 Open Access Article
Open Access Article3D quantitative phase imaging via the transport of intensity equation: applications for studying red blood cells
Anusha Pillaia,
Saritha Kamath U.*b,
Sushma Belurkarc,
Anand K. Asundid and
Ajeetkumar Patil *a
*a
aManipal Institute of Applied Physics (MIAP), Manipal Academy of Higher Education, Manipal, 576104, India. E-mail: ajeetkumar.p@manipal.edu
bDepartment of Medical Laboratory Technology, Manipal College of Health Professionals, Manipal Academy of Higher Education, Manipal, 576104, India. E-mail: saritha.kamath@manipal.edu
cDepartment of Pathology, Kasturba Medical College, Manipal Academy of Higher Education, Manipal, 576104, India
dd'Optron Pte Ltd, 8 CleanTech Loop, 637145, Singapore
First published on 14th April 2025
Abstract
Quantitative phase imaging (QPI) is a versatile, label-free technique for investigating the morphological and biophysical properties of biological cells. Here, we used 3D QPI to study the behavior of human red blood cells (RBCs) across a saline gradient from 1% to 0%. RBCs are osmotic-sensitive, and their phase profiles provide important measures of membrane integrity, shape transitions, and lumen hemolysis. Deducing phase alterations across each concentration allowed us to quantify the osmotic impacts on RBC morphology and assess the corresponding biophysical changes. Other studies on osmotic shock contributed to understanding the behavior of RBCs in hypotonic environments and their applications in hematological and biomedical fields.
Introduction
Red blood cells, or erythrocytes, are essential for the transportation of oxygen from the lungs to tissues and the return of carbon dioxide to the lungs. This knowledge of RBCs is crucial for diagnosing and treating disorders, such as sickle cell disease and anaemia. In these diseases, RBC count, size, shape, and haemoglobin content are often abnormal.1 Moreover, there is also potential for exploring the biophysical properties of RBCs, such as membrane deformability, which is essential for examining their function in oxygen delivery and identifying disease markers. RBCs are analysed in clinical laboratories using automated haematology analysers for complete blood counts and a peripheral blood smear for visual observation of cell shape. However, such methods are unable to capture single-cell idiosyncrasies or structural and biophysical components, thus falling short of providing a thorough analysis of individual RBCs necessary for interpreting complex blood diseases.1 Given the complexity of these cells under different conditions, research into RBCs continues to be a key area of study. This will only be possible once we gain a deeper understanding of their behaviour and function. Imaging of RBCs, combined with the analysis of their cellular function, enables the acquisition of new insights into RBC characteristics with potential therapeutic benefits. The predominant research approaches today are based on light microscopy, including bright field,2 phase contrast,3 and differential interference contrast (DIC) microscopy,4 which are used to visualize RBC size, shape, and count.Fluorescence microscopy,5 such as standard fluorescence and confocal laser scanning microscopy,6 offers detailed views of cellular components and dynamics. It provides insights into specific cellular components through labelling and is one of the most widely used methods. By using various fluorescent agents, such as organic dyes (DAPI, FITC, rhodamine, etc.), it offers specific information on cellular components. Genetically encoded fluorescent proteins (GFP, mCherry, etc.)7 and nanoparticle-based probes, such as quantum dots,8 enable targeted imaging of specific cellular structures. However, these agents have major limitations. Organic dyes may photobleach over time, leading to a loss of signal intensity, while genetically encoded fluorescent proteins can disrupt normal cellular functions. The use of external labelling agents can also degrade cell viability and may not successfully label all types of cell compartments, rendering them unsuitable for live-cell imaging over extended periods.
Super-resolution microscopy techniques, such as STED,9,10 SIM, PALM, and STORM,11 provide molecular-level insights beyond the diffraction limit. Holographic microscopy, particularly digital holographic microscopy (DHM),12 offers non-invasive 3D imaging and provides label-free 3D imaging by capturing phase shifts of light. While the term “non-invasive” is often applied to DHM, it primarily emphasizes DHM's ability to obtain quantitative phase data without the need for dyes or physical contact, similar to the label-free nature of other optical techniques. Optical coherence tomography (OCT) provides high-resolution 3D images based on optical scattering,13 revealing structural details; however, its lateral resolution is lower than those of the super-resolution methods. Raman microscopy14,15 combined with optical imaging allows the chemical mapping of RBCs without labelling. While bright field microscopy is commonly used to assess RBC size and shape, its inability to provide contrast without staining limits its application for transparent samples like RBCs. Phase contrast microscopy improves the visualization of such samples but introduces halo artefacts that obscure fine details and does not yield absolute quantitative data. DIC microscopy, though effective for edge contrast, struggles with isotropic structures, offering a pseudo-3D view that may lead to the misinterpretation of cellular height.
To address these challenges, we present an imaging method grounded on the principles of the Transport of Intensity Equation (TIE). TIE is a high-resolution, label-free imaging technique to visualize nanoscale cellular structures while keeping the integrity of the cell. In contrast to fluorescence imaging, TIE-based techniques yield quantitative phase information, enabling the measurement of cellular morphological features, dry mass distribution and dynamic processes within living cells without an exogenous contrast agent. This allows for its wide application in non-invasive imaging, such as the characterization of cancer cells or the enhancement of image contrast using nanoparticles,16 as well as other applications, such as tissue engineering and regenerative medicine.17
In this study, we present 3D quantitative phase imaging (QPI) via the Transport of Intensity Equation (TIE), enabling label-free 3D characterization of single RBCs at high detail. TIE reconstructs phase maps from defocused images,18 which encode rich information about the morphology, membrane dynamics and internal constituents of the RBC without any staining or labelling.19 This method allows for in situ imaging of living cells at high resolution for a detailed understanding of the mechanism and structural properties of RBCs. A system used to study RBCs must be sensitive, maintain single-cell assessment capabilities and not alter morphology. This granular-level analysis of individual cells is critical to system efficacy for it to be considered a diagnostic and research tool, especially for blood disorders that might have vague symptoms but can be life-threatening to patients of any age. We demonstrate the use of our, 3D QPI system that employs the TIE to study red blood cells with high precision at various saline concentrations, allowing us to detect small, differential changes in RBC volume and morphology. This system fulfils the minimum requirements of sensitivity and specificity and enables the evaluation of single cells without impacting their physiological state. By detecting slight differences in the shape of cells, this method demonstrates the potential for broad biomedical applications. Overall, its capacity to provide accurate, label-free data about cell dynamics has potential value in clinical diagnostics and research, which can help explore, for instance, cell responses to changes in the environment, disease progression, or the effects of a drug on the morphology of cells.20
Materials and methods
In this study, we have employed 3D QPI using TIE to study red blood cells under various saline concentrations. To optimize the system, polystyrene-based microparticles of size 8 ± 0.2 μm from Sigma-Aldrich were purchased initially. For the RBC studies, 3 mL of blood was collected from healthy volunteers at Kasturba Medical College, Manipal, India, with informed consent (IEC No: IEC-651-2020). To avoid clotting of the blood, we used ETDA vacutainers. For the erythrocyte study, saline solutions of different concentrations ranging from 1% to 0% were used. About 3 μL of the blood was added to each solution and allowed for about 30 minutes before imaging. After 30 minutes, about 1 μL each of the solutions at different concentrations were suspended in glass slides and imaged. Using the same samples, the UV-visible spectroscopic study was performed.Instrumentation
Fig. 1a and b show the optical setup for phase imaging based on the Transport of Intensity Equation (TIE). We used an upright microscope (Nikon JAPAN model ci-L plus). The illumination source was a white LED, and the light then passed through a condenser lens, which collected and focused the light onto the sample. The slide was mounted on a stage for accurate placement and motion. After examining the sample and reading information, the light was next collected using a 60× objective lens, which is important to generate a high-magnification image. The light path could then be manipulated with a series of extra optical elements, even by passing through an additional tube lens in this whole setup. The lenses used here served to focus and direct the light to accurately pass through an optical system. A critical part of this system was a phase modulation element, which introduced controlled defocusing in the light path. The TIE algorithm requires this defocusing since it enables recording the intensity variations at different focal planes, which can then be employed in the phase retrieval of samples.21 The light finally reached a detector (d'Bioimager camera from d'Optron Pvt. Ltd), and the intensity images were captured. Finally, these phase images recorded at different defocused positions were then analysed through computational algorithms built on the TIE to recover the quantitative phase map of the sample.18,22 The resulting phase image was thereby decoded into a three-dimensional view of the slice. TIE offers the advantage of obtaining structural information and is non-invasive, which makes it a powerful technique for 3D imaging.23,24 | ||
| Fig. 1 (a) 3D quantitative phase imaging using TIE instrumentation; (b) schematic of the microscope and camera components. | ||
Fig. 2 shows a focused image of the polystyrene beads. Using TIE and the d'Optron Bioimager camera, we could image all the polystyrene beads in a single capture in real-time and extract information about all the beads, thus demonstrating a real-time, high-resolution, high-throughput and robust 3D imaging technique.
Results and discussion
The measurements of polystyrene beads using the optimized 3D Quantitative Phase Imaging (QPI) system revealed distinct quantitative parameters. 2D images of the polystyrene beads were captured using a Nikon DS-Fi3 camera. These images served as a valuable reference for our research. Following precise measurements of the diameters of 50 beads, the average diameter was determined to be approximately 7.653 μm, with a standard deviation of ±0.32 μm. Fig. 3a displays the image of a polystyrene bead captured with the Nikon camera, while Fig. 3b shows the focused image obtained using the d'Bioimager camera. Additionally, measurements using the d'Bioimager camera yielded an average diameter of 7.73 μm, with a standard deviation of ±0.25 μm. A key distinction between the two cameras is that the Nikon camera could provide only the diameter, whereas the d'Bioimager camera enabled the measurement of additional parameters, such as the thickness and volume of the beads, as discussed in subsequent sections. | ||
| Fig. 3 (a) Image of the polystyrene bead captured using a Nikon camera. (b) Image of the polystyrene bead captured using a d'Bioimager camera. | ||
This measurement, along with the specifications provided by Sigma-Aldrich, served as a baseline to validate our data and confirmed the accuracy of our quantitative phase imaging method. One focused image and two slightly defocused images of the polystyrene beads were captured with a defocus distance of ±3.5 μm, as shown in Fig. 4a–c.
These three images were combined using the Transport of Intensity Equation, followed by the reconstruction of the phase image. From the phase data, quantitative features, such as height, surface area, and volume, of the bead were extracted. Fig. 4d shows the intensity derivative image, Fig. 4e displays the phase image, Fig. 4f depicts the height profile, and Fig. 4g and h illustrate the 3D profile of a single polystyrene bead. The average height of the 100 measured polystyrene beads was determined to be 7.9 micrometres, with a standard deviation of about ±0.25 μm. The volume of the beads was calculated to be 138 cubic micrometres on average, showing minimal variability across the sample set, with a standard deviation of about ±5.85 μm. To calculate the volume, the region of interest (ROI) was first identified. The height of each pixel within the ROI was measured and multiplied by the area of the pixel to evaluate the volume contribution of each pixel. The total volume of the ROI was then determined by summing the volumes of all individual pixels within the selected region.
Quantitative analysis of erythrocyte morphology
We performed a quantitative morphological analysis of the 3D TIE QPM images of erythrocytes under different saline concentrations. Under hypertonic conditions, at an elevated concentration (1% NaCl), the erythrocytes had a shrivelled appearance, and a marked decrease in cell volume, indicative of cell dehydration. On the other hand, concentrations of isotonic saline between 0.9% and 0.65% showed erythrocyte mean cell volumes matching the values reported for isotonic saline in the literature,25 confirming the stable cellular morphology and osmotic balance. Bulging of the erythrocytes was noted from 0.6% NaCl, indicating a mild hypotonicity (an increase in cell volume by the influx of water). As the concentration of the hypotonic solution increased, a greater proportion of erythrocytes exhibited signs of membrane deformation and compromised structural integrity. Notably, at 0.4% NaCl concentration, a significant number of erythrocytes underwent haemolysis, characterized by rupture of the cell membrane and the release of haemoglobin into the surrounding medium, leaving behind ghost cells devoid of intracellular contents, as shown in Fig. 5. Additionally, phase imaging using 3D TIE QPM enabled the visualization of cellular variations of erythrocytes under different osmotic conditions. The height profile maps revealed the bulging of the cell membrane by hypotonic stress, as shown in Fig. 5.A quantitative analysis of the erythrocyte images provided insights into changes in key morphological parameters, such as cell height and cell volume. These parameters were computed across different saline concentrations to characterize the osmotic fragility profile of erythrocytes.
Fig. 6 shows the average volume of about 100 RBCs for each NaCl concentration along with the standard deviation. Under hypotonic stress, we observed that as the saline concentrations decreased and water concentrations increased, the erythrocytes started swelling, leading to an increase in their average volume. Beyond 0.4% saline concentration, the cells ruptured, and no cells were observed, indicating haemolysis. Therefore, in Fig. 7, we have extrapolated up to 0% concentrations and considered the volume to be zero as no cells were observed. Fig. 7 also shows the trend of the increase in volume with the increase in water concentration. This rupture and subsequent decrease in volume further support the occurrence of haemolysis.
UV-visible absorption spectral studies with saline at various concentrations were conducted to support the above findings. The absorption of beta and alpha heme at 540 nm and 575 nm, respectively, increased rapidly beyond 0.4% NaCl concentration, indicating hemolysis, as shown in Fig. 8.
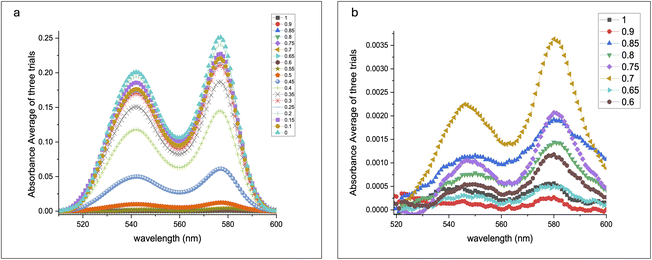 | ||
| Fig. 8 (a) UV-visible absorption of beta and alpha haemoglobins under various concentrations of saline; (b) zoomed-out scale athigher concentrations. | ||
Fig. 9 illustrates that the volume obtained from the 3D QPI system aligns closely with the normalized absorption data of beta and alpha haemoglobins from the UV-visible spectra. The graph in Fig. 9b provides a zoomed-in view of the plots of alpha and beta Hb. Since the errors were minimal, a 10-time expanded scale was required to observe the error bars for the alpha band, and for the beta band, about a 3-time expanded scale was required to visualize the error bars, which highlighted the accuracy of the data. The observed trends demonstrate that haemoglobin absorption increased after 0.4% saline concentration, stabilizing at 0%. Simultaneously, the RBC volume expanded up to 0.5% saline, signifying cellular bulging, and subsequently decreased beyond 0.45%, indicating cell rupture.
 | ||
| Fig. 9 (a) Combined UV-visible and volume data; (b) inset of 9 (a) showing a 3× zoomed-out scale for normalized beta Hb band. | ||
Here, we have used 3D Quantitative Phase Imaging (QPI) based on the Transport of Intensity Equation (TIE) to quantify the volume of a single RBC across a continuum of sodium chloride concentrations (1% to 0%) and study morphological changes with high precision. No previous study has reported this type of imaging analysis across all concentrations, thus our work is unique in its comprehensiveness.
Current literature has investigated theoretical modeling approaches and 3D imaging techniques to provide insights into the morphology of RBCs. One of the studies has evaluated RBC shape at specific saline concentrations (0%, 0.15%, 0.3%, 0.45%, 0.6%, and 0.75%), relying on cDOT-based qualitative analysis. While this work highlights the stabilizing role of glucose and mannitol, our study provides direct volume measurements, allowing for precise quantification of morphological variations across a finer concentration gradient.26 In another study, the use of spherical harmonic parameterization allowed theoretical predictions to be compared with 3D images of live cells, emphasizing the mechanical role of the membrane skeleton (MS) in stabilizing the shape of RBCs.27 Moreover, Shape-From-Shading (SFS) in combination with holographic reconstruction has been explored to estimate the concave RBC shape, and the advantage of this method is in recovering the concave surfaces versus traditional SFS.28 While these studies yield important insights into RBC structural dynamics, they either focus exclusively on specific shape transformations or examine discrete NaCl concentrations, rather than a systematic exploration of morphological changes across a fine gradient. Although 3D refractive index tomography (3D-RIT), confocal microscopy and holographic interference microscopy (HIM) enable high-accuracy morphological analysis, they usually depend on complex instrumentation and computational resources, as well as advanced sample preparation. In comparison, our TIE-based method is a straightforward, low-cost, and label-free method, and enables fast real-time volume extraction, with minimal installation effort. As such, it is very useful for regular laboratory activity and diagnostic use.
Significantly, no studies prior to this have performed a detailed imaging analysis across all concentrations of a concentration gradient. This approach allows for the observation of subtle morphological changes that may have been missed in discrete sampling methods. Our results validate the importance of osmolyte-induced stabilization of RBCs and can assist in the formulation of blood storage solutions and intravenous infusion protocols. This suggests that future work on long-term RBC viability and deformability under these conditions can lead to a richer understanding of membrane mechanics and cytoskeletal interactions.
Conclusion
In this work, we applied 3D Quantitative Phase Microscopy (QPM) based on the Transport of Intensity Equation (TIE) to investigate the morphology of erythrocytes at different saline concentrations. We found differential changes in the cell volume and function under different osmotic conditions. However, hypertonic NaCl concentrations (1% NaCl) caused dehydration of erythrocytes, marked by a decline in cell volume, indicating osmotic stress. At isotonic conditions (0.9–0.65% NaCl), the mean cell volume matched with the literature values,29 demonstrating a stable morphology and equilibrium. Yet, as with other solutes, at low concentrations of saline (lower than 0.6% NaCl), the cells continuously swelled in osmolyte-reduced media until water-induced membrane deformation occurred. At 0.4% NaCl, extensive haemolysis was observed, which was further confirmed by imaging and UV spectroscopic analysis. At this critical concentration, the UV spectra emphasized a significant enhancement of the β and α haemoglobin absorption bands, corroborating with the rupture of membranes and haemoglobin release. This work elucidates the dynamics of erythrocyte responses to different magnitudes of osmotic stress and highlights the potential of TIE-based QPM for cellular applications. These findings show that 3D QPI exhibits strong sensitivity in the non-invasive detection of small changes in red blood cell volume in saline. Our study demonstrates the capability of the 3D QPI system in detecting minor fluctuations in cell volume in response to hypotonic stress and determining critical points for cell swelling and rupture. Finally, the ability of the system to correlate the large changes in volume with haemolysis, as confirmed using UV-visible absorption spectra, further highlights the potential of this bio-imaging system for the detection and monitoring of cellular abnormalities. The Transport of Intensity Equation (TIE) also has strong potential for applications in cellular imaging, which are outside the scope of this work. Due to its non-invasive, label-free nature and its capacity to extract quantitative phase information, this technique provides a unique nanometre-scale spatial–temporal resolution of sub-cellular dynamics in a variety of biological situations. TIE can be very useful for the early identification of pathological changes in cells, such as those observed in blood diseases, cancer, and neurodegenerative diseases. It is adaptable for high-throughput and real-time imaging, extending its applicability to drug screening and personalized medicine. On the other hand, cellular imaging with TIE is a relatively less explored domain, however, with limitless opportunities for addressing future needs and even new developments.Ethical statement
For the RBC studies, we collected 3 mL of blood from healthy volunteers who visited Kasturba Hospital, Manipal, India for routine check-ups, with informed consent. All experiments were conducted in accordance with the guidelines of the Kasturba Hospital Institutional Ethics Committee and were approved by the Institutional Ethics Committee (IEC No: IEC-651-2020).Data availability
The data supporting the findings of this study are part of a patent application submitted to the Indian Patent Office (Application No. 202441046450, dated 17th June 2024) and may be available upon request.Conflicts of interest
The present work has been filed for intellectual property rights with the Indian Patent Office (Application No. 202441046450, dated 17th June 2024).Acknowledgements
Ms. Anusha Pillai gratefully acknowledges the Dr T. M. A Pai Scholarship provided by Manipal Academy of Higher Education. The authors also thank Manipal Academy of Higher Education for the INTRAMURAL GRANT –MAHE/CDS/PHD/IMF/2019.References
- L. Barbalato and L. S. Pillarisetty, Histology, Red Blood Cell, 2019 Search PubMed.
- D. Lazari, J. K. Freitas Leal, R. Brock and G. Bosman, Front. Physiol., 2020, 11, 288 Search PubMed.
- G. Popescu, Y. K. Park, W. Choi, R. R. Dasari, M. S. Feld and K. Badizadegan, Blood Cells, Mol., Dis., 2008, 41, 10–16, DOI:10.3389/FPHYS.2020.00288/BIBTEX.
- A. Nardo-Marino, T. H. Braunstein, J. Petersen, J. N. Brewin, M. N. Mottelson, T. N. Williams, J. A. L. Kurtzhals, D. C. Rees and A. Glenthøj, Front. Physiol., 2022, 13, 859906 Search PubMed.
- M. Bardyn, J. Allard, D. Crettaz, B. Rappaz, G. Turcatti, J. D. Tissot and M. Prudent, Int. J. Mol. Sci., 2021, 22, 4293 CrossRef CAS PubMed.
- A. Zeidan and D. Yelin, Biomed. Opt. Express, 2015, 6, 4335–4343, DOI:10.1364/BOE.6.004335.
- Y. Sung, W. Choi, C. Fang-Yen, K. Badizadegan, R. R. Dasari and M. S. Feld, Opt. Express, 2009, 17, 266–277, DOI:10.1364/OE.17.000266.
- L. Liu, S. Jin, Y. Hu, Z. Gu and H. C. Wu, J. Nanomater., 2011, 2011, 834139, DOI:10.1155/2011/834139.
- J. G. Schloetel, J. Heine, A. F. Cowman and M. Pasternak, Sci. Rep., 2019, 9(1), 1–10, DOI:10.1038/s41598-019-40718-z.
- G. Krampert, I. Kleppe, T. Kalkbrenner, K. Weisshart, R. Wolleschensky and M. Kempe, Biophotonics, 2010, 7715, 771503, DOI:10.1117/12.854633.
- J. Liu, Y. Y. Tan, W. Zheng, Y. Wang, L. A. Ju and Q. P. Su, J. Nanobiotechnol., 2024, 22, 363, DOI:10.1186/S12951-024-02605-2.
- D. Kucharski and M. Bartczak, Opt. Laser Technol., 2021, 142, 107262, DOI:10.1016/J.OPTLASTEC.2021.107262.
- A. Dubois, L. Vabre, A.-C. Boccara and E. Beaurepaire, Appl. Opt., 2002, 41, 805–812 CrossRef PubMed.
- M. A. Toderi, G. E. Galizzi, B. D. Riquelme and D. Dumas, Study of the red blood cell aggregation by coherent anti-Stokes Raman spectroscopy, Proc. SPIE, 2020, vol. 11251, DOI:10.1117/12.2543073.
- J. Dybas, F. C. Alcicek, A. Wajda, M. Kaczmarska, A. Zimna, K. Bulat, A. Blat, T. Stepanenko, T. Mohaissen, E. Szczesny-Malysiak, D. Perez-Guaita, B. R. Wood and K. M. Marzec, TrAC, Trends Anal. Chem., 2022, 146, 116481, DOI:10.1016/J.TRAC.2021.116481.
- D. A. Fernandes, S. Appak-Baskoy, E. Berndl and M. C. Kolios, RSC Adv., 2021, 11, 4906–4920, 10.1039/D0RA08009H.
- D. A. Fernandes and M. C. Kolios, Opt. Mater. Express, 2019, 9, 4532 CrossRef CAS.
- C. Zuo, J. Li, J. Sun, Y. Fan, J. Zhang, L. Lu, R. Zhang, B. Wang, L. Huang and Q. Chen, Opt. Lasers Eng., 2020, 135, 106187, DOI:10.1016/J.OPTLASENG.2020.106187.
- H. Park, S. Lee, M. Ji, K. Kim, Y. Son, S. Jang and Y. Park, Sci. Rep., 2016, 6(1), 1–10, DOI:10.1038/srep34257.
- S. Carney, T. C. Khoo, A. Sheikhsofla, S. Ghazanfarpour, A. Sharikova, S. D. Mahajan, A. Khmaladze and J. C. Petruccelli, Opt. Lasers Eng., 2023, 166, 107581 Search PubMed.
- C. Zuo, W. Qu, Q. Chen and A. Asundi, Opt. Express, 2013, 21, 24060–24075 CrossRef PubMed.
- T.-C. Poon, U. Abeywickrema, M. Basunia, H. Zhang and P. P. Banerjee, Appl. Opt., 2016, 55(33), 9546–9554 CrossRef PubMed.
- S. Sarangapani, A. Patil, Y. K. Ngeow, R. Elsa Mohan, A. Asundi and M. J. Lang, Integr. Biol., 2018, 10, 313–324, DOI:10.1364/AO.55.009546.
- A. Pillai, S. Kamath U, S. Belurkar, A. K. Asundi and A. Patil, Proc. SPIE, 2024, vol. 13069, DOI:10.1117/12.3023411.
- P. M. S. Roma, L. Siman, F. T. Amaral, U. Agero and O. N. Mesquita, Appl. Phys. Lett., 2014, 104, 7 Search PubMed.
- M. Son, Y. S. Lee, M. J. Lee, Y. K. Park, H. R. Bae, S. Y. Lee, M. G. Shin and S. Yang, PLoS One, 2021, 16, e0262106, DOI:10.1371/JOURNAL.PONE.0262106.
- K. Khairy, J. Foo and J. Howard, Cell. Mol. Bioeng., 2010, 1, 173, DOI:10.1007/S12195-008-0019-5.
- P. Memmolo, L. Miccio, F. Merola, O. Gennari, P. A. Netti and P. Ferraro, Cytometry, Part A, 2014, 85, 1030–1036, DOI:10.1002/CYTO.A.22570.
- M. Son, Y. S. Lee, M. J. Lee, Y. K. Park, H. R. Bae, S. Y. Lee, M. G. Shin and S. Yang, PLoS One, 2021, 16, e0262106, DOI:10.1371/JOURNAL.PONE.0262106.
| This journal is © The Royal Society of Chemistry 2025 |





