Open Access Article
Open Access ArticleDesign and computational screening of high-energy, low-sensitivity bistetrazole-based energetic molecules†
Peihao Cheng,
Yunhe Jin ,
Dongqi Wang and
Shengyang Tao
,
Dongqi Wang and
Shengyang Tao *
*
School of Chemistry, Dalian University of Technology, Dalian, 116024, Liaoning, China. E-mail: taosy@dlut.edu.cn
First published on 14th April 2025
Abstract
Bistetrazole-based compounds are novel high-nitrogen energetic molecules that have garnered attention in recent years. They possess a higher energy density and lower sensitivity, and are less challenging to synthesize than complex cage-like molecules. This study employed a molecular auto-generation mechanism to generate 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322 bistetrazole-based molecules with 20 bridgeheads and 29 side substituents. A combination of quantum chemical calculations and machine learning models was used to sequentially screen the molecules based on their oxygen balance index, synthesis difficulty, density, and detonation pressure, thus rapidly narrowing the search scope. Three bistetrazole-based energetic molecules with high potential were identified. The theoretical enthalpy of the formation of the designed molecules was as high as 854.76 kJ mol−1 and their detonation velocity reached 9.58 km s−1. Further calculations also demonstrated that these molecules have better macroscopic stability than trinitrotoluene, making them promising candidates for practical applications in developing energetic materials.
322 bistetrazole-based molecules with 20 bridgeheads and 29 side substituents. A combination of quantum chemical calculations and machine learning models was used to sequentially screen the molecules based on their oxygen balance index, synthesis difficulty, density, and detonation pressure, thus rapidly narrowing the search scope. Three bistetrazole-based energetic molecules with high potential were identified. The theoretical enthalpy of the formation of the designed molecules was as high as 854.76 kJ mol−1 and their detonation velocity reached 9.58 km s−1. Further calculations also demonstrated that these molecules have better macroscopic stability than trinitrotoluene, making them promising candidates for practical applications in developing energetic materials.
1 Introduction
Energetic materials are a special class of active substances that can release energy through intense redox reactions when stimulated externally. Such materials have significantly contributed to human progress and prosperity, and they are widely used in developing natural resources, mining, demolition, rocket launches, and other fields. Energy, sensitivity, and density are the three characteristics of energetic materials most focused on.1 However, there is always a contradictory and restrictive relationship between them. Generally, the high energy of energetic materials is always accompanied by increased sensitivity and density.2 Therefore, developing new energetic materials simultaneously possessing high energy, low sensitivity, and higher density remains a considerable challenge.Over the past two hundred years, chemists have developed and widely used only a few high-performance energetic molecules such as trinitrotoluene (TNT), nitroglycerin, cyclotetramethylene tetranitramine (HMX), cyclotrimethylenetrinitramine (RDX), and 2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane (CL-20).3–5 The development of energetic molecules has progressed from blunt-type to cage-type nitro compounds to high-energy nitrogen-rich molecules. The currently used highest-performance energetic molecule, CL-20, is a typical caged nitrogen-rich energetic compound. However, synthesizing cage molecules is relatively tricky, requiring precious metal catalysts like palladium,6–8 significantly increasing the synthesis cost. High-nitrogen compounds have attracted widespread attention from researchers in recent years due to their ease of synthesis and environmentally friendly explosive products.9 Modulated energetic ionic salts and high-nitrogen heterocyclic compounds have been developed. Bistetrazole-based energetic molecules have received attention due to their simple structure, high energy density, and low sensitivity. The bistetrazole-based skeleton has two tetrazole rings, providing good conjugation performance and being combined with good high-energy groups. Dihydroxylammonium 5,5′-bBistetrazole-1,1′-diolate (TKX-50)10–17 is one of the representative molecules. Therefore, constructing new energetic molecules based on the bistetrazole-based structure has become necessary in developing energetic materials.18,19
Traditional development of energetic molecules depends on experienced chemists designing molecular structures and conducting synthesis verification. This process is fraught with explosion riskshighly, and many of the reagents used are toxic, so chemists have always tried to find and predict high-performance energetic molecules from a theoretical design perspective to reduce the dangers of developing energetic molecules. Empirical models used to guide the design of energetic materials have been developed, such as the Kamlet–Jacobs20 equation for predicting detonation properties and the nitro group charge method (NGCM)21 for predicting mechanical sensitivity. With subsequent improvements, quantum chemical calculation methods have been able to predict some electronic properties and performance of energetic materials. However, these empirical models are rarely used for large-scale pre-screening of energetic materials before experimental synthesis due to the considerable time and money costs associated with quantum mechanical calculations. For a long time, the discovery of new energetic materials has heavily relied on scientific intuition through experimental and traditional trial-and-error processes, which are inefficient, have a small screening range, and are highly uncertain.
With the advent of the big data era, the research paradigm of energetic materials has undergone profound changes. Compared to empirical models, machine learning models usually have various advantages in accuracy, generalizability, and handling nonlinear problems and are, therefore, widely used in various fields of materials science. However, simultaneously, the accuracy of machine learning models highly depends on collecting a large amount of effective data. However, in the field of energetic materials, due to the complex synthesis process, high safety risks, and military secrecy involved, the publicly available data are limited and not highly reliable. This results in the trained machine-learning models being inaccurate or limited to molecules with specific features.
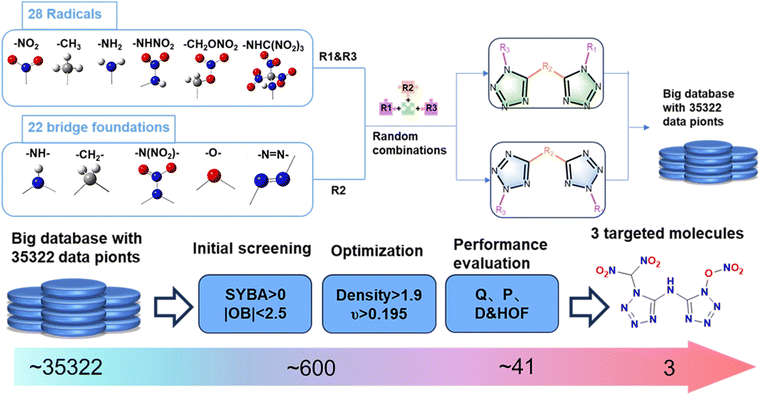
Here, we demonstrate a high-throughput virtual screening method that combines domain knowledge, existing machine learning models, and quantum chemical calculations to accelerate the discovery of new bistetrazole-based energetic materials with well-balanced energy safety properties. This method quickly screens promising target molecules from 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322 exhaustively generated molecular structures. The screened compounds also have higher energy density, lower sensitivity, and better detonation performance. After further evaluating the feasibility of the synthesis, we provide possible synthesis schemes for the relevant compounds. These findings demonstrate the effectiveness of the proposed method and the great potential of applying quantitative calculation methods in the pre-screening design of high-performance energetic materials.
322 exhaustively generated molecular structures. The screened compounds also have higher energy density, lower sensitivity, and better detonation performance. After further evaluating the feasibility of the synthesis, we provide possible synthesis schemes for the relevant compounds. These findings demonstrate the effectiveness of the proposed method and the great potential of applying quantitative calculation methods in the pre-screening design of high-performance energetic materials.
2 Experimental and computational section
2.1 Generation of molecules to be screened
In order to use a heuristic exhaustive method to expand the chemical space of molecules to be screened, we collected structures of energetic materials containing tetrazole structures from the literature over the past decade and extracted 29 common high-energy groups as side groups in the tetrazole structure. At the same time, we identified 20 common bridging groups from the reviewed energetic materials as linking groups and connected these groups with two types of tetrazole rings. Using a Python script based on the RDKit software package, the 29 high-energy groups, 20 bridging groups, and 2 types of bistetrazole rings were exhaustively combined into SMILES representations. The SMILES data cleaning process consists of two parts: one involves SMILES data that cannot be read by RDKit, which are considered to represent unreasonable molecular structures and are excluded; the other part involves removing duplicate SMILES strings.22,23 After cleaning, it was found that all 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322 generated SMILES were readable by RDKit with no completely identical duplicate SMILES. Therefore, we retained this dataset of molecules for subsequent screening.
322 generated SMILES were readable by RDKit with no completely identical duplicate SMILES. Therefore, we retained this dataset of molecules for subsequent screening.
2.2 Virtual screening process
The key to the entire high-throughput virtual screening method lies in the logical arrangement of the sequence in which molecular properties are predicted. By first calculating the properties that require less computational time, we can quickly reduce the number of molecules for subsequent screening. This reduction in number decreases the computational load for later, more precise, and time-consuming calculations, thereby ensuring computational accuracy while increasing speed.Utilizing previously cleaned molecular structure data, we first calculate each molecule's zero oxygen balance index (OB) using a Python program, selecting those with an OB between −0.25% and 0.25% for the next screening step. OB is a value that measures whether the oxygen content in an explosive is balanced with the oxygen required to completely oxidize the combustible elements. The data that meet the zero oxygen balance criteria are then evaluated for synthetic difficulty using the SYnthetic Bayesian Accessibility (SYBA)24 model based on functional groups and skeleton structures. In SYBA, a positive score indicates an easily synthesizable molecule, while a negative score indicates difficulty in synthesis, with larger numerical values representing lower synthetic difficulty. We select molecules with a SYBA score greater than 0 for further screening. After this initial screening, the number of molecules is reduced from 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322 to 600. The molecular data from the initial screening, represented in SMILES format, is converted into “.sdf” files through the RDkit platform. A Python program is then written to convert these “.sdf” files into submission files for Gaussian 16 software, where batch computations are submitted using DFT25,26 methods at the B3LYP/6-31G** level for molecular structure optimization and frequency calculation tasks. Using the optimized chemical structures, the Multiwfn27 program reads the computation results and automatically calculates the density values and electrostatic balance parameters ν, a measure of the balance between positive and negative regions. Those with a density value28–30 greater than 1.90 and an electrostatic balance parameter (ν) greater than 0.195 are selected for the next round of more accurate screening, reducing the data to 41 molecules.
322 to 600. The molecular data from the initial screening, represented in SMILES format, is converted into “.sdf” files through the RDkit platform. A Python program is then written to convert these “.sdf” files into submission files for Gaussian 16 software, where batch computations are submitted using DFT25,26 methods at the B3LYP/6-31G** level for molecular structure optimization and frequency calculation tasks. Using the optimized chemical structures, the Multiwfn27 program reads the computation results and automatically calculates the density values and electrostatic balance parameters ν, a measure of the balance between positive and negative regions. Those with a density value28–30 greater than 1.90 and an electrostatic balance parameter (ν) greater than 0.195 are selected for the next round of more accurate screening, reducing the data to 41 molecules.
Furthermore, we submit these 41 geometrically optimized structures for another round of geometric optimization and frequency calculation tasks in Gaussian 16 at a higher accuracy level using B3LYP/6-311G**.31,32 We also use the G4 (ref. 33) method combined with the B3LYP/Def2-TZVP basis set level to calculate energy, successively computing the enthalpy of formation (HOF), the heat of detonation (Q), detonation pressure (P), and detonation velocity (D) of these molecules.19,34–36 By analyzing the predicted performance data, we identify three bistetrazole-based nitrogen-rich energetic molecules promising for laboratory synthesis, with high density, high energy, and good stability. We also predict their synthetic routes and electronic structures.37 A Dell 7920T tower workstation equipped with two Gold 6238R CPUs and 192GB of RAM was used during the calculations. The generation and initial screening of the energetic molecules took about 30 minutes; the secondary screening involved DFT calculations at the B3LYP/6-31G** level for structural optimization and frequency analysis of 612 molecules, combined with density calculation formulas to compute molecular density and electrostatic balance parameter (ν), taking about two weeks for batch submission of computation tasks. Finally, the higher precision calculation and performance prediction for the selected 41 molecules took one week.
3 Results and discussion
3.1 High-throughput virtual screening method
The entire methodology flow is shown in Fig. 1.In this high-throughput virtual screening method, eight properties of candidate molecules were evaluated to screen for suitable energetic molecules. These properties include the oxygen balance index (OB), SYBA scores, density, electrostatic balance parameters (ν), the enthalpy of formation (HOF), heat of detonation Q, detonation pressure P, and detonation velocity D. The formulas or methods used to calculate the properties above are elaborated in detail in the ESI (p4–p7).†
In generating energetic molecules, 20 bridging groups, 29 high-energy groups, and two tetrazole rings were utilized. Through exhaustive group combination, 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322 molecule SMILES codes were generated in only 1 hour, which far exceeds the speed of manual molecular design (corresponding data are given in Table S1 in the ESI†).
322 molecule SMILES codes were generated in only 1 hour, which far exceeds the speed of manual molecular design (corresponding data are given in Table S1 in the ESI†).
The preliminary screening data following the OB is shown in Fig. 1. The screening process for OB took 120 seconds and resulted in 612 molecules that met the criteria, reducing the number of molecules to be screened by 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710. It significantly reduces the computational workload for subsequent steps. Although few molecules are near the OB = 0 position, selecting molecules with a high OB means that the designed energetic materials can oxidize more completely during detonation, potentially providing better performance.
710. It significantly reduces the computational workload for subsequent steps. Although few molecules are near the OB = 0 position, selecting molecules with a high OB means that the designed energetic materials can oxidize more completely during detonation, potentially providing better performance.
The machine learning model of SYBA, based on a Bernoulli Naïve Bayes classifier, can quickly rank large molecular data sets. Subsequently, the SYBA model was used to screen for the synthetic difficulty of the molecules, taking 1.5 hours. The data shows that most of the selected molecules are defined by the SYBA model as easy to synthesize. The distribution of synthetic difficulty between the two types of tetrazole rings is not completely consistent, with slightly more molecules defined as challenging to synthesize in the category with an ortho position nitrogen substitutable.
By eliminating molecules with a SYBA score below zero, 600 remaining molecular structures (corresponding data are given in Table S2 in the ESI†) were input in batches into Gaussian 16 software for optimization to obtain rational chemical structures and energy calculations, predicting density and electrostatic balance parameters (ν). At this stage, computational time increases noticeably. However, with 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 722 fewer molecules than the initial 35
722 fewer molecules than the initial 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322, the computation time is relatively short since these two properties do not require high precision basis sets. The results of the secondary screening are shown in Fig. 2.
322, the computation time is relatively short since these two properties do not require high precision basis sets. The results of the secondary screening are shown in Fig. 2.
In Fig. 2, the vertical axis represents molecular density, and the horizontal axis represents the sensitivity-related electrostatic balance parameters (ν). From the data distribution, most molecules have a predicted density greater than 1.9 g cm−3, proving that our previous strategy was effective. However, the prediction method for density is highly dependent on the structure. Subsequent use of high-precision calculation methods will make the structure more rational and the predicted density data more accurate. The secondary screening primarily uses the electrostatic balance parameters (ν) as a reference, selecting molecules with more uniform electron distribution and more stable structures.
After the secondary screening, the number of molecules was reduced from 600 to 41 (Fig. 3). These 41 molecules were input into Gaussian 16 software for higher precision calculations. The results, combined with empirical formulas for detonation performance and enthalpy of formation energy data, are used for deeper screening (Fig. 4 and 5). This step takes the longest time, averaging 3 hours per molecule. The optimized structures, molecular formulas, and corresponding data of the 41 molecules that entered the deep screening are shown in Table S4.†
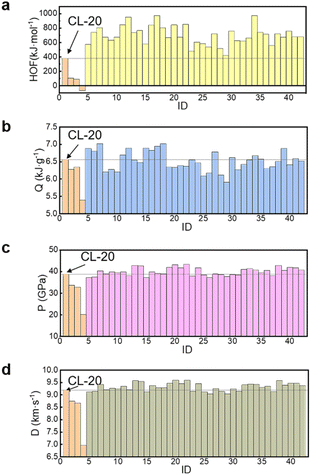 | ||
| Fig. 5 (a) Plot of energy data for candidate molecule enthalpy of formation (HOF); (b) heat of detonation (Q); (c) detonation pressure (P); (d) detonation velocity (D). | ||
3.2 Comparison between designed molecules and reference molecules
The performance of three designed molecules was compared against four well-known energetic molecules, TNT, RDX, HMX, and CL-20, using them as reference molecules.As demonstrated in Fig. S1 and Table S3,† the quantum chemical calculation methods employed for predicting the properties of the four classic energetic molecules resulted in outcomes that did not significantly deviate from the measured values. Furthermore, the overall trends observed for the four molecules were consistent. Thus, the predicted results are considered reasonable within an acceptable error range. The prediction closest to experimental values was the molecules' solid enthalpy of formation (HOF), with an average deviation of 1.51 kJ mol−1, consistent with experimental results and less than 6.87% off. The detonation velocity (D) also showed minimal deviation, with an average difference of 0.2 kJ mol−1. The calculations showed a relatively larger deviation for the CL-20 molecule. The comparison results for density and detonation pressure (P) showed the largest relative error, but the absolute deviation was not significantly different from other molecules. These results show that the chosen calculation methods and high-precision basis sets are reasonable and feasible for predicting molecules' density and detonation pressure. Therefore, the calculated values can screen out molecules with lower density and detonation pressure. The final three molecules screened were molecules #7, #22, and #33. The properties of the 4 reference and 38 design molecules are shown in Table S5.†
The weakest bond dissociation energy, the nitrocharge method, and the corresponding reaction kinetics simulation can analyze the thermal stability of the molecule and correspond one-to-one. In addition, the molecular macroscopic stability, or mechanical sensitivity, is related to the molecular surface charge distribution, the maximal ESP analysis, etc., and the HOMO–LUMO gap energy reflects the molecular reactivity and macroscopic stability.9,21,41,42 The discussion of energy-containing material stability is broad in scope, encompassing three aspects: thermal sensitivity, mechanical sensitivity, and electrostatic sensitivity. In the case of molecules, the general macroscopic stability is initiated by the overall electron distribution of individual molecules, such as ν and maximum ESP values, which reflect the mechanical sensitivity of the energy-containing molecule. Concurrently, the local nitro energies of bond-breaking energy and nitro charge reflect thermal stability related to the thermal decomposition temperature. The nitro charge method and the nitro connected to the atom on the nitro charge have a significant impact. The nitro charge is generally positive when connected to the oxygen atom. However, a different situation arises when connected to the C, N, and O atoms.43,44 For the N atom or C atom connected to the case, the overall display of the negative charge depends on the position of the C and N atoms relative to the benzene ring and the aliphatic ring. The connecting atoms determine the ability to donate electrons. However, when the O atom is connected to the nitro charge, the nitro energy is reflected in the thermal stability and thermal decomposition temperature. In this case, the charge of the nitro group connected to the O atom will be negative overall.
The thermal stability of a molecule can be predicted by calculating its bond dissociation energy (BDE). A lower BDE of the weakest bond indicates lower thermal stability and a lower thermal decomposition temperature.
Similarly, the mechanical stability of a molecule can be evaluated based on its electronic distribution, such as the electrostatic equilibrium parameter or the maximum electrostatic potential (ESP) value. These parameters serve as indicators of mechanical sensitivity, where a larger electrostatic equilibrium parameter and a lower maximum ESP value correspond to improved mechanical stability.
A detailed analysis of the electrostatic potential distribution of three molecules was conducted (ESI Section 9, Fig S2–4†). In terms of the distribution of positive and negative potential regions, the positive potential region of molecule #7 accounts for approximately 57% of the total area, while for molecules #22 and #33, the positive potential regions account for approximately 62% and 67%, respectively. Based on this data, molecule #7 has a smaller proportion of positive potential area and a larger ν value, indicating better overall mechanical sensitivity. As for the average values of positive and negative potentials, for molecules #7, #33, and #22, the average of the positive potential region gradually increases, while the ν value decreases correspondingly. Overall, in the electrostatic potential distribution of the molecules, the larger the average positive potential and the greater the proportion of the positive potential region, the worse the mechanical stability of the molecule.
3.3 Further analysis of candidate molecules' performance
| CL-20 | HMX | RDX | TNT | #7 | #22 | #33 | |
|---|---|---|---|---|---|---|---|
| Density (g cm−3) | 1.913 | 1.781 | 1.753 | 1.635 | 1.884 | 1.999 | 1.944 |
| ν | 0.059 | 0.136 | 0.172 | 0.202 | 0.241 | 0.213 | 0.221 |
| Q (kJ g−1) | 6.561 | 6.28 | 6.341 | 5.396 | 7.015 | 6.362 | 6.551 |
| D (km s−1) | 9.186 | 8.745 | 8.67 | 6.957 | 9.419 | 9.586 | 9.437 |
| P (GPa) | 38.78 | 33.7 | 32.81 | 20.23 | 40.43 | 43.3 | 41.32 |
| SYBA | 1.26 | 2.08 | 2.08 | 34.53 | 9.93 | 7.69 | 16.08 |
| ESP_Max (kJ mol−1 e−1) | 266.14 | 235.14 | 206.35 | 163.85 | 169.83 | 286.06 | 258.74 |
| BDE (kJ mol−1) | 165.79 | 190.59 | 171.25 | 259.96 | 55.74 | 53.03 | 122.81 |
The data clearly show that bond dissociation energies gradually increase from CL-20 to TNT, indicating enhanced structural stability. By comparing the bond dissociation energy, nitro charge, and thermal decomposition temperature of the reference molecule, along with the calculated BDE and nitro charge values for molecules #7, #22, and #33, the following conclusions can be drawn: molecules #7 and #33, which contain the O–NO2 bond, exhibit lower BDE values than the reference molecule, indicating reduced thermal stability. Consequently, their thermal decomposition temperatures are unlikely to exceed 200 °C, the decomposition temperature of the reference molecule. In contrast, molecule #33 has a BDE and nitro charge value similar to those of the reference molecule, suggesting comparable high thermal stability (the data are presented in Table S6†).
The prediction of thermal stability for energetic molecules can be approached from the bond dissociation energy (BDE) and predicted thermal decomposition temperature. The experimentally obtained thermal decomposition temperature can intuitively reflect the thermal stability of energetic molecules. The calculated BDE value is positively correlated with the molecular thermal stability. The larger the BDE value, the higher the thermal decomposition temperature and the better the molecular thermal stability. Wu47 et al. used machine learning (ML) methods to build multiple models for predicting thermal decomposition temperature based on a dataset composed of 1022 different compounds, achieving good results. The three candidate molecules in this paper were predicted for their thermal decomposition temperatures using the model, and the corresponding results are shown in Table S6.†
 | ||
| Fig. 6 (a) Predicted synthetic routes for the #7 molecule; (b) predicted synthetic routes for the #22 molecule; (c) predicted synthetic routes for the #33 molecule. | ||
The design of the synthetic routes was based on a linear synthetic strategy, in which the intermediates to be synthesised in the process were deduced backwards from the final molecular formula, and the reactions were deduced backwards from the molecular groups, basically according to the establishment of the bridging and bis-tetrazolium skeleton, followed by the substitution of high-energy groups and the associated nitration reactions.48–51
In several rounds of screening, each round eliminates certain molecules. The first round of screening primarily involves OB value selection and synthetic difficulty screening. These two molecular properties eliminate most molecules. The OB value eliminates molecules that do not contain oxygen (O) elements or those with excessive nitro and hydroxyl groups. The former lacks high-energy functional groups, while the latter has too many high-energy groups, making them sensitive and unstable. Only molecules with a certain proportion of –CHO groups are retained, as these molecules ensure more complete oxidation during explosions. The synthetic difficulty screening eliminates molecules with inappropriate or unreasonable functional group connections because such molecules are difficult to synthesize and are discarded. After the initial screening, the remaining molecules, aside from those with a bis-azole ring, generally consist of organic molecules containing two or more nitro groups, with relatively simple molecular structures. The second screening involves density and ν selection. Molecules with higher density values and better mechanical stability are typically those with simple bridged groups and side groups containing high-energy groups, such as nitro groups, with a total of 2–6 such groups in the molecule.
In summary, the three molecules #7, #22, and #33 selected fulfill the initial concept of high-energy, low-sensitivity bistetrazole-based energetic materials. Molecule #7 exhibits the best overall performance and, according to the retrosynthetic analysis, is promising for laboratory synthesis and has significant application potential.52
4 Conclusions
In this work, we developed and applied existing quantitative calculation methods combined with machine learning models to accelerate high-throughput virtual screening to guide the selection and discovery of bistetrazole-based energetic materials. This high-throughput virtual screening method accounts for the time differences in property screening processes, sensibly conducts molecular property selection, significantly reducing the number of molecules in time-consuming computations, thereby notably enhancing the screening speed and reducing computational costs. Subsequent high-precision quantum chemical calculations demonstrated that the computational tasks and basis sets used in this screening process were effective and reliable. Using this method, we were able to quickly select three high-energy, low-sensitivity molecules with good performance from a potential pool of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 322 bistetrazole molecular structures in just three weeks. Further electronic and experimental studies showed that these molecules exhibited the expected excellent performance and superior detonation characteristics. This work demonstrates the potential of our high-throughput virtual screening method in rapidly finding high-performance new energetic materials. Moreover, the proposed systematic approach can be extended to the discovery of other novel organic functional materials.
322 bistetrazole molecular structures in just three weeks. Further electronic and experimental studies showed that these molecules exhibited the expected excellent performance and superior detonation characteristics. This work demonstrates the potential of our high-throughput virtual screening method in rapidly finding high-performance new energetic materials. Moreover, the proposed systematic approach can be extended to the discovery of other novel organic functional materials.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge the support of this research by the National Natural Science Foundation of China (No. 22372025), the Fundamental Research Funds for the Central Universities (No. DUT22LAB607, DUT22QN226), and the Projects of International Cooperation and Exchanges NSFC-BRFFR (No. 22211530456).References
- K. Zhong and C. Zhang, Chem. Eng. J., 2024, 483, 149202 CrossRef CAS.
- Y. Wen, L. Wen, B. Tan, J. Dou, M. Xu, Y. Liu, B. Wang and N. Liu, J. Mater. Chem. A, 2024, 12, 9427–9437 RSC.
- R.-b. Lv, J.-y. Zhou, L. He, T.-w. Wang, H.-z. Li and Q. Zhang, Energ. Mater. Front., 2024, 5, 17–26 CrossRef CAS.
- V. D. Ghule, P. M. Jadhav, R. S. Patil, S. Radhakrishnan and T. Soman, J. Phys. Chem. A, 2010, 114, 498–503 CrossRef CAS PubMed.
- J.-y. Zhang, H.-c. Du, F. Wang, X.-d. Gong and Y.-s. Huang, J. Mol. Model., 2012, 18, 165–170 CrossRef CAS PubMed.
- Y. Wang, L. Hu, S. Pang and J. n. M. Shreeve, J. Mater. Chem. A, 2023, 11, 13876–13888 RSC.
- L. Zhu, Q. Zhou, W. Wang, H. Li, B. Li, Y. Zhang and J. Luo, Energ. Mater. Front., 2024 DOI:10.1016/j.enmf.2024.06.005.
- L. Wen, T. Yu, W. Lai, J. Shi, M. Liu, Y. Liu and B. Wang, J. Phys. Chem. Lett., 2021, 12, 11591–11597 CrossRef CAS PubMed.
- J. Liu, S. Zhao, B. Duan, X. He, C. Yang, X. Pu, X. Zhang, Y. Xiao, F. Nie, W. Qian, G. Li and C. Zhang, J. Mater. Chem. A, 2023, 11, 25031–25044 RSC.
- Q. Lai, L. Pei, T. Fei, P. Yin, S. Pang and J. n. M. Shreeve, Nat. Commun., 2022, 13, 6937 CrossRef CAS PubMed.
- H. Niu, S. Chen, Q. Shu, L. Li and S. Jin, J. Hazard. Mater., 2017, 338, 208–217 CrossRef CAS PubMed.
- W. Pang, T. M. Klapötke, L. T. DeLuca, D. OuYang, Z. Qin, Y. Hu, X. Fan and F. Zhao, in Nano and Micro-Scale Energetic Materials, 2023, pp. 149–190, DOI:10.1002/9783527835348.ch6.
- T. M. Klapötke, in Materials Research and Applications, 2021, ch. 1, pp. 1–91, DOI:10.1007/978-981-15-9223-2_1.
- T. M. Klapötke, S. Cudziło, W. A. Trzciński, J. Paszula, L. Bauer, C. Riedelsheimer and J. T. Lechner, Propellants, Explos., Pyrotech., 2023, 48, e202380611 CrossRef.
- T. M. Klapötke, S. Cudziło and W. A. Trzciński, Propellants, Explos., Pyrotech., 2022, 47, e202100358 CrossRef.
- V. K. Golubev and T. M. Klapötke, J. Phys.: Conf. Ser., 2022, 2154 Search PubMed.
- N. Fischer, D. Fischer, T. M. Klapötke, D. G. Piercey and J. Stierstorfer, J. Mater. Chem., 2012, 22, 20418 Search PubMed.
- N. V. Muravyev, D. R. Wozniak and D. G. Piercey, J. Mater. Chem. A, 2022, 10, 11054–11073 RSC.
- T. Wang, H. Gao and J. n. M. Shreeve, Z. Anorg. Allg. Chem., 2021, 647, 157–191 CrossRef CAS.
- M. J. Kamlet and S. J. Jacobs, J. Chem. Phys., 1968, 48, 23–35 CrossRef CAS.
- C. Zhang, Propellants, Explos., Pyrotech., 2008, 33, 139–145 Search PubMed.
- N. Zohari and F. Ghiasvand Mohammadkhani, Cent. Eur. J. Energ. Mater., 2020, 17, 31–48 Search PubMed.
- RDKit: Open-source cheminformatics, https://www.rdkit.org, DOI:10.5281/zenodo.591637.
- M. Vorsilak, M. Kolar, I. Cmelo and D. Svozil, J. Cheminf., 2020, 12, 35 Search PubMed.
- C. T. Lee, W. T. Yang and R. G. Parr, Phys. Rev. B:Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- A. D. Becke, Phys. Rev. A:At., Mol., Opt. Phys., 1988, 38, 3098–3100 CrossRef CAS.
- T. Lu and F. W. Chen, J. Comput. Chem., 2012, 33, 580–592 CrossRef CAS.
- L. Wilbraham, S. H. M. Mehr and L. Cronin, Acc. Chem. Res., 2021, 54, 253–262 CrossRef CAS PubMed.
- J. S. Murray, T. Brinck, P. Lane, K. Paulsen and P. Politzer, J. Mol. Struct.:THEOCHEM, 1994, 307, 55–64 CrossRef.
- C. Yang, J. Chen, R. Wang, M. Zhang, C. Zhang and J. Liu, J. Chem. Inf. Model., 2021, 61, 2582–2593 Search PubMed.
- R. Krishnan, J. S. Binkley, R. Seeger and J. A. Pople, J. Chem. Phys., 1980, 72, 650–654 CrossRef CAS.
- S. Grimme, J. Antony, S. Ehrlich and H. Krieg, J. Chem. Phys., 2010, 132, 19 CrossRef PubMed.
- B. Chan, A. Karton and K. Raghavachari, J. Chem. Theory Comput., 2019, 15, 4478–4484 CrossRef CAS PubMed.
- W. Pang, C. Deng, H. Li, L. T. DeLuca, D. Ouyang, H. Xu and X. Fan, Nanomaterials, 2021, 12(1), 133 CrossRef PubMed.
- H. Lin, S.-G. Zhu, L. Zhang, X.-H. Peng, P.-Y. Chen and H.-Z. Li, Struct. Chem., 2013, 24, 1655–1663 CrossRef CAS.
- F. Wang, H. Du, J. Zhang and X. Gong, J. Phys. Chem. A, 2011, 115, 11788–11795 CrossRef CAS.
- J. y. Zhang, F. Wang and X. d. Gong, Struct. Chem., 2013, 24, 1339–1346 CrossRef CAS.
- B. Chen, H. Lu, J. Chen, Z. Chen, S.-F. Yin, L. Peng and R. Qiu, Top. Curr. Chem., 2023, 381, 25 CrossRef CAS PubMed.
- T. Zhao, L. Li, Z. Dong, Y. Zhang, G. Zhang, M. Huang and H. Li, Chin. J. Org. Chem., 2014, 34, 304–315 Search PubMed.
- I. Chi-Duran, J. Enriquez, C. Manquian, R. A. Fritz, A. Vega, D. Serafini, F. Herrera and D. P. Singh, ACS Omega, 2019, 4, 14398–14403 Search PubMed.
- C. Zhang, J. Hazard. Mater., 2009, 161, 21–28 CrossRef CAS.
- G. Li and C. Zhang, J. Hazard. Mater., 2020, 398, 122910 CrossRef CAS.
- Q. Sun, N. Ding, C. Zhao, Q. Zhang, S. Zhang, S. Li and S. Pang, Sci. Adv., 2022, 8, eabn3176 CrossRef CAS.
- Q. Wu, L. Tan, Z. Hang, J. Wang, Z. Zhang and W. Zhu, RSC Adv., 2015, 5, 93607–93614 RSC.
- J.-y. Zhang, H.-c. Du, F. Wang, X.-d. Gong and Y.-s. Huang, J. Phys. Chem. A, 2011, 115, 6617–6621 CrossRef CAS.
- G.-z. Zhao and M. Lu, J. Mol. Model., 2013, 19, 57–64 CrossRef CAS PubMed.
- J.-n. Wu, S.-w. Song, X.-l. Tian, Y. Wang and X.-j. Qi, Energ. Mater. Front., 2023, 4, 254–261 CrossRef CAS.
- J.-y. Zhang, F. Wang and X.-d. Gong, Struct. Chem., 2013, 24, 2163–2172 CrossRef CAS.
- D. Xiang, H. Chen, W. Zhu and H. Xiao, Can. J. Chem., 2016, 94, 667–673 CrossRef CAS.
- Q. Wu, W. Zhu and H. Xiao, J. Mol. Model., 2014, 20, 2483 CrossRef PubMed.
- F. Yang, P. Lu, T. B. Ren, X. B. Zhang and L. Yuan, Smart Mol., 2023, 1, e20220002 CrossRef.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, and D. J. Fox, Gaussian 16, Revision C.02, Gaussian, Inc., Wallingford CT, 2019 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra01604e |
| This journal is © The Royal Society of Chemistry 2025 |