Open Access Article
Open Access ArticleMulti-target antibacterial Ru-based metallodrugs containing phenylsulfonyl indole derivatives: synthesis and their efficacy against Gram-positive and -negative bacteria†
Shijie Lin‡
a,
Guangying Yu‡c,
Liu Zhang‡c,
Xiangwen Liao c,
Guijuan Jiang
c,
Guijuan Jiang *c and
Lianghong Liu*b
*c and
Lianghong Liu*b
aDepartment of Pharmacy, Hainan General Hospital (Hainan Affiliated Hospital of Hainan Medical University), Haikou, 570311, China
bSchool of Pharmaceutical Sciences, Hunan University of Medicine, Huaihua, 418000, China. E-mail: llhfe@qq.com
cSchool of Pharmacy, Jiangxi Science & Technology Normal University, Nanchang, 330013, China. E-mail: jgjchem@163.com
First published on 23rd June 2025
Abstract
The escalating crisis of antimicrobial resistance (AMR) underscores the critical need for the development of novel antibacterial drugs with unique mechanisms of action. Herein, a series of phenylsulfonyl indole-modified Ru-based metallodrugs with multi-target antibacterial mechanisms were prepared and evaluated. Notably, all these complexes showed strong bacteriostatic efficacy against Staphylococcus aureus (S. aureus), and the most active complex, RuS2, showed a lower MIC than that of many common antibiotics. Importantly, RuS2 also showed robust bactericidal efficacy against Gram-negative bacteria (Escherichia coli) in the presence of subinhibitory concentrations of polymyxin B. In addition, complex RuS2 can reduce bacterial pathogenicity by inhibiting hemolysin secretion and biofilm formation. More importantly, RuS2 can curb the production of drug-resistant bacteria and has significant activity against clinically isolated resistant bacteria. Mechanism studies have demonstrated that RuS2 can destroy the bacterial membrane, cause membrane depolarization, and induce the production of reactive oxygen species (ROS). Finally, the G. mellonella wax worms and mouse infection models confirm that RuS2 has low toxicity and significant anti-infective potency in vivo. Taken together, the results presented herein pave a promising way for combating Gram-positive and -negative bacterial infections.
1 Introduction
The overuse and misuse of antibiotics in humans, agriculture, and livestock have accelerated the rapid spread of drug-resistant bacteria.1–3 Nowadays, antimicrobial resistance (AMR) has gradually become a major cause of death worldwide.4–6 It is reported that about 1.27 million people died directly from AMR and that 4.95 million deaths were related to antibiotic-resistant infections in 2019. The World Health Organization (WHO) has identified AMR as one of the top 10 global public health threats, with projections estimating 10 million annual deaths by 2050 if no action is taken.7,8 Worse yet, the development of new antibiotics lags far behind the emergence of drug-resistant bacteria. In the last two decades, only six new antibacterial drugs have been approved, and none of them are effective against Gram-negative bacteria.9 Thus, there is an urgent need for novel antibiotics that are hard to develop resistance to and have strong efficacy against Gram-positive and -negative bacteria. However, this remains a significant challenge.Metallodrugs play an important role in medicine owing to their diverse biochemical properties, versatile redox states, and ability to interact with biological targets.10–14 Platinum-based drugs remain an important drug in the clinical treatment of malignant tumors.15,16 Gold(I) complexes, such as auranofin, were used for rheumatoid arthritis by inhibiting thioredoxin reductase and modulating immune responses.17 In addition, metal-based agents act as powerful candidates in combating bacterial infections.18,19 Silver sulfadiazine and silver nanoparticles showed robust efficacy in combating antibiotic-resistant pathogens in wound dressings and coatings.20–22 Bismuth compounds (bismuth citrate) are the cornerstone drug in Helicobacter pylori infection treatment.23 It is worth noting that the ruthenium-based antibacterial agents were reported to have robust bactericidal potency in vitro and in vivo by targeting bacterial membrane integrity.24 They showed a low frequency of resistance and inhibited the exotoxin secretion, with a great advantage in combating drug-resistant bacteria.25–28
Indole-containing derivatives have been reported to have excellent activity against both Gram-positive and -negative bacteria.29,30 For example, eudistomin U has robust potency against Staphylococcus aureus (S. aureus), Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa), with MIC ranging from 3.4 to 27.7 μg mL−1. Dragmacidin G exhibited a broad-spectrum antibacterial activity, including S. aureus (MIC, 0.62 μg mL−1), MRSA (MIC, 0.62 μg mL−1) and Mycobacterium tuberculosis (MIC, 21 μM) (Scheme 1A).31 In addition to indole, phenylsulfonyl-containing drugs also have significant antimicrobial activity, such as sulfapyridine and sulfamethoxazole (Scheme 1A). Here, to develop some novel multi-target antibacterial Ru-based metallodrugs with good activity against Gram-positive and -negative bacteria, a phenylsulfonyl indole moiety was linked to the Ru-based structure, and three novel complexes were obtained (Scheme 1B). Subsequently, their antibacterial potency against S. aureus and E. coli was investigated. Next, the ability of the most active complex, RuS2, to reduce bacterial pathogenicity and combat bacterial resistance was further confirmed. In addition, its multi-target antibacterial mechanism was verified using a series of methods. Finally, two animal infection models were employed to investigate the toxicity and anti-infective potency of RuS2 in vivo.
2 Results and discussion
2.1 Antibacterial activity of Ru-based agents in vitro
The synthesis routes of three Ru-based complexes containing phenylsulfonyl indole derivatives are illustrated in Scheme 1C. The structure of the three complexes was characterized by HRMS spectroscopy, 1H NMR, and 13C NMR spectra, and their purity was confirmed by HPLC (Fig. S1–S15†). In addition, the stabilities of the three complexes were investigated using NMR spectroscopy. The results indicate that all complexes are stable at room temperature (Fig. S16†).Next, the minimum inhibitory concentration (MIC) of each compound against S. aureus and E. coli was first determined. The results indicated that three complexes, RuS1, RuS2 and RuS3, showed low MIC values (1.56–6.25 μg mL−1) against S. aureus, indicating their robust antibacterial efficacy. Importantly, the most active agent RuS2 has the same potency against S. aureus (1.56 μg mL−1) as vancomycin, a clinically used antibiotic as the last resort for intractable Gram-positive bacterial infections32 (Fig. 1). Next, the biocompatibility of the three complexes was explored. The results suggested that RuS1, RuS2 and RuS3 showed low toxicity against fresh rabbit blood cells, with all HC50 above 200 μg mL−1 (Fig. 2).
 | ||
| Fig. 1 Minimum inhibitory concentration (MIC) of three Ru-based complexes containing phenylsulfonyl indole derivatives in this work and clinically used antibiotics against S. aureus. | ||
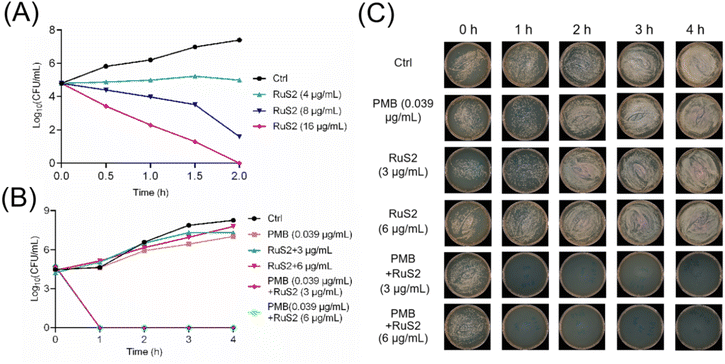
It is noteworthy that all the three complexes showed relatively poor efficacy against E. coli, with MIC lying at or above 100 μg mL−1. We speculated that the unique lipopolysaccharide (LPS) outer membrane of Gram-negative bacteria may limit the potency of Ru-based complexes.33 Notably, polymyxin B (PMB) binds to the LPS of the bacterial outer cell membrane and then disrupts the integrity of the cell membrane.34 Thus, the combination with PMB significantly enhances their antibacterial potency against Gram-negative bacteria. It turned out that the checkerboard assay (Fig. 3A) confirmed that there was an obvious synergistic effect between RuS2 and PMB, with FICI values lying at 0.25 (synergistic effects defined by FICIs ≤ 0.5) (Fig. 3B). In addition, the antibacterial potency of RuS2 against E. coli was increased 32 times, with the MIC decreasing from 100 to 3.125 μg mL−1 in the presence of sub-inhibitory concentrations of polymyxin B (0.039 μg mL−1, 0.1 × MIC) (Fig. 3C).
Time-kill curves are a common method for directly visualizing the speed and extent of sterilization of antimicrobial agents.35 Therefore, to investigate the bactericidal effect of RuS2, its time–kill curves against S. aureus or E. coli were explored. As shown in Fig. 4, RuS2 killed >99.9% S. aureus within 2 hours at 16 μg mL−1. In addition, although RuS2 had no bactericidal effect on E. coli when used alone, its bactericidal efficacy was significantly elevated in the presence of 0.039 μg mL−1(0.1 × MIC) polymyxin B. RuS2 at 3 μg mL−1 killed >99.9% E. coli within only 1 h. Overall, these results suggest that Ru-based agents containing phenylsulfonyl indole derivatives showed good activity against Gram-positive and -negative bacteria in vitro.
2.2 Ability of RuS2 to combat bacterial resistance
Antimicrobial resistance refers to the phenomenon in which pathogenic bacteria gradually decrease or even completely lose their sensitivity to antibiotics under long-term exposure, which poses a serious threat to human health.36 Thus, overcoming the emergence of bacterial resistance is a required ability for new antibacterial drugs. Here, the drug-resistance development of S. aureus against RuS2 was explored. In brief, S. aureus was exposed to RuS2 or clinical antibiotics for 20 consecutive generations, and changes in the MIC of RuS2 and antibiotics were recorded. As shown in Fig. 5A, the MIC of RuS2 increased only 2-fold after 20 generations. However, under the same conditions, the MICs of clinically used antibiotics vancomycin, paromomycin, gentamicin and cefalexin were increased by 64, 256, 320 and 512 fold, respectively. These results indicate that RuS2 can effectively avoid the generation of bacterial resistance. Next, a cross-resistance study was performed to confirm the efficacy of RuS2 against a series of drug-resistant S. aureus. As shown in Fig. 5B, the antibiotics had lost their efficacy against the corresponding resistance strains. However, RuS2 still has strong antimicrobial efficacy against all drug-resistant S. aureus, showing low MIC values. Notably, RuS2 also showed robust potency against the clinical isolates of MRSA (Table S1†). Overall, these results confirm that RuS2 can efficiently combat bacterial resistance.2.3 RuS2 reduces the pathogenicity of bacteria
Biofilm and hemolytic toxins (such as α-hemolysin) are two core factors of bacterial pathogenicity.37,38 They work in concert during the infection process, significantly enhancing the bacteria's invasive ability, immune evasion, and host damage. Among them, α-hemolysin is the core virulence factor of S. aureus, which is involved in various diseases, such as skin and soft tissue infections, necrotizing pneumonia, and sepsis.38 In recent years, antitoxin strategies that inhibit the secretion of α-hemolysin have emerged as a new way to fight against drug-resistant infections. To further investigate the effect of RuS2 on toxin secretion in S. aureus, the sub-inhibitory concentrations of RuS2 were co-incubated with S. aureus for 12 hours; then, the amount of α-hemolysin in the supernatant was detected using rabbit red blood cells. The results confirmed that RuS2 can efficiently inhibit the secretion of α-hemolysin in S. aureus at sub-inhibitory concentrations (Fig. 6). | ||
| Fig. 6 Inhibition effect of RuS2 on hemolytic toxin secretion from S. aureus. (A) Quantitative analysis of hemolytic toxin secretion in the presence of RuS2. (B) Photos of red blood cell rupture. | ||
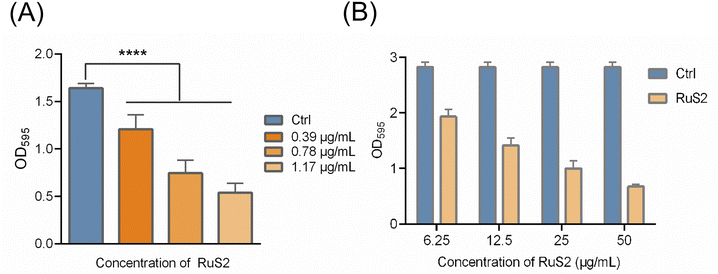
As other core factors of bacterial pathogenicity, biofilm formation provides a stable growth environment, protects bacteria from host immune defenses and facilitates bacterial dissemination,39 posing a great challenge to clinical treatment. Next, the effect of RuS2 on bacterial biofilms was further explored. As shown in Fig. 7A, the formation of bacterial biofilm was gradually reduced upon co-incubated with the sub-inhibitory concentration of RuS2, confirming its inhibition effect on S. aureus biofilm formation. It is reported that bacterial resistance in the mature biofilm can be significantly enhanced by 10 to 1000 times.40 Thus, it is necessary to further explore the eradication effect of RuS2 on mature biofilms. In brief, bacteria were incubated for 24 hours to form mature biofilms and then co-incubated with different doses of RuS2, and the viable bacteria in mature biofilms were detected with MTT. The results indicated that RuS2 can effectively eradicate the mature biofilm, resulting in a significant decrease in the number of viable bacteria in the biofilm (Fig. 7B). Collectively, the data presented here demonstrate that RuS2 can reduce bacterial pathogenicity by inhibiting hemolysin secretion and biofilm formation, which is very advantageous for the effective control of complex infections.
2.4 Antibacterial mechanism studies of RuS2
Encouraged by the excellent antibacterial properties of RuS2 in vitro, we set out our efforts to further uncover its underlying multi-target mechanism. First, the destructive effects of RuS2 on the cell membranes of S. aureus and E. coli were explored through nucleic acid leakage experiments. The results showed that the content of nucleic acid in the supernatant increased significantly when S. aureus was co-incubated with RuS2, as indicated by the higher absorbance at both 260 and 280 nm than the PBS-treated group (Fig. 8A and B). In addition, in the presence of sub-inhibitory concentrations of polymyxin B (0.039 μg mL−1, 0.1 × MIC), RuS2 exhibited a similar effect on E. coli, leading to a significant increase in the absorbance of the bacterial supernatant at both 260 and 280 nm as well (Fig. 8C and D). Next, two fluorescence dyes (DAPI and PI) were used to further explore the action of RuS2 on the bacterial cell membrane. DAPI can label all bacterial cells blue, while PI can only label the membrane-damaged cells red. As shown in Fig. 9, red fluorescence was observed only in the RuS2-treated group. In addition, the number of red-labeled bacteria significantly increased after incubation with a higher dose of RuS2, as indicated by the red fluorescence intensity (Fig. S17†). These results demonstrate that RuS2 can efficiently disrupt the bacterial cell membrane. | ||
| Fig. 9 Membrane damage effect of RuS2 and kanamycin on S. aureus was evaluated by DAPI and PI staining. | ||
It is reported that an antibacterial drug binds to the negatively charged components of the bacterial cell membrane, which can increase membrane permeability.41 Therefore, the effect of RuS2 on bacterial membrane potential was further explored. As shown in Fig. 10A, the zeta potential of S. aureus gradually appeared to shift from negative to positive values with an increase in the concentration of RuS2, indicating that RuS2 can disrupt the charge distribution on the surface of the bacterial cell membrane. In addition, in the presence of sub-inhibitory concentrations of polymyxin B (0.039 μg mL−1, 0.1 × MIC), RuS2 exhibited a similar effect on E. coli, leading to significant changes in membrane potentials as well (Fig. 10B). Next, we further investigated whether RuS2 could also result in the depolarization of the bacterial membrane. In brief, bacteria were co-incubated with RuS2 for 4 hours; then, a green fluorescence dye DiSC3(5) for depolarization monitoring was added. As shown in Fig. 11A, strong green fluorescence was observed in the RuS2-treated group, indicating the depolarization of the bacterial membrane. In addition, the intensity of green fluorescence induced by RuS2 is concentration-dependent (Fig. 11B). Notably, the same dose of vancomycin failed to alter the green fluorescence intensity (Fig. 11B). These data confirm that RuS2 can result in changes in bacterial membrane potential and lead to depolarization.
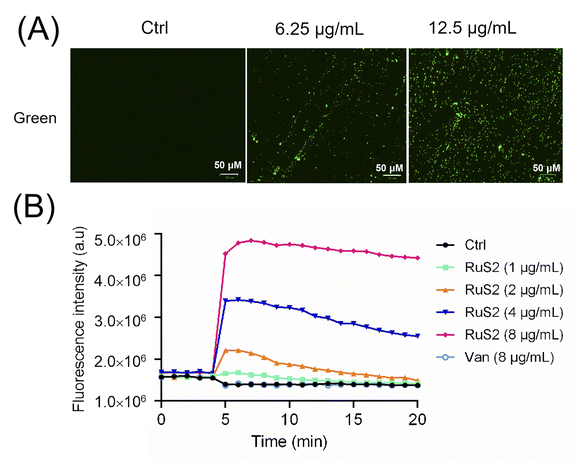 | ||
| Fig. 11 Membrane depolarization of S. aureus was evaluated by DiSC3(5) staining. Fluorescence imaging (A) and fluorescence intensity (B) of S. aureus after co-incubation with RuS2. | ||
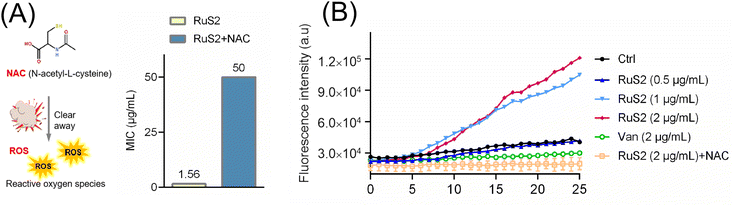
In addition to targeting the bacterial membrane, Ru-based antibacterial agents can efficiently kill bacteria by inducing ROS production as well.24 Therefore, the ROS scavenger (N-acetyl-cysteine, NAC)24 was employed to verify the important role of ROS in the bactericidal process of RuS2. The results indicated that NAC significantly attenuated the antibacterial potency of RuS2; the MIC of RuS2 against S. aureus increased from the initial 1.56 to 50 μg mL−1 in the presence of NAC (Fig. 12A). In addition, the production of ROS in bacteria after co-incubated with RuS2 was further monitored using a green fluorescent probe (2,7-dichlorodihydrofluorescein diacetate, DCFH-DA).24 As shown in Fig. 12B, a strong intensity of green fluorescence was detected in the RuS2-treated group, suggesting the accumulation of ROS. In addition, the intensity of green fluorescence induced by RuS2 is concentration-dependent. Notably, no ROS accumulation was detected in the vancomycin-treated group. As expected, the addition of NAC eliminated the accumulation of ROS induced by RuS2 (Fig. 12B). Collectively, these results demonstrated that RuS2 killed bacteria in many ways: destroying the bacterial membrane, causing membrane depolarization, and inducing the production of ROS.
2.5 In vivo toxicity of RuS2
Given that RuS2 showed good antimicrobial effects against both S. aureus and E. coli, its toxicity was further investigated using Galleria mellonella (G. mellonella). Generally, G. mellonella is widely used for toxicity testing because its immune system is similar to that of mammals.42 In brief, high doses of RuS2 (ranging from 16 to 128 mg kg−1) were injected into the G. mellonella larvae, followed by observation of their survival and physiological responses, and vancomycin was selected as the control. The results indicated that RuS2 had no significant effect on the survival rate of G. mellonella larvae. Even though the tested dose of RuS2 reached 128 mg kg−1, the survival rate of G. mellonella was still as high as 80%, indicating its low acute toxicity. Notably, RuS2 did not show higher toxicity compared with the antibiotic vancomycin (Fig. 13). These data confirmed that RuS2 has similar acute toxicity to vancomycin, at least in the case of G. mellonella.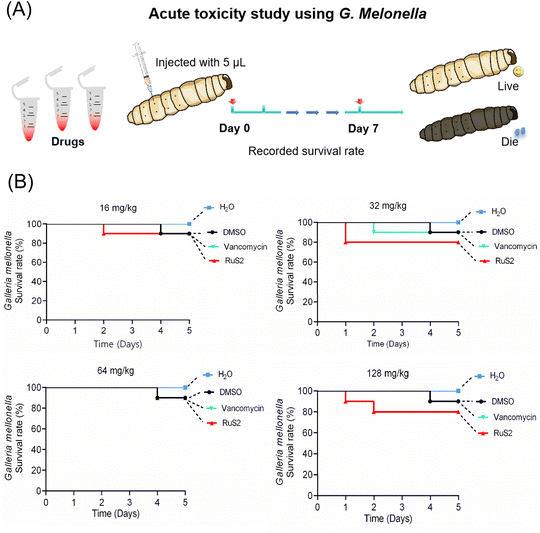 | ||
| Fig. 13 (A) Acute toxicity study of RuS2 using G. mellonella. (B) Survival rate of G. mellonella wax worms after treatment with different doses of RuS2 or vancomycin. | ||
2.6 Anti-infective efficacy of RuS2 in vivo
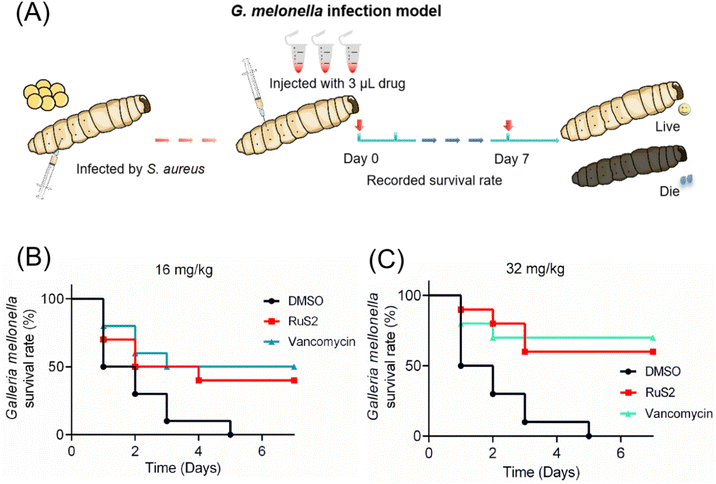
Two animal infection models were established to further investigate the anti-infective efficacy of RuS2 in vivo. To begin, 5 μL bacteria (OD600 = 0.3) were injected into the body of G. mellonella larvae with a microsyringe. After 30 min, the S. aureus-infected G. mellonella was treated with RuS2 or vancomycin; then, the survival rate of G. mellonella was recorded (Fig. 14A). The results indicated that the therapeutic efficacy of RuS2 was comparable to that of vancomycin, resulting in a 60% survival rate of S. aureus-infected G. mellonella at a dose of 32 mg kg−1 (Fig. 14). Next, a mouse skin infection model (Fig. 15A) was further established to further confirm the anti-infective efficacy of RuS2. In brief, a circular wound was cut on the back of the mouse, and a bacterial suspension was added to the wound. Next, creams containing RuS2 or vancomycin were uniformly applied to the wounds. During the treatment, the size of the wounds was photographed and documented daily, and the body weight data of the mice were recorded. On the 11th day, the infected tissue was collected for abrasive coating to monitor the number of viable bacteria. The changes in the size and the photograph of the infected wound clearly confirmed that the wound healing rate in the RuS2-treated group was better than that in the PBS-treated groups, indicating a wound that was almost completely healed on the 11th day (Fig. 15B and C). In addition, RuS2 showed a stronger therapeutic effect than vancomycin at the same dose, resulting in a lower viable bacterial load in the infected tissue than in the vancomycin-treated group (Fig. 15D). Notably, the weight of the mice did not change significantly throughout the treatment (Fig. 15E). Collectively, the results presented here demonstrate that RuS2 has robust anti-infective efficacy against S. aureus in animal infection models as well.3 Conclusion
In summary, three novel Ru-based antibacterial agents bearing phenylsulfonyl indole derivatives were successfully prepared and evaluated in this work. The antibacterial potency investigation in vitro indicated that all the three complexes showed strong bactericidal activity against S. aureus. The most active agent, RuS2, completely inhibited bacterial growth at only 1.56 μg mL−1, a lower MIC value than that of the most commonly used antibiotics. These data confirm that the incorporation of the indole moiety can enhance the antibacterial activity of ruthenium compounds against S. aureus. Moreover, RuS2 showed a lower lipophilicity/hydrophilicity ratio (C![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 0.524) and displayed the most potent antibacterial capacity, indicating that the fine-tuned lipophilicity/hydrophilicity ratio governed the antibacterial potency of the compounds. Although RuS2 was less effective against Gram-negative bacteria (E. coli) when used alone, its potency against E. coli can be increased 32 times using polymyxin B, with the MIC decreasing from 100 to 3.125 μg mL−1. Notably, RuS2 at 3 μg mL−1 killed >99.9% E. coli within only 1 hour in the presence of 0.1 × MIC polymyxin B. More importantly, RuS2 exhibits numerous advantages over conventional antibiotics. First, RuS2 can effectively avoid the generation of bacterial resistance and has no cross-resistance with traditional antibiotics. Second, RuS2 can reduce bacterial pathogenicity by inhibiting hemolysin secretion and biofilm formation. Finally, RuS2 showed many antibacterial mechanisms: destroying the bacterial membrane, causing membrane depolarization, and inducing the production of ROS. These advantages were partially verified by the good anti-infective potency of RuS2 in vivo as well. In the mouse infection model, RuS2 showed a stronger therapeutic effect than vancomycin at the same dose. Collectively, these results presented here pave a promising way in combating Gram-positive and -negative bacterial infection.
P = 0.524) and displayed the most potent antibacterial capacity, indicating that the fine-tuned lipophilicity/hydrophilicity ratio governed the antibacterial potency of the compounds. Although RuS2 was less effective against Gram-negative bacteria (E. coli) when used alone, its potency against E. coli can be increased 32 times using polymyxin B, with the MIC decreasing from 100 to 3.125 μg mL−1. Notably, RuS2 at 3 μg mL−1 killed >99.9% E. coli within only 1 hour in the presence of 0.1 × MIC polymyxin B. More importantly, RuS2 exhibits numerous advantages over conventional antibiotics. First, RuS2 can effectively avoid the generation of bacterial resistance and has no cross-resistance with traditional antibiotics. Second, RuS2 can reduce bacterial pathogenicity by inhibiting hemolysin secretion and biofilm formation. Finally, RuS2 showed many antibacterial mechanisms: destroying the bacterial membrane, causing membrane depolarization, and inducing the production of ROS. These advantages were partially verified by the good anti-infective potency of RuS2 in vivo as well. In the mouse infection model, RuS2 showed a stronger therapeutic effect than vancomycin at the same dose. Collectively, these results presented here pave a promising way in combating Gram-positive and -negative bacterial infection.
4 Experimental section
4.1 Synthesis of compounds RuS1, RuS2 and RuS3
The ammonium acetate (20.0 mmol, 1.541 g), phenylsulfonyl indole carboxaldehyde (1.0 mmol, 0.476 g), 1,10-phthalophenanthroline-5,6-dione (1.1 mmol, 0.231 g) and glacial acetic acid (10.0 mL) were mixed and then reacted at 120 °C for 4 hours. After cooling to room temperature, ddH2O was added; then, the pH of the reaction solution was adjusted to neutral with NH3·H2O to obtain a light yellow precipitate. Next, recrystallization was carried out with anhydrous ethanol to produce a light pink needle-like solid (the ligand, CHNOS). Next, the obtained ligand CHNOS (0.2 mmol, 0.095 g), [Ru(bpy)2Cl2]2H2O (0.2 mmol, 0.108 g) and ethylene glycol (10 mL) were mixed and then heated at 150 °C for 8 hours under argon. After cooling to room temperature, the KPF6 aqueous solution (15 mL) was added, and a dark red precipitate was obtained. Finally, the crude product was collected and purified by column chromatography (neutral alumina, acetonitrile/xylene = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) to prepare compound RuS1. The synthesis methods of compounds RuS2 and RuS3 were similar to that of compound RuS1 (Scheme 1).
1, v/v) to prepare compound RuS1. The synthesis methods of compounds RuS2 and RuS3 were similar to that of compound RuS1 (Scheme 1).
CHNOS (Ligand): 1H NMR (400 MHz, DMSO-d6) δ 13.86 (s, 1H), 9.05 (dd, J = 9.6, 2.6 Hz, 2H), 8.99 (d, J = 7.4 Hz, 1H), 8.80 (t, J = 8.3 Hz, 2H), 8.74 (s, 1H), 8.08 (d, J = 7.5 Hz, 2H), 8.04 (d, J = 4.9 Hz, 1H), 7.91 (s, 1H), 7.82 (s, 1H), 7.73 (d, J = 7.2 Hz, 1H), 7.65 (d, J = 7.8 Hz, 2H), 7.51 (d, J = 8.3 Hz, 2H).13C NMR (101 MHz, DMSO-d6) δ 148.12 (s), 145.96 (s), 143.44 (s), 137.16 (s), 135.50 (s), 135.04 (s), 130.57 (s), 128.27 (s), 127.25 (s), 126.35 (s), 125.92 (d, J = 13.2 Hz), 124.86 (s), 123.72 (d, J = 47.2 Hz), 113.75 (d, J = 20.7 Hz). HRMS (ESI): m/z calcd for [M + H]+ C27H18N5O2S 476.1181; found: 476.1393.
[Ru(bpy)2(CHONS)](PF6)2 (RuS1): 1H NMR (400 MHz, DMSO-d6) δ 8.99 (t, J = 9.0 Hz, 3H), 8.86 (dd, J = 17.2, 7.8 Hz, 4H), 8.22 (dd, J = 24.4, 16.0 Hz, 3H), 8.14–7.95 (m, 5H), 7.88 (d, J = 5.2 Hz, 2H), 7.84–7.65 (m, 5H), 7.59 (t, J = 6.5 Hz, 6H), 7.43 (s, 2H), 7.36 (s, 2H), 1.22 (s, 1H). 13C NMR (151 MHz, DMSO-d6) δ 157.34 (s), 157.14 (s), 151.83 (s), 151.53 (s), 147.13 (s), 143.89 (s), 138.17 (s), 137.99 (s), 137.37 (s), 135.23 (d, J = 17.1 Hz), 130.35 (d, J = 7.5 Hz), 128.26 (s), 127.20 (s), 126.06 (d, J = 12.7 Hz), 125.43 (s), 124.84 (s), 124.21 (s), 123.57 (s), 113.52 (s). HRMS (ESI): m/z calcd for [M-2PF6]2+ C47H34O2N9RuS, 444.5767; found: 444.5800.
[Ru(dmb)2(CHONS)](PF6)2 (RuS2): 1H NMR (400 MHz, DMSO-d6) δ 9.03 (d, J = 8.0 Hz, 2H), 8.89 (s, 1H), 8.72 (d, J = 20.4 Hz, 4H), 8.46 (s, 1H), 7.99 (d, J = 17.0 Hz, 5H), 7.89–7.81 (m, 2H), 7.68 (d, J = 5.7 Hz, 3H), 7.59 (t, J = 7.6 Hz, 2H), 7.53–7.36 (m, 6H), 7.16 (d, J = 5.7 Hz, 2H), 2.56 (s, 6H), 2.45 (s, 6H), 1.20 (s, 1H). 13C NMR (151 MHz, DMSO-d6) δ 156.84 (s), 156.76 (d, J = 21.0 Hz), 150.87 (s), 149.88 (s), 149.72 (s), 144.95 (s), 137.07 (s), 135.40 (s), 135.05 (s), 130.65–130.56 (m), 130.33 (d, J = 40.0 Hz), 128.87 (d, J = 22.3 Hz), 127.23 (s), 126.06 (s), 125.32 (s), 124.61 (s), 113.61 (s), 21.24(s). HRMS (ESI): m/z calcd for [M-2PF6]2+ C51H42O2N9RuS, 472.6080; found: 472.6091.
[Ru(dtb)2(CHONS)](PF6)2 (RuS3): 1H NMR (400 MHz, DMSO-d6) δ 8.93 (dd, J = 41.6, 20.7 Hz, 6H), 8.32 (s, 1H), 8.01 (d, J = 5.7 Hz, 2H), 7.80 (s, 4H), 7.74–7.53 (m, 7H), 7.41 (dd, J = 42.8, 5.7 Hz, 6H), 1.42 (s, 18H), 1.32 (s, 18H), 1.20 (s, 2H), 0.82 (d, J = 8.1 Hz, 1H). 13C NMR (151 MHz, DMSO-d6) δ 162.07 (s), 161.89 (s), 157.09 (s), 156.90 (s), 151.45–151.25 (m), 151.06 (d, J = 31.1 Hz), 147.29 (s), 144.26 (s), 137.30 (s), 135.22 (s), 130.39 (s), 130.09 (s), 129.47 (s), 127.19 (s), 125.64 (s), 125.18 (s), 124.94 (s), 124.25 (d, J = 12.8 Hz), 122.19 (s), 113.53 (s), 35.99 (s), 30.45 (s). HRMS (ESI): m/z calcd for [M-2PF6]2+ C63H66O2N9RuS, 556.7021; found: 556.7043.
4.2 Determination of the minimum inhibition concentration
The tested complexes, which dissolved in DMSO, were added into a 96-well plate and then diluted with sterile water to obtain a concentration gradient of 0.2–200 μg mL−1. Next, 200 μL bacterial suspension was added into the 96-well plate and then further incubated in a constant temperature incubator at 37 °C for 20 hours. Finally, the turbidity of the bacteria in the 96-well plate was observed to determine the MIC value. Each set of experiments was repeated at least 3 times.4.3 Inhibiting the formation of bacterial biofilms
Bacterial suspension and the tested drugs were mixed in a 24-well plate and then incubated for 48 hours at 37 °C. After washing away the floating bacteria with PBS (phosphate-buffered saline), the biofilm was stained with 0.1% crystal violet for 20 minutes. After washing away the crystal violet with PBS, 50% glacial acetic acid was added to dissolve the stained crystal violet. Finally, the absorbance at 595 nm was determined using a microplate reader.4.4 Bacterial biofilm eradication assay
Bacteria at the logarithmic phase were diluted 1000-fold, and then 200 μL bacterial solution was added into a 96-well plate. After incubation at 37 °C for 24 hours, the plate was washed with PBS and dried at 37 °C for 10 minutes. Next, the tested drug solutions were added and further incubated at 37 °C for 24 hours. Afterwards, an equal amount of MTT (thiazolyl blue, 500 μg mL−1) was added and further incubated for 30 minutes. Finally, the excess MTT was removed, and the absorbance of the solution at 595 nm was measured using a microplate reader after adding dimethyl sulfoxide (DMSO).4.5 Checkerboard experiments
Checkerboard experiments were performed according to the reported protocol.25 In brief, the two drugs tested together with 200 μL bacterial suspensions were added into a 96-well plate and then incubated at 37 °C for 20 hours. Next, the growth of the bacteria was observed, and the FICI of the tested drug was calculated to analyze the combination effect of the Ru-based complexes with antibiotics. The FICI values were calculated based on the following formula: FICI = FICIA + FICIB = CA/MICA + CB/MICB.4.6 DAPI/PI staining assay
S. aureus was co-incubated with antibiotics or RuS2 at 37 °C for 4 hours; then, bacteria were collected by centrifugation (5000 rpm, 2 min) and washed with PBS. Next, 30 μM DAPI and 30 μM PI were added into the bacteria suspension and further incubated for 15 minutes in the dark. Finally, the bacteria were observed using a fluorescence microscope and photographed for recording.4.7 ROS detection through DCFH-DA
Bacteria in the logarithmic phase were collected and then suspended in sterile PBS to OD600 = 0.3. Then, bacteria suspension together with different doses of RuS2 was added into a 96-well black enzyme labeling plate. Afterwards, 30 μL (DCF-DA, 2′,7′-dichlorodihydrofluorescein diacetate) was further added with the final concentration at 30 μM. Finally, the fluorescence intensity over time was monitored using a microtiter plate reader (excitation wavelength at 488 nm and emission wavelength at 530 nm).4.8 Membrane depolarization monitored with DiSC3(5)
Bacteria were co-incubated with RuS2 at 37 °C for 1 hour. Then, the bacteria was collected via centrifugation (5000 rpm, 2 min) and further washed with PBS. Next, 30 μL DiSC3(5) (50 μM) was added into the bacterial resuspension solution (300 μL) and incubated for 30 minutes in the dark. Next, 10 μL of bacteria was placed on the slide and observed under a fluorescence microscope. For quantitative detection, the bacterial suspension was processed as above and added into a 96-well black enzyme labeling plate. Then, the fluorescence intensity over time was monitored using a microtiter plate reader (excitation wavelength at 622 nm and emission wavelength at 670 nm).4.9 Time-kill kinetics assays
Bacteria at the logarithmic phase were diluted 1000-fold and then co-incubated with RuS2 in a shaker at 37 °C. Then, 50 μL bacteria were collected at different times and then spread on an agar plate. Next, the plate was incubated at 37 °C for 20 hours; then, the number of colonies was recorded. Finally, the time-kill curve was plotted according to the number of viable bacteria over time.4.10 HC50 detection
First, the rabbit blood erythrocytes were washed 3 times with sterile PBS. Next, 150 μL rabbit blood cells and 50 μL Ru-based complexes were added into 1 mL PBS and incubated at 37 °C for 30 minutes. The supernatant was collected via centrifugation (2000 rpm, 2 minutes), and the absorbance value at 543 nm was determined. TriX-100 was used as a positive control, and the PBS group was used as a negative control. Each group of experiments was independently repeated at least three times. The hemolysis rate was calculated as follows: hemolysis rate = [OD543(experimental group) − OD543(PBS)/OD543(Triton-100) − OD543(PBS)]%.4.11 Resistance development assays
First, the MIC of the tested drug was determined using a 96-well plate. Then, bacteria from 0.5 × MIC wells were collected and diluted with fresh TSB medium. After incubation at 37 °C for 20 hours, the MIC values of the tested drug were determined once again. This step was repeated consecutively up to the 20th generation, and the changes in the MIC value over the generation were recorded.4.12 Inhibiting the secretion of hemolytic toxins
The bacteria were inoculated into a fresh TSB medium, and then RuS2 was added. After incubation for 10 hours at 37 °C, the supernatant was collected by centrifugation (5000 rpm, 2 min). Meanwhile, rabbit erythrocytes were washed 3 times with PBS buffer (2000 rpm, 2 min). In 1 mL PBS solution, 50 μL bacterial supernatant and 50 μL erythrocyte suspension were sequentially added and further incubated at 37 °C for 30 minutes. Finally, the supernatant was collected by centrifugation (2000 rpm, 2 min), and the absorbance at 543 nm was measured to monitor the hemolytic activity.4.13 DNA leakage
Bacteria in the logarithmic phase were diluted with fresh PBS to OD600 = 0.3; then, RuS2 was added. Next, 2 mL bacterial suspension was transferred into 24-well plates and then incubated at 37 °C for 2 hours. Afterwards, the bacterial supernatant was collected by centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 5 min) and filtered through a microporous membrane. The absorbance of the supernatant at 260 or 280 nm was measured using a UV spectrophotometer. Untreated bacteria were used as the control group.
000 rpm, 5 min) and filtered through a microporous membrane. The absorbance of the supernatant at 260 or 280 nm was measured using a UV spectrophotometer. Untreated bacteria were used as the control group.
4.14 Zeta potential studies
Bacteria in the logarithmic phase were collected and diluted with fresh ultrapure water. Then, 6 mL bacterial suspension was mixed with the tested drug and pipetted into a 10 mL shaker tube. After incubating in a shaker at 37 °C for 1 hour, the zeta potential of the bacterial suspension was measured using a particle size meter, and the water-treated bacterial suspension was used as a blank control. Each set of experiments was repeated three times.4.15 G. mellonella infection model
For acute toxicity screening, the injection site of G. mellonella larvae was sterilized with ethanol. Then, RuS2 or antibiotics (6–128 mg kg−1) was injected from the hind foot of the larvae using a 50 μL microsyringe. Afterwards, the survival of the larvae was observed and recorded every 24 hours. There were 10 larvae in each group, and DMSO was used as a negative control. For anti-infective efficacy exploration, bacteria were first incubated in fresh TSB to the logarithmic stage and then suspended in sterile PBS to OD600 = 0.3. Next, 5 μL bacterial suspension was injected into the body of G. mellonella larvae from the left hind foot. After 30 minutes, RuS2 or antibiotics (16 or 32 mg kg−1) were injected from the right hind foot of the G. mellonella larvae. Finally, the survival rate of the infected larvae was observed and recorded at 24-hour intervals over 7 days.4.16 Mouse skin infection models
Female KM mice (25–30 g) were selected for the mouse skin infection model. In brief, the wounds on the back of the mice were infected with bacteria for 12 hours to induce abscess formation. Afterwards, the infected mice were randomly divided into 4 groups, and RuS2 containing ointment was administered three times daily. The weight change of the mice and the recovery rate of the wounds were recorded daily. On the 11th day, the tissue of the infected wound in each group was collected for viable bacteria counting.Ethical statement
The animal study was reviewed and approved by Institutional Animal Care and use Committee of Jiangxi Science & Technology Normal University.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors have declared that there is no conflict of interest.Acknowledgements
We gratefully acknowledge the generous support provided by the Health Research Project of Hainan Province (Grant No. 22A200010) and Jiangxi Science & Technology Normal University (2021QNBJRC001).References
- M. William and A. Cesar, ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics, Nat. Rev. Microbiol., 2024, 22(10), 598–616 CrossRef PubMed.
- I. Marchal, A synthetic antibiotic overcomes antimicrobial resistance, Nat. Biotechnol., 2024, 42(3), 379 Search PubMed.
- A. L. K. Morgan, D. Moran and T. P. Van Boeckel, Taxation of veterinary antibiotics to reduce antimicrobial resistance, One Health, 2023, 17, 100650 CrossRef PubMed.
- X. He, Y. Qian, C. Wu, J. Feng, X. Sun, Q. Zheng, X. Li and J. Shen, Entropy-mediated high-entropy MXenes nanotherapeutics: NIR-II-enhanced intrinsic oxidase mimic activity to combat Methicillin-resistant Staphylococcus aureus infection, Adv. Mater., 2023, 35(26), e2211432 CrossRef PubMed.
- X. He, Y. Lv, Y. Lin, H. Yu, Y. Zhang, Y. Tong and C. Zhang, Platinum nanoparticles regulated V2C MXene nanoplatforms with NIR-II enhanced nanozyme effect for photothermal and chemodynamic anti-infective therapy, Adv. Mater., 2024, 36(25), e2400366 CrossRef PubMed.
- G. Qi, X. Liu, H. Li, Y. Qian, C. Liu, J. Zhuang, L. Shi and B. Liu, A dual-mechanism luminescent antibiotic for bacterial infection identification and eradication, Sci. Adv., 2025, 11(15), eadp9448 CrossRef CAS PubMed.
- A. Acosta, W. Tirkaso, F. Nicolli, T. P. Van Boeckel, G. Cinardi and J. Song, The future of antibiotic use in livestock, Nat. Commun., 2025, 16(1), 2469 CrossRef CAS PubMed.
- E. Mahase, AMR: Increased vaccine use could cut global antibiotic use by 22%, WHO estimates, BMJ, 2024, 387, q2248 CrossRef PubMed.
- J. K. Martin, J. P. Sheehan, B. P. Bratton, G. M. Moore, A. Mateus, S. H. Li, H. Kim, J. D. Rabinowitz, A. Typas, M. M. Savitski, M. Z. Wilson and Z. Gitai, A Dual-mechanism antibiotic kills Gram-negative bacteria and avoids drug resistance, Cell, 2020, 181(7), 1518–1532 CrossRef CAS PubMed.
- S. Abdolmaleki, A. Aliabadi and S. Khaksar, Riding the metal wave: A review of the latest developments in metal-based anticancer agents, Coord. Chem. Rev., 2024, 501, 215579 CrossRef CAS.
- K. Almagambetova, K. Murugesan and M. Rueping, Transition-Metal and Photocatalyst-Free, Redox-Neutral Heteroarylation of C(sp3)–H Bonds, ACS Catal., 2024, 14(16), 12664–12670 CrossRef CAS.
- H. Y. Khan, M. F. Ansari, S. Tabassum and F. Arjmand, A review on the recent advances of interaction studies of anticancer metal-based drugs with therapeutic targets, DNA and RNAs, Drug Discovery Today, 2024, 29(7), 104055 CrossRef CAS PubMed.
- G. Moreno-Alcántar, P. Picchetti and A. Casini, Gold Complexes in Anticancer Therapy: From New Design Principles to Particle-Based Delivery Systems, Angew Chem. Int. Ed. Engl., 2023, 62(22), e202218000 CrossRef PubMed.
- M. He, Z. Ma, L. Zhang, Z. Zhao, Z. Zhang, W. Liu, R. Wang, J. Fan, X. Peng and W. Sun, Sonoinduced Tumor Therapy and Metastasis Inhibition by a Ruthenium Complex with Dual Action: Superoxide Anion Sensitization and Ligand Fracture, J. Am. Chem. Soc., 2024, 146(37), 25764–25779 CrossRef CAS PubMed.
- F. Song and X. Cheng, CLO25-070: Effects and Mechanisms of CD16+/−NK Cells on Ovarian Malignant Tumor Cells Platinum-Based Drug Responsiveness in Tumor Microenvironment, J. Natl. Compr. Cancer Network, 2025, 23(3.5), CLO25–CLO70 Search PubMed.
- P. S. Starha and R. Krikavová, Platinum(IV) and platinum(II) anticancer complexes with biologically active releasable ligands, Coord. Chem. Rev., 2024, 501, 215578 CrossRef CAS.
- R. Venkatesan, H. Xiong, Y. J. Yao, J. R. Nakkala, T. Zhou, S. F. Li, C. Y. Fan and C. Y. Gao, Immuno-modulating theranostic gold nanocages for the treatment of rheumatoid arthritis, Chem. Eng. J., 2022, 446, 136868 CrossRef CAS.
- J. Cebula, K. Fink, W. Goldeman, B. Szermer-Olearnik, A. Nasulewicz-Goldeman, M. Psurski, M. Cuprych, A. Kędziora, B. Dudek, G. Bugla-Płoskońska, M. Chaszczewska-Markowska, M. Gos, P. Migdał and T. M. Goszczyński, Structural Patterns Enhancing the Antibacterial Activity of Metallacarborane-Based Antibiotics, J. Med. Chem., 2023, 66(21), 14948–14962 CrossRef CAS PubMed.
- Y. Lin, H. Jung, C. A. Bulman, J. Ng, R. Vinck, C. O'Beirne, S. Zhong, M. S. Moser, N. Tricoche, R. Peguero, R. W. Li, J. F. Urban Jr, P. Le Pape, F. Pagniez, M. Moretto, T. Weil, S. Lustigman, K. Cariou, M. Mitreva, J. A. Sakanari and G. Gasser, Discovery of New Broad-Spectrum Anti-Infectives for Eukaryotic Pathogens Using Bioorganometallic Chemistry, J. Med. Chem., 2023, 66(23), 15867–15882 CrossRef CAS PubMed.
- S. Shrestha, B. Wang and P. K. Dutta, Commercial Silver-Based Dressings: In Vitro and Clinical Studies in Treatment of Chronic and Burn Wounds, Antibiotics, 2024, 13(9), 910 CrossRef CAS PubMed.
- M. A. Mofazzal Jahromi, P. Sahandi Zangabad, S. M. Moosavi Basri, K. Sahandi Zangabad, A. Ghamarypour, A. R. Aref, M. Karimi and M. R. Hamblin, Nanomedicine and advanced technologies for burns: Preventing infection and facilitating wound healing, Adv. Drug Delivery Rev., 2018, 123, 33–64 CrossRef CAS PubMed.
- L. M. Stabryla, K. A. Johnston, N. A. Diemler, V. S. Cooper, J. E. Millstone, S. J. Haig and L. M. Gilbertson, Role of bacterial motility in differential resistance mechanisms of silver nanoparticles and silver ions, Nat. Nanotechnol., 2021, 16(9), 996–1003 CrossRef CAS PubMed.
- R. Wang, T. P. Lai, P. Gao, H. Zhang, P. L. Ho, P. C. Woo, G. Ma, R. Y. Kao, H. Li and H. Sun, Bismuth antimicrobial drugs serve as broad-spectrum metallo-β-lactamase inhibitors, Nat. Commun., 2018, 9(1), 439 CrossRef PubMed.
- Y. Song, J. Wang, Y. Sun, S. Dong, G. Yu, W. Lin, Y. Xiong, Y. Tan, Y. Xiong, G. Jiang, J. Wang, X. Liao and L. Liu, Targeting bacterial efflux pump effectively enhances the efficacy of Ru-based antibacterial agents against Gram-negative pathogen, J. Inorg. Biochem., 2025, 263, 112772 CrossRef CAS PubMed.
- J. Wang, Y. Song, Z. Huang, W. Lin, G. Yu, Y. Xiong, G. Jiang, Y. Tan, J. Wang and X. Liao, Coupling a Virulence-Targeting Moiety with Ru-Based AMP Mimics Efficiently Improved Its Anti-Infective Potency and Therapeutic Index, J. Med. Chem., 2023, 66(18), 13304–13318 CrossRef CAS PubMed.
- A. Jabłońska-Wawrzycka, P. Rogala, G. Czerwonka, K. Gałczyńska, M. Drabik and M. Dańczuk, Ruthenium Complexes with 2-Pyridin-2-yl-1H-benzimidazole as Potential Antimicrobial Agents: Correlation between Chemical Properties and Anti-Biofilm Effects, Int. J. Mol. Sci., 2021, 22(18), 10113 CrossRef PubMed.
- F. Li, J. G. Collins and F. R. Keene, Ruthenium complexes as antimicrobial agents, Chem. Soc. Rev., 2015, 44(8), 2529–2542 RSC.
- R. Tao, Y. Lu, W. Xia, C. Zhang and C. Wang, Characterization and antibacterial activity of ruthenium-based shikimate cross-linked chitosan composites, Int. J. Biol. Macromol., 2022, 217, 890–901 CrossRef CAS PubMed.
- Q. Kong, W. Pan, H. Xu, Y. Xue, B. Guo, X. Meng, C. Luo, T. Wang, S. Zhang and Y. Yang, Design, Synthesis, and Biological Evaluation of Novel Pyrimido[4,5-b]indole Derivatives Against Gram-Negative Multidrug-Resistant Pathogens, J. Med. Chem., 2021, 64(12), 8644–8665 CrossRef CAS PubMed.
- P. Dhiman, S. Das, V. Pathania, S. Rawat, H. S. Nandanwar, K. G. Thakur and V. D. Chaudhari, Discovery of Conformationally Constrained Dihydro Benzo-Indole Derivatives as Metallo-β-Lactamase Inhibitors to Tackle Multidrug-Resistant Bacterial Infections, J. Med. Chem., 2025, 68(7), 7062–7081 CrossRef CAS PubMed.
- Y. Liu, Y. Cui, L. Lu, Y. Gong, W. Han and G. Piao, Natural indole-containing alkaloids and their antibacterial activities, Arch. Pharm., 2020, 353(10), e2000120 CrossRef PubMed.
- W. Cheng, T. Xu, L. Cui, Z. Xue, J. Liu, R. Yang, S. Qin and Y. Guo, Discovery of Amphiphilic Xanthohumol Derivatives as Membrane-Targeting Antimicrobials against Methicillin-Resistant Staphylococcus aureus, J. Med. Chem., 2023, 66(1), 962–975 CrossRef CAS PubMed.
- E. R. Rojas, G. Billings, P. D. Odermatt, G. K. Auer, L. Zhu, A. Miguel, F. Chang, D. B. Weibel, J. A. Theriot and K. C. Huang, The outer membrane is an essential load-bearing element in Gram-negative bacteria, Nature, 2018, 559(7715), 617–621 CrossRef CAS PubMed.
- X. Hu, D. Li, H. Li, Y. Piao, H. Wan, T. Zhou, M. Karimi, X. Zhao, Y. Li, L. Shi and Y. Liu, Reaction-Induced Self-Assembly of Polymyxin Mitigates Cytotoxicity and Reverses Drug Resistance, Adv. Mater., 2024, 36(36), e2406156 CrossRef PubMed.
- M. Sumini, C. R. Souza, G. J. S. Andrade, I. R. C. Oliveira, S. Scandorieiro, C. A. Tischer, R. K. T. Kobayashi and G. Nakazato, Cellulose Hydrogel with Hyaluronic Acid and Silver Nanoparticles: Sustained-Release Formulation with Antibacterial Properties against Pseudomonas aeruginosa, Antibiotics, 2023, 12(5), 873 CrossRef CAS PubMed.
- G. Kalın, E. Alp, A. Chouaikhi and C. Roger, Antimicrobial Multidrug Resistance: Clinical Implications for Infection Management in Critically Ill Patients, Microorganisms, 2023, 11(10), 2575 CrossRef PubMed.
- J. A. Finbloom, P. Raghavan, M. Kwon, B. N. Kharbikar, M. A. Yu and T. A. Desai, Codelivery of synergistic antimicrobials with polyelectrolyte nanocomplexes to treat bacterial biofilms and lung infections, Sci. Adv., 2023, 9(3), eade8039 CrossRef PubMed.
- L. Azizova, A. Al Dalaty, E. Brousseau, J. Birchall, T. Wilkinson, A. Sloan and W. N. Ayre, Antimicrobial release from a lipid bilayer titanium implant coating is triggered by Staphylococcus aureus alpha-haemolysin, Appl. Surf. Sci., 2024, 665, 160337 CrossRef CAS.
- Q. Peng, X. Tang, W. Dong, N. Sun and W. Yuan, A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism, Antibiotics, 2022, 12(1), 12 CrossRef PubMed.
- F. Khan, D. T. N. Pham, S. F Oloketuyi and Y. Kim, Antibiotics Application Strategies to Control Biofilm Formation in Pathogenic Bacteria, Curr. Pharm. Biotechnol., 2020, 21(4), 270–286 CAS.
- H. Liu, S. Li, C. S. Brennan and Q. Wang, Antimicrobial activity of Arg–Ser–Ser against the food-borne pathogen Pseudomonas aeruginosa, Int. J. Food Sci. Technol., 2020, 55(1), 379–388 CrossRef CAS.
- M. W. De Lima Gualque, C. O. Vaso, K. S. Dos Santos, M. C. Galeane, P. C. Gomes, M. S. Palma, M. J. Soares Mendes Giannini, A. Moroz and A. M. Fusco Almeida, Peptides from Galleria mellonella against Cryptococcus spp: toxicity in three-dimensional cell cultures and G. mellonella, Future Microbiol., 2025, 20(1), 11–21 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra03098f |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |