Open Access Article
Open Access ArticleDesign and synthesis of terephthalic dihydrazide analogues as dual inhibitors of glycation and urease†
Hazrat Bilalab,
Saeed Ullahc,
Sobia Ahsan Halimc,
Momin Khan d,
Satya Kumar Avula
d,
Satya Kumar Avula c,
Aftab Alame,
Eman Serry Zayedf,
Sabah H. El-Ghaieshgj,
Hanan A. Ogaly
c,
Aftab Alame,
Eman Serry Zayedf,
Sabah H. El-Ghaieshgj,
Hanan A. Ogaly h,
Zarbad Shah
h,
Zarbad Shah *a,
Ajmal Khan*ci and
Ahmed Al-Harrasi
*a,
Ajmal Khan*ci and
Ahmed Al-Harrasi *c
*c
aDepartment of Chemistry, Bacha Khan University Charsadda, Charsadda-24420, Khyber Pakhtunkhwa, Pakistan. E-mail: zshej@hotmail.com
bDepartment of Chemistry, Government Postgraduate College Dargai Malakand, Pakistan
cNatural and Medical Sciences Research Center, University of Nizwa, PO Box 33, 616 Birkat Al Mauz, Nizwa, Sultanate of Oman. E-mail: ajmalkhan@unizwa.edu.om; aharrasi@unizwa.edu.om; Tel: +968-98957352 Tel: +968 25446328
dDepartment of Chemistry, Abdul Wali Khan University, Mardan, Mardan, 23200, Pakistan
eDepartment of Biochemistry, Abdul Wali Khan University, Mardan, Mardan, 23200, Pakistan
fDepartment of Clinical Biochemistry, Faculty of Medicine, University of Tabuk, Tabuk 71491, Saudi Arabia
gDepartment of Pharmacology, Faculty of Medicine, University of Tabuk, Tabuk 71491, Saudi Arabia
hChemistry Department, College of Science, King Khalid University, Abha 61421, Saudi Arabia
iDepartment of Chemical and Biological Engineering, College of Engineering, Korea University, Seongbuk-gu, 02841, Republic of Korea
jDepartment of Pharmacology, Faculty of Medicine, Tanta University, Tanta, Egypt
First published on 28th March 2025
Abstract
The overexpression of urease is the root cause of peptic ulcers and gastritis. Therefore, introducing new inhibitors against urease is a possible therapeutic approach to overcoming the pathogenesis; for instance, limiting the risk of development of urinary calculi. Moreover, glycation is the leading cause of several complications. Thus, in this study, we synthesized novel terephthalic dihydrazide analogues and evaluated their biological importance. These terephthalic dihydrazide analogues were characterized using advanced spectroscopic techniques, such as 1H NMR, 13C NMR, 19F NMR and HRMS (ESI+), and FT-IR. Fortunately, 6 of the 11 synthesized compounds exhibited urease inhibitory capability, and 8 compounds exhibited anti-glycation capability. Compounds 13–14, 20 and 23 showed significant urease inhibition with IC50 values of 63.12 ± 0.28, 65.71 ± 0.40, 49.2 ± 0.49 and 51.45 ± 0.39 μM, respectively. Meanwhile, they exhibited potent anti-glycation activity with IC50 values of 67.53 ± 0.46, 68.06 ± 0.43, 48.32 ± 0.42 and 54.36 ± 0.40 μM, respectively. Molecular docking of active urease inhibitors showed their good binding at the entrance of the active site and good correlation with our in vitro results.
1. Introduction
Compounds containing the azomethine (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–) functional group are called Schiff bases. They are produced when a carbonyl compound reacts with a primary amine. The aliphatic aldehydes involved in the formation of Schiff bases are comparatively more reactive and easily polymerized than the Schiff bases of aromatic aldehydes, which usually contain an active conjugated system. There are numerous applications of Schiff bases, such as the synthesis, identification, and recognition of carbonyl compounds, as well as the refinement of ketones, aldehydes and compounds containing amino groups. In addition, the above-mentioned groups can also be protected during the formation of complexes or some sensitive compounds. In certain dyes, this functional group is used as a basic unit.
N–) functional group are called Schiff bases. They are produced when a carbonyl compound reacts with a primary amine. The aliphatic aldehydes involved in the formation of Schiff bases are comparatively more reactive and easily polymerized than the Schiff bases of aromatic aldehydes, which usually contain an active conjugated system. There are numerous applications of Schiff bases, such as the synthesis, identification, and recognition of carbonyl compounds, as well as the refinement of ketones, aldehydes and compounds containing amino groups. In addition, the above-mentioned groups can also be protected during the formation of complexes or some sensitive compounds. In certain dyes, this functional group is used as a basic unit.
Schiff bases have bi-or tri-dentate ligands that can form highly stable transition-metal complexes. Many of these complexes are used as liquid crystals, which have numerous applications in electronic devices like displays and LCDs. The reactions of Schiff bases are favourable in organic synthesis to form a bond between carbon and nitrogen.1 The characteristic functional group of Schiff bases is the imine group (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–), which is significant to interpreting the mechanisms of their reactions like racemization and transamination in living organisms; they also have significant antitumor, antimicrobial, herbicidal and antifungal activities.2,3 Schiff bases have held an important position as ligands in transition metal coordination chemistry for nearly a century since their discovery.4
N–), which is significant to interpreting the mechanisms of their reactions like racemization and transamination in living organisms; they also have significant antitumor, antimicrobial, herbicidal and antifungal activities.2,3 Schiff bases have held an important position as ligands in transition metal coordination chemistry for nearly a century since their discovery.4
Schiff bases with hydroxyl substituents formed by the reaction of aniline and substituted aromatic aldehydes are reported for their biological importance.5,6 Their antioxidant effects on the stable galvinoxyl radical in methanol and ethyl ethanoate, as well as 2,2′-azobis (2-amidinopropane hydrochloride)-induced deoxyribonucleic acid (DNA) helix breakdown and its anti-proliferative effect on human hepatoma (HepG-2) cells, have also been checked. A structure–activity relationship study revealed that the o-dihydroxyl group on one benzene ring and the p-hydroxyl group present on the second benzene ring contribute chiefly to their antiproliferative and antioxidant activities.7
Some other Schiff bases like naphthalene-amine phenols and naphthalene-Schiff bases have been explored in the development of cheaper anti-malarial medicines to target CQ-resistant species.8 Among all the prepared compounds, only one exhibited an important biological activity with an inhibitory concentration (IC50) of 1.70 μM against the CQ-resistant Dd-2 species.8 Earlier, some terephthalic dihydrazide Schiff bases have been reported as poly (ADP-ribose) glycopyrrolate (PARG) inhibitors; the structures and activities of these Schiff bases are shown in Fig. 1 .2,3,7,9,10
As urease is a very common enzyme responsible for the hydrolysis of urea in living organisms to form carbon dioxide and ammonia,11,12 high concentrations of ammonia are obtained with urease hyperactivity, which leads to a rise in the pH of the stomach, thus resulting in complications like peptic and gastric ulcers,13 hepatic coma,14 pyelonephritis and kidney stones.15,16 Such clinical complications demand the use of inhibitors that can regulate the activity of urease.12,16–18 Studies have revealed that a variety of compounds, especially Schiff base hydrazones, can act as urease inhibitors.19,20 Urease inhibitors amplify urea N uptake in plants21 and hence reduce environmental issues.16,22–24 Additionally, they play a role as strong antiulcer drugs.25,26
Furthermore, we extended our study to obtain insights into the antiglycation capability of these derivatives. Therefore, we investigated the antiglycation activity. First, the reducing sugars react with the amino (-NH2) group of the proteins and form a reversible Schiff base, which upon reordering gives Amadori products. Then, the glycated proteins in the presence of transition metals and molecular oxygen give dicarbonyl intermediates, such as 3-deoxyglucosone (3-DG), glyoxal (GO) and methylglyoxal (MG), which regulate the process of glycation, leading to the second step or an advanced stage to produce AGEs as the end products.27,28 Several glycation inhibitors have been used in the past, including flavonoids, aspirin, vitamin B6, and aminoguanidine. Many of them have shown good results when explored as potential therapeutic targets.29
Urease inhibition and anti-glycation activity are linked to their effects on inflammation and oxidative stress, which contribute to aging, diabetes, and metabolic diseases.30–33 Urease inhibitors like thiourea block urea breakdown, and aid the treatment of H. pylori infections, kidney stones, and UTIs.34 Glycation is the process in which sugars form advanced glycation end-products (AGEs), which damage tissues in conditions like diabetes.35 Urease inhibition reduces ammonia production and lowers oxidative stress and inflammation, which may decrease AGE formation.36–38 This can improve metabolic balance in diabetics, protect kidneys from damage, and reduce AGE accumulation, thus helping manage diabetic nephropathy and chronic infections. Overall, urease inhibitors may lower inflammation and oxidative stress, thereby reducing AGE formation and managing diabetes and kidney disease complications.
The structural motifs of terephthalic dihydrazide and its derivatives are ideal for interaction with important biological targets related to urease and glycation activities. It is well-known that the hydrazide functionality can interrupt the glycation process by forming stable hydrazone derivatives with reducing sugars.39 Furthermore, the rigid aromatic core of terephthalic acid can enhance urease activity through further modifications. Terephthalic dihydrazide is a perfect option for dual inhibition because of its bifunctionality, which targets both urease and glycation pathways.
The current study focuses on the synthesis of novel terephthalic dihydrazide analogues to gain insights into their medicinal importance by evaluating their urease and glycation inhibitory performance.
2. Methodology
2.1. Chemicals used
All experiments were carried out in dry reaction vessels under a dry nitrogen atmosphere. All reagents were purchased from Sigma-Aldrich, Germany. The solvents were purified and dried using standard procedures before being used in experiments. All reactions were monitored by Thin-Layer-Chromatography (TLC) using silica gel F254. Visualization was accomplished using UV light and I2 staining. The solvents used for the catalytic reaction were of technical grade and dried using standard procedures. The solvents used for column chromatography (EtOAc, n-hexane) were of technical grade and distilled prior to use. Organic extracts were dried over anhydrous MgSO4.2.2. Instrumentation
The mass spectra were recorded using the following HR-ESI-MS instrument: Agilent Technologies, 6530 Accurate Mass. The 1H and 13C NMR spectra were recorded on 600 MHz and 150 MHz spectrometers using the solvent peak as the internal reference (CDCl3, δ H: 7.26; δ C: 77.0). The 19F NMR spectra were recorded on a 564 MHz spectrometer. Data are reported in the following order: chemical shift (δ) in ppm; multiplicities are indicated as m = multiplet; q = quadrate, dd = doublet of doublet, t = triplet, d = doublet, s = singlet; coupling constants (J) are in hertz (Hz). The HPLC analysis was performed with a variable wavelength detector. Column chromatography was carried out by using silica gel of selected particle size (100–200 mesh). The HRMS (ESI+) spectra of the prepared products were recorded on a Waters Quattro Premier XE mass spectrometer (Waters, Milford, MA).The melting points of all the prepared compounds were determined on a STUART melting point SMP-10; the UV spectra were recorded on a Shimadzu (UV-1800) UV-visible spectrophotometer with a version 2.32 UV probe. The ATR-FTIR data were recorded on a PerkinElmer Spectrum version 10.4.00.
2.3. General experimental procedures
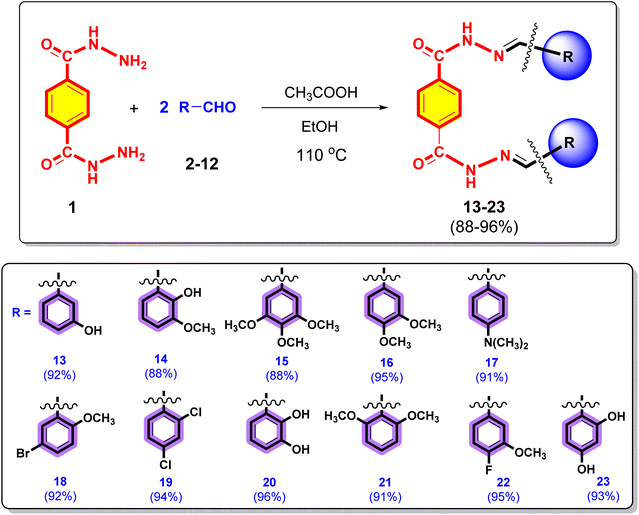
In a 150 mL conical flask, terephthalic dihydrazide (1) (1.0 equiv.) and different commercially available substituted aromatic aldehydes (2–12) (2.0 equiv.) were reacted in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio. Then, 20 mL of EtOH was added as the solvent. The reaction was then catalysed by adding two to three drops of ethanoic acid. The reaction mixture was refluxed at 110 °C for 3–4 h until the completion of the reaction (monitored by TLC analysis). After the completion of the reaction, the product was extracted with EtOAc (3 times). The combined organic layer was dried over anhydrous MgSO4 and concentrated under reduced pressure on a rotary evaporator to obtain the crude product, which was purified by flash column chromatography to obtain several target terephthalic dihydrazide derivatives (13–23) in their pure form and high yields (88–96%). The purity of the terephthalic dihydrazide analogues was characterized by advanced spectroscopic techniques, such as 1H NMR, 13C NMR, 19F NMR and HRMS (ESI+).
2 ratio. Then, 20 mL of EtOH was added as the solvent. The reaction was then catalysed by adding two to three drops of ethanoic acid. The reaction mixture was refluxed at 110 °C for 3–4 h until the completion of the reaction (monitored by TLC analysis). After the completion of the reaction, the product was extracted with EtOAc (3 times). The combined organic layer was dried over anhydrous MgSO4 and concentrated under reduced pressure on a rotary evaporator to obtain the crude product, which was purified by flash column chromatography to obtain several target terephthalic dihydrazide derivatives (13–23) in their pure form and high yields (88–96%). The purity of the terephthalic dihydrazide analogues was characterized by advanced spectroscopic techniques, such as 1H NMR, 13C NMR, 19F NMR and HRMS (ESI+).
White solid powder; yield = 92% (360 mg); m.p. 293–295 °C; UV (λmax): 309 nm; IR νmax (cm−1): 3467 (O–H), 3248 (N–H), 3100 (–C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H), 1631 (C
C–H), 1631 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1612 (C
O), 1612 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1586 (Ar–C
N), 1586 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1H NMR: (600 MHz, d6-DMSO) δ 11.92 (s, 2H, NH, amide), 9.66 (s, 2H, OH, phenolic), 8.38 (s, 2H, CH, 7), 8.04 (s, 4H, CH, 2/3/5/6), 7.26 (t, 2H, J = 7.8 Hz, CH, 6′), 7.21 (s, 2H, CH, 5′), 7.11 (s, 2H, CH, 2′), 6.87–6.81 (m, 2H, CH, 4′); 13C NMR: (150 MHz, d6-DMSO): δ 165.63, 162.85, 158.17, 148.91, 136.58, 135.96, 130.41, 128.25, 127.42, 119.40, 118.08, 113.16; HRMS (ESI+) calcd for C22H19N4O4 [M + H]+ 403.1368; found 403.1366. Elemental analysis: C22H18N4O4 calcd: C, 65.68; H, 4.53; N, 13.89; O, 13.90; found: C, 65.66; H, 4.51; N, 13.92; O, 15.90.
C), 1H NMR: (600 MHz, d6-DMSO) δ 11.92 (s, 2H, NH, amide), 9.66 (s, 2H, OH, phenolic), 8.38 (s, 2H, CH, 7), 8.04 (s, 4H, CH, 2/3/5/6), 7.26 (t, 2H, J = 7.8 Hz, CH, 6′), 7.21 (s, 2H, CH, 5′), 7.11 (s, 2H, CH, 2′), 6.87–6.81 (m, 2H, CH, 4′); 13C NMR: (150 MHz, d6-DMSO): δ 165.63, 162.85, 158.17, 148.91, 136.58, 135.96, 130.41, 128.25, 127.42, 119.40, 118.08, 113.16; HRMS (ESI+) calcd for C22H19N4O4 [M + H]+ 403.1368; found 403.1366. Elemental analysis: C22H18N4O4 calcd: C, 65.68; H, 4.53; N, 13.89; O, 13.90; found: C, 65.66; H, 4.51; N, 13.92; O, 15.90.Off-white solid powder; yield = 88% (390 mg); m.p. 294–297 °C, UV (λmax): 318 nm, IR νmax (cm−1): 3564 (O–H), 3391 (N–H), 3178 (Ar–C–H), 3003 (Al–C–H), 1626 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1604 (C
O), 1604 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1574 (Ar–C
N), 1574 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1H NMR: (600 MHz, d6-DMSO): δ 12.19 (s, 2H, OH, phenolic), 10.85 (s, 2H, NH, amide), 8.68 (s, 2H, CH, 7), 8.07 (s, 4H, CH, 2/3/5/6), 7.17 (d, 2H, J = 7.8 Hz, CH, 6′), 7.04 (d, 2H, J = 8.0 Hz, CH, 5′), 6.87 (t, 2H, J = 7.9 Hz, CH, 4′), 3.81 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.57, 148.94, 148.44, 147.64, 136.22, 128.33, 121.12, 119.59, 119.43, 114.37, 56.31; HRMS (ESI+) calcd for C24H23N4O6 [M + H]+ 463.1539; found 463.1542. Elemental analysis: C24H22N4O6 calcd: C, 62.35; H, 4.78; N, 12.10; O, 20.78; found: C, 62.33; H, 4.80; N, 12.12; O, 20.76.
C), 1H NMR: (600 MHz, d6-DMSO): δ 12.19 (s, 2H, OH, phenolic), 10.85 (s, 2H, NH, amide), 8.68 (s, 2H, CH, 7), 8.07 (s, 4H, CH, 2/3/5/6), 7.17 (d, 2H, J = 7.8 Hz, CH, 6′), 7.04 (d, 2H, J = 8.0 Hz, CH, 5′), 6.87 (t, 2H, J = 7.9 Hz, CH, 4′), 3.81 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.57, 148.94, 148.44, 147.64, 136.22, 128.33, 121.12, 119.59, 119.43, 114.37, 56.31; HRMS (ESI+) calcd for C24H23N4O6 [M + H]+ 463.1539; found 463.1542. Elemental analysis: C24H22N4O6 calcd: C, 62.35; H, 4.78; N, 12.10; O, 20.78; found: C, 62.33; H, 4.80; N, 12.12; O, 20.76.Off-white solid powder; yield = 88% (459 mg); m.p. more than 300 °C, UV (λmax): 328 nm, IR νmax (cm−1): 3222 (N–H), 2942 (Ar–C–H), 2841 (Al–C–H), 1649 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1607 (C
O), 1607 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1575 (Ar–C
N), 1575 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 11.96 (s, 2H, NH, amide), 8.40 (s, 2H, CH, 7), 8.03 (s, 4H, CH, 2/3/5/6), 7.04 (s, 4H, CH, 2′/6′), 3.84 (s, 12H, CH3, methoxy), 3.71 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.93, 153.70, 148.87, 139.83, 136.63, 130.19, 128.26, 104.89, 60.62, 56.46; HRMS (ESI+) calcd for C28H31N4O8 [M + H]+ 551.1783; found 551.1780. Elemental analysis: C28H30N4O8 calcd: C, 61.10; H, 5.52; N, 10.20; O, 23.23; found: C, 61.08; H, 5.49; N, 10.18; O, 23.25.
C); 1H NMR: (600 MHz, d6-DMSO): δ 11.96 (s, 2H, NH, amide), 8.40 (s, 2H, CH, 7), 8.03 (s, 4H, CH, 2/3/5/6), 7.04 (s, 4H, CH, 2′/6′), 3.84 (s, 12H, CH3, methoxy), 3.71 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.93, 153.70, 148.87, 139.83, 136.63, 130.19, 128.26, 104.89, 60.62, 56.46; HRMS (ESI+) calcd for C28H31N4O8 [M + H]+ 551.1783; found 551.1780. Elemental analysis: C28H30N4O8 calcd: C, 61.10; H, 5.52; N, 10.20; O, 23.23; found: C, 61.08; H, 5.49; N, 10.18; O, 23.25.Off-white Solid powder; yield = 95% (465 mg); m.p. 283–285 °C, UV (λmax): 324 nm, IR νmax (cm−1): 3237 (N–H), 3077 (Ar–C–H), 2839 (Al–C–H), 1644 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1601 (C
O), 1601 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1577 (Ar–C
N), 1577 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 11.86 (s, 2H, NH, amide), 9.86 (s, 2H, CH, 7), 8.02 (s, 4H, CH, 2/3/5/6), 7.36 (s, 2H, CH, 6′), 7.22 (d, 2H, J = 8.2 Hz, CH, 2′), 7.03 (d, 2H, J = 8.2 Hz, CH, 5′), 3.81 (d, 12H, J = 9.7 Hz, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 165.62, 162.75, 151.35, 149.57, 149.06, 136.65, 135.93, 128.19, 127.42, 122.52, 111.97, 108.75, 56.06, 55.95; HRMS (ESI+) calcd for C26H27N4O6 [M + H]+ 491.2011; found 491.2014. Elemental analysis: C26H26N4O6 calcd: C, 63.68; H, 5.31; N, 11.40; O, 19.54; found: C, 63.66; H, 5.34; N, 11.42; O, 19.57.
C); 1H NMR: (600 MHz, d6-DMSO): δ 11.86 (s, 2H, NH, amide), 9.86 (s, 2H, CH, 7), 8.02 (s, 4H, CH, 2/3/5/6), 7.36 (s, 2H, CH, 6′), 7.22 (d, 2H, J = 8.2 Hz, CH, 2′), 7.03 (d, 2H, J = 8.2 Hz, CH, 5′), 3.81 (d, 12H, J = 9.7 Hz, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 165.62, 162.75, 151.35, 149.57, 149.06, 136.65, 135.93, 128.19, 127.42, 122.52, 111.97, 108.75, 56.06, 55.95; HRMS (ESI+) calcd for C26H27N4O6 [M + H]+ 491.2011; found 491.2014. Elemental analysis: C26H26N4O6 calcd: C, 63.68; H, 5.31; N, 11.40; O, 19.54; found: C, 63.66; H, 5.34; N, 11.42; O, 19.57.Buttery solid powder; yield = 91% (413 mg); m.p. more than 300 °C, UV (λmax): 319 nm, IR νmax (cm−1): 3233 (N–H), 3081 (Ar–C–H), 2900 (Al–C–H), 1642 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1608 (C
O), 1608 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1594 (Ar–C
N), 1594 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 11.65 (s, 2H, NH, amide), 8.32 (s, 2H, CH, 7), 8.00 (s, 4H, CH, 2/3/5/6), 7.55 (d, 4H, J = 8.3 Hz, CH, 3′/5′), 6.76 (d, 4H, J = 8.4 Hz, CH, 2′/6′), 2.98 (s, 12H, CH3, methyl); 13C NMR: (150 MHz, d6-DMSO): δ 162.42, 152.09, 149.60, 136.70, 129.02, 128.05, 127.48, 121.92, 112.29, 48.92; HRMS (ESI+) calcd for C26H29N6O2 [M + H]+ 457.2083; found 457.2080. Elemental analysis: C26H28N6O2 calcd: C, 68.43; H, 6.20; N, 18.39; O, 7.03; found: C, 68.40; H, 6.18; N, 18.41; O, 7.01.
C); 1H NMR: (600 MHz, d6-DMSO): δ 11.65 (s, 2H, NH, amide), 8.32 (s, 2H, CH, 7), 8.00 (s, 4H, CH, 2/3/5/6), 7.55 (d, 4H, J = 8.3 Hz, CH, 3′/5′), 6.76 (d, 4H, J = 8.4 Hz, CH, 2′/6′), 2.98 (s, 12H, CH3, methyl); 13C NMR: (150 MHz, d6-DMSO): δ 162.42, 152.09, 149.60, 136.70, 129.02, 128.05, 127.48, 121.92, 112.29, 48.92; HRMS (ESI+) calcd for C26H29N6O2 [M + H]+ 457.2083; found 457.2080. Elemental analysis: C26H28N6O2 calcd: C, 68.43; H, 6.20; N, 18.39; O, 7.03; found: C, 68.40; H, 6.18; N, 18.41; O, 7.01.White solid powder; yield = 92% (518 mg); m.p. 300 °C, UV (λmax): 312 nm, IR νmax (cm−1): 3550 (N–H), 3288 (Ar–C–H), 2835 (Al–C–H), 1652 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1601 (C
O), 1601 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1553 (Ar–C–H); 1H NMR: (600 MHz, d6-DMSO): δ 12.08 (s, 2H, NH, amide), 9.85 (s, 2H, CH, 7), 7.97–7.82 (m, 4H, CH, 2/3/5/6), 7.11 (d, 2H, J = 8.9 Hz, CH, 6′), 5.74 (s, 2H, CH, 4′), 4.51 (s, 2H, CH, 3′), 3.87 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 165.61, 142.54, 139.67, 136.39, 135.92, 128.29, 127.96, 127.41, 114.98, 112.99, 56.63; HRMS (ESI+) calcd for C24H2179Br2N4O4 [M + H]+ 589.0448 found 589.0450. HRMS (ESI+) calcd for C24H2181Br2N4O4 [M + H]+ 591.0368; found 591.0365. Elemental analysis: C24H20Br2N4O4 calcd: C, 49.02; H, 3.40; N, 9.54; O, 10.86; found: C, 49.00; H, 3.43; Br, 27.17; N, 9.52; O, 10.88.
N), 1553 (Ar–C–H); 1H NMR: (600 MHz, d6-DMSO): δ 12.08 (s, 2H, NH, amide), 9.85 (s, 2H, CH, 7), 7.97–7.82 (m, 4H, CH, 2/3/5/6), 7.11 (d, 2H, J = 8.9 Hz, CH, 6′), 5.74 (s, 2H, CH, 4′), 4.51 (s, 2H, CH, 3′), 3.87 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 165.61, 142.54, 139.67, 136.39, 135.92, 128.29, 127.96, 127.41, 114.98, 112.99, 56.63; HRMS (ESI+) calcd for C24H2179Br2N4O4 [M + H]+ 589.0448 found 589.0450. HRMS (ESI+) calcd for C24H2181Br2N4O4 [M + H]+ 591.0368; found 591.0365. Elemental analysis: C24H20Br2N4O4 calcd: C, 49.02; H, 3.40; N, 9.54; O, 10.86; found: C, 49.00; H, 3.43; Br, 27.17; N, 9.52; O, 10.88.Off-white solid powder; yield = 94% (476 mg); m.p. more than 300 °C, UV (λmax): 306 nm, IR νmax (cm−1): 3182 (N–H), 3045 (Ar–C–H), 1646 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 (C
O), 1587 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1551 (Ar–C
N), 1551 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 12.26 (s, 2H, NH, amide), 8.84 (s, 2H, CH, 7), 8.08 (s, 4H, CH, 2/3/5/6), 8.04 (d, 2H, J = 8.5 Hz, CH, 6′), 7.74 (s, 2H, CH, 3′), 7.54 (d, 2H, J = 8.7 Hz, CH, 5′); 13C NMR: (150 MHz, d6-DMSO): δ 162.93, 154.07, 152.42, 148.00, 136.58, 131.71, 128.28, 127.41, 121.56, 116.83, 111.37; HRMS (ESI+) calcd for C22H15Cl4N4O2 [M + H]+ 509.0202; found 509.0205. Elemental analysis: C22H14Cl4N4O2 calcd: C, 52.02; H, 2.81; N, 11.01; O, 6.28; found: C, 52.00; H, 2.78; Cl, 27.90; N, 11.03; O, 6.30.
C); 1H NMR: (600 MHz, d6-DMSO): δ 12.26 (s, 2H, NH, amide), 8.84 (s, 2H, CH, 7), 8.08 (s, 4H, CH, 2/3/5/6), 8.04 (d, 2H, J = 8.5 Hz, CH, 6′), 7.74 (s, 2H, CH, 3′), 7.54 (d, 2H, J = 8.7 Hz, CH, 5′); 13C NMR: (150 MHz, d6-DMSO): δ 162.93, 154.07, 152.42, 148.00, 136.58, 131.71, 128.28, 127.41, 121.56, 116.83, 111.37; HRMS (ESI+) calcd for C22H15Cl4N4O2 [M + H]+ 509.0202; found 509.0205. Elemental analysis: C22H14Cl4N4O2 calcd: C, 52.02; H, 2.81; N, 11.01; O, 6.28; found: C, 52.00; H, 2.78; Cl, 27.90; N, 11.03; O, 6.30.White solid powder; yield = 419 mg (96%); m.p. more than 300 °C, UV (λmax): 316 nm, IR νmax (cm−1): 3230 (O–H), 3180 (N–H), 2900 (Ar–C–H), 1630 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1616 (C
O), 1616 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1579 (Ar–C
N), 1579 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H-NMR: (600 MHz, d6-DMSO): δ 12.23 (s, 2H, OH, phenolic), 11.03 (s, 2H, NH, amide), 9.27 (s, 2H, OH, phenolic), 8.62 (s, 2H, CH, 7), 8.08 (s, 4H, CH, 2/3/5/6), 6.99 (d, 2H, J = 7.8 Hz, CH, 6′), 6.86 (d, 2H, J = 7.8 Hz, CH, 4′), 6.75 (t, 2H, J = 7.8 Hz, CH, 5′); 13C NMR: (150 MHz, d6-DMSO): δ 165.46, 162.55, 149.84, 146.63, 146.09, 136.70, 136.17, 135.39, 128.35, 128.17, 127.62, 120.45, 119.69, 119.26, 117.97; HRMS (ESI+) calcd for C22H19N4O6 [M + H]+ 435.0978; found 435.0976. Elemental analysis: C22H18N4O6 calcd: C, 60.81; H, 4.16; N, 12.93; O, 22.14; found: C, 60.83; H, 4.18; N, 12.90; O, 22.10.
C); 1H-NMR: (600 MHz, d6-DMSO): δ 12.23 (s, 2H, OH, phenolic), 11.03 (s, 2H, NH, amide), 9.27 (s, 2H, OH, phenolic), 8.62 (s, 2H, CH, 7), 8.08 (s, 4H, CH, 2/3/5/6), 6.99 (d, 2H, J = 7.8 Hz, CH, 6′), 6.86 (d, 2H, J = 7.8 Hz, CH, 4′), 6.75 (t, 2H, J = 7.8 Hz, CH, 5′); 13C NMR: (150 MHz, d6-DMSO): δ 165.46, 162.55, 149.84, 146.63, 146.09, 136.70, 136.17, 135.39, 128.35, 128.17, 127.62, 120.45, 119.69, 119.26, 117.97; HRMS (ESI+) calcd for C22H19N4O6 [M + H]+ 435.0978; found 435.0976. Elemental analysis: C22H18N4O6 calcd: C, 60.81; H, 4.16; N, 12.93; O, 22.14; found: C, 60.83; H, 4.18; N, 12.90; O, 22.10.White solid powder; yield = 91% (445 mg); m.p. 295 °C, UV (λmax): 320 nm, IR νmax (cm−1): 3209 (N–H), 3100 (Ar–C–H), 2834 (Al–C–H), 1645 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1606 (C
O), 1606 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1593 (Ar–C
N), 1593 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 11.76 (s, 2H, NH, amide), 8.63 (s, 2H, CH, 7), 8.02 (s, 4H, CH, 2/3/5/6), 7.35 (t, 2H, J = 8.4 Hz, CH, 4′), 6.71 (dd, 4H, J = 17.9, CH, 3′/5′), 3.81 (s, 12H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.57, 159.24, 159.13, 144.48, 136.66, 131.82, 130.66, 128.15, 127.14, 111.45, 104.90, 56.46; HRMS (ESI+) calcd for C26H27N4O6 [M + H]+ 491.1733; found 491.1731. Elemental analysis: C26H26N4O6 calcd: C, 63.68; H, 5.32; N, 11.45; O, 19.60; found: C, 63.66; H, 5.34; N, 11.42; O, 19.57.
C); 1H NMR: (600 MHz, d6-DMSO): δ 11.76 (s, 2H, NH, amide), 8.63 (s, 2H, CH, 7), 8.02 (s, 4H, CH, 2/3/5/6), 7.35 (t, 2H, J = 8.4 Hz, CH, 4′), 6.71 (dd, 4H, J = 17.9, CH, 3′/5′), 3.81 (s, 12H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.57, 159.24, 159.13, 144.48, 136.66, 131.82, 130.66, 128.15, 127.14, 111.45, 104.90, 56.46; HRMS (ESI+) calcd for C26H27N4O6 [M + H]+ 491.1733; found 491.1731. Elemental analysis: C26H26N4O6 calcd: C, 63.68; H, 5.32; N, 11.45; O, 19.60; found: C, 63.66; H, 5.34; N, 11.42; O, 19.57.White solid powder; yield = 95% (439 mg); m.p. more than 300 °C, UV (λmax): 313 nm, IR νmax (cm−1): 3228 (N–H), 3082 (Ar–C–H), 2986 (Al–C–H), 1644 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1613 (C
O), 1613 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1590 (Ar–C
N), 1590 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 11.99 (s, 2H, NH, amide), 8.45 (s, 2H, CH, 7), 8.04 (s, 4H, CH, 5′/6′), 7.52 (d, 2H, J = 8.3 Hz, CH, 2′), 7.35–7.27 (m, 4H, CH, 2/3/5/6), 3.91 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.93, 154.07, 152.42, 148.11, 148.00, 136.58, 131.71, 128.28, 121.51, 116.83, 116.70, 111.37, 56.50; 19F NMR (564 MHz, d6-DMSO) δ −58.84; HRMS (ESI+) calcd for C24H20F2N4O4Na [M + Na]+ 489.1776; found 489.1778. Elemental analysis: C24H20F2N4O4 calcd: C, 61.82; H, 4.30; N, 12.03; O, 13.75; found: C, 61.80; H, 4.32; N, 12.01; O, 13.72.
C); 1H NMR: (600 MHz, d6-DMSO): δ 11.99 (s, 2H, NH, amide), 8.45 (s, 2H, CH, 7), 8.04 (s, 4H, CH, 5′/6′), 7.52 (d, 2H, J = 8.3 Hz, CH, 2′), 7.35–7.27 (m, 4H, CH, 2/3/5/6), 3.91 (s, 6H, CH3, methoxy); 13C NMR: (150 MHz, d6-DMSO): δ 162.93, 154.07, 152.42, 148.11, 148.00, 136.58, 131.71, 128.28, 121.51, 116.83, 116.70, 111.37, 56.50; 19F NMR (564 MHz, d6-DMSO) δ −58.84; HRMS (ESI+) calcd for C24H20F2N4O4Na [M + Na]+ 489.1776; found 489.1778. Elemental analysis: C24H20F2N4O4 calcd: C, 61.82; H, 4.30; N, 12.03; O, 13.75; found: C, 61.80; H, 4.32; N, 12.01; O, 13.72.Yellow solid powder; yield = 93% (401 mg); m.p. 297 °C, UV (λmax): 320 nm, IR νmax (cm−1): 3632 (O–H), 3507 (N–H), 3191 (Ar–C–H), 3056 (Al–C–H), 1651 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1625 (C
O), 1625 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1586 (Ar–C
N), 1586 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 1H NMR: (600 MHz, d6-DMSO): δ 12.02 (s, 2H, OH, phenolic), 11.39 (s, 2H, NH, amide), 9.97 (s, 2H, CH, 7), 8.53 (s, 2H, CH, 3′), 8.04 (s, 4H, CH, 2/3/5/6), 7.33 (d, 2H, J = 8.4 Hz, CH, 6′), 6.36 (dd, 2H, J = 8.4 Hz, CH, 5′); 6.32 (t, 2H, J = 1.7 Hz, OH, phenolic); 13C NMR: (150 MHz, d6-DMSO): δ 162.26, 161.34, 160.00, 150.02, 136.22, 131.80, 128.21, 110.98, 108.26, 103.14; HRMS (ESI+) calcd for C22H19N4O6 [M + H]+ 435.0988; found 435.0985. Elemental analysis: C22H18N4O6 calcd: C, 60.85; H, 4.21; N, 12.93; O, 22.13; found: C, 60.83; H, 4.18; N, 12.90; O, 22.10.
C); 1H NMR: (600 MHz, d6-DMSO): δ 12.02 (s, 2H, OH, phenolic), 11.39 (s, 2H, NH, amide), 9.97 (s, 2H, CH, 7), 8.53 (s, 2H, CH, 3′), 8.04 (s, 4H, CH, 2/3/5/6), 7.33 (d, 2H, J = 8.4 Hz, CH, 6′), 6.36 (dd, 2H, J = 8.4 Hz, CH, 5′); 6.32 (t, 2H, J = 1.7 Hz, OH, phenolic); 13C NMR: (150 MHz, d6-DMSO): δ 162.26, 161.34, 160.00, 150.02, 136.22, 131.80, 128.21, 110.98, 108.26, 103.14; HRMS (ESI+) calcd for C22H19N4O6 [M + H]+ 435.0988; found 435.0985. Elemental analysis: C22H18N4O6 calcd: C, 60.85; H, 4.21; N, 12.93; O, 22.13; found: C, 60.83; H, 4.18; N, 12.90; O, 22.10.2.4. In vitro urease inhibition assay
The urease inhibition capability of all compounds was determined using our previously reported procedure.40 Initially, 25 μL of urease enzyme from Canavalia ensiformis (Jack bean), 5 μL of the synthesised compounds at different concentrations and 55 μL of urea (100 mM) were incubated for 15 mints at 30 °C in 96-well plates. To each vial, 45 μL of a phenolic reagent (A) and 70 μL of an alkali reagent (B) were added, making up a total volume of 200 μL. The phenolic reagent (A) was a combination of 1% w/v phenol and 0.005% w/v sodium nitroprusside. The alkali reagent (B) was composed of 0.5% w/v NaOH and 0.1% w/v NaOCl. To investigate the urease inhibition action of the new derivatives, the method published by Weatherburn based on the release of ammonia after hydrolysis was used. The absorbance values were recorded before and after 50 minutes using a microplate reader (xMark™ Microplate Spectrophotometer, BIO-RAD) to calculate the difference. Each reaction was performed in triplicate using a final volume of 200 μL. Thiourea was used as the standard urease inhibitor.412.5. Statistical analysis
To analyse the obtained results and determine biological activity, the SoftMax Pro package and Microsoft Excel were utilized. The below formula (eqn (1)) was used to calculate percentage inhibition.
 | (1) |
EZ-FIT (Perrella Scientific, Inc., USA) was used to calculate the IC50 values of all compounds. To avoid expected errors, all experiments were performed in triplicate, and variations in the results are reported as Standard Error of Mean values (SEM) (calculated using eqn (2)).
 | (2) |
2.6. In vitro anti-glycation assay
A buffer of sodium phosphate (pH = 7.4) was prepared by reacting NaH2PO4 and Na2HPO4 to obtain a final concentration of 67 mM and 3 mM sodium azide. Phosphate buffer saline (PBS) was prepared through the reaction of Na2HPO4 (8.1 mM), NaCl (137 mM), KH2PO4 (1.47 mM) and KCl (2.68 mM), while pH was adjusted to 10 using NaOH (0.25 mM). Anhydrous glucose (50 mg mL−1) and BSA (10 mg mL−1) solutions were prepared using the sodium phosphate buffer. The samples were dissolved in DMSO (1 mM mL−1) for testing. In the 96-well-plate activity assay, each well contained 60 μL of the reaction mixture (20 μL of anhydrous glucose (50 mg mL−1), 20 μL BSA (10 mg mL−1), and 20 μL of the test sample). The glycation control contained 20 mL glucose, 20 mL BSA and 20 mL buffer of sodium phosphate (Na2SO4), while in the blank control, 40 mL sodium phosphate buffer and 20 mL BSA were present. Then, the reaction mixture was incubated for seven days at 37 °C. After that, 6 μL of TCA (100%) was added to all wells, and the plate was centrifugated at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for five minutes at 4 °C. Then, the obtained pellets were washed with 50 μL TCA (5%). The upper layer that contained the standard inhibitors, glucose and interfering substances was removed, and the pellet containing AGE-BSA was redissolved in 60 mL of PBS in each cell. The fluorescence spectra (ex. 370 nm) and changes in the intensity of fluorescence (360–440 nm) based on AGE synthesis were recorded using a Spectro-fluorimeter (RF-1500, Shimadzu, Japan). The percentage inhibition was then determined using the formula:
000 rpm for five minutes at 4 °C. Then, the obtained pellets were washed with 50 μL TCA (5%). The upper layer that contained the standard inhibitors, glucose and interfering substances was removed, and the pellet containing AGE-BSA was redissolved in 60 mL of PBS in each cell. The fluorescence spectra (ex. 370 nm) and changes in the intensity of fluorescence (360–440 nm) based on AGE synthesis were recorded using a Spectro-fluorimeter (RF-1500, Shimadzu, Japan). The percentage inhibition was then determined using the formula:| % Inhibition = (1 − fluorescence of test sample/fluorescence of glycated sample) × 100 |
2.7. Molecular docking of urease inhibitors
The active urease inhibitors found in this study were docked into the binding site of jack bean urease by MOE (Molecular Operating Environment version 2022.02).42 The structure of jack bean urease (PDB code: 4H9M) was obtained from the Protein Databank and optimized using the QuickPrep module of MOE with the addition of hydrogen atoms and partial charges via OPLSAA forcefield. The nickel atoms in the active site were assigned +2 charge with default parameters (mass = 58.6930, q = 2.0, R = 1.4170, Eps = 0.1225, m = 12 and n = 6). After preparing the protein structure, the docking capacity of MOE was analysed by re-docking thiourea into its binding site using the Alpha triangle docking algorithm of MOE with a cutoff of 100 docked conformations scored by the London dG scoring function.During re-docking, thiourea produced almost a similar binding pattern to its X-ray crystal structure (4H9M) with an RMSD of 1.19 Å compared to its X-ray conformation and a score of −12.20 kcal mol−1. The re-docking results (Fig. S1†) showed that the given docking parameters can allow the docking of our compounds, thus we applied Alpha triangle along with the London dG scoring function for docking our urease inhibitors.
The structures of the active urease inhibitors were generated by Chem Draw and converted into their 3D form using MOE with energy minimization (RMS gradient = 0.5 kcal mol−1 Å−1, charges = AM1-BCC). Afterwards, docking was conducted with a cutoff of 50 solutions, and the docked conformations were analysed on the MOE interface.
3. Results and discussion
3.1. Synthesis of terephthalic dihydrazide analogues (13–23)
The synthesis of terephthalic dihydrazide analogues (13–23) is depicted in the Scheme 1. Terephthalic dihydrazide (1) was reacted with different commercially available substituted aromatic aldehydes (2–12) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, and EtOH was used as the solvent. The reaction mixture was then catalysed by adding 2 or 3 drops of ethanoic acid and refluxed at 110 °C for 4 h to produce the target terephthalic dihydrazide derivatives (13–23) with high yields (88–96%).
2 ratio, and EtOH was used as the solvent. The reaction mixture was then catalysed by adding 2 or 3 drops of ethanoic acid and refluxed at 110 °C for 4 h to produce the target terephthalic dihydrazide derivatives (13–23) with high yields (88–96%).
By analysing their spectral data obtained using UV, ATR-FTIR, 1H NMR, HRMS (ESI+) spectroscopy techniques and elemental analysis, the structures of all newly synthesized compounds 13–23 were confirmed.
3.2. Biological evaluation
The general structural features of the synthesized terephthalic dihydrazide analogues (13–23) are shown in Fig. 2. A rigid motif of the terephthalic dihydrazide base group is attached to two variable moieties (different aromatic groups), which decide the varied degree of activity.All the synthesized compounds 13–23 were screened for urease inhibitory function. Among them, six compounds exhibited urease inhibition potential, with IC50 values in the range of 49.20–81.74 μM, while comparing our results with the available standard inhibitor thiourea (IC50 = 21.00 ± 0.28 μM) (Table 1). Compounds 20 (IC50 = 49.20 ± 0.28 μM) and 23 (IC50 = 51.45 ± 0.39 μM) exhibited almost similar urease inhibition performance and were the most active compounds. This inhibitory effect might be due to the better binding of the hydroxyl groups found in both compounds to the enzyme pocket.
| Compound | IC50 (μM ± SEM) | Compound | IC50 (μM ± SEM) |
|---|---|---|---|
| a SEM = standard error of the mean, NA = not active; Thiourea is the standard inhibitor used for the urease activity assay. | |||
| 13 | 63.12 ± 0.28 | 19 | NA |
| 14 | 65.71 ± 0.40 | 20 | 49.2 ± 0.49 |
| 15 | 74.42 ± 0.57 | 21 | NA |
| 16 | 81.74 ± 0.60 | 22 | NA |
| 17 | NA | 23 | 51.45 ± 0.39 |
| 18 | NA | Thiourea | 21.00 ± 0.28 |
Compounds 13 (IC50 = 63.12 ± 0.28 μM) and 14 (IC50 = 65.71 ± 0.40 μM) also exhibited good inhibitory capability with almost similar potency. Compound 15 exhibited slightly lower urease inhibitory capability (IC50 = 74.42 ± 0.57 μM) than the other compounds. Similarly, the urease inhibitory effect of compound 16 was also decreased (IC50 = 81.74 ± 0.60 μM). The overall structure–activity relationship demonstrates that hydroxyl groups might be involved in the urease inhibition.
Compounds 13–23 were also evaluated for their antiglycation capability. Interestingly, eight compounds exhibited anti-glycation performance in the range of 48.32–652.06 μM. Compounds 13 and 14 with hydroxyl and methoxy substituents exhibited similar anti-glycation capability, with IC50 values 67.53 ± 0.46 and 68.06 ± 0.43 μM, respectively, which are comparable to the standard rutin (IC50 = 70.00 ± 0.50 μM) (Table 2). On the other hand, compounds 15 and 16 exhibited comparatively weaker anti-glycation capability, with IC50 values 132.11 ± 1.26 and 149.03 ± 1.18 μM, respectively. Similarly, compounds 21 and 22 also exhibited weak glycation inhibitory capability, with IC50 values 354.58 ± 1.53 and 652.06 ± 1.60 μM, respectively. In contrast, compounds 20 and 23 exhibited the best anti-glycation performance, with IC50 values of 48.32 ± 0.42 and 54.36 ± 0.40 μM, respectively.
| Compound | IC50 (μM ± SEM) | Compound | IC50 (μM ± SEM) |
|---|---|---|---|
| a SEM = standard error of the mean, NA = not active, Rutin = common standard for the anti-glycation. | |||
| 13 | 67.53 ± 0.46 | 19 | NA |
| 14 | 68.06 ± 0.43 | 20 | 48.32 ± 0.42 |
| 15 | 132.11 ± 1.26 | 21 | 354.58 ± 1.53 |
| 16 | 149.03 ± 1.18 | 22 | 652.06 ± 1.60 |
| 17 | NA | 23 | 54.36 ± 0.40 |
| 18 | NA | Rutin | 70.00 ± 0.50 |
3.3. Molecular docking of active urease inhibitors
The active urease inhibitors (compounds 13–16, 20, and 23) were docked at the binding site of urease to elucidate their binding patterns (Table 3) and predict the structure–activity relationship. The most active molecule, namely compound 20, exhibited excellent interactions with a highly negative docking score (−9.05 kcal mol−1) compared with the other inhibitors. Compound 20 showed good fit near the entrance of the active site, with one of its hydrazide moieties mediating a hydrogen bond with the Arg439 side chain at 3.36 Å, and Asp494 providing a bidentate interaction with the ortho-substituted –OH group. Moreover, the other hydrazide group displayed interaction with the carbonyl of Ala636 (2.90 Å). These interactions stabilize compound 20 at the ligand binding site, resulting in the highest docking score among all the docked compounds. The binding mode of compound 20 is shown in Fig. 3.| Compounds | Score (kcal mol−1) | Inhibitor atoms | Urease atoms | Interactions | Distance (Å) |
|---|---|---|---|---|---|
| 20 | −9.05 | N27 | O-ALA636 | H–donor | 2.90 |
| O29 | OD1-ASP494 | H–donor | 2.88 | ||
| O29 | OD2-ASP494 | H–donor | 3.20 | ||
| O23 | NH2-ARG439 | H–acceptor | 3.36 | ||
| 23 | −8.66 | N31 | SG-CME592 | H–donor | 4.37 |
| O32 | OD1-ASP494 | H–donor | 3.18 | ||
| O25 | NH2-ARG439 | H–acceptor | 2.72 | ||
| 13 | −7.77 | O30 | O-ALA440 | H–donor | 2.83 |
| O23 | NH2-ARG439 | H–acceptor | 3.41 | ||
| 6-Ring | CB-ARG439 | π–H | 3.47 | ||
| 14 | −7.75 | O29 | SD-CME592 | H–donor | 3.63 |
| O30 | O-ARG439 | H–donor | 2.13 | ||
| 6-Ring | CB-ARG439 | π–H | 3.40 | ||
| 6-Ring | CA-GLN635 | π–H | 3.71 | ||
| 15 | −6.50 | N39 | SG-CME592 | H–donor | 3.43 |
| 6-Ring | CZ-CME592 | π–H | 3.77 | ||
| 16 | −6.32 | N41 | SG-CME592 | H–donor | 3.24 |
| O35 | NH2-ARG439 | H–acceptor | 1.66 | ||
| Thiourea | −12.20 (RMSD = 1.19 Å) | O2 | NE2-HIS492 | H–acceptor | 3.17 |
| O9 | NI | Metal | 2.31 | ||
| O2 | NI | Ionic | 3.28 | ||
| O9 | NI | Ionic | 2.38 | ||
| O9 | NI | Ionic | 2.31 |
 | ||
| Fig. 3 Binding mode of compound 20 (green ball and sticks) to the active site of urease. The residues are represented by yellow sticks, and hydrogen bonds are shown as black lines. | ||
Furthermore, compound 23 showed good inhibition of urease. In this compound, again an ortho-substituted OH group mediates a hydrogen bond with Asp494, whereas one of its hydrazides is linked to Cme592 and Arg439, and the other hydrazide interacts with Met588 through hydrogen bonds. Compound 13 possesses a meta-substituted phenol group, which interacts with Ala440 via hydrogen bonding. Moreover, Arg439 mediates a hydrogen bond with its hydrazide moiety, contributing to the hydrophobic interactions of compound 13. Similar to compound 13, compounds 14–16 also displayed a tilted position at the active site entrance, where the ortho-substituted hydroxyl groups at both the methoxyphenyl rings formed hydrogen bonds with the thiol of Cme592 (3.63 Å) and the carbonyl of Arg439 (2.13 Å). Additionally, Arg439 and Gln635 mediate hydrophobic interactions with compound 14. In compound 15, only one hydrazide group interacts with the thiol of Cme592 through hydrogen bonding and hydrophobic interaction. Meanwhile, its trimethoxy benzene ring does not bind with the surrounding residues. Again, in compound 16, Cme592 and Arg439 form hydrogen bonds with its hydrazide moiety, while its dimethoxy-substituted rings do not participate in binding. The docking observations indicate that the substitution of –OH groups at the ortho-, meta-, para-position of the rings produces beneficial inhibitory effects, whereas the substitution of the methoxy group reduces the inhibitory potential of the compounds. The docking scores of compounds 23 and 13 to 16 were in the range of −8.66 to −6.32 kcal mol−1. The docking scores and interactions of the molecules are tabulated in Table 3, while their 3D interactions are given in ESI (Fig. S1).†
A comparison of the binding modes of our compounds with that of thiourea revealed a distinct binding behaviour. The thiourea molecule is a small fragment; due to its smaller size, it gets deep into the core of the active site, where it directly binds with the bi-nickel atoms through ionic and metallic interactions. Moreover, His492 provides hydrogen bonding to the thiourea molecule, thus inhibiting the urease activity (Fig. S1†). In contrast, our inhibitors do not bind with nickel atoms and remain at the active site entrance. Moreover, thiourea also exhibited a higher docking score (−12.20 kcal mol−1) than our inhibitors.
4. Conclusion
In this research work, eleven (11) new terephthalic dihydrazide analogues were synthesized by the condensation of terephthalic dihydrazide and eleven different aromatic aldehydes under reflux conditions. Ethyl alcohol was used as the solvent, and all reactions were catalysed by ethanoic acid. The purity of the synthesized compounds (13–23) was confirmed through TLC. Structure elucidation of the synthesized terephthalic dihydrazide derivatives was carried out using FT-IR, UV, ESI-MS and 1H NMR. All the prepared products (13–23) were evaluated for their in vitro anti-glycation and urease inhibition activities. In the antiglycation assay, compounds 13, 14, 20 and 23 showed better activity than the standard drug. Thus, these compounds are considered excellent drug candidates for future work. On the other hand, the rest showed moderate to low activity, and some of the synthesized compounds were inactive. Similarly, all the synthesized compounds were screened for urease inhibition activity in vitro. Compounds 20 and 23 showed excellent inhibitory activity, comparable to the common standard. Thus, these compounds are excellent drug candidates for future work. Molecular docking of urease inhibitors indicated the significance of –OH moieties in the phenyl rings in the binding mechanisms. Further, the results evidence that the active molecules bind at the entrance of the urease binding site, and the docking study correlates with our in vitro findings.Data availability
The data supporting this article have been included in the ESI.†Author contributions
Hazrat Bilal: methodology, data curation, and writing – original draft. Saeed Ullah: methodology, data curation, and writing – original draft. Sobia Ahsan Halim: methodology, formal analysis, and writing – review & editing. Momin Khan: methodology, conceptualization, and writing – original draft. Satya Kumar Avula: software, data curation, and formal analysis, Aftab Alam: data curation, formal analysis, and investigation. Eman Serry Zayed: investigation, formal analysis, and writing – review & editing. Sabah H El-Ghaiesh: data curation, investigation, formal analysis and writing – review & editing. Hanan A. Ogaly: formal analysis, resources, and funding acquisition. Zarbad Shah: conceptualization, project administration, supervision, and writing – review & editing. Ajmal Khan: data curation, formal analysis, resources, and writing – review & editing. Ahmed Al Harrasi: conceptualization, project administration, supervision, and writing – review & editing.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through the Large Research Project under grant number RGP2/152/45.References
- H. I. Ugras, I. Basaran, T. Kilic and U. Cakir, J. Heterocycl. Chem., 2006, 43, 1679–1684 CAS.
- A. H. El-masry, H. Fahmy and S. Ali Abdelwahed, Molecules, 2000, 5, 1429–1438 CAS.
- E. Aguilar-Llanos, S. E. Carrera-Pacheco, R. González-Pastor, J. Zúñiga-Miranda, C. Rodríguez-Pólit, A. Mayorga-Ramos, O. Carrillo-Naranjo, L. P. Guamán, J. C. Romero-Benavides and C. Cevallos-Morillo, ACS Omega, 2023, 8, 42632–42646 CrossRef CAS.
- M. A. Ashraf, K. Mahmood, A. Wajid, M. J. Maah and I. Yusoff, Int. Proc. Chem., Biol. Environ. Eng., 2011, 10, 185 Search PubMed.
- F. Nworie, F. Nwabue, N. Elom and S. Eluu, J. Basic Appl. Res. Biomed, 2016, 2, 295–305 Search PubMed.
- I. Mushtaq, M. Ahmad, M. Saleem and A. Ahmed, FJPS, 2024, 10, 16 Search PubMed.
- L.-X. Cheng, J.-J. Tang, H. Luo, X.-L. Jin, F. Dai, J. Yang, Y.-P. Qian, X.-Z. Li and B. Zhou, Bioorg. Med. Chem. Lett., 2010, 20, 2417–2420 CrossRef CAS PubMed.
- S. E. Harpstrite, S. D. Collins, A. Oksman, D. E. Goldberg and V. Sharma, Curr. Top. Med. Chem., 2008, 4, 392–395 CAS.
- R. Islam, F. Koizumi, Y. Kodera, K. Inoue, T. Okawara and M. Masutani, Bioorg. Med. Chem. Lett., 2014, 24, 3802–3806 CAS.
- S. E. Harpstrite, S. D. Collins, A. Oksman, D. E. Goldberg and V. Sharma, Med. Chem., 2008, 4, 392–395 CAS.
- P. Kafarski and M. Talma, J. Adv. Res., 2018, 13, 101–112 CAS.
- P. Kosikowska and Ł. Berlicki, Expert Opin. Ther. Pat., 2011, 21, 945–957 CAS.
- J. G. Kusters, A. H. M. van Vliet and E. J. Kuipers, Clin. Microbiol. Rev., 2006, 19, 449–490 CAS.
- R. Rai, V. A. Saraswat and R. K. Dhiman, J. Clin. Exp. Hepatol., 2015, 5, S29–S36 CrossRef.
- I. J. Rosenstein, J. M. Hamilton-Miller and W. Brumfitt, Infect. Immun., 1981, 32, 32–37 CrossRef CAS.
- B. Krajewska, J. Mol. Catal. B: Enzym., 2009, 59, 9–21 CAS.
- J. Bremner, Nitrogen Economy in Tropical Soils, 1995, pp. 321–329 Search PubMed.
- H. Beraldo and D. Gambino, Mini-Rev. Med. Chem., 2004, 4, 31–39 CAS.
- A. Hameed, Z. Shafiq, M. Yaqub, M. Hussain, M. A. Hussain, M. Afzal, M. N. Tahir and M. M. Naseer, New J. Chem., 2015, 39, 9351–9357 CAS.
- L. S. B. Upadhyay, Indian J. Biotechnol., 2012, 11, 381–388 CAS.
- Y. F. Rego, M. P. Queiroz, T. O. Brito, P. G. Carvalho, V. T. de Queiroz, Â. de Fátima and F. Macedo Jr, J. Adv. Res., 2018, 13, 69–100 CrossRef CAS.
- R. D. Hauck, Nitrogen in Crop Production: Proceedings of a Symposium Held 25-27 May 1982 at Sheffield, Alabama, American Society of Agronomy, 1984 Search PubMed.
- G. McCarty, J. Bremner and J. Lee, Plant Soil, 1990, 127, 269–283 CrossRef CAS.
- K. M. Khan, S. Iqbal, M. A. Lodhi, G. M. Maharvi, M. I. Choudhary and S. Perveen, Bioorg. Med. Chem., 2004, 12, 1963–1968 CAS.
- M. A. S. Aslam, S.-u. Mahmood, M. Shahid, A. Saeed and J. Iqbal, Eur. J. Med. Chem., 2011, 46, 5473–5479 CAS.
- Y. Onoda, H. Iwasaki, T. Magaribuchi and H. Tamaki, Arzneim. Forsch., 1991, 41, 546–548 CAS.
- C.-H. Wu and G.-C. Yen, J. Agric. Food Chem., 2005, 53, 3167–3173 CAS.
- N. Sattarahmady, F. Khodagholi, A. A. Moosavi-Movahedi, H. Heli and G. H. Hakimelahi, Int. J. Biol. Macromol., 2007, 41, 180–184 CrossRef CAS.
- H. Shahzadi, M. A. Sheikh, A. Hameed and A. Jamil, Int. J. Agric. Biol., 2015, 17, 539–546 CrossRef CAS.
- D. Evstafeva, F. Ilievski, Y. Bao, Z. Luo, B. Abramovic, S. Kang, C. Steuer, E. Montanari, T. Casalini and D. Simicic, Nat. Commun., 2024, 15, 2226 CrossRef CAS.
- C. He, Z. Yang and N.-H. Lu, World J. Gastroenterol., 2014, 20, 4607 Search PubMed.
- C. D. Mărginean, C. O. Mărginean and L. E. Meliț, Children, 2022, 9, 1352 Search PubMed.
- V. J. Shetty, H. K. Prasad, A. S. Konamme, K. J. Shetty and G. Kulamarva, BBRJ, 2020, 4, 41–44 Search PubMed.
- I. Khan, S. Ali, S. Hameed, N. H. Rama, M. T. Hussain, A. Wadood, R. Uddin, Z. Ul-Haq, A. Khan and S. Ali, Eur. J. Med. Chem., 2010, 45, 5200–5207 CrossRef CAS PubMed.
- R. Singh, A. Barden, T. Mori and L. Beilin, Diabetologia, 2001, 44, 129–146 CrossRef CAS PubMed.
- J. Zhang, E. C. Hoedt, Q. Liu, E. Berendsen, J. J. Teh, A. Hamilton, A. W. O’Brien, J. Y. Ching, H. Wei and K. Yang, Gastroenterology, 2021, 160, 317–330 CrossRef CAS PubMed.
- R. Ryvchin, V. Dubinsky, K. Rabinowitz, N. Wasserberg, I. Dotan and U. Gophna, J. Crohn's Colitis, 2021, 15, 2066–2077 CrossRef PubMed.
- K. C. Richards-Corke, Y. Jiang, V. Yeliseyev, Y. Zhang, E. A. Franzosa, Z. A. Wang, M. Yapa Abeywardana, P. A. Cole, C. Huttenhower and L. Bry, ACS Chem. Biol., 2025, 20, 48–55 CrossRef CAS PubMed.
- N. Ahmed, Glycation and the Maillard Reaction in Vitro: Implications for Diabetes Mellitus, Open University (United Kingdom), 1992 Search PubMed.
- J. Uddin, S. Ullah, S. A. Halim, M. Waqas, A. Ibrar, I. Khan, A. Bin Muhsinah, A. Khan and A. Al-Harrasi, ACS Omega, 2023, 8, 31890–31898 CAS.
- M. Islam, A. Khan, M. T. Shehzad, A. Hameed, N. Ahmed, S. A. Halim, M. Khiat, M. U. Anwar, J. Hussain and R. Csuk, Bioorg. Chem., 2019, 87, 155–162 CAS.
- CULC, Chemical Computing Group ULC, McGill University: Montreal, QC, Canada, 2020 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Binding modes of compounds are given in S1. Spectroscopic spectra for compounds 13–23 are given in S2–S35. See DOI: https://doi.org/10.1039/d5ra00459d |
| This journal is © The Royal Society of Chemistry 2025 |