Open Access Article
Open Access ArticleIntegrating gene knockout, GC-O-MS and multivariate statistical analysis: revealing the impact of fadA and bkdB genes on the aroma characteristics of Bacillus subtilis BJ3-2 fermented soybeans†
Feng Wen,
Yongjun Wu *,
Jing Jin,
Lincheng Zhang
*,
Jing Jin,
Lincheng Zhang ,
Shuoqiu Tong,
Cen Li,
Li Ran and
Yuzhang Zhu
,
Shuoqiu Tong,
Cen Li,
Li Ran and
Yuzhang Zhu
Key Laboratory of Plant Resource Conservation and Germplasm Innovation in Mountainous Region (Ministry of Education), College of Life Sciences/Institute of Agro-Bioengineering, Guizhou University, Guiyang 550025, China. E-mail: wyjbio@163.com
First published on 11th July 2025
Abstract
Fermented soybean products are an integral part of the Chinese diet, valued for their distinctive aroma. Bacillus subtilis (B. subtilis) BJ3-2 is a highly effective fermentation strain widely used in the industrial production of bacterial-type douchi. However, the specific aroma characteristics of soybeans fermented by this strain remain unclear. This study utilized gene knockout, GC-O-MS, and multivariate statistical analysis to comprehensively investigate the aroma components of soybeans fermented by B. subtilis BJ3-2. For the first time, 12 key aroma skeleton compounds were identified, including guaiacol, 2,5-dimethylpyrazine, and 2-methylbutyric acid, which primarily contribute to the smoky, roasted, and sweaty aroma attributes of fermented soybeans. Additionally, 17 compounds were considered to have important coordinating effects on soybean aroma, such as 3-hydroxy-2-butanone (buttery, creamy). Partial least squares regression (PLSR) analysis demonstrated a good correlation between key aroma compounds and sensory attributes. Gene knockout of fadA and bkdB resulted in a significant increase in 2,5-dimethylpyrazine and 1-octen-3-ol levels, along with a marked reduction in 2-methylbutyric acid levels. These changes enhanced the roasted and beany notes, reduced the sweaty aroma, and ultimately improved the overall aroma profile. These findings provide insights into the genetic regulation of aroma in fermented soybean products and form a foundation for reducing undesirable aromas and improving the overall aroma quality of industrially produced douchi.
1 Introduction
Soybean (Glycine max [L.] Merrill) is one of the most important crops globally, valued for its long cultivation history, high yield, and rich nutritional content. It is widely processed into various food products.1 Among the many soybean processing techniques, fermentation has been extensively utilized worldwide as a simple and cost-effective method that not only enhances the nutritional value of soybeans but also imparts unique aroma characteristics.2 Fermented soybean products are highly diverse, with notable examples including douchi (China), used as a seasoning; doenjang and cheonggukjang (Korea), commonly used in soups and stews; natto (Japan), consumed as a side dish; and kanjang (Korea and Japan), used as a liquid condiment.3 During fermentation, the proteins, fats, and carbohydrates in soybeans are broken down by microorganisms into various metabolites, which significantly influence the aroma and overall sensory qualities of these fermented foods.Aroma is a critical factor influencing the quality and consumer acceptance of fermented soybean products.4 Gas chromatography-olfactometry-mass spectrometry (GC-O-MS), which integrates gas chromatography-olfactometry (GC-O) and gas chromatography-mass spectrometry (GC-MS), is a powerful tool for studying food aromas and has been widely used in the aroma analysis of various foods.5 To comprehensively understand the changes in volatile compounds during fermentation and their relationships with aroma quality, various statistical methods are commonly employed, including Quantitative Descriptive Analysis (QDA),6 Relative Odor Activity Value (ROAV),7 Principal Component Analysis (PCA),8 Partial Least Squares-Discriminant Analysis (PLS-DA),9 and Partial Least Squares Regression (PLSR).10 These methods not only facilitate the quantification and classification of volatile compounds but also uncover their potential correlations with aroma attributes in fermented products.
B. subtilis is a key functional strain in the production of fermented soybean products, including soy sauce, natto, and douchi.3,11,12 During fermentation, B. subtilis produces various enzymes, such as nattokinase, proteases, and lipases, which break down complex macromolecules like proteins and fats into smaller compounds, including amino acids, fatty acids, and organic acids. These compounds are further metabolized into volatile compounds that contribute to the distinctive aromas of fermented soybeans.11,13 Currently, B. subtilis has been widely applied in food aroma regulation. For example, Studies have shown that B. subtilis NRCZ144 generates rich and pleasant aroma compounds through solid-state fermentation of soybean meal combined with L-lysine and L-threonine.14 However, during its metabolic processes, B. subtilis can also produce off-aroma compounds, such as 2-methylbutyric acid and 2-methylpropanoic acid, which may negatively impact the aroma of fermented foods.15 This underscores the importance of developing strategies to optimize the aroma profiles of fermented products.
In recent years, the rapid development of gene-editing technologies has provided powerful tools for the precise regulation of metabolic pathways in B. subtilis. By knocking out or modifying specific aroma-related genes, it is possible to alter metabolic fluxes and optimize the aroma profiles of fermented foods. For example, knocking out the rocF gene in B. subtilis significantly increased tetramethylpyrazine production and improved soybean aroma, but its impact on other key aroma components was not clarified.16 Studies have shown that 2-methylbutyric acid, a key volatile compound in soybeans fermented by B. subtilis, is primarily produced through the degradation of isoleucine.13 Our previous research analyzed the transcriptomic data of B. subtilis BJ3-2 under different fermentation temperatures. In the 53 °C vs. 37 °C comparison, fadA and bkdB were identified as significantly downregulated genes enriched in the valine, leucine, and isoleucine degradation pathway, suggesting that these two genes contribute to the stronger aroma of fermented soybeans at 37 °C.17 However, the roles of these two genes in the aroma profiles of fermented soybeans have not been fully elucidated. Moreover, aroma is a multidimensional and complex characteristic; although potential key compounds have been identified in previous studies, they remain insufficient to fully explain the aroma profile of soybeans.
Therefore, this study employed GC-O-MS to detect volatile compounds in four types of fermented soybeans and applied QDA, ROAV, and PLSR statistical methods to comprehensively evaluate the effects of the fadA and bkdB genes on the aroma of fermented soybeans. For the first time, the key aroma skeleton components of soybeans fermented by B. subtilis BJ3-2 were identified, along with the supplementation of the types and quantities of potential key aroma components. Furthermore, relevant literature was reviewed to summarize the sources of key aroma compounds, and the potential regulatory roles of the fadA and bkdB genes were hypothesized. These findings provide new insights into the analysis and genetic-level regulation of soybean aroma.
2 Materials and methods
2.1. Microorganisms, plasmids, and medium
All microorganisms and plasmids used in this study were listed in Table S1.† B. subtilis BJ3-2, which was isolated from fermented soybeans, has had its genome sequence deposited in the NCBI database (GI: CP025941). Both B. subtilis BJ3-2 and Escherichia coli DH5α were cultured in Luria-Bertani (LB) broth medium (5 g per L yeast extract, 10 g per L tryptone, and 10 g per L NaCl). Antibiotics were used at the following final concentrations: ampicillin (100 μg mL−1), chloramphenicol (5 μg mL−1), and erythromycin (1 μg mL−1).2.2. Chemicals and reagents
Reference standards were obtained from the following sources. 2-Octanol (≥99.0%), 2,3-butanedione (≥99.0%), and 3-ethyl-2,5-dimethylpyrazine (98%) were purchased from Macklin Biochemical Technology Corporation (Shanghai, China). 3-Hydroxy-2-butanone (98%) was purchased from Aladdin Industries Corporation (Shanghai, China). 2-Ethylfuran (>98.0%), 2-heptanone (>98.0%), 2-pentylfuran (>98.0%), 1-octen-3-ol (>98.0%), 2,3,5-trimethylpyrazine (>98.0%), 2,4,5-trimethyloxazole (>98.0%), 2,5-dimethylpyrazine (>98.0%), methyl 2-methylbutyrate (>98.0%), 2-methylbutyric acid (>98.0%), and guaiacol (>98.0%) were purchased from TCI Co. Ltd (Shanghai, China). N-Alkanes (C7–C30) (Supelco, Bellefonte, PA, USA) was employed to calculate the retention indices (RI). All primers used in this work were synthesized by Beijing Tsingke Biotech Co., Ltd (Beijing, China) and were shown in Table S2.†2.3. Reverse transcription-quantitative real-time PCR (RT-qPCR)
To evaluate the reliability of RNA sequencing (RNA-seq), total RNA was extracted from bacteria cultured at 37 °C and 53 °C using a column-based bacterial total RNA extraction and purification kit (Sangon Biotech, Shanghai, China). High-purity cDNA was synthesized using the StarScript Pro All-in-One RT Mix with gDNA Remover (Genstar, Beijing, China). RT-qPCR was performed on a CFX96™ Real-Time System (Bio-Rad, USA). The reaction mixture consisted of 1 μL of cDNA, 5 μL of 2× RealStar Fast SYBR qPCR Mix (Genstar, Beijing, China), 0.5 μL of primer pairs (10 mM), and 3.5 μL of sterile water. Reaction conditions were set according to the manufacturer's instructions. The 16S rRNA gene served as the internal reference, and relative expression levels were calculated using the 2−ΔΔCT method.18 The primers used for each gene were listed in Table S2.† All RT-qPCR experiments were performed in triplicate with a significance level of p < 0.05.2.4. Construction of mutant strains
The single knockout strains (BJ3-2ΔfadA and BJ3-2ΔbkdB) were constructed using the pre-laboratory serA knockout vector (pUC18-HLarm(serA)-cm-HRarm(serA)).19 First, the upstream (HLarm) and downstream (HRarm) flanking regions of the target genes were amplified from B. subtilis BJ3-2 genomic DNA, digested with restriction endonucleases (EcoRI/KpnI for HLarm and BamHI/SalI for HRarm), and ligated into the vector using T4 DNA ligase (Promega, Beijing, China). The ligation products were then transformed into E. coli DH5α competent cells and verified. Finally, the verified recombinant plasmid was transformed into B. subtilis BJ3-2 receptor cells, and positive transformants were screened on LB solid plates containing chloramphenicol.20The double knockout strains were obtained by following the method of Du et al.,21 with minor modifications. The bkdB fragment and pUC18 vector were digested with KpnI/HindIII and ligated to form the pUC18-bkdB plasmid. An erythromycin resistance gene (erm), amplified from the pMarA plasmid, was inserted into the pUC18-bkdB plasmid using the Hieff Clone® Plus One Step Cloning Kit (Yeasen, Shanghai, China). The successfully constructed pUC18-bkdB::erm recombinant plasmid was then transformed into BJ3-2ΔfadA receptor cells, and positive strains were screened on media containing chloramphenicol and erythromycin. A final series of validation steps was conducted, and the primers used for validation were listed in Table S2.†
2.5. Preparation and sensory evaluation of fermented soybean samples
Fermented soybean samples were prepared as described by Tao et al.,22 with appropriate modifications. BJ3-2, BJ3-2ΔfadA, BJ3-2ΔbkdB, and BJ3-2ΔfadAΔbkdB were individually inoculated into 5 mL of LB medium and incubated at 37 °C with shaking at 180 rpm for 12 h. The bacterial suspension was then adjusted to an OD600 value of approximately 1.20. Subsequently, the adjusted bacterial suspensions were individually inoculated into sterilized soybeans (soaked overnight and sterilized at 121 °C for 20 min) at an inoculation volume of 1% (vol/M ratio). The mixtures were thoroughly mixed and collectively placed in an incubator at 37 °C for fermentation over 72 h. The resulting fermented soybean samples were designated as FS1, FS2, FS3, and FS4, corresponding to BJ3-2, BJ3-2ΔfadA, BJ3-2ΔbkdB, and BJ3-2ΔfadAΔbkdB, respectively.The sensory evaluation of fermented soybeans was conducted using quantitative descriptive analysis (QDA).23,24 Sensory evaluation team was made up of 10 (five men and five women, aged 23 to 30) experienced members. A 10 g fermented soybean sample was placed in a 50 mL odorless transparent PET bottle and provided to the sensory panel for evaluation at room temperature (25 °C). All panel members underwent a week-long training prior to the analysis. Firstly, members independently sniffed the samples and recorded aroma descriptors. Secondly, based on the frequency and intensity of the descriptors, a consensus was reached to select six aroma attributes: smoky, buttery, beany, sweaty, roasted, and caramel. Six attributes were defined as the following aroma references: guaiacol for “smoky” attribute, 3-hydroxy-2-butanone for “buttery” attribute, 1-octen-3-ol for “beany” attribute, 2-methylbutyric acid for “sweaty” attribute, 2,5-dimethylpyrazine for “roasted” attribute, 2-ethylfuran for “caramel” attribute. Finally, the samples were labeled with random three-digit numbers and distributed to the panel members, who evaluated the intensity of each aroma attribute by sniffing. The intensity was rated on a 0–5 scale, with higher scores indicating stronger aroma intensity. Each sample was tested three times, and the average intensity of each aroma attribute was calculated.
2.6. Volatile compounds analysis by gas chromatography-olfactometry-mass spectrometry analysis (GC-O-MS)
Gas chromatography-olfactometry-mass spectrometry (GC-O-MS) was performed using a 8890/5977B GC-MS (Agilent Technologies, Inc., Santa Clara, CA, USA) equipped with an ODP4 olfactory detector (Gerstel, Mulheim, Germany), following the method of Zhao et al., with slight modifications.25 Before first measurement, a 50/30 μm DVB/CAR/PDMS fiber tip (Supelco, Bellefonte, PA, USA) was aged at 270 °C for 30 min to remove residual substances. Then, the 3 g sample was accurately weighed and placed into a 15 mL headspace vial, added 5 μL, 2-octanol (0.18 mg mL−1) as an internal standard, quickly tightened the cap, and placed in the autosampler tray. The samples were equilibrated at 50 °C for 20 minutes, followed by inserting the extraction fiber into the headspace vial for 40 minutes. Finally, the extraction head was inserted into the GC injection port, and desorbed at 250 °C for 5 min. GC conditions: sample separation was achieved using a highly polar DB-WAX capillary column (60 m × 0.25 mm × 0.25 μm, Agilent Technologies); the sample injection was conducted in a splitless mode to ensure optimal sensitivity and precision; ramp-up procedure: inlet port temperature 250 °C, initial column temperature 40 °C, hold for 3 min, ramp up to 130 °C at 3 °C min−1, hold for 5 min, ramp up to 200 °C at 4 °C min−1, hold for 5 min; the samples were separated by GC and analyzed by an olfactory detector and a mass spectrometer with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 split ratio. MS conditions: the mass spectrometer operated with a scan range of 35–500 m/z, 70 eV electron energy, 230 °C ion source, 150 °C quadrupole, and a 3 min solvent delay. Linear retention indices were calculated using n-alkane standards (C7–C30) under identical GC-MS conditions.
1 split ratio. MS conditions: the mass spectrometer operated with a scan range of 35–500 m/z, 70 eV electron energy, 230 °C ion source, 150 °C quadrupole, and a 3 min solvent delay. Linear retention indices were calculated using n-alkane standards (C7–C30) under identical GC-MS conditions.
GC-O analysis: GC-O was performed using a GC-MS system equipped with an ODP4 olfactory detection port. The olfactory evaluations were conducted by three trained assessors (one male and two females), who recorded the retention times, odor characteristics, and intensity of aroma compounds by sniffing at the olfactory detection port. Intensity of each aroma was rated on a 0–5 scale, with higher scores indicating stronger odor intensities.26 The GC-O-MS analysis was performed in triplicate.
2.7. Qualitative and quantitative analysis of the volatile compounds
Volatile compounds were identified with reference to previously reported literature.27 Initial characterization was performed using MS. Then, a series of homologous n-alkane standards (C7–C30) were used to calculate the retention indices (RIs) of the volatile compounds under the same chromatographic conditions as described by Yang et al.28 These calculated RIs were further compared with theoretical RIs from the NIST 20.0 database for confirmation. Olfactory descriptions (O) were used as supplementary characterization, and certain key compounds were verified using authentic standards (S) to complete the final identification. The content of volatile compounds was calculated using a semi-quantitative method with 2-octanol as the internal standard.29 All analyses were performed in triplicate, with results expressed as mean ± standard deviation (SD). One-way ANOVA was used to determine significant differences between samples at p < 0.05.2.8. Relative odor activity values (ROAV) calculation
ROAV was calculated as the ratio of the relative concentration of each compound to its absolute odor threshold in water. The calculation followed the approach of Yue et al.30| ROAV = Ci/OTi |
2.9. Statistical analysis
All experiments were independently repeated three times, with results reported as mean ± SD. Excel 2019 (Redmond, WA, USA) was used for statistical analysis. Radar plots, line plots, bar graphs, and Venn plots were created using Origin 2021 (OriginLab Corporation, Northampton, MA, USA). One-way ANOVA was performed to analyze the data at a significance level of p < 0.05. Heatmaps were produced by TBtools v1.068 (Heatmap Illustrator, Wuhan, China). PCA and PLS-DA were conducted using SIMCA-P 14.1 (Umetrics, Umeå, Sweden). PLSR analysis was produced by The Unscrambler 9.7 (CAMO Software, Oslo, Norway).3 Results and discussion
3.1. RT-qPCR validation
2-Methylbutyric acid, a key aroma compound in B. subtilis-fermented soybeans, was typically produced through isoleucine degradation.13,33 Previous studies in our laboratory analyzed the transcriptomic data of B. subtilis BJ3-2 under different temperatures. In the 53 °C vs. 37 °C comparison, fadA and bkdB were identified as significantly downregulated genes enriched in the valine, leucine, and isoleucine degradation pathways. These genes were therefore selected as key candidates for further investigation.17 Transcriptomic analysis revealed that the log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FoldChange (log2
FoldChange (log2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) FC) values of −2.24 and −2.09 for fadA and bkdB, respectively (Table S3†), indicating lower expression levels at 53 °C compared to 37 °C. To validate these results, RT-qPCR was performed (Table S4†). As shown in Fig. 1A, the log2FC (53 °C/37 °C) values for both genes were negative in transcriptome sequencing (RNA-Seq) and RT-qPCR, confirming higher gene expression at 37 °C. Additionally, no significant differences were observed between the RNA-Seq and RT-qPCR results, demonstrating consistency between the two methods. These findings suggested that the expression of fadA and bkdB might play a critical role in modulating food aroma.
FC) values of −2.24 and −2.09 for fadA and bkdB, respectively (Table S3†), indicating lower expression levels at 53 °C compared to 37 °C. To validate these results, RT-qPCR was performed (Table S4†). As shown in Fig. 1A, the log2FC (53 °C/37 °C) values for both genes were negative in transcriptome sequencing (RNA-Seq) and RT-qPCR, confirming higher gene expression at 37 °C. Additionally, no significant differences were observed between the RNA-Seq and RT-qPCR results, demonstrating consistency between the two methods. These findings suggested that the expression of fadA and bkdB might play a critical role in modulating food aroma.
3.2. Morphological characteristics and growth curves of the strains
In this study, we successfully constructed two single-gene knockout vectors and one double-gene knockout vector, with detailed validation results shown in Fig. S1–S5.† Using chemical transformation, we generated two single-gene knockout strains (BJ3-2ΔfadA and BJ3-2ΔbkdB) and one double-gene knockout strain (BJ3-2ΔfadAΔbkdB), with corresponding validation results presented in Fig. S6–S8.† Recent studies have revealed that deleting key genes in microorganisms can influence strain growth. For instance, the deletion of bkdAB genes in certain B. subtilis strains has been reported to promote growth, whereas the knockout of gbsAB genes in other strains negatively affects growth.34,35 To investigate the growth characteristics of the mutant strains, morphological observations and growth capacity measurements were conducted. As shown in Fig. 1B–E, the colony morphology of the three mutant strains on LB solid plates was similar to that of the wild-type (WT) strain (BJ3-2), characterized by a grayish-white color with a rough, opaque surface, numerous elevations and wrinkles, and irregular edges. Additionally, microscopic morphology was observed using Gram staining (Fig. 1F–I). The results showed that both the mutant and WT strains stained purple, with no noticeable differences in cell morphology or size, indicating that the knockout of fadA and bkdB genes had no effect on the morphological characteristics of the strains. Furthermore, growth curves were measured to assess growth ability (Fig. 1J). The results showed that the growth trend of the mutant strain was consistent with that of BJ3-2, and the strain exhibited normal growth after the knockout of fadA and bkdB (Table S5†).3.3. Sensory evaluation of fermentation samples
During microbial fermentation, macromolecules in soybeans (proteins, carbohydrates, and lipids) were broken down into small-molecule substrates (amino acids, sugars, and fatty acids), which were subsequently converted into volatile compounds that contributed to the unique aroma of fermented foods.36 To evaluate the overall aroma characteristics of the four fermented soybean samples (FS1–FS4), we performed QDA using a 0–5 point scale, with the scoring criteria as follows: 0 = no odor, 1 = very weak, 2 = weak, 3 = moderate, 4 = strong, and 5 = very strong. The results (Table 1) revealed that the aroma characteristics of fermented soybeans were dominated by smoky, beany, sweaty, and roasted attributes, which exhibited higher aroma intensities, while caramel and buttery attributes were relatively weaker. Smoky and beany aromas were typically associated with phenolics, alcohols, or aldehydes,37,38 whereas roasted aromas were primarily linked to the formation of pyrazine compounds during fermentation.39–41 In our study, all four samples exhibited strong roasted aromas, indicating that a substantial amount of pyrazine compounds was likely produced during the fermentation process. Notably, significant differences in sweaty odor, a generally undesirable off-aroma, were observed among the samples. FS1 displayed the highest sweaty odor intensity (3.66 ± 0.19), followed by FS2 (3.00 ± 0.24) and FS3 (2.83 ± 0.37), while FS4 exhibited the lowest intensity (2.00 ± 0.26). This unpleasant odor in natto has been attributed to branched-chain short-chain fatty acids (BCFAs).42 Based on these findings, we hypothesized that fadA and bkdB were involved in BCFAs production. Further analysis revealed that the FS4 sample exhibited the strongest beany aroma (3.33 ± 0.21), roasted aroma (4.00 ± 0.33), and caramel aroma (2.17 ± 0.49). Overall, FS4 was the most favored by panelists due to its more balanced and appealing aroma profile.| Aroma attributes | QDA scores (mean ± SD) | |||
|---|---|---|---|---|
| FS1 | FS2 | FS3 | FS4 | |
| Smoky | 2.00 ± 0.27 | 3.17 ± 0.24 | 3.33 ± 0.27 | 2.17 ± 0.19 |
| Buttery | 1.33 ± 0.25 | 1.83 ± 0.22 | 1.67 ± 0.33 | 1.00 ± 0.28 |
| Beany | 2.00 ± 0.00 | 2.67 ± 0.22 | 3.00 ± 0.30 | 3.33 ± 0.21 |
| Sweaty | 3.66 ± 0.19 | 3.00 ± 0.24 | 2.83 ± 0.37 | 2.00 ± 0.26 |
| Roasted | 3.00 ± 0.48 | 3.17 ± 0.32 | 3.33 ± 0.18 | 4.00 ± 0.33 |
| Caramel | 1.17 ± 0.25 | 1.50 ± 0.21 | 1.67 ± 0.31 | 2.17 ± 0.49 |
3.4. GC-O-MS analysis of volatile compounds in fermented soybeans
To explore the sources of aroma in fermented soybeans, we analyzed the volatile compounds in four types of fermented soybeans using GC-O-MS, identifying a total of 100 volatile compounds (Table S6†). These compounds included 12 pyrazines, 27 alcohols, 23 ketones, 4 acids, 11 esters, and 23 others. As shown in Fig. 2A, FS4 contained the highest number of volatile compounds (73), suggesting a richer aroma profile. Additionally, Fig. S9† illustrated the composition and differences in volatile compounds among the samples. Pyrazines were the most abundant, with an average concentration of 8970.72 μg kg−1, followed by ketones (2800.64 μg kg−1), alcohols (1125.69 μg kg−1), and acids (889.24 μg kg−1). Compared to FS1, FS2 showed significantly higher levels of pyrazines, ketones, and alcohols. FS3 exhibited a notable increase in alcohols but a significant decrease in acids, while FS4 showed elevated levels of pyrazines and alcohols, accompanied by a substantial decrease in acids.Cluster analysis was performed to visualize the relative distribution of volatile compounds and the similarities in aroma profiles among the four fermented soybean samples (Fig. 2B). The samples were divided into two distinct clusters: FS1–FS3 were categorized into cluster 1, whereas FS4 was assigned to cluster 2. Furthermore, the dendrogram classified the volatiles into two groups, each comprising 50 compounds. Group I was dominated by pyrazines, alcohols, ketones, and acids, while Group II mainly consisted of pyrazines, alcohols, and ketones. Interestingly, all four samples contained high levels of pyrazine compounds, including 2,3,5-trimethylpyrazine and 2,5-dimethylpyrazine. Pyrazines, key aroma compounds in fermented soybeans, were found to provide rich nutty, cocoa, and chocolate-like aromas, and were typically produced through non-enzymatic browning reactions and microbial metabolism.39 Higher levels of 1-octen-3-ol were detected in FS3 and FS4. This compound was identified as a common volatile substance in fermented soybeans and as one of the sources of the mushroom-like or beany aroma in the samples.43 Although acetic acid was present in the samples, its high odor threshold limited its contribution to the overall aroma. FS1 contained the highest level of 2-methylbutyric acid, a compound with a sweaty odor. This compound was considered an unpleasant off-aroma in Bacillus-fermented foods and has continued to hinder the industrial development of fermented soybeans.44 Fortunately, many esters and ketones, such as methyl 2-methylbutyrate (apple) and 2-heptanone (banana), have been reported to produce pleasant fruity aromas that can mask the undesirable aromas in fermented foods.45–47 Additionally, phenolic compounds (e.g., guaiacol and maltol) and furan compounds (e.g., 2-ethylfuran and 2-pentylfuran) were detected in fermented soybeans, contributing malty, smoky, caramel, and grassy aromas to the samples.48–50 The above results indicated that knocking out the related genes increased the variety of aroma compounds in soybeans, thereby enhancing the complexity of the overall aroma profile in fermented soybeans.
3.5. GC-O and ROAV identification of key aroma compounds
Studies have shown that only a portion of the volatile compounds detected by GC-MS significantly affect the perception of overall aroma.51 Additionally, volatile aroma compounds have different thresholds for different people, so the contribution of each compound to the aroma profile cannot be assessed by their relative content.38,52 Therefore, a combined GC-O and ROAV analysis was utilized in this study to further explore the key aroma components of the four fermented soybean samples.GC-O has been widely used to identify potential key aroma components, utilizing the human nose as a detector.53,54 In this study, GC-O analysis detected 19 volatile compounds from four fermented soybean samples (Table S7†). Among these, 2,5-dimethylpyrazine (3.00–4.00), 2,3,5-trimethylpyrazine (2.33–3.33), 1-octen-3-ol (2.00–2.83), 2-methylbutyric acid (2.00–3.67), and guaiacol (2.00–3.33) exhibited higher odor intensities and were thus considered potential key aroma compounds. Additionally, compounds with ROAV > 1 were also identified as potential contributors to the aroma profile.55,56 In our study, only 18 compounds exhibited ROAV > 1, contributors to the aroma profile (Table S8†).57 Notably, 2,3-butanedione (49.75–221.88), 2,5-dimethylpyrazine (93.44–289.06), 1-octen-3-ol (40.54–86.58), 2-methylbutyric acid (233.27–1294.69), and guaiacol (65.35–171.61) exhibited relatively high ROAV values across different samples, indicating their significant contributions to the overall aroma. We further used GC-O and ROVA > 1 for joint analysis, and the results were shown in Table 2. A total of 13 volatile compounds were screened by both methods, including 2,5-dimethylpyrazine, 2,3,5-trimethylpyrazine, 3-ethyl-2,5-dimethylpyrazine, 1-octen-3-ol, 2,3-butanedione, 2-heptanone, acetoin, 2-methylbutyric acid, methyl 2-methylbutyrate, 2-ethylfuran, 2-pentylfuran, guaiacol, and 2,4,5-trimethyloxazole. Among them, 2,5-dimethylpyrazine, 2,3,5-trimethylpyrazine, 2-methylbutyric acid, and guaiacol exhibited both high aroma intensity and ROAV, proving their crucial role in the composition of soybean aroma. Meanwhile, other compounds with lower GC-O intensity and ROAV might still have contributed to the overall aroma through synergistic effects. The results of the GC-O and ROAV analyses were consistent with the sensory evaluation results, further confirming the importance of these key aroma compounds in the overall aroma of fermented soybeans.
| No. | Aroma compounds | Aroma description | Odor intensity of GC-O | Thresholda (μg kg−1) | ROAV > 1 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FS1 | FS2 | FS3 | FS4 | FS1 | FS2 | FS3 | FS4 | ||||
| a Odor thresholds in water. | |||||||||||
| Pyrazines | |||||||||||
| 1 | 2,5-Dimethylpyrazine | Roasted, nutty | 3.00 | 3.17 | 3.33 | 4.00 | 20 (ref. 31) | 93.44 | 122.97 | 96.39 | 289.06 |
| 2 | 2,3,5-Trimethylpyrazine | Roasted, peanut | 2.67 | 3.00 | 3.33 | 2.33 | 71 (ref. 31) | 65.57 | 69.61 | 72.69 | 53.51 |
| 3 | 3-Ethyl-2,5-dimethylpyrazine | Stir-fried cocoa | 1.00 | 1.67 | 1.00 | 1.00 | 25 (ref. 19) | 2.54 | 4.54 | 2.58 | 1.96 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||||
| Alcohols | |||||||||||
| 4 | 1-Octen-3-ol | Mushroom | 2.00 | 2.33 | 2.50 | 2.83 | 7 (ref. 31) | 40.54 | 60.73 | 77.60 | 86.58 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||||
| Ketones | |||||||||||
| 5 | 2,3-Butanedione | Buttery | 1.00 | 1.83 | 1.33 | 1.00 | 3 (ref. 31) | 87.73 | 221.88 | 97.07 | 49.75 |
| 6 | 2-Heptanone | Banana | 1.83 | 1.00 | 2.00 | 1.83 | 70 (ref. 31) | 1.92 | 0.96 | 2.46 | −2.23 |
| 7 | Acetoin | Buttery, creamy | 1.50 | 2.00 | 1.67 | 1.00 | 40 (ref. 31) | 12.27 | 49.08 | 16.30 | 1.02 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||||
| Acids | |||||||||||
| 8 | 2-Methylbutyric acid | Sweaty, stink | 3.67 | 3.00 | 2.83 | 2.00 | 0.7 (ref. 32) | 1294.69 | 1032.69 | 950.03 | 233.27 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||||
| Esters | |||||||||||
| 9 | Methyl 2-methylbutyrate | Apple | 1.00 | 1.00 | 1.17 | 2.00 | 0.25 (ref. 31) | 28.00 | 24.46 | 42.32 | 102.87 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||||
| Others | |||||||||||
| 10 | 2-Ethylfuran | Caramel | 1.00 | 1.50 | 1.50 | 2.17 | 2.3 (ref. 19) | 1.88 | 3.87 | 4.39 | 8.01 |
| 11 | 2-Pentylfuran | Green, buttery | 1.00 | 1.50 | 1.00 | 1.67 | 4.8 (ref. 31) | 1.64 | 4.45 | 3.42 | 5.13 |
| 12 | Guaiacol | Smoky, woody | 2.00 | 3.17 | 3.33 | 2.17 | 0.75 (ref. 31) | 65.35 | 156.16 | 171.61 | 73.54 |
| 13 | 2,4,5-Trimethyloxazole | Earthy, cucumbers | 1.67 | 2.00 | 1.50 | 1.00 | 5 (ref. 13) | 14.24 | 27.16 | 13.99 | 6.73 |
3.6. Multivariate statistical analysis of key aroma compounds in fermented soybean samples
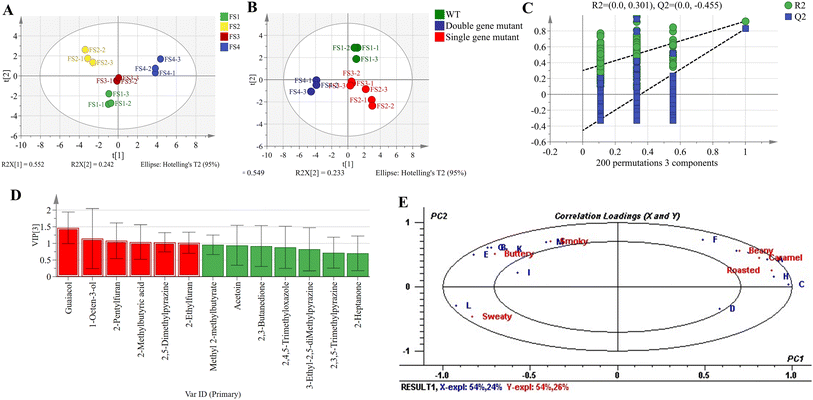
To investigate the differences among fermented soybean samples, we performed PCA and PLS-DA analyses on the key aroma compounds. The PCA score plot showed that the cumulative variance contribution rate of PC1 and PC2 was 79.4%, indicating that these two principal components effectively captured the major differences among the samples (Fig. 3A). Moreover, the four groups of samples exhibited clear separation, with samples within each group clustered tightly, demonstrating good reproducibility. Specifically, FS1 and FS3 samples were both distributed in the third quadrant, suggesting certain similarities between them, which was consistent with the results of hierarchical clustering analysis. To further elucidate the differences between groups, PLS-DA was applied to analyze the key aroma compounds in the four groups of fermented soybean samples. The PLS-DA score plot (Fig. 3B) revealed that the WT, single-gene knockout strains (Single Gene Mutant), and double-gene knockout strains (Double Gene Mutant) samples were distributed in different quadrants, highlighting significant differences among the groups. Through 200 cross-validations (Fig. 3C), the results showed that the intersection of the Q2 regression line with the vertical axis was less than 0, demonstrating that the PLS-DA model was not overfitted, thereby confirming the validity of the model and the reliability of sample classification.55 The Variable Importance in Projection (VIP) values were used to evaluate the importance of variables in distinguishing the samples, with larger VIP values indicating a greater contribution of the variable to sample differentiation.58 As shown in Fig. 3D, compounds with VIP values greater than 1 were marked in red, including guaiacol, 1-octen-3-ol, 2-pentylfuran, 2-methylbutyric acid, 2,5-dimethylpyrazine, and 2-ethylfuran. Therefore, these six key aroma compounds were considered to play an important role in differentiating the fermented soybean samples produced by different strains.To explore the correlation between key aroma compounds and sensory attributes, we performed PLSR analysis. As shown in Fig. 3E, the PLSR loading plot indicated that two principal components explained 78% of the variance in the 13 key aroma compounds (X variables) and 80% of the variance in the 6 sensory attributes (Y variables). The inner and outer ellipses in the plot represent the ranges explaining 50% and 100% of the variance, respectively. Except for D (2-heptanone) and I (2,3,5-trimethylpyrazine), the remaining 11 aroma-active compounds and all sensory attributes were located outside the inner ellipse but within the outer ellipse, indicating that the PLSR model had strong explanatory power. Meanwhile, the loading plot clearly demonstrated close correlations between different aroma compounds and specific sensory attributes. Two variables in the same quadrant were positively correlated, and the correlation became stronger when a sensory attribute was closer to a specific aroma compound.59 For instance, A (2-ethylfuran), H (2,5-dimethylpyrazine), and J (1-octen-3-ol) were closely associated with caramel, roasted, and beany sensory attributes, respectively. M (guaiacol) was highly correlated with smoky aroma; B (2,3-butanedione) and G (3-hydroxy-2-butanone) were linked to creamy aroma; and L (2-methylbutyric acid) was associated with sweaty aroma. Studies have reported that 2-pentylfuran is a key compound contributing to the beany and grassy notes in soybean aroma,50 a finding consistent with our results. Additionally, the compounds closely related to these six sensory attributes have also been documented in the literature. For instance, 2,5-dimethylpyrazine (roasted, nutty), 2-methylbutyric acid (sweaty, stink), 3-hydroxy-2-butanone (buttery, creamy), and 2-pentylfuran (green bean, buttery) were identified as major volatile compounds in soybean fermented by Bacillus species.13,60 Similarly, 1-octen-3-ol (mushroom) was one of the primary components in soy sauce.61 Furthermore, 2,3-butanedione and guaiacol were key aroma-active compounds in Yongchuan Douchi, where 2,3-butanedione contributed buttery and milky aromas, while guaiacol imparted smoky attributes.37
3.7. Determination of key aroma skeleton components
In a previous study, sdaAA and katX genes of B. subtilis BJ3-2 were knocked out, and soybeans were fermented. A total of 20 aroma compounds were detected by GC-O analysis, while 15 compounds with ROAV > 1 were identified through ROAV analysis. Among these, 13 aroma compounds were simultaneously screened in all fermented soybean samples through both GC-O and ROAV > 1 analysis.13 The detailed compound information from previous studies mentioned above can be found in Table S9.† As shown in Fig. 4A, our findings were compared with previous studies to identify the key aroma skeleton components of B. subtilis BJ3-2 fermented soybeans. The compounds contained in SGR1, SGR2, GR1, and GR2 were listed in Table S9.† The Venn diagram showed that 12 identical aroma compounds were detected in both the GR1 and GR2 groups, including 2-ethylfuran, 2-pentylfuran, 2,4,5-trimethyloxazole, 1-octen-3-ol, 2-methylbutyric acid, methyl 2-methylbutyrate, 2-heptanone, 2,3-butanedione, 2,5-dimethylpyrazine, 2,3,5-trimethylpyrazine, 3-ethyl-2,5-dimethylpyrazine, and guaiacol. Previous studies have indicated that compounds detected by GC-O are widely considered potential key aroma components, while those with ROAV > 1 are similarly regarded as key contributors to aroma profiles.53,55 These 12 compounds not only satisfied both GC-O detection and ROAV > 1 criteria but were also consistently identified as key aroma compounds in both previous and current studies. Therefore, these compounds can be regarded as the key aroma skeleton of soybeans fermented by B. subtilis BJ3-2. In addition to these key aroma skeleton components, 17 additional compounds were identified in the SGR1 and SGR2 groups (Table S9†) and were classified as key auxiliary aroma components of B. subtilis-fermented soybeans due to their GC-O detection and ROAV > 1. KEGG analysis indicated that the knockout of sdaAA and katX primarily affected serine and tryptophan metabolism, while the knockout of fadA and bkdB mainly influenced isoleucine metabolism. These metabolic changes, together with raw materials and fermentation conditions, contributed to the observed results.3.8. Sources and changes of key aroma skeleton components
The key aroma skeleton components detected in this study have been previously reported in the literature, further confirming their significant roles in the aroma formation of fermented soybean products. To understand the effects of fadA and bkdB on key aroma compounds, we compared the levels of these compounds across four fermented soybean samples (Table 3, p < 0.05). Additionally, we inferred the potential formation pathways of these compounds (Fig. 4B). 2-Methylbutyric acid, characterized by a sweaty odor, was primarily derived from isoleucine degradation.33,44 Based on the KEGG database, bkdB in Bacillus subtilis encodes the lipoamide acyltransferase component of the branched-chain alpha-keto acid dehydrogenase complex, which primarily mediates the conversion of branched-chain alpha-keto acids to 2-methylbutyryl-CoA during isoleucine degradation. fadA encodes acetyl-CoA acyltransferase, which primarily facilitates the conversion of 2-methylacetoacetyl-CoA to acetyl-CoA during isoleucine degradation and also plays a role in generating acetyl-CoA through fatty acid β-oxidation. 2-Methylbutyryl-CoA and acetyl-CoA are key precursor metabolites for the synthesis of 2-methylbutyric acid.62–64 Thus, the deletion of these two genes significantly reduced 2-methylbutyric acid content in FS2–FS4, weakening the sweaty odor. Additionally, methyl 2-methylbutyrate could be synthesized from 2-methylbutyric acid, contributing to a fruity aroma.65,66 Ketones can be generated through microbial enzymatic activity acting on amino acids or lipids.67 B. subtilis produced α-acetolactate under aerobic conditions, which was decarboxylated to 3-hydroxy-2-butanone (acetoin) and further oxidized to 2,3-butanedione.68 Their levels were higher in FS2 and FS3 than in FS1 and FS4, indicating richer buttery aromas in FS2 and FS3. Moreover, 2,3-butanedione reacted with cysteine or alanine to form 2,4,5-trimethyloxazole.13 2-Heptanone, contributing a banana-like aroma, was detected in soybean oil and derived mainly from the carboxyl group of linoleic acid.69 Pyrazines, key aroma compounds in fermented soybean products, contributed to their roasted aroma. Their production depended on microbial metabolism using precursors like α-acetolactate, acetoin, amino acids, and ammonia.68 L-Threonine was converted through dehydrogenation, decarboxylation, and non-enzymatic reactions into alkylpyrazines.70 According to the KEGG database, fadA and bkdB were involved in the isoleucine degradation pathway. Knocking out these genes likely disrupted the pathway, causing L-threonine accumulation and increased production of 2,3,5-trimethylpyrazine, 2,5-dimethylpyrazine, and 3-ethyl-2,5-dimethylpyrazine in mutant strains. 1-Octen-3-ol, with a mushroom-like aroma, was derived from the enzymatic oxidation of linoleic and linolenic acids.43,50 2-Pentylfuran, contributing beany and grassy notes, likely formed via linolenic acid autoxidation and played a similar role in Korean doenjang.50 Both compounds were associated with the beany aroma, showing higher levels in FS2-FS4 than in FS1, with FS4 exhibiting the strongest beany aroma. 2-Ethylfuran, a marker of lipid oxidation, contributed caramel-like attributes and was mainly produced through lipid degradation.71 Its content varied across samples, with FS4 showing the highest level, enhancing its caramel-like aroma. Guaiacol, with its smoky aroma, was generated from the thermal degradation of lignin-related phenolic acids.37 Its content was higher in FS2 and FS3, aligning with their stronger smoky aromas, while FS1 and FS4 had lower levels, resulting in milder smoky aromas. These results were consistent with the QDA findings, confirming the impact of gene knockouts on aroma characteristics.| No. | Aroma compounds | RIca | RIrb | Compound contentc (μg kg−1) | |||
|---|---|---|---|---|---|---|---|
| FS1 | FS2 | FS3 | FS4 | ||||
| a Retention indices of the compounds calculated on a DB-WAX column (60 m × 0.25 mm × 0.25 μm) with a homologous series of n-alkanes (C7–C30).b Retention index obtained from a specific database (https://webbook.nist.gov/chemistry/name-ser/).c Different superscript letters (a–c) indicate significant differences in compound content (p < 0.05). | |||||||
| 1 | 2-Ethylfuran | 942.78 | 944 | 4.33 ± 2.47c | 8.91 ± 0.44bc | 10.09 ± 2.30b | 18.43 ± 4.09a |
| 2 | 2,3-Butanedione | 959.45 | 954 | 263.20 ± 27.18b | 665.64 ± 55.86a | 291.21 ± 69.68b | 149.24 ± 15.31c |
| 3 | Methyl 2-methylbutyrate | 1017.87 | 1004 | 7.00 ± 1.97c | 6.12 ± 0.87c | 10.58 ± 1.37b | 25.72 ± 0.20a |
| 4 | 2-Heptanone | 1181.68 | 1184 | 134.73 ± 19.13b | 67.43 ± 5.74c | 172.20 ± 5.34a | 156.27 ± 6.40a |
| 5 | 2,4,5-Trimethyloxazole | 1197.31 | 1197 | 71.21 ± 9.84b | 135.81 ± 6.81a | 69.95 ± 6.54b | 33.66 ± 3.07c |
| 6 | 2-Pentylfuran | 1226.42 | 1229 | 7.86 ± 2.16b | 21.37 ± 3.67a | 16.40 ± 3.94ab | 24.64 ± 8.77a |
| 7 | Acetoin | 1278.58 | 1286 | 490.60 ± 89.67b | 1963.38 ± 188.10a | 652.11 ± 174.58b | 40.63 ± 4.75c |
| 8 | 2,5-Dimethylpyrazine | 1333.13 | 1323 | 1868.70 ± 41.22c | 2459.33 ± 108.38b | 1927.81 ± 323.07c | 5781.23 ± 268.62a |
| 9 | 2,3,5-Trimethylpyrazine | 1408.95 | 1416 | 4655.50 ± 368.80ab | 4942.39 ± 1343.98ab | 5160.84 ± 154.00a | 3799.16 ± 277.16b |
| 10 | 1-Octen-3-ol | 1437.04 | 1435 | 283.77 ± 71.33c | 425.08 ± 112.61b | 543.22 ± 18.78ab | 606.03 ± 56.61a |
| 11 | 3-Ethyl-2,5-dimethylpyrazine | 1444.29 | 1447 | 63.55 ± 22.79b | 113.41 ± 23.50a | 64.53 ± 9.25b | 48.96 ± 7.51b |
| 12 | 2-Methylbutyric acid | 1653.67 | 1670 | 906.28 ± 88.38a | 722.88 ± 14.39b | 665.02 ± 27.36b | 163.29 ± 76.6c |
| 13 | Guaiacol | 1853.74 | 1859 | 49.01 ± 0.72b | 117.12 ± 24.32a | 128.70 ± 7.26a | 55.16 ± 0.70b |
In summary, the fadA and bkdB genes primarily influenced the formation of key aroma compounds by participating in the degradation pathways of valine, leucine, isoleucine, and fatty acid metabolism. The knockout of fadA and bkdB blocked isoleucine and fatty acid metabolism, reducing the production of 2-methylbutyryl-CoA and acetyl-CoA, which significantly decreased the levels of 2-methylbutyric acid. Moreover, the inhibition of isoleucine degradation redirected L-threonine into alternative metabolic pathways, affecting the production of 2,3-butanedione, 3-hydroxy-2-butanone, and 2,4,5-trimethyloxazole, while leading to the accumulation of aminoacetone. As a critical precursor, elevated aminoacetone levels directly increased alkylpyrazine concentrations. Additionally, 1-octen-3-ol, 2-heptanone, and 2-pentylfuran were derived from the metabolism of unsaturated fatty acids (linoleic acid or linolenic acid), while 2-ethylfuran was produced through lipid metabolism, indicating a close association between these aroma compounds and fatty acid metabolism. However, due to the complexity of biological metabolic regulation, the precise mechanisms by which fadA and bkdB knockout affect key aroma compounds remain unclear and require further study.
4 Conclusion
Current research on the aroma of fermented soybeans is abundant, but studies exploring soybean aroma from a genetic level are relatively scarce. This study systematically investigated the overall aroma profile and key aroma compounds of soybeans fermented by B. subtilis BJ3-2 and its mutant strains. Using multiple approaches—such as gene knockout, QDA, GC-O-MS, and PLSR—it specifically focused on the effects of the fadA and bkdB genes on the fermentation aroma. The fermented soybeans exhibited strong smoky, buttery, beany, and sweaty attributes. A total of 100 volatile compounds were detected by GC-MS. PLSR analysis revealed a good correlation between key aroma compounds and sensory attributes. By comparing previous studies, 12 key aroma skeleton compounds—such as 2,5-dimethylpyrazine (roasted, nutty), guaiacol (smoky, woody), and 2-methylbutyric acid (sweaty, stinky)—and 17 auxiliary aroma compounds were identified, collectively shaping the characteristic aroma of B. subtilis-fermented soybeans. The knockout of the fadA and bkdB genes improved the aroma of fermented soybeans primarily by influencing metabolic pathways involved in fatty acid metabolism and the degradation of branched-chain amino acids (valine, leucine, and isoleucine). Specifically, the gene knockout significantly reduced the production of 2-methylbutyric acid, thereby decreasing off-flavors, while promoting the formation of pyrazine compounds (e.g., 2,5-dimethylpyrazine and other roasted aroma substances), ultimately enhancing the overall aroma characteristics of fermented soybeans. Although the impact of gene knockout on key aroma components has been demonstrated, the accumulation of intermediate metabolites still requires analysis through metabolomics, and the molecular regulatory mechanisms remain to be further elucidated. The findings of this study provide new insights into the aroma profile of soybeans fermented by B. subtilis BJ3-2, offering valuable guidance for aroma control in future fermented soybean products. These strains will be applied in industrial fermentation to further validate their role in douchi production. Additionally, metabolic engineering will be employed to regulate precursor substances, aiming to enhance aroma consistency and improve product quality stability.Ethical statement
In this study, all participants provided informed consent before the experiments involving human subjects were conducted. All participants were in good health and had the freedom to withdraw from the study at any time.Data availability
All data supporting the conclusions of this study are included in the manuscript and ESI.†Author contributions
Feng Wen: investigation, methodology, experiment, data analysis, writing–original draft. Yongjun Wu: funding acquisition, supervision, project administration, and writing – review & editing. Jing Jin: supervision, project administration. Lincheng Zhang: writing – review & editing. Shuoqiu Tong: conceptualization, writing – review & editing. Cen Li: writing – review & editing. Li Ran: investigation, experiment, data analysis. Yuzhang Zhu: writing – review & editing.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was funded by the National Natural Science Foundation of China (Grant No. 32060585) and the Guizhou University Project (No. [2020]40).References
- Y. Qiao, K. Zhang, Z. Zhang, C. Zhang, Y. Sun and Z. Feng, Food Res. Int., 2022, 158, 111575 CrossRef CAS PubMed.
- L. Liu, X. Chen, L. Hao, G. Zhang, Z. Jin, C. Li, Y. Yang, J. Rao and B. Chen, Crit. Rev. Food Sci. Nutr., 2022, 62, 1971–1989 CrossRef CAS PubMed.
- N. K. Lee, W. S. Kim and H. D. Paik, Food Sci. Biotechnol., 2019, 28, 1297–1305 CrossRef CAS PubMed.
- Y. Feng, G. Su, H. Zhao, Y. Cai, C. Cui, D. Sun-Waterhouse and M. Zhao, Food Chem., 2015, 167, 220–228 CrossRef CAS PubMed.
- H. Song and J. Liu, Food Res. Int., 2018, 114, 187–198 CrossRef CAS PubMed.
- W. He and H. Y. Chung, Food Res. Int., 2019, 125, 108548 CrossRef CAS PubMed.
- Y. Xiao, H. Chen, Y. Wang, J. Ma, A. Hou, Y. Wang, Y. Chen and X. Lu, Curr. Res. Food Sci., 2024, 9, 100854 CrossRef CAS PubMed.
- Y. Yang, B. Wang, Y. Fu, Y. G. Shi, F. L. Chen, H. N. Guan, L. L. Liu, C. Y. Zhang, P. Y. Zhu, Y. Liu and N. Zhang, Food Chem., 2021, 346, 128880 CrossRef CAS PubMed.
- M. K. Park and Y. S. Kim, Food Chem., 2020, 305, 125461 CrossRef CAS PubMed.
- Y. Jia, B. Yuan, Y. Yang, C. Zheng and Q. Zhou, Food Chem., 2023, 423, 136290 CrossRef CAS.
- T. Gopikrishna, H. K. Suresh Kumar, K. Perumal and E. Elangovan, Ann. Microbiol., 2021, 71, 30 CrossRef PubMed.
- X. Chen, Y. Lu, A. Zhao, Y. Wu, Y. Zhang and X. Yang, Food Chem., 2022, 374, 131725 CrossRef CAS PubMed.
- Z. Chen, Y. Wu, S. Tong, J. Jin, L. Zhang, C. Li, Q. Tan, F. Wen and Y. Tao, Fermentation, 2024, 10, 409 CrossRef CAS.
- A. M. Abd-Alrahman, M. M. Ramadan, M. F. Maraay, R. Salem, F. M. Saleh, M. A. Hashim, A. Zhernyakova and T. M. El-Messery, Front. Nutr., 2023, 10, 1280209 CrossRef PubMed.
- Y. Gao, M. Hu, W. Meng, W. Wen, P. Zhang, B. Fan, F. Wang and S. Li, Food Chem., 2024, 443, 138523 CrossRef CAS PubMed.
- Z. Liu, Y. Wu, L. Zhang, S. Tong, J. Jin, X. Gong and J. Zhong, BMC Biotechnol., 2022, 22, 18 CrossRef CAS PubMed.
- Y. Wu, Y. Tao, J. Jin, S. Tong, S. Li and L. Zhang, BMC Microbiol., 2022, 22, 142 CrossRef CAS PubMed.
- Z. Yao, Y. Meng, H. G. Le, S. J. Lee, H. S. Jeon, J. Y. Yoo and J. H. Kim, J. Microbiol. Biotechnol., 2021, 31, 833–839 CrossRef CAS PubMed.
- Q. Tan, Y. Wu, C. Li, J. Jin, L. Zhang, S. Tong, Z. Chen, L. Ran, L. Huang and Z. Zuo, Foods, 2024, 13, 2731 CrossRef CAS PubMed.
- X. Gong, Master's thesis, Guizhou University, 2020.
- W. X. Du, M. Zhang and R. Ban, Chem. Biochem. Eng., 2007, 52–55 CAS.
- Y. Tao, X. Huang, S. Tong, C. Li and Y. Wu, J. Food Ferment. Sci. Technol., 2022, 58, 9–15 Search PubMed.
- H. Luo, Y. Wu, J. Jin, L. Zhang, S. Tong, C. Li, Q. Tan and Q. Han, RSC Adv., 2024, 14, 16368–16378 RSC.
- F. Tan, P. Wang, P. Zhan and H. Tian, Food Chem., 2022, 366, 130604 CrossRef CAS PubMed.
- H. Zhao, J. Xu, R. Wang, X. Liu, X. Peng and S. Guo, Foods, 2023, 12, 329 CrossRef CAS PubMed.
- X. Yin, Y. Xiao, K. Wang, W. Wu, J. Huang, S. Liu and S. Zhang, Food Res. Int., 2023, 174, 113515 CrossRef CAS PubMed.
- P. Yang, H. Song, L. Wang and H. Jing, J. Agric. Food Chem., 2019, 67, 7926–7934 CrossRef CAS.
- P. Yang, H. Song, Y. Lin, T. Guo, L. Wang, M. Granvogl and Y. Xu, Food Funct., 2021, 12, 4797–4807 RSC.
- X. H. Huang, B. S. Fu, L. B. Qi, L. D. Huo, Y. Y. Zhang, M. Du, X. P. Dong, B. W. Zhu and L. Qin, Food Funct., 2019, 10, 6473–6483 RSC.
- Y. Yue, C. Wang, Y. Chen, M. Zheng, Y. Zhang, Q. Deng and Q. Zhou, Food Chem., 2024, 432, 137095 CrossRef CAS PubMed.
- L. J. Van Gemert, Odour Thresholds : Compilations of Odour Threshold Values in Air, Water and Other Media, Oliemans Punter, 2011 Search PubMed.
- A. Giri, K. Osako, A. Okamoto and T. Ohshima, Food Res. Int., 2010, 43, 1027–1040 CrossRef CAS.
- D. I. Serrazanetti, M. Ndagijimana, S. L. Sado-Kamdem, A. Corsetti, R. F. Vogel, M. Ehrmann and M. E. Guerzoni, Appl. Environ. Microbiol., 2011, 77, 2656–2666 CrossRef CAS PubMed.
- E. Huang, J. S. Yan, R. G. Gicana, Y. R. Chiang, F. I. Yeh, C. C. Huang and P. H. Wang, Chemosphere, 2023, 322, 138200 CrossRef CAS PubMed.
- J. Boch, B. Kempf, R. Schmid and E. Bremer, J. Bacteriol., 1996, 178, 5121–5129 CrossRef CAS PubMed.
- J. G. Baek, S. M. Shim, D. Y. Kwon, H. K. Choi, C. H. Lee and Y. S. Kim, Biosci., Biotechnol., Biochem., 2010, 74, 1860–1868 CrossRef CAS PubMed.
- S. Wang, Y. Chang, B. Liu, H. Chen, B. Sun and N. Zhang, Molecules, 2021, 26, 3048 CrossRef CAS PubMed.
- G. Wei, J. M. Regenstein and P. Zhou, Food Res. Int., 2021, 147, 110473 CrossRef CAS PubMed.
- W. Fan, Y. Xu and Y. Zhang, J. Agric. Food Chem., 2007, 55, 9956–9962 CrossRef CAS PubMed.
- C. Larroche, I. Besson and J.-B. Gros, Process Biochem., 1999, 34, 667–674 CrossRef CAS.
- I. Besson, C. Creuly, J. Gros and C. Larroche, Appl. Microbiol. Biotechnol., 1997, 47, 489–495 CrossRef CAS.
- C. Hong, Y. Chen, L. Li, S. Chen and X. Wei, J. Agric. Food Chem., 2017, 65, 1592–1597 CrossRef CAS PubMed.
- S. M. Lee, S. B. Kim and Y. S. Kim, J. Food Sci., 2019, 84, 2758–2776 CrossRef CAS PubMed.
- Y. Hong, H.-J. Jung and H.-Y. Kim, Food Sci. Biotechnol., 2012, 21, 1163–1172 CrossRef CAS.
- Y. Xu, J. Zhao, X. Liu, C. Zhang, Z. Zhao, X. Li and B. Sun, Food Chem., 2022, 369, 130920 CrossRef CAS PubMed.
- M. Flores, M. A. Durá, A. Marco and F. Toldrá, Meat Sci., 2004, 68, 439–446 CrossRef CAS PubMed.
- Z. Xiao, S. Tingting, J. Hang, G. Chunfeng, W. Zhouli, Y. Tianli and Y. Yahong, LWT, 2022, 155, 112936 CrossRef.
- Y. Suezawa and M. Suzuki, Biosci., Biotechnol., Biochem., 2007, 71, 1058–1062 CrossRef CAS PubMed.
- A. Adams, C. Bouckaert, F. Van Lancker, B. De Meulenaer and N. De Kimpe, J. Agric. Food Chem., 2011, 59, 11058–11062 CrossRef CAS PubMed.
- Q. C. Chen, Y. X. Xu, P. Wu, X. Y. Xu and S. Y. Pan, Int. J. Food Sci. Technol., 2011, 46, 1823–1829 CrossRef CAS.
- A. M. Vidal, S. Alcalá, A. De Torres, M. Moya, J. M. Espínola and F. Espínola, Molecules, 2019, 24, 3587 CrossRef CAS PubMed.
- R. Ni, P. Wang, P. Zhan, H. Tian and T. Li, Food Chem., 2021, 357, 129786 CrossRef CAS PubMed.
- M. Q. Wang, W. J. Ma, J. Shi, Y. Zhu, Z. Lin and H. P. Lv, Food Res. Int., 2020, 130, 108908 CrossRef CAS PubMed.
- C. M. Delahunty, G. Eyres and J. P. Dufour, J. Sep. Sci., 2006, 29, 2107–2125 CrossRef CAS.
- S. Duan, Z. Tian, X. Zheng, X. Tang, W. Li and X. Huang, Food Chem., 2024, 458, 139422 CrossRef CAS PubMed.
- Y. Huang, X. Peng, Y. Chen, Y. Wang, J. Ma, M. Zhu, Z. Liu and Y. Xiao, LWT, 2025, 117765 CrossRef CAS.
- D. Meng, D. Zhao, Z. Zhao, X. Wang, Y. Wu, Y. Li, Z. Lv and Q. Zhong, Food Chem., 2025, 476, 143430 CrossRef CAS PubMed.
- J. Zhang, W. Zhang and L. Xing, Food Chem., 2021, 365, 130411 CrossRef CAS PubMed.
- S. Bao, Q. An, Y. Yang, X. Li, G. Chen, Y. Chen, J. Chen, Z. Liu and J. Huang, Food Res. Int., 2025, 208, 116278 CrossRef CAS PubMed.
- A. Leejeerajumnean, S. C. Duckham, J. D. Owens and J. M. Ames, J. Sci. Food Agric., 2001, 81, 525–529 CrossRef CAS.
- S.-J. Lee and B. Ahn, Food Chem., 2009, 114, 600–609 CrossRef CAS.
- J. Qi, X. Xiao, L. Ouyang, C. Yang, Y. Zhuang and L. Zhang, Microb. Cell Fact., 2022, 21, 238 CrossRef CAS PubMed.
- A. Matich and D. Rowan, J. Agric. Food Chem., 2007, 55, 2727–2735 CrossRef CAS PubMed.
- B. Ganesan, P. Dobrowolski and B. C. Weimer, Appl. Environ. Microbiol., 2006, 72, 4264–4273 CrossRef CAS PubMed.
- Y. Zhang, M. A. Fraatz, F. Birk, M. Rigling, A. Hammer and H. Zorn, Food Chem., 2018, 266, 475–482 CrossRef CAS PubMed.
- X. Zhang, X. Lu, C. He, Y. Chen, Y. Wang, L. Hu, Q. Qing, M. Zhu, Z. Liu and Y. Xiao, Food Res. Int., 2025, 209, 116279 CrossRef CAS PubMed.
- Y. Xiao, H. Chen, Y. Chen, C. T. Ho, Y. Wang, T. Cai, S. Li, J. Ma, T. Guo, L. Zhang and Z. Liu, Food Res. Int., 2024, 197, 115219 CrossRef CAS PubMed.
- Y. Liu, H. Song and H. Luo, Food Res. Int., 2018, 112, 175–183 CrossRef CAS PubMed.
- S. Ghorbani Gorji, M. Calingacion, H. E. Smyth and M. Fitzgerald, J. Food Sci. Technol., 2019, 56, 4076–4090 CrossRef CAS PubMed.
- L. Zhang, Y. Cao, J. Tong and Y. Xu, Appl. Environ. Microbiol., 2019, 85, e01807 CAS.
- K. Kantono, N. Hamid, D. Chadha, Q. Ma, I. Oey and M. M. Farouk, Foods, 2021, 10, 1148 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S5: construction of homologous recombination knockout plasmids; Fig. S6–S8: validation of BJ3-2ΔfadA, BJ3-2ΔbkdB, and BJ3-2ΔfadAΔbkdB; Fig. S9: determination of volatile content in fermented soybeans; Table S1: the strains and plasmids used for this study; Table S2: primers and PCR details used in this study; Table S3: transcriptome sequencing results; Table S4: RT-qPCR results; Table S5: results of variance analysis of OD600 values at each period in the growth curves of the four strains; Table S6: identification and quantification of volatile compounds in different fermented soybeans; Table S7: GC-O analysis; Table S8: ROAV values of all aroma compounds; Table S9: screening of key aroma skeleton components and auxiliary aroma components in combination with previous findings. See DOI: https://doi.org/10.1039/d5ra02302e |
| This journal is © The Royal Society of Chemistry 2025 |