 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efficacy of micro–nano bubble enhanced immobilized Chlorella vulgaris in the removal of typical antibiotics†
Tao Zhua,
Mengyao Jingbcd,
Jianping Zhangbcd,
Hui Lie,
Min Zhou *fg and
Guijuan Libcd
*fg and
Guijuan Libcd
aHenan College of Transportation, Zhengzhou 450008, Henan, China
bSchool of Water and Environment, Chang'an University, Xi'an 710054, China
cKey Laboratory of Subsurface Hydrology and Ecology in Arid Areas, Ministry of Education, Chang'an University, Xi'an 710054, China
dKey Laboratory of Eco-hydrology and Water Security in Arid and Semi-arid Regions of Ministry of Water Resources, Chang'an University, Xi'an 710054, China
eHenan Transport Investment Group Co., Ltd, Zhengzhou, China
fOcean University of China, Qingdao 266100, Shandong, China. E-mail: zhoumin2023@126.com
gHenan Provincial Department of Transport, Zhengzhou 45000, Henan, China
First published on 16th June 2025
Abstract
Antibiotic pollution poses a global environmental challenge, with effective removal technologies for different antibiotic types still lacking. This study investigates an innovative micro–nano bubble (MNB)-augmented immobilized Chlorella vulgaris system for remediating groundwater contaminated with sulfadiazine (SD) and chloramphenicol (CAP) antibiotics. Key parameters, including initial concentration (5–30 mg L−1), algal bead density (0.25–4 beads per mL), aeration time (5–30 min), and coexisting ions, were evaluated. SEM and FT-IR analyses revealed removal mechanisms. Results showed MNBs significantly improved microalgal biomass and removal efficiency (SD: 79.97%; CAP: 93.92%). SD elimination primarily depends on initial concentration and aeration, while CAP removal shows stronger ionic environment dependence. FT-IR confirmed stronger interactions (electrostatic attraction, surface adsorption) between algae and CAP. The technology showed particular effectiveness for CAP, achieving over 90% removal through MNB-algae synergy, providing valuable insights for targeted antibiotic remediation strategies.
1 Introduction
The increasing prevalence of antibiotic contamination and associated resistance genes has become a critical environmental issue.1–4 Multiple factors contribute to this problem, including partial metabolic breakdown in organisms5,6 and inadequate removal by conventional wastewater treatment.7 These contaminants migrate to groundwater through various pathways, 8,9 threatening aquatic ecosystems and potable water resources.10,11 Current remediation approaches face substantial challenges: 12,13 biological methods risk inducing resistance, 14 chemical treatments often prove costly and generate hazardous byproducts, 2 while physical techniques suffer from interference and high operational expenses.15 This situation demands innovative solutions for groundwater remediation that are simultaneously effective, economical, and environmentally sustainable.Microalgae technology has the potential to remove pollutants and nutrients, reduce biochemical oxygen demand, efficiently capture CO2, be eco-friendly, and produce high-value products, so it has become a promising alternative.16,17 Frascaroli et al. (2024)18 used three microalgae (Auxenochlorella protothecoides, Tetradesmus obliquus, and Chlamydomonas acidophila) to effectively remove the mixture of seven antibiotics (ciprofloxacin, clarithromycin, erythromycin, metronidazole, ofloxacin, sulfamethoxazole and trimethoprim). At the same time, since microalgae are non-target organisms of antibiotics,19 microalgae show significant adaptability during antibiotic treatment.20 In particular, heterotrophic microalgae have higher biomass production, extracellular polymeric substances (EPS) accumulation, and pollutant removal efficiency than photosynthetic autotrophic microalgae.21 Similar situations exist in other microorganisms, such as heterotrophic denitrifying bacteria.22 In this sense, microalgae can fully adapt to the dark environment of groundwater.
However, despite the excellent performance of microalgae in antibiotic removal, its removal efficiency is still affected by many factors, such as microalgae species, antibiotic species and concentration, dissolved oxygen (DO) concentration, light intensity, etc.23 Therefore, in practical applications, the selection of appropriate microalgae species and optimization of process conditions are key to improving the removal efficiency of antibiotics. To further increase the efficiency of antibiotic removal by microalgae, researchers have begun to explore new technological tools in recent years.24 For instance, micro–nanobubble (MNB) technology has emerged as a prospective support, with potential to enhance performance across various bioprocesses.25,26 Miao et al. (2024)27 demonstrated that the application of MNBs by using novel oxygenated MNBs loaded with micropore biochar to stimulate indigenous aerobic denitrifying bacteria could effectively alleviate hypoxia, enhance the overall collaboration among microorganisms, and promote the expression of relevant genes. Temesgen et al. (2023)28 applied nanobubbles to the aeration mechanism of a semi-intermittent bioreactor and improved the biodegradation rate of organic matter in wastewater from 5.83 to 17.5 mg L−1 h−1. Compared to conventional bubbles, MNBs exhibit faster mass transfer rates and lower rise velocities due to their small diameter and high internal pressure.29 Introducing MNBs into microalgae culture systems significantly enhances the efficiency of oxygen transfer from internal bubbles to the external water column, generating higher DO concentrations to improve the low-oxygen environment of groundwater and promoting the growth and metabolic activities of microalgae, which can accelerate their degradation of antibiotics.
In addition, the application of immobilization technology also provides innovative approaches for antibiotic removal by microalgae. Immobilizing microalgae onto carriers improves their stability and reusability in wastewater treatment.30,31 Xie et al. (2020)17 showed that immobilized Chlorella outperformed the free state of Chlorella in antibiotic removal.
Sulfonamides and amphenicols account for 12% and 8% of global antibiotic usage, respectively, and their environmental occurrence concentrations and associated ecological risks exhibit significant differences.8 This study selected sulfadiazine (SD, representing sulfonamides) and chloramphenicol (CAP, representing amphenicols) as representative antibiotics. The aqueous residual concentration of SD can reach the μg L−1 level, while CAP is more prone to accumulation in sediments.32 The strong lipophilicity of CAP facilitates its adsorption onto algal cell membranes, whereas the high water solubility of SD enables its diffusion primarily through the aqueous phase. The two antibiotics exhibit significant differences in chemical structure, physicochemical properties, and degradation pathways, making them effective indicators for assessing the system's removal characteristics for different types of antibiotics. In this study, the widely used green algae genus Chlorella vulgaris (C. vulgaris) was employed as the experimental material to investigate the effect of MNBs on the removal of SD and CAP by immobilized C. vulgaris.33,34 We examined the effects of various factors on removing antibiotics from groundwater using MNBs-enhanced immobilized C. vulgaris and determined the dominant factors through correlation analysis. Afterward, two characterization methods, scanning electron microscopy (SEM) and Fourier transform infrared spectroscopy (FT-IR), were used to analyze the antibiotic removal pathway and determine the ESP content to reveal the antibiotic removal mechanism further. This work provides new insights for applying immobilized microalgae technology in groundwater pollution remediation, contributing to upgrading traditional biotechnology.
2 Materials and methods
2.1 Preparation of micro–nano bubbles
The micro–nano bubble generator (ZJC-NM-200L, Shanghai Zhongjing Environmental Technology Co., Ltd) utilizes multiphase vortex gas–liquid shear and swirl countercurrent technology (Fig. SI1†). During the experiment, the operating pressure of the device was maintained at 0.3–0.35 MPa, with an air intake of approximately 100 mL min−1. The dissolution rate of the generated MNBs exceeded 95%. The resulting bubbles exhibited a size distribution of 100–1000 nm in diameter and demonstrated remarkable stability, persisting for over one week in an aqueous solution (Fig. SI2 and SI3†).2.2 Materials
The Chlorella vulgaris strain used in the experiment, with the strain number FACHB-8, was purchased from the Freshwater Algae Culture Collection at the Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan. Antibiotics were chromatographically pure and purchased from Shanghai Yuanye Biotechnology Co., Ltd, China, and their physicochemical properties are shown in Table 1. The simulated groundwater samples were prepared in the proportion of 0.1665 g per L CaCl2, 0.1952 g per L MgSO4, 0.3884 g per L NaHCO3, and 0.0160 g per L KNO3, referring to the literature.352.3 Immobilization and de-immobilization of C. vulgaris
C. vulgaris was inoculated into BG-11 liquid medium (containing NaNO3 1.5 g L−1; other components are listed in Table SI1†) and maintained at pH7–7.5. The cultures were continuously subculture for 2–3 cycles in an illuminated incubator (25 °C, 3000–4000 lux, light–dark cycle of 12 h![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 h). OD680 was monitored daily, and the standard for activity recovery was defined as a stable biomass growth rate (ΔOD680 per day > 0.15).
12 h). OD680 was monitored daily, and the standard for activity recovery was defined as a stable biomass growth rate (ΔOD680 per day > 0.15).
The algae cells were collected by centrifugation and resuspended in sterile deionized water, and the absorbance of the algae solution was adjusted to 0.3. The algae solution was mixed with 5% sodium alginate solution at a volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The mixed droplets were added to 2% CaCl2 (w/v) solution at a rate of 3.6 mL min−1 using a peristaltic pump to form immobilized algal beads with a diameter of about 3–4 mm. The beads were hardened for 4 h, separated using filter paper, and rinsed with sterile water for subsequent use. Each algal bead contained approximately 0.0524 mL of algal solution and weighed about 0.05 g. For de-immobilization, the algal beads were placed in a 10% (w/v) sodium citrate solution and gently shaken in an oscillator until the beads were dissolved entirely.17
1. The mixed droplets were added to 2% CaCl2 (w/v) solution at a rate of 3.6 mL min−1 using a peristaltic pump to form immobilized algal beads with a diameter of about 3–4 mm. The beads were hardened for 4 h, separated using filter paper, and rinsed with sterile water for subsequent use. Each algal bead contained approximately 0.0524 mL of algal solution and weighed about 0.05 g. For de-immobilization, the algal beads were placed in a 10% (w/v) sodium citrate solution and gently shaken in an oscillator until the beads were dissolved entirely.17
2.4 Experimental design
The initial conditions were set with an antibiotic concentration of 10 mg L−1, algal bead concentration of 1 bead per mL, pH 7, DO 5 mg L−1, and aeration frequency of 10 min every 48 h. Antibiotic concentrations were measured before each aeration cycle. Unless otherwise specified, all experiments were conducted under these standardized conditions and repeated in triplicate.
Effect of initial antibiotic concentration: tested concentrations: 5, 10, 15, 20, and 30 mg L−1 of SD and CAP.
Effect of algal bead concentration: tested concentrations: 0.25, 0.5, 1, 2, and 4 beads per mL.
Effect of aeration duration: tested aeration durations: 5, 10, 15, 20, and 30 min every 48 h.
Effect of coexisting ions: prepared 2 L of simulated groundwater, with deionized water as a blank control. Added ion solutions (K+, Na+, Mg2+, Ca2+, HCO3−, NO3−, and SO42−) at concentrations of 0, 2, and 5 mM.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 v/v) via ultrasonication (40 kHz, 2.2 kW, 1 h) followed by centrifugation.36
2 v/v) via ultrasonication (40 kHz, 2.2 kW, 1 h) followed by centrifugation.362.5 Measurement methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90, v/v). The column temperature was 30 °C. The detection wavelengths of SD and CAP were 260 nm and 278 nm, respectively. LOQs were 0.03 mg per L (SD) and 0.05 mg per L (CAP), while LODs were 0.01 mg per L (SD) and 0.02 mg per L (CAP).
90, v/v). The column temperature was 30 °C. The detection wavelengths of SD and CAP were 260 nm and 278 nm, respectively. LOQs were 0.03 mg per L (SD) and 0.05 mg per L (CAP), while LODs were 0.01 mg per L (SD) and 0.02 mg per L (CAP).The removal process of antibiotics were analyzed by using pseudo-first-order kinetic model, which can be expressed as:37
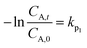 | (1) |
The half-life equation:
 | (2) |
 | (3) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 15 min). The supernatant was filtered (0.22 μm) to obtain a crude EPS solution. Protein content was determined by the Coomassie Brilliant Blue method, while polysaccharide content was measured using the anthrone–sulfuric acid assay.39
000 rpm, 15 min). The supernatant was filtered (0.22 μm) to obtain a crude EPS solution. Protein content was determined by the Coomassie Brilliant Blue method, while polysaccharide content was measured using the anthrone–sulfuric acid assay.393 Results and discussion
3.1 Algae performance
As shown in Fig. 1(a), C. vulgaris growth stabilized and reached its maximum on Day 6. When SD and CAP were added, the growth remained stable on Day 6. However, in the CAP + MNBs group, the maximum was reached on Day 8, possibly due to the synergistic effect of MNBs, which prolonged the growth cycle. This delay suggests that MNBs alleviated the toxic inhibition caused by CAP. At the end of the 12-day culture period, the number of algae in the SD + MNBs group was 3.235 × 109, and the CAP + MNBs group was 5.745 × 109. Compared to the SD group and the CAP group, the increases were 1.61 times and 2.4 times, respectively. MNBs significantly promoted the growth of C. vulgaris because MNBs not only increased the content of DO in water (Fig. SI4†), provided oxygen for the development of C. vulgaris, but also accelerated the transfer of nutrients in the water medium to organisms, thereby indirectly stimulating enzyme activity42 and promoting the growth and development of microorganisms.43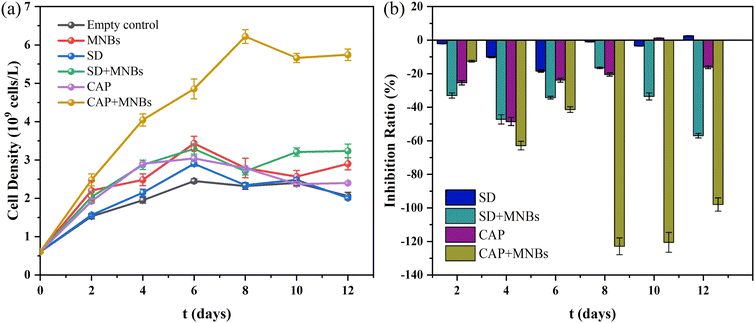 | ||
| Fig. 1 Growth curves of C. vulgaris under different conditions (a), and growth inhibition of antibiotic on C. vulgaris (b). | ||
As can be seen from Fig. 1(b), the growth inhibition of C. vulgaris was negative in the first 8 days of the exposure period, and both antibiotics promoted algae growth to varying degrees. The CAP group demonstrated a promotional effect after a temporary inhibition observed on the 10th day, attributed to the degradation of antibiotics and the acclimatization of C. vulgaris. The SD group showed the weakest promotion effect and finally showed an inhibition effect. Because microalgae respond differently to different antibiotics,36 SD induces toxicity and exceeds the tolerance limit of the algal cells, destroying and disintegrating the cellular structure. In contrast, MNBs resulted in much higher algae populations when antibiotic concentrations were relatively low and acted as an exogenous carbon source to promote algae growth. Ma et al. (2024)38 showed that higher initial algal biomass alleviated the growth inhibition of Chlorella sp. by SDZ-induced stress.
3.2 Removal efficiency and kinetics
Fig. 2 illustrates the removal process and kinetic fitting of the SD and CAP in MNBs, IC, and MNBs-IC. The enhanced system shows the highest antibiotic removal rates, achieving 79.97% and 93.92%, respectively. Compared with the single IC system, the removal rate was significantly improved, increasing to 2.36 and 1.89 times, respectively. The collapsed MNBs can form a high-speed jet with high energy density, and the surrounding water molecules are pyrolyzed to produce hydroxyl radicals to degrade organic matter.44 However, Fig. 2 shows that the degradation effect of MNBs is significantly smaller than that of C. vulgaris. Combined with 3.1, we can conclude that MNBs primarily contribute to the growth of C. vulgaris. | ||
| Fig. 2 SD (left) and CAP (right) are removed over time in different systems (a) and kinetic fitting (b). | ||
The Table 2 below displays the kinetic parameters related to the antibiotic removal process. The pseudo-first-order model is typically applicable to diffusion-dominated rapid adsorption processes.45 The high water solubility of SD facilitates its adsorption onto the microalgae cell surface via hydrophobic interactions, resulting in liquid film diffusion acting as the rate-limiting step, which aligns well with the assumptions of the pseudo-first-order model.46 In contrast, the strong hydrophobicity of CAP leads to its removal relying on multi-step mechanisms, such as surface adsorption, chemical bonding, and biodegradation. These complex processes cannot be adequately described by the pseudo-first-order model. The removal efficiency of CAP is significantly higher than that of SD due to the strong specificity of algae in removing different antibiotics.36 Research indicates that the antibiotic removal rate is highly relevant to the alcohol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow).47 The log
Kow).47 The log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow values of CAPs and SD are 1.14 and −0.09, respectively. The larger the log
Kow values of CAPs and SD are 1.14 and −0.09, respectively. The larger the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kow value is, the higher the concentration of organic compounds in octanol is, indicating that the material is susceptible to passing through the phospholipid bilayer structure of the cell membrane and the easier it is to enter the cell. Moreover, the adsorption of antibiotics onto microalgae is affected by electrostatic attraction, which links to the ionization properties of the antibiotics.48 In this experiment, the pH value was maintained at around 7, making CAP primarily positively charged and SD predominantly negatively charged. Due to the presence of carboxyl, hydroxyl and amino groups on the cell membrane, the surface of Chlorella is usually considered to be negatively charged. MNBs are also negatively charged due to the presence of hydroxyl radicals at the interface and the electric double layer effect.49,50 Therefore, the electrostatic attraction between MNBs-IC system and CAP is stronger than that of SD, which further explains the higher removal rate of CAP.
Kow value is, the higher the concentration of organic compounds in octanol is, indicating that the material is susceptible to passing through the phospholipid bilayer structure of the cell membrane and the easier it is to enter the cell. Moreover, the adsorption of antibiotics onto microalgae is affected by electrostatic attraction, which links to the ionization properties of the antibiotics.48 In this experiment, the pH value was maintained at around 7, making CAP primarily positively charged and SD predominantly negatively charged. Due to the presence of carboxyl, hydroxyl and amino groups on the cell membrane, the surface of Chlorella is usually considered to be negatively charged. MNBs are also negatively charged due to the presence of hydroxyl radicals at the interface and the electric double layer effect.49,50 Therefore, the electrostatic attraction between MNBs-IC system and CAP is stronger than that of SD, which further explains the higher removal rate of CAP.
| SD | CAP | |||||||
|---|---|---|---|---|---|---|---|---|
| K (d−1) | t1/2 (d) | R2 | Removal rate | K (d−1) | t1/2 (d) | R2 | Removal rate | |
| IC | 0.033 | 20.482 | 0.982 | 33.87% | 0.054 | 12.914 | 0.991 | 49.67% |
| MNBs | 0.004 | 168.066 | 0.997 | 4.90% | 0.007 | 103.315 | 0.975 | 8.19% |
| MNBs-IC | 0.140 | 4.972 | 0.995 | 79.97% | 0.238 | 3.479 | 0.939 | 93.92% |
3.3 Analysis of influencing factors and correlations
The effectiveness of antibiotic removal based on microalgae varies with the type of antibiotics and microalgae species.60 Microalgae are more effective in removing macrolide antibiotics than β-lactam and sulfonamide antibiotics, with an overall removal rate of 62.3%.61 Chen et al. (2020)62 studied that the removal rate of 10 mg per L SD by C. vulgaris was only 29%. Wang et al. (2023)46 used Chlorella pyrenoidosa to remove SD at different initial concentrations (100, 200 and 500 μg L−1). The removal efficiency is 65.9–67.6%. The method used in this study has obvious advantages in removal efficiency.
3.4 Antibiotic removal mechanisms
SEM analysis revealed distinct morphological differences in immobilized C. vulgaris after 12-day cultivation (Fig. SI5†). Control group cells exhibited extensive rupture and mortality, with wrinkled and distorted surfaces. In contrast, antibiotic-exposed cells maintained smoother surfaces and intact morphology, particularly in CAP-treated samples, which showed optimal cellular integrity and higher cell density. These observations suggest that nutrient limitation (carbon/nitrogen sources) in the control group induced algal senescence, while SD and CAP potentially served as supplemental nutrients promoting algal growth. These findings are consistent with the growth trends observed in Section 3.1.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations. The peak at 1430 cm−1, assigned to –COO– symmetric stretching,33 exhibited minimal shift, suggesting limited participation in antibiotic removal. Significant peak shifts were observed at 1640 cm−1 (amide I–C
O stretching vibrations. The peak at 1430 cm−1, assigned to –COO– symmetric stretching,33 exhibited minimal shift, suggesting limited participation in antibiotic removal. Significant peak shifts were observed at 1640 cm−1 (amide I–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching) and 1100 cm−1 (–C–O–C stretching).65 A new peak appeared at 1360 cm−1 in all antibiotic groups, which was attributed to the symmetric stretching vibration of –NO2 in CAP group, 66 and may reflect the C–N stretching of the degradation product 2-aminopyrimidine in SD group.67 The peak at 1271.8 cm−1 may be the stretching vibration peak of the C–N of the amide III band. These findings suggest that functional groups such as –NH, –OH, –C
O stretching) and 1100 cm−1 (–C–O–C stretching).65 A new peak appeared at 1360 cm−1 in all antibiotic groups, which was attributed to the symmetric stretching vibration of –NO2 in CAP group, 66 and may reflect the C–N stretching of the degradation product 2-aminopyrimidine in SD group.67 The peak at 1271.8 cm−1 may be the stretching vibration peak of the C–N of the amide III band. These findings suggest that functional groups such as –NH, –OH, –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N, and –C–O–C in C. vulgaris likely participate in the enhanced removal of antibiotics by MNBs, but the specific mechanisms require further investigation.
O, C–N, and –C–O–C in C. vulgaris likely participate in the enhanced removal of antibiotics by MNBs, but the specific mechanisms require further investigation.
 | ||
| Fig. 7 Mechanism of antibiotic removal: (a) percentage of removal pathways for different concentrations of antibiotics (b) EPS content of C. vulgaris. | ||
In addition to the nature of antibiotics, biosorption is also affected by hydrophobic mechanisms, such as EPS.69 ESP is mainly composed of proteins and polysaccharides. There are many forces in the process of binding with antibiotics, including the van der Waals force, hydrogen bond, and electrostatic attraction.70 With the increase in antibiotic concentration, the ESP of C. vulgaris in the SD group increased to varying degrees, while that in the CAP group increased first and then decreased. This may be caused by the combined effect of the biomass of C. vulgaris and the stress of antibiotics on C. vulgaris.10 Wang et al. (2019)71 studies have shown that high concentrations of antibiotics can stimulate Chlorella to secrete ESP to protect and maintain cell activity. We found that the content of EPS well correlated with the biosorption ratio of algae cells combined with the removal pathway, especially in the CAP group. Because of its strong hydrophobicity and chemical makeup, CAP facilitated chemical bonding and physical adsorption by algal cell walls and EPS.72 According to the literature report,73 EPS affects the adsorption of antibiotics by Chlorella mainly through the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, –NH2, and –OH of proteins and the C–O–C functional groups of polysaccharides, which can provide more active sites to adsorb antibiotics, corroborated by FT-IR results in Section 3.4.1.
O, –NH2, and –OH of proteins and the C–O–C functional groups of polysaccharides, which can provide more active sites to adsorb antibiotics, corroborated by FT-IR results in Section 3.4.1.
4 Conclusions
MNBs can promote C. vulgaris growth by increasing the amount of DO in water and significantly enhancing the removal efficiency of antibiotics by the IC. The system has a firmer enhancement effect on the SD removal process. The removal process followed the first-order kinetic model. The experimental results of various influencing factors showed that the toxic effect of SD on C. vulgaris may be greater than that of CAP; algae concentration is the most significant factor affecting the removal rate of antibiotics. Aeration time affects the removal effect by influencing the DO value in water, and the coexisting ions in the groundwater promote the removal of antibiotics. Initial antibiotic concentration and aeration time had a more pronounced effect on SD removal, and coexisting ions had a more potent impact on CAP. Both antibiotic removal pathways include adsorption, biosorption, bioaccumulation, and degradation (biodegradable and non-biodegradable) of immobilized carriers. Degradation was the primary mechanism among them, and the amount of adsorption was the primary determinant of antibiotic degradation efficiency. ESP and the antibiotics' nature worked together to give CAP a higher degradation efficacy than SD.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Tao Zhu: writing – original draft, funding acquisition, formal analysis. Mengyao Jing: writing – original draft, supervision, methodology. Jianping Zhang: writing – review & editing, supervision, data curation. Hui Li: writing – review & editing, data curation. Min Zhou: writing – review & editing, data curation. Guijuan Li: writing – review & editing, supervision, methodology, conceptualization.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Henan Province Transportation Science and Technology Project (No. 2020G-2-10); the National Natural Science Foundation of China (No. 41977163).Notes and references
- H. B. Hawash, A. A. Moneer, A. A. Galhoum, A. M. Elgarahy, W. A. A. Mohamed, M. Samy, H. R. El-Seedi, M. S. Gaballah, M. F. Mubarak and N. F. Attia, Occurrence and spatial distribution of pharmaceuticals and personal care products (PPCPs) in the aquatic environment, their characteristics, and adopted legislations, J. Water Process Eng., 2023, 52, 103490 CrossRef.
- M. Kumar, S. Sridharan, A. D. Sawarkar, A. Shakeel, P. Anerao, G. Mannina, P. Sharma and A. Pandey, Current research trends on fmerging contaminants pharmaceutical and personal care products (PPCPs): A comprehensive review, Sci. Total Environ., 2023, 859, 160031 CrossRef CAS PubMed.
- M. Narayanan, M. El-sheekh, Y. Ma, A. Pugazhendhi, D. Natarajan, G. Kandasamy, R. Raja, R. M. Saravana Kumar, S. Kumarasamy, G. Sathiyan, R. Geetha, B. Paulraj, G. Liu and S. Kandasamy, Current status of microbes involved in the degradation of pharmaceutical and personal care products (PPCPs) pollutants in the aquatic ecosystem, Environ. Pollut., 2022, 300, 118922 CrossRef CAS PubMed.
- C. F. Ottosen, P. L. Bjerg, S. Kümmel, H. H. Richnow, P. Middeldorp, H. Draborg, G. G. Lemaire and M. M. Broholm, Natural attenuation of sulfonamides and metabolites in contaminated groundwater – Review, advantages and challenges of current documentation techniques, Water Res., 2024, 254, 121416 CrossRef CAS PubMed.
- J. Li, Z. Min, W. Li, L. Xu, J. Han and P. Li, Interactive effects of roxithromycin and freshwater microalgae, chlorella pyrenoidosa: Toxicity and removal mechanism, Ecotoxicol. Environ. Saf., 2020, 191, 110156 CrossRef CAS.
- Y. Fang, Y. Liu and J. Zhang, Mechanisms for the increase in lipid production in cyanobacteria during the degradation of antibiotics, Environ. Pollut., 2023, 322, 121171 CrossRef CAS PubMed.
- S. Rodriguez-Mozaz, I. Vaz-Moreira, S. Varela Della Giustina, M. Llorca, D. Barceló, S. Schubert, T. U. Berendonk, I. Michael-Kordatou, D. Fatta-Kassinos, J. L. Martinez, C. Elpers, I. Henriques, T. Jaeger, T. Schwartz, E. Paulshus, K. O'Sullivan, K. M. M. Pärnänen, M. Virta, T. T. Do, F. Walsh and C. M. Manaia, antibiotic residues in final effluents of european wastewater treatment plants and their impact on the aquatic environment, Environ. Int., 2020, 140, 105733 CrossRef CAS.
- L. Ma, Y. Liu, Q. Yang, L. Jiang and G. Li, Occurrence and distribution of pharmaceuticals and ppersonal care products (PPCPs) in wastewater related riverbank groundwater, Sci. Total Environ., 2022, 821, 153372 CrossRef CAS.
- K. Zhang, Y. Li, Z. Yu, T. Yang, J. Xu, L. Chao, J. Ni, L. Wang, Y. Gao, Y. Hu and Z. Lin, Xin’anjiang nested experimental watershed (XAJ-NEW) for understanding multiscale mwater cycle: Scientific objectives and experimental design, Engineering, 2022, 18, 207–217 CrossRef.
- T. Fayaz, N. Renuka and S. K. Ratha, Antibiotic occurrence, environmental risks, and their removal from aquatic environments using microalgae: Advances and future perspectives, Chemosphere, 2024, 349, 140822 CrossRef CAS PubMed.
- S. M. Zainab, M. Junaid, N. Xu and R. N. Malik, Antibiotics and antibiotic resistant genes (ARGs) in groundwater: A global review on dissemination, sources, interactions, environmental and human health risks, Water Res., 2020, 187, 116455 CrossRef CAS.
- J. P. Bavumiragira, I. Eheneden, H. Yin, A. W. Mumbi, G. D. S. Quoie, P. Uyisaba, R. Wang and J. Zhao, Insight on prioritization of antibiotics in China, their occurrence, and removal by different wastewater treatment technologies, Discovery Environ., 2024, 2, 28 CrossRef.
- J. Peng, K.-L. Cao, S.-B. Lv, Y.-X. Hu, J. Lin, Q.-Z. Zhou and J.-H. Wang, Algal strains, treatment systems and removal mechanisms for treating antibiotic wastewater by microalgae, J. Water Process Eng., 2023, 56, 104266 CrossRef.
- L. Cheng, X. Li, S. An, Z. Liu, Y. Liu and D. Ren, Preparation and characterization of polyethylene-based composite films coated with carboxymethyl chitosan/sodium alginate/nisin and application in the packaging of Rosa roxburghii Tratt, Food Packag. Shelf Life, 2024, 43, 101295 CrossRef CAS.
- S. Zheng, Y. Wang, C. Chen, X. Zhou, Y. Liu, J. Yang, Q. Geng, G. Chen, Y. Ding and F. Yang, Current progress in natural degradation and enhanced removal techniques of antibiotics in the environment: A Review, Int. J. Environ. Res. Publ. Health, 2022, 19, 10919 CrossRef CAS.
- Y. Chu, X. Chen, S. Li, X. Li, N. Ren and S.-H. Ho, Novel insights into revealing the intrinsic degradation mechanism of ciprofloxacin by Chlorella sorokiniana: Removal efficiency, pathways and metabolism, Chem. Eng. J., 2024, 500, 157015 CrossRef CAS.
- B. Xie, X. Tang, H. Y. Ng, S. Deng, X. Shi, W. Song, S. Huang, G. Li and H. Liang, Biological sulfamethoxazole degradation along with anaerobically digested centrate treatment by immobilized microalgal-bacterial consortium: Performance, mechanism and shifts in bacterial and microalgal communities, Chem. Eng. J., 2020, 388, 124217 CrossRef CAS.
- G. Frascaroli, J. Roberts, C. Hunter and A. Escudero, Removal efficiencies of seven frequently detected antibiotics and related physiological responses in three microalgae species, Environ. Sci. Pollut. Res., 2024, 31, 14178–14190 CrossRef CAS.
- A. Escudero, C. Hunter, J. Roberts, K. Helwig and O. Pahl, Pharmaceuticals removal and nutrient recovery from wastewaters by Chlamydomonas acidophila, Biochem. Eng. J., 2020, 156, 107517 CrossRef CAS.
- Y. Wang, J. Zheng, J. Cheng, R. Zhou, X. Li, J. Hu and J. Lü, Nanobubbles can modulate microbial communities and sedimentary ecosystem during the treatment of pond water, Environ. Sci.: Water Res. Technol., 2023, 9, 1804–1812 RSC.
- J. Peng, Y.-Y. He, Z.-Y. Zhang, X.-Z. Chen, Y.-L. Jiang, H. Guo, J.-P. Yuan and J.-H. Wang, Removal of levofloxacin by an oleaginous microalgae Chromochloris zofingiensis in the heterotrophic mode of cultivation: Removal performance and mechanism, J. Hazard. Mater., 2022, 425, 128036 CrossRef CAS PubMed.
- Y. Zhu, H. Dai and S. Yuan, The competition between heterotrophic denitrification and DNRA pathways in hyporheic zone and its impact on the fate of nitrate, J. Hydrol., 2023, 626, 130175 CrossRef CAS.
- D. Liberti, F. Pinheiro, B. Simões, J. Varela and L. Barreira, Beyond bioremediation: the untapped potential of microalgae in wastewater treatment, Water, 2024, 16, 2710 CrossRef CAS.
- S. Dubey, C.-W. Chen, D. Haldar, V. S. Tambat, P. Kumar, A. Tiwari, R. R. Singhania, C.-D. Dong and A. K. Patel, Advancement in algal bioremediation for organic, inorganic, and emerging pollutants, Environ. Pollut., 2023, 317, 120840 CrossRef CAS.
- J. Silva, L. Arias-Torres, C. Carlesi and G. Aroca, Use of nanobubbles to improve mass transfer in bioprocesses, Processes, 2024, 12, 1227 CrossRef CAS.
- W. Xiao, G. Xu and G. Li, Effect of nanobubble application on performance and structural characteristics of microbial aggregates, Sci. Total Environ., 2021, 765, 142725 CrossRef CAS.
- X. Miao, J. Xu, B. Yang, J. Luo, Y. Zhi, W. Li, Q. He and H. Li, Indigenous mixotrophic aerobic denitrifiers stimulated by oxygen micro/nanobubble-loaded microporous biochar, Bioresour. Technol., 2024, 391, 129997 CrossRef CAS.
- T. Temesgen and M. Han, Advancing aerobic digestion efficiency using ultrafine bubbles in wastewater treatment, J. Water Process Eng., 2023, 55, 104072 CrossRef.
- N. Evode, X. Zhang, X. Chai, J. Gu, S. Zhao and L. Yutao, Performance test and analysis of tetracycline degradation using a Micro-Nano Bubble system, J. Environ. Manage., 2024, 363, 121328 CrossRef CAS.
- M. Han, C. Zhang and S.-H. Ho, Immobilized microalgal system: An achievable idea for upgrading current microalgal wastewater treatment, Environ. Sci. Ecotechnol., 2023, 14, 100227 CrossRef CAS PubMed.
- M. Han, P. Xie, N. Ren and S.-H. Ho, Cytoprotective alginate microcapsule serves as a shield for microalgal encapsulation defensing sulfamethoxazole threats and safeguarding nutrient recovery, J. Hazard. Mater., 2024, 465, 133454 CrossRef CAS PubMed.
- Q. Zhong and J. Xoing, A Globally Distributed Cyanobacterial Nitroreductase Capable of Conferring Biodegradation of Chloramphenicol, Research, 2025, 8, 0692 CrossRef PubMed.
- Y. He, J. Lian, L. Wang, H. Su, L. Tan, Q. Xu, H. Wang, Y. Li, M. Li, D. Han and Q. Hu, Enhanced brewery wastewater purification and microalgal production through algal-bacterial synergy, J. Clean. Prod., 2022, 376, 134361 CrossRef CAS.
- Y. Cao, S. Yang, J. Wang, W. Kong, B. Guo, Y. Xi, A. Zhang and B. Yue, Metabolomic exploration of the physiological regulatory mechanism of the growth and metabolism characteristics of Chlorella vulgaris under photoautotrophic, mixotrophic, and heterotrophic cultivation conditions, Biomass Bioenergy, 2023, 173, 106775 CrossRef CAS.
- H. Lee, D. Jeong, S. Im and A. Jang, Optimization of alginate bead size immobilized with Chlorella vulgaris and Chlamydomonas reinhardtii for nutrient removal, Bioresour. Technol., 2020, 302, 122891 CrossRef CAS PubMed.
- C. Song, Y. Wei, Y. Qiu, Y. Qi, Y. Li and Y. Kitamura, Biodegradability and mechanism of florfenicol via Chlorella Sp. UTEX1602 and L38: Experimental study, Bioresour. Technol., 2019, 272, 529–534 CrossRef CAS PubMed.
- S.-F. Yang, C.-F. Lin, C.-J. Wu, K.-K. Ng, A. Yu-Chen Lin and P.-K. Andy Hong, Fate of sulfonamide antibiotics in contact with activated sludge – Sorption and biodegradation, Water Res., 2012, 46, 1301–1308 CrossRef CAS.
- Y. Ma, S. Lin, T. Guo, C. Guo, Y. Li, Y. Hou, Y. Gao, R. Dong and S. Liu, Exploring the influence of sulfadiazine-induced stress on antibiotic removal and transformation pathway using microalgae Chlorella sp, Environ. Res., 2024, 256, 119225 CrossRef CAS.
- J. Jia, D. Han, H. G. Gerken, Y. Li, M. Sommerfeld, Q. Hu and J. Xu, Molecular mechanisms for photosynthetic carbon partitioning into storage neutral lipids in Nannochloropsis oceanica under nitrogen-depletion conditions, Algal Res., 2015, 7, 66–77 CrossRef.
- J. Zhen, Z.-B. Wang, B.-J. Ni, S. Ismail, A. El-Baz, Z. Cui and S.-Q. Ni, Synergistic Integration of Anammox and Endogenous Denitrification Processes for the Simultaneous Carbon, Nitrogen, and Phosphorus Removal, Environ. Sci. Technol., 2024, 58, 10632–10643 CrossRef CAS PubMed.
- A. Ahmad, A. H. Bhat and A. Buang, Biosorption of transition metals by freely suspended and Ca-alginate immobilised with Chlorella vulgaris: Kinetic and equilibrium modeling, J. Clean. Prod., 2018, 171, 1361–1375 CrossRef CAS.
- J. Wang and R. Zhuan, Degradation of antibiotics by advanced oxidation processes: An overview, Sci. Total Environ., 2020, 701, 135023 CrossRef CAS PubMed.
- W. Xiao and G. Xu, Mass transfer of nanobubble aeration and its effect on biofilm growth: Microbial activity and structural properties, Sci. Total Environ., 2020, 703, 134976 CrossRef CAS PubMed.
- S. Zhou, S. Nazari, A. Hassanzadeh, X. Bu, C. Ni, Y. Peng, G. Xie and Y. He, The effect of preparation time and aeration rate on the properties of bulk micro-nanobubble water using hydrodynamic cavitation, Ultrason. Sonochem., 2022, 84, 105965 CrossRef CAS PubMed.
- J. P. Vareda, On validity, physical meaning, mechanism insights and regression of adsorption kinetic models, J. Mol. Liq., 2023, 376, 121416 CrossRef CAS.
- H. Wang, C. Hu, Y. Wang, Y. Zhao, C. Jin and L. Guo, Elucidating microalgae-mediated metabolism for sulfadiazine removal mechanism and transformation pathways, Environ. Pollut., 2023, 327, 121598 CrossRef CAS PubMed.
- S. Chen, W. Zhang, J. Li, M. Yuan, J. Zhang, F. Xu, H. Xu, X. Zheng and L. Wang, Ecotoxicological effects of sulfonamides and fluoroquinolones and their removal by a green alga (Chlorella vulgaris) and a cyanobacterium (Chrysosporum ovalisporum), Environ. Pollut., 2020, 263, 114554 CrossRef CAS PubMed.
- S.-F. Yang, C.-F. Lin, A. Yu-Chen Lin and P.-K. Andy Hong, Sorption and biodegradation of sulfonamide antibiotics by activated sludge: Experimental assessment using batch data obtained under aerobic conditions, Water Res., 2011, 45, 3389–3397 CrossRef CAS PubMed.
- A. Lal and D. Das, Biomass production and identification of suitable harvesting technique for Chlorella sp. MJ 11/11 and Synechocystis PCC 6803, 3 Biotech, 2016, 6, 41 CrossRef PubMed.
- H. Li, L. Hu, D. Song and F. Lin, Characteristics of Micro Nano Bubbles and Potential Application in Groundwater Bioremediation, Water Environ. Res., 2014, 86, 844–851 CrossRef CAS.
- P. Wu, T. Hu, L. Sun and J. Fan, Efficient degradation of sulfadiazine by photosynthetic cyanobacteria Synechocystis Sp. PCC 6803 coupling with recombinant laccase strategy, Chem. Eng. J., 2023, 474, 145974 CrossRef CAS.
- J.-Q. Xiong, S.-J. Kim, M. B. Kurade, S. Govindwar, R. A. I. Abou-Shanab, J.-R. Kim, H.-S. Roh, M. A. Khan and B.-H. Jeon, Combined effects of sulfamethazine and sulfamethoxazole on a freshwater microalga, Scenedesmus obliquus: toxicity, biodegradation, and metabolic fate, J. Hazard. Mater., 2019, 370, 138–146 CrossRef CAS PubMed.
- X. You, L. Yang, H. Chu, L. Zhang, Y. Hong, Y. Lin, X. Zhou and Y. Zhang, Micro-nano-bubbles and their application in microalgae production: Wastewater treatment, carbon capture and microalgae separation, Algal Res., 2024, 78, 103398 CrossRef.
- X. R. Feng, J. Hu, N. Liu, W. K. Tong, M. Gao, C. Dai, Y. Han and J. Li, Response model between nanobubble preparation parameters and properties: Controllable preparation & its application example, J. Water Process Eng., 2024, 64, 105660 CrossRef.
- A. M. Zafar, M. A. Javed, A. Aly Hassan, K. Mehmood and E. Sahle-Demessie, Recent updates on ions and nutrients uptake by halotolerant freshwater and marine microalgae in conditions of high salinity, J. Water Process Eng., 2021, 44, 102382 CrossRef.
- M. G. Mostofa, M. M. Rahman, T. K. Ghosh, A. H. Kabir, M. Abdelrahman, M. A. Rahman Khan, K. Mochida and L.-S. P. Tran, Potassium in plant physiological adaptation to abiotic stresses, Plant Physiol. Biochem., 2022, 186, 279–289 CrossRef CAS PubMed.
- H. Ji, Y. Gong, J. Duan, D. Zhao and W. Liu, Degradation of petroleum hydrocarbons in seawater by simulated surface level atmospheric ozone: Reaction kinetics and effect of oil dispersant, Mar. Pollut. Bull., 2018, 135, 427–440 CrossRef CAS PubMed.
- M. A. Farha, S. French, J. M. Stokes and E. D. Brown, Bicarbonate Alters Bacterial Susceptibility to Antibiotics by Targeting the Proton Motive Force, ACS Infect. Dis., 2018, 4, 382–390 CrossRef CAS PubMed.
- G. Pérez-Lucas, A. E. Aatik, M. Aliste, G. Navarro, J. Fenoll and S. Navarro, Removal of Contaminants of Emerging Concern from a Wastewater Effluent by Solar-Driven Heterogeneous Photocatalysis: A Case Study of Pharmaceuticals, Water Air Soil Pollut., 2023, 234, 55 CrossRef.
- S. Li, P. Show, H. Ngo and S.-H. Ho, Algae-mediated antibiotic wastewater treatment: A critical review, Environ. Sci. Ecotechnol., 2022, 9, 100145 CrossRef CAS PubMed.
- W. Lu, C. Xu, F. Liu, M. Su, S. Cheng and Y. Zhang, Antibiotic removal efficiency by microalgae: A systematic analysis combined with meta-analysis, Process Saf. Environ. Prot., 2023, 174, 912–920 CrossRef CAS.
- S. Chen, L. Wang, W. Feng, M. Yuan, J. Li, H. Xu, X. Zheng and W. Zhang, Sulfonamides-induced oxidative stress in freshwater microalga Chlorella vulgaris: Evaluation of growth, photosynthesis, antioxidants, ultrastructure, and nucleic acids, Sci. Rep., 2020, 10(1), 8243 CrossRef CAS PubMed.
- D. Kowalczuk and M. Pitucha, Application of FTIR Method for the Assessment of Immobilization of Active Substances in the Matrix of Biomedical Materials, Materials, 2019, 12, 2972 CrossRef CAS PubMed.
- M. Cheng, C. Shi, B.-H. Zhao, T.-Y. Wang, N. Zhang, R.-B. Liu, D.-Q. Cao and X.-D. Hao, Distribution characteristics of sulfonamide antibiotics between water and extracellular polymeric substances in municipal sludge, Environ. Res., 2024, 259, 119576 CrossRef CAS PubMed.
- X. Zhao, X. Wang and T. Lou, Preparation of fibrous chitosan/sodium alginate composite foams for the adsorption of cationic and anionic dyes, J. Hazard. Mater., 2021, 403, 124054 CrossRef CAS PubMed.
- Q. Li, X. Li, C. Zhou, C. Lu, B. Liu and G. Wang, Insight into oxidation and adsorption treatment of algae-laden water: Algal organic matter transformation and removal, Chem. Eng. J., 2021, 420, 129887 CrossRef CAS.
- D. Elkobrosy, A. A. AI-Askar, H. El-Gendi, Y. Su, R. Nabil, A. Abdelkhalek and S. Behiry, Nematocidal and Bactericidal Activities of Green Synthesized Silver Nanoparticles Mediated by Ficus sycomorus Leaf Extract, Life, 2023, 13, 1083 CrossRef CAS PubMed.
- L. Leng, L. Wei, Q. Xiong, S. Xu, W. Li, S. Lv, Q. Lu, L. Wan, Z. Wen and W. Zhou, Use of microalgae based technology for the removal of antibiotics from wastewater: A review, Chemosphere, 2020, 238, 124680 CrossRef CAS PubMed.
- Y. Chu, C. Zhang, R. Wang, X. Chen, N. Ren and S.-H. Ho, Biotransformation of sulfamethoxazole by microalgae: Removal efficiency, pathways, and mechanisms, Water Res., 2022, 221, 118834 CrossRef CAS PubMed.
- Q. Cheng, Y. Jiang, Z. Jin, C. Hui, L. Xu, Q. Zhou, Y. Zhao, L. Du and H. Jiang, Enhanced excretion of extracellular polymeric substances associated with nonylphenol tolerance in Dictyosphaerium sp, J. Hazard. Mater., 2020, 395, 122644 CrossRef CAS PubMed.
- C. Wang, D. Dong, L. Zhang, Z. Song, X. Hua and Z. Guo, Response of Freshwater Biofilms to Antibiotic Florfenicol and Ofloxacin Stress: Role of Extracellular Polymeric Substances, Int. J. Environ. Res. Publ. Health, 2019, 16, 715 CrossRef CAS PubMed.
- L. M. Nguyen, N. T. T. Nguyen, T. T. T. Nguyen, T. T. Nguyen, D. T. C. Nguyen and T. V. Tran, Occurrence, toxicity and adsorptive removal of the chloramphenicol antibiotic in water: a review, Environ. Chem. Lett., 2022, 20, 1929–1963 CrossRef CAS PubMed.
- M. Hejna, D. Kapuścińska and A. Aksmann, Pharmaceuticals in the Aquatic Environment: A Review on Eco-Toxicology and the Remediation Potential of Algae, Int. J. Environ. Res. Publ. Health, 2022, 19, 7717 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra02082d |
| This journal is © The Royal Society of Chemistry 2025 |







