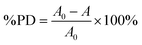Open Access Article
Open Access ArticleHarnessing nature for dual action: silver nanoparticles synthesized from guava leaf extract for photocatalytic degradation of methyl red and antibacterial applications†
Quoc-An Trieu *ab,
Quynh Nu Ai Unga,
Phung Ngoc Thaia,
Tam Minh Maia and
Dong Van Nguyencd
*ab,
Quynh Nu Ai Unga,
Phung Ngoc Thaia,
Tam Minh Maia and
Dong Van Nguyencd
aInstitute of Applied Technology and Sustainable Development, Nguyen Tat Thanh University, 331 National Highway 1A, An Phu Dong Ward, District 12, Ho Chi Minh City 700000, Vietnam. E-mail: tqan@ntt.edu.vn
bFaculty of Applied Science and Technology (FAST), Nguyen Tat Thanh University, 331 National Highway 1A, An Phu Dong Ward, District 12, Ho Chi Minh City 700000, Vietnam
cFaculty of Chemistry, University of Science, 227 Nguyen Van Cu Street, Ward 4, District 5, Ho Chi Minh City, Vietnam
dVietnam National University Ho Chi Minh City (VNU-HCM), Linh Trung Ward, Thu Duc, Ho Chi Minh City, Vietnam
First published on 25th April 2025
Abstract
This study revealed novel insights into key parameters affecting the biogenic synthesis of silver nanoparticles (AgNPs) with a dual-faceted application via a green route utilizing aqueous guava (Psidium guajava L.) leaf extract as both a reducing and stabilizing agent. The formation of AgNPs was visually confirmed by a color change of the reaction mixture from pale yellow to reddish-brown. Characterization of the synthesized AgNPs revealed a surface plasmon resonance band at 415–420 nm in the Ultraviolet-Visible (UV-Vis) spectra, confirming the presence of AgNPs. Dynamic light scattering (DLS) analysis indicated an average particle size of approximately 29 nm, while X-ray diffraction (XRD) analysis confirmed the crystalline nature and high purity of the synthesized AgNPs. Transmission electron microscopy (TEM) images displayed a primarily spherical morphology with an average size of about 12 nm. Fourier transform infrared (FTIR) spectroscopic analysis further supported the role of phytochemicals, such as phenolic acids and flavonoids, in the bioreduction and stabilization of the AgNPs. The synthesized AgNPs exhibited significant antibacterial activity against Gram-positive (S. aureus, E. faecalis) and Gram-negative (P. aeruginosa) bacterial strains, as demonstrated by the disc diffusion method. Furthermore, the AgNPs demonstrated promising photocatalytic activity, achieving approximately 95–96% degradation of methyl red (MR) within 72 hours under sunlight exposure. The dual functionality of the as-synthesized AgNPs opens up exciting avenues in both environmental remediation and biomedical fields.
1. Introduction
The global silver nanoparticles (AgNPs) market has experienced substantial growth, expanding from approximately $1 billion in 2015 to a projected $3 billion by 2024.1 Medical applications, particularly in healthcare and life sciences, are expected to be the largest sector, surpassing $1 billion. This is followed by the textile sector, with an estimated market value of over $750 million, and the food and beverage sector, which was forecasted to reach over $300 million by 2024.2 The unique physical, chemical, and biological properties of silver nanoparticles (AgNPs) arise from their nanoscale size (<100 nm) and high surface-to-volume ratio. The “quantum size effect” plays a critical role in determining these properties, as it explains how the behavior of AgNPs changes significantly with variations in their size.3 Unlike bulk silver, where quantum effects are averaged out, the smaller size of AgNPs allows these forces to have a more pronounced impact on the behavior of the nanoparticles. As a result, AgNPs exhibit distinct properties, including enhanced catalytic and biopharmaceutical activities, therapeutic potential, and improved sensing capabilities.Silver nanoparticles can be synthesized using a multitude of techniques that are fundamentally classified into two main approaches: top-down and bottom-up methods.4 While physical methods, such as mechanical thermal evaporation/condensation milling, laser ablation, arc discharge, and milling, predominantly rely on the top-down approach via breaking down bulk material into nanoparticles, chemical and biological methods predominantly employ the bottom-up approach.5 Chemical synthesis of silver nanoparticles (AgNPs) involves the transformation of silver ionic precursors into elemental silver (Ag0) atoms through chemical reduction. This process leads to the formation of nanoscale Ag0 atomic clusters, which are stabilized by capping agents. Various types of chemical reactions have been utilized for the synthesis of AgNPs, including the reduction reaction with borohydride,6 the reduction reaction using ethylene glycol,7 the Turkevich method,8 and the Tollens reaction.9 Nevertheless, these approaches frequently exhibit limitations, including elevated energy consumption, substantial costs, and reliance on potentially hazardous chemicals. Therefore, the green synthesis of AgNPs is increasingly being developed, exploiting natural biological sources, including bacteria,10 fungi,11 yeast,12 algae,13 plants,14 and natural biopolymers.15 Biological synthetic routes offer several notable advantages, including cost-effectiveness, compatibility with environmental and biological systems, rapid synthesis rates, scaling-up potential, and ease of manipulation.
Plants have captured considerable interest among biological agents owing to their rich diversity of biomolecules, including flavonoids, terpenoids, alkaloids, polyphenols, saccharides, amino acids, proteins, and others,16 which can be utilized for AgNP synthesis. This wealth of naturally occurring compounds makes them ideal candidates for various biotechnological applications. The versatility of plant-based materials is evident in the green synthesis of AgNPs, with diverse plant parts such as fruits, roots, flowers, leaves, and peels effectively utilized. Veerasamy et al.17 successfully synthesized AgNPs using a green approach, employing Garcinia mangostana leaf extract as a reducing agent. This study highlighted the potential of green chemistry for the development of effective antimicrobial agents against multi-drug resistant pathogens. Banerjee et al.18 fabricated silver nanoparticles (AgNPs) using leaf extracts from medicinal plants such as neem (Azadirachta indica), banana (Musa balbisiana), and black tulsi (Ocimum tenuflorum). The synthesized AgNPs exhibited potent antibacterial activity against E. coli and Bacillus sp., surpassing the antimicrobial effects of both silver nitrate (AgNO3) and the plant extracts alone. Importantly, the study demonstrated minimal toxicity of these green-synthesized AgNPs on the germination and oxidative stress response of Moong Bean and Chickpea seeds. Sharifi-Rad et al.19 reported the use of Lallemantia royleana leaf extract for the green synthesis of silver nanoparticles (AgNPs). The synthesized AgNPs demonstrated promising antimicrobial activity against a range of bacterial and fungal pathogens and exhibited significant cytotoxicity against human cancer cell lines (Hep-G2 and MCF-7). Moreover, the AgNPs displayed catalytic activity, highlighting their possibilities for various applications. In the work of Varghese Alex et al.,20 the sensing and photocatalytic properties of green-synthesized silver nanoparticles (AgNPs) produced using different plant leaf extracts were explored. Among the extracts tested, neem leaf extract, rich in diterpenoids as revealed by GC-MS analysis, yielded AgNPs with optimal Surface Plasmon Resonance (SPR) behavior. The study further demonstrates the potential of these green-synthesized AgNPs for biosensing applications, with a linear response observed between mancozeb (MCZ) concentration and SPR peak position. Moreover, the AgNPs exhibited photocatalytic activity towards MCZ degradation under UV-visible illumination, attributed to the formation of reactive oxygen species. Wahid et al.21 studied the ability of wheat leaf extract-synthesized silver nanoparticles (AgNPs) to enhance salt tolerance in wheat plants (Triticum aestivum L. cv. Pusa Kiran). Their findings revealed that AgNPs played a crucial role in mitigating the negative impacts of salt stress by modulating key physiological processes such as seed germination, K+ ion balance, abscisic acid (ABA) levels, antioxidants, proline accumulation, nitrogen metabolism, chlorophyll content, and stomatal function. These findings highlighted the potentiality of AgNPs in improving crop productivity and contributing to more sustainable agricultural practices. A study by Karthiga et al.22 showcased the selective colorimetric detection of hazardous heavy metal ions—specifically Hg2+, Pb2+, and Zn2+—in aqueous solutions utilizing silver nanoparticles (AgNPs) synthesized through green chemistry methods. These AgNPs, derived from plant extracts of neem, mango, and pepper, demonstrated distinct preferences for different metal ions. Notably, these sensors demonstrated robust performance over a wide pH range (2.0–11), emphasizing the potential use of plant-based AgNPs for practical environmental monitoring applications. In a study by Ahn et al.,23 thirty plant extracts were evaluated for their ability to synthesize silver nanoparticles (AgNPs). Among these, seven extracts—Cratoxylum formosum, Phoebe lanceolata, Scurrula parasitica, Ceratostigma minus, Mucuna birdwoodiana, Myrsine africana, and Lindera strychnifolia—demonstrated successful AgNP synthesis and exhibited superior antioxidant activity compared to the other extracts tested. The synthesized AgNPs demonstrated promising biological activities, including cytotoxicity against human lung cancer cells, particularly those synthesized from Cratoxylum formosum and Mucuna birdwoodiana extracts, and wound healing activity observed with Lindera strychnifolia extract-synthesized AgNPs. Recent studies have identified Uvaria narum leaf extract,24 Rubus discolor leaf extract,25 Premna integrifolia root extract,26 Croton macrostachyus bark extract, Aerva lanata flower extract,27 Azadirachta indica fruit extract,28 Phoenix dactylifera seed extract29 as promising green sources for the synthesis of silver nanoparticles (AgNPs) that exhibit enhanced medicinal and catalytic properties. It should be emphasized that the principles of green chemistry underpin the plant-mediated synthesis of AgNPs,30 minimizing the use of hazardous chemicals and waste generation. This approach presents a commitment to sustainable development by aligning with SDGs 3, 6, 9, and 12, prioritizing environmental sustainability and human health. Through maximizing atom economy and utilizing eco-friendly solvents, green nanotechnology can significantly contribute to achieving these SDGs.31
The guava (Psidium guajava L.) tree, a common fruit tree in tropical regions such as India, Indonesia, Bangladesh, and Vietnam, belongs to the Myrtaceae family. Guava leaves are rich in phytochemicals, including quercetin, avicularin, apigenin, and various flavonoids and phenolic acids. Previous studies have demonstrated the diverse biological activities of guava leaf extracts, encompassing anticancer, antidiabetic, antioxidant, antidiarrheal, antimicrobial, lipid-lowering, and hepatoprotective properties.32 Numerous reports have highlighted the significance of guava leaf aqueous extract in the synthesis of silver nanoparticles.20,33,34 While extensive research exists on the use of nature-derived AgNPs for dye removal from water bodies, no study has specifically examined guava leaf extract-synthesized AgNPs (AgNPs-GuLE) for this application. This study seeks to fill this important gap in knowledge and to explore a dual-faceted application of AgNPs-GuLE in wound healing and dye-contaminated water treatment, which has not been addressed in preceding studies. Dyes, frequently used in industries like food, pharmaceuticals, cosmetics, paper, textiles, plastics, ink, and paint, are common water contaminants. Plant-mediated AgNPs exhibit not only dye removal capabilities (via adsorption and photocatalysis) but also emerging multifunctional properties. This has paved the way for the development of AgNP-based nanohybrids with diverse applications in areas such as environmental remediation, diagnostics, and therapeutics.
Building upon prior research on the antimicrobial properties of AgNPs synthesized from guava leaf extract, this study aimed to optimize the synthesis process, to assess their antibacterial effectiveness against Staphylococcus aureus, Enterococcus faecalis, and Pseudomonas aeruginosa, and to investigate their photocatalytic degradation of methyl red (MR). To the best of our knowledge, this study marks the inaugural application of AgNPs-GuLE for dye treatment, underscoring the dual antibacterial and photocatalytic capabilities of AgNPs-GuLE.
2. Experimental
2.1. Materials
Dried guava leaves were procured from a local store in Vietnam, while silver nitrate (AgNO3) was sourced from China. Analytical-grade sodium hydroxide (NaOH) and hydrochloric acid (HCl) were employed in the study.2.2. Methods
 . The Scherrer formula relates the average crystallite size (Dp) to several key parameters, including the FWHM (β) of the diffraction peak, the X-ray wavelength (λ), and the Bragg angle (θ). The Scherrer constant (K), which has a value of 0.94 for spherical crystals with cubic symmetry, is an important factor in this equation.
. The Scherrer formula relates the average crystallite size (Dp) to several key parameters, including the FWHM (β) of the diffraction peak, the X-ray wavelength (λ), and the Bragg angle (θ). The Scherrer constant (K), which has a value of 0.94 for spherical crystals with cubic symmetry, is an important factor in this equation.or
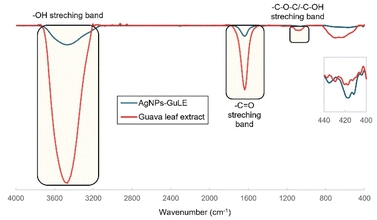
This equation establishes a relationship between the initial state of the MR solution (A0 and C0) and its state following photocatalytic degradation (A and C). This quantitative assessment is crucial for evaluating the efficiency of the photocatalytic process.
To account for the disc diameter itself, a normalization step was employed following the procedure by Serrano-Aroca et al.35 The assessment involved measuring both the diameter of the inhibition zone (diz) and the diameter of the disc (d). The normalized width (nw) of the inhibition zone was then calculated based on these measurements.
3. Results and discussion
3.1. Observation of the formation of silver nanoparticles (AgNPs) through visual analysis and subsequent examination of ultraviolet-visible (UV-vis) spectral data
The synthesis of silver nanoparticles (AgNPs) using plant extracts is known to produce solutions with characteristic yellowish-brown or reddish-brown colors.36 The characteristic color change from colorless to orangish-brown upon the addition of guava leaf extract to an aqueous AgNO3 solution provides a visual cue for monitoring the progress of AgNP formation, highlighting the potential for real-time observation and optimization of the green synthesis process.32,33 This color change indicates the formation of AgNPs, which is further supported by their ability to exhibit a localized surface plasmon resonance (SPR) band. The ability of UV-vis spectroscopy to provide information on AgNPs size, shape, and concentration highlights its significance as a valuable analytical tool in the field of nanotechnology.37–39 The increased production of AgNPs is typically indicated by a rise in maximum absorbance of the SPR band.4,40 The Full-Width at Half Maximum (FWHM) of the SPR peaks serves as a crucial parameter for assessing the size distribution of AgNPs, distinguishing between monodisperse, polydisperse, and highly polydisperse size distributions.41 Furthermore, the position of the SPR peak, as well as shifts toward longer (red shift) or shorter (blue shift) wavelengths, offers insights into the size and stability of the nanoparticles.42 In Fig. 1, the spectrum of AgNPs synthesized using guava leaf extract displays a distinct SPR band peaking at 417 nm, which is characteristic of well-formed silver nanoparticles. A proposed mechanism for the synthesis of AgNPs via guava leaf extract involves the action of quercetin and flavonoids, which serve as both reducing and stabilizing agents. These biomolecules facilitate the reduction of Ag+ ions to metallic silver (Ag0) and simultaneously stabilize the resulting nanoparticles by undergoing a tautomeric shift from the enol to the keto form (Fig. 2).433.2. The impact of pH on the synthesis of AgNPs suspensions using guava leaf extract
The sensitivity of AgNPs synthesis to pH underscores the need for meticulous pH control to achieve predictable and reproducible nanoparticle properties.58 Fig. 3 depicts the UV-vis spectra of AgNP suspensions synthesized at varying pH. Characteristic SPR bands of AgNPs were observed within the 400–420 nm range. A red-shift in the SPR peak was observed with increasing pH, accompanied by a decrease in peak intensity at pH 12, suggesting AgNP aggregation. Enhanced basicity likely increased the ionization of flavonoids and phenolics in the guava extract, thereby accelerating the reduction of silver ions and AgNPs formation.59 Furthermore, elevated pH levels may facilitate the conversion of Ag+ ions into Ag–O− groups that strongly interact with hydroxyl in plant extracts, thereby effectively covering the surface of AgNPs, and stabilizing them in the aqueous media.44 Consequently, a pH range of 8 to 10 within reaction mixtures was found to be optimal for the synthesis of AgNPs. | ||
| Fig. 3 UV-vis Spectra of guava leaf extract-mediated AgNPs unveiling the role of pH in AgNPs synthesis. | ||
DLS measurements show that AgNPs reached their smallest size, approximately 29 nm, within a pH range of 8 to 10 (Fig. S1 and S2, ESI†). Beyond pH 10, the size significantly increased. The observed trends were consistent with the UV-vis spectral analysis, providing further evidence for the relationship between pH, AgNPs size, and the observed spectral characteristics. TEM images (Fig. 7) indicate a primarily spherical morphology with an average size of about 12 nm, which was much smaller than that of the DLS results. This difference arises because TEM measures the size of the dry particles as they appear on the grid, while DLS (Dynamic Light Scattering) determines the hydrodynamic size of the nanoparticles, which includes the surrounding hydration layers. Essentially, TEM provides a “naked” particle size, whereas DLS captures the particles with their “coats of phytochemicals and water” on, leading to a larger apparent size in DLS measurements.34 The observed organic layers surrounding the AgNPs (Fig. 7), likely derived from biomolecules within the guava leaf extract, play a crucial role in stabilizing the nanoparticles by preventing aggregation and promoting their dispersion in the suspension.45
3.3. The role of AgNO3 concentration in the green synthesis of silver nanoparticles using guava leaf extract
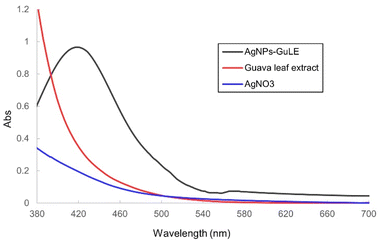
As the concentration of AgNO3 increased (Fig. 4), the UV-vis spectra display a shift of the absorption maxima to longer wavelengths (red-shift), indicating that the size of the silver nanoparticles (AgNPs) increased with higher AgNO3 concentrations. Unlike the pH-dependent study, this red shift did not coincide with a decrease in maximum absorbance. Instead, the absorbance consistently grew with increasing AgNO3 concentration, confirming that the amount of synthesized AgNPs was also rising. The SPR peaks were observed within the range of 417–448 nm, and the resulting AgNPs suspensions exhibited reddish-brown colors that became more intense as the concentration of AgNO3 was elevated. The SPR bands widened significantly with the increase in AgNO3 concentration, suggesting a broader range of sizes and possibly more varied particle distribution in the produced AgNPs. This is probably due to the higher reduction rate of AgNO3 at elevated concentrations, speeding up the nucleation and growth processes. Such quick synthesis can lead to differences in nanoparticle sizes and a higher chance of aggregation due to weak van der Waals forces. These results are in accordance with what has been observed in earlier research.3.4. Characterization of silver nanoparticles through X-ray diffraction analysis
XRD plays an integral role in the characterization of plant-extract synthesized silver nanoparticles (AgNPs), providing valuable information about their crystalline structure and purity. The Debye–Scherrer equation provides a method for estimating the particle size of AgNPs by correlating the broadening of diffraction peaks with reduced particle dimensions. In Fig. 5, the XRD pattern displays distinctive peaks for metallic silver (Ag0) at angles of 37.9°, 44.2°, 64.5°, and 77.6°, which correspond to the (111), (200), (220), and (311) crystal planes of metallic silver (Ag0). These peaks match the reference data from the ICDD Powder Diffraction File (PDF) no. 03-065-2871 and confirm the face-centered cubic (fcc) structure of the synthesized AgNPs. A comparison with standard XRD patterns for Ag2O (ICDD PDF no. 00-041-1104)46 and AgCl (ICDD PDF no. 00-001-1013) reveals that the nanoparticles consist purely of metallic silver, with no detectable impurities from other silver compounds. Utilizing Scherrer's formula, the average crystallite size of the AgNPs was calculated to be 9.6 nm.3.5. FITR characterization
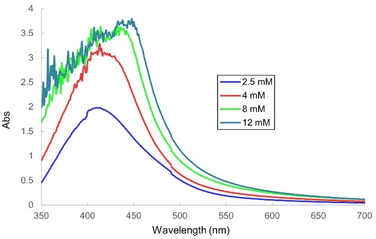
The FTIR spectrum analysis identified functional groups present in both the guava leaf extract and the AgNPs formed after the extract interacted with Ag+ ions. The spectral analysis of both the guava leaf extract and the synthesized AgNPs (Fig. 6) revealed several notable similarities, including absorption peaks at 3471 cm−1. These peaks are indicative of the stretching vibrations of the hydroxyl (–OH) group. Additionally, peaks at 1638 cm−1 were attributed to the stretching vibrations of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group, while bands at 1061 cm−1 and 1079 cm−1 indicated the vibrations of the C–O–C bond. These functional groups suggest the presence of biomolecules acting as capping and stabilizing agents on the surface of the AgNPs-GuLE. Specially, a unique absorption peak at 417 cm−1, observed only in the AgNPs spectrum, was characteristic of the vibrations associated with silver metal.47 These results are in accord with previous studies on the synthesis of AgNPs using plant extracts.48–50
O group, while bands at 1061 cm−1 and 1079 cm−1 indicated the vibrations of the C–O–C bond. These functional groups suggest the presence of biomolecules acting as capping and stabilizing agents on the surface of the AgNPs-GuLE. Specially, a unique absorption peak at 417 cm−1, observed only in the AgNPs spectrum, was characteristic of the vibrations associated with silver metal.47 These results are in accord with previous studies on the synthesis of AgNPs using plant extracts.48–50
3.6. TEM imaging analysis
As illustrated in Fig. 7, the majority of the silver nanoparticles (AgNPs) exhibit a spherical morphology and demonstrate a relatively polydisperse particle size distribution. It is noteworthy that aggregates of AgNPs can be observed in certain regions of the figure, while individual particles also exist independently. The average size of the particles was found to be 12 nm, with a standard deviation of 6 nm. This indicates that the sizes of the particles could range from 4 nm to 18 nm (SD = ± 9.0 nm). This broad range highlights the polydispersity in particle sizes (Fig. S3†), which is an important factor to consider when analyzing their properties and potential applications.51 These findings are consistent with those of prior research conducted by Kordy et al.52 and González-Pedroza et al.533.7. Photocatalytic degradation of MR using AgNPs-GuLE
As illustrated in Fig. 10b, the UV-vis absorption decreased progressively with an increase in the AgNPs-GuLE dosage, indicating that a higher concentration of AgNPs-GuLE enhanced the removal efficiency of MR from the solution. Sufficient AgNPs concentration ensures better dispersion and greater availability at active sites, thereby optimizing the photocatalytic degradation of MR.59 Fig. 10a indicates that at a volume of 1 mL of AgNPs-GuLE, the maximum percentage of photodegradation (% PD) achieved was 95 ± 4% (±SD), corroborating the findings of prior research.60
In the investigation of room light with an intensity of 230 Lux, it was observed that sunlight penetrated the room during daylight hours, whereas samples exposed to the LED light condition were shielded from natural illumination. The decrease in absorbance at 518 nm was more pronounced when samples were exposed to the 230 Lux room light compared to the 353 Lux LED light, which produces a small amount of UV compared to that generated by sunlight. This resulted in a higher percentage of photodegradation (%PD) for the 230 Lux room light relative to the LED intensities of 353 Lux. Furthermore, as the LED light intensity was increased to 588 Lux and 764 Lux, a gradual decrease in absorbance was noted, with % PD values of 68 ± 0.2% (±SD) and 79 ± 1.2% (±SD), respectively (Fig. 11a). These findings underscore the significant impact of the light nature and intensity on the efficiency of MR decomposition.
Under continuous sunlight exposure from sunrise to sunset, with an average intensity of 2394 Lux, the removal of methyl red (MR) displayed the highest efficiency among the three light types, achieving a % PD of 96 ± 0.2% (±SD). This interaction with sunlight, which is a full-spectrum light and contains a significant amount of UV radiation, promotes the excitation of electrons in silver nanoparticles (AgNPs), facilitating the generation of electron–hole pairs.56,61 The generated photoelectrons interacting with atmospheric oxygen form superoxide radicals (O2˙−), while the photogenerated holes produce highly reactive hydroxyl radicals (˙OH). The adsorbed MR can inject electrons into positive holes and be reduced,20,61 thereby preventing the recombination process. Free radicals oxidize MR molecules and cause the mineralization of MR. These reactive oxygen species are capable of decomposing MR into carbon dioxide, water, and various ions.55,62 While the intensity of illumination does influence the photocatalytic degradation of MR, the most rapid and marked effects are observed under solar radiation, aligning with the findings of prior studies.58,63
3.8. Antibacterial activity
Research indicates that phytochemicals found in guava leaf extracts possess selective antibacterial properties against Gram-positive bacterial strains.65,66 Furthermore, the antibacterial efficacy and the biocompatibility of green-synthesized AgNPs, particularly the safety of plant-synthesized AgNPs, have been extensively documented, mitigating concerns regarding AgNP toxicity.67 Assessments of the cytotoxicity of these green-synthesized AgNPs across various cell lines demonstrate robust antiproliferative activity against cancerous cell lines, e.g., lungs, pancreas, and prostate,68 with minimal impact on normal cell lines.25This study evaluates guava leaf extract-mediated silver nanoparticles (AgNPs-GuLE) for their antibacterial activity against three bacterial strains: Staphylococcus aureus (S. aureus) and Enterococcus faecalis (E. faecalis), which are Gram-positive, alongside Pseudomonas aeruginosa (P. aeruginosa), a Gram-negative bacterium. These pathogens are frequently implicated in infected wounds.69–71 These findings underline the enhanced antibacterial activity of AgNPs-GuLE compared to the guava leaf extract, highlighting the potential of green-synthesized nanoparticles for developing novel antimicrobial agents (Fig. 13 and S6, ESI†).
 | ||
| Fig. 13 Measurement of normalized inhibition zone widths for guava leaf extract and AgNPs against S. aureus, E. faecalis, and P. aeruginosa. | ||
The specific mechanisms underlying the antibacterial selectivity of AgNPs towards Gram-negative and Gram-positive bacteria remain unclear. Various factors may influence this selectivity, including the size,72 shape,73,74 surface charge,75 and surface chemistry76 of AgNPs, as well as the distinctive composition of bacterial cell walls.77–79 Certain key factors must be considered as a rule of thumb in the fabrication of AgNPs for robust antibacterial activity, high biocompatibility, and controlled toxicity. The surface chemistry should be engineered for dispersion stability, preventing aggregation while simultaneously improving tailored Ag+ release through optimized chemical instability.80 A positive surface charge strengthens adhesion to bacterial cell membranes. Furthermore, minimizing particle size74 and a spherical or platelet-like shape maximizes specific surface area and increases the Ag+ release rate from the AgNPs dissolution, thereby amplifying antibacterial efficacy. Despite substantial research on the antimicrobial properties of plant-mediated AgNPs, the precise mechanisms are not yet fully elucidated. Two main mechanisms have been proposed: (i) AgNPs self-affix to the bacterial cell wall, penetrate the membrane, and subsequently damage intracellular biomolecules such as DNA and proteins,81 and (ii) Ag+ ions released from AgNPs (“Trojan horse” mechanistic pathway82) leads to the disruption of cellular functions by interacting with essential proteins and enzymes. Moreover, the production of reactive oxygen species (ROS) by both AgNPs and the released Ag+ ions83–85 can interfere with vital cellular processes, including respiration and cell division, ultimately compromising microbial survival and function.
4. Conclusions
This study demonstrates the effects of critical parameters, including pH and AgNO3 concentration, on the synthesis of silver nanoparticles (AgNPs) using guava leaf extract as a reductant and a stabilizer (AgNPs-GuLE). Various characterization techniques, such as UV-vis, DLS, XRD, TEM, FTIR were employed to confirm the successful synthesis of AgNPs via guava leaf extract. The UV-vis spectrum displayed surface plasmon resonance (SPR) bands in the 415–420 nm range, confirming the successful synthesis of silver nanoparticles (AgNPs) under optimized conditions. These conditions included a pH range of 8–10, a silver nitrate to guava leaf extract volume ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and a reaction time of 30 minutes. The resulting AgNPs, produced through this green synthesis approach, had an average particle size of approximately 12 nm. The XRD results confirmed the pure crystallinity of the AgNPs derived from guava extract. The FTIR characterization demonstrated the presence of capping agents on the surface of AgNPs-GuLE, partly shedding light on the mechanism of the AgNPs-GuLE synthesis. Furthermore, the AgNPs-GuLE exhibited significant antibacterial activity against both Gram-positive (S. aureus and E. faecalis) and Gram-negative (P. aeruginosa) bacterial strains, highlighting their potential applications in the pharmaceutical, cosmetic, and agricultural sectors. The demonstration of efficient photocatalytic dye degradation by AgNPs-GuLE highlights their potential as an innovative approach to wastewater treatment, offering a promising alternative for environmental remediation. The potential toxicity of AgNPs has prompted widespread safety concerns regarding cell selectivity, dosage, short-term and long-term exposure, etc. Continued in vitro and in vivo research can help tackle these issues to deploy the full potential of AgNPs and accelerate clinical studies. Moreover, understanding the exact impacts of size, shape, and phytochemicals of plant extracts on the antibacterial mechanism could result in fit-for-purpose AgNPs, which can be accomplished through precise optimization of the nanosynthesis. Research into the application of green-synthesized AgNPs in agriculture, food safety and security is expected to achieve promising outcomes.
1, and a reaction time of 30 minutes. The resulting AgNPs, produced through this green synthesis approach, had an average particle size of approximately 12 nm. The XRD results confirmed the pure crystallinity of the AgNPs derived from guava extract. The FTIR characterization demonstrated the presence of capping agents on the surface of AgNPs-GuLE, partly shedding light on the mechanism of the AgNPs-GuLE synthesis. Furthermore, the AgNPs-GuLE exhibited significant antibacterial activity against both Gram-positive (S. aureus and E. faecalis) and Gram-negative (P. aeruginosa) bacterial strains, highlighting their potential applications in the pharmaceutical, cosmetic, and agricultural sectors. The demonstration of efficient photocatalytic dye degradation by AgNPs-GuLE highlights their potential as an innovative approach to wastewater treatment, offering a promising alternative for environmental remediation. The potential toxicity of AgNPs has prompted widespread safety concerns regarding cell selectivity, dosage, short-term and long-term exposure, etc. Continued in vitro and in vivo research can help tackle these issues to deploy the full potential of AgNPs and accelerate clinical studies. Moreover, understanding the exact impacts of size, shape, and phytochemicals of plant extracts on the antibacterial mechanism could result in fit-for-purpose AgNPs, which can be accomplished through precise optimization of the nanosynthesis. Research into the application of green-synthesized AgNPs in agriculture, food safety and security is expected to achieve promising outcomes.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI†. These data are available from the corresponding author upon request.Author contributions
Quynh Nu Ai Ung: investigation, data curation, formal analysis, visualization, writing – original draft. Phung Ngoc Thai: investigation, data curation, formal Analysis, visualization, writing – original draft. Tam Minh Mai: data curation, formal analysis, visualization. Dong Van Nguyen: methodology, validation. Quoc-An Trieu: conceptualization, methodology, investigation, data curation, resources, supervision, validation, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam.References
- S. Temizel-Sekeryan and A. L. Hicks, Resour., Conserv. Recycl., 2020, 156, 104676 CrossRef.
- A. Syafiuddin, Salmiati, M. R. Salim, A. Beng Hong Kueh, T. Hadibarata and H. Nur, J. Chin. Chem. Soc., 2017, 64, 732–756 CrossRef CAS.
- A. Campos, N. Troc, E. Cottancin, M. Pellarin, H.-C. Weissker, J. Lermé, M. Kociak and M. Hillenkamp, Nat. Phys., 2019, 15, 275–280 Search PubMed.
- N. S. Alharbi, N. S. Alsubhi and A. I. Felimban, J. Radiat. Res. Appl. Sci., 2022, 15, 109–124 Search PubMed.
- S. Iravani, H. Korbekandi, S. V. Mirmohammadi and B. Zolfaghari, Res. Pharm. Sci., 2014, 9, 385–406 Search PubMed.
- L. Mulfinger, S. D. Solomon, M. Bahadory, A. V. Jeyarajasingam, S. A. Rutkowsky and C. Boritz, J. Chem. Educ., 2007, 84, 322 CrossRef.
- L. Pourzahedi and M. J. Eckelman, Environ. Sci.: Nano, 2015, 2, 361–369 RSC.
- S. H. Lee and B.-H. Jun, Int. J. Mol. Sci., 2019, 20, 865 CrossRef CAS PubMed.
- M. A. AbuDalo, I. R. Al-Mheidat, A. W. Al-Shurafat, C. Grinham and V. Oyanedel-Craver, PeerJ, 2019, 7, e6413 CrossRef PubMed.
- P. Singh, S. Pandit, C. Jers, A. S. Joshi, J. Garnæs and I. Mijakovic, Sci. Rep., 2021, 11, 12619 CrossRef CAS PubMed.
- D. Wang, B. Xue, L. Wang, Y. Zhang, L. Liu and Y. Zhou, Sci. Rep., 2021, 11, 10356 CrossRef CAS PubMed.
- B. Skóra, U. Krajewska, A. Nowak, A. Dziedzic, A. Barylyak and M. Kus-Liśkiewicz, Sci. Rep., 2021, 11, 13451 CrossRef PubMed.
- R. Algotiml, A. Gab-Alla, R. Seoudi, H. H. Abulreesh, M. Z. El-Readi and K. Elbanna, Sci. Rep., 2022, 12, 2421 CrossRef CAS PubMed.
- M. Ghaedi, M. Yousefinejad, M. Safarpoor, H. Z. Khafri and M. K. Purkait, J. Ind. Eng. Chem., 2015, 31, 167–172 CrossRef CAS.
- H. E. Emam and H. B. Ahmed, Carbohydr. Polym., 2016, 135, 300–307 CrossRef CAS PubMed.
- M. A. Huq, M. Ashrafudoulla, M. M. Rahman, S. R. Balusamy and S. Akter, Polymers, 2022, 14, 742 CrossRef CAS PubMed.
- R. Veerasamy, T. Z. Xin, S. Gunasagaran, T. F. W. Xiang, E. F. C. Yang, N. Jeyakumar and S. A. Dhanaraj, J. Saudi Chem. Soc., 2011, 15, 113–120 CrossRef CAS.
- P. Banerjee, M. Satapathy, A. Mukhopahayay and P. Das, Bioresour. Bioprocess., 2014, 1, 3 CrossRef.
- M. Sharifi-Rad, H. S. Elshafie and P. Pohl, J. Photochem. Photobiol., A, 2024, 448, 115318 CrossRef CAS.
- K. Varghese Alex, P. Tamil Pavai, R. Rugmini, M. Shiva Prasad, K. Kamakshi and K. C. Sekhar, ACS Omega, 2020, 5, 13123–13129 CrossRef CAS PubMed.
- I. Wahid, S. Kumari, R. Ahmad, S. J. Hussain, S. Alamri, M. H. Siddiqui and M. I. R. Khan, Biomolecules, 2020, 10, 1506 CrossRef CAS PubMed.
- D. Karthiga and S. P. Anthony, RSC Adv., 2013, 3, 16765–16774 RSC.
- E.-Y. Ahn, H. Jin and Y. Park, Mater. Sci. Eng. C, 2019, 101, 204–216 CrossRef CAS PubMed.
- A. P. Ajaykumar, A. Mathew, A. P. Chandni, S. R. Varma, K. N. Jayaraj, O. Sabira, V. A. Rasheed, V. S. Binitha, T. R. Swaminathan, V. S. Basheer, S. Giri and S. Chatterjee, Antibiotics, 2023, 12, 564 CrossRef CAS PubMed.
- S. Ghasemi, S. Dabirian, F. Kariminejad, D. E. Koohi, M. Nemattalab, S. Majidimoghadam, E. Zamani and F. Yousefbeyk, Sci. Rep., 2024, 14, 4130 CrossRef CAS PubMed.
- C. Singh, S. K. Anand, R. Upadhyay, N. Pandey, P. Kumar, D. Singh, P. Tiwari, R. Saini, K. N. Tiwari, S. K. Mishra and R. Tilak, Mater. Chem. Phys., 2023, 297, 127413 CrossRef CAS.
- S. Palithya, S. A. Gaddam, V. S. Kotakadi, J. Penchalaneni, N. Golla, S. B. N. Krishna and C. V. Naidu, Part. Sci. Technol., 2022, 40, 84–96 CrossRef CAS.
- N. S. Alharbi and N. S. Alsubhi, J. Radiat. Res. Appl. Sci., 2022, 15, 335–345 CAS.
- N. N. Farshori, M. M. Al-Oqail, E. S. Al-Sheddi, S. M. Al-Massarani, Q. Saquib, M. A. Siddiqui, R. Wahab and A. A. Al-Khedhairy, J. Drug Delivery Sci. Technol., 2022, 70, 103260 CrossRef CAS.
- H. Duan, D. Wang and Y. Li, Chem. Soc. Rev., 2015, 44, 5778–5792 RSC.
- V. O. Nyandoro, H. K. Masioge and Z. L. Malago, Clean Technol. Environ. Policy, 2024, 27, 517–530 CrossRef.
- N. T. T. Le, B. T. D. Trinh, D. H. Nguyen, L. D. Tran, C. H. Luu and T. T. Hoang Thi, J. Cluster Sci., 2021, 32, 601–611 CrossRef CAS.
- P. R. Sougandhi and S. Ramanaiah, Inorg. Nano-Met. Chem., 2020, 50, 1290–1294 CrossRef CAS.
- F. N. Eze, C. Ovatlarnporn, S. Nalinbenjapun and S. Sripetthong, Arabian J. Chem., 2022, 15, 104167 CrossRef CAS.
- M. Marti, B. Frigols and A. Serrano-Aroca, J. Vis. Exp., 2018, 57710 Search PubMed.
- S. Ahmed, M. Ahmad, B. L. Swami and S. Ikram, J. Adv. Res., 2016, 7, 17–28 CrossRef CAS PubMed.
- A. Amirjani, N. N. Koochak and D. F. Haghshenas, J. Chem. Educ., 2019, 96, 2584–2589 CrossRef CAS.
- G. A. Martínez-Castañón, N. Niño-Martínez, F. Martínez-Gutierrez, J. R. Martínez-Mendoza and F. Ruiz, J. Nanopart. Res., 2008, 10, 1343–1348 CrossRef.
- V. Amendola, O. M. Bakr and F. Stellacci, Plasmonics, 2010, 5, 85–97 CrossRef CAS.
- S. Jain and M. S. Mehata, Sci. Rep., 2017, 7, 15867 CrossRef PubMed.
- A. W. Orbaek, M. M. McHale and A. R. Barron, J. Chem. Educ., 2015, 92, 339–344 CrossRef CAS.
- S. L. Smitha, K. M. Nissamudeen, D. Philip and K. G. Gopchandran, Spectrochim. Acta, Part A, 2008, 71, 186–190 CrossRef CAS PubMed.
- N. Ahmad, S. Sharma, M. K. Alam, V. N. Singh, S. F. Shamsi, B. R. Mehta and A. Fatma, Colloids Surf., B, 2010, 81, 81–86 CrossRef CAS PubMed.
- J. Jacob, T. Mukherjee and S. Kapoor, Mater. Sci. Eng. C, 2012, 32, 1827–1834 CrossRef CAS PubMed.
- Q.-A. Trieu, C. T. B. Le, C. M. Pham and T. H. Bui, J. Exp. Nanosci., 2023, 18, 2225759 CrossRef.
- N. Jahan Tamanna, M. Sahadat Hossain, N. Mohammed Bahadur and S. Ahmed, Results Chem., 2024, 7, 101313 CrossRef CAS.
- W. K. A. Wan Mat Khalir, K. Shameli, S. D. Jazayeri, N. A. Othman, N. W. Che Jusoh and N. M. Hassan, Front. Chem., 2020, 8, 620 CrossRef PubMed.
- O. Pawar, N. Deshpande, S. Dagade, S. Waghmode and P. Nigam Joshi, J. Exp. Nanosci., 2016, 11, 28–37 CrossRef CAS.
- V. K. Vidhu and D. Philip, Micron, 2014, 56, 54–62 CrossRef CAS PubMed.
- N. K. R. Bogireddy, H. A. Kiran Kumar and B. K. Mandal, J. Environ. Chem. Eng., 2016, 4, 56–64 CrossRef CAS.
- M. Danaei, M. Dehghankhold, S. Ataei, F. Hasanzadeh Davarani, R. Javanmard, A. Dokhani, S. Khorasani and M. R. Mozafari, Pharmaceutics, 2018, 10, 57 CrossRef PubMed.
- M. G. M. Kordy, M. Abdel-Gabbar, H. A. Soliman, G. Aljohani, M. BinSabt, I. A. Ahmed and M. Shaban, Nanomaterials, 2022, 12, 373 CrossRef CAS PubMed.
- M. G. González-Pedroza, A. R. T. Benítez, S. A. Navarro-Marchal, E. Martínez-Martínez, J. A. Marchal, H. Boulaiz and R. A. Morales-Luckie, Sci. Rep., 2023, 13, 790 CrossRef PubMed.
- G. Prasad Tajpuriya, P. Shah, N. Shahi and A. Bhattarai, Spectrochim. Acta, Part A, 2021, 255, 119646 CrossRef CAS PubMed.
- V. Railean-Plugaru, P. Pomastowski, M. Wypij, M. Szultka-Mlynska, K. Rafinska, P. Golinska, H. Dahm and B. Buszewski, J. Appl. Microbiol., 2016, 120, 1250–1263 CrossRef CAS PubMed.
- P. Kumar, M. Govindaraju, S. Senthamilselvi and K. Premkumar, Colloids Surf., B, 2013, 103, 658–661 CrossRef CAS PubMed.
- H. D. Tran, D. Q. Nguyen, P. T. Do and U. N. P. Tran, RSC Adv., 2023, 13, 16915–16925 RSC.
- R. Mohammadzadeh Kakhki, S. Hedayat and K. Mohammadzadeh, J. Mater. Sci.: Mater. Electron., 2019, 30, 8788–8795 CrossRef CAS.
- K. Tahir, S. Nazir, B. Li, A. U. Khan, Z. U. H. Khan, A. Ahmad and F. U. Khan, Sep. Purif. Technol., 2015, 150, 316–324 CrossRef CAS.
- A. S. Santhosh, S. Sandeep and N. Kumara Swamy, Surf. Interfaces, 2019, 14, 50–54 CrossRef CAS.
- S. Kaviya and E. Prasad, RSC Adv., 2015, 5, 17179–17185 RSC.
- P. P. A. Jose, M. S. Kala, N. Kalarikkal and S. Thomas, Res. Chem. Intermed., 2018, 44, 5597–5621 CrossRef CAS.
- Z. Z. Vasiljevic, M. P. Dojcinovic, J. D. Vujancevic, I. Jankovic-Castvan, M. Ognjanovic, N. B. Tadic, S. Stojadinovic, G. O. Brankovic and M. V. Nikolic, R. Soc. Open Sci., 2020, 7, 200708 CrossRef CAS PubMed.
- D. A. Reddy, J. Choi, S. Lee, R. Ma and T. K. Kim, RSC Adv., 2015, 5, 67394–67404 RSC.
- B. Biswas, K. Rogers, F. McLaughlin, D. Daniels and A. Yadav, Int. J. Microbiol., 2013, 2013, 746165 CrossRef PubMed.
- L. A. Worku, M. Zebeaman, R. K. Bachheti, A. Bachheti, Y. S. Rawat, A. Husen and R. G. Shiferaw, J. Food Qual., 2024, 2024, 6076403 CrossRef CAS.
- D. Somda, J. L. Bargul, J. M. Wesonga and S. W. Wachira, Sci. Rep., 2024, 14, 29273 CrossRef CAS PubMed.
- K. Jadhav, S. Deore, D. Dhamecha, R. H R, S. Jagwani, S. Jalalpure and R. Bohara, ACS Biomater. Sci. Eng., 2018, 4, 892–899 CrossRef CAS PubMed.
- L. J. Bessa, P. Fazii, M. Di Giulio and L. Cellini, Int. Wound J., 2015, 12, 47–52 CrossRef PubMed.
- K. K. L. Chong, W. H. Tay, B. Janela, A. M. H. Yong, T. H. Liew, L. Madden, D. Keogh, T. M. S. Barkham, F. Ginhoux, D. L. Becker and K. A. Kline, J. Infect. Dis., 2017, 216, 1644–1654 CrossRef CAS PubMed.
- B. Dai, C. Gao, J. Guo, M. Ding, Q. Xu, S. He, Y. Mou, H. Dong, M. Hu, Z. Dai, Y. Zhang, Y. Xie and Z. Lin, Nano Lett., 2024, 24, 4816–4825 CAS.
- A. Ivask, I. Kurvet, K. Kasemets, I. Blinova, V. Aruoja, S. Suppi, H. Vija, A. Käkinen, T. Titma, M. Heinlaan, M. Visnapuu, D. Koller, V. Kisand and A. Kahru, PLoS One, 2014, 9, e102108 CrossRef PubMed.
- A. Alshareef, K. Laird and R. B. M. Cross, Appl. Surf. Sci., 2017, 424, 310–315 CrossRef CAS.
- A. Menichetti, A. Mavridi-Printezi, D. Mordini and M. Montalti, J. Funct. Biomater., 2023, 14, 244 CrossRef CAS PubMed.
- A. M. El Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2011, 45, 283–287 CrossRef CAS PubMed.
- B. Le Ouay and F. Stellacci, Nano Today, 2015, 10, 339–354 CrossRef CAS.
- P. R. More, S. Pandit, A. D. Filippis, G. Franci, I. Mijakovic and M. Galdiero, Microorganisms, 2023, 11, 369 CrossRef CAS PubMed.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef CAS PubMed.
- F. Mirzajani, A. Ghassempour, A. Aliahmadi and M. A. Esmaeili, Res. Microbiol., 2011, 162, 542–549 CrossRef CAS PubMed.
- S. Tang and J. Zheng, Adv. Healthcare Mater., 2018, 7, 1701503 CrossRef PubMed.
- Y. Qing, L. Cheng, R. Li, G. Liu, Y. Zhang, X. Tang, J. Wang, H. Liu and Y. Qin, Int. J. Nanomed., 2018, 13, 3311–3327 CrossRef CAS PubMed.
- E.-J. Park, J. Yi, Y. Kim, K. Choi and K. Park, Toxicol. in Vitro, 2010, 24, 872–878 CrossRef CAS PubMed.
- E. Z. Gomaa, J. Gen. Appl. Microbiol., 2017, 63, 36–43 CrossRef CAS PubMed.
- M. A. Quinteros, V. Cano Aristizábal, P. R. Dalmasso, M. G. Paraje and P. L. Páez, Toxicol. in Vitro, 2016, 36, 216–223 CrossRef CAS PubMed.
- Y. M. Long, L. G. Hu, X. T. Yan, X. C. Zhao, Q. F. Zhou, Y. Cai and G. B. Jiang, Int. J. Nanomed., 2017, 12, 3193–3206 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra02503f |
| This journal is © The Royal Society of Chemistry 2025 |