Open Access Article
Open Access ArticleSynthesis of SGLT2 inhibitor O-glucoside derivatives for enhancing anti-heart failure activity†
Yu Liua,
Xiaodong Fanga,
Meng Suna,
Xutong Wanga,
Kejing Maa,
Bing Wanga,
Qiming Lia,
Jan Mohammad Omarb,
Yihui Liu*b,
Heng Liu*c and
Weina Han *a
*a
aDepartment of Medicinal Chemistry and Natural Medicine Chemistry, College of Pharmacy, Harbin Medical University, Harbin, People's Republic of China. E-mail: hanweina@hrbmu.edu.cn
bSecond Affiliated Hospital of Harbin Medical University, Harbin, 150081, China. E-mail: 502986473@qq.com
cPharmaceutical Experiment Teaching Center, College of Pharmacy, Harbin Medical University, Harbin, China. E-mail: 2454802160@qq.com
First published on 27th June 2025
Abstract
SGLT2 inhibitors were derived from the natural product phlorizin and were used clinically to treat heart failure with preserved ejection fraction (HFpEF). In this study, based on phlorizin as the lead compound, tyramine moiety was introduced while retaining the pharmacophoric glucoside, thereby enhancing SGLT2 protein inhibitory activity and improving gastrointestinal absorption properties. We synthesized 11 new O-glucoside derivatives and evaluated their anti-heart failure activities. Results showed that compound D4 significantly increased the inhibition rate of the SGLT2 protein and anti-heart failure activity compared with empagliflozin and phlorizin and increased the ATP content in the impaired cardiomyocytes, which exhibited the best inhibition effect on SOD enzymes. According to the docking scores, it was speculated that compound D4 could treat heart failure by inhibiting cellular oxidative stress and reducing ROS and lipid peroxidation. The pharmacokinetic prediction results showed that compound D4 had good plasma protein binding force and hydrophilicity.
1. Introduction
Heart failure (HF) is a cardiovascular disease caused by structural or functional abnormalities owing to a variety of reasons, but it is the final stage in the progression of heart diseases.1–3 The treatment for heart failure remains an uphill battle, and the clinical treatment for heart failure with preserved ejection fraction (HFpEF) still lacks good drugs.4 According to research studies, two antidiabetic sodium-glucose cotransporter 2 (SGLT2) inhibitors, namely, dapagliflozin (DAPA) and empagliflozin (EMPA), significantly reduce mortality in HFpEF patients, both with and without diabetes.5,6 Currently, most SGLT2 inhibitors used in clinical practice are C-glucoside derivatives. Long-term use of these compounds may impose a considerable burden on the kidneys, particularly in patients with pre-existing renal insufficiency.7,8 Therefore, the design and synthesis of novel O-glucoside SGLT2 derivatives may offer a new therapeutic strategy for the treatment of heart failure. By enhancing SGLT2 inhibition, these compounds not only improve the clinical symptoms of heart failure patients but also reduce renal burden, delay the progression of heart failure, and ultimately improve the quality of life of patients.SGLT2 inhibitors, derived from O-glucoside phlorizin,9,10 lower blood glucose by inhibiting renal glucose reabsorption. Their mechanism also promotes urinary sodium excretion and osmotic diuresis, while modulating sodium–hydrogen exchange in the kidneys and heart. These effects collectively reduce body weight, blood pressure, plasma volume, and cardiac preload/afterload, thereby contributing to heart failure treatment.11–15 The aim of this study was to enhance the inhibition of the SGLT2 protein and improve the gastrointestinal absorption and pharmacological efficacy of O-glucoside derivatives for the treatment of heart failure. We introduced tyramine fragments into structure of compound while maintaining the key phlorizin glucoside pharmacophore. The introduction of the tyramine fragment could promote the synthesis and release of norepinephrine, potentially reversing the symptoms of myocardial hypertrophy in patients with heart failure and contributing to the treatment of chronic heart failure.16–18 Additionally, the introduction of the tyramine fragment provided an amide group, which enhanced hydrogen bonding between the compound and the SGLT2 protein, thereby stabilizing the interaction. This design was further validated by the molecular docking results (Fig. 4 and S41†). Simultaneously, we examined the 11 new synthesized compounds for cytotoxicity, anti-heart failure activity, SGLT2 inhibition rate, ATP content and SOD enzyme inhibition. Meanwhile, 13 proteins associated with the treatment of heart failure were individually docked using Autodock software to investigate the potential therapeutic targets of these compounds. In addition, the pharmacokinetic prediction results showed that compound D4 improved the gastrointestinal absorption, plasma protein binding ability and hydrophilicity (Fig. 1).
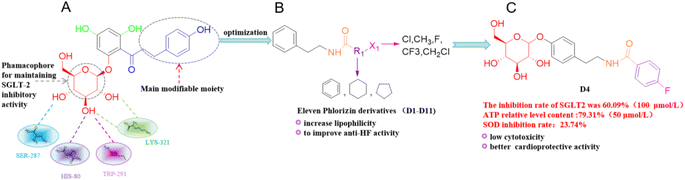 | ||
| Fig. 1 Synthesis of O-glucoside derivatives and anti-heart failure activity. (A) Interactions of phlorizin with SGLT2 (PDB:7VSi) in the binding domain of hydroxyl groups; (B) structural modification of the distal benzene ring (group R1 and X1) of phlorizin; and (C) structure of the optimal derivative D4. | ||
2. Results and discussion
2.1. Synthesis
Compound III-1–III-11 was obtained by the reaction of 4-methoxyphenylethylamine and compound II in dichloromethane (DCM) under alkaline conditions. Compounds III-1–III-11 were dissolved in anhydrous DCM, and BBr3 was slowly added at 0 °C under argon conditions. The reaction was performed for 1–2 hours and quenched by saturated Na2CO3 aqueous solution. Compounds IV-1–IV-11 and acetyl bromide-α-D-glucose were dissolved in DCM, and sodium hydroxide (5 M) and the catalyst tetrabutylammonium bromide were added, and the reaction was performed for 12–28 hours to obtain compounds V-1–V-11, which were dissolved in anhydrous MeOH. At room temperature, lithium hydroxide was added, and the reaction was carried out for 1–2 hours to obtain compound D-1–D-11. All the O-glucoside derivatives were characterized by 1H NMR, 13C NMR and ESI-MS analyses.The presence of unsubstituted hydrogen on the amide group identified by 1H NMR confirmed the substitution of the hydroxyl group on the benzene ring by the glucose group, and the resulting product was the O-glucoside derivatives (Table 1). The designed synthetic route was reasonable, and all target compounds were successfully obtained. The purity of the compounds was no less than 95%, as determined by HPLC (ESI†), which meets the requirements for biological experiments.
2.2. Survival rate of cardiomyocytes in vitro
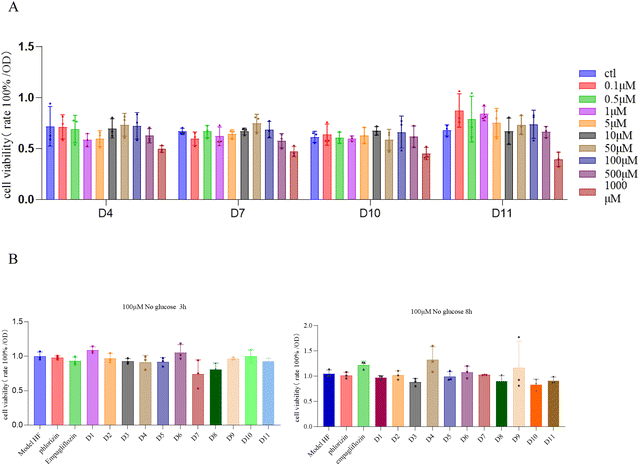
The survival rate of cardiomyocytes was assessed using the CCK-8 assay. It was shown that compounds D4, D7, D10, and D11 exhibited lower cytotoxicity compared to the control group at multiple concentrations (Fig. 2A). The survival rates of cardiomyocytes in other compound groups are provided in the ESI (Fig. S34†). When cultured in a glucose-free DMEM medium for 3 h, compounds D1 and D6 showed better protection than phlorizin at 100 μM (Fig. 2B). Specific dosing concentrations are shown in the ESI.† It was noteworthy that the survival rate of cardiomyocytes treated with compound D4 for 8 h in a glucose-free medium was higher than that of cardiomyocytes treated with empagliflozin (Fig. 2B). Based on the results from the above experiment, 100 μM of compound D4 was selected and administered in the subsequent experiments. Due to the presence of a nitrosamine structure in the synthesized compound, there is a potential risk of nitrosamine formation. However, based on our cytotoxicity and therapeutic activity experiments on damaged cardiomyocytes, we believe that the risk of nitrosamine formation is negligible.2.3. SGLT2 protein content and relative inhibition rate
The results of the SGLT2 inhibition experiment showed that the inhibition rate of compound D4 on SGLT2 was significantly better than that of phlorizin (Fig. 3A). It was noted that the inhibition rate of compound D4 on SGLT2, with over 60%, was double that of empagliflozin at the same dose concentration (100 μM) (Fig. 3A and B). Inhibition of SGLT2 protein reduces the reabsorption of glucose and Na+, thereby decreasing blood pressure, plasma volume, and loading of the heart, which contributes to the treatment of heart failure.2.4. ATP content and SOD inhibition rate
The experimental results showed that the compound D4 increased the amount of ATP in cardiomyocyte injury and was slightly higher than empagliflozin (Fig. 4A). In the SOD inhibition assay, compound D4 exhibited stronger inhibitory activity on SOD compared to empagliflozin and demonstrated a concentration-dependent enhancement of SOD inhibition (Fig. 4B). It is preliminarily speculated that the reason why compound D4 was more effective than empagliflozin in cardiomyocyte injury was that it is related to the inhibition of cellular oxidative stress, reduction of ROS and lipid peroxidation.2.5. Molecular docking
Based on the docking results of the compounds with SGLT2 protein, we deduced that when phlorizin's pyranose ring is substituted at the ortho-benzene, pyran at the 5-position affects the formation of the hydrogen bond between the hydroxyl group and the SGLT2 protein due to its relative spatial hindrance. However, the substitution of compound D4 and empagliflozin with para and meta glycosides on the benzene ring precisely solved this spatial problem (Fig. 5A and S41†). The binding affinity of compound D4 to the SGLT2 protein was −10.2 kcal mol−1, which was higher than that of empagliflozin. This is because the hydrogen of the amide group in compound D4 forms hydrogen bonding with the THR-87 residue of the SGLT2 protein, along with π–π stacking interactions, which contribute to its higher binding stability (Fig. S41†). This likely contributes to the higher inhibition rate of D4 on SGLT2 compared to empagliflozin. In addition, the heatmap results showed that the docking index of the compounds with proteins, including SIRT1 (PDB:Q9VK34), CAT (PDB:6vko) and Nrf2 (PDB:7K2F), was ≤−10.0. Especially with the CAT protein, the docking score of compound D4 was higher than that of empagliflozin (Fig. 5B). The reason for the better effect of compound D4 may also be that, besides targeting the SGLT2 protein, it may also alleviate cardiomyocyte injury by activation of the above three proteins and thus reducing the ROS production. It could be useful to further optimize its structure in future studies.2.6. Pharmacokinetic prediction
The synthesis of 11 O-glucoside derivatives were predicted using SwissADME 2.0 (https://admetmesh.scbdd.com/explanation/index) (https://www.swissadme.ch/) and ADMET. Some of the predicted parameters are presented in Table 2. The predicted results showed that compared with phloridin, the synthesized target compounds improved the absorption degree and permeability of phloridin in the human gastrointestinal tract. Structurally, D4 incorporates an O-glucoside moiety, which enhances hydrophilicity and may improve cellular uptake and bioavailability compared to the C-glucoside structure of empagliflozin. The pharmacokinetic predictions further corroborate the aforementioned findings.| Name | PPB | Caco-2 permeability | GI absorption | BBB | P-gp substrate | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) S (ESOL) |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w (iLOGP) Po/w (iLOGP) |
|---|---|---|---|---|---|---|---|
| Phlorizin | 75.02% | −6.381 | Low | NO | Yes | Soluble | 1.25 |
| EMPD | 95.407% | −5.191 | High | NO | Yes | Soluble | 3.18 |
| D1 | 94.40% | −5.429 | High | NO | Yes | Moderately soluble | 3.03 |
| D2 | 94.42% | −5.221 | High | NO | Yes | Moderately soluble | 2.32 |
| D3 | 93.26% | −5.346 | High | NO | Yes | Soluble | 2.73 |
| D4 | 93.58% | −5.409 | High | NO | Yes | Soluble | 1.56 |
| D5 | 94.84% | −5.292 | High | NO | Yes | Moderately soluble | 2.68 |
| D6 | 92.97% | −5.321 | High | NO | Yes | Soluble | 2.43 |
| D7 | 87.22% | −5.467 | High | NO | Yes | Soluble | 2.76 |
| D8 | 80.53% | −5.539 | High | NO | Yes | Soluble | 2.02 |
| D9 | 95.92% | −5.268 | High | NO | Yes | Moderately soluble | 2.57 |
| D10 | 92.49% | −5.328 | High | NO | Yes | Soluble | 2.67 |
| D11 | 95.35% | −5.330 | High | NO | Yes | Moderately soluble | 3.04 |
3. Conclusion
In this study, 11 O-glucoside derivatives were designed and synthesized, including D4, which showed low toxicity and high SGLT2 protein inhibition rate, and is a promising compound for the treatment of heart failure. The active hydrogen of the amide group was identified from the 1H NMR and the hydroxyl group on the benzene ring was replaced by the glucose group. In the in vitro experiments, at the same treatment dose, the effect of D4 on the ATP content and SOD enzyme in cardiomyocyte injury was significantly stronger than the positive drug empagliflozin, and the different therapeutic effects may be related to the structure of the compound. The compound D4 incorporates the tyramine group, and the hydrogen on the amide group enhances hydrogen bonding with the SGLT2 protein, thereby increasing the binding affinity and stabilizing the interaction with the protein. Further mechanistic studies showed that the synthesized target compounds showed good binding activity to SIRT1, CAT and Nrf2. However, the prediction of the above mechanism is from computer simulations, and its reliability needs to be further verified in subsequent experiments.4. Experimental
4.1. General information
All chemicals and reagents were of commercial grade. The progress of the reactions was followed using pre-coated aluminum sheet thin layer chromatography (TLC) plates. 1H NMR (600 MHz, DMSO-d6), 13C NMR (151 MHz, DMSO-d6), chemical shifts (δ), and coupling constants (J) were shown in ppm and Hz, respectively. All reagents were chemically pure or purified using standard methods. Solvents and anhydrous reagents were pure analytes and dried according to conventional protocols. To monitor the reaction process, thin layer chromatography (TLC) was performed on silica gel TLC plates fabricated by Shaoyuan Chemical Technology Co. The spots were observed under UV illumination (254 nm) using a triple-duty UV analyser from Shanghai Jiapeng Technology Co. Column chromatography was performed using a silica gel column (silica gel with a mesh size of 200 to 300) manufactured by Qingdao Ocean Chemical Co. After the reaction, the product was concentrated in a rotary evaporator from Shanghai Yarong Biochemical Instrument Factory under a specific atmospheric pressure.Purities of the representative compounds were determined by HPLC on an Agilent eclipse plus C18 column (4.6 cm × 150 mm, 5 μm, λ = 254/365 nm, 1 mL min−1) using MeOH/H2O (80/20, v/v) as an eluent.
4.2. Chemistry
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol, 60
methanol, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
4.2.1.1 4-(Chloromethyl)-N-(4-methoxyphenethyl)benzamide (III-1). White solid with 92.52% yield.1H NMR (600 MHz, DMSO-d6) δ 8.55 (t, J = 5.6 Hz, 1H), 7.82 (d, J = 8.3 Hz, 2H), 7.51 (d, J = 8.3 Hz, 2H), 7.15 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.5 Hz, 2H), 4.80 (s, 2H), 3.71 (s, 3H), 3.44 (q, J = 5.8 Hz, 2H), 2.78 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.67, 157.68, 140.49, 134.50, 131.37, 129.60, 129.60, 128.68, 128.68, 127.47, 127.47, 113.77, 113.77, 54.96, 45.46, 41.14, 34.18.
4.2.1.2 4-Chloro-N-(4-methoxyphenethyl)benzamide (III-2). White solid with 90.34% yield. 1H NMR (600 MHz, DMSO-d6) δ 8.61 (t, J = 5.6 Hz, 1H), 7.84 (d, J = 8.5 Hz, 2H), 7.53 (d, J = 8.6 Hz, 2H), 7.15 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 3.71 (s, 3H), 3.44 (q, J = 6.7, 2H), 2.77 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.06, 157.69, 135.89, 133.35, 131.32, 129.60, 129.60, 129.05, 129.05, 128.35, 128.35, 113.77, 113.77, 54.96, 41.17, 34.13.
4.2.1.3 N-(4-Methoxyphenethyl)-4-methylbenzamide (III-3). White solid with 92.52% yield.1H NMR (600 MHz, DMSO-d6) δ 8.44 (t, J = 5.6 Hz, 1H), 7.73 (d, J = 8.2 Hz, 2H), 7.25 (d, J = 7.9 Hz, 2H), 7.15 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 3.71 (s, 3H), 3.43 (q, 7.6 Hz, 2H), 2.77 (t, J = 7.5 Hz, 2H), 2.34 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 165.99, 157.67, 140.86, 131.85, 131.45, 129.59, 129.59, 128.77, 128.77, 127.12, 127.12, 113.76, 113.76, 54.96, 41.08, 34.27, 20.91.
4.2.1.4 4-Fluoro-N-(4-methoxyphenethyl)benzamide (III-4). White solid with 96.51% yield. 1H NMR (600 MHz, DMSO-d6) δ 8.55 (t, J = 5.6 Hz, 1H), 7.89 (d, J = 14.3 Hz, 2H), 7.28 (d, J = 17.6 Hz, 2H), 7.15 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 3.71 (s, 3H), 3.43 (q, J = 6.6 Hz, 2H), 2.77 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.07, 157.69, 131.36, 129.75, 129.69, 129.59, 129.59, 115.23, 115.23, 115.09, 115.09, 113.77, 113.77, 54.96, 41.17, 34.19.
4.2.1.5 2-Chloro-N-(4-methoxyphenethyl)benzamide (III-5). White solid with 89.86% yield.1H NMR (600 MHz, chloroform-d) δ 7.60 (d, J = 7.5 Hz, 1H), 7.36 (d, J = 7.9 Hz, 1H), 7.33 (t, J = 7.5 Hz, 1H), 7.29 (t, J = 7.4 Hz, 1H), 7.17 (d, J = 8.3 Hz, 2H), 6.85 (d, J = 8.5 Hz, 2H), 3.79 (s, 3H), 3.71 (q, J = 6.6 Hz, 2H), 2.89 (t, J = 6.9 Hz, 2H). 13C NMR (151 MHz, CDCl3) δ 166.59, 158.48, 136.04, 131.33, 130.77, 130.73, 130.32, 130.21, 129.92, 129.92, 127.17, 114.22, 114.22, 55.40, 41.54, 34.69.
4.2.1.6 N-(4-Methoxyphenethyl)-2-methylbenzamide (III-6). White solid with 90.65% yield. 1H NMR (600 MHz, chloroform-d) δ 7.28 (d, J = 7.5 Hz, 1H), 7.25 (t, J = 8.6 Hz, 1H), 7.21 (d, J = 7.56 Hz, 1H), 7.17 (t, J = 8.6 Hz, 1H), 7.15 (d, J = 8.6 Hz, 2H), 6.86 (d, J = 8.6 Hz, 2H), 3.79 (s, 3H), 3.68 (q, J = 6.6 Hz, 2H), 2.87 (t, J = 6.9 Hz, 2H), 2.39 (s, 3H). 13C NMR (151 MHz, CDCl3) δ 170.19, 158.47, 136.68, 136.14, 131.10, 130.85, 129.90, 129.86, 129.86, 126.73, 125.82, 114.24, 114.24, 55.41, 41.10, 34.91, 22.10.
4.2.1.7 N-(4-Methoxyphenethyl)cyclohexanecarboxamide (III-7). White solid with 88.63% yield. 1H NMR (600 MHz, chloroform-d) δ 7.09 (d, J = 8.5 Hz, 2H), 6.85 (d, J = 8.5 Hz, 2H), 3.79 (s, 3H), 3.46 (q, J = 6.7 Hz, 2H), 2.74 (t, J = 6.9 Hz, 2H), 2.00 (tt, J = 11.8, 3.5 Hz, 1H), 1.78 (m, 5H), 1.38 (m, 2H), 1.20 (m, 3H). 13C NMR (151 MHz, CDCl3) δ 176.61, 158.91, 131.12, 129.85, 129.85, 114.63, 114.63, 55.39, 46.13, 40.65, 34.91, 29.79, 29.79, 25.85, 25.85, 25.85.
4.2.1.8 N-(4-Methoxyphenethyl)cyclopentanecarboxamide (III-8). White solid with 94.41% yield.1H NMR (600 MHz, chloroform-d) δ 7.10 (d, J = 8.6 Hz, 2H), 6.84 (d, J = 8.6 Hz, 2H), 3.79 (s, 3H), 3.47 (q, J = 6.7 Hz, 2H), 2.75 (t, J = 6.9 Hz, 2H), 2.43 (p, J = 7.9 Hz, 1H), 1.80 (q, J = 6.9, 5.9 Hz, 2H), 1.73 (m, 4H), 1.54 (q, J = 5.64 Hz, 2H). 13C NMR (151 MHz, CDCl3) δ 176.29, 158.37, 131.13, 129.85, 129.85, 114.14, 114.14, 55.39, 46.05, 40.83, 34.94, 30.53, 30.53, 25.99, 25.99.
4.2.1.9 3-Chloro-N-(4-methoxyphenethyl)benzamide (III-9). White solid with 94.78% yield.1H NMR (600 MHz, DMSO-d6) δ 8.65 (t, J = 5.6 Hz, 1H), 7.85 (s, 1H), 7.78 (d, J = 7.6 Hz, 1H), 7.58 (d, J = 7.9 Hz, 1H), 7.49 (t, J = 7.9 Hz, 1H), 7.15 (d, J = 8.6 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 3.71 (s, 3H), 3.44 (dd, J = 14.7, 5.8 Hz, 2H), 2.77 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 164.67, 157.70, 136.60, 133.14, 131.28, 130.91, 130.30, 129.59, 129.59, 126.94, 125.88, 113.77, 113.77, 54.96, 41.19, 34.06.
4.2.1.10 N-(4-Methoxyphenethyl)-3-methylbenzamide (III-10). White solid with 97.23% yield. 1H NMR (600 MHz, DMSO-d6) δ 8.46 (t, J = 5.6 Hz, 1H), 7.64 (s, 1H), 7.60 (d, J = 5.2 Hz, 1H), 7.33 (t, J = 7.32 Hz, 1H) 7.32 (d, J = 6.7 Hz, 1H), 7.15 (d, J = 8.8 Hz, 2H), 6.85 (d, J = 8.6 Hz, 2H), 3.71 (s, 3H), 3.43 (q, J = 6.9 Hz, 2H), 2.77 (t, J = 7.6 Hz, 2H), 2.35 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 166.24, 157.67, 137.47, 134.66, 131.57, 131.42, 129.58, 129.58, 128.12, 127.69, 124.23, 113.76, 113.76, 54.95, 41.09, 34.23, 20.94.
4.2.1.11 N-(4-Methoxyphenethyl)-3-(trifluoromethyl)benzamide (III-11). White solid with 84.66% yield.1H NMR (600 MHz, DMSO-d6) δ 8.80 (t, J = 5.6 Hz, 1H), 8.15 (s, 1H), 8.12 (d, J = 7.8 Hz, 1H), 7.89 (d, J = 7.8 Hz, 1H), 7.71 (t, J = 7.8 Hz, 1H), 7.16 (d, J = 8.6 Hz, 2H), 6.86 (d, J = 8.6 Hz, 2H), 3.71 (s, 3H), 3.47 (dd, J = 13.6, 6.8 Hz, 2H), 2.79 (t, J = 7.5 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 164.64, 157.72, 135.46, 131.26, 131.24, 129.63, 129.61, 129.21, 129.00, 129.00, 127.66, 123.71, 113.78, 113.78, 54.95, 41.24, 34.07.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate, 5
ethyl acetate, 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–1
1–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v).
2, v/v).
4.2.3.1 2-(Acetoxymethyl)-6-(4-(2-(4-(chloromethyl)benzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-1). White solid with a yield of 26.01%.1H NMR (600 MHz, DMSO-d6) δ 8.60 (s, 1H), 7.80 (t, J = 11.4 Hz, 2H), 7.51 (d, J = 5.3 Hz, 2H), 7.19 (d, J = 8.0 Hz, 2H), 6.92 (d, J = 6.6 Hz, 2H), 5.51 (d, J = 6.5 Hz, 1H), 5.40 (t, J = 10.6 Hz, 1H), 5.01 (dt, J = 30.0, 12.0 Hz, 2H), 4.77 (d, J = 39.7 Hz, 2H), 4.22 (t, J = 18.0 Hz, 2H), 4.05 (d, J = 12.0 Hz, 1H), 3.45 (s, 2H), 2.82 (d, J = 30.5 Hz, 2H), 2.01 (s, 6H),1.99 (s, 3H),1.97 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 170.38, 170.01, 169.74, 169.53, 166.05, 155.26, 140.91, 134.80, 134.42, 130.19, 130.00, 129.57, 129.10, 127.89, 127.86, 122.03, 116.72, 97.62, 72.29, 71.10, 68.41, 62.00, 45.84, 41.37, 34.53, 33.95, 20.85, 20.78, 20.73, 20.68.
4.2.3.2 2-(Acetoxymethyl)-6-(4-(2-(4-chlorobenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-2). White solid with yield 23.96%.1H NMR (600 MHz, DMSO-d6) δ 8.63 (t, J = 5.6 Hz, 1H), 7.83 (d, J = 8.5 Hz, 2H), 7.53 (d, J = 8.5 Hz, 2H), 7.18 (d, J = 8.4 Hz, 2H), 6.91 (d, J = 8.6 Hz, 2H), 5.49 (d, J = 8.0 Hz, 1H), 5.39 (t, J = 9.6 Hz, 1H), 5.03 (t, J = 8.1 Hz, 1H), 4.98 (t, J = 9.6 Hz, 1H), 4.20 (q, J = 2.3 Hz, 2H), 4.05 (d, J = 10.0 Hz, 1H), 3.44 (q, J = 6.8 Hz, 3H), 2.79 (t, J = 7.3 Hz, 2H), 2.01 (s, 3H), 2.00 (s, 3H), 1.99 (s, 3H), 1.96 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 170.02, 169.65, 169.38, 169.18, 165.13, 154.93, 136.44, 134.03, 133.32, 129.82, 129.82, 129.08, 129.08, 127.82, 127.82, 116.39, 116.39, 97.31, 71.97, 70.78, 70.75, 68.09, 61.66, 41.04, 34.13, 20.49, 20.42, 20.37, 20.32.
4.2.3.3 2-(Acetoxymethyl)-6-(4-(2-(4-methylbenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-3). White solid with yield 25.76%.1H NMR (600 MHz, DMSO-d6) δ 8.48 (t, J = 5.6 Hz, 1H), 7.72 (d, J = 7.9 Hz, 2H), 7.25 (d, J = 7.9 Hz, 2H), 7.18 (d, J = 8.2 Hz, 2H), 6.91 (d, J = 8.2 Hz, 2H), 5.50 (d, J = 8.0 Hz, 1H), 5.40 (t, J = 9.6 Hz, 1H), 5.03 (dd, J = 9.8, 8.0 Hz, 1H), 4.98 (t, J = 9.6 Hz, 1H), 4.22 (dd, J = 13.8, 5.8 Hz, 1H), 4.18 (t, J = 6.0 Hz, 1H), 4.05 (dd, J = 12.2, 2.1 Hz, 1H), 3.42 (q, J = 6.9 Hz, 2H), 2.78 (t, J = 7.4 Hz, 2H), 2.34 (s, 3H), 2.01 (s, 3H), 2.00 (s, 3H), 1.99 (s, 3H), 1.96 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 170.09, 169.72, 169.44, 169.24, 166.05, 154.93, 140.99, 131.82, 129.87, 129.87, 128.86, 128.86, 127.20, 127.20, 116.39, 116.39, 97.29, 71.97, 70.78, 70.75, 68.08, 61.68, 61.68, 41.01, 34.30, 21.00, 20.55, 20.49, 20.43, 20.38.
4.2.3.4 2-(Acetoxymethyl)-6-(4-(2-(4-fluorobenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-4). White solid with yield 25.87%. 1H NMR (600 MHz, DMSO-d6) δ 8.57 (t, J = 5.6 Hz, 1H), 7.88 (dd, J = 8.7, 5.7 Hz, 2H), 7.28 (t, J = 8.9 Hz, 2H), 7.19 (d, J = 8.6 Hz, 2H), 6.91 (d, J = 8.6 Hz, 2H), 5.49 (d, J = 8.0 Hz, 1H), 5.39 (t, J = 9.6 Hz, 1H), 5.03 (t, J = 9.7 Hz, 1H) 4.98 (t, J = 9.6 Hz, 1H), 4.20 (q, J = 4.4 Hz, 2H), 4.05 (dd, J = 11.9, 1.9 Hz, 1H), 3.44 (q, J = 6.6 Hz, 2H), 2.79 (t, J = 7.4 Hz, 2H), 2.01 (s, 3H), 2.00 (s, 3H), 1.99 (s, 3H), 1.96 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 170.03, 169.65, 169.38, 169.17, 165.13, 163.01, 154.92, 134.07, 131.07, 129.82, 129.82, 129.79, 129.72, 116.40, 116.40, 115.28, 115.14, 97.33, 71.98, 70.79, 70.76, 68.10, 61.66, 41.04, 34.19, 20.48, 20.41, 20.37, 20.31.
4.2.3.5 2-(Acetoxymethyl)-6-(4-(2-(2-chlorobenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-5). White solid with yield 26.67%.1H NMR (600 MHz, chloroform-d) δ 7.60 (d, J = 7.5 Hz, 1H), 7.36 (t, J = 4.71 Hz, 1H), 7.33 (d, J = 7.8 Hz, 1H), 7.30 (t, J = 7.7 Hz, 1H), 7.18 (d, J = 8.2 Hz, 2H), 6.94 (d, J = 8.1 Hz, 2H), 5.29 (t, J = 9.2 Hz, 1H), 5.25 (t, J = 8.4 Hz, 1H), 5.16 (t, J = 9.4 Hz, 1H), 5.05 (d, J = 7.4 Hz, 1H), 4.28 (dd, J = 12.3, 5.3 Hz, 1H), 4.16 (dd, J = 12.3, 2.5 Hz, 1H), 3.85 (dd, J = 4.8, 2.5 Hz, 1H), 3.70 (q, J = 6.7 Hz, 2H), 2.91 (t, J = 7.0 Hz, 2H), 2.06 (s, 3H), 2.05 (s, 3H), 2.04 (s, 3H), 2.03 (s, 3H). 13C NMR (151 MHz, CDCl3) δ 170.73, 170.40, 169.56, 169.46, 166.62, 155.75, 135.17, 133.84, 131.44, 130.68, 130.68, 130.35, 130.22, 130.07, 127.23, 117.39, 117.39, 99.36, 72.84, 72.15, 71.28, 68.40, 62.10, 41.47, 34.82, 20.83, 20.77, 20.77, 20.73.
4.2.3.6 2-(Acetoxymethyl)-6-(4-(2-(2-methylbenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-6). White solid with yield 22.78%.1H NMR (600 MHz, chloroform-d) δ 7.28 (d, J = 9.0 Hz, 1H), 7.24 (t, J = 14.8 Hz, 1H), 7.18 (d, J = 7.6 Hz, 1H), 7.15 (t, J = 8.3 Hz, 1H), 7.15 (t, J = 8.3 Hz, 2H), 6.93 (d, J = 8.5 Hz, 2H), 5.75 (d, J = 11.9 Hz, 1H), 5.28 (t, J = 9.4 Hz, 1H), 5.24 (t, J = 7.3 Hz, 1H), 5.15 (t, J = 9.4 Hz, 1H), 5.04 (d, J = 7.4 Hz, 1H), 4.27 (dd, J = 12.3, 5.3 Hz, 1H), 4.15 (dd, J = 12.4, 2.5 Hz, 1H), 3.66 (q, J = 6.7 Hz, 2H), 2.88 (t, J = 6.9 Hz, 2H), 2.38 (s, 3H), 2.05 (s, 3H), 2.04 (s, 3H), 2.03 (s, 3H), 2.02 (s, 3H). 13C NMR (151 MHz, CDCl3) δ 170.72, 170.39, 170.21, 169.55, 169.45, 155.73, 136.55, 136.16, 133.91, 131.15, 130.01, 130.01, 126.69, 125.85, 117.38, 117.38, 99.35, 72.83, 72.15, 71.28, 68.39, 62.08, 41.02, 35.06, 20.82, 20.77, 20.74, 20.72, 19.84.
4.2.3.7 2-(Acetoxymethyl)-6-(4-(2-(cyclohexanecarboxamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-7). White solid with yield 27.29%.1H NMR (600 MHz, chloroform-d) δ 7.10 (d, J = 8.1 Hz, 2H), 6.93 (d, J = 8.1 Hz, 2H), 5.44 (t, J = 5.7 Hz, 1H), 5.29 (t, J = 9.3 Hz, 1H), 5.25 (t, J = 8.4 Hz, 1H), 5.16 (t, J = 9.4 Hz, 1H), 5.05 (d, J = 7.4 Hz, 1H), 4.28 (dd, J = 12.4, 5.4 Hz, 1H), 4.17 (dd, J = 12.2, 2.4 Hz, 1H), 3.46 (q, J = 7.0 Hz, 2H), 2.75 (t, J = 6.9 Hz, 2H), 2.07 (s, 3H), 2.06 (s, 3H), 2.04 (s, 3H), 2.03 (s, 3H), 1.77 (t, J = 16.2 Hz, 5H), 1.38 (m, 2H), 1.21 (m, 4H). 13C NMR (151 MHz, CDCl3) δ 176.61, 171.25, 170.39, 169.55, 169.45, 155.64, 134.16, 130.00, 130.00, 117.29, 117.29, 99.36, 72.83, 72.15, 71.29, 68.40, 62.08, 45.66, 40.59, 35.07, 29.80, 29.80, 25.83, 25.83, 25.83, 20.83, 20.77, 20.74, 20.72.
4.2.3.8 2-(Acetoxymethyl)-6-(4-(2-(cyclopentanecarboxamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-8). White solid with yield 22.60%.1H NMR (600 MHz, DMSO-d6) δ 7.79 (t, J = 5.7 Hz, 1H), 7.15 (d, J = 8.7 Hz, 2H), 6.92 (d, J = 8.6 Hz, 2H), 5.50 (d, J = 8.0 Hz, 1H), 5.42 (t, J = 9.6 Hz, 1H), 5.06 (t, J = 7.98 Hz, 1H), 5.00 (t, J = 9.6 Hz, 1H), 4.23 (dd, J = 21.12 Hz, 7.74 Hz, 2H), 4.08 (dd, J = 12.0, 2.1 Hz, 1H), 3.23 (q, J = 7.2 Hz, 2H), 2.66 (t, J = 7.3 Hz, 2H), 2.49 (d, J = 7.4 Hz, 1H), 2.03 (s, 6H), 2.02 (s, 3H), 1.98 (s, 3H), 1.69 (t, J = 9.5 Hz, 2H), 1.59 (m, 4H), 1.49 (m, 2H).13C NMR (151 MHz, DMSO-d6) δ 175.08, 169.97, 169.59, 169.32, 169.10, 154.86, 134.11, 129.75, 129.75, 116.29, 116.29, 97.34, 71.95, 70.77, 70.72, 68.08, 61.65, 44.30, 40.24, 34.32, 29.93, 29.93, 25.56, 25.56, 20.46, 20.37, 20.32, 20.28.
4.2.3.9 2-(Acetoxymethyl)-6-(4-(2-(3-chlorobenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-9). White solid with yield 28.48%.1H NMR (600 MHz, DMSO-d6) δ 8.66 (t, J = 5.6 Hz, 1H), 7.85 (s, 1H), 7.78 (d, J = 7.8 Hz, 1H), 7.59 (d, J = 5.8 Hz, 1H), 7.49 (t, J = 7.8 Hz, 1H), 7.19 (d, J = 8.2 Hz, 2H), 6.92 (d, J = 8.2 Hz, 2H), 5.50 (d, J = 7.9 Hz, 1H), 5.40 (t, J = 9.6 Hz, 1H), 5.04 (t, J = 7.9 Hz, 1H), 4.99 (t, J = 9.6 Hz, 1H), 4.21 (dd, J = 17.2 Hz, 7.7 Hz, 2H), 4.06 (dd, J = 12.1, 2.1 Hz, 1H), 3.45 (q, J = 6.8 Hz, 2H), 2.80 (t, J = 7.2 Hz, 2H), 2.01 (d, J = 3.0 Hz, 6H), 1.99 (s, 3H), 1.96 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 169.96, 169.59, 169.33, 169.11, 164.70, 154.91, 136.55, 133.97, 133.15, 130.94, 130.31, 129.78, 129.78, 126.94, 125.88, 116.37, 116.37, 97.31, 71.95, 70.77, 70.73, 68.09, 61.64, 41.03, 34.04, 20.44, 20.37, 20.33, 20.28.
4.2.3.10 2-(Acetoxymethyl)-6-(4-(2-(3-methylbenzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-10). White solid with yield 25.87%.1H NMR (600 MHz, DMSO-d6) δ 8.47 (t, J = 5.2 Hz, 1H), 7.63 (s, 1H), 7.60 (d, J = 6.1 Hz, 1H), 7.35 (t, J = 6.4 Hz, 1H), 7.32 (d, J = 5.0 Hz, 1H), 7.19 (d, J = 8.1 Hz, 2H), 6.92 (d, J = 8.1 Hz, 2H), 5.50 (d, J = 8.1 Hz, 1H), 5.40 (t, J = 9.6 Hz, 1H), 5.04 (t, J = 8.9 Hz, 1H), 4.99 (t, J = 9.6 Hz, 1H), 4.20 (dd, J = 20.8, 8.8 Hz, 2H), 4.06 (d, J = 11.9 Hz, 1H), 3.44 (q, J = 7.0 Hz, 2H), 2.80 (t, J = 7.5 Hz, 2H), 2.35 (s, 3H), 2.01 (d, J = 3.5 Hz, 6H), 1.99 (s, 3H), 1.96 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 169.96, 169.58, 169.32, 169.11, 166.24, 154.88, 137.47, 134.61, 134.10, 131.59, 129.76, 129.76, 128.12, 127.68, 124.22, 116.37, 116.37, 97.32, 71.95, 70.76, 70.72, 68.08, 61.63, 40.91, 34.20, 20.93, 20.43, 20.37, 20.32, 20.27.
4.2.3.11 2-(Acetoxymethyl)-6-(4-(2-(3-(trifluoromethyl)benzamido)ethyl)phenoxy)tetrahydro-2H-pyran-3,4,5-triyl triacetate (V-11). White solid with yield 24.94%.1H NMR (600 MHz, DMSO-d6) δ 8.81 (t, J = 5.6 Hz, 1H), 8.15 (s, 1H), 8.12 (d, J = 8.0 Hz, 1H), 7.89 (d, J = 7.8 Hz, 1H), 7.71 (t, J = 7.8 Hz, 1H), 7.20 (d, J = 8.1 Hz, 2H), 6.92 (d, J = 8.1 Hz, 2H), 5.49 (d, J = 8.0 Hz, 1H), 5.40 (t, J = 9.6 Hz, 1H), 5.04 (t, J = 8.9 Hz, 1H), 4.99 (t, J = 9.6 Hz, 1H), 4.20 (t, J = 12.2 Hz, 2H), 4.06 (d, J = 11.9 Hz, 1H), 3.48 (q, J = 7.0 Hz, 2H), 2.82 (t, J = 7.5 Hz, 2H), 2.01 (s, 6H), 1.99 (s, 3H), 1.96 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 169.94, 169.58, 169.32, 169.10, 164.65, 154.92, 135.41, 133.94, 131.24, 129.78, 129.78, 129.63, 129.22, 129.00, 127.70, 123.72, 116.38, 116.38, 97.31, 71.94, 70.76, 70.73, 68.08, 61.63, 41.08, 34.05, 20.41, 20.37, 20.30, 20.27.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol, 30
methanol, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–5
1–5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) (Scheme 1).
1, v/v) (Scheme 1).
4.2.4.1 4-(Chloromethyl)-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D1). White solid with yield 90.2%.1H NMR (600 MHz, DMSO-d6) δ 8.58 (dt, J = 25.7, 5.6 Hz, 1H), 7.81 (t, J = 8.2 Hz, 2H), 7.45 (dd, J = 78.1, 8.1 Hz, 2H), 7.14 (d, J = 8.4 Hz, 2H), 6.95 (d, J = 8.4 Hz, 2H), 5.31 (d, J = 5.2 Hz, 1H), 5.11 (d, J = 4.8 Hz, 1H), 5.05 (d, J = 5.3 Hz, 1H), 4.81 (t, J = 6.0 Hz, 2H), 4.58 (t, J = 5.8 Hz, 1H), 4.46 (s, 1H), 3.67 (dd, J = 11.9, 5.3 Hz, 1H), 3.46 (t, J = 6.0 Hz, 2H), 3.42 (q, J = 6.0 Hz, 1H), 3.25 (td, J = 8.8, 4.9 Hz, 1H), 3.20 (td, J = 9.0, 8.4, 5.1 Hz, 1H), 3.14 (td, J = 9.1, 5.3 Hz, 1H), 2.78 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.74, 155.96, 141.53, 140.59, 134.51, 133.71, 132.79, 129.59, 128.81, 127.57, 127.16, 116.18, 100.49, 77.04, 76.66, 73.29, 73.12, 69.72, 60.71, 57.76, 45.54, 41.19, 34.30. MS (ESI+): m/z: calculated for C22H26ClNO7+ ([M + H]+) 452.35; found 452.14. Purity >99% by HPLC.
4.2.4.2 4-Chloro-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phenethyl)benzamide (D2). White solid with yield 77.4%.1H NMR (600 MHz, DMSO-d6) δ 8.63 (t, J = 5.6 Hz, 1H), 7.83 (d, J = 8.3 Hz, 2H), 7.53 (d, J = 8.2 Hz, 2H), 7.14 (d, J = 8.2 Hz, 2H), 6.95 (d, J = 8.4 Hz, 2H), 5.28 (d, J = 5.1 Hz, 1H), 5.07 (d, J = 4.8 Hz, 1H), 5.01 (d, J = 5.3 Hz, 1H), 4.80 (d, J = 7.6 Hz, 1H), 4.55 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 10.7, 4.8 Hz, 1H), 3.45 (q, J = 6.1 Hz, 2H), 3.42 (d, J = 6.3 Hz, 1H), 3.35 (dd, J = 15.3, 9.5 Hz, 1H), 3.24 (dd, J = 8.7, 4.9 Hz, 1H), 3.21 (dd, J = 7.8, 5.1 Hz, 1H), 3.16 (dd, J = 9.4, 4.4 Hz, 1H), 2.78 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.15, 155.96, 135.95, 133.36, 132.73, 129.55, 129.55, 129.10, 129.10, 128.42, 128.42, 116.20, 116.20, 100.53, 77.03, 76.66, 73.28, 69.75, 60.74, 41.86, 34.21. MS (ESI+): m/z: calculated for C21H24ClNO7+ ([M]+) 437.23; found 437.12. Purity >99% by HPLC.
4.2.4.3 4-Methyl-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D3). White solid with yield 84.19%. 1H NMR (600 MHz, DMSO-d6) δ 8.46 (t, J = 5.6 Hz, 1H), 7.72 (d, J = 7.9 Hz, 2H), 7.25 (d, J = 7.8 Hz, 2H), 7.14 (d, J = 8.2 Hz, 2H), 6.95 (d, J = 8.3 Hz, 2H), 5.28 (d, J = 5.0 Hz, 1H), 5.07 (d, J = 4.8 Hz, 1H), 5.01 (d, J = 5.3 Hz, 1H), 4.80 (d, J = 7.5 Hz, 1H), 4.56 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 10.8, 4.7 Hz, 1H), 3.46 (t, J = 6.1 Hz, 1H), 3.42 (q, J = 6.7 Hz, 2H), 3.29 (dd, J = 14.9, 10.8 Hz, 1H), 3.24 (dd, J = 8.8, 4.8 Hz, 1H), 3.20 (dd, J = 13.0, 7.7 Hz, 1H), 3.17 (dd, J = 10.2, 5.1 Hz, 1H), 2.77 (t, J = 7.4 Hz, 2H), 2.34 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 166.06, 155.93, 140.93, 132.85, 131.85, 129.53, 129.53, 128.83, 128.83, 127.17, 127.17, 116.20, 116.20, 100.54, 77.02, 76.65, 73.29, 69.75, 60.73, 41.55, 34.75, 20.97. MS (ESI+): m/z: calculated for C22H27NO7+ ([M + H]+) 418.92; found 418.18. Purity >92% by HPLC.
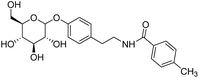
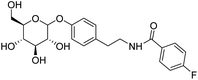
4.2.4.4 4-Fluoro-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D4). White solid with yield 70.31%. 1H NMR (600 MHz, DMSO-d6) δ 8.57 (t, J = 5.6 Hz, 1H), 7.88 (d, J = 14.0 Hz, 2H), 7.28 (d, J = 17.4 Hz, 2H), 7.14 (d, J = 8.4 Hz, 2H), 6.95 (d, J = 8.2 Hz, 2H), 5.29 (d, J = 4.9 Hz, 1H), 5.08 (d, J = 4.6 Hz, 1H), 5.02 (d, J = 5.2 Hz, 1H), 4.81 (d, J = 7.5 Hz, 1H), 4.56 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 11.4, 6.0 Hz, 1H), 3.47 (t, J = 6.0 Hz, 2H), 3.41 (q, J = 6.1 Hz, 1H), 3.30 (dd, J = 15.6, 9.8 Hz, 1H), 3.25 (dd, J = 8.8, 4.8 Hz, 1H), 3.208 (dd, J = 13.0, 7.9 Hz 1H), 3.14 (dd, J = 11.3, 5.2 Hz, 1H), 2.78 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 165.14, 163.01, 155.95, 132.77, 131.11, 129.80, 129.74, 129.54, 129.54, 116.20, 115.30, 115.15, 100.53, 77.02, 76.65, 73.28, 69.75, 60.73, 41.15, 34.26. MS (ESI+): m/z: calculated for C21H24FNO7+ ([M]+) 421.30; found 421.15. Purity >92% by HPLC.
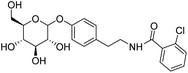
4.2.4.5 2-Chloro-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D5). White solid with yield 78.63%. 1H NMR (600 MHz, DMSO-d6) δ 8.49 (t, J = 5.7 Hz, 1H), 7.47 (d, J = 7.9 Hz, 1H), 7.42 (t, J = 6.8 Hz, 1H), 7.37 (t, J = 7.4 Hz, 1H), 7.33 (d, J = 6.0 Hz, 1H), 7.17 (d, J = 8.1 Hz, 2H), 6.96 (d, J = 8.2 Hz, 2H), 5.29 (d, J = 5.1 Hz, 1H), 5.07 (d, J = 4.9 Hz, 1H), 5.01 (d, J = 5.3 Hz, 1H), 4.81 (d, J = 7.6 Hz, 1H), 4.56 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 10.5, 5.5 Hz, 1H), 3.46 (t, J = 5.8 Hz, 1H), 3.42 (q, J = 6.8 Hz, 2H), 3.30 (dd, J = 9.4, 4.7 Hz, 1H), 3.25 (dd, J = 8.7, 4.8 Hz, 1H), 3.21 (dd, J = 13.9, 5.3 Hz, 1H), 3.16 (dd, J = 14.5 Hz, 9.2 Hz, 1H), 2.77 (t, J = 7.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 166.30, 156.00, 137.15, 132.62, 130.69, 129.90, 129.62, 129.62, 129.62, 128.81, 127.10, 116.20, 116.20, 100.58, 77.03, 76.67, 73.30, 69.76, 60.75, 40.81, 34.11. MS (ESI+): m/z: calculated for C21H24ClNO7+ ([M]+) 437.34; found 437.12. Purity >98% by HPLC.
4.2.4.6 2-Methyl-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D6). White solid with yield 95.18%. 1H NMR (600 MHz, DMSO-d6) δ 8.27 (t, J = 5.6 Hz, 1H), 7.29 (d, J = 6, 3 Hz, 1H), 7.29 (t, J = 7.2 Hz, 1H), 7.23 (d, J = 7.2 Hz, 1H), 7.21 (d, J = 7.7 Hz, 2H), 7.16 (d, J = 8.2 Hz, 2H), 6.96 (d, J = 8.1 Hz, 2H), 5.29 (d, J = 5.1 Hz, 1H), 5.07 (d, J = 4.8 Hz, 1H), 5.01 (d, J = 5.4 Hz, 1H), 4.80 (d, J = 7.6 Hz, 1H), 4.56 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 11.4, 5.8 Hz, 1H), 3.46 (q, J = 5.6 Hz, 1H), 3.42 (t, J = 7.0 Hz, 2H), 3.30 (dd, J = 9.5, 6.0 Hz, 1H), 3.25 (dd, J = 8.7, 4.8 Hz, 1H), 3.21 (dd, J = 13.6 Hz, 5.9 Hz, 1H), 3.16 (dd, J = 14.5 Hz, 9.1 Hz, 1H), 2.77 (t, J = 7.3 Hz, 2H), 2.26 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 169.06, 155.97, 137.39, 135.14, 132.77, 130.39, 129.59, 129.59, 129.16, 126.95, 125.48, 116.20, 116.20, 100.61, 77.03, 76.67, 73.30, 69.76, 60.75, 40.60, 34.22, 19.36. MS (ESI+): m/z: calculated for C22H27NO7+ ([M + H]+) 418.82; found 418.18. Purity >99% by HPLC.
4.2.4.7 N-(4-((3,4,5-Trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)cyclohexanecarboxamide (D7). White solid with yield 89.19%.1H NMR (600 MHz, DMSO-d6) δ 7.71 (t, J = 5.6 Hz, 1H), 7.08 (d, J = 8.6 Hz, 2H), 6.93 (d, J = 8.5 Hz, 2H), 5.28 (d, J = 5.1 Hz, 1H), 5.07 (d, J = 4.8 Hz, 1H), 5.01 (d, J = 5.3 Hz, 1H), 4.79 (d, J = 7.6 Hz, 1H), 4.55 (t, J = 5.5 Hz, 1H), 3.68 (dd, J = 9.7, 5.3 Hz, 1H), 3.45 (t, J = 6.0 Hz, 1H), 3.28 (dd, J = 5.7, 2.1 Hz, 1H), 3.24 (dd, J = 8.8, 4.9 Hz, 1H), 3.20 (dd, J = 12.7 Hz, 7.3 Hz, 3H), 3.15 (dd, J = 9.2 Hz, 3.8 Hz, 1H), 2.62 (t, J = 7.3 Hz, 2H), 2.03 (tt, J = 11.6, 3.4 Hz, 1H), 1.68 (m, 2H), 1.61 (dd, J = 16.4, 12.9 Hz, 3H), 1.29 (m, 2H), 1.17 (dd, J = 25.2, 12.3 Hz, 3H). 13C NMR (151 MHz, DMSO-d6) δ 175.12, 155.89, 132.85, 129.52, 129.52, 116.13, 116.13, 100.56, 77.02, 76.65, 73.28, 69.75, 60.73, 44.03, 40.21, 34.37, 29.25, 25.51, 25.51, 25.32, 25.32. MS (ESI+): m/z: calculated for C21H31NO7+ ([M]+) 409.28; found 409.21. Purity >99% by HPLC.
4.2.4.8 N-(4-((3,4,5-Trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)cyclopentanecarboxamide (D8). White solid with yield 84.08%. 1H NMR (600 MHz, DMSO-d6) δ 7.82 (t, J = 5.6 Hz, 1H), 7.09 (d, J = 8.1 Hz, 2H), 6.93 (d, J = 8.1 Hz, 2H), 5.29 (d, J = 5.0 Hz, 1H), 5.10 (d, J = 4.8 Hz, 1H), 5.04 (d, J = 5.2 Hz, 1H), 4.79 (d, J = 7.6 Hz, 1H), 4.57 (t, J = 5.8 Hz, 1H), 3.67 (dd, J = 13.0, 4.3 Hz, 1H), 3.45 (dd, J = 11.8, 6.0 Hz, 2H), 3.29 (dd, J = 9.5, 5.6 Hz, 1H), 3.25 (dd, J = 8.8, 4.7 Hz, 1H), 3.21 (dd, J = 11.2, 5.9 Hz, 3H), 3.16 (dd, J = 9.1, 5.2 Hz, 1H), 2.63 (t, J = 7.4 Hz, 2H), 1.68 (t, J = 9.4 Hz, 2H), 1.58 (m, 4H), 1.47 (t, J = 9.4 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 175.21, 155.91, 132.87, 132.87, 129.52, 129.52, 116.16, 100.57, 77.02, 76.66, 73.29, 69.75, 60.73, 44.35, 40.44, 34.40, 30.01, 25.64, 30.01, 25.64. MS (ESI+): m/z: calculated for C20H29NO7+ ([M + H]+) 396.83; found 396.19. Purity >86% by HPLC.
4.2.4.9 3-Chloro-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D9). White solid with yield 84.57%.1H NMR (600 MHz, DMSO-d6) δ 8.67 (t, J = 5.6 Hz, 1H), 7.85 (s, 1H), 7.78 (d, J = 7.8 Hz, 1H), 7.59 (d, J = 6.1 Hz, 1H), 7.49 (t, J = 7.9 Hz, 1H), 7.15 (d, J = 8.2 Hz, 2H), 6.95 (d, J = 8.0 Hz, 2H), 5.26 (d, J = 5.1 Hz, 1H), 5.05 (d, J = 4.8 Hz, 1H), 4.99 (d, J = 5.3 Hz, 1H), 4.81 (d, J = 7.6 Hz, 1H), 4.53 (t, J = 5.9 Hz, 1H), 3.68 (dd, J = 12.0, 5.7 Hz, 1H), 3.47 (dd, J = 11.6, 6.0 Hz, 1H), 3.44 (t, J = 6.4 Hz, 2H), 3.30 (t, J = 5.0 Hz, 1H), 3.25 (dd, J = 8.7, 4.6 Hz, 1H), 3.21 (dd, J = 13.0, 4.9 Hz, 1H), 3.16 (dd, J = 14.3, 9.0 Hz, 1H), 2.78 (t, J = 7.5 Hz, 2H). 13C NMR (151 MHz, DMSO-d6) δ 164.71, 155.95, 136.59, 133.16, 132.67, 130.93, 130.33, 129.50, 129.50, 126.96, 125.90, 116.19, 116.19, 100.53, 77.00, 76.64, 73.26, 69.72, 60.70, 41.16, 34.13. MS (ESI+): m/z: calculated for C21H24ClNO7+ ([M]+) 437.36; found 437.12. Purity >99% by HPLC.
4.2.4.10 3-Methyl-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D10). White solid with yield 67.17%.1H NMR (600 MHz, DMSO-d6) δ 8.47 (t, J = 5.6 Hz, 1H), 7.63 (s, 1H), 7.60 (d, J = 6.3 Hz, 1H), 7.32 (d, J = 6.1 Hz, 2H), 7.14 (d, J = 8.1 Hz, 2H), 6.95 (d, J = 8.1 Hz, 2H), 5.26 (d, J = 5.1 Hz, 1H), 5.05 (d, J = 4.7 Hz, 1H), 4.99 (d, J = 5.2 Hz, 1H), 4.81 (d, J = 7.5 Hz, 1H), 4.54 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 11.2, 4.6 Hz, 1H), 3.47 (dd, J = 12.1, 6.0 Hz, 1H), 3.42 (s, 2H), 3.30 (dd, J = 9.4, 5.9 Hz, 1H), 3.25 (dd, J = 8.7, 4.8 Hz, 1H), 3.22 (dd, J = 7.8, 5.1 Hz, 1H), 3.16 (dd, J = 9.8, 5.3 Hz, 1H), 2.78 (t, J = 7.4 Hz, 2H), 2.35 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 166.25, 155.91, 137.48, 134.65, 132.80, 131.59, 129.48, 129.48, 128.14, 127.69, 124.24, 116.17, 116.17, 100.53, 76.99, 76.63, 73.26, 69.72, 60.70, 41.04, 34.27, 20.95. MS (ESI+): m/z: calculated for C22H27NO7+ ([M + H]+) 418.87; found 418.18. Purity >99% by HPLC.
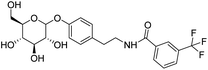
4.2.4.11 3-(Trifluoromethyl)-N-(4-((3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)phenethyl)benzamide (D11). White solid with yield 67.17%.1H NMR (600 MHz, DMSO-d6) δ 8.47 (t, J = 5.6 Hz, 1H), 7.63 (s, 1H), 7.60 (d, J = 6.3 Hz, 1H), 7.32 (d, J = 6.1 Hz, 2H), 7.14 (d, J = 8.1 Hz, 2H), 6.95 (d, J = 8.1 Hz, 2H), 5.26 (d, J = 5.1 Hz, 1H), 5.05 (d, J = 4.7 Hz, 1H), 4.99 (d, J = 5.2 Hz, 1H), 4.81 (d, J = 7.5 Hz, 1H), 4.54 (t, J = 5.8 Hz, 1H), 3.68 (dd, J = 11.2, 4.6 Hz, 1H), 3.47 (dd, J = 12.1, 6.0 Hz, 1H), 3.42 (s, 2H), 3.30 (dd, J = 9.4, 5.9 Hz, 1H), 3.25 (dd, J = 8.7, 4.8 Hz, 1H), 3.22 (dd, J = 7.8, 5.1 Hz, 1H), 3.16 (dd, J = 9.8, 5.3 Hz, 1H), 2.78 (t, J = 7.4 Hz, 2H), 2.35 (s, 3H). 13C NMR (151 MHz, DMSO-d6) δ 166.25, 155.91, 137.48, 134.65, 132.80, 131.59, 129.48, 129.48, 128.14, 127.69, 124.24, 116.17, 116.17, 100.53, 76.99, 76.63, 73.26, 69.72, 60.70, 41.04, 34.27, 20.95. MS (ESI+): m/z: calculated for C22H27NO7+ ([M + H]+) 418.87; found 418.18. Purity >99% by HPLC.
4.3. Cell culture and isolation of neonatal rat cardiomyocytes
Neonatal mice of the C57BL/6 strain (1–3 days) were provided by the Laboratory Animal Center of the Second Affiliated Hospital of Harbin Medical University. The use of animals in this study and the experimental process were strictly in accordance with the National Institutes of Health laboratory animal care and use guidelines and were approved by the Ethics Committee of Harbin Medical University.Neonatal rats (within 3 days of birth) were alcohol-sterilized. Using sterile scissors, an incision was made on the left chest to expose and extract the heart, which was then placed in ice-cold D-Hank's solution. The heart was carefully cleaned of surface blood vessels, residual blood, and attached lung tissue, then transferred to another Petri dish containing fresh D-Hank's solution for further washing. Afterward, D-Hank's solution and collagenase were added, and the heart was placed in a 4 °C shaker for 10–12 hours for digestion. Type II collagenase was added again to continue the digestion process. After digestion, the solution was collected in a 15 mL centrifuge tube and centrifuged at 1000 rpm for 5 minutes. The supernatant was discarded, and the cell pellet was resuspended in a DMEM medium containing 10% FBS. After gentle pipetting, the cell suspension was transferred to a culture flask and incubated at 37 °C with 5% CO2 for 1.5–2 hours. Differential adhesion was performed, and the floating cardiomyocytes were aspirated. The cardiomyocytes were then plated onto a 96-well plate for further cultivation. After 48 hours, the cells were used for subsequent experiments when they exhibited optimal conditions.
4.4. Cytotoxicity assay
The primary cardiomyocytes were digested and then resuspended in 96-well plates with an appropriate concentration, 200 μL per well. After 48 hours of continuous culture, the liquid was changed, and the medicine was added when the condition were favorable. The compounds D1–D11 were prepared with concentrations of 0.1 μM, 0.5 μM, 1 μM, 5 μM, 10 μM, 50 μM, 100 μM, 500 μM and 1000 μM, and were cultured in three multiple wells for 24 hours at each concentration. The CCK-8 solution was diluted with the DMEM medium under the condition of avoiding light, and the dilution ratio was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. After incubation at 37 °C for 1–4 hours, it was shaken in an enzyme label for 10 seconds, and the Optical Density (OD450) at 450 nm was recorded. This experiment assessed cell viability using the CCK-8 assay. CCK-8 is a cell proliferation and viability assay based on the WST-8 reagent. WST-8 is a water-soluble tetrazolium salt that is reduced by dehydrogenases in viable cells (such as succinate dehydrogenase) to form an orange, water-soluble formazan dye, which exhibits a maximum absorbance at 450 nm.
10. After incubation at 37 °C for 1–4 hours, it was shaken in an enzyme label for 10 seconds, and the Optical Density (OD450) at 450 nm was recorded. This experiment assessed cell viability using the CCK-8 assay. CCK-8 is a cell proliferation and viability assay based on the WST-8 reagent. WST-8 is a water-soluble tetrazolium salt that is reduced by dehydrogenases in viable cells (such as succinate dehydrogenase) to form an orange, water-soluble formazan dye, which exhibits a maximum absorbance at 450 nm.
4.5. Derivatives screen assay
Cultured cardiomyocytes were placed in 96-well plates and further cultured for 48 hours. When in good condition, the fluid was changed and the drug was added. The cells were cultured in the DMEM medium without glucose and fetal bovine serum for 3 h and 8 h, respectively, to induce different degrees of cell damage. Compounds D1–D11 were prepared at a concentration of 100 μM; the same concentration of phlorizin and empagliflozin was used as the control group and incubated for 24 h. After incubation, the culture medium was removed, cells were washed with PBS once, and cardiomyocyte activity was measured at 450 nm by the CCK-8 method.4.6. Determination of SGLT2 protein
HK-2 cells were purchased from Hibo Biotechnology Co., Ltd, Qingdao, China. The cultured HK-2 cells were inoculated in 6-well plates and incubated for 24 hours. The next day, the culture medium was removed and the cells were washed multiple times with a PBS buffer solution. The concentrations of D4 were set at 50 μM, 100 μM and 150 μM, while the concentrations of phlorizin and empagliflozin in the control group were set at 100 μM, and the culture was continued for 24 hours. The renal epithelial cells were collected, rinsed twice with PBS, and lysed in a RIPA buffer containing protease and phosphatase inhibitors. The cell debris was removed by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes at 4 °C. The horseradish peroxidase (HRP) labeled detection antibody was added, the plate membrane was sealed, and it was kept in a constant temperature box at 37 °C for 60 minutes. The waste liquid was discarded, the plate was washed and patted dry, a color-developing agent was added for color development, and the Optical Density (OD450) at 450 nm was recorded.
000 rpm for 15 minutes at 4 °C. The horseradish peroxidase (HRP) labeled detection antibody was added, the plate membrane was sealed, and it was kept in a constant temperature box at 37 °C for 60 minutes. The waste liquid was discarded, the plate was washed and patted dry, a color-developing agent was added for color development, and the Optical Density (OD450) at 450 nm was recorded.
4.7. ATP levels of O-glucoside derivatives were determined in vitro
The kit is based on the fact that the enzyme firefly luciferase (also known as luciferase) requires ATP energy to catalyze luciferin to produce fluorescence. The assay system containing saturating concentrations of luciferin and luciferase produces luminescence linearly proportional to ATP concentration. The cultured cardiomyocytes were spread on a 6-well plate and cultured for 48 hours without DMEM damage, and the concentrations of D4 were set at 50 μM and 100 μM, while the concentrations of phlorizin and empagliflozin in the control group were set at 100 μM, and the culture was continued for 24 hours. The culture medium was sucked out, and 200 μL lysate was added to the cells according to the enzyme pore of the 6-well plate. The lysate was collected, centrifuged at 4 °C at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 minutes, and the supernatant was discarded. The ATP content in cardiomyocytes was detected using a luminometer.
000 rpm for 5 minutes, and the supernatant was discarded. The ATP content in cardiomyocytes was detected using a luminometer.
4.8. SOD inhibition rate of O-glucoside derivatives determined in vitro
Superoxide Dismutase SOD (Superoxide Dismutase SOD) can catalyze the disproportionation of superoxide anions to produce hydrogen peroxide and oxygen and is an important antioxidant enzyme in living organisms. It plays a vital role in the oxidation and antioxidant balance of the body. This enzyme can remove superoxide anion free radicals and protect cells from damage. The activity of SOD was determined by the WST-1 method. Cultured cardiomyocytes were spread in a 10 cm dish and cultured for 48 hours without DMEM damage, and the concentrations of D4 were 50 μM and 100 μM, while the concentrations of phlorizin and empagliflozin in the control group were 100 μM, and the culture was continued for 24 hours. Primary cardiomyocytes were collected, washed with PBS, digested with trypsin, and finally collected in PBS buffer. The solution was centrifuged at 4 °C at 1000 rpm/10 min, and the supernatant was discarded. The SOD sample preparation solution was added, centrifuged for 5 minutes at 4 °C, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, and the supernatant was discarded. The Optical Density (OD450) at 450 nm and 650 nm was recorded with an enzymograph.
000 rpm, and the supernatant was discarded. The Optical Density (OD450) at 450 nm and 650 nm was recorded with an enzymograph.
4.9. Molecular docking
Using Pubmed literature search software, we selected 13 heart failure-related proteins. Moreover, the target compound was molecularly docked with 13 proteins by downloading PDB (https://www.rcsb.org/) and UniProt (https://www.uniprot.org/) and using software such as Autodock, PyMOL and Ligplot, their docking index was calculated. The heat map analyzes the binding strength, and the deep color represents the strong binding force between the two components. The interaction of phlorizin and D4 with SGLT2(PDB:7VSi) was explored using molecular docking by Autodock and PyMOL to predict the binding model of the two compounds towards SGLT2. The parameters of the docking process were set as default.4.10. Pharmacokinetic prediction
SwissADME and ADMET 2.0 were used to predict 11 synthetic O-glucoside derivatives. Some predicted parameters are presented, including plasma protein binding rate (PPB), Caco-2 cell permeability, blood–brain barrier, gastrointestinal absorptivity, solubility, and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp lipid–water distribution coefficient. The chemotoxicity of the compounds were investigated.
Kp lipid–water distribution coefficient. The chemotoxicity of the compounds were investigated.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this study.Acknowledgements
We thank the Natural Science Foundation of Heilongjiang Province (LH2022H013) for providing the financial support for this study.References
- G. Savarese and L. H. Lund, Global Public Health Burden of Heart Failure, Card. Fail. Rev., 2017, 3(1), 7–11 CrossRef.
- E. Braunwald, Heart failure, JACC: Heart Failure, 2013, 1, 1–20 Search PubMed.
- G. A. Mensah, G. A. Roth and V. Fuster, The Global Burden of Cardiovascular Diseases and Risk Factors: 2020 and Beyond, J. Am. Coll. Cardiol., 2019, 74(20), 2529–2532 CrossRef PubMed.
- A. M. Ilieşiu and A. S. Hodorogea, Advances in Experimental Medicine and Biology, 2018, vol. 1067, pp. 67–87 Search PubMed.
- J. J. V. McMurray, S. D. Solomon and S. E. Inzucchi, et al., N. Engl. J. Med., 2019, 381(21), 1995–2008 CrossRef PubMed.
- M. Packer, S. D. Anker and J. Butler, et al., N. Engl. J. Med., 2020, 383(15), 1413–1424 CrossRef PubMed.
- A. Gajewska, J. Wasiak, N. Sapeda, E. Młynarska, J. Rysz and B. Franczyk, Int. J. Mol. Sci., 2024, 25(9), 4959 CrossRef CAS PubMed.
- A. Szalat, A. Perlman, M. Muszkat, M. Khamaisi, Z. Abassi and S. N. Heyman, Drug Saf., 2018, 41(3), 239–252 CrossRef.
- J. R. Ehrenkranz, N. G. Lewis, C. R. Kahn and J. Roth, Diabetes/Metab. Res. Rev., 2005, 21(1), 31–38 CrossRef CAS.
- R. A. DeFronzo, L. Norton and M. Abdul-Ghani, Nat. Rev. Nephrol., 2017, 13(1), 11–26 CrossRef CAS PubMed.
- A. Fathi, K. Vickneson and J. S. Singh, Heart Failure Rev., 2021, 26(3), 623–642 CrossRef CAS.
- L. Pinto, D. Rados and L. Remonti, Diabetol. Metab. Syndr., 2015, 7, A58 CrossRef.
- C. C. J. Dekkers, C. D. Sjöström, P. J. Greasley, V. Cain, D. W. Boulton and H. J. L. Heerspink, Diabetes, Obes. Metab., 2019, 21(12), 2667–2673 CrossRef CAS.
- S. Mudaliar, D. Polidori, B. Zambrowicz and R. R. Henry, Diabetes Care, 2015, 38(12), 2344–2353 CrossRef CAS.
- C. G. Santos-Gallego, J. A. Requena-Ibanez and R. San Antonio, et al., J. Am. Coll. Cardiol., 2019, 73(15), 1931–1944 CrossRef CAS.
- T. Obata and M. Miyashita, Eur. J. Pharmacol., 2013, 700(1–3), 60–64 CrossRef PubMed.
- A. J. Scriven, C. T. Dollery, M. B. Murphy, I. Macquin and M. J. Brown, Clin. Pharmacol. Ther., 1983, 33(6), 710–716 CrossRef PubMed.
- V. S. Krishnamurty and A. Grollman, J. Pharmacol. Exp. Ther., 1972, 182(2), 264–272 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra00235d |
| This journal is © The Royal Society of Chemistry 2025 |