Open Access Article
Open Access ArticleExplanation of the potential mechanism of Sophora flavescens Ait and processed products treating ulcerative colitis based on the analysis method of “chemical composition characterization-target prediction”
Xiaoting Wang a,
Yue Liuc,
Tianni Jiangb and
Hui Gao*b
a,
Yue Liuc,
Tianni Jiangb and
Hui Gao*b
aLiaoning University of Traditional Chinese Medicine, Shenyang, 110847, Liaoning, China
bSchool of Pharmacy, Liaoning University of Traditional Chinese Medicine, Dalian, 116600, China. E-mail: gaohuitcm@163.com
cLiaoning Agricultural Vocational and Technical College, Yingkou, 115009, China
First published on 14th April 2025
Abstract
Processing is often used to prepare decoctions of traditional Chinese medicine (TCM) with reduced toxicity and enhanced efficacy. While Sophora flavescens Ait (SFA) is often used directly, processing with rice-washed water (RSFA) was performed in ancient times, and processing with wheat bran (WSFA) is a more modern method. Processing has been shown to enhance the anti-inflammatory and antibacterial effects of SFA, though the mechanisms underlying this change are unclear. In this study, a total of 106 active components of SFA, RSFA, and WSFA, mostly alkaloid and flavonoid derivatives, were identified using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry, and a total of 159 potential molecular targets in the treatment of ulcerative colitis were identified by network pharmacology. Relationships among key targets, including epidermal growth factor receptor, heat shock protein 90, SRC, and p100α, were identified through development of a protein–protein interaction network. GO enrichment indicated that peptidyl-tyrosine phosphorylation, peptidyl-tyrosine modification, and cellular response to chemical stress are important in the action of SFA against ulcerative colitis, and KEGG enrichment showed that the phosphoinositide 3-kinase-AKT signaling pathway is another key target. Molecular docking showed that the active components have strong affinities for phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha and protein kinase C alpha. In vitro cell experiments have demonstrated that five active components in SFA can exert anti-inflammatory effects by modulating IL-6 and IL-10. We found that processing results in changes in the chemical composition of SFA that influence the treatment of UC. This study provides a reference for further research into the pharmacodynamic basis for the enhanced efficacy of processed SFA in the treatment of ulcerative colitis.
1. Introduction
The clinical manifestations of ulcerative colitis (UC) include abdominal pain, diarrhea and mucopurulent and bloody stools, as well as urgency and heaviness. In traditional Chinese medicine (TCM), the specific symptoms “bloody stool” and “chronic dysentery” are both associated with UC.1 Damp heat is one of the main syndromes associated with UC in TCM. Damp heat is an important factor underlying the onset and development of UC, and it is also the main factor leading to its recurrence and difficulty in treatment.2,3TCMs are derived from plants, animals, and minerals. Many of these sources are hard and thick in texture, some contain impurities and sands, and some are associated with significant side effects when used in unprocessed forms. Accordingly, the native materials generally cannot be directly used in clinical practice and instead must be processed before application.4 Many changes can occur during processing of TCM with excipients, including alterations to efficacy, flavor, and toxicity. Liquid excipients include wine, vinegar, honey, and rice-washed water, while solid excipients include wheat bran, rice, and river sand. Rice-washed water alone can invigorate qi, relieve vexation and thirst, and resolve toxins, and as an excipient, it can decrease a drug's pungent flavor, mitigate extreme properties, and strengthen the functions of replenishing the spleen and strengthening the middle energizer. Processing the drug with wheat bran can also moderate a drug's drastic properties and improve its therapeutic effectiveness.
TCM represents a potentially useful tool in the treatment of UC. In particular, lightyellow sophora root, also known as “kushen” in China, is the dried roots of Sophora flavescens Ait (SFA), which is associated with both bitterness and cold and is a candidate treatment for UC due to its ability to eliminate heat and dampness, its stimulation of water excretion, its effectiveness at treating carbuncles and other types of sores, and its activity against enterorrhagia and bloody stool.5 Accordingly, several processed products derived from SFA are widely used as an important component of TCM in the treatment of UC. Due to its unique manufacturing method and active ingredients, it has unique advantages in the treatment of UC. In TCM, SFA is usually used to treat heat dysentery, bloody stool, jaundice with anuria, red or white vaginal discharge, pudendal swelling and itch, eczema, dampness sore, itching of skin, scabies and tinea leprosy.6 Extracts of SFA have demonstrated several pharmacological activities, including anti-inflammatory, antioxidant, and antitumor effects. Its main components, such as alkaloids and flavonoids, have activity.
The processing of SFA has been studied in depth. For example, based on ancient books, the Chinese Pharmacopoeia, and modern literature, a research group conducted a comparative study on the chemical and pharmacological properties of 8 processed products of SFA.7 The results showed that processing with wheat bran to produce WSFA and with rice-washed water to produce RSFA led to significant changes to the activities of SFA, and the products exhibited stronger anti-inflammatory and antibacterial effects in vitro and stronger anti-UC effects in vivo than did the raw products, supporting the rationality of the historical use of WSFA and RSFA. The study also found that some components of SFA undergo transformations during the processing, leading to changes in their amounts. On this basis, we speculate that the enhanced therapeutic effect of processed SFA may be related to changes to the molecular composition of these materials. At present, however, few reports have investigated the mechanisms of SFA or its processed products in treating UC by combining the pharmacological components of SFA after processing with target prediction.8
The integration of network pharmacology and molecular docking methods can serve as an alternative to screening compounds before animal experiments. It focuses on the interaction between components and target proteins, providing strong support for elucidating the “multi-component, multi-target” nature of TCM.9,10 In this study, we performed a UPLC-Q-TOF-MS/MS-based analysis of the chemical composition of SFA and its processed products. The resulting information, combined with network pharmacology analysis of the correlation between targets, disease, and drugs, allowed a preliminary verification of the material basis and possible molecular mechanisms involved in the treatment of UC by SFA and its processed products. In addition, an in silico molecular docking analysis provided a theoretical basis for further research on the synergistic mechanisms underlying the clinical treatment of UC by SFA and its processed products.
2. Materials and methods
2.1. Identification of the chemical constituents in SFA and the processed products by UPLC-Q-TOF-MS/MS
To prepare processed SFA with rice-washed water, SFA decoction pieces were soaked with rice-washed water overnight at room temperature (25 ± 5 °C).11 The pieces were removed, rinsed twice with clean water, and steamed for 8 h. The processed decoction pieces were dried in the sun, crushed, and sifted through a No. 4 sieve.
The Waters Xevo G2-XS system (Waters Corp., MA, USA), capable of performing electrospray ionization (ESI), was used in positive and negative ionization modes. High purity N2 was used as an auxiliary spray ionization and desolvation gas: N2 temperature, 450 °C; desolventizing nitrogen flow rate, 800 L h; taper hole counter blowing nitrogen, 50 L h; capillary ionization voltage, 3 KV; and full scan range, 100–1500 m/z.
2.2. Network pharmacology analysis
2.3 Molecular docking
Computer-assistant molecular docking technology was utilized to investigate the physical interactions between SFA compounds and their targets. The 10 most significant ingredients were selected as ligands to analyze against their respective potential pharmacodynamic targets. The SDF files of these 10 compounds were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and converted to three-dimensional structures using Chem3D (22.0, PerkinElmer informatics Inc). The molecular structures of target proteins were obtained from the Protein Data Bank (PDB, http://www.rcsb.org/). Discover Studio 2019 was used for molecular docking, and RStudio was used to sort the interaction strengths by binding free energy.2.4 Molecular dynamics simulation
The complex was analyzed via 100 ns MD simulations using Gromacs 2023. The protein was modeled with CHARMM 36 force field parameters, while the ligand topology was generated using GAFF2 force field parameters. The protein-ligand complex is placed in a cubic box with periodic boundary conditions, and the box is filled with water molecules using the TIP3P water model. Electrostatic interactions are handled using Particle Mesh Ewald and the Verlet algorithm. Subsequently, we performed 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps of isothermal isovolumetric ensemble equilibrium followed by isothermal isobaric ensemble equilibrium, utilizing a coupling constant of 0.1 ps and a simulation duration of 100 ps. Both van der Waals and Coulomb interactions were computed using a cutoff distance of 1.0 nm. Finally, the system underwent molecular dynamics simulation using Gromacs 2023, maintaining a constant temperature of 300 K and a constant pressure of 1 bar, with a total simulation duration of 100 ns.
000 steps of isothermal isovolumetric ensemble equilibrium followed by isothermal isobaric ensemble equilibrium, utilizing a coupling constant of 0.1 ps and a simulation duration of 100 ps. Both van der Waals and Coulomb interactions were computed using a cutoff distance of 1.0 nm. Finally, the system underwent molecular dynamics simulation using Gromacs 2023, maintaining a constant temperature of 300 K and a constant pressure of 1 bar, with a total simulation duration of 100 ns.
2.5 In vitro cell experiments
3. Results
3.1 Identification of the chemical constituents of SFA and its processed products by UPLC-Q-TOF-MS/MS
According to the standard total ion chromatograms (TICs) of the nonvolatile ingredients extracted from SFA, RSFA, and WSFA (Fig. 1), these preparations contain multiple types of compounds. Notably, the positive ESI mode was found to be more sensitive than the negative ESI mode for detecting compounds in the SFA; therefore, the positive ion mode was chosen for component analysis (Table 1). Changes to the composition of SFA during stir-frying with wheat bran were analyzed using Masslynx V4.1 and by comparing the results to chemical in the UNIFI database. Through this analysis, a total of 106 chemical components were determined to be present in SFA both before and after processing.| No. | tR/min | Components | Chemical formula | Ion mode | m/z | Source | ||
|---|---|---|---|---|---|---|---|---|
| SFA | WSFA | RSFA | ||||||
| 1 | 1.78 | Baptifoline | C15H20N2O2 | [M + H]+ | 261.15 | + | + | + |
| 2 | 2.96 | Kaempferol | C15H10O6 | [M + H]+ | 287.05 | + | + | + |
| 3 | 3.88 | Isokuraramine | C12H18N2O2 | [M + H]+ | 223.14 | + | + | + |
| 4 | 4.04 | Cytisine | C11H14N2O | [M + H]+ | 191.11 | + | + | + |
| 5 | 4.19 | Anagyrine | C15H20N2O | [M + H]+ | 245.16 | + | + | + |
| 6 | 4.49 | Oxysophocarpine | C15H22N2O2 | [M + H]+ | 263.17 | + | − | + |
| 7 | 4.55 | Isomatrine | C15H24N2O | [M + H]+ | 249.19 | + | + | − |
| 8 | 4.70 | Umbelliferone | C9H6O3 | [M + H]+ | 163.03 | + | + | + |
| 9 | 4.83 | Isosophocarpine | C15H22N2O | [M + H]+ | 247.18 | + | + | + |
| 10 | 4.88 | 7,11-Dehydromatrine | C15H22N2O | [M + H]+, [M + Na]+ | 247.18 | + | + | + |
| 11 | 4.97 | 2,4-Dihydroxybenzoic acid | C7H6O4 | [M + H]+ | 155.03 | + | + | + |
| 12 | 5.02 | 13,14-Dehydrosophoridine | C15H22N2O | [M + H]+ | 247.18 | + | + | + |
| 13 | 5.08 | N-Methylcytisine | C12H16N2O | [M + H]+ | 205.13 | + | + | + |
| 14 | 5.20 | Lupanine | C15H24N2O | [M + H]+ | 249.19 | + | + | + |
| 15 | 5.37 | Lehmannine | C15H22N2O | [M + H]+, [M + Na]+ | 247.18 | + | − | + |
| 16 | 5.48 | Mamanine | C15H22N2O2 | [M + H]+ | 263.17 | + | + | + |
| 17 | 5.51 | Rhombifoline | C15H20N2O | [M + H]+ | 245.16 | + | + | + |
| 18 | 5.57 | Oxymatrine | C15H24N2O2 | [M + H]+ | 265.19 | + | + | + |
| 19 | 5.69 | Matrine | C15H24N2O | [M + H]+ | 249.19 | + | + | + |
| 20 | 6.02 | Glabrol | C25H28O4 | [M + H]+ | 393.20 | + | + | + |
| 21 | 6.07 | Sophoranol N-oxide | C15H24N2O3 | [M + H]+, [M + Na]+ | 281.18 | + | + | + |
| 22 | 6.15 | Sophoranol | C15H24N2O2 | [M + H]+ | 265.19 | + | + | + |
| 23 | 6.19 | Sophoramine | C15H20N2O | [M + H]+ | 245.16 | − | + | − |
| 24 | 6.29 | Rutin | C27H30O16 | [M + Na]+, [M + H]+ | 633.14 | + | − | − |
| 25 | 6.47 | 12α-Hydroxysophocarpine | C15H22N2O2 | [M + H]+, [M + Na]+ | 263.17 | + | + | + |
| 26 | 6.58 | Isorhamnetin-3-O-β-rutinoside | C28H32O16 | [M + H]+ | 625.17 | + | + | + |
| 27 | 7.05 | Kosamol A | C30H38O8 | [M + H]+ | 527.26 | + | + | + |
| 28 | 7.13 | Biochanin A | C16H12O5 | [M + H]+ | 285.07 | + | + | + |
| 29 | 7.47 | Kushenol S | C20H20O5 | [M + H]+ | 341.13 | + | + | + |
| 30 | 7.58 | Resokaempferol | C15H10O5 | [M + H]+ | 271.06 | + | + | + |
| 31 | 8.20 | Kushenol J | C27H32O14 | [M + H]+ | 581.18 | + | + | + |
| 32 | 8.36 | Genistein | C15H10O5 | [M + H]+ | 271.05 | + | + | + |
| 33 | 8.53 | Kushenin | C16H14O5 | [M + H]+ | 287.09 | + | + | + |
| 34 | 9.28 | Kushenol O | C27H30O13 | [M + H]+, [M + Na]+ | 563.17 | + | + | + |
| 35 | 9.93 | Quercetin | C15H10O7 | [M + H]+ | 303.04 | + | + | − |
| 36 | 9.95 | Kushenol V | C21H22O7 | [M + H]+ | 387.14 | + | − | + |
| 37 | 10.03 | Kushecarpins A | C17H18O6 | [M + H]+ | 319.11 | − | + | − |
| 38 | 10.37 | Formononetin | C16H12O4 | [M + H]+ | 269.08 | + | + | + |
| 39 | 10.47 | Sophoraflacoside | C59H96O27 | [M + H]+, [M + Na]+ | 1237.62 | + | + | + |
| 40 | 10.49 | Soyasaponin I | C48H78O18 | [M + H]+, [M + Na]+ | 943.52 | + | + | + |
| 41 | 10.50 | Azukisaponin I | C42H68O13 | [M + H]+ | 781.47 | + | + | + |
| 42 | 10.82 | Pterocarpin | C17H14O5 | [M + H]+, [M + Na]+ | 299.09 | + | + | + |
| 43 | 11.05 | Kaikasaponin II | C48H78O17 | [M + Na]+, [M + H]+ | 949.51 | − | + | − |
| 44 | 11.46 | Kushenol H | C26H32O8 | [M + H]+, [M + Na]+ | 473.21 | + | + | + |
| 45 | 11.71 | 1,8-Dihydroxy-3-methylanthraquinone | C15H10O4 | [M + H]+ | 255.06 | + | + | + |
| 46 | 11.75 | Trifolirhizin | C22H22O10 | [M + H]+ | 447.12 | + | + | + |
| 47 | 11.81 | Isorhamnetin | C16H12O7 | [M + H]+ | 317.06 | + | + | + |
| 48 | 11.87 | Kushenol K | C26H32O8 | [M + H]+, [M + Na]+ | 473.21 | + | + | + |
| 49 | 12.46 | Glucuronic acid | C6H10O7 | [M + H]+ | 195.0 | + | + | + |
| 50 | 12.96 | Maackiain | C16H12O5 | [M + H]+, [M + Na]+ | 285.07 | + | + | + |
| 51 | 13.04 | Azukisaponin V | C48H77O18 | [M + H]+ | 943.52 | + | + | + |
| 52 | 14.02 | 4,7,2′-Trihydroxy-4′-methoxyisoflavanol | C15H12O6 | [M + H]+ | 289.10 | + | + | + |
| 53 | 14.07 | Flavenochromane C | C21H20O6 | [M + H]+, [M + Na]+ | 369.13 | + | − | − |
| 54 | 14.26 | Trifolirhizin 6′-monoacertate | C24H25O11 | [M + H]+, [M + Na]+ | 489.13 | + | + | + |
| 55 | 14.79 | Kushecarpins C | C17H16O7 | [M + Na]+ | 355.07 | + | + | + |
| 56 | 14.97 | Kuraridinol | C26H32O7 | [M + H]+, [M + Na]+ | 457.22 | + | + | + |
| 57 | 15.02 | Noranhyoicaritin | C20H34O6 | [M + H]+, [M + Na]+ | 371.14 | + | + | + |
| 58 | 15.16 | Kushenol N | C26H30O7 | [M + H]+, [M + Na]+ | 455.20 | + | + | + |
| 59 | 15.71 | Kushenol Q | C26H32O7 | [M + H]+, [M + Na]+ | 457.22 | + | + | + |
| 60 | 16.49 | Ethylparaben | C9H10O3 | [M + H]+ | 167.06 | + | + | + |
| 61 | 17.24 | Xanthohumol | C21H22O5 | [M + H]+, [M + Na]+ | 355.15 | + | + | + |
| 62 | 17.41 | Azukisaponin II | C42H68O14 | [M + H]+ | 797.46 | + | + | + |
| 63 | 17.80 | Neokurarinol | C27H34O7 | [M + H]+ | 471.23 | + | + | + |
| 64 | 17.94 | Flavenochromane B | C25H26O6 | [M + H]+ | 423.17 | + | + | + |
| 65 | 17.98 | Kushenol G | C25H28O8 | [M + H]+ | 457.18 | + | + | + |
| 66 | 17.99 | Kaikasaponin III | C48H78O17 | [M + H]+, [M + Na]+ | 949.51 | + | + | + |
| 67 | 18.00 | Kushenol T | C25H30O6 | [M + H]+ | 427.21 | + | + | + |
| 68 | 18.05 | Kushenol W | C21H22O7 | [M + H]+ | 387.14 | + | + | + |
| 69 | 18.07 | Leachianone G | C20H20O6 | [M + H]+ | 357.13 | + | + | + |
| 70 | 18.09 | 3′-Methoxydaidzein | C16H12O5 | [M + H]+ | 285.07 | + | + | + |
| 71 | 18.37 | Kushenol A | C25H28O5 | [M + H]+ | 409.19 | + | + | + |
| 72 | 18.41 | Kushenol I | C26H30O7 | [M + H]+, [M + Na]+ | 455.20 | + | + | + |
| 73 | 18.74 | 3,4′,5-Trihydroxy-7-methoxy-8-isopentenylflavone | C21H20O6 | [M + H]+ | 369.13 | + | + | + |
| 74 | 19.12 | Isokurarinone | C26H30O6 | [M + H]+, [M + Na]+ | 439.21 | + | + | + |
| 75 | 19.48 | Kushenol P | C26H32O7 | [M + H]+, [M + Na]+ | 457.22 | + | + | + |
| 76 | 19.59 | Kushenol X | C25H28O7 | [M + H]+, [M + Na]+ | 441.19 | + | + | + |
| 77 | 19.93 | Kushenol L | C25H28O7 | [M + H]+ | 441.19 | + | + | + |
| 78 | 20.37 | 8-C-Prenylkaempferol | C20H18O6 | [M + H]+ | 355.11 | + | + | + |
| 79 | 20.60 | Neorkurainol | C27H34O7 | [M + H]+ | 441.19 | − | − | + |
| 80 | 20.74 | Glucose | C6H12O6 | [M + H]+ | 181.06 | + | − | + |
| 81 | 21.03 | Kushenol R | C26H30O5 | [M + H]+, [M + Na]+ | 423.21 | + | + | + |
| 82 | 21.45 | Diphenyl sulfone | C12H10O2S | [M + H]+ | 219.04 | − | + | + |
| 83 | 21.46 | 2-Methoxykurarinone | C27H32O6 | [M + H]+, [M + Na]+ | 453.22 | + | + | + |
| 84 | 21.97 | Kushenol F | C25H28O6 | [M + H]+ | 425.19 | + | + | + |
| 85 | 21.75 | Kushenquinone A | C17H22O4 | [M + H]+ | 291.16 | + | + | + |
| 86 | 22.08 | Lupinifolin | C25H26O5 | [M + H]+ | 407.18 | + | + | + |
| 87 | 22.33 | Kushenol E | C25H28O6 | [M + H]+ | 425.19 | + | + | + |
| 88 | 22.46 | Isoanhyoicaritin | C22H24O6 | [M + H]+ | 385.16 | + | + | + |
| 89 | 22.60 | Kushenol C | C25H26O7 | [M + H]+ | 439.17 | + | + | + |
| 90 | 23.00 | Kushecarpins B | C18H18O7 | [M + Na]+ | 360.09 | + | + | + |
| 91 | 24.22 | Geranyl acetate | C12H20O2 | [M + H]+ | 197.15 | − | + | + |
| 92 | 24.51 | Isoxanthohumol | C21H22O5 | [M + H]+ | 355.15 | − | − | + |
| 93 | 25.02 | Sophoraisoflavone A | C20H16O6 | [M + H]+ | 353.10 | + | + | + |
| 94 | 25.44 | Naringenin | C15H12O5 | [M + H]+ | 273.07 | − | + | + |
| 95 | 26.05 | Linolenic acid | C18H30O2 | [M + H]+ | 279.23 | + | + | − |
| 96 | 26.16 | β-Sitosterol | C29H50O | [M + Na]+, [M + H]+ | 437.37 | + | − | − |
| 97 | 26.29 | Kuraridin | C26H30O6 | [M + H]+ | 439.21 | + | + | + |
| 98 | 26.95 | Kushenol M | C30H36O7 | [M + H]+, [M + Na]+ | 509.25 | + | + | + |
| 99 | 29.50 | Kushenol D | C27H32O6 | [M + H]+, [M + Na]+ | 453.22 | + | + | + |
| 100 | 29.65 | Kushenol B | C30H36O6 | [M + H]+ | 493.25 | + | + | + |
| 101 | 29.80 | Tonkinensisol | C25H24O6 | [M + H]+ | 421.16 | + | − | + |
| 102 | 29.85 | Hexadecanoic acid | C16H32O2 | [M + H]+ | 257.24 | + | + | + |
| 103 | 30.12 | Myristic acid | C14H28O2 | [M + H]+ | 229.21 | + | − | − |
| 104 | 31.53 | Sophoranodichromane B | C25H28O5 | [M + H]+ | 409.19 | − | + | − |
| 105 | 32.07 | Soyasaponin III | C42H67O14 | [M + H]+ | 813.50 | + | − | − |
| 106 | 32.55 | Betulin | C30H50O2 | [M + H]+ | 443.38 | − | − | + |
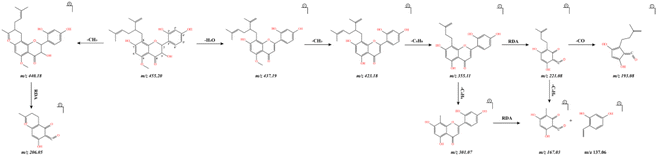
The identification of the alkaloids utilized the MS data, as described here for matrine and lupanine as representative compounds. The molecular ion of matrine was identified at m/z 249.19 [M + H]+. The characteristic fragment ions at m/z 248.18 [M + H − H]+, m/z 218.17 [M + H − H–CH2O]+, m/z 263.13 [M + H − CH2O–NH3]+were also found in the MS/MS spectrum. These ions are derived from the successive loss of H, CH2O and NH3 from the molecular ions. In addition, another ions was observed at m/z 189.15; this ion resulted from the loss of CH2 and rearrangement of the product at m/z 263.13. The fragmentation produced ions at m/z 137.12, m/z 121.10, and m/z 97.08. The major fragmentation ions and the proposed fragmentation pathway of matrine are shown in Fig. 2.
The molecular ion of lupanine was observed at m/z 249.19 [M + H]+. The generation of an ion observed at m/z 248.18 [M + H − 2H]+ was due to the loss of two H+ ions from the molecular ion. Due to their structural characteristics, the C7–17 and C9–11 rings of lupanine are prone to cleavage upon fragmentation due to the McDonnell rearrangement; two ions consistent with this cleavage were observed, at m/z 151.09 and 99.10. The major fragmentation ions and the proposed fragmentation pathway of lupanine are shown in Fig. 3.
We describe here the process of identification of formononetin, kushenol I, and trifolrhizin as representative flavonoids. The major fragment ions and the proposed fragmentation pathway of these compounds are shown in Fig. 4, 5, and 6. The molecular ion of formononetin was found at m/z 269.08 [M + H]+, and formononetin yielded ions at m/z 199.07 [M + H − CH3–CO − CHO]+, m/z 227.07 [M + H − CH3–CO]+, and m/z 255.06 [M + H − CH3]+, which were attributed to the successive losses of CHO and then CO from the parent compound. Fragment ions observed at m/z 237.02 and 119.05 were found to have derived from a retro-Diels–Alder reaction, which is a characteristic reaction of flavonoids. The ions at m/z 109.02 and 82.04 were attributed to the continuous losses of CO and in the ion at m/z 237.02.
For kushenol I, the product ions at m/z 440.18 and m/z 437.19 were formed by the neutral losses of CH3 and H2O, respectively, from the molecular ion [M + H]+ at m/z 455.20. Similarly, the fragment ion at m/z 423.18 was derived from the loss of CH3 from the product ion at m/z 437.19. The product ions at m/z 355.11 were attributed to the loss of C5H8 from the fragment ion at m/z 423.18. The fragment ions at m/z 237.02 and 119.05, 137.06, 167.03, 206.05, and 221.08 were found to derive from the retro-Diels–Alder reaction.
The parent nucleus of trifolrhizin is different from that of ordinary flavonoids in that it cannot undergo the retro-Diels–Alder reaction and due to the presence of a glucosyl group. The molecular ion of trifolrhizin was identified at m/z 447.12 [M + H]+, and a fragment ion was produced at m/z 285.07 [M + H–C6H10O5]+ due to the loss of glucose. The fragment ions m/z 148.05 and 163.04 in the structure of trifolrhizin were considered to be indicative of the dihydrofuran and dihydropyran rings.
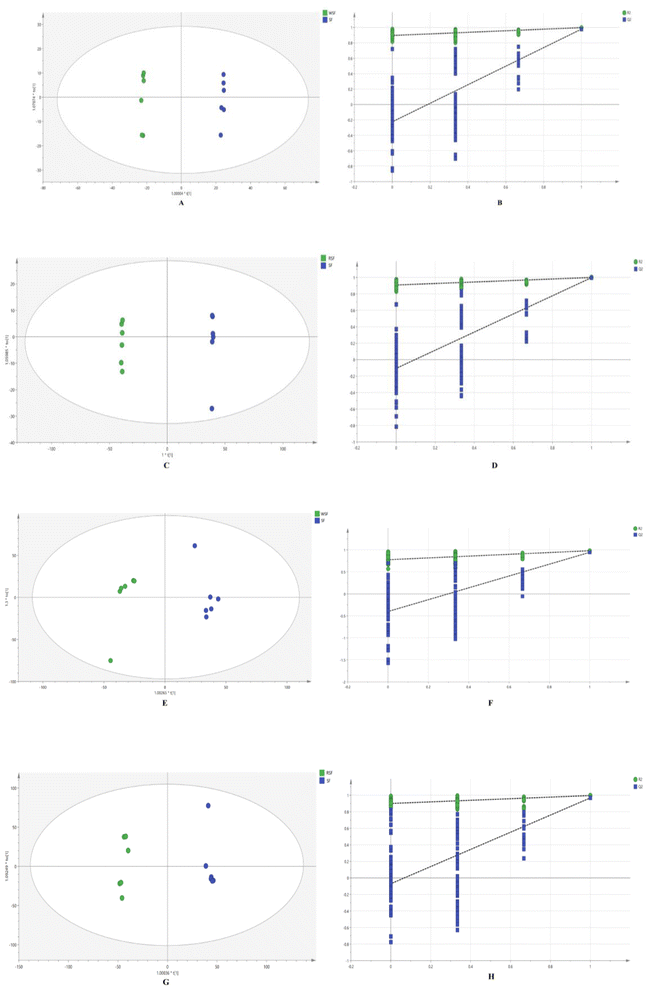
PCA analysis was conducted on SFA, WSFA, and RSFA, with orthogonal partial least squares discriminant analysis (OPLS-DA) models identified and validated through internal and external methods (Fig. 8). In the positive ion mode, the internal validation results for the SFA and WSFA showed an R2Y of 0.998 and a Q2 of 0.979. Similarly, the internal validation results for the SFA and RSFA showed an R2Y of 1.000 and a Q2 of 0.998. The external validation of the model comparing SFA and WSFA gave values of R2 of 0.897 and Q2 of −0.225, while the values of the model comparing Sophora flavescens made from SFA and RSFA were an R2 of 0.909 and a Q2 of −0.106. These results suggest that the evaluation model is reliable and effective with no overfitting. In negative ion mode, the results of internal validation of the model comparing SFA and WSFA were an R2Y of 0.966 and a Q2 of 0.590. The internal validation results for the comparison of SFA and RSFA were R2Y = 0.996 and Q2 = 0.967, indicating the model has good predictive ability. The results of external validation of the comparison of SFA and WSFA was R2 = 0.777 and Q2 = −0.400, and the results of external validation of the comparison of SFA and RSFA was R2 = 0.900 and Q2 = −0.0707. These values indicate that the evaluation model is reliable and effective, and there is no overfitting.
3.2 Network pharmacology analysis
| No. | Compound | Formula | PubChem ID |
|---|---|---|---|
| C1 | Formononetin | C16H12O4 | 5280378 |
| C2 | Isoxanthohumol | C21H22O5 | 513197 |
| C3 | Kurarinone | C26H30O6 | 11982640 |
| C4 | Kushenol I | C26H30O7 | 20832634 |
| C5 | Matrine | C15H24N2O | 91466 |
| C6 | Oxymatrine | C15H24N2O2 | 114850 |
| C7 | Sophocarpine | C15H22N2O | 115269 |
| C8 | Sophoraflavanone G | C25H28O6 | 72936 |
| C9 | Sophoridine | C15H24N2O | 165549 |
| C10 | Trifolrhizin | C22H22O10 | 442827 |
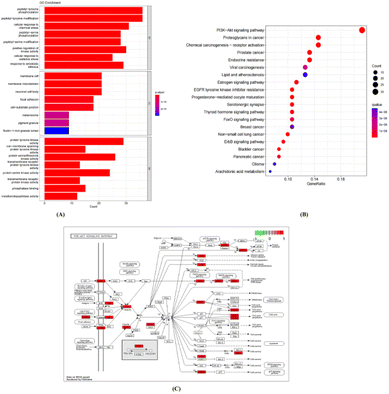
A KEGG pathway analysis was also conducted to explore the significance of the putative targets. Among the top 20 signaling pathways as identified using R 4.1.0 (Fig. 11B) were the phosphoinositide 3-kinase (PI3K)-AKT signaling pathway, signaling pathways involving proteoglycans in cancer, and signaling pathways of importance to prostate cancer. A schematic diagram of the PI3K-AKT signaling pathway is shown in Fig. 11C.
3.3 Molecular docking of active components with potential targets
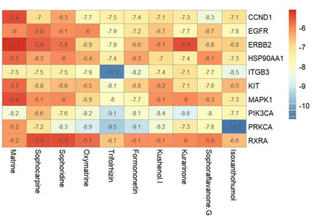
In silico molecular docking analyses were performed in order to further investigate the interactions between SFA compounds and the potential targets of SFA in the treatment of UC. In receptor-ligand docking analyses, more highly negative changes in free energy indicate stronger binding. As shown in Fig. 12, in the present study, the protein-compound pairs that exhibited the strongest binding were protein kinase C alpha (PRKCA) with isoxanthohumol, integrin beta 3 with trifolrhizin, PRKCA with trifolrhizin, the p110α subunit of PI3K with trifolrhizin, and PRKCα with formononetin. The binding energies were less than −9 kcal mol−1. The structures that resulted from the docking of each of the five receptor-ligand pairs were visualized using Discovery studio (Fig. 13).3.4 Molecular dynamics simulations
Molecular dynamics simulation was utilized to ascertain the binding capabilities of small molecule compounds with key target proteins exhibiting optimal binding in molecular docking. Isoxanthohumol and PRKCA were selected for molecular dynamics simulations, given they demonstrated the strongest binding force in molecular docking procedures. The stability of the simulation system was assessed using root mean square deviation (RMSD). As shown in Fig. 14a, the complex system reaches equilibrium after 40 ns, and finally fluctuates around 3.3 Å. This indicates that the small molecule demonstrates high stability when bound to the target protein. Further analysis revealed that the composite system exhibited an initial upward trend from 60 ns to 95 ns, followed by a decline that gradually stabilized. This indicates that the composite system initially expanded, then contracted during the simulation process, ultimately achieving a stable state (Fig. 14b). Solvent Accessible Surface Area (SASA) serves as an important metric for assessing the surface area of a protein. In this simulation, the SASA was calculated for the interaction between the target protein and the small molecule (Fig. 14c). The results indicated that the complex system exhibited minor fluctuations during movement, eventually stabilizing over time. This suggests that the binding of small molecules influences the binding microenvironment, resulting in notable changes in SASA. The number of hydrogen bonds formed between the small molecule and the target protein during the dynamic process ranges from 0 to 4. In most instances, the complex system exhibits approximately 2 hydrogen bonds, suggesting that it has favorable hydrogen bond interactions (Fig. 14d). The Root mean square fuctuation (RMSF) value of the (Fig. 14e). | ||
| Fig. 14 Molecular dynamics simulation between isoxanthohumol and PRKCA. (a) RMSD. (b) Rg. (c) SASA. (d) HBond Numners. (e) RMSF. | ||
3.5 In vitro cell experiments
4. Discussion
UC is a non-specific, chronic, recurrent, and inflammatory disease of the intestinal tract. Its pathogenesis involves the destruction of the intestinal mucosal barrier, resulting in abnormalities in the stability and gut microbiota of the colon; these changes lead to activation of macrophages and T cells, further altering the levels of immune-related cytokines and inducing a series of inflammatory reactions.12,13Currently, the most commonly used drugs for the treatment of UC derive from Western medicine, including aminosalicylates, corticosteroids, and immunosuppressants. However, these drugs are often accompanied by adverse reactions, making the discovery of safe and effective drugs to prevent and treat UC an important research goal. Notably, TCM may have advantages in treating UC, though the complexity of the chemical components in such medicinal preparations means that the material basis for TCM action and its molecular mechanisms often remain unclear. In this study, UPLC-Q-TOF-MS/MS was employed to identify the chemical components of SFA and its processed products, and its key targets and pharmacodynamic mechanisms in the treatment of UC were analyzed using network pharmacology and molecular docking techniques, providing a reference for elucidating the mechanisms underlying the efficacy of SFA.
In previous studies, SFA was found to be able to treat UC, and the processed products showed more significant therapeutic effects. Still, the pharmacological material basis has yet to be thoroughly explored, and the specific mechanisms of action in its prevention and treatment of UC have yet to be elucidated. Here, UPLC-Q-TOF-MS/MS combined with network pharmacology allowed us to identify 159 potential targets of SFA in the treatment of UC, including epidermal growth factor receptor (EGFR), the protein-tyrosine kinase SRC, PIK3CA, and heat shock protein 90 alpha (HSP90ɑ).
Pathways related to these targets have been associated with UC development. For example, the EGFR signaling pathway alters the rates of apoptosis and proliferation, and its activity is involved in the occurrence of colon mucosal injury. Damage to the intestinal mucosa in ulcerative colitis has been shown to affect the expression of EGFR, thereby reducing its protective and reparative effects on the colon mucosa. Accordingly, EGFR levels have been found to correlate closely with UC recurrence. Therefore, the EGFR signaling pathway is a potential target in the treatment of UC.14,15 Similarly, HSP90ɑ is closely linked with inflammation, and inhibiting the expression of HSP90 can alleviate the inflammatory response. HSP90ɑ is known to regulate the activation of inflammasomes and the secretion of interleukin (IL) lβ via its interactions with NLR family pyrin domain containing 3 (NLRP3).16
Inflammation serves as a defensive response of the organism to injury, infection, or other harmful stimuli, characterized by a dynamic equilibrium between pro-inflammatory and anti-inflammatory factors. Pro-inflammatory factors initiate and amplify the inflammatory response, while anti-inflammatory factors suppress inflammation and facilitate tissue repair. An imbalance between these two types of factors can lead to excessive or persistent inflammatory responses, potentially resulting in disease. PRKCA, a serine/threonine-specific protein kinase, plays a pivotal role in cellular signaling and regulates various biological processes, including cell proliferation, differentiation, and survival. The role of PRKCA in inflammation has been extensively studied. Research indicates that PRKCA is also crucial in modulating the production of reactive oxygen species (ROS), which are key mediators in numerous inflammatory responses. By inhibiting ROS production, PRKCA can alleviate cellular oxidative stress, thereby mitigating the inflammatory response.17 Furthermore, the activation of PRKCA has been shown to suppress LPS-induced inflammatory responses, reducing the release of pro-inflammatory cytokines.18 This reduction in pro-inflammatory cytokine release can, to some extent, upregulate anti-inflammatory factors, thereby inhibiting the onset of inflammation.
The active ingredients of SFA for the treatment of UC were identified through a joint analysis of components and targets. These ingredients mainly include alkaloids and flavonoids, including matrine, oxymatrine, formononetin, and kushenol I. Among the alkaloids, oxymatrine has been determined to be one of the main active ingredients of SFA, as it exhibits a wide range of pharmacological effects. It exerts therapeutic effects on organ fibrosis, protective effects on myocardial ischemia and infarction, and anti-inflammatory, antiviral, bactericidal, and anti-tumor effects.19 Sophocarpine also has been demonstrated to have strong anti-inflammatory and analgesic effects, which may be related to calcium and inhibition of the secretion of inflammatory factors.20
The flavonoids in SFA have also been demonstrated to exert anti-inflammatory effects through interactions with modulation of NF-κB and other related pathways, which lead to inhibition of the expression of inflammatory factors.21 Sophoraflavanone G can inhibit the neuroinflammation of lipopolysaccharide-activated microglia by regulating mitogen-activated protein kinases (MAPKs) and a signaling pathway involving nuclear factor erythroid 2-related factor and heme oxygenase-1.22 Kushenol F can inhibit the production of IL-1 by inhibiting the activation of PI3K, AKT, and the p38 MAPK proteins β, IL-6, and tumor necrosis factor α. The inhibition of release of pro-inflammatory factors promotes, the secretion of anti-inflammatory factor IL-10, reduces inflammation damage to colon tissue, and thus improves related symptoms in UC mice.23
Based on the previous work suggesting the effectiveness of SFA components in inflammatory diseases, we conducted in silico analyses of molecular docking between the active ingredients of SFA and core targets in UC. The results showed that although each of the components was predicted to interact with each target, the binding affinities were different due to their different structures. The wide breadth of binding activities means that the active ingredients of SFA may synergistically act on multiple protein targets and signaling pathways to exert its anti-inflammatory effects. However, after processing, the degree of the therapeutic effect may change due to changes in chemical composition.
5. Conclusions
We determined that the active compounds of SFA act on multiple core targets, including EGFR, SRC, PIK3CA, and HSP90AA1, and they regulate multiple signaling pathways, including PI3K/AKT, lipid and atherosclerosis, and proteoglycans in cancer. Thus, by combining the characterization of chemical composition with network pharmacology, the mechanisms of action of SFA in the treatment of UC were determined to involve multiple targets, multiple components, and multiple paths. This information also provides a reference for clinical medication. However, the reasons for the increased efficacy after processing still need to be clarified.Abbreviations
| TCM | Traditional Chinese medicine |
| RSFA | Sophora flavescens Ait processed with rice-washed water |
| WSFA | Sophora flavescens Ait processed with wheat bran |
| SFA | Sophora flavescens Ait |
| UPLC-Q-TOF-MS/MS | Ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry |
| UC | Ulcerative colitis |
| ESI | Electrospray ionization |
| BP | Biological process |
| MF | Molecular function |
| CC | Cellular component |
| TIC | Total ion chromatogram |
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This research was supported by the Provincial Traditional Chinese Medicine Processing Technology Inheritance Base Project of the Liaoning Provincial Administration of Traditional Chinese Medicine (CCJDXM2020).References
- D. G. Li, Q. Yang, Y. R. Cai, B. L. Li, D. Y. Zhao, P. L. Du, Y. X. Guo, T. Guo, B. T. Guo and X. M. Jia, Professor Li Diangui's Experience in the Treatment of Ulcerative Colitis with a Combination of Traditional Chinese and Western Medicine, Chin. J. Integr. Tradit. West. Med. Dig., 2019, 27(04), 244–246 Search PubMed.
- Y. Q. An and H. J. Wang, Research progress on the distribution of Traditional Chinese Medicine syndrome and its correlation with enteroscopy in ulcerative colitis, Chin. J. Integr. Tradit. West. Med. Dig., 2021, 29(05), 373–376 Search PubMed.
- T. H. Zhang and H. Shen, Ulcerative colitis and its TCM pattern-differentiation classification and inflammatory activity indexes: a correlation analysis, J. Beijing Univ. Tradit. Chin. Med., 2019, 42(08), 685–690 Search PubMed.
- N. Li and L. Y. Zhong, Processing Of Chinese Materia Medica, China Medical Science Press, 2020, pp. 57–58 Search PubMed.
- G. R. Ding, Summary Of Medicinal Properties, 1917, pp. 19–20 Search PubMed.
- Chinese Pharmacopoeia Commission, Pharmacopoeia Of The People's Republic Of China, China Medical Science and Technology Press, Beijing, 2020, vol. I, pp. 211 Search PubMed.
- Y. Liu, L. Q. Shan and H. Gao, Comparison of the effects of water extract of Sophora flavescens Ait and its different processed products on dextran sulfate induced ulcerative colitis in mice, Chin. Tradit. Pat. Med., 2021, 43(05), 1323–1328 Search PubMed.
- X. L. Pei, T. N. Jiang, Y. Liu and H. Gao, Regulatory effects of Sophora flavescens and its processed products on TLR4/MyD88/NF-κB signaling pathway in mice with damp-heat Ulcerative Colitis, Chin. Arch. Tradit. Chin. Med., 2023, 1–11 CAS.
- L. Mouad, H. El-Idrissi, S. Taoufiq, I. Bouchaîb, N. Saffaj, R. Mamouni and Y. Kandri Rodi, QSAR modeling, molecular docking and molecular dynamic simulation of phosphorus-substituted quinoline derivatives as topoisomerase I inhibitors, Arabian J. Chem., 2023, 16(6), 104783–104783 CrossRef.
- M. Lahyaoui, A. Diane, H. El-Idrissi, T. Saffaj, Y. K. Rodi and B. Ihssane, QSAR modeling and molecular docking studies of 2-oxo-1, 2-dihydroquinoline-4- carboxylic acid derivatives as p-glycoprotein inhibitors for combating cancer multidrug resistance, Heliyon, 2023, 9(1), e13020, DOI:10.1016/j.heliyon.2023.e13020.
- Y. Q. Zhang and Q. F. Gong, Research on the ancient and modern application of rice swill in the processing of traditional Chinese medicine, J. Tradit. Chin. Med. Sci., 2011, 42(04), 64–66 Search PubMed.
- B. Chassaing, J. D. Aitken, M. Malleshappa and M. Vijay-Kumar, Dextran sulfate sodium (DSS)-induced colitis in mice, Curr. Protoc. Immunol., 2014, 104, 15, DOI:10.1002/0471142735.im1525s104.
- R. Toumi, I. Soufli, H. Rafa, M. Belkhelfa, A. Biad and C. Touil-Boukoffa, Probiotic bacteria lactobacillus and bifidobacterium attenuate inflammation in dextran sulfate sodium-induced experimental colitis in mice, Int. J. Immunopathol. Pharmacol., 2014, 27(4), 615–627, DOI:10.1177/039463201402700418.
- P. Y. Chen, C. Yuan, Z. C. Hong, Y. Zhang, X. G. Ke, B. Yu, C. Wang, X. C. Xiao, H. Z. Wu and Y. F. Yang, Revealing the mechanism of “Huai Hua San” in the treatment of ulcerative colitis based on network pharmacology and experimental study, J. Ethnopharmacol., 2021, 281, 114321, DOI:10.1016/j.jep.2021.114321.
- T. He, W. Wang, L. Li, S. L. Qiu, S. M. Sun and L. Zong, PCNA,EGFR,MUC2 expression levels and clinical significance in ulcerative colitis, J. Med. Res., 2022, 51(03), 104–107 Search PubMed.
- M. Zhang, L. Liu, X. Lin, Y. Wang, Y. Li, Q. Guo, S. Li, Y. Sun, X. Tao, D. Zhang, X. Lv, L. Zheng and L. Ge, A Translocation Pathway for Vesicle-Mediated Unconventional Protein Secretion, Cell, 2020, 181(3), 637–652.e15, DOI:10.1016/j.cell.2020.03.031.
- Q. J. Zhu, J. Wang, Y. Li, Z. J. Bai, X. B. Guo and T. Pan, PRKCA Promotes Mitophagy through the miR-15a-5p/PDK4 Axis to Relieve Sepsis-Induced Acute Lung Injury, Infect. Immun., 2023, 91(1), e0046522, DOI:10.1128/iai.00465-22.
- M. Wang, H. Zhong, X. Zhang, X. Huang, J. Wang, Z. Li, M. Chen and Z. Xiao, EGCG promotes PRKCA expression to alleviate LPS-induced acute lung injury and inflammatory response, Sci. Rep., 2021, 11(1), 11014, DOI:10.1038/s41598-021-90398-x.
- X. Lan, J. Zhao, Y. Zhang, Y. Chen, Y. Liu and F. Xu, Oxymatrine exerts organ- and tissue-protective effects by regulating inflammation, oxidative stress, apoptosis, and fibrosis: From bench to bedside, Pharmacol. Res., 2020, 151, 104541, DOI:10.1016/j.phrs.2019.104541.
- F. L. Wang, H. Wang, J. H. Wang, D. X. Wang, Y. Gao, B. Yang, H. J. Yang, Y. B. Ji and G. S. Xin, Analgesic and Anti-Inflammatory Activities of Sophocarpine from Sophora viciifolia Hance, BioMed Res. Int., 2021, 2021, 8893563, DOI:10.1155/2021/8893563.
- X. D. He, Z. Y. Fu, Y. Huang, J. B. He, X. M. Pu, J. Li, Y. J. Li, J. Yu and X. X. Yang, Research progress on the chemistry and bioactivity of isopentenyl flavonoids from Sophora flavescens, Chin. J. Clin. Pharmacol. Ther., 2022, 27(08), 899–907 Search PubMed.
- C. Guo, L. Yang, C. X. Wan, Y. Z. Xia, C. Zhang, M. H. Chen, Z. D. Wang, Z. R. Li, X. M. Li, Y. D. Geng and L. Y. Kong, Anti-neuroinflammatory effect of Sophoraflavanone G from Sophora alopecuroides in LPS-activated BV2 microglia by MAPK, JAK/STAT and Nrf2/HO-1 signaling pathways, Phytomedicine, 2016, 23(13), 1629–1637, DOI:10.1016/j.phymed.2016.10.007.
- X. D. He, H. Y. Ni, J. B. He, M. Li, Y. K. Hu, D. H. Gong, J. L. Yao, J. Yu and X. X. Yang, Intervention effect of kushenol F on ulcerative colitis mice, China Pharm., 2024, 35(04), 419–424 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |