Open Access Article
Open Access ArticleDecarburization, denitrification characteristics and microbial community analysis of a full-scale two-stage anoxic–oxic process for treating refractory coking wastewater†
Jie Hu *a,
Bing Xu
*a,
Bing Xu *a,
Jiabao Yan
*a,
Jiabao Yan b and
Guozhi Fan
b and
Guozhi Fan a
a
aSchool of Chemical and Environmental Engineering, Wuhan Polytechnic University, Wuhan 430023, China. E-mail: hujie9231@whpu.edu.cn; xubing200806@163.com
bHubei Province Key Laboratory of Coal Conversion and New Carbon Materials, School of Chemistry and Chemical Engineering, Wuhan University of Science and Technology, Wuhan 430081, China
First published on 27th March 2025
Abstract
Coking wastewater is a representative intractable industrial wastewater, which contains plenty of organic pollutants and nutrient nitrogen and needs to be treated effectively. The decarburization, denitrification characteristics and microbial community composition and structure of coking wastewater treated by a full-scale two-stage anoxic–oxic (A/O) process were systematically investigated. The results showed that the full-scale two-stage A/O process exhibited outstanding decarburization and denitrification capability with a removal efficiency above 90% for chemical oxygen demand (COD), ammonium nitrogen (NH4+–N), and total nitrogen (TN) in coking wastewater. Different biological reaction tanks in the two-stage A/O process played various roles in coking wastewater treatment. COD was mainly removed in the first stage anoxic tank (A1), TN was mainly removed in A1 and the second stage anoxic tank (A2), and NH4+–N was mainly removed in the first stage oxic tank (O1). The function of different biological reaction tanks was highly associated with the composition and structure of the microbial community. The differential microorganisms in different biological reaction tanks were determined by multidimensional analysis. Thiobacillus, Thauera, Thioalkalispira, Pedomicrobium, Azoarcus, etc, were the key differential microorganisms in A1. Mycobacterium, Nitrospira, Acinetobacter, Pseudomonas, Nitrosomonas, etc, were the key differential microorganisms in O1. Bacillus, Thiobacillus, Mesorhizobium, Pusillimonas, etc, were the key differential microorganisms in A2. Truepera, Legionella, Sphingobium, Pseudomonas, etc, were the key differential microorganisms in the second stage oxic tank (O2). Augmenting the key microorganisms in different biological reaction tanks is crucial for boosting the treatment effect of actual coking wastewater.
1. Introduction
Coking wastewater largely originates from the procedures of high-temperature coal destructive distillation, coal oven gas decontamination and coal tar processing, which contains a large amount of intractable organic contaminants, e.g. phenols, polyaromatic hydrocarbons, nitrogenous heterocyclic compounds, as well as nutrient nitrogen.1–6 Excessive organic pollutants and nitrogen emissions will cause water environment pollution and destroy the ecological balance.7–10 Therefore, an effective coking wastewater treatment process is of critical significance for the stable discharge of wastewater and protecting the water ecological environment.Biological treatment technology is the prevalent method in coking wastewater treatment, which mainly utilizes the metabolism of microorganisms to convert pollutants into harmless substances.11–14 However, the traditional biological treatment processes, like sequencing batch reactor (SBR), and anoxic–oxic (A/O) treatment process, have limited effectiveness in treating coking wastewater and the effluent is difficult to meet the more stringent discharge standards.7,15–17 Recently, some novel biological treatment processes, including oxic–hydrolytic–oxic (O/H/O) process, anaerobic–anoxic–oxic (A/A/O) process, and two-stage anoxic–oxic (A/O) process, have been gradually utilized in coking wastewater disposal to enhance the treatment efficacy of coking wastewater.8,18–23 For example, Wei et al. investigated the effect of A/A/O and O/H/O processes in coking wastewater disposal and discovered that A/A/O process was suitable for wastewater with low-load chemical oxygen demand (COD) while O/H/O process was favorable for wastewater with high-load COD.19 Two-stage A/O treatment process is an effective method for coking wastewater, which has been successfully applied in a large amount of coking wastewater treatment plants (CWWTP) throughout China, such as Shanghai, Guangdong Province, and Hubei Province, etc., to boost the removal efficacy of carbonaceous and nitrogenous pollutants.
Microbial community is the critical factor that ascertains the operation performance of CWWTP and has a significant impact on the biological treatment process of coking wastewater.24–28 The microbial community can be controlled by changing the environmental parameters, microbial inoculation and bioaugmentation, and nutrient management. Microorganisms in the two-stage A/O coking wastewater treatment process are highly sensitive to temperature and dissolved oxygen. Most heterotrophic microorganisms thrive in a temperature range of 25–35 °C and effectively remove organic pollutants and nitrogen in coking wastewater. In the oxic tanks, the dissolved oxygen level of 2–4 mg L−1 is typically suitable for aerobic bacteria. In the anoxic tanks, maintaining a low dissolved oxygen level (<0.5 mg L−1) is essential for denitrifying bacteria. In coking wastewater treatment process, when the organic carbon source is insufficient, the growth of heterotrophic bacteria may be severely inhibited. Pseudomonas, Thauera, Thiobacillus, and Nitrospira have been reported to be widely present in the coking wastewater treatment systems.7,12,27–29 Different microorganisms play various roles in the process of treating coking wastewater. It is highly significant to examine the impact of microbial community structure on the treatment efficacy of coking wastewater to optimize the biological treatment process, boost the removal efficiency and reduce the operating cost.26,30–32 At present, most of the reported two-stage A/O coking wastewater treatment processes are based on the experiment-scale studies. These experiment-scale reactors often simplify the complexity of actual two-stage A/O CWWTP. For instance, the hydraulic conditions in the experiment-scale reactors are far from those in the full-scale CWWTP. The flow patterns, mixing intensities, and residence times in the actual CWWTP can be more intricate due to the large volume and complex structure of the treatment tanks. As a result, the treatment efficiency and pollutant removal performance observed in experiment-scale studies may not reflect the actual operation performance of full-scale two-stage A/O CWWTP. In addition, the current microbial community analysis in two-stage A/O process mainly focuses on the composition of microbial communities in each single stage. There is a dearth of research on the change and regulation of microbial communities and the differential microorganisms during the alternate stages of anoxic and oxic processes. Microbial communities are highly dynamic and respond rapidly to changes in environmental conditions. During the transition from anoxic to oxic and vice versa, different microbial species will thrive or decline based on their metabolic capabilities.
A CWWTP with full-scale two-stage A/O process was built in Baowu Steel Group Co., Ltd (Wuhan, China) to treat coking wastewater. In this work, the decarbonization and denitrification performance of the actual full-scale two-stage A/O process were investigated systematically. Continuous monitoring over a period was carried out to accurately capture the dynamic changes of COD, ammonium nitrogen (NH4+–N), total nitrogen (TN), nitrite nitrogen (NO2−–N), and nitrate nitrogen (NO3−–N) in the influent, effluent, and each biological reaction tanks of actual full-scale two-stage A/O process. The microscopic morphology of activated sludge samples in each anoxic and oxic biological reaction tank was observed. Moreover, the metagenomic sequencing was conducted to investigate the composition of microbial community in each biological reaction tank. High-throughput sequencing technologies were applied to sequence the whole microbial genomes and identify the dominant microorganisms that played key roles in the treatment process. The change and regulation of microbial communities and the differential microorganisms in the alternate anoxic and oxic biological reaction tanks were investigated by principal component analysis (PCA), hierarchical cluster analysis (HCA), and orthogonal partial least squares discriminant analysis (OPLS-DA). PCA was used to reduce the dimensionality of the large-scale metagenomic data, identifying the most significant variables that contributed to the variation in microbial communities between different biological reaction tanks. HCA was employed to group samples based on their similarity in microbial composition, revealing distinct clusters that corresponded to specific stages of the two-stage A/O process. OPLS-DA, a powerful statistical method, was utilized to find the differential microorganisms that were most responsible for the treatment performance of different biological reaction tank. Overall, the systematic analysis of the operation efficiency of full-scale two-stage A/O CWWTP and the change and regulation of microbial communities in the alternate anoxic and oxic biological reaction tanks provided valuable insights into the actual coking wastewater treatment systems.
2. Materials and methods
2.1. Full-scale two-stage A/O coking wastewater treatment system
The full-scale two-stage A/O CWWTP was built in Baowu Steel Group Co., Ltd (Wuhan, China). The schematic diagram of this CWWTP is represented in Fig. 1 and the relative parameters of main structures are shown in Table S1.† The influent for the biological treatment process was obtained following water quality regulation of regulation tank to maintain the stable water quality parameters, such as temperature 30–35 °C, pH 6–8, salinity < 1%, etc. The biological treatment process of two-stage A/O consisted of a conventional anoxic–oxic (A/O) process followed by another A/O process. In the first stage A/O process, there are the first stage anoxic tank (A1), first stage oxic tank (O1), and first stage sedimentation tank (S1). The nitrate returning from O1 to A1 was set as 200–300% and the sludge return from S1 to A1 was set as 80–100%. The second stage A/O process included the second stage anoxic tank (A2), second stage oxic tank (O2), and second stage sedimentation tank (S2). The tanks including anoxic tanks, oxic tanks, and sedimentation tanks were constructed of reinforced concrete. The sludge returning from S2 to A2 was set as 80–100% as well. In A2, sodium acetate was utilized as supplementary carbon source to provide nutrients for the denitrification of microorganisms and ensure the nitrogen removal efficiency of A2. After the biological treatment process, the effluent was further treated by the subsequent advanced treatment process.2.2. Decarburization and denitrification performance of two-stage A/O process
The coking wastewater samples were obtained from the influent, A1, O1, A2, O2, and effluent of biological treatment process to assess the decarburization and denitrification performance of full-scale two-stage A/O process. Samples were taken continuously for 20 days at each location and immediately stored at 4 °C until measurement of water quality parameters. The COD, NH4+–N, NO2−–N, NO3−–N, and TN of each sample were determined by the standard methods.33 The decarbonization and denitrification performance of each biological reaction tank were evaluated according to the results of water quality parameters. The statistical analysis and significant differences analysis were conducted by T-tests and the mean and standard deviation were calculated to reflect the variation of each indicator in different biological reaction tanks.2.3. Microscopic morphology of activated sludge samples
The activated sludge samples were taken from the biological reaction tanks of A1, O1, A2, and O2 for microscopic morphology analysis. The microscopic morphology analysis of activated sludge samples was performed according to the reported procedures.12,34,35 Samples were centrifuged at 4000×g to obtain the concentrated sludge samples. The concentrated sludge samples were immobilized by 2.5% glutaraldehyde solution for 24 h. The fixed activated sludge samples were washed three times using phosphate buffer solution, followed by gradient dehydration with ethanol solution of 50%, 70%, 80%, 90%, 100% concentrations. The dewatered activated sludge samples were subjected to supercritical drying and coated with gold by sputtering process, and then observed by scanning electron microscope (SEM, Hitachi, SU8010).2.4. Metagenomic sequencing and microbial community analysis
The activated sludge samples in different biological reaction tanks were taken 6 times respectively at various sites and then used for metagenomic sequencing and microbial community analysis. The total DNA of activated sludge samples was extracted with the PowerSoil DNA Kit (MoBio, California, USA). Metagenomic sequencing was performed on the Illumina MiSeq platform (Illumina, California, USA) by SangonBiotech (Shanghai, China). The sequences were trimmed and assembled for further analysis. The taxonomic classification and Alpha diversity analysis were performed by Mothur program (https://www.mothur.org/).2.5. Differential microorganism analysis in two-stage A/O process
The change and regulation of microbial communities in the alternate anoxic and oxic biological reaction tanks were investigated by PCA, HCA, and OPLS-DA to determine the differential microorganisms in different biological reaction tanks. The PCA and OPLS-DA models were generated by SIMCA software. The HCA was constructed by HEML software. PCA and HCA were utilized to uncover the differences and clusters of microbial communities in the activated sludge samples collected from each biological reaction tank. Pairwise comparisons of microbial communities in the alternate anoxic and oxic biological reaction tanks were performed by using OPLS-DA to identify the differential microorganisms in different biological reaction tanks. The differential microorganisms were obtained by the variable importance for the projection (VIP) values that were determined according to the OPLS-DA model.3. Results and discussion
3.1. Operation performance of two-stage A/O process
The operation performance of full-scale two-stage A/O process was evaluated by determining the changes of COD, NH4+–N, NO2−–N, NO3−–N, and TN in coking wastewater (Fig. 2). As presented in Fig. 2A, the COD of the influent was 2056 ± 208 mg L−1. After the treatment in the A1, more than 60% of COD was removed and the COD of coking wastewater was reduced to 755 ± 124 mg L−1, indicating that the A1 was of great importance in COD removal. In the two-stage A/O wastewater treatment process, COD was mainly removed in the first anoxic biological reaction tank.36–38 After the treatment in the O1, COD was further reduced to 461 ± 59 mg L−1, corresponding to the removal efficiency of 77.2%. Considering that insufficient carbon sources may affect the nitrogen removal effect of microorganisms, sodium acetate was added into the A2 as organic carbon source to provide nutrients for the growth and nitrogen removal of microorganisms. Therefore, the COD of coking wastewater was slightly elevated after the treatment in the A2. Subsequently, the COD of the coking wastewater was further reduced to 279 ± 38 mg L−1 after the treatment in the O2. Finally, the COD of the effluent was < 200 mg L−1 and the removal efficiency reached above 90%. The outstanding COD removal performance is a testament to the high efficiency of the actual two-stage A/O process for coking wastewater treatment.Nutrient nitrogen is one of the important pollution indicators of wastewater and excessive nitrogen discharge will lead to the water eutrophication.39,40 Therefore, it is imperative to eliminate nitrogen from the coking wastewater. The nitrogen removal performances are presented in Fig. 2B–E. NH4+–N in the influent was 216.4 ± 29.4 mg L−1. After the treatment in the A1, NH4+–N was decreased to 138.7 ± 17.5 mg L−1. The increase of NO2−–N and NO3−–N was mainly due to the reflux of nitrate from O1 to A1. TN was reduced from 334.7 ± 26.7 mg L−1 to 177.7 ± 20.0 mg L−1 and the removal efficiency was around 47%. In the O1, microbial nitrification was carried out. NH4+–N was converted into NO2−–N under oxic condition and further to NO3−–N.7,38,41 Therefore, NH4+–N decreased dramatically to 21.8 ± 6.6 mg L−1, corresponding to the increase of NO2−–N and NO3−–N to 3.5 ± 0.75 and 96.2 ± 7.4 mg L−1. TN was slightly reduced to 150.9 ± 19.3 mg L−1, which may be the assimilation of microorganisms. Part of the generated NO2−–N and NO3−–N were removed under anoxic condition by the nitrate reflux into the A1 and others were removed by the second stage A/O process.
In the second stage A/O process, NH4+–N remained nearly unchanged after the treatment in the A2. NO2−–N and NO3−–N were removed rapidly to 0.33 ± 0.13 and 11.9 ± 2.6 mg L−1. Meanwhile, TN was reduced to 46.2 ± 7.1 mg L−1 and the removal rate reached over 85%. It can be found that TN was mainly removed in two anoxic biological reaction tanks of A1 and A2. After the treatment in the O2, the concentrations of various nitrogen were further reduced to ensure the effluent quality. NH4+–N and TN were 4.9 ± 2.4 and 17.1 ± 4.6 mg L−1 after the two-stage A/O biological treatment process, corresponding to the removal efficiency of 97.7% and 94.8%. NO2−–N, NO3−–N were below 0.1 and 6 mg L−1. The low nitrogen concentration in the final effluent indicates that the actual two-stage A/O coking wastewater treatment process is highly effective in removing various forms of nitrogen.
3.2. Characteristics of activated sludge samples
The activated sludge samples were obtained from different biological reaction tanks to measure the sludge characteristics. The mixed liquor suspended solids (MLSS) concentration of activated sludge in A1, O1, A2, O2 biological reaction tanks were 4.5 ± 0.3, 3.3 ± 0.4, 3.6 ± 0.4, and 2.9 ± 0.2 g L−1. The higher organic compounds content in A1 biological reaction tank is more conducive to the growth of microorganisms, making the higher MLSS concentration. The settling velocity at 30 minutes (SV30) of activated sludge samples were 38 ± 6, 25 ± 3, 29 ± 4, and 21 ± 3%, corresponding to the sludge volume index (SVI) of 84, 76, 81, and 72 mL g−1. According to the relevant parameters of the activated sludge samples, it can be found that the activated sludge samples in A1, O1, A2, and O2 biological reaction tanks had great settling performance.As shown in Fig. 3, the microscopic morphology of activated sludge taken from various biological reaction tanks of A1, O1, A2, and O2 was observed by SEM. These figures provided the high-resolution view, allowing for the detailed insights into the structure of the activated sludge samples. A large amount of rod and globular bacteria were observed from the activated sludge samples taken from the biological reaction tanks of A1 and O1 (Fig. 3A and B). The rod-shaped bacteria are often known to play crucial roles in various metabolic processes such as degradation of complex organic compounds within the activated sludge system. The globular bacteria may involve in the process of nutrient cycling. In addition, due to the more organic carbon source in A1 and O1 biological reaction tanks, plenty of extracellular secretions were produced by microbial growth and metabolism and attached to the surface of bacteria. In the A2 and O2, the low content of organic carbon sources resulted in relatively low extracellular secretions produced by microorganisms (Fig. 3C and D).
3.3. Analysis of microbial community
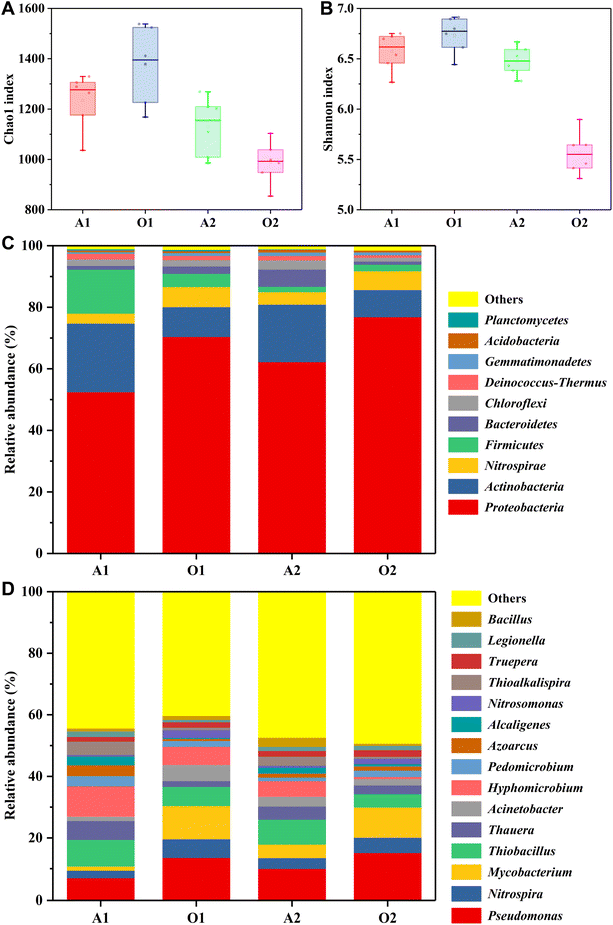
The composition and structure of microbial community in activated sludge samples are intimately connected with the disposal effect of coking wastewater.26,30,42 Therefore, it is vital to analyze the composition and structure of microbial community in activated sludge samples. As shown in Fig. 4A and B, the Alpha diversity indexes including Chao1 index and Shannon index were different in activated sludge samples taken from various biological reaction tanks, indicating that the diversity and richness of microbial community are different. Generally, the larger Chao1 index indicates the higher species richness and the larger Shannon index indicates the more diverse species.5,12,18,43 The Chao1 index and Shannon index of microbial communities in A1, O1, A2, and O2 biological reaction tanks were 1233, 1375, 1131, 988 and 6.57, 6.73, 6.48, 5.56, respectively. It can be found that the diversity and richness were higher in the O1, comparable in two anoxic biological reaction tanks of A1 and A2, and lower in the O2.The taxonomic classification of microbial community at phylum level is shown in Fig. 4C. Among 10 primary phyla, Proteobacteria was the dominant phylum in the activated sludge samples, accounting for 52.5%, 70.4%, 62.3%, and 76.9% in the biological reaction tanks of A1, O1, A2, and O2, respectively. It has been reported that Proteobacteria are mainly involved in biological nitrogen removal and degradation of organic pollutants in coking wastewater disposal.7,12,17 The relative abundance of Actinobacteria in the anoxic biological reaction tanks of A1 and A2 was higher than that in the oxic biological reaction tanks of O1 and O2. Inversely, the relative abundance of Nitrospirae in the oxic biological reaction tanks of O1 and O2 was higher than that in the anoxic biological reaction tanks of A1 and A2. Interestingly, the relative abundance of Firmicutes in the A1 (14.3%) was higher than that in other biological reaction tanks of O1, A2, and O2.
Fig. 4D shows the dominant bacteria in full-scale two-stage A/O process at genus level. As the dominant bacteria, Pseudomonas accounted for the higher proportion in the four biological reaction tanks. Especially in the oxic biological reaction tanks of O1 and O2, the relative abundance of Pseudomonas was more than 10%. Nitrospira is an important nitrifying bacterium, which can oxidize nitrite to nitrate under oxic condition and plays a crucial role in biological nitrogen removal of wastewater.7,36,38,44 The relative abundance of Nitrospira in two oxic biological reaction tanks of O1 and O2 was 6.1% and 5.0%, which was higher than that in two anoxic biological reaction tanks of A1 and A2. Mycobacterium was the subdominant bacteria in the oxic biological reaction tanks of O1 and O2 with a relative abundance of nearly 10%. In the anoxic biological reaction tanks of A1 and A2, the relative abundances of Thiobacillus and Thauera were higher than that in oxic biological reaction tanks of O1 and O2. As reported, Thauera was the dominant nitrate-reducing bacteria in the biological nitrogen removal of coking wastewater.7,8,43,45 Moreover, the relative abundance of Hyphomicrobium in the A1 was much higher than that in other three biological reaction tanks of O1, A2, and O2. The microbial community in the final effluent was similar to that in the biological reaction tank of O2, mainly including Pseudomonas, Mycobacterium, Nitrospira, and Thiobacillus, etc.
3.4. Differential microorganism analysis in two-stage A/O process
To further analyze the differential microorganisms in the full-scale two-stage A/O process, PCA, HCA, and OPLS-DA were used to reveal the change and regulation of microorganisms in four biological reaction tanks. Here top 40 dominant microorganisms in each biological reaction tank were selected for PAC, HCA, and OPLS-DA. As an unsupervised multivariate statistical analysis, PCA was utilized to analyze the clustering trend of multidimensional data. The PCA score plot shows that microbial communities are clearly divided into 4 clusters according to the different biological reaction tanks of A1, O1, A2, and O2 (Fig. S1†). The samples collected from the same biological reaction tank were clustered, demonstrating the excellent reproducibility and uniformity of the replicates. The principal component of PCA model can account for 41.1% and 25.4% of the overall variance among the samples, indicating that microbial communities in each group were different in various biological reaction tanks of two-stage A/O process.Fig. 5 shows the differences in the relative abundance of microorganisms in different biological reaction tanks. From the HCA heatmap, the composition and relative abundances of microorganisms in the same biological reaction tank were similar, while the relative abundances of microorganisms in different biological reaction tanks were significantly different. According to the decarbonization and denitrification characteristics of two-stage A/O process, different biological reaction tanks played different roles, e.g. the COD of coking wastewater was mainly removed in the A1, TN was mainly removed in two anoxic biological reaction tanks of A1 and A2, and NH4+–N was mainly removed in the O1. Therefore, the differential microorganisms in different biological reaction tanks were the key functional microorganisms that played different roles in the treatment of coking wastewater.
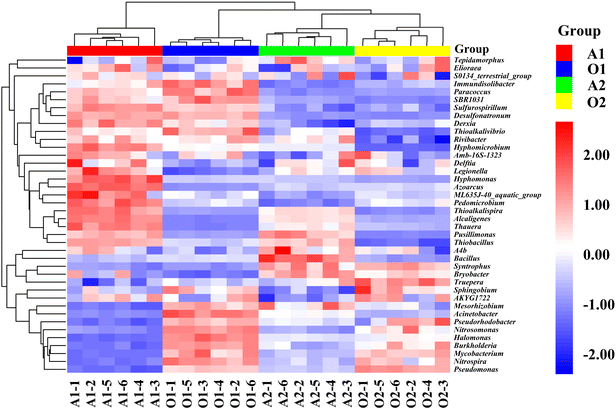 | ||
| Fig. 5 HCA heatmap constructed from the relative abundance of microorganisms in different biological reaction tanks of two-stage A/O process. | ||
To determine the differential microorganisms in different biological reaction tanks, the OPLS-DA model was constructed by pairwise comparisons of microbial communities in the alternate anoxic and oxic biological reaction tanks. The VIP values were determined according to the OPLS-DA model and utilized to identify the differential microorganisms that make the greatest contribution to the categorization. The microorganisms with VIP values ≥1 were chosen as the key differential microorganisms (Tables S2–S5†). In the S-plots generated by the OPLS-DA model (Fig. 6), the key differential microorganisms with significantly different relative abundances were marked as red pentagrams and green diamonds in different biological reaction tanks, respectively.
For the first stage A/O process, the microbial communities in the two biological reaction tanks of A1 and O1 were compared (Fig. 6A). It can be found that 9 microorganisms had higher relative abundance in the O1, including Halomonas, Acinetobacter, Nitrospira, Burkholderia, Mycobacterium, Nitrosomonas, Pseudorhodobacter, Mesorhizobium, and Pseudomonas, which were mainly aerobic bacteria. Thereinto, Nitrosomonas and Nitrospira were the key nitrifying bacteria that could convert NH4+–N into NO2−–N and NO3−–N under oxic condition and played a vital role in biological removal of NH4+–N.46–49 Other 9 microorganisms including Azoarcus, Thauera, Alcaligenes, Legionella, Thioalkalispira, Hyphomonas, Pedomicrobium, Pusillimonas, and Thiobacillus had higher relative abundance in the A1 and could effectively remove organic pollutants under anoxic condition. The microbial communities in two anoxic biological reaction tanks of A1 and A2 were compared as well (Fig. 6B). Six microorganisms had higher relative abundance in the A2 and other 12 microorganisms had higher relative abundance in the A1. In the biological reaction tank of A1, the content of organic carbon source was higher, which was more conducive to the growth and metabolism of microorganisms.
For the second stage A/O process, the microbial communities in two biological reaction tanks of A2 and O2 were compared (Fig. 6C). Eight microorganisms had higher relative abundance in biological reaction tank of O2, such as Sphingobium, Pseudomonas, Mycobacterium, AKYG1722, Nitrosomonas, Pedomicrobium, Truepera, and Nitrospira. Nine microorganisms had higher relative abundance in biological reaction tank of A2, including Bacillus, Thiobacillus, Pusillimonas, Rivibacter, Thioalkalispira, Hyphomicrobium, Alcaligenes, Thioalkalivibrio, Thauera, most of which were reported to have denitrification capability.7,50,51 In the coking wastewater treatment process, sodium acetate was used as organic carbon source and added into the A2 for microbial growth and denitrification. Comparison of microbial communities in two oxic biological reaction tanks of O1 and O2, 4 microorganisms had higher relative abundance in the O2 and 12 microorganisms had higher relative abundance in the O1. The composition, structure, and differential microorganisms of microbial communities in the full-scale two-stage A/O process were closely related to the function of wastewater biological treatment. It is of great significance to augment the key microorganisms in each biological reaction tank to improve the disposal efficacy of actual coking wastewater.
4. Conclusions
The operation performance and microbial community structure of full-stage two-stage A/O process for treating intractable coking wastewater were investigated systematically. More than 90% of COD, ammonium nitrogen, and total nitrogen were removed by the two-stage A/O process, exhibiting outstanding decarburization and denitrification capability. COD was mainly removed in the first anoxic biological reaction tank, ammonium nitrogen was mainly removed in the first oxic biological reaction tank, and total nitrogen was mainly removed in two anoxic biological tanks. Microbial community analysis showed that differential microorganisms existed in different biological reaction tanks and played different roles in the disposal of coking wastewater. Enhancing the dominant microorganisms in various biological reaction tanks is of crucial importance in boosting the treatment efficacy of actual coking wastewater.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its ESI.†Author contributions
Jie Hu: conceptualization, methodology, investigation, formal analysis, writing – original draft, writing – review & editing, funding acquisition. Bing Xu: conceptualization, resources, supervision, writing – review & editing, funding acquisition. Jiabao Yan: conceptualization, methodology, supervision, funding acquisition. Guozhi Fan: methodology, investigation, formal analysis.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by Nature Science Foundation of Hubei Province (2023AFB385), Key Laboratory of Hubei Province for Coal Conversion and New Carbon Materials (Wuhan University of Science and Technology) (WKDM202301), Research and Innovation Initiatives of WHPU (2023Y27), Research Funding of Wuhan Polytechnic University (2024R2006) and Research Program Project of Hubei Provincial Department of Education (F2023009).References
- X. Gao, H. Zhang, Y. Wang, H. Wang, Y. Tang, Y. Hu, Y. Lv and J. Bai, Chem. Eng. J., 2023, 455, 140696 CAS.
- S. Chen, S. Li, W. Wu, H. Ye, H. Liu, S. Ma and Q. Chen, RSC Adv., 2024, 14, 32389 CAS.
- L. Gao and J. L. Goldfarb, RSC Adv., 2011, 9(9), 16018 Search PubMed.
- C. Wang, M. Zheng, G. Chen, Z. Yan, B. Xie, W. Wang and H. Han, J. Water Process Eng., 2023, 56, 104492 Search PubMed.
- W. Xu, H. Zhao, H. Cao, Y. Zhang, Y. Sheng, T. Li, S. Zhou and H. Li, Bioresour. Technol., 2020, 300, 122667 CAS.
- C. Zhao, Y. Ran, Y. Gong, C. Hong, Y. Xing, Y. Sun, H. Wang, W. Ling, Y. Wang, W. Feng, J. Hou, X. Zhai and C. Liu, J. Environ. Chem. Eng., 2024, 12, 114591 CAS.
- H. Chu, X. Liu, J. Ma, T. Li, H. Fan, X. Zhou, Y. Zhang, E. Li and X. Zhang, Chem. Eng. J., 2021, 417, 129204 CrossRef CAS.
- Z. Hou, X. Zhou, Z. Zhao, W. Dong, H. Wang, H. Liu, Z. Zeng and J. Xie, J. Environ. Manage., 2022, 322, 116140 CrossRef CAS.
- G. Meng, N. Jiang, Y. Wang, H. Zhang, Y. Tang, Y. Lv and J. Bai, J. Water Process Eng., 2022, 45, 102482 CrossRef.
- X. Zhou, Z. Zhang and Y. Li, RSC Adv., 2017, 7, 23714 RSC.
- J. Wang, X. Wang, Z. Yu, S. Huang, D. Yao, J. Xiao, W. Chen, Z. Wang and F. Zan, J. Cleaner Prod., 2022, 334, 130269 CrossRef CAS.
- K. Yuan, S. Li and F. Zhong, J. Hazard. Mater., 2020, 400, 123117 CrossRef CAS PubMed.
- J. Yue, Y. Zhao, Y. Sheng, H. Cao and H. Wen, Ind. Eng. Chem. Res., 2020, 59, 5022–5031 CrossRef.
- X. Zhang, Z. Song, Q. Tang, M. Wu, H. Zhou, L. Liu and Y. Qu, J. Environ. Sci., 2021, 101, 373–381 CAS.
- C. Na, Y. Zhang, X. Quan, S. Chen, W. Liu and Y. Zhang, J. Hazard. Mater., 2017, 338, 186–193 CAS.
- G. Ren, M. Zhou, Q. Zhang, X. Xu, Y. Li, P. Su, M. Paidar and K. Bouzek, Water Res., 2019, 154, 336–348 CAS.
- C. Wang, Y. Liu, M. Huang, W. Xiang, Z. Wang, X. Wu, F. Zan and T. Zhou, Bioresour. Technol., 2022, 363, 127897 CAS.
- Q. Ban, L. Zhang and J. Li, Chemosphere, 2022, 286, 131724 CAS.
- C. Wei, J. Wei, Q. Kong, D. Fan, G. Qiu, C. Feng, F. Li, S. Preis and C. Wei, Sci. Total Environ., 2020, 742, 140400 CAS.
- G. Wei, T. Wei, Z. Li, C. Wei, Q. Kong, X. Guan, G. Qiu, Y. Hu, C. Wei, S. Zhu, Y. Liu and S. Preis, Chem. Eng. J., 2023, 466, 143257 CAS.
- H. Zhang, C. Wei, A. Chen, X. Ke, Z. Li, Z. Qin, Y. Tian, H. Wu, G. Qiu and S. Zhu, Bioresour. Technol., 2025, 416, 131754 CAS.
- W. Zhao, Q. Sui and X. Huang, Sci. Total Environ., 2018, 635, 716–724 CAS.
- S. Zhu, H. Wu, C. Wu, G. Qiu, C. Feng and C. Wei, Water Res., 2019, 164, 114963 CAS.
- M.-H. Cai, Y.-C. Tian, A.-M. Li, Y. Li, Y.-Z. Han, J. Li, H.-F. Sun, X. Wang, Q. Zhou and W.-T. Li, J. Environ. Chem. Eng., 2023, 11, 109043 CAS.
- W. Li, Y. Xia, N. Li, J. Chang, J. Liu, P. Wang and X. He, J. Environ. Sci., 2024, 137, 455–468 CAS.
- Y. Li, Q. Wang, H. Chen, C. Song, Y. Zheng, Z. Chai and M. Zheng, Bioresour. Technol., 2024, 411, 131271 CAS.
- Z. Tan, W. Chen, Z. Guo, X. Xu, J. Xie, J. Dai, Y. Lin, B. Sheng, S. Preis, C. Wei and S. Zhu, Appl. Microbiol. Biotechnol., 2024, 108, 490 CAS.
- S. Zhu, Z. Tan, Z. Guo, H. Zheng, B. Zhang, Z. Qin, J. Xie, Y. Lin, B. Sheng, G. Qiu, S. Preis and C. Wei, Water Res., 2024, 257, 121741 CAS.
- S. Zhu, H. Wu, C. Wei, L. Zhou and J. Xie, Appl. Microbiol. Biotechnol., 2016, 100, 949–960 CAS.
- X. Bai, M. Nie, Z. Diwu, L. Wang, H. Nie, Y. Wang, Q. Yin and B. Zhang, Bioresour. Technol., 2022, 347, 126377 CrossRef CAS PubMed.
- B. Cui, S. Fu, X. Hao and D. Zhou, Chemosphere, 2023, 318, 137956 CrossRef CAS.
- P. Xiang, P. Ma, Q. He, Z. Song and Z. Miao, Bioresour. Technol., 2024, 394, 130207 CrossRef CAS PubMed.
- APHA, AWWA, WEF, Standard Methods for the Examination of Water and Wastewater, APHA, Washington, DC, 22th edn, 2012 Search PubMed.
- Z. Li, S. Cun, G. Han, X. Guo, B. Liu, T. Huang, D. Hou, R. Liu and X. Liu, Environ. Res., 2023, 219, 115161 CrossRef CAS.
- Y. Yang, M. Li, Z. Hu, H. Shim, J.-G. Lin, X.-Y. Li and J.-D. Gu, J. Cleaner Prod., 2020, 276, 124176 CAS.
- Y. Feng, S. Wang and Y. Peng, Bioresour. Technol., 2022, 361, 127693 CAS.
- X. Huang, J. Zhu, W. Duan, J. Gao and W. Li, Bioresour. Technol., 2020, 300, 122595 CAS.
- Y. Yang, S. Zhang, A. Yang, J. Li, L. Zhang and Y. Peng, Bioresour. Technol., 2020, 310, 123468 CAS.
- J. Hu, J. Yan, L. Wu, Y. Bao, D. Yu and J. Li, Bioresour. Technol., 2022, 351, 126925 CAS.
- J. Hu, J. Yan, L. Wu, Y. Bao, D. Yu and J. Li, Bioresour. Technol., 2021, 341, 125818 CAS.
- Y. Feng, Y. Peng, B. Wang, B. Liu and X. Li, Sci. Total Environ., 2021, 771, 145387 CrossRef CAS.
- D. R. Joshi, Y. Zhang, H. Zhang, Y. Gao and M. Yang, J. Environ. Sci., 2018, 63, 105–115 CrossRef CAS PubMed.
- X. Zhou, G. Wang, Z. Yin, J. Chen, J. Song and Y. Liu, Chemosphere, 2020, 243, 125382 CrossRef CAS PubMed.
- X. Yan, C. Zhu, B. Huang, Q. Yan and G. Zhang, Bioresour. Technol., 2018, 247, 157–164 CrossRef CAS PubMed.
- Q. Ma, Y. Qu, W. Shen, Z. Zhang, J. Wang, Z. Liu, D. Li, H. Li and J. Zhou, Bioresour. Technol., 2015, 179, 436–443 CAS.
- I. Cotto, K. J. Vilardi, L. Huo, E. C. Fogarty, W. Khunjar, C. Wilson, H. De Clippeleir, K. Gilmore, E. Bailey, S. Lücker and A. J. Pinto, Water Res., 2023, 229, 119497 CAS.
- M. Oshiki, H. Netsu, K. Kuroda, T. Narihiro, N. Fujii, T. Kindaichi, Y. Suzuki, T. Watari, M. Hatamoto, T. Yamaguchi, N. Araki and S. Okabe, Environ. Microbiol., 2022, 24, 3735–3750 CAS.
- Y.-H. Shao, J.-H. Wu and H.-W. Chen, Water Res., 2024, 257, 121698 CrossRef CAS.
- S. Siripong and B. E. Rittmann, Water Res., 2007, 41, 1110–1120 CrossRef CAS PubMed.
- D. Chen, L. Wei, Z. Zou, K. Yang and H. Wang, Appl. Microbiol. Biotechnol., 2016, 100, 6805–6813 CrossRef CAS PubMed.
- X. Zhao, Y. Xie, B. Sun, Y. Liu, S. Zhu, W. Li, M. Zhao and D. Liu, Environ. Res., 2023, 239, 117402 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ra00218d |
| This journal is © The Royal Society of Chemistry 2025 |