.25 | Flavonoids | 440![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735 735 |
0.620 | 0.591 | 0.439 |
| 14 | Naringenin | C15H12O5 | [M–H]− | 1.95 | 7.521 | 271.05845 | 272.25 | Flavonoids | 439![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 246 246 |
0.584 | 0.591 | 0.584 |
| 15 | Luteolin | C15H10O6 | [M–H]− | 1.39 | 6.880 | 285.03643 | 286.24 | Flavonoids | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 445 445 |
0.566 | 0.647 | 0.553 |
| 16 | Norstictic acid | C18H12O9 | [M–H]− | 1.78 | 9.336 | 371.03360 | 372.30 | Phenylpropanoids and polyketides | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 379 379![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 540 540 |
0.598 | 0.415 | 0.799 |
| 17 | α-Linolenic acid | C18H30O2 | [M–H]− | 1.23 | 11.985 | 293.20690 | 294.42 | Fatty acid derivative | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 280![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 934 934 |
0.786 | 0.646 | 0.713 |
| 18 | FA 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2+1° 2+1° |
C18H32O3 | [M–H]− | 2.14 | 12.606 | 003![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 296.4 296.4 |
295.22 | Fatty acid derivative | 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 921 921![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 871 871 |
0.772 | 0.624 | 0.645 |
| 19 | Suberic acid | C8H14O4 | [M–H]− | 1.02 | 5.019 | 173.0807 | 174.19 | Fatty acid derivative | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 457 457 |
0.472 | 0.476 | 0.751 |
| 20 | (Z)-5,8,11-Trihydroxyoctadec-9-enoic acid | C18H34O5 | [M–H]− | 1.45 | 8.585 | 329.2276 | 330.50 | Fatty acid derivative | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 096![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 399 |
0.611 | 0.416 | 0.572 |
Flavonoids, alkaloids, and other secondary metabolites such as phenylpropanoids, polyketides, and terpenoids appeared to play important roles in the antioxidant activity observed in T. crispa extracts. Several of these compounds consistently showed positive correlations across all three antioxidant assays. Which aligns with previous reports supporting their pharmacological activities.2,3
To further explore their abundance and expression patterns across different extraction solvents, box plots were generated for 16 representative metabolites: d-tetrahydropalmatine, n-methylcorydaldine, piperine, reticuline, 13-oxoODE, α-linolenic acid, FA 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 + 1°, suberic acid, apigenin, naringenin chalcone, eriodictyol, naringenin, luteolin, trans-ferulic acid, vanillin and loliolide. These visualizations highlight the variation in metabolite abundance among extracts and reinforce their potential contribution to the antioxidant activity observed in the experimental assays (Fig. 4).
2 + 1°, suberic acid, apigenin, naringenin chalcone, eriodictyol, naringenin, luteolin, trans-ferulic acid, vanillin and loliolide. These visualizations highlight the variation in metabolite abundance among extracts and reinforce their potential contribution to the antioxidant activity observed in the experimental assays (Fig. 4).
To complement the selection of bioactive candidate metabolites of T. crispa extracts, a Circos plot was constructed to visualize the relative abundance and solvent-specificity of these compounds across ethyl acetate, ethanol, and water extracts (Fig. 5).
In the positive ion mode (Fig. 5A), distinct patterns of metabolite abundance were observed among the solvents. For the representative metabolites in the ethanol extract, TC0183 (D-tetrahydropalmatine) and TC0162 (piperine) were more abundant than in the ethyl acetate extract, with AUCs of 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510 vs. 21
510 vs. 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760 (2.4-fold) and 43
760 (2.4-fold) and 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920 vs. 6586 (6.7-fold), respectively. Both metabolites were detected only at trace levels in the water extract. For the ethyl acetate extract, the representative metabolites TC0052 (apigenin), TC0148 (n-methylcorydaldine), and TC0187 (trans-ferulic acid) showed higher abundances compared with the ethanol extract, with AUCs of 74
920 vs. 6586 (6.7-fold), respectively. Both metabolites were detected only at trace levels in the water extract. For the ethyl acetate extract, the representative metabolites TC0052 (apigenin), TC0148 (n-methylcorydaldine), and TC0187 (trans-ferulic acid) showed higher abundances compared with the ethanol extract, with AUCs of 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470 vs. 50
470 vs. 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 210 (1.48-fold), 41
210 (1.48-fold), 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 030 vs. 19
030 vs. 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510 (2.10-fold), and 122
510 (2.10-fold), and 122![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 vs. 42
100 vs. 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (2.91-fold), respectively. These metabolites were either undetected or present at negligible levels in the water extract. For the other detected metabolites, although the differences between the ethanol and ethyl acetate extracts were not pronounced enough to suggest clear solvent-specific separation, their abundances were markedly higher than in the water extract, where these compounds were barely detectable. Additionally, the water extract contained only a limited number of identifiable metabolites, and the representative metabolite TC0171 (reticuline) was still lower in abundance than in the ethanol extract, with AUCs of 26
000 (2.91-fold), respectively. These metabolites were either undetected or present at negligible levels in the water extract. For the other detected metabolites, although the differences between the ethanol and ethyl acetate extracts were not pronounced enough to suggest clear solvent-specific separation, their abundances were markedly higher than in the water extract, where these compounds were barely detectable. Additionally, the water extract contained only a limited number of identifiable metabolites, and the representative metabolite TC0171 (reticuline) was still lower in abundance than in the ethanol extract, with AUCs of 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 vs. 33
400 vs. 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 940 (1.3-fold).
940 (1.3-fold).
In the negative ion mode (Fig. 5B), the representative metabolite in the ethanol extract, TC0224 (α-linolenic acid), showed a higher abundance compared with the ethyl acetate extract, with AUC values of 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 330 vs. 33
330 vs. 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 (1.18-fold). Conversely, for the ethyl acetate extract, the representative metabolite TC0239 (norstictic acid) was more abundant than in the ethanol extract, with AUCs of 25
230 (1.18-fold). Conversely, for the ethyl acetate extract, the representative metabolite TC0239 (norstictic acid) was more abundant than in the ethanol extract, with AUCs of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 750 vs. 6512 (3.96-fold). For the other detected metabolites, the differences between the ethanol and ethyl acetate extracts were present but not sufficiently pronounced to indicate clear solvent-specific separation. Nonetheless, their abundances remained noticeably higher than those found in the water extract. In the water extract, no prominent metabolites were observed. Although TC0199 ((Z)-5,8,11-trihydroxyoctadec-9-enoic acid) was detectable, its abundance remained substantially lower than in both the ethyl acetate and ethanol extracts, by 1.8-fold and 1.4-fold, respectively.
750 vs. 6512 (3.96-fold). For the other detected metabolites, the differences between the ethanol and ethyl acetate extracts were present but not sufficiently pronounced to indicate clear solvent-specific separation. Nonetheless, their abundances remained noticeably higher than those found in the water extract. In the water extract, no prominent metabolites were observed. Although TC0199 ((Z)-5,8,11-trihydroxyoctadec-9-enoic acid) was detectable, its abundance remained substantially lower than in both the ethyl acetate and ethanol extracts, by 1.8-fold and 1.4-fold, respectively.
These findings indicate that most bioactive metabolites were preferentially extracted by semi-polar solvents, particularly ethanol and ethyl acetate.
These candidate metabolites were classified into flavonoids, phenylpropanoids, alkaloids, fatty acid derivatives, and terpenoids. Flavonoids such as apigenin, luteolin, and naringenin displayed strong positive correlations, consistent with their well-documented radical scavenging and metal-chelating activities.39–45 Phenylpropanoids, including vanillin and trans-ferulic acid, also contributed significantly, reflecting their ability to modulate oxidative stress through hydrogen donation and ROS neutralization.46,47 Alkaloids such as piperine and reticuline showed moderate correlations and have been implicated in neuroprotection, anti-inflammatory activity, and reactive oxigen species regulation.48–50 Fatty acids including linoleic and α-linolenic acid correlated with antioxidant activity by stabilizing cell membranes and supporting inflammatory resolution.51 Additionally, terpenoids such as betulinic acid contributed to antioxidant and anticancer activities.18 Additional pharmacological effects of these compound classes are summarized in Table 2.
| Compound class | Representative metabolites | Major pharmacological effects |
|---|---|---|
| Flavonoids | Apigenin, naringenin chalcone, eriodictyol | Antioxidant, anti-inflammatory, anticancer, and neuroprotective activities |
| Naringenin, luteolin | ||
| Phenylpropanoids and polyketides | N-trans-P-coumaroyltyramine | Antioxidant, antimicrobial and anti-inflammatory and anticancer activities |
| Feruloyl dehydrotyramine, trans-ferulic acid | ||
| Vanillin, norstictic acid | ||
| Alkaloids and derivatives | N-methylcorydaldine, piperine, reticuline | Antioxidant, anti-inflammatory, neuroactive, anticancer, and antidiabetic effects |
| D-tetra-hydropalmatine | ||
| Fatty acid derivatives | 13-OxoODE, α-linolenic acid, FA 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 + 1° 2 + 1° |
Anti-inflammatory, wound-healing, neuroprotective, and anticancer effects |
| Suberic acid, (Z)-5,8,11-trihydroxyoctadec-9-enoic acid | ||
| Terpenoids | Loliolide | Antioxidant, anti-inflammatory and neuro-protective effects |
Taken together, these results suggested that the antioxidant potential of T. crispa was not solely dependent on phenolic content but rather arises from a complex interplay of multiple classes of bioactive metabolites. The identification of these candidated compounds underscores the therapeutic potential of T. crispa as a multifunctional medicinal plant with applications in oxidative stress reduction, inflammation control, and disease prevention.
4 Conclusion
Our study emphasises the critical role of solvent polarity in shaping the phytochemical profile and antioxidant potential of T. crispa extracts. Semi-polar solvents, particularly ethyl acetate and ethanol, exhibited superior extraction efficiency compared to water, resulting in higher yields of diverse bioactive metabolites and enhanced antioxidant activity. Metabolomic analysis, supported by multivariate and correlation approaches, identified flavonoids, alkaloids, and other secondary metabolites as key contributors to antioxidant capacity. These findings highlight the importance of solvent selection in maximizing phytochemical recovery and reinforce the potential of T. crispa as a promising natural source of antioxidant agents.Author contributions
Methodology, investigation, data curation, formal analysis, visualization, writing—original draft, writing – review and editing, N. M.; methodology, investigation, formal analysis, data curation, writing—original draft, writing – review & editing, P. P.; methodology, resources, writing—review and editing, N. N.; methodology, resources, writing – review and editing, P. K.; methodology, resources, writing—review & editing, A. W.; methodology, investigation, formal analysis, data curation, writing – original draft, writing—review and editing, H. D.; methodology, writing – review and editing, S. S.; supervision, writing—review and editing, A. T.; conceptualization, supervision, project administration, funding acquisition, writing—review & editing, W. L. All the authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Abbreviations
| CV | Cross-validation |
| DPPH | 2,2-Diphenyl-1-picrylhydrazyl |
| ESI | Electrospray ionization |
| FRAP | Ferric reducing antioxidant power |
| LC-Q-TOF/MS | Liquid chromatography-quadrupole-time-offlight mass spectrometry |
| PCA | Principal component analysis |
| PLS-DA | Partial least-squares discriminant analysis |
| TPC | Total phenolic content |
| VIP | Variable importance in projection |
Data availability
The raw MS/MS spectra data are available in ProteomeXchange: JPST004023 and PXD067528 (preview URL for Reviewers: https://repository.jpostdb.org/preview/200271828368a6e21b0033c, Access key: 3098).Supplementary Information (SI) is available. See DOI: https://doi.org/10.1039/d5ra07211e.
Acknowledgements
We would like to thank Professor Ross H. Andrews for editing the manuscript. We also acknowledge that this work was supported by the Research Program from the Research Department of Khon Kaen University and the National Research Council of Thailand through the Hub of Knowledge Grant to WL. A. T. and N. M. were partially supported by the Program Management Unit for Human Resources & Institutional Development, Research and Innovation (PMU-B) [grant number B13F660135]. We confirm that the funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.References
- M. A. Z. Benjamin, R. A. Mohd Mokhtar, M. Iqbal, A. Abdullah, R. Azizah, L. Sulistyorini, N. Mahfudh and Z. A. Zakaria, J. Ethnopharmacol., 2024, 330, 118239 CrossRef CAS PubMed.
- W. Ahmad, W. Ahmad, I. Jantan and S. N. A. Bukhari, Front. Pharmacol., 2016, 7, 59 Search PubMed.
- E. Haque, M. S. Bari, L. Khandokar, J. Anjum, I. Jantan, V. Seidel and M. A. Haque, Phytochem. Rev., 2023, 22, 211–273 CrossRef CAS PubMed.
- H. Noor and S. J. H. Ashcroft, J. Ethnopharmacol., 1989, 27, 149–161 CrossRef CAS PubMed.
- N. Yusof, M. P. Y. Goh and N. Ahmad, Nat. Prod. Sci., 2022, 28, 105–114 CrossRef CAS.
- A. Chaudhary, R. Das, K. Mehta and D. K. Mehta, Heliyon, 2024, 10, e31229 CrossRef CAS PubMed.
- A. Thomas, E. K. Rajesh and D. S. Kumar, Phytother Res., 2016, 30, 357–366 CrossRef CAS PubMed.
- U. M. Zuhri, N. D. Yuliana, F. Fadilah, L. Erlina, E. H. Purwaningsih and A. Khatib, J. Ethnopharmacol., 2024, 319, 117296 CrossRef CAS PubMed.
- D. Joladarashi, N. D. Chilkunda and P. V. Salimath, J. Nutr. Sci., 2012, 1, e7 CrossRef PubMed.
- M. Srinivasan, A. R. Sudheer and V. P. Menon, J. Clin. Biochem. Nutr., 2007, 40, 92–100 CrossRef CAS PubMed.
- N. Maliyakkal, A. Appadath Beeran, S. A. Balaji, N. Udupa, S. Ranganath Pai and A. Rangarajan, Integr. Cancer Ther., 2015, 14, 156–171 CrossRef PubMed.
- A. Kumar and V. Jaitak, Eur. J. Med. Chem., 2019, 176, 268–291 CrossRef CAS PubMed.
- C. Bitwell, S. S. Indra, C. Luke and M. K. Kakoma, Sci. Afr., 2023, 19, e01585 CAS.
- Q. D. Do, A. E. Angkawijaya, P. L. Tran-Nguyen, L. H. Huynh, F. E. Soetaredjo, S. Ismadji and Y. H. Ju, J. Food Drug Anal., 2014, 22, 296–302 CrossRef CAS PubMed.
- T. Lefebvre, E. Destandau and E. Lesellier, J. Chromatogr. A, 2021, 1635, 461770 CrossRef CAS PubMed.
- N. N. Azwanida, Med. Aromat. Plants, 2015, 4, 196 Search PubMed.
- J. Dai and R. J. Mumper, Molecules, 2010, 15, 7313–7352 CrossRef CAS PubMed.
- P. Elliott, J. M. Posma, Q. Chan, I. Garcia-Perez, A. Wijeyesekera, M. Bictash, T. M. D. Ebbels, H. Ueshima, L. Zhao, L. van Horn, M. Daviglus, J. Stamler, E. Holmes and J. K. Nicholson, Sci. Transl. Med., 2015, 7, 285ra262 Search PubMed.
- P. Suwanchaikasem, K. Rattanapisit, R. Strasser and W. Phoolcharoen, Sci. Rep., 2024, 14, 9629 CrossRef CAS PubMed.
- M. Yu and V. Philip, Anal. Chem., 2025, 97, 17309–17314 CrossRef CAS PubMed.
- K. Hajnajafi and M. A. Iqbal, Proteome Sci., 2025, 23, 5 CrossRef PubMed.
- L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W. M. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, T. Hankemeier, N. Hardy, J. Harnly, R. Higashi, J. Kopka, A. N. Lane, J. C. Lindon, P. Marriott, A. W. Nicholls, M. D. Reily, J. J. Thaden and M. R. Viant, Metabolomics, 2007, 3, 211–221 CrossRef CAS PubMed.
- L. M. Quiros-Guerrero, P. M. Allard, L. F. Nothias, B. David, A. Grondin and J. L. Wolfender, Sci. Data, 2024, 11, 415 CrossRef CAS PubMed.
- Z. Pang, Y. Lu, G. Zhou, F. Hui, L. Xu, C. Viau, A. F. Spigelman, P. E. MacDonald, D. S. Wishart, S. Li and J. Xia, Nucleic Acids Res., 2024, 52, W398–W406 CrossRef PubMed.
- V. Shulaev, Briefings Bioinf., 2006, 7, 128–139 CrossRef CAS PubMed.
- S. N. Nakorn, H. Dokduang, N. Namwat, P. Klanrit, A. Wangwiwatsin, B. Promraksa, S. Sitthirak, T. Seaban and W. Loilome, Heliyon, 2024, 10 CrossRef CAS PubMed.
- L. Shi, W. Zhao, Z. Yang, V. Subbiah and H. A. R. Suleria, Environ. Sci. Pollut. Res., 2022, 29, 81112–81129 CrossRef CAS PubMed.
- F. Imtiaz, D. Ahmed, O. A. Mohammed, U. Younas and M. Iqbal, Results Chem., 2025, 13, 101960 CrossRef CAS.
- S. Baliyan, R. Mukherjee, A. Priyadarshini, A. Vibhuti, A. Gupta, R. P. Pandey and C. M. Chang, Molecules, 2022, 27, 1326 CrossRef CAS PubMed.
- J. Zheng, W. Tian, C. Yang, W. Shi, P. Cao, J. Long, L. Xiao, Y. Wu, J. Liang, X. Li, S. Zhao, K. Zhang, H. Zhi and P. Sun, Ind. Crops Prod., 2019, 127, 36–45 CrossRef CAS.
- B. Singh, S. Nathawat and R. A. Sharma, Arab. J. Chem., 2021, 14, 103381 CrossRef CAS.
- H. Abd Hamid, R. Mutazah, I. H. Yahya and S. S. Zeyohannes, Mater. Sci. Forum, 2021, 1025, 163–168 Search PubMed.
- M. Tourabi, K. Faiz, R. Ezzouggari, B. Louaste, M. Merzouki, M. Dauelbait, M. Bourhia, K. S. Almaary, F. Siddique, B. Lyoussi and E. Derwich, Bioresour Bioprocess, 2025, 12, 24 CrossRef PubMed.
- W. C. Chen, S. W. Wang, C. W. Li, H. R. Lin, C. S. Yang, Y. C. Chu, T. H. Lee and J. J. Chen, Antioxidants, 2022, 11, 398 CrossRef CAS PubMed.
- W. C. Chen, S. W. Wang, C. W. Li, H. R. Lin, C. S. Yang, Y. C. Chu, T. H. Lee and J. J. Chen, Antioxidants, 2022, 11, 398 CrossRef CAS PubMed.
- R. Paranthaman, K. Sureshkumar and P. Muthukumaran, Pharmacogn. J., 2018, 10, 457–462 CrossRef.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, Nucleic Acids Res., 2025, 53, D1516–D1525 CrossRef PubMed.
- P. Schober, C. Boer and L. A. Schwarte, Anesth. Analg., 2018, 126, 1763–1768 CrossRef PubMed.
- S. Tang, B. Wang, X. Liu, W. Xi, Y. Yue, X. Tan, J. Bai and L. Huang, Food Front., 2025, 6, 218–247 CrossRef CAS.
- E. Middleton, C. Kandaswami and T. C. Theoharides, Pharmacol. Rev., 2000, 52, 673–751 CrossRef CAS PubMed.
- V. Naponelli, M. T. Rocchetti and D. Mangieri, Int. J. Mol. Sci., 2024, 25, 5569 CrossRef CAS PubMed.
- G. Li, J. Zhou, M. Sun, J. Cen and J. Xu, J. Funct. Foods, 2020, 74, 104196 CrossRef CAS.
- M. Mahwish, M. Imran, H. Naeem, M. Hussain, S. A. Alsagaby, W. Al Abdulmonem, A. Mujtaba, M. A. Abdelgawad, M. M. Ghoneim, A. H. El-Ghorab, S. Selim, S. K. Al Jaouni, E. M. Mostafa and T. F. Yehuala, Food Sci. Nutr., 2025, 13, e4682 CrossRef CAS PubMed.
- S. M. Nabavi, D. Šamec, M. Tomczyk, L. Milella, D. Russo, S. Habtemariam, I. Suntar, L. Rastrelli, M. Daglia, J. Xiao, F. Giampieri, M. Battino, E. Sobarzo-Sanchez, S. F. Nabavi, B. Yousefi, P. Jeandet, S. Xu and S. Shirooie, Biotechnol. Adv., 2020, 38, 107316 CrossRef CAS PubMed.
- X. Liu, N. Wang, S. Fan, X. Zheng, Y. Yang, Y. Zhu, Y. Lu, Q. Chen, H. Zhou and J. Zheng, Sci. Rep., 2016, 6, 39735 CrossRef CAS PubMed.
- S. Liga, C. Paul and F. Péter, Plants, 2023, 12, 2732 CrossRef CAS PubMed.
- C. Iannuzzi, M. Liccardo and I. Sirangelo, Int. J. Mol. Sci., 2023, 24, 1817 CrossRef CAS PubMed.
- S. Sirin, S. Nigdelioglu Dolanbay and B. Aslim, Health Sci. Rev., 2023, 6, 100071 Search PubMed.
- Y. R. Kong, K. C. Tay, Y. X. Su, C. K. Wong, W. N. Tan and K. Y. Khaw, Molecules, 2021, 26, 728 CrossRef CAS PubMed.
- F. Al-Mamoori and A. M. A. Qasem, in Medicinal Plants–Chemical, Biochemical, and Pharmacological Approaches, ed. M. S. de Oliveira, E. Helena De Aguiar Andrade, R. Kumar and S. N. Mali, IntechOpen, London, 2023 Search PubMed.
- P. C. Calder, Biochem. Soc. Trans., 2017, 45, 1105–1115 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |