 Open Access Article
Open Access ArticleAntidiabetic potential of Abelmoschus esculentus leaves and fruits: a comparative study assisted by chemical profiling, in vitro and in silico studies
Ahlam Hashem Elwekeel *a,
Elham Amin
*a,
Elham Amin b,
Ahmed M. Sayed
b,
Ahmed M. Sayed cd and
Marwa H. A. Hassan
cd and
Marwa H. A. Hassan *a
*a
aDepartment of Pharmacognosy, Faculty of Pharmacy, Beni-Suef University, Beni-Suef, 62514, Egypt. E-mail: ahlam_hasham2000@yahoo.com; mh_elsefy@yahoo.com; marwa.hassan@pharm.bsu.edu.eg; Tel: +201278982288 Tel: +201122803247
bDepartment of Medicinal Chemistry and Pharmacognosy, College of Pharmacy, Qassim University, Buraydah 51452, Saudi Arabia. E-mail: El.saleh@qu.edu.sa
cDepartment of Pharmacognosy, Faculty of Pharmacy, Nahda University, Beni-Suef, 62513, Egypt. E-mail: ahmed.mohamed.sayed@nub.edu.eg
dDepartment of Pharmacognosy, College of Pharmacy, Almaaqal University, 61014 Basrah, Iraq
First published on 2nd June 2025
Abstract
Abelmoschus esculentus (L.) is an edible plant from the Malvaceae family known for its nutritional value. The phytochemical content and medicinal usefulness of the fruits of this plant have been extensively discussed, but the leaves have not been adequately investigated. The present research provides a comparative study of leaves and fruits in terms of phytochemical content and biological potential. Interestingly, the current findings highlight the higher contents of phenolics and flavonoids in the leaf extracts than in the fruit extracts. The results of GC-MS and LC-HRMS/MS analyses indicated the rich and diverse content of both organs. LC-HRMS/MS analysis allowed the annotation of seventy-four metabolites, with the leaf extract being richer (60 annotated metabolites) than the fruit extract (32 metabolites). Flavonoids, phenolics, and fatty acids were the most predominant classes of the detected metabolites. Remarkably, fatty acid derivatives, coumarins, iridoids, and lignans were reported for the first time in the genus Abelmoschus. The investigation of the potential activities of the two organs concluded that the antioxidant activity of leaves (9.9 ± 0.71 mg AAE per g) is better than that of fruits (7.32 ± 0.91 mg AAE per g). Similarly, the IC50 values for the anti-enzymatic activity of the leaf extract (4.47 ± 0.1 mg mL−1, 3.54 ± 0.08 mg mL−1, 0.385 ± 0.019 μg mL−1, and 1.044 ± 0.05 mg mL−1 against α-glucosidase, α-amylase, DPP-4, and lipase enzymes, respectively) were lower than those for the anti-enzymatic activity of the fruit extract (10.4 ± 0.2 mg mL−1, 6.53 ± 0.15 mg mL−1, 2.669 ± 0.132 μg mL−1, and 14.66 ± 0.67 mg mL−1, respectively). Additionally, the molecular docking simulation study concluded the distinct role of flavonoids in the observed bioactivities. In conclusion, A. esculentus leaves, which are considered agriculture waste, show richer metabolic content and more potent activities than the fruits. Therefore, okra leaves should be valued for these results.
1 Introduction
Human well-being is directly related to nutrition and food. At the health level, dietary habits play a valuable and certain role. In recent years, there has been an increase in evidence-based research focused on functional foods and its various effects on human health, preventing numerous health disorders and blocking degenerative diseases.1–3 Growing research has proven the protective effect of a plant-rich diet against various health disorders, while a diet poor in plants might lead to human disorders.2 A huge number of publications and research have emphasized that vegetables and fruits act as a bioactive barrier against cancer and are identified as “chemo-preventers” owing to presence of plant phytochemicals.3 Therefore, numerous edible plants have been explored for their nutritional values and biological properties.Diabetes mellitus (DM) is a metabolic disease commonly associated with alterations in carbohydrate, protein and fat metabolism.4 DM has been categorized as one of the five directing causes of death worldwide, with statistics indicating that more than 450 million people between the ages of 20 and 79 have been affected by diabetes.5 It is a progressive disease in which inflammation and oxidative stress have been recognized as its most common causes.6 Diabetes occurrence has been closely related to disorders in functional enzymes such as α-glucosidase and α-amylase. These enzymes are closely related to type 2 diabetes as they are involved in the digestion process of carbohydrates.7 Moreover, a high lipid profile and obesity have been accompanied by type 2 diabetes.8
Abelmoschus esculentus (L.) is a member of the Malvaceae family and it is commonly known as lady finger or okra. Okra is an edible plant that can be eaten fresh, cooked and used in salads, stews and soups. Okra is native to Africa, and is now widely cultivated throughout the world: Southern Europe, Middle East, Asia, and America.9 Okra has been used traditionally as a diuretic and in the management of acute inflammation, diarrhea, stomach irritation, dysuria, and gonorrhea infection, bronchitis and pneumonia.10 Also, it was reported that okra can be used as an antihyperlipidemic as it decreases lipid and cholesterol absorption. Moreover, it was found that the polysaccharide contents of okra fruits can lower the glucose levels in the body and improve the glucose tolerance,11 while the polysaccharide content of okra leaves showed potent antioxidant potential.12 The seeds and the fleshy part of okra are rich in polyphenolic constituents such as isoquercetin, quercetin, quercetin-3-O-gentibioside, rutin, and catechin derivatives, hydroxycinnamic derivatives which are known to be main bioactive metabolites in the plant.13–15 Therefore, okra extract may be used to develop novel products with different applications in the nutraceutical section, which could include functional foods with significant antioxidant, antidiabetic, and other health-promoting bioactive properties.16,17 Additionally, the ingestion of okra extract reduces peroxidation of lipids, boosts the levels of antioxidant enzymes such as catalase, superoxide dismutase and glutathione peroxidase and reduces glutathione levels in diabetes, which is a chronic disease characterized by organ damage induced by oxidative stress.18
Based on the previous data, the aim of this work is based on a comparative study between the leaves and fruits of okra (A. esculentus). The study includes phytochemical analysis, involving the determination of total phenolic and total flavonoids as well as GC-MS and LC-HRMS/MS. Biological investigations include in vitro determination of the antioxidant capacity and the inhibitory potential of the extracts against α-glucosidase, α-amylase, DPP-4 (dipeptidyl-peptidase-4) and lipase enzymes. In addition, in silico studies were performed to elucidate the mode of interactions between the identified metabolites in the leaves and fruit extracts against the tested enzymes.
2 Materials and methods
2.1. Plant materials
The leaves and fruits of A. esculentus were collected from the cultivated plants in Beni-Suef governate, South Egypt. The plant parts were air dried and separately ground into a fine powder to yield 500 g of the powdered leaves and fruits, then kept separately in an airtight container until extraction.2.2. Solvents and chemicals
The solvents used during this work (methanol, n-hexane and ethanol) were purchased from El-Nasr Company for chemicals; solvents used in LC/MS/MS analysis were obtained from Sigma-Aldrich. Chemicals used for total phenolic (TPC) and flavonoid (TFC), and antioxidants were Folin–Ciocalteu (F–C), sodium carbonate, gallic acid, aluminum chloride, sodium hydroxide, sodium nitrite, quercetin, ascorbic acid and DPPH, which were purchased from Sigma-Aldrich.2.3. Preparation of the plant extracts
For LC/MS analysis and biology, 100 g of the leaves and fruits was extracted with methanol (300 mL × 3) till complete extraction and then filtered and dried to obtain 5 g and 1 g of dry extract, respectively. One hundred mg of the dried extract was used for LC-MS/MS analysis and biological evaluation. For GC-MS analysis, 10 g of the dried plant powders were extracted with 50 mL n-hexane, then filtered and dried. The n-hexane extract was used for GC-MS analysis. For TPC, TFC and antioxidant, one gram of the dried powder of leaves and fruits was extracted with 50 mL methanol twice using an orbital shaker for 2 h, filtered and the collected filtrates were adjusted to 100 mL in a volumetric flask.2.4. Total phenolic content (TPC)
The TPC of the leaves and fruits was determined using F–C reagent as follows: 100 μL of the extract was mixed with 0.75 mL of F–C reagent (diluted tenfold with distilled water); after 5 min, 0.75 mL of 6% sodium carbonate was added, the mixture was kept for 90 min in the dark at room temperature, and the developed color was measured at 725 nm using a spectrophotometer (Jenway 6300). Different concentrations of gallic acid (25–200 μg mL−1) were used to plot the standard curve and the results were calculated as mg gallic acid equivalents/g dry plant powder.19,202.5. Total flavonoid content (TFC)
The TFC of the leaves and fruits was measured using 10% AlCl3 as follows: in a test tube, 0.5 mL of the extract was diluted with 2.25 mL of distilled water and 0.15 mL of 5% NaNO2 solution was added. After 6 min, 0.3 mL of 10% AlCl3·6H2O solution was added to the mixture and allowed to stand for another 5 min. Finally, 1.0 mL of 1 M NaOH was added. The mixture was mixed well with a vortex. The absorbance was immediately measured at 510 nm using a spectrophotometer. The results were expressed as mg quercetin equivalents in 1 g of dried sample.19,202.6. GC-MS profiling of the n-hexane extract of the leaves and fruits of A. esculentus
GC-MS analysis of the n-hexane extracts was performed using a TRACE GC Ultra gas chromatograph (Thermo Scientific Corp., USA) coupled with a Thermo Scientific mass spectrometer detector (ISQ single quadrupole mass spectrometer). The GC-MS apparatus was outfitted with a TR-5 MS column (30 m × 0.32 mm i.d., 0.25 μm film thicknesses). Helium was used as the carrier gas with flow rate of 1.0 mL min−1 and a split ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 using the following temperature program: 60 °C for 1 min, rising at 4.0 °C min−1 to 240 °C and held for 1 min. The injector and detector were held at 210 °C. Mass spectra were obtained by electron ionization (EI) at 70 eV using a spectral range of 40–450 m/z. The chemical constituents were identified bases on Wiley and NIST libraries as well as comparison of the retention indices. The compounds were identified after comparison with the available data in the computer library (NIST and Wiley) attached to the GC-MS instrument.
10 using the following temperature program: 60 °C for 1 min, rising at 4.0 °C min−1 to 240 °C and held for 1 min. The injector and detector were held at 210 °C. Mass spectra were obtained by electron ionization (EI) at 70 eV using a spectral range of 40–450 m/z. The chemical constituents were identified bases on Wiley and NIST libraries as well as comparison of the retention indices. The compounds were identified after comparison with the available data in the computer library (NIST and Wiley) attached to the GC-MS instrument.
2.7. Metabolites profiling of the alcoholic extract of the leaves and fruits of A. esculentus using LC-MS/MS
Separation was performed using a Thermo Scientific C18 column (Acclaim™ Polar Advantage II, 3 × 150 mm, 3 μm particle size) on an UltiMate 3000 UHPLC system (Dionex). Gradient elution was performed at a flow rate of 0.4 mL min−1 and a column temperature of 40 °C using H2O + 0.1% formic acid (A) and 100% acetonitrile (B) with a total run time of 22 minutes. The injection volume of the sample was 3 μL. The gradient started at 5% B to 80% B in 15 min, followed by a reverse gradient back to 5% B at 22 min. High resolution mass spectrometry was carried out using a MicroTOF QIII Bruker Daltonic with ESI positive ionization and the following settings: capillary voltage: 4500 V; nebulizer pressure: 2.0 bar; drying gas: 8 L min−1 at 300 °C. The mass range was 50–1000 m/z. The accurate mass data of the molecular ions provided by the TOF analyzer were processed by Compass Data Analysis software (Bruker Daltonik GmbH).2.8. Biological activities
| Scavenging effect (%) = [(absorbance of control − absorbance of sample)/absorbance of control] × 100 |
The calibration curve was plotted using different concentrations of ascorbic acid. The result was calculated as mg ascorbic acid equivalent antioxidant capacity in 1 g of sample (mg AEAC per g).
2.8.2.1. In vitro α-glucosidase inhibitory assay. The assay was performed according to the instructions in the kit protocol (Bio-vision, catalog # K938) as follows: in a 96 well clear plate, 10 μL of the sample (plant extract, Acarbose) was mixed with 10 μL of the assay buffer and 10 μL of the enzyme (dilute 2 μL of α-glucosidase with 38 μL of α-glucosidase assay buffer), adjust the volume to 80 μL with the buffer, mix well and incubate at room temperature for 15–20 min. Twenty μL of the enzyme substrate (p-nitrophenyl-α-D-glucopyranoside (PNPG)) was mixed and the absorbance was measured at 410 nm. The IC50 for the extracts and Acarbose were calculated.19,21
2.8.2.2. In vitro α-amylase inhibitory assay. The inhibitory potential of the extracts and the reference drug Acarbose was evaluated in vitro according to the instructions in the kit protocol (Bio-vision, catalog # K482): in a 96 well microplate, 50 μL of the extract or the standard was mixed with 50 μL of the enzyme solution, mixed well and incubated at room temperature for 10 min, followed by the addition of 50 μL of the starch solution and incubated for 3 min. Then, 50 μL of DNS reagent (3,5-dinitrosalicylic acid) was added to stop the reaction the reaction and boiled for 10 min at 85–90 °C in a water bath. The mixture was cooled to room temperature and the absorbance was measured at 405 nm.22
2.8.2.3. In vitro DPP-4 inhibitory assay. The activities of the extracts as inhibitors of DPP-4 were measured using a DPP-4 inhibitor screening assay kit (RayBio Quantichrom DPP-4 inhibitor screening kit) and compared with Sitagliptin (reference drug) according to the manufacturer's protocol. In a 96 well plate, 50 μL of the diluted DPP-4 enzyme solution was mixed with 25 μL of the sample (extract or control), mixed well, and incubated for 10 minutes at 37 °C. Twenty-five μL of the substrate was added to each well and incubated for 30 minutes at 37 °C. After incubation, the fluorescence was measured at Ex/Em = 360/460 nm.23,24
2.8.2.4. In vitro lipase inhibitory assay. Lipase inhibition activity of the leaves, fruit extracts and the reference drug (orlistat) was evaluated according to.25 In this method, the activity was assayed using p-nitrophenyl butyrate (PNB) as a substrate. Fifty μL of the sample (extract or standard) was added to 20 μL of the enzyme (pancreatic lipase, type II, ≥125 units per mg protein from Sigma-Aldrich), diluted with 120 μL of Tris-base buffer solution, and incubated for 25 minutes at 37 °C. Then, 20 μL of the substrate was added, and the amount of p-nitrophenol released in the reaction was measured using a Robonik p2000 ELISA reader at 450 nm.
2.9. Molecular docking study
AutoDock Vina software was used in all molecular docking experiments.26 All dereplicated compounds were docked against the active sites of human α-amylase, α-glucosidase, and dipeptidyl peptidase-4 (DPP4) (with PDB codes 4W9, 3L4W, and 2ONC, respectively).27,28 The docking sites were determined according to the enzyme's co-crystallized ligands. The co-ordinates of the grid boxes were x = −9.682; y = 4.274; z = −23.145; and x = 45.424; y = 92.375; z = 34.811; and x = −10.456; y = 17.557; z = −76.278, respectively. The size of the grid box was set to 20 Å3. The exhaustion was set to 24. Ten poses were generated for each docking experiment. Docking poses were analyzed and visualized using Pymol software.262.10. Statistical analysis
All the data was expressed as means ± standard deviation (SD) from three experiments. The data of TPC, TFC and antioxidant was calculated from the linear calibration curve plotted by Excel software using different concentrations of the standards. In addition, the difference for α-glucosidase, α-amylase, DDP-4 and lipase was considered significant at a P value < 0.05 using one-way analysis of variance (ANOVA) for comparison of the group's differences followed by Tukey's test for multiple comparisons using GraphPad Prism 8 (La Jolla, CA, USA).3 Results and discussion
A. esculentus, known as okra, is a species in the Malvaceae family whose fruit is widely consumed in the human diet. The nutritional value of different parts of okra have been thoroughly investigated and found to be rich in proteins, fibers, polysaccharides, vitamins, and minerals.29 Its medicinal value has also been discussed in several studies that reported its antioxidant, antimicrobial and cytotoxic effects.12,30–32 The current research aims to value okra leaves that are considered as agriculture waste and to compare their phytochemical content and biological efficacy with those of fruits (the edible part of the plant). The metabolic content of the two organs was explored using GC-MS and LC-MS/MS analyses, then the DPPH scavenging effect and the antidiabetic activity of the two organs extracts were compared. It is noteworthy that this study is the first report of the metabolic profiling and antidiabetic potential of an alcoholic extract of A. esculentus (okra) leaves. Moreover, an in silico study was conducted to figure out which of the dereplicated metabolites is effective in inhibiting the tested enzymes.3.1. Total phenolics and total flavonoids
The TPC of the leaves and fruits were evaluated using Folin–Ciocalteu (FC) reagent. The phenolic compounds reduce the FC reagent, forming a blue complex measured at 725 nm, revealing that the leaves showed higher content of phenolics (6.84 ± 0.19 mg GAE per g dry weight (DW)) than fruits, which showed 3.89 ± 0.034 mg GAE per g DW. A previous study of the phenolic content of the leaves, fruits and seeds of okra indicated higher content of phenolics in leaves;33 these results support the current findings. TFC was also evaluated using 10% aluminum chloride and the results were expressed as quercetin equivalent (QE) per g dry weight. The flavonoid content in leaves was quantified as 2.04 ± 0.2 mg QE per g DW while the fruits exhibited 1.4 ± 0.08 mg QE per g DW. Similar results were previously reported by Wu et al. 2020 who investigated the flavonoid content of the fruits in different okra cultivars and concluded variable contents of flavonoids ranging from 1.75 to 3.39 mg RE per g DW.343.2. GC-MS profiling of the n-hexane extract of leaves and fruits of A. esculentus
The constituents in the n-hexane extracts from leaves and fruits were analyzed using GC-MS and the identified compounds were recorded in Table 1. The results noted that the linoleic acid, palmitic acid and oleic acid fatty acids with peak areas 14.52%, 10.73% and 8.83%, respectively are the major components in the fruit extract. Oleic acid, 2-methylhexacosane, and hexadecanoic acid ethyl ester with peak areas of 12.16%, 10.58% and 7.13%, respectively were the major identified compounds in the leaf extract. Reviewing the relevant literature, several studies were found discussing GC-MS analysis of okra.35,36 Osman et al., analyzed okra fruit extracts using GC-MS and concluded that hexadecanoic acid methyl ester and 9,12-octadecadienoic acid methyl ester were among the major detected components.36 Interestingly, the current findings stated the identification of hexadecanoic acid methyl ester (7.13%) and 9,12-octadecadienoic acid ethyl ester (4.98%) in the leaf extract, while 9,12-octadecadienoic acid (14.52%) was detected in the fruit extract.| Compound name | Rt | Fruit | Leaves | Molecular formula | |
|---|---|---|---|---|---|
| 1 | Tridecanol | 4.28 | 2.19 | — | C13H28O |
| 2 | 2,7-Dimethyl-1-octanol | 4.86 | 3.94 | — | C10H22O |
| 3 | 1-Octadecanethiol | 5.26 | 1.56 | — | C18H38S |
| 4 | Propane, 1-(dodecyloxy)-2,3-epoxy- | 5.49 | 1.40 | — | C15H30O2 |
| 5 | Trans-3-decen-1-ol | 5.86 | 3.60 | — | C10H20O |
| 6 | 4-Tridecene, (Z)- | 6.76 | 4.89 | — | C13H26 |
| 7 | 1-Octadecyne | 7.12 | 1.08 | — | C18H34 |
| 8 | 1-Hexadecyne | 7.38 | 1.41 | — | C16H30 |
| 9 | 1-Chlorooctadecane | 8.01 | 3.64 | — | C18H37Cl |
| 10 | 1-Octene | 8.95 | 2.22 | — | C8H16 |
| 11 | 1-Octanol, 2-butyl- | 9.13 | 2.42 | — | C12H26O |
| 12 | n-Heptadecylcyclohexane | 9.53 | 2.27 | — | C23H46 |
| 13 | 1-Chlorohexadecane | 9.95 | 1.21 | — | C16H33Cl |
| 14 | Benzene, (2-decyldodecyl) | 10.06 | 1.76 | — | C28H50 |
| 15 | 10-Heneicosene, 11-phenyl- | 10.57 | 3.74 | — | C27H46 |
| 16 | Oxirane, dodecyl | 10.97 | 1.49 | — | C14H28O |
| 17 | Neophytadiene | 19.98 | — | 3.58 | C20H38 |
| 18 | Hexahydrofarnesyl acetone | 20.06 | — | 1.71 | C18H36O |
| 19 | 1-Heptatriacotanol | 21.19 | — | 2.46 | C37H76O |
| 20 | Hexadecanoic acid, methyl ester | 21.29 | — | 2.16 | C17H34O2 |
| 21 | Hexadecanoic acid, ethyl ester | 22.31 | — | 7.13 | C18H36O2 |
| 22 | Methyl octadecanoate | 21.36 | 1.24 | — | C19H38O2 |
| 23 | Tetradecanoic acid | 23.11 | 3.94 | — | C14H28O2 |
| 24 | Phytol | 24.05 | 1.16 | 2.59 | C20H40O |
| 25 | Dodecanoic acid | 24.15 | 1.46 | — | C12H24O2 |
| 26 | 9,12-Octadecadienoic acid (9Z,12Z)-, ethyl ester | 24.68 | — | 4.98 | C20H36O2 |
| 27 | Hexadecanoic acid | 24.85 | 10.37 | — | C16H32O2 |
| 28 | Heptadecanoic acid, ethyl ester | 25.12 | — | 1.68 | C19H38O2 |
| 29 | Oleic acid | 25.51 | 8.83 | 12.16 | C18H34O2 |
| 30 | 9,12-Octadecadienoic acid (Z,Z)- | 27.41 | 14.52 | — | C18H32O2 |
| 31 | Stigmasterol | 29.36 | — | 3.74 | C29H48O |
| 32 | Di-n-octyl phthalate | 30.19 | 1.87 | 6.16 | C24H38O4 |
| 33 | Cholest-22-ene-21-ol, 3,5-dehydro-6-methoxy-, pivalate | 31.74 | — | 6.45 | C33H54O3 |
| 34 | 2,6,10-Trimethyltetradecane | 31.83 | — | 1.89 | C17H36 |
| 35 | 2-Methylhexacosane | 32.00 | 1.68 | 10.58 | C27H56 |
| 36 | Squalene | 32.71 | — | 4.70 | C30H50 |
| 37 | (9E)-8-Methyl-9-tetradecenyl acetate | 32.82 | — | 1.05 | C17H32O2 |
| 38 | Methyl 4,4-difluororetinoate | 33.01 | — | 3.29 | C21H28F2O2 |
| 39 | Methoprene | 33.24 | — | 3.59 | C19H34O3 |
| 40 | Octadecanoic acid | 33.47 | — | 1.14 | C18H36O2 |
| 41 | Cholest-22-ene-21-ol, 3,5-dehydro-6-methoxy-, pivalate | 34.76 | — | 1.10 | C33H54O3 |
| 42 | Cholest-4-en-3-one | 35.15 | — | 1.58 | C27H44O |
| 43 | Cedryl propyl ether | 35.23 | — | 2.34 | C18H32O |
| 44 | Cholesta-4,6-dien-3-ol | 35.48 | — | 1.91 | C27H44O |
3.3. Metabolomic profiling of the alcoholic extract of leaves and fruits of A. esculentus
Metabolic profiling using LC-HRMS/MS of A. esculentus led to the annotation of 74 metabolites with variable chemical structures: 60 metabolites were identified in the leaf extract while 32 metabolites were identified in the fruit extract. The annotated metabolites (Table 2 and Fig. 1–3) could be classified according to their chemical identity as follows.| No. | Tentative identification | Rt | m/za | Molecular formula | MS/MS-fragments | Chemical class | Plant organ | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Leaves | Fruits | ||||||||
| a All the compounds were detected as M + H. | |||||||||
| 1 | Shikimic acid | 1.3 | 175.1186 | C7H10O5 | 116.0699, 130.0970, 175.1181 | Hydroxy carboxylic acid | − | + | 37 |
| 2 | Azelaic acid | 1.6 | 189.1118 | C9H16O4 | 130.0871, 189.1247 | Fatty acid | − | + | 38 and 39 |
| 3 | 5-Hydroxyferulic acid | 1.7 | 211.0719 | C10H10O5 | 211.0719 | Phenolic acid | + | − | 40 |
| 4 | Paeonolide | 1.9 | 461.2013 | C20H28O12 | 461.2013 | Phenolic compound | + | − | 41 |
| 5 | Salicylic acid (organic acid) | 1.9 | 138.0548 | C7H6O3 | 120.0825, 138.0545 | Phenolic acid | + | + | 42 |
| 6 | Agnuside | 2.0 | 467.1908 | C22H26O11 | 467.1908 | Iridoid glycoside | + | − | 43 and 44 |
| 7 | Linifolin A | 2.2 | 305.1330 | C17H20O5 | 215.1339, 227.1079, 304.1513, 305.1378 | Sesquiterpene lactones | + | + | 45 |
| 8 | Vernolic acid | 2.3 | 297.0977 | C18H32O3 | 297.0977 | Fatty acid | + | + | 46 |
| 9 | Iridotrial glucoside | 2.8 | 345.1357 | C16H24O8 | 165.0549, 280.1175, 308.1125, 326.1238 | Iridoid glycoside | + | − | 44 |
| 10 | Luteoliflavan | 2.9 | 275.0945 | C15H14O5 | 130.0497, 192.0652, 226.0715, 238.0708 | Flavonoid | + | − | 47 |
| 11 | Davidigenin | 3.0 | 259.0922 | C15H14O4 | 130.0495, 131.0523, 171.0776, 223.0984 | Flavonoid | + | + | 48 |
| 12 | Methyl 2-(2-hydroxyphenyl) acetate | 3.6 | 167.0891 | C9H10O3 | 120.0809, 131.0485, 166.0831 | Phenolic compound | + | − | 49 |
| 13 | p-Courmaryol-hexose | 4.1 | 328.1380 | C15H18O8 | 166.0859, 264.1225, 292.1171, 310.1277 | Phenolic compound | + | + | 13 |
| 14 | 37(3β)-9,18-Dihydroxyolean-12-en-3-yl acetate | 4.7 | 485.2498 | C32H52O3 | 485.2498 | Triterpene | + | − | 50 |
| 15 | 7-Hydroxydodecanoate | 5.0 | 217.1725 | C12H24O3 | 114.103, 132.1133, 216.1704 | Fatty acid ester | + | − | 51 |
| 16 | Quercetin | 7.0 | 303.1318 | C15H10O7 | 195.0906, 285.1224, 303.1329 | Flavonoid | + | − | 13 and 52 |
| 17 | 17a-Kaempferol | 7.1 | 287.1299 | C15H9O6 | 210.0777, 215.5759, 286.1290 | Flavonoid | + | − | 13 |
| 17b-Luteolin | 53 and 54 | ||||||||
| 18 | Traumatic acid | 7.1 | 229.1551 | C12H20O4 | 229.1551 | Fatty acid | − | + | 55 |
| 19 | (3β,21β)-19,21-Epoxylup-20(29)-en-3-yl acetate | 7.4 | 483.1977 | C32H52O | 483.1977 | Triterpene | + | − | 50 |
| 20 | Isobergapten | 7.7 | 217.1725 | C12H8O4 | 144.0805, 217.0959 | Furanocoumarin | + | + | 56 |
| 21 | 5-Caffeoylshikimic acid | 7.8 | 337.1060 | C16H16O8 | 337.1060 | Phenolic acid | + | − | 57 and 58 |
| 22 | Quercetin 3-O-(6-O-acetyl-β-D-glucopyranoside) | 7.9 | 507.2122 | C23H22O13 | 507.2122 | Flavonoid glycoside | + | − | 59 |
| 23 | Cycloart-23-ene-3β,25-diol or cycloart-25-en-3,24-diol | 8.0 | 443.2240 | C30H50O2 | 132.1125, 177.0542, 443.2242 | Triterpene | + | − | 60 and 61 |
| 24 | Delphinidin 3-O-sambubioside | 8.2 | 599.2448 | C26H30O16 | 599.2448 | Flavonoid glycoside | + | − | 62 |
| 25 | Abscisic acid | 8.3 | 265.0961 | C15H20O4 | 206.0830, 247.0867, 265.0961 | Sesquiterpene | + | − | 63 |
| 26 | Floramanoside F | 8.4 | 523.1986 | C23H21O14 | 523.1986 | Flavonoid glycoside | + | − | 52 |
| 27 | Apigenin 7-O-neohesperidoside | 8.6 | 579.1621 | C27H30O14 | 313.0691, 397.0888, 415.0998, 433.1100 | Flavonoid glycoside | + | − | 64 |
| 28 | 28a Quercetin 3,7 diglucoside, 28b-Quercetin 3-O-sophoroside | 8.7 | 627.2373 | C27H30O17 | 127.0379, 145.0492, 303.0496 | Flavonoid glycoside | − | + | 37 |
| 29 | Calanolide A | 8.8 | 371.2023 | C22H26O5 | 133.1005, 137.0946, 209.1531 | Coumarin | + | − | 65 and 66 |
| 30 | Quercetin 3-O-[β-D-xylosyl-(1→2)-β-D-glucoside] | 8.8 | 597.1400 | C26H28O16 | 127.0384, 303.0486 | Flavonoid glycoside | − | + | 59 |
| 31 | (−)-Pinoresinol glucoside | 9.1 | 521.2143 | C26H32O11 | 177.0543, 191.0691 | Lignan | + | − | 67 |
| 32 | Feruloylquinic acid | 9.1 | 369.1162 | C17H21O9 | 145.0276, 177.0541, 178.0567 | Phenolic acid | + | − | 57 and 58 |
| 33 | Quercetin-3-O-glucoside | 9.2 | 465.1011 | C21H20O12 | 127.0374, 303.0494 | Flavonoid glycoside | − | + | 52 and 68 |
| 34 | Herniarin | 9.2 | 177.0537 | C10H8O3 | 117.0339, 145.0269 | Coumarin | + | − | 69 |
| 35 | Acetosyringone | 9.4 | 197.1164 | C10H12O4 | 145.0277, 149.0583, 177.0541 | Phenolic compound | + | + | 42 |
| 36 | Gallic acid | 9.6 | 171.0994 | C7H6O5 | 171.1492 | Phenolic acid | + | + | 70 and 71 |
| 37 | Coniferyl ferulate | 9.6 | 357.1456 | C20H20O6 | 357.1456 | Lignan | + | − | 72 |
| 38 | Hispidulin | 9.7 | 300.1219 | C16H12O6 | 121.0645, 163.0385, 300.1204 | Flavonoid | + | − | 42 |
| 39 | (+)-Epicatechin | 9.7 | 291.0967 | C15H14O6 | 159.0911, 188.0695, 227.0794 | Flavonoid | + | − | 73 |
| 40 | N-E-Feruloyltyramine (moupinamide) | 9.9 | 314.0853 | C18H19NO4 | 121.0638, 177.0540, 255.2301 | Nitrogenous metabolite | + | + | 49 and 74 |
| 41 | Myricetin 3-O-rutinose | 10.1 | 627.2658 | C27H30O17 | 177.0539, 314.1378 | Flavonoid glycoside | + | + | 59 |
| 42 | Floramanoside D | 10.2 | 625.2477 | C28H32O16 | 201.0537, 460.1715, 488.1653 | Flavonoid glycoside | + | + | 59 and 75 |
| 43 | Ferulic acid | 10.7 | 195.1375 | C10H10O4 | 135.1174, 194.1164 | Phenolic acid | + | − | 70 and 71 |
| 44 | Caffeic acid | 10.9 | 181.121 | C9H8O4 | 135.1151, 163.1107, 181.1210 | Phenolic acid | + | − | 37 |
| 45 | Allamandin | 11.1 | 309.0819 | C15H16O7 | 263.0803, 281.0908, 309.0852 | Iridoid | + | − | 76 |
| 46 | 9,12,13-Trihydroxy-octadecenoic acid | 11.2 | 331.2463 | C18H34O5 | 277.2149, 295.2245 | Fatty acid | + | + | 77 |
| 47 | Syringic acid | 11.3 | 199.1336 | C9H10O5 | 199.1336 | Phenolic acid | + | − | 37 |
| 48 | Kaempferol-3-O-glucoside (astragalin) | 11.7 | 449.3742 | C21H19O11 | 186.1231, 225.1974, 449.2699 | Flavonoid glycoside | + | + | 42 |
| 49 | Apigenin-7-glucoside | 11.9 | 433.2750 | C21H20O10 | 137.0446, 415.2666 | Flavonoid glycoside | − | + | 54 and 42 |
| 50 | β-Sitosterol | 12.0 | 415.2082 | C29H50O | 119.0851, 120.0878 | Sterol | + | − | 60 and 61 |
| 51 | Pinoresinol | 12.1 | 359.2367 | C20H22O6 | 359.2367 | Lignan | + | − | 67 |
| 52 | Scopoletin | 12.3 | 193.1471 | C10H8O4 | 135.0791, 191.1415 | Coumarin | + | − | 42 |
| 53 | Hibiscetin | 12.5 | 335.2147 | C15H10O9 | 333.2022, 335.2168 | Flavonoid | − | + | 42, 53 and 78 |
| 54 | (12Z)-9,10-Dihydroxyoctadec-12-enoic acid | 12.6 | 315.2493 | C18H34O4 | 133.0993, 147.1153, 279.2298 | Fatty acid | − | + | 79 |
| 55 | Stigmast-4,22-dien-3,6-dione | 12.9 | 425.3736 | C29H44O2 | 137.1312, 407.3639 | Sterol | + | + | 60 and 61 |
| 56 | Stearidonic acid (6,9,12,15-octadecatetraenoic acid) | 13.0 | 277.126 | C18H28O2 | 121.1003, 135.1150, 277.2066 | Fatty acid | + | + | 80 |
| 57 | Rosmarinic acid | 13.0 | 361.2195 | C18H16O8 | 129.0146, 361.2341 | Phenolic acid | + | − | 70 |
| 58 | Ammoresinol | 13.1 | 383.1963 | C24H30O4 | 281.1330, 327.1381, 383.2028 | Coumarin | + | − | 81 and 82 |
| 59 | Quercetin-4′-O-methy-3-O-β-D-glucopyranoside | 13.2 | 479.2969 | C22H22O12 | 337.2713, 479.3372 | Flavonoid glycoside | − | + | 83 |
| 60 | Yamogenin 3-O-neohesperidoside | 13.3 | 723.4957 | C39H62O12 | 723.4957 | Saponin glycoside | + | − | 84 |
| 61 | Tiliroside | 13.5 | 595.3724 | C30H26O13 | 578.3660, 595.3750 | Flavonoid glycoside | + | + | 49 and 42 |
| 62 | Linolenic acid | 13.7 | 279.2291 | C18H29O2 | 123.1162, 137.1305, 279.2275 | Fatty acid | + | + | 77 |
| 63 | Isorhamnetin | 13.8 | 317.1164 | C16H12O7 | 317.2073 | Flavonoid | + | + | 85 and 86 |
| 64 | Isorhamnetin 3-O-glucoside-7-O-xyloside | 13.9 | 611.4198 | C27H30O16 | 317.2074, 318.2095, 358.2330 | Flavonoid glycoside | − | + | 62 |
| 65 | Scutellarin | 13.9 | 463.2982 | C21H18O12 | 231.0635, 341.0973, 463.2981 | Flavonoid | + | − | 87 |
| 66 | Oleic acid | 14.1 | 283.2604 | C18H34O2 | 283.2604 | Fatty acid | + | − | 77 |
| 67 | Esculentoside | 14.3 | 331.2801 | C13H18N2O8 | 239.2366, 313.2694 | Nitrogenous metabolites (pyridine-imidazole derivative) | + | − | 49 and 74 |
| 68 | (4Z,7Z,10Z,13Z,16Z,19Z)-Docosahexaenoic acid ethyl ester | 14.5 | 357.2955 | C24H36O2 | 135.1162, 247.2425, 265.2481 | Fatty acid ester | + | − | 88 |
| 69 | 69a-Myricetin (syn.: Myricetol or cannabiscetin) | 14.6 | 319.2203 | C15H10O8 | 318.3111, 319.2224 | Flavonoid | − | + | 52 |
| 69b-Gossypetin (syn.: articulatidin and equisporol) | 59, 89 and 90 | ||||||||
| 70 | (−)-Isoamijiol | 14.6 | 305.2447 | C20H32O2 | 121.1004, 135.1147, 305.2424 | Diterpene | + | − | 91 |
| 71 | Linoleic acid | 14.8 | 281.2451 | C18H32O2 | 133.0975, 135.1124, 45.2270 | Fatty acid | − | + | 92 |
| 72 | Gallocatechin | 15.6 | 307.2546 | C15H14O7 | 123.1153, 261.2210 | Flavonoid | + | + | 73 |
| 73 | Chlorogenic acid | 16.0 | 355.281 | C16H18O9 | 245.2238, 263.2341, 337.2715 | Phenolic acid | + | + | 71 |
| 74 | Quercetin-3-O-(malonyl) glucoside | 16.1 | 551.3482 | C24H22O15 | 534.3579, 549.2513 | Flavonoid glycoside | + | + | 13 |
 | ||
| Fig. 1 LC-HRMS chromatogram of dereplicated metabolites from the alcoholic extract of the leaves of Abelmoschus esculentus (positive mode). | ||
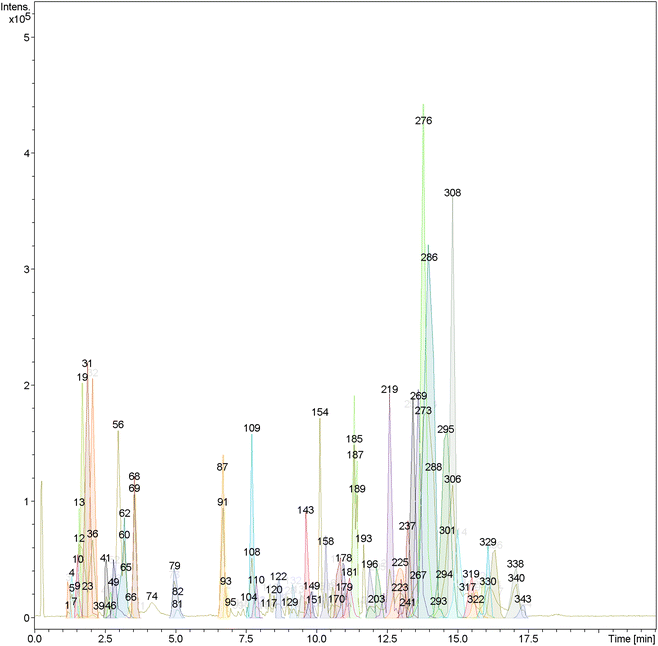 | ||
| Fig. 2 LC-HRMS chromatogram of dereplicated metabolites from the alcoholic extract of the fruits of Abelmoschus esculentus (positive mode). | ||
 | ||
| Fig. 3 Structure of the major identified metabolites in the alcoholic extract of the leaves and fruits of Abelmoschus esculentus. | ||
In conclusion, compounds 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 15, 18, 17b, 20, 21, 27, 29, 31, 32, 34, 35, 37, 38, 45, 46, 48, 49, 51, 52, 53, 54, 56, 58, 60, 62, 65, 66, 68, 70, and 71 have been identified for the first time in the leaves and fruits extract of the genus Abelmoschus. Compounds 12, 13, 14, 16, 17a, 19, 22, 23, 24, 25, 26, 36, 39, 40, 41, 42, 43, 47, 57, 63, 67, 72, and 74 have been discovered for the first time in the leaves extract of A. esculentus. On the other hand, compounds 28, 30, 33, 36, 41, 42, 63, 64 and 73 have been detected for the first time in the fruit extract of A. esculentus.
3.4. Biological activities
3.4.2.1. In vitro α-glucosidase inhibitory assay. α-Glucosidase is an enzyme that catalyzes the liberation of glucose from low molecular weight carbohydrates. Inhibition of the enzyme decreases the blood glucose level after a carbohydrate meal.95 α-Glucosidase inhibitors represent important candidates in diabetes treatment. The present results indicated a potent activity for the leaf extract with an IC50 of 4.47 ± 0.1 mg mL−1 compared with Acarbose standard which exhibits an IC50 of 3.2 ± 0.07 mg mL−1, while the fruit extract showed a higher IC50 value of 10.4 ± 0.2 mg mL−1 (Table 3). Reviewing the relevant literature, a previous study investigating α-glucosidase inhibition of okra seeds, outer skin and inner skin concluded that there was inhibitory activity for the seeds while the outer and inner skin showed no activity against α-glucosidase.15
| Extract | α-Glucosidase | α-Amylase | DPP-4 | Lipase |
|---|---|---|---|---|
| IC50 mg mL−1 | IC50 mg mL−1 | IC50, μg mL−1 | IC50 mg mL−1 | |
| a The data in the table represent means ± SD, where **P < 0.01 and ****P < 0.0001 are considered statistically significant when comparing group differences of Acarbose (for α-glucosidase and α-amylase), Sitagliptin (for DDP-4), and Orlistat (for lipase), using one-way analysis of variance (ANOVA) followed by Tukey's test for multiple comparisons. | ||||
| Leaves | 4.47 ± 0.1**** | 3.54 ± 0.08**** | 0.385 ± 0.019** | 1.044 ± 0.05**** |
| Fruits | 10.4 ± 0.2**** | 6.53 ± 0.15**** | 2.669 ± 0.132**** | 14.66 ± 0.67**** |
| Acarbose® | 3.2 ± 0.07 | 2.21 ± 0.05 | — | — |
| Sitagliptin® | — | — | 0.082 ± 0.004 | — |
| Orlistat® | — | — | — | 5.05 ± 0.23 |
3.4.2.2. In vitro α-amylase inhibitory assay. α-Amylase is an enzyme which catalyzes the hydrolysis of starch into low-molecular-weight dextrin and sugars. Inhibition of the enzyme might retard the digestion of carbohydrates and decrease the rate of glucose absorption, which may contribute to glycemic control in type-2 diabetes.96 Therefore, the inhibition of α-amylase is considered as a good approach for the development of antidiabetic drugs. Herein, the results revealed a characteristic activity for the leaves extract with an IC50 of 3.54 ± 0.08 mg mL−1 compared with Acarbose standard which reported an IC50 of 2.21 ± 0.05 mg mL−1, while the fruit extract showed a higher IC50 value expressed as 6.53 ± 0.15 mg mL−1 (Table 3). Remarkably, previous research have stated considerable α-glucosidase and α-amylase inhibitory activities of okra peel and seeds,97 while another study also revealed that okra seed protein hydrolysate exhibits potent inhibition of carbohydrate hydrolyzing enzymes together with lipase enzyme.98 An additional study exploring the antidiabetic activity of okra fruits unveiled the potential inhibitory activity of the methanolic extract against α-glucosidase and α-amylase with 14.36% to 19.23% inhibition of α-glucosidase and 15.89 to 37.19% inhibition of α-amylase at concentrations of 50–200 mg mL−1.32
3.4.2.3. In vitro DPP-4 inhibitory assay. Dipeptidyl peptidase (DPP-4) is a serine protease enzyme which acts via cleavage and inactivation of peptides as incretin hormones, glucose-dependent insulinotropic polypeptide (GIP) and glucagon-like peptide-1 (GLP-1). Therefore, inhibition of DPP-4 could be a useful approach for treating type 2 diabetes.99 Results unveiled a considerable activity for the leaf extract with an IC50 of 0.385 ± 0.019 μg mL−1 compared with Sitagliptin standard (IC50: 0.082 ± 0.004 μg mL−1) (Table 3). Meanwhile, fruit extract displayed an IC50 of 2.669 ± 0.132 μg mL−1. The antidiabetic activity of okra fruit was previously reported to be attributed to the suppression of DPP-4 signaling.100,101
3.4.2.4. In vitro lipase inhibitory assay. Lipase enzyme is one of the important enzymes in the breakdown of triglycerides; therefore, inhibition of pancreatic lipase is a crucial strategy for managing obesity and hyperlipidemia.102 The current findings highlighted remarkable potent activity for the leaf extract with an IC50 of 1.044 ± 0.05 mg mL−1 compared with the Orlistat standard (IC50: 5.05 ± 0.23 mg mL−1), while the fruit extract exhibited an IC50 of 14.66 ± 0.67 mg mL−1 (Table 3). Interestingly, the results indicated a better activity of the leaf extract than the Orlistat standard. Previous in vivo studies revealed that the dichloromethane and methanol extracts of okra fruits reduced the levels of triglyceride and cholesterol in mice taking tyloxapol to induce hyperlipidemia mice.103 Also, okra peel and the seeds extracts showed antihyperlipidemic activity in STZ induced diabetic rats.104 Another report revealed that okra fruits exhibited remarkable antioxidant potential and potent inhibitory effects on α-glucosidase, α-amylase and lipase digestive enzymes.105
3.5. Molecular docking analysis
To gain insights into the potential bioactive metabolites in the alcoholic extracts of leaves and fruits of okra, all the dereplicated compounds were subjected to molecular docking against human α-amylase, α-glucosidase, and dipeptidyl peptidase-4 (DPP-4) with PDB codes 4W9, 3L4W, and 2ONC, respectively. As illustrated in Fig. 4, the docking scores for all the dereplicated compounds ranged from approximately −4 to −12 kcal mol−1 with the three enzymes. The majority of compounds scored less than 7 kcal mol−1 with the three enzymes: α-amylase, α-glucosidase, and DPP-4. Considering α-amylase, the top-scoring compounds which achieved docking scores below −10 kcal mol−1 were quercetin diglucoside (28) (detected in fruits), tiliroside (61) (detected in leaves-fruits), and isorhamnetin 3-O-glucoside-7-O-xyloside (64) (detected in fruits). They exhibited various binding modes within the enzyme's active site comparable with that of the co-crystalized inhibitor montbretin A (Fig. 5A–D), where they established multiple H-bonds, particularly with ASP-197, GLU-233, and ASP-356. In addition, they established further hydrophobic interactions with TRP-59. Previous reports on tiliroside revealed its effectiveness in inhibiting pancreatic α-amylase in vitro and its effective inhibition for plasma glucose levels increase in an oral glucose tolerance test, but not in an intraperitoneal glucose tolerance test. Tiliroside also show inhibitory effects against glucose uptake which is mediated by both sodium-dependent glucose transporter 1 (SGLT1) and glucose transporter 2 (GLUT2) inhibitors (phlorizin and phloretin, respectively).106 | ||
| Fig. 4 Docking score distribution of dereplicated metabolites in the leaf and fruit extracts of A. esculentus against human α-amylase (A), α-glucosidase (B), and (C) DPP-4 (PDB 4w9, 3l4w and 2onc, respectively). | ||
 | ||
| Fig. 5 Docking poses of compounds with docking scores < −10.0 kcal mol−1 (i.e., 28, 61, and 64) along with the co-crystalized inhibitor montbretin A inside human α-amylase ((A–D), respectively). | ||
Regarding α-glucosidase, the best scoring compounds with scores < −10 kcal mol−1 were luteoliflavan (10) (leaves), quercetin (16) (leaves), kaempferol (17a) (leaves), (+)-epicatechin (39) (leaves), hibiscetin (53) (fruits), myricetin (69a) (fruits), and gossypetin (69b) (fruits) which showed different binding interactions inside the enzyme's active site (Fig. 6). Compounds (10) and (39) established interactions highly similar to that of the co-crystalized inhibitor miglitol, where H-bonds were the predominant with ASP-203, ASP-327, TRP-406, ASP-443, ASN-449, ARG-526, ASP-542, and HIS-600 (Fig. 6A, C and F). On the other hand, their major hydrophobic interactions were with TRP-406, and PHE-450. The flavonol derivatives (16, 53, 69a and 69b) showed fewer H-bonds with only ASP-327 and ASP-203, in addition to three hydrophobic interactions with TYR-299, TRP-406, and PHE-450 (Fig. 6B, D and E).
In 2016, Meng et al. investigated the inhibitory activity of myricetin and quercetin previously isolated from Hovenia dulcis against α-glucosidase. The two compounds showed a noncompetitive inhibition against the enzyme. The IC50 results of both compounds could be attributed to the presence of a –OH group at the 5′-position of B ring which boosts the inhibitory activity against α-glucosidase.107
A group of flavonoids previously isolated from Saccharomyces cerevisiae, including quercetin, kaempferol, catechin and epicatechin, were chosen for the assessment of their inhibitory activity versus α-glucosidase. The results revealed that the most active compound was quercetin. Analyzing the collected results suggests that the absence of a double bond at C2–C3 in (catechin and epicatechin) may decrease the inhibitory activity. Meanwhile, the occurrence of the catechol group in B ring may improve the inhibitory activity of the flavonoids which was observed in the results of quercetin and kaempferol.108 Generally, it has been observed that flavonoids are effective in blocking glucose transporters by suppressing the activities of α-amylase and α-glucosidase. A previous study revealed a relationship between the hydroxyl group number on rings A and B in the flavonoid skeleton and the capacity of inhibition.109 Considering the α-amylase enzyme, the presence of the 4-keto group, the double bond between C2 and C3, and the hydroxyl groups of rings A (at position C6 or C7) and B (at position C4′ or C5′) potentiate the inhibitory potential of the flavonoid against α-amylase, while the occurrence of an –OH group at C3 and the glycosylation or the methylation of –OH groups in rings A and B may reduce the inhibition activity against α-amylase.109 Regarding the favorable features of flavonoids for inhibition of α-glucosidase, the existence of the carbonyl group at C4, the double bond at C2 and C3, and the –OH groups at ring C (C3), ring B (C3′, 4′ and/or 5′ positions), and at ring A (5, 6, 7 and/or at 8 positions) enhances the activity of flavonoids against α-glucosidase. The replacement of the hydroxyl groups by the alkyl or glycosyl moiety in rings A and B may reduce the inhibition activity of the flavonoids.109
Finally, only two compounds exhibited docking scores below −10 kcal mol−1 with human DPP4: davidigenin (11) (leaves-fruits) and N-E-feruloyltyramine (syn.: moupinamide) (40) (leaves-fruits). As depicted in Fig. 7, both compounds shared a common hydrogen bond with TYR-662, along with the co-crystallized inhibitor. Additionally, compound 11 formed a hydrogen bond with GLU-206, similar to the co-crystallized inhibitor.
 | ||
| Fig. 7 Docking poses of compounds with docking scores < −10.0 kcal mol−1 (i.e., 11 and 40, along with the co-crystalized inhibitor) inside human DPP-4 ((A–C), respectively). | ||
4 Conclusion
The study of the phytochemical content and biological activities of the leaves and fruits extracts of the edible plant A. esculentus revealed higher content of bioactive metabolites in the leaves than in the fruits of the plant. A total of 74 metabolites were dereplicated in the extracts from the two organs, among which flavonoids were the most prevailing chemical class. Investigation of the potential biological activities indicated superior antioxidant and antidiabetic efficacy of the leaves extract compared with the fruit extract. A molecular docking simulation study noted the role of flavonoids in the antidiabetic activity of okra extracts. This study highlighted the medicinal potential of okra leaves. However, due to the limitations of these in vitro studies, future research is required to target the in vivo validation, bioavailability studies and toxicity assessment to bridge these gaps and provide stronger evidence of the importance of okra leaves that are often treated as waste.Data availability
All data generated or analyzed during this study are included in this published article.Author contributions
Conceptualization, A. H. E., and M. H. A. H.; investigation, A. H. E., M. H. A. H., and E. A.; methodology, A. H. E., M. H. A. H., and E. A.; software, A. H. E., M. H. A. H., and A. S.; writing original draft preparation, A. H. E., M. H. A. H., and A. S.; writing, review and editing, A. H. E., M. H. A. H., and E. A.; all authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
The authors would like to thank Dr Esam Rashwan, Head of the Confirmatory Diagnostic Unit, VACSERA-EGYPT, for carrying out the in vitro α-glucosidase, α-amylase, DPP-4, and lipase inhibitory assays. The authors would also like to thank the Institut Biologi Sistem, Universiti Kebangsaan Malaysia, Liquid Chromatography-Time of Flight-Mass Spectrometry, LC-TOF-MS laboratory for carrying out the LC-HRMS/MS analysis.References
- J. C. Espín, M. T. García-Conesa and F. A. Tomás-Barberán, Phytochemistry, 2007, 68, 2986–3008 CrossRef PubMed.
- M. T. Murray and J. Pizzorno, The Encyclopedia of Healing Foods, Simon and Schuster, 2010 Search PubMed.
- N. Martins, L. Barros and I. C. Ferreira, Trends Food Sci. Technol., 2016, 48, 1–12 CrossRef CAS.
- T. Vetrichelvan, M. Jegadeesan and B. A. U. Devi, Biol. Pharm. Bull., 2002, 25, 526–528 CrossRef CAS PubMed.
- C. Ning, Y. Jiao, J. Wang, W. Li, J. Zhou, Y.-C. Lee, D.-L. Ma, C.-H. Leung, R. Zhu and H.-M. D. Wang, Food Sci. Hum. Wellness, 2022, 11, 1121–1133 CrossRef CAS.
- P. Zimmet, Diabetologia, 1999, 42, 499–518 CrossRef CAS PubMed.
- S.-H. Lee, M.-H. Park, S.-J. Heo, S.-M. Kang, S.-C. Ko, J.-S. Han and Y.-J. Jeon, Food Chem. Toxicol., 2010, 48, 2633–2637 CrossRef CAS PubMed.
- M. Tomoda, N. Shimizu, R. Gonda, M. Kanari, H. Yamada and H. Hikino, Carbohydr. Res., 1989, 190, 323–328 Search PubMed.
- A. Durazzo, M. Lucarini, E. Novellino, E. B. Souto, P. Daliu and A. Santini, Molecules, 2018, 24(1), 38 Search PubMed.
- S. Habtemariam, Medicinal Foods as Potential Therapies for Type-2 Diabetes and Associated Diseases, Elsevier, Amsterdam, The Netherlands, 2019, pp. 307–332 Search PubMed.
- P. Dubey and S. Mishra, J. Med. Plants Stud., 2017, 5, 23–26 Search PubMed.
- I. F. Olawuyi and W. Y. Lee, Food Chem., 2021, 354, 129437 CrossRef CAS PubMed.
- P. Arapitsas, Food Chem., 2008, 110, 1041–1045 CrossRef CAS PubMed.
- F. Xia, Y. Zhong, M. Li, Q. Chang, Y. Liao, X. Liu and R. Pan, Nutrients, 2015, 7, 8846–8858 CrossRef CAS PubMed.
- E. S. Ong, C. L. Y. Oh, J. C. W. Tan, S. Y. Foo and C. H. Leo, Plants, 2021, 10, 1645 CrossRef CAS PubMed.
- S. Petropoulos, Â. Fernandes, L. Barros, A. Ciric, M. Sokovic and I. C. Ferreira, Food Funct., 2017, 8, 4733–4743 RSC.
- C.-H. Leo and O. L. Woodman, J. Cardiovasc. Pharmacol., 2015, 65, 532–544 CrossRef CAS PubMed.
- V. Sabitha, S. Ramachandran, K. R. Naveen and K. Panneerselvam, J. Ayurveda Integr. Med., 2012, 3, 188 CrossRef PubMed.
- A. Elwekeel, D. El Amir, E. I. Mohamed, E. Amin, M. H. Hassan and M. A. Zaki, Plants, 2022, 11, 208 CrossRef CAS PubMed.
- M. F. A. Bakar, M. Mohamed, A. Rahmat and J. Fry, Food Chem., 2009, 113, 479–483 CrossRef.
- A. Bhatia, B. Singh, R. Arora and S. Arora, BMC Compl. Alternative Med., 2019, 19, 74 CrossRef PubMed.
- M. N. Wickramaratne, J. Punchihewa and D. Wickramaratne, BMC Complementary Altern. Med., 2016, 16, 1–5 Search PubMed.
- Z. Wang, L. Yang, H. Fan, P. Wu, F. Zhang, C. Zhang, W. Liu and M. Li, PeerJ, 2017, 5, e3283 CrossRef PubMed.
- S. S. El-Hawary, A. Elwekeel, S. O. A. El-Ela, U. R. Abdelmohsen and A. I. Owis, RSC Adv., 2023, 13, 14855–14862 RSC.
- R. M. Abdallah, H. M. Hammoda, N. S. El-Gazzar, R. S. Ibrahim and S. M. Sallam, RSC Adv., 2023, 13, 27167–27173 RSC.
- D. Seeliger and B. L. de Groot, J. Comput.-Aided Mol. Des., 2010, 24, 417–422 CrossRef CAS PubMed.
- L. K. Williams, X. Zhang, S. Caner, C. Tysoe, N. T. Nguyen, J. Wicki, D. E. Williams, J. Coleman, J. H. McNeill and V. Yuen, Nat. Chem. Biol., 2015, 11, 691–696 CrossRef CAS PubMed.
- L. Sim, K. Jayakanthan, S. Mohan, R. Nasi, B. D. Johnston, B. M. Pinto and D. R. Rose, Biochemistry, 2010, 49, 443–451 CrossRef CAS PubMed.
- M. E. E. Caluête, L. M. P. de Souza, E. dos Santos Ferreira, A. P. de França, C. De Akneuda Gadelha, J. de Souza Aquino and T. Santi-Gadelha, Afr. J. Biotechnol., 2015, 14, 683–687 CrossRef.
- M. Hafeez, S. M. Hassan, S. S. Mughal, M. Munir and M. K. Khan, Chemical and Biomolecular Engineering, 2020, 5, 69 CrossRef.
- S. Guebebia, C. Espinosa-Ruiz, L. Zourgui, A. Cuesta, M. Romdhane and M. Á. Esteban, Fish Shellfish Immunol., 2023, 138, 108799 CrossRef CAS PubMed.
- M. H. Siddique, A. Ashraf, S. Hayat, B. Aslam, M. Fakhar-e-Alam, S. Muzammil, M. Atif, M. Shahid, S. Shafeeq and M. Afzal, J. Saudi Chem. Soc., 2022, 26, 101418 CrossRef CAS.
- S. Guebebia, A. Gharsallaoui, E. Dumas, F. Baghi, L. Zourgui, M. Romdhane, G. Agusti and S. Ghnimi, Appl. Sci., 2023, 13, 12273 CrossRef CAS.
- D.-T. Wu, X.-R. Nie, D.-D. Shen, H.-Y. Li, L. Zhao, Q. Zhang, D.-R. Lin and W. Qin, Molecules, 2020, 25, 1276 CrossRef CAS PubMed.
- D. Selvaraj, A. Subramanian and T. Samuel, World J. Adv. Res. Rev., 2020, 5, 067–079 CrossRef CAS.
- A. I. A. Osman, Nat. Volatiles Essent. Oils, 2021, 9578–9587 CAS.
- M. H. Romdhane, H. Chahdoura, L. Barros, M. I. Dias, R. C. G. Corrêa, P. Morales, M. Ciudad-Mulero, G. Flamini, H. Majdoub and I. C. F. R. Ferreira, Molecules, 2020, 25, 4739 CrossRef CAS PubMed.
- M. Nazzaro-Porro, J. Am. Acad. Dermatol., 1987, 17, 1033–1041 CrossRef CAS PubMed.
- J. Y. Cho, H. K. Kim, S. J. Ma, J. H. Moon and K. H. Park, Food Sci. Biotechnol., 2000, 9, 313–316 Search PubMed.
- N. Misawa, T. Hosoya, S. Yoshida, O. Sugimoto, T. Yamada-Kato and S. Kumazawa, Phytother. Res., 2018, 32, 1304–1310 CrossRef CAS PubMed.
- L. Sun, C. Kong, H. Zhao, D. Wu and F. Xu, Rec. Nat. Prod., 2022, 1–6 Search PubMed.
- H. R. Bajes, S. A. Oran and Y. K. Bustanji, Res. J. Pharm. Technol., 2021, 14, 6447–6454 Search PubMed.
- S. Arokiyaraj, K. Perinbam, P. Vivek and N. K. Udaya Prakash, J. Pharma Res., 2012, 5, 2548–2552 CAS.
- Y. Deng, Chin. Pharm. J., 2017, 2146–2150 CAS.
- J. Jodynis-Liebert, M. Murias and E. Błoszyk, Planta Med., 1999, 65, 320–324 CrossRef CAS PubMed.
- M. L. Wang, B. Morris, B. Tonnis, J. Davis and G. A. Pederson, J. Agric. Food Chem., 2012, 60, 6620–6626 CrossRef CAS PubMed.
- D. T. H. Trang, P. T. Trang, D. M. Phuong, D. H. Anh, N. Q. Anh, P. Van Kiem and P. H. Viet, Vietnam J. Chem., 2023, 61, 1–7 CrossRef.
- O. Desire, C. Rivière, R. Razafindrazaka, L. Goossens, S. Moreau, J. Guillon, S. Uverg-Ratsimamanga, P. Andriamadio, N. Moore and A. Randriantsoa, J. Ethnopharmacol., 2010, 130, 320–328 CrossRef CAS PubMed.
- S. Mi, R. Guo, C. Xie and X. Huang, Asian J. Tradit. Med., 2021, 16, 269–275 Search PubMed.
- Y. Zhou, X. Jia, J. Shi, Y. Xu, L. Jing and L. Jia, Helv. Chim. Acta, 2013, 96, 533–537 CrossRef CAS.
- S. Tahara, K. Hosokawa and J. Mizutani, Agric. Biol. Chem., 1980, 44, 193–197 CAS.
- S.-Y. Yang, Y.-M. Zhao, Z.-L. Li, Q.-Y. Zuo and S.-H. Qian, Chem. Nat. Compd., 2018, 54, 257–260 CrossRef CAS.
- V. Vadivel, Int. J. Green Pharm., 2016, 10(1), S33–S45 CAS.
- E. H. Naser, L. S. Mahdiand and R. T. Alasadi, Sci. J. Med. Res., 2022, 6(23), 35–44 Search PubMed.
- D. C. Zimmerman and C. A. Coudron, Plant Physiol., 1979, 63, 536–541 Search PubMed.
- H. Saleem, M. Sarfraz, H. M. Ahsan, U. Khurshid, S. A. J. Kazmi, G. Zengin, M. Locatelli, I. Ahmad, H. H. Abdallah and M. F. Mahomoodally, Processes, 2020, 8, 336 CrossRef CAS.
- R. A. El-Shiekh, R. M. Ashour, E. A. Abd El-Haleim, K. A. Ahmed and E. Abdel-Sattar, Biomed. Pharmacother., 2020, 128, 110303 CrossRef CAS PubMed.
- J. Zhen, T. S. Villani, Y. Guo, Y. Qi, K. Chin, M.-H. Pan, C.-T. Ho, J. E. Simon and Q. Wu, Food Chem., 2016, 190, 673–680 CrossRef CAS PubMed.
- S. Yin, Z. Cai, C. Chen, Y. Mei, L. Wei, S. Liu, L. Zou, N. Wu, J. Yuan and X. Liu, Horticulturae, 2022, 8, 317 CrossRef.
- L. Jia, D. Li, L. Jing and M. Guo, J. Chin. Med. Mater., 2010, 33, 1262–1265 CAS.
- L. Jia, M. Guo, D. Li and L. Jing, China J. Chin. Mater. Med., 2011, 36, 891–895 CAS.
- Y. Zhang, T. Zhang, Q. Zhao, X. Xie, Y. Li, Q. Chen, F. Cheng, J. Tian, H. Gu and J. Huang, Foods, 2021, 10, 2180 Search PubMed.
- P. Daliu, G. Annunziata, G. C. Tenore and A. Santini, Nat. Prod. Res., 2020, 34, 3–9 CrossRef CAS PubMed.
- I. Matlawska, Acta Pol. Pharm., 2001, 58, 127–132 CAS.
- A. Mandal, D. Biswas and B. Hazra, Stud. Nat. Prod. Chem., 2020, 66, 225–271 CAS.
- L. Nahar, A. D. Talukdar, D. Nath, S. Nath, A. Mehan, F. M. D. Ismail and S. D. Sarker, Molecules, 2020, 25, 1–23 Search PubMed.
- S.-J. Lee, Y.-S. Yun, I.-K. Lee, I.-J. Ryoo, B.-S. Yun and I.-D. Yoo, Planta Med., 1999, 65, 658–660 CrossRef CAS PubMed.
- N. Li, H. Tang, L. Wu, H. Ge, Y. Wang, H. Yu, X. Zhang, J. Ma and H. F. Gu, Phytother. Res., 2021, 35, 198–206 Search PubMed.
- A. Ahmad and L. Misra, Int. J. Pharmacogn., 1997, 35, 121–125 CAS.
- S. Ray, S. Mishra, K. Bisen, S. Singh, B. K. Sarma and H. B. Singh, Microbiol. Res., 2018, 207, 100–107 CrossRef CAS PubMed.
- I. F. Olawuyi, J. J. Park and W. Y. Lee, Food Sci. Preserv., 2020, 27, 476–486 Search PubMed.
- C. Chen, C. Wu, X. Lu, Z. Yan, J. Gao, H. Zhao and S. Li, J. Evidence-Based Complementary Altern. Med., 2013, 2013 Search PubMed.
- P. Khomsug, W. Thongjaroenbuangam, N. Pakdeenarong, M. Suttajit and P. Chantiratikul, Res. J. Biol. Sci., 2010, 5, 310–313 CrossRef.
- I. O. Kehinde, O. E. Olatunji and A. Adegoke, World J. Adv. Res. Rev., 2022, 14, 104–111 Search PubMed.
- Y. Zhang, W. He, C. Li, Q. Chen, L. Han, E. Liu and T. Wang, J. Nat. Med., 2013, 67, 78–85 CrossRef CAS PubMed.
- S. M. Kupchan, A. L. Dessertine, B. T. Blaylock and R. F. Bryan, J. Org. Chem., 1974, 39, 2477–2482 Search PubMed.
- F. C. Rodrigues, A. T. L. dos Santos, R. P. da Cruz, J. W. Almeida-Bezerra, H. D. M. Coutinho, P. R. V. Ribeiro, E. S. de Brito, M. F. B. Morais-Braga and A. F. M. de Oliveira, S. Afr. J. Bot., 2022, 147, 286–293 Search PubMed.
- A. Ismailov, A. Karimdzhanov, S. Y. Islambekov and Z. Rakhimkhanov, Chem. Nat. Compd., 1994, 30, 1–14 CrossRef.
- D. M. Harris, F. Berrue, R. Kerr and C. L. Patten, Rhizosphere, 2018, 6, 98–111 CrossRef.
- J. L. Guil-Guerrero, Eur. J. Lipid Sci. Technol., 2007, 109, 1226–1236 CrossRef CAS.
- A. Abedini, V. Roumy, S. Mahieux, A. Gohari, M. Farimani, C. Rivière, J. Samaillie, S. Sahpaz, F. Bailleul and C. Neut, Lett. Appl. Microbiol., 2014, 59, 412–421 CrossRef CAS PubMed.
- H.-R. Adhami, J. Lutz, H. Kählig, M. Zehl and L. Krenn, Sci. Pharm., 2013, 81, 793–806 CrossRef CAS PubMed.
- H. Liao, H. Liu and K. Yuan, Pharmacogn. Mag., 2012, 8, 12 CrossRef CAS PubMed.
- M. K. Aadesariya, V. R. Ram and P. N. Dave, Int. J. Adv. Res. Chem. Sci., 2017, 4, 1–14 Search PubMed.
- J. Guo, L. Du, E. Shang, T. Li, Y. Liu, D. Qian, Y. Tang and J. Duan, Pharm. Biol., 2016, 54, 595–603 CrossRef CAS PubMed.
- J. Li, G.-y. Ye, H.-l. Liu and Z.-h. Wang, J. Plant Biochem. Biotechnol., 2022, 31, 351–360 CrossRef CAS.
- N. M. E. Satti, M. B. Suliman and A. S. Jagdouj, Int. J. Botany Stud., 2020, 5(4), 108–111 Search PubMed.
- R. Rayssan and M. S. Shawkat, J. Pharmaceut. Sci. Res., 2019, 11, 70–74 CAS.
- M. T. Islam, Phytother. Res., 2019, 33, 72–80 CrossRef PubMed.
- G. Rak, P. Fodor and L. Abrankó, Int. J. Mass Spectrom., 2010, 290, 32–38 CrossRef CAS.
- S. Rodríguez-Morales, B. Ocampo-Medina, N. Romero-Ceronio, C. Alvarado-Sánchez, M. Á. Vilchis-Reyes, L. F. Roa de la Fuente, R. Ortiz-Andrade and O. Hernández-Abreu, Chem. Biodiversity, 2021, 18, e2000820 CrossRef PubMed.
- V. Tešević, V. Vajs, S. Lekic, I. Đorđević, M. Novaković, L. V. Vujisić and M. Todosijevic, Arch. Biol. Sci., 2012, 64, 221–227 CrossRef.
- O. H. Abdelhafez, M. A. Fawzy, J. R. Fahim, S. Y. Desoukey, M. Krischke, M. J. Mueller and U. R. Abdelmohsen, PLoS One, 2018, 13, e0202362 CrossRef PubMed.
- H. Faisal and S. Handayani, Indonesian Journal of Pharmaceutical and Clinical Research, 2019, 2, 6–13 CrossRef.
- H. E. Lebovitz, Endocrinol. Metab. Clin. North Am., 1997, 26, 539–551 Search PubMed.
- H. Zeng, Q. Liu, J. Yu, M. Wang, M. Chen, R. Wang, X. He, M. Gao and X. Chen, J. Sep. Sci., 2015, 38, 3897–3904 CrossRef CAS PubMed.
- V. Sabitha, K. Panneerselvam and S. Ramachandran, Asian Pac. J. Trop. Biomed., 2012, 2, S162–S164 CrossRef.
- O. S. Ijarotimi, A. O. Akinola-Ige and T. D. Oluwajuyitan, Meas.: Food, 2023, 11, 100101 Search PubMed.
- C.-N. Huang, C.-J. Wang, Y.-J. Lee and C.-H. Peng, PLoS One, 2017, 12, e0180285 Search PubMed.
- C.-H. Peng, Y.-B. Ker, H.-H. Li, S.-H. Tsou, C.-L. Lin and C.-N. Huang, PLoS One, 2022, 17, e0265444 CrossRef CAS PubMed.
- C.-N. Huang, C.-J. Wang, C.-L. Lin, A.-T. Yen, H.-H. Li and C.-H. Peng, PLoS One, 2019, 14, e0217400 CrossRef CAS PubMed.
- B. Zhang, Z. Deng, D. D. Ramdath, Y. Tang, P. X. Chen, R. Liu, Q. Liu and R. Tsao, Food Chem., 2015, 172, 862–872 Search PubMed.
- T. H. Ngoc, Q. N. Ngoc, A. Tran and N. V. Phung, J. Pharm. Sci., 2008, 35, 42–46 Search PubMed.
- V. Sabitha, S. Ramachandran, K. Naveen and K. Panneerselvam, J. Pharm. BioAllied Sci., 2011, 3, 397–402 CrossRef CAS PubMed.
- D.-D. Shen, X. Li, Y.-L. Qin, M.-T. Li, Q.-H. Han, J. Zhou, S. Lin, L. Zhao, Q. Zhang and W. Qin, J. Food Sci. Technol., 2019, 56, 1275–1286 CrossRef CAS PubMed.
- T. Goto, M. Horita, H. Nagai, A. Nagatomo, N. Nishida, Y. Matsuura and S. Nagaoka, Mol. Nutr. Food Res., 2012, 56, 435–445 CrossRef CAS PubMed.
- Y. Meng, A. Su, S. Yuan, H. Zhao, S. Tan, C. Hu, H. Deng and Y. Guo, Plant Foods Hum. Nutr., 2016, 71, 444–449 CrossRef CAS PubMed.
- M. N. Sarian, Q. U. Ahmed, S. Z. Mat So'ad, A. M. Alhassan, S. Murugesu, V. Perumal, S. N. A. Syed Mohamad, A. Khatib and J. Latip, BioMed Res. Int., 2017, 2017, 8386065 Search PubMed.
- T. Hussain, B. Tan, G. Murtaza, G. Liu, N. Rahu, M. S. Kalhoro, D. H. Kalhoro, T. O. Adebowale, M. U. Mazhar and Z. ur Rehman, Pharmacol. Res., 2020, 152, 104629 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |

