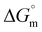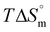Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bio-physical insights into the interaction of biocompatible iron oxide nanoparticles with biomolecules: microcalorimetric and spectroscopic evaluation
Aditi
Pandey
,
Vishakha
Choudhary
,
Bhawna
Sharma
 and
Achal
Mukhija
and
Achal
Mukhija
 *
*
Department of Chemistry, Banasthali Vidyapith, Tonk, Rajasthan 304022, India. E-mail: mukhija18@gmail.com
First published on 8th October 2025
Abstract
Drug delivery systems (DDSs), despite extensive research, have yet to achieve optimal therapeutic outcomes. In this study, we synthesized amino acid-coated iron oxide nanoparticles (IONPs) to investigate a nano-bio interface through thermodynamic analysis and assess how surface coating influences DDS efficiency. The synthesized systems were characterized using FTIR, XRD, BET, SEM, and DLS techniques. Isothermal titration calorimetry in combination with spectroscopy was employed for interaction studies and to obtain data on the binding and thermodynamics of interaction. Thermodynamic parameters and therapeutic efficiency were correlated with the functional groups of the coating material of the IONPs. Experimental findings imply that coating IONPs with amino acids improves their interactions with DNA and drug-loading efficiency. Comparison of the efficiency of different amino acid-coated IONPs based on the functional group of the coating material reveals the importance of the –OH group over other functional groups. Additionally, results demonstrated how the efficiency of the DDSs changes in the homologous series of amino acids and highlight how the size and length of the side substituent as well as the type of amino acid impact their interaction with DNA and drug loading efficiency with 5-fluorouracil. Correlating the energetics of the interactions with the structure and physical characteristics of amino acids enabled quantitative structure–activity relationship (QSAR) studies and will facilitate the AI-based design of efficient DDSs.
1. Introduction
Drug delivery systems (DDSs) have gained significant interest in the diagnosis and treatment of cancer, yet despite numerous formulations explored, requisite results have still not been attained.1–5 For the application of nanoformulation techniques for diagnosis and therapeutics, crucial parameters such as coating material, formulation size, and interactions with nucleic acids need to be precisely correlated with the efficiency of nanoformulations. These correlations provide an understanding of the nano-bio interface, through which quantitative structure–activity relationships (QSARs) can be obtained to guide AI-driven design of effective DDSs.6–8Iron oxide nanoparticles (IONPs), such as ferumoxytol (Feraheme™), ferucarbotran, and Feridex®, are clinically approved DDSs, due to their biocompatibility and non-toxic nature, which has led to a breakthrough in cancer treatment.9–13 Thus, in the present study, six different amino acid-coated IONPs, namely, glycine-, alanine-, aspartic acid-, tryptophan-, serine-, and threonine-coated IONPs, along with bare IONPs, are synthesized and used as DDSs. Coating IONPs with amino acids offers several benefits by making the system more biocompatible, boosting colloidal stability, and preventing agglomeration. Literature reports suggest that amino acids are promising candidates for surface coating of IONPs and can be used in different biomedical applications. A report by Barick et al. suggested that the carboxylate group of glycine chemisorbs onto IONPs, leaving the amine group free for further functionalization; glycine-coated IONPs thus have strong potential to be used for drug delivery and magnetic hyperthermia applications.14 A similar report by Salehiabar et al. also demonstrated that aspartic acid-coated IONPs exhibit promising characteristics for drug delivery applications.15 Additionally, amino acid-coated IONPs further conjugated with anti-cancer drugs have been shown to exhibit remarkable anti-cancer effects. A report by Nosrati et al. showed that lysine-coated and methotrexate-conjugated IONPs are excellent candidates for drug delivery applications.16
Thus, in this study, amino acids from different structural categories have been chosen as coating materials to assess the effect of functional groups on the therapeutic efficacy of DDSs. Glycine and alanine are small and non-polar amino acids with –H and –CH3 α-substituents, respectively (Fig. S1(a) and (b)). Aspartic acid, serine, and threonine are polar amino acids with –CH2–COOH, –CH2–OH, and –CH(OH)–CH3 α-substituted R groups, respectively (Fig. S1(c), (e) and (f)). Tryptophan was chosen from the neutral amino acid category, with an indole side chain as the β-substituent (Fig. S1(d)). Correlation of these structural properties of different amino acids with the energetics of their interactions with biomolecules is a topic of interest to derive QSAR.
To understand and gain quantitative insights into the nano-bio interface, it is imperative to study how synthesized DDSs interact with biomolecules when administered intravenously.4,5,17–20 Accordingly, to understand the thermodynamics of the interaction of the synthesized DDSs with the biomolecule DNA, isothermal titration calorimetry (ITC) and fluorescence spectroscopy techniques are used. ITC offers a very precise measurement of the heat change in the system and is used to address NP-biomolecule interactions. Analysis of the heat obtained can provide necessary insights into the mechanism and mode of interaction. Quantitative aspects of interactions in terms of a binding constant (Kb), stoichiometry (n), and complete thermodynamics of interaction, i.e., enthalpy change ( ), free energy change (
), free energy change (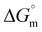 ), and entropy change (
), and entropy change (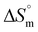 ) are obtained from ITC studies.21 Fluorescence spectroscopy is utilized as an additional method to study NP-biomolecule interactions. Using the modified Stern–Volmer equation, the Kb value for interactions between the synthesized systems and DNA is investigated.22 Furthermore, the essential prerequisites and fundamental standards for an effective DDS are summarized as: retain, evade, target, and release. Therefore, drug loading experiments with anticancer drugs, i.e., 5-fluorouracil (5-FU) (Fig. S1(g)), are carried out to investigate the therapeutic effectiveness of the synthesized nanoformulations. 5-FU hampers nucleoside metabolism of DNA/RNA, leading to cancer cell death.23,24
) are obtained from ITC studies.21 Fluorescence spectroscopy is utilized as an additional method to study NP-biomolecule interactions. Using the modified Stern–Volmer equation, the Kb value for interactions between the synthesized systems and DNA is investigated.22 Furthermore, the essential prerequisites and fundamental standards for an effective DDS are summarized as: retain, evade, target, and release. Therefore, drug loading experiments with anticancer drugs, i.e., 5-fluorouracil (5-FU) (Fig. S1(g)), are carried out to investigate the therapeutic effectiveness of the synthesized nanoformulations. 5-FU hampers nucleoside metabolism of DNA/RNA, leading to cancer cell death.23,24
Thus, in the present study, bare and structurally different amino acids, i.e., glycine, alanine, serine, threonine, aspartic acid, and tryptophan-coated IONPs, have been synthesized and systematically characterized with different spectroscopic and microscopic techniques. Their interaction with DNA was studied using a combination of ITC and fluorescence spectroscopic techniques, and their structural characteristics were correlated with the interaction thermodynamics for the development of QSAR. The study focused on understanding how the functional groups present in the amino acids influence interactions with DNA. Additionally, in relation to their biomedical applications, the therapeutic efficacy of the synthesized nanoformulations was investigated and correlated with the amino acid structure for future AI-based designs of efficient DDSs.
2. Materials and methods
Ferric (Fe3+) chloride (97%), ferrous (Fe2+) chloride tetrahydrate (≥99%) from Sigma-Aldrich Chemicals Pvt. Ltd, were used as reaction precursors to synthesize bare IONPs, and aqueous ammonia solution (25%) from Molychem Chemicals Pvt. Ltd was used as a precipitating agent. Glycine (≥99%, mol. wt 75.07 g mol−1), L-alanine (≥98%, mol. wt 89.09 g mol−1), L-serine (≥99%, mol. wt 105.09 g mol−1), L-threonine (≥98%, mol. wt 119.12 g mol−1), L-aspartic acid (≥98%, mol. wt 133.103 g mol−1), and L-tryptophan (≥98%, mol. wt 204.23 g mol−1) were used as coating materials, sourced from Sigma-Aldrich Chemicals Pvt. Ltd, to synthesize different amino acid-coated IONPs. For interaction and drug loading studies on the synthesized nanosystems, all solutions were prepared in phosphate-buffered saline (PBS ACS reagent ≥99.0%), at pH 7.4. Calf thymus DNA (ct-DNA), intercalating dye ethidium bromide (EtBr, Bioreagent), and anti-cancer drug 5-fluorouracil (≥99%), were also obtained from Sigma-Aldrich Chemicals Pvt. Ltd. All reagents and chemicals utilized in the experimental analysis were of analytical grade purity and used as received without purification.2.1. Synthesis of bare IONPs and different amino acid-coated IONPs
Water-dispersible, biomolecule-coated IONPs were prepared by a one-pot synthesis method.14 Attempts were made to synthesize different amino acid-coated IONPs with a similar method to make it feasible to assess the effect of coating material on the efficacy of IONPs, as synthesis methods affect the physical properties of nanoformulations significantly. In brief, Fe(III) and Fe(II) salts were dissolved in 100 mL of water in a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and mechanically stirred in a nitrogen atmosphere to prevent further oxidation. After maintaining the temperature at 70 °C for 30 min, 30 mL of 25% aqueous ammonia was gradually introduced to the reaction mixture, and the reaction was allowed to proceed for 30 min under similar conditions. After the reaction, the formed black precipitates of magnetic bare IONPs were magnetically separated from the supernatant, washed, and dried in an oven.
1 and mechanically stirred in a nitrogen atmosphere to prevent further oxidation. After maintaining the temperature at 70 °C for 30 min, 30 mL of 25% aqueous ammonia was gradually introduced to the reaction mixture, and the reaction was allowed to proceed for 30 min under similar conditions. After the reaction, the formed black precipitates of magnetic bare IONPs were magnetically separated from the supernatant, washed, and dried in an oven.
For surface functionalization of the IONPs with different amino acids, a similar method was used to evaluate the effect of the coating material. The ratio (w/w) of Fe3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe2+ was taken as 2
Fe2+ was taken as 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and ammonia solution was used to precipitate the iron salts. The obtained black precipitate indicated successful synthesis of the IONPs. The selected amino acids were then introduced to the reaction with precise Fe3+
1, and ammonia solution was used to precipitate the iron salts. The obtained black precipitate indicated successful synthesis of the IONPs. The selected amino acids were then introduced to the reaction with precise Fe3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe2+: amino acid (w/w) as 2
Fe2+: amino acid (w/w) as 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n, where n was varied as 0.5, 0.25, 0.5, 0.25, 0.125, and 0.25 for glycine, alanine, serine, threonine, aspartic acid, and tryptophan, respectively. Thereafter, all the finally prepared amino acid-coated systems were separated magnetically, washed with double-distilled water several times to remove any uncoordinated amino acid residues, and dried in an oven.
n, where n was varied as 0.5, 0.25, 0.5, 0.25, 0.125, and 0.25 for glycine, alanine, serine, threonine, aspartic acid, and tryptophan, respectively. Thereafter, all the finally prepared amino acid-coated systems were separated magnetically, washed with double-distilled water several times to remove any uncoordinated amino acid residues, and dried in an oven.
2.2. Characterization of bare IONPs and different amino-acid-coated IONPs
To confirm the successful synthesis of the nanoformulations, the following characterization techniques were employed.The Zetasizer instrument uses the DLS technique to determine hydrodynamic diameter (size) and zeta potential values of dispersed nanosystems. A Zetasizer from Malvern (B.2590 nano series), equipped with a 4 mW He–Ne laser (λ = 633 nm), was used to perform DLS measurements with a dispersant (water) refractive index of 1.330, ensuring accurate data acquisition. These parameters provide insights into the particle size distribution and colloidal stability of the synthesized IONPs. For analysis, all the sample solutions were prepared in a buffer at pH 7.4. Studies were performed at 25 °C with a quartz cuvette with a 3 mL sample volume. The hydrodynamic diameters of bare IONPs and different amino acid-coated IONPs were observed to study changes in the hydrodynamic diameter of the IONPs upon coating. Furthermore, zeta potential measurement studies were conducted to assess the effect of coating material on the colloidal stability of the synthesized systems. Comparative studies between bare IONPs and amino acid-coated IONPs highlight the impact of the coating materials on the overall stability of the nanoparticles.
In addition to this, studies related to the specific surface area of the synthesized formulations provide information about their effectiveness as DDSs. BET analysis enables specific surface area measurements of materials by employing a completely automated analyzer to evaluate nitrogen multilayer adsorption as a function of relative pressure. A BET instrument (Quanta Chrome version 5.0 – Autosorb iQ) was used to study the specific surface area of the synthesized systems and to examine how the surface coating affects their specific surface area. Prior to measurement, samples were degassed under vacuum for at least 8 hours at 60 °C, to remove any adsorbed moisture or contaminants, ensuring the accuracy of the results. The specific surface area was determined by N2 adsorption–desorption isotherm analysis under cryogenic conditions utilizing the multi-point BET method as a function of relative pressure (p/p0). A graph was plotted showing the amount of nitrogen gas adsorbed by the particles (cm3 g−1 at STP, on the y-axis) against relative pressure of the gas (P/P0, on the x-axis), with an R2 value of 0.99. The study examined the effects of surface coatings on specific surface area, specifically the presence of amino acids in the synthesized systems. This data is essential for evaluating the potential effects of surface modifications on the functional characteristics of IONPs, such as drug loading efficiency and interaction with biomolecules.
Furthermore, to understand the morphology and topographical patterns, i.e., information about the surface features of the bare IONPs and the amino acid-coated IONPs, SEM analysis was performed. For analysis, a field emission scanning electron microscope (MIRA3 TESCAN) was employed. For sample preparation, a small amount of sample was simply dropped on a copper grid that had been coated with carbon to ensure good conductivity and reduce charging effects during imaging. Excess sample was wiped away to create a thin film of the sample, and images were then recorded.
2.3. Interaction studies of the synthesized nanoformulations with biomolecules
Knowledge of nano-bio interfaces is linked to the intelligent design of safe and effective DDSs. Therefore, it is important to study how nanoformulations interact with biomolecules. Calorimetric and spectroscopic investigations have been performed to gain a comprehensive understanding of the interactions taking place at the nano-bio interface.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 bp cm−1 M−1).25 Purity of DNA was ascertained by studying the absorbance ratio at 260 and 280 nm. An absorbance ratio (i.e., A260/A280) of 1.85 signifies the essence of protein-free DNA.22
200 bp cm−1 M−1).25 Purity of DNA was ascertained by studying the absorbance ratio at 260 and 280 nm. An absorbance ratio (i.e., A260/A280) of 1.85 signifies the essence of protein-free DNA.22
To determine the thermodynamic parameters of the interactions, a DNA solution was prepared in PBS of 20 mM concentration, and the same buffer solution was filled in a reference cell. The sample cell was loaded with DNA at set concentrations of 100 μM, and a syringe was filled with variously produced IONP solutions. Different IONP solutions were also prepared in the same buffer solution and injected into the sample cell containing 200 μL of biomolecules using a computer-operated 40 μL syringe. The syringe concentration was optimized for each experiment to obtain saturated heat on completion of each titration. Each experimental analysis involved nineteen sequential injections, starting with an initial 0.4 μL injection, followed by eighteen, 2 μL injections with an interval of 150 s between successive injections. All titrations were carried out at a controlled stirring speed of 750 rpm throughout the experiments, to ensure proper mixing of the solutions.
Calorimetric data analysis was performed by using the ITC analysis software provided with the instrument. By integration of the area under the peaks of the power-versus-time curve over the entire course of the experiment, the enthalpy change associated with each injection was determined. Dilution experiments were carried out for each experimental analysis, and dilution-corrected heats of interaction were plotted against molar ratio and fitted to a single-site binding model to obtain thermodynamic interaction parameters. To test the reproducibility of results, all experiments were performed in duplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. For this, DNA was used at a concentration of 50 μg mL−1, while EtBr was maintained at 10 μg mL−1. The scanning rate was fixed at 900 nm min−1, and both the emission and excitation slit widths were precisely set at 10 nm. Excitation was performed at 470 nm, and the emission spectra were recorded over the range of 500 to 700 nm. Bare IONPs and different amino acid-coated IONPs were incrementally added to the DNA-EtBr solution, and the IONP concentration was systematically varied to achieve significant quenching. The fluorescence quenching spectra of DNA both in the absence and in the presence of quencher molecules were recorded, and the binding constant of interactions of the synthesized bare IONPs and different amino acid-coated IONPs with DNA was calculated by using a modified Stern–Volmer equation.26,27
1. For this, DNA was used at a concentration of 50 μg mL−1, while EtBr was maintained at 10 μg mL−1. The scanning rate was fixed at 900 nm min−1, and both the emission and excitation slit widths were precisely set at 10 nm. Excitation was performed at 470 nm, and the emission spectra were recorded over the range of 500 to 700 nm. Bare IONPs and different amino acid-coated IONPs were incrementally added to the DNA-EtBr solution, and the IONP concentration was systematically varied to achieve significant quenching. The fluorescence quenching spectra of DNA both in the absence and in the presence of quencher molecules were recorded, and the binding constant of interactions of the synthesized bare IONPs and different amino acid-coated IONPs with DNA was calculated by using a modified Stern–Volmer equation.26,27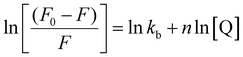 | (1) |
2.4. Therapeutic efficiency determination of the synthesized DDSs
DDSs offer the potential to improve therapeutic efficacy, decrease toxicity, increase patient compliance, and facilitate the development of novel medical therapies. Drug loading studies in this regard can explain the efficiency of synthesized nanoformulations as DDSs. Therefore, to ascertain the efficiency of the synthesized IONPs, drug-loading studies with the anti-cancer drug 5-FU were conducted. For this, a Shimadzu UV-VIS spectrophotometer was employed, and the absorbance of both 5-FU and 5-FU-loaded IONP samples was recorded. These experimental analyses were based on the determinations of the absorbance of 5-FU-loaded nanoparticles compared with free 5-FU.25 For this, different aqueous solutions of IONPs and 5-FU were prepared in a 20 mM phosphate buffer solution in four different ratios, i.e., 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.75, 1
0.75, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, and 1
0.5, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25. The absorbance maximum for 5-FU was recorded at 266 nm. To assess the drug-loading efficiency over time, the absorbance of the supernatant of 5-FU-loaded IONP samples was studied at different time intervals, i.e., 0 h, 2 h, 8 h, and 24 h of mixing, and the percentage 5-FU loading efficiency was calculated by using the equation:
0.25. The absorbance maximum for 5-FU was recorded at 266 nm. To assess the drug-loading efficiency over time, the absorbance of the supernatant of 5-FU-loaded IONP samples was studied at different time intervals, i.e., 0 h, 2 h, 8 h, and 24 h of mixing, and the percentage 5-FU loading efficiency was calculated by using the equation: | (2) |
3. Results and discussion
3.1. Comprehensive physiochemical characterization of bare IONPs and amino-acid-coated IONPs
All the nanoformulations, i.e., bare IONPs and amino acid-coated IONP-based DDSs, were systematically prepared by co-precipitation of Fe3+ and Fe2+ (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio) in an alkaline medium followed by in situ functionalization with the amino acids. The formation of a black precipitate indicated the completion of the reaction and the formation of IONPs. For confirmation of the successful synthesis of the nanoformulations, different spectroscopic and surface analysis techniques were used.
1 molar ratio) in an alkaline medium followed by in situ functionalization with the amino acids. The formation of a black precipitate indicated the completion of the reaction and the formation of IONPs. For confirmation of the successful synthesis of the nanoformulations, different spectroscopic and surface analysis techniques were used.
The FTIR spectra of bare IONPs are shown in Fig. 1(a)-(i). The FTIR vibrational peak observed at around 570 cm−1 is ascribed to the Fe–O vibrational mode of Fe3O4 and further suggests the successful synthesis of the IONPs. The coating of amino acids on the surface of the IONPs is confirmed by comparing the spectra of bare IONPs, pure amino acids, and amino acid-coated IONPs. On comparing the FTIR spectra of pure glycine and glycine-coated IONPs (Fig. 1((a)-ii, iii)), it was observed that the FTIR spectrum of glycine-coated IONPs exhibits vibrational modes primarily corresponding to the characteristic functional groups of glycine, with little shift.31,32 The asymmetric and symmetric stretching vibrations of the COO− group in glycine, which are initially observed at 1600 cm−1 and 1406 cm−1, respectively, exhibited a shift to 1583 cm−1 and 1393 cm−1 upon surface coating of the IONPs with glycine.14 Therefore, the wavenumber separation between the shifted asymmetric and symmetric stretching bands of the COO− group, i.e., Δ = 190 cm−1, is in accordance with the reported value and implies a bridging bidentate type linkage of the COO− group to the Fe3O4.14 This complexation imparts a partial single-bond character to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, due to which a slight shift in frequency value was observed. In addition to this, the absorption bands observed for pure glycine were sharp and well-defined, whereas those corresponding to glycine-coated IONPs were rather broad and shifted, suggesting interactions between glycine molecules and the nanoparticle surface. A broad band observed at around 3000 cm−1 and a peak at around 3118 cm−1 correspond to the νs and νas vibration modes of the NH3+ group. These peaks were also observed in the spectra of glycine-coated IONPs, without any change in their position. These observations suggest that glycine is linked to the IONP surface primarily through the COO− group, resulting in the amine (–NH2) group being freely exposed. Similarly, on comparing the FTIR spectra of pure glycine and pure alanine, it was observed that both amino acids exhibit similar FTIR spectra. For this reason, alanine-coated IONPs (Fig. S3(a)) also showed similarity in their spectra with glycine-coated IONPs.
O, due to which a slight shift in frequency value was observed. In addition to this, the absorption bands observed for pure glycine were sharp and well-defined, whereas those corresponding to glycine-coated IONPs were rather broad and shifted, suggesting interactions between glycine molecules and the nanoparticle surface. A broad band observed at around 3000 cm−1 and a peak at around 3118 cm−1 correspond to the νs and νas vibration modes of the NH3+ group. These peaks were also observed in the spectra of glycine-coated IONPs, without any change in their position. These observations suggest that glycine is linked to the IONP surface primarily through the COO− group, resulting in the amine (–NH2) group being freely exposed. Similarly, on comparing the FTIR spectra of pure glycine and pure alanine, it was observed that both amino acids exhibit similar FTIR spectra. For this reason, alanine-coated IONPs (Fig. S3(a)) also showed similarity in their spectra with glycine-coated IONPs.
Similar patterns of peaks were observed with aspartic acid, serine, threonine, and tryptophan-coated IONPs, which suggests the coating of different sets of amino acids onto the surface of IONPs by bidentate chelation (Fig. S3). As an illustration, distinctive groups of aspartic acid are –COOH and NH2, therefore its spectrum shows peaks at around 3300 cm−1, 2994 cm−1, 1685 cm−1, 1400 cm−1, and 1112 cm−1 assigned to N–H (νN–H), O–H (νO–H), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (νC
O (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), COO– (νCOOH), and C–NH2 (νC–N), respectively.33,34 These peaks are also observed in the FTIR spectra of aspartic acid-coated IONPs with a slight shift in their absorption frequency and width of peaks (Fig. S3(b)). Equivalently, for tryptophan-coated IONPs, in addition to the characteristic bands of amino acids, modes associated with pyrrole and benzene rings (1464 cm−1, 1371–797 cm−1), are also observed in the spectra of tryptophan-coated IONPs (Fig. S3(c)).35 This again suggests the successful coating of the amino acid tryptophan on the surface of the IONPs. In a continuation of this, serine- and threonine-coated IONPs display all the distinctive peaks of the amino acids, and a vibrational mode with the frequency of the –O–H group was also seen in the spectra of both serine- and threonine-coated IONPs at around 3300 cm−1 (Fig. S3(d) and (e)). Thus, the FTIR results confirm the coating of amino acids onto the surface of the IONPs through the carboxylate group, whereas the amine group is freely exposed. From the obtained FTIR spectra, it can be concluded that the respective amino acids are linked to the IONP surface.
O), COO– (νCOOH), and C–NH2 (νC–N), respectively.33,34 These peaks are also observed in the FTIR spectra of aspartic acid-coated IONPs with a slight shift in their absorption frequency and width of peaks (Fig. S3(b)). Equivalently, for tryptophan-coated IONPs, in addition to the characteristic bands of amino acids, modes associated with pyrrole and benzene rings (1464 cm−1, 1371–797 cm−1), are also observed in the spectra of tryptophan-coated IONPs (Fig. S3(c)).35 This again suggests the successful coating of the amino acid tryptophan on the surface of the IONPs. In a continuation of this, serine- and threonine-coated IONPs display all the distinctive peaks of the amino acids, and a vibrational mode with the frequency of the –O–H group was also seen in the spectra of both serine- and threonine-coated IONPs at around 3300 cm−1 (Fig. S3(d) and (e)). Thus, the FTIR results confirm the coating of amino acids onto the surface of the IONPs through the carboxylate group, whereas the amine group is freely exposed. From the obtained FTIR spectra, it can be concluded that the respective amino acids are linked to the IONP surface.
| S. no. | Synthesized systems | Hydrodynamic diameter (nm) | Specific surface area (m2 g−1) | Zeta potential (mV) |
|---|---|---|---|---|
| 1 | Bare IONPs | 101.4 ± 4.1 | 205.27 ± 14 | −11.7 ± 1.1 |
| 2 | Glycine IONPs | 130.4 ± 3.2 | 152.47 ± 11 | −29.6 ± 1.3 |
| 3 | Alanine IONPs | 138.1 ± 3.1 | 128.58 ± 8 | −26.2 ± 1.9 |
| 4 | Aspartic acid IONPs | 170.9 ± 2.2 | 112.27 ± 7 | −37.4 ± 1.1 |
| 5 | Tryptophan IONPs | 181.5 ± 3.5 | 111.44 ± 9 | −21.5 ± 3.7 |
| 6 | Serine IONPs | 155.5 ± 4.3 | 122.46 ± 5 | −35.5 ± 2.1 |
| 7 | Threonine IONPs | 167.5 ± 3.3 | 118.68 ± 4 | −33.9 ± 1.5 |
Furthermore, results of zeta potential studies, listed in Table 1, indicate that the zeta potential values of the amino acid-coated IONPs were more negative than that of the bare IONPs. This suggests that surface coating IONPs with amino acids increases their colloidal stability. The obtained data also suggest that the zeta potential values of polar amino acid-coated IONPs, i.e., aspartic acid, serine, and threonine-coated IONPs, were more negative than non-polar amino acid-coated IONPs, i.e., glycine, alanine, and tryptophan-coated IONPs.
Thus, comprehensive statistical analysis of the hydrodynamic diameter, specific surface area, and zeta potential demonstrates that amino acids used as coating material significantly influence the physicochemical properties of IONPs. Coating IONPs with larger amino acids markedly increases their particle size while reducing their specific surface area, and polar coatings further enhance negative surface charge, boosting colloidal stability and effectively preventing aggregation. These trends provide compelling evidence for the successful and controlled coating of IONPs, highlighting how small variations in coating chemistry significantly influence the physicochemical properties. Collectively, results offer critical insights for optimizing IONPs for highly efficient drug delivery applications.
Additionally, images of the sample surface were obtained with a targeted electron beam under SEM examination to study the surface topography and composition. The acquired images of bare IONPs (Fig. 2(a)) indicate that there is a positive signature of agglomeration. In contrast, IONPs with glycine and other amino acid coatings are all well dispersed and have a spherical form in comparison to bare IONPs (Fig. 2(b)). From the acquired images, it is also clear that the synthesized amino acid-coated nanoformulations exhibit improved stability as clearer images are obtained. Therefore, the observed SEM images of bare IONPs and IONPs coated with amino acids indicate the increased stability of IONPs on surface coating, supporting the zeta potential results.
3.2. MALDI/TOF analysis
The exact molecular weight of amino acid-coated IONPs is fundamental to calculating the binding and thermodynamic parameters of interactions in ITC and fluorescence quenching studies. Therefore, MALDI/TOF analysis was used to obtain the accurate molecular weight of amino acid-coated IONPs and facilitate their direct comparison with bare IONPs. Fig. 3 shows the normalized signal intensity systematically plotted against the molecular weight-to-charge ratio (m/z) of alanine-coated IONPs. Previous studies demonstrated that the molar mass of bare IONPs is 231.5 g mol−1.36,37 Results obtained from MALDI analysis suggested that the molecular weight of alanine (Fig. 3), glycine, aspartic acid, tryptophan, serine, and threonine-coated IONPs were 453.3, 414.2, 697.7, 717.6, 485.1, and 525.2 g mol−1, respectively (Fig. S4). The increased molecular weight of all the amino acid-coated IONPs further confirms the coating of the respective amino acids on the IONPs. Subsequently, ITC and fluorescence quenching studies were performed using the obtained molecular weight of the synthesized amino acid-coated IONPs.3.3. Calorimetric and spectroscopic insights into the nano-bio-interface: comparative analysis of the effect of coating material on DNA binding affinity of amino acid-coated IONPs and bare IONPs
Understanding the nano-bio interface and the interaction mechanism is important for determining the applicability of the synthesized DDSs for biomedical applications. To develop QSAR by quantitatively understanding the nano-bio interface, it is crucial to correlate interaction parameters with the functional groups of the coating material. ITC and fluorescence emission techniques were employed to study the interaction parameters of the different amino acid-coated IONPs and bare IONPs with DNA. Comparison of the interaction parameters between structurally different amino acid-coated IONPs and bare IONPs with DNA can provide guidelines for developing efficient coating materials for IONPs for different biomedical applications. Fig. 4(a) represents ITC profiles illustrating the binding interaction of the synthesized bare IONPs with DNA. Raw data for heat rate (μW min−1) is shown in the top panel of the figure, whereas the bottom panel shows dilution-corrected heat as a function of gradually increasing molar ratio of IONPs to DNA. The solid bold line shows that the data points are fitted with a single binding site interaction model, and quantitative thermodynamic parameters are obtained. | ||
| Fig. 4 (a) ITC profile for interactions of bare IONPs with 100 μM DNA. (b) Fluorescence emission quenching graph of the complex formed by DNA-EtBr with increasing bare IONP concentration. | ||
Associated quantitative insights into the interactions of bare IONPs with DNA are summarised in Table 2. The obtained Kb = 1.3 × 10 4 M−1, with positive 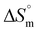 and negative
and negative 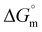 , suggests intercalative binding of bare IONPs with DNA, which is both entropically favorable and spontaneous in nature.38 The obtained
, suggests intercalative binding of bare IONPs with DNA, which is both entropically favorable and spontaneous in nature.38 The obtained  and stoichiometry value (n) suggest endothermic binding of approximately five molecules of bare IONPs per DNA base pair. Further, fluorescence quenching studies using the DNA-EtBr complex also validated the intercalative mode of binding of bare IONPs with DNA (Fig. 4b).
and stoichiometry value (n) suggest endothermic binding of approximately five molecules of bare IONPs per DNA base pair. Further, fluorescence quenching studies using the DNA-EtBr complex also validated the intercalative mode of binding of bare IONPs with DNA (Fig. 4b).
| (i) Results obtained from isothermal titration calorimetry studies at pH 7.4 and 298 K | ||||||
|---|---|---|---|---|---|---|
| S. no. | Synthesized iron oxide nanoparticles | K b (M−1) | (kJ mol−1) | (kJ mol−1) | (kJ mol−1) | n |
| 1 | Bare IONPs | (1.3 ± 0.1) × 104 | (0.15 ± 0.01) | (−23.5 ± 0.2) | (23.6 ± 0.2) | (4.6 ± 0.31) |
| 2 | Glycine IONPs | (1.4 ± 0.05) × 105 | (−1.02 ± 0.01) | (−29.3 ± 0.11) | (28.3 ± 0.21) | (2.7 ± 0.1) |
| 3 | Alanine IONPs | (1.2 ± 0.03) × 105 | (−0.56 ± 0.02) | (−28.9 ± 0.13) | (28.4 ± 0.11) | (2.5 ± 0.2) |
| 4 | Aspartic acid IONPs | (5.1 ± 0.02) × 105 | (−0.55 ± 0.6) | (−32.6 ± 0.22) | (32.0 ± 0.14) | (3.9 ± 0.5) |
| 5 | Tryptophan IONPs | (2.3 ± 0.9) × 104 | (0.88 ± 0.09) | (−25.0 ± 0.41) | (25.9 ± 0.35) | (1.9 ± 0.1) |
| 6 | Serine IONPs | (6.4 ± 0.2) × 105 | (−0.63 ± 0.6) | (−33.2 ± 0.21) | (32.5 ± 0.16) | (2.7 ± 0.6) |
| 7 | Threonine IONPs | (5.6 ± 0.09) × 105 | (−1.60 ± 0.5) | (−32.9 ± 0.12) | (31.3 ± 0.21) | (1.8 ± 0.2) |
| (ii) Results obtained from fluorescence quenching studies at pH 7.4 and 298 K | ||
|---|---|---|
| S. no. | Synthesized iron oxide nanoparticles | K b (M−1) |
| 1 | Bare IONPs | (1.8 ± 0.2) × 104 |
| 2 | Glycine IONPs | (2.0 ± 0.3) × 105 |
| 3 | Alanine IONPs | (1.4 ± 0.1) × 105 |
| 4 | Aspartic acid IONPs | (5.0 ± 0.4) × 105 |
| 5 | Tryptophan IONPs | (3.0 ± 0.6) × 104 |
| 6 | Serine IONPs | (6.4 ± 0.2) × 105 |
| 7 | Threonine IONPs | (5.3 ± 0.2) × 105 |
The observed quantitative aspects of interactions of serine and threonine-coated IONPs with DNA are listed in Table 2. The obtained Kb value of the order of 105 indicates that both systems bind to DNA strongly through an intercalative mode, similar to that observed for standard intercalators.39 Among all the synthesized systems, serine-coated IONPs were found to exhibit the strongest interaction with DNA. This could be due to the presence of the hydroxyl group in the serine side chain, which contributes to their highest binding with DNA. Moreover, the binding constant obtained for threonine-coated IONPs was somewhat lower than that for serine-coated IONPs. This can be explained by serine and threonine having similar functional groups, –CH2–OH and –CH(OH)–CH3, respectively, but the presence of one extra –CH3 group in threonine causes a slight decrease in the Kb value. Furthermore, the energy values with positive 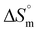 suggest entropically favorable interactions of the two formulations with DNA. Exothermic enthalpy indicates the presence of H-bonding, and the negative
suggest entropically favorable interactions of the two formulations with DNA. Exothermic enthalpy indicates the presence of H-bonding, and the negative 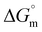 value directly relates to the binding affinity for spontaneous binding between the synthesized systems and DNA. Binding of around 3 molecules of serine-coated IONPs and 2 molecules of threonine-coated IONPs per base pair of DNA was suggested by the obtained stoichiometry (n) values (Fig. 5(e) and (f)).
value directly relates to the binding affinity for spontaneous binding between the synthesized systems and DNA. Binding of around 3 molecules of serine-coated IONPs and 2 molecules of threonine-coated IONPs per base pair of DNA was suggested by the obtained stoichiometry (n) values (Fig. 5(e) and (f)).
On a similar line, glycine and alanine are also structural analogs. The Kb value observed for glycine-coated IONPs was slightly greater than that observed for alanine-coated IONPs (Table 2). This indicates that a similar effect of one extra –CH3 group on the Kb value was observed for glycine- and alanine-coated IONPs with DNA, as observed with serine and threonine-coated IONPs. Lower Kb values for glycine- and alanine-coated IONPs in comparison to serine- and threonine-coated IONPs can be understood by considering that glycine and alanine are devoid of any polar groups at the α-position, and instead have non-polar –H and –CH3, respectively, and belong to the class of aliphatic non-polar amino acids. Furthermore, the Kb value for glycine- and alanine-coated IONPs was found to be greater than that of bare IONPs with DNA, indicating that even though glycine and alanine are non-polar amino acids, the interactions of the corresponding amino acid-coated formulations with DNA were stronger than those of bare IONPs. Entropic support and the spontaneous nature of the binding process are demonstrated by the positive 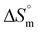 and negative
and negative 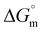 values. The obtained exothermic
values. The obtained exothermic  values for both glycine- and alanine-coated IONPs indicate the presence of hydrophilic contacts during the binding process. The obtained n values suggest that around 3 molecules of both glycine- and alanine-coated IONPs bind to each base pair of DNA (Fig. 5(a) and (b)).
values for both glycine- and alanine-coated IONPs indicate the presence of hydrophilic contacts during the binding process. The obtained n values suggest that around 3 molecules of both glycine- and alanine-coated IONPs bind to each base pair of DNA (Fig. 5(a) and (b)).
Continuing the series, the thermodynamic parameters obtained from the calorimetric data of aspartic acid- and tryptophan-coated IONPs with DNA are presented in Table 2. The Kb value for aspartic acid- and tryptophan-coated IONPs with DNA is higher than that of the bare IONPs. The obtained thermodynamic parameters also suggest that, since aspartic acid is a polar amino acid, the binding of aspartic acid-coated IONPs was greater than that of non-polar glycine- and alanine-coated IONPs. In addition, the obtained energy values suggest the presence of hydrophilic, entropically supported, and spontaneous interactions. The stoichiometry value indicates binding of 4 molecules of aspartic acid-coated IONPs per base pair of DNA (Fig. 5(c)). Moreover, tryptophan, with one aromatic residue (i.e., the indole moiety) at the β-carbon, is a non-polar amino acid and has a bulky β-substituent. Therefore, tryptophan-coated IONPs show a much lower Kb value that is approximately the same as that of bare IONPs, which also indicates the presence of hydrophobic interactions, with a positive  value during the binding process. The obtained positive
value during the binding process. The obtained positive 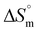 and negative
and negative 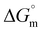 , together with the stoichiometry value, point towards entropically supported and spontaneous binding of 2 molecules of tryptophan-coated IONPs for each base pair of DNA (Fig. 5(d)).
, together with the stoichiometry value, point towards entropically supported and spontaneous binding of 2 molecules of tryptophan-coated IONPs for each base pair of DNA (Fig. 5(d)).
The findings of the calorimetric investigations on the DNA binding constants for various amino acid-coated IONPs and bare IONPs are as follows:
Serine-coated IONPs > threonine-coated IONPs > aspartic acid-coated IONPs > glycine-coated IONPs > alanine-coated IONPs > tryptophan-coated IONPs ≥ bare IONPs.
To further elucidate the binding mode of bare IONPs and the different amino acid-coated IONPs with DNA, and to complement the ITC results, EtBr displacement studies were performed with the DNA-EtBr complex (Fig. 4(b) and 6). For analysis, the synthesized IONPs were incrementally titrated into the DNA-EtBr solution in aliquots, which caused quenching of fluorescence emission. This suggests that intercalated EtBr molecules on the DNA are replaced by the synthesized IONPs.40–42Fig. 4(b) and 6 show the fluorescence emission quenching spectra of the DNA-EtBr complex upon incremental addition of bare and amino acid-coated IONPs, along with a Stern–Volmer plot of log [(F0 − F)/F] as a function of the log of concentration of nanoparticles. Binding constant (Kb) values for all the synthesized systems were calculated by applying the modified Stern–Volmer equation (eqn (1)). The Kb value obtained from the ITC studies, of the order of 104 and 105, points to strong binding of the synthesized IONPs with DNA. Similarly, complete quenching was observed for the DNA-EtBr complex as the concentration of IONPs increased, and the Kb value of the order of 104 for bare IONPs with DNA (Fig. 4(b)) confirms the intercalation (Table 2). From the obtained Kb values, it was also concluded that IONPs coated with amino acids show even stronger interactions with DNA compared to bare IONPs (Table 2). Moreover, the trend of binding constant values for all the amino acid-coated IONPs in the fluorescence studies complements the trend obtained in the ITC studies. IONPs coated with polar amino acids show higher Kb values in comparison to IONPs coated with non-polar amino acids. The Kb values of threonine- and alanine-coated IONPs, which were seen to be smaller than their structural counterparts serine- and glycine-coated IONPs, also show the impact of inclusion of one extra –CH3 group. Due to its bulky side chain (indole group), tryptophan-coated IONPs were consistently shown to have the lowest Kb values, which were roughly equivalent to that of bare IONPs with DNA (Table 2).
Conclusively, the results obtained from the calorimetric and fluorescence studies were observed to complement each other and have significant clinical relevance. Interaction studies demonstrated that polar amino acids exhibit greater binding affinities when coated on the surface of IONPs than non-polar amino acids. Thus, polar amino acids, by enhancing the binding strength, offer a pathway to the design of DDSs with superior DNA delivery efficiency. It was also noted that the inclusion of one extra –CH3 group in threonine and alanine compared to their respective structural counterparts, serine and glycine, lowers the Kb value. This again highlights how even small structural differences in amino acid coatings significantly influence the therapeutic performance. Likewise, tryptophan, with the largest side chain, has the lowest DNA binding affinity, about equivalent to that of bare IONPs, which demonstrates that steric hindrance from large substituents significantly impairs biomolecular interactions. Such molecular-level insights offer a blueprint for designing next-generation nanomedicines that are safer and more effective in cancer therapeutics. From all these obtained results, it is possible to infer that surface functionalization of IONPs with amino acids increases their ability to bind DNA, making them more effective DDSs than IONPs on their own.
3.4. Quantitative determination of drug loading efficiency to evaluate the efficacy of the synthesized DDSs
Drug loading efficiency of the synthesized IONPs was determined at four different ratios of nanoparticle to drug, as detailed in the experimental section. The loading efficiency of bare IONPs with 5-FU was observed to be around 20% at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.25 ratio.38 Different amino acid-coated formulations, including serine, threonine, aspartic acid, glycine, alanine, and tryptophan-coated IONPs, show drug loading efficiencies of 71%, 62%, 60%, 45%, 40%, and 22%, respectively (Fig. 7). These obtained values suggest that amino acid-coated nanoformulation enables significantly better drug loading efficiency compared to bare IONPs,38 demonstrating their improved efficiency as DDSs over bare IONPs. Among the different amino acid-coated formulations, serine-coated IONPs showed the highest loading efficiency, whereas threonine-coated IONPs showed the second-highest drug-loading efficiency. This difference can be ascribed to the fact that threonine has an extra –CH3 group in comparison to serine, which may hinder its interactions. The loading efficiency of aspartic acid-coated IONPs was also observed to be very close to that of the threonine-coated IONPs. However, the loading efficiency of glycine, alanine, and tryptophan-coated IONPs was found to be on the lower side. Lower drug loading efficiency of alanine-coated IONPs as compared to glycine-coated IONPs can again be understood by considering the presence of one extra -CH3 group. Furthermore, the lowest drug loading efficiency of the tryptophan-coated IONPs, which is almost equivalent to the loading efficiency of bare IONPs, signifies the influence of the bulky side substituents of the coating material on their loading efficiency. Drug loading data further suggest that polar amino acids, when functionalized on the surface of the IONPs, increase the loading efficiency of IONPs to a greater extent than the non-polar amino acids. This is primarily attributed to enhanced hydrogen-bonding interactions as well as stronger electrostatic attractions.43 These findings imply that the surface coating of IONPs with different sets of amino acids not only enhances their interaction with the biological environment but also increases their efficiency for theranostics applications. Thus, the results have significant clinical implications since they show that amino acid-coated IONPs can achieve significantly higher drug loading efficiencies than bare IONPs, allowing for the administration of higher drug doses. Various literature reports suggested that different ligand-coated IONPs exhibit excellent loading efficiency and hold strong therapeutic potential. A report by Prabha et al. demonstrated that IONPs coated with different ligands, such as chitosan, polyethylene glycol, and polyvinylpyrrolidone, show very high Curcumin loading efficiency and, hence, are potential candidates for cancer therapeutics applications.44 In addition to this, a report by Kumar et al. demonstrated that glycine-coated IONPs exhibited significant efficacy in the treatment of other diseases such as visceral leishmaniasis, underscoring their therapeutic potential.45 Thus, coating IONPs with an appropriate ligand improves treatment efficiency, reduces systemic toxicity, and opens the door for next-generation, personalized theranostics platforms that combine effective drug delivery with better therapeutic outcomes.
0.25 ratio.38 Different amino acid-coated formulations, including serine, threonine, aspartic acid, glycine, alanine, and tryptophan-coated IONPs, show drug loading efficiencies of 71%, 62%, 60%, 45%, 40%, and 22%, respectively (Fig. 7). These obtained values suggest that amino acid-coated nanoformulation enables significantly better drug loading efficiency compared to bare IONPs,38 demonstrating their improved efficiency as DDSs over bare IONPs. Among the different amino acid-coated formulations, serine-coated IONPs showed the highest loading efficiency, whereas threonine-coated IONPs showed the second-highest drug-loading efficiency. This difference can be ascribed to the fact that threonine has an extra –CH3 group in comparison to serine, which may hinder its interactions. The loading efficiency of aspartic acid-coated IONPs was also observed to be very close to that of the threonine-coated IONPs. However, the loading efficiency of glycine, alanine, and tryptophan-coated IONPs was found to be on the lower side. Lower drug loading efficiency of alanine-coated IONPs as compared to glycine-coated IONPs can again be understood by considering the presence of one extra -CH3 group. Furthermore, the lowest drug loading efficiency of the tryptophan-coated IONPs, which is almost equivalent to the loading efficiency of bare IONPs, signifies the influence of the bulky side substituents of the coating material on their loading efficiency. Drug loading data further suggest that polar amino acids, when functionalized on the surface of the IONPs, increase the loading efficiency of IONPs to a greater extent than the non-polar amino acids. This is primarily attributed to enhanced hydrogen-bonding interactions as well as stronger electrostatic attractions.43 These findings imply that the surface coating of IONPs with different sets of amino acids not only enhances their interaction with the biological environment but also increases their efficiency for theranostics applications. Thus, the results have significant clinical implications since they show that amino acid-coated IONPs can achieve significantly higher drug loading efficiencies than bare IONPs, allowing for the administration of higher drug doses. Various literature reports suggested that different ligand-coated IONPs exhibit excellent loading efficiency and hold strong therapeutic potential. A report by Prabha et al. demonstrated that IONPs coated with different ligands, such as chitosan, polyethylene glycol, and polyvinylpyrrolidone, show very high Curcumin loading efficiency and, hence, are potential candidates for cancer therapeutics applications.44 In addition to this, a report by Kumar et al. demonstrated that glycine-coated IONPs exhibited significant efficacy in the treatment of other diseases such as visceral leishmaniasis, underscoring their therapeutic potential.45 Thus, coating IONPs with an appropriate ligand improves treatment efficiency, reduces systemic toxicity, and opens the door for next-generation, personalized theranostics platforms that combine effective drug delivery with better therapeutic outcomes.
 | ||
| Fig. 7 Drug loading efficiency profile of 5-FU on the synthesized iron oxide nanoparticles at pH 7.4 and 37 °C. | ||
4. Conclusion
In conclusion, in this study, we synthesized and characterized different amino acid-coated IONPs along with bare IONPs to quantitatively understand the nano-bio interface through thermodynamics of interactions and therapeutic efficiency. The interaction studies were carried out using ITC, complemented by fluorescence spectroscopy, to assess the binding interactions and the mode of binding. Drug loading studies with 5-FU were performed to evaluate the efficiency of IONPs in drug delivery applications. Thermodynamic parameters of interaction and therapeutic efficiency show that coating materials with polar groups leads to better binding and loading efficiency as compared to non-polar groups for IONPs, and the addition of one CH3 group slightly reduces the therapeutic efficiency. The results highlight the impact of size, length of the side substituent, and type of amino acid utilized on their interaction with biomolecules and drug loading efficiency. The findings suggest that surface coating improves both interaction with biomolecules and drug loading efficiency, with the choice of coating material being a key factor influencing the overall efficacy and performance of the IONPs. The correlation of functional groups with the energetics of interaction and activity will help developing guidelines for AI-based creation of effective DDSs.Conflicts of interest
The authors declare no competing financial interest.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5pm00182j.Acknowledgements
The authors acknowledge Banasthali Vidyapith, Rajasthan, for providing access to Isothermal Titration Calorimetry and other facilities.References
- M. Kumar, P. A. Chawla, A. Faruk and V. Chawla, RSC Pharm., 2025, 2(2), 318–332 RSC.
- R. Singh, F. R. Long, A. Saini, N. Joma, A. Basu, M. Mahmoudi, H. Vali and A. Kakkar, RSC Pharm., 2025, 2(1), 44–58 RSC.
- E. Myrovali, A. T. Chatzitaki, K. Papadopoulos and D. G. Fatouros, RSC Pharm., 2025, 2(2), 292–302 RSC.
- N. Ahmed, H. Fessi and A. Elaissari, Drug Discovery Today, 2012, 17(17–18), 928–934 CrossRef CAS PubMed.
- J. Naskar, G. Palui and A. Banerjee, J. Phys. Chem. B, 2009, 113(35), 11787–11792 CrossRef CAS PubMed.
- J. E. Gagner, S. Shrivastava, X. Qian, J. S. Dordick and R. W. Siegel, J. Phys. Chem. Lett., 2012, 3(21), 3149–3158 CrossRef CAS PubMed.
- T. E. Mallouk and P. Yang, J. Am. Chem. Soc., 2009, 131(23), 7937–7939 CrossRef CAS PubMed.
- J. Subbotina and V. Lobaskin, J. Phys. Chem. B, 2022, 126(6), 1301–1314 CrossRef CAS PubMed.
- N. Ajinkya, X. Yu, P. Kaithal, H. Luo, P. Somani and S. Ramakrishna, Materials, 2020, 13(20), 4644 CrossRef CAS PubMed.
- C. Bai, P. Hu, N. Liu, G. Feng, D. Liu, Y. Chen, M. Ma, N. Gu and Y. Zhang, ACS Appl. Nano Mater., 2020, 3(4), 3585–3595 CrossRef CAS.
- A. Kamal, M. Saba and M. Farooq, J. Basic Microbiol., 2023, 63(2), 156–167 CrossRef CAS PubMed.
- Y. Wu, Z. Lu, Y. Li, J. Yang and X. Zhang, Nanomaterials, 2020, 10(8), 1441 CrossRef CAS PubMed.
- A. Lazaro-Carrillo, M. Filice, M. J. Guillén, R. Amaro, M. Viñambres, A. Tabero, K. O. Paredes, A. Villanueva, P. Calvo, M. D. P. Morales and M. Marciello, Mater. Sci., Eng. C, 2020, 107, 110262 CrossRef CAS PubMed.
- K. C. Barick and P. A. Hassan, J. Colloid Interface Sci., 2012, 369(1), 96–102 CrossRef CAS PubMed.
- M. Salehiabar, H. Nosrati, S. Davaran, H. Danafar and H. K. Manjili, Drug Res., 2018, 68(05), 280–285 CrossRef CAS PubMed.
- H. Nosrati, M. Salehiabar, S. Davaran, H. Danafar and H. K. Manjili, Drug Dev. Ind. Pharm., 2018, 44(6), 886–894 CrossRef CAS PubMed.
- P. Gehr, Colloids Surf., B, 2018, 172, 395–399 CrossRef CAS PubMed.
- C. Auría-Soro, T. Nesma, P. Juanes-Velasco, A. Landeira-Viñuela, H. Fidalgo-Gomez, V. Acebes-Fernandez, R. Gongora, M. J. A. Parra, R. Manzano-Roman and M. Fuentes, Nanomaterials, 2019, 9(10), 1365 CrossRef PubMed.
- S. Roy, K. A. Aastha, K. Deo, A. K. Dey, A. Gaharwar and A. Jaiswal, ACS Appl. Mater. Interfaces, 2023, 15(30), 35753–35787 CrossRef CAS PubMed.
- M. S. Ural, J. M. Joseph, F. Wien, X. Li, M. A. Tran, M. Taverna, C. Smadja and R. Gref, Drug Delivery Transl. Res., 2023, 1–15 Search PubMed.
- Biocalorimetry 2: applications of calorimetry in the biological sciences, ed. J. E. Ladbury and M. L. Doyle, John Wiley & Sons, 2004 Search PubMed.
- Principles of fluorescence spectroscopy, ed. J. R. Lakowicz, springer US, Boston, MA, 2006 Search PubMed.
- A. Mukhija and N. Kishore, J. Mol. Liq., 2019, 283, 558–572 CrossRef CAS.
- N. Zhang, Y. Yin, S. J. Xu and W. S. Chen, Molecules, 2008, 13(8), 1551–1569 CrossRef CAS PubMed.
- K. C. Barick, S. Singh, N. V. Jadhav, D. Bahadur, B. N. Pandey and P. A. Hassan, Adv. Funct. Mater., 2012, 22(23), 4975–4984 CrossRef CAS.
- G. Felsenfeld and S. Z. Hirschman, J. Mol. Biol., 1965, 13(2), 407–427 CrossRef CAS PubMed.
- M. Liu, W. Zhang, L. Qiu and X. Lin, J. Biochem., 2011, 149(1), 27–33 CrossRef CAS PubMed.
- M. Arakha, S. Pal, D. Samantarrai, T. K. Panigrahi, B. C. Mallick, K. Pramanik, B. Mallick and S. Jha, Sci. Rep., 2015, 5(1), 14813 CrossRef CAS PubMed.
- M. Mahdavi, M. B. Ahmad, M. J. Haron, F. Namvar, B. Nadi, M. Z. A. Rahman and J. Amin, Molecules, 2013, 18(7), 7533–7548 CrossRef CAS PubMed.
- K. Song, Y. Lee, M. R. Jo, K. M. Nam and Y. M. Kang, Nanotechnology, 2012, 23(50), 505401 CrossRef PubMed.
- R. K. Khanna, M. Horak and E. R. Lippincott, Spectrochim. Acta, 1966, 22(10), 1759–1771 CrossRef CAS.
- S. Kumar, A. K. Rai, V. B. Singh and S. B. Rai, Spectrochim. Acta, Part A, 2005, 61(11–12), 2741–2746 CrossRef PubMed.
- Z. Wang, X. Xiao, Y. Yang, T. Zou, X. Xing, R. Zhao, Z. Wang and Y. Wang, Nanomaterials, 2019, 9(8), 1165 CrossRef CAS PubMed.
- G. Marinescu, L. Patron, D. C. Culita, C. Neagoe, C. I. Lepadatu, I. Balint, L. Bessais and C. B. Cizmas, J. Nanopart. Res., 2006, 8, 1045–1051 CrossRef CAS.
- A. Barth, Prog. Biophys. Mol. Biol., 2000, 74(3–5), 141–173 CrossRef CAS PubMed.
- T. T. D. Pham, Y. H. Seo, D. Lee, J. Noh, J. Chae, E. Kang, J. H. Park, T. J. Shin, S. Kim and J. Park, Polymer, 2019, 161, 205–213 CrossRef CAS.
- I. M. Obaidat, C. Nayek, K. Manna, G. Bhattacharjee, I. A. Al-Omari and A. Gismelseed, Nanomaterials, 2017, 7(12), 415 CrossRef PubMed.
- A. Pandey, B. Sharma, V. Choudhary and A. Mukhija, J. Mol. Liq., 2024, 126741 Search PubMed.
- P. B. Dervan and M. M. Becker, J. Am. Chem. Soc., 1978, 100(6), 1968–1970 CrossRef CAS.
- J. Olmsted III and D. R. Kearns, Biochemistry, 1977, 16(16), 3647–3654 CrossRef PubMed.
- C. Qiao, S. Bi, Y. Sun, D. Song, H. Zhang and W. Zhou, Spectrochim. Acta, Part A, 2008, 70(1), 136–143 CrossRef PubMed.
- A. Banerjee, J. Singh and D. Dasgupta, J. Fluoresc., 2013, 23, 745–752 CrossRef CAS PubMed.
- R. A. Harris, J. Mol. Liq., 2022, 360, 119515 CrossRef CAS.
- G. Prabha and V. Raj, J. Biomed. Mater. Res., Part B, 2016, 104(4), 808–816 CrossRef CAS PubMed.
- R. Kumar, K. Pandey, G. C. Sahoo, S. Das, V. N. R. Das, R. K. Topno and P. Das, Mater. Sci. Eng., C, 2017, 75, 1465–1471 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |