Open Access Article
Open Access ArticleLiver magnetic resonance spectroscopy as an alternative for evaluating Niemann-Pick C disease progression†
Aline Xavier ab,
Juan E. Oyarzun
ab,
Juan E. Oyarzun bc,
Flavia Zacconi
bc,
Flavia Zacconi de,
Silvana Zanlungof and
Marcelo E. Andia*bc
de,
Silvana Zanlungof and
Marcelo E. Andia*bc
aFaculty of Engineering, Universidad de Santiago de Chile, Santiago, 9170124, Chile
bMillennium Institute for Intelligent Healthcare Engineering - IHEALTH, Santiago, Chile
cRadiology Department & Biomedical Imaging Center, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile. E-mail: meandia@uc.cl
dFaculty of Chemistry and Pharmacy, Pontificia Universidad Católica de Chile, Santiago, Chile
eInstitute for Biological and Medical Engineering, Schools of Engineering, Medicine, and Biological Sciences, Pontificia Universidad Católica de Chile, Santiago, Chile
fGastroenterology Department, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile
First published on 6th February 2025
Abstract
Niemann-Pick disease (NP) is a group of rare genetic disorders that affect normal lipid metabolism and cause an accumulation of lipids in the liver, spleen, brain, and bone marrow. NP patients develop brain alterations and a very fast progression of liver damage. The purpose of this study is to characterize the changes in liver lipid composition during the progression of this disease using ex vivo magnetic resonance spectroscopy (MRS) in mouse models with the aim of identifying potential biomarkers to support a future non-invasive technique to follow-up these patients. NP type C (NPC) and wild-type (WT) mice were fed a chow diet and euthanized at 5 weeks of age (n = 5 per group) and 9 weeks of age (n = 5 per group). We extracted lipids from their livers and analyzed them with Gas Chromatography-Mass Spectrometry (GC-MS) and MRS. With the GC-MS analysis, 7 main fatty acids (FA) and cholesterols were quantified. Using MRS, we identified 5 metabolite peaks that correspond to FA only, 3 peaks that correspond to cholesterol only, and 2 peaks that correspond to FA and cholesterol. Our results show that the increase in liver cholesterol is the key biomarker for liver damage in NPC, which is consistent with a bad liver disease prognosis due to the association of increased cholesterol levels and liver inflammation. Additionally, we identified a difference in the pool of FA stored in the NPC compared to the WT mouse livers. Those different liver spectra could provide potential biomarkers for the non-invasive follow-up of NPC patients.
1. Introduction
Niemann-Pick disease (NP) is a group of rare genetic disorders. Patients who have this disease are characterized by having an abnormal lipid metabolism which causes the accumulation of lipids in the cells of the body, particularly in the brain, liver, spleen, and bone marrow.1 This disease was first described by Albert Niemann in 1914 (ref. 2) and histologically characterized by Ludwig Pick in 1926.3 In 1961, NP disease was classified into four types according to their clinical form (A, B, C, and D) characterized by the accumulation of sphingomyelin and cholesterol in the cell's lysosomes. The cause of types A and B was identified in 1966 with the discovery of the acid sphingomyelinase lysosomal enzyme, whose deficiency results in lysosomal accumulation of sphingomyelin.4 Later, in the 1990s, it was determined that types C and D are the result of mutations in genes related to lysosomal efflux of cholesterol (NPC1 and/or NPC2).5,6 NP type D is considered an allelic variant of type C and has only been found in the French-Canadian population of Yarmouth County, Nova Scotia.7Patients with Niemann-Pick disease type C (NPC) exhibit rapid and progressive neurodegeneration, growth retardation, and liver damage. The prevalence of NPC is around 1 in 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 live births, making it a rare disease, with a life expectancy of between days to decades.8 The life expectancy depends on the onset of the disease which ranges from the perinatal period to adulthood.9
000 live births, making it a rare disease, with a life expectancy of between days to decades.8 The life expectancy depends on the onset of the disease which ranges from the perinatal period to adulthood.9
Unfortunately, NPC disease has no cure; however, in some cases, the symptoms can be treated with a multidisciplinary approach. The main treatment focuses on helping people live with the symptoms, such as delaying neurological symptoms and physical therapy to preserve mobility as much as possible. NPC patients present with considerable damage in the liver.8 Some aspects of the pathophysiology associated with the NPC liver have been investigated using the NPC mouse model. In particular, Beltroy et al.1 performed a study which found that NPC mice have a similar liver disease to that seen in NPC infants, making this a useful model for exploring the molecular events. Those studies with a mouse model reported cholesterol accumulation, progressive apoptosis, inflammation, oxidative stress, and fibrosis in the NPC mouse liver10–12.
Nowadays, the most common way to evaluate the accumulation of lipids in the liver is with ultrasound, however, it is not quantitative, and it is a very operator-dependent technique13,14. A better non-invasive way to quantify the fat in the liver is using Magnetic Resonance Spectroscopy (MRS) which is the most accurate method to determine the exact percentage of fat in the liver, commonly termed the “hepatic fat fraction”15,16. This modality has been proposed for studying patients with non-alcoholic fatty liver disease (NAFLD).17–20 In the case of NPC patients, it could be used to evaluate the response to liver treatment.
Magnetic resonance technique has emerged as the ideal technique to quantify the fat infiltration in solid organs. Considering that most of the acquired MR signal comes from water and fat protons, Fat Fraction (FF, the proportion of the acquired signal derived from fat protons) has been proposed as non-invasive biomarker of different metabolic diseases.21
MRS uses the magnetic property of an atom's nucleus when it is exposed to a static magnetic field and then excited by a radio-frequency pulse to produce a spectrum. This spectrum provides chemical information about a tissue, by showing the resonance frequency of protons from this tissue (in parts per million, ppm) versus its amplitude when exposed to an external magnetic field. Each set of chemically equivalent protons originates a signal (or peak). In this way, MRS makes it possible to infer the chemical structure of a molecular species and, in turn, obtain metabolic information.22,23
In MRS, the fat fraction is calculated by a ratio between the water (W) peak, present in 4.7 ppm, and the main lipid (L) peak, present at 1.3 ppm, given by (L/L + W). This occurs since water peak represents the protons in water molecules. As water is abundant in biological tissues, this peak is usually dominant. Lipid peak corresponds to the protons in fat molecules, particularly in triglycerides, which resonate at a different frequency than water. However, lipid content originates not only one peak, but many other peaks in the MRS spectrum.24 All those peaks can contribute to characterizing the type of lipid accumulated in the liver. Some studies have already correlated the fatty acids (FA) profile with the progression of NAFLD25–28.
Regarding NPC disease, a study analyzing the lipidomics of this disease found differences in FA composition between NPC and healthy mice29 by using gas chromatography, which is the gold standard method to quantify fatty acids and cholesterol. Therefore, our hypothesis is that this invasive analysis could be done using MRS with the possibility of progress towards a non-invasive technique.
To the best of our knowledge, the liver lipids of Niemann-Pick disease have never been analyzed with ex vivo magnetic resonance spectroscopy. The attraction of this method is the possibility to control not only the fat fraction of a NPC liver with MRS, but also to characterize the composition of fatty acids and cholesterol in the liver by doing a non-invasive magnetic resonance exam.
The purpose of this study is to characterize the composition of liver lipids in NPC and wild-type mouse models using ex-vivo spectroscopy with the aim of providing a follow-up technique for NPC patients. This study showed for the first time the uses of MRS spectra of hepatic lipids to evaluate the progression of NPC disease.
2. Subjects and methods
2.1. Animal model
In this study, the protocols used with the mouse models were in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals in the Institute for Laboratory Animal Research Guide for Care and Use of Laboratory Animals and approved by the review board for animal studies at our institution (Comité Etica Bienestar Animal, CEBA-MedUC; ID#3-2009).NPC and wild-type (WT) mice, hereinafter referred to as control mice, were fed a chow diet since their weaning and euthanized at 5 weeks of age and at 9 weeks of age to obtain their livers (n = 5 per group). These time points were chosen based on reports indicating that liver damage begins around 5 weeks (evidenced by changes in aminotransferase levels), and by 9 weeks, the damage is consolidated.1,30–33 We extracted lipids from these livers and then analyzed them with Gas Chromatography-Mass Spectrometry (GC-MS) and Magnetic Resonance Spectroscopy (MRS).
2.2. Lipid extraction and esterification
Intracellular lipids were extracted using a protocol adapted from Folch et al.,34 followed by esterification to obtain more stable fatty acid methyl esters (FAME).35 Each liver was weighed and homogenized for 5 min at 0 °C with a known amount of CHCl3/MeOH (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), NaCl, CH3COOH, and C19 fatty acid (as standard), and the organic phase was extracted with n-hexane. The solvent was evaporated to dryness under a stream of nitrogen gas. Once completely dry, the final mass of lipids extracted from the initial liver mass was obtained.
1), NaCl, CH3COOH, and C19 fatty acid (as standard), and the organic phase was extracted with n-hexane. The solvent was evaporated to dryness under a stream of nitrogen gas. Once completely dry, the final mass of lipids extracted from the initial liver mass was obtained.
The esterification process was carried out by adding 1 mL of 10% acetyl chloride in anhydrous methanol and 0.5 mL of dry toluene as a solvent and bringing it up to 55 °C overnight. Each sample was neutralized with 7 mL of 6% sodium bicarbonate. n-hexane (2 ml) was added for the final extraction of the FAME, to which, after a centrifugation process to remove residual water, the solvent was displaced with nitrogen gas, and the final methyl esters mass was obtained. From previous studies,36,37 we found that 300 mg of the liver from healthy mice was sufficient to obtain 5 mg of FAME, which is the minimum amount of sample needed to perform a MRS (400 MHz) with good signal-to-noise ratio (SNR) while only 2 mg of FAME is required for GC-MS.
2.3. Gas chromatography with a mass spectrometer
The lipids were analyzed using GC-MS (PerkinElmer, Clarus 680) equipped with a HP-Innowax capillary column (length 25 m, internal diameter 0.2 mm, film 0.2 mm). The injector temperature was kept at 220 °C, and the oven ramp was 150 °C for 1 min, changed to 200 °C at a 15 °C min−1 rate and maintained for 5 min and then increased from 200 °C to 260 °C at a 12 °C min−1 rate. Sample injection was done in split mode (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) with helium as the carrier gas with a flow of 1 mL min−1. Electron impact ionization mode of the mass detector was set with the ionization potential of 70 eV. The data register operated in SCAN mode, and peak integration was achieved using TurboMass Training 2016 PRO software. FAME and cholesterol were determined through the comparison of retention times to established standards and by aligning them with mass spectra from the NIST library (National Institute of Standards and Technology, USA) to ensure originality.37 FA and cholesterol compositions were defined as the percentage of individual compounds (fatty acids or cholesterol) with respect to the total.
20) with helium as the carrier gas with a flow of 1 mL min−1. Electron impact ionization mode of the mass detector was set with the ionization potential of 70 eV. The data register operated in SCAN mode, and peak integration was achieved using TurboMass Training 2016 PRO software. FAME and cholesterol were determined through the comparison of retention times to established standards and by aligning them with mass spectra from the NIST library (National Institute of Standards and Technology, USA) to ensure originality.37 FA and cholesterol compositions were defined as the percentage of individual compounds (fatty acids or cholesterol) with respect to the total.
2.4. Magnetic resonance spectroscopy
MRS spectra of the lipid samples were obtained using a Bruker Avance spectrometer (9.4 T) with the Zg30 acquisition protocol. A known mass of lipids from each sample was dissolved in 0.7 mL of deuterated chloroform containing tetramethylsilane as an internal reference. The sample was introduced into a 5 mm diameter tube. The spectrum was acquired with Topspin V3.0 at 25 °C, a 1s relaxation delay, a spectral width of 8012.8 Hz, number of scans equal to 16, flip angle of 30° (to avoid T1 relaxation effects), and a total acquisition time of 48.7 s. The data were analyzed with MestReNova V12, and the area under the curve was calculated. The metabolite composition was defined as the percentage of individual peaks with respect to its total. Oostendorf et al.24 was used as a reference to match the peaks with the corresponding metabolite.2.5. Statistical analysis
Prism 6 (GraphPad Software Inc, La Jolla, CA) was used to perform statistical analyses. All graphs were generated with mean values and their corresponding standard deviation (SD). The data was first analyzed with the ROUT method to eliminate outliers, and a Shapiro–Wilk test was performed to confirm the normality of the data. To compare the four groups (Control 5 weeks, Control 9 weeks, NPC 5 weeks, and NPC 9 weeks), the ANOVA test was performed followed by a Bonferroni's multiple comparison test. To compare NPC 5 weeks and NPC 9 weeks to evaluate the progression, a T-test was performed.3. Results
3.1. Control mice and Niemann-Pick C mice show differences in the total amount of fat in the liver
After the lipid extraction, the percentage of fat was calculated as total lipids divided by liver weight, obtaining the percentage of fat in the liver (Fig. 1). As expected, the total amount of fat in the NPC mouse livers was higher than in the control mouse livers. However, there was no significant difference between NPC mouse livers at 5 weeks and 9 weeks of age.3.2. MRS spectra showed changes in the metabolite peaks between control and Niemann-Pick C mice
By using MRS, it was possible to identify five metabolite peaks that correspond to FA only, three peaks that correspond to cholesterol only, and two peaks that correspond to an overlap between the FA peak and cholesterol peak. Fig. 2 shows a comparison between the spectrum corresponding to a control group (red) and a NPC group (blue). The peaks highlighted with a black arrow are the fatty acid peaks, the ones with an orange arrow are a mixture of FA and cholesterol, and the others in green are cholesterol only. Note that although it is possible to appreciate some changes in the peaks with only fatty acids, the main changes are due to the presence of a larger amount of cholesterol in NPC mice.Table 1 shows the correspondence between each peak and the metabolite. Peaks 2 to 7 and peaks 9 and 10 overlap. The peak at 3.7 ppm corresponds to the ester radical, which was expected since we esterified the sample.
| Peak | Peak frequency (ppm) | Assignment |
|---|---|---|
| 1 | 0.68 | Total cholesterol protons |
| 2 | 0.86/0.87 | Total cholesterol protons |
| 3 | 0.88 | Fatty acid protons |
| 4 | 0.91 | Total cholesterol protons |
| 5 | 1.01 | Free cholesterol protons |
| 6 | 1.02 | Esterified cholesterol protons |
| 7 | 1.05–1.19 | Multiple cholesterol protons |
| 8 | 1.24–1.37 | Fatty acid protons |
| 9 | 1.42–1.55 | Multiple cholesterol protons |
| 10 | 1.55–1.65 | Fatty acid protons |
| 11 | 1.79–1.88 | Multiple cholesterol protons |
| 12 | 1.98–2.09 | Fatty acid protons |
| 13 | 2.24–2.35 | Fatty acid protons |
| 14 | 2.77–2.87 | Fatty acid protons |
| 15 | 3.48–3.57 | Free cholesterol protons |
| 16 | 5.29–5.43 | Fatty acid protons |
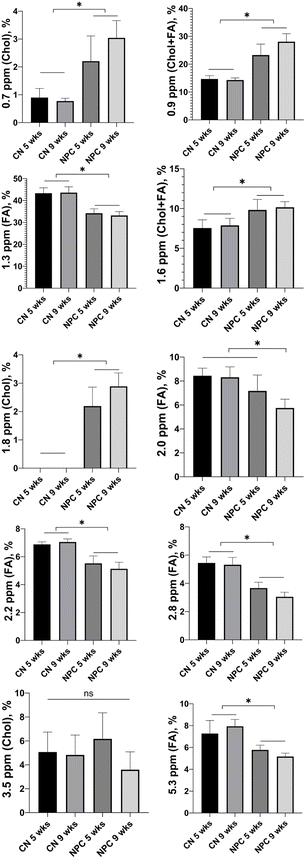
The area under the curve (AUC) of each peak was calculated and normalized by the total area of the spectrum to obtain the relative quantity of each metabolite. Fig. 3 shows a graph with all metabolites found in the spectra for each group. In all of them, it is possible to observe differences between control and NPC mice, except for the peak at 3.5 ppm. This result could indicate that MRS is a good technique to classify a NP liver versus a healthy liver.
3.3. With MRS, it was possible to identify the progression of the disease by comparing NPC mice at 5 and 9 weeks of age
By analyzing the NPC mice at two points of their life (Fig. 4), one can see the progression of their liver disease. The peaks at 2.8 ppm and 5.3 ppm decrease. These results are in accordance with the disease progression since the decrease in those peaks means a decrease in polyunsaturated fatty acids (PUFAs).3.4. Control mice and Niemann-Pick C mice show differences in the amount of cholesterol in the liver
With the GC-MS analysis, we were able to quantify 7 main fatty acid peaks and 2 peaks of cholesterol. We calculated the percentage of cholesterol with respect to the total lipids (Fig. 5). We found that there is a huge difference between control mouse and NPC mouse livers. This is in accordance with the results of Fig. 3, as the peaks at 0.7 ppm, 0.9 ppm, 1.6 ppm, and 1.8 ppm, all related to cholesterol, also show significant differences.3.5. With GC-MS, it was possible to observe changes in the fatty acid content during the disease progression as observed with MRS
As the NPC disease progressed, a reduction in some PUFAs, such as arachidonic and linoleic acid (Fig. 6A), was detected in the mouse livers. The combination of arachidonic and linoleic acid (Fig. 6B) shows a significant difference between NPC mice at 5 and 9 weeks of age which is related to the double bounds correspondent to the peaks at 2.8 ppm and 5.3 ppm in the MRS.4. Discussion
In this study, we have analyzed the fatty acid and cholesterol distributions in the liver of Niemann-Pick type C mice by using magnetic resonance spectroscopy. Those results were compared with the gold standard method to quantify fatty acids and cholesterol. In total, we have compared four groups of study: NPC mice at 5 weeks of age, NPC mice at 9 weeks of age, WT mice at 5 weeks of age, and WT mice at 9 weeks of age.Mice with NPC have more cholesterol in their liver than WT mice because of their genetic condition that affects proteins related to cholesterol transport.5 FA composition in the liver also differs between WT and NPC mice. As expected, the total amount of fat in the NPC mouse livers was higher than in the control mouse livers. However, there was no significant difference in the livers of NPC mice at 5 weeks and 9 weeks of age.
With MRS, it was possible to detect significant differences in the liver spectra. Regarding the fatty acid peaks, we notice a significant decrease in the AUC of the diallylic (2.8 ppm) and olefinic (5.3 ppm) peaks in NPC mouse livers when compared to the WT group. We also see a decrease of the same peaks between 5 weeks and 9 weeks of age in the mice with NPC, showing a progression of the disease. Those peaks correspond to polyunsaturated fatty acids (PUFAs), and the reduction in PUFAs is related to an increase in liver inflammation, since some of them are precursors of anti- and pro-inflammatory mediators.38 By analyzing the same results with GC-MS, we found a significant decrease in the amount of linoleic and arachidonic acids which also correlates with the progression of the disease, confirming the results obtained with MRS. A previous study conducted by members of our group31 showed that inflammation increased in NPC mice from 6 to 9 weeks of age. To analyze inflammation, the authors of that study evaluated CD68, a marker of inflammatory macrophages. Their evaluation showed that, from 6 to 9 weeks of age, there is an increase in the CD68 marker in the liver of NPC mice compared to control animals, and this increase is progressive over time.
In the present study, when analyzing the cholesterol peaks with MRS, we also found significant differences between the NPC group and the control group, corroborated by the GC-MS analysis. According to Van Rooyen et al.,39 accumulation of free cholesterol facilitates the transition from steatosis to nonalcoholic steatohepatitis (NASH) which is a more advanced stage of the liver disease. However, there is no knowledge about the reason that the cholesterol generates this damage in the liver. When the peak of cholesterol was analyzed to study the progression of the disease (from week 5 to week 9), we found no significant difference in the cholesterol peak. Similar results were found by Beltroy et al.;1 they studied mouse models with NPC disease across many weeks, and they also found no significant difference in cholesterol concentration between 5 and 9 weeks of age. In that study, the authors observed an increase in the cholesterol content only in week 10.
Our study has shown that the changes in fatty acids and cholesterol could be evaluated using magnetic resonance spectroscopy. An interesting application of this could be to evaluate treatment responses.40 Although the spectral resolution of 1.5 and 3 T MR scanners probably does not allow adequate visualization of these peaks in vivo, there have been some satisfactory experiences using 7 T machines.41
Even though NPC is a rare metabolic disease, the rapid progression of liver damage to cirrhosis it causes can shed light on possible mechanisms of liver damage in other common liver pathologies such as NAFLD. Indeed, the accumulation of cholesterol seems to be a key marker in the rapid progression of this disease and could be an interesting marker to assess in the NAFLD population.
Our study has some limitations. First, by using MRS, we have the overlap of cholesterol and FA peaks, which do not allow those metabolites to be studied separately and have a better classification. Another limitation is that the entire liver was used for the fatty acid extraction, therefore we do not have the histology results which would provide more information on the relationship between the inflammation process and the PUFA reduction. However, as this is a known model, we can infer from other papers the stage of the disease.1,31
5. Conclusions
Our results, using ex vivo MRS, show that an increase in the liver cholesterol concentration is the main difference of NPC mouse liver manifestation. There is also a different pool of fatty acids stored in the NPC and WT mouse livers. Both the increased concentration of cholesterol in the liver and the decrease in some FA could be biomarkers for tracking the progression of the disease and the response to treatment in this group of patients. Those changes also have the potential to be detected non-invasively using in vivo MRS.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
FZ is grateful to FONDEQUIP EQM120021 and EQM150020 from Pontificia Universidad Católica de Chile. This work was funded by ANID – Millennium Science Initiative Program – ICN2021_004, FONDECYT #1220922, #1190334 #11250733, FONDECYT Postdoctorado 2023 #3230777.Notes and references
- E. P. Beltroy, J. A. Richardson, J. D. Horton, S. D. Turley and J. M. Dietschy, Hepatology, 2005, 42, 886–893 CrossRef CAS PubMed.
- A. Niemann, Jahrb. Kinderheilkd., 1914, 79, 1–10 Search PubMed.
- L. Pick, Ergebenisse Der Inneren Medizin Und Kingderheilkunde, 1926, vol. 29, pp. 519–527 Search PubMed.
- R. O. Brady, J. N. Kanfer, M. B. Mock and D. S. Fredrickson, Proc. Natl. Acad. Sci., 1966, 55, 366–369 CrossRef CAS PubMed.
- E. D. Carstea, J. A. Morris, K. G. Coleman, S. K. Loftus, D. Zhang, C. Cummings, J. Gu, M. A. Rosenfeld, W. J. Pavan and D. B. Krizman, Science, 1997, 277, 228–231 CrossRef CAS PubMed.
- S. Naureckiene, D. E. Sleat, H. Lackland, A. Fensom, M. T. Vanier, R. Wattiaux, M. Jadot and P. Lobel, Science, 2000, 290, 2298–2301 CrossRef CAS PubMed.
- M. T. Vanier, Handbook of Clinical Neurology, 2013, vol. 113, pp. 1717–1721 Search PubMed.
- M. C. Patterson, C. J. Hendriksz, M. Walterfang, F. Sedel, M. T. Vanier and F. Wijburg, Mol. Genet. Metab., 2012, 106, 330–344 CrossRef CAS PubMed.
- T. Geberhiwot, A. Moro, A. Dardis, U. Ramaswami, S. Sirrs, M. P. Marfa, M. T. Vanier, M. Walterfang, S. Bolton and C. Dawson, Orphanet J. Rare Dis., 2018, 13, 1–19 CrossRef PubMed.
- V. M. Rimkunas, M. J. Graham, R. M. Crooke and L. Liscum, Hepatology, 2008, 47, 1504–1512 CrossRef CAS PubMed.
- M. C. Vázquez, T. del Pozo, F. A. Robledo, G. Carrasco, L. Pavez, F. Olivares, M. González and S. Zanlungo, PLoS One, 2011, 6, e28777 CrossRef PubMed.
- R. Fu, C. A. Wassif, N. M. Yanjanin, D. E. Watkins-Chow, L. L. Baxter, A. Incao, L. Liscum, R. Sidhu, S. Firnkes and M. Graham, Hum. Mol. Genet., 2013, 22, 3508–3523 CrossRef CAS PubMed.
- M. V. Machado and H. Cortez-Pinto, J. Hepatol., 2013, 58, 1007–1019 CrossRef PubMed.
- R. Williams and S. D. Taylor-Robinson, Clinical Dilemmas in Non-alcoholic Fatty Liver Disease, John Wiley & Sons, 2016 Search PubMed.
- L. S. Szczepaniak, P. Nurenberg, D. Leonard, J. D. Browning, J. S. Reingold, S. Grundy, H. H. Hobbs and R. L. Dobbins, Am. J. Physiol. Endocrinol. Metab., 2005, 288, E462–E468 CrossRef CAS PubMed.
- S. B. Reeder, I. Cruite, G. Hamilton and C. B. Sirlin, J. Magn. Reson. Imag., 2011, 34, 729–749 CrossRef PubMed.
- D. Pasanta, K. T. Htun, J. Pan, M. Tungjai, S. Kaewjaeng, H. Kim, J. Kaewkhao and S. Kothan, Diagnostics, 2021, 11(5), e842 CrossRef PubMed.
- B. Lavin, T. R. Eykyn, A. Phinikaridou, A. Xavier, S. Kumar, X. Buque, P. Aspichueta, C. Sing-Long, M. Arrese, R. M. Botnar and M. E. Andia, NMR Biomed., 2023, 36, e4932 CrossRef CAS PubMed.
- S. Qadri, E. Vartiainen, M. Lahelma, K. Porthan, A. Tang, I. S. Idilman, J. H. Runge, A. Juuti, A. K. Penttila, J. Dabek, T. E. Lehtimaki, W. Seppanen, J. Arola, P. Arkkila, J. Stoker, M. Karcaaltincaba, M. Pavlides, R. Loomba, C. B. Sirlin, T. Tukiainen and H. Yki-Jarvinen, JHEP Rep., 2024, 6, 100928 CrossRef PubMed.
- J. H. Runge, L. P. Smits, J. Verheij, A. Depla, S. D. Kuiken, B. C. Baak, A. J. Nederveen, U. Beuers and J. Stoker, Radiology, 2018, 286, 547–556 CrossRef PubMed.
- T. J. Bray, M. D. Chouhan, S. Punwani, A. Bainbridge and M. A. Hall-Craggs, Br. J. Radiol., 2018, 91, 20170344 CrossRef PubMed.
- M. van der Graaf, Eur. Biophys. J., 2010, 39, 527–540 CrossRef CAS PubMed.
- D. E. Befroy and G. I. Shulman, Diabetes, 2011, 60, 1361–1369 CrossRef CAS PubMed.
- M. Oostendorp, U. F. Engelke, M. A. Willemsen and R. A. Wevers, Clin. Chem., 2006, 52, 1395–1405 CrossRef CAS PubMed.
- K. Yamada, E. Mizukoshi, H. Sunagozaka, K. Arai, T. Yamashita, Y. Takeshita, H. Misu, T. Takamura, S. Kitamura and Y. Zen, Liver Int., 2015, 35, 582–590 CrossRef CAS PubMed.
- X. Wang, Y. Cao, Y. Fu, G. Guo and X. Zhang, Lipids Health Dis., 2011, 10, 1–7 CrossRef CAS PubMed.
- P. Puri, R. A. Baillie, M. M. Wiest, F. Mirshahi, J. Choudhury, O. Cheung, C. Sargeant, M. J. Contos and A. J. Sanyal, Hepatology, 2007, 46, 1081–1090 CrossRef CAS PubMed.
- A. Xavier, F. Zacconi, C. Gainza, D. Cabrera, M. Arrese, S. Uribe, C. Sing-Long and M. E. Andia, RSC Adv., 2019, 9, 42132–42139 RSC.
- M. R. Pergande, F. Serna-Perez, S. B. Mohsin, J. Hanek and S. M. Cologna, Proteomics, 2019, 19, 1800285 CrossRef PubMed.
- E. Balboa, T. Marin, J. E. Oyarzun, P. S. Contreras, R. Hardt, T. van den Bosch, A. R. Alvarez, B. Rebolledo-Jaramillo, A. D. Klein, D. Winter and S. Zanlungo, Cells, 2021, 10 CrossRef CAS PubMed.
- A. D. Klein, J. E. Oyarzun, C. Cortez and S. Zanlungo, Int. J. Mol. Sci., 2018, 19 Search PubMed.
- M. C. Vazquez, T. del Pozo, F. A. Robledo, G. Carrasco, L. Pavez, F. Olivares, M. Gonzalez and S. Zanlungo, PLoS One, 2011, 6, e28777 CrossRef CAS PubMed.
- N. L. Sayre, V. M. Rimkunas, M. J. Graham, R. M. Crooke and L. Liscum, J. Lipid Res., 2010, 51, 2372–2383 CrossRef CAS PubMed.
- J. Folch, M. Lees and G. H. Sloane Stanley, J. Biol. Chem., 1957, 226, 497–509 CrossRef CAS PubMed.
- B. Tang and K. H. Row, Monatsh. Chem., 2013, 144, 1427–1454 CrossRef CAS.
- A. Xavier, F. Zacconi, D. Cabrera, K. Fuenzalida and M. Andia, Singapore, 2019 Search PubMed.
- A. Xavier, F. Zacconi, F. Santana-Romo, T. R. Eykyn, B. Lavin, A. Phinikaridou, R. Botnar, S. Uribe, J. E. Oyarzun, D. Cabrera, M. Arrese and M. E. Andia, Ann. Hepatol., 2021, 25, 100358 CrossRef CAS PubMed.
- G. Bannenberg, M. Arita and C. N. Serhan, TheScientificWorldJOURNAL, 2007, 7, 1440–1462 CrossRef CAS PubMed.
- D. M. Van Rooyen, C. Z. Larter, W. G. Haigh, M. M. Yeh, G. Ioannou, R. Kuver, S. P. Lee, N. C. Teoh and G. C. Farrell, Gastroenterology, 2011, 141, 1393–1403.e5 CrossRef CAS PubMed.
- H. D. Le, J. A. Meisel, V. E. De Meijer, E. M. Fallon, K. M. Gura, V. Nose, B. R. Bistrian and M. Puder, J. Parenter. Enteral Nutr., 2012, 36, 431–441 CrossRef CAS PubMed.
- A. Xavier, C. Arteaga de Castro, M. E. Andia, P. R. Luijten, D. W. Klomp, A. Fillmer and J. J. Prompers, NMR Biomed., 2020, 33, e4343 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06781a |
| This journal is © The Royal Society of Chemistry 2025 |