Open Access Article
Open Access ArticleSynthesis, molecular docking, and biological investigations of new pyrazolone chalcones†
Ahmed A. Noser *a,
Maha M. Salem
*a,
Maha M. Salem b,
Esraa M. ElSaftya,
Mohamed H. Barena,
Adel I. Selima and
Hamada S. A. Mandoura
b,
Esraa M. ElSaftya,
Mohamed H. Barena,
Adel I. Selima and
Hamada S. A. Mandoura
aOrganic Chemistry, Chemistry Department, Faculty of Science, Tanta University, Tanta 31527, Egypt. E-mail: ahmed.nosir@science.tanta.edu.eg; esraa_elsafty@science.tanta.edu.eg; chemistnoser2010@yahoo.com; adel.saleem@science.tanta.edu.eg; hamada.mandour@science.tanta.edu.eg
bBiochemistry Division, Chemistry Department, Faculty of Science, Tanta University, Tanta 31527, Egypt. E-mail: maha_salem@science.tanta.edu.eg
First published on 25th April 2025
Abstract
Heterocyclic compounds are essential to the drug development and discovery processes. Herein, we synthesized new pyrazolone chalcones (3a–g) through the reaction of azopyrazolone (2) with different aromatic aldehydes in a basic medium. Numerous techniques including elemental analysis, 1H-NMR, 13C-NMR, and FT-IR spectroscopies, were used to characterize pyrazolone chalcone derivatives. Compound 3b exhibited the highest binding energy towards YAP/TEAD protein with a value of −8.45 kcal mol−1 in in silico studies. This observation suggested that compound 3b inhibits the YAP/TEAD Hippo signaling pathway. In addition, compound 3b offered a prospective anticancer effect against various cancer cell lines, such as HepG-2, MCF-7, and HCT-116, among the other synthesized compounds, with IC50 values equal to 5.03 ± 0.4, 3.92 ± 0.2, and 6.34 ± 0.5 μM, respectively. These results validated our findings regarding the in silico suppression of the YAP/TEAD protein. Its pharmacokinetic properties were theoretically observed using ADMET. Additionally, compound 3b demonstrated a potent antioxidant scavenging action (in vitro) against DPPH free radicals. Thus, based on our findings, compound 3b may act as a potential anticancer scaffold owing to its inhibitory impact towards the YAP/TEAD-mediated Hippo signaling pathway with a safe toxic profile on normal cells.
Introduction
Cancer is a worldwide civic health concern, and its diagnosis in early stage is of great importance.1 Currently, the primary treatment strategies for cancer are chemotherapy, surgery, and targeted therapy. Targeted therapy has become a standard treatment for cancer patients and has significantly increased the efficacy of cancer therapy.2 According to a previous report, regulating organ growth and inhibiting tumors depend on the Hippo signaling target pathway's control over cell division and proliferation.3 Because of its significant influence on patient prognosis and chemotherapeutic drug resistance, the Hippo pathway is a desirable therapeutic target.4 Pyrazoles have attracted significant attention owing to their effective pharmaceutical properties such as antibiotic, antifungal, antibacterial, pesticidal, antioxidant, anti-inflammatory, antiviral, and antitumor effects.5–11 Chalcones are α,β-unsaturated ketones with a broad range of pharmacological actions, making them interesting synthons and bioactive scaffolds with great therapeutic potential. It is commonly known that they have significant biological effects such as antibacterial, antioxidant, and anticancer effects.12–15 Chalcones facilitate the synthesis of novel heterocyclic compounds including pyrimidinones, azepines, pyridine, and pyran, which have strong biological activities.16,17 There have been numerous reports on marketed anticancer drugs containing pyrazolone and a chalcone core, such as metochalcone, sofalcone, butein, metamizole, and propyphenazone,18–22 as shown in Fig. 1.The goal of our study was to develop a new class of pyrazolone chalcone derivatives, examine their radical scavenging activities and investigate their anticancer effects by inhibiting the YAP/TEAD Hippo signaling pathway (in silico). our findings confirmed their impact on cancer cell lines (in vitro). Additionally, absorption–distribution–metabolism–excretion–toxicity (ADMET) pharmacokinetic features were used to assess their drug-likeness and bioavailability.
Results and discussion
Chemistry of the novel compounds
Scheme 1 describes the reaction of ethyl cyanoacetate with phenyl hydrazine, which leads to the formation of compound 1, as described previously by Weissberger.23 Further modification of compound 1 with p-acetyl phenyl diazonium chloride offered azo pyrazolone 2 with 74% yield (Scheme 1). The chemical structures of compounds 3a–g were established according to their elemental analysis and different spectral data. Elemental analysis was used to indicate the exact ratio of C, H, and N. Fourier transform infrared (FT-IR) spectroscopy was used to illustrate the presence of the main functional groups. Nuclear magnetic resonance (NMR) was used to indicate the different types of hydrogen and carbon atoms in the synthesized compounds. The FT-IR analysis of compound 2 showed different absorption bands at 1600 cm−1 and 1540 cm−1 due to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and N
O and N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N groups, respectively. Its 1H NMR spectrum showed a new singlet signal at δ 2.20 ppm attributed to the CH3 group.
N groups, respectively. Its 1H NMR spectrum showed a new singlet signal at δ 2.20 ppm attributed to the CH3 group.
Modifying azo compound (2) with a different aromatic aldehyde in a basic medium led to the formation of pyrazolone chalcone 3a–g (Scheme 2).24 The FT-IR analysis of chalcones (3a–g) showed absorption bands at 1610–1670 cm−1, 1520–1600 cm−1, and 1500–1560 cm−1, for C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, and N
N, and N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N groups. Compound 3b–d showed absorption bands at 3100–3310 cm−1 for the OH group. The 1H NMR spectrum of compound 3a revealed two new signals resonated at δ 7.54, and 7.68 ppm attributed to two olefinic protons. Compound 3b showed four new signals at δ 7.00, 7.40, 3.86, and 2.08 ppm attributed to two olefinic protons, OCH3 protons, and OH proton, respectively. Additionally, compounds 3c and 3d showed three new signals at δ 6.92–7.45, 7.55–7.56, and 10.12–10.26 ppm attributed to two olefinic protons and OH proton, respectively. Finally, the 1H NMR spectrum of compound 3g revealed four new signals resonated at δ 7.62, 7.68, 8.02, and 8.07 ppm attributed to olefinic protons.
N groups. Compound 3b–d showed absorption bands at 3100–3310 cm−1 for the OH group. The 1H NMR spectrum of compound 3a revealed two new signals resonated at δ 7.54, and 7.68 ppm attributed to two olefinic protons. Compound 3b showed four new signals at δ 7.00, 7.40, 3.86, and 2.08 ppm attributed to two olefinic protons, OCH3 protons, and OH proton, respectively. Additionally, compounds 3c and 3d showed three new signals at δ 6.92–7.45, 7.55–7.56, and 10.12–10.26 ppm attributed to two olefinic protons and OH proton, respectively. Finally, the 1H NMR spectrum of compound 3g revealed four new signals resonated at δ 7.62, 7.68, 8.02, and 8.07 ppm attributed to olefinic protons.
The suggested mechanism for the synthesis of compounds 3a–g is illustrated in Scheme 3.
Antioxidant capacity
Fig. 2 and 3 show that the antioxidant activity enhanced with the increase in chalcone concentration using the stable DPPH radical. Compared to normal L-ascorbic acid (IC50 = 16.81 ± 0.10 μM), compound 3b exhibited the greatest DPPH scavenging activity with an IC50 value of 19.95 ± 0.14 μM. Compounds 3a and 3f exhibited a strong antioxidant activity with IC50 values of 21.40 ± 0.17 and 27.25 ± 0.19 μM. Moreover, compounds 3e and 3d had moderate effects on scavenging with the activity of 32.79 ± 0.21 and 48.59 ± 0.28 μM, respectively, while compounds 3g and 3c showed a limited ability to quench free radicals with IC50 values of 73.63 ± 0.39 and 94.18 ± 0.45 μM, respectively. | ||
| Fig. 2 Antioxidant scavenging activity of chalcone derivatives (3a–g). Results are reported as mean ± SE; where n = 3. | ||
 | ||
| Fig. 3 Inhibitory concentrations (IC50) of chalcones 3a–g via DPPH. Results are reported as mean ± SE, where n = 3. | ||
Docking studies
For creating anticancer drugs, the novel chalcone ligands 3a–g were docked into YAP/TEAD, a well-known and alluring therapeutic target protein in the Hippo signaling pathway. Numerous studies in the fields of oncology and regenerative medicine have examined the Hippo pathway, and these studies have also suggested that the route may be important as a regulatory variable in human biology or as an inhibitory target for drug development. Generally, the Hippo pathway consists of two constitutive modules: the upstream serine/threonine kinase cascade, which comprises MST1/2 and LATS1/2, and the downstream transcriptional modules, which mainly include the YAP/TEAD complex.25The YAP/TEAD genetic and pharmacological interaction significantly decreased carcinogenesis in YAP-dependent cancer models.26 As a result, the YAP/TEAD complex has become a desirable target for the creation of anti-cancer medications.27 Table 1 and Fig. 4 summarize all new chalcone interactions with the target YAP/TEAD protein. According to our findings, compound 3b had the highest binding energy (−8.45 kcal mol−1) against the target YAP/TEAD protein. Compound 3a had high binding energy (−7.86 kcal mol−1) with the target protein. With binding energies of −7.11, −7.13, and −7.7.22 kcal mol−1, respectively, compounds 3d, 3e, and 3f also demonstrated a mild inhibitory effect. Conversely, compounds 3c and 3g showed low binding energies of −6.11 and −6.09 kcal mol−1, respectively. Compound 3b was therefore highly recommended for use as an anticancer agent because of its potential to inhibit the YAP/TEAD mediated Hippo signaling pathway.
| Compounds | YAP/TEAD target Hippo signaling protein | |
|---|---|---|
| Docking score (ΔGbind) | Docked complex (amino acid–ligand) interactions | |
| 3a | −7.86 | H-acceptor |
| LYS344 | ||
| CYS367 | ||
| Electrostatic interactions | ||
| PHE229 | ||
| LLE395 | ||
| MET370 | ||
| LEU366 | ||
| 3b | −8.45 | H-donor |
| PRO365 | ||
| π-Hydrogen | ||
| LYS344 | ||
| VAL345 | ||
| Electrostatic interactions | ||
| LEU366 | ||
| MET370 | ||
| CYS367 | ||
| LLE395 | ||
| GLU343 | ||
| CYS367 | ||
| 3c | −6.11 | Electrostatic interactions |
| CYS367 | ||
| LEU366 | ||
| MET370 | ||
| LYS344 | ||
| LLE395 | ||
| 3d | −7.11 | H-acceptor |
| LYS344 | ||
| Electrostatic interactions | ||
| CYS367 | ||
| THR332 | ||
| PHE229 | ||
| LLE395 | ||
| PHE415 | ||
| VAL342 | ||
| 3e | −7.13 | H-acceptor |
| LYS344 | ||
| Electrostatic interactions | ||
| CYS367 | ||
| THR332 | ||
| PHE229 | ||
| LLE395 | ||
| PHE415 | ||
| VAL342 | ||
| 3f | −7.22 | H-acceptor |
| LYS344 | ||
| Electrostatic interactions | ||
| CYS367 | ||
| VAL342 | ||
| LLE395 | ||
| MET370 | ||
| LEU366 | ||
| 3g | −6.09 | Electrostatic interactions |
| LEU366 | ||
| GLU368 | ||
| VAL342 | ||
| CYS367 | ||
| LYS344 | ||
| LLE395 | ||
| PHE229 | ||
 | ||
| Fig. 4 Two- and three-dimensional molecular interaction networks for all chalcone derivatives (3a–g) with the target YAP/TEAD-mediated Hippo signaling pathway. | ||
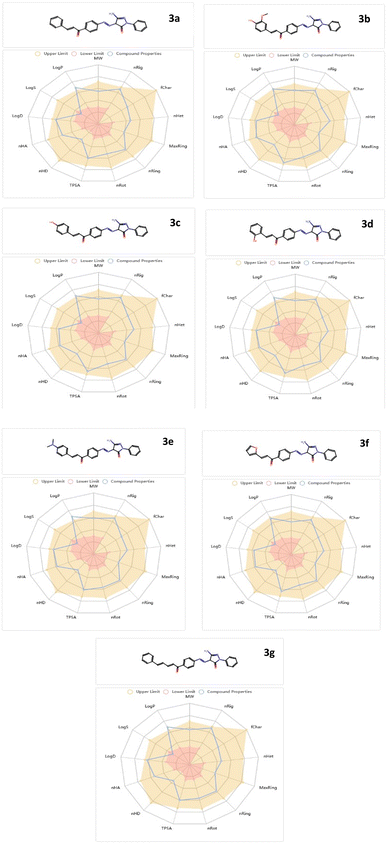
ADMET features
The bioavailability and drug-like characteristics of the studied chalcone derivatives (3a–g) were determined, and are shown in Fig. 5. With a total polar surface area (TPSA) ranging from 102.25 to 149.35 Å2, the compounds 3a–g met all the requirements for exceptional permeability and had sufficient oral bioavailability. To further demonstrate flexibility, they also used rotatable bonds between 0 and 10. They were more soluble in cellular membranes because their hydrogen-bond accepted (HBA) and hydrogen-bond donated (HBD) values fell within the fulfilled range. Table 2 demonstrates that octanol/water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p) values less than 5 were indicative of good lipophilicity features. The chalcone derivatives also showed a higher percentage of human intestinal absorption (HIA) ratings in accordance with the ADMET criteria, indicating that the human intestine could absorb them more effectively. The studied chalcones have a great safety profile for the central nervous system (CNS) since they do not penetrate the blood–brain barrier. They are safe because every AMES toxicity and carcinogenicity test result was negative.
p) values less than 5 were indicative of good lipophilicity features. The chalcone derivatives also showed a higher percentage of human intestinal absorption (HIA) ratings in accordance with the ADMET criteria, indicating that the human intestine could absorb them more effectively. The studied chalcones have a great safety profile for the central nervous system (CNS) since they do not penetrate the blood–brain barrier. They are safe because every AMES toxicity and carcinogenicity test result was negative.
| Molecular weight (g mol−1) | Blood–brain barrier (BBB) | % human intestinal absorption (HIA+) | TPSA A2 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p p |
nHA | nHD | N rotatable | AMES toxicity | Carcinogenicity | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3a | 409.15 | No | 89.6 | 100.48 | 3.39 | 7 | 2 | 6 | Nontoxic | Noncarcinogenic |
| 3b | 455.16 | No | 92.1 | 129.94 | 2.98 | 9 | 3 | 7 | Nontoxic | Noncarcinogenic |
| 3c | 425.15 | No | 84.3 | 120.71 | 3.08 | 8 | 3 | 6 | Nontoxic | Noncarcinogenic |
| 3d | 425.15 | No | 75.9 | 120.71 | 3.43 | 8 | 3 | 6 | Nontoxic | Noncarcinogenic |
| 3e | 452.20 | No | 83.4 | 103.72 | 3.61 | 8 | 2 | 7 | Nontoxic | Noncarcinogenic |
| 3f | 399.13 | No | 79.1 | 113.62 | 2.90 | 8 | 2 | 6 | Nontoxic | Noncarcinogenic |
| 3g | 435.17 | No | 77.3 | 100.48 | 3.47 | 7 | 2 | 7 | Nontoxic | Noncarcinogenic |
In vitro anticancer study
The antitumor effect of chalcone derivatives (3a–g) on HCT-116, HepG2, MCF-7, and WI-38 normal cell lines was assessed in this study using the MTT assay (Fig. 6). Compound 3b demonstrated noteworthy antitumor effects on the cancer cell lines HepG2, MCF-7, and HCT-116, with IC50 values of 5.03 ± 0.4, 3.92 ± 0.2, and 6.34 ± 0.5 μM, respectively. Compounds 3a and 3f demonstrated impressive antitumor effects with HCT-116, HepG2, and MCF-7 cell lines. Compounds 3e and 3d demonstrated a moderate effect on all cancer cell lines. Conversely, Compounds 3g and 3c demonstrated a weak effect on all cancer cell lines when compared to the reference chemotherapeutic drug (DOX). Moreover, all new derivatives (3a–g) showed lower cytotoxic effects on WI-38 normal cells except 3c. Our finding was in line with the previous study.24 This suggests that compound 3b worked against proliferative cancer by blocking the Hippo signaling pathway mediated by YAP/TEAD. | ||
| Fig. 6 Antitumor and cytotoxic impacts of chalcones 3a–g. Results are reported as mean ± SE of three independent triplicates. | ||
Structure–anti-tumor activity relationship (SAR)
The synthesized chalcones demonstrated their anticancer effect for the following reasons (Fig. 7):• The presence of different substituents in the aryl group enhanced cytotoxic activity against different cancer cell lines (HepG-2, MCF-7, and HCT-116).
• The presence of hetero atoms such as nitrogen as well as carbonyl groups in the chemical structure of the pyrazolone chalcones enhanced the value of binding energy through the interaction between the pyrazolone chalcone and the target protein via hydrogen bonding.28
• The azo linker enhanced the hydrogen bonding with the target protein 29.
• Compound 3b was considered the most active compound due to the presence of the phenyl group and olefinic protons, which enhanced the hydrogen bonding and π–π interaction with the target protein and, hence, increased the value of the binding energy.30
Conclusion
We synthesized and characterized new pyrazolone chalcones (3a–g) using various spectral data. Among all the synthesized compounds, chalcone 3b demonstrated the strongest antioxidant activity. Furthermore, it has a high docking score because of hydrogen bonds, hydrophobic interactions, and electrostatic interactions with important residues in the binding pocket of the YAP/TEAD target protein. For that, molecular docking suggested that it may be a targeted anticancer agent. Additionally, the in vitro anti-cancer effect against different cancer cell lines was explained by the suppressive effect of chalcone 3b, which inhibited the YAP/TEAD-mediated Hippo signaling pathway to cause apoptosis and obstruct cell growth and survival. According to these results, chalcone 3b may be a newly targeted anticancer scaffold that targets a variety of cancer cells.Experimental section
Materials and instrumentation
Section S1 in the ESI† includes the data of chemicals and instrumentation.Synthesis of 3-amino-1-phenyl-1H-pyrazol-5(4H)-one (1)
Weissberger lab was previously preparing compound 1 (ref. 23)Synthesis of 4-((4-acetylphenyl)diazenyl)-3-amino-1-phenyl-1H-pyrazol-5(4H)-one (2)
A sodium nitrite solution (12.7 mmol) was added slowly to p-amino acetophenone (13.7 mmol) in concentrated HCl. The resulting diazonium chloride was added to a solution of compound 1 (8.5 mmol) in pyridine. Then, the product was filtered and dried.Orange powder; yield 74%; mp 89–91 °C; 1H NMR (400 MHz, DMSO-d6) δ (ppm): 9.14 (s, 2H, NH2), 7.08–8.11 (m, 9H, Ar–H), 2.59 (s, 1H, CH–pyrazolone), 2.20 (s, 3H, CH3); IR (KBr) ν: 1600 (CO), 1540 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C17H15N5O2 (321.33): C, 63.54%; H, 4.71%; N, 21.79%. Found: C, 63.34%; H, 4.57%; N, 21.67%.
N); anal. calcd for C17H15N5O2 (321.33): C, 63.54%; H, 4.71%; N, 21.79%. Found: C, 63.34%; H, 4.57%; N, 21.67%.
General procedure for the synthesis of pyrazolone chalcones (3a–g)
An ethanolic solution of compound 2 (10.0 mmol), aromatic aldehyde (10.0 mmol), and sodium hydroxide (20.0 mmol) was stirred at 25 °C for 10 h, put into freezing water, filtered, and then dried.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.54 (d, 1H, COCH
CH), 7.54 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.54–8.40 (m, 14H, Ar–H), 2.60 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm): 171.72 (C
), 6.54–8.40 (m, 14H, Ar–H), 2.60 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm): 171.72 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O chalcone), 168.04 (C
O chalcone), 168.04 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O pyrazolone), 146.95 (C
O pyrazolone), 146.95 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyrazolone), 113.20–133.22 (Ar–C), 127.60, 135.25 (C–olefinic), 74.43 (CH–pyrazolone); IR (KBr) ν: 3405 (NH2), 1650 (C
N pyrazolone), 113.20–133.22 (Ar–C), 127.60, 135.25 (C–olefinic), 74.43 (CH–pyrazolone); IR (KBr) ν: 3405 (NH2), 1650 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1597 (C
O), 1597 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1550 (N
N), 1550 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C24H19N5O2 (409.15): C, 70.40%; H, 4.68%; N, 17.10%. Found: C, 70.25%; H, 4.46%; N, 17.02%.
N); anal. calcd for C24H19N5O2 (409.15): C, 70.40%; H, 4.68%; N, 17.10%. Found: C, 70.25%; H, 4.46%; N, 17.02%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.00 (d, 1H, COCH
CH), 7.00 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.89–8.17 (m, 12H, Ar–H), 3.86 (s, 3H, OCH3), 2.61 (s, 1H, CH–pyrazolone), 2.08 (s, 1H, OH); 13C NMR (101 MHz, DMSO-d6) δ (ppm):153.80 (C
), 6.89–8.17 (m, 12H, Ar–H), 3.86 (s, 3H, OCH3), 2.61 (s, 1H, CH–pyrazolone), 2.08 (s, 1H, OH); 13C NMR (101 MHz, DMSO-d6) δ (ppm):153.80 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O chalcone), 150.94 (C
O chalcone), 150.94 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O pyrazolone), 149.03 (C
O pyrazolone), 149.03 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyrazolone), 145.49 (C–OH), 111.54–130.62 (Ar–C), 122.71, 130.93 (C–olefinic), 70.29 (CH–pyrazolone), 55.98 (OCH3); IR (KBr) ν: 3400 (NH2), 3310 (OH), 1610 (C
N pyrazolone), 145.49 (C–OH), 111.54–130.62 (Ar–C), 122.71, 130.93 (C–olefinic), 70.29 (CH–pyrazolone), 55.98 (OCH3); IR (KBr) ν: 3400 (NH2), 3310 (OH), 1610 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1520(N
N), 1520(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C25H21N5O4 (455.16): C, 65.93%; H, 4.65%; N, 15.38%. Found: C, 65.77%; H, 4.47%; N, 15.26%.
N); anal. calcd for C25H21N5O4 (455.16): C, 65.93%; H, 4.65%; N, 15.38%. Found: C, 65.77%; H, 4.47%; N, 15.26%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.92 (d, 1H, COCH
CH), 6.92 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.84–8.21 (m, 13H, Ar–H), 2.59 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm):163.65 (C
), 6.84–8.21 (m, 13H, Ar–H), 2.59 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm):163.65 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O chalcone), 160.64 (C
O chalcone), 160.64 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O pyrazolone), 159.35 (C
O pyrazolone), 159.35 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyrazolone), 135.57 (C–OH), 98.54–131.58 (Ar–C), 123.23, 132.56 (C–olefinic), 74.42 (CH–pyrazolone); IR (KBr) ν: 3400 (NH2), 3200 (OH), 1620 (C
N pyrazolone), 135.57 (C–OH), 98.54–131.58 (Ar–C), 123.23, 132.56 (C–olefinic), 74.42 (CH–pyrazolone); IR (KBr) ν: 3400 (NH2), 3200 (OH), 1620 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1590 (C
O), 1590 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1520(N
N), 1520(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C24H19N5O3 (425.15): C, 67.76%; H, 4.50%; N, 16.46%. Found: C, 67.66%; H, 4.36%; N, 16.38%.
N); anal. calcd for C24H19N5O3 (425.15): C, 67.76%; H, 4.50%; N, 16.46%. Found: C, 67.66%; H, 4.36%; N, 16.38%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.45 (d, 1H, COCH
CH), 7.45 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.79–8.22 (m, 13H, Ar–H), 2.60 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm): 167.74 (C
), 6.79–8.22 (m, 13H, Ar–H), 2.60 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm): 167.74 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O chalcone), 156.32 (C
O chalcone), 156.32 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O pyrazolone), 146.51 (C
O pyrazolone), 146.51 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyrazolone), 143.65 (C–OH), 116.98–131.27 (Ar–C), 122.70, 133.51 (C–olefinic), 63.92 (CH–pyrazolone); IR (KBr) ν: 3200 (NH2), 3100 (OH), 1670 (C
N pyrazolone), 143.65 (C–OH), 116.98–131.27 (Ar–C), 122.70, 133.51 (C–olefinic), 63.92 (CH–pyrazolone); IR (KBr) ν: 3200 (NH2), 3100 (OH), 1670 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1540 (N
N), 1540 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C24H19N5O3 (425.15): C, 67.76%; H, 4.50%; N, 16.46%. Found: C, 67.66%; H, 4.42%; N, 16.38%.
N); anal. calcd for C24H19N5O3 (425.15): C, 67.76%; H, 4.50%; N, 16.46%. Found: C, 67.66%; H, 4.42%; N, 16.38%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 6.74 (d, 1H, COCH
CH), 6.74 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 7.09–8.35 (m, 13H, Ar–H), 3.04 (s, 6H, N(CH3)2), 2.84 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm):154.73 (C
), 7.09–8.35 (m, 13H, Ar–H), 3.04 (s, 6H, N(CH3)2), 2.84 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm):154.73 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O chalcone), 152.51 (C
O chalcone), 152.51 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O pyrazolone), 145.51 (C
O pyrazolone), 145.51 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyrazolone), 111.52–137.28 (Ar–C), 122.98, 138.22 (C–olefinic), 79.78 (CH–pyrazolone), 55.34 (N(CH3)2); IR (KBr) ν: 3400 (NH2), 1660 (C
N pyrazolone), 111.52–137.28 (Ar–C), 122.98, 138.22 (C–olefinic), 79.78 (CH–pyrazolone), 55.34 (N(CH3)2); IR (KBr) ν: 3400 (NH2), 1660 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1520 (C
O), 1520 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1500 (N
N), 1500 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C26H24N6O2 (452.20): C, 69.01%; H, 5.35%; N, 18.57%. Found: C, 68.91%; H, 5.55%; N, 18.39%.
N); anal. calcd for C26H24N6O2 (452.20): C, 69.01%; H, 5.35%; N, 18.57%. Found: C, 68.91%; H, 5.55%; N, 18.39%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH), 7.13 (d, 1H, COCH
CH), 7.13 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 6.71–8.17 (m, 12H, Ar–H), 2.17 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm): 151.58 (C
), 6.71–8.17 (m, 12H, Ar–H), 2.17 (s, 1H, CH–pyrazolone); 13C NMR (101 MHz, DMSO-d6) δ (ppm): 151.58 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O chalcone), 147.45 (C
O chalcone), 147.45 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O pyrazolone), 146.80 (C
O pyrazolone), 146.80 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyrazolone), 112.83–136.01 (Ar–C), 124.28, 136.37 (C–olefinic), 75.72 (CH–pyrazolone); IR (KBr) ν: 3420 (NH2), 1630 (C
N pyrazolone), 112.83–136.01 (Ar–C), 124.28, 136.37 (C–olefinic), 75.72 (CH–pyrazolone); IR (KBr) ν: 3420 (NH2), 1630 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1600 (C
O), 1600 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1530 (N
N), 1530 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C22H17N5O3 (399.13): C, 66.16%; H, 4.29%; N, 17.53%. Found: C, 66.06%; H, 4.21%; N, 17.35%.
N); anal. calcd for C22H17N5O3 (399.13): C, 66.16%; H, 4.29%; N, 17.53%. Found: C, 66.06%; H, 4.21%; N, 17.35%.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
 –CH), 8.02 (d, 1H, COCH
–CH), 8.02 (d, 1H, COCH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 7.68 (t, 1H,
), 7.68 (t, 1H, 
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH–Ar), 7.62 (d, 1H, CH
CH–Ar), 7.62 (d, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif)
 –Ar), 6.74–8.60 (m, 14H, Ar–H), 2.08 (s, 1H, CH–pyrazolone); IR (KBr) ν: 3400 (NH2), 1620 (C
–Ar), 6.74–8.60 (m, 14H, Ar–H), 2.08 (s, 1H, CH–pyrazolone); IR (KBr) ν: 3400 (NH2), 1620 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1590 (C
O), 1590 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1560 (N
N), 1560 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N); anal. calcd for C26H21N5O2 (435.17): C, 71.71%; H, 4.86%; N, 16.08%. Found: C, 71.55%; H, 4.68%; N, 15.92%.
N); anal. calcd for C26H21N5O2 (435.17): C, 71.71%; H, 4.86%; N, 16.08%. Found: C, 71.55%; H, 4.68%; N, 15.92%.Biological investigations
Anticancer assessments (in vitro)
Abbreviations
| FT-IR | Fourier transform infrared |
| NMR | nuclear magnetic resonance |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| ADMET | absorption–distribution–metabolism–excretion–toxicity |
| HBA | hydrogen bond accepted |
| HBD | hydrogen bond donated |
| HIA | human intestinal absorption |
| CNS | central nervous system |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P | octanol/water partition coefficient |
Data availability
The cell lines were provided by the American Type Culture Collection (ATCC) via VACSERA, Cairo, Egypt, and all accession codes were added: mammary gland (MCF-7; #ATCC HTB-22 (https://www.atcc.org/products/htb-22)), colorectal adenocarcinoma (HCT-116; #ATCC CCL-247 (https://www.atcc.org/products/htb-37)), hepatocellular carcinoma (HepG-2; #ATCC HB-8065 (https://www.atcc.org/products/hb-8065)), and human lung fibroblast (WI-38; #ATCC CCL-75 (https://www.atcc.org/products/ccl-75)). The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The macromolecule protein structure is deposited in the worldwide protein data bank repository (https://www.rcsb.org/structure/3KYS).Author contributions
Ahmed A. Noser, Maha M. Salem, and Esraa M. ElSafty: writing – review & editing, writing – original draft, and methodology. Maha M. Salem: formal analysis. Adel I. Selim, Ahmed A. Noser, Mohamed H. Baren, and Hamada S. A. Mandour: supervision.Conflicts of interest
The authors have no relevant financial or non-financial interests to disclose.References
- P. Das, G. Sayan, A. Tarun, M. Pritiprasanna, G. Sabyasachi, C. Sumita and G. Subhashis, Mater. Chem. Phys., 2019, 237, 121860 CrossRef CAS.
- W. Cao, H. D. Chen, Y. W. Yu, N. Li and W. Q. Chen, Chin. Med., 2020, 134, 783e791 Search PubMed.
- L. Zhang, F. Ren, Q. Zhang, Y. Chen, B. Wang and J. Jiang, Dev. Cell, 2008, 14, 377–387 CrossRef CAS PubMed.
- Y. Zhao and X. Yang, Int. J. Cancer, 2015, 137, 2767–2773 CrossRef CAS PubMed.
- J. V. Patil, S. S. Shubhangi, U. Shweta, G. Pankaj and B. Suresh, ChemistrySelect, 2024, 9, e202303391 CrossRef CAS.
- S. M. Emam, B. Samir and A. M. Ahmed, Results Chem., 2023, 5, 100725 CrossRef CAS.
- A. N. Ahmed, H. A. Aboubakr and M. S. Maha, Bioorg. Chem., 2023, 131, 106299 CrossRef PubMed.
- A. N. Ahmed, A. S. Ihsan, H. A. Aboubakr and M. S. Maha, ACS Omega, 2022, 7, 25265–25277 CrossRef PubMed.
- M. Islam, M. Abdullah, N. Essam, Y. Sammer, A. Muhammad, N. Asif and W. Abdul, J. Mol. Struct., 2022, 1269, 133843 CrossRef CAS.
- S. Dabhade, P. Manjushri, S. Lala, A. Sachin, A. Shweta, Y. Somdatta and N. Santosh, Chem. Biodiversity, 2024, 21, e202400015 CrossRef PubMed.
- F. Rizk, N. Ahmed, A. Seham and K. Amira, J. Heterocycl. Chem., 2022, 59, 2190–2206 CrossRef.
- H. Şenol, G. Mansour, Ö. Gülbahar, T. Alim and G. Uğur, J. Mol. Struct., 2024, 1295, 136804 CrossRef.
- A. N. Ahmed, A. A. El-Barbary, M. S. Maha, A. Hayam and S. Mohamed, Sci. Rep., 2024, 14, 3530 CrossRef PubMed.
- K. J. Setshedi, M. B. Richard, I. Kayhan, M. Dorien, C. Guy and J. L. Lesetja, Med. Chem. Res., 2024, 3, 977–988 CrossRef.
- A. N. Ahmed, A. I. Saham, A. Hayam, M. A. Nora and S. A. Hamada, J. Iran. Chem. Soc., 2023, 20, 2963–2976 CrossRef.
- S. S. Shafi, Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem., 2021, 60, 1132–1136 Search PubMed.
- S. K. Ramadan and A. R. Sameh, J. Iran. Chem. Soc., 2022, 19, 187–201 CrossRef CAS.
- J. Zhou, W. Feng, X. Bin, L. Xin, P. Cheng and F. U. Peng, Oncol. Res., 2024, 32, 943 CrossRef PubMed.
- H. Tanaka, N. Seito, O. Kenji, T. Tomoya and H. Toshihiko, Biochem. Biophys. Res. Commun., 2009, 381, 566–571 CrossRef CAS PubMed.
- G. Padmavathi, K. R. Nand, B. Devivasha, A. Frank, M. Srishti, S. Gautam, B. Anupam and B. K. Ajaikumar, Phytomedicine, 2017, 25, 118–127 CrossRef CAS PubMed.
- A. Jasiecka, T. Maslanka and J. J. Jaroszewski, Pol. J. Vet. Sci., 2014, 17, 207–214 CAS.
- H. G. Kraetsch, T. Hummel, J. Lötsch, R. Kussat and G. Kobal, Eur. J. Clin. Pharmacol., 1996, 49, 377–382 CrossRef CAS PubMed.
- A. Weissberger, H. D. Porter and W. A. Gregory, J. Am. Chem. Soc., 1944, 66, 1851–1855 CrossRef CAS.
- J. Dong, H. Guang, Z. Qijing, W. Zengtao, C. Jiahua, W. Yan, M. Qingqing and L. Shaoshun, MedChemComm, 2019, 10, 1606–1614 RSC.
- B. Zhao, X. Ye, J. Yu, L. Li, W. Li, S. Li, J. Yu, J. D. Lin, C. Y. Wang, A. M. Chinnaiyan, Z. C. Lai and K. L. Guan, Genes Dev., 2008, 22, 1962–1971 CrossRef CAS PubMed.
- Y. Liu-Chittenden, B. Huang, J. S. Shim, Q. Chen, S. J. Lee, R. A. Anders, J. O. Liu and D. Pan, Genes Dev., 2012, 26, 1300–1305 CrossRef CAS PubMed.
- H. Zhang, S. K. Ramakrishnan, D. Triner, B. Centofanti, D. Maitra, B. Gyorffy, J. S. Sebolt-leopold, M. K. Dame, J. Varani, D. E. Brenner, E. R. Fearon, M. B. Omary and Y. M. Shah, Sci. Signaling, 2015, 8, 98 Search PubMed.
- A. N. Ahmed, E. Mohamed, D. Thoria and H. A. Aboubakr, Molecules, 2020, 25, 4780 CrossRef PubMed.
- S. K. Liew, M. Sharan, M. A. Norhafiza and H. N. Noor, Biomolecules, 2020, 10, 138 CrossRef CAS PubMed.
- Y. Ali, S. A. Hamid and R. Umer, Mini-Rev. Med. Chem., 2018, 18, 1548–1558 CrossRef CAS PubMed.
- W. M. Hamada, M. N. El-Nahass, A. A. Noser, T. A. Fayed, M. El-Kemary, M. M. Salem and E. Bakr, Sci. Rep., 2023, 13, 15420 CrossRef CAS PubMed.
- A. Noser, H. Mohamed, A. Saham, M. Rekaby and M. Maha, ChemistrySelect, 2023, 8, e202204670 CrossRef CAS.
- Y. Son, J. Kim, Y. Kim, S. G. Chi, T. Kim and J. Yu, Bioorg. Chem., 2023, 131, 106274 CrossRef CAS PubMed.
- H. A. Hekal, O. M. Hammad, N. R. El-Brollosy, M. M. Salem and A. K. Allayeh, Bioorg. Chem., 2024, 147, 107353 CrossRef CAS PubMed.
- M. N. El-Nahass, E. A. Bakr, T. A. Fayed, W. M. Hamada, M. M. Salem and A. M. Radwan, J. Iran. Chem. Soc., 2024, 21, 699–718 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and instruments used during the synthesis and the spectral data of the new compounds. See DOI: https://doi.org/10.1039/d5ra01233c |
| This journal is © The Royal Society of Chemistry 2025 |