Open Access Article
Open Access ArticleIntegrative network pharmacology and molecular dynamics analysis of Ceiba pentandra compounds with experimental support for colorectal cancer therapy
Menaga Suriyakanthan and
Geetha Natesan *
*
Department of Botany, Bharathiar University, Coimbatore, Tamil Nadu 641 046, India. E-mail: ngeethaptc@gmail.com
First published on 11th December 2025
Abstract
This study investigated the anticancer potential of Ceiba pentandra leaf extract against colorectal cancer (CRC) through integrated computational and experimental approaches. Twenty-nine phytochemicals were identified, of which ten compounds with high gastrointestinal absorption and drug-like properties were shortlisted. SwissTargetPrediction and network pharmacology analyses (GeneMANIA, STRING, Cytoscape) revealed 267 common targets between C. pentandra compounds and CRC-associated genes. Key hub genes included AKT1, TP53, and STAT3. Molecular docking using Schrödinger followed by MM-GBSA calculations and 100 ns molecular dynamics (MD) simulations identified kaempferol, linarin, and β-sitosterol-β-D-glucoside as the most active compounds. The 3CQU–kaempferol complex showed the greatest structural stability and favorable binding free energy, supporting strong ligand–protein interactions. KEGG and GO enrichment analyses indicated that these targets were predominantly involved in the PI3K-Akt and colorectal cancer signaling pathways. In vitro validation using the MTT assay demonstrated a dose-dependent cytotoxic effect of C. pentandra extract on HCT-116 colon cancer cells with an IC50 value of 36.9 µg mL−1, while showing good biocompatibility toward 3T3L1 fibroblast cells. Morphological changes consistent with apoptosis were observed in treated cells. Overall, C. pentandra exhibits potent, multi-target anticancer activity against colorectal cancer by modulating key oncogenic pathways. These findings integrate in silico predictions with in vitro validation, highlighting C. pentandra as a promising natural therapeutic candidate warranting further mechanistic and preclinical investigations.
Introduction
Cancer remains one of the most pressing global health challenges, with colorectal cancer (CRC) ranking among the top causes of cancer-related morbidity and mortality worldwide.1 Although conventional treatments such as chemotherapy and radiotherapy have improved survival rates, they are often associated with significant toxicity, resistance, and high financial burden. In light of these limitations, there is increasing interest in exploring natural products, particularly plant-derived compounds, as alternative or complementary anticancer therapies.2 Medicinal plants have historically been a rich source of bioactive molecules, and several plant-based compounds, such as paclitaxel and vincristine, have already been successfully translated into clinically approved anticancer drugs.3 Colorectal cancer (CRC) management primarily relies on a combination of surgery, chemotherapy, radiotherapy, and targeted or immunotherapeutic approaches, depending on disease stage and molecular characteristics. Surgical resection remains the cornerstone for localized tumors, often combined with adjuvant chemotherapy using agents such as 5-fluorouracil, oxaliplatin, and irinotecan to prevent recurrence and improve survival. Targeted therapies, including anti-EGFR (cetuximab, panitumumab) and anti-VEGF (bevacizumab) monoclonal antibodies, have further enhanced clinical outcomes in metastatic CRC.4 However, these treatments are frequently accompanied by severe adverse effects, including gastrointestinal toxicity, myelosuppression, and neuropathy, and their efficacy is often limited by the emergence of drug resistance and tumor heterogeneity. Moreover, the high cost and limited accessibility of these therapies in developing regions underscore the urgent need for safer, more affordable, and multitargeted therapeutic alternatives such as those derived from medicinal plants in order to improve global CRC management5C. pentandra, a tropical tree widely used in traditional medicine, has attracted scientific attention due to its diverse pharmacological properties, including antioxidant, anti-inflammatory, and anticancer activities.6 Recent studies have demonstrated that C. pentandra extracts exhibit significant antiproliferative and pro-apoptotic effects against various cancer cell lines, including breast, liver, and lung cancers, supporting its potential relevance in oncology.7 Moreover, emerging evidence highlights the modulation of oxidative stress and inflammatory pathways by C. pentandra phytoconstituents, suggesting a plausible mechanistic basis for its activity against colorectal cancer. To assess the drug potential of these phytochemicals, drug-likeness and ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) profiling using in silico tools such as SwissADME is essential. These predictive models help identify compounds with favourable pharmacokinetic properties and minimize the risk of adverse effects, thereby streamlining the drug discovery process.8 A network pharmacology approach offers a systems-level perspective on the multitarget nature of plant-based compounds, which is particularly relevant for complex diseases like cancer. Tools such as SwissTargetPrediction enable the identification of putative molecular targets, while protein–protein interaction (PPI) networks constructed using the STRING database can highlight key regulatory genes involved in cancer pathogenesis.9–11 To gain further mechanistic insight, KEGG pathway enrichment analysis is performed to identify the signalling pathways modulated by the target proteins. In the context of colorectal cancer, critical pathways such as PI3K-Akt, MAPK, p53 signalling, and apoptosis are often implicated.12
Molecular docking is a widely used computational technique in drug discovery and development that predicts the interaction between small molecules (ligands) and their biological targets, typically proteins or enzymes.13 The primary goal of docking studies is to estimate the binding affinity and orientation (pose) of a ligand within the active site of a target protein,14 providing insight into the molecular mechanisms underlying therapeutic activity. By simulating these interactions, molecular docking helps identify potential drug candidates, prioritize compounds for synthesis or testing, and understand structure–activity relationships.15 The process involves two key components: sampling, which explores possible conformations and positions of the ligand, and scoring, which evaluates the stability and strength of the ligand–receptor complex based on binding energies. Docking studies are especially valuable in the early stages of drug discovery as they are cost-effective, time-efficient, and capable of screening large compound libraries.16 In vitro cytotoxicity assays serve as a fundamental approach in cancer research, enabling the preliminary evaluation of the anticancer potential of various natural and synthetic compounds.17 The HCT-116 cell line, derived from human colorectal carcinoma, is widely used for studying colorectal cancer. Among various methods, the MTT assay is commonly employed to determine cell viability by measuring mitochondrial metabolic activity, offering a reliable estimate of cytotoxic effects.18,19 Utilizing HCT-116 cells for cytotoxicity screening of plant extracts provides valuable insights into their potential as natural anticancer agents and supports the scientific validation of traditional medicinal practices.20 Despite the traditional use of C. pentandra, its anticancer potential against colorectal cancer remains underexplored. Previous studies lack an integrated approach combining computational predictions with experimental validation. Specifically, there is limited evidence on its phytochemical interactions with key oncogenic targets such as AKT1, TP53, and STAT3. This highlights the need for a comprehensive study linking molecular docking, dynamics, and in vitro cytotoxicity to uncover its therapeutic potential.
While a few prior studies have investigated C. pentandra through basic docking or MTT assays, these efforts have been largely isolated, lacking an integrative systems-level interpretation or comparative evaluation across major CRC-related targets. Furthermore, no previous work has comprehensively combined network pharmacology, enrichment analysis, and molecular dynamics to delineate the multi-target anticancer mechanism of C. pentandra compounds. The present study uniquely advances this field by bridging computational predictions with experimental validation, thereby revealing how specific phytoconstituents modulate central oncogenic regulators, AKT1, TP53, and STAT3 within critical signalling cascades. This integrative approach not only defines new mechanistic insights into C. pentandra's action against CRC but also distinguishes this research as the first to establish a consolidated pharmacology-to-validation framework for this plant.
Materials and methods
Selection of phytocompounds
C. pentandra, a medicinal plant traditionally used in ethnomedicine, exhibits a broad spectrum of pharmacological activities, including anticancer, anti-inflammatory, antioxidant, and antimicrobial effects. Recent studies have highlighted its potential in oncology, particularly through modulation of pathways involved in cancer progression. To explore its therapeutic promise, the phytochemical constituents of C. pentandra were identified using multiple phytochemical and ethnobotanical databases, including IMPPAT (Indian Medicinal Plants, Phytochemistry and Therapeutics), KNAPSACK, and RePharm, facilitating comprehensive retrieval of 29 known bioactive compounds. These compounds also consist of compounds previously identified from our earlier study through GC-MS of C. pentandra leaf extracts were also incorporated to ensure experimental relevance and continuity between computational and in vitro analyses.21 These compounds were further evaluated for drug-likeness and pharmacokinetic properties using the SwissADME tool. Lipinski's rule of five was applied, allowing at most one violation, while Veber and Egan criteria were used to assess molecular flexibility, polar surface area (TPSA ≤ 140 Å2), and oral bioavailability. Additionally, compounds were screened for high gastrointestinal (GI) absorption and acceptable water solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S within −6 to 0).22 Based on these filters, 7 compounds failed the Lipinski criteria, 5 did not satisfy the Veber/Egan thresholds, and 7 exhibited either low GI absorption or poor solubility, resulting in the exclusion of 19 compounds in total. The remaining 10 phytocompounds showed favourable drug-like and ADMET properties. The potential protein targets for each selected compound were then predicted using the SwissTargetPrediction database, and these compounds were further analysed for their molecular interactions with colorectal cancer-associated proteins.23
S within −6 to 0).22 Based on these filters, 7 compounds failed the Lipinski criteria, 5 did not satisfy the Veber/Egan thresholds, and 7 exhibited either low GI absorption or poor solubility, resulting in the exclusion of 19 compounds in total. The remaining 10 phytocompounds showed favourable drug-like and ADMET properties. The potential protein targets for each selected compound were then predicted using the SwissTargetPrediction database, and these compounds were further analysed for their molecular interactions with colorectal cancer-associated proteins.23
Identification of colorectal cancer related targets
The keywords “colorectal cancer” were used to search three databases: GeneCards (https://www.genecards.org/), OMIM (https://omim.org/), and the GEO database (https://www.ncbi.nlm.nih.gov/geo/) to identify relevant target genes.24 From GEO, the microarray dataset GSE110224 was obtained, based on the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array platform, which includes 17 colorectal cancer (CRC) and 17 matched normal tissue samples.25 Differentially expressed genes (DEGs) between CRC and normal samples were identified using the LIMMA package with a paired t-test, and genes with adjusted P < 0.05 and |log2 fold-change| ≥ 1 were considered significant. Based on adjusted P values, the top 10 upregulated and top 10 downregulated genes were selected for further analysis. In total, 2580 genes from GeneCards, 27 from OMIM, and 20 from GEO were collected, and all genes were merged and labelled as colorectal cancer-associated genes, resulting in 2672 genes for subsequent analyses.Construction and analysis of the protein–protein interactions (PPI) network
To identify overlapping genes between the compound target dataset and colorectal cancer-associated genes, a Venn diagram was generated. The intersecting gene set was then subjected to a protein–protein interaction (PPI) analysis using the STRING database version 12.0 (https://string-db.org/).26 The analysis was conducted under the following parameters: organism set to Homo sapiens and a medium confidence score threshold of ≥0.4, which is commonly used for moderately reliable interaction predictions. The resulting PPI network was exported and visualized using Cytoscape software (version 3.10.3). Topological parameters, including degree centrality, betweenness centrality (BC), and closeness centrality (CC), were computed to assess the influence and connectivity of each node within the network.27 These metrics help identify key regulatory proteins by evaluating how centrally or strategically they are positioned within the network.Hub genes were identified by applying the CytoHubba plugin (version 0.1), which ranks nodes based on maximum degree values, indicating their potential as critical network regulators. Additionally, Molecular Complex Detection (MCODE) (version 2.0.2) analysis was performed to identify densely connected modules within the network, representing potential functional clusters or biological complexes.28
Gene–gene interactions
To investigate potential interactions among target genes, gene–gene interaction networks were constructed using the GeneMANIA prediction tool (https://genemania.org/).29 This online bioinformatics platform integrates multiple genomic and proteomic datasets to predict functional relationships based on co-expression, physical interactions, shared protein domains, co-localization, and genetic interactions.KEGG pathway and GO gene enrichment analysis
This study utilized multiple biological model databases to explore the roles of cellular components (CC), molecular functions (MF), and biological processes (BP) within large-scale gene datasets. Gene Ontology (GO) enrichment analysis, performed using the STRING 12.0 platform, provided in-depth understanding of the biological roles, pathways, and subcellular localizations of enriched genes.30 In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was employed to systematically examine gene functions and annotate relevant signaling pathways. By integrating GO and KEGG enrichment analyses, the research offered a broad functional perspective on common target genes, enabling the identification of critical drug–disease interaction pathways. Genes filtered through STRING were subjected to both GO and KEGG analyses, producing detailed insights into CC, MF, BP categories, and associated pathways. The top 10 GO terms and top 20 KEGG pathways-ranked by statistical significance (FDR) were selected for visualization using bar and bubble charts via the bioinformatics visualization platform (https://www.bioinformatics.com.cn/). Additionally, the interactive network of active compounds, corresponding targets, and involved pathways was constructed and illustrated using Cytoscape software (version 3.8).In silico docking of compounds against colorectal cancer proteins
Ligand structure preparation
Ligand structure preparation using LigPrep involves a comprehensive toolset tailored to generate accurate, all atom three dimensional (3D) models of drug-like compounds. It processes both two-dimensional (2D) and 3D molecular inputs in formats such as SDF or Maestro to produce geometrically refined ligand structures suitable for computational analyses.Receptor grid generation
Receptor grid generation is a crucial preliminary step in molecular docking, carried out using the receptor grid generation module. Docking simulations cannot proceed without completing this step. A properly prepared protein structure-with accurate bond orders and formal charges is required for grid construction. The process involves navigating through four key configuration tabs to define and generate the docking grid around the active site. The grid box coordinates were obtained from PrankWeb (https://prankweb.cz/) and subsequently validated before being used for docking simulations. The grid box was centered on the top-ranked predicted binding pocket, with dimensions of 20 Å in all directions around the site center.32Ligand docking
In the next step, the grids generated were used to dock the ligands prepared from LigPrep performed. Ligands were docked in extra precision mode for better results. The resulting docking score was analyzed, and hydrogen bonding was also visualized using 2D and 3D diagrams.Molecular dynamics (MD) simulation analysis
The Optimized Potentials for Liquid Simulations 2005 (OPLS_2005) force field was used for all simulations. Prior to production runs, the systems were energy-minimized using Desmond's built-in relaxation protocol (convergence threshold: 1 kcal mol−1; 2000 iterations), followed by pre-equilibration under NVT and NPT ensembles. During equilibration, positional restraints were applied to heavy atoms, and constraints on all bond lengths involving hydrogen atoms were maintained using the M-SHAKE algorithm.33Simulations were performed in Desmond (Schrödinger 2023-1) using the OPLS_2005 force field and TIP3P water model within an orthorhombic box (10 Å buffer). Systems were neutralized with Na+/Cl− and adjusted to 0.15 M ionic strength. Energy minimization and NVT/NPT equilibration preceded 100 ns production runs (310 K, 1 bar, 2 fs time step) with Nosé–Hoover thermostat and Martyna–Tobias–Klein barostat. Post-simulation analyses included evaluation of the root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), solvent-accessible surface area (SASA), and intermolecular hydrogen bond profiles to assess structural stability, compactness, and interaction persistence of the protein–ligand complexes throughout the trajectories.
The trajectories for RMSD and RMSF of both protein and ligand were computed using standard mathematical expressions, which quantify atomic displacement relative to a reference structure over time.
where, T represents the total duration of the trajectory over which the RMSF is calculated, while tref is the reference time point (time t = 0). Here, r corresponds to the position of an atom at the reference time tref, and r′ is the position of the atom at time t after alignment to the reference frame.
Prime MM-GBSA analysis
The most stable docking poses, based on docking scores and visual inspection of key interactions, were selected for subsequent analysis. The binding free energies (ΔG-bind) of the docked ligand–protein complexes were estimated using the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) approach implemented in the Prime module of Schrödinger Maestro (Schrödinger Release 2023-4). The MM/GBSA method provides an approximate free energy of binding by combining molecular mechanics energies with solvation and surface area contributions.34The total binding free energy was calculated using the following equation:
| ΔGbind = Gcomplex − (Gprotein + Gligand) |
| G = EMM + Gsolv + GSA |
The Prime MMGBSA calculation was executed using the default OPLS4 force field, with the VSGB 2.0 solvation model applied for implicit solvent representation. The protein–ligand complexes obtained from docking studies were minimized before energy evaluation. Both solvated (ΔG_bind) and no-solvent (ΔG_bind(NS)) binding energies were computed to assess the influence of solvation effects on complex stability. The coulombic and van der Waals energy components were extracted to analyze the electrostatic and hydrophobic contributions to binding.35
Plant collection and extraction
Plant materials were collected from the Coimbatore district and authenticated by the Botanical Survey of India, Coimbatore (BSI/SRC/5/23/2024-25/Tech./514). The leaves were thoroughly cleaned, shade-dried for six weeks, and then powdered using a mixer grinder. A total of 50 g of the powdered leaf material was subjected to sequential Soxhlet extraction using five solvents of differing polarity in a Soxhlet apparatus. Finally, methanolic leaf extract was chosen and the extracts obtained were concentrated to dryness under reduced pressure using a rotary evaporator and, for cell-based assays, the extracts were dissolved in dimethyl sulfoxide (DMSO) to prepare stock solutions, ensuring that the final DMSO concentration in the culture medium did not exceed 0.5%, and vehicle controls were included.Cell line propagation and treatment
HCT-116, a human colorectal carcinoma cell line, and 3T3L1, a normal fibroblast cell line, were procured from the National Centre for Cell Science (NCCS), Pune, India, and cultured in T25 flasks containing Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% antibiotics (100 µg per mL penicillin and 100 µg per mL streptomycin) at 37 °C in a humidified incubator with 5% CO2.36 Once the cells reached confluency, they were detached using trypsin and sub-cultured for experimental use, and following treatment, cell viability was determined and expressed as a percentage relative to the untreated control.Extract preparation for anti-cancer activity
The antiproliferative effect of C. pentandra leaf extract was assessed on HCT-116 and 3T3L1 fibroblast cells using doxorubicin (1.5 µM) as a positive control. Methanol Soxhlet extracts of C. pentandra leaves were used for all cell-based assays. Extraction procedures and phytochemical quantification (including yield, compound profiling, and characterization) followed the previously published protocol by the same author, ensuring reproducibility and methodological transparency. All experiments included vehicle controls, using DMSO at a final concentration not exceeding 0.1% (v/v). Cells were cultured in T25 flasks using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of DMEM and Leibovitz's L-15 medium, supplemented with 10% FBS and 1% antibiotics (100 U per mL penicillin, 100 µg per mL streptomycin), and maintained at 37 °C with 5% CO2. Upon reaching confluency, cells were trypsinized and seeded into 96 well plates at a density of 5000 cells per well. Once 50% confluent, cells were treated with C. pentandra extract at concentrations of 50–300 µg mL−1 for HCT-116 cells and 50–500 µg mL−1 for 3T3L1 fibroblast cells for 24 hours. Cell viability was determined using the MTT assay, with absorbance measured at 450 nm using a microplate reader (EnSpire, PerkinElmer, USA).
1 mixture of DMEM and Leibovitz's L-15 medium, supplemented with 10% FBS and 1% antibiotics (100 U per mL penicillin, 100 µg per mL streptomycin), and maintained at 37 °C with 5% CO2. Upon reaching confluency, cells were trypsinized and seeded into 96 well plates at a density of 5000 cells per well. Once 50% confluent, cells were treated with C. pentandra extract at concentrations of 50–300 µg mL−1 for HCT-116 cells and 50–500 µg mL−1 for 3T3L1 fibroblast cells for 24 hours. Cell viability was determined using the MTT assay, with absorbance measured at 450 nm using a microplate reader (EnSpire, PerkinElmer, USA).
Assessment of cell proliferation
The antiproliferative activity of C. pentandra leaf extract on HCT-116 and 3T3L1 fibroblast cells was evaluated using the MTT assay. Cells were seeded in 96 well plates (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) and incubated at 37 °C with 5% CO2. At 60–70% confluency, cells were treated with C. pentandra extract. After 24 hours, MTT solution (2 mg mL−1) was added and incubated for 3 hours. Formazan crystals were solubilized in 200 µL DMSO, and absorbance was measured at 570 nm. Reduced absorbance indicated decreased viability. Data are presented as mean ± SD from triplicate experiments.37
000 cells per well) and incubated at 37 °C with 5% CO2. At 60–70% confluency, cells were treated with C. pentandra extract. After 24 hours, MTT solution (2 mg mL−1) was added and incubated for 3 hours. Formazan crystals were solubilized in 200 µL DMSO, and absorbance was measured at 570 nm. Reduced absorbance indicated decreased viability. Data are presented as mean ± SD from triplicate experiments.37
Statistical analysis
Statistical analysis was performed using GraphPad Prism software. Data were expressed as mean ± standard deviation (SD) of three independent experiments. One-way analysis of variance (ANOVA) was conducted to assess significant differences among groups, followed by Tukey's post hoc test for multiple comparisons.Results
Analysis of bioactive compounds present in the sample
In this study, 29 bioactive compounds of C. pentandra were retrieved from various databases. From these, 10 phytocompounds exhibiting high gastrointestinal (GI) absorption and favourable drug-like characteristics were shortlisted using the SwissADME tool. Furthermore, the potential targets for each selected compound were identified using the SwissTargetPrediction database. A probability cutoff of ≥0.1 was applied to retain reliable target predictions, with the species restricted to Homo sapiens. After all filtering steps, a total of 519 ligand target genes were selected for subsequent analysis. The details of these 10 compounds are presented in Table 1.| S. no. | Compound name | GI absorption | H-bond donor | H-bond acceptor | Molecular weight | TPSA | Rotatable bond |
|---|---|---|---|---|---|---|---|
| 1 | (+)-Catechin | High | 5 | 6 | 290.27 | 74.33 | 1 |
| 2 | (−)-Beta-sitosterol | Low | 1 | 1 | 414.71 | 133.2 | 6 |
| 3 | Vavain | High | 2 | 7 | 344.32 | 91.44 | 4 |
| 4 | D-Glucuronic acid | Low | 5 | 7 | 194.14 | 36.35 | 1 |
| 5 | D-Galactose | Low | 5 | 6 | 180.16 | 35.74 | 1 |
| 6 | 2,7-Dimethoxy-5-isopropyl-3-methyl-8,1-naphthalene carbolactone | High | 0 | 4 | 286.32 | 81.44 | 3 |
| 7 | Kaempferol | High | 4 | 6 | 286.24 | 76.01 | 1 |
| 8 | Linarin | Low | 7 | 14 | 592.55 | 141.8 | 7 |
| 9 | Beta-sitosterol-beta-D-glucoside | Low | 4 | 6 | 576.85 | 165.6 | 9 |
| 10 | Caffeic acid | High | 3 | 4 | 180.16 | 47.16 | 2 |
Therapeutic targets of C. pentandra bioactive compounds for colorectal cancer treatment
To identify genes associated with colorectal cancer, the keyword “Colorectal Cancer” was used to search multiple databases. A total of 2672 disease-related genes were collected, of which 2580 were retrieved from GeneCards, representing literature and database supported targets, 27 were from OMIM and 20 were obtained from the GEO database, reflecting experimentally validated genes. These disease-associated genes were then compared with 519 ligand target genes, resulting in 267 overlapping genes. These common genes, representing potential therapeutic targets supported by both disease relevance and ligand targeting, were selected for further analyses, as illustrated in Fig. 1.Gene–gene interactions
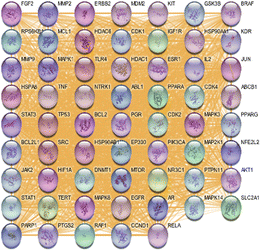
A Gene MANIA-derived interaction network, dominated by physical (49.34%) and genetic (22.45%) interactions, revealed TP53, AKT1, EGFR, STAT3, and SRC as central hub genes interconnected through diverse functional associations, highlighting their key roles in apoptosis, cell signalling, and oncogenic regulation. Fig. 2.PPI network visualization and analysis
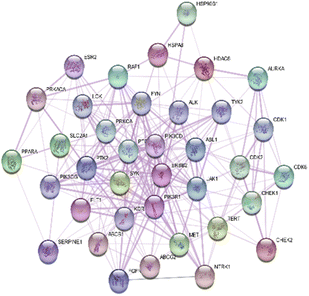
To elucidate the molecular interactions among the 267 predicted therapeutic targets of C. pentandra, a protein–protein interaction (PPI) network was constructed using the STRING database and visualized in Cytoscape version 3.10.3. The resulting network comprised 267 nodes and 5528 edges, with an average node degree of 45.853, indicating extensive interconnectivity among the targets (Fig. 3).Topological analysis revealed AKT1, TP53, STAT3, TNF, EGFR, BCL2, HSP90AA1, ESR1, JUN, and MAPK3 as the top 10 hub genes based on degree centrality (Table 2). Further assessment of betweenness and closeness centralities confirmed the prominent regulatory positions of AKT1, TP53, STAT3, TNF, and EGFR, all of which are crucial mediators in colorectal cancer (CRC) progression. These findings suggest that C. pentandra may exert therapeutic effects by targeting multiple central nodes within cancer-associated signalling pathways. Functional module analysis using the MCODE plugin identified four major clusters within the PPI network (Table 3).
| Target name | Degree | Closeness centrality | Betweenness centrality | ASPL |
|---|---|---|---|---|
| AKT1 | 171 | 0.76 | 0.06 | 1.31 |
| TP53 | 170 | 0.74 | 0.04 | 1.33 |
| TNF | 169 | 0.74 | 0.53 | 1.34 |
| EGFR | 160 | 0.72 | 0.03 | 1.37 |
| STAT3 | 151 | 0.70 | 0.27 | 1.41 |
| BCL2 | 144 | 0.69 | 0.03 | 1.43 |
| HSP90AA1 | 143 | 0.69 | 0.02 | 1.44 |
| ESR1 | 142 | 0.69 | 0.04 | 1.46 |
| JUN | 138 | 0.68 | 0.02 | 1.49 |
| MAPK3 | 122 | 0.65 | 0.01 | 1.52 |
Cluster 1 (score = 42.0), the most significant, contained key proteins such as AKT1, TP53, STAT3, EGFR, TNF, and MTOR, predominantly enriched in PI3K/AKT/mTOR, MAPK, and NF-κB signaling pathways. These are central to cell survival, apoptosis, and proliferation, suggesting that C. pentandra compounds may simultaneously modulate multiple oncogenic cascades. Cluster 2, with a score of 12.5 comprised regulators of cell cycle, JAK/STAT, and VEGF signaling (e.g., JAK1, TYK2, CDK1, RAF1), indicating a role in inhibiting tumor growth and angiogenesis. Cluster 3 (score = 5.3) included MMP1, MMP9, TGFBR1, ROCK1, and NOS2, associated with extracellular matrix remodelling and metastasis, implying potential anti-invasive effects of C. pentandra constituents. Cluster 4 (score = 5.2) was enriched with CYP family members and prostaglandin-related enzymes, suggesting modulation of lipid metabolism and inflammatory mediators relevant to tumor microenvironment regulation. These results indicate that C. pentandra compounds act on multiple functional modules and hub proteins involved in key oncogenic pathways, providing a mechanistic basis for their potential therapeutic role in colorectal cancer.
KEGG enrichment analysis and GO ontology
KEGG pathway enrichment analysis identified 127 signaling pathways, with the top 20 (Fig. 4A) prominently associated with pathways in cancer, colorectal cancer, PI3K-Akt signaling, ErbB signaling, estrogen signaling, and IL-17 signaling. Key targets such as PIK3CA, TP53, BCL2, CDK4, STAT3, AR, and ESR1 were enriched in these networks, indicating that C. pentandra bioactives may influence multiple oncogenic and survival pathways. Mechanistically, the modulation of the PI3K-Akt and MAPK pathways may restore apoptotic signaling and inhibit abnormal cell proliferation, while regulation of IL-17 and NF-κB pathways could suppress the pro-inflammatory microenvironment that drives colorectal cancer progression. The involvement of estrogen and ERBB signalling further suggests potential interference with hormone receptor-mediated pathways, highlighting the multi-pathway synergistic action of C. pentandra phytochemicals. This KEGG pathway map titled Pathways in Cancer illustrates the complex network of signalling pathways and molecular interactions that drive cancer development and progression. The red-highlighted genes and proteins such as PIK3CA, TP53, BCL2, CDK4, STAT3, and hormone receptors like AR and ESR1 represent key molecules that are upregulated in the dataset being analyzed. These genes play critical roles in regulating cell proliferation, inhibiting apoptosis, promoting cell cycle progression, enabling genomic instability, and modulating hormone responses. The KEGG pathway analysis (Fig. 4B) revealed that the identified target genes regulated by C. pentandra compounds are predominantly involved in key cancer-associated pathways governing apoptosis, cell proliferation, and survival signalling.Gene Ontology (GO) analysis revealed significant enrichment in biological processes such as positive regulation of gene expression, suppression of apoptosis, and response to xenobiotic stimuli; cellular components like the nucleus and cytosol; and molecular functions including protein binding, enzyme regulation, and ubiquitin ligase activity (Fig. 4C). These results indicate that C. pentandra compounds may modulate key intracellular regulatory systems affecting protein stability, transcriptional control, and stress responses. Collectively, the KEGG and GO findings suggest that C. pentandra exerts anticancer effects through coordinated modulation of cell survival, inflammatory, apoptotic, and hormonal signaling networks, providing a mechanistic basis for its therapeutic potential against colorectal cancer.
Relationship between active compounds-potential therapeutic target, and critical pathways involved
The interaction between bioactive compounds, target genes/proteins, and KEGG pathways relevant to cancer are shown in Fig. 5. The yellow nodes are phytochemicals (e.g., catechin, kaempferol, beta-sitosterol), the green ellipses are their associated protein targets, and the pink triangles indicate the KEGG pathways (e.g., hsa05200: pathways in cancer, hsa04151: PI3K-Akt signalling pathway). The numerous connecting edges show how each compound influences multiple targets, and how these targets are involved in multiple cancer-related pathways. This indicates that the phytochemicals may exert their anticancer effects through multi-target, multi-pathway mechanisms. Among them the most interactive compounds are vavain, linarin, beta-sitosterol-beta-D-glucoside, caffeic acid beta-sitosterol, and kaempferol with a score of 100 and the most interactive genes are AKT1, MAPK3, Tp53, and MTOR with a score of 21. These compounds are further chosen for molecular docking using Schrodinger software.Anti-COAD hub gene expression and prognosis of colorectal cancer patients
The study also aimed to determine whether the hub targets of C. pentandra, which act on colorectal cancer (CRC), could serve as potential molecular markers for predicting CRC prognosis. To achieve this, we employed Kaplan–Meier survival analysis to evaluate the association between the expression levels of ten hub target genes and the overall survival of colorectal cancer patients Fig. 6. According to the investigation, patients with high expression of TP53 and MMP9 showed better overall survival (OS) than those with low expression of these genes (p < 0.05). In contrast, patients with low expression of AKT1, TNF, and EGFR had significantly better OS compared to those with high expression levels (p < 0.01). Meanwhile, the expression levels of STAT3, HSP90AA1, HIF1A, and MAPK3 did not show any statistically significant effect on the overall survival of colorectal cancer patients (p > 0.05). | ||
| Fig. 6 KM plotter analysis of the relationship between hub target gene expression and colorectal cancer patient survival. | ||
Molecular docking
The top genes were selected, and key compounds such as linarin, beta-sitosterol, kaempferol, caffeic acid, vavain, beta-sitosterol-beta-D-glucoside, and (+)-catechin were identified from the compound–ligand target interaction network. The crystal structure 1XQH represents the TP53 macromolecule co crystallized with SET7/9 lysine methyltransferase and the cofactor S-adenosyl-L-homocysteine (SAH). The structure was resolved at 1.75 Å and contains chains A, B, E, and F. It corresponds to the wild-type enzyme, with no engineered mutations reported. Several residues and loop regions are missing, particularly in flexible surface-exposed regions as indicated in the PDB entry.The 6TLC structure corresponds to human STAT3 in complex with a designed monobody inhibitor (MS3-6). It was determined at a resolution of 2.90 Å, containing multiple chains (B–D) representing STAT3 dimers and monobody units. The structure represents unphosphorylated wild-type STAT3, with no small-molecule cofactors. Minor missing residues are reported in surface-exposed loop regions and N-/C-terminal ends, which are typical for flexible domains not resolved in electron density.
The 3CQU structure represents the active form of human AKT1 kinase domain in complex with a substrate peptide and a spiroindoline inhibitor. The structure includes chain A, covering residues 144–480 of AKT1, and was determined at 2.70 Å resolution. The protein carries a S473D mutation to mimic phosphorylation and contains phosphorylated Thr308 (pT308), corresponding to its active conformation. The N-terminal regulatory region (residues 1–143) and a few surface loops are missing from the model due to lack of interpretable density.
The analysis demonstrated the strongest binding affinities between the selected ligands and target proteins: kaempferol with AKT1 (binding affinity: −7.307), linarin with TP53 (binding affinity: −5.177), and beta-sitosterol-beta-D-glucoside with STAT3 (binding affinity: −5.324), as illustrated in Fig. 7, and their 2D and 3D structures are shown in Fig. 8A–C.
 | ||
| Fig. 7 Clustering heatmap of binding energies (kcal mol−1) from molecular docking. The darker the color, the higher the free energy for the phytocompounds to bind to the hub targets. | ||
 | ||
| Fig. 8 (A–C) Shows the binding affinity and their 2D and 3D images of 1XQH with linarin, 3CQU with kaempferol and 6TLC with beta-sitosterol-beta-D-glucoside. | ||
Molecular dynamics simulation studies
Three distinct cancer-related receptors such as 1XQH, 3CQU, and 6TLC, were selected for molecular dynamics simulation studies. Each receptor was paired with its top-performing ligand based on docking scores: linarin for 1XQH, kaempferol for 3CQU, and beta-sitosterol-β-D-glucoside for 6TLC.Root mean square deviation (RMSD) plot over a 100 nanosecond molecular dynamics (MD) simulation
The root-mean-square deviation (RMSD) profiles over a 100 ns molecular dynamics simulation provided valuable insights into the conformational stability of the protein–ligand complexes. The 1XQU with linarin complex (Fig. 9A) displayed a gradual structural deviation, with the protein backbone RMSD increasing from approximately 2.5 Å to 5.6 Å, indicating moderate conformational flexibility. Meanwhile, the ligand RMSD rose significantly from ∼1.0 Å to over 12.0 Å, peaking around 13.5 Å, suggesting notable ligand repositioning within the binding pocket. In contrast, the 3CQU–kaempferol complex (Fig. 9B) demonstrated higher structural stability, with protein RMSD maintained between 1.4 and 2.8 Å and the ligand RMSD remaining stable around 2.5–3.0 Å, apart from minor fluctuations near 50 ns, indicating a well-maintained binding conformation. However, the 6TLC–beta-sitosterol-β-D-glucoside complex (Fig. 9C) exhibited substantial fluctuations, with the protein RMSD increasing from ∼2.0 Å to 6.5 Å and the ligand RMSD peaking at 17 Å, reflecting pronounced ligand movement and potential binding instability. Overall, the 3CQU–kaempferol complex showed the greatest stability among the three, while the other complexes displayed higher deviations, indicating more dynamic interactions. | ||
| Fig. 9 (A–C) Shows the RMSD plot of the receptor (A) 1XQU with linarin (B) 3CQU with kaempferol and (C) 6TLC–beta-sitosterol-β-D-glucoside complex. | ||
Root mean square fluctuation (RMSF)
The RMSF analysis reveals distinct flexibility profiles across the three protein–ligand complexes. For the 1XQU-Linarin complex (Fig. 10A), moderate fluctuations are observed overall, with pronounced peaks reaching approximately 8.0 Å around residues 240–260 and 470–490, indicating highly flexible loop regions, while the majority of residues remain within a stable range of 1.0–3.0 Å. In the case of the 3CQU–kaempferol complex (Fig. 10B), residue flexibility is predominantly low, ranging from ∼0.5 to 1.5 Å, with isolated peaks near residues 50, 200, and 300 showing fluctuations up to 4.3 Å, suggesting limited localized flexibility within an otherwise rigid structure. The 6TLC–beta-sitosterol-β-D-glucoside complex (Fig. 10C) displays a wider range of fluctuations, with multiple peaks occurring between residues 500 and 1200, particularly around residues 600, 800, and 1000. Where RMSF values exceed 6.0 Å, indicating substantial structural flexibility; in contrast, the region spanning residues 100–400 exhibits relatively stable behaviour, with fluctuations ranging from 1.0 to 2.5 Å. | ||
| Fig. 10 (A–C) Shows the RMSF plot of the receptor (A) 1XQU with linarin (B) 3CQU with kaempferol and (C) 6TLC–beta-sitosterol-β-D-glucoside complex. | ||
Molecular docking interaction profiles of selected ligands with target protein residues
The interaction profiles of the ligand–protein complexes reveal that linarin, when bound to 1XQU, forms strong hydrogen bonds with HIS297 (57%), ILE223, and TRP352 (61%), with a notable interaction between the carbonyl oxygen and a hydroxyl group (96%), indicating substantial binding stability (Fig. 11A). Kaempferol in complex with 3CQU exhibits prominent interactions with PHE161, ASP292 (73%), LEU156 (47%), GLU228, and ALA230 (69%), along with water-mediated hydrogen bonds, suggesting a robust binding affinity (Fig. 11B). In contrast, the 6TLC–beta-sitosterol-β-D-glucoside complex shows a single dominant interaction with TRP510, reflecting a relatively weaker but specific binding, primarily driven by hydrophobic interactions (Fig. 11C). | ||
| Fig. 11 (A–C) Shows the interaction profiles of the ligand–protein complexes (A) 1XQU with linarin (B) 3CQU with kaempferol and (C) 6TLC–beta-sitosterol-β-D-glucoside complex. | ||
Protein–ligand interaction analysis of selected cancer receptors
The protein–ligand interaction analysis revealed that 1XQU–linarin and 3CQU–kaempferol complexes exhibited extensive hydrogen bonding and water bridge interactions, with TRP351 in 1XQU and ASP292 in 3CQU contributing significantly to ligand binding through high interaction fractions Fig. 12A and B. Notably, 3CQU–kaempferol also displayed strong hydrophobic and ionic interactions at residues such as GLU234 and TYR229, suggesting a highly stable and diverse binding mode. In contrast, the 6TLC–beta-sitosterol-β-D-glucoside complex demonstrated fewer and weaker interactions, predominantly water bridges and hydrophobic contacts, with a prominent but isolated hydrophobic peak at TRP510 Fig. 12C. These findings indicate that linarin and kaempferol form more stable and functionally relevant interactions with their respective receptors compared to beta-sitosterol-β-D-glucoside, supporting their potential as promising anticancer agents. | ||
| Fig. 12 (A–C) Shows the interaction profiles of the protein–ligand contacts of (A) 1XQU with linarin (B) 3CQU with kaempferol and (C) 6TLC–beta-sitosterol-β-D-glucoside complex. | ||
Prime MMGBSA calculations
Prime MMGBSA calculations were performed to evaluate the binding affinities of the docked complexes. Among the compounds, 3CQH exhibited the most favorable total binding free energy (ΔG_bind = −129.47 kcal mol−1), indicating the strongest interaction with the target protein. 1XQH showed moderate binding (−63.56 kcal mol−1), while 6TLC had the weakest affinity (−25.17 kcal mol−1). The coulombic energy supported these results, with 3CQH (−64.64 kcal mol−1) showing strong electrostatic stabilization, compared to 1XQH (−15.09 kcal mol−1) and 6TLC (−14.87 kcal mol−1). Similarly, no-solvent (NS) binding energies followed the same trend, where 3CQH (−172.28 kcal mol−1) demonstrated superior stability relative to 1XQH (−67.33 kcal mol−1) and 6TLC (−35.22 kcal mol−1).Overall, 3CQH formed the most stable and energetically favorable complex, mainly due to strong electrostatic and van der Waals interactions, while 6TLC showed the least stable binding among the tested ligands (Table 4).
| Compound | ΔG_bind (kcal mol−1) | ΔG_Coulomb (kcal mol−1) | ΔG_bind(NS) (kcal mol−1) | ΔG(NS)_Coulomb (kcal mol−1) |
|---|---|---|---|---|
| 3CQH | −129.47 | −64.64 | −172.28 | −80.53 |
| 1XQH | −63.56 | −15.09 | −67.33 | −21.04 |
| 6TLC | −25.17 | −14.87 | −35.22 | −14.07 |
Effects of C. pentandra leaf extract and doxorubicin on the cytotoxicity of HCT-116 cell line
The cytotoxic activity of the leaf extract of C. pentandra against the HCT-116 colorectal cancer cell line was evaluated using the MTT assay. Cells were treated with increasing concentrations of the extract (50–300 µg mL−1) for 24 hours, and the percentage of cell inhibition was calculated. The results revealed a clear dose-dependent increase in cytotoxicity, where the extract showed approximately 49% inhibition at 50 µg mL−1, 60% at 100 µg mL−1, 75% at 150 µg mL−1, 79% at 200 µg mL−1, 87% at 250 µg mL−1, and reached 93% inhibition at 300 µg mL−1. The calculated IC50 value of the extract was 36.9 µg mL−1, indicating potent anticancer activity at higher concentrations.For comparison, the standard drug doxorubicin (positive control) also exhibited a concentration-dependent cytotoxic effect, with percentage inhibitions of 18.09%, 38.39%, 57.89%, 71.09%, and 80.07% at concentrations 1–5 µg mL−1, respectively. These findings suggest that while doxorubicin demonstrated strong cytotoxic potential as expected, the C. pentandra leaf extract also exerted a significant inhibitory effect on HCT-116 cells, supporting its potential as a natural anticancer agent Fig. 13A–C.
Effects of C. pentandra leaf extract on the viability of 3T3L1 fibroblast cells
The cytotoxic effects of the plant extract were evaluated on 3T3-L1 fibroblast cells using a cytotoxicity assay. Cells were treated with increasing concentrations of the plant extract (0–500 µg mL−1), and cell viability was assessed after the treatment period. The findings revealed a dose-dependent reduction in cell viability shows in Fig. 14A and B.At the lowest concentration (50 µg mL−1), cell survival was slightly reduced to 94.54% compared to the control (100%). As the concentration increased, a gradual decline in viability was observed, with 84.05% at 100 µg mL−1, 79.70% at 200 µg mL−1, and 79.19% at 300 µg mL−1. Notably, a more pronounced cytotoxic effect was evident at higher concentrations, with viability decreased to 59.78% at 400 µg mL−1 and 44.79% at 500 µg mL−1. These findings suggest that C. pentandra leaf extract exhibits dose-dependent cytotoxicity in 3T3-L1 cells, with significant reductions observed at concentrations ≥ 400 µg mL−1. The IC50 value of the cells are 446.3 µg mL−1.
Discussion
Twenty-nine bioactive compounds were initially retrieved from multiple databases, of which ten were shortlisted based on likable pharmacokinetic profiles, including high gastrointestinal absorption and compliance with drug-likeness filters assessed using SwissADME. These parameters including Lipinski's rule of five, bioavailability, and GI permeability support the selection of compounds with potential for oral systemic activity. The gene–gene interaction network generated through GeneMANIA revealed a highly interconnected module dominated by physical (49.34%) and genetic (22.45%) interactions, underscoring the biological complexity of the identified targets. Hub genes such as TP53, AKT1, EGFR, STAT3, and SRC emerged as key regulators of apoptosis, cell proliferation, and oncogenic signaling. The centrality of TP53 (a tumor suppressor) and AKT1/EGFR (master regulators of the PI3K/AKT and EGFR/ERK pathways) aligns with their well-documented roles in colorectal cancer (CRC) progression and therapeutic resistance.38 Protein–protein interaction (PPI) analysis further identified AKT1, TP53, STAT3, TNF, and EGFR as major hub genes based on high degree, betweenness, and closeness centrality values. These genes collectively influence cellular survival, DNA repair, and apoptotic regulation core mechanisms implicated in CRC.39 KEGG pathway enrichment highlighted their involvement in cancer-associated signaling, including PI3K-Akt, ErbB, estrogen, and IL-17 pathways. Rather than implying redundancy, the recurrence of these pathways emphasizes their convergence on cell survival and inflammatory mechanisms, which may synergistically or competitively influence therapeutic outcomesS.40 Potential off-target or toxicity effects arising from multi-pathway interactions should also be considered in future studies. Phytochemicals such as linarin, β-sitosterol, and kaempferol demonstrated multitarget interactions with these hub genes, suggesting a polypharmacological mode of action. However, molecular docking and dynamics simulations are hypothesis-generating tools and not definitive proof of efficacy. The observed stable binding affinities, such as kaempferol with AKT1 (−7.307 kcal mol−1), linarin with TP53 (−5.177 kcal mol−1), and β-sitosterol-β-D-glucoside with STAT3 (−5.324 kcal mol−1) suggest plausible biochemical interactions that warrant validation through target-engagement assays. Future experimental validation will involve western blot analyses of p-AKT and p-ERK to confirm pathway modulation, along with TP53 functional assays such as p53 reporter or phosphorylation assays to substantiate the computational predictions. In addition, qRT-PCR will be conducted to assess the expression of key target genes (AKT1, TP53, BCL2, and CASP3), while flow cytometry and immunofluorescence analyses will be employed to evaluate apoptosis, cell cycle arrest, and AKT phosphorylation, thereby providing comprehensive biochemical confirmation of the in silico findings. Similar approaches have been reported that β-sitosterol suppressed the PI3K/AKT and MAPK signaling pathways through decreased p-AKT and p-ERK expression in HT-29 colon cancer cells.41Kaplan–Meier survival analysis indicated that elevated expression of TP53 and MMP9 correlates with improved CRC patient survival, while high levels of AKT1, TNF, and EGFR associate with poorer prognosis. These results should be interpreted cautiously, as TP53 mutation status and expression level dynamics can produce variable clinical outcomes. Therefore, further integration of mutation data with expression-based survival analysis is recommended for more accurate prognostic insights. The cytotoxic potential of C. pentandra leaf extract against HCT-116 colorectal cancer cells revealed a dose-dependent reduction in cell viability, with an IC50 of 35.77 µg mL−1 after 24 hours. This activity is comparable to reports from other medicinal plants such as Annona muricata (IC50 = 11.43 ± 1.87 µg mL−1) and Eclipta alba (IC50 = 179 ± 0.81 µg mL−1), highlighting the moderate but significant cytotoxicity of C. pentandra.42,43 The induction of apoptosis-like morphological features at IC50 suggests possible engagement of intrinsic apoptotic pathways. Overall, these findings support C. pentandra as a promising candidate for anticancer exploration through multi-target modulation. Nevertheless, the in silico and network-based results should be considered as preliminary hypotheses requiring confirmation via targeted biochemical assays, such as phosphorylation analysis of AKT/ERK, TP53 activation studies, and validation in normal versus cancer cell lines to substantiate the predicted molecular mechanisms and assess potential off-target effects.
Conclusion
This study highlights the anticancer potential of C. pentandra against colorectal cancer using a combination of computational, network pharmacology, and in vitro approaches. Out of 29 identified phytocompounds, 10 showed favourable ADME profiles, with kaempferol, linarin, and β-sitosterol-β-D-glucoside showing strong interactions with key cancer targets like AKT1, TP53, and STAT3. Molecular docking and MD simulations confirmed stable binding, especially in the 3CQU–kaempferol complex, which showed minimal fluctuations. Network and pathway analyses revealed the involvement of multiple cancer-related signalling pathways, including PI3K-Akt and colorectal cancer pathways. The MTT assay demonstrated dose-dependent cytotoxicity of C. pentandra against HCT-116 cells (IC50 = 36.9 µg mL−1), with morphological signs of apoptosis. C. pentandra acts through a multi-target, multi-pathway mechanism, supporting its potential as a natural therapeutic agent for colorectal cancer.Author contributions
MS contributed to writing the original draft, investigation, formal analysis, data curation and conceptualization. GN: initial ideology, editing, workflow, supervision, and project administration of the study.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Data availability
All data generated or analysed during this study are included in this published article. Additional in silico data are available from the corresponding author upon reasonable request.References
- G. G. Dagnaw and H. Dejene, Semin. Oncol., 2025, 52, 19–26 CrossRef PubMed.
- C. A. Dehelean, I. Marcovici, C. Soica, M. Mioc, D. Coricovac, S. Iurciuc, O. M. Cretu and I. Pinzaru, Molecules, 2021, 26, 1109 CrossRef CAS PubMed.
- A. Esmeeta, S. Adhikary, V. Dharshnaa, P. Swarnamughi, Z. U. Maqsummiya, A. Banerjee, S. Pathak and A. K. Duttaroy, Biomed. Pharmacother., 2022, 153, 113384 CrossRef CAS PubMed.
- W. C. Da Silva, V. E. De Araujo, E. M. E. A. Lima, J. B. R. Dos Santos, M. R. R. Da Silva, P. H. R. F. Almeida, F. De Assis Acurcio, B. Godman, A. Kurdi, M. L. Cherchiglia and E. I. G. Andrade, BioDrugs, 2018, 32, 585–606 CrossRef CAS.
- N. J. Jacobo-Herrera, F. E. Jacobo-Herrera, A. Zentella-Dehesa, A. Andrade-Cetto, M. Heinrich and C. Pérez-Plasencia, J. Ethnopharmacol., 2016, 179, 391–402 CrossRef PubMed.
- S. Menaga and N. Geetha, Next Research, 2025, 2, 100500 CrossRef.
- R. Kumar, N. Kumar, G. V. Ramalingayya, M. M. Setty and K. S. R. Pai, Cytotechnology, 2016, 68, 1909–1923 CrossRef.
- P. Ojuka, N. M. Kimani, S. Apollo, J. Nyariki, R. S. Ramos and C. B. R. Santos, South Afr. J. Bot., 2023, 157, 106–114 CrossRef CAS.
- Y. Zhai, L. Liu, F. Zhang, X. Chen, H. Wang, J. Zhou, K. Chai, J. Liu, H. Lei, P. Lu, M. Guo, J. Guo and J. Wu, Chin. Med., 2025, 20, 1–20 CrossRef.
- L. Yan, Z. Zhang, Y. Liu, S. Ren, Z. Zhu, L. Wei, J. Feng, T. Duan, X. Sun, T. Xie and X. Sui, Front. Mol. Biosci., 2022, 9, 862932 CrossRef CAS PubMed.
- M. K. Elbashir, M. Mohammed, H. Mwambi and B. Omolo, Appl. Sci., 2023, 13, 2403 CrossRef CAS.
- S. A. Ruban, F. J. Raj and P. Thangaraj, Biochim. Biophys. Acta, Rev. Cancer, 2025, 1880, 189349 CrossRef PubMed.
- F. Stanzione, I. Giangreco and J. C. Cole, Prog. Med. Chem., 2021, 60, 273–343 Search PubMed.
- R. G. Coleman, M. Carchia, T. Sterling, J. J. Irwin and B. K. Shoichet, PLoS One, 2013, 8, e75992 CrossRef CAS PubMed.
- P. G. R. Achary, Mini-Rev. Med. Chem., 2020, 20, 1375–1388 CrossRef PubMed.
- L. Patel, T. Shukla, X. Huang, D. W. Ussery and S. Wang, Molecules, 2020, 25, 5277 CrossRef CAS PubMed.
- P. Chunarkar-Patil, M. Kaleem, R. Mishra, S. Ray, A. Ahmad, D. Verma, S. Bhayye, R. Dubey, H. N. Singh and S. Kumar, Biomedicines, 2024, 12, 201 CrossRef CAS.
- L. Xiang, B. He, Q. Liu, D. Hu, W. Liao and R. Li, Oncol. Rep., 2020, 44, 1997–2008 CAS.
- A. M. Surin, R. R. Sharipov, I. A. Krasil'nikova, D. P. Boyarkin, O. Y. Lisina, L. R. Gorbacheva, A. V. Avetisyan and V. G. Pinelis, Biochemistry, 2017, 82, 737–749 CAS.
- V. K. Nelson, N. K. Sahoo, M. Sahu, H. H. Sudhan, C. P. Pullaiah and K. S. Muralikrishna, BMC Complementary Med. Ther., 2020, 20, 1–8 CrossRef.
- S. Menaga and N. Geetha, Next Research, 2025, 2, 100500 CrossRef.
- L. P. Ruan, S. Chen, B. Y. Yu, D. N. Zhu, G. A. Cordell and S. X. Qiu, Eur. J. Med. Chem., 2006, 41, 605–610 CrossRef CAS PubMed.
- F. Mayr, G. Möller, U. Garscha, J. Fischer, P. R. Castaño, S. G. Inderbinen, V. Temml, B. Waltenberger, S. Schwaiger, R. W. Hartmann, C. Gege, S. Martens, A. Odermatt, A. V. Pandey, O. Werz, J. Adamski, H. Stuppner and D. Schuster, Int. J. Mol. Sci., 2020, 21, 7102 CrossRef CAS.
- X. Qi, H. Xu, P. Zhang, G. Chen, Z. Chen, C. Fang and L. Lin, Evid. Base Compl. Alternative Med., 2021, 2021, 3905367 Search PubMed.
- M. Pawlak, Ż. Kałuzińska-Kołat, Z. W. Pasieka, D. Kołat and E. Płuciennik, Comput. Biol. Med., 2025, 190, 110116 CrossRef CAS PubMed.
- M. Al-Mustanjid, S. M. H. Mahmud, M. R. I. Royel, M. H. Rahman, T. Islam, M. R. Rahman and M. A. Moni, Genomics, 2020, 112, 3416–3426 CrossRef CAS.
- F. T. J. Fariha, M. Fuad, C. S. Saha, S. Hossen and M. J. Hossain, Sci. Rep., 2025, 15(1), 34490 CrossRef CAS.
- H. Ma, Z. He, J. Chen, X. Zhang and P. Song, Sci. Rep., 2021, 11, 1–11 CrossRef.
- S. Mostafavi, D. Ray, D. Warde-Farley, C. Grouios and Q. Morris, Genome Biol., 2008, 9, 1–15 CrossRef PubMed.
- D. Mutailifu, A. Aini and A. Maimaitiaili, Biomed. Technol., 2025, 10, 100087 CrossRef CAS.
- M. A. Binmujlli, Processes, 2024, 12, 1659 CrossRef CAS.
- A. A. T. Naqvi, T. Mohammad, G. M. Hasan and M. I. Hassan, Curr. Top. Med. Chem., 2019, 18, 1755–1768 CrossRef.
- M. Askarzadeh, H. Azizian, M. Adib, M. Mohammadi-Khanaposhtani, S. Mojtabavi, M. A. Faramarzi, S. M. Sajjadi-Jazi, B. Larijani, H. Hamedifar and M. Mahdavi, Sci. Rep., 2022, 12, 1–16 CrossRef.
- Z. Zaman, S. Khan, F. Nouroz, U. Farooq and A. Urooj, Life Sci., 2021, 264, 118621 CrossRef CAS PubMed.
- K. W. Wang, J. Lee, H. Zhang, D. Suh and W. Im, J. Phys. Chem. B, 2022, 126, 7354–7364 CrossRef CAS PubMed.
- N. Bharadwaj, M. S. Manimuthu, S. Vimal and N. Radhakrishnan, J. Pharm. BioAllied Sci., 2024, 16, S1181–S1185 CrossRef PubMed.
- L. Benov, J. A. Gralnick, A. Hoc, J. Oh and S. Olsen, Microbiol. Spectr., 2021, 9(3), e0163721 CrossRef PubMed.
- Z. Zhang, R. Hao, Q. Guo, S. Zhang and X. Wang, Front. Cell Dev. Biol., 2021, 9, 759154 CrossRef PubMed.
- M. Xiang, Y. Gao, Y. Zhou, M. Wang and X. Yao, BMC Cancer, 2023, 23, 1–19 CrossRef PubMed.
- M. Signore, M. Buccarelli, E. Pilozzi, G. De Luca, M. Cappellari, M. Fanciulli, F. Goeman, E. Melucci, M. Biffoni and L. Ricci-Vitiani, Oncotarget, 2016, 7, 44113 CrossRef PubMed.
- S. Nandi, A. Nag, S. Khatua, S. Sen, N. Chakraborty, A. Naskar, K. Acharya, D. Calina and J. Sharifi-Rad, Phytother Res., 2024, 38, 592–619 CrossRef CAS PubMed.
- S. Z. Moghadamtousi, H. Karimian, E. Rouhollahi, M. Paydar, M. Fadaeinasab and H. A. Kadir, J. Ethnopharmacol., 2014, 156, 277–289 CrossRef PubMed.
- V. K. Nelson, N. K. Sahoo, M. Sahu, H. H. Sudhan, C. P. Pullaiah and K. S. Muralikrishna, BMC Complementary Med. Ther., 2020, 20, 1–8 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |