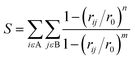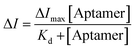Open Access Article
Open Access ArticleAccelerated aptamer selection via SELEX and molecular simulations for lipopolysaccharide detection
Thuy-Duong Thi Tran†
a,
Hai Ly Nguyen†b,
Phan Thi Ngoc Hoa a,
Thu Thao Phamc,
Nguyen Thi Thanh Huyena,
Giang Dau Huonga,
Hoa Thi Hoanga,
Patrick Wagnerd,
Anne Charriere,
Truong T. N. Lien
a,
Thu Thao Phamc,
Nguyen Thi Thanh Huyena,
Giang Dau Huonga,
Hoa Thi Hoanga,
Patrick Wagnerd,
Anne Charriere,
Truong T. N. Lien *a,
Phuong H. Nguyenf and
Nguyen The Toan
*a,
Phuong H. Nguyenf and
Nguyen The Toan *g
*g
aVietnam-Korea Institute of Science and Technology, Hoa Lac High-Teck Park, Thach That, Hanoi, Vietnam. E-mail: ttnlien@mst.gov.vn
bInstitute of Theoretical and Applied Research, Duy Tan University, Hanoi, 100000, Vietnam
cGraduate University of Science and Technology, Vietnam Academy of Science and Technology, Hanoi, 100000, Vietnam
dLaboratory for Soft Matter and Biophysics, ZMB, Department of Physics and Astronomy, KU Leuven, Celestijnenlaan 200 D, B-3001 Leuven, Belgium
eAix Marseille Univ, CNRS, CINaM, Marseille, France
fUniversité Paris Cité, CNRS, Laboratoire de Biochime Théorique, 13 Rue Pierre et Marie Curie, Paris, F-75005, Frances
gKey Laboratory for Multiscale Simulation of Complex Systems and Faculty of Physics, University of Science, Vietnam National University, Hanoi, Vietnam
First published on 27th November 2025
Abstract
Lipopolysaccharides (LPS), the major components of Gram-negative bacterial outer membranes, are critical virulence factors that can trigger severe immune responses, leading to sepsis and other inflammatory diseases. Rapid and accurate LPS detection is therefore essential in clinical diagnosis, pharmaceutical safety, and food monitoring. Aptamers represent a promising alternative to antibodies due to their high specificity, stability, and cost-effectiveness. However, the conventional SELEX process for aptamer selection is labor-intensive and time-consuming, often generating numerous candidates requiring costly experimental validation. Herein, we present a computationally assisted post-SELEX refinement pipeline that integrates molecular docking and molecular dynamics simulations to efficiently prioritize and validate aptamer candidates obtained from SELEX. Using LPS from Klebsiella pneumoniae ATCC 15380 as the model target, ten rounds of SELEX yielded candidate ssDNA sequences that were subsequently evaluated through our computational workflow. Among the refined candidates, aptamer seq. 5 exhibited the highest predicted and experimental affinity, with a dissociation constant (Kd) of 6.68 nM, supported by both in vitro assays and molecular simulations. These finding demonstrate that the proposed SELEX-simulation pipeline significantly accelerates post-SELEX aptamer refinement, effectively reducing the number of candidates requiring expensive experimental screening and validation.
1. Introduction
Lipopolysaccharides (LPSs), also known as endotoxins, are essential components of the outer membrane of Gram-negative bacteria and are major contributors to sepsis. Even at low concentrations, LPSs can trigger strong immune responses in the human circulatory system, leading to the release of pro-inflammatory cytokines (e.g., IL-6, IL-1β, and TNF-α). This immune cascade may cause tissue damage, organ failure, and, if untreated, death.1 Structurally, LPSs constitute about 75% of the bacterial outer membrane and are composed of three domains: the hydrophilic O-antigen, the hydrophobic lipid A, and the core polysaccharide bridging the two.2 As LPSs are continuously shed during bacterial growth, division, and lysis, they are commonly found in air, water, soil, and surfaces. Their high thermal stability and ability to pass through 0.2 µm filters make their removal particularly challenging. Therefore, identifying LPS-binding molecules is crucial for sensitive and reliable detection, supporting the diagnosis, monitoring, and prevention of endotoxin-related health risks.Several methods have been established for endotoxin detection. Traditional approaches, including the rabbit pyrogen test (RPT), Limulus amoebocyte lysate (LAL), monocyte activation test (MAT), and recombinant factor C (rFC), are widely applied in regulatory and industrial settings. However, these assays are often costly, time-consuming, technically complex, and raise ethical concerns due to animal testing.3 More recently, EndoLISA has been introduced as a less labor-intensive alternative, but it still relies on expensive reagents (capture proteins, recombinant factor C, fluorescent substrates) and requires specialized equipment.4
To address these challenges, aptamers have emerged as promising alternatives, through the SELEX process, which typically involves multiple rounds of experimental screening. Aptamers are short single-stranded nucleic acids that bind specifically and with high affinity to target molecules. They are selected through the systematic evolution of ligands by exponential enrichment (SELEX) process and offer advantages such as low cost, stability, rapid detection, and potential integration into point-of-care devices.5 Compared with conventional assays, aptamer-based platforms represent simpler, faster, and more versatile approaches for endotoxin detection. Several aptamer-based biosensors for LPS detection have demonstrated impressive performance. For example, Kim et al. (2012) reported an electrochemical aptasensor detecting LPS in the range of 0.01–1 ng mL−1 using a thiol-modified aptamer assembled on a gold electrode.5 Similarly, Su et al. (2013) developed an impedance-based aptasensor employing a polymerized self-assembled monolayer (SAM) of thiol-functionalized 3,4-ethylenedioxythiophene (EDOT) and AuNPs, achieving an ultra-low detection limit of 0.1 pg mL−1.6 However, previous studies have mainly focused on aptamer selection for LPS from Escherichia coli.5,7,8 Klebsiella pneumoniae especially multidrug-resistant (MDR) or carbapenem-resistant (CRKP) strains, is one of the leading nosocomial pathogens, causing high morbidity and mortality.9 Existing studies have mainly focused on selecting aptamers targeting whole-cell or outer membrane proteins (OMPs) of K. pneumoniae, instead of LPS.10,11 Klebsiella pneumoniae ATCC 15380 strain carries O1 antigen on LPS and K2 antigen on capsule which are strongly associated with K. pneumoniae strains causing hypervirulent invasive infections such as liver abscesses.12,13 Aptamers which could recognize LPS-O1 of K. pneumoniae would therefore be highly valuable for differential diagnosis between dangerous K. pneumoniae infections and other Gram-negative bacteria.
Despite these advances, the SELEX process itself remains time-consuming, labor-intensive, and costly, generating a large pool of potential sequences that must be experimentally screened and validated to identify high-affinity candidates. It requires multiple iterative rounds of binding, partitioning, and amplification, followed by affinity evaluation of multiple candidate sequences.14 To streamline the post-SELEX phase, molecular modeling techniques have been introduced as powerful complementary tools to assist aptamer refinement and candidate prioritization.15 These computational approaches allow researchers to predict aptamer conformations, simulate aptamer–target interactions, and evaluate binding stability in silico, thereby shortening the candidate validation process. Furthermore, molecular simulations provide valuable mechanistic insights that support rational aptamer design and optimization. For instance, for post-SELEX analysis and optimization, Rabal et al. (2016) proposed a workflow integrating Rosetta 3D modeling and 3dRPC docking for aptamer–protein interactions,16 while Vu et al. applied computational methods to analyze aptamer–PDGF-B binding, highlighting structural roles and suggesting therapeutic applications.17
Aptamers are an ideal research subject for molecular simulation. Unlike rigid drug targets, single-stranded nucleic acids like ssDNA and ssRNA are inherently flexible molecules with numerous configurations.18 This complexity makes Molecular Dynamics (MD) simulation a non-dispensable tool required to accurately sample this vast conformational space, understand their true structure and function, and model their dynamic interaction with biomolecules such as LPS. Thus, our computational approach is broadly applicable, offering a critical approach for investigating numerous other biological systems where ssDNA and ssRNA are central players. In this study, we integrated the SELEX process with a computational post-SELEX refinement pipeline based on molecular docking and molecular dynamics simulations. This integrated strategy accelerates aptamer refinement and candidate validation after SELEX, reducing the number of sequences requiring costly experimental testing. Using LPS from Klebsiella pneumoniae ATCC 15380 as a model target, we demonstrate how this hybrid workflow facilitates the identification of optimized aptamers with superior binding properties.
2. Experimental
2.1 Materials and reagents
A 91-nucleotide single-stranded DNA library containing 55 bases of randomized sequence 5′-ATCCGTCACACCTGCTCT–N55–TGGTGTTGGCTCCCGTAT-3′ was obtained from IDT, Inc. (California, USA). The DNA library was amplified using a forward primer 5′-Biotin-ATC CGT CAC ACC TGC TCT-3′ and a phosphorylated reverse primer 5′-/5Phos/ATA CGG GAG CCA ACA CCA-3′, also synthesized by IDT, Inc., California, USA. The LPS from Klebsiella pneumoniae ATCC 15380 was purchased from L4268, Sigma, MO, USA. All chemicals needed for the aptamer selection process, including the buffers, solution, and materials were obtained from Sigma (Germany), Merck (Germany), or Thermo Fisher (USA), and the experiment was performed on a Costar 96-well EIA/RIA plate (Immunochemistry Technologies, CA, USA).2.2 SELEX procedure combined with molecular simulations
2.2.1.1 SELEX procedure. The aptamer selection process was performed with minor modifications from a previously reported method.19 In the first round, lipopolysaccharide (LPS) from Klebsiella pneumoniae ATCC 15380 was diluted in carbonate, bicarbonate buffer (pH 9.6) and immobilized on a Costar 96-well EIA/RIA plate overnight at 4 °C. Wells were then blocked with 3% (w/v) bovine serum albumin (BSA) at 37 °C for 2 h. A 10 µM ssDNA library in binding buffer (20 mmol L−1 HEPES, pH 7.35; 1 mmol L−1 MgCl2; 1 mmol L−1 CaCl2; 150 mmol L−1 NaCl) was heated at 95 °C for 5 min and rapidly cooled on ice for 10 min prior to incubation. After blocking, the LPS-coated wells were washed three times with washing buffer (binding buffer + 0.05% Tween-20) and incubated with the ssDNA pool at 37 °C for 1 h with shaking. Unbound sequences were removed by six washing steps. Bound ssDNA was eluted by adding nuclease-free water and heating at 95 °C for 15 min, then collected and amplified by PCR. PCR products were analyzed by 2% agarose gel electrophoresis. In subsequent rounds, the selection procedure was maintained with gradually increased stringency. From the fourth round onward, a negative selection step was introduced: the ssDNA library was first incubated in BSA-blocked blank wells at 37 °C for 1 h with shaking to eliminate non-specific binders before exposure to LPS-coated wells.
2.2.1.2 Symmetric PCR optimization. PCR optimization was carried out to determine the optimal annealing temperature, cycle number, and template concentration. Each 50 µL reaction contained 25 µL DreamTaq PCR Master Mix, 1 µL of each primer (10 µM), the DNA template, and nuclease-free water. The thermocycling program included an initial denaturation at 95 °C for 3 min, followed by cycles of 95 °C for 10 s (denaturation), annealing at variable temperatures for 10 s, and 72 °C for 10 s (extension), with a final extension at 72 °C for 3 min. PCR products were analyzed by 2% agarose gel electrophoresis, and the condition that produced strong specific bands with minimal non-specific amplification was selected. Following optimization, large-scale PCR was performed to generate sufficient DNA for ssDNA regeneration. The PCR products were purified using a Microcon-30 kDa centrifugal filter unit (Ultracel-30 membrane) to remove residual primers.
2.2.1.3 ssDNA regeneration. To obtain ssDNA as the enriched library for the next selection round, PCR products were used as templates for asymmetric PCR (aPCR). The variables optimized for aPCR were the optimal annealing temperature, forward/reverse primer ratio, cycle number, and template concentration. The thermocycler program was set as follows: initial denaturation at 95 °C for 3 minutes, followed by 20 seconds for each of three steps (denaturation at 95 °C, annealing at variable temperatures, and extension at 72 °C), and 3 minutes at 72 °C for final extension. The aPCR products were analyzed on a 4% agarose gel, and the condition that produced an intense ssDNA band with minimal non-specific amplification was selected for further use.
To assess library enrichment during the selection process, lambda was used to digest the 5′−phosphorylated reverse strand of double-stranded DNA (dsDNA). Symmetric PCR products were first purified using the PureLink™ Quick Gel Extraction Kit to obtain dsDNA labelled with a biotinylated forward primer and a phosphate-labeled reverse primer. Then, 1 µg of purified dsDNA was incubated with 10 U of lambda exonuclease at 37 °C for 60 minutes. The reaction was terminated by heating at 80 °C for 10 minutes, and the digestion products were analyzed on a 4% agarose gel.
The SELEX procedure is presented in Fig. S8.
A total of 45 white colonies were picked, and plasmids were extracted and sequenced. The 45 valid sequences were aligned using MegaX, and phylogenetic analysis by Mfold was performed to group them into families. The representative sequences given are those with the most repeat frequency and the highest secondary structure stability.
Molecular docking of the aptamers to LPS was carried out using the Schrödinger 2016.4 software package23 with a flexible docking protocol that permitted limited local relaxation. Docking poses were scored based on docking energy and the number of intermolecular hydrogen bonds (H-bonds). Three representative aptamer–LPS complexes were selected for further MD simulations: two with the lowest docking energies and one with the highest number of H-bonds.
To analyze the structural and energetic properties of aptamer–LPS binding, trajectories were examined for RMSD, RMSF, H-bond formation, coordination numbers, and binding free energies. The coordination number between molecules A and B was calculated as:
Binding free energies were estimated from equilibrated snapshots using the MM/GBSA method32 as implemented in the gmx_MMPBSA Python library.33,34 Visualization and figure generation were performed using VMD.
2.3 Fabrication of the electrochemical aptasensor
Commercial disposable electrochemical printed (DEP) chips were purchased from BioDevice Technology Ltd, Japan (https://www.biodevicetech.com). These chips were fabricated using screen-printing technology and configured as a three-electrode system, consisting of a carbon working electrode, a carbon counter electrode, and an Ag/AgCl reference electrode. The working electrode had a surface area of 2.64 mm2.For surface modification, gold nanoparticles (AuNPs) were electrodeposited onto the working electrode using cyclic voltammetry (CV). Tetrachloroauric acid (HAuCl4) was diluted in 10× phosphate-buffered saline (PBS) to a final concentration of 165 µM. A 35 µL aliquot of this solution was applied to the DEP chip, covering all three electrodes. CV was performed within a potential window of −650 to +650 mV versus Ag/AgCl for 20 cycles at a scan rate of 50 mV s−1. After deposition, the AuNP-modified electrodes were rinsed with deionized water and dried under a nitrogen (N2) stream. To remove any residual gold oxide, the electrodes were scanned in 1 M H2SO4 over a potential range of −0.2 to +1.4 V versus Ag/AgCl for 10 cycles. Finally, the AuNP-coated SPCEs were washed twice with distilled water and dried under N2 before use.
For aptamer immobilization, aptamers were commercially synthesized with a 5′-biotin modification (Integrated DNA Technologies, IDT). The AuNP-coated electrodes were first incubated with 0.5 mg mL−1 streptavidin at 37 °C for 1 h to allow streptavidin binding. The streptavidin-modified electrodes were then immersed in biotinylated aptamer solutions at varying concentrations for 30 min at 37 °C. To minimize nonspecific binding, the electrodes were blocked with 1 mM mercaptosuccinic acid for 2 h at 37 °C.
For target recognition, the aptamer-modified electrodes were exposed to different concentrations of lipopolysaccharide (LPS) from Klebsiella pneumoniae ATCC 15380 in 1× PBS. Specificity tests were performed using bovine serum albumin (BSA), glucose, and LPS from Escherichia coli O127:B8, chosen as structural analogues containing lipid, polysaccharide, and polypeptide components similar to the target LPS. After each step, electrodes were thoroughly washed with buffer and deionized water, and dried under N2 gas.
2.4 Characterization and analytical techniques
Biotinylated aptamer sequences were denatured at 95 °C for 5 min and rapidly cooled on ice for at least 10 min in binding buffer before use. After blocking, 50 µL of 200 nM biotinylated aptamer solution was added to each well, followed by gentle shaking at 37 °C for 1 h. Unbound sequences were removed with three washes of PBS-T. Subsequently, 50 µL of streptavidin-poly HRP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10.000 dilution) was added and incubated at 37 °C for 30 min. After five additional washes with 200 µL PBS-T, 100 µL of TMB substrate solution was added and incubated in the dark at 37 °C for 10 min. The reaction was terminated with 25 µL of 0.5 M H2SO4, and absorbance was measured at 450 nm using an Epoch Microplate Spectrophotometer (Agilent BioTek, USA).
10.000 dilution) was added and incubated at 37 °C for 30 min. After five additional washes with 200 µL PBS-T, 100 µL of TMB substrate solution was added and incubated in the dark at 37 °C for 10 min. The reaction was terminated with 25 µL of 0.5 M H2SO4, and absorbance was measured at 450 nm using an Epoch Microplate Spectrophotometer (Agilent BioTek, USA).
Gold nanoparticles (AuNPs) were synthesized via the citrate reduction of chloroauric acid (HAuCl4) using the Turkevich–Frens method, yielding monodisperse spherical particles with an average diameter of ∼15 ± 2 nm.36 To construct the sensing platform, AuNPs (OD 10) were first conjugated with streptavidin (91 µg mL−1), followed by incubation with biotinylated aptamers (430 µg mL−1) for 1 h at room temperature. LPS (71.5 µg mL−1) was subsequently introduced and incubated for 30 min before SERS measurements. Streptavidin–biotin coupling ensured stable immobilization of aptamers on the AuNP surface, while the subsequent interaction with LPS induced conformational rearrangements that altered the local molecular environment. These changes affected the vibrational modes of the aptamer–LPS complex, leading to distinct shifts and variations in SERS peak intensities. Comparison of spectra before and after LPS addition provided direct evidence of specific aptamer–LPS binding and offered insights into their molecular recognition mechanism. A schematic representation of the functionalization and binding process is shown in Fig. S2.
In this study, an aptamer-based sensor was fabricated using a gold nanoparticles (AuNPs)-modified electrode to improve surface area and conductivity. The aptamer was immobilized through a streptavidin–biotin linkage. Upon LPS binding, conformational changes at the electrode interface hindered electron transfer between the electrode and the electrolyte.
EIS measurements were carried out with a potentiostat/galvanostat (CS350M, Cortest Instruments, China) using a 10 mV AC voltage over the frequency range of 0.05 Hz–10 kHz, with a Ag/AgCl reference electrode. Impedance spectra were displayed as Nyquist plots, and data were fitted using CS Studio6 software. The electrolyte consisted of 5 mM K3[Fe(CN)6]/K4[Fe(CN)6] (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) in 0.1 M KCl. Binding of LPS to the aptamer increased the charge transfer resistance (RCT), confirming reduced electron transfer efficiency. Analysis of these EIS responses enabled evaluation of the sensitivity, selectivity, and overall performance of the sensor, highlighting its potential for rapid and reliable LPS detection in biomedical and environmental applications.
1 molar ratio) in 0.1 M KCl. Binding of LPS to the aptamer increased the charge transfer resistance (RCT), confirming reduced electron transfer efficiency. Analysis of these EIS responses enabled evaluation of the sensitivity, selectivity, and overall performance of the sensor, highlighting its potential for rapid and reliable LPS detection in biomedical and environmental applications.
The resulting peak current variations (ΔI) were plotted against aptamer concentration to construct a binding curve. The dissociation constant (Kd) was obtained by fitting the data to the Langmuir isotherm model, which describes the equilibrium between free and bound aptamer–LPS complexes, according to the equation:
3. Results and discussion
3.1 Aptamer selection by SELEX
3.1.1.1 Optimizing PCR conditions. PCR amplification is a crucial step in the SELEX process.39 However, amplifying random DNA libraries differs significantly from conventional homogeneous DNA amplification.40 Improper amplification of the random DNA library can lead to the loss of the main product and the accumulation of unwanted by-products, potentially causing SELEX failure. In this study, we investigated the impact of several factors on PCR efficiency, including annealing temperature, cycle number, and template concentration, during the first and second rounds of SELEX. The tested conditions for each factor are summarized in Table S2. As shown in Fig. S3A and D, within the annealing temperature range of 50–59.5 °C, the strongest PCR product band with minimal smearing was observed at 56.3 °C. At both higher and lower annealing temperatures, a decrease in band intensity (Fig. S3A) and increased smearing (Fig. S3D) were noted, although the differences were not significant. This observation aligns with previous findings that annealing temperature is more critical for long PCR templates than for short ones, such as DNA aptamers.39
Overamplification of random DNA pools can lead to opportunistic mispairing, resulting in aberrant PCR products. Therefore, optimizing the PCR cycle number and template concentration is essential. In the first SELEX round, unwanted by-products appeared from the 10th PCR cycle (Fig. S3B) and at an ssDNA template concentration of 0.1 ng µL−1 (Fig. S3C). In the second round, by-products emerged from the 12th PCR cycle (Fig. S3E) and at an ssDNA template concentration of 1 ng µL−1 (Fig. S3F). These differences could be attributed to the reduction in ssDNA pool size or changes in ssDNA purity across selection rounds.41 Thus, while an annealing temperature of 56.3 °C was found to be optimal, cycle number and template concentration should be carefully adjusted in each round within minimal variation ranges to prevent undesired amplification artifacts.
3.1.1.2 Optimizing asymmetric PCR conditions. Following the optimization of PCR parameters, we further investigated conditions for asymmetric PCR, which was employed to regenerate ssDNA for subsequent SELEX rounds. Several methods for ssDNA regeneration were considered, such as magnetic separation using streptavidin-coated beads, lambda exonuclease digestion of the phosphorylated strand, size separation on denaturing polyacrylamide gel electrophoresis based on unequal strand lengths, and asymmetric PCR. Among these, aPCR was chosen due to its straightforward procedure, low input requirement, effectiveness, and cost-efficiency.39 Furthermore, aPCR products could be used directly for the next round of SELEX without additional purification.42
The primer-removed PCR products were used for asymmetric PCR optimization. Factors that could influence ssDNA yield, including annealing temperature, template concentration, forward/reverse primer ratio, and cycle number, were systematically examined. As summarized in Table S3, ssDNA bands migrated to lower positions compared to dsDNA bands (91 bp). As shown in Fig. S4A, the optimal annealing temperature was determined to be 59 °C, which was then applied in subsequent rounds. In Fig. S4B, higher template concentrations (>0.1 ng µL−1) caused smearing and by-product formation. Interestingly, when sufficient template was used, variations in forward/reverse primer ratios or cycle numbers did not significantly affect the relative yield of ssDNA compared to dsDNA (Fig. S4C and D). However, consistent with previous reports, the optimal aPCR conditions in the first SELEX round differed from those in later rounds, likely due to decreasing purity and size of ssDNA pools throughout the selection process.41 Therefore, except for maintaining an annealing temperature of 59 °C, other parameters should be carefully optimized in each round within minimal variation ranges.
Taken together, the optimized PCR and aPCR conditions ensured high-quality ssDNA library regeneration with minimal by-products, providing a robust foundation for the subsequent SELEX rounds.
The enrichment was clearly evidenced by the enzyme-linked oligonucleotide assay: while the initial library showed only a weak absorbance at 450 nm, the signal rose significantly from round 8 onward and nearly plateaued by round 10 (Fig. 1). This progressive increase confirmed that LPS-binding aptamers were being enriched and that maximal binding capacity had nearly been reached. Consequently, the selection was terminated after the tenth round. The enriched products from round 10 were amplified by PCR, cloned into E. coli TOP10, and sequenced. A total of 15 unique sequences, each appearing at least twice, were identified. These sequences represented the dominant aptamer candidates and were subsequently subjected to molecular docking analysis to evaluate their predicted binding affinity to LPS (Table S1). Therefore, the successful enrichment and identification of 15 dominant aptamer candidates provided a solid foundation for subsequent in silico docking, molecular dynamics simulations, and experimental validation, enabling the rational selection of the most promising sequences for LPS recognition.
3.2 In silico evaluation of aptamer candidates
To complement the experimental SELEX results, a systematic computational screening pipeline was established, consisting of molecular docking, molecular dynamics (MD) simulations, and coordination number (CN) analysis. This multi-step approach not only enabled the prediction and refinement of aptamer–LPS binding modes, but also provided a quantitative framework to discriminate among candidate sequences. In particular, the CN analysis served as a critical highlight of the pipeline, offering a robust metric for ranking aptamer performance and thereby underscoring the strength of our integrative in silico strategy.| Aptamer seq. | Docking energy (kcal mol−1) | Aptamer seq | Docking energy (kcal mol−1) |
|---|---|---|---|
| 1 | −45.80 | 9 | −44.27 |
| 2 | −44.20 | 10 | −46.73 |
| 3 | −46.38 | 11 | −47.11 |
| 4 | −49.05 | 12 | −45.60 |
| 5 | −50.13 | 13 | −45.99 |
| 6 | −49.10 | 14 | −44.60 |
| 7 | −49.30 | 15 | −49.20 |
| 8 | −47.09 |
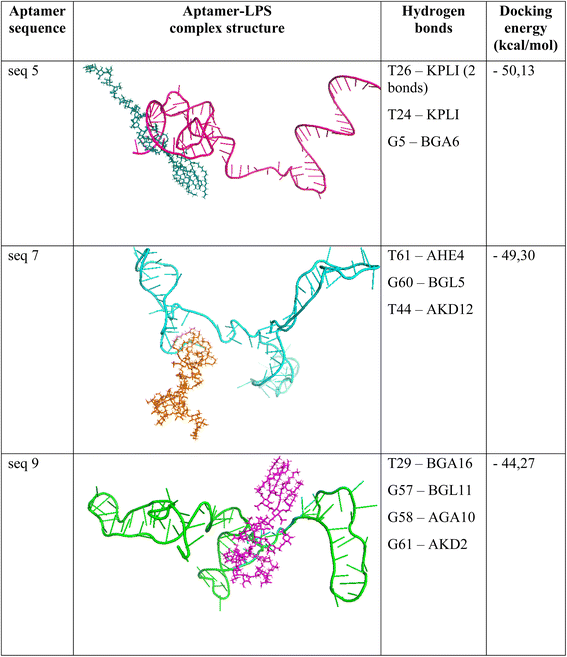
Notably, aptamers seq. 5 and seq. 7 exhibited the lowest docking energies, suggesting particularly strong initial binding propensities toward LPS. Interestingly, although seq. 9 showed a slightly higher average docking energy (−44.27 kcal mol−1), it established four distinct hydrogen bonds with LPS, the highest number among all candidates. This finding implies that binding affinity may not be determined solely by docking energy, but also by the quality and distribution of non-covalent interactions. Aptamer seq. 5 also formed four hydrogen bonds; however, two of these involved the same residue pair (T26 and the KPLI moiety of LPS), which may limit the diversity of its interaction network compared with aptamer seq. 9.
Taken together, these docking results highlight aptamer seq. 5, aptamer seq. 7, and aptamer seq. 9 as promising candidates that balance favorable binding energies and meaningful interaction profiles. The representative docking poses and hydrogen bond interactions of these three aptamers are shown in Fig. 2. Therefore, they were prioritized for further evaluation through molecular dynamics simulations to assess the stability and robustness of their binding under dynamic conditions.
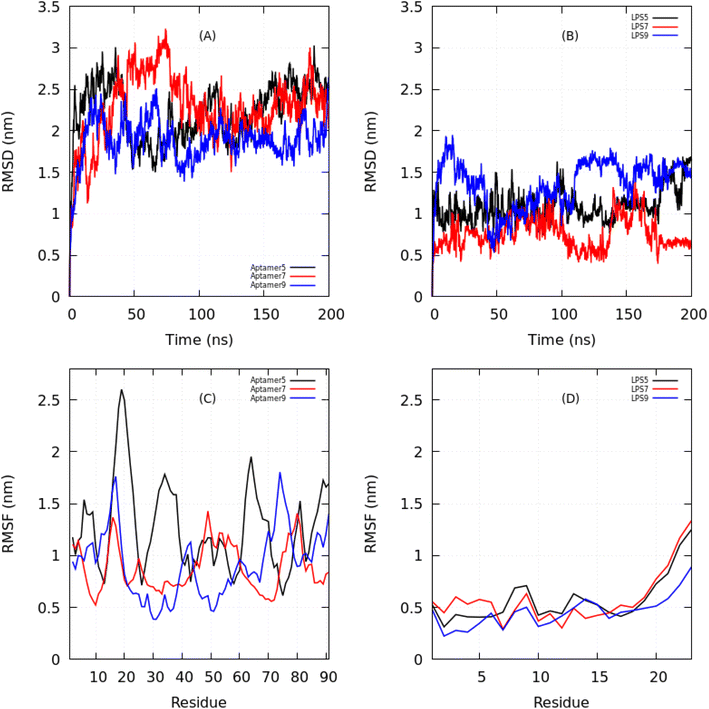
3.2.2.1 Global conformational rearrangements. The RMSD analysis provided an overview of structural stability over the 200 ns simulations (Fig. 3A and B). All aptamers showed an initial rise in RMSD up to ∼2.0 nm within the first 30 ns, reflecting conformational adaptation. LPS molecules exhibited a similar equilibration phase, followed by distinct stabilization patterns. Among the complexes, LPS9 fluctuated the most (∼1.8 nm, with persistent oscillations), indicating high conformational flexibility. LPS7 stabilized at ∼1.0–1.3 nm after 30 ns, whereas LPS5 maintained the lowest RMSD (<1.0 nm after 50 ns), suggesting the most stable complex formation.
RMSF analysis revealed residue-specific flexibility of the aptamers and LPS molecules (Fig. 3C and D). Aptamer seq. 5 was the most flexible, with pronounced peaks (>2.5 nm) around residues 20–25 and additional fluctuations near residues 35, 50, and 75–85. Aptamer seq. 7 showed the lowest overall fluctuations (<1.5 nm), indicating a relatively rigid structure. Aptamer seq. 9 displayed intermediate flexibility, with moderate peaks at residues 15, 35–40, and 75–80, and reduced mobility at residues involved in H-bonding (29, 57, 58, 61). For the LPS molecules, all complexes showed weak flexibility (0.2–1.3 nm), with enhanced motion at the C-terminal region (residues 18–22). Notably, LPS bound to aptamer seq. 7 was the most flexible, whereas LPS5 remained the most stable. These results suggest that aptamer binding differentially modulates the local dynamics of LPS.
3.2.2.2 Hydrogen bond formation. The number of intra- and intermolecular hydrogen bonds was analyzed to assess structural stability and interaction strength (Fig. 4A–C). Aptamer seq. 9 maintained the highest number of intramolecular H-bonds (45–60), indicating compact folding, while aptamer seq. 7 showed intermediate stability (38–55), and seq. 5 the lowest (35–50), consistent with its extended conformation. For LPS molecules, complexes with aptamer seq. 5 and aptamer seq. 7 retained 10–20 intramolecular H-bonds, while LPS9 showed fewer (7–14), implying stronger perturbation of its structure upon binding. Importantly, intermolecular H-bond analysis revealed that seq. 9 consistently formed the largest number (15–22), surpassing aptamer seq. 5 (10–15) and aptamer seq. 7 (7–13).
Overall, MD simulations highlight aptamer seq. 9 as the strongest interactor with LPS, characterized by a compact aptamer conformation, the highest number of intermolecular H-bonds, and pronounced modulation of LPS structure. Aptamer seq. 5 exhibited intermediate binding, while aptamer seq. 7 maintained structural rigidity but engaged in relatively weak interactions.
Because CN is a structural descriptor involving distance, not energy, the data can be noisy and dependent on the selected cutoff distance. This is why we employed a multitude of analyses to investigate the problem at hand. Additionally, the initial equilibration portion can be inferred from other analyses, such as RMSD or free energy time series. During molecular dynamics simulations, the coordination number, defined as the number of aptamer atoms within a cutoff distance of 8 Å from the LPS molecule, serves as a sensitive descriptor of molecular recognition. For computational tractability, only the C5′ atom of each nucleotide residue was considered. Fig. 5 depicts the time evolution of the coordination numbers between LPS and aptamers seq. 5, 7, and 9.
 | ||
| Fig. 5 Time evolution of the coordination number between the aptamers and LPS molecules during the MD simulations. Results are shown for three aptamer–LPS complexes (seq. 5, seq. 7, and seq. 9). | ||
Aptamer seq. 9 exhibited dynamic but consistently high coordination, fluctuating between 20 and 40 atoms, with distinct peaks around 75, 150, and 180 ns, although a transient drop occurred at ∼45 ns. Aptamer seq. 5 showed a comparable coordination profile, also maintaining values between 20 and 40, but with less dramatic fluctuations, indicating steadier contacts. In contrast, aptamer seq. 7 consistently displayed much lower coordination numbers (0–15) with only minor variations, pointing to weaker and less intimate contact with LPS. Notably, these patterns mirror the hydrogen bond analysis, where aptamer seq. 9 and aptamer seq. 5 formed more numerous and persistent interactions, whereas aptamer seq. 7 engaged in weaker and more transient binding.
3.2.3.1 Binding free energy analysis. To complement the contact-based analysis, we calculated the binding free energy (ΔG) for the aptamer–LPS complexes over 200 ns (Fig. 6). Due to its limited stability, aptamer seq. 7 was excluded from this analysis. Both aptamers seq. 5 and seq. 9 exhibited persistently negative ΔG values, reflecting thermodynamically favorable binding. Aptamer seq. 9 displayed markedly low ΔG in the early and mid-stages (as low as −85 kcal mol−1 around 60 ns), followed by stabilization in the range of −40 to −60 kcal mol−1, suggesting strong yet dynamic binding. Aptamer seq. 5 initially showed weak binding (ΔG ∼0 kcal mol−1 within the first 50 ns), but then stabilized into a favorable range (−30 to −50 kcal mol−1), reflecting a slower but ultimately stable interaction process.
 | ||
| Fig. 6 Time evolution of the binding energy between LPS and the aptamers for two representative aptamer–LPS complexes (aptamers seq. 5 and seq. 9). | ||
Decomposition of the binding energies (Table 2) revealed contrasting driving forces. For aptamer seq. 5, binding was dominated by van der Waals interactions (−46.1 ± 7.2 kcal mol−1), whereas its large positive electrostatic contribution (3188.8 ± 376.8 kcal mol−1) was nearly canceled by the solvation energy (−3175.0 ± 384.1 kcal mol−1). seq. 9 exhibited even stronger van der Waals interactions (−93.8 ± 11.1 kcal mol−1) but also much larger electrostatic/solvation compensation (4510.7 ± 213.3 vs. −4445.6 ± 200.5 kcal mol−1), yielding a net ΔG of −29.5 ± 19.3 kcal mol−1. This comparison highlights that aptamer seq. 5, despite weaker instantaneous contacts, forms a more energetically balanced and favorable complex, whereas aptamer seq. 9 achieves tight binding through stronger non-bonded interactions but suffers from large enthalpy-entropy trade-offs.
| Aptamer seq | ΔGelec (kcal mol−1) | ΔGvdW (kcal mol−1) | ΔGsurf+GB (kcal mol−1) | ΔGtotal (kcal mol−1) |
|---|---|---|---|---|
| 5 | 3188.8 ± 376.8 | −46.1 ± 7.2 | −3175.0 ± 384.1 | −41.8 ± 8.6 |
| 9 | 4510.7 ± 213.3 | −93.8 ± 11.1 | −4445.6 ± 200.5 | −29.5 ± 19.3 |
3.2.3.2 Methodological significance. Taken together, the coordination number, hydrogen bond dynamics, and free energy decomposition provide a multi-dimensional view of aptamer–LPS recognition. Importantly, this pipeline demonstrates that binding affinity cannot be inferred from a single descriptor (e.g., docking energy or H-bond count) but instead arises from a nuanced interplay of van der Waals, electrostatics, solvation, and entropic effects. Such an integrative workflow is precisely what enables the reconciliation of computational predictions with experimental observations: while aptamer seq. 9 appears superior in terms of contact frequency and structural persistence, aptamer seq. 5 ultimately achieves the more favorable binding free energy and experimentally validated recognition performance.
This highlights the strength of our computational–experimental pipeline, which moves beyond static docking toward dynamic, energy-resolved insights. By systematically layering structural, energetic, and dynamic descriptors, the pipeline provides a robust framework for aptamer selection and validation. We believe this integrative methodological perspective, highlighting the importance of combining diverse simulation readouts with experimental validation, provides a broadly applicable framework not only for rational aptamer selection and characterization but also for guiding molecular recognition studies across a wide range of biomolecular systems.
3.3 Experimental validation of selected aptamers
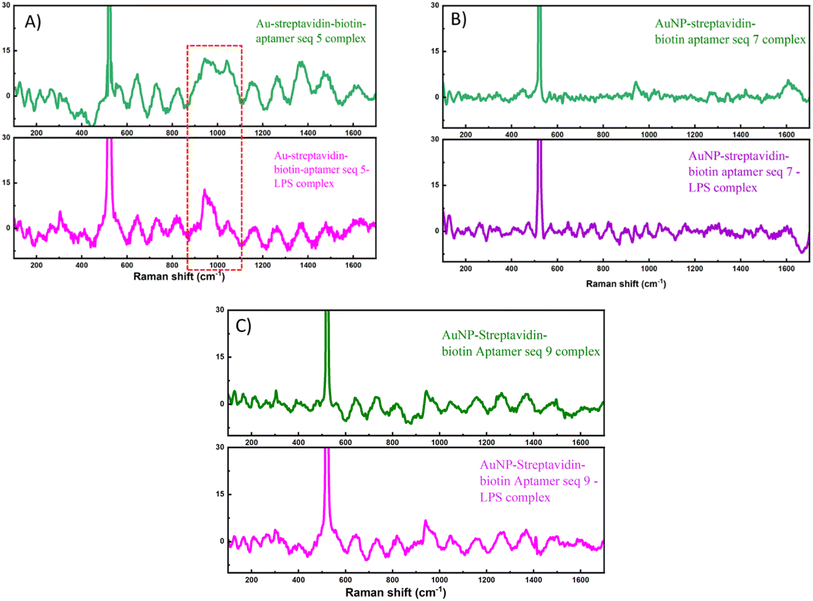
To corroborate the computational predictions and assess their practical relevance, we performed experimental validation of the selected aptamer candidates, focusing on their binding performance against LPS under laboratory conditions.After confirming surface charge variations through zeta potential analysis, SERS was performed to gain deeper insight into the molecular-level interactions. SERS is a sensitive method that provides spectral information that allows label-free identification of the molecules. This method has proved its ability to detect traces of various kinds of analytes ranging from metallic ions or pollutants to more complex compounds such as proteins or even microorganisms such as bacteria.43 Specifically, SERS has been used as a powerful tool for the analysis of aptamer and aptamer–protein interactions,35 prompted us to investigate the changes in SERS spectra of our selected sequences with the presence of LPS from Klebsiella pneumoniae ATCC 15380. To demonstrate the validity of the functionalization process, the full set of SERS spectra recorded at each modification step (citrate-stabilized AuNPs, AuNP-streptavidin, AuNP-streptavidin–biotin aptamer, and AuNP-streptavidin–biotin aptamer–LPS) is provided in Fig. S6. Meanwhile, the main text focuses on the SERS spectra of the three representative aptamers in the presence of LPS, as shown in Fig. 7.
Aptamers are single-stranded DNA molecules and therefore exhibit strong, characteristic SERS signatures of oligonucleotides. However, differences in nucleotide sequence and base composition can lead to variations in their SERS spectra, as the signal is highly sensitive to the molecular configuration of the aptamer and its binding characteristics with the metallic surface used for enhancement.44 In Fig. 7, the three aptamers display variations in spectral intensity, yet all show characteristic DNA-related vibrational features. Specifically, the bands at 640–646 cm−1, 724–733 cm−1, and 1360–1367 cm−1 correspond to the ring-breathing modes of nucleobases. The bands at 942–950 cm−1, 1152–1158 cm−1, 1261–1273 cm−1, 1470–1472 cm−1, and 1606–1607 cm−1 are assigned to stretching vibrations of C–C, C–N, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds in the bases, while the bands at 1042–1044 cm−1 are attributed to sugar vibrations (C–O and N-sugar stretching). In addition, the bands at 814–826 cm−1 correspond to vibrations of the DNA phosphate backbone.45 Notably, aptamer seq. 7 exhibits significantly weaker intensities at these bands reduction is compared with seq. 5 and seq. 9. This most likely due to its secondary structure and orientation when immobilized via the streptavidin–biotin linkage, which may place the nucleobases farther from the gold surface, thereby diminishing the chemical enhancement effect.
C bonds in the bases, while the bands at 1042–1044 cm−1 are attributed to sugar vibrations (C–O and N-sugar stretching). In addition, the bands at 814–826 cm−1 correspond to vibrations of the DNA phosphate backbone.45 Notably, aptamer seq. 7 exhibits significantly weaker intensities at these bands reduction is compared with seq. 5 and seq. 9. This most likely due to its secondary structure and orientation when immobilized via the streptavidin–biotin linkage, which may place the nucleobases farther from the gold surface, thereby diminishing the chemical enhancement effect.
After the addition of LPS, a selective decrease in the intensity of DNA-related peaks together with slight peak shifts was observed, most prominently for aptamer seq. 5 (Fig. 7A). In the range of 856–1106 cm−1 in the SERS spectrum of gold nanoparticles functionalized with aptamer seq. 5 through the streptavidin–biotin linkage, three partially overlapping peaks were detected. Upon interaction with LPS, these peaks exhibited a clear reduction in signal intensity. This spectral region is generally assigned to vibrations of the DNA sugar-phosphate backbone, including C–O/C–C stretching of ribose (∼930–1060 cm−1) and the symmetric stretching of the phosphate group (PO2−, ∼1085–1100 cm−1). Binding of seq. 5 to LPS likely alters the orientation and spatial arrangement of the nucleobases and backbone segments, causing them to reorient relative to the gold surface or become shielded by the bulky dielectric environment of LPS. Consequently, both chemical enhancement from direct base-Au interactions and electromagnetic enhancement at the hotspots are reduced, leading to weaker DNA-related bands. Moreover, due to the abundance of sugar and phosphate groups in LPS, complex formation can further occupy space around the aptamer, decrease hotspot density, and modify the local dielectric constant, thereby intensifying the attenuation of DNA vibrational signals in this region. Taken together, these observations strongly indicate that the spectral changes originate from the specific formation of aptamer–LPS complexes, which alter the molecular conformation and local environment of the aptamer on the AuNP surface.
In summary, Zeta potential measurements confirmed that all three aptamers interacted with LPS, as reflected by the significant changes in surface charge after complex formation. However, the SERS results revealed that only aptamer seq. 5 exhibited clear spectral changes characteristic of specific binding to LPS, while seq. 7 and seq. 9 did not show comparable alterations. This discrepancy suggests that although all three aptamers can electrostatically associate with LPS, only aptamer seq. 5 establishes a stable and structurally favorable interaction that brings its nucleobases into an orientation and proximity suitable for SERS detection. These findings highlight the importance of both aptamer sequence and conformational compatibility in determining the strength and detectability of aptamer–target interactions, underscoring seq. 5 as the most promising candidate for LPS recognition in this system.
This system was subsequently employed to investigate the binding affinity of each selected sequence to LPS from Klebsiella pneumoniae ATCC 15380 by exposing the aptamer-modified electrodes to different concentrations of LPS in 1× PBS. As expected, changes in electrical impedance were observed upon LPS binding. Specifically, seq. 5 exhibited a clear increase in impedance, whereas seq. 7 showed a decrease (Fig. 8B and C). In contrast, the impedance response of seq. 9 remained almost unchanged in the presence of its target (Fig. 8D), indicating a weak interaction with LPS. These findings suggest that among the tested sequences, seq. 5 demonstrated the strongest and most consistent binding affinity toward LPS, while seq. 7 displayed an atypical response and seq. 9 showed negligible affinity. This evidence highlights seq. 5 as the most promising aptamer candidate for sensor development.
Interestingly, the ΔRCT values exhibited opposite trends between aptamer seq. 5 and seq. 7 upon exposure to LPS. For aptamer seq. 5, a positive ΔRCT was observed, which can be rationalized by the formation of a stable aptamer–LPS complex on the electrode surface that hinders the charge transfer of the redox probe, thereby increasing the interfacial resistance. In contrast, aptamer seq. 7 displayed a negative ΔRCT, indicating a decrease in charge-transfer resistance after LPS binding. This atypical behavior may arise from a surface rearrangement induced by the binding event, where the aptamer–LPS interaction alters the interfacial structure in a way that facilitates electron transfer. Alternatively, the strong negative charge of LPS might have partially neutralized or redistributed charges at the electrode interface in the presence of aptamer seq. 7, reducing the electrostatic barrier for the [Fe(CN)6]3−/4− redox couple. These differences can be further explained by the molecular dynamics (MD) analysis. Aptamer seq. 5 was found to establish stable hydrogen bonds with LPS, including T26-KPLI (two bonds), T24-KPLI, and G5-BGA6, which provide strong anchoring points for complex formation and thereby account for the consistent impedance increase. In contrast, aptamer seq. 7 mainly formed hydrogen bonds with residues T61-AHE4, G60-BGL5, and T44-AKD12, which likely resulted in weaker or less stable interactions, consistent with its atypical ΔRCT decrease. Together, these results suggest that aptamer seq. 5 engages in more favorable molecular interactions with LPS, leading to reliable electrochemical responses, while aptamer seq. 7 exhibits unstable binding behavior that compromises its potential as a sensing element. In addition, the nearly unchanged EIS response of aptamer seq. 9 across LPS concentrations is consistent with the MD findings, which revealed only a sparse and weakly cooperative hydrogen-bond network (T29-BGA16, G57-BGL11, G58-AGA10, and G61-AKD2). These bonds are distributed across the aptamer and likely occur at peripheral O-antigen/core sites, leading to a shallow binding energy minimum and high conformational mobility. Such transient and unstable interactions prevent the formation of a compact blocking layer or the close positioning of LPS to the electrode surface, thereby failing to hinder the access of the [Fe(CN)6]3−/4− redox couple. As a result, RCT and other interfacial parameters remain essentially unchanged. In other words, aptamer seq. 9 engages LPS only weakly and transiently, yielding low surface coverage and negligible perturbation of charge transfer. This explains the flat EIS dose–response and confirms that aptamer seq. 9 is not a suitable candidate for sensitive LPS detection.
To further evaluate the specificity of the aptamers toward LPS, we examined their responses to potential interfering substances, including LPS from E. coli, BSA, and glucose, each at a 100-fold higher concentration compared with LPS from Klebsiella pneumoniae ATCC 15380. The results revealed that aptamer seq. 5 exhibited minimal responses to these non-target analytes, as reflected by negligible changes in RCT (Fig. 9A). In contrast, seq. 7 displayed low affinity with LPS from E. coli but unexpectedly showed high responses to BSA and glucose (Fig. 9B), with ΔRCT values being negative. This indicates that its EIS signal mainly originated from nonspecific interactions and environmental effects rather than target-specific binding. BSA, a negatively charged protein (pI 4.7), may adsorb onto the aptamer-functionalized electrode surface and reorganize the local electrostatic environment, thereby reducing repulsion against the [Fe(CN)6]3−/4− redox couple and effectively lowering the interfacial resistance. Glucose, at elevated concentrations, can modulate the dielectric constant and viscosity of the medium and form weak hydrogen bonds with exposed aptamer bases or phosphate groups, as well as with the streptavidin/biotin interface, promoting surface rearrangements that favor charge transfer. By contrast, the distinct O-antigen and core motifs of LPS from E. coli appear incompatible with the recognition sites of aptamer seq. 7, consistent with MD simulations showing a sparse and weakly cooperative hydrogen-bond network, leading to poor binding stability. Taken together, these results suggest that aptamer seq. 7 is more sensitive to protein fouling and solute-induced interfacial effects than to specific recognition of LPS, thereby limiting its applicability as a reliable sensing probe.
To describe the strength of binding affinity between aptamer seq. 5 and LPS, the dissociation constant (Kd) of the aptamer was calculated from the change in peak current (ΔI) measured using Differential Pulse Voltammetry (DPV). The Kd was determined by fitting the data to the Langmuir isotherm model, as described in the Experimental section. The binding affinity of aptamer seq. 5 was 6.68 nM (Fig. 9C). This value was comparable to high affinity antibodies typically showing nanomolar ranges of Kd for small targets.47 Compared to aptamers selected for LPS from E. coli 055:B5 using NECEEM-based non-SELEX method, Kd of aptamer seq. 5 also showed a comparable value.5 The aptamer with the highest binding affinity (Kd = 12 nM) for LPS from E. coli 055:B5 in the study of Kim et al. (2012) has been used in several studies to develop biosensors with detection limit of up to 1 pg mL−1 of LPS, suggesting that our aptamer (seq. 5) is highly promising in the development of good aptasensors.5,48,49
4. Conclusion
This work demonstrates a synergistic strategy that integrates computational modeling with conventional SELEX for the rational selection of aptamers against lipopolysaccharides (LPS). Starting from SELEX-derived candidates, docking and molecular dynamics (MD) simulations were employed to support the post-SELEX refinement and candidate validation process. Aptamer seq. 5 ultimately identified as the most stable and selective aptamer. Importantly, the use of coordination number analysis provided a quantitative measure of hydrogen-bonding and noncovalent contacts, enabling a more reliable assessment of binding stability than docking scores alone. This methodological innovation serves as a pipeline that bridges the gap between large-scale in silico screening and targeted experimental validation.Electrochemical impedance spectroscopy (EIS) and surface-enhanced Raman scattering (SERS) experiments further corroborated the MD predictions, confirming seq. 5 as a high-affinity and specific probe for LPS detection. The strong agreement between simulations and experiments not only validates the computational workflow but also highlights its potential to reduce trial-and-error in aptamer discovery. Beyond the current case, this combined framework-particularly the incorporation of coordination number as a stability descriptor-offers a broadly applicable tool for accelerating aptamer-based biosensor development, with implications for diagnostics, food safety, and environmental monitoring.
Author contributions
This article was produced with equal contributions from two groups: experimental: Thuy-Duong Thi Tran: writing – original draft, visualization, validation, methodology, investigation, formal analysis, conceptualization, Phan Thi Ngoc Hoa: writing – review & editing, visualization, validation, methodology, investigation, conceptualization, Thu Thao Pham: investigation, Nguyen Thi Thanh Huyen: investigation, Hoa Thi Hoang: investigation, Patrick Wagner: methodology, Anne Charrier: methodology, and Truong TN Lien: supervision, resources, funding acquisition. computational stimulation: Hai Ly Nguyen: writing – original draft, visualization, validation, methodology, investigation, formal analysis, conceptualization, software, formal analysis, data curation, Phuong H. Nguyen: writing – review & editing, formal analysis, and Nguyen The Toan: writing – review & editing. formal analysis.Conflicts of interest
The authors declare that there is no conflict of interest.Data availability
All the raw experimental and computational data supporting the findings of this study are available from the corresponding author upon reasonable request.The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra06759f.
Acknowledgements
The authors gratefully acknowledge the financial support provided by the Institute of Big Data, Vingroup, under the Vingroup Innovation Foundation – VINIF Scholar (grant number VINIF.2022.DA00032). The authors also express their sincere gratitude to Dr Kasper Eersels from the Sensor Engineering group, Faculty of Science and Engineering, Maastricht University, for his invaluable contribution and thoughtful guidance during the revision of this work. His support has been deeply appreciated, and the authors wish him strength and recovery during this challenging time.References
- D. Esmaeilli, A. M. Mobarez, A. H. Salmanian and A. Z. Hosseini, Bioactivity and immunological evaluation of LPS from different serotypes of Helicobacter pylori, Iran. J. Microbiol., 2013, 5(2), 142 Search PubMed , Accessed: Jun. 19, 2025. [Online]. Available: https://pmc.ncbi.nlm.nih.gov/articles/PMC3696850/.
- A. Gorman and A. P. Golovanov, Lipopolysaccharide Structure and the Phenomenon of Low Endotoxin Recovery, Eur. J. Pharm. Biopharm., 2022, 180, 289–307, DOI:10.1016/j.ejpb.2022.10.006.
- B. Tindall, D. Demircioglu and T. Uhlig, Recombinant bacterial endotoxin testing: a proven solution, Biotechniques, 2021, 70(5), 290–300, DOI:10.2144/btn-2020-0165.
- H. Grallert, S. Leopoldseder, M. Schuett, P. Kurze and B. Buchberger, EndoLISA®: a novel and reliable method for endotoxin detection, Nat. Methods, 2011, 8(10), 3–5, DOI:10.1038/nmeth.f.350.
- S. E. Kim, W. Su, M. Cho, Y. Lee and W. S. Choe, Harnessing aptamers for electrochemical detection of endotoxin, Anal. Biochem., 2012, 424, 12–20, DOI:10.1016/j.ab.2012.02.016.
- W. Su, M. Cho, J. Do Nam, W. S. Choe and Y. Lee, Aptamer-Assisted Gold Nanoparticles/PEDOT Platform for Ultrasensitive Detection of LPS, Electroanalysis, 2013, 25, 380–386, DOI:10.1002/elan.201200453.
- J. L. Ding, S. T. Gan and B. Ho, Single-stranded DNA oligoaptamers: molecular recognition and LPS antagonism are length- and secondary structure-dependent, J. Innate Immun., 2009, 1(1), 46–58, DOI:10.1159/000145542.
- Y. J. Lee, S. R. Han, J.-S. Maeng, Y.-J. Cho and S.-W. Lee, In vitro selection of Escherichia coli O157:H7-specific RNA aptamer, Biochem. Biophys. Res. Commun., 2012, 417(1), 414–420, DOI:10.1016/j.bbrc.2011.11.130.
- T. Karampatakis, K. Tsergouli and P. Behzadi, Carbapenem-Resistant Klebsiella pneumoniae: Virulence Factors, Molecular Epidemiology and Latest Updates in Treatment Options, Antibiotics, 2023, 12(2), 234, DOI:10.3390/antibiotics12020234.
- B. C. Kim, Y. S. Kim, J. Y. Chung, J. S. Jurng and M. Y. Song, Single-stranded nucleic acid aptamers specifically binding to Klebsiella pneumoniae and method for detecting K. Pneumonia Using the Same, US Pat., US20150141270A1, 2016 Search PubMed.
- A. Deb, M. Gogoi, T. K. Mandal, S. Sinha and P. S. G. Pattader, Specific Instantaneous Detection of Klebsiella pneumoniae for UTI Diagnosis with a Plasmonic Gold Nanoparticle Conjugated Aptasensor, ACS Appl. Bio Mater., 2023, 6(8), 3309–3318, DOI:10.1021/acsabm.3c00369.
- M. K. Paczosa and J. Mecsas, Klebsiella pneumoniae: Going on the Offense with a Strong Defense, Microbiol. Mol. Biol. Rev., 2016, 80(3), 629–661, DOI:10.1128/MMBR.00078-15.
- P.-F. Hsieh, et al., Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess, PLoS One, 2012, 7(3), e33155, DOI:10.1371/journal.pone.0033155.
- K. Sefah, D. Shangguan, X. Xiong, M. B. O'Donoghue and W. Tan, Development of DNA aptamers using cell-selex, Nat. Protoc., 2010, 5, 1169–1185, DOI:10.1038/nprot.2010.66.
- J. Li, et al., Novel p22 and p30 dual-proteins combination based indirect ELISA for detecting antibodies against African swine fever virus, Front. Vet. Sci., 2023, 10 DOI:10.3389/fvets.2023.1093440.
- O. Rabal, F. Pastor, H. Villanueva, M. M. Soldevilla, S. Hervas-Stubbs and J. Oyarzabal, In Silico Aptamer Docking Studies: From a Retrospective Validation to a Prospective Case Study-TIM3 Aptamers Binding, Mol. Ther. Nucleic Acids, 2016, 5, e376, DOI:10.1038/mtna.2016.84.
- C. Q. Vu, P. Rotkrua, B. Soontornworajit and Y. Tantirungrotechai, Effect of PDGF-B aptamer on PDGFRβ/PDGF-B interaction: molecular dynamics study, J. Mol. Graph. Model., 2018, 82, 145–156, DOI:10.1016/j.jmgm.2018.04.012.
- A. Pal and Y. Levy, Structure, stability and specificity of the binding of ssDNA and ssRNA with proteins, PLOS Comput. Biol., 2019, 15(4), e1006768, DOI:10.1371/journal.pcbi.1006768.
- H. Li, et al., Aptamer selection for the detection of Escherichia coli K88, Can. J. Microbiol., 2011, 57(6), 453–459, DOI:10.1139/w11-030.
- J. Sarzynska, M. Popenda, M. Antczak and M. Szachniuk, RNA tertiary structure prediction using RNAComposer in CASP15, Proteins: Struct., Funct., Bioinf., 2023, 91, 1790–1799, DOI:10.1002/prot.26578.
- M. Zuker, Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Res., 2003, 31, 3406–3415, DOI:10.1093/nar/gkg595.
- J. Lee, et al., CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans, J. Chem. Theory Comput., 2019, 15(1), 775–786, DOI:10.1021/acs.jctc.8b01066.
- L. Schrödinger, Schrödinger 2016, 4, 2016 Search PubMed.
- M. J. Abraham, et al., Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25, DOI:10.1016/j.softx.2015.06.001.
- J. Huang and A. D. Mackerell, CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data, J. Comput. Chem., 2013, 34, 2135–2145, DOI:10.1002/jcc.23354.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926–935, DOI:10.1063/1.445869.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092, DOI:10.1063/1.464397.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, LINCS: A Linear Constraint Solver for molecular simulations, J. Comput. Chem., 1997, 18, 1463–1472, DOI:10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 126(1), 014101, DOI:10.1063/1.2408420.
- H. J. C. Berendsen, J. P. M. Postma, W. F. Van Gunsteren, A. Dinola and J. R. Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys., 1984, 81, 3684–3690, DOI:10.1063/1.448118.
- Q. Ke, X. Gong, S. Liao, C. Duan and L. Li, Effects of thermostats/barostats on physical properties of liquids by molecular dynamics simulations, J. Mol. Liq., 2022, 365, 120116, DOI:10.1016/j.molliq.2022.120116.
- S. Genheden and U. Ryde, The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities, Expert Opin. Drug Discovery, 2015, 10, 449–461, DOI:10.1517/17460441.2015.1032936.
- B. R. Miller, T. D. McGee, J. M. Swails, N. Homeyer, H. Gohlke and A. E. Roitberg, MMPBSA.py: an efficient program for end-state free energy calculations, J. Chem. Theory Comput., 2012, 8, 3314–3321, DOI:10.1021/ct300418h.
- M. S. Valdés-Tresanco, M. E. Valdés-Tresanco, P. A. Valiente and E. Moreno, Gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS, J. Chem. Theory Comput., 2021, 17, 6281–6291, DOI:10.1021/acs.jctc.1c00645.
- R. Gillibert, M. N. Triba and M. Lamy de la Chapelle, Surface enhanced Raman scattering sensor for highly sensitive and selective detection of ochratoxin A, Analyst, 2017, 143(1), 339–345, 10.1039/c7an01730h.
- T. T. Pham, et al., Staining-Enhanced Peroxidase-Mimicking Gold Nanoparticles in Nano-ELISA for Highly Sensitive Detection of Klebsiella pneumoniae, ACS Omega, 2023, 8, 49211–49217, DOI:10.1021/acsomega.3c07503.
- X. Li, et al., Electrochemical impedance spectroscopy for study of aptamer-thrombin interfacial interactions, Biosens. Bioelectron., 2008, 23(11), 1624–1630, DOI:10.1016/j.bios.2008.01.029.
- M. Ebrahimi, M. Johari-Ahar, H. Hamzeiy, J. Barar, O. Mashinchian and Y. Omidi, Electrochemical impedance spectroscopic sensing of methamphetamine by a specific aptamer, Bioimpacts, 2012, 2(2), 91–95, DOI:10.5681/bi.2012.013.
- C. Marimuthu, T.-H. Tang, J. Tominaga, S.-C. Tan and S. C. B. Gopinath, Single-stranded DNA (ssDNA) production in DNA aptamer generation, Analyst, 2012, 137(6), 1307–1315, 10.1039/C2AN15905H.
- M. U. Musheev and S. N. Krylov, Selection of aptamers by systematic evolution of ligands by exponential enrichment: addressing the polymerase chain reaction issue, Anal. Chim. Acta, 2006, 564(1), 91–96, DOI:10.1016/j.aca.2005.09.069.
- M. Heiat, R. Ranjbar, A. M. Latifi, M. J. Rasaee and G. Farnoosh, Essential strategies to optimize asymmetric PCR conditions as a reliable method to generate large amount of ssDNA aptamers, Biotechnol. Appl. Biochem., 2017, 64(4), 541–548, DOI:10.1002/bab.1507.
- S. Y. Low, J. E. Hill and J. Peccia, A DNA aptamer recognizes the Asp f 1 allergen of Aspergillus fumigatus, Biochem. Biophys. Res. Commun., 2009, 386(3), 544–548, DOI:10.1016/j.bbrc.2009.06.089.
- D. Cialla-May, et al., Recent advances of surface enhanced Raman spectroscopy (SERS) in optical biosensing, TrAC, Trends Anal. Chem., 2024, 181 DOI:10.1016/j.trac.2024.117990.
- S. Rubin, P. H. L. Nguyen, and Y. Fainman, The effect of DNA bases permutation on surface-enhanced Raman scattering spectrum, 2021, vol. 10, pp. 1581–1593, DOI:10.1515/nanoph-2021-0021.
- W. Safar, A. Azziz, M. Edely and M. Lamy de la Chapelle, Conventional Raman, SERS and TERS Studies of DNA Compounds, Chemosensors, 2023, 11(7), 399, DOI:10.3390/chemosensors11070399.
- A. R. Harris, P. Carter, R. Cowan and G. G. Wallace, Impact of Protein Fouling on the Charge Injection Capacity, Impedance, and Effective Electrode Area of Platinum Electrodes for Bionic Devices, ChemElectroChem, 2021, 8(6), 1078–1090, DOI:10.1002/celc.202001574.
- J. Zhou and J. Rossi, Aptamers as targeted therapeutics: current potential and challenges, Nat. Rev. Drug Discovery, 2017, 16(3), 181–202, DOI:10.1038/nrd.2016.199.
- W. Su, M. Lin, H. Lee, M. S. Cho, W. S. Choe and Y. Lee, Determination of endotoxin through an aptamer-based impedance biosensor, Biosens. Bioelectron., 2012, 32, 32–36, DOI:10.1016/j.bios.2011.11.009.
- J. Niu, et al., Femtomolar Detection of Lipopolysaccharide in Injectables and Serum Samples Using Aptamer-Coupled Reduced Graphene Oxide in a Continuous Injection-Electrostacking Biochip, Anal. Chem., 2019, 91, 2360–2367, DOI:10.1021/acs.analchem.8b05106.
Footnote |
| † These authors contribute equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2025 |