 Open Access Article
Open Access ArticleExploiting nano-in-micro-technologies to couple PLGA-hydroxyl-FK866 nanoparticles to a hydrogel network for local drug release†
Eugenia
Spessot‡
 a,
Xue
Bai‡§
b,
Daniel
Moranduzzo
a,
Chen
Zhao
b,
Sam
Butterworth
b,
Devid
Maniglio
a and
Annalisa
Tirella
a,
Xue
Bai‡§
b,
Daniel
Moranduzzo
a,
Chen
Zhao
b,
Sam
Butterworth
b,
Devid
Maniglio
a and
Annalisa
Tirella
 *ab
*ab
aBIOtech-Center for Biomedical Technologies, Department of Industrial Engineering, University of Trento, Via delle Regole 101, 38123 Trento, Italy. E-mail: annalisa.tirella@unitn.it
bDivision of Pharmacy and Optometry, School of Health Science, Faculty of Biology, Medicine and Health, University of Manchester, Oxford Road, Manchester M13 9PT, UK. E-mail: annalisa.tirella@manchester.ac.uk
First published on 2nd April 2025
Abstract
Technological advancements in the formulation and delivery strategies of potent chemotherapeutic agents have been exploited to direct a site-specific drug delivery for the local treatment of tumours. Of these, new generations of nanoparticles are engineered to control the release of therapeutic agents, but they still possess off-target and overall systemic delivery. Injectable hydrogels have unique physico-chemical properties enabling their use as carriers to ensure site-specific targeting. Based on such observations, nanoparticle-loaded hydrogels represent an optimal candidate to both make use of controlled release chemotherapeutic agents (nanoparticles) and local delivery agents (hydrogels) using minimally invasive procedures to reach the target site. Here, we explore the interaction of drug-polymer conjugated nanoparticles with an alginate-based hydrogel network to confine and release a highly cytotoxic compound (hydroxyl-FK866). Specifically, chitosan coating was used to covalently link poly(lactic-co-glycolic acid) nanoparticles to oxidised alginate: confinement and interaction of nanoparticles within alginate-based hydrogels were evaluated using atomic force microscopy measurements, confirming the nanoparticle/hydrogel interaction. Deployment of composite injectable hydrogels in 3D printing was finally investigated. Rheological characterisation and printability tests were performed to assess the printability of alginate-based drug delivery systems to match site-specific geometrical requirements. Then, alginate hydrogels loaded with nanoparticles were ionically crosslinked to match the properties of soft tissues (e.g. breast tissue). The efficacy of 3D printed hydrogels loaded with a known dose of hydroxyl-FK866 was tested using human breast cancer MDA-MB-231 cells. Results confirmed the expected cytotoxicity, showing approx. 52% toxicity of the hydrogel loaded, after 48 hours of incubation, whereas lower viability (approx. 36%) was measured in cells treated with free nanoparticles (control).
1. Introduction
Systemic administration of anticancer drugs is the most used therapeutic plan, but new strategies are required to tackle significant problems related to poor intra-tumoral drug delivery and off-target effects, as shown in the use of injected nanoparticles.1 Technological advancements in the formulation and delivery strategies of potent drugs have been exploited to design new therapeutic approaches for the local treatment of tumours, when the surgical procedure and tumour type and size permit it. Local treatment after surgery shows potential to reduce the risk of cancer recurrence and achieve long-term remission.2 Local drug delivery platforms, for example, peritumoral injections/implants, provide an alternative and reliable approach for cancer therapy, allowing drugs to be delivered directly to the target site at a known dose, bypassing physiological barriers and related issues.3 In this, it is essential to precisely dose potent drugs and control their release in the surrounding environment.4,5Injectable and transplantable hydrogels have recently received much attention thanks to their biocompatibility, low toxicity, biodegradability and easy synthesis processes, showing great promise as carriers for local drug administration.6,7 However, when small drugs are loaded in hydrogels, rapid diffusion is observed due to the poor interaction with the hydrogel network, hence limiting their use for local delivery.8 Drug-loaded nanoparticles (NPs) offer the opportunity not only to load a known dose of therapeutic agent, but also most importantly, they can be engineered to control its release.9 Exploiting the use of composite hydrogels encapsulating known concentrations of drug-loaded NPs is ground-breaking, offering the unique characteristics of loading compounds at a known dose, confining NPs within hydrogels at known concentrations, matching the mechanical properties of the target tissue, enhancing adhesion to the tissue and most importantly, enabling the localized and sustained release with known kinetics.10
In this study, we confined different types of poly(lactic-co-glycolic acid) (PLGA) NPs in an alginate-based hydrogel network based on the following two hypotheses: first, positively charged NPs should form ‘weak’ interactions with the negatively charged alginate chains; secondly, the coating of NPs with primary amines could form covalent and stable bonds with the hydrogel network. Based on our recent work (Bai et al.),11 we used PLGA-based NPs for the sustained release of the highly toxic therapeutic agent hydroxyl-FK866. Chitosan (CS), a linear polysaccharide composed of β(1-4) linked D-glucosamine and N-acetyl-D-glucosamine units,12 was selected to coat PLGA NPs and obtain a homogeneous positively charged surface, also exposing primary amines.
Alginate was selected as it is a non-toxic and biocompatible linear polysaccharide, derived from brown algae, composed of irregular blocks of β-D-mannuronic acid and 1–4 linked α-L-guluronic residues able to form stable ionically cross-linked hydrogels using divalent cations that cannot degrade in vivo.13 Modifications of alginate are used to promote hydrolysis, hence controlling degradation of alginate hydrogels under physiological conditions.14 Oxidation of alginate is one of the most used approaches to induce degradation of alginate-derived hydrogels, both in vitro and in vivo.15 We reported the preparation of oxidized alginates (OAs) with known degrees of oxidation (DOs) and exhibiting an inverse correlation between the degree of oxidation and the molecular weight.16 We also showed that combinations of alginate, oxidized alginates with varying DOs, and crosslinking strategies can be selected to form hydrogels with known mechanical properties and degradation profiles.17 Moreover, as OAs presents aldehyde groups proportional to DOs, excess or unreacted groups are still available to react with primary amines (Schiff's base reaction) and form covalent links.18
In this study, we used an unmodified alginate and OA at a low degree of oxidation (DO = 5%) loaded with PLGA NPs as injectable technology for the local and sustained release of a potent chemotherapeutic agent: hydroxyl-FK866. Optimization of the chitosan coating of PLGA NPs was performed to direct confinement of PLGA NPs (uncoated, negatively charged) or CS/PLGA NPs (chitosan-coated, positively charged) within the alginate-based hydrogel network. The feasibility of using such formulations as biomaterial inks for the 3D printing of drug delivery technologies was then assessed, and the printability of biomaterial inks was tested at different NP concentrations and without/with chitosan-coating. Interactions between alginate, OA, PLGA NPs and CS/PLGA NPs were investigated using atomic force microscopy (AFM) imaging. AFM allowed us to assess whether positively charged NPs exposing primary amines on their surface could be linked within the negatively charged alginate-based hydrogel network, distinguishing between electrostatic interactions and covalent crosslinks (Schiff's base reaction).19,20 We found that CS-coated NPs can be effectively linked to the hydrogel network, with high control over their homogeneous distribution across the 3D-printed scaffolds.
Finally, toxicity studies using the human triple-negative breast cancer cell line sensitive to hydroxyl-FK866 (MDA-MB 231) confirmed the expected release of hydroxyl-FK866 with toxicity comparable to NPs dispersed in cell culture media, slightly lower due to diffusion through the hydrogel, as per NPs suspended in cell culture media. We herein show new strategies to load and constrain NPs in degradable hydrogels for local and sustained release of chemotherapeutic agents, further opening the possibility of deploying this formulation for 3D printing of implantable devices with a known geometry, mechanical properties, and in situ drug release profiles.
This study demonstrates the broad ramifications of the use of nanoparticles and hydrogels for personalized medicine and shows new strategies to better control the local release of chemotherapeutic agents at the target site. Injectable and 3D printed drug delivery technologies are promising alternatives to increase drug efficacy and reduce inflammatory processes in situ, using minimally invasive delivery procedures to limit discomfort to patients.
2. Materials and methods
2.1. Materials
Poly(lactic-co-glycolic acid) (PLGA, RG752H, product no. 719919, lactide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycolide 75
glycolide 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, Mw 4000–15
25, Mw 4000–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), poly(vinyl alcohol) (PVA, product no. 360627-25G), citric acid (product no. 251275), Dulbecco's modified Eagle's medium (DMEM, product no. D6429), fetal bovine serum (FBS, product no. F9665), trypsin (product no. T3924), L-glutamine (product no. G7513), antibiotics (penicillin–streptomycin, product no. P0781), trypan blue (product no. 15250061), cell proliferation reagent WST-1 (product no. CELLPRO-RO), chitosan (CS, product no. 448869-50G), and sodium alginate (ALG, product no. 71238-250G) were all purchased from Sigma-Aldrich (UK). Acetonitrile (HPLC-grade, 34851-2.5L) and methanol (HPLC-grade, product no. 34966-2.5L) were purchased from Honeywell. Hydroxyl-FK866-PLGA was prepared and characterized as described in our previous work.11 Oxidized alginate with a target degree of oxidation of 5% (OA5) was prepared and characterized using protocols developed by our group and as described by Zhao et al.16
000), poly(vinyl alcohol) (PVA, product no. 360627-25G), citric acid (product no. 251275), Dulbecco's modified Eagle's medium (DMEM, product no. D6429), fetal bovine serum (FBS, product no. F9665), trypsin (product no. T3924), L-glutamine (product no. G7513), antibiotics (penicillin–streptomycin, product no. P0781), trypan blue (product no. 15250061), cell proliferation reagent WST-1 (product no. CELLPRO-RO), chitosan (CS, product no. 448869-50G), and sodium alginate (ALG, product no. 71238-250G) were all purchased from Sigma-Aldrich (UK). Acetonitrile (HPLC-grade, 34851-2.5L) and methanol (HPLC-grade, product no. 34966-2.5L) were purchased from Honeywell. Hydroxyl-FK866-PLGA was prepared and characterized as described in our previous work.11 Oxidized alginate with a target degree of oxidation of 5% (OA5) was prepared and characterized using protocols developed by our group and as described by Zhao et al.16
2.2. Preparation of FK866-loaded nanoparticles using a microfluidic system
PLGA-based NPs used in this work were prepared using an automated Dolomite microfluidic system (Dolomite, Royston UK) equipped with a 5-input Chip (part no. 3200735), a compressor (part no. 3200117), a Mitos P-Pump (part no. 3200016), Mitos P-Pump Remote Chamber 30 (part no. 3200178), a Mitos Flow Rate Sensor 1–50 μL min−1 (part no. 3200098), a Mito Flow Rate Sensor 30–1000 μL min−1 (part no. 3200097), a Meros Temperature Control Unite (part no. 3200428), an In-line Valve (part no. 3200087), and a T-Connector (part no. 3000397). Pure HPLC-grade H2O and a 1% (w/v) PVA solution (aq.) were used as the aqueous (or continuous) phase in all the experiments. The organic (or dispersed) phases were prepared by dissolving PLGA (and derivatives) in acetonitrile at 5.0 mg mL−1. In specific, rhodamine labelled PLGA NPs were used for imaging (Rho-PLGA NPs, ESI SI.1†) and hydroxyl-FK866 loaded NPs for drug delivery studies (hydroxyl-FK866-PLGA NPs, ESI SI.2†). NPs were all prepared using the microfluidic configuration and settings previously optimized: a flow rate ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and a total flow rate of 200 μL min−1 at a temperature of 25 °C and a back pressure of 2 bars.11 Prior to each experiment, aqueous and organic solutions were filtered twice through 0.22 μm PVDF filters (Millex-CV, Merck Millipore Ltd) and 0.2 μm PTFE filters (code 1514-1499, Fisherbrand), respectively. Of note, hydroxyl-FK866-PLGA NPs were used immediately after purification; PLGA and Rho-PLGA NPs were discarded after 2 weeks of storage at 4 °C. All NPs from now on are referred to as PLGA NPs for simplicity unless necessary to specify the formulation.
3 and a total flow rate of 200 μL min−1 at a temperature of 25 °C and a back pressure of 2 bars.11 Prior to each experiment, aqueous and organic solutions were filtered twice through 0.22 μm PVDF filters (Millex-CV, Merck Millipore Ltd) and 0.2 μm PTFE filters (code 1514-1499, Fisherbrand), respectively. Of note, hydroxyl-FK866-PLGA NPs were used immediately after purification; PLGA and Rho-PLGA NPs were discarded after 2 weeks of storage at 4 °C. All NPs from now on are referred to as PLGA NPs for simplicity unless necessary to specify the formulation.
2.3. Coating of nanoparticles with chitosan
Prior to chitosan (CS) coating, PLGA NPs were concentrated to 40 mg mL−1 with dialysis against a 10% w/v PEG 20 kDa (81298, Sigma-Aldrich) solution (aq.) overnight at RT using a dialysis tube (Flot-A-Lyzer G2 dialysis tubes, 3.5–5 kDa membrane cut-off, G235029, Spectra/Por®). CS solutions at a concentration of 0.1% w/v and 0.01% w/v in 4.6 mM HCl (aq.) were mixed with concentrated PLGA NPs in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution and incubated overnight while stirring (RT, 70 rpm) allowing coating. As a control, PLGA NPs were diluted in 4.6 mM HCl solution (aq.) at the same ratio and incubated overnight at RT. Of note, plain PLGA NPs and hydroxyl-FK866-PLGA NPs coated with CS were diluted to final concentrations of 1 mg mL−1 and 20 mg mL−1 and used to formulate biomaterial inks (Table 1) and hydrogels for imaging (Table 2).
1 dilution and incubated overnight while stirring (RT, 70 rpm) allowing coating. As a control, PLGA NPs were diluted in 4.6 mM HCl solution (aq.) at the same ratio and incubated overnight at RT. Of note, plain PLGA NPs and hydroxyl-FK866-PLGA NPs coated with CS were diluted to final concentrations of 1 mg mL−1 and 20 mg mL−1 and used to formulate biomaterial inks (Table 1) and hydrogels for imaging (Table 2).
| Biomaterial ink ID | ALG (10% w/v) | OA5 (10% w/v) | HBS | PLGA NPs (1 mg mL−1) | PLGA NPs (20 mg mL−1) | CS/PLGA NPs (1 mg mL−1) | CS/PLGA NPs (20 mg mL−1) | Hydroxyl-FK866-PLGA (20 mg mL−1) | CS/hydroxyl-FK866-PLGA NPs (20 mg mL−1) |
|---|---|---|---|---|---|---|---|---|---|
| Abbreviations: ALG: unmodified alginate; OA5: oxidized alginate, 5% degree of oxidation; HBS: HEPES buffer solution and CS: chitosan. | |||||||||
| 1 | 800 μL | 100 μL | 100 μL | — | — | — | — | ||
| 2 | 800 μL | 100 μL | — | 100 μL | — | — | — | — | — |
| 3 | 800 μL | 100 μL | — | — | 100 μL | — | — | — | — |
| 4 | 800 μL | 100 μL | — | — | — | 100 μL | — | — | — |
| 5 | 800 μL | 100 μL | — | — | — | — | 100 μL | — | — |
| 6 | 800 μL | 100 μL | — | — | — | — | — | 100 μL | — |
| 7 | 800 μL | 100 μL | — | — | — | — | — | — | 100 μL |
| Sample_ID | ALG (10% w/v) | OA5 (10% w/v) | HBS | PLGA NPs (20 mg mL−1) | CS/PLGA NPs (20 mg mL−1) | PLGA NPs (20 mg mL−1) | Rho-PLGA NPs (1 mg mL−1) | Rho-PLGA NPs (20 mg mL−1) |
|---|---|---|---|---|---|---|---|---|
| ALG | 5 μL | — | 5 μL | — | — | — | — | — |
| OA5 | — | 5 μL | 5 μL | — | — | — | — | — |
| ALG – NPs | 5 μL | — | — | 5 μL | ||||
| OA5 – NPs | 5 μL | 5 μL | ||||||
| ALG – CS/NPs | 5 μL | — | — | 5 μL | ||||
| OA5 – CS/NPs | — | 5 μL | — | — | 5 μL | |||
| ALG-OA5-NPs | 800 μL | 100 μL | 100 μL | |||||
| ALG-OA5-RhoL | 800 μL | 100 μL | — | — | — | 100 μL | ||
| ALG-OA5-RhoH | 800 μL | 100 μL | — | — | — | 100 μL |
2.4. Physico-chemical characterisation of FK866-nanoparticles
Dynamic light scattering. Measurements of PLGA NP concentration were performed using Zetasizer software v8.02. Samples scoring a “good particle size distribution sample” (determined automatically by the Zetasizer software when a good quality signal is detected) were used, recording the size and intensity of the sample. To calculate the concentration of NPs, the “concentration utilities calculator” under “tools” – “calculator” of the software was used and the settings were as follows: the final volume was set to 10, the instrument was set to Zetasizer S, and the attenuator was set to 11 (following manufacturers’ procedures). Then, the measured sample radius and the refractive index of the material were added to the software, and the nanoparticle concentration was calculated.21
Tracking analysis. PLGA NPs’ tracking analysis was performed using a NanoSight NS300 instrument and NanoSight NTA 3.4 software, build 3.4.003 (Malvern Panalytical, Malvern, UK). Prior measurements, PLGA NPs were diluted 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 with deionized water. The diluted samples were then introduced to the NanoSight NS300 instrument. The settings of the camera (sCMOS, laser type: green) were modified to the level of 16. Post-acquisition settings were kept constant between samples and any video files that experienced uncorrected vibration and/or poor tracking analysis were excluded from the final analysis report.
500 with deionized water. The diluted samples were then introduced to the NanoSight NS300 instrument. The settings of the camera (sCMOS, laser type: green) were modified to the level of 16. Post-acquisition settings were kept constant between samples and any video files that experienced uncorrected vibration and/or poor tracking analysis were excluded from the final analysis report.
2.5. Nanoparticle-loaded biomaterial ink formulation and characterization
2.6. Assessment of printability
Printability assessment was performed by printing bidimensional geometries (n = 3) at variable pressures (130, 140, and 150 kPa) and printing velocities (5, 10, and 15 mm s−1). Straight lines and grids were printed to quantitatively determine the shape fidelity of the constructs and thus determine the optimal printing parameters.
The second parameter investigated was the spreading ratio (SR), calculated on the same images acquired for UF by using eqn (2), where tav is the average width of the filament and dnozzle is the internal nozzle diameter (i.e., for the 25 G nozzle dnozzle = 0.250 mm). SR is helpful in determining whether an ink is suitable to print structures with a certain degree of precision and with limited collapse.
The last parameter selected for printability assessment was the printability index (Pr), used to predict the shape retention of an ink and calculated using eqn (3) in which C is the circularity of an enclosed area. Pr is evaluated using a printed geometry 20 × 20 mm squared in the CAD model and with 15% infill density considering the 16 central squares of the grids.22 If perfectly squared holes are printed, then Pr is equal to 1, while Pr < 1 is found when more circular “collapsed” pores are detected and Pr > 1 indicates irregular shapes and uneven extrusion.
Biomaterial ink 1 was used for preliminary tests to identify optimal printing parameters, and then biomaterial inks 2–4 were assessed, using the printability index Pr to confirm the quality of printed objects.
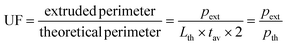 | (1) |
 | (2) |
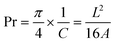 | (3) |
2.7. Laser scanning confocal microscopy (LSCM)
An A1 Laser Microscope (Nikon Instruments Europe BV) was used to acquire volumetric datasets of alginate-based hydrogels with/without rhodamine-labelled PLGA NPs (i.e., Rho-PLGA NPs). PLGA was conjugated with rhodamine following what was described in the literature;23 then Rho-PLGA NPs were fabricated with microfluidics and characterized in size (ESI, SI.1†). Sections and high-resolution (3D-HR) volumetric datasets were acquired to precisely determine Rho-PLGA NPs’ localisation within printed alginate-based hydrogels. For 3D-HR acquisitions, the confocal settings were set as follows: 1 Airy unit, a scan speed of 0.25 frame per s, average line ×2, and 20 μm z-step. Datasets were collected with a Plan Apo 20× DIC M N2 objective and with a 561/595 nm laser. Sections were then processed and analysed using NIS-Element AR 5.30.05, and 3D rendering was performed using NIS-Element AR 5.30.05. Before acquisitions, the bottom and the top levels were determined using the Rho-PLGA NP signal. To compare the amount of particles dispersed in the hydrogels between conditions, images were imported in ImageJ (version 1.51, NIH) and binarised using the Otsu threshold (range 30–240) and the percentage of area occupied by Rho-PLGA NPs was determined with the “analyze particles” tool.2.8. Atomic force microscopy (AFM)
Surface topography was studied using an NT-MDT Solver Pro system equipped with an S7 scanner. Samples (reported in Table 2) were imaged in semi-contact mode using silicon tips (NSG-11, NT-MDT, 10 nm nominal tip radius, with a resonance frequency of 181 kHz). 500 × 500 nm topography and phase-contrast maps (512 × 512 pixels) were collected on different regions of each sample. Alginate-based solutions (aq.) and NP suspensions (aq.) were mixed in a specified volume ratio for a few minutes, then pipetted over the AFM sample holder and left to air dry at room temperature for 24 h in a fume hood and scanned thereafter. AFM data were analysed with the support of Gwyddion analysis software (v. 2.64).242.9. Cell experiments
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2 and left to adhere overnight. Alginate-based hydrogels (Table 1) with a volume of 100 μL were prepared under sterile conditions, washed with HBS and included in each well; all the samples were crosslinked with 50 mM CaCl2 (aq.) for 10 min. Hydroxyl-FK866-PLGA NPs were prepared as described in our previous work and characterized in size and surface charge without and with CS coating (ESI SI.2†). Biomaterial inks 6 and 7 (Table 1) were immersed in the cell culture medium and incubated for 48 h; biomaterial inks 1 and 5 (Table 1) and untreated cells were used as positive controls, whereas cells treated with 30% (v/v) methanol were used as negative controls. Of note, the amount of hydroxyl-FK866-PLGA NPs loaded in alginate hydrogels was calculated based on our recent study and determined as a final 20 mg mL−1 concentration of hydroxyl-FK866-PLGA NPs.11 After incubation for 48 h, the hydrogel and cell culture media were removed from each well, and cells were washed with PBS (n = 3) and then incubated with WST-1 reagent (1 h, 37 °C). Experiments were performed with n = 3 replicates and repeated (N = 2) with biological independent experiments.
000 cells per cm2 and left to adhere overnight. Alginate-based hydrogels (Table 1) with a volume of 100 μL were prepared under sterile conditions, washed with HBS and included in each well; all the samples were crosslinked with 50 mM CaCl2 (aq.) for 10 min. Hydroxyl-FK866-PLGA NPs were prepared as described in our previous work and characterized in size and surface charge without and with CS coating (ESI SI.2†). Biomaterial inks 6 and 7 (Table 1) were immersed in the cell culture medium and incubated for 48 h; biomaterial inks 1 and 5 (Table 1) and untreated cells were used as positive controls, whereas cells treated with 30% (v/v) methanol were used as negative controls. Of note, the amount of hydroxyl-FK866-PLGA NPs loaded in alginate hydrogels was calculated based on our recent study and determined as a final 20 mg mL−1 concentration of hydroxyl-FK866-PLGA NPs.11 After incubation for 48 h, the hydrogel and cell culture media were removed from each well, and cells were washed with PBS (n = 3) and then incubated with WST-1 reagent (1 h, 37 °C). Experiments were performed with n = 3 replicates and repeated (N = 2) with biological independent experiments.
2.10. Statistical analysis
All data represent the mean of a minimum of three independent experiments ± standard deviation (st. dev.), unless otherwise stated. Statistical analyses were performed by one-way ANOVA, followed by Sidak's multiple comparison tests in GraphPad Prism 9. Differences were considered significant at p < 0.05 (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001).3. Results and discussion
3.1. Nanoparticle characterisation
As reported in our recent work, acid-terminated PLGA in acetonitrile (organic phase) and PVA solution (aq.) (aqueous phase) were used to prepare PLGA NPs with the Dolomite microfluidic system.11 PLGA NPs have a mean Z-average size of 207 ± 15 nm and negative ζ-potential (−21.90 ± 2.13 mV) due to the presence of carboxylic end groups in PLGA. CS-coated PLGA NPs have larger hydrodynamic size, showing an increase in NP size (218 ± 4 nm and 245 ± 20 nm) with increased concentrations of CS (0.01% w/v vs. 0.1% w/v) (Table S1†), as reported in similar studies.25,26 As expected, non-coated NPs have a lower PDI (0.118 ± 0.007) when compared to CS-coated NPs (0.207 ± 0.024 and 0.256 ± 0.025, respectively for 0.01% w/v CS and 0.1 w/v CS). Of note, all NPs have PDI < 0.3, which indicates a monodispersed particle size in all formulations tested (Table S1†).PLGA NPs with low concentrations of CS (0.01% w/v) showed an increase in the ζ-potential (23.00 ± 4.32 mV), indicating the successful coating and the presence of positive charge on NPs’ surface due to the protonation of amine groups in chitosan. When a higher concentration of CS is used to coat NPs (0.1% w/v), an increase in the ζ-potential (49.33 ± 1.25 mV) was observed and found to be proportional to the CS concentration used. For this reason, and as the ζ-potential impacts on particle stability, cell adhesion and interaction with negatively charged biomaterials,18 it was decided to further use CS-coated PLGA NPs obtained with higher concentrations of CS (i.e., 0.1% w/v, Fig. S2†).
TEM images confirmed that the coating process did not impact the morphology of PLGA NPs, with all NPs showing a spherical morphology (Fig. 1E and F). Furthermore, TEM images do not demonstrate variations in the size of PLGA NPs without/with CS coating; the slight increase in size measured by DLS could be caused by the presence of a hydrated thin layer of CS (proportional to CS concentration used in the NP coating step) and not detected in dry particles analysed by TEM (Fig. S3†).
NanoSight tracking analysis was further used to determine the concentration of CS/PLGA NPs (obtained with the optimised protocol, 0.1% w/v CS concentration) and the uncoated control in the dispersed phase. The precise quantification of NP concentration is considered essential to further dose hydroxyl-FK866-PLGA NPs in the alginate-based hydrogels for further drug delivery studies.27 Herein, we compared two methods to determine the concentrations of PLGA nanoparticles: NanoSight and DLS. The concentrations of uncoated PLGA NPs measured in both methods are similar, that is, (2.68 ± 0.61) × 107 particles and (2.46 ± 0.34) × 107 particles, measured respectively with DLS and NanoSight. In the case of CS/PLGA NPs, variations in the measured concentrations are observed (in the range of 15–20%). Again, differences in the detection method (light scattering vs. image analysis) could affect the returned measure in the presence of the CS layer: a concentration of (3.98 ± 0.17) × 108 CS/PLGA NPs is measured with DLS being much higher than the (3.30 ± 0.02) × 107 CS/PLGA NPs detected by the Nanosight, and more similar to the controls.
3.2. Rheology
The rheological properties were evaluated on the materials before (biomaterial ink) and after printing (crosslinked hydrogel), as previously reported.28–30 Firstly, the viscosity and viscoelasticity of biomaterial inks (i.e., aqueous polymeric solution) before printing were analysed to assess their flow properties and suitability to be processed with extrusion-based bioprinters. Of note, this characterisation could be used to evaluate their injectability. Then, the viscoelastic properties of the crosslinked hydrogels were analysed to compare the mechanical properties of the hydrogels to those of breast tissues, both healthy and tumoral. | ||
| Fig. 2 (A) Viscosity average curves for all the tested conditions acquired with a rotational shear rate sweep. (B) Oscillatory stress sweep tests for biomaterial ink 3 before extrusion, as representative curves for all the conditions. (C) Oscillatory stress sweep tests for biomaterial ink 3 crosslinked, as representative curves for all the conditions. (D) Mean storage and loss moduli acquired from oscillatory stress sweep tests in the linear viscoelastic region of the crosslinked inks. For biomaterial inks composition refer to Table 1. Data presented as average ± st. dev. (n = 3). | ||
The viscoelastic properties of the biomaterial inks before extrusion have been assessed; an example of the flowing properties of the biomaterial ink 3 (Table 1) is shown in Fig. 2B, highlighting the presence of yield stresses and showing that the biomaterial ink behaves as a viscous fluid (G′′ > G′). This was as expected since the solutions tested were not crosslinked before extrusion. Moreover, the yield stress, which delimits the linear viscoelastic region, occurred at around 400 Pa, predicting the possibility of printing these solutions.29 In fact, previous studies reported that values of yield stress higher than 100 Pa led to acceptable printing outcomes.30–33
Additionally, results showed that the presence of PLGA NPs (without/with CS coating) did not impact the hydrogel mechanical properties due to a low dispersion of particles in the hydrogel network. All hydrogels showed G′ > G′′, denoting the dominant elastic character of hydrogels, which suggested a completely gelled and elastic structure driven only by the presence of physical ionic bonds, and with the presence of viscous components.36
3.3. Printability assessment
Three-dimensional (3D) printability of a biomaterial ink is defined as the ability of a hydrogel to form and maintain a reproducible 3D structure with dimensional integrity.37 Rheological properties and printing parameters, such as extrusion pressure and speed, can affect print quality. For example, a low pressure would lead to biomaterial ink extrusion failure, a high pressure could cause poor printability, and a low printing speed would result in a high quantity of extruded material per unit time, resulting in poor printability. High-speed print nozzles can also result in discontinuous filaments.38In this study, firstly we defined the optimal printing parameters for the biomaterial ink 1 (without NPs) as we did not find differences in the rheological properties of the biomaterial inks with PLGA NPs (Fig. 2A). Then, we validated the optimal printing parameters for the biomaterial ink 3 with the highest concentration of PLGA NPs (20 mg mL−1) and the biomaterial ink 5 with 20 mg mL−1 CS/PLGA NPs (Table 1) as shown in Fig. 3F.
The UF and the SR of biomaterial ink 1 at different pressures/velocities were evaluated on a simple printed line (Fig. 3A). UF values close to 1 (Fig. 3B) were found for most of the tested parameters, which is the ideal value as UF = 1 is an indication of a smooth extrusion, while UF > 1 represents over-gelated hydrogels.
This result was as expected since the biomaterial ink 1 has a viscous-like behaviour with G′′ > G′. From the analysis of the SR, it is possible to observe that all the tested combinations have SR > 1 (Fig. 3C). Thus, they have a higher width than the one designed with the CAD. However, SR values below 2 are rarely present in the literature due to the viscoelastic nature of the material used, due to the relaxation of the polymeric chains. SR is lower with the decrease of pressure and increase in velocity.
To quantitatively evaluate the shape fidelity of biomaterial ink 1, we evaluated the Pr values and grid morphology under different pressures and speeds (Fig. 3D and E). In specific, biomaterial ink 1 ensured adequate flow conditions, with smooth and uniform filaments being continuously extruded, whilst having perfect printability state of geometries with square-shaped interconnecting channels and Pr values of 1; literature reports that when Pr is in the range of 0.8–1.1, the 3D printed hydrogel structures will exhibit good filament morphology and mechanical stability.39
We used biomaterial ink 1 for all printability tests and selected the combination of pressure and velocity which had the lowest SR and a Pr value close to 1 as the optimal printing parameters. Thus, the optimal printing parameters were 140 kPa and 10 mm s−1.
Then, we evaluated the shape fidelity with the measure of the printability index of different biomaterial inks (i.e., biomaterial ink 1, biomaterial ink 3 and biomaterial ink 5, Table 1) printed with the optimal parameters; as shown in Fig. 3F, all biomaterial inks used have good printability and the presence of the NPs (regardless of the CS coating) does not affect the printability of the biomaterial ink.
3.4. Composite alginate-based hydrogels: interaction and distribution of PLGA-NPs in the hydrogel
Two distinctive imaging acquisition methods were used: atomic force microscopy (AFM) to interrogate the interaction between PLGA and CS/PLGA NPs and the alginate network and understand if any physical and/or chemical interaction may occur and laser scanning confocal microscopy (LSCM) to assess volumetric distribution of Rho-PLGA NPs within alginate-based hydrogels (Table 2). Of note, and due to the relatively low concentration of PLGA NPs used in the formulation of biomaterial inks, higher concentrations of PLGA NPs and different alginate formulations were selected to better evaluate the interactions occurring between positive (CS/PLGA NPs) and negative (PLGA NPs) charges within the hydrogel network (supposedly negative, due to the presence of hydrated alginate chains), and the concentration of PLGA NPs was increased only for sample preparation for AFM studies (Table 2). | ||
| Fig. 4 AFM images of topography and phase contrast images of a 500 × 500 nm scan area of: (A) ALG; (B) OA5; (C) ALG-PLGA NPs; (D) OA5-PLGA NPs; (E) ALG-CS/PLGA NPs; and (F) OA5-CS/PLGA NPs. | ||
Both alginate films (ALG and OA5) were characterised by a granular structure, as expected. While in the case of ALG, grains are characterised by an elongated shape ranging several hundred microns, and OA5 grains are more circular, with a diameter of a few tenths microns (Fig. 4B, phase contrast). This confirms our previous study that the oxidation of alginate causes a reduction of the molecular weight.16
The loading of uncoated PLGA NPs (207 ± 20 nm; −21.90 ± 2.73 mV) showed different interactions as a function of the polysaccharide used. An evident segregation of NPs at the surface with ALG with poor interfacial affinity is observed, possibly due to the negative charge of both PLGA NPs and ALG, which is emphasized by the film retraction caused by the polymer drying (Fig. 4C). Differently, when PLGA NPs are embedded in OA5 (Fig. 4D), a higher interaction is observed between NPs and the modified polysaccharide matrix, with no evidence of surface segregation and no significant difference in morphology from that of a pure OA5 film (Fig. 4B) with a slight enhancement of the globular edges.
The loading of CS/PLGA NPs (245 ± 20 nm, +49.33 ± 1.25 mV) within alginate-based solutions showed different levels of interactions with the alginates. In the case of ALG (Fig. 4E), there is still some level of segregation between particles and matrix; however, the nanoparticles are visible at the surface and appear to be coated by an alginate layer. This phenomenon is probably due to the opposite charges that characterise ALG (negatively charged) and CS/PLGA NPs (positively charged), which resulted in an overall improvement of the interfacial interactions. Interestingly, OA5 and CS/PLGA NPs show no evidence of NP segregation at the surfaces (Fig. 4F), suggesting a further degree of interaction between the primary amine group on CS chains (hence on the NP surface) and the aldehyde groups on oxidized alginate. Moreover, and as observed for the OA5 and PLGA NPs composite hydrogel (Fig. 4D), the globular morphology of the OA5 matrix is enhanced with respect to ALG, leading to a slight increase in the Z topography profile.
3.5. Cytotoxicity of hydrogels loaded with hydroxyl-FK866-PLGA NPs
Hydroxyl-FK866, a cytotoxic compound, is a highly specific inhibitor of the biosynthesis of the coenzyme nicotinamide adenine dinucleotide, which could be used for cancer treatment. In our previous work, we have shown that hydroxyl-FK866-PLGA NPs are able to release hydroxyl-FK866 by a hydrolytically cleavable linkage for up to 3 months under physiological conditions with effective toxicity to cancer cells (Fig. S2A†).11The hydroxyl-FK866-PLGA NPs used in this work have an encapsulation efficiency of 98.6 ± 5.8% (data not shown). The presence of CS coating slightly increases the Z-average size of NPs (Fig. S2B†), as well as switching the ζ-potential from negative (hydroxyl-FK866-PLGA NPs) to positive (CS/hydroxyl-FK866-PLGA NPs) values (Fig. S2C†).
Prior to loading in biomaterial inks, the toxicity of hydroxyl-FK866-PLGA NPs was assessed using PLGA NPs as controls (Fig. 6A). As expected, the drug-loaded NPs were toxic to the cells regardless of the presence of CS coating (no statistical difference between hydroxyl-FK866-PLGA NPs and CS/hydroxyl-FK866-PLGA NPs); in specific, the release of hydroxyl-FK866 impacted cell viability more than the charge of NPs (typically positively charged NPs are reported to be more toxic40). Of note, the concentration of CS used to coat NPs is well reported in the literature and found to not impact cell viability;41,42 our study aligns with the literature, showing no significant difference in viability between CS/PLGA NPs and untreated controls (Fig. 6A).
Then, sterile biomaterial inks 1, 5, 6 and 7 were formulated (Table 1), crosslinked with 50 mM sterile CaCl2 solution (aq.) and hydrogels incubated with MDA-MB-231 cells.
All nanoparticles used (i.e., CS/PLGA NPs, hydroxyl-FK866-PLGA NPs and CS/hydroxyl-FK866-PLGA NPs) were at a concentration of 20 mg mL−1, concentration previously selected as ensuring release of hydroxyl-FK866 at toxic doses in the observed time point (48 h). Alginate only hydrogel (biomaterial ink 1) and CS/PLGA NP loaded alginate hydrogels (biomaterial ink 5) were well tolerated by cells, showing viability of 90–95% and similar to untreated control (100%, Fig. 6B). As expected, and regardless of the presence of CS coating, both hydrogels loading hydroxyl-FK866-PLGA NPs were found to be toxic (biomaterial inks 6 and 7), without statistically significant difference at the observed timepoint.
These results showed that the viability of cells treated with drug-loaded NPs dispersed in a medium (hydroxyl-FK866-PLGA NPs and CS/hydroxyl-FK866-PLGA NPs, Fig. 6A) was lower than the one observed when the same NPs were loaded in alginate hydrogels (Fig. 6B), being 36 ± 1% vs. 52 ± 2%. Such different cell responses could be explained considering that hydroxyl-FK866 released by NPs in a hydrogel should diffuse throughout the polysaccharide matrix and, thus, is made available to cells with a delay.
In this work, we used a small hydrophilic cytotoxic compound (hydroxyl-FK866) with molecular weight (MW) well below the one of alginate.16 Considering that the diffusion coefficient of molecules with MW < 2 × 104 in alginate hydrogels is similar to those in the water system,43 it was possible to assume that no difference was present in the diffusive profiles of hydroxyl-FK866 within alginate hydrogels. Therefore, the only driving force for hydroxyl-FK866 release resides in the hydrolysis from PLGA in hydroxyl-FK866-PLGA NPs, hence depending on the pH of the microenvironment. Since alginate hydrogels can immediately reach an equilibrium with the environment pH,43,44 it is possible to assume that the hydroxyl-FK866-PLGA NPs are exposed to a constant pH. In our previous study,11 the effective release of hydroxyl-FK866 was characterized at different pH values, mimicking both the tumoral (pH = 6.4) and healthy (pH 7.4) environmental pH of breast tissues and with results displaying a prolonged release of low doses of the cytotoxic compound up to two months.
Overall, results show that alginate-based hydrogels could act as depot drug delivery systems for different treatments, such as cancer. Based on our previous work, we can corroborate that hydroxyl-FK866-PLGA NPs could provide sustained release even when loaded and interlinked in the polysaccharide network, constraining NPs at a given location and ensuring the release of the therapeutic for prolonged periods of time, thereby minimizing systemic toxicity and reducing dosing frequency.
4. Conclusions
A novel alginate-based biomaterial ink loading and confining PLGA NPs for the localised and sustained release of potent cytotoxic compounds for cancer treatment has been presented in this study. CS/PLGA NPs were found homogeneously dispersed and confined within oxidized alginate (OA5) hydrogels via covalent crosslinks (primary amines and aldehydes). Composite biomaterial inks (9% w/v ALG-OA5; 20 mg mL−1 PLGA NPs) showed shear thinning properties with excellent printability (mean Pr in the range of 0.80–0.95). Rheological characterization of composite hydrogels showed mechanical properties compatible with soft tissues (e.g., breast tissue) and, overall, in line with the maintenance of cell phenotype and not triggering inflammatory responses due to mechanical mismatch. Combinations of hydroxyl-FK866-PLGA NPs and alginates were used to treat triple-negative human breast cancer (MDA-MB-231), showing toxicity of all drug-loaded systems (30–40% viability at 48 h) when compared to the controls. This study demonstrates the loading and confinement of highly toxic compounds (CS/hydroxyl-FK866-PLGA NPs) within injectable alginate-based hydrogels for the local treatment of solid tumours. Such a hybrid system can facilitate the delivery of toxic small chemotherapeutics at the tumour site, improving the efficacy of treatments against cancer cells. Further long-term in vitro studies are required to clearly demonstrate the effectiveness in the local and sustained release of hydroxyl-FK866, as well as the confinement of CS/PLGA NPs in modified alginate hydrogels for both in vitro and in vivo applications.Author contributions
Eugenia Spessot and Xue Bai are responsible for conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, and writing – review & editing. Daniel Moranduzzo is responsible for methodology, formal analysis and visualization. Chen Zhao is responsible for methodology. Sam Butterworth is responsible for conceptualization, supervision, funding acquisition and writing – review & editing. Devid Maniglio is responsible for formal analysis, methodology, visualization and writing – review & editing. Annalisa Tirella is responsible for conceptualization, data curation, formal analysis, funding acquisition, supervision, visualisation, and writing – review & editing.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
All the authors would like to thank Prof. Michela Denti and Miss Ilaria Brentari (Department of Cellular, Computational and Integrative Biology, University of Trento, Italy) for their help in the acquisition and interpretation of NanoSight data. XB is grateful to Dr Francesca Agostinacchio (Biotech – Department of Industrial Engineering, University of Trento, Italy) for the help in the acquisition of confocal microscopy images. ES and AT are grateful for the support of Fondazione VRT. ES, DM, and AT acknowledge the SHIFT Project, funded by the European Union's Horizon 2020 Research and Innovation Programme under the Maria Sklodowska-Curie grant agreement no. 101008041.References
- C. Pacheco, A. Baião, T. Ding, W. Cui and B. Sarmento, Adv. Drug Delivery Rev., 2023, 194, 114724 CrossRef CAS PubMed.
- Q. Leng, Y. Li, P. Zhou, K. Xiong, Y. Lu, Y. Cui, B. Wang, Z. Wu, L. Zhao and S. Fu, Mater. Sci. Eng., C, 2021, 129, 112390 CrossRef CAS PubMed.
- A. Spadea, A. Tirella, J. M. Rios de la Rosa, E. Lallana, M. Mehibel, B. Telfer, N. Tirelli, M. J. Lawrence, K. J. Williams, I. J. Stratford and M. Ashford, Pharmaceutics, 2024, 16, 1286 CrossRef CAS PubMed.
- Y. Xie, M. Liu, C. Cai, C. Ye, T. Guo, K. Yang, H. Xiao, X. Tang and H. Liu, Front. Oncol., 2023 DOI:10.3389/fonc.2023.1027254.
- J. Zheng, X. Song, Z. Yang, C. Yin, W. Luo, C. Yin, Y. Ni, Y. Wang and Y. Zhang, J. Controlled Release, 2022, 350, 898–921 CrossRef CAS PubMed.
- X. Bai and A. Tirella, Int. J. Drug Discovery Pharmacol., 2022, 10–10 CrossRef.
- F. Perin, E. Spessot and A. Motta, in Multiscale Cell-Biomaterials Interplay in Musculoskeletal Tissue Engineering and Regenerative Medicine, ed. J. M. Oliveira, R. L. Reis and S. Pina, Academic Press, 2024, pp. 219–240 Search PubMed.
- J. Min Jung, Y. Lip Jung, S. Han Kim, D. Sung Lee and T. Thambi, J. Colloid Interface Sci., 2023, 636, 328–340 CrossRef CAS PubMed.
- I. V. Zelepukin, O. Y. Griaznova, K. G. Shevchenko, A. V. Ivanov, E. V. Baidyuk, N. B. Serejnikova, A. B. Volovetskiy, S. M. Deyev and A. V. Zvyagin, Nat. Commun., 2022, 13, 6910 CrossRef CAS PubMed.
- Y. Jiang, N. Krishnan, J. Heo, R. H. Fang and L. Zhang, J. Controlled Release, 2020, 324, 505–521 CrossRef CAS PubMed.
- X. Bai, S. Tang, S. Butterworth and A. Tirella, Biomater. Adv., 2023, 154, 213649 CrossRef CAS PubMed.
- Y. A. Khan, K. Ozaltin, A. Bernal-Ballen and A. Di Martino, J. Drug Delivery Sci. Technol., 2021, 61, 102126 CrossRef CAS.
- F. Abasalizadeh, S. V. Moghaddam, E. Alizadeh, E. Akbari, E. Kashani, S. M. B. Fazljou, M. Torbati and A. Akbarzadeh, J. Biol. Eng., 2020, 14, 8 CrossRef CAS PubMed.
- C. Gao, M. Liu, J. Chen and X. Zhang, Polym. Degrad. Stab., 2009, 94, 1405–1410 CrossRef CAS.
- T. Boontheekul, H.-J. Kong and D. J. Mooney, Biomaterials, 2005, 26, 2455–2465 CrossRef CAS PubMed.
- C. Zhao, A. Latif, K. J. Williams and A. Tirella, React. Funct. Polym., 2022, 175, 105292 CrossRef CAS.
- A. Bucciarelli, C. Zhao, X. Bai, A. Latif, K. Williams and A. Tirella, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-7bt78.
- I. A. Duceac and S. Coseri, Gels, 2022, 8, 779 CrossRef CAS PubMed.
- N. Amiryaghoubi, M. Fathi, A. Safary, Y. Javadzadeh and Y. Omidi, Int. J. Biol. Macromol., 2024, 256, 128335 Search PubMed.
- S. E. Samuel, T. Ghosh, A. Damodar Nayak, R. Deveswaran and B. V. Basavaraj, J. Drug Delivery Sci. Technol., 2024, 97, 105746 CrossRef CAS.
- Zetasizer family software update v8.02, https://www.malvernpanalytical.com/en/support/product-support/software/zetasizer-family-software-update-v8-02, (accessed 25 October 2024).
- F. Perin, E. Spessot, A. Famà, A. Bucciarelli, E. Callone, C. Mota, A. Motta and D. Maniglio, ACS Biomater. Sci. Eng., 2023, 9, 1320–1331 CrossRef CAS PubMed.
- J. M. Rios De La Rosa, A. Spadea, R. Donno, E. Lallana, Y. Lu, S. Puri, P. Caswell, M. J. Lawrence, M. Ashford and N. Tirelli, Sci. Rep., 2020, 10, 14505 CrossRef CAS PubMed.
- D. Nečas and P. Klapetek, Open Phys., 2012, 10, 181–188 CrossRef.
- B. Lu, X. Lv and Y. Le, Polymers, 2019, 11, 304 CrossRef PubMed.
- H. M. Aldawsari, N. A. Alhakamy, R. Padder, M. Husain and S. Md, Coatings, 2020, 10, 439 CrossRef CAS.
- N. G. Khlebtsov, Anal. Chem., 2008, 80, 6620–6625 CrossRef CAS PubMed.
- M. Chopin-Doroteo, E. A. Mandujano-Tinoco and E. Krötzsch, Biochim. Biophys. Acta, Gen. Subj., 2021, 1865, 129782 CrossRef CAS PubMed.
- A. Schwab, R. Levato, M. D'Este, S. Piluso, D. Eglin and J. Malda, Chem. Rev., 2020, 120, 11028–11055 Search PubMed.
- D. A. Rau, M. J. Bortner and C. B. Williams, Addit. Manuf., 2023, 75, 103745 CAS.
- C. Minas, D. Carnelli, E. Tervoort and A. R. Studart, Adv. Mater., 2016, 28, 9993–9999 CrossRef CAS PubMed.
- S. S. L. Chan, R. M. Pennings, L. Edwards and G. V. Franks, Addit. Manuf., 2020, 35, 101335 Search PubMed.
- D. Kokkinis, M. Schaffner and A. R. Studart, Nat. Commun., 2015, 6, 8643 CrossRef PubMed.
- A. M. Teixeira and P. Martins, Front. Bioeng. Biotechnol., 2023 DOI:10.3389/fbioe.2023.1161815.
- N. Shpaisman, L. Sheihet, J. Bushman, J. Winters and J. Kohn, Biomacromolecules, 2012, 13, 2279–2286 CrossRef CAS PubMed.
- F. Perin, C. Mota, I. Mancini, A. Motta and D. Maniglio, Polymer, 2022, 252, 124941 CrossRef CAS.
- S. Naghieh, M. D. Sarker, N. K. Sharma, Z. Barhoumi and X. Chen, Appl. Sci., 2020, 10, 292 CrossRef CAS.
- S. Naghieh and X. Chen, J. Pharm. Anal., 2021, 11, 564–579 CrossRef PubMed.
- L. Ouyang, R. Yao, Y. Zhao and W. Sun, Biofabrication, 2016, 8, 035020 Search PubMed.
- H. Chen, W. Yang, H. Chen, L. Liu, F. Gao, X. Yang, Q. Jiang, Q. Zhang and Y. Wang, Colloids Surf., B, 2009, 73, 212–218 CrossRef CAS PubMed.
- C. Lee, J. S. Choi, I. Kim, K. T. Oh, E. S. Lee, E.-S. Park, K. C. Lee and Y. S. Youn, Int. J. Nanomed., 2013, 8, 2975–2983 Search PubMed.
- M. Trif, P. E. Florian, A. Roseanu, M. Moisei, O. Craciunescu, C. E. Astete and C. M. Sabliov, J. Biomed. Mater. Res., Part A, 2015, 103, 3599–3611 Search PubMed.
- H. Tanaka, M. Matsumura and I. A. Veliky, Biotechnol. Bioeng., 1984, 26, 53–58 CrossRef CAS PubMed.
- A. Tirella, M. La Marca, L.-A. Brace, G. Mattei, J. W. Aylott and A. Ahluwalia, J. Biomed. Nanotechnol., 2015, 11, 1451–1460 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4pm00334a |
| ‡ First co-authors. |
| § Present address: Institute of Food Science and Technology, Chinese Academy of Agricultural Sciences/Key Laboratory of Agro-Products Processing, Ministry of Agriculture, Beijing 100193, China. |
| This journal is © The Royal Society of Chemistry 2025 |




