Open Access Article
Open Access ArticleEnzyme-catalyzed C(sp3)–H aminations for the highly enantioselective construction of chiral 2-oxazolidinones†
Hanzi Gao ,
Miao Li and
Guojun Zheng
,
Miao Li and
Guojun Zheng *
*
State Key Laboratory of Chemical Resources Engineering, Beijing University of Chemical Technology, Beijing, 100029, People's Republic of China. E-mail: zhenggj@buct.edu.cn
First published on 10th June 2025
Abstract
As a rapidly growing field, C(sp3)–H functionalization is being used to access a wide range of important molecular targets. The enzymatic activity of C(sp3)–H is a powerful synthetic tool to develop valuable building blocks. In this study, engineered myoglobin variants were found to be capable of C(sp3)–H activation under mild conditions via mediated nitrene transfer. Using this approach, 2-oxazolidinones and γ-lactams with high enantioselectivity were obtained through intramolecular cyclization using readily available and stable N-acetoxyamides as substrates.
Introduction
One of the most prevalent chemical bonds in organic chemistry and biochemistry is the carbon–nitrogen bond. Nitrogen-containing heterocycles, which are important structural units,1–4 are frequently found in natural products and medications such as antibiotics,5 antiepileptic drugs,6 analgesics,7 and antimicrobials8 (Scheme 1a). However, how to directly convert C–H bonds into C–N bonds is still a challenging research topic. In previous studies, transition metal-catalyzed amino transfer has shown advantages of high regioselectivity and stereoselectivity.9–11 In 1983, Breslow carried out pioneering work using a (tetraphenylporphyrin)FeIII catalyst (5 mol%) and cytochrome P450 to achieve nitrene transfer to obtain cyclosulfonamides.12,13 Recently, using dioxazolones as nitrene precursors, Chang's group reported the formation of lactams via an Ir complex-catalyzed C–H amidation reaction.14,15 These transformations are mediated by reactive metal-nitrenoid species generated via the reaction of transition metal catalysts with nitrene precursor reagents (Scheme 1b). Using this strategy, various cyclic amines, including oxazolidinones, sulfamates, and pyrrolidines, can be obtained. The development of more cost-effective and environmentally friendly catalysts for efficient C–N bond formation is a goal of ongoing research.Heme-containing enzymes, such as cytochrome P450, can naturally accomplish regio- and enantioselective C–H functionalization, such as hydroxylation and epoxidation.16–18 In recent years, it has been found that hemoproteins can also catalyze reactions that differ from their widely known catalytic reactions, known as enzyme promiscuity.19 In 2013, Arnold's group and Fasan's group achieved cytochrome P450 variant-mediated nitrene transfer,20,21 which is one of the major breakthroughs in the area of C–H functionalization. Over the subsequent decade, combined with directed evolution,22 heme-containing enzyme-mediated nitrene transfer was extended to many types of reactions, realizing the great potential of C–H functionalization on a wide range of substrates23–27 while maintaining high reaction rates and enantioselectivity, as demonstrated.
In 2015, P450-catalyzed conversion of carbonazidate substrates into oxazolidinones was achieved28 by Fasan's group (Scheme 1c). However, the poor safety of azide substrates, a low total turnover number (TTN) and the absence of enantioselectivity in this method limited its further development. In 2023, a strategy for the asymmetric synthesis of β-, γ- and δ-lactams via hemoprotein-catalyzed intramolecular C–H amidation of dioxazolones was reported.29 This approach enables precise control over regioselectivity and stereoselectivity. Mechanistic and computational studies were conducted to elucidate the nature of C–H amidation and enantio-determining steps and provide insights into the protein-mediated control of regioselectivity and stereoselectivity.
In this study, we report an intramolecular C–H amidation reaction catalyzed by a hemoprotein to yield 2-oxazolidinones and γ-lactams (Scheme 1d). Engineered myoglobin variants containing heme were used to catalyze the C(sp3)–H amination reaction using easily accessible N-acyloxyamides as nitrene precursors to obtain the target products through directed evolution and optimization of a series of conditions. This method enables the enzyme-catalyzed synthesis of optically active 2-oxazolidinone products and can also be applied to the generation of γ-lactams.
Note added
During the revision of this manuscript, a related study on highly enantioselective enzymatic C(sp3)–H amination for constructing chiral oxazolidinone rings was published (https://doi.org/10.1021/acscatal.4c06066). It should be noted that while some conclusions may overlap, the two studies were carried out independently, and no conflicts of interest exist between them.Results and discussion
Inspired by the work of the Meggers group,28,30 we envisioned the possibility of using N-acyloxyamides for the enzyme-catalyzed γ-C–H amidation reaction via nitrene transfer. First, N-benzoyloxycarbamate (1-1) was used as the initial substrate. In order to identify the initial biocatalyst for this reaction, a number of various heme-containing and iron-containing non-heme enzymes were screened, including the myoglobin variant Mb-H64V/V68A (Mb1),26,27,29,31 and variants of cytochrome P450 (ref. 32–35) and Az1.36 Among all the enzymes, only myoglobin showed the desired activity, and the target product 4-phenyl-2-oxazolidinone (2a) was detected with excellent enantioselectivity (96% e.e.) (Scheme 2). Meanwhile, a 78% yield of the carbamate byproduct (3) was also obtained, presumably resulting from the reduction of the nitrenoid intermediate. This result provides a starting point to validate our study.Subsequently, several substrates with different acyl groups were tested. It was found that the pivaloyl substrate (1-2) exhibited a better yield (30%) and similar enantioselectivity (98% e.e.) compared with the benzoyl substrate (1-1). Therefore, it was decided to use phenethyl N-pivaloyloxycarbamate (1-2) as a model substrate and the myoglobin variant Mb1 as the initial catalyst for subsequent studies.
Stronger ligand–enzyme interactions will lead to an increase in enzyme activity and simultaneously can inhibit the production of byproducts. In order to further improve the reaction yield, attempts were made to obtain enzyme mutants with higher catalytic activity through directed evolution. Based on the interaction model between myoglobin and ligands,27 a total of 16 amino acid sites, including H93, which is on the heme axial ligand, H97, S92, and R45, which can interact with heme, as well as others around the substrate pocket and adjacent flexible loops, were selected for mutagenesis. In each round of site saturation mutagenesis, the mutants were assayed by HPLC to determine the transformation yields.
Finally, three mutants, L104F, L104H, and L104Y, which can lead to a pronounced increase in yields, were obtained. Among them, the mutant L104F can achieve transformation with a 42% yield and 98% e.e., which may result from the introduction of larger phenylalanine compared to the original hydrophobic leucine (see ESI Fig. S2†). This mutation can maintain the hydrophobic environment of the active pocket while intensifying protein stacking,37,38 resulting in a more rigid protein structure and a strict conformational landscape and retaining the high enantioselectivity of the product.
The saturation mutation screening for the other 15 sites yielded no beneficial mutations (see ESI Table S3†). A few mutants, such as L32M, L89V, and R45K, were comparable to the wild type in terms of yield. Before and after these mutations, the amino acids were similar in size and characteristics, and the original activity was maintained without disrupting the hydrophobic environment of the active pocket. To explore how the enzyme regulates product selectivity, we constructed a mutant Mb1–L104F protein structure using homology modeling and simulated the protein–ligand interaction by molecular docking. The molecular docking results (see ESI Fig. S3†) corroborate the findings of previous studies.29 Analysis of amino acid residues within the active pocket revealed that Ile107, Val64, and Ala68 are located on either side of the substrate, and the aromatic ring of the substrate extends into the protein core between Ile107 and Ala68, facilitating the immobilization of the substrate-binding site. Due to the high resistance of the left side, the substrate adopts a conformation that favors loop closure from the right side, resulting in the R-conformation of the 2-oxazolidinone product. Thus, mutation of all three positions resulted in a significant decrease in yield.
Based on previous studies on the mechanism of C–H amination by metal-mediated nitrene transfer, it was hypothesized that an alkaline environment might be beneficial for the C–H amination reaction; hence, a series of optimizations were performed on the type of buffer and pH for the reaction. Potassium phosphate buffer with pH = 11 yielded the best result. However, a high pH would lead to a decrease in enzyme activity. According to the previous report,29 an organic co-solvent could favor the desired C–H amidation reaction and correspondingly inhibit the formation of the amide byproduct 3. Screening of various organic co-solvents was conducted, which revealed that 7.5% (v/v) acetonitrile was beneficial in increasing the yield of the main product while reducing the formation of compound 3. The optimized reaction conditions were determined, and finally, 2a was obtained with a yield of 55%, TTN of 223 and enantioselectivity of 98% e.e. (Fig. 1).
Under optimized reaction conditions, the adaptation of the mutant Mb1–L104F to different N-pivaloyloxycarbamate substrates was tested. The results are shown in Scheme 3, which reveal that the enzyme is well-tolerant to aryl ring para-substitution and generates 2-oxazolidinone products with excellent enantioselectivity (97–99% e.e.). However, the yields are significantly affected by the size of the substituent groups, with larger substituents leading to a significant decrease in yield (2h–j). Both electron-withdrawing and electron-donating substituents have no effect on the reaction yields (2b, c, k). Para-fluorine substitution produced the corresponding 2d with high yield (76%) and high enantiopurity (>99% e.e.), which may be attributed to the hydrogen bonding interaction between fluorine and the enzyme. Furthermore, the enzymatic synthesis of 2l with >99% e.e. and 74% yield also demonstrated the tolerance of the enzyme towards aromatic heterocycles. The yields of the ortho- and meta-substituted compounds decreased slightly compared to the para-substituted compound but still maintained a high enantioselectivity (2e–g). Collectively, these results reveal the remarkable universality of enzyme catalysts for the stereoselective synthesis of 2-oxazolidinones of different sizes and substitutions. The wide scope of the substrate indicates the practicality of this biocatalytic system.
 | ||
| Scheme 3 Substrate scope of 2-oxazolidinone. CDI, carbonyldiimidazole; DCC, N,N′-dicyclohexylcarbodiimide. | ||
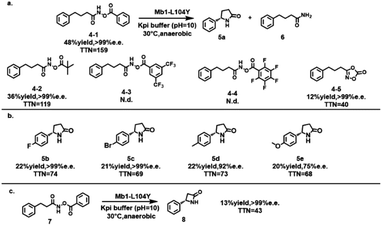
To further extend the adaptability of this mutant, different N-acyloxyamides were tested in this catalytic process. Encouragingly, several substrates were found to be capable of intramolecular C–H amination. Optimization of these reaction conditions allowed the catalytic conversion of (1-oxo-4-phenylbutyl)azanyl ester (4-1) to 5-phenyl-γ-lactam (5a) with the Mb1–L104Y mutant, with an enhanced yield of 48% and excellent S-configured enantioselectivity (>99% e.e.) (Scheme 4a). In previous studies, it has been found that the key acyl nitrenoid intermediates generated in the reaction are prone to Curtius rearrangements, which can hamper the desired nitrene insertion and lead to the formation of isocyanate byproducts.39,40 Under the conditions we optimized, only the γ-lactam product and the byproduct 4-phenylbutyramide (6) were detected in the reaction solution. To assess the substrate scope of the method, Mb1–L104F was tested against a range of para-substituted substrates (5b–e) (Scheme 4b). The enzyme tolerated both electron-withdrawing and electron-donating substituted substrates well; however, the enantiomeric excess of the electron-donating substituent products was reduced (5d, 5e), probably due to the larger para-substituents that prevent the substrate from approaching the enzyme. Attempts to catalyze (1-oxo-3-phenylpropyl)azanyl ester (7) with the Mb1–L104Y mutant were successful in obtaining a highly enantioselective β-lactam product (8) (Scheme 4c). This approach further expands the scope of enzyme-catalyzed reactions, demonstrating the strong ability of the enzyme to control stereoselectivity and regioselectivity.
Conclusions
In summary, we have developed a potential biocatalytic strategy for the asymmetric synthesis of N-heterocyclic compounds through nitrene transfer, starting from readily available and stable N-acyloxycarbamate compounds. During this process, engineered myoglobin variants were utilized as a biocatalyst to produce a variety of 2-oxazolidinones and γ-lactams with high enantioselectivity. Saturation mutagenesis indicated that several amino acid residues, such as L104, V64, and A68, were crucial for enzyme activity. This enzymatic approach demonstrates its potential application in asymmetric synthesis. This paves the way for the development of other asymmetric enzyme-catalyzed nitrene transfer reactions using carbamates as nitrene precursors.Data availability
The authors confirm that all data are true and reliable. The data that support the findings of this study are available within the main text and the ESI.† Details about materials, methods, experimental procedures, characterization data, and NMR and HPLC spectra are available in the ESI,† and all other data are available from the corresponding author upon request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Key R&D Program of China (2021YFC2102900 and 2021YFC2101503).Notes and references
- J. Bariwal and E. Van der Eycken, Chem. Soc. Rev., 2013, 42, 9283–9303 RSC.
- A. Trowbridge, S. M. Walton and M. J. Gaunt, Chem. Rev., 2020, 120, 2613–2692 CrossRef CAS PubMed.
- B. Eftekhari-Sis, M. Zirak and A. Akbari, Chem. Rev., 2013, 113, 2958–3043 CrossRef CAS PubMed.
- Y. Wang, W. X. Zhang and Z. Xi, Chem. Soc. Rev., 2020, 49, 5810–5849 RSC.
- K. Tahlan and S. E. Jensen, J. Antibiot., 2013, 66, 401–410 CrossRef CAS PubMed.
- C. Palleria, G. Cozza, R. Khengar, V. Libri and G. De Sarro, Curr. Pharm. Des., 2017, 23, 5606–5624 CrossRef CAS PubMed.
- Y. Zhao, B. Huang, C. Yang, Q. Chen and W. Xia, Org. Lett., 2016, 18, 5572–5575 CrossRef CAS PubMed.
- K. Tsai, V. Stojković, D. J. Lee, I. D. Young, T. Szal, D. Klepacki, N. Vázquez-Laslop, A. S. Mankin, J. S. Fraser and D. G. Fujimori, Nat. Struct. Mol. Biol., 2022, 29, 162–171 CrossRef CAS PubMed.
- H. M. Davies and J. R. Manning, Nature, 2008, 451, 417–424 CrossRef CAS PubMed.
- Y. Kim, S. Chang and Y. Park, Chem. Rev., 2017, 117, 9247–9301 CrossRef PubMed.
- N. Sauermann, T. H. Meyer, Y. Qiu and L. Ackermann, ACS Catal., 2018, 8, 7086–7103 CrossRef CAS.
- R. Breslow and S. H. Gellman, J. Am. Chem. Soc., 1983, 105, 6728–6729 CrossRef CAS.
- E. W. Svastits, J. H. Dawson, R. Breslow and S. H. Gellman, J. Am. Chem. Soc., 2002, 107, 6427–6428 CrossRef.
- S. Y. Hong, Y. Park, Y. Hwang, Y. B. Kim, M. H. Baik and S. Chang, Science, 2018, 359, 1016–1021 CrossRef CAS PubMed.
- S. Huh, S. Y. Hong and S. Chang, Org. Lett., 2019, 21, 2808–2812 CrossRef CAS PubMed.
- N. Ma, Z. Chen, J. Chen, J. Chen and Z. Cong, Angew. Chem., 2018, 130, 7628–7633 CrossRef PubMed.
- Y. Wei, E. L. Ang and H. Zhao, Curr. Opin. Chem. Biol., 2018, 43, 1–7 CrossRef CAS PubMed.
- K. Geisler, R. K. Hughes, F. Sainsbury, G. P. Lomonossoff, M. Rejzek, S. Fairhurst, C. E. Olsen, M. S. Motawia, R. E. Melton, A. M. Hemmings, S. Bak and A. Osbourn, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3360–3367 Search PubMed.
- R. Chen, B. Gao, X. Liu, F. Ruan, Y. Zhang, J. Lou, K. Feng, C. Wunsch, S. M. Li, J. Dai and F. Sun, Nat. Chem. Biol., 2017, 13, 226–234 CrossRef CAS PubMed.
- J. A. McIntosh, P. S. Coelho, C. C. Farwell, Z. J. Wang, J. C. Lewis, T. R. Brown and F. H. Arnold, Angew Chem. Int. Ed. Engl., 2013, 52, 9309–9312 CrossRef CAS PubMed.
- R. Singh, M. Bordeaux and R. Fasan, ACS Catal., 2014, 4, 546–552 CrossRef CAS PubMed.
- R. Breslow and S. H. Gellman, J. Chem. Soc., 1982, 24, 1400–1401 Search PubMed.
- T. K. Hyster, C. C. Farwell, A. R. Buller, J. A. McIntosh and F. H. Arnold, J. Am. Chem. Soc., 2014, 136, 15505–15508 CrossRef CAS PubMed.
- N. W. Goldberg, A. M. Knight, R. K. Zhang and F. H. Arnold, J. Am. Chem. Soc., 2019, 141, 19585–19588 CrossRef CAS PubMed.
- Z. Y. Qin, S. Gao, Y. Zou, Z. Liu, J. B. Wang, K. N. Houk and F. H. Arnold, ACS Cent. Sci., 2023, 9, 2333–2338 CrossRef CAS PubMed.
- M. Bordeaux, R. Singh and R. Fasan, Bioorg. Med. Chem., 2014, 22, 5697–5704 CrossRef CAS PubMed.
- M. Pott, M. Tinzl, T. Hayashi, Y. Ota, D. Dunkelmann, P. R. E. Mittl and D. Hilvert, Angew Chem. Int. Ed. Engl., 2021, 60, 15063–15068 CrossRef CAS PubMed.
- R. Singh, J. N. Kolev, P. A. Sutera and R. Fasan, ACS Catal., 2015, 5, 1685–1691 CrossRef CAS PubMed.
- S. Roy, D. A. Vargas, P. Ma, A. Sengupta, L. Zhu, K. N. Houk and R. Fasan, Nat. Catal., 2023, 7, 65–76 CrossRef PubMed.
- Z. Zhou, Y. Tan, X. Shen, S. Ivlev and E. Meggers, Sci. China: Chem., 2021, 64, 452–458 CrossRef CAS.
- M. Bordeaux, V. Tyagi and R. Fasan, Angew Chem. Int. Ed. Engl., 2015, 54, 1744–1748 CrossRef CAS PubMed.
- C. K. Prier, R. K. Zhang, A. R. Buller, S. Brinkmann-Chen and F. H. Arnold, Nat. Chem., 2017, 9, 629–634 CrossRef CAS PubMed.
- R. K. Zhang, K. Chen, X. Huang, L. Wohlschlager, H. Renata and F. H. Arnold, Nature, 2019, 565, 67–72 CrossRef CAS PubMed.
- Z. J. Jia, S. Gao and F. H. Arnold, J. Am. Chem. Soc., 2020, 142, 10279–10283 CrossRef CAS PubMed.
- O. F. Brandenberg, K. Chen and F. H. Arnold, J. Am. Chem. Soc., 2019, 141, 8989–8995 CrossRef CAS PubMed.
- J. Rui, Q. Zhao, A. J. Huls, J. Soler, J. C. Paris, Z. Chen, V. Reshetnikov, Y. Yang, Y. Guo, M. Garcia-Borràs and X. Huang, Science, 2022, 376, 869–874 CrossRef CAS PubMed.
- P. J. Fleming and F. M. Richards, J. Mol. Biol., 2000, 299, 487–498 CrossRef CAS PubMed.
- J. Pontius, J. Richelle and S. J. Wodak, J. Mol. Biol., 1996, 264, 121–136 CrossRef CAS PubMed.
- S. Bräse, C. Gil, K. Knepper and V. Zimmermann, Angew Chem. Int. Ed. Engl., 2005, 44, 5188–5240 CrossRef PubMed.
- D. Li, T. Wu, K. Liang and C. Xia, Org. Lett., 2016, 18, 2228–2231 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra01905b |
| This journal is © The Royal Society of Chemistry 2025 |