Open Access Article
Open Access ArticleInhibitory mechanism of pterostilbene and pinostilbene on aldose reductase and α-glucosidase: a new insight from inhibition kinetics and molecular docking studies
Lili Jiang,
Yanyan Jia,
Hang Yin and
Yong Liu *
*
School of Chemical Engineering, Ocean and Life Sciences, Dalian University of Technology, No. 2 Dagong Road, Liaodongwan New District, Panjin 124221, Liaoning, China. E-mail: yliu@dlut.edu.cn; Tel: +86-04272631433
First published on 18th July 2025
Abstract
Pterostilbene, a dietary polyphenol in blueberries, grapes and wine, possesses an anti-diabetic efficacy. However, the mechanism of anti-diabetic efficacy for pterostilbene is poorly understood. The purpose of the present study was to systematically investigate inhibitory effects and inhibition mechanisms of pterostilbene or its metabolite (pinostilbene) against aldose reductase and α-glucosidase. In the present study, inhibition kinetics and molecular docking were applied to investigate the effect of pterostilbene and pinostilbene on the activity of aldose reductase and α-glucosidase. The results showed that pinostilbene exhibited stronger inhibition on aldose reductase (IC50 = 55.41 ± 4.31 μM) and α-glucosidase (IC50 = 11.69 ± 0.888 μM) in noncompetitive-type manner. Whereas, pterostilbene displayed the weak effect on aldose reductase and α-glucosidase (IC50 = 1569 ± 177.858 μM) inhibition. Moreover, pinostilbene interacted with amino acid residues outside of aldose reductase and α-glucosidase active site to inhibit its activities by van der Waals, π–anion or π–alkyl interactions. The current study has provided comprehensive reference not only for the anti-diabetic mechanism of pterostilbene, but also offered a basis to develop and design adjuvant antidiabetic drugs or novel functional foods.
1 Introduction
Diabetes mellitus is characterized by hyperglycemia and associated with disorders in metabolism resulting from insufficient insulin. It is a group of metabolic chronic diseases, affecting millions of individuals globally.1,2 Long-term uncontrolled hyperglycemia can cause cellular and tissue damage, and lead to major complications like retinopathy, atherosclerosis, neuropathy, nephropathy.3,4Aldose reductase plays a crucial role in the polyol pathway, converting glucose to sorbitol. It is considered to be associated with the development of diabetic complications.5 α-Glucosidase, a cell membrane bound enzyme, situates in epithelium cells of small intestines, whose function is to hydrolyze polysaccharides.6 Thus, due to their significant role in diabetic complications, carbohydrate digestion, and postprandial hyperglycemia, aldose reductase and α-glucosidase have received a great deal of attention. Inhibitions of them are considered to be a crucial diabetes management by delaying the digestion and absorption of disaccharides and starch, preventing and attenuating diabetic complications. In addition, aldose reductase and α-glucosidase inhibitors, for example epalrestat, acarbose, voglibose, and miglitol, have been selected to control blood glucose levels and prevent complications in diabetes mellitus patients.7–9 However, the extensive usage of these drugs has been associated with various side effects, such as abdominal pain, flatulence, hepatic injury, acute hepatitis, abdominal fullness, renal tumors, or diarrhea.5,10,11 Therefore, foodborne aldose reductase and α-glucosidase inhibitors, known to be safe, could provide an alternative and complementary treatment for diabetes mellitus.
Natural polyphenol pterostilbene (Fig. 1A) is widely occurring in our daily diets, for instance huckleberries, grapes, blueberries, peanuts, or red wine.12 As with its well-known precursor, resveratrol, pterostilbene exhibited numerous beneficial biological activities, such as antiaging, antiinflammation, anti-cancer, anti-diabetic, anti-obesity, anti-oxidation, and cholesterol lowering.13–18 Moreover, pterostilbene proved to be higher oral bioavailability than resveratrol, making it a promising dietary supplement or potential therapeutic agent that could combat diabetes mellitus in humans.19
Animal studies have demonstrated that pterostilbene could significantly reduce glycosylated hemoglobin and blood glucose concentration in rats with diabetes mellitus.20,21 Additionally, it was also demonstrated that pterostilbene improved glycemic control of rats fed an obesogenic diet by regulating hepatic glucokinase activities.22 However, the mechanism of anti-diabetic efficacy for pterostilbene is poorly understood, especially regarding the inhibitory effects of pterostilbene against aldose reductase and α-glucosidase activities. In addition, pinostilbene (Fig. 1B), a major bioactive metabolite of pterostilbene,23,24 underwent complex physiological and biochemical processes, also played an important role in overall biological activities of small intestine.
Thus, the purpose of present study was to investigate the inhibitory effects and inhibition mechanisms of pterostilbene and pinostilbene against the activities of aldose reductase and α-glucosidase relevant to diabetes mellitus. The enzyme kinetic analysis as well as molecular docking methods were used in this study. The current study provides comprehensive reference not only for the anti-diabetic mechanism of pterostilbene, but also offers a basis to develop and design new functional foods or adjuvant antidiabetic drugs.
2 Materials and methods
2.1 Reagents
The 4-nitrophenyl-β-D-glucopyranosiduronic acid (pNPG), α-glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20), DL-glyceraldehyde and acarbose were purchased from Sigma-Aldrich (St, Louis, USA). Pterostilbene and pinostilbene were obtained from Selleck Chemicals (USA). Epalrestat, lithium sulfate and p-nitrophenol (pNP) were from Aladdin Biotechnology (Shanghai, China). Nicotinamide adenine dinucleotide phosphate (NADPH) was from Yuanye Bio-Technology Company (Shanghai, China). The analytical reagent grade was used for all other chemicals and reagents. Aqueous solutions were prepared through freshly ultrapure water.2.2 Enzyme inhibition assay
| Relative activity (%) = (1 − Asample/Ablank) × 100% |
| Relative activity (%) = (1 − Asample/Ablank) × 100% |
The positive controls were acarbose and epalrestat, which are both known aldose reductase and α-glucosidase inhibitors. The half maximal inhibitory concentration of inhibitor (IC50) was measured by nonlinear regression analysis of relative activity (%) of enzymes versus inhibitor concentration ([I]).
2.3 Inhibitory kinetic assay
The underlying inhibition mechanisms were probed by the inhibitory kinetic assay. Numerous concentrations of pNPG and DL-glyceraldehyde as substrate in the absence or presence of pinostilbene were measured. The inhibition mode against aldose reductase and α-glucosidase was explored by kinetic parameters analysis. Briefly, the data were analyzed using the nonlinear regression Michaelis–Menten enzyme kinetics of the Lineweaver–Burk plot and Dixon plot. The kinetic parameters were calculated using formulas for competitive inhibition (eqn (1)), noncompetitive inhibition (eqn (2)), uncompetitive (eqn (3)), or mixed inhibition (eqn (4)).| v = (VmaxS)/(Km(1 + I/Ki) + S) | (1) |
| v = (VmaxS)/(Km + S)(1 + I/Ki) | (2) |
| v = (VmaxS)/(Km+(1 + I/Ki)S) | (3) |
| v = (VmaxS)/[Km(1 + I/Ki) + S(1 + I/αKi)] | (4) |
Secondary plots can be calculated from
| Slop = (Km/Vmax) + (Km[I]/VmaxKi) | (5) |
2.4 Molecular docking analysis
Molecular docking was conducted to explore the effects of pinostilbene against α-glucosidase. 3D structure of pinostilbene (3-methoxy-4′,5-dihydroxy-trans-stilbene, CAS: 42438-89-1) were prepared from PubChem database, and further optimized in AutoDockTools software. The crystal structures of α-glucosidase (PDB ID: 3TOP) and aldose reductase (PDB ID: 1US0) were obtained from Protein Data Bank (https://www.rcsb.org/). Before docking, water molecules and original ligands were removed from the target proteins. Then, charge calculation, hydrogenation, and distribution as well as atom type specification were performed using AutoDockTools. AutoDock and Discovery Studio 2016 were used to estimate binding affinity between pinostilbene and enzyme.2.5 Statistical analysis
The data were subjected to nonlinear regression, the results were expressed as the mean ± standard deviation (SD). In addition, the data were analyzed by one-way analysis of variance. p < 0.05 was considered as statistically significant.3 Results
3.1 Inhibition of pterostilbene and pinostilbene on α-glucosidase activity
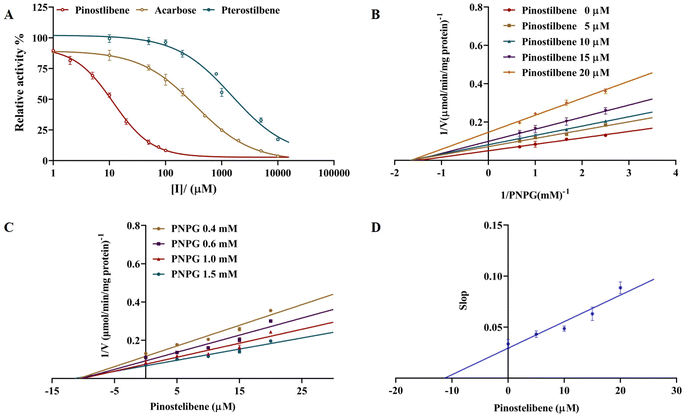
The inhibition of three compounds (pterostilbene, pinostilbene and acarbose) against α-glucosidase activity was shown in Fig. 2A. As presented in Fig. 2A, significant differences on the inhibitions of α-glucosidase in each compound were observed. Pinostilbene exhibited the high effect on α-glucosidase inhibition with IC50 value of 11.69 ± 0.888 μM (95% CI: 9.593 to 15.01), and its inhibitory effect was better than acarbose (IC50 = 342.8 ± 20.702 μM) (95% CI: 265.6 to 402.8). Whereas pterostilbene displayed the weak effect on α-glucosidase inhibition with IC50 value of 1569 ± 177.858 μM (95% CI: 939.2 to 1861). Our results revealed that not pterostilbene but its metabolite (pinostilbene) exhibited a more potent inhibition against α-glucosidase activities than that of acarbose.3.2 Enzymatic kinetics of α-glucosidase inhibition
The inhibitory type of pinostilbene against α-glucosidase was performed by the calculation of Km and Vmax via using the nonlinear regression Michaelis–Menten enzyme kinetics of Dixon plots and Lineweaver–Burk plots. As observed Fig. 2B and C (Lineweaver–Burk and Dixon plots), all lines possessed the different slopes, and dropped with the increase in pinostilbene concentration. Vmax value decreased significantly, while the Km value was fixed. These results suggested that pinostilbene exhibited a noncompetitive-type inhibition against α-glucosidase. The Ki value was estimated to be 14.28 ± 1.492 μM by the eqn (2). Meanwhile, the secondary re-plot of the apparent slop versus pinostilbene (R2 = 0.9210) was linearly fitted, suggesting that pinostilbene probably bound to α-glucosidase at a single site or a single inhibition-site class outside of the active sites, but not directly in the active site pockets (Fig. 2D).3.3 Inhibition of pterostilbene and pinostilbene on aldose reductase activity
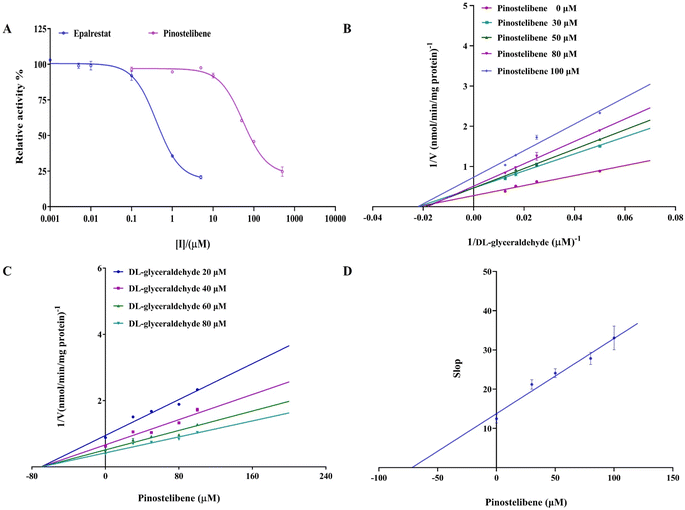
The inhibitory effects of pinostilbene and epalrestat on aldose reductase activity were detected and the results were shown in Fig. 3A. As seen in Fig. 3A, significantly different inhibitions of pinostilbene and pterostilbene on aldose reductase were observed. Pinostilbene exhibited a high effect on aldose reductase inhibition with IC50 value of 55.41 ± 4.31 μM (95% CI: 45.99 to 64.99), though its inhibitory effect was lower than that of epalrestat (IC50 = 0.413 ± 0.004 μM) (95% CI: 0.3055 to 0.5514). Whereas, pterostilbene displayed a weak effect on aldose reductase inhibition (the data was not shown). Our results revealed that pinostilbene (a metabolite of pterostilbene) exhibited a potent inhibition against aldose reductase activities.3.4 Enzymatic kinetics of aldose reductase inhibition
In order to deepen the knowledge of pinostilbene's mode of inhibition against aldose reductase, the nonlinear regression Michaelis–Menten enzyme kinetics of Dixon plots and Lineweaver–Burk plots were performed for calculating values of apparent kinetic parameters (Km and Vmax). As observed in Fig. 3B and C (Lineweaver–Burk and Dixon plots), all lines possessed the different slopes, and dropped with the increase in pinostilbene concentration. Vmax value decreased significantly, while the Km value was fixed. Based on these results, it was evident that the inhibition of pinostilbene against aldose reductase occurred through a noncompetitive-type inhibition mechanism, with a Ki value of 60.02 ± 4.636 μM. Meanwhile, as it can be noted in Fig. 3D, the linear secondary re-plot of the apparent slop versus pinostilbene (R2 = 0.9718) was observed, suggesting that pinostilbene probably bound to aldose reductase at a single site or a single inhibition-site class outside of the active sites, but not directly in the active site pockets.3.5 The binding site prediction
To predict the binding mode for pinostilbene on aldose reductase and α-glucosidase, molecular docking was used. As shown in Fig. 4, pinostilbene apparently showed good fit and strong binding to the reactive catalytic pocket which lied outside of the active site pockets of enzymes, surrounded by parts of the catalytic residues Val1035, Asp1056, Lys1059, Arg1061, Tyr1062, Asp1117, Gly1209 for α-glucosidase (binding energy = −6.72 kcal mol−1), Phe161, Glu193, Lys194, Leu195, Arg296, Pro310 for aldose reductase (binding energy = −9.38 kcal mol−1), respectively. van der Waals were formed between the hydroxyl group, methoxy group of pinostilbene and the active site the residue Lys1059 (5.07 Å), Tyr1062 (2.08 Å) of α-glucosidase or Glu193 (2.0 Å), Arg296 (2.94 Å), Pro310 (2.01 Å) of aldose reductase respectively (Fig. 4B and D). Furthermore, pinostilbene also interacted with Val1035 (5.14 Å), Asp1056 (4.17 Å), Arg1061 (5.07 Å), Asp1117 (4.62 Å) of α-glucosidase and Lys194 (4.63 Å), Leu195 (4.76 Å) of aldose reductase by π–anion or π–alkyl interactions (Fig. 4B and D), indicating that the π–anion and π–alkyl also played important roles in the binding of pinostilbene with aldose reductase or α-glucosidase. The docking prediction suggested that pinostilbene might interact with the amino acid residues outside of the aldose reductase or α-glucosidase active sites to inhibit their activities, which was accordance with noncompetitive-type inhibitory modes of the experimental results shown above.4 Discussion
In past decades, there have been increasing interests in natural polyphenols as therapeutic compounds especially in the prevention or treatment of diabetes.30–32 Pterostilbene, one of the most abundant natural polyphenols, is a common constituent of the average everyday diet, which is known to have diverse pharmacological benefits for the prevention and treatment of various diseases, for instance inflammation, cancer, diabetes mellitus, and dyslipidemia.18,33 Among various beneficial effects, influence on the controlling hyperglycemia associated with diabetes mellitus was through a variety of mechanisms in vivo and in vitro studies. In vivo, pterostilbene exerted its hypoglycemic effects by ether increasing glycolysis or decreasing gluconeogenesis.21 Additionally, pterostilbene applied its hypoglycemic activity by controlling insulin secretion of the residual islet β-cells34 or inhibition of the islet β-cells apoptosis.35 Our data offer in vitro evidence that pterostilbene may be connected with the inhibition of aldose reductase and α-glucosidase leading to diabetes mellitus.Interestingly, our results showed that not pterostilbene but its metabolite (pinostilbene) exhibited more potent inhibitory activity against α-glucosidase than that of acarbose which is a clinical drug used in diabetes mellitus treatment. Importantly, pinostilbene, a major demethylated metabolite of pterostilbene, possessed a relatively high abundance which was even comparable to that of pterostilbene in colonic content and colonic mucosa.24 Therefore, after oral administration of pterostilbene, its metabolite (pinostilbene) may significantly contribute to the anti-diabetic efficacy, such as inhibition against aldose reductase or α-glucosidase activity. This study also inspired that we could combine the pterostilbene metabolites with its bioactivities in a follow-up study.
Our molecular docking demonstrated that the binding sites of pinostilbene were at hydrophobic pockets outside of the catalytic sites of aldose reductase or α-glucosidase. This result was consistent with that from enzymatic kinetics of aldose reductase and α-glucosidase inhibition assay. Our data give an in-depth understanding of the mechanisms, which can help us to determine the potential therapeutic application of pinostilbene.
In addition, our results found that pinostilbene bound directly to aldose reductase or α-glucosidase outside of the active sites, which was different from the binding pockets of acarbose or epalrestat.36 These different active sites can provide an insight into the inhibition of aldose reductase or α-glucosidase, allowing identification of new target sites which was different from that of other polyphenols.32,37
Taken together, the results complied herein can serve as a comprehensive reference for the anti-diabetic mechanisms of pinostilbene. We also advance it as a future adjuvant therapeutic agent for diabetes mellitus. Meanwhile, the findings in present study will be useful to develop or design novel alpha a-glucosidase or aldose reductase inhibitors. Of course, in vitro cellular experiments and further molecular dynamics simulations are needed to elucidate its in-depth mechanisms or to adjust its dose dependent effects in vivo.
5 Conclusions
The present study focused on the mechanism of pterostilbene controlling hyperglycemia associated with diabetes mellitus in vitro, and provided useful information for researchers, i.e., not pterostilbene but its metabolite (pinostilbene) exhibited more potent inhibitory effect against aldose reductase or α-glucosidase activity. Enzymatic kinetics of aldose reductase and α-glucosidase inhibition assay showed that pinostilbene led to a noncompetitive-type inhibition and likely bound directly to aldose reductase or α-glucosidase outside of the active site, which was consistent with that from the molecular docking. The current results provided comprehensive reference not only for the anti-diabetic mechanism of pterostilbene, but also offered a basis to develop and design new functional foods or adjuvant antidiabetic drugs.Data availability
All data included in this study are available upon request by contact with the corresponding authors (Yong Liu, E-mail: yliu@dlut.edu.cn).Author contributions
Formal Analysis: Lili Jiang. Funding acquisition: Yong Liu. Investigation: Lili Jiang, Yanyan Jia. Methodology: Lili Jiang, Yanyan Jia, Hang Yin. Project administration: Yong Liu. Writing – original draft: Lili Jiang. Writing – review & editing: Yong Liu.Conflicts of interest
The authors declare that there is no conflict of interests regarding the publication of this article.Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (82274006).References
- M. Nauck, C. Gerdes, A. Petersmann, D. Mueller-Wieland, U. A. Mueller, G. Freckmann, L. Heinemann, E. Schleicher and R. Landgraf, Diabetologe, 2021, 17(4), 404–410 CrossRef.
- World Health Organization, 2016, available from: https://www.who.int/diabetes/global-report, accessed February 11, 2019.
- A. Mitra, D. Dewanjee and B. Dey, World J. Diabetes, 2012, 3, 201–207 CrossRef PubMed.
- D. M. Nathan, N. Engl. J. Med., 1993, 328, 1676–1685 CrossRef CAS PubMed.
- A. M. Warren, S. T. Knudsen and M. E. Cooper, Expert Opin. Ther. Targets, 2019, 23, 579–591 CrossRef PubMed.
- W. F. Caspary, Lancet, 1978, 1, 1231–1233 CrossRef CAS PubMed.
- K. Kaku, Expet Opin. Pharmacother., 2014, 15, 1181–1190 CrossRef CAS PubMed.
- L. J. Scott and C. M. Spencer, Drugs, 2000, 59, 521–549 CrossRef CAS PubMed.
- A. E. Martin and P. A. Montgomery, Am. J. Health-Syst. Pharm., 1996, 53, 2277–2290 CrossRef CAS PubMed.
- R. Sudhir and V. Mohan, Treat. Endocrinol., 2002, 1, 105–116 CrossRef PubMed.
- K. R. Rengasamy, M. A. Aderogba, S. O. Amoo, W. A. Stirk and J. Van Staden, Food Chem., 2013, 141, 1412–1415 CrossRef CAS PubMed.
- A. M. Rimando, W. Kalt, J. B. Magee, J. Dewey and J. R. Ballington, J. Agric. Food Chem., 2004, 52, 4713–4719 CrossRef CAS PubMed.
- M. Dvorakova and P. Landa, Pharmacol. Res., 2017, 124, 126–145 CrossRef CAS PubMed.
- Y. R. Li, S. Li and C. C. Lin, Biofactors, 2018, 44, 69–82 CrossRef CAS PubMed.
- M. H. Pan, J. C. Wu, C. T. Ho and C. S. Lai, Biofactors, 2018, 44, 50–60 CrossRef CAS PubMed.
- K. W. Lange and S. Li, Biofactors, 2018, 44, 83–90 CrossRef CAS PubMed.
- W. Nawaz, Z. Zhou, S. Deng, X. Ma, X. Ma, C. Li and X. Shu, Nutrients, 2017, 9(11), 1188 CrossRef PubMed.
- H. Y. Tsai, C. T. Ho and Y. K. Chen, J. Food Drug Anal., 2017, 25, 134–147 CrossRef CAS PubMed.
- R. Kosuru, U. Rai, S. Prakash, A. Singh and S. Singh, Eur. J. Pharmacol., 2016, 789, 229–243 CrossRef CAS PubMed.
- M. Manickam, M. Ramanathan, M. A. F. Jahromi, J. P. N. Chansouria and A. B. Ray, J. Nat. Prod., 1997, 60, 609–610 CrossRef CAS PubMed.
- L. Pari and M. A. Satheesh, Life Sci., 2006, 79, 641–645 CrossRef CAS PubMed.
- S. Gomez-Zorita, A. Fernandez-Quintela, L. Aguirre, M. T. Macarulla, A. M. Rimando and M. P. Portillo, Food Funct., 2015, 6, 1968–1976 RSC.
- X. Shao, X. Chen, V. Badmaev, C. T. Ho and S. Sang, Rapid Commun. Mass Spectrom., 2010, 24, 1770–1778 CrossRef CAS PubMed.
- Y. Sun, X. Wu, X. Cai, M. Song, J. Zheng, C. Pan, P. Qiu, L. Zhang, S. Zhou, Z. Tang and H. Xiao, Mol. Nutr. Food Res., 2016, 60, 1924–1932 CrossRef CAS PubMed.
- J. Q. Zhao, Y. M. Wang, Y. L. Yang, Y. Zeng, Q. L. Wang, Y. Shao, L. J. Mei, Y. P. Shi and Y. D. Tao, Food Chem., 2017, 227, 93–101 CrossRef CAS PubMed.
- E. Apostolidis, Y. I. Kwon and K. Shetty, Innov. Food Sci. Emerg. Technol., 2007, 8, 46–54 CrossRef CAS.
- S. Hayman and J. Kinoshita, J. Biol. Chem., 1965, 240, 877 CrossRef CAS PubMed.
- M. N. Islam, I. J. Ishita, H. A. Jung and J. S. Choi, Food Chem. Toxicol., 2014, 69, 55–62 CrossRef CAS PubMed.
- A. R. Rao, C. Veeresham and K. Asres, Phytother Res., 2013, 27, 753–760 CrossRef CAS PubMed.
- J. B. Yang, J. Y. Tian, Z. Dai, F. Ye, S. C. Ma and A. G. Wang, Fitoterapia, 2017, 117, 65–70 CrossRef CAS PubMed.
- M. Li, X. Wu, X. Wang, T. Shen and D. Ren, Nat. Prod. Res., 2018, 32, 36–42 CrossRef CAS PubMed.
- L. Jiang, Z. Wang, X. Wang, S. Wang, J. Cao and Y. Liu, RSC Adv., 2020, 10, 4529–4537 RSC.
- Y. Liu, Y. You, J. Lu, X. Chen and Z. Yang, Molecules, 2020, 25(21), 5166 CrossRef CAS PubMed.
- B. Elango, S. Dornadula, R. Paulmurugan and K. M. Ramkumar, Chem. Res. Toxicol., 2016, 29, 47–57 Search PubMed.
- S. Paul, A. M. Rimando, H. J. Lee, Y. Ji, B. S. Reddy and N. Suh, Cancer Prev. Res., 2009, 2, 650–657 Search PubMed.
- K. Bharatham, N. Bharatham, K. H. Park and K. W. Lee, J. Mol. Graphics Modell., 2008, 26, 1202–1212 CrossRef CAS PubMed.
- X. Wu, H. Ding, X. Hu, J. Pan, Y. Liao, D. Gong and G. Zhang, J. Funct.Foods, 2018, 48, 200–209 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2025 |