Open Access Article
Open Access ArticlePolymer-lipid hybrid nanoparticles incorporating green coffee extract: enhancing treatment for hepatic steatosis and fibrosis through metabolomic insights
Marco A. Uriostegui-Camposa,
Shaula A. Castro-Murrietaa,
Ximena I. Lopez-Cesatia,
Ricardo Colín-Delgadoa,
Gabriela I. Carballo-Lópeza,
Karla Cervantes-Luevanoa,
Aldo Moreno-Ulloaa and
Ana B. Castro-Ceseña *ab
*ab
aDepartamento de Innovación Biomédica, Centro de Investigación Científica y de Educación Superior de Ensenada, Baja California (CICESE), Carretera Ensenada-Tijuana No. 3918, Zona Playitas, C.P. 22860, Ensenada, Baja California, Mexico. E-mail: acastro@cicese.mx
bSECIHTI-Departamento de Innovación Biomédica, Centro de Investigación Científica y de Educación Superior de Ensenada, Baja California (CICESE), Carretera Ensenada-Tijuana No. 3918, Zona Playitas, C.P. 22860, Ensenada, Baja California, Mexico
First published on 27th November 2025
Abstract
Metabolic dysfunction-associated steatotic liver disease (MASLD) affects approximately 30% of adults globally. Its severe form, metabolic dysfunction-associated steatohepatitis (MASH), results in liver damage due to a combination of steatosis and fibrosis. Green coffee extract (GCE) derived from unroasted coffee beans demonstrates therapeutic potential for MASLD/MASH attributed to its rich profile of bioactive compounds. However, challenges remain regarding bioavailability and understanding of mechanisms. In this study, we investigated the bioactive potential of GCE encapsulated in polymer-lipid hybrid nanoparticles (PLNs) as a therapeutic option for MASLD and associated fibrosis. Encapsulation of GCE into PLNs enhanced its physicochemical properties, including reduced particle size (191.5 ± 3.6 nm) and improved polydispersity index (PDI) (0.291), as confirmed by dynamic light scattering (DLS) and transmission electron microscopy (TEM) (145.4 ± 26 nm). The encapsulation efficiency (55%) and release kinetics of GCE-PLNs were optimized for sustained bioavailability over 24 h. Metabolomic and chemoinformatic analyses identified 1942 metabolites, including GCE-derived bioactive compounds with structural similarities to therapeutic agents like mesalamine, idebenone, and cysteamine, highlighting potential antifibrotic and hepatoprotective effects. In vitro assays, using HepG2 cells, demonstrated that GCE-PLNs modulate the expression of steatosis- and fibrosis-related genes, such as PLIN1, CPT1A, ACTA2, and COL1A1, in HepG2 and HHSC-N cellular models, suggesting reduced lipid accumulation and fibrotic activity. These findings suggest that GCE-PLNs may represent a promising nanotechnological strategy for the treatment of MASLD.
Introduction
Metabolic dysfunction-associated steatotic liver disease (MASLD), previously referred to as NAFLD, affects approximately 30% of the global adult population.1,2 MASLD is a spectrum of liver conditions that occur in the presence of at least one cardiometabolic risk factor and in the absence of excessive alcohol consumption (≥20–30 g per day) or other chronic liver diseases. This spectrum includes simple steatosis and more severe stages, such as cirrhosis and hepatocellular carcinoma (HCC).3,4 A characteristic feature of MASLD is impaired hepatic metabolism of carbohydrates and fatty acids, leading to triglycerides accumulation (TGs), oxidative stress, inflammation, and fibrogenesis. Over time, these processes contribute to genomic instability and progressive liver dysfunction. Additionally, the pathogenesis of MASLD is complex and heterogeneous, involving various pathophysiological mechanisms with significant inter-individual variability.5,6Metabolic dysfunction-associated steatohepatitis (MASH), a severe form of MASLD, is marked by hepatocyte inflammation and fibrogenesis, frequently resulting in irreversible cirrhosis. Fibrosis occurs due to the excessive deposition of type I collagen, predominantly driven by activated hepatic stellate cells (HSCs). The interaction between steatosis and fibrosis intensifies liver damage, underscoring the necessity for therapies aimed at reducing TGs and inhibiting mechanisms that promote fibrosis, such as the activation of HSCs.7 Present treatments emphasize lifestyle modifications, including diet and exercise; however, low adherence rates and the absence of effective pharmacological options highlight the need for innovative therapeutic strategies.8,9
The development of new therapies for MASLD from natural products is a promising avenue. Among natural products, green coffee extract (GCE), derived from unroasted coffee beans, is a rich source of bioactive compounds like polyphenols and caffeine. These constituents have demonstrated significant antioxidant, anti-inflammatory, and lipid-lowering effects in MASLD patients, positioning GCE as a promising natural therapy for managing MASLD and its advanced stage, MASH.10–13 However, the clinical application of GCE is limited by challenges such as low bioavailability, insufficient hepatic accumulation and limited understanding of its mechanisms, which hinder its efficacy in targeting liver pathologies.14,15 Nanotechnology offers a promising solution to overcome the limitations associated with bioavailability and efficacy of bioactive substances through nanoencapsulation in hybrid polymer-lipid nanoparticles (PLNs).16,17 PLNs combine the structural advantages of polymers, like poly-lactic-co-glycolic acid (PLGA), with the biocompatibility of lipid-based systems, such as lecithin, enabling controlled drug release and optimized therapeutic efficacy. This kind of nanoparticles offers a versatile and efficient platform for the encapsulation and controlled delivery of phytochemicals with potential antioxidant, anti-inflammatory, anti-fibrotic or even anti steatotic properties. These hybrid systems combine the structural stability of polymers with the biocompatibility and high loading capacity of lipids, enabling a more effective transport of compounds with limited solubility or stability, and also can protect sensitive molecules from enzymatic degradation and premature release, prolonging their circulation time and enhancing their pharmacokinetic profile.18–20
This study involves the extraction and metabolomic profiling of GCE, followed by its encapsulation in PLNs composed of PLGA and soy lecithin (HSPC), to assess their therapeutic potential against MASLD/MASH. Using an in vitro model of steatotic HepG2 cells and activated HSCs from a MASH donor (HHSC-N), the effects of GCE-PLNs on steatosis and fibrosis-related gene expression were evaluated. Metabolomic and chemoinformatic analyses were also conducted to characterize the extract composition and identify potential therapeutic compounds for MASLD/MASH.
Materials and methods
Green coffee extract
To obtain green coffee extract (GCE), unroasted coffee beans of the species Coffea canephora (San Felipe coffee company, Mexico) grown and harvested in Cordoba, Veracruz, Mexico were used. The unroasted coffee beans were ground using a burr mill. Ground coffee was added to absolute methanol at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio and the mixture was subjected to magnetic stirring at 1000 rpm for 1 h at room temperature. Following this, the mixture was filtered with a 0.20 µM syringe filter and concentrated in a vacuum evaporator Rocket Synergy (Genevac, UK) at 50 °C and <30 mBar for 2 h. The resulting solid was stored protected from light at −20 °C.
10 ratio and the mixture was subjected to magnetic stirring at 1000 rpm for 1 h at room temperature. Following this, the mixture was filtered with a 0.20 µM syringe filter and concentrated in a vacuum evaporator Rocket Synergy (Genevac, UK) at 50 °C and <30 mBar for 2 h. The resulting solid was stored protected from light at −20 °C.
Determination of total phenolic content
The total phenolic content (TPC) of GCE was quantified using the Folin-Ciocalteu assay.21 Briefly, 100 µL of 0.2 N Folin-Ciocalteu reagent was combined with 20 µL of the sample, and the mixture was incubated at room temperature for 5 minutes. After incubation, 80 µL of 700 mM Na2CO3 was added, and the reaction was incubated in the dark for 2 h at room temperature. Following this, the plate was read at 760 nm in an Epoch plate reader. The TPC of the samples was calculated based on a gallic acid (GA) standard curve and the results were expressed in mg of GA equivalents (GAE) per g of GCE.Determination of antioxidant activity
The antioxidant activity of GCE was evaluated by 2,2-Diphenyl-1-Picrylhydrazyl (DPPH). A DPPH radical (DPPH˙) solution (60 µM) was prepared in absolute ethanol; then, 150 µL of the solution was transferred to a 96-well plate with 50 µL of GCE. The mixture was incubated in the dark at room temperature for 30 min, after, absorbance was measured at 517 nm in an Epoch plate reader. The DPPH˙ inhibition percentage was calculated using the following formula:where Abscontrol, Abssample and Absblank are the absorbance values of the control, sample, and sample blank at 517 nm, respectively. The DPPH˙ inhibition of GCE, was compared to a citric acid (CA) standard curve and the results were expressed in µM of CA equivalents (CAE).
Ultra performance liquid chromatography (UPLC) and tandem mass spectrometry (MS/MS) data acquisition
The widely targeted metabolomic profiling was performed by MetWare Biotechnology Inc (Massachusetts, USA) according to standard procedures as described below. For the sample processing, GCE was ground using a ball mill grinder (30 Hz, 1.5 min) (MM 400, Retsch). Fifty milligrams of the powdered sample were extracted with 1200 µL of pre-cooled 70% methanol (−20 °C) containing internal standards. The mixture was vortexed for 30 seconds every 30 minutes over a 3 hours period (6 cycles total) and centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 minutes at 4 °C. The supernatant was filtered through a 0.22 µm membrane and stored for UPLC-MS/MS analysis.
000 rpm for 3 minutes at 4 °C. The supernatant was filtered through a 0.22 µm membrane and stored for UPLC-MS/MS analysis.
Metabolites were analyzed using an Ultra-Performance Liquid Chromatography (UPLC) system ExionLC™ AD (Sciex, Canada), coupled to tandem mass spectrometry (MS/MS). Chromatographic separation was achieved on an Agilent SB-C18 column (2.1 mm × 100 mm, 1.8 µm) at 40 °C. The mobile phases consisted of ultrapure water with 0.1% formic acid (phase A) and acetonitrile with 0.1% formic acid (phase B). The gradient began at 5% B, linearly increased to 95% B over 9 minutes, held at 95% B for 1 minute, decreased to 5% B over 1.1 minutes, and equilibrated at 5% B for 4 minutes (total runtime: 14 minutes). The flow rate was 0.35 mL min−1, and the injection volume was 2 µL.
Mass spectrometry was conducted using a TripleTOF 6600+ system (Sciex, Canada) and a QTRAP® 6500+ system (Sciex, Canada) equipped with an ESI Turbo ion spray interface operating in both positive and negative ion modes. For the TripleTOF system, source temperature was set to 500 °C, ion spray voltage to 5500 V (positive mode) or −4500 V (negative mode), and the curtain gas (CUR), gas I (GSI), and gas II (GSII) were set to 25, 50, and 60 psi, respectively. The declustering potential (DP) was 80 V, and the collision energy (CE) was 30 V, with a collision energy spread of 15 V.
For the QTRAP system, the source temperature was 550 °C, and the ion spray voltage was 5500 V (positive mode) or −4500 V (negative mode). CUR, GSI, and GSII were maintained at 25, 50, and 60 psi, respectively. Collision-induced dissociation was set to high, and metabolite detection was performed in MRM mode using nitrogen as the collision gas. Data acquisition was managed with Analyst 1.6.3 software (Sciex, Canada), monitoring MRM ion pairs according to the metabolites' elution times.
UPLC-MS/MS data processing and analysis
The metabolite analysis was performed using a standardized workflow with quality control measures to ensure reliability. The raw data were processed to remove isotope signals, repeated signals containing K+, Na+, and NH4+ ions, as well as fragment signals of larger molecular weight compounds. Missing values in the raw data file (S1) were imputed using one-fifth of the minimum value of each metabolite row, and metabolites with a coefficient of variation (CV) value below 0.5 in QC samples were retained. Once the annotation and elimination of contaminants such as parabens of the metabolites were made, SMILES strings and chemical classes were assigned to the features using ClassyFire tool (S2).22Mass spectrometry data were processed using Analyst 1.6.3 software. Total ion chromatograms (TIC) and extracted ion chromatograms (XIC) of mixed QC samples were generated, where the X-axis represented the retention time (Rt), and the Y-axis represented ion flow intensity. Each chromatographic peak, distinguished by color, corresponded to a detected metabolite, with signal intensity (CPS) measured for characteristic ions by triple quadrupole mass spectrometry. Chromatographic peaks were integrated and corrected using MultiQuant software.
To ensure accurate qualitative and quantitative analysis, metabolite peaks were corrected for retention time and peak distribution. Quality control samples, prepared as mixtures of all sample extracts, were analyzed every ten test samples to monitor reproducibility. Overlapping TIC diagrams from different QC samples demonstrated high reproducibility and stability of the analytical process.
Prediction of potential antisteatotic and antifibrotic molecules by molecular fingerprints-based analysis
To explore the potential bioactivity of GCE metabolites against MASLD and MASH, an in silico analysis was performed by comparing their molecular fingerprints with those of drugs approved or indicated for MASLD treatment. These drugs, sourced from the ChEMBL database, included agonists, inhibitors, antagonists, and modulators of known therapeutic targets for the disease, as well as small molecules used in MASLD/MASH treatment without identified targets. ChEMBL IDs, target proteins, and SMILES of these drugs were collected for comparison.A comparative analysis was performed using Similarity Chart as the dimensionality reduction method, implemented in the DataWarrior program (version 5.5.0) with the following parameters: descriptor, FragFp and a 90% similarity limit. The analysis incorporated the molecular fingerprints of the metabolites and drugs, which were computed using the FragFP descriptor. FragFP is a binary molecular fingerprint based on a dictionary of 512 predefined substructural fragments.
Lipid-polymer nanoparticles synthesis
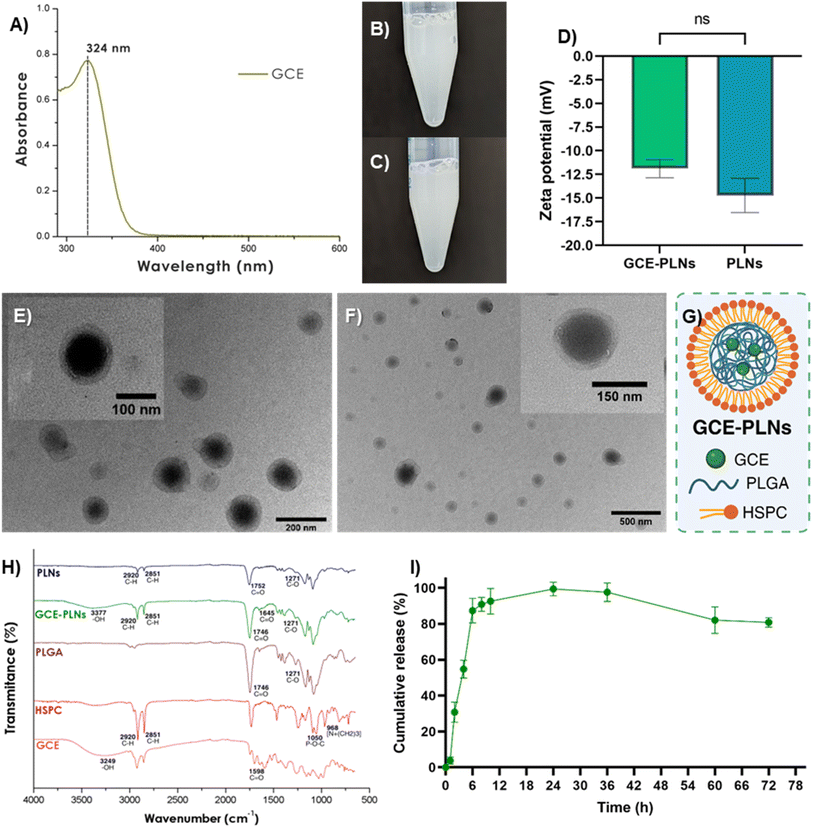
The synthesis of polymer-lipid hybrid nanoparticles (PLNs) was carried out using a nanoprecipitation method combined with self-assembly23,24 with some modifications (Fig. 1). First, stock solutions of PLGA (Nanosoft Polymers, USA; 11088-20-50K) at 20 mg mL−1 in acetonitrile and soybean lecithin (HSPC) (Nanosoft Polymers, USA; 26372) at 10 mg mL−1 in 4% ethanol (EtOH) were prepared at 65 °C to achieve proper homogenization of HSPC. An organic phase loaded with either 1 mg of GCE or empty (without GCE) was prepared. The organic phase, was gradually added at a rate of 1 mL min−1 to an aqueous phase containing 200 µL of 10 mg mL−1 HSPC stock and 3.8 mL of 4% EtOH, under constant stirring at 180 rpm and 65 °C. Subsequently, 5 mL of Milli-Q water was added, and the resulting mixture was sonicated for 7 minutes at 42 kHz in an MH 2800 series ultrasonic bath (Branson Ultrasonics, USA). Finally, the solution was subjected to a series of centrifugation washes at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 3, 6, 12, and 18 min using Microsep™ Advance 10 kDa MWCO filters (Cytiva/Pall Life Sciences, USA) to remove free molecules and purify the nanoparticles. The filtered volume was recovered and stored protected from light to determine the encapsulation efficiency. PLNs loaded with GCE were labeled as GCE-PLNs and empty PLNs only as PLNs.
000×g for 3, 6, 12, and 18 min using Microsep™ Advance 10 kDa MWCO filters (Cytiva/Pall Life Sciences, USA) to remove free molecules and purify the nanoparticles. The filtered volume was recovered and stored protected from light to determine the encapsulation efficiency. PLNs loaded with GCE were labeled as GCE-PLNs and empty PLNs only as PLNs.
GCE-PLNs characterization
Encapsulation efficiency
The encapsulation efficiency (EE%) of GCE-PLNs was assessed using a direct method. Lyophilized nanoparticles were dissolved in 95% methanol and vortexed vigorously to disrupt their structure, followed by incubation at room temperature for 2 h, protected from light. Encapsulated GCE was quantified using its characteristic absorbance of chlorogenic acids (CGAs) at 324 nm25 with an Epoch microplate reader, referencing an GCE standard curve. The EE% was calculated using the following equation:where GCEtotal is the total mass of GCE used for GCE-PLNs synthesis and GCEfree is the mass of GCE detected at 324 nm.
Release kinetics
The release profile of GCE-PLNs was evaluated using a mini dialysis kit (Cytiva, USA; 80648432). Briefly, freshly prepared GCE-PLNs were carefully recovered and placed inside an 8 kDa MWCO dialysis membrane in 1 mL of PBS 1X (pH 7.4). The release solution consisted of 6 mL of the same buffer. The assay was carried out in an incubator with constant shaking at 37 °C and 120 rpm. At specific intervals, from 1 to 72 hours, samples were taken from the release solution to quantify the amount of GCE released, the sample volume (0.6 mL) was replaced by the same volume of PBS 1X to maintain a constant volume in the release solution. Quantification of GCE was performed using the same spectrophotometric detection method as for EE%. The cumulative release of GCE was calculated using the following equations:where Mn is the µg of GCE detected in the release solution at time “n”; Cn is the concentration in µg mL−1 of GCE detected in the release solution at time “n”; Vt is the volume in mL of the release solution; Cn−1 is the concentration in µg mL−1 of GCE before time “n” and Vm is the volume in mL of the sample.
Cell culture
Human hepatocellular carcinoma HepG2 cells (Addexbio Technologies, USA; C0015002) and activated hepatic stellate cells (HHSC-N), derived from a patient with MASH (iX Cells Biotechnologies, USA; 10HU-210N) were employed to establish the in vitro model of hepatic steatosis and fibrosis, respectively. Both cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) with high glucose (4.5 g L−1) (Gibco, USA; 10569010), supplemented with 10% fetal bovine serum (FBS) (Gibco, USA; 26140-079) and 1% antibiotic/antimycotic solution (A/A) (Gibco, USA; 15140-122). Cultures were incubated under standard conditions at 37 °C in a humidified atmosphere with 5% CO2.Cells were grown in 100 mm culture plates, and the medium was replaced every 48 hours. Once 70–80% confluence was reached, cells were detached with 0.25% trypsin–EDTA 1X (Gibco, USA; 25200056) and used for further experiments. HepG2 cells were used between passages 14 and 22, while HHSC-N cells were limited to passage 7, as per the supplier's recommendations.
Cell viability assays
Cell viability of HepG2 cells was assessed using the MTT assay. HepG2 cells (2 × 104 cells per well) were cultured in 96-well plates with high-glucose DMEM and treated with GCE, GCE-PLNs, and PLNs at equivalent concentrations (100 µg mL−1 of GCE or its equivalent in PLNs) for 24 h. Post-treatment, 90 µL of DMEM with 10 µL of MTT (5 mg mL−1) was added, incubated, and then solubilized with HCl/SDS for absorbance measurement at 570 nm. Viability was calculated as a percentage relative to untreated controls using the following equation:where ODsample is the absorbance value of the wells with treated cells and ODcontrol corresponds to the absorbance of the wells with untreated cells.
The viability of HHSC-N cells was assessed using the trypan blue exclusion method to evaluate cellular proliferation in response to treatments. Cells were seeded at a density of 2.5 × 105 cells per well in 6-well plates and incubated for 48 h with complete DMEM containing GCE, GCE-NPLs, or PLNs. After treatment, cells were detached using 0.25%-EDTA trypsin, and a 20 µL aliquot of the cell suspension was mixed with 20 µL of 0.4% trypan blue solution. Viable cells were manually counted using a hemocytometer under an EVOS XL inverted microscope at 20X magnification. Results were expressed as cells per mL and compared with the initial cell count (0 h).
Steatosis induction in HepG2
To induce steatosis in HepG2 hepatocytes, cells were exposed to a mixture of free fatty acids (FFAs) comprising oleic acid (OA) (Sigma-Aldrich, USA; O1008) and palmitic acid (PA) (MedChemExpress, USA; HY-N0830) at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio.26 The FFAs were conjugated with fatty acid-free bovine serum albumin (BSA) (MP Biomedicals, USA; 152401) by dissolving OA and PA in molecular biology-grade ethanol at concentrations of 500 mM and 250 mM, respectively. A stock solution of 7.5 mM (5 mM OA and 2.5 mM PA) was prepared in complete DMEM supplemented with 10% BSA. The conjugation was performed by incubating the FFA-BSA mixture at 37 °C for 1 h. HepG2 cells were then treated for 24 h with various concentrations (0.1–1 mM) of the FFA mixture in complete DMEM containing 1% BSA. The impact of the FFA treatment on cell viability was evaluated using the MTT assay as previously described. A final concentration of 1 mM OA-PA 2
1 molar ratio.26 The FFAs were conjugated with fatty acid-free bovine serum albumin (BSA) (MP Biomedicals, USA; 152401) by dissolving OA and PA in molecular biology-grade ethanol at concentrations of 500 mM and 250 mM, respectively. A stock solution of 7.5 mM (5 mM OA and 2.5 mM PA) was prepared in complete DMEM supplemented with 10% BSA. The conjugation was performed by incubating the FFA-BSA mixture at 37 °C for 1 h. HepG2 cells were then treated for 24 h with various concentrations (0.1–1 mM) of the FFA mixture in complete DMEM containing 1% BSA. The impact of the FFA treatment on cell viability was evaluated using the MTT assay as previously described. A final concentration of 1 mM OA-PA 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was employed to achieve steatotic conditions in HepG2.27
1 was employed to achieve steatotic conditions in HepG2.27
Analysis of steatotic gene expression in HepG2 and fibrotic gene expression in HHSC-N
To investigate the effects of GCE-PLNs on the expression of genes associated with hepatic steatosis and fibrosis, steatotic HepG2 and activated HHSC-N cells were treated with GCE, GCE-PLNs, and PLNs for 24 and 48 hours, respectively. Correspondingly, HepG2 and HHSC-N cells were seeded in 6-well plates at densities of 4 × 105 and 2.5 × 105 cells per well. After treatment, total RNA was extracted using the PureLink™ RNA Mini Kit (Invitrogen, USA).Gene expression analysis was conducted using real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR) with RNA extracted from both cell lines. The analysis was performed on a 7500 Real-Time PCR System (Applied Biosystems, USA) with the GoTaq® 1-Step RT-qPCR kit (Promega, USA). Forward and reverse oligonucleotides were designed based on published literature (Table 1) and validated using SnapGene software (GSL Biotech LLC, USA) before being acquired from T4 OLIGO (Mexico). Data normalization was achieved using GAPDH as the endogenous control for HepG2 cells and HPRT1 for HHSC-N cells. Relative expression was calculated using the 2−ΔΔCT method.28 The expression of the genes carnitine palmitoyltransferase I (CPT1A) and perilipin 1 (PLIN1) was quantified in HepG2 cells. In contrast, in HHSC-N cells, the analyzed genes were smooth muscle alpha-actin (ACTA2) and alpha-1 collagen chain type I (COL1A1).
| Gene | ID | F (5′ 3′) | R (5′ 3′) | |
|---|---|---|---|---|
| a HHSC-N: activated HSCs from a MASH donor. | ||||
| HepG2 | CPT1A | NM_001876.4 | GATCCTGGACAATACCTCGGAG | CTCCACAGCATCAAGAGACTGC |
| PLIN1 | NM_002666.5 | GCGGAATTTGCTGCCAACACTC | AGACTTCTGGGCTTGCTGGTGT | |
| GAPDH | NM_002046.7 | GCATCTTCTTGTGCAGTGCC | GAGAAGGCAGCCCTGGTAAC | |
| HHSC-N | ACTA2 | NM_001613.4 | CTGTTCCAGCCATCCTTCAT | TCATGATGCTGTTGTAGGTGGT |
| COL1A1 | NM_000088.4 | GCCTCAAGGTATTGCTGGAC | ACCTTGTTTGCCAGGTTCAC | |
| HPRT1 | NM_000194.3 | TGGTCAGGCAGTATAATCCAAAGA | TTCAAATCCAACAAAGTCTGGCT | |
Statistical analysis
The data were analyzed using GraphPad Prism software, version 9.4.1 (Dotmatics, UK). A one-way or two-way analysis of variance (ANOVA) with Tukey or Dunnett's multiple comparisons tests was used to evaluate the results. Vertical bars in the graphs represent standard deviation (SD). All assays were performed in triplicate (n = 3). Statistical significance was defined as p < 0.05, with significance levels indicated as follows: not significant (ns), p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****).Results and discussion
Total phenolic content and antioxidant activity of CGE
The Total phenolic content (TPC) and antioxidant activity of GCE were evaluated using the Folin-Ciocalteu colorimetric method and DPPH˙ inhibition assay, respectively. Results showed a TPC of 180.51 ± 30.26 mgEAG g−1 of GCE and an antioxidant activity of 30.86 ± 4.3 µM EAC in 100 µg mL−1 of GCE, equivalent to a 40% inhibition of DPPH˙. These findings align with prior studies on Robusta coffee bean extracts, where phenolic compounds were strongly linked to antioxidant capacity.29,30 Notably, Robusta beans, known for their elevated phenolic content and antioxidant properties,31 highlight their suitability as a phytochemical-rich source for obtaining GCE.Untargeted metabolomics and chemoinformatics
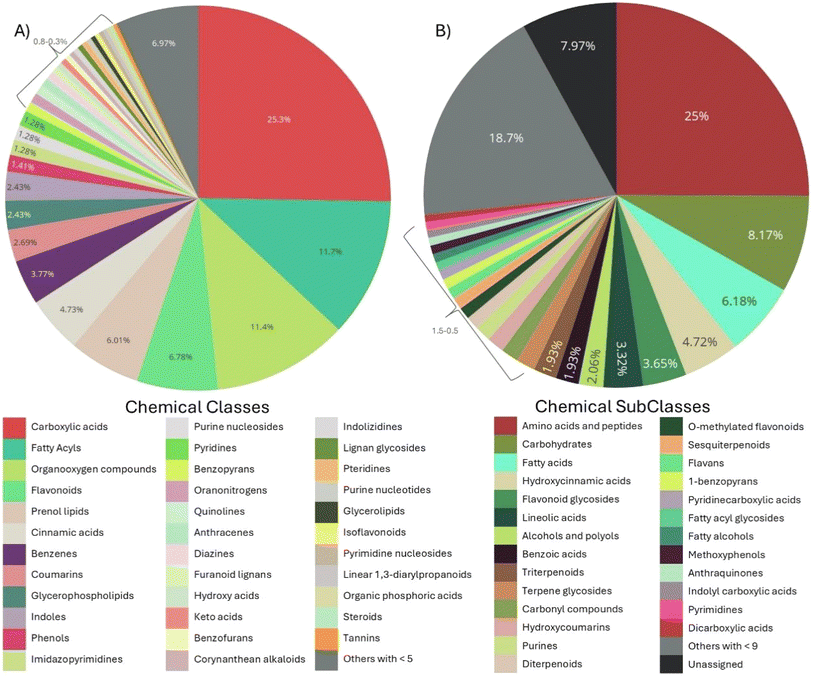
The metabolomic analysis was made by UPLC-MS/MS and after the filtering steps such as eliminating contaminants, 1942 molecules from 96 different classes and 145 subclasses were identified. However, to enhance confidence in the presence of chemical classes in GCE, those represented by fewer than 5 metabolites at the class level and fewer than 9 at the subclass level were grouped as “others with <5” and “others with <9”, respectively. This refinement resulted in a final list of 35 chemical classes and 26 subclasses (Fig. 2A and B).The use of chemoinformatic tools revealed the possible bioactivity of the metabolites by comparing them with indicated drugs that help reduce the progression of MASLD and or MASH. These chemoinformatic analyses identified a metabolite from GCE structurally like mesalamine, as shown in Fig. 3A. Mesalamine is a PPAR-gamma agonist, a receptor that promotes lipid storage in adipocytes, enhancing insulin sensitivity and reducing free fatty acid flux to the liver. Consequently, hepatic fatty acid uptake and steatosis are decreased upon PPAR-gamma activation.32,33 Moreover, PPAR-gamma is a marker of differentiated HSCs, and its expression diminishes as these cells transdifferentiate into myofibroblasts. Agonism of PPAR-gamma has been shown to attenuate HSC activation,34,35 suggesting a potential antifibrotic effect.
Moreover, the in silico analysis also revealed a metabolite from GCE structurally similar to idebenone, as shown in Fig. 3B. Idebenone is a Shc inhibitor known for its hepatoprotective effects, improvement of steatosis, and reduction of pro-fibrotic markers like COL1A1 in mouse models. These effects are attributed to its antioxidant properties and modulation of Shc, which is more active in MASH.36,37 Additionally, a GCE metabolite cysteamine bitartrate-like was identified (Fig. 3C). Cysteamine, a cystine-reducing agent approved for cystinosis treatment, has demonstrated promising effects in NAFLD, including liver enzyme improvement, oxidative stress reduction, and antifibrotic activity. Although its precise mechanism is not fully understood, its influence on oxidative stress modulation and adiponectin levels suggests a potential role in metabolic and hepatic health.38,39
Synthesis and characterization of polymer-lipid hybrid nanoparticles
| Sample | Hydrodynamic diameter (nm) | PDI |
|---|---|---|
| GCE-PLNs | 191.5 ± 3.6 | 0.291 ± 0.016 |
| PLNs | 218.2 ± 8.0 | 0.394 ± 0.021 |
The formation and size of the nanoparticles were confirmed through TEM. Both GCE-PLNs and PLNs exhibited a spherical morphology with a core–shell structure, characterized by two distinct phases: a high-contrast core corresponding to PLGA and a thin, low-contrast outer layer attributed to HSPC (Fig. 4E and F).44 Also, TEM analysis corroborated the hydrodynamic diameter measurements obtained by DLS, showing that GCE-PLNs (145.4 ± 26 nm) were smaller than PLNs (179 ± 36.7 nm). Meanwhile, ζ analysis revealed similar negative values for both formulations, with no significant differences (Fig. 4D). These findings are consistent with those previously reported for PLNs coated with HSPC, emphasizing the anionic nature imparted by the lipid coating.23,45 In drug delivery, the surface charge of nanoparticles plays a critical role in determining their efficacy and safety. A near-neutral charge (−10 to +10 mV) is optimal for systemic delivery, as it minimizes rapid clearance, enhances compatibility with biological systems, and improves therapeutic outcomes.46,47 Negatively charged nanoparticles are particularly advantageous for in vivo applications due to their lower cytotoxicity and hemolytic activity compared to positively charged counterparts. Additionally, their biodistribution favors liver-targeted therapies, as they naturally accumulate in the liver, unlike positively charged nanoparticles, which are rapidly cleared via the hepatobiliary system.48,49
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester and C-O stretching, respectively.50,51 HSPC presented peaks at 2920 and 2851 cm−1, corresponding to C–H bond stretches of its hydrophobic chains, which were also observable in the spectra of GCE-PLNs and PLNs. Additionally, HSPC showed signals for the P–O–C bond at 1050 cm−1 and the choline moiety [-N+(CH2)3] at 968 cm−1.52,53 GCE displayed peaks at 3249 cm−1 and 1598 cm−1, related to C
O ester and C-O stretching, respectively.50,51 HSPC presented peaks at 2920 and 2851 cm−1, corresponding to C–H bond stretches of its hydrophobic chains, which were also observable in the spectra of GCE-PLNs and PLNs. Additionally, HSPC showed signals for the P–O–C bond at 1050 cm−1 and the choline moiety [-N+(CH2)3] at 968 cm−1.52,53 GCE displayed peaks at 3249 cm−1 and 1598 cm−1, related to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and –OH functional groups from phenolic and other phytochemical compounds.54,55 In GCE-PLNs, these signals shifted to 3377 cm−1 and 1645 cm−1, suggesting the adsorption of some extract compounds on the nanoparticle surface. However, the absence of other distinctive GCE signals and the dominance of PLGA and HSPC signals in GCE-PLNs indicate a successful encapsulation of GCE within the nanoparticles.56,57
O and –OH functional groups from phenolic and other phytochemical compounds.54,55 In GCE-PLNs, these signals shifted to 3377 cm−1 and 1645 cm−1, suggesting the adsorption of some extract compounds on the nanoparticle surface. However, the absence of other distinctive GCE signals and the dominance of PLGA and HSPC signals in GCE-PLNs indicate a successful encapsulation of GCE within the nanoparticles.56,57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 for its encapsulation, as this polymer ratio generally favors hydrophilic compounds. Some studies suggest that PLGA 65
50 for its encapsulation, as this polymer ratio generally favors hydrophilic compounds. Some studies suggest that PLGA 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 could enhance EE% for bioactive compounds from plant extracts.59,60
35 could enhance EE% for bioactive compounds from plant extracts.59,60As illustrated in Fig. 4I, the release profile of GCE from the nanoparticle system was monitored over 72 hours quantifying the content of CGAs in the release medium. The data revealed a rapid initial release (∼85%) within the first 6 hours, followed by a sustained release phase that extended to 24 hours, at which point complete GCE release was achieved. Subsequently, starting at 36 hours, a decrease in GCE accumulation in the release solution was observed. The burst-like initial release observed in GCE-PLNs likely stems from phytochemical compounds near the nanoparticle surface, which rapidly diffuse into the solution due to nanoparticle degradation mechanisms.25,61 Besides, CGAs exhibit a moderate hydrophilic nature (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.42),62 facilitating their dispersion in aqueous environments like PBS and contributing to this rapid release. The subsequent decline in GCE levels after 36 hours suggests potential CGAs degradation.60
P = 1.42),62 facilitating their dispersion in aqueous environments like PBS and contributing to this rapid release. The subsequent decline in GCE levels after 36 hours suggests potential CGAs degradation.60
Cell viability of HepG2 and HHSC-N
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
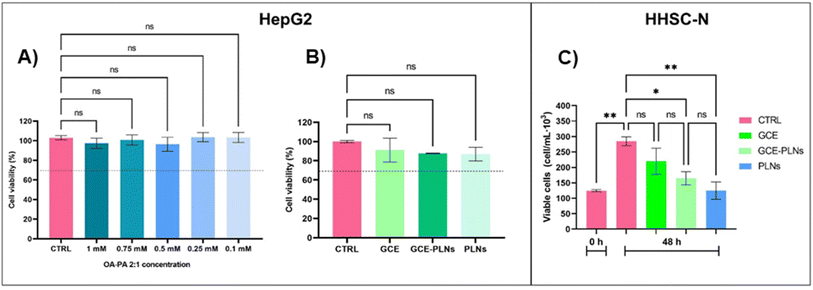
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture. MTT assay results showed that none of the concentrations were cytotoxic to HepG2 after 24 h of incubation, and no significant differences in cell viability were observed between the different concentrations tested (Fig. 5A). Oleic and palmitic acid are commonly used in experimental models of hepatic steatosis because they promote TGs accumulation in hepatocytes, mimicking conditions seen in patients with metabolic liver diseases.64,65 Some in vitro studies have shown that concentrations of ≥1 mM of OA-PA effectively induce a steatotic state in hepatocytes without compromising cell viability.66–68 These findings are consistent with the cell viability results obtained from this work for HepG2 cells exposed to OA-PA mixture.
1 mixture. MTT assay results showed that none of the concentrations were cytotoxic to HepG2 after 24 h of incubation, and no significant differences in cell viability were observed between the different concentrations tested (Fig. 5A). Oleic and palmitic acid are commonly used in experimental models of hepatic steatosis because they promote TGs accumulation in hepatocytes, mimicking conditions seen in patients with metabolic liver diseases.64,65 Some in vitro studies have shown that concentrations of ≥1 mM of OA-PA effectively induce a steatotic state in hepatocytes without compromising cell viability.66–68 These findings are consistent with the cell viability results obtained from this work for HepG2 cells exposed to OA-PA mixture.
On the other hand, HHSC-N cells treated for 48 hours exhibited a significant increase in cell number in the untreated control and GCE-treated groups (Fig. 5C). Remarkably, GCE-PLNs and PLNs maintained cell numbers like the baseline observed in freshly seeded untreated cells (0 h). These results indicate that both GCE-PLNs and PLNs controlled cell growth of HHSC-N, maintaining cell numbers near baseline levels. Upon activation, HSCs adopt a proliferative phenotype, and their increased presence contributes to fibrosis progression in MASH.70,71 Thus, it is important that the treatment controls HHSC-N proliferation. To clarify this mechanism, further assays assessing apoptosis markers and proliferation-associated gene expression would be necessary.
Effect of treatments on steatosis-associated genes in HepG2
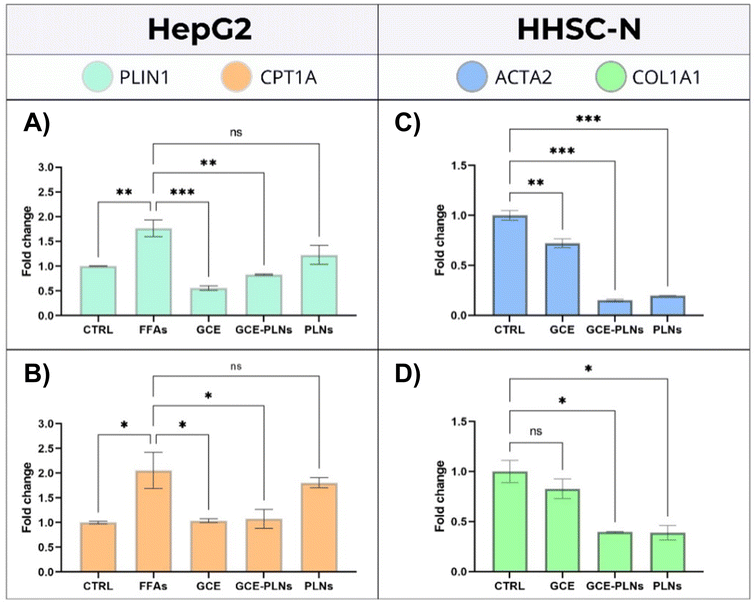
After 24 h of incubation with the FFAs mixture and 24 h of treatment with GCE, GCE-PLNs and PLNs, the expression of steatosis-related genes in HepG2 cells was quantified by RT-qPCR. As shown in Fig. 6A and B, the steatotic control group (FFAs) exhibited a significant upregulation of PLIN1 and CPT1A, with increases of 76% and 94%, respectively, compared to the untreated cells (CTRL). The upregulation PLIN1 validates the accumulation of lipids within hepatocytes, as it plays a critical role in lipid droplet (LD) formation and stability, facilitating lipid storage.63,72 Treatments with GCE and GCE-PLNs significantly reduced the expression of both genes, however, there is no significant difference between free GCE and encapsulated GCE. Likewise, PLNs alone did not alter gene expression relative to the steatotic control, suggesting that the observed reductions are specifically attributed to the bioactive compounds of GCE.During MASH, PLIN1 is markedly overexpressed, contributing to macrovesicular steatosis by promoting lipid droplet formation and activating genes involved in lipid synthesis and storage.73 The observed reduction in PLIN1 expression in cells treated with both free and encapsulated GCE suggests its potential role in decreasing TGs accumulation, which could mitigate hepatic steatosis. To corroborate this lipid reduction, it would be beneficial to quantify intracellular TGs levels through the expression of enzymes pivotal in TGs synthesis.74 Such analyses would provide more comprehensive evidence of lipid modulation and offer a stronger validation of GCE's impact on steatosis.
CPT1A is a key enzyme in the β-oxidation of FFAs and plays a crucial role in regulating hepatic lipid metabolism. Its expression and activity are intricately linked to the progression of MASLD. While elevated CPT1A levels can be advantageous by enhancing FFAs oxidation and reducing hepatic inflammation, a deficiency in certain contexts may confer a protective effect, mitigating diet-induced liver damage.75,76 Studies indicate that a moderate increase in CPT1A activity effectively reduces intracellular TGs accumulation in both in vitro and in vivo models.77,78 Thus, it is important to maintain a balanced CPT1A expression to ensure lipid metabolism homeostasis and prevent exacerbation of the disease. Notably, GCE and GCE-PLNs appear to support this equilibrium by not significantly altering CPT1A expression.
Effect of treatments on fibrosis-associated genes in HHSC-N
The expression of profibrotic genes in HHSC-N cells was evaluated after 48 h of incubation with GCE, GCE-PLNs, and PLNs. As illustrated in Fig. 6C and D, both GCE-PLNs and PLNs significantly downregulated COL1A1 and ACTA2 genes. Free GCE also markedly reduced ACTA2 expression; however, its effect on COL1A1 was less pronounced, showing only a downward trend compared to the untreated HHSC-N control. These findings suggest that PLNs enhance the impact of GCE on ACTA2 and COL1A1 expression, demonstrating an additive or synergistic effect attributable to the nanoparticle delivery system, specifically to HSPC.79The downregulation of ACTA2 and COL1A1 genes suggests an antifibrotic effect of GCE-PLNs, as ACTA2 expression is closely related to the activation of HSCs.80 Previous studies,81,82 have shown that ACTA2 deficiency significantly reduces type I collagen production, which is consistent with the results of this work, where down-regulation of ACTA2 and COL1A1 was observed in HHSC-N after 48 h of treatment with GCE-PLNs. Additionally, these findings align with the cheminformatics analysis results, where the presence of compounds like mesalamine and idebenone in the GCE suggests an antifibrotic effect related to the activation of HSCs and the production of type I collagen. However, further studies are warranted to validate these results, including direct measurements of type I collagen synthesis and a more comprehensive characterization of the profibrotic phenotype in HHSC-N cells. Such investigations would provide deeper insight into the mechanism of action of GCE-PLNs and strengthen their potential as a therapeutic option for liver fibrosis.
Conclusions
A green coffee extract (GCE) with high phenolic content and antioxidant activity was successfully obtained and encapsulated into hybrid polymer-lipid nanoparticles (GCE-PLNs) formulated from PLGA and HSPC. The incorporation of GCE enhanced the physicochemical properties of the nanoparticles, resulting in smaller and more stable particles. The nanoparticles ranged in size from 140 to 200 nm, with the incorporation of GCE reducing particle size by 20%. By cheminformatics analysis, we identified 1942 molecules from 96 different classes and 145 subclasses, revealing structural similarities between GCE metabolites and three distinct therapeutic compounds for MASLD and MASH: mesalamine, a PPAR-gamma agonist involved in lipid metabolism and fibrosis regulation; cysteamine bitartrate, a cystine-reducing agent with antioxidant and antifibrotic properties; and idebenone, a Shc inhibitor known for its hepatoprotective effects. These findings highlight the possible bioactivity of GCE metabolites structurally like known drugs, suggesting additional pathways through which GCE-PLNs could exert their beneficial effects.An in vitro MASLD model was employed using human cells. Hepatic steatosis was modeled by inducing lipid accumulation in HepG2 cells using a free fatty acid mixture for 24 hours, while hepatic fibrosis was modeled with activated hepatic stellate cells (HHSC-N) from a MASH patient. None of the treatments exhibited cytotoxicity in the in vitro model and GCE-PLNs controlled cell growth of HHSC-N after 48 hours. Moreover, GCE-PLNs modulated the expression of genes associated with steatosis and fibrosis in liver cells. Specifically, treatments with GCE and GCE-PLNs reduced lipid accumulation and the expression of fibrosis-related genes, indicating their potential therapeutic effects in liver diseases. Both free GCE and GCE-PLNs reduced PLIN1 expression in HepG2 cells without altering CPT1A, suggesting an anti-steatotic effect that preserves lipid metabolism homeostasis. Likewise, GCE-PLNs and PLNs significantly decreased the expression of COL1A1 and ACTA2 in HHSC-N after 48 hours of treatment, demonstrating an enhanced effect attributed to PLNs.
Our findings highlight the therapeutic potential of GCE and its encapsulation in addressing the dual challenges of hepatic steatosis and fibrosis. Further studies are necessary to confirm these results in more complex models and assess additional mechanisms. Overall, our results support the use of GCE-PLNs as a promising approach for liver therapy, with the potential for clinical application in MASLD treatment.
Author contributions
Marco A. Uriostegui-Campos: conceptualization, methodology, formal analysis, investigation, writing – original draft & visualization. Shaula A. Castro-Murrieta: methodology, formal analysis & writing. Ximena I. Lopez-Cesati: methodology, review & editing. Ricardo Colín-Delgado: methodology, review & editing. Gabriela I. Carballo-López: methodology, review & editing. Karla Cervantes-Luevano: methodology. Aldo Moreno-Ulloa: formal analysis, writing – review & editing. Ana B. Castro-Ceseña: conceptualization, formal analysis, writing – review & editing, project administration, funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial or personal relationships that could have appeared to influence the work reported in this paper.Data availability
Data supporting this article have been included as part of the supplementary information (SI), that contains a list of all the identified metabolites in samples, as well as a list of processed identified metabolites in samples. The raw datasets supporting the metabolomics results are available in the GNPS/MassIVE public repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp?redirect=auth) under the accession number: MSV000097479. Direct link: MassIVE Private Dataset (https://massive.ucsd.edu/ProteoSAFe/private-dataset.jsp?task=f3efb5e8d8224c599c11d0f627da5484). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra06552f.Acknowledgements
This work was supported by the Secretaría de Ciencia, Humanidades, Tecnología e Innovación (SECIHTI) (Project CF-2023-I-2458), and CICESE (Project 685112). Marco A. Uriostegui-Campos acknowledges SECIHTI for his scholarship. Prof. Norma Cortez-Lemus is thanked for her help with DLS measurements. During the preparation of this work, the authors used Microsoft 365 Copilot to improve readability and language of the work. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.References
- W. K. Chan, K. H. Chuah, R. B. Rajaram, L. L. Lim, J. Ratnasingam and S. R. Vethakkan, Metabolic Dysfunction-Associated Steatotic Liver Disease (MASLD): A State-of-the-Art Review, J. Obes. Metab. Syndr., 2023, 32(3), 197–213 CrossRef PubMed.
- L. Miao, G. Targher, C. D. Byrne, Y. Y. Cao and M. H. Zheng, Current status and future trends of the global burden of MASLD, Trends Endocrinol. Metab., 2024, 35(8), 697–707 CrossRef CAS PubMed.
- E. E. Powell, V. W. Wong and M. Rinella, Non-alcoholic fatty liver disease, Lancet, 2021, 397(10290), 2212–2224 CrossRef CAS PubMed.
- European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD) and European Association for the Study of Obesity (EASO), EASL-EASD-EASO Clinical Practice Guidelines on the management of metabolic dysfunction-associated steatotic liver disease (MASLD), J. Hepatol., 2024, 81(3), 492–542 CrossRef PubMed.
- S. L. Friedman, B. A. Neuschwander-Tetri, M. Rinella and A. J. Sanyal, Mechanisms of NAFLD development and therapeutic strategies, Nat. Med., 2018, 24(7), 908–922 CrossRef CAS PubMed.
- P. Golabi, S. Owrangi and Z. M. Younossi, Global perspective on nonalcoholic fatty liver disease and nonalcoholic steatohepatitis – prevalence, clinical impact, economic implications and management strategies, Aliment. Pharmacol. Ther., 2024, 59(Suppl 1), S1–S9 Search PubMed.
- C. Y. Zhang, W. G. Yuan, P. He, J. H. Lei and C. X. Wang, Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets, World J. Gastroenterol., 2016, 22(48), 10512–10522 CrossRef CAS PubMed.
- K. C. Lee, P. S. Wu and H. C. Lin, Pathogenesis and treatment of non-alcoholic steatohepatitis and its fibrosis, Clin. Mol. Hepatol., 2023, 29(1), 77–98 CrossRef PubMed.
- J. Zeng, J. G. Fan and S. M. Francque, Therapeutic management of metabolic dysfunction associated steatotic liver disease, United Eur. Gastroenterol. J., 2024, 12(2), 177–186 CrossRef CAS PubMed.
- L. Barrea, G. Pugliese and E. Frias-Toral, et al., Coffee consumption, health benefits and side effects: a narrative review and update for dietitians and nutritionists, Crit. Rev. Food Sci. Nutr., 2023, 63(9), 1238–1261 CrossRef PubMed.
- S. Hosseinabadi, M. Rafraf, S. Asghari, M. Asghari-Jafarabadi and S. Vojouhi, Effect of green coffee extract supplementation on serum adiponectin concentration and lipid profile in patients with non-alcoholic fatty liver disease: A randomized, controlled trial, Complement. Ther. Med., 2020, 49, 102290 CrossRef PubMed.
- O. Nikpayam, A. H. Faghfouri, O. M. Tavakoli-Rouzbehani, S. M. Jalali, M. Najafi and G. Sohrab, The effect of green coffee extract supplementation on lipid profile: A systematic review of clinical trial and in-vivo studies, Diabetes Metab. Syndr., 2020, 14(5), 1521–1528 CrossRef PubMed.
- H. A. Shahmohammadi, S. A. Hosseini, E. Hajiani, A. S. Malehi and M. Alipour, Effects of Green Coffee Bean Extract Supplementation on Patients with Non-Alcoholic Fatty Liver Disease: A Randomized Clinical Trial, Hepatitis Mon., 2017, 17(4), e12299 Search PubMed.
- M. Á. Seguido, R. M. Tarradas and S. González-Rámila, et al., Sustained Consumption of a Decaffeinated Green Coffee Nutraceutical Has Limited Effects on Phenolic Metabolism and Bioavailability in Overweight/Obese Subjects, Nutrients, 2022, 14(12), 2445 CrossRef CAS PubMed.
- O. Asbaghi, S. Kashkooli and M. Mardani, et al., Effect of green coffee bean extract supplementation on liver function and inflammatory biomarkers: A meta-analysis of randomized clinical trials, Complement. Ther. Clin. Pract., 2021, 43, 101349 CrossRef PubMed.
- L. Pachuau, N. Laldinchhana, P. K. Roy, J. H. Zothantluanga, S. Ray and S. Das, Encapsulation of bioactive compound and its therapeutic potential, in Bioactive Natural Products for Pharmaceutical Applications, 2020, pp. 687–714 Search PubMed.
- A. Mukherjee, A. K. Waters, P. Kalyan, A. S. Achrol, S. Kesari and V. M. Yenugonda, Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: state of the art, emerging technologies, and perspectives, Int. J. Nanomed., 2019, 14, 1937–1952 CrossRef CAS PubMed.
- N. Tahir, M. T. Haseeb, A. Madni, et al., Lipid polymer hybrid Nanoparticles: a novel approach for drug delivery, in Role of Novel Drug Delivery Vehicles in Nanobiomedicine, 2019 Search PubMed.
- J. Ghitman, E. I. Biru, R. Stan and H. Iovu, Review of hybrid PLGA nanoparticles: Future of smart drug delivery and theranostics medicine, Mater. Des., 2020, 193, 108805 CrossRef CAS.
- S. E. Varghese, M. K. Fariya, G. S. Rajawat, F. Steiniger, A. Fahr and M. S. Nagarsenker, Lecithin and PLGA-based self-assembled nanocomposite, Lecithmer: preparation, characterization, and pharmacokinetic/pharmacodynamic evaluation, Drug Delivery Transl. Res., 2016, 6(4), 342–353 CAS.
- I. Rubio-Elizalde, J. Bernáldez-Sarabia and A. Moreno-Ulloa, et al., Scaffolds based on alginate-PEG methyl ether methacrylate-Moringa oleifera-Aloe vera for wound healing applications, Carbohydr. Polym., 2019, 206, 455–467 CrossRef CAS PubMed.
- Y. Djoumbou Feunang, R. Eisner and C. Knox, et al., ClassyFire: automated chemical classification with a comprehensive, computable taxonomy, J. Cheminf., 2016, 8, 61 Search PubMed.
- P. Ahmaditabar, A. A. Momtazi-Borojeni, A. H. Rezayan, M. Mahmoodi, A. Sahebkar and M. Mellat, Enhanced entrapment and improved in vitro controlled release of N-Acetyl cysteine in hybrid PLGA/Lecithin nanoparticles prepared using a Nanoprecipitation/Self-Assembly method, J. Cell. Biochem., 2017, 118(12), 4203–4209 CrossRef CAS PubMed.
- R. H. Fang, S. Aryal, C. M. Hu and L. Zhang, Quick synthesis of lipid-polymer hybrid nanoparticles with low polydispersity using a single-step sonication method, Langmuir, 2010, 26(22), 16958–16962 CrossRef CAS PubMed.
- Y. A. Moussa, M. H. Teaima, D. Attia, M. M. Elmazar and M. A. El-Nabarawi, Unroasted Green Coffee Extract-Loaded Solid Lipid Nanoparticles for Enhancing Intestinal Permeation, ACS Omega, 2023, 8(23), 20251–20261 CrossRef CAS PubMed.
- W. E. Knight, Y. Cao, P. Dillon and K. Song, A simple protocol to produce mature human-induced pluripotent stem cell-derived cardiomyocytes, STAR Protoc., 2021, 2(4), 100912 CrossRef CAS PubMed.
- M. Ricchi, M. R. Odoardi and L. Carulli, et al., Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes, J. Gastroenterol. Hepatol., 2009, 24(5), 830–840 CrossRef CAS PubMed.
- S. C. Taylor, K. Nadeau, M. Abbasi, C. Lachance, M. Nguyen and J. Fenrich, The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time, Trends Biotechnol., 2019, 37(7), 761–774 CrossRef CAS PubMed.
- S. Pergolizzi, V. D'Angelo and M. Aragona, et al., Evaluation of antioxidant and anti-inflammatory activity of green coffee beans methanolic extract in rat skin, Nat. Prod. Res., 2020, 34(11), 1535–1541 CrossRef CAS PubMed.
- E. Suryanti, N. D. Retnowati and N. M. E. Prastya, et al., Chemical Composition, Antioxidant, Antibacterial, Antibiofilm, and Cytotoxic Activities of Robusta Coffee Extract (Coffea canephora), HAYATI J. Biosci., 2023, 30(4), 632–642 CrossRef.
- U. H. A. Hasbullah and D. R. Umiyati, Antioxidant activity and total phenolic compounds of arabica and robusta coffee at different roasting levels, J. Phys. Conf. Ser., 2021, 1764(1), 012033 CrossRef CAS.
- M. Schwab, V. Reynders and S. Loitsch, et al., PPARgamma is involved in mesalazine-mediated induction of apoptosis and inhibition of cell growth in colon cancer cells, Carcinogenesis, 2008, 29(7), 1407–1414 CrossRef CAS PubMed.
- C. Linard, O. Grémy and M. Benderitter, Reduction of peroxisome proliferation-activated receptor gamma expression by gamma-irradiation as a mechanism contributing to inflammatory response in rat colon: modulation by the 5-aminosalicylic acid agonist, J. Pharmacol. Exp. Ther., 2008, 324(3), 911–920 CrossRef CAS PubMed.
- B. Staels, L. Butruille and S. Francque, Treating NASH by targeting peroxisome proliferator-activated receptors, J. Hepatol., 2023, 79(5), 1302–1316 CrossRef CAS PubMed.
- J. Pan, W. Zhou, R. Xu, L. Xing, G. Ji and Y. Dang, Natural PPARs agonists for the treatment of nonalcoholic fatty liver disease, Biomed. Pharmacother., 2022, 151, 113127 CrossRef CAS PubMed.
- J. X. Jiang, A. Tomilov, C. Montgomery, C. K. Hui, N. J. Török and G. Cortopassi, Shc inhibitor idebenone ameliorates liver injury and fibrosis in dietary NASH in mice, J. Biochem. Mol. Toxicol., 2021, 35(10), e22876 CrossRef CAS PubMed.
- A. Tomilov, S. Allen, C. K. Hui, A. Bettaieb and G. Cortopassi, Idebenone is a cytoprotective insulin sensitizer whose mechanism is Shc inhibition, Pharmacol. Res., 2018, 137, 89–103 CrossRef CAS PubMed.
- J. Tonascia, Cysteamine Bitartrate Delayed-Release for the Treatment of Nonalcoholic Fatty Liver Disease (NAFLD) in Children, NIDDK Central Repository Resources for Research (NIDDK-CR R4R), 2023 Search PubMed.
- R. Dohil, L. Meyer, S. Schmeltzer, B. L. Cabrera, J. E. Lavine and S. A. Phillips, The effect of cysteamine bitartrate on adiponectin multimerization in non-alcoholic fatty liver disease and healthy subjects, J. Pediatr., 2012, 161(4), 639–645 CrossRef CAS PubMed.
- S. Abdelghany, T. Parumasivam and A. Pang, et al., Alginate modified-PLGA nanoparticles entrapping amikacin and moxifloxacin as a novel host-directed therapy for multidrug-resistant tuberculosis, J. Drug Delivery Sci. Technol., 2019, 52, 642–651 CrossRef CAS.
- J. W. Hickey, J. L. Santos, J. M. Williford and H. Q. Mao, Control of polymeric nanoparticle size to improve therapeutic delivery, J. Controlled Release, 2015, 219, 536–547 CrossRef CAS PubMed.
- V. Rosiuk, A. Runser, A. Klymchenko and A. Reisch, Controlling Size and Fluorescence of Dye-Loaded Polymer Nanoparticles through Polymer Design, Langmuir, 2019, 35(21), 7009–7017 CrossRef CAS PubMed.
- N. Tahir, A. Madni and A. Correia, et al., Lipid-polymer hybrid nanoparticles for controlled delivery of hydrophilic and lipophilic doxorubicin for breast cancer therapy, Int. J. Nanomed., 2019, 14, 4961–4974 CrossRef CAS PubMed.
- J. Ismail, L. C. Klepsch and P. Dahlke, et al., PEG-Lipid-PLGA Hybrid Particles for Targeted Delivery of Anti-Inflammatory Drugs, Pharmaceutics, 2024, 16(2), 187 CrossRef CAS PubMed.
- Y. Yang, Y. Meng, E. Zhang and J. Ding, A Facile Way to Increase the Cellular Uptake Efficiency of Hybrid Nanoparticles, J. Nanosci. Nanotechnol., 2018, 18(7), 4559–4564 CrossRef CAS PubMed.
- S. A. Dilliard, Q. Cheng and D. J. Siegwart, On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(52), e2109256118 CrossRef CAS PubMed.
- C. Martin, E. Dolmazon and K. Moylan, et al., A charge neutral, size tuneable polymersome capable of high biological encapsulation efficiency and cell permeation, Int. J. Pharm., 2015, 481(1–2), 1–8 CrossRef CAS PubMed.
- S. Hirn, M. Semmler-Behnke and C. Schleh, et al., Particle size-dependent and surface charge-dependent biodistribution of gold nanoparticles after intravenous administration, Eur. J. Pharm. Biopharm., 2011, 77(3), 407–416 CrossRef CAS PubMed.
- M. Xie, Y. Xu, H. Shen, S. Shen, Y. Ge and J. Xie, Negative-charge-functionalized mesoporous silica nanoparticles as drug vehicles targeting hepatocellular carcinoma, Int. J. Pharm., 2014, 474(1–2), 223–231 CAS.
- L. M. d. A. C. Gaspar, A. C. S. Dórea and D. Droppa-Almeida, et al., Development and characterization of PLGA nanoparticles containing antibiotics, J. Nanopart. Res., 2018, 20(11), 289 CrossRef.
- S. Kumar, P. Sangwan, V. Lather and D. Pandita, Biocompatible PLGA-oil hybrid nanoparticles for high loading and controlled delivery of resveratrol, J. Drug Delivery Sci. Technol., 2015, 30, 54–62 CrossRef CAS.
- S. Biswas, P. K. Mukherjee, A. Kar, S. Bannerjee, R. Charoensub and T. Duangyod, Optimized piperine-phospholipid complex with enhanced bioavailability and hepatoprotective activity, Pharm. Dev. Technol., 2021, 26(1), 69–80 CrossRef CAS PubMed.
- F. V. Leimann, M. H. Biz and K. C. Kaufmann, et al., Characterization of progesterone loaded biodegradable blend polymeric nanoparticles, Cienc. Rural, 2015, 45(11), 2082–2088 CrossRef CAS.
- A. Masek, M. Latos-Brozio, J. Kałużna-Czaplińska, A. Rosiak and E. Chrzescijanska, Antioxidant properties of green coffee extract, Forests, 2020, 11(5), 557 CrossRef.
- C. M. Topala and L. D. Tataru, Infrared Spectra of Green Arabica Coffee Extraction using Supercritical Carbon Dioxide and Soxhlet Technique, Rev. Chim., 2015, 66(8), 1128–1131 CAS.
- S. E. Garcia-Solis, V. Perez-Perez and D. Tapia-Maruri, et al., Microencapsulation of the green coffee waste extract with high antioxidant activity by spray-drying, J. Food Process. Preserv., 2022, 46(10), e16864 CAS.
- M. Kesente, E. Kavetsou and M. Roussaki, et al., Encapsulation of Olive Leaves Extracts in Biodegradable PLA Nanoparticles for Use in Cosmetic Formulation, Bioengineering, 2017, 4(3), 75 CrossRef PubMed.
- N. M. Desai, J. Gilbert Stanley and P. S. Murthy, Green coffee nanoparticles: optimisation, in vitro bioactivity and bio-release property, J. Microencapsulation, 2020, 37(1), 52–64 CrossRef CAS PubMed.
- B. A. Oseni, C. P. Azubuike, O. O. Okubanjo, C. I. Igwilo and J. Panyam, Encapsulation of Andrographolide in poly(lactide-co-glycolide) Nanoparticles: Formulation Optimization and in vitro Efficacy Studies, Front. Bioeng. Biotechnol., 2021, 9, 639409 CrossRef PubMed.
- M. C. Pereira, D. A. Oliveira and L. E. Hill, et al., Effect of nanoencapsulation using PLGA on antioxidant and antimicrobial activities of guabiroba fruit phenolic extract, Food Chem., 2018, 240, 396–404 CrossRef CAS PubMed.
- B. Mandal, N. K. Mittal, P. Balabathula, L. A. Thoma and G. C. Wood, Development and in vitro evaluation of core-shell type lipid-polymer hybrid nanoparticles for the delivery of erlotinib in non-small cell lung cancer, Eur. J. Pharm. Sci., 2016, 81, 162–171 CrossRef CAS PubMed.
- D. Scherbl, S. Muentnich and E. Richling, In vitro absorption studies of chlorogenic acids from coffee using the Ussing chamber model, Food Res. Int., 2014, 63, 456–463 CrossRef CAS.
- O. Khalifa, N. S. Al-Akl, K. Errafii and A. Arredouani, Exendin-4 alleviates steatosis in an in vitro cell model by lowering FABP1 and FOXA1 expression via the Wnt/-catenin signaling pathway, Sci. Rep., 2022, 12(1), 2226 CrossRef CAS PubMed.
- F. A. Müller and S. J. Sturla, Human in vitro models of nonalcoholic fatty liver disease, Curr. Opin. Toxicol., 2019, 16, 9–16 CrossRef.
- S. Sharma, J. E. Mells, P. P. Fu, N. K. Saxena and F. A. Anania, GLP-1 analogs reduce hepatocyte steatosis and improve survival by enhancing the unfolded protein response and promoting macroautophagy, PLoS One, 2011, 6(9), e25269 CrossRef CAS PubMed.
- R. A. Egnatchik, A. K. Leamy, Y. Noguchi, M. Shiota and J. D. Young, Palmitate-induced activation of mitochondrial metabolism promotes oxidative stress and apoptosis in H4IIEC3 rat hepatocytes, Metabolism, 2014, 63(2), 283–295 CrossRef CAS PubMed.
- P. J. Giraudi, V. J. Becerra, V. Marin, N. C. Chavez-Tapia, C. Tiribelli and N. Rosso, The importance of the interaction between hepatocyte and hepatic stellate cells in fibrogenesis induced by fatty accumulation, Exp. Mol. Pathol., 2015, 98(1), 85–92 CrossRef CAS PubMed.
- A. Moravcová, Z. Červinková, O. Kučera, V. Mezera, D. Rychtrmoc and H. Lotková, The effect of oleic and palmitic acid on induction of steatosis and cytotoxicity on rat hepatocytes in primary culture, Physiol. Res., 2015, 64(Suppl 5), S627–S636 Search PubMed.
- D. Lim, W. Kim, M. G. Lee, H. J. Heo, O. K. Chun and D. O. Kim, Evidence for protective effects of coffees on oxidative stressinduced apoptosis through antioxidant capacity of phenolics, Food Sci. Biotechnol., 2012, 21(6), 1735–1744 CrossRef CAS.
- O. Nunez Lopez, F. J. Bohanon and X. Wang, et al., STAT3 Inhibition Suppresses Hepatic Stellate Cell Fibrogenesis: HJC0123, a Potential Therapeutic Agent for Liver Fibrosis, RSC Adv., 2016, 6(102), 100652–100663 RSC.
- A. Zisser, D. H. Ipsen and P. Tveden-Nyborg, Hepatic Stellate Cell Activation and Inactivation in NASH-Fibrosis-Roles as Putative Treatment Targets?, Biomedicines, 2021, 9(4), 365 CrossRef CAS PubMed.
- D. Greco, A. Kotronen and J. Westerbacka, et al., Gene expression in human NAFLD, Am. J. Physiol.: Gastrointest. Liver Physiol., 2008, 294(5), G1281–G1287 CrossRef CAS PubMed.
- R. M. Carr and R. S. Ahima, Pathophysiology of lipid droplet proteins in liver diseases, Exp. Cell Res., 2016, 340(2), 187–192 CrossRef CAS PubMed.
- N. L. Gluchowski, K. R. Gabriel and C. Chitraju, et al., Hepatocyte Deletion of Triglyceride-Synthesis Enzyme Acyl CoA: Diacylglycerol Acyltransferase 2 Reduces Steatosis Without Increasing Inflammation or Fibrosis in Mice, Hepatology, 2019, 70(6), 1972–1985 CrossRef CAS PubMed.
- C. W. Lin, Y. J. Peng, Y. Y. Lin, H. J. Mersmann and S. T. Ding, LRRK2 Regulates CPT1A to Promote β-Oxidation in HepG2 Cells, Molecules, 2020, 25(18), 4122 CrossRef CAS PubMed.
- W. Sun, T. Nie and K. Li, et al., Hepatic CPT1A facilitates Liver–Adipose cross talk via induction of FGF21 in mice, Diabetes, 2021, 71(1), 31–42 CrossRef PubMed.
- N. Da Silva Lima, M. F. Fondevila and E. Nóvoa, et al., Inhibition of ATG3 ameliorates liver steatosis by increasing mitochondrial function, J. Hepatol., 2021, 76(1), 11–24 CrossRef PubMed.
- M. Stefanovic-Racic, G. Perdomo, B. S. Mantell, I. J. Sipula, N. F. Brown and R. M. O'Doherty, A moderate increase in carnitine palmitoyltransferase 1a activity is sufficient to substantially reduce hepatic triglyceride levels, Am. J. Physiol.: Endocrinol. Metab., 2008, 294(5), E969–E977 CrossRef CAS PubMed.
- Q. Cao, K. M. Mak and C. S. Lieber, DLPC decreases TGF-beta1-induced collagen mRNA by inhibiting p38 MAPK in hepatic stellate cells, Am. J. Physiol.: Gastrointest. Liver Physiol., 2002, 283(5), G1051–G1061 CrossRef CAS PubMed.
- Y. Tsuchiya, T. Seki and K. Kobayashi, et al., Fibroblast growth factor 18 stimulates the proliferation of hepatic stellate cells, thereby inducing liver fibrosis, Nat. Commun., 2023, 14(1), 6304 CrossRef CAS PubMed.
- I. N. Chomsy, M. S. Rohman and H. Khotimah, et al., Effect of the ethanolic extract of green tea and green coffee on cardiac fibrosis attenuation by suppressing activin-a and collagen-1 gene expression, AIP Conf. Proc., 2022, 2543, 020002 CrossRef.
- D. C. Rockey, Q. Du, N. D. Weymouth and Z. Shi, Smooth Muscle α-Actin Deficiency Leads to Decreased Liver Fibrosis via Impaired Cytoskeletal Signaling in Hepatic Stellate Cells, Am. J. Pathol., 2019, 189(11), 2209–2220 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |