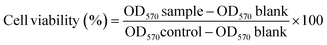DOI:
10.1039/D5RA02118A
(Paper)
RSC Adv., 2025,
15, 22006-22017
Mn and Cu complexes of a novel malic acid–cysteine ligand with remarkable ROS scavenging activity†
Received
26th March 2025
, Accepted 10th June 2025
First published on 4th July 2025
Abstract
The scientific community is very interested in investigating antioxidant activity using various assays to treat oxidative stress and reduce the harmful effects of free radicals. Cysteine, a sulfur-containing compound can treat oxidative stress. The primary objective of this study was to create a novel ligand through the combination of L-cysteine with malic acid and its metal complexes with copper and manganese. The following discussion focuses on how the complexes mimic natural antioxidant enzymes and explains how they could be used to remove reactive oxygen species. Various methods were used to verify the synthesis, including FT-IR, UV-vis, TGA, CHNS, EDX, SEM, mass, and fluorescence. These techniques indicated structural changes upon complexation and integration with β-CD. Spectrophotometric tests were used to evaluate the compounds' peroxidase-, catalase-, and superoxide dismutase-like activities under different conditions. Cytotoxicity was evaluated using HFF-2 cells and the MTT test. The amounts of carbon, hydrogen, nitrogen, and sulfur were 39.48, 4.7, 6.58, and 15.23, and the ligand formula is C7H11NSO6. Changes in infrared spectral signals upon metal interaction, particularly in the carboxylate region, indicate significant metal–ligand bonding. The results showed that the β-CD-modified complexes, especially Cu-complex/β-CD and Mn-complex/β-CD, had better antioxidant activity (91% and 78%, respectively). They were better at scavenging O2˙− radicals and breaking down H2O2 into safe byproducts. Additionally, Mn-complex/β-CD was less cytotoxic. In conclusion, the produced complexes are promising antioxidants and potential treatments for diseases linked to oxidative stress, especially the β-CD derivatives.
1 Introduction
Nowadays, numerous food and health products incorporate advantageous phytochemical compounds. Among these compounds, malic acid is a specific type of α-hydroxy acid in fruits like apples and pears. Malic acid is utilized to manage microbial contamination and inhibit the proliferation of pathogens in food before and after food items are processed.1,2 It assumes a crucial function in the metabolic pathways of both plants and animals, participating in various fundamental biochemical processes, such as the Krebs cycle.3 The Krebs cycle comprises a sequence of enzymatic reactions aimed at acquiring energy through the breakdown of carbohydrates. Malic acid plays a crucial role in energy metabolism within the muscle system by promoting the synthesis of adenosine triphosphate (ATP), a molecule essential for providing energy to muscle cells. Furthermore, malic acid exhibits metal chelating properties, enabling it to bind to harmful metals accumulated in the liver and facilitate their removal.4 It has also been identified as a versatile ligand in coordination chemistry. A study conducted in 2013 indicated that malic acid might have therapeutic benefits in treating melasma.5 Furthermore, research demonstrated that the combination of malic acid and magnesium could potentially alleviate pain and sensitivity in individuals with fibromyalgia.6 A recent study demonstrated that malic acid exhibits good antioxidant properties and significant antibacterial effects.7 Malic acid has been found to interact with specific amino acids, including arginine.8 Furthermore, L-histidine s-malate is a bioactive chemical. Cysteine stands out from other naturally occurring amino acids due to its thiol-containing side chain, which plays a crucial role in oxidation–reduction reactions. This amino acid exhibits antioxidant characteristics, such as binding to metals and eliminating free radicals.9 Amino acids are capable of generating metal–organic framework (MOF) hydrogels.10 Cysteine is instrumental in the formation of these hydrogels, especially those based on peptides, as it promotes self-assembly and cross-linking via disulfide bonds. Cyclodextrins have a non-polar hole within and hydroxyl groups on the outside. The main mechanism via which hydrophobic compounds are included is through hydrophobic interactions between the cyclodextrin cavity walls and guest molecules.11 Hydrophobic interactions between the guest molecules and the cyclodextrin cavity walls are the primary mechanism by which hydrophobic compounds undergo inclusion complexation. The guest binding may also be related to other forces like van der Waals and dipole–dipole interaction.12–14
Stress oxidative in aerobic organisms produces reactive oxygen species (ROS). Production and removal of ROS are in a reasonably balanced state under normal conditions. However, oxidative stress can result from a large increase in ROS production under internal or external environmental stress.15 Antioxidant compounds have been widely recognized for their potential therapeutic benefits in addressing various disorders linked to oxidative stress caused by free radicals. The detrimental impact of oxidative stress processes has been implicated in a range of health conditions, including diabetes, neurodegenerative diseases, cancer, and aging.16 ROS scavenging natural and synthetic antioxidants have become increasingly popular due to the search for new medicinal agents. In this viewpoint, synthetic versions of antioxidant enzymes, such as superoxide dismutase (SOD) or catalase, are proposed as potential novel medications to avert damage caused by oxidative stress. Transition metals are essential for the biological functions linked to bioinorganic particles.
Until now, there have been no reports of a combination that includes cysteine and malic acid. The conjunction of cysteine and malic acid is a viable ligand for metal coordination while enhancing their antioxidant properties. The utilization of multiple antioxidant efficacy assays such as (SOD)-like, catalase-like, and peroxidase-like activities enable a comprehensive understanding of the antioxidant activity exhibited by the ligand and mononuclear complexes of manganese and copper.
2 Experimental
2.1. Materials and equipment
L-Cysteine (C3H7NO2S, 99%), DL-malic acid (C4H6O5, 98%), manganese(II) chloride dihydrate (MnCl2·2H2O, 98%), copper(II) acetate monohydrate (Cu(CH3COO)2·H2O, 99%), ammonium iron(II) sulfate (FeSO4(NH4)2SO4·6H2O, 99.99%), tris(hydroxylmethyl)aminomethane (Tris-buffer, 99%), sodium acetate (CH3COONa, 98.5%) and 3,3′,5,5′-tetramethylbenzidine dihydrochloride (TMB, 99%) were obtained from Merck Company (Germany), and pyrogallol (C6H3(OH)3, 98%), 1,10-phenanthroline monohydrate (99%), and sulfuric acid (H2SO4, 95%) from Sigma-Aldrich without further purification. Methanol (99.8%), ethanol (96%), hydrogen peroxide (H2O2, 30%), and β-Cyclodextrin (98%) were purchased from Mojallali Company (Saveh, Iran) and S. D. Fine chemicals in Mumbai, respectively. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, 99%), and dimethyl sulfoxide (DMSO, 99.99%) were sourced from Sigma/Aldrich without the requirement for further purification. The Roswell Park Memorial Institute 1640 (RPMI) and penicillin/streptomycin were obtained from Bio-idea in Tehran, Iran. The fetal bovine serum (FBS) was sourced from Capricorn Scientific GmbH. HFF-2 cells were purchased from the National Cell Bank of Iran (NCBI) in Tehran, Iran, at the Pasteur Institute.
Elemental analysis (C, H, N, and S) of the ligand was performed using a model FLASH EA 1112 SERIES, Thermo Finnigan analyzer. The mass spectrum of the ligand was recorded on an Agilent Technologies G708113 550D. Fourier transform infrared (FTIR) spectroscopy was conducted using a Bruker TENSOR 27 model FTIR spectrophotometer from Germany, examining the 400–4000 cm−1 range. Sample preparation for FTIR analysis involved thoroughly mixing compounds and KBr powders to create pellets, which were subsequently compressed. Electronic spectroscopy employed an ultraviolet-visible (UV-vis) spectrophotometer in deionized water (Analytik Jena, Specord 250) for electronic spectra. The fluorescent characteristics of compound solutions were measured using a JASCO Corp. FP-750 spectrofluorometer. Energy dispersive X-ray (EDX) spectra and Scanning Electron Microscopy (SEM) were carried out using a Tescan MIRA3 FEG-SEM instrument from the Czech, providing comprehensive analytical data. The optical absorbance of the wells at a wavelength of 570 nm was quantitatively determined using a microplate reader obtained from BioTek Instruments, Inc., USA. Thermogravimetric analysis (TGA) was carried out on the samples using a TG/SDTA 851 Mettler Toledo apparatus. Around 10 milligrams of the specimen underwent heating at a rate of 10 °C min−1 until reaching 700 °C in an air atmosphere.
2.2. Synthesis of CMA ligand: [4-((1-carboxy-2-mercaptoethyl)amino)-3-hydroxy-4-oxobutanoic acid]
The ligand was synthesized by dissolving a mixture of 0.01 mol of cysteine and 0.01 mol of malic acid in deionized water. After an hour of stirring, the mixture was heated for 9 hours at 135 °C. The resulting precipitate had a mustard color (1.6 gr, yield: 67%); m. p.: 165 °C; Anal calcd. for C7H11NSO6 (MW: 237.07): C, 39.48; H, 4.7; N, 6.58; and S, 15.23. Found: C, 40.76; H, 4.28; N, 7.38; and S, 15.23. FT-IR data (ν/cm−1): 1719 ν(carboxylic acid group), 1561 ν(amide group), 1538 and 1097 ν(C–N), 2571 ν(S–H), 2947 ν(C–H). Mass (m/z): 236.1 [C7H11NSO6].
2.3. Synthesis of metal complexes
To a ligand solution in 10 mL of methanol (0.3 g, 125 × 10−5 mol), a heated methanolic solution (5 mL) of MnCl2·2H2O (0.2 g, 125 × 10−5 mol) was added. Subsequently, the reaction mixture was refluxed at 65 °C and stirred continuously for 6 hours before being cooled in an ice bath to induce precipitation. The resulting solid product was filtered, washed multiple times with methanol, and then dried. The complexes were prepared in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio. In the synthesis of the Cu(II) complex, the ligand (0.3 g, 125 × 10−5 mol) was dissolved in 10 mL of ethanol and mixed with an ethanolic solution of metal(II) acetate (0.25 g, 125 × 10−5 mol). The resulting mixture was refluxed at 75 °C for 12 hours. After the completion of the reaction, the precipitate was separated by centrifugation and cleaned with ethanol, and dried at room temperature.
1 stoichiometric ratio. In the synthesis of the Cu(II) complex, the ligand (0.3 g, 125 × 10−5 mol) was dissolved in 10 mL of ethanol and mixed with an ethanolic solution of metal(II) acetate (0.25 g, 125 × 10−5 mol). The resulting mixture was refluxed at 75 °C for 12 hours. After the completion of the reaction, the precipitate was separated by centrifugation and cleaned with ethanol, and dried at room temperature.
2.4. Synthesis of metal complexes/β-CD
0.15 grams of the metal complexes were introduced into 5 mL of deionized water and heated at 70 °C for 30 minutes. Subsequently, 0.255 g of β-CD was dissolved in 5 mL of distilled water. The pH of the solution was raised to 9 by adding NaOH and integrated into the solution through vigorous stirring for 9 hours. Following this, ethanol was gradually added until a solid precipitate formed. The resulting precipitate was filtered and left to dry under normal atmospheric conditions.
2.5. Quantum yield (QY)
The QY of the prepared samples was determined by using quinine sulfate as the standard fluorescent molecule, which has a quantum yield of 54% in a water solution. Four different samples and quinine sulfate concentrations were prepared to determine the QY to achieve absorbance levels below 0.1. Following this, the fluorescence emissions were measured at these concentrations. The quantum yield of the nanoparticle can be calculated utilizing the following equation:
In this formula, the symbols ‘S’ and ‘R’ represent the samples and Q.S, respectively. The variables F, A, and η represent the area of the fluorescence spectrum, absorbance, and refractive index, respectively.17
2.6. Antioxidant assay
2.6.1. In vitro SOD-like assay. In an alkaline solution, pyrogallol can self-autoxidize; during this process, O2˙– is produced. SOD exhibits the ability to degradation of O2˙–, and finally prevent the autoxidation process of pyrogallol.18 The specific experimental protocol followed: 3 mL of Tris–HCl buffer (50 mM, pH 8.2) was mixed with 2 mg of complexes at 37 °C. The reaction mixtures were incubated for 10 minutes before being combined with pyrogallol (9 mM) in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio (Tris–HCl buffer
1 volume ratio (Tris–HCl buffer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pyrogallol). After that, a UV-vis spectrophotometer operating at wavelengths of 325 and 420 nm was used to monitor the reaction mixture. After that, the capacity to scavenge superoxide radicals (O2˙–) was determined as:
pyrogallol). After that, a UV-vis spectrophotometer operating at wavelengths of 325 and 420 nm was used to monitor the reaction mixture. After that, the capacity to scavenge superoxide radicals (O2˙–) was determined as:
A0 represents the absorbance value of pyrogallol while A1 represents the absorbance value of the sample after a 10 minute reaction.19
2.6.2. In vitro catalase-like assay. To assess the H2O2 scavenging activity of antioxidants, ferrous ammonium sulfate and 1,10-phenanthroline were utilized.20 In each test tube, 5 mg of the complex and 1 mL of H2O2 solution (10 mM) were combined and subsequently incubated at room temperature for 30 minutes. After the incubation period, 3 mL of 1,10-phenanthroline (1 mM) and 0.5 mL of ferrous ammonium sulfate (1 mM) were introduced into the test tube, and thoroughly mixed. Finally, the absorption was measured at 510 nm. A reference solution was prepared without any active compounds, consisting of the same levels of 1,10-phenanthroline and ferrous ammonium sulfate. The solution with the highest absorbance acted as a benchmark to compare the effectiveness of other solutions in removing hydrogen peroxide, which was evaluated using a formula calculating the percentage of H2O2 removal. This formula was as follows:
In this formula, Atest represents the absorbance of the solution containing the mentioned compounds. By comparing the absorbance of the test solution to the absorbance of the blank solution, the percentage of H2O2 scavenging activity could be determined, providing insight into the antioxidative capabilities of the compounds under investigation.
2.6.3. In vitro peroxidase-like assay. The peroxidase-like activity of compounds was measured by an oxidation reaction of substrate TMB in the presence of H2O2. A mixture containing 100 μL of catalysts (5 mg mL−1), 100 μL of 4.16 mM 3,3′,5,5′-tetramethylbenzidine (TMB) dissolved in ethyl acetate, and 100 μL of 4.98 M H2O2 was added to 2 mL of HAcNaAc buffer (0.1 M, pH 5.0). After a 15-minute incubation at 30 °C, the absorbance of the samples was measured between 500–750 nm, and the absorbance intensity at 652 nm of the blue solution from TMB oxidation was determined with a UV-vis spectrophotometer.21 The percentage of the samples' relative activity was calculated using the formula:
A0 and A represent the absorbance values in the presence and absence of a catalyst, respectively.
2.6.3.1. Optimization conditions for the reaction. Various experimental factors like the pH of the buffer, temperature, incubation time, and Cu-complex/β-CD concentration influenced the catalytic activity of the samples. Therefore, it is necessary to examine their impacts. The pH of the buffer was studied within the range of 3.5 to 5.5, the incubation time from 1 to 15 minutes, the temperature ranged from 20 °C to 45 °C, and the concentration of Cu-complex/β-CD (0, 43.47, 86.95, 130.43, 173.91, 217 μg mL−1) in the reaction medium was determined.
2.6.3.2. Kinetic assay of the peroxidase-like activity. The investigation was focused on the steady-state kinetic property of Cu-complex/β-CD. In brief, experiments were incubated under optimal conditions with Cu-complex/β-CD (173.91 μg mL−1), a fixed H2O2 concentration of 0.109 M, and various TMB concentrations in acetate buffer. In contrast, other mixtures contained catalyst (173.91 μg mL−1), a fixed TMB concentration of 0.18 mM, and various H2O2 concentrations in acetate buffer at pH 5, were incubated for 15 min at 30 °C. All solutions were analyzed using a UV-vis spectrophotometer. Subsequently, the kinetic parameters were determined using Lineweaver–Burk plots and the Michaelis–Menten equation.21,22
Vmax, V, Km, and [S] represent the maximum reaction rate, initial reaction rate, the Michaelis constant, and substrate concentration, respectively. The values of Vmax and Km were obtained from the Lineweaver–Burk double reciprocal plots.
2.7. Cell viability assay
In this particular investigation, the utilization of HFF-2 cells was employed to assess the potential toxicity of the compounds under examination. The approach taken in this methodology hinges upon the reduction of a tetrazolium salt known as MTT.23 To culture and maintain the HFF-2 cells, a specific medium known as RPMI was utilized. To enhance the growth and viability of the cells, 10% FBS, and 1% penicillin/streptomycin were added to the medium as well. The cells were cultured in a controlled environment with a specific temperature of 37 °C and a carbon dioxide (CO2) concentration of 5%. A particular procedure was followed to evaluate the prepared samples' cytotoxic effects on the MCF-7 cells. Firstly, a total of 1.0 × 104 cells per well were seeded into a 96-well plate. These cells were then allowed to incubate for approximately 24 hours. Following this incubation period, the cells were subjected to various concentrations of compounds ranging from 0 to 700 μg mL−1. The cells were further incubated for 48 hours. After the abovementioned incubation, each well was treated with 20 μL of MTT solution with a 3.0 mg mL−1 concentration. These treated wells were then subjected to an additional 4 hours of incubation at 37 °C. Once the 4-hour incubation period had concluded, the resultant purple formazan crystals were solubilized. This solubilization process was done by replacing the RPMI media in each well with 200 μL of DMSO.24 The following formula was used to determine cell viability:25
3 Results and discussion
3.1. FT-IR spectra
In the 400–4000 cm−1 vibration band, the racemic form of malic acid was described and discussed (Fig. S1†): 3447 cm−1 (υOH of CHOH), 3061 cm−1 (υOH of COOH), 1443 cm−1 (υC–O, δOH of COOH), 1408 cm−1 (δCH2), 1360 cm−1 ((δCH2)scis, (δCH2)wag), 1293 cm−1 (δOH, υC–O of COOH), 1220 cm−1 ((δCH2)wag), 1181 cm−1 ((δCH2)twist, (δCH2)scis), 1101 cm−1 (δCH2, υC–O, δOH), 1028 cm−1 (υC–C, γCH), 960 cm−1 (υC–C, δC–O), 885 cm−1 (δC–O, υC–CH2, δC–H), 664 cm−1 (δC–H, δC–CH2, δOH, δCH2, υC–C), 608 cm−1 (hydrogen bonded) and 478 cm−1 ((δCOO)wag, (δCOO)rock).26 Fig. 1 illustrates the FTIR spectrum of ligand, Mn-complex, Mn-complex/β-CD, Cu-complex, and Cu-complex/β-CD. The IR spectra of the ligand exhibited numerous significant distinctions when compared to cysteine and malic acid. The different sharp FTIR peaks represent the ligand's functional groups. The thiol stretch at 2571 cm−1 shows free thiol in the ligand. The appearance of a new peak at 1651 cm−1 could be linked to the presence of amide groups within the ligand. Furthermore, the prominent peaks at 1719 cm−1 can be associated with the carboxylic acid functional group present in the ligand. The vibrational frequencies observed at 1397, 1523, and 3368 cm−1 can be attributed to the symmetric and asymmetric stretching of the carboxylate group, and the stretching motion of the OH stretching, respectively. The absorption peaks at 1180 and 1097 cm−1 show the C–O stretching bonds.
 |
| | Fig. 1 FT-IR spectra of prepared compounds. | |
According to Pearson's principle,27 the reduction in both asymmetric and symmetric stretches of the carboxylate functional group within manganese and copper complexes compared to the free ligand is attributed to their coordination with the metal through oxygen atoms originating from the carboxylic acid groups present in the ligand. The observed decrease in the asymmetric stretch vibration indicates the coordination of Mn with the carboxylate group. As the metal and ligand bond strengthens, the frequency shifts move towards lower or higher values.28 Moreover, the absence of the thiol group implies the potential coordination of values.28 Moreover, the absence of the thiol group implies the potential coordination of copper with sulfur atoms, highlighting the likelihood of a distinct chemical bonding interaction between copper and sulfur within the complexes. Within the spectrum of β-CD inclusion complexes, a broad peak at 3380 cm−1 signifies the stretching vibration of O–H, while the asymmetric stretching vibration of the –CH aliphatic group is evident at 2930 cm−1. A prominent peak at 1028 cm−1 is associated with the C–O–C stretching mode, with peaks at 1156 cm−1 and 1418 cm−1 denoting the asymmetric C–O stretching and CH2 bending modes, respectively. The intensity of the amide group disappeared and decreased in Mn-complex/β-CD and Cu-complex/β-CD, respectively. The absorption peak at 1646 cm−1 is attributed to the bending vibration of H–O–H in βCD (Fig. 1).
3.2. Electronic transition and fluorescence studies
UV-vis absorption and fluorescence spectra of the ligand and its related complexes are shown in Fig. 2a and b. The n–σ* and π–π* transitions, as well as the n–π* transitions of the lone pair electrons from the amine and sulfur to the π* orbital of a carbonyl group, have been observed at 220 nm, a shoulder at 255 nm, and at 350–360 nm.29 The absence of n–π* transitions associated with the ligand in Cu-complexes validates the coordination of copper with the ligand and the formation of copper complexes. Moreover, the observation of ligand-to-metal transfer (LMCT) bands at 235 nm, which overlap with the ligand peak at 255 nm, confirms metal coordination with the oxygen atoms of Cys and malic.
 |
| | Fig. 2 (a) UV-vis spectra and (b) fluorescence spectra of ligand and its metal compounds. | |
Fig. 2b displays the fluorescence spectra of Cys, malic acid, and the synthesized compounds in deionized water at natural pH. While cysteine and malic acid showed low fluorescence, the ligand and its complexes showed strong fluorescence with a significant emission peak at 464 nm under excitation at 370 nm. Therefore, it can be suggested that the fluorescence property was associated with an organic one-pot reaction between malic acid and Cys, induced by the electronic excitation of n–π*. A reduction in the fluorescence intensity of the complexes due to the addition of metal to the ligand is observed, which confirms the metal-to-ligand coordination, as Mn(II) and Cu(II) ions quench the intrinsic fluorescence of the ligand.30,31 The fluorescence quenching observed in metal-containing samples is attributed to the static effect of metal ion coordination (Mn2+, Cu2+) with the amino acid groups (amino and carboxyl groups) of the ligand. The fluorescence absorbance and quantum yields of Mn-complex/β-CD and Cu-complex/β-CD decreased more significantly in comparison to Mn-complex and Cu-complex, respectively, due to the interactions between (Mn-complex) and (Cu-complex) with β-CD.
3.3. Energy-dispersive X-ray analysis
To assess the existence of C, O, N, S, Mn, and Cu elements, a Field Emission Scanning Electron Microscope (FESEM) equipped with an energy-dispersive X-ray (EDX) was utilized Fig. S2,† Table 1. The presence of Mn and Cu confirms the coordination of ligand to metal. Through the EDX data analysis, it was deduced that adding complexes to cyclodextrin decreases the metal content within the complexes, while the percentage of carbon and oxygen increases in both samples. The determined ratios closely corresponded with the stoichiometric ratios.
Table 1 Analytical data and some physical properties of the ligand and their complexes
| Compounds |
Formula |
Mp (°C) |
Colour |
ɸ |
Wta (%) |
| C |
O |
N |
S |
Metal |
| The prepared compounds' elemental content was determined using EDX analysis. |
| Ligand |
C7H11NSO6 |
165 |
Mustard |
44.4% |
40.84 |
37.22 |
9.81 |
12.14 |
— |
| Mn-complex |
[Mn(C7H10NO6S)2(H2O)2] |
110 |
Cucao |
39.9% |
40.58 |
24.67 |
9.08 |
15.35 |
Mn (10.38) |
| Mn-complex/β-CD |
— |
208 |
Pine |
36.1% |
42.76 |
44.23 |
3.99 |
1.83 |
Mn (2.60) |
| Cu-complex |
[Cu(C7H10NO6S)(CH3COO)] |
235 |
Olive |
12.8% |
36.79 |
22.2 |
12.93 |
11.61 |
Cu (16.48) |
| Cu-complex/β-CD |
— |
280 |
Almand |
2.7% |
46.15 |
43.75 |
3.52 |
2.66 |
Cu (3.92) |
3.4. Scanning electron microscopy
The morphology and surface features of both the ligand and its metal complexes are shown in Fig. 3, as determined by SEM analysis. The ligand demonstrates a sheet-like structure. After the formation of Mn-complex and Cu-complex, the sheet-like structure of the ligand was deformed to a spherical structure. The SEM images indicated that the addition of cyclodextrin led to a growth in particle size.
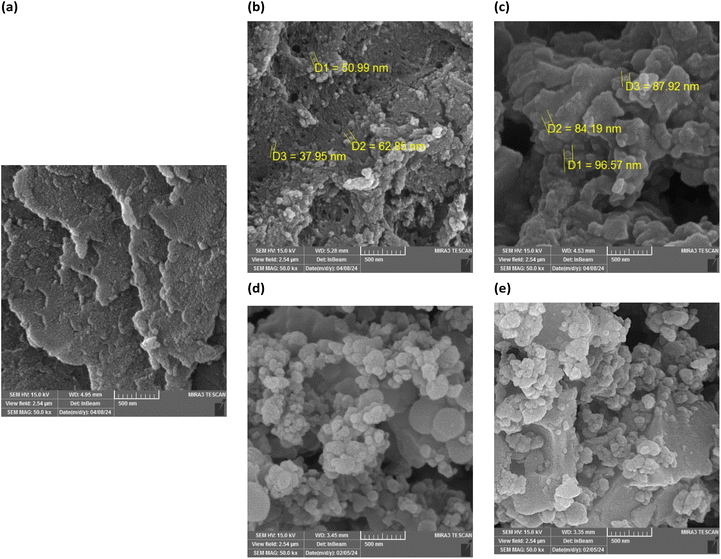 |
| | Fig. 3 SEM image of (a) ligand; (b) Mn-complex; (c) Mn-complex/β-CD; (d) Cu-complex and (e) Cu-complex/β-CD. | |
3.5. Thermogravimetric analysis
Fig. 4 illustrates the TGA diagrams of Mn-complex/β-CD and Cu-complex/β-CD, conducted at a temperature range of 0 –700 °C with a heating rate of 10 °C min−1 under an air atmosphere. The distinct stages of degradation were identified through an analysis of weight loss values related to temperature. βCD demonstrates two distinct weight losses at 95 and 310 °C, attributed to the evaporation of water molecules (11.75% weight loss) and the decomposition of βCD (66.7% weight loss), respectively.32 Analysis of the pure cysteine revealed a single-step decomposition process, with the removal of organic groups and adsorbed complex/β-CD exhibits a total of four distinct steps: the first one, the loss of water from within the β-CD cavity occurs between 26–183 °C, followed by the second step at 183–235 °C attributed to cysteine and lastly, the temperature of 105 °C, associated with water loss, is considerably lessened in the Cu-complex/β-CD in contrast to the Mn-complex/β-CD. The second step occurs at 190 °C due to cysteine decomposition, and the third step at 361 °C relates to β-CD. The reduced water loss in Cu-complex/β-CD compared to Mn-complex/β-CD suggests stronger water molecule interactions in the Cu-complex/β-CD. The weight loss in the fourth step from 361 °C for both compounds is due to metal oxide formation. Cysteine and βCD decomposition temperatures are higher in Mn-complex/β-CD and Cu-complex/β-CD, indicating increased thermal stability. The compound is confirmed to be highly pure and free of inorganic impurities when the weight fraction drops to zero after continued heating.33
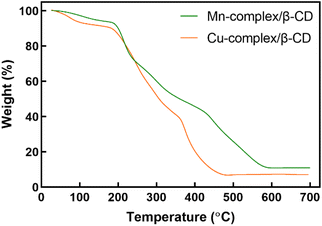 |
| | Fig. 4 TGA curves of Mn-complex/β-CD and Cu-complex/β-CD. | |
3.6. Antioxidant assay
3.6.1. SOD-like activity. A metalloenzyme known as superoxide dismutase catalyzes superoxide radicals (O2˙–) into H2O2 autoxidation in a pH 8.2 tris buffer solution was measured (Fig. 5). During the autoxidation process, UV–vis spectra exhibit three absorption peaks at 270–275 nm, 310–320 nm, and 420–440 nm. Li demonstrated that the absorbance at 325 nm exhibited greater sensitivity than that at 420 nm under pH conditions of 7.4 and 8.2.34 The antioxidant can deactivate O2˙–, resulting in a reduction of the absorbance of the reaction mixture at 325 nm. Theoretically, an oxidized metal ion can be reduced by O2˙– through an outer-sphere (eqn (1)–(3)) or inner-sphere (eqn (4) and (5)) pathway. O2˙– becomes a ligand in the oxidized metal ion's first coordination sphere at the active site before electron transfer and O2 release in an inner-sphere mechanism. Superoxide must enter the first coordination sphere by a ligand exchange process if the metal ion does not have an open coordination site. An electron transfer with O2˙– would occur outside the first coordination sphere. Thus farther away from the metal ion, in an outer-sphere mechanism, but a previous ligand exchange reaction is unnecessary. Proton donors alone are used in an outer-sphere process to stabilize peroxide. Thus, O2˙– needs to be docked at a location with one or more proton donors that can donate one or two protons to O22− as it forms and creates a hydrogen bond with O2˙–.35,36 The rapid exchange of ligands that are axially coordinated to the central metal ion, along with minimal steric hindrance for the O2˙– anion approach, is essential for the effective binding of the O2˙−species. Furthermore, the ability of the metal complex to undergo geometrical rearrangements during the redox cycling of Mn+/Mn−1 significantly aids in the interaction with O2˙–. Additionally, the characteristics of the ligands coordinated with the metal ion are crucial in augmenting the SOD-like catalytic activity of the SOD functional models. The enzymatic activity oxidation and reduction of the Mn ion between the Mn(II) and Mn(III) oxidation states. SOD takes two molecules of superoxide and transfers the excess electron from one to the other. Thus, one forms normal oxygen with fewer electrons, whereas the other forms with more electrons. The one with the additional electron quickly takes up two hydrogen ions to create H2O2.37,38 Eqn (1) and (2) are combined to get a complete reaction eqn (3):| | |
Mn2+ (H+) + O2˙− → Mn3+ + H2O2
| (1) |
| | |
Mn3+ + O2˙− → Mn2+ (H+) + O2
| (2) |
| | |
O2˙− + O2˙− + 2H+ → O2 + H2O2
| (3) |
| | |
Mn+(O2−) + O2− → Mn+(O22−) + 2H+ → Mn + H2O2 + O2
| (5) |
 |
| | Fig. 5 UV-vis spectrum of pyrogallol autoxidation and SOD-like activity of prepared samples. | |
Compounds containing carboxyl groups can create H+, speeding up the catalytic process shown in eqn (3) through a positive reaction. If the redox potential of the complexes falls within the oxidation and reduction potentials range for superoxide [E°′(O2/O2˙−) = −0.160 V, and E°′ (O2˙−/H2O2) = 0.890 V, these complexes could function as efficient functional models of the enzyme accountable for decompose superoxide, similar to the natural enzyme.18,39 According to this, Cu-complexes might theoretically decompose O2˙– according to thermodynamic principles (E°′ (Cu2+/Cu+) = 0.153 V). Additionally, enzymes that possess a Cu2+ state convert to stable Cu+ upon receiving an electron from the superoxide radical anion, transforming the radical anion into oxygen.
| | |
Cu2+ + O2˙− + H+ → Cu+ + O2
| (6) |
| | |
Cu+ + O2˙− + H+ → Cu2+ + H2O2
| (7) |
| | |
2O2˙− + 2H+ → O2 + H2O2
| (8) |
The rate of O2˙− decomposition increased with the addition of synthesized samples. The assessment outcomes on superoxide scavenging activity indicated that all produced compounds displayed greater efficacy than that of scavenging H2O2 (Cu-complex/β-CD (6 mg/10 mL) > Cu-complex (2 mg/10 mL) > Mn-complex/β-CD (4 mg/10 mL) > Mn-complex (1 mg/10 mL) > Ligand (1 mg/10 mL); 92% > 80% > 77% > 74% > 70%). By cycling between the (Cu2+/Cu+) and (Mn2+/Mn3+) oxidation states, the synthesized complexes may function as electron transport mediators and promote the decomposition of O2˙–. It was observed that the coordination of metal to CD resulted in a notable enhancement in the SOD activity of β-CD. Vecchio and his colleagues previously observed this. They developed several SOD mimetics utilizing metal complexes of β-cyclodextrin conjugates and found that in certain instances, the SOD activity of these complexes was notably amplified by the presence of the β-CD cavity.40,41 These investigations defense. The concentration of a substance needed to cause 50% inhibition is known as the IC50. Antioxidant concentrations that result in 50% inhibition of the superoxide anion radical are known as inhibitory concentrations or IC50 values.19 When substrate concentrations rise, for instance, the IC50 of a competitive inhibitor will also increase. The concentration that to 50% suppression of superoxide anion radical is lower in more effective complexes. In the presence of ligand/Mn-complex/Mn-complex/β-CD/Cu-complex/Cu-complex/β-CD, the IC50 (μg mL−1) (Table 2).
Table 2 IC50 (μg mL−1) values with the pyrogallol autoxidation method
| Sample |
Ligand |
Mn-complex |
Mn-complex/β-CD |
Cu-complex |
Cu-complex/β-CD |
| IC50 |
91.48 |
80.04 |
40.9 |
49.36 |
27.33 |
3.6.2. H2O2 scavenging process. To evaluate the catalase-like activity of prepared samples, their ability to degrade H2O2 molecules was assessed. The experiments were conducted multiple times to ensure the reliability and reproducibility of the findings. The results of the antioxidant properties of the compounds have been visually represented in Fig. 6, providing a clear depiction of their efficacy. The Mn-complex/β-CD and Cu-complex/β-CD showed H2O2 removal abilities of 86% and 72%, respectively. The metal complexes showed a significant protective effect, possibly due to their ability to act as SOD and CAT mimetics under in vitro conditions. Notably, due to the transfer of electrons from the metal to H2O2, metal complexes exhibited better reduction results than the ligand (Mn-complex/β-CD > Cu-complex/β-CD > Mn-complex > Cu-complex > Ligand; 86% > 72% > 66% > 51% > 30%). Moreover, the Mn-complex/β-CD demonstrates a higher efficacy than the Cu-complex/β-CD in decomposing H2O2. The primary factor contributing to this phenomenon is the high suitability of Mn(II) complexes as potential candidates for the process of H2O2 dismutation.
 |
| | Fig. 6 Catalase-like activity of prepared samples. | |
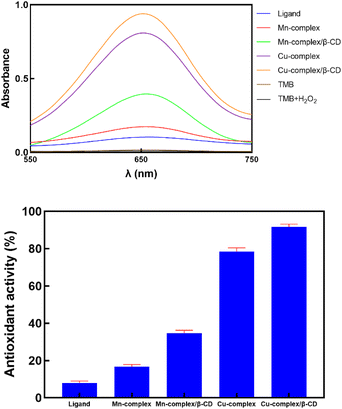
3.6.3. Peroxidase-like activity. The primary function of the peroxidase enzyme is to decompose H2O2 into non-toxic substances. When oxygen is breathed, a hazardous byproduct called H2O2 is produced.42 The colorimetric reaction with the color shift from clear to blue utilizing 3,3′,5,5′-tetramethylbenzidine (TMB) as the enzyme-substrate and H2O2 were used in this investigation to assess the enzyme-like activity of the produced materials. The oxidized product of TMB, a common peroxidase substrate, has a high molar extinction coefficient (εTMBox= 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 at 652 nm).43 The decomposition of H2O2 absorbed on the substrate most likely produces hydroxyl radicals, which initiate the catalytic reaction of TMB. As the H2O2 decomposition process continues, TMB's blue hue occurs.44 This is a two-stage response. During the first stage, TMB donates an electron to form TMB+ in the presence of H2O2. The TMB becomes deep blue instead of colorless due to this reaction. Subsequently, the solution changes from blue to yellow, TMB+ loses its second electron, and a peak is seen at about 450 nm. Using UV-vis spectroscopy, the color shift results in an absorption band at 655 nm, which shows an inc It's also important to note that, under the same reaction conditions, the control reaction, which was conducted with neither the prepared samples nor H2O2, did not exhibit any discernible color change, suggesting that the peroxidase-like activity was exclusively originating from the prepared materials. Cu-complex/β-CD exhibits the highest catalytic rease in absorbance with time. Activity among all five samples, whereas all prepared samples showed peroxidase-like activity as predicted (Fig. 7). The pH of the buffer solution, incubation time, temperature, concentration of catalyst, H2O2 concentration, and TMB concentration were among the Fig. 7 peroxidase-like activity of prepared samples to investigate how they affect the absorbance of oxidized TMB, as shown in Fig. 8. The following ideal conditions have been achieved to maximize the Cu-complex/β-CD's catalytic activity: pH 5, 10 min, 30 °C, and 173.91 μg mL−1 of Cu-complex/β-CD (in the reaction medium). In the presence of antioxidants, Fig. 8 illustrates the effect of buffer solution pH varies from 3.5 to 5.5 on the absorbance of oxidized TMB. The absorbance was at its highest at a pH of 5. Antioxidant incubation duration also affected TMB changes at 655 nm. Absorbance increased as incubation time increased from 0 to 15 min. As TMB was added, the absorbance increased immediately and stayed constant, according to the data. But the absorbance started to drop after 10 minutes. The ideal incubation period was thus 10 minutes. The results indicate that the molar absorptivity of the proposed system was markedly impacted by variations in the Cu-complex/β-CD concentration. The color shift of the solution is mainly attributed to TMB and H2O2. For this reason, the TMB and H2O2 concentrations were examined.
000 M−1 cm−1 at 652 nm).43 The decomposition of H2O2 absorbed on the substrate most likely produces hydroxyl radicals, which initiate the catalytic reaction of TMB. As the H2O2 decomposition process continues, TMB's blue hue occurs.44 This is a two-stage response. During the first stage, TMB donates an electron to form TMB+ in the presence of H2O2. The TMB becomes deep blue instead of colorless due to this reaction. Subsequently, the solution changes from blue to yellow, TMB+ loses its second electron, and a peak is seen at about 450 nm. Using UV-vis spectroscopy, the color shift results in an absorption band at 655 nm, which shows an inc It's also important to note that, under the same reaction conditions, the control reaction, which was conducted with neither the prepared samples nor H2O2, did not exhibit any discernible color change, suggesting that the peroxidase-like activity was exclusively originating from the prepared materials. Cu-complex/β-CD exhibits the highest catalytic rease in absorbance with time. Activity among all five samples, whereas all prepared samples showed peroxidase-like activity as predicted (Fig. 7). The pH of the buffer solution, incubation time, temperature, concentration of catalyst, H2O2 concentration, and TMB concentration were among the Fig. 7 peroxidase-like activity of prepared samples to investigate how they affect the absorbance of oxidized TMB, as shown in Fig. 8. The following ideal conditions have been achieved to maximize the Cu-complex/β-CD's catalytic activity: pH 5, 10 min, 30 °C, and 173.91 μg mL−1 of Cu-complex/β-CD (in the reaction medium). In the presence of antioxidants, Fig. 8 illustrates the effect of buffer solution pH varies from 3.5 to 5.5 on the absorbance of oxidized TMB. The absorbance was at its highest at a pH of 5. Antioxidant incubation duration also affected TMB changes at 655 nm. Absorbance increased as incubation time increased from 0 to 15 min. As TMB was added, the absorbance increased immediately and stayed constant, according to the data. But the absorbance started to drop after 10 minutes. The ideal incubation period was thus 10 minutes. The results indicate that the molar absorptivity of the proposed system was markedly impacted by variations in the Cu-complex/β-CD concentration. The color shift of the solution is mainly attributed to TMB and H2O2. For this reason, the TMB and H2O2 concentrations were examined.
 |
| | Fig. 7 Peroxidase-like activity of prepared samples. | |
 |
| | Fig. 8 Peroxidase-like activity dependence on (a) pH; (b) time; (c) temperature; and (d) Cu-complex/β-CD concentration. | |
Fig. 9 displays the double-reciprocal Lineweaver–Burk plots and the Michaelis–Menten curves connected to velocity and substrates for Cu-complex/β-CD, are shown in Table 3. The kinetics of H2O2 degradation and TMB oxidation revealed a non-linear connection between substrate concentration and reaction rate. In contrast, the Lineweaver–Burk plot showed a linear connection between substrate concentration and response velocity. The kinetic data were obtained by adjusting one substrate's concentration while maintaining the other substrate's concentration. The highest activity of Cu-complex/β-CD, compared to HRP at higher H2O2 concentration, can be attributed to their larger Km value for H2O2, which fits in with the pattern.
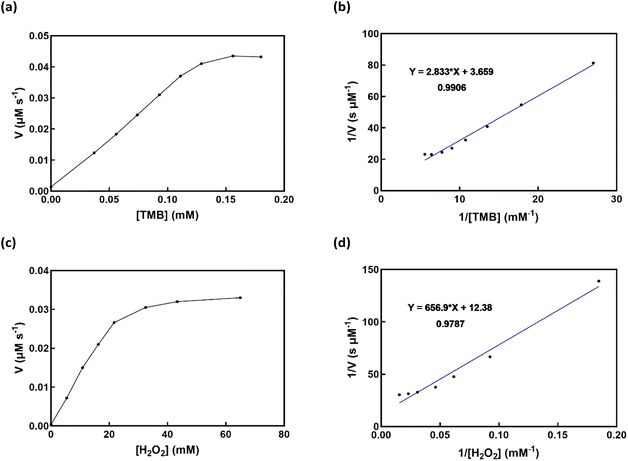 |
| | Fig. 9 The Cu-complex/β-CD Michaelis–Menten curve and Lineweaver–Burk plot (insets), (a and b) in addition to varying the TMB concentration, the H2O2 concentration was 109 mM. (c and d) TMB was present at a concentration of 0.18 mM, whereas the concentration of H2O2 was adjusted. | |
Table 3 Comparison of Km and Vmax values of Cu-complex/β-CD with HRP and other peroxidase mimics
| Catalysts |
Km (mM) |
Vmax (10−8 M s−1) |
Ref. |
| TMB |
H2O2 |
TMB |
H2O2 |
| HRP |
0.434 |
3.7 |
10.0 |
8.7 |
45 |
| Cu NPs |
0.648 |
29.16 |
5.96 |
4.22 |
46 |
| Cu(OH)2 |
1.335 |
0.379 |
42.1 |
39.1 |
47 |
| Cu NPs/CoO/CNFs |
0.26 |
0.14 |
12.32 |
42.24 |
22 |
| Cu-N-C |
3.76 |
19.94 |
75.05 |
20.07 |
48 |
| Cu-SAC |
0.077 |
0.155 |
5.97 |
6.14 |
49 |
| Cu-complex/β-CD |
53 |
0.76 |
8.07 |
27.32 |
This work |
3.7. Cytotoxicity results of ligand and complexes
In the widely used colorimetric test called the MTT assay, MTT is reduced by the succinate dehydrogenase enzyme, which is found in the mitochondria of live cells. Measurements of the materials' cytotoxicity were conducted using this assay.12 The MTT assay works by MTT entering cells and moving through mitochondria to become insoluble purple formazan. If cell metabolism is low, MTT reduction is also low. Using HFF-2 cells and the MTT assay, the study tested the viability of all compounds and demonstrated that even at concentrations of 400 μg mL−1, there is no notable toxic effect after 48 hours of incubation, and cell viability remains above 75% (Fig. 10). The IC50 of these compounds is presented in Table 4.
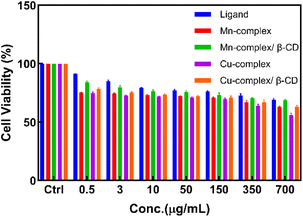 |
| | Fig. 10 MTT cell viability assay of compounds on HFF-2 normal cell line (measured at 48 h). | |
Table 4 IC50 (μg mL−1) values
| Sample |
Ligand |
Mn-complex |
Mn-complex/β-CD |
Cu-complex |
Cu-complex/β-CD |
| IC50 |
1002.6 |
694.4 |
895.6 |
549.6 |
694.6 |
Conclusions
In conclusion, a novel CMA ligand was synthesized by combining malic acid and L-cysteine. Its metal complexes with manganese and copper were also created and investigated. To increase these complexes' solubility, stability, and bioavailability, β-CD was added. To evaluate the ligand and metal complexes, the study used a variety of spectroscopic methods, including mass spectrometry, fluorescence spectroscopy, FT-IR, UV-vis, TGA, CHNS, EDX, and SEM. The study clarified how metals coordinate with oxygen and sulfur atoms in the ligand, highlighting the changes in vibrational frequencies and electronic transitions during complexation. TGA was used to investigate the complexes in detail to see how the addition of β-CD caused significant changes in thermal stability and spectrum shifts. Metal–ligand coordination was further confirmed by fluorescence tests, which showed notable emission peaks. Interestingly, the metal complexes' observed fluorescence quenching suggested that the metal ions were effectively interacting with the functional groups of the ligand. The study demonstrates how these recently synthesized compounds may be used as multifunctional agents, especially as antioxidants and potential treatments for illnesses where oxidative stress plays a significant role. It has been observed that the Mn-complex/β-CD and Cu-complex/β-CD have essential SOD-like activity and can catalytically scavenge O2˙– into O2 and H2O2. They proceed to convert H2O2 into O2 and H2O. The catalytic performance was demonstrated to be efficient by the IC50 value and catalytic dynamics. They can exhibit SOD-like activity by cycling between the Mn(II) and Mn(III) oxidation states due to the proper redox potential of Mn-complex/β-CD. Kinetic studies confirmed these results, showing that the metal complexes effectively defend against oxidative stress by performing reliable mimics of natural antioxidant enzymes. Additionally, adding β-CD greatly increased the metal complexes' antioxidant activity, demonstrating the importance of including complex formation for boosting biological efficiency.
Data availability
All data generated or analyzed during this study are included in this published article and its ESI.†
Author contributions
Saeedeh Khadivi-Derakhshan: methodology, writing – original draft, validation, investigation, project administration. Mahtab Pirouzmand: conceptualization, methodology, writing – original draft, supervision, project administration. Jafar Soleymani: investigation, project administration. Mehdi D. Esrafili: investigation, project administration.
Conflicts of interest
The authors have no conflict of interest.
Acknowledgements
The authors thank the Research Affairs of the University of Tabriz for financial support.
Notes and references
- A. Almasoud, N. Hettiarachchy, S. Rayaprolu, D. Babu, Y. M. Kwon and A. Mauromoustakos, Lwt, 2016, 66, 560–564 CrossRef CAS
 .
. - S. Suriyaprom, P. Mosoni, S. Leroy, T. Kaewkod, M. Desvaux and Y. Tragoolpua, Antioxidants, 2022, 11, 602 CrossRef CAS
 .
. - Y. P. Wang, A. Sharda, S. N. Xu, N. van Gastel, C. H. Man, U. Choi, W. Z. Leong, X. Li and D. T. Scadden, Cell Metab., 2021, 33, 1027–1041.e8 CrossRef CAS
 .
. - M. Riri, M. Hor, F. Serdaoui and M. Hlaibi, Arab. J. Chem., 2016, 9, S1478–S1486 CrossRef CAS
 .
. - M. B. Taylor, J. S. Yanaki, D. O. Draper, J. C. Shurtz and M. Coglianese, J. Drugs Dermatol., 2013, 12, 45–50 CAS
 .
. - I. Ferreira, Á. Ortigoza and P. Moore, Medwave, 2019, 19, e7632–e7638 Search PubMed
 .
. - D. Zhang, S. Nie, M. Xie and J. Hu, Food Sci. Biotechnol., 2020, 29, 17–25 CrossRef CAS
 .
. - I. Szãm, J. Szentner, I. Hegedüs-Wein and A. Vass, Int. J. Clin. Pharmacol., 1972, 6, 260–265 Search PubMed
 .
. - J. H. Kim, H. J. Jang, W. Y. Cho, S. J. Yeon and C. H. Lee, Arab. J. Chem., 2020, 13, 1678–1684 CrossRef CAS
 .
. - S. Biswas, M. Haouas, C. Freitas, C. Vieira Soares, A. Al Mohtar, A. Saad, H. Zhao, G. Mouchaham, C. Livage, F. Carn, N. Menguy, G. Maurin, M. L. Pinto and N. Steunou, Chem. Mater., 2022, 34, 9760–9774 CrossRef CAS
 .
. - A. R. Hedges, W. J. Shieh and C. T. Sikorski, Use of cyclodextrins for encapsulation in the use and treatment of food products, in Encapsulation and Controlled Release of Food Ingredients, ed. S. J. Risch and G. A. Reineccius, ACS Publications, Washington, 1995, pp. 60–71 Search PubMed
 .
. - S. Azizi, J. Soleymani and N. Shadjou, J. Mol. Recognit., 2020, 33, e2871 CrossRef CAS
 .
. - A. Cid-Samamed, J. Rakmai, J. C. Mejuto, J. Simal-Gandara and G. Astray, Food Chem., 2022, 384, 132467 Search PubMed
 .
. - K. Mishra and B. Singh, Liq. Cryst., 2021, 48, 980–990 CrossRef CAS
 .
. - Y. Zhu, K. Wang, X. Jia, C. Fu, H. Yu and Y. Wang, Med. Res. Rev., 2024, 44, 275–364 Search PubMed
 .
. - I. Liguori, G. Russo, F. Curcio, G. Bulli, L. Aran, D. Della-Morte, G. Gargiulo, G. Testa, F. Cacciatore and D. Bonaduce, Clin. Interventions Aging, 2018, 757–772 CrossRef CAS
 .
. - X. Tang, H. Yu, B. Bui, L. Wang, C. Xing, S. Wang, M. Chen, Z. Hu and W. Chen, Bioact. Mater., 2021, 6, 1541–1554 CAS
 .
. - J. Mu, X. Zhao, J. Li, E. C. Yang and X. J. Zhao, J. Mater. Chem. B, 2016, 4, 5217–5221 RSC
 .
. - Q. A. Zhang, X. Wang, Y. Song, X. H. Fan and J. F. García-Martín, J. AOAC Int., 2016, 99, 504–511 CrossRef CAS
 .
. - D. Mukhopadhyay, P. Dasgupta, D. Sinha Roy, S. Palchoudhuri, I. Chatterjee, S. Ali and S. Ghosh Dastidar, Free Radicals Antioxid., 2016, 6, 124–132 CrossRef CAS
 .
. - M. Shojaei, M. Pirouzmand and M. Khatamian, Polyhedron, 2023, 232, 116294 CrossRef CAS
 .
. - P. B. Niu, Y. Q. Wang, R. Hu and T. Yang, ACS Appl. Nano Mater., 2024, 7, 5996–6004 CrossRef CAS
 .
. - S. Khadivi-Derakhshan, M. Pirouzmand, M. D. Esrafili and J. Soleymani, J. Mol. Struct., 2024, 1306, 137950 Search PubMed
 .
. - D. Alhashmialameer, G. G. Mohamed, Y. Al-hawamy, A. Abdou, H. A. H. Alshehri, F. Alkhatib and A. M. Abu-Dief, Appl. Organomet. Chem., 2024, 38, e7667 CrossRef CAS
 .
. - A. M. Abu-Dief, R. M. El-Khatib, T. El-Dabea, A. Abdou, F. S. Aljohani, E. S. Al-Farraj, I. O. Barnawi and M. A. E. A. A. Ali, J. Mol. Liq., 2023, 386, 122353 CrossRef CAS
 .
. - H. Barańka, J. Kuduk-Jaworska, R. Szostak and A. Romaniewska, J. Raman Spectrosc., 2003, 34, 68–76 Search PubMed
 .
. - T. L. Ho, Chem. Rev., 1975, 75, 1–20 CrossRef CAS
 .
. - K. Nakamoto, J. Fujita, S. Tanaka and M. Kobayashi, J. Am. Chem. Soc., 1957, 79, 4904–4908 CrossRef CAS
 .
. - R. W. Newberry and R. T. Raines, Acc. Chem. Res., 2017, 50, 1838–1846 CrossRef CAS
 .
. - S. Zhang, T. Yu, M. Sun, H. Yu, Z. Zhang, S. Wang and H. Jiang, Talanta, 2014, 126, 185–190 CrossRef CAS
 .
. - S. Chakraborty, A. Patra, A. Mondal, S. Lohar, E. Zangrando and P. Chattopadhyay, Polyhedron, 2021, 203, 115226 Search PubMed
 .
. - I. L. Sousa, C. M. Porto, K. C. Bassani, M. H. Martins, F. B. T. Pessine and N. H. Morgon, J. Braz. Chem. Soc., 2020, 31, 1585–1596 CAS
 .
. - A. Dehghani, G. Bahlakeh and B. Ramezanzadeh, Composites, Part B, 2020, 197, 108152 CrossRef CAS
 .
. - X. Li, J. Agric. Food Chem., 2012, 60, 6418–6424 CrossRef CAS
 .
. - S. J. Lippard and J. M. Berg, Principles of bioinorganic chemistry: By S J Lippard and J M Berg. pp 411. University Science Books, Mill Valley, California. 1994. $30 ISBN 0-935702-73-3 (paper), Wiley Online Library, 1995, vol. 23 Search PubMed
 .
. - Y. Sheng, I. A. Abreu, D. E. Cabelli, M. J. Maroney, A. F. Miller, M. Teixeira and J. S. Valentine, Chem. Rev., 2014, 114, 3854–3918 CrossRef CAS
 .
. - M. E. McAdam, R. A. Fox, F. Lavelle and E. M. Fielden, Biochem. J., 1977, 165, 71–79 CrossRef CAS
 .
. - A. S. Hearn, M. E. Stroupe, D. E. Cabelli, J. R. Lepock, J. A. Tainer, H. S. Nick and D. N. Silverman, Biochemistry, 2001, 40, 12051–12058 CrossRef CAS
 .
. - G. Csire, J. Demjén, S. Timári and K. Várnagy, Polyhedron, 2013, 61, 202–212 CrossRef CAS
 .
. - V. Oliveri, A. Puglisi and G. Vecchio, Dalton Trans., 2011, 40, 2913–2919 RSC
 .
. - A. Puglisi, G. Tabbì and G. Vecchio, J. Inorg. Biochem., 2004, 98, 969–976 CrossRef CAS
 .
. - S. Thangudu and C. H. Su, Biomolecules, 2021, 11, 1015 CrossRef CAS
 .
. - S. Ghosh, P. Singh, S. Roy, K. Bhardwaj and A. Jaiswal, ChemBioChem, 2022, 23, e202100691 CrossRef CAS
 .
. - M. Pirouzmand, B. Nikzad-Kojanag, S. A. Hosseini-Yazdi and S. Khadivi-Derakhshan, Inorg. Chem. Commun., 2024, 162, 112298 CrossRef CAS
 .
. - L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS
 .
. - L. Hu, Y. Yuan, L. Zhang, J. Zhao, S. Majeed and G. Xu, Anal. Chim. Acta, 2013, 762, 83–86 CrossRef CAS
 .
. - R. Cai, D. Yang, X. Chen, Y. Huang, Y. Lyu, J. He, M. Shi, I. T. Teng, S. Wan, W. Hou and W. Tan, J. Mater. Chem. B, 2016, 4, 4657–4661 RSC
 .
. - X. Liu, F. Wu, X. Zheng, H. Liu, F. Ren, J. Sun, H. Ding, R. Yang and L. Jin, ACS Appl. Nano Mater., 2023, 6, 10303–10311 CrossRef CAS
 .
. - Y. Wu, J. Wu, L. Jiao, W. Xu, H. Wang, X. Wei, W. Gu, G. Ren, N. Zhang, Q. Zhang, L. Huang, L. Gu and C. Zhu, Anal. Chem., 2020, 92, 3373–3379 CrossRef CAS
 .
.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Jafar Soleymani
*a,
Jafar Soleymani b and
Mehdi D. Esrafilic
b and
Mehdi D. Esrafilic
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio. In the synthesis of the Cu(II) complex, the ligand (0.3 g, 125 × 10−5 mol) was dissolved in 10 mL of ethanol and mixed with an ethanolic solution of metal(II) acetate (0.25 g, 125 × 10−5 mol). The resulting mixture was refluxed at 75 °C for 12 hours. After the completion of the reaction, the precipitate was separated by centrifugation and cleaned with ethanol, and dried at room temperature.
1 stoichiometric ratio. In the synthesis of the Cu(II) complex, the ligand (0.3 g, 125 × 10−5 mol) was dissolved in 10 mL of ethanol and mixed with an ethanolic solution of metal(II) acetate (0.25 g, 125 × 10−5 mol). The resulting mixture was refluxed at 75 °C for 12 hours. After the completion of the reaction, the precipitate was separated by centrifugation and cleaned with ethanol, and dried at room temperature.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio (Tris–HCl buffer
1 volume ratio (Tris–HCl buffer![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pyrogallol). After that, a UV-vis spectrophotometer operating at wavelengths of 325 and 420 nm was used to monitor the reaction mixture. After that, the capacity to scavenge superoxide radicals (O2˙–) was determined as:
pyrogallol). After that, a UV-vis spectrophotometer operating at wavelengths of 325 and 420 nm was used to monitor the reaction mixture. After that, the capacity to scavenge superoxide radicals (O2˙–) was determined as:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 at 652 nm).43 The decomposition of H2O2 absorbed on the substrate most likely produces hydroxyl radicals, which initiate the catalytic reaction of TMB. As the H2O2 decomposition process continues, TMB's blue hue occurs.44 This is a two-stage response. During the first stage, TMB donates an electron to form TMB+ in the presence of H2O2. The TMB becomes deep blue instead of colorless due to this reaction. Subsequently, the solution changes from blue to yellow, TMB+ loses its second electron, and a peak is seen at about 450 nm. Using UV-vis spectroscopy, the color shift results in an absorption band at 655 nm, which shows an inc It's also important to note that, under the same reaction conditions, the control reaction, which was conducted with neither the prepared samples nor H2O2, did not exhibit any discernible color change, suggesting that the peroxidase-like activity was exclusively originating from the prepared materials. Cu-complex/β-CD exhibits the highest catalytic rease in absorbance with time. Activity among all five samples, whereas all prepared samples showed peroxidase-like activity as predicted (Fig. 7). The pH of the buffer solution, incubation time, temperature, concentration of catalyst, H2O2 concentration, and TMB concentration were among the Fig. 7 peroxidase-like activity of prepared samples to investigate how they affect the absorbance of oxidized TMB, as shown in Fig. 8. The following ideal conditions have been achieved to maximize the Cu-complex/β-CD's catalytic activity: pH 5, 10 min, 30 °C, and 173.91 μg mL−1 of Cu-complex/β-CD (in the reaction medium). In the presence of antioxidants, Fig. 8 illustrates the effect of buffer solution pH varies from 3.5 to 5.5 on the absorbance of oxidized TMB. The absorbance was at its highest at a pH of 5. Antioxidant incubation duration also affected TMB changes at 655 nm. Absorbance increased as incubation time increased from 0 to 15 min. As TMB was added, the absorbance increased immediately and stayed constant, according to the data. But the absorbance started to drop after 10 minutes. The ideal incubation period was thus 10 minutes. The results indicate that the molar absorptivity of the proposed system was markedly impacted by variations in the Cu-complex/β-CD concentration. The color shift of the solution is mainly attributed to TMB and H2O2. For this reason, the TMB and H2O2 concentrations were examined.
000 M−1 cm−1 at 652 nm).43 The decomposition of H2O2 absorbed on the substrate most likely produces hydroxyl radicals, which initiate the catalytic reaction of TMB. As the H2O2 decomposition process continues, TMB's blue hue occurs.44 This is a two-stage response. During the first stage, TMB donates an electron to form TMB+ in the presence of H2O2. The TMB becomes deep blue instead of colorless due to this reaction. Subsequently, the solution changes from blue to yellow, TMB+ loses its second electron, and a peak is seen at about 450 nm. Using UV-vis spectroscopy, the color shift results in an absorption band at 655 nm, which shows an inc It's also important to note that, under the same reaction conditions, the control reaction, which was conducted with neither the prepared samples nor H2O2, did not exhibit any discernible color change, suggesting that the peroxidase-like activity was exclusively originating from the prepared materials. Cu-complex/β-CD exhibits the highest catalytic rease in absorbance with time. Activity among all five samples, whereas all prepared samples showed peroxidase-like activity as predicted (Fig. 7). The pH of the buffer solution, incubation time, temperature, concentration of catalyst, H2O2 concentration, and TMB concentration were among the Fig. 7 peroxidase-like activity of prepared samples to investigate how they affect the absorbance of oxidized TMB, as shown in Fig. 8. The following ideal conditions have been achieved to maximize the Cu-complex/β-CD's catalytic activity: pH 5, 10 min, 30 °C, and 173.91 μg mL−1 of Cu-complex/β-CD (in the reaction medium). In the presence of antioxidants, Fig. 8 illustrates the effect of buffer solution pH varies from 3.5 to 5.5 on the absorbance of oxidized TMB. The absorbance was at its highest at a pH of 5. Antioxidant incubation duration also affected TMB changes at 655 nm. Absorbance increased as incubation time increased from 0 to 15 min. As TMB was added, the absorbance increased immediately and stayed constant, according to the data. But the absorbance started to drop after 10 minutes. The ideal incubation period was thus 10 minutes. The results indicate that the molar absorptivity of the proposed system was markedly impacted by variations in the Cu-complex/β-CD concentration. The color shift of the solution is mainly attributed to TMB and H2O2. For this reason, the TMB and H2O2 concentrations were examined.

.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.