Open Access Article
Open Access ArticleExploring the potential molecular targets and therapeutic mechanisms of Cyperi Rhizoma in treating diabetic cardiomyopathy: a computational approach†
Su-Rui Lu‡
a,
Zi-Xiang Fu‡a,
Yan Xia‡a,
Yan-Mei Xua,
Feng-Liang Wangc,
Zhao-Wen Pana,
Yi-Wei Tua,
Wen-Jing Penga,
Ming-Li Hana,
Wei Yu*a and
Wen-Liang Zha *ab
*ab
aSchool of Pharmacy, Xianning Medical College, Hubei University of Science and Technology, Xianning 437100, China. E-mail: xyzwl800@163.com; yuwei0805@163.com
bSecond Affiliated Hospital, Xianning Medical College, Hubei University of Science and Technology, Xianning 437100, China
cMinda Hospital of Hubei Minzu University, En Shi 445000, China
First published on 8th July 2025
Abstract
Diabetic cardiomyopathy (DCM) is a major contributor to heart failure in diabetic patients, characterized by complex pathophysiological mechanisms and a lack of effective targeted therapies. Cyperi Rhizoma (CR), a traditional Chinese medicinal herb, has shown potential in regulating Qi and improving metabolic disorders, yet its therapeutic mechanisms in DCM remain unclear. This study systematically explored the bioactive components and underlying mechanisms of CR in the treatment of DCM by integrating network pharmacology, molecular docking, molecular dynamics simulation (MDS), and in vitro experiments. Active compounds of CR were identified, and DCM-related targets were obtained from GEO, DisGeNET, OMIM, and GeneCards databases. Quercetin, luteolin, and patchoulenone were identified as key compounds, while AKT1, IL6, TNF, IL1B, and TP53 emerged as core targets. Functional enrichment analysis indicated that the AGE-RAGE and TNF signaling pathways may play critical roles in the protective effects of CR against DCM. Molecular docking and MDS demonstrated a stable binding interaction between luteolin and AKT1. Furthermore, in vitro validation confirmed the protective effect of luteolin against high glucose and palmitic acid-induced injury in H9c2 cardiomyocytes. Collectively, this study elucidates the multi-level pharmacological basis and molecular mechanisms of CR in mitigating DCM, providing a theoretical and experimental foundation for its potential therapeutic application.
1. Introduction
Diabetes cardiomyopathy (DCM) is a condition that affects the heart structurally and functionally as a result of diabetes.1 The hallmark of DCM is abnormal functioning of the left ventricle, which is distinct from traditional cardiovascular disorders such as coronary artery disease or hypertension.2 With the escalating global prevalence of diabetes, DCM has emerged as a major contributor to diabetes-related morbidity and mortality.3 The disease typically manifests initially as diastolic dysfunction, progressing to systolic dysfunction in advanced stages, which significantly exacerbates mortality and disability rates.4 Although substantial progress has been made in the development of therapeutic strategies aimed at glycemic control and the symptomatic management of heart failure, targeted pharmacological interventions that effectively halt or reverse the progression of DCM remain elusive. This underscores the critical imperative to explore novel therapeutic strategies and elucidate the underlying molecular mechanisms to address this unmet medical need.As a part of traditional Chinese medicine (TCM), Cyperi Rhizoma (CR) has been extensively employed for centuries to address a diverse spectrum of health conditions, owing to its multifaceted pharmacological properties.5 Rooted in TCM theory, CR is traditionally ascribed with the functions of alleviating pain, promoting blood circulation, harmonizing Qi flow, and exerting hepatoprotective effects.6 Its therapeutic applications span a range of disorders, including but not limited to, menstrual irregularities, gastric discomfort, and thoracic and abdominal distension.7 Lately, CR has garnered extensive attention in contemporary pharmacological investigations in light of its diverse therapeutic potential, with emerging evidence highlighting its regulatory potential in the management of diabetes.8 Notably, emerging studies have suggested that CR may hold therapeutic promise in diabetes management, with evidence pointing to its role in improving glucose metabolism, reducing insulin resistance, and mitigating diabetic complications. However, despite preliminary findings suggesting cardioprotective benefits, the precise molecular mechanisms through which CR exerts its effects on DCM remain inadequately understood. This identified knowledge gap underscores the need for systematic investigation of CR's pharmacodynamics, molecular targets, and mechanistic pathways in DCM pathophysiology to facilitate its translation into evidence-based clinical applications.
Modern drug target research increasingly relies on advanced computational methodologies, with network pharmacology, molecular docking, as well as molecular dynamics simulations (MDS) serving as complementary and indispensable tools for elucidating drug mechanisms, identifying therapeutic targets, and analyzing molecular interactions.9 The multi-target drug analysis platform integrates comprehensive data on drugs, targets, diseases, and associated signaling pathways, facilitating the construction of drug-target-disease network models. This systematic approach enables the exploration of the multi-target effects of drugs and the prediction of potential therapeutic pathways and biological outcomes through large-scale data analysis.10 Molecular docking, a key simulation technology, predicts the interaction patterns, binding sites, and binding affinity between drug molecules and target proteins, thereby providing a critical foundation for drug discovery and development.11 Building on molecular docking, MDS further simulates the dynamic interactions and kinetic properties of drug-target complexes under varying environmental conditions, offering deeper structural insights and validating the stability of predicted interactions.12 These computational technologies collectively represent a paradigm shift in drug research, enabling a more precise and comprehensive understanding of drug action mechanisms on a molecular scale.
To shed light on the bioactive constituents and therapeutic targets of CR within the context of DCM, this study employs an integrative research strategy that synergistically combines computational pharmacology approaches, ligand-target interaction modeling, MDS, and in vitro cellular assays. Computational pharmacology will be utilized to identify bioactive compounds and their corresponding targets, while ligand-target interaction modeling and MDS will provide detailed insights into the binding mechanisms and dynamic stability of these interactions. Additionally, in vitro experiments using H9c2 cardiomyocytes will be conducted to validate the cardioprotective effects of the identified bioactive compound under pathological conditions. The overarching objective of this research is to uncover novel drug candidates and therapeutic strategies for the clinical management of DCM. Fig. 1 describes a schematic illustration of the integrated methodology outlined in the preceding sections.
2. Experimental procedures
2.1. Database URLs and software
TCMSP (https://old.tcmsp-e.com/tcmsp.php); BATMAN-TCM (https://bionet.ncpsb.org.cn/batman-tcm/index.php/Home/Index/target/jobId/batman-I2024-05-26-86050-1716690714/cutoff/20/pVal/0.05); Swiss Target Prediction (https://swisstargetprediction.ch/); Uniprot (https://www.uniprot.org/); GEO (https://www.ncbi.nlm.nih.gov/geo/); DisGenet (https://www.disgenet.org/); Gene Cards (https://www.genecards.org/); OMIM (https://www.omim.org/); jvenn (https://www.bioinformatics.com.cn/static/others/jvenn/example.html); STRING (https://cn.string-db.org/cgi/); David (https://david.ncifcrf.gov/home.jsp); Wechat (https://www.bioinformatics.com.cn/); PubChem (https://pubchem.ncbi.nlm.nih.gov/); Protein Data Bank (PDB) (https://www.rcsb.org/); gmx_mmpbsa (https://jerkwin.github.io/gmxtool); Software Cytoscape3.9.1; AutoDock Vina1.1.2; PyMOL2.4.1; PyMOL2.5.0; VMD1.9.3.2.2 Network-based pharmacological investigation
2.3 Molecular docking
Three-dimensional structures of target proteins and small molecules were retrieved from the PubChem and PDB databases.23 Molecular docking was carried out with the help of AutoDock Vina to determine the optimal binding conformation exhibiting a minimum binding energy. The resulting complexes were displayed and analyzed using PyMol.242.4 MDS
MDS were conducted using GROMACS 2020.3 to investigate protein–ligand interactions.25 The amber99sb-ildn and GAFF force fields were utilized in the generation of parameters and topologies for the protein and ligand, respectively. A simulation box was created with protein atoms positioned at least 1.0 nm from the edges, filled with SPC216 water molecules, and neutralized using Na+ and Cl− counterions. The system was energy-minimized using the steepest descent method and equilibrated through 100-ps NVT and NPT ensemble simulations at 300 K and 1 bar. A 50 ns production simulation was performed under periodic boundary conditions, with temperature and pressure maintained using the V-rescale and Parrinello-Rahman algorithms.26 A 2 fs time step was considered for leapfrog integration, while long-range electrostatic interactions were computed using the Particle Mesh Ewald (PME) method. Bond lengths were constrained by LINCS algorithm. Trajectories were visualized and analyzed with VMD 1.9.3 and PyMOL 2.4.1, and binding free energies were measured using gmx_mmpbsa.272.5 Cell viability assay using CCK-8
Well-cultured H9c2 cardiomyocytes were harvested, digested, centrifuged, and resuspended, followed by cell counting and seeding into 96-well culture plates at a density of 5 × 103 cells per well.28 After cell adherence, the medium was replaced with drug-containing medium at predetermined concentrations for each experimental group. The cells were incubated for 48 hours under standardized conditions. Subsequently, CCK-8 solution (10%) was added to each well and exposed for an additional 2 to 4 hours. A 450 nm absorbance was measured using a multifunctional microplate reader to assess cell viability.2.6 Detection of reactive oxygen species (ROS) by DHE staining
H9c2 cells were grown in culture and supplemented with drug-containing medium at specified concentrations for 48 hours. Following three washes with PBS, the cells were fixed in 300 liters of chilled methanol for ten minutes, followed by another three PBS washes. To facilitate the permeabilization of the cell membranes, 0.1% Triton X-100 was added for five minutes, followed by additional washes with PBS. Next, 500 μL of 0.1% DIPA staining solution was applied and incubated for 5 minutes to pre-stain the cells. After PBS washing, 500 μL of 0.1% DHE solution was added, followed by incubation in the dark for 30 min. As a final step, intracellular ROS levels were evaluated by using a fluorescence microscope.3. Results
3.1 Effective active components and related targets of CR
A total of eighteen bioactive compounds were identified by searching the TCMSP database using the keyword “Cyperi Rhizoma,” as summarized in Table 1. The corresponding molecular targets of these active ingredients were mapped to human gene names using the UniProt database, and duplicate entries were removed, resulting in the final identification of 10 active compounds associated with 172 potential targets. Similarly, querying the Bat ANT-TCM database with the same keyword yielded seven active compounds and 142 potential targets, as shown in Table 2. Further target prediction was performed using the Swiss Target Prediction tool, applying a probability threshold of >0.7, which led to the identification of 93 additional potential targets. After merging and eliminating redundancies among the targets obtained from the three databases, a total of 376 unique targets were identified (Fig. 2).| Molecular ID | Molecule name | OB (%) | DL |
|---|---|---|---|
| 003044 | Chryseriol | 35.85 | 0.27 |
| 000354 | Isorhamnetin | 49.6 | 0.31 |
| 003542 | 8-Isopentenyl-kaempferol | 38.04 | 0.39 |
| 000358 | Beta-sitosterol | 36.91 | 0.75 |
| 000359 | Sitosterol | 36.91 | 0.75 |
| 004027 | 1,4-Epoxy-16-hydroxyheneicos-1,3,12,14,18-pentaene | 45.1 | 0.24 |
| 004053 | Isodalbergin | 35.45 | 0.2 |
| 004058 | Khell | 33.19 | 0.19 |
| 004059 | Khellol glucoside | 74.96 | 0.72 |
| 010489 | Resivit | 30.84 | 0.27 |
| 004068 | Rosenonolactone | 79.84 | 0.37 |
| 004071 | Hyndarin | 73.94 | 0.64 |
| 004074 | Stigmasterol glucoside_qt | 43.83 | 0.76 |
| 004077 | Sugeonyl acetate | 45.08 | 0.2 |
| 000422 | Kaempferol | 41.88 | 0.24 |
| 000449 | Stigmasterol | 43.83 | 0.76 |
| 000006 | Luteolin | 36.16 | 0.25 |
| 000098 | Quercetin | 46.43 | 0.28 |
| Compound | Score |
|---|---|
| Copadiene | 48 |
| Patchoulenone | 80.882 |
| Cyperolone | 48 |
| Alpha-cyperone | 48 |
| Cyperol | 55.444 |
| Isocyperol | 48 |
| Isokobusone | 48 |
3.2 Screening of targets for DCM
The GEO repository identified 418 qualifying therapeutic targets, with DisGeNET generating 220 cardiomyopathy-associated candidates. GeneCards screening revealed 1413 potential targets, complemented by 505 OMIM-curated entries. Subsequent consolidation of these multi-source datasets through deduplication yielded 2277 unique DCM-related target genes.3.3 CR and DCM intersection targets and PPI network construction
Intersection analysis through jvenn visualization tool determined 139 shared molecular targets between CR and dilated cardiomyopathy (Fig. 3A). Bioinformatics interrogation of these overlapping entries using STRING established a curated PPI network (139 nodes, 2424 edges) with robust interactome topology metrics: mean node degree of 34.9 and clustering coefficient of 0.664 (Fig. 3B). The network was further visualized using Cytoscape 3.9.1, where nodes with higher degree values were represented by larger and darker red circles, with size and color gradients indicating node significance (Fig. 3C). By ranking nodes based on their degree values, the top 10 hub genes were identified, including AKT1, IL6, TNF, IL1B, TP53, SRC, CASP3, BCL2, ESR1, and PTGS2 (Fig. 3D). These hub genes may play critical roles in the therapeutic effects of CR against DCM.3.4 CR major active ingredients-intersection targets-diabetic myocardial injury network construction
A network targeting the intersection of Cyperi Rhizoma (CR) and diabetic myocardial injury was constructed based on 139 shared targets and 17 active compounds from CR to elucidate the potential therapeutic mechanisms of CR against DCM (Fig. 4). Among the active compounds, luteolin (degree = 27), quercetin (degree = 33), and patchoulenone (degree = 13) were identified as the most influential components, exhibiting the highest degree values within the network. These findings indicate that the bioactive constituents of CR may exert significant therapeutic effects in the treatment of diabetic myocardial injury by modulating key target proteins and related biological processes.3.5 Analysis of GO biological functions and KEGG pathways
In total, 139 key targets were entered into the DAVID database for GO enrichment analysis, with results ranked in ascending order based on P-value. The top ten most significant terms were selected for visualization (Fig. 5A–C). GO enrichment analysis identified 673 biological processes (BP), predominantly involving positive regulation of gene expression, inflammatory response, apoptotic process, and oxidative stress response. The 74 cellular components (CC) were mainly associated with the extracellular space, membrane raft, and receptor complex, while the 132 molecular functions (MF) were primarily associated with enzyme binding, cytokine receptor binding, and identical protein binding.To identify the signaling pathways associated with the active components of CR in addressing DCM, a KEGG enrichment pathway analysis was conducted, revealing the top 10 pathways that exhibited the strongest correlations (Fig. 5D). The analysis predominantly highlighted the AGE-RAGE signaling pathway in diabetic complications (Fig. S1†) and the TNF signaling pathway (Fig. S2†) as the most significantly enriched pathways among others. The primary objective of this herb is to modulate these pathways to facilitate the treatment of diabetic myocardial injury.
3.6 CR-active ingredient-DCM-disease target-pathway network analysis
In order to determine the active constituents of CR-targeted DCM and their related KEGG pathways, a diagram illustrating the relationships among the active components, the top 10 enriched pathways, and potential therapeutic targets was created using Cytoscape 3.9.1 (Fig. 6).3.7 Molecular docking
Based on the quantity of effective component targets, the three most active components were chosen for docking with the five top-ranked targets according to their degree value. As illustrated in Fig. 7A, patchoulenone interacts with multiple targets, exhibiting binding energies ranging from −5.4 kcal mol−1 to −7.6 kcal mol−1, reflecting favorable binding; in contrast, luteolin and quercetin engage with several targets at energies lower than −7.0 kcal mol−1, demonstrating significant binding strength. Among these, the docking configurations that were the most stable for luteolin with AKT1, IL6, TNF, IL1B, and TP53 were depicted in Fig. 7B–F. The binding between luteolin and AKT1 was the most stable, even more so than that between AKT1 and its natural ligand PIP3 (Fig. S3†). This further supports the idea that the active components of CR target essential factors involved in diabetic myocardial injury, contributing to the repair of affected tissues and organs. | ||
| Fig. 7 Calculation of binding energy and visualization of molecular docking between effective components of CR and key targets. (A) A heatmap illustrating the binding energies of docking molecular engagements among the activated components of CR and their principal targets. (B) AKT1 (PDB ID: 3O96) in relation to luteolin. (C) The association of luteolin with IL6 (PDB ID: 1ALU). (D) Luteolin's interaction with TNF (PDB ID: 7JRA). (E) The connection between luteolin and IL1B (PDB ID: 2I1B). (F) Luteolin's relationship with TP53 (PDB ID: 1ULO). | ||
3.8 MDS
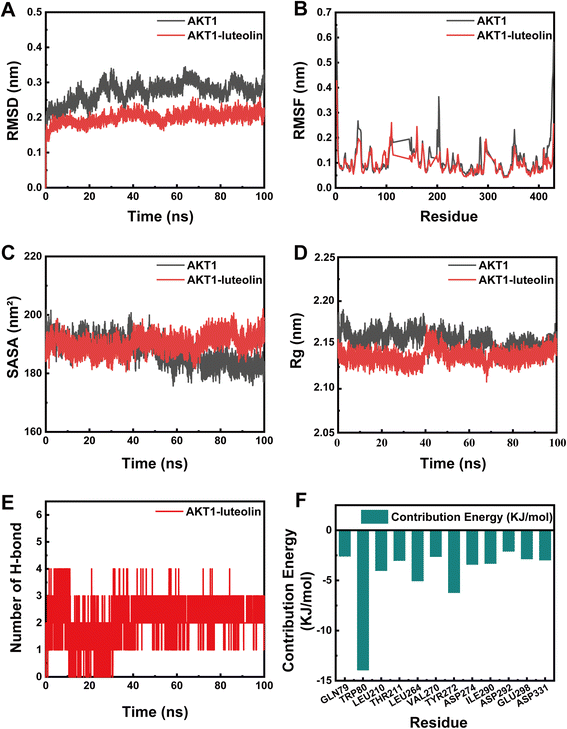
Following molecular docking, the AKT1-luteolin complex with the highest binding affinity, as well as the AKT1-quercetin complex with a comparable binding energy, were selected for molecular dynamics simulation (MDS) analysis. The simulation results indicated that both apo-AKT1 (unbound protein) and the AKT1-luteolin complex reached system equilibrium within 25 nanoseconds, demonstrating the robustness and reliability of the simulation protocol (Fig. 8A). Quantitative analysis revealed that the average root-mean-square deviation (RMSD) values for apo-AKT1 and the AKT1-luteolin complex were 0.2725 nm and 0.1992 nm, respectively, suggesting that luteolin binding enhanced the structural stability of AKT1. Further root-mean-square fluctuation (RMSF) analysis (Fig. 8B) showed average values of 0.1126 nm for apo-AKT1 and 0.0940 nm for the complex, providing additional evidence for the stabilizing effect of luteolin on the protein structure.In terms of solvent accessible surface area (SASA), no significant difference was observed between apo-AKT1 and the AKT1-luteolin complex (Fig. 8C), indicating that the structural integrity of the complex was maintained upon binding. Analysis of the radius of gyration (Rg) revealed that the AKT1-luteolin complex had a lower Rg value than apo-AKT1 (Fig. 8D), suggesting that luteolin binding promoted a more compact protein structure. Protein–ligand interaction analysis showed that an average of 1.96 hydrogen bonds were formed between AKT1 and luteolin (Fig. 8E), indicating the presence of stable and specific polar interactions at the binding interface. Binding free energy analysis (Table 3) demonstrated that van der Waals interactions were the predominant driving force in the AKT1-luteolin complex, with a total binding free energy of −94.547 kJ mol−1, confirming the formation of a stable complex. Energy decomposition analysis further identified TRP80, LEU264, and TYR272 as key residues contributing significantly to the binding energy (Fig. 8F). Collectively, these findings indicate that luteolin forms specific and stable interactions with AKT1, providing a potential molecular basis for its pharmacological effects in CR.
| Name | Degree |
|---|---|
| Pathways in cancer | 49 |
| Lipid and atherosclerosis | 35 |
| Fluid shear stress and atherosclerosis | 31 |
| AGE-RAGE signaling pathway in diabetic complications | 29 |
| Kaposi sarcoma-associated herpesvirus infection | 28 |
| Non-alcoholic fatty liver disease | 27 |
| Hepatitis B | 27 |
| Influenza A | 26 |
| C-type lectin receptor signaling pathway | 23 |
| TNF signaling pathway | 23 |
In comparison, MDS analysis of the AKT1-quercetin complex revealed an average RMSD of 0.2437 nm for the protein (Fig. S4A†), indicating relatively high structural stability. However, its average RMSF value was 0.0999 nm (Fig. S4B†), slightly higher than that of the AKT1-luteolin complex, suggesting marginally reduced structural stability. The SASA of the AKT1-quercetin complex was greater than that of apo-AKT1 (Fig. S4C†), indicating increased solvent exposure due to ligand binding. Additionally, the Rg of the AKT1-quercetin complex was higher than that of apo-AKT1 (Fig. S4D†), implying reduced structural compactness upon quercetin binding. Although the average number (Table 4) of hydrogen bonds formed in the AKT1-quercetin complex was 2.95—greater (Fig. S4E†) than the 1.96 observed in the AKT1-luteolin complex—its total binding free energy was −85.053 kJ mol−1 (Fig. S4F and Table S1†), lower than that of the luteolin complex. These results suggest that the AKT1-luteolin complex possesses superior overall structural stability and binding affinity.
| Energy | Complex |
|---|---|
| van der Waals energy (kJ mol−1) | −157.706 |
| Electrostatic energy (kJ mol−1) | −68.827 |
| Polar solvation energy (kJ mol−1) | 142.235 |
| Nonpolar solvation energy (kJ mol−1) | −19.011 |
| Total binding energy (kJ mol−1) | −103.309 |
| −TΔS (kJ mol−1) | 8.762 |
| Total binding free energy (kJ mol−1) | −94.547 |
3.9 Cytoprotective effects of luteolin on H9c2 cardiomyocytes under diabetic conditions
In diabetic models, we observed a significant spike in intracellular ROS compared with controls using DHE staining (P < 0.05). Nuclear staining with DAPI confirmed cellular localization. Notably, luteolin treatment at concentrations of 5, 10, and 20 μM effectively attenuated ROS production, with all treatment groups showing significantly lower fluorescence intensity than the DM group (P < 0.05) (Fig. 9A and B).Cell viability analysis showed a significant decline in the diabetic cohort as compared with the control group (P < 0.05). However, luteolin treatment at concentrations of 5, 10, and 20 μM exhibits marked higher cell survival rates than the DM group (P < 0.05) (Fig. 9C). These results delineates that luteolin exerts potential cardioprotective properties by mitigating oxidative stress in cardiomyocytes under diabetic conditions.
To more comprehensively elucidate the potential effects of CR components on diabetic cardiomyopathy, H9c2 cells subjected to high-glucose and high-lipid stimulation were treated with quercetin at concentrations of 5, 10, and 20 μM. Cell viability analysis revealed that the survival rates in all treatment groups were significantly higher than that of the DM group (P < 0.05), with the 20 μM quercetin group showing the most pronounced improvement (94.44%), slightly lower than the effect observed with 20 μM luteolin (97.92%) (Fig. S5†).
4. Discussion
DM is defined by an elevated blood glucose level resulting from metabolic disorders that contribute to its development.29 China ranks as the second most populous nation affected by DM.30 DCM represents a prevalent and serious cardiovascular complication among patients with diabetes, markedly heightening the risk of mortality.31 Despite notable progress in contemporary medical treatments for diabetes, specific pharmacological interventions for DCM remain insufficient. Traditional Chinese medicine, particularly CR, has been found to exhibit anti-diabetic properties as a herbal remedy.32 Despite this, the exact molecular mechanisms underlying the effects of CR on DCM have not been fully elucidated. In this study, we conducted network pharmacology, molecular docking, MDS, along with in vitro cellular assays to elucidate the therapeutic targets and effects of CR in the context of DCM, thereby furnishing novel theoretical and therapeutic perspectives.The TCMSP and BATMAN-TCM platforms indicate that the principal bioactive constituents identified in CR are quercetin, luteolin, and patchoulenone, which are widely acknowledged as efficacious constituents. Extensive research has been conducted on these compounds, demonstrating their considerable biological activity across a range of diseases. Quercetin, a flavonoid, exhibits antioxidant, anti-inflammatory, and anti-apoptotic pharmacological actions particularly under the framework of diabetes and cardiovascular diseases.33 Luteolin, another flavonoid commonly found in various herbal extracts, has demonstrated anti-diabetic properties in several studies.34 Additionally, patchoulenone possesses both anti-inflammatory and antioxidant capabilities, helping to minimize cardiac damage.35 It is possible that these compounds function synergistically through different pathways to aid as a therapeutic strategy for DCM.
The establishment of the PPI network for the key targets related to CR in DCM revealed the most important targets including AKT1, IL6, TNF, IL1B, TP53, SRC, CASP3, BCL2, ESR1, PTGS2, among others. Positioned downstream of PI3K within the insulin (INS) signaling pathway,36 AKT exerts a criticaleffect in decreasing apoptosis of myocardial cells by inhibiting the transcription of apoptotic genes such as BAX and caspase-3.37 IL-6 and TNF-α serve as biomarkers circulating levels in DCM individuals and can worsen myocardial cellular apoptosis by exacerbating oxidative burden and activating the ligand-receptor interaction death signaling pathway, which heightens the risk of heart failure.38 As a well-known tumor suppressor, TP53 oversees various signal transduction pathways to avert malignant transformation of cells.39 Nonetheless, under pathological conditions, abnormal activation of the TP53 signaling pathway can lead to detrimental effects such as cardiac fibrosis, cell death, heart failure, and premature mortality.40
An analysis of pathway enrichment utilizing GO and KEGG pathways indicated that CR might influence its effects through the regulation of various biological processes. The findings from the GO analysis suggest that CR could impact DCM by modulating gene expression and downregulating cell apoptosis. Furthermore, the KEGG pathway assessment reveals that CR might oppose DCM by affecting key pathways, in particular the TNF and AGE-RAGE signaling pathways. Both AGE-RAGE and TNF signaling pathways play pivotal roles in the pathogenesis of diabetic cardiomyopathy (DCM). The AGE-RAGE pathway contributes to myocardial damage in diabetes by amplifying oxidative stress, fostering inflammatory responses, and promoting extracellular matrix deposition, which collectively lead to myocardial fibrosis and impaired cardiac contractility.41 Persistent hyperglycemia accelerates the formation of advanced glycation end-products (AGEs), which bind to their receptor RAGE on cardiomyocytes and endothelial cells, initiating a cascade that exacerbates tissue damage.42 Similarly, the TNF signaling pathway intensifies inflammatory processes through the activation of NF-κB and downstream cytokines, contributing to cardiomyocyte apoptosis, mitochondrial dysfunction, and eventual cardiac remodeling.43 The involvement of these two pathways underscores a multifactorial mechanism in DCM progression that intertwines inflammation, oxidative stress, and metabolic dysregulation. Therefore, targeting these pathways may provide a strategic therapeutic approach for preventing or attenuating DCM.
The findings from the molecular docking experiments offered additional support that the key active constituents of CR, which consist of quercetin, luteolin, and patchoulenone, possess a significant capacity to associate with essential targets such as AKT1, IL6, TNF, IL1B, and TP53. Numerous studies have indicated a robust correlation between these targets and conditions like inflammatory heart disease, oxidative stress, and cell apoptosis, all of which are believed to be crucial in the context of DCM.44 Moreover, MDS further validated the strong binding affinity of luteolin with AKT1, highlighting its importance in the regulation of metabolism and survival of myocardial cells post interaction with the target.
In the in vitro cellular study, luteolin was employed to assess its impact on an H9c2 cardiomyocyte injury model triggered by elevated glucose and PA conditions. The findings revealed that luteolin showed considerable therapeutic potential in alleviating this injury. Additionally, further examination utilizing DHE staining suggested that CR might deliver its therapeutic benefits for DCM by decreasing the levels of ROS. This observation aligns well with earlier conclusions obtained from network pharmacology, molecular docking, and MDS, thereby offering a more thorough experimental basis for clarifying the mechanism through which CR addresses DCM.
5. Study limitations and future directions
Although the current investigation has offered valuable understanding of the mechanisms of CR in addressing DCM through integrated computational biology, network pharmacology, and in vitro experiments, several limitations must be acknowledged. While MDS and network pharmacology have shed light on the multi-target and multi-component nature of CR, the complexity of biological systems requires further experimental validation to fully elucidate these mechanisms. Additionally, the study primarily focused on computational and in vitro approaches, which may not fully capture the systemic effects of CR in a physiological context. Future research should include comprehensive clinical trial to evaluate the safety, efficacy, and feasibility of CR as a therapeutic intervention for DCM. Moreover, exploring the interactions between CR and other therapeutic strategies, as well as investigating the potential role of gut microbiota and metabolic pathways, could provide a more holistic understanding of its effects. Addressing these limitations and expanding the scope of research will be critical to translating these findings into clinically relevant interventions for diabetic patients.It is important to note that this study employed the H9c2 cell line under high glucose and palmitic acid conditions as an in vitro model to investigate the cardioprotective effects of CR active compounds. While this model provides a simplified and controllable environment for preliminary mechanistic studies, it cannot fully replicate the complex physiological and pathological environment in vivo, which involves multiple cell types, systemic interactions, and metabolic factors. Furthermore, although multiple bioactive compounds were identified, experimental validation primarily focused on luteolin due to limited experimental resources and our previous research on quercetin. We have supplemented molecular dynamics simulations and cellular experiments for the quercetin-AKT1 complex, but the synergistic effects of multiple compounds remain to be further explored.
6. Conclusion
This study employed an integrative approach combining network pharmacology, molecular docking, MDS, and in vitro experiments to systematically elucidate the multi-target mechanisms of CR and its active components in DCM, highlighting the scientific value of CR as a potential therapeutic strategy. Although luteolin has been previously studied in the context of DCM, the novelty of this work lies in the first comparative MDS analysis of luteolin and quercetin, revealing that both compounds stably bind to the core target AKT1. This finding not only addresses the limitations of earlier studies that focused on a single compound but also provides new molecular evidence for the synergistic mechanisms of multiple components in CR. The constructed PPI network further identified AKT1, IL6, TNF, TP53, and other key targets as central to CR's therapeutic effects on DCM, supporting its multi-pathway regulatory and systemic therapeutic potential. In vitro experiments using H9c2 cardiomyocytes under high glucose and palmitic acid conditions preliminarily validated the cardioprotective effects of luteolin, while the MDS data on the quercetin-AKT1 complex enhanced the credibility of the findings. The key targets and pathways identified in this study may serve as a theoretical foundation for future drug development and targeted intervention strategies. Further exploration of these mechanisms could lead to novel therapeutic approaches to alleviate cardiovascular complications in diabetic patients and ultimately improve clinical outcomes.Abbreviations
| DM | Diabetes mellitus |
| DCM | Diabetic cardiomyopathy |
| CR | Cyperi Rhizoma |
| TCM | Traditional Chinese medicine |
| PA | Palmitic acid |
| DAPI | 4′,6-Diamidino-2-phenylindole |
| ROS | Reactive oxygen species |
| RMSF | Root mean square fluctuation |
| SASA | Solvent accessible surface area |
| OB | Oral bioavailability |
| MDS | Molecular dynamics simulation |
| PBS | Phosphate buffered saline |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| PPI | Protein–protein interaction |
| BP | Biological processes |
| CC | Cellular component |
| MF | Molecular function |
| RMSD | Root mean square deviation |
| RG | Radius of gyration |
| HUNBM | Number of hydrogen bonds |
| DL | Drug-like properties |
| DHE | Dihydroethidium |
| DAVID | The Database for Annotation, Visualization and Integrated Discovery |
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. All relevant data generated or analyzed during this study are included in this published article.Author contributions
Su-Rui Lu: carried out visualization, conducted investigations, and performed formal analysis. Zixiang Fu: conducted investigations and handled formal analyses. Yan Xia: engaged in investigations and validation processes. Yanmei Xu: data curation and handled formal analyses. Fengliang Wang: participated in investigations. Zhaowen Pan: focused on visualization, software development, and data curation. Yiwei Tu: engaged in both investigations and validation. Wenjing Peng: focused on investigations and formal analysis. Mingli Han: conducted investigations and formal analyses. Wei Yu: developed the concept, created the methodology. Wenliang Zha: oversaw the project, performed formal analysis, managed data curation, participated in the assessment and refinement of the manuscript., and assisted in financial backing acquisition.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The research of our groups has been assisted by Hubei Provincial Natural Science Project (2024AFD064), Scientific Research Projects of Hubei Health Commission (WJ2023M119), Xianning Natural Science Foundation (2024RCFW005), Hubei Research Innovation Team (T20201022).References
- K. S. Spector, Clin. Cardiol., 1998, 21, 885–887, DOI:10.1002/clc.4960211205.
- J. L. Huo, Q. Feng, S. Pan, W. J. Fu, Z. Liu and Z. Liu, Cell Death Discovery, 2023, 9, 256, DOI:10.1038/s41420-023-01553-4.
- S. Dandamudi, J. Slusser, D. W. Mahoney, M. M. Redfield, R. J. Rodeheffer and H. H. Chen, J. Card. Failure, 2014, 20, 304–309, DOI:10.1016/j.cardfail.2014.02.007.
- A. Fiordelisi, F. A. Cerasuolo, R. Avvisato, A. Buonaiuto, M. Maisto, A. Bianco, V. D'Argenio, P. Mone, C. Perrino, S. D'Apice, R. Paolillo, A. Pezone, F. Varzideh, G. Santulli, D. Sorriento, G. Iaccarino and J. Gambardella, Cardiovasc. Diabetol., 2024, 23, 450, DOI:10.1186/s12933-024-02490-x.
- S. Wu, T. Zhao, L. Jin and M. Gong, J. Ethnopharmacol., 2024, 335, 118608, DOI:10.1016/j.jep.2024.118608.
- J. Lu, W. Li, N. Xu, P. Yao, S. Wang, C. Fu, J. Pei, H. Chen and S. Wang, J. Evidence-Based Complementary Altern. Med., 2022, 2022, 8309832, DOI:10.1155/2022/8309832.
- B. X. Xue, R. S. He, J. X. Lai, N. A. Mireku-Gyimah, L. H. Zhang and H. H. Wu, Phytochem. Rev., 2023, 1–46, DOI:10.1007/s11101-023-09870-3.
- P. Singh, R. L. Khosa, G. Mishra and K. K. Jha, J. Pharm. BioAllied Sci., 2015, 7, 289–292, DOI:10.4103/0975-7406.168028.
- M. Ren, J. Ma and M. Qu, Medicine, 2024, 103, e40929, DOI:10.1097/md.0000000000040929.
- H. Liang, C. Fang and M. Qiu, Sci. Rep., 2025, 15, 159, DOI:10.1038/s41598-024-83921-3.
- Z. H. Shen, T. Ye, B. Chen, C. Wan, X. Lu, T. H. Chen, S. Lin, J. X. Ye, L. Xie and Y. S. Fu, Sci. Rep., 2025, 15, 594, DOI:10.1038/s41598-024-84259-6.
- M. Naveed, I. Ali, T. Aziz, A. Saleem, Z. Rajpoot, S. Khaleel, A. A. Khan, M. Al-Harbi and T. H. Albekairi, Sci. Rep., 2025, 15, 131, DOI:10.1038/s41598-024-83908-0.
- H. Yan, Z. Li and Z. Zhang, Medicine, 2024, 103, e39138, DOI:10.1097/md.0000000000039138.
- X. Xu, W. Zhang, C. Huang, Y. Li, H. Yu, Y. Wang, J. Duan and Y. Ling, Int. J. Mol. Sci., 2012, 13, 6964–6982, DOI:10.3390/ijms13066964.
- UniProt Consortium, Nucleic Acids Res., 2023, 51, D523–D531, DOI:10.1093/nar/gkac1052.
- Z. Liu, F. Guo, Y. Wang, C. Li, X. Zhang, H. Li, L. Diao, J. Gu, W. Wang, D. Li and F. He, Sci. Rep., 2016, 6, 21146, DOI:10.1038/srep21146.
- S. Shi, K. Ding, F. Chen, M. Yang, L. Ni and X. Wu, Adipocyte, 2024, 13, 2423723, DOI:10.1080/21623945.2024.2423723.
- Z. Yue, L. Li, H. Fu, Y. Yin, B. Du, F. Wang, Y. Ding, Y. Liu, R. Zhao, Z. Zhang and S. Yu, J. Cell. Mol. Med., 2021, 25, 7500–7512, DOI:10.1111/jcmm.16786.
- H. Heberle, G. V. Meirelles, F. R. da Silva, G. P. Telles and R. Minghim, BMC Bioinf., 2015, 16, 169, DOI:10.1186/s12859-015-0611-3.
- Y. Yao, J. Wang, H. Zhang, T. Peng, Y. Sun, R. Zhang, X. Meng, X. Lu, Y. Gao, Y. Jin, Y. Zhang and L. Chen, J. Ethnopharmacol., 2025, 338, 118974, DOI:10.1016/j.jep.2024.118974.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. T. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Genome Res., 2003, 13, 2498–2504, DOI:10.1101/gr.1239303.
- L. Xin, H. C. Feng, Q. Zhang, X. L. Cen, R. R. Huang, G. Y. Tan and Q. Zhang, J. Ethnopharmacol., 2025, 338, 119023, DOI:10.1016/j.jep.2024.119023.
- M. Liu, Y. Wang, W. Deng, J. Xie, Y. He, L. Wang, J. Zhang and M. Cui, Front. Cell. Infect. Microbiol., 2024, 14, 1453529, DOI:10.3389/fcimb.2024.1453529.
- C. Wu, W. Zheng, J. Zhang and X. He, J. Evidence-Based Complementary Altern. Med., 2022, 2022, 2407462, DOI:10.1155/2022/2407462.
- J. Wu, F. Ge, L. Zhu and N. Liu, Environ. Sci. Technol., 2023, 57, 4852–4862, DOI:10.1021/acs.est.2c09352.
- M. Kawata and U. Nagashima, Chem. Phys. Lett., 2001, 340, 165–172 Search PubMed.
- R. Carretero-González, P. G. Kevrekidis, I. G. Kevrekidis, D. Maroudas and D. J. Frantzeskakis, Phys. Lett. A, 2005, 341, 128–134, DOI:10.1016/j.physleta.2005.04.046.
- W. Li, Y. Yao, X. Weng, X. Yue and F. Li, Anim. Biotechnol., 2022, 33, 1582–1587, DOI:10.1080/10495398.2021.1919130.
- V. V. Lupu, I. Miron, L. M. Trandafir, E. Jechel, I. M. Starcea, I. Ioniuc, O. E. Frasinariu, A. Mocanu, F. D. Petrariu, C. Danielescu, A. H. Nedelcu, D. L. Salaru, N. Revenco and A. Lupu, Front. Pharmacol., 2024, 15, 1472670, DOI:10.3389/fphar.2024.1472670.
- NCD Risk Factor Collaboration (NCD-RisC), Lancet, 2024, 404, 2077–2093, DOI:10.1016/s0140-6736(24)02317-1.
- Z. Zhuang, G. Ye and B. Huang, Med. Sci. Monit., 2017, 23, 3925–3931, DOI:10.12659/msm.902491.
- Y. Lei, M. Du, G. Zhang, L. Chen, Y. Fu, Y. Zhong and E. Zhang, J. Evidence-Based Complementary Altern. Med., 2021, 2021, 8614963, DOI:10.1155/2021/8614963.
- K. Ferenczyova, B. Kalocayova and M. Bartekova, Int. J. Mol. Sci., 2020, 21, 1585, DOI:10.3390/ijms21051585.
- Y. J. Wang, Y. L. Wang, X. F. Jiang and J. E. Li, World J. Diabetes, 2023, 14, 1659–1671, DOI:10.4239/wjd.v14.i11.1659.
- C. Thebtaranonth, Y. Thebtaranonth, S. Wanauppathamkul and Y. Yuthavong, Phytochemistry, 1995, 40, 125–128, DOI:10.1016/0031-9422(95)00260-e.
- B. Boezio, K. Audouze, P. Ducrot and O. Taboureau, Mol. Inf., 2017, 36(10), 1700048, DOI:10.1002/minf.201700048.
- N. Zhu, B. Huang, L. Zhu and Y. Wang, J. Diabetes Res., 2021, 2021, 9944589, DOI:10.1155/2021/9944589.
- H. Saad, H. A. Soliman, B. Mahmoud, A. A. Moneim and M. Y. Zaky, Inflammation, 2023, 46, 146–160, DOI:10.1007/s10753-022-01718-w.
- H. K. Punja, D. P. Nanjappa, N. Babu, K. Kalladka, B. Shanti Priya Dias, G. Chakraborty, S. M. Rao and A. Chakraborty, Mol. Biol. Rep., 2021, 48, 5093–5097, DOI:10.1007/s11033-021-06505-8.
- L. Rouhi, S. Fan, S. M. Cheedipudi, A. Braza-Boïls, M. S. Molina, Y. Yao, M. J. Robertson, C. Coarfa, J. R. Gimeno, P. Molina, P. Gurha, E. Zorio and A. J. Marian, Cardiovasc. Res., 2022, 118, 1466–1478, DOI:10.1093/cvr/cvab197.
- M. Snelson, E. Lucut and M. T. Coughlan, Int. J. Mol. Sci., 2022, 23, 1766, DOI:10.3390/ijms23031766.
- N. Wang and C. Zhang, Antioxidants, 2024, 13, 455, DOI:10.3390/antiox13040455.
- C. Jin, Z. Zhong, L. Gao, X. Wu, C. Zhou, G. Zhou and S. Liu, Mol. Rev. Cardiovasc. Med., 2024, 25, 110, DOI:10.31083/j.rcm2504110.
- J. Wu, S. Xia, B. Kalionis, W. Wan and T. Sun, BioMed Res. Int., 2014, 2014, 615312, DOI:10.1155/2014/615312.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra02505b |
| ‡ Su-Rui Lu, Zi-Xiang Fu, and Yan Xia are the co-first authors of this paper. |
| This journal is © The Royal Society of Chemistry 2025 |