Open Access Article
Open Access ArticleEnzyme graphene oxide interaction: the case system of β-lactamases†
Alessandra Piccirilli a,
Mariagrazia Perilli
a,
Mariagrazia Perilli a,
Fabrizia Brisdelli
a,
Fabrizia Brisdelli a,
Dario Mastrippolito
a,
Dario Mastrippolito *b,
Gianluca D'Olimpiob,
Soheil Ershadradc,
Biplab Sanyal
*b,
Gianluca D'Olimpiob,
Soheil Ershadradc,
Biplab Sanyal c and
Luca Ottaviano*b
c and
Luca Ottaviano*b
aDepartment of Biotechnological and Applied Clinical Sciences, University of L'Aquila, L'Aquila, Italy
bDepartment of Physical and Chemical Sciences, University of L'Aquila, L'Aquila, Italy. E-mail: mastrippolito@insp.upmc.fr; luca.ottaviano@aquila.infn.it
cDepartment of Physics and Astronomy, Uppsala University, Uppsala, Sweden
First published on 23rd June 2025
Abstract
We report an extensive survey of the inhibition effect of graphene oxide (GO) on the enzymatic activity of β-lactamases (BLs), namely: serine β-lactamases (SBLs) SHV-1, CTX-M-15, GES-1, KPC-3, OXA-23, and metallo-β-lactamases (MBLs) (NDM-1, VIM-1, CphA, and L1). The inhibition study has been performed with GO dose-dependent kinetic assays (using nitrocefin as a substrate). On all the SBLs, a formidable inhibition activity is demonstrated already at a GO concentration of 0.5 µg ml−1. Inhibition is observed for all the MBLs in the monomeric phase. For MBLs, the interaction mechanism with GO has been directly investigated with X-ray photoemission spectroscopy, focusing on the interaction of NDM-1 with GO. Data do not show any specific interaction with GO of the Zn atoms at the active site of the enzyme, while a detailed analysis of the C 1s and N 1s core levels points to the interaction of GO with the enzyme via its positive residues, indicating a non-competitive inhibition mechanism. Together with the modeling of the nitrocefin–GO system with density functional theory, we demonstrate that the GO basal plane links the substrate molecule via oxidation of one of the two sulfur atoms of the substrate, strongly reducing the inhibition effectiveness if GO is pre-incubated with the substrate.
Introduction
The interaction of surfaces with organic or biological matter in liquid environments is a subject of pivotal relevance. Indeed, surfaces, offering a platform for catalysis, adhesion, and self-organization, are considered to be at the very origin of the evolution of life.1 Two-dimensional materials, being the ultimate “all-surface” systems, offer a unique platform to study the interaction of organic, soft, and living matter with surfaces, and among them, graphene oxide (GO) is unique.2 As reviewed in ref. 3, many features determine such uniqueness. GO is soluble in water, it appears in the form of single-layer two-dimensional sheets of highly oxidized graphene having dimensions ranging from tens of nanometers to hundreds of micrometers, it is made of the same light elements of living matter (carbon, oxygen, hydrogen), it is very prone to functionalization, it is flexible in water, its reduction degree can be tuned, and, accordingly, its transition from being hydrophilic to hydrophobic can also be tuned. The interaction of GO with proteins has been the subject of increasing interest for the last two decades. To this aim, the research has been mostly focused on globular proteins and enzymes like α-chymotrypsin,4 trypsine,5 β-galactosidase,6 Horse-Radish peroxidase,7 lysozyme,7 human ubiquitin,8 hemoglobin,9 myoglobin,10 human serum albumin,11 or catalase.12 A good summary review of the interaction of proteins with graphene oxide is reported in ref. 13, where, overall, so far, fourteen protein–GO systems have been reported and examined. In most of the above-mentioned studies, the research was focused not only on adhesion adsorption itself but also on its consequences for the enzymes' biochemical activity. Their adsorption on GO is likely via linking with the built-in functional groups on both sides of its surface. The proposed adhesion mechanism is via π–π interaction, electrostatic interactions, hydrogen bonding, or even covalent bonding.13 As a result of such interaction, the ternary structure of the enzymes can likely be perturbed, leading to the inhibition of the enzymatic activity.5,14,15Among enzymes, some of bacterial genesis are involved in the breakdown of antibiotics, playing a key role in the development of antibiotic resistance. β-Lactams resistant bacteria have evolved by developing specific enzymes, the β-lactamases (BLs), that enable antibiotic inactivation via hydrolysis of the amide bond of the typical lactam ring of the drug, rendering the product biologically inactive16,17 (among antibiotics, β-lactams are the most widely used due to their effectiveness, low cost, and minimal side effects18). As a matter of fact, bacterial evolution has led to a large family of BLs. To date, several thousand of these enzymes, classified by molecular point of view in classes A, B, C, and D, have been discovered.19 The enzymes included in classes A, C, and D are serine β-lactamases (SBLs) that hydrolyze β-lactams by forming a transient or stable acyl-enzyme through an active site serine, whereas class B BLs are metallo-enzymes that utilize one or two zinc ions for their catalysis.20 Metallo-β-lactamases (MBLs) are further categorized into three subclasses (B1, B2, B3) based on amino acid sequence, structure, and catalytic mechanism.21 The most common BLs in the clinical environment are MBLs such as NDM- and VIM-types and extended-spectrum BLs including KPC, CTX-M, and OXA families enzymes.22,23 Only quite recently, attention has been focused on GO/BLs systems with a study on the VIM-2 one that clearly demonstrated the occurrence of a significant inhibition.14 However, a systematic survey of the interaction of GO with A, B, C, and D BLs is still lacking.
In this study, we surveyed the enzymatic activity of GO via kinetic assays on a very representative choice of BLs. Nine different GO–enzyme systems have been considered, producing a leap contribution to the pre-existing literature.13 We selected the most representative BL with a narrow β-lactams spectrum that is SHV-1 (found mainly in K. pneumoniae and E. coli clinical isolates), and also extended spectrum BLs such as CTX-M-15 and GES-1. KPC-3, known as carbapenemase, was also considered for its ability to hydrolyze very efficiently carbapenems such as imipenem, meropenem, and ertapenem.24 The OXA-23 enzyme, a class D BL frequently isolated in A. baumanni and sometimes in Enterobacterales strains,25 was also chosen due to its strong hydrolytic activity against oxacillin (a semisynthetic penicillin) and carbapenems. All the above are SBLs. To complete the survey, MBLs such as NDM-1 and VIM-1 (subclass B1), CphA (subclass B2), and L1 (subclass B3) were included. All BLs used in this study, except L1, which is a tetrameric enzyme, are monomeric.
For a well-rounded methodological approach, X-ray Photoelectron Spectroscopy (XPS) has been used to study the specific interaction of GO with the nitrocefin substrate; the same interaction has been studied with first principles density functional theory (DFT) calculations. Both experimental data and theoretical predictions clearly show the chemisorption of the antibiotic on the GO basal plane via one of the S atoms of the drug. This cross-talk between the GO inhibitor and the substrate provides a rationale to explain the results of the kinetic assays, where the GO, pre-incubated with the substrate, exhibits a marked reduction of its inhibition activity. The GO/BL interaction was then studied by performing GO dose-dependent kinetic assays (using nitrocefin as a substrate), and, after the GO concentration was optimized via incubation time-dependent kinetic assays. On all the SBLs, we demonstrated a formidable inhibition activity at GO concentration values almost one order of magnitude lower than in previous reports.14 The inhibition is universally effective on all monomeric enzymes (including MBLs) and is observed to be markedly reduced only for L1, which is a tetramer BL. As a representative and very relevant case system, the interaction of NDM-1 with GO has been studied with XPS. Data indicate no chemical shifts of the Zn core level of the zinc atoms of NDM-1 in the interaction with GO, while a detailed analysis of the C 1s XPS spectra gives an indication consistent with covalent bonding of amino groups of the enzymes with the oxidized basal plane of the GO. The evidences are consistent with a model interaction where enzyme–GO linking preferentially occurs via the bonding of lysine residues.
Results
GO morphological characterization
Graphene oxide is an ill-defined material. In particular, it does not have a specific molecular weight as it appears in the form of individual flakes, whose size distribution is strongly dependent on its synthesis methods and post-treatment procedures.3 To this purpose, to assess its effects on biological systems, as size-dependent effects are well known to play a role,26,27 it is mandatory to characterize it in terms of its flake size distribution. In particular, it has already demonstrated that the lateral size distribution of GO always obeys a lognormal statistic (see ref. 3), where the main effective parameter is the modal flake size. Therefore, bearing this general occurrence in mind, even a qualitative estimate of the modal size of dispersion of GO flakes can give a clear idea of the size distribution. In this work, GO from the same solution used for the kinetic essays illustrated in this manuscript has been spin-coated onto 100 nm SiO2/Si(100) substrates and then investigated with scanning electron microscopy (SEM). Results are reported in Fig. 1 in panels (a) and (b) at two different magnifications. The images show very large individual GO flakes with a typical lateral size exceeding few microns, while the smallest flakes (see Fig. 1(b)) have a typical lateral size of few hundreds nanometers. With a conservative estimate of the GO lateral flake size of 2 µm, as estimated by SEM analysis (see size distribution in Fig. S1a†). We can assess, using the same approach proposed in ref. 3, that only a mass fraction less than 10−5 of GO flakes in our solution corresponds to flakes having less than 100 nm lateral size. This is important as the occurrence of edge effect might come into play only for GO flakes sizes of the order of 40 nm, as evidenced in ref. 27 and in consistency with our quantitative estimate of edge-to-surface effects as reported in the ESI (Fig. S1).† Therefore, we can ensure with perfect confidence that the results reported hereafter are inherently related to the interaction of enzymes (having a typical size of 2–3 nm) with the basal plane of comparatively extremely larger GO flakes, where no edge effects come into play. | ||
| Fig. 1 SEM images of GO flakes from a spin-coated dispersion from the same GO solution used in the kinetic assays. Panels (a) and (b) refer to different area samples and different magnifications. | ||
Substrate–GO interaction: DFT investigation, XPS analysis, and kinetic assays
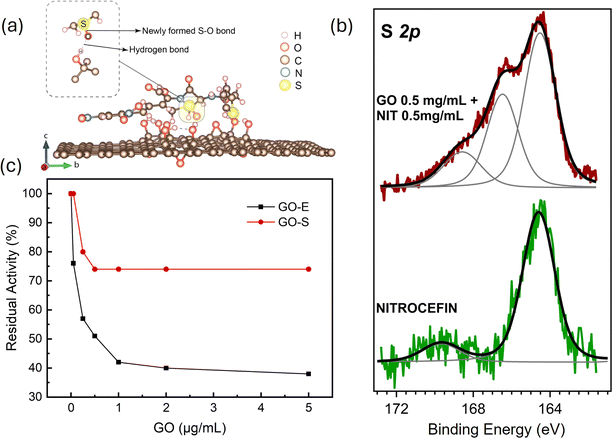
To set up a right protocol in the kinetic assays, considering that the GO flake size is orders of magnitude larger than the linear dimensions of the substrate, we have to verify the occurrence of substrate–GO interaction, and therefore appropriately disentangle such effect in the analysis of kinetic assays of the GO–enzyme interaction. To this purpose, we have addressed this problem with a combined theoretical and experimental approach. The GO interaction with the substrate has been modeled with DFT simulations, and compared with XPS experiments and kinetic assays as illustrated in Fig. 2. For DFT, we explored fourteen different initial “landing” configurations of the nitrocefin molecule near the GO basal plane, in order to mimic the most likely mutual spatial arrangements of the two systems, with the long axis of the nitrocefin molecule either perpendicular or parallel to the GO surface. Thirteen (out of fourteen) optimized GO–nitrocefin configurations are reported in Fig. S2–S14 of the ESI.† Out of the fourteen optimized structures, the one reported in Fig. 2, panel (a) is the one with the lowest energy. The unique feature of this structure is that one oxygen atom of the GO has created a bond to one sulfur atom of nitrocefin and a hydrogen bond on the GO side. Most importantly, this chemisorbed configuration is, on average, 2.7 eV (3.3 eV) more energetically favorable with respect to all the other horizontal (vertical) landing modes (see ESI†). We have thus strong DFT evidences of S oxidation of one of the two S atoms in nitrocefin as a result of the interaction with GO. An accurate XPS characterization of the substances in the pure phase and in the mixed one supports, on the experimental side, the simulated outcome. The observation of the S 2p core level signal, which is only specific to nitrocefin, brings a great deal of information. Fig. 2(b) reports such spectra for the pure nitrocefin and a one-to-one mass mixture of GO and nitrocefin (for the analysis of the C 1s core level, see Fig. S15 in ESI†). The spectral S 2p line shape of the pure phase is dramatically changed once the nitrocefin is mixed with GO. In particular, in the S 2p core level related to pure nitrocefin, the spectrum is characterized by a single component centered at 164.0 eV binding energy. In the mixed phase with GO, two other broad peaks appear with an almost 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 spectral weight ratio with the main nitrocefin component at low binding energy. In this case, the low binding energy component indicates the presence of S atoms unperturbed by the adsorption to GO, but almost half of the whole spectral weight of the core level spectrum has to be assigned to S atoms forming sulfite (S
1 spectral weight ratio with the main nitrocefin component at low binding energy. In this case, the low binding energy component indicates the presence of S atoms unperturbed by the adsorption to GO, but almost half of the whole spectral weight of the core level spectrum has to be assigned to S atoms forming sulfite (S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds) and sulfone (SO2), detected at 166.2 eV and 168.2 eV.28 The same spectral weight of such components indicates that, if nitrocefin alone is let to interact with GO then a chemisorption with its basal plane is very likely, confirming the oxidization of the sulfur-active spot of the nitrocefin as predicted by DFT simulations. These evidences provide a rationale for understanding the results of the kinetic assay reported in Fig. 2(c). Data refer to GO tested as an inhibitor of the enzymatic activity of one BL (NDM-1). In the figure, the data labeled as GO–E refer to tests where GO at various concentrations is submitted to a pre-incubation (2 min) with 15 nM of NDM-1, whereas GO–S are data taken after GO was instead 5 min pre-incubated with the substrate (100 µM nitrocefin). When GO is pre-incubated with NDM-1, the residual activity (at 1.0 µg ml−1) is about 42%, but when GO is pre-incubated with nitrocefin then the residual activity stays above 73%. Evidently, the pre-incubation of GO with nitrocefin favors the formation of nitrocefin–grafted GO and hampers the inhibition activity of GO itself.
O bonds) and sulfone (SO2), detected at 166.2 eV and 168.2 eV.28 The same spectral weight of such components indicates that, if nitrocefin alone is let to interact with GO then a chemisorption with its basal plane is very likely, confirming the oxidization of the sulfur-active spot of the nitrocefin as predicted by DFT simulations. These evidences provide a rationale for understanding the results of the kinetic assay reported in Fig. 2(c). Data refer to GO tested as an inhibitor of the enzymatic activity of one BL (NDM-1). In the figure, the data labeled as GO–E refer to tests where GO at various concentrations is submitted to a pre-incubation (2 min) with 15 nM of NDM-1, whereas GO–S are data taken after GO was instead 5 min pre-incubated with the substrate (100 µM nitrocefin). When GO is pre-incubated with NDM-1, the residual activity (at 1.0 µg ml−1) is about 42%, but when GO is pre-incubated with nitrocefin then the residual activity stays above 73%. Evidently, the pre-incubation of GO with nitrocefin favors the formation of nitrocefin–grafted GO and hampers the inhibition activity of GO itself.
Determination of β-lactamase activity: GO concentration and time dependent experiments
To study the interaction of GO with BLs, in order to further refine the experimental protocol, attention has first been focused on SBLs. Summarizing results are reported in Fig. 3. To determine the response to GO at varying concentrations, inhibition assays were performed by incubating 0.05, 0.25, 0.5, 1, 2, and 5 µg ml−1 of GO for 1 min, for each BL. The optimal concentration for each enzyme is selected based on the kinetic parameters (Km and Vmax) obtained with nitrocefin as substrate. The inset in Fig. 3(a) shows the result for KPC-3 as a representative example. The residual activity (%) of the enzymes was then calculated as a reduction of nitrocefin hydrolysis. Our data analysis showed that, from GO concentration values of 0.5 µg ml−1 and above, the residual activity values remain constant. Fig. 3(a) reports then, the time-dependent kinetic response of all the SBLs considered in our evaluation class A (KPC-3, CTX-M-15, GES-1, and SHV-1) and class D (OXA-23). In this protocol, the time-dependent inhibition assays between SBLs and GO were performed by pre-incubating the enzymes with 0.5 µg per ml GO, with a pre-incubation time varying from zero to 60 min. Each reaction was then monitored by spectrophotometric assay for 5 min. Both the concentration and time-dependent experiments bring very relevant information. As far as the GO concentration-dependent experiments are concerned, we observe a marked inhibition activity of GO already at concentration values, which are one order of magnitude lower than the values reported for the inhibition survey of GO onto VIM-1 (see Fig. 3b in ref. 14). We will discuss this occurrence later on in the manuscript, but the result is itself notable because it quantitatively lowers by one order of magnitude the minimum GO concentration value necessary to observe inhibition effectiveness. The second observation is that, in any case, for all the SBLs considered, although the time response variation curves are quite enzyme specific, there is a clear general trend to a substantial inhibition of the residual activity below 10% after 60 min of pre-incubation. This is a remarkable result as it clearly and convincingly demonstrates the inhibition effectiveness of GO (even at a relatively low concentration value) on all the SBLs (to this aim, the robustness of our statement is given by the specific evaluation of the response to the OXA-23 enzyme, which is representing a very large class of more than four hundred of similar enzymes29). To grasp a general trend, the time-dependent graphs of each specific enzyme have been averaged and plotted in panel (b) of Fig. 3. In this case, the data more nicely accommodates onto a smooth experimental trend that we have fitted with a phenomenological logarithmic curve (solid line in Fig. 3(b)). Since the logarithmic function is undefined at x = 0, we numerically replaced the value with a small positive 0+, corresponding to 10−6. This is a commonly used technique in numerical fitting to avoid singularities while preserving the functional form of the logarithmic function. The model curve interpolates very well the experimental data, and it allows for quantitatively extrapolating to the value of 90 ± 5 min the pre-incubation time required for complete enzymatic inhibition by GO of SBLs. Back to a more detailed analysis of Fig. 3(a), one can also observe that each specific enzyme has its own time-dependent variation of the residual activity. In particular, if the pre-incubation time is set to 5 min, the response spectrum in terms of residual activity is clearly and nicely opened. This suggests the potential use of such parameters in enzyme-selective sensors of SBLs.The summary of the kinetic assay investigations on MBLs is reported in Fig. 4. Once considering MBLs, due to their methodological relevance in this specific case, a preliminary kinetic assay was performed to evaluate the effect of ZnCl2 on NDM-1, VIM-1, CphA, and L1 enzymes activity. We observed that the four MBLs all showed similar residual activity values in the presence and absence of 50 µM of ZnCl2, therefore, only data with the averaged response are reported in panel (a) of Fig. 4. All kinetic experiments (summarized in Fig. 4(b) and (c)) were then performed using Hepes buffer without the addition of ZnCl2 to limit the possible interferences with kinetic assays. In panel (b) of Fig. 4 the NDM-1, VIM-1, CphA, and L1 time-dependent residual activity is reported. Also in this case, as for SBLs, for NDM-1, CphA, and VIM-1, we observed a very significant GO inhibition already within the first 10 min with a loss of about 70–80% of the enzymatic activity. After 60 min of pre-incubation, the residual activity drops to 16–18% for NDM and CphA and to 4% in the case of VIM-1. It is interesting to note that L1 was only partially inhibited by GO, with a significant residual activity of 52% after 60 min of pre-incubation. We stress that, in the case of L1 enzyme, the incubation was prolonged until 120 min, but its residual activity remained constant at 50%. Consistent with the analysis approach of Fig. 3(b), in panel (c) of Fig. 4, we plotted (black squares) the averaged time response of NDM-1, CphA, and VIM-1 and fitted the data with the same model phenomenological curve adopted for SBLs. The MBL-related black curve in Fig. 4(c) is reported in direct comparison (red curve) with the phenomenological best-fitting curve of SBLs of Fig. 3(b). We observe a very similar time-dependent behavior. This is strong evidence that GO offers a universally effective inhibiting potential toward all BLs, either serine or metallo ones, and, once the anomalous behavior of the L1 response is considered, is a strong indication that their occurrence in a monomeric phase is the only relevant factor that comes into play.
 | ||
| Fig. 4 Pre-incubation time-dependent inhibition of 0.5 µg per ml GO towards metallo-β-lactamases (MBLs). Panel (a) averaged response with or without 50 µM ZnCl2 in the buffer solution. Panel (b) individual time response of NDM-1, VIM-1, CphA, and L1 in the presence of 0.5 µg per ml GO and no ZnCl2. Panel (c) (black circles) averaged the response of NDM-1, VIM-1, and CphA (from panel (b)). The black line is a fitted phenomenological logarithmic curve. The red line is a similarly derived fitting curve for SBL from panel (b) of Fig. 3. Panel (d) BL model structures of NDM-1, VIM-1, CphA, and L1. The former three BLs are colored consistently with the corresponding graphs of panel (b). | ||
Core-level XPS analysis of the enzyme–GO interaction
To elucidate the GO–BL interaction mechanism better, direct information can be accessed with XPS. To this aim, given the observed universality of the GO inhibition activity, we decided to focus the attention on one specific GO–BL interaction, namely the NDM–GO one, as we believe it can be reasonably representative of the general trends. The rationale for this choice is twofold: (i) the specific relevance of this MBL due to its presence in super-resistant bacteria,30 and, (ii) the presence of a metal, namely zinc, in the enzyme active site whose electronic environment (and its perturbations) can be directly probed with XPS via the analysis of the metal core level spectra.The results of the XPS analysis of the interaction of GO with NDM-1 are reported in Fig. 5–7. In Fig. 5 we focus on the Zn 2p3/2 core level. The upper curve (violet) reports the Zn signal relative to pure NDM-1, while the lower curve (red) is related to the Zn signal of NDM-1 in the mixed phase with GO. Data are vertically displaced for clarity and are reported on the same intensity scale. We note that, apart from an obvious intensity decrease in the mixed phase, the Zn signal does not change, within the limits of the experimental accuracy, neither in the energy position nor line shape. This points to no alterations of the outer electronic charge distribution of the Zn metal atom of the enzyme, either in the pure phase or while the enzyme is exposed to GO. In other words, there is no direct evidence of preferential interaction (either oxidation or reduction) of the Zn of the MBL with the GO basal plane.
An enlightening detailed analysis of the C 1s and N 1s spectra is reported in Fig. 6 and 7. In Fig. 6 (left panel), we report the XPS C 1s spectra for pure NDM 0.5 mg ml−1 (violet), pure GO 0.5 mg ml−1 (green), and GO 0.5 mg ml−1 mixed with NDM-1 0.5 mg ml−1 (red). Grey curves are the fitting and background lines. Close to the red curve, the sum of the GO + NDM-1 isolated spectra is reported in dotted grey. From bottom to top: the C 1s of the NDM-1 is nicely fitted with three main contributions, corresponding to the graphitic one due to C–C and C–H bonds (centered at 284.7 eV), an intermediate energy component (at 286.2 eV) that we assign to C–O and C–N bonds, and a third significant contribution at 288.2 eV assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds. This spectral analysis is a very plausible result for the XPS C 1s spectrum of proteins. The GO spectrum, green curve in Fig. 6 on the left, is, instead, very much typical of pure non-reduced GO. Its spectral line shape and its corresponding detailed analysis are consistent with our previous reports.31 Once GO is mixed with NDM-1, the XPS spectrum reported in red in Fig. 6 is obtained. This spectrum, at a very first visual inspection, shows a close resemblance to the one that can be obtained by just summing the spectra (green and violet curves) from the non-interacting compounds. Indeed, once the agreement, between the XPS C 1s signal of the mixed phase (red in both panels in Fig. 6) and one of the isolated phases, is let maximize by taking the best-fitting linear combination of the two pure phases (blue dotted curve in Fig. 6) there is, in any case, a residual mismatch between the two curves. This indicates that a non-interacting model between GO and NDM in the mixed phase does not explain the experimental evidence. There is a residual curve, well above the noise level and peaked at 284.6 eV, 286.1 eV, and 288.0 eV. Complementary information is obtained from the XPS spectra of N 1s. In Fig. 7 are reported the N 1s spectra for pure NDM-1 (violet) and the GO–NDM-1 mixed phase (red). In this case, the analysis is more straightforward, as the N 1s signal arises solely from N atoms belonging to the structure of the NDM-1 enzyme, as nitrogen is not present in GO. For pure NDM-1 the signal is centered at 400.8 eV, while for the mixed phase, the signal is reduced to the lower binding energy side (reduction) and centered at 400.3 eV. We elaborate on the interpretation of such residual spectrum in the following section.
O bonds. This spectral analysis is a very plausible result for the XPS C 1s spectrum of proteins. The GO spectrum, green curve in Fig. 6 on the left, is, instead, very much typical of pure non-reduced GO. Its spectral line shape and its corresponding detailed analysis are consistent with our previous reports.31 Once GO is mixed with NDM-1, the XPS spectrum reported in red in Fig. 6 is obtained. This spectrum, at a very first visual inspection, shows a close resemblance to the one that can be obtained by just summing the spectra (green and violet curves) from the non-interacting compounds. Indeed, once the agreement, between the XPS C 1s signal of the mixed phase (red in both panels in Fig. 6) and one of the isolated phases, is let maximize by taking the best-fitting linear combination of the two pure phases (blue dotted curve in Fig. 6) there is, in any case, a residual mismatch between the two curves. This indicates that a non-interacting model between GO and NDM in the mixed phase does not explain the experimental evidence. There is a residual curve, well above the noise level and peaked at 284.6 eV, 286.1 eV, and 288.0 eV. Complementary information is obtained from the XPS spectra of N 1s. In Fig. 7 are reported the N 1s spectra for pure NDM-1 (violet) and the GO–NDM-1 mixed phase (red). In this case, the analysis is more straightforward, as the N 1s signal arises solely from N atoms belonging to the structure of the NDM-1 enzyme, as nitrogen is not present in GO. For pure NDM-1 the signal is centered at 400.8 eV, while for the mixed phase, the signal is reduced to the lower binding energy side (reduction) and centered at 400.3 eV. We elaborate on the interpretation of such residual spectrum in the following section.
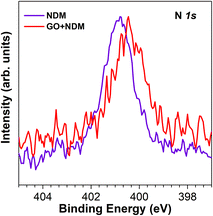 | ||
| Fig. 7 Normalized N 1s XPS core-level spectra of pure NDM-1 0.5 mg ml−1 (violet) and GO 0.5 mg ml−1 mixed with NDM-1 0.5 mg ml−1 (red). | ||
Discussion
The experimental evidences from the kinetic essays demonstrate that there is a strong inhibition of the catalytic activity of all (but the L1 one) of the BLs considered in our study. Although indirect, inhibition is an unquestionable evidence of adsorption/interaction. With this in mind, from the results of the kinetic assays reported in Fig. 3, one can determine that there is an increasing interaction strength for the various SBLs with the following order: SHV-1, OXA-23, CTX-M-15, KPC-3, GES-1. For MBLs (see Fig. 4), the inhibition response is comparable for NDM-1 and CphA, while the interaction strength is higher in the case of VIM-1. A rationale proposed in the past to explain the GO enzyme interaction is that it is driven by electrostatic interaction. At the zeroth order of such interaction is the attraction (repulsion) between molecules bearing opposite (equal) signs of net electric charge. This model to explain interaction and loss of enzymatic activity by controlling the charge disproportionation between GO and the enzymes has been proposed in ref. 32, where α-chymotrypsin, with an isoelectric point (pI = 8.75), has been incubated with positive GO (GO grafted with ethylenediamine, EDA, GO–EDA) observing no loss of enzymatic activity, while the strongest inhibition of the enzyme has been observed for pristine (“negative”) GO by De et al.4 The same rationale was proposed to explain the interaction of GO8 and reduced GO33 with human ubiquitin via titration experiments. To this aim, in order to predict an overall attraction/repulsion, one has to consider the pH-dependent surface charge of GO and the one of the enzymes considered. GO carries a negative net charge in a wide pH range (from 4.0 to 10.0),34 while the net charge per protein depends on the isoelectric point (pH value corresponding to charge neutrality of the enzyme).In Table 1, together with other relevant information, the measured isoelectric point (pI) is reported for each enzyme studied in this work. At solution, pH that is above (below) the pI, the surface of a protein bears a negative (positive) charge. According to this simple rationale, considering that all the kinetic essays have been performed and physiological conditions (pH = 7). KPC-3 and OXA-23 are expected to be slightly negative. GES-1, NDM-1, VIM-1, and L1 are even more negatively charged, while CTX-M-15, SHV-1, and CphA bear a positive charge at pH = 7. Under this crude hypothesis, these latter positive enzymes should manifest a stronger interaction with GO (stronger inhibition), but this is not consistent with what was observed. For example, SHV-1 (positively charged) is the least interacting enzyme among SBLs, and GES-1 (negatively charged) shows the strongest interaction. Moreover, VIM-1 (pI 5.1, negative) and CphA (pI 8.0, positive) have extreme values for the pIs under consideration, but exhibit an almost identical response in the kinetic assays. Evidently, this very simple model interpretation for the enzyme GO interaction is contradicted by experimental evidences.
| BL | Protein chains | PDB ID | Weight (kDa) | Modelled residue count | Isoelectric point | Arg | Lys |
|---|---|---|---|---|---|---|---|
| KPC-3 | 1 | 6QWD | 31.5 | 268 | 6.7 (ref. 35) | 15 | 10 |
| CTX-M-15 | 1 | 4HBT | 29.15 | 263 | 8.7 (ref. 36) | 15 | 14 |
| GES-1 | 1 | 2QPN | 31.0 | 270 | 6.0 (ref. 37) | 18 | 13 |
| SHV-1 | 1 | 1SHV | 29.42 | 265 | 7.7 (ref. 38) | 23 | 7 |
| OXA-23 | 1 | 4K0X | 27.81 | 243 | 6.7 (ref. 39) | 7 | 20 |
| NDM-1 | 1 | 3ZR9 | 24.81 | 231 | 5.2 (ref. 40) | 7 | 8 |
| VIM-1 | 1 | 5N5G | 27.26 | 233 | 5.1 (ref. 41) | 9 | 5 |
| CphA | 1 | 1X8G | 25.44 | 224 | 8.0 (ref. 42) | 9 | 15 |
| L1 | 4 | 2HB9 | — | — | 5.9 (ref. 43) | 60 | 32 |
Indeed, the same assumption that GO bears a net negative charge has to be more clearly discussed. In ref. 44 Li et al. have demonstrated that the net negative charge of GO flakes, under the pH values where it has been examined in ref. 34, is dependent almost exclusively on the ionization of the carboxylic groups COOH that are found at the edges of the GO flakes. Therefore, once large (larger than 100 nm) GO flakes are considered, the net surface charge per unit area of a GO flake is negligible, due to negligible edge effects (see Fig. S1†). However, GO with its surface hydroxyl and epoxy groups has a polar nature, and it can therefore interact with the electric dipoles that emerge at the surface of a protein. In other words, the surface of GO bears almost charge neutrality, but it is polar in nature. Bearing that in mind, one can examine in order of relevance the potential interaction mechanism of GO with those amino acids that, in one way or another, can interact with GO. Glutamic acid (Glu) and aspartic acid (Asp) are acidic amino-acids (negatively charged) although GO is negatively charged, negative–negative interactions can still occur due to local variations in charge distribution, and although these interactions are usually weaker than the positive–negative interactions, nonetheless, the carboxyl groups in these amino acids can still form hydrogen bonds with the surface of GO. Phenylalanine (Phe), tyrosine (Tyr), and tryptophan (Trp) are, instead, aromatic amino acids that can interact with graphene oxide through π–π hydrophobic interactions with the pure graphene patches that are always present on the GO surface.45 Glutamine (Gln) and asparagine (Asn) have polar amide groups in their side chains that can form hydrogen bonds with the hydroxyl groups on the GO surface. In this respect, for example, a detailed molecular dynamics (MD) study has demonstrated the important role of asparagine in the immobilization of α-chymotrypsin5 on GO via the formation of hydrogen bonding. However, all of the above potential interactions of residues with GO are relatively weak, in comparison with the interaction with inherently positively charged residues like lysine and arginine. These interactions are typically quite strong and stable at physiological pH. In particular, at physiological pH (around 7.0), the side chain of lysine, which contains an amino group (–NH3+), remains protonated and positively charged. The pKa of the amino group on lysine's side chain is around 10.5, which means it remains protonated (and thus positively charged) at pH 7.0 (at pH 7.0, the lysine side chain will be in its +1 charged state). Arginine has a side chain with a guanidino group that has a pKa around 12.5, meaning it remains protonated and positively charged at physiological pH (pH 7.0). At pH 7.0, the guanidino group will retain its proton, giving arginine a +1 charge. In summary, we are guided in our model interpretation of the experimental results by the presence (and position) of lysine and arginine on the enzyme surface. To support this work hypothesis, we observe that, in another MD work where the interaction of cytochrome c (Cyt c) with GO was simulated, it has emerged that Cyt c is adsorbed onto the GO residues mainly through lysine residues.46 Indeed, once individual interaction of amino-acids is considered with GO, recent works have demonstrated that GO can be easily drafted with those two amino-acids47–52 and in particular via hydrogen bonding of the ε-aminic group of lysine with the oxidized part of GO.50
This interaction model is consistent with the outcomes of the XPS analysis on NDM-1. The Gaussian surface of this enzyme is reported in Fig. 8. In the figure the enzyme is represented by six different images obtained after a left-handed rotation of 72° around the vertical axis (in the figure). The top left image shows the enzyme surface exposing the active catalytic site (in white), while the surface lysine (arginine) is represented in magenta (mustard). There, the lysine residues (magenta) protrude significantly with their amine group ε at the end of the lateral aliphatic chain. These protrusions are very likely anchoring points with the GO surface. NDM-1 has an almost equivalent number of arginine (seven) and lysine (eight) residues (see Table 1). Such residues, especially the lysine ones, are close to the active catalytic site of the enzyme (see Fig. 8 top left). The XPS residual signal in Fig. 6 (green curve right panel), as already stressed, shows clear components peaked at 284.6 eV, 286.1 eV, and 288.0 eV. The most intense one, at 284.6 eV, is related to aromatic carbon. This clearly indicates a reduction of the GO, which can occur with the opening of epoxide rings. Indeed, in ref. 50 the epoxide ring opening reaction with the concomitant partial reduction of GO is the chemical route for grafting lysine or arginine onto GO. The residual signal at 286.1 eV is the characteristic binding energy of C–N bonds. In ref. 47 the XPS analysis of Arg/GO clearly shows, in agreement with our results, the increase of a C–N component resulting from the nucleophilic interaction of NH2 groups of Arg and GO epoxy groups. The residual signal at 288.0 eV is indicative of the formation of amidic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O again, in agreement with the XPS results in ref. 47. As far as the N 1s signal (see Fig. 7), our data are in perfect agreement, in terms of energy position and line shapes, with the XPS study of lysine functionalized GO recently reported in ref. 51, while this spectroscopic evidence is not in agreement with arginine modified GO.50 Thus, we have significant XPS evidences that the NDM-1 GO interaction is mediated preferentially through lysine linking with GO, while also arginine interaction cannot be excluded. We propose this model interaction based on the adsorption interaction of all the other BLs enzymes examined in the work. According to this model, interaction strength has to be mediated by the number of arginine and lysine residues in the enzyme. Indeed, from Table 1, one can extract the average total number of Arg and Lys residues per unit mass (in kDa) for SLBs and MBLs (with the reasonable exclusion of L1), which is 142/149 ≈ 0.95 kDa−1 and 53/77.5 ≈ 0.68 kDa−1, respectively. The MBL examined in this study possesses, on average, only 70% of the positive residues that are present in the SBLs. Interestingly, this is in line with the observation in Fig. 4 of a more efficient inhibition activity of SBLs with respect to MBLs. Some further discussion, based on the specific arrangements of the positive residues in the enzymes, can be reasonably proposed to further explain other details in the kinetic assays. Let us consider the following: although all the SBL are substantially immobilized after 60 min of pre-incubation, with a comparable residual activity, nevertheless there are remarkable differences between the response of SHV-1 (the least interacting) with the one of GES-1 at intermediate pre-incubation times (the residual activity of SHV-1 after 5 min pre-incubation is almost one order of magnitude higher than the one of GES-1).
O again, in agreement with the XPS results in ref. 47. As far as the N 1s signal (see Fig. 7), our data are in perfect agreement, in terms of energy position and line shapes, with the XPS study of lysine functionalized GO recently reported in ref. 51, while this spectroscopic evidence is not in agreement with arginine modified GO.50 Thus, we have significant XPS evidences that the NDM-1 GO interaction is mediated preferentially through lysine linking with GO, while also arginine interaction cannot be excluded. We propose this model interaction based on the adsorption interaction of all the other BLs enzymes examined in the work. According to this model, interaction strength has to be mediated by the number of arginine and lysine residues in the enzyme. Indeed, from Table 1, one can extract the average total number of Arg and Lys residues per unit mass (in kDa) for SLBs and MBLs (with the reasonable exclusion of L1), which is 142/149 ≈ 0.95 kDa−1 and 53/77.5 ≈ 0.68 kDa−1, respectively. The MBL examined in this study possesses, on average, only 70% of the positive residues that are present in the SBLs. Interestingly, this is in line with the observation in Fig. 4 of a more efficient inhibition activity of SBLs with respect to MBLs. Some further discussion, based on the specific arrangements of the positive residues in the enzymes, can be reasonably proposed to further explain other details in the kinetic assays. Let us consider the following: although all the SBL are substantially immobilized after 60 min of pre-incubation, with a comparable residual activity, nevertheless there are remarkable differences between the response of SHV-1 (the least interacting) with the one of GES-1 at intermediate pre-incubation times (the residual activity of SHV-1 after 5 min pre-incubation is almost one order of magnitude higher than the one of GES-1).
 | ||
| Fig. 8 NDM-1 enzyme structure (Gaussian surface) as generated from https://www.rcsb.org/, (ID: 3ZR9) of the NDM-1 metallo BL. Lysine (arginine) residues are colored in purple (mustard). The active catalytic pocket is indicated in white. To have a complete view of the enzyme surface, from top left to bottom right, the 3D generated model is left-handed, turned 72° around the vertical axis in the figure. Similarly generated images for all the other enzymes studied in this work are reported in the ESI.† | ||
If the total number of lysine and arginine residues is considered, then (see Table 1), there seems to be no reason to justify the evidently strongest interaction of GES-1 with GO (the two enzymes have respectively 31 and 30 Lys + Arg residues for GES-1 and SHV-1 respectively). However, if we focus on their distribution with respect to the active site, as visible in Fig. 9, we observe that for SHV-1 the surface of the molecule exposing the active site has two arginine and one lysine, while the GES-1 has four lysine and four arginine residues, making the anchoring more plausible in one case rather than the other. Moreover, in consistency with the XPS results on NDM-1, which point to a more likely interaction with lysine, we observe that GES-1 has six more Lys residues than SHV-1. These are, of course, non-conclusive arguments, and only a microscopic molecular dynamics simulation can substantiate the strength of such statements.
As far as the enzyme inhibition mechanism of GO, which we have directly demonstrated with kinetic assays, all the arguments and evidences illustrated above point to a non-competitive inhibition. On the one hand, the active catalytic site is not directly interested in interaction with GO (we verified no chemical shifts in the Zn core level spectrum of NDM-1), but on the other hand there is a significant adhesion of the enzymes on the GO basal plane via positive residues with a consequent more than a plausible distortion of the enzyme's ternary structure and of the active site that forces the inhibition of the catalytic activity of the enzymes, in analogy with what directly proved with MD for the immobilization/inhibition of α-chymotrypsin/GO.5
Conclusions
In summary, we have studied the interaction/inhibition of five SBLs (KPC-3, CTX-M-15, GES-1, SHV1, and OXA-23) and four MBLs (NDM-1, VIM-1, CphA, and L1) with graphene oxide. The study adds significantly to the previous literature of studies of the interaction of proteins with GO that have been considered so far, to the best of our literature review, fourteen other proteins. We demonstrated directly with kinetic assays a universal inhibition, with the notable exception of the L1 enzyme, which is significantly more resilient to the inhibition itself. The average inhibition is more effective for SBLs than for MBLs. With the support of XPS evidences (carried out for the NDM-1/GO system) which is fully consistent with a model interpretation where the enzymes are anchored to the oxidized patches of the GO basal plane via grafting of positive residues (lysine, and arginine), we propose a non-competitive inhibition mechanism where the ternary structure of the enzyme protein is altered upon adsorption of GO with consequent deformation of the catalytic site. A complementary study has been performed on the interaction of the nitrocefin molecule (used as a substrate in the kinetic assays) with GO, via DFT calculations and XPS. Both simulations and experiments directly indicate the chemisorption of nitrocefin on the oxidized part of GO, with the oxidation of one of the two sulfur atoms of the molecule. This explains the marked reduction of the inhibition of GO once its surface is previously grafted with nitrocefin, as directly demonstrated with kinetic assays.Experimental and DFT simulations
Chemicals and vectors
Graphene oxide (4 mg ml−1 dispersion in water) was purchased from Ossila and washed in deionized water following the procedure of ref. 53. Sodium phosphate buffer, and Hepes-sodium salt were purchased from Sigma-Aldrich. The isopropyl-β-thiogalactoside (IPTG) was from Euroclone (Milan, Italy). Nitrocefin (Sigma-Aldrich, Milan, Italy) is a chromogenic cephalosporin used as a substrate in the kinetic assay involving BLs. The enzymes used in the present study were SBLs MBLs that are able to hydrolyze β-lactams with two different catalytic mechanisms. Among MBLs, the metalloenzymes of subclasses B1 (i.e. NDM-1 and VIM-1), B2 (CphA), and B3 (L1) were used for kinetic assays. Concerning SBLs, the study was focused on molecular Class A (SHV-1, KPC-3, CTX-M-15, GES-1) and Class D enzymes (i.e. OXA-23). Except for OXA-23, which was purified in this study, all enzymes were from our laboratory collection.54–58Cloning, expression, and purification of OXA-23 serine-β-lactamase
The blaOXA-23 gene was isolated from A. baumannii clinical isolates59 and cloned into pET-24a (+) vector using NdeI and BamHI restriction enzymes. The authenticity of OXA-23 was confirmed by sequencing the recombinant plasmid using an automated sequencer (ABI PRISM3500, Life Technologies, Monza, Italy). E. coli NovaBlue strain was used as non-expression host for the initial cloning, and E. coli BL21(DE3) cells were used for enzyme expression.OXA-23 was extracted from 1 l of an overnight culture of E. coli BL21(DE3)/pET24-OXA-23. The growth curve was performed to overproduce the OXA-23 enzyme, which is the only tested enzyme that we purified ourselves. The other enzymes were already purified and were from our laboratory collection. The growth curve for the recombinant strain expressing OXA-23 was generated using the pET system protocol. The IPTG, 0.4 mM, was added when the culture reached an OD of 0.7, measured at a wavelength of 600 nm. After the addition of IPTG, the culture was incubated at 22 °C for 16 h. Cells were harvested by centrifugation at 8000 rpm for 10 min at 4 °C, washed twice with 25 mM sodium phosphate buffer (pH 7.0). The cells were then suspended in 100 ml of 30 mM Tris–HCl buffer (pH 8.0) containing 27% sucrose. The crude enzyme was obtained by sonication in ice (5 cycles at 60 W for 1 min and 2 min of break). The lysate was centrifuged at 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 60 min, dialyzed overnight at 4 °C in 25 mM sodium acetate buffer (pH 5.2), and loaded onto an SP Sepharose FF (GE Healthcare, Life Sciences, Milan, Italy) equilibrated with the same buffer. The column was extensively washed to remove unbound proteins, and BLs were eluted with a linear gradient of NaCl (0 to 1 M) in the same buffer. Active fractions were pooled and dialyzed in 25 mM sodium phosphate buffer, pH 7.0, and loaded onto a Sephacril-100 column (GE Healthcare, Life Sciences, Milan, Italy) equilibrated with 25 mM sodium phosphate buffer pH 7.0 + 0.15 M NaCl.
000 rpm for 60 min, dialyzed overnight at 4 °C in 25 mM sodium acetate buffer (pH 5.2), and loaded onto an SP Sepharose FF (GE Healthcare, Life Sciences, Milan, Italy) equilibrated with the same buffer. The column was extensively washed to remove unbound proteins, and BLs were eluted with a linear gradient of NaCl (0 to 1 M) in the same buffer. Active fractions were pooled and dialyzed in 25 mM sodium phosphate buffer, pH 7.0, and loaded onto a Sephacril-100 column (GE Healthcare, Life Sciences, Milan, Italy) equilibrated with 25 mM sodium phosphate buffer pH 7.0 + 0.15 M NaCl.
GO and β-lactamases interaction
The effect of GO on BLs activity was evaluated at different concentrations of GO (0.05, 0.25, 0.5, 1, 2, and 5 µg ml−1) using pure enzymes at concentrations ranging from 15 nM to 30 nM. This experiment was performed by (a) incubating for 2 min each concentration of GO with enzymes, and (b) incubating for 2 min each concentration of GO with 100 µM of substrate (nitrocefin). In the type (a) assay, the reaction starts with the addition of substrate, whereas in the type (b) assay, the reaction starts with enzyme addition. Time-dependent interaction of GO towards BLs was determined by measuring nitrocefin residual activity after incubation of GO and enzymes, at 25 °C, for 0, 1, 2, 5, 10, 15, 20, 30, 60 min. Kinetic experiments involving SBLs were carried out in sodium phosphate buffer (25 mM, pH 7.0), whereas for MBLs Hepes 20 mM, pH 7.0 with and without 50 µM ZnCl2 was used. For all experiments, the concentration of GO was 0.5 µg mL−1. Residual activity was calculated relative to the activity of the corresponding enzyme towards nitrocefin, which was set at 100%.The data were collected with a Lambda 25 spectrophotometer (PerkinElmer) using 100 µM nitrocefin as reporter substrate (λ = 482 nm; ΔεM482 = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1·cm−1).
000 M−1·cm−1).
SEM experiments
SEM was performed using a Gemini SEM 500 instrument. The sample for SEM analysis was prepared by depositing 30 µl of a GO solution (0.5 mg ml−1) by spin-coating at 2200 rpm on a 300 nm SiO2/Si(100) substrate. SEM images were analyzed to determine the lateral dimension of the flakes.XPS experiments
A detailed chemical investigation of the systems under consideration was carried out via XPS. Samples were prepared in ambient atmosphere conditions via spin coating onto 100 nm Au/Si(100) substrates: (a) GO 0.5 mg ml−1, (b) NMD 0.5 mg ml−1, (c) nitrocefin 0.5 mg ml−1, and mixtures of equal weights of (a) + (b), or (a) + (c). Samples were then readily inserted into ultra-high vacuum (UHV) chamber for the XPS investigation. XPS was performed using a PHI 1257 spectrometer equipped with an Al Kα source (hν = 1486.6 eV) operated at a spectral resolution of 100 meV, using a pass energy of 9.16 eV. Spectra were acquired at room temperature and a base pressure of 2.0 × 10−9 mbar. All XPS data were calibrated to the Au 4f peak of a clean gold reference, set to 84.0 eV binding energy. The analysis of the S 2p and C 1s core-level spectra was carried out after Shirley background subtraction. In the fitting procedure, where shown, spectra were decomposed using the product of Gaussian and Lorentzian line shapes.DFT simulations
Density functional theory simulations of the interaction of GO with nitrocefin have been performed using the Vienna Ab Initio Simulation Package (VASP). Nitrocefin, C21H16N4O8S2, is a chromogenic cephalosporin substrate typically used to detect the presence of β-lactamase enzymes via kinetic assays. Its molecular structure has been downloaded from the PubChem database and then optimized separately in VASP. GO was modeled with a 9 × 9 graphene platelet, functionalized with epoxy oxygen atoms (on both sides of the graphene plane), one carbonyl oxygen, and six hydroxyl (–OH) groups. Such structure was optimized separately in VASP. The optimized structure of nitrocefin was then put in proximity to the graphene oxide monolayer, assuming fourteen different initial “landing” configurations, with the long axis of the molecule either perpendicular or parallel to the graphene oxide plane. All fourteen structures were optimized using the vdW correction (Grimme method).60 The structures presented (thirteen out of fourteen in the ESI†) are the final (optimized) configurations of the GO–nitrocefin system. The “normalized” energy values for each optimized configuration are given in terms of the energy offset with respect to the “ground state” adsorption configuration (reported in the main manuscript).Data availability
The data that support the findings of this study are available on request from the corresponding author.Author contributions
AP, MP, and LO designed the project and methodology. MP and LO supervised the project. SE and BS conducted the DFT analysis. GD acquired and analyzed, together with DM, the XPS and SEM data. AP, MP, and FB acquired and analyzed, together with DM, the kinetic assay data. DM and LO wrote the manuscript, which has been proofread by all the authors.Conflicts of interest
The authors declare no competing interests.References
- R. M. Hazen and D. A. Sverjensky, Mineral surfaces, geochemical complexities, and the origins of life, Cold Spring Harbor Perspect. Biol., 2010, 2, a002162 CrossRef PubMed.
- S. Priyadarsini, S. Mohanty, S. Mukherjee, S. Basu and M. Mishra, Graphene and graphene oxide as nanomaterials for medicine and biology application, J. Nanostruct. Chem., 2018, 8, 123–137 CrossRef CAS.
- F. Perrozzi, S. Prezioso and L. Ottaviano, Graphene oxide: from fundamentals to applications, J. Phys.: Condens. Matter, 2014, 27, 013002 CrossRef PubMed.
- M. De, S. S. Chou and V. P. Dravid, Graphene oxide as an enzyme inhibitor: modulation of activity of α-chymotrypsin, J. Am. Chem. Soc., 2011, 133, 17524–17527 CrossRef CAS PubMed.
- X. Sun, Z. Feng, T. Hou and Y. Li, Mechanism of graphene oxide as an enzyme inhibitor from molecular dynamics simulations, ACS Appl. Mater. Interfaces, 2014, 6, 7153–7163 CrossRef CAS PubMed.
- Z. Li, H. Wang, S. Chen, Y. Liu and C. Hao, Molecular insights into the interaction between graphene oxide and β-galactosidase: From multispectral experiment, enzyme kinetics experiment to computational simulations, J. Mol. Liq., 2024, 414, 126116 CrossRef CAS.
- J. Zhang, F. Zhang, H. Yang, X. Huang, H. Liu, J. Zhang and S. Guo, Graphene oxide as a matrix for enzyme immobilization, Langmuir, 2010, 26, 6083–6085 CrossRef CAS PubMed.
- S. Mondal, R. Thirupathi, L. P. Rao and H. S. Atreya, Unraveling the dynamic nature of protein–graphene oxide interactions, RSC Adv., 2016, 6, 52539–52548 RSC.
- Y. Wang, Z. Zhu, H. Zhang, J. Chen, B. Tang and J. Cao, Investigation on the conformational structure of hemoglobin on graphene oxide, Mater. Chem. Phys., 2016, 182, 272–279 CrossRef CAS.
- Z. Zhu, Y. Wang, Y. Kang, H. Zhang, Z. Zhang, Z. Fei and J. Cao, Graphene oxide destabilizes myoglobin and alters its conformation, Carbon, 2017, 114, 449–456 CrossRef CAS.
- Z. Ding, H. Ma and Y. Chen, Interaction of graphene oxide with human serum albumin and its mechanism, RSC Adv., 2014, 4, 55290–55295 RSC.
- P. Bolibok, M. Wiśniewski, K. Roszek and A. P. Terzyk, Controlling enzymatic activity by immobilization on graphene oxide, Sci. Nat., 2017, 104, 1–10 CrossRef CAS PubMed.
- K. Chaudhary, K. Kumar, P. Venkatesu and D. T. Masram, Protein immobilization on graphene oxide or reduced graphene oxide surface and their applications: Influence over activity, structural and thermal stability of protein, Adv. Colloid Interface Sci., 2021, 289, 102367 CrossRef CAS PubMed.
- P.-J. J. Huang, R. Pautler, J. Shanmugaraj, G. Labbé and J. Liu, Inhibiting the VIM-2 Metallo-β-Lactamase by Graphene Oxide and Carbon Nanotubes, ACS Appl. Mater. Interfaces, 2015, 7, 9898–9903 CrossRef CAS PubMed.
- S. R. Ali, S. Pandit and M. De, 2D-MoS2-based β-lactamase inhibitor for combination therapy against drug-resistant bacteria, ACS Appl. Bio Mater., 2018, 1, 967–974 CrossRef CAS PubMed.
- K. Bush and P. A. Bradford, β-Lactams and β-lactamase inhibitors: an overview, Cold Spring Harbor Perspect. Med., 2016, 6, a025247 CrossRef PubMed.
- R. A. Bonomo, β-Lactamases: a focus on current challenges, Cold Spring Harbor Perspect. Med., 2017, 7, a025239 CrossRef PubMed.
- K. Bush, β-Lactam antibiotics: penicillins, Antibiotic and Chemotherapy: Expert Consult, 2010, pp. 200–225 Search PubMed.
- R. P. Ambler, The structure of β-lactamases, Philos. Trans. R. Soc., B, 1980, 289, 321–331 CAS.
- K. Bush and G. A. Jacoby, Updated functional classification of β-lactamases, Antimicrob. Agents Chemother., 2010, 54, 969–976 CrossRef CAS PubMed.
- G. Garau, I. García-Sáez, C. Bebrone, C. Anne, P. Mercuri, M. Galleni, J.-M. Frère and O. Dideberg, Update of the standard numbering scheme for class B β-lactamases, Antimicrob. Agents Chemother., 2004, 48, 2347–2349 CrossRef CAS PubMed.
- L. Chen, B. Mathema, K. D. Chavda, F. R. DeLeo, R. A. Bonomo and B. N. Kreiswirth, Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding, Trends Microbiol., 2014, 22, 686–696 CrossRef CAS PubMed.
- K. Bush, Past and present perspectives on β-lactamases, Antimicrob. Agents Chemother., 2018, 62, e01076–18 CrossRef CAS PubMed.
- J. Alba, Y. Ishii, K. Thomson, E. S. Moland and K. Yamaguchi, Kinetics Study of KPC-3, a Plasmid-Encoded Class A Carbapenem-Hydrolyzing β-Lactamase, Antimicrob. Agents Chemother., 2005, 49, 4760–4762 CrossRef CAS PubMed.
- B. A. Evans and S. G. Amyes, OXA β-lactamases, Antimicrob. Agents Chemother., 2014, 27, 241–263 Search PubMed.
- L. De Marzi, L. Ottaviano, F. Perrozzi, M. Nardone, S. Santucci, J. De Lapuente, M. Borras, E. Treossi, V. Palermo and A. Poma, Flake size-dependent cyto and genotoxic evaluation of graphene oxide on in vitro A549, CaCo2 and vero cell lines, J. Biol. Regul. Homeostatic Agents, 2014, 28, 281–289 CAS.
- A. Kobayashi, Y. Yamaguchi, S. Maruki, S. Kim, Y. Arakawa and S. Kurihara, Size, functional group effects and inhibition mode of graphene oxide for IMP-1 metallo-β-lactamase, Mater. Lett., 2022, 315, 131901 CrossRef CAS.
- K. S. Siow, L. Britcher, S. Kumar and H. J. Griesser, XPS study of sulfur and phosphorus compounds with different oxidation states, Sains Malays., 2018, 47, 1913–1922 CrossRef CAS.
- E.-J. Yoon and S. H. Jeong, Class D β-lactamases, J. Antimicrob. Chemother., 2021, 76, 836–864 CrossRef CAS PubMed.
- M. T. Zmarlicka, M. D. Nailor and D. P. Nicolau, Impact of the New Delhi metallo-beta-lactamase on beta-lactam antibiotics, Infect. Drug Resist., 2015, 8, 297 CAS.
- F. Perrozzi, S. Prezioso, M. Donarelli, F. Bisti, P. De Marco, S. Santucci, M. Nardone, E. Treossi, V. Palermo and L. Ottaviano, Use of optical contrast to estimate the degree of reduction of graphene oxide, J. Phys. Chem. C, 2013, 117, 620–625 CrossRef CAS.
- J. Li, L.-J. Wu, S.-S. Guo, H.-E. Fu, G.-N. Chen and H.-H. Yang, Simple colorimetric bacterial detection and high-throughput drug screening based on a graphene-enzyme complex, Nanoscale, 2013, 5, 619–623 RSC.
- S. A. Malik, Z. Mohanta, C. Srivastava and H. S. Atreya, Modulation of protein–graphene oxide interactions with varying degrees of oxidation, Nanoscale Adv., 2020, 2, 1904–1912 RSC.
- T. Szabó, E. Tombácz, E. Illés and I. Dékány, Enhanced acidity and pH-dependent surface charge characterization of successively oxidized graphite oxides, Carbon, 2006, 44, 537–545 CrossRef.
- G. Li, Q. Wei, Y. Wang, X. Du, Y. Zhao and X. Jiang, Novel genetic environment of the plasmid-mediated KPC-3 gene detected in Escherichia coli and Citrobacter freundii isolates from China, Eur. J. Clin. Microbiol. Infect. Dis., 2011, 30, 575–580 CrossRef CAS PubMed.
- E. M. Valenzuela de Silva, J. R. Mantilla Anaya, M. T. Reguero Reza, E. B. Gonzalez Mejia, I. Y. Pulido Manrique, I. Dario Llerena and D. Velandia, Detection of CTX-M-1, CTX-M-15, and CTX-M-2 in clinical isolates of Enterobacteriaceae in Bogota, Colombia, J. Clin. Microbiol., 2006, 44, 1919–1920 CrossRef CAS PubMed.
- L. Poirel, I. Le Thomas, T. Naas, A. Karim and P. Nordmann, Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae, Antimicrob. Agents Chemother., 2000, 44, 622–632 CrossRef CAS PubMed.
- G. Vedel, B. Picard, G. Paul, L. Gilly, P. Goullet and P. Nevot, Titration curve (pH gradient electrophoresis) of SHV-1, SHV-2 and of a new SHV-2 like betalactamase, Pathol. Biol., 1988, 36, 366–369 CAS.
- J. Walther-Rasmussen and N. Høiby, OXA-type carbapenemases, J. Antimicrob. Chemother., 2006, 57, 373–383 CrossRef CAS PubMed.
- G. Peirano, J. Ahmed-Bentley, N. Woodford and J. D. Pitout, New Delhi metallo-β-lactamase from traveler returning to Canada, Emerging Infect. Dis., 2011, 17, 242 CrossRef CAS PubMed.
- N. Franceschini, B. Caravelli, J.-D. Docquier, M. Galleni, J.-M. Frere, G. Amicosante and G. M. Rossolini, Purification and biochemical characterization of the VIM-1 metallo-β-lactamase, Antimicrob. Agents Chemother., 2000, 44, 3003–3007 CrossRef CAS PubMed.
- B. Segatore, O. Massidda, G. Satta, D. Setacci and G. Amicosante, High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance, Antimicrob. Agents Chemother., 1993, 37, 1324–1328 CrossRef CAS PubMed.
- A. Felici, G. Amicosante, A. Oratore, R. Strom, P. Ledent, B. Joris, L. Fanuel and J.-M. Frère, An overview of the kinetic parameters of class B β-lactamases, Biochem. J., 1993, 291, 151–155 CrossRef CAS PubMed.
- M.-J. Li, C.-M. Liu, Y.-B. Xie, H.-B. Cao, H. Zhao and Y. Zhang, The evolution of surface charge on graphene oxide during the reduction and its application in electroanalysis, Carbon, 2014, 66, 302–311 CrossRef CAS.
- C. Gómez-Navarro, J. C. Meyer, R. S. Sundaram, A. Chuvilin, S. Kurasch, M. Burghard, K. Kern and U. Kaiser, Atomic structure of reduced graphene oxide, Nano Lett., 2010, 10, 1144–1148 CrossRef PubMed.
- D. Zhao, L. Li and J. Zhou, Simulation insight into the cytochrome c adsorption on graphene and graphene oxide surfaces, Appl. Surf. Sci., 2018, 428, 825–834 CrossRef CAS.
- M. Haghshenas, M. Mazloum-Ardakani, L. Amiri-Zirtol and F. Sabaghian, Arginine-functionalized graphene oxide for green and high-performance symmetric supercapacitors, Int. J. Hydrogen Energy, 2021, 46, 30219–30229 CrossRef CAS.
- S. Khabnadideh, E. Mirzaei and L. Amiri-Zirtol, L-arginine modified graphene oxide: A novel heterogeneous catalyst for synthesis of benzo[b]pyrans and pyrano[3, 2-c]chromenes, J. Mol. Struct., 2022, 1261, 132934 CrossRef CAS.
- S. Mantovani, S. Khaliha, T. D. Marforio, A. Kovtun, L. Favaretto, F. Tunioli, A. Bianchi, G. Petrone, A. Liscio, V. Palermo and others, Facile high-yield synthesis and purification of lysine-modified graphene oxide for enhanced drinking water purification, Chem. Commun., 2022, 58, 9766–9769 RSC.
- G. Moro, S. Khaliha, A. Pintus, S. Mantovani, M. Feltracco, A. Gambaro, T. D. Marforio, M. Calvaresi, V. Palermo and M. Melucci, others Amino acid modified graphene oxide for the simultaneous capture and electrochemical detection of glyphosate, Mater. Today Chem., 2024, 36, 101936 CrossRef CAS.
- S. Sibt-e Hassan, K. A. Hasan, N. Adil, S. N. Khan, M. Usman, D. Hussain and S. G. Musharraf, Lysine-functionalized graphene oxide on cost-effective microporous support: A hydrophilic and fouling resistant membrane for safe lead filtration, Carbon, 2024, 223, 118974 CrossRef CAS.
- S. Mantovani, A. Pintus, A. Kovtun, A. Gondolini, S. Casadio, A. Sanson, T. D. Marforio, M. Calvaresi, M. Rancan and L. Armelao, others Graphene Oxide-Arginine Composites: Efficient Dual Function Materials for Integrated CO2 Capture and Conversion, ChemSusChem, 2024, 17, e202301673 CrossRef CAS PubMed.
- I. Barbolina, C. Woods, N. Lozano, K. Kostarelos, K. Novoselov and I. Roberts, Purity of graphene oxide determines its antibacterial activity, 2D Mater., 2016, 3, 025025 CrossRef.
- C. Bottoni, M. Perilli, F. Marcoccia, A. Piccirilli, C. Pellegrini, M. Colapietro, A. Sabatini, G. Celenza, F. Kerff, G. Amicosante, M. Galleni and P. S. Mercuri, Kinetic studies on CphA mutants reveal the role of the P158-P172 loop in activity versus carbapenems, Antimicrob. Agents Chemother., 2016, 60, 3123–3126 CrossRef CAS PubMed.
- A. Sabatini, F. Brisdelli, G. Celenza, F. Marcoccia, M. Colapietro, M. M. Tavío, A. Piccirilli, G. Amicosante and M. Perilli, Interaction of carbapenems and β-lactamase inhibitors towards CTX-M-15 and CTX-M-15G238C mutant, J. Global Antimicrob. Resist., 2017, 10, 95–100 CrossRef PubMed.
- A. Piccirilli, P. S. Mercuri, M. Galleni, M. Aschi, A. Matagne, G. Amicosante and M. Perilli, P174E Substitution in GES-1 and GES-5 β-Lactamases Improves Catalytic Efficiency toward Carbapenems, Antimicrob. Agents Chemother., 2018, 62, e01851–17 CAS.
- A. Piccirilli, F. Brisdelli, M. Aschi, G. Celenza, G. Amicosante and M. Perilli, Kinetic profile and molecular dynamic studies show that Y229W substitution in an NDM-1/L209F variant restores the hydrolytic activity of the enzyme toward penicillins, cephalosporins, and carbapenems, Antimicrob. Agents Chemother., 2019, 63, e02270–18 CrossRef CAS PubMed.
- A. Piccirilli, F. Brisdelli, J. D. Docquier, M. Aschi, S. Cherubini, F. De Luca, A. Matagne, G. Amicosante and M. Perilli, Amino Acid Replacement at Position 228 Induces Fluctuation in the Ω-Loop of KPC-3 and Reduces the Affinity against Oxyimino Cephalosporins: Kinetic and Molecular Dynamics Studies, Catalysts, 2020, 10, 1–14 Search PubMed.
- M. Perilli, A. Sabatini, E. Pontieri, G. Celenza, B. Segatore, C. Bottoni, P. Bellio, A. Mancini, F. Marcoccia, F. Brisdelli and G. Amicosante, OXA-23 Carbapenemase in multidrug-resistant Acinetobacter baumannii ST2 type: first identification in L'Aquila Hospital (Italy), Microb. Drug Resist., 2015, 21, 97–101 CrossRef CAS PubMed.
- S. Grimme, Semiempirical GGA-type density functional constructed with a long-range dispersion correction, J. Comput. Chem., 2006, 27, 1787–1799 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra01697e |
| This journal is © The Royal Society of Chemistry 2025 |