Open Access Article
Open Access ArticleMicrofluidic chip designs and their application for E antigen typing on red blood cells
Pornsuda Maraminga,
Nang Noon Shean Ayea,
Pattarawarin Panyakakaewa,
Patcharaporn Tippayawata,
Sakda Daduangb,
Kiattawee Choowongkomonc,
Nichada Jearanaikoond,
Kriettisak Srisomd,
Rungrueang Phatthanakun*d and
Jureerut Daduang *a
*a
aCentre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University, Khon Kaen 40002, Thailand. E-mail: pornsma@kku.ac.th; nangnoonshean.aye@kkumail.com; papattaravarin@kkumail.com; patchatip@kku.ac.th; jurpoo@kku.ac.th
bDivision of Pharmacognosy and Toxicology, Faculty of Pharmaceutical Sciences, Khon Kaen University, Khon Kaen 40002, Thailand. E-mail: sakdad@kku.ac.th
cDepartment of Biochemistry, Faculty of Science, Kasetsart University, Bangkok 10900, Thailand. E-mail: kiattawee.c@ku.ac.th
dSynchrotron Light Research Institute (Public Organization), Nakhon Ratchasima 30000, Thailand. E-mail: kriettisak@slir.or.th; nchada@slri.or.th; rungrueang@slri.or.th
First published on 24th February 2025
Abstract
The E antigen is one of the main Rh antigens on red blood cells (RBCs). The high ability of the E antigen to trigger an immune response along with the presence of anti-E, can cause serious issues such as hemolytic transfusion reactions and hemolytic disease in newborns. In this study, we developed a microfluidic biochip for Rh typing (E antigen). Three different micromixer types were compared for simulation, mixing tests, and optimal interactions of blood typing reagents and RBCs to provide an accurate agglutination reaction. Using separate syringe pumps, a blood sample and anti-E reagent were introduced into the microfluidic device through the respective inlet channels. Then, the fluids underwent thorough mixing within the micromixers before reaching reaction reservoirs where RBC agglutination was observed. The 8-shape micromixer design showed the most agglutination and indicated the best performance for E antigen detection and was able to differentiate blood clinical samples with E-positive and -negative RBCs. The microfluidic chip could also be applied for RBC antigen detection in blood bank procedures during blood typing and compatibility testing.
1 Introduction
Blood group antigens are surface markers of red blood cells (RBCs) that can evoke immune antibodies and elicit an immune response after a transfusion or a pregnancy event.1 The blood group system is essential for blood transfusion and organ transplant matching.2 In addition to ABO blood groups, the Rh blood group system has a secondary clinical importance in safe transfusion practice because of the Rh antigens.3,4 Rh antigens are highly immunogenic. Alloimmunization to Rh antigens, where recipients can produce the corresponding antibodies, results in hemolytic transfusion reactions (HTRs) and hemolytic disease of the fetus and newborn (HDFN).5,6In general, there are more than three hundred genetically different blood groups.7 Blood group system antigenic differences occur between the donor and recipient as well as the immune-modulatory effect of allogenic blood transfusion on the recipient's immune system are effective factors in alloimmunization.8 Previous studies have shown that multitransfused patients and multiparous women produce most antibodies directly against Rh, Duffy, Kell, and Kidd blood group antigens, which results in HTRs or HDFNs.9,10 In addition to the D antigen in the Rh blood group system, C, c, E, and e antigens are common and important antigens of the system. Therefore, RBC phenotyping is also performed for detecting other antigens of the Rh blood group system including C, c, E, and e antigens. These antigens are not routinely detected in patients. RBC phenotypes are required for the blood transfusion of antigen-matched units for patients who have alloantibodies or receive multiple transfusions to prevent alloimmunization such as patients with thalassemia, myelodysplasia, and sickle cell disease.9
There are several blood typing techniques that differ from each other in terms of sensitivity, the reagents and equipment needed, the time of operation, and throughput analysis.11 The tube test is the gold standard for identifying ABO and Rh phenotype blood groups. For cell grouping of ABO and Rh typing (D, C, c, E, and e), two drops of each antibody are mixed with blood adjusted to a concentration of approximately 3–5% in a test tube for 5 min at room temperature, centrifuged, and gently resuspended and examined for agglutination.12 In general, the tube method is much more sensitive than the slide test but it also requires centrifugation and expertise to read and grade the agglutination reactions.11 The slide test method involves mixing a specific antiserum with the blood of the patient or donor on a clean glass slide or ceramic tile. Antigen-positive RBCs react with their corresponding antibody, which results in strong agglutination. The main advantage of the slide test method is that it is a rapid and inexpensive test that can be executed without the need for specific equipment such as a centrifuge. However, this test cannot detect a low quantity of antigen and results in a weak reaction.12 Microplate technology is a more sensitive and faster blood typing analysis with the feasibility of automation. This technique can detect both antibodies in blood plasma and antigens on RBCs.13,14 Typically, microplates consist of many small tubes that contain a few μL of reagents, which are used to treat blood samples. Dispenser devices, microplate readers, and microplate centrifuges are needed for this method, which is laborious and inconvenient for field applications.15 The column or gel test has been developed to examine RBC grouping and serum grouping. The specific pore size of the gel matrix is packed in the microtube column to discriminate the strength of agglutinated cells after centrifugation.16,17 The cost and centrifugation are the main disadvantages of this test. The ABO-Rh blood group and antigen phenotyping of donors and patients provide important information required for compatibility testing before blood transfusion. However, current methods still have limitations, such as being expensive and requiring complicated and time-consuming processes.
Microfluidic systems have many benefits, such as cost-effectiveness, shorter reaction times, parallel operation on a single chip, and less cross-contamination through precise fluid control elements. Microfluidics has evolved into a new area of research in biomedical diagnostics, drug delivery, and the food and chemical industries.18,19 However, adequate, homogeneous, and rapid mixing of liquids is a major challenge in the development of lab-on-a-chip devices and micro total analysis systems (μ-TASs).20–22 Recently, paper-based and polymeric microfluidic chips for blood grouping have been established.17,23–26 In paper-based microfluidics devices, the immunoreaction between RBCs and antibodies is limited by the finite diffusion and incubation times of blood samples passing through the paper network and results in the inability to detect weakly interacting blood groups. The microfluidic thread-based analytical devices have been applied for low-cost and simple red blood cell typing. The fundamental principle of the device is that the blood sample is placed on the fibers that have been pre-treated with antibodies. This allows large agglutinated RBCs to remain on threads, which can be observed with the naked eye.27–30 Despite the validation of using threads for blood typing, the transport mechanism of RBCs in threads is still unspecified. Careful consideration of the thread's properties is required.31 On the other hand, polymeric microfluidic devices can provide rapid and high-throughput RBC agglutination detection, low sample volume, and reagent consumption.32 Kim et al. fabricated a disposable microfluidic biochip for blood typing that is comprised of microchannels, chaotic micromixers, reaction microchambers, and detection microfilters.23
In most microfluidic systems, the mixing of two or more reagents relies on diffusion, and consequently, the mixing reaction is sluggish. Micromixers are an essential component of lab-on-a-chip and micro total analysis systems applied for a wide range of chemical and biological applications.33 Different kinds of micromixers have been introduced to reduce the mixing time, microchannel length, and pressure inside. The micromixers are divided into active and passive micromixers depending on whether moving parts or external energy is provided or not.34 For example, active mixing requires external energy, such as magnetohydrodynamic energy,35 pulse flow,36 electro-osmosis,37 and thermal distribution,38 to agitate and mix the fast fluid. However, there are disadvantages, such as high energy consumption and difficulty in fabricating and integrating with other microfluidic components, whereas passive micromixers enhance mixing by designing or modifying microchannels of various shapes and structures without requiring external energy.
A serpentine laminating F-shape micromixer was designed by DS Kim et al. (2005) to characterize the fluid mixing efficiency. The initial F-shaped design involved a double-layer microchannel, which was effective but burdensome to fabricate. Therefore, we designed a single-layer F-micromixer to create a simple and efficient mixing reaction.39 A tesla J-shape micromixer was shown to provide efficient mixing performance via passive micromixing.40 The tesla 8-shape micromixer with a circular obstacle was observed to be very effective in terms of mixing and energy savings due to the synergistic effect of transverse dispersion and chaotic advection.41 In this study, we aimed to develop a low-cost, lab-on-a-chip integrated microfluidic biochip of compact size for E-antigen typing, which requires a very small sample blood volume. We introduced the design and operation process of the microfluidic biochip and the components integrated into the biochip as follows: microchannels, chaotic micromixers, and reaction microchambers. We performed Attenuated total reflectance-Fourier transform infrared (ATR-FTIR) analysis of PDMS surface chemistry before and after O2 plasma treatment. Three different micromixer prototypes (F, J, and 8 shapes) were investigated for mixing efficiency using numerical simulation and dye testing. In addition, RBC agglutinations were compared among these chips. The best micromixer was selected for testing with clinical samples. The optimal interaction of blood typing reagents and RBCs provides an accurate agglutination reaction. Scheme 1 shows a schematic of the overall concept of microfluidic devices for the detection of the E antigen on RBCs.
 | ||
| Scheme 1 A schematic illustration of the concept of microfluidic device for red cell antigen typing. | ||
2 Materials and methods
2.1 Reagents and materials
A blood-typing reagent (monoclonal anti-E antibody) was purchased from the National Blood Centre, Thai Red Cross Society. SUEX® thick dry film sheets were purchased from DJ MicroLaminates HQ (Tokyo, Japan). 2-(1-Methoxy) propyl acetate (99%, Thermo Scientific Acros, Oslo, Norway) was applied as an SU8 developer. Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corporation, Wiesbaden, Germany) was used to fabricate the microfluidic devices. PTFE (Teflon) tubing (1/16′′ OD) was obtained from Darwin Microfluidics, Paris, France. Ethanol (70%), phosphate-buffered saline (PBS), and sodium chloride were obtained from Sigma-Aldrich (St. Louis, MO). The micromixer design and fabrication of microfluidic chips were performed in Microsystems, the Electronics and Control System Laboratory (MECS), and the Synchrotron Light Research Institute (Nakhon Ratchasima, Thailand).2.2 Fabrication of the PDMS microchannel
UV lithography technique was used for the fabrication of the PDMS microchannel. Processes for lithography include photoresist coating, soft baking, photomasking, UV exposure, and postexposure baking (Fig. 1). The SU8 dry film (100 μm) was mounted on a glass substrate followed by hot-roll lamination at 70 °C twice. The photoresist was prebaked by heating at 65 °C for 5 min followed by heating at 95 °C for 10 min and cooling to room temperature. The UV mask was used to cover the resist with a UV exposure dose of 450 mJ cm−2 for 45 s. The substrate was subjected to a postexposure bake (65 °C for 5 min and 95 °C for 10 min) to remove the solvent and promote adhesion. After cooling, the pattern was developed in SU8 developer for 5–10 min followed by a simultaneous rinsing with isopropyl alcohol. For PDMS lithography, PDMS was mixed with the curing agent at a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Degassing was conducted by placing the PDMS in a vacuum desiccator for 30 min. After degassing, the mold was placed in a Petri dish, and the PDMS was poured over the mold. Then, the mixture was cured at 70 °C for 1 h to solidify it. After it cooled, it was peeled off from the mold, and the inlet and outlet holes were punched.
1. Degassing was conducted by placing the PDMS in a vacuum desiccator for 30 min. After degassing, the mold was placed in a Petri dish, and the PDMS was poured over the mold. Then, the mixture was cured at 70 °C for 1 h to solidify it. After it cooled, it was peeled off from the mold, and the inlet and outlet holes were punched.
2.3 Assembly of the microfluidic chip
The irreversible bonding of PDMS to glass slides was achieved through oxygen plasma bonding, which is a method that enhances the surface properties of both materials. In this process, the PDMS replica, which was oriented with the patterned surface facing upward, was placed alongside a glass substrate in an oxygen plasma cleaner. The oxidation occurred at a pressure of 0.5 Mbar using 40 W of power for 1 minute, which effectively activated the surface by generating silanol groups. The bonded surfaces were subsequently subjected to thermal treatment at 80 °C for 1 hour, which facilitated the formation of strong covalent bonds between the PDMS and glass through siloxane linkages. This combination of plasma treatment and thermal curing not only ensures robust and permanent bonding but also minimizes the risk of delamination during subsequent use in microfluidic applications.2.4 Overall design and operating process of microfluidic biochips
The microfluidic chip for blood antigen testing consisted of three major parts as follows: (1) fluid stream microchannels for test samples and reagents, (2) micromixer parts, and (3) a reaction chamber (Fig. 2). The blood sample and blood typing reagents were injected into the microfluidic device through the separated inlet channels via two syringe pumps at an appropriate flow rate, which was followed by complete mixing within micromixers and finally RBC agglutination in the reaction reservoirs. The agglutination of RBCs was initially observed within the micromixers, but the larger scale of agglutinates was clearly investigated in the reaction microchamber. Finally, the fluid flow passed through the outlet channel into the waste reservoir. Results of blood agglutination were further observed by the naked eye and an inverted Olympus IX71 microscope (Olympus, Shinjuku, Tokyo, Japan) with an 10× objective lens for image acquisition. | ||
| Fig. 2 Designs and dimensions of (A) F-shaped micromixer, (B) Tesla (J)-shaped micromixer and (C) 8-shaped micromixer. | ||
2.5 ATR-FTIR analysis of PDMS surface chemistry
ATR-FTIR spectra for PDMS (untreat and O2 plasma treated) samples were acquired using a Bruker Tensor 27 spectrometer equipped with Hyperion 3000 FTIR microscope (20× objective) and a mercury cadmium telluride (MCT) detector (Bruker Optik GmbH, Ettlingen, Germany) Spectra were recorded at wavenumber 4000–600 cm−1 with spectral resolution of 4 cm−1. The FTIR spectra were analyzed using Unscrambler X 10.4 software (Camo, Oslo, Norway) to classify the differences between untreated and treated PDMS. The spectra were preprocessed by performing second derivatives with Savitzky–Golay 13-points smoothing and normalized using Extended Multiplicative Signal Correction (EMSC). Principal Component Analysis (PCA) was conducted on mean-centered data with leverage correction validation using spectral ranges containing bands between 3020–2900 cm−1 and 1300–650 cm−1. The resulting scores plots were used to visualize data clustering, while loading plots identified the spectral regions that contributed most to the variance in the data.2.6 Simulation of micromixers
The mixing efficiency of the micromixer design was evaluated through 2D simulation via SOLIDWORKS Flow Simulation software. This allowed us to observe how the two fluids mix and easily assess the micromixer performance for each design. In the SolidWorks simulation, the fluid used in the first inlet was blood, and the solution in the second inlet was slurry, which are both classified as non-Newtonian fluids because it is necessary to simulate both inlets to have similar density values to those of the test in practice (the RBCs and antibody fluids). We achieved this by simulating various flow ratios between blood and slurry, such as 200 μL min−1 and 100 μL min−1 for each fluid, with configurations including blood at 80 μL min−1 and slurry at 20 μL min−1 as well as a mixture of blood at 70 μL min−1 with slurry at 30 μL min−1. The properties of the fluids used in the simulation are detailed in Table 1. The simulation focused on flow rates and their effects.| Properties | Blood | Slurry | Unit |
|---|---|---|---|
| Density | 1003 | 1647.2 | kg m−3 |
| Specific heat | 4182 | 4000 | J kg−1 K−1 |
| Thermal conductivity | 0.6 | 0.6 | W m−1 K−1 |
| Viscosity | Power-law model | Herschel–Vulkley model | |
| Consistency coefficient | 0.012171 | 0.05546 | Pa s |
| Power-law index | 0.7991 | 0.86523 |
2.7 Dye testing
To characterize the realized micromixer, blue- and yellow-dyed DI water was injected into the micromixer at equal flow rates (100 μL min−1) via a syringe pump to characterize the mixing performance. A CCD camera was used to record and visualize the mixing performance according to the color intensity of the dyed water. The mixing reaction was observed within the micromixing units as well as the reaction chamber.2.8 RBC antigen typing via a microfluidic chip
The blood typing process using the biochip in this study was performed as follows. A constant volume of 1 mL of a RBC suspension (3%) in normal saline or standard anti-E serum (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was loaded into the corresponding inlets and injected into the biochip through flow-splitting microchannels with a syringe pump as depicted in Scheme 1. The bloodstream and corresponding serum stream were then introduced into a chaotic micromixer for efficient mixing to induce the effective agglutination of RBCs with the antibody.
1) was loaded into the corresponding inlets and injected into the biochip through flow-splitting microchannels with a syringe pump as depicted in Scheme 1. The bloodstream and corresponding serum stream were then introduced into a chaotic micromixer for efficient mixing to induce the effective agglutination of RBCs with the antibody.
For the selection of micromixer types, a mixture of Rh E-positive blood samples and anti-E was flowed through the reaction area of the microfluidics chip at a flow rate of 200 μL min−1 for 10 s, followed by a flow rate of 1 μL min−1 for 30 s. The flow was then stopped. After 3 min, agglutination in the reaction area was observed in the reaction chambers of the F, J, and 8 micromixers. The micromixer with the best agglutination was selected to optimize flow rates. Different flow rates of E-positive RBCs and anti-E (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 80
30, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 90
20, 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, 100
10, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and 200
100, and 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 μL min−1) were tested. After each test, the flow rate was reduced to 1 μL min−1 for 30 s and then stopped to eliminate the driving pressure applied to the biochip and thereby keeping the mixtures inside the reaction microchambers during the reaction time. RBC agglutinations were observed in the reaction microchambers after the flow rate was stopped for 3 min. The optimal micromixer type and flow rate were selected for clinical sample testing. The clinical performance of the proposed microfluidics chip was determined by testing with clinical EDTA-containing blood samples. Blood samples were collected from the Blood Bank, Faculty of Medicine, Khon Kaen University, Thailand, following the Declaration of Helsinki and the ICH Good Clinical Practice Guidelines (HE674003). E-negative and E-positive RBCs (3 samples in each group) were obtained for the detection of E antigens via the microfluidic chip. After anti-E was mixed with the clinical samples, the microfluidic chip measurements were performed as described in the previous section.
200 μL min−1) were tested. After each test, the flow rate was reduced to 1 μL min−1 for 30 s and then stopped to eliminate the driving pressure applied to the biochip and thereby keeping the mixtures inside the reaction microchambers during the reaction time. RBC agglutinations were observed in the reaction microchambers after the flow rate was stopped for 3 min. The optimal micromixer type and flow rate were selected for clinical sample testing. The clinical performance of the proposed microfluidics chip was determined by testing with clinical EDTA-containing blood samples. Blood samples were collected from the Blood Bank, Faculty of Medicine, Khon Kaen University, Thailand, following the Declaration of Helsinki and the ICH Good Clinical Practice Guidelines (HE674003). E-negative and E-positive RBCs (3 samples in each group) were obtained for the detection of E antigens via the microfluidic chip. After anti-E was mixed with the clinical samples, the microfluidic chip measurements were performed as described in the previous section.
2.9 Image processing and agglutination analysis
RBC images were obtained in the reaction chambers with an inverted Olympus IX71 microscope (Olympus, Shinjuku, Tokyo, Japan) with an 10× objective lens. Images were captured three minutes after a flow rate of 1 μL min−1 was reached.Each image was first converted to grayscale, and a Gaussian blur (kernel size of 3 × 3) was applied to each image to reduce noise. The blurred images were then binarized using a fixed threshold of 155 for the binary inversion method, which allowed the image to be more distinctly black and white, which facilitated further analysis. The detection of agglutination was performed by identifying significant contours in the binary images. Which was determined based on the image resolution and the typical size of RBC clusters observed in this study. The contour area thresholds correspond to the pixel resolution of the images and were empirically chosen to capture the majority of RBC aggregates while excluding noise and non-agglutinated cells. Additionally, a contour was considered indicative of agglutination if it exhibited specific shape characteristics as defined by aspect ratio thresholds and the presence of gaps within the contour. Examples of image processing procedures portraying the original, binary, and contour images are depicted for non-agglutinated (Fig. 3A) and agglutinated RBCs (Fig. 3B).
 | ||
| Fig. 3 Process of agglutination analysis showing the original, binary and contour area of red cell images in (A) negative (non-agglutinated) and (B) positive (agglutinated) RBCs. | ||
2.10 Ethical statement
The study was reviewed and approved by the Khon Kaen University Ethics Committee for Human Research in accordance with the Declaration of Helsinki and the ICH Good Clinical Practice Guidelines (HE674003).3 Results and discussion
3.1 Design of the micromixer
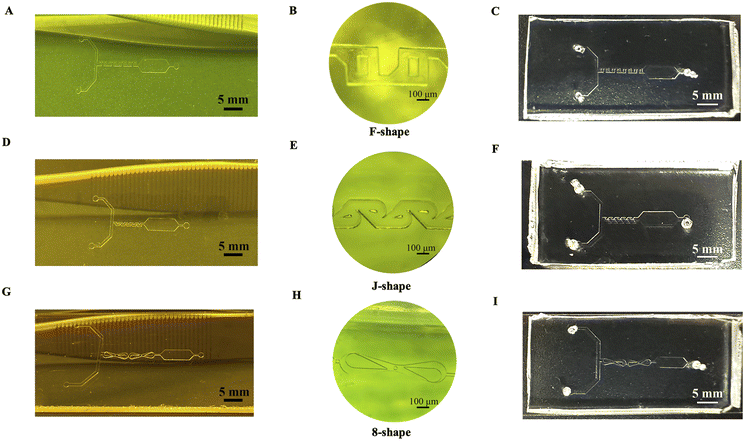
The micromixers were designed in the Layout Editor's full version. Three micromixer models have been designed for testing the agglutination reaction including the F-shape, the Tesla (J)-shape, and the 8-shape with an obstacle. The first micromixer type contains eight small “F-shape” mixing units (250 μm width) comprising a total mixing length of 10.5 mm. Moreover, the Tesla (J) micromixer was composed of ten small “J-shape” units and had a mixing length of 6.5 mm. A Tesla micromixer works by utilizing the Coanda effect to split an approaching stream and then redirecting one of the diverging flows so that it will meet the second tributary. Thus, the alternating flow within the transverse direction improved the mixing performance. Two “8-shape” micromixer units comprising a total mixing length of 12.3 mm in the last micromixer type were used. The main microchannels of all the micromixers were 100 μm high, and the reaction chamber width was 2500 μm. The detailed dimensions of each micromixer can be observed in Fig. 2. Despite 8-shaped micromixer has only two mixing nodes, the total mixing length is the highest, compared to F-shaped and J-shaped designs. It was proven that the integration of circular obstruction in the fluid flow path of an 8-shaped micromixer significantly enhances the chaotic advection, thus improving the mixing efficacy within the first micromixer unit.1 The Tesla (J)-shaped micromixer reported in previous studies indicated the enhanced mixing efficiency by applying five mixing pairs and a mixing length of ∼7 mm.2,3 F-shaped micromixers typically consist of four mixing pairs, which provide efficient mixing performance.4,5 Since the results of our fluid simulation are consistent with previous studies, the mixing units for each type of micromixer are set at certain values.The proposed microfluidic device contains two inlet channels, one micromixer unit, a reaction chamber, and an outlet. PDMS mold (4 mm high and 7.5 mm long) was used for the fabrication of a microfluidic device. Prior to plasma bonding, two inlets and one outlet hole were punched by a PDMS puncher, and 1.6 mm (OD) Tygon tubing was inserted for fluid flow. Fig. 4 shows the overall and microscopic structures of the master mold and fabricated microfluidic chips: F-shape (A–C), J-shape (D–F), and 8-shape (G–I) micromixers. Normal RBCs have a biconcave-discoidal shape with a diameter of 6–8 μm. The center of the RBC is relatively narrower (∼1 μm) than the surrounding borders (∼3 μm) under normal conditions.42,43 According to the rheological characteristics of RBCs, a fluidic channel lower than the thickness of the RBC's outer edge will prevent agglutinated RBCs. A large, low-aspect-ratio fluidic channel was necessary to further reduce the chance of false-positive results from channel obstruction.44 Therefore, our micromixers were specifically designed to prevent the effect of RBCs clogging inside the mixing units.
3.2 ATR-FTIR analysis of PDMS surface chemistry
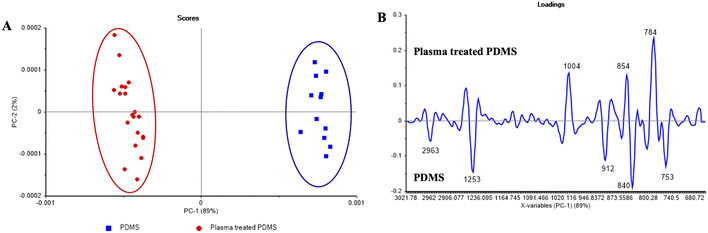
Using ATR-FTIR, the surface chemistry of PDMS and O2 plasma-treated PDMS was characterized (see Fig. 5). PCA score plot revealed that untreated and oxygen plasma-treated PDMS spectra were separated along PC1 (see Fig. 5a). All bands detected on the FTIR spectrum confirm the PDMS membrane polymeric structure (Fig. 5b). The absorption peak of PDMS at 2958 cm−1 can be assigned to symmetric stretching of –CH3 in Si–CH3.9 The peaks at 1251 cm−1 in untreated PDMS and 1270 cm−1 in treated PDMS represent the CH3 symmetric bending in Si–CH3.8 The bands of 1020 and 1004 cm−1 were associated with Si–O–Si asymmetric stretching,7 whereas a band at 867 cm−1 is associated with stretching Si–C in Si–CH3.4,5 Moreover, the peak at 753 cm−1 represents CH3 rocking in Si–CH3.1–3 The presence of 784 and 854 cm−1 bands in treated PDMS indicates the Si–O symmetric and asymmetric stretching from SiO4, respectively. The loading plot revealed at approximately 2963 cm−1 and 1251 cm−1 are higher in untreated PDMS. This is due to the replacement of methyl groups with oxygen atom upon oxidation10 and the decrease in methyl (–CH3) functional group at the PDMS surface with the plasma treatment, signifying the hydrophilic characteristics of the PDMS surface.11 After plasma exposure, the methyl groups of the siloxane oligomers are trapped, thus forming a SiOx structure. Therefore, the change of CH3–Si–O– to –O–Si–O– enhances the affinity with water molecules, thereby increasing the hydrophilicity of the PDMS surface.11,123.3 Evaluating the mixing efficiency of micromixers
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, 100
200, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 80
100, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, and 70
20, and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30. The objective was to observe the behavior of the fluid in the micromixing region and evaluate the mixing performance. In the micromixing unit and reaction chamber region, the focus was on assessing the mixing performance. Higher performance, which is characterized by homogeneous mixing, was achieved, particularly in the early stages of the micromixing unit.
30. The objective was to observe the behavior of the fluid in the micromixing region and evaluate the mixing performance. In the micromixing unit and reaction chamber region, the focus was on assessing the mixing performance. Higher performance, which is characterized by homogeneous mixing, was achieved, particularly in the early stages of the micromixing unit.
3.3.1.1 F-shape model. The F-shape model results in the best mixing performance among the tested shapes. It achieved a homogeneous mix with just 2 units of the micromixing unit, which indicates efficient mixing across all flow ratios (refer to Fig. 6A for visualization). The result obtained is similar to the previous report for simulation of F-mixing units, where there are two turns of fluid flow into the micromixer and chaotic convection has occurred.45
 | ||
| Fig. 6 2D Simulations of all three micromixer types: (A) F-shape, (B) Tesla (J)-shape and (C) 8-shape. | ||
3.3.1.2 Tesla (J)-shape model. The Tesla model demonstrated moderate mixing performance. Three units of the micromixing region were required to achieve a homogeneous mixture of the two fluids (Fig. 6B). The changes in the flow ratio indicated homogeneous mixing efficiency with different mixing intensities. A study reported that the tesla micromixer achieved full mixing after passing four mixing units according to the fluidic simulation.46 Our simulation results are consistent with the previous reports showing the transverse dispersion of fluids at the end of each mixing unit.
3.3.1.3 8-Shape model. This model exhibited diverse mixing performances among the three shapes. This can be attributed to its design, which includes only 2 micro mixing regions (see Fig. 6C). When equal flow rates of blood and slurry were applied, there was evidence of multilayered mixed fluid, which indicated incomplete mixing in the reaction microchamber. However, the application of a lower volume of slurry seemed to improve the mixing efficacy.
These results suggest that the design and configuration of the micromixing unit and reaction chambers significantly impact the mixing performance. The F-shape model, with its optimized design, shows promise for efficient and effective mixing under various flow conditions.
The Tesla (J)-shape micromixer, which was by far the most common, exhibited excellent mixing performance in both simulated and practical dye testing. Owing to the alternative fluid flow and transverse dispersion, complete mixing was observed after the first four mixing units were passed (Fig. 7B). However, the amount of air trapped inside the curved area of the micromixer obstructs fluid flow and hence interferes with RBC agglutination. The 8-shape micromixer illustrated an alternative fluid stream providing efficient mixing around the circular obstruction and inside the reaction cell (Fig. 7C). It was proven that integration of circular obstruction in fluid flow path enhance the chaotic advection significantly and thus improving the mixing efficacy.41 The resulting green color was introduced at the fluid interfaces, and the dispersion of color intensity in the reaction well indicated the different levels of mixing. In conclusion, the mixing efficiencies of all three proposed micromixers are promising for application in RBC antigen typing. Nevertheless, the divergent mixing reactions and the dispersion of fluid streams would have affected RBC antigen and antibody interactions.
3.4 RBC testing in microfluidic chips
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 80
30, 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, 100
20, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, and 200
100, and 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 μL min−1) at the same time.
200 μL min−1) at the same time.The agglutination strength changed with different flow rates. As shown in Fig. 9A, weak agglutination was observed at 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 μL min−1. At a flow rate of 80
30 μL min−1. At a flow rate of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 μL min−1, the agglutinates were uniformly distributed with no unreacted red cells in the detection region (see Fig. 9B). Higher flow rates led to larger agglutinates but there were greater variations in agglutination sizes (Fig. 9C and D). An 80
20 μL min−1, the agglutinates were uniformly distributed with no unreacted red cells in the detection region (see Fig. 9B). Higher flow rates led to larger agglutinates but there were greater variations in agglutination sizes (Fig. 9C and D). An 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 μL min−1 flow rate was selected as optimal for red cell testing.
20 μL min−1 flow rate was selected as optimal for red cell testing.
3.5 Clinical sample testing
To validate the potential application of the microfluidic chips for E-antigen typing, blood samples containing both positive and negative E antigens were tested. Agglutination was observed in the positive E-antigen samples, which presented clusters of RBCs, whereas only single red cells were visible in the negative samples (Fig. 10A and B). Red cell agglutination is typically interpreted by visual inspection (naked eyes or microscopically). Automation of analysis has been used recently to offset bias and human error in manual methods.47,48 However, absolute quantitation of red cell agglutination is still lacking. In this study, an image processing algorithm, as outlined in Section 2.8, was applied to quantify the extent of agglutination. This analysis utilized thresholding and contour detection to identify and measure clusters and led to a quantifiable agglutination index. Fig. 10C shows a greater agglutination index for E-positive RBCs than for E-negative RBCs and confirms the efficacy of the image processing approach for distinguishing between positive and negative E-antigen samples.4 Conclusions
The novel microfluidic chip and optimal conditions developed in this work have been successfully employed for detecting E antigens on RBCs. We validated the successful O2 plasma treatment of PDMS by using ATR-FTIR analysis of PDMS surface chemistry. The PDMS surface demonstrated enhanced hydrophilicity, indicating that the microfluidic chip is ready to use. Simulations and dye mixing studies were performed in microfluidic chips with F-, J-, and 8-shape designs to understand and prove that the physical capture hypothesis aligns with the experimental results. These microfluidic chips also showed different agglutination indices when E + RBCs and anti-E serum were tested. The 8-shape design emerged as the best micromixer for the detection of E antigen. The microfluidic chip could be integrated into blood bank procedures and streamlining E antigen detection during blood typing and compatibility testing. This would potentially reduce the risk of HTRs and HDFNs by ensuring better blood type matching. The chip also has the potential to be adapted for multiplexing and allow simultaneous detection of other blood group antigens alongside the E antigen in the future.Data availability
The data collected from human participants, described in Fig. 10 are not available for confidentiality reasons.Author contributions
Conceptualization, P. M. and R. P.; methodology, N. N. S. and P. M.; software, R. P.; validation, P. M. and P. P.; formal analysis, N. N. S. and P. M.; investigation, P. P., N. N. S., and P. M.; resources, J. D., P. T., K. S., and R. P.; data curation, N. J. and S. D.; writing—original draft preparation, N. N. S. and P. M.; writing—review and editing, all authors; visualization, N. N. S. and P. M.; supervision, K. C., R. P., and J. D.; project administration, P. M.; funding acquisition, P. M. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have no conflicts of interest.Acknowledgements
This project is funded by the National Research Council of Thailand (NRCT) grant no. N42A660918 and the Centre of Research and Development of Medical Diagnostic Laboratories (CMDL), Faculty of Associated Medical Sciences, Khon Kaen University, Thailand.References
- J. Poole and G. Daniels, Transfus. Med. Rev., 2007, 21, 58–71 CrossRef PubMed.
- L. Dean, The ABO Blood Group, National Center for Biotechnology Information (US), 2005 Search PubMed.
- N. D. Avent and M. E. Reid, Blood, 2000, 95, 375–387 CrossRef CAS PubMed.
- C. M. Westhoff, Semin. Hematol., 2007, 44, 42–50 CrossRef CAS PubMed.
- B. Michalewska, A. Ejduk and K. Pniewska, Vox Sang., 2005, 89, 61 CrossRef CAS PubMed.
- V. Chatziantoniou, N. Heeney, T. Maggs, C. Rozette, C. Fountain, T. Watts, C. Harrison, D. Pasupathy, S. Sankaran, P. Kyle and S. Robinson, Transfus. Med., 2017, 27, 275–285 CrossRef CAS PubMed.
- Z. Liu, M. Liu, T. Mercado, O. Illoh and R. Davey, Transfus. Med. Rev., 2014, 28, 177–186 CrossRef PubMed.
- M. Vaziri, H. JavadzadehShahshahani, M. Moghaddam and N. Taghvaee, Iran J. Pediatr. Hematol. Oncol., 2015, 5, 93–99 CAS.
- S. Kulkarni, B. Choudhary, H. Gogri, J. Sharma and M. Madkaikar, Indian J. Med. Res., 2020, 152, 273 CrossRef CAS PubMed.
- S. T. Chou, T. Jackson, S. Vege, K. Smith-Whitley, D. F. Friedman and C. M. Westhoff, Blood, 2013, 122, 1062–1071 CrossRef CAS PubMed.
- A. Mujahid and F. Dickert, Sensors, 2015, 16, 51 CrossRef PubMed.
- W. Malomgré and B. Neumeister, Anal. Bioanal. Chem., 2009, 393, 1443–1451 CrossRef PubMed.
- J. H. Spindler, H. Klüter and M. Kerowgan, Transfusion (Paris), 2001, 41, 627–632 CrossRef CAS PubMed.
- W. Xu, F. Wan, Y. Lou, J. Jin and W. Mao, Clin. Lab., 2014, 60, 241–244 Search PubMed.
- Y.-J. Chang, C.-Y. Ho, X.-M. Zhou and H.-R. Yen, Biosens. Bioelectron., 2018, 102, 234–241 CrossRef CAS PubMed.
- S. Chomean, S. Ingkananth, M. Kiatchaipar and C. Kaset, Anal. Chim. Acta, 2021, 1180, 338884 CrossRef CAS PubMed.
- T. Songjaroen and W. Laiwattanapaisal, Anal. Chim. Acta, 2016, 921, 67–76 CrossRef CAS PubMed.
- S. Hossain, A. Fuwad, K.-Y. Kim, T.-J. Jeon and S. M. Kim, Ind. Eng. Chem. Res., 2020, 59, 3636–3643 CrossRef CAS.
- C. T. Culbertson, T. G. Mickleburgh, S. A. Stewart-James, K. A. Sellens and M. Pressnall, Anal. Chem., 2014, 86, 95–118 CrossRef CAS PubMed.
- V. Hessel, H. Löwe and F. Schönfeld, Chem. Eng. Sci., 2005, 60, 2479–2501 CrossRef CAS.
- C. J. Ingham and J. E. T. van Hylckama Vlieg, Lab Chip, 2008, 8, 1604–1616 RSC.
- T. H. Schulte, R. L. Bardell and B. H. Weigl, Clin. Chim. Acta, 2002, 321, 1–10 CrossRef CAS PubMed.
- D. S. Kim, S. H. Lee, C. H. Ahn, J. Y. Lee and T. H. Kwon, Lab Chip, 2006, 6, 794–802 RSC.
- H. Ashiba, M. Fujimaki, K. Awazu, T. Tanaka and M. Makishima, Sens. Bio. Sens. Res., 2016, 7, 121–126 CrossRef.
- S. H. Choi, D. S. Kim and T. H. Kwon, Microsyst. Technol., 2009, 15, 309–316 CrossRef CAS.
- J.-H. Lin, T.-T. Tsai, Q. Zeng, C.-Y. Chang, J.-Y. Guo, C.-J. Lin and C.-F. Chen, ACS Sens., 2020, 5, 3082–3090 CrossRef CAS PubMed.
- S. Min, T. Zhan, Y. Lu, D. Pan, X. Chen and B. Xu, Lab Chip, 2023, 23, 4680–4689 RSC.
- X. Wu, S. Min, T. Zhan, Y. Huang, H. Niu and B. Xu, Lab Chip, 2024, 24, 4379–4389 RSC.
- B. Xu, J. Zhang, D. Pan, J. Ni, K. Yin, Q. Zhang, Y. Ding, A. Li, D. Wu and Z. Shen, Lab Chip, 2022, 22, 4382–4392 RSC.
- H. Wang, S. Duan, M. Wang, S. Wei, Y. Chen, W. Chen, Y. Li and S. Ding, Sens. Actuators, B, 2020, 320, 128376 CrossRef CAS.
- A. Nilghaz, L. Zhang, M. Li, D. R. Ballerini and W. Shen, ACS Appl. Mater. Interfaces, 2014, 6, 22209–22215 CrossRef CAS PubMed.
- J. Park and J.-K. Park, Anal. Chem., 2019, 91, 11636–11642 CrossRef CAS PubMed.
- A. Afzal and K.-Y. Kim, in Analysis and Design Optimization of Micromixers, ed. A. Afzal and K.-Y. Kim, Springer, Singapore, 2021, pp. 11–34 Search PubMed.
- G. D. Xia, Y. F. Li, J. Wang and Y. L. Zhai, Int. Commun. Heat Mass Tran., 2016, 71, 188–196 CrossRef CAS.
- H. H. Bau, J. Zhong and M. Yi, Sens. Actuators, B, 2001, 79, 207–215 CrossRef CAS.
- I. Glasgow and N. Aubry, Lab Chip, 2003, 3, 114–120 RSC.
- R. H. Vafaie, M. Mehdipoor, A. Pourmand, E. Poorreza and H. B. Ghavifekr, Biotechnol. Bioprocess Eng., 2013, 18, 594–605 CrossRef CAS.
- S.-J. Kim, F. Wang, M. A. Burns and K. Kurabayashi, Anal. Chem., 2009, 81, 4510–4516 CrossRef CAS PubMed.
- D. S. Kim, S. H. Lee, T. H. Kwon and C. H. Ahn, Lab Chip, 2005, 5, 739–747 RSC.
- M. Juraeva and D.-J. Kang, Micromachines, 2022, 13, 1375 CrossRef PubMed.
- S. Hossain, A. Fuwad, K.-Y. Kim, T.-J. Jeon and S. M. Kim, Ind. Eng. Chem. Res., 2020, 59, 3636–3643 CrossRef CAS.
- Y. Park, C. A. Best, T. Auth, N. S. Gov, S. A. Safran, G. Popescu, S. Suresh and M. S. Feld, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1289–1294 CrossRef CAS PubMed.
- M. Kinnunen, A. Kauppila, A. Karmenyan and R. Myllylä, Biomed. Opt. Express, 2011, 2, 1803 CrossRef PubMed.
- J.-Y. Chen, Y.-T. Huang, H.-H. Chou, C.-P. Wang and C.-F. Chen, Lab Chip, 2015, 15, 4533–4541 RSC.
- X. Chen and J. Shen, Anal. Methods, 2017, 9, 1885–1890 RSC.
- C.-C. Hong, J.-W. Choi and C. H. Ahn, Lab Chip, 2004, 4, 109–113 RSC.
- M. Huet, M. Cubizolles and A. Buhot, Biosens. Bioelectron., 2017, 93, 110–117 CrossRef CAS PubMed.
- M. Nguyen, K. Fries, R. Khoury, L. Zheng, B. Hu, S. W. Hildreth, R. Parkhill and W. Warren, SLAS Technol., 2016, 21, 287–296 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |