Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efficient constitution of a library of rotenoid analogs active against Trypanosoma cruzi from a digitalized plant extract collection†
Arnaud Gaudry *ab,
Laurence Marcourt
*ab,
Laurence Marcourt ab,
Marcel Kaiser
ab,
Marcel Kaiser cd,
Julien Flückigerab,
Bruno David
cd,
Julien Flückigerab,
Bruno David e,
Antonio Grondin
e,
Antonio Grondin e,
Jean-Robert Iosetf,
Pascal Mäser
e,
Jean-Robert Iosetf,
Pascal Mäser cd,
Emerson Ferreira Queiroz
cd,
Emerson Ferreira Queiroz ab,
Pierre-Marie Allard
ab,
Pierre-Marie Allard abg and
Jean-Luc Wolfender
abg and
Jean-Luc Wolfender *ab
*ab
aInstitute of Pharmaceutical Sciences of Western Switzerland, University of Geneva, 1211 Geneva 4, Switzerland. E-mail: arnaud.gaudry@unige.ch; jean-luc.wolfender@unige.ch; laurence.marcourt@unige.ch; fluckiger.julien@gmail.com; emerson.ferrerira@unige.ch
bSchool of Pharmaceutical Sciences, University of Geneva, 1211 Geneva 4, Switzerland
cDepartment of Medical and Parasitology and Infection Biology, Swiss Tropical and Public Health Institute, 4123 Allschwil, Switzerland. E-mail: marcel.kaiser@swisstph.ch; pascal.maeser@swisstph.ch
dFaculty of Science, University of Basel, 4002 Basel, Switzerland
eGreen Mission Pierre Fabre, Institut de Recherche Pierre Fabre, 3 Avenue Hubert Curien, 31562 Toulouse, France. E-mail: brunoxdavid@gmail.com; antonio.grondin@pierre-fabre.com
fDrugs for Neglected Diseases Initiative (DNDi), 1202 Geneva, Switzerland. E-mail: jrioset@dndi.org
gDepartment of Biology, University of Fribourg, 1700 Fribourg, Switzerland. E-mail: pierre-marie.allard@unifr.ch
First published on 9th May 2025
Abstract
Natural products (NP) have proven to be a rich source of potentially bioactive compounds, and metabolomics is the current method of choice for characterizing natural extracts. To integrate the vast amount of data and information produced by modern metabolomics workflows, we recently developed a sample-centric approach for the semantic enrichment and alignment of metabolomics datasets. The resulting Experimental Natural Products Knowledge Graph (ENPKG) is queryable and integrates both newly acquired digitalized experimental data and information, and previously reported knowledge. It allows the highlighting of putative bioactive compounds at the extract level by comparing, for example, the occurrence of compounds of a given chemical class with bioactivity results. Using this approach, we recently described potent anti-Trypanosoma cruzi activity of two rotenoids, deguelin and rotenone. These compounds were identified in six active extracts from four plant species: Cnestis palala (Connaraceae), Chadsia grevei, Pachyrhizus erosus, and Desmodium heterophylum (Fabaceae). In this work, we present the results of the phytochemical investigation of four of these extracts and the establishment of a library of structural analogs for in vitro bioactivity testing. This work led to the isolation, characterization, and biological evaluation of the anti-T. cruzi potential of 41 compounds, including 11 rotenoids and seven compounds reported for the first time. Thanks to modern metabolite annotation and single-step isolation procedures, this work also demonstrates the possibility of considering natural extract libraries as a reservoir of rapidly accessible pure NPs. This perspective could increase the options for NP research and help accelerate NP drug discovery efforts.
Introduction
Chagas disease, or American trypanosomiasis, is a potentially fatal infection caused by Trypanosoma cruzi.1 Six to seven million people are estimated to be chronically infected with T. cruzi, mostly in Latin America, and are at risk of developing severe complications such as arrhythmia, dilated heart, or dilated colon, making Chagas disease an important public health issue.2–4 Current treatments – benznidazole and nifurtimox – are sometimes associated with severe adverse effects and suboptimal efficacy.5 There is therefore a need for new drugs targeting asymptomatic infections before complications appear.6Plants are an important resource for discovering new antiparasitic molecules, with the well-known example of artemisinin isolated from Artemisia annua.7 Because of their particular properties as distinct from synthetic compounds (generally with more sp3 carbons, chiral centers, or oxygen atoms), NPs are more pertinent to biological targets and have higher hit rates in drug discovery screening programs compared to compounds of synthetic origin.8–13 However, obtaining new drug candidates from natural sources also comes with challenges: their structural complexity makes NPs difficult to synthesize, and the quantity and diversity of molecules in natural extracts (NE) make them challenging to comprehensively characterize.
Modern metabolomics methods have been effectively used to decipher NE content.14 These methods are usually based on ultra-high-performance liquid chromatography coupled with data-dependant acquisition (DDA) tandem high-resolution mass spectrometry (UHPLC-HRMS2) analysis, typically followed by molecular networking (MN) and spectral annotation.15,16 Different spectral annotation tools exist for labelling unknown spectra at the structural level, such as SIRIUS/CSI:FingerID or spectral matching, or CANOPUS at the chemical class level, for example.16–19 On the other hand, the same spectral and structural datasets can also be leveraged within research projects for the discovery of bioactive compounds. Several computational approaches have been developed, such as multi-informative MN, bioactivity-based MN, or Compound Activity Mapping, which allow the integration of bioassay screening results and metabolomics data to highlight potentially active compounds in NE prior to any physical isolation processes.20–22
These computational developments led to increasing data volumes in NP research and triggered the need for advanced data analytics methods. We, therefore, recently developed a sample-centric framework to semantically enrich and align LC-MS data from many samples into a single knowledge graph, the Experimental Natural Products Knowledge Graph (ENPKG). We used this framework to explore the results from antiparasitic screening and metabolomic profiling of 1600 plant extracts, and anticipated two putative structural scaffolds that could be responsible for the anti-T. cruzi bioactivities observed, namely rotenoids and quinolones.23–27 Among the eight extracts active against T. cruzi, the activity of one of them (Melochia umbellata (Houtt.) Stapf stems) could be explained by the presence of quinolone derivatives (query). Due to the extensive work already done on the anti-T. cruzi activity of this scaffold, it was not deemed necessary to carry out phytochemical work on this extract.25,26,28,29 For six other active extracts, the activity was linked to rotenoid derivatives, particularly deguelin and rotenone. Biological evaluation of the commercial standards of both compounds showed nanomolar IC50 against the amastigote form of the parasite without toxicity to the host cell.27 Rotenoid derivatives demonstrated significant potential as antiparasitic agents in other studies, particularly against nematodes and protozoan parasites. Deguelin and rotenone, exhibit potent nematocidal activity against Haemonchus contortus, with deguelin showing selective inhibition of larval motility without high cytotoxicity.30 In another study, modified rotenone derivatives have also displayed antiplasmodial activity against Plasmodium falciparum (EC50 values <50 μM) and moderate effects against Leishmania panamensis, though cytotoxicity remains a challenge for some compounds.31 Since, to the best of our knowledge, the anti-T. cruzi activity of rotenoids had not been reported, we went on to investigate the phytochemistry of the six rotenoid-containing active extracts. We used an optimized single-step chromatography isolation workflow to efficiently and rapidly generate a library of rotenoid and related isoflavone analogs and to test them in vitro. This approach enabled an efficient translation from metabolite profiling results to targeted isolation of selected compounds and assessment of their in vitro bioactivity.
Results and discussion
By integrating metabolite profiling and bioactivity data into the ENPKG (https://enpkg.commons-lab.org/graphdb/), we were able to discover that six of the eight extracts active against T. cruzi in the initial screening contained many potential rotenoid derivatives. This was done by analyzing the CANOPUS annotations of the different active extracts at the NPClassifier class level (query: https://enpkg.commons-lab.org/graphdb/sparql?savedQueryName=count_features_canopus_rotenoids_in_samples&owner=admin) and the individual structural annotations (query: https://enpkg.commons-lab.org/graphdb/sparql?savedQueryName=count_annotation_occurence_in_selection_vs_dataset&owner=admin).19,27,32 These extracts were obtained from the following species: Desmodium heterophyllum (Willd.) DC., Chadsia grevei Drake, Pachyrhizus erosus (L.) Urb. (Leguminosae), and Cnestis palala (Lour.) Merr. (Connaraceae).27 For two species, C. grevei and C. palala, two extracts from two different plant parts were active (Fig. 1A). The ENPKG allowed us to highlight the chemical similarity between these six extracts that are not closely related taxonomically: three Fabaceae from different genera and one Connaraceae.27 | ||
| Fig. 1 Experimental workflow of the present study. (A) Identification of rotenoids as responsible for the activity of six extracts against T. cruzi using the Experimental Natural Products Knowledge Graph framework.27 (B) Plant material corresponding to the six active extracts containing rotenoids [Desmodium heterophyllum underground parts, Chadsia grevei roots, and bark, Pachyrhizus erosus leaves, and Cnestis palala woody stems and roots (see ref. 27 for details)] was reextracted by maceration. The obtained EtOAc extracts were prepared using SPE and profiled on a UHPLC-PDA-HRMS2-CAD platform. (C) The LC-HRMS2 data were processed using MZmine 3, SIRIUS-CANOPUS, and GNPS to generate an annotated Ion-Identity Feature-Based Molecular Network (IIN-FBMN). LC-MS peaks corresponding to rotenoids were retrieved using the CANOPUS chemical class annotations. Using the PDA and CAD data, we could confirm they were UV-reactive and present in a relative amount sufficient for isolation. (D) The HPLC gradient was optimized for better separating the targeted compounds for the four selected extracts, one by species. (E) Finally, targeted compounds were obtained using semi-preparative HPLC in a single chromatography step. Pure compounds' structural elucidation was performed using NMR, HRMS, and chiroptical methods, and finally (F) their bioactivity against T. cruzi was assessed in vitro. | ||
As the extracts used in the initial screening were not available in sufficient quantity for further fractionation, dried plant material from the six active plant parts was extracted at a larger scale using maceration with solvents of increasing polarities (hexane, ethyl acetate, and methanol). Ethyl acetate extracts were used for the remainder of the work, as this was the solvent used to generate the initial 1600-extract library.23 To confirm the presence of rotenone and deguelin analogs in the newly obtained extracts, we used UHPLC-DDA-HRMS2-based profiling for metabolite annotation. The analysis of the new extracts was performed using a better chromatographic resolution than was used when obtaining the initial screening metabolomics data (100 mm vs. 50 mm column, longer elution gradient), and charged aerosol detector (CAD) detection was added to obtain semi-quantitative information about the composition of the extracts (Fig. 1B).33,34 The data obtained from this improved metabolite profiling were subjected to Ion-Identity (IIN) Feature-Based Molecular Networking (FBMN) combined with CANOPUS chemical class annotation to highlight compounds of interest9–13 (Fig. 1C). Four extracts covering a maximum of the chemical diversity from the six active extracts, one per species, were selected to further isolate the targeted compounds. These were isolated using one-step high-resolution semi-preparative HPLC and characterized using HRMS, NMR, and chiroptical methods (Fig. 1D and E).35 Finally, the cytotoxicity of the compounds and their activity against T. cruzi were evaluated in vitro (Fig. 1F).
Ion-identity molecular networking and SIRIUS-CANOPUS annotation of the selected extracts
The extracts UHPLC-HRMS2 profiling data in positive ionization (PI) mode were processed using the Ion Identity-FBMN workflow36 and SIRIUS/CSI:FingerID and CANOPUS17–19 were used to annotate the content of the extracts and confirm the presence of rotenoids. The IIN-FBMN allows both (1) the grouping and annotating of features corresponding to the same molecular species (adducts, in-source fragments, etc., for each chromatographic peak) using the MS1 feature peak-shape correlation and the difference in m/z and (2) visualization of spectral similarities among features.36 CANOPUS, on the other hand, is a computational tool that allows the annotation of unknown MS2 spectra at the pathway, superclass, and class level following the NPClassifier taxonomy.32 Because a chemical class can be assigned even if the structure is not reported in any database, it is a particularly relevant tool for annotating analogs of a given chemical class, such as, in this case, rotenoids.In the annotated IIN-FBMN from the six newly obtained extracts, after feature alignment a total of 2639 features were deconvoluted among the extracts, and this number was reduced to 1990 collapsed features after IIN grouping, with 680 of them corresponding to an IIN unique molecular species (i.e., at least two correlated adducts/in-source fragments detected). The IIN-MN clustering (cosine and adducts links) returned a main cluster of 1014 nodes containing most of the compounds of interest (see Fig. 2 for this main cluster and ESI Fig. 7† for the whole MN). Among the 1990 features of the whole MN, at the NPClassifier superclass level, 391 were annotated as flavonoids, 297 as isoflavonoids, and 134 as coumarins by CANOPUS (Fig. 2A and ESI Fig. 7A†).19,32 At the more scaffold-specific defined NPClassifier class level, the main classes are flavanones (141 features), chalcones (123), and rotenoids (116) (Fig. 2B and ESI Fig. 7B†), with coumarins not appearing in the top 3. This analysis at the chemical class level confirms that the selected extracts are rich in rotenoids and other (iso)flavone derivatives, as expected from the ENPKG results. Using standards, we could also unambiguously identify rotenone and deguelin in the MN (Fig. 2C). By visualizing the relative feature intensity mapping on the FBMN nodes (Fig. 2C), we observed that deguelin was detected in all extracts except P. erosus leaf extract, and rotenone in all extracts except the two C. palala extracts. Other potential derivatives were highlighted by their close relationship in the MN, and the relative feature intensity mapping shows that they were either specific to a given species or shared among different extracts (Fig. 2C). These data confirm that the newly obtained extracts are rich in isoflavone and rotenoid derivatives and, therefore, were used to generate a library of isolated rotenoid analogs.
Targeted isolation of rotenoids and isoflavonoids derivatives
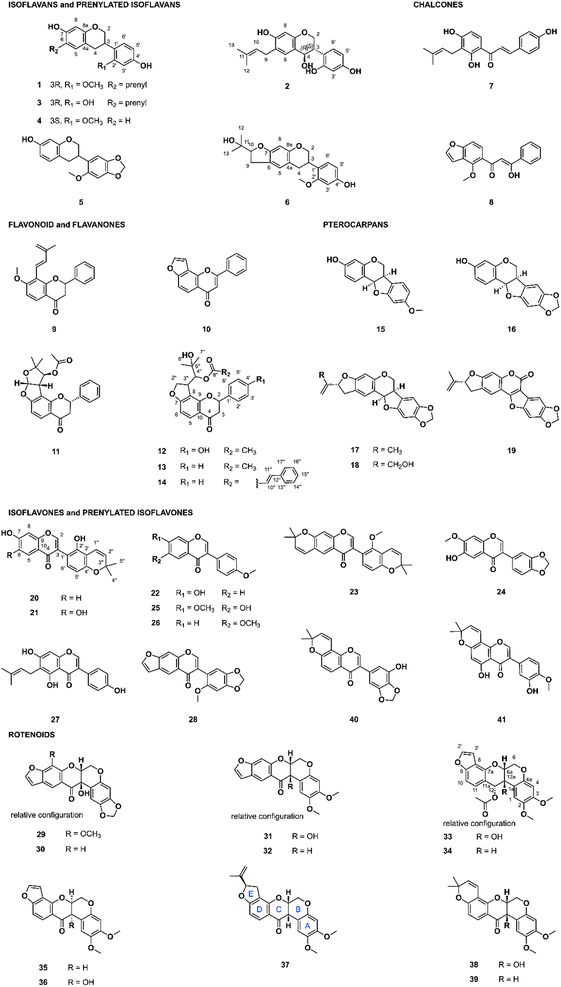
The FBMN indicated that the retention time (RT) of the above-mentioned features annotated as rotenoid and isoflavonoid derivatives was mainly between 4.5 and 9.5 min. In addition, the semi-quantitative CAD signal confirmed that targeted compounds were apparently present in sufficient quantity to isolate amounts suitable for characterization and bioactivity evaluation starting from about 120 mg of extracts (ESI Fig. 1–6†). Because both Chadsia grevei and Cnestis palala extracts presented similar chromatographic profiles, only one extract from each species (C. grevei roots bark and C. palala roots, respectively) was selected for isolation. We performed gradient optimization to maximize the resolution in the chromatographic area of interest using analytical HPLC coupled to a photodiode array (PDA) detector (4.6 × 250 mm I.D., 5 μm, C18 column). The obtained conditions were transferred to semi-preparative HPLC (19 × 250 mm I.D., 5 μm, C18 column) coupled to dry-load injection (see ESI Fig. 8 and 9†) using a geometrical gradient transfer.35 Since all targeted compounds share a characteristic UV-PDA chromophore, their isolation could be easily monitored using UV detection alone. Following this process, compounds of interest were isolated in a single high-resolution semi-preparative chromatography step. This led to the isolation of 9 compounds from Cnestis palala, 11 from Chadsia grevei, 17 from Pachyrhizus erosus, and 9 from Desmodium heterophyllum. This represents a total of 41 different pure NPs (Fig. 3), some being isolated in multiple extracts (see ESI Table 5† for the details about the originating extract(s) for each compound).Of the 41 isolated compounds, 34 had already been reported and identified through their NMR and chiroptical data, while seven (1, 6, 12, 13, 14, 21, and 33) were not reported to our knowledge. Regarding scaffolds, 6 were isoflavans or prenylated isoflavans, 2 were chalcones, 6 were flavonoids or flavanones, 5 were pterocarpans, 11 were isoflavones or prenylated isoflavones, and 11 were rotenoids. The chemical classes of the isolated compounds were consistent with that expected from the CANOPUS annotations.
The 34 known compounds were identified based on their HRMS and NMR data as bavaisoflavanol (2), manuifolin H (3), 7,4′-dihydroxy-2′-methoxyisoflavan (4), astraciceran (5), isobavachalcone (7), pongamol (8), dehydroisoderricin (9), lanceolatin B (10), (−)-purpurin (11), medicarpin (15), maackiain (16), emoroidocarpan (17), (2′R)-4′-hydroxyemoroidocarpan (18), tephcalostan (19), glabrone (20), formononetin (22), munetone (23), acicerone (24), alfalone (25), afrormosin (26), wighteone (27), dehydroneotenone (28), 12a-hydroxypachyrhizone (29), 12a-hydroxydolineone (30), 12a-hydroxyerosone (31), erosone (32), 12-deoxo-12α-acetoxyelliptone (34), elliptone (35), 12a-hydroxyelliptone (36), rotenone (37), tephrosin (38), deguelin (39), norisojamaicin (40) and 3′-hydroxy-4′-O-metyhlerrone (41). The original references used for identification are available in ESI.†
The seven unreported compounds were identified as 2′-O-methylmanuifolin H (1), 4-(2-(2-hydroxypropan-2-yl)-2,3,6,7-tetrahydro-5H-furo[3,2-g]chromen-6-yl)-3-methoxyphenol (6), 2-hydroxy-1-(2-(4-hydroxyphenyl)-4-oxo-3,4,8,9-tetrahydro-2H-furo[2,3-h]chromen-9-yl)-2-methylpropyl acetate (12), 2-hydroxy-2-methyl-1-(4-oxo-2-phenyl-3,4,8,9-tetrahydro-2H-furo[2,3-h]chromen-9-yl)propyl acetate (13), 2-hydroxy-2-methyl-1-(4-oxo-2-phenyl-3,4,8,9-tetrahydro-2H-furo[2,3-h]chromen-9-yl)propyl cinnamate (14), 5′,6,7-trihydroxy-2′,2′-dimethyl-2′H,4H-[3,6′-bichromen]-4-one (21), and 12-deoxo-12α-acetoxy-12a-β-hydroxyelliptone (33). Details about the structural elucidation are shown in ESI.†
Among these isolated compounds, deguelin (39)37 had already been reported in C. grevei and rotenone (37),38 erosone (32),39 12a-hydroxypachyrrhizone (29),38,40 12a-hydroxydolineone (30)40,41 and dehydroneotenone (28)41 in P. erosus. To our knowledge, none of these isolated compounds had been reported for D. heterophyllum and C. palala. While the occurrence of these flavone derivatives in Leguminosae species was expected,42 the Connaraceae family (C. palala) has not been subject to extensive phytochemical investigation and this is the first reported occurrence of rotenoids in this family.
Bioactivity of isolated compounds
The bioactivity of isolated compounds against intracellular T. cruzi amastigotes grown in rat L6 cardiomyocytes is presented in Table 1. Only tephrosin (38) showed activity in the nanomolar range (IC50 of 0.04 μM) comparable to that of deguelin (39, 0.02 μM) and rotenone (37, 0.01 μM). Interestingly, the eight other rotenoids tested (compounds 29–36) were not active against T. cruzi, demonstrating that the activity is very specific even for compounds of the same chemical class. Rotenone and deguelin are known to be strong mitochondrial complex I (also known as NADH:ubiquinone oxidoreductase) inhibitors,43–45 while different activities between closely related rotenoids (rotenone, 5′-α-epirotenone, and 5′-β-epirotenone) has already been shown. The activity of rotenone on mitochondrial complex I was found to be dependent on the bent form of the rotenoid at the B/C ring junction, E ring substitution, ligand flexibility, and 2,3 dimethoxy substitution.46–49 Evaluation of the activity of a series of rotenone and deguelin analogs on the NADH:ubiquinone oxidoreductase also demonstrated important differences in activity.50 It is evident from the various examples presented that the activities of rotenoids on mitochondrial complex I can vary significantly, even with minor structural changes. The variation in activity observed on our rapidly generated series of structural analogs points in the same direction. As shown in Fig. 3 and Table 1, the cyclic and bent form of the rotenoid at the B/C ring junction is essential for activity, since compounds 40 and 41 were not active. The lack of activity of compounds 33–36 highlights the central role of an aliphatic substitution on the E ring for bioactivity.| Compound | IC50a (μM) | Selectivity indexc | |
|---|---|---|---|
| T. cruzi amastigotes | Cytotoxicityb | ||
| a The IC50 are the means of two independent assays.b Rat skeletal myoblast (L6 cells).c Selectivity index (SI) = IC50 cytotoxicity/IC50 against T. cruzi. | |||
| 1 | 28.19 | 99.00 | 3.5 |
| 2 | 43.08 | 49.51 | 1.1 |
| 3 | 28.31 | 106.01 | 3.7 |
| 4 | 247.34 | 358.06 | 1.4 |
| 5 | 94.07 | 75.42 | 0.8 |
| 6 | 202.58 | 153.62 | 0.8 |
| 7 | 60.12 | 103.59 | 1.7 |
| 8 | 26.83 | 73.56 | 2.7 |
| 9 | 66.17 | 95.36 | 1.4 |
| 10 | 9.58 | 130.21 | 13.6 |
| 11 | 1.78 | 6.25 | 3.5 |
| 12 | 58.80 | 99.41 | 1.7 |
| 13 | 122.72 | 130.54 | 1.1 |
| 14 | 15.97 | 81.62 | 5.1 |
| 15 | 201.83 | 226.25 | 1.1 |
| 16 | 118.91 | 155.84 | 1.3 |
| 17 | 47.81 | 84.77 | 1.8 |
| 18 | 40.12 | 118.19 | 2.9 |
| 19 | 65.59 | 118.70 | 1.8 |
| 20 | 149.40 | 186.27 | 1.2 |
| 21 | 56.76 | 76.63 | 1.4 |
| 22 | 216.58 | 224.04 | 1.0 |
| 23 | 7.35 | 38.82 | 5.3 |
| 24 | 86.78 | 186.38 | 2.1 |
| 25 | 240.87 | 263.17 | 1.1 |
| 26 | 205.84 | 255.46 | 1.2 |
| 27 | 46.55 | 103.59 | 2.2 |
| 28 | 3.59 | 18.27 | 5.1 |
| 29 | 93.25 | 124.63 | 1.3 |
| 30 | 195.01 | 141.08 | 0.7 |
| 31 | 19.74 | 113.35 | 5.7 |
| 32 | 39.31 | 114.52 | 2.9 |
| 33 | 65.11 | 98.21 | 1.5 |
| 34 | 27.12 | 103.94 | 3.8 |
| 35 | 18.25 | 99.05 | 5.4 |
| 36 | 5.42 | 100.18 | 18.5 |
| 37 | 0.01 | 0.48 | 51.7 |
| 38 | 0.04 | 72.37 | 1937.0 |
| 39 | 0.02 | 19.40 | 1148.5 |
| 40 | 21.83 | 123.37 | 5.7 |
| 41 | 8.04 | 55.68 | 6.9 |
| Benznidazole | 3.14 | ||
| Podophyllotoxin | 0.02 | ||
In T. cruzi epimastigotes, rotenone has been observed to have a weak effect on NADH-fumarate reductase, NADH-cytochrome c reductase, succinate–cytochrome c reductase, and sn-glycerol-phosphate cytochrome c reductase.51 In addition, T. cruzi kDNA mutants with deletions in complex I subunits demonstrated no changes in mitochondrial bioenergetics, production of ROS, or redox state when compared to their wild-type counterparts, implying a restricted role for complex I in the functioning of T. cruzi epimastigotes.52 However, no studies have been performed on the amastigote form of the parasite to our knowledge; therefore, the role of complex I in the metabolism of this form remains unclear.53 The very selective anti-T. cruzi activity observed for compounds 37–39, known complex I inhibitors, compared to that of the large set of analogs tested, suggests that the observed activity may be linked to complex I inhibition. However, because of the phenotypic nature of the assay used, we can not exclude that the activity observed is due to action on one or more different target(s). Further target deconvolution experiments are needed to confirm the target identity.
Material and methods
General experimental procedures
UV spectra were recorded on a JASCO J-815 spectrometer (Loveland, CO, United States) in MeOH, using a 1 cm cell. The optical rotations were measured in acetonitrile on a JASCO P-1030 polarimeter (Loveland, CO, United States) in a 1 ml, 10 cm tube. NMR data were recorded on a Bruker Avance Neo 600 MHz NMR spectrometer equipped with a QCI 5 mm cryoprobe and a SampleJet automated sample changer (Bruker BioSpin, Rheinstetten, Germany). 1D and 2D NMR experiments (1H, COSY, ROESY, HSQC, and HMBC) were recorded in CD3OD or CDCl3. The residual CD3OD or CDCl3 signals (δH 3.31; δC 49.0 and δH 7.26; δC 77.2, respectively) were used as internal standards for 1H and 13C NMR, respectively. Chemical shifts are reported in parts per million (δ) and coupling constants (J) in Hz.Plant material
The plants investigated are part of the Pierre Fabre Laboratories (PFL) collection, which is among one of the largest collections of plant samples worldwide with over 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 unique samples, and was registered on April 1, 2020, at the European Commission under the accession number 03-FR-2020.23,54 This registration certifies that the collection meets the criteria set out in the EU ABS Regulation which implements at EU level the requirements of the Nagoya Protocol regarding access to genetic resources and the fair and equitable sharing of benefits arising from their utilization. The PFL supplied all the vegetal material (grounded dry material). Precise localization of the initial collection, unique ID (VXXXXX) and barcode are stored in databases. The plant material was dried, and then grounded and stored in plastic pots at a controlled temperature and humidity in the PFL facilities.
000 unique samples, and was registered on April 1, 2020, at the European Commission under the accession number 03-FR-2020.23,54 This registration certifies that the collection meets the criteria set out in the EU ABS Regulation which implements at EU level the requirements of the Nagoya Protocol regarding access to genetic resources and the fair and equitable sharing of benefits arising from their utilization. The PFL supplied all the vegetal material (grounded dry material). Precise localization of the initial collection, unique ID (VXXXXX) and barcode are stored in databases. The plant material was dried, and then grounded and stored in plastic pots at a controlled temperature and humidity in the PFL facilities.
Plant material extraction
Plant material was extracted with three solvents of increasing polarity (hexane, ethyl acetate, and methanol). For each solvent, the dried plant material was macerated 3 consecutive times for 12 hours with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (m/v) solvent ratio. The macerate was filtered, and the filtrate was dried using a rotavapor. For the rest of the work, EtOAc extracts were used. The different PFL identifiers of the plant parts used in this work are: Desmodium heterophyllum (Willd.) DC. underground parts: V114311, Chadsia grevei Drake roots: V113889, C. grevei bark: V113890, Pachyrhizus erosus (L.) Urb. leaves: V112311, and Cnestis palala (Lour.) Merr. woody stems: V113330, C. palala roots: V113332.
10 (m/v) solvent ratio. The macerate was filtered, and the filtrate was dried using a rotavapor. For the rest of the work, EtOAc extracts were used. The different PFL identifiers of the plant parts used in this work are: Desmodium heterophyllum (Willd.) DC. underground parts: V114311, Chadsia grevei Drake roots: V113889, C. grevei bark: V113890, Pachyrhizus erosus (L.) Urb. leaves: V112311, and Cnestis palala (Lour.) Merr. woody stems: V113330, C. palala roots: V113332.
UHPLC-PDA-HRMS2-CAD analysis
Ethyl acetate extracts were analyzed using UHPLC-HRMS/MS-CAD-PDA. Prior to analysis, too apolar compounds of the extracts were removed by C18 solid phase extraction (SPE 1000 mg/12 ml, Finisterre, Teknokroma, Barcelona, Spain). About 10 mg of extract was loaded on the cartridge, followed by elution with 10 ml of methanol (MeOH). The prepared extracts were dried under nitrogen flux. For UHPLC-PDA/HRMS2/CAD analysis, extracts were diluted in MeOH at a 5 mg mL−1 concentration. Chromatographic separation was performed on a Waters Acquity UPLC system interfaced with a Q-Exactive Focus mass spectrometer (Thermo Scientific, Bremen, Germany), using a heated electrospray ionization (HESI-II) source. Thermo Scientific Xcalibur 3.1 software was used for instrument control. The LC conditions were as follows: column, Waters BEH C18 100 × 2.1 mm I.D., 1.7 μm; mobile phase, (A) water with 0.1% formic acid; (B) acetonitrile with 0.1% formic acid; flow rate, 600 μl min−1; injection volume, 5 μl; gradient, a linear gradient of 5–100% B over 13.5 min and isocratic at 100% B for 2 min. The optimized HESI-II parameters were as follows: source voltage, 3.5 kV (pos); sheath gas flow rate (N2), 55 units; auxiliary gas flow rate, 15 units; spare gas flow rate, 3.0; capillary temperature, 350.00 °C, S-Lens RF level, 45. The mass analyzer was calibrated using a mixture of caffeine, methionine–arginine–phenylalanine–alanine–acetate (MRFA), sodium dodecyl sulfate, sodium taurocholate, and Ultramark 1621 in an acetonitrile/methanol/water solution containing 1% formic acid by direct injection. The data-dependent MS/MS events were performed on the three most intense ions detected in full scan MS (Top3 experiment). The MS/MS isolation window width was 1 Da, and the stepped normalized collision energy was set to 15, 30, and 45 units. In data-dependent MS/MS experiments, full scans were acquired at a resolution of 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 FWHM (at m/z 200) and MS/MS scans at 17
000 FWHM (at m/z 200) and MS/MS scans at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 FWHM with an automatically determined maximum injection time. After being acquired in an MS/MS scan, parent ions were placed in a dynamic exclusion list for 2.0 s.
500 FWHM with an automatically determined maximum injection time. After being acquired in an MS/MS scan, parent ions were placed in a dynamic exclusion list for 2.0 s.
UHPLC-MS/MS data-treatment
The MS data were converted from RAW (Thermo) standard data format to mzML format using the MSConvert software, part of the ProteoWizard package.55 The converted files were treated using the MZmine software suite v. 3.1.0.56,57 The parameters were adjusted as follows: the centroid mass detector was used for mass detection with the noise level set to 1.0 × 105 for MS level set to 1, and to 0 for MS level set to 2. The ADAP chromatogram builder was used and set to a minimum group size of scans of 5, minimum group intensity threshold of 1.0 × 105, minimum highest intensity of 5.0 × 105 and m/z tolerance of 10 ppm. For chromatogram deconvolution, the algorithm used was the wavelets (ADAP).58 The intensity window S/N was used as S/N estimator with a signal-to-noise ratio set at 15, a minimum feature height at 5.0 × 105, a coefficient area threshold at 110, a peak duration ranges from 0.1 to 0.5 min and the RT wavelet range from 0.02 to 0.06 min. Isotopes were detected using the isotopes peaks grouper with a m/z tolerance of 15 ppm, a RT tolerance of 0.05 min (absolute), the maximum charge set at 2 and the representative isotope used was the most intense. Individual feature lists were aligned using a RT tolerance of 0.05 min, a m/z tolerance of 10 ppm, a weight for m/z of 2 and a weight for RT of 1. At this point, only features with a corresponding MS/MS spectrum, at least 2 features in the 13C isotope pattern and a RT between 0.5 and 15.5 min were kept. For IIN, correlated features were grouped using the metaCorrelate module (RT tolerance of 0.05 min, no minimal feature height, and an intensity correlation threshold of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). Finally, adducts were annotated for correlated features using the IIN module with a m/z tolerance of 10 ppm and no minimal feature height. Files (.mgf spectra file, .csv features quantification table and .csv IIN edges file) were exported for IIFBMN using the export to GNPS – FBMN module.
000). Finally, adducts were annotated for correlated features using the IIN module with a m/z tolerance of 10 ppm and no minimal feature height. Files (.mgf spectra file, .csv features quantification table and .csv IIN edges file) were exported for IIFBMN using the export to GNPS – FBMN module.
Ion-identity molecular networking
Exported files (spectra, feature quantification table and IIN edges) were uploaded on GNPS to perform IIFBMN.15,36,59 Parameters were set as follows: precursor ion and fragment mass tolerance were set to 0.02, the modified-cosine score threshold was set to 0.7, the minimum matched fragments ions were set to 6 and the maximum component size was set to 100. The resulting IIFBMN is available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=bef399e77f86411ab6857bce924f4b07.Semi-preparative HPLC-PDA purification
To purify the targeted compounds, the chromatographic conditions were first optimized using an HP 1260 Agilent High-Performance liquid chromatography equipped with a photodiode array detector (HPLC-PDA) (Agilent Technologies, Santa Clara, CA, United States). The chromatographic separation was performed on an XBridge C18 column (250 × 4.6 mm I.D., 5 μm; Waters) equipped with a C18 pre-column at 1 mL min−1, with H2O (A) and MeCN or MeOH (B) both containing 0.1% formic acid as solvents. MeCN was used for Cnestis palala roots, Chadsia grevei roots bark, and Pachyrhizus erosus leaves extracts, while MeOH was used for Desmodium heterophyllum underground parts. The UV absorbance was measured at 254 nm, and the UV-PDA spectra were recorded between 190 and 600 nm (step 2 nm).The geometrically transferred gradients were used at the semi-preparative scale on a Shimadzu system equipped with an LC-20 A module pumps, an SPD-20 A UV/VIS, a 7725I Rheodyne® valve, and an FRC-40 fraction collector (Shimadzu, Kyoto, Japan). The separation was performed on an XBridge C18 column (250 mm × 19 mm I.D., 5 μm; Waters) equipped with a C18 precolumn cartridge holder (10 mm × 19 mm I.D., 5 μm; Waters) at 17 mL min−1, with H2O (A) and MeOH or MeCN (B) (same solvent as analytical HPLC optimization) both containing 0.1% formic acid as solvents. The UV detection was set at 210 and 254 nm. The mixtures were injected on the semi-preparative HPLC column using a dry-load methodology.35
Isolated compounds
For details on isolated compounds and structural elucidation, see ESI.†Cytotoxicity assay: L-6 cells
The pure compounds cytotoxicity was assessed against L6 cells as described in ref. 27.Activity against Trypanosoma cruzi
The pure compounds activity was assessed against T. cruzi amastigotes as described in ref. 27.Conclusion
Applying the ENPKG sample-centric and semantic enrichment methodology to anti-parasitic screening results and the associated metabolomics dataset of 1600 plant extracts, we quickly identified rotenoids as being responsible for anti-T. cruzi activity. These derivatives were located in six different plant extracts from four different botanical species: Desmodium heterophyllum, Chadsia grevei, Pachyrhizus erosus, and Cnestis palala. Deguelin or rotenone, two highly active rotenoids, were identified in all six active extracts. Using IIN-FBMN, we found several rotenoid analogs in active extracts from the four botanical species considered. We targeted the isolation of these potentially active analogs using a streamlined single chromatographic-step semi-preparative HPLC procedure. This resulted in a library of 41 compounds, seven of which, to our knowledge, have not been previously reported. Their bioactivity was assessed against intracellular T. cruzi amastigotes, and only tephrosin displayed an activity range similar to deguelin and rotenone. In this work, we demonstrate how modern annotation strategies and mining tools, coupled with state-of-the-art isolation and structural characterization techniques, can help to efficiently generate a library of structural analogs for bioactivity assessment. While only having limited amounts of botanical material and isolated compounds present certain difficulties for complete and detailed structural analysis, the strategy enables fast deconvolution of the compounds responsible for the bioactivity of the crude extracts. This method allows for identification of metabolites responsible for bioactivity in individual extracts, and in the entire NE library, thanks to the connection possible through ENPKG and its multimodal alignment.27 As showcased in this study, and since plants often produce various structurally related scaffolds, the compounds obtained can be used for structure–activity relationship evaluation. This workflow demonstrates how modern NP research methods can facilitate the transition from an NE library to a digitalized NP library. Together with recent advances in computational metabolomics and data analysis, these advances show how these tools can allow researchers to efficiently explore the potential for natural products drug discovery.Data availability
The ENPKG containing data information of the original screening on 1600 plant extracts is available at https://enpkg.commons-lab.org/graphdb/. The IIFBMN obtained from the newly generated extracts of selected species is available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=bef399e77f86411ab6857bce924f4b07.Author contributions
Conceptualization: AGa, P-MA and J-LW. Data curation: AGa, MK and LM. Funding acquisition: J-RI, J-LW and P-MA. Investigation: AGa, JF, MK, LM, EFQ and P-MA. Project administration: J-LW and P-MA. Resources: MK, PM, J-RI, BD, AGr and J-LW. Supervision: J-LW and P-MA. Visualization: AGa. Writing – original draft: AGa. Writing – review & editing: AGa, LM, MK, PM, JR, AGr, BD, J-RI, EFQ, J-LW and P-MA.Conflicts of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.Acknowledgements
The authors are grateful to Green Mission Pierre Fabre, Pierre Fabre Research Institute, Toulouse, France, for establishing and sharing this unique library of extracts. They also thank the Drugs for Neglected Diseases initiative (DNDi) for the received support to fund the bioassays from UK International Development, UK; the Dutch Ministry of Foreign Affairs (DGIS), the Netherlands; and the Federal Ministry of Education and Research (BMBF) through KfW, Germany. J-LW and P-MA thank the Swiss National Science Foundation for receiving support for the project (SNF No. CRSII5_189921/1). J-LW is thankful to the Swiss National Science Foundation for the support in the acquisition of the NMR 600 MHz (SNF Research Equipment grant 316030_164095). P-MA is thankful for the Swiss Open Research Data Grants (CHORD) in Open Science I, a program coordinated by swissuniversities.References
- J. A. Pérez-Molina and I. Molina, Chagas disease, Lancet, 2018, 391, 82–94 CrossRef PubMed.
- M. C. Field, D. Horn, A. H. Fairlamb, M. A. Ferguson, D. W. Gray and K. D. Read, et al., Anti-trypanosomatid drug discovery: an ongoing challenge and a continuing need, Nat. Rev. Microbiol., 2017, 15, 217–231 CrossRef CAS PubMed.
- Centers for Disease Control and Prevention, CDC – Chagas Disease, 2009, https://www.cdc.gov/parasites/chagas/biology.html Search PubMed.
- World Health Organization, Chagas disease, https://www.who.int/news-room/fact-sheets/detail/chagas-disease-(american-trypanosomiasis) Search PubMed.
- A. M. Mejia, B. S. Hall, M. C. Taylor, A. Gómez-Palacio, S. R. Wilkinson and O. Triana-Chávez, et al., Benznidazole-resistance in Trypanosoma cruzi is a readily acquired trait that can arise independently in a single population, J. Infect. Dis., 2012, 206 DOI:10.1093/infdis/jis331.
- J. Martín-Escolano, E. Medina-Carmona and R. Martín-Escolano, Chagas Disease: Current View of an Ancient and Global Chemotherapy Challenge, ACS Infect. Dis., 2020, 6, 2830–2843 CrossRef PubMed.
- J. Wang, C. Xu, Y. K. Wong, Y. Li, F. Liao and T. Jiang, et al., Artemisinin, the magic drug discovered from traditional Chinese medicine, Engineering, 2019, 5, 32–39 CrossRef CAS.
- M. Feher and J. M. Schmidt, Property Distributions: Differences between Drugs, Natural Products, and Molecules from Combinatorial Chemistry, J. Chem. Inf. Comput. Sci., 2003, 43, 218–227 CrossRef CAS PubMed.
- P. A. Clemons, N. E. Bodycombe, H. A. Carrinski, J. A. Wilson, A. F. Shamji and B. K. Wagner, et al., Small molecules of different origins have distinct distributions of structural complexity that correlate with protein-binding profiles, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 18787–18792 CrossRef CAS PubMed.
- K. U. Bindseil, J. Jakupovic, D. Wolf, J. Lavayre, J. Leboul and D. van der Pyl, Pure compound libraries; a new perspective for natural product based drug discovery, Drug Discovery Today, 2001, 6, 840–847 CrossRef CAS PubMed.
- S. C. K. Sukuru, J. L. Jenkins, R. E. J. Beckwith, J. Scheiber, A. Bender and D. Mikhailov, et al., Plate-based diversity selection based on empirical HTS data to enhance the number of hits and their chemical diversity, J. Biomol. Screening, 2009, 14, 690–699 CrossRef CAS PubMed.
- C. F. Stratton, D. J. Newman and D. S. Tan, Cheminformatic comparison of approved drugs from natural product versus synthetic origins, Bioorg. Med. Chem. Lett., 2015, 25, 4802–4807 CrossRef CAS PubMed.
- B. David, A. Grondin, P. Schambel, M. Vitorino and D. Zeyer, Plant natural fragments, an innovative approach for drug discovery, Phytochem. Rev., 2020, 19, 1141–1156 CrossRef CAS.
- J.-L. Wolfender, M. Litaudon, D. Touboul and E. F. Queiroz, Innovative omics-based approaches for prioritisation and targeted isolation of natural products - new strategies for drug discovery, Nat. Prod. Rep., 2019, 36, 855–868 RSC.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg and Y. Peng, et al., Sharing and community curation of mass spectrometry data with GNPS, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- P.-M. Allard, T. Péresse, J. Bisson, K. Gindro, L. Marcourt and V. C. Pham, et al., Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication, Anal. Chem., 2016, 88, 3317–3323 CrossRef CAS PubMed.
- K. Duhrkop, M. Fleischauer, M. Ludwig, A. A. Aksenov, A. V. Melnik and M. Meusel, et al., SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information, Nat. Methods, 2019, 16, 299–302 CrossRef PubMed.
- K. Duhrkop, H. Shen, M. Meusel, J. Rousu and S. Bocker, Searching molecular structure databases with tandem mass spectra using CSI:FingerID, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12580–12585 CrossRef PubMed.
- K. Dührkop, L.-F. Nothias, M. Fleischauer, R. Reher, M. Ludwig and M. A. Hoffmann, et al., Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra, Nat. Biotechnol., 2021, 39, 462–471 CrossRef PubMed.
- F. Olivon, P.-M. Allard, A. Koval, D. Righi, G. Genta-Jouve and J. Neyts, et al., Bioactive Natural Products Prioritization Using Massive Multi-informational Molecular Networks, ACS Chem. Biol., 2017, 12, 2644–2651 CrossRef CAS PubMed.
- L.-F. Nothias, M. Nothias-Esposito, R. da Silva, M. Wang, I. Protsyuk and Z. Zhang, et al., Bioactivity-Based Molecular Networking for the Discovery of Drug Leads in Natural Product Bioassay-Guided Fractionation, J. Nat. Prod., 2018, 81, 758–767 CrossRef CAS PubMed.
- K. L. Kurita, E. Glassey and R. G. Linington, Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11999–12004 CrossRef CAS PubMed.
- P.-M. Allard, A. Gaudry, L.-M. Quirós-Guerrero, A. Rutz, M. Dounoue-Kubo and T. W. N. Walker, et al., Open and reusable annotated mass spectrometry dataset of a chemodiverse collection of 1,600 plant extracts, GigaScience, 2022, 12 DOI:10.1093/gigascience/giac124.
- A. Gaudry, F. Huber, L.-F. Nothias, S. Cretton, M. Kaiser and J.-L. Wolfender, et al., MEMO: Mass Spectrometry-Based Sample Vectorization to Explore Chemodiverse Datasets, Front. Bioinform., 2022, 2 DOI:10.3389/fbinf.2022.842964.
- S. Cretton, S. Dorsaz, A. Azzollini, Q. Favre-Godal, L. Marcourt and S. N. Ebrahimi, et al., Antifungal Quinoline Alkaloids from Waltheria indica, J. Nat. Prod., 2016, 79, 300–307 CrossRef CAS PubMed.
- S. Cretton, L. Breant, L. Pourrez, C. Ambuehl, L. Marcourt and S. N. Ebrahimi, et al., Antitrypanosomal quinoline alkaloids from the roots of Waltheria indica, J. Nat. Prod., 2014, 77, 2304–2311 CrossRef CAS PubMed.
- A. Gaudry, M. Pagni, F. Mehl, S. Moretti, L.-M. Quiros-Guerrero and L. Cappelletti, et al., A sample-centric and knowledge-driven computational framework for natural products drug discovery, ACS Cent. Sci., 2024 DOI:10.1021/acscentsci.3c00800.
- S. Cretton, L. Bréant, L. Pourrez, C. Ambuehl, R. Perozzo and L. Marcourt, et al., Chemical constituents from Waltheria indica exert in vitro activity against Trypanosoma brucei and T. cruzi, Fitoterapia, 2015, 105, 55–60 CrossRef CAS PubMed.
- S. Cretton, M. Kaiser, S. Karimou, S. N. Ebrahimi, P. Mäser and M. Cuendet, et al., Pyridine-4(1H)-one Alkaloids from Waltheria indica as Antitrypanosomatid Agents, J. Nat. Prod., 2020, 83, 3363–3371 CrossRef CAS PubMed.
- H. M. P. Dilrukshi Herath, S. Preston, A. Hofmann, R. A. Davis, A. V. Koehler and B. C. H. Chang, et al., Screening of a small, well-curated natural product-based library identifies two rotenoids with potent nematocidal activity against Haemonchus contortus, Vet. Parasitol., 2017, 244, 172–175 CrossRef CAS PubMed.
- Y. Upegui, J. F. Gil, W. Quinones, F. Torres, G. Escobar and S. M. Robledo, et al., Preparation of rotenone derivatives and in vitro analysis of their antimalarial, antileishmanial and selective cytotoxic activities, Molecules, 2014, 19, 18911–18922 CrossRef PubMed.
- H. W. Kim, M. Wang, C. A. Leber, L.-F. Nothias, R. Reher and K. B. Kang, et al., NPClassifier: A Deep Neural Network-Based Structural Classification Tool for Natural Products, J. Nat. Prod., 2021, 84, 2795–2807 CrossRef CAS PubMed.
- M. Ligor, S. Studzińska, A. Horna and B. Buszewski, Corona-Charged Aerosol Detection: An Analytical Approach, Crit. Rev. Anal. Chem., 2013, 43, 64–78 CrossRef CAS.
- A. Rutz and J.-L. Wolfender, Automated composition assessment of natural extracts: Untargeted mass spectrometry-based metabolite profiling integrating semiquantitative detection, J. Agric. Food Chem., 2023, 71, 18010–18023 CrossRef CAS PubMed.
- E. F. Queiroz, A. Alfattani, A. Afzan, L. Marcourt, D. Guillarme and J. L. Wolfender, Utility of dry load injection for an efficient natural products isolation at the semi-preparative chromatographic scale, J. Chromatogr. A, 2019 DOI:10.1016/j.chroma.2019.03.042.
- R. Schmid, D. Petras, L.-F. Nothias, M. Wang, A. T. Aron and A. Jagels, et al., Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment, Nat. Commun., 2021, 12, 3832 CrossRef CAS PubMed.
- F. Manjary, A. Petitjean, J. Y. Conan, M. T. Martin, F. Frappier and P. Rasoanaivo, et al., Degueline: A Rotenoid Constituent of Chadsia grevei, Planta Med., 1994, 60, 602 CrossRef CAS PubMed.
- E. A. Estrella-Parra, J. C. Gomez-Verjan, I. González-Sánchez, E. R. Vázquez-Martínez, E. Vergara-Castañeda and M. A. Cerbón, et al., Rotenone isolated from Pachyrhizus erosus displays cytotoxicity and genotoxicity in K562 cells, Nat. Prod. Res., 2014, 28, 1780–1785 CrossRef CAS PubMed.
- L. B. Norton and R. Hansberry, Constituents of the Insecticidal Resin of the Yam Bean (Pachyrrhizus erosus), J. Am. Chem. Soc., 1945, 67, 1609–1614 CrossRef CAS.
- M. Krishnamurti, Y. R. Sambhy and T. R. Seshadri, Chemical study of indian yam beans (Pachyrrhizus erosus): Isolation of two new rotenoids: 12a-hydroxydolineone and 12a-hydroxypachyrrhizone, Tetrahedron, 1970, 26, 3023–3027 CrossRef CAS.
- A. Phrutivorapongkul, V. Lipipun, N. Ruangrungsi, T. Watanabe and T. Ishikawa, Studies on the constituents of seeds of Pachyrrhizus erosus and their anti herpes simplex virus (HSV) activities, Chem. Pharm. Bull., 2002, 50, 534–537 CrossRef CAS PubMed.
- N. C. Veitch, Isoflavonoids of the leguminosae, Nat. Prod. Rep., 2009, 26, 776–802 RSC.
- P. Caboni, T. B. Sherer, N. Zhang, G. Taylor, H. M. Na and J. T. Greenamyre, et al., Rotenone, deguelin, their metabolites, and the rat model of Parkinson's disease, Chem. Res. Toxicol., 2004, 17, 1540–1548 Search PubMed.
- P. Lümmen, Complex I inhibitors as insecticides and acaricides, Biochim. Biophys. Acta, 1998, 1364, 287–296 CrossRef PubMed.
- J. Schiller and V. Zickermann, Binding of natural inhibitors to respiratory complex I, Pharmaceuticals, 2022, 15, 1088 CrossRef CAS PubMed.
- H. Ueno, H. Miyoshi, K. Ebisui and H. Iwamura, Comparison of the inhibitory action of natural rotenone and its stereoisomers with various NADH-ubiquinone reductases, Eur. J. Biochem., 1994, 225, 411–417 CrossRef CAS PubMed.
- H. Ueno, H. Miyoshi, M. Inoue, Y. Niidome and H. Iwamura, Structural factors of rotenone required for inhibition of various NADH-ubiquinone oxidoreductases, Biochim. Biophys. Acta, 1996, 1276, 195–202 CrossRef PubMed.
- H. Miyoshi, Structure-activity relationships of some complex I inhibitors, Biochim. Biophys. Acta, 1998, 1364, 236–244 CrossRef CAS PubMed.
- C. S. Pereira, M. H. Teixeira, D. A. Russell, J. Hirst and G. M. Arantes, Mechanism of rotenone binding to respiratory complex I depends on ligand flexibility, Sci. Rep., 2023, 13, 6738 CrossRef CAS PubMed.
- N. Fang and J. E. Casida, Cubé resin insecticide: Identification and biological activity of 29 rotenoid constituents, J. Agric. Food Chem., 1999, 47, 2130–2136 CrossRef CAS PubMed.
- F. R. Hernandez and J. F. Turrens, Rotenone at high concentrations inhibits NADH-fumarate reductase and the mitochondrial respiratory chain of Trypanosoma brucei and T. cruzi, Mol. Biochem. Parasitol., 1998, 93, 135–137 CrossRef CAS PubMed.
- J. C. Carranza, A. J. Kowaltowski, M. A. G. Mendonça, T. C. de Oliveira, F. R. Gadelha and B. Zingales, Mitochondrial bioenergetics and redox state are unaltered in Trypanosoma cruzi isolates with compromised mitochondrial complex I subunit genes, J. Bioenerg. Biomembr., 2009, 41, 299–308 CrossRef PubMed.
- Z. Liu, R. Ulrich vonBargen and L.-I. McCall, Central role of metabolism in Trypanosoma cruzi tropism and Chagas disease pathogenesis, Curr. Opin. Microbiol., 2021, 63, 204–209 CrossRef CAS PubMed.
- European Commission, Official European Commission register of collections, 2020, https://ec.europa.eu/environment/nature/biodiversity/international/abs/pdf/Register%20of%20Collections.pdf Search PubMed.
- M. C. Chambers, B. Maclean, R. Burke, D. Amodei, D. L. Ruderman and S. Neumann, et al., A cross-platform toolkit for mass spectrometry and proteomics, Nat. Biotechnol., 2012, 30, 918–920 CrossRef CAS PubMed.
- T. Pluskal, S. Castillo, A. Villar-Briones and M. Orešič, MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinf., 2010, 11, 395 CrossRef PubMed.
- R. Schmid, S. Heuckeroth, A. Korf, A. Smirnov, O. Myers and T. S. Dyrlund, et al., Integrative analysis of multimodal mass spectrometry data in MZmine 3, Nat. Biotechnol., 2023, 41, 447–449 CrossRef CAS PubMed.
- O. D. Myers, S. J. Sumner, S. Li, S. Barnes and X. Du, One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks, Anal. Chem., 2017, 89, 8696–8703 CrossRef CAS PubMed.
- L.-F. Nothias, D. Petras, R. Schmid, K. Dührkop, J. Rainer and A. Sarvepalli, et al., Feature-based molecular networking in the GNPS analysis environment, Nat. Methods, 2020, 17, 905–908 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Spectral and HRMS data, as well as references, for known isolated compounds. Structural elucidation, including NMR spectra, for new compounds. Occurrence of compounds in each extract. UHPLC and HPLC chromatographic profiles. See DOI: https://doi.org/10.1039/d4ra08652j |
| This journal is © The Royal Society of Chemistry 2025 |