Open Access Article
Open Access ArticleSpecific phosphorylation patterns control the interplay between aggregation and condensation of Tau-R4 peptides†
Shachar Guy
Bressler‡
ae,
Dana
Grunhaus‡
ae,
Amit
Aviram
a,
Stefan G. D.
Rüdiger
 bcd,
Mattan
Hurevich
bcd,
Mattan
Hurevich
 *ae and
Assaf
Friedler
*ae and
Assaf
Friedler
 *ae
*ae
aThe Institute of Chemistry, The Hebrew University of Jerusalem, Edmond J. Safra Campus, Givat Ram, Jerusalem, 91904, Israel. E-mail: assaf.friedler@mail.huji.ac.il; mattan.hurevich@mail.huji.ac.il
bProtein Chemistry of Disease, Department of Chemistry, Utrecht University, Padualaan 8, 3584CH Utrecht, The Netherlands
cCellular Protein Chemistry, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584CH Utrecht, The Netherlands
dScience for Life, Utrecht University, Padualaan 8, 3584CH Utrecht, The Netherlands
eCenter for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem, Jerusalem 9190401, Israel
First published on 19th June 2025
Abstract
Specific phosphorylation patterns regulate the activity of proteins and play a central role in protein self-assembly. In Tau, such patterns drive the formation of disease-related condensates and aggregates. Understanding their functional impact is essential for studying Tauopathies such as Alzheimer's Disease. Here we show how specific phosphorylation patterns regulate Tau self-assembly and control the interplay between its aggregation and condensation, using a peptide-based approach that allows systematic analysis of libraries of specific phosphorylation patterns at the domain level and is complementary to the current protein-level methods. We applied our methodology to study the effect of specific phosphorylations on the aggregation and condensation of the R4 domain of Tau that is pivotal for its self-assembly, forming the β-helix motif that is common to various Tau patient fibrils. Using advanced phosphopeptide synthesis methods developed in our labs, we generated a library of multi-phosphorylated peptides derived from Tau R4. We found that phosphorylation at Ser341 promotes aggregation, while Ser352 enhances condensation. Phosphorylation at Ser356 inhibits both processes. The source of these different outcomes is the distinct microenvironments around each phosphorylated site. Our results provide a residue-level resolution of how the decision between Tau condensation and aggregation is being made. This was possible by using our peptide-based approach, which is complementary to protein-level method and enables efficient identification of active phosphorylation patterns. These can later be studied at the protein level.
Introduction
Phosphorylation is one of the most abundant posttranslational modifications (PTM) in eukaryotes.1,2 Phosphorylation has a key role in regulating protein–protein interactions (PPI) and self-assembly of monomeric protein to oligomers.3–5 The most prevalent phosphorylation sites occur on serine and threonine, and most phosphoproteins are phosphorylated at multiple sites.4,6 Proteins can oligomerize to different types of assemblies, ranging from coacervates formed via liquid–liquid phase separation (LLPS) to aggregates of different types such as amorphous, native-like, or amyloid fibrils.7 In cells, coacervates function as membrane-less organelles, which have several possible functions such as bioreactors that catalyze protein interactions and cellular processes.8–10 Aggregation, however, is frequently associated with pathological pathways that lead to disease.11Different phosphorylation patterns can regulate protein activity and dictate which pathway the protein will undergo. They regulate PPI and may control the oligomerization type of proteins, selecting between aggregation and condensation.3,12 The existence of multiple phosphorylation sites in proteins means that there are many possible phosphorylation patterns, each with a unique effect on oligomerization. The ideal way for studying the effect of each such pattern is by obtaining a library of multi-phosphorylated derivatives of the protein of interest, each with a unique and specific phosphorylation pattern. Such a library would cover all relevant patterns. However, obtaining such a diverse multi-phosphorylated protein library in sufficient quantities for extensive biophysical, biochemical and structural studies is difficult. The methods for obtaining recombinant phosphorylated proteins for in vitro biophysical, biochemical and structural studies include kinase-mediated phosphorylation, the use of “phosphomimetic” amino acids and semisynthetic methods. These methods are complementary, each having its pros and cons, raising the need for using more than one method to obtain the full picture. There is thus a need for additional methods to be added to the existing toolbox.
Replacing phosphorylated amino acids with other negatively charged residues is very popular.13,14 This common substitution of phosphoserine by negatively charged glutamate or aspartate (commonly referred to as phosphomimic residues) allows for performing large scale studies at the protein level.15 This is an easy and fast method to make libraries. However, using this method results in a significant compromise as there is a crucial difference between a phosphate group and a carboxylic group. Replacing a phosphoserine or phosphothreonine by glutamate or aspartate retains only one negative charge at the specific site but overlooks the unique chemical and physical properties of the phosphate group.13 A phosphate has two negative charges at physiological pH, while glutamate has only one. Moreover, the pKa of phosphate is 5.58, while the pKa of glutamate is 4.25.16 The phosphate also occupies much larger volume that is derived from its tetrahedral geometry compared to the planar carboxylate group. These factors govern the distinctive interplay between the phosphorylated residue and its surroundings, dictating the role of the phosphate at each specific microenvironment within the protein. Comparative studies showed that Glu substitution of the phosphorylated Ser did not necessarily result in the same protein structure or function.17,18 It was demonstrated for α-synuclein that while phosphorylation of Ser129 inhibited fibrillation, mutation to Asp or Glu did not reproduce this activity.19 This difference is prevalent in the study of condensation, as it is a highly charge-dependent process.20In vitro phosphorylation of recombinant proteins by kinases although widely used, introduces constraints, as it is usually results in a mixture of proteins with different phosphorylation patterns that are difficult to isolate and analyze, limiting our understanding of their precise functional roles.21 Enzymatic phosphorylation by different kinases is not totally selective.21 A prior knowledge of each specific kinase is needed for achieving the desired specific phosphorylation is required.18 The above limits the access to many possible phosphorylation patterns that can be of high importance. To obtain the specific phosphorylation pattern with high selectivity and high yields, chemical peptide synthesis should rather be used.
Semisynthetic strategies, which include the chemical synthesis of a phosphorylated domain followed by its ligation to a recombinant protein (such as native chemical ligation), have the advantage of introducing the actual phosphorylation pattern into the protein. However, these approaches are time consuming, low yielding processes that provide only small quantities of the desired proteins and are completely impractical for the preparation of libraries with phosphorylation patterns.22,23 Thus, while excellent for studying a specific pattern at the protein level, these approaches are not suitable for identifying which pattern is the active one since it is practically impossible to produce libraries and systematically study them using the mentioned semisynthetic methods. Most importantly, protein synthesis depends on the availability of the corresponding multiphosphopeptides which is still a significant problem. Since the peptides are later incorporated into the proteins using NCL, the successful synthesis of the peptides is crucial for the success of the NCL later and is another layer in the process. If the peptides are difficult to make, the proteins could not be made either. The synthesis of proteins hence depends on the development of synthetic methods to prepare multi-PTM peptides and demonstrate their relevance in studies such as the one presented here and as described below.
The synthesis of long, multiphosphorylated peptides remains challenging due to the steric bulk and lability of protected phospho-serine and phospho-threonine residues under standard Fmoc conditions. These factors limit coupling efficiency and overall yield. Despite these hurdles, we have established robust synthetic method that enables the efficient production of phosphorylated peptide libraries derived from aggregation-prone domains.24–27 The method is based on a combination of high temperature and fast mixing for accelerating each coupling step while minimizing the formation of side products and maximizing the efficiency.25 Using this method, we demonstrated the synthesis of peptides containing up to eight phosphorylation sites. The above strategy is ideal for synthesizing libraries of multiphosphorylated peptides. These libraries can then be used for systematic studies of the effect of specific phosphorylation patterns on the biological activity of proteins. Here we applied our method for studying the aggregation processes of the Tau protein.28,29
The Tau protein forms amyloid fibrils in neurons as part of the pathology of neurodegenerative diseases (ND). in vivo, fibrils of Tau are hyperphosphorylated,30 an event that is linked to aggregation and condensate formation by the protein.21,31–34 However, the role of the specific phosphorylation patterns in controlling the initial molecular events leading to amyloid aggregates or condensates is mostly unclear. Tau is a highly disordered protein that is composed of an N-terminal domain, a proline-rich domain, microtubule binding domain (RD) with five homologous repeats (R1–R4, R′) and a C-terminal part (Fig. 1A). In Tauopathies such as Alzheimer's disease (AD) and chronic traumatic encephalopathy (CTE), Tau undergoes aggregation into amyloid fibrils via its RD. Fibrils from AD and CTE patients share a hairpin with a hydrophobic core that is formed by the fourth repeat (R4) of Tau-RD (β-helix; Fig. 1B). In fibrils, R4 has three beta-sheets, interconnected by disordered linkers. In addition, Tau-RD is essential for the condensation of Tau.34,35 Although it is known that phosphorylation of the full-length Tau initiates aggregation and condensation,21,36 there is almost no information about the effect of specific phosphorylations, mostly due to the difficulty in obtaining Tau derivatives with specific phosphorylation patterns. For example, single phosphorylations at Ser293 in R2 or Ser305 in R3 promote amyloid fibril formation in vitro.37 However, the effect of specific phosphorylation patterns on oligomerization of R4 is yet unknown. R4 has three sites that were found to be phosphorylated in ND: Ser341, Ser352 and S356. Each of these serine residues is located at a different chemical environment. Ser341 is located between two charged residues, Lys340 and Glu342, within the disordered linker. Ser352 is positioned next to Gln351 and Lys353 and is located within a loop structure, while Ser 356 is located between two aliphatic residues, Gly355 and Leu357 at the end of the loop of R4. The side chains of Ser341 and Ser352 are facing toward the core of the hairpin, while Ser356 is facing out (Fig. 1C).
 | ||
| Fig. 1 The Tau R4 domain (residues 336–358). Tau 336–358 is derived from the R4 repeat and forms a hairpin structure within native Tau fibrils. (A) Domain structure of Tau: the N-terminal part (N-ter), the proline-rich domain (PD), the repeat domain (RD) with its five repeats R1–R4, R′ (highlighted), and the C-terminus part (C-ter). The sequence of Tau R4 (serine 341 in red, 352 in magenta and 356 in purple). (B) Cryo-EM structures of Tau filaments isolated from patients of different Tauopathies: AD amyloid filament (PDB ID: 5O3T),38 CTE amyloid filaments (PDB: 6NWQ);39 Tau R4 that adopts a hairpin structure is framed. (C) The hairpin structure that Tau R4 forms in AD PHF patients. Serine 341 labeled in red, 352 in magenta and 356 in purple. hydrophobic residues in black. | ||
Due to the challenges associated with synthesizing full-length Tau protein, studies of Tau self-assembly often rely on the use of Tau-derived fragments. These fragments vary in length, from short hexapeptides such as those derived from key amyloidogenic motifs in the R3 and R2 domains40 to larger constructs of ∼100 residues, including the First Intermediate Amyloid (FIA) fragment41 and well-established aggregation models such as K18 and K19.42 Peptides are vastly used to explore molecular mechanisms of canonical domains that undergo phase separation.43,44 Peptides have several advantages in bridging the gap between the authentic yet inaccessible multiphosphorylated proteins to the much-needed biophysical and biochemical input. First, synthesis of peptides can provide each specific pattern at high purity, specificity and reliability. Second, synthetic processes guarantee that the peptides can be obtained reproducibly and at a sufficient quantity. Third, peptides can be labeled by different chemical tags to enable analysis via diverse biochemical assays. Most importantly: they provide an easy and accessible route to libraries.
Here we harnessed our novel approaches for multiphosphorylated peptide synthesis12,24,45 to study systematically how specific phosphorylation pattern affect aggregation and condensation of the Tau 336–358 (Tau R4 domain). We designed and synthesized a library of peptides representing the R4 domain and containing all the possible phosphorylation patterns. Then, we determined whether each peptide undergoes condensation or aggregation by fluorescence and electron microscopy. We found that the fully phosphorylated peptide can condensate or aggregate depending on the conditions, while the non-phosphorylated peptide cannot oligomerize at all. Phosphorylation of Ser341 promoted aggregation, while phosphorylation of Ser352 promoted condensation. Our research emphasizes the importance of a methodological study that explores the effects of phosphorylation patterns on oligomerization using phosphorylated peptide libraries, as a complementary method for the other protein-level methods that are currently in use.
Results
Synthesis of a phosphorylated peptide library derived from Tau R4
To study the effect of phosphorylation on the condensation and aggregation of R4, we synthesized a library of peptides with patterns including all combinations of phosphorylations in Ser341, Ser352, and Ser356. The peptides were synthesized using our newly developed Accelerated MPP synthesis,24 which enabled us to synthesize the library in a short time and in sufficient amounts (see Table 1 for peptide sequences and ESI Fig. 1 and 2† for the analytical data of the peptides). The synthesis was performed with a constant overhead stirring of 1200 rpm, at 90 °C. The assembly of each peptide was completed in less than 35 min. Sixteen fluorescent and non-fluorescent peptides were obtained in over 95% purity (ESI Fig. 1 and 2†) and multi-mg scale in less than two weeks of synthesis. The peptides were then studied for their aggregation and condensation using biophysical analyses as described below.Full phosphorylation of Tau R4 promotes its aggregation and condensation
To test if the phosphorylation of Tau R4 affects its aggregation and condensation, we first compared the fully phosphorylated (3P) and the non-phosphorylated (0P) peptides. We performed turbidity assays as a quick screening method to test first if the peptides undergo any type of high-order oligomerization. This may indicate possible condensation or aggregation of the peptides. Mixtures of 5% Fluorescein labeled (FL) FL-0P and FL-3P with 95% non-labeled peptides were tested at NaCl concentrations of 0, 150, 500 and 1000 mM, and peptide concentrations of 50, 300 and 1000 μM. Testing the dependence of the peptide oligomerization on the ionic strength was performed to examine whether the process is charge-dependent. This is important since the phosphorylation adds extra negative charges to the peptides. 3P showed high turbidity in 1 mM peptide at all NaCl concentrations. It also showed high turbidity values in 1 M NaCl at all peptide concentrations. In contrast, 0P showed low turbidity at all tested conditions except 1 M NaCl (Fig. 2A).To test if the high turbidity of 3P resulted from aggregation or condensation, we analyzed the oligomers using fluorescence microscopy. For 3P, we observed condensates formation in 50 μM and 300 μM of peptide at all NaCl concentrations and aggregation of the peptide at 1000 μM of peptide in all NaCl concentrations. For 0P we did not observe aggregation nor condensation of peptide except at very high salt concentration (Fig. 2B and C). This also confirmed that the fluorescein labeling itself did not promote condensation or aggregation.
An important feature of self-assembly processes is their dynamics, meaning the ability of the peptides to diffuse in and out of the condensates. This defines the rate of condensate maturation which is the conversion of condensates into a more viscous gel-like phase.46 To evaluate the dynamics of the condensate formation by FL-3P and test whether they have similar properties to the condensates of the recombinant Tau,46 we used fluorescence recovery after photobleaching (FRAP). This method allows monitoring the diffusion of molecules such as peptides in and out of the condensates. Higher recovery means a higher diffusion rate and less viscous condensates. We bleached the entire 3P condensate areas 15 and 40 minutes after they were formed. ∼80% of the condensates that were bleached after 15 min recovered after 30 seconds. More mature condensates that were bleached 40 min after formation showed only partial recovery of ∼35%. In both cases, a plateau was reached at approximately 40 seconds (Fig. 2D). These results indicate that after 15 minutes the condensates are liquid-like, which allows fast diffusion of 3P in and out of the condensates. The partial recovery of the mature condensates can indicate a solidification process that led to a more gel-like character (Fig. 2D and E).
TEM and SEM images of 3P taken after 10 days (Fig. 3A and B) showed that it formed spherical aggregates of 10–60 nm. These units together formed higher-order oligomeric structures with a flower-like morphology. Under the same conditions, 0P did not aggregate even after this long incubation period (Fig. 3A and B).
Specific phosphorylation patterns control Tau-R4 condensation and aggregation
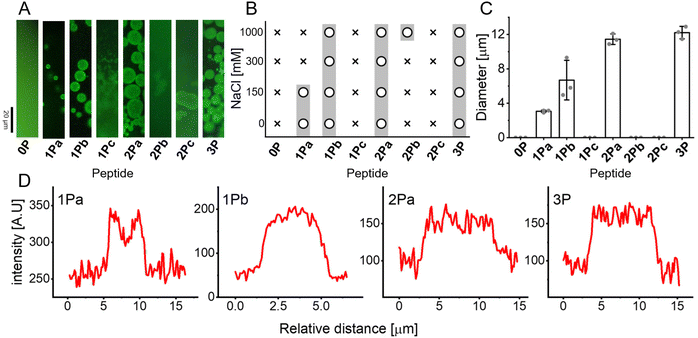
Controlling the condensation and aggregation of Tau R4, we characterized the ability of the mono and di-phosphorylated peptides to form condensates at NaCl concentrations of 0, 150, 300 and 1000 mM (Fig. 4A and B) using fluorescence microscopy. Mixtures of 5% fluorescein-labeled and 95% of non-labeled peptides were prepared. 1Pa, 1Pb and 2Pa formed condensates, while 1Pc, 2Pb and 2Pc did not form condensates. Like 3P, 1Pb and 2Pa formed condensates in 0, 150, 300 and 1000 mM NaCl. In contrast, 1Pa formed condensates only at lower salt concentrations of 0 and 150 mM. 2Pb is the only peptide that underwent condensation only at 1000 mM NaCl. These findings suggest that: (1) phosphorylation is required for condensation of Tau R4; (2) phosphorylations of Ser341 and Ser352 facilitate condensation; (3) phosphorylation of Ser356 inhibits condensation. A closer investigation of the size of the condensates showed that while 1Pa had an average diameter of ∼3.5 μm, 1Pb formed condensates with diameters that ranged between 4 to 12 μm. The largest condensates of 1Pb were in the same range as the mean diameter of 2Pa ∼12 μm, and 3P ∼12.3 μm. Condensates of 1Pa, 2Pa and 3P were uniform in their size, with relatively small standard deviations, while 1Pb showed a large range of condensate size (Fig. 4C). The intensity of the fluorescence in each condensate was examined since it is an indication of peptide concentration. We showed that while the peptide concentration of 1Pb, 2Pa and 3P was homogeneous across the condensate droplets. The edge of 1Pa condensates had higher fluorescence intensity (Fig. 4D) suggesting that peptides accumulated on the surface of the condensate, which can stop the growth of the condensates to give smaller entities.SEM was used to characterize the aggregates formed by the phosphorylated peptides after 10 days of incubation (Fig. 5A). 2Pa, 1Pb, 1Pc, 2Pb and 2Pc did not aggregate (ESI Fig. S2†). Like 3P, 1Pa formed spherical aggregates with flower-like morphology. These results suggest that phosphorylation of Ser341 contributes to the aggregation behavior of Tau R4, while phosphorylation of Ser352 and Ser356 did not contribute to the phenomena.
 | ||
| Fig. 5 (A) SEM of mono- and di-phosphorylated Tau336–358 peptides without Tau-RD (B) SEM of 0P, 1Pa, 1Pb, 2Pc and 3P Tau336–358 peptides with Tau-RD. Scale bar = 500 nm. | ||
The effect of specific R4 phosphorylations and their combinations on the Tau-RD protein (244–372) aggregation
Since it is technically impossible to study the effect of phosphorylation patterns at the protein level via a library of phosphoproteins, we decided to study the effect of phosphorylation patterns on Tau oligomerization by testing the effect of the phosphorylated Tau R4 peptide library on aggregation the full R domain of Tau (Tau-RD, residues 244–372), which is responsible for its amyloid aggregation. In vitro Tau forms amyloids via its MTBR only at the presence of an inducer such as heparin, which is highly negatively charged. Here, we tested if the phosphorylated peptides have a similar effect, due to their negative charges. When Tau-RD was incubated for 10 days with 3P, spherical aggregates with a diameter of 20–60 nm were formed, interconnected by larger amorphous aggregates. This suggests that aggregation is indeed dependent on the interactions between 3P and Tau-RD.To test the effects of the specific phosphorylation patterns at the protein level, 0P, 1Pa, 1Pb, 1Pc and 2Pc were incubated with the recombinant Tau-RD for 10 days at 37 °C and the samples were tested using SEM (Fig. 5B). 3P and 1Pb induced Tau-RD aggregation, resulting in aggregates with string-like morphology. 1Pa, 1Pc, and 2Pc incubation with Tau-RD did not result in oligomer formation. Tau-RD also did not aggregate by itself (ESI Fig. 3†).
Specific phosphorylation patterns control Tau-R4 condensation and aggregation
Charge and steric hindrance alone cannot explain the different effects of the three phosphorylations on the oligomerization of Tau R4. We used the PEP-FOLD3 predictor to examine the conformations and secondary structures of Tau R4.47 The most probable 29 models that were calculated and generated (ESI Fig. 4†) shared the same closed hairpin-like conformation as was found in AD amyloids. All these models contain a combination of disordered and structured domains (Fig. 1C). A notable difference is that according to the models, the Tau R4 fragment has a helical domain whereas the same region in the full-length protein has a beta-sheet structure (Fig. 7A and B). Indeed, peptides usually adopt helical rather than a β-sheet structures since the stabilizing partner strands are missing. In all the calculated models, Ser341 is in the disordered region of the peptide, from Q356 to Phe346. Ser352 is in the helical domain that includes 7 residues from Lys347 to Lys353, and Ser356 is located in the IDR tail from Ile354 to Asp358 (Fig. 6A), similar to their locations in the AD amyloid structure. The CD spectrum of 0P, had a minimum at ∼200 nm and ∼225 nm, corresponding to disordered and helical conformations (Fig. 6B). 0P and 3P had similar CD spectra suggesting they obtain the same secondary structure (Fig. 6B). Using the PEPstrMOD server,48 we generated a de novo structure prediction of Tau R4 β-helix motif phosphorylated at all three p-sites. The results show that the phosphorylations do not affect the secondary structure (Fig. 6C). Thus, we added the phosphate groups to the backbone of 0P using a PyTM plugin (version 1.8.4.0).49 Using PyMol measurement wizard,50 we found that Ser341 and Ser356 are more solvent-exposed than Ser352 (Fig. 6D). Calculated distances between the p-site and their nearest entity showed that: (1) pSer352 may interact with the amine group on the backbone of Val337 via hydrogen bond of 1.5 Å, (2) pSer341 had a 4.3 Å distance from the most adjacent positive charged Lys351, and (3) pSer356 had a 4.7 Å distance from the adjacent amide of the side chain of Gln351 (Fig. 6E). Altogether, pSer 352 is the least exposed of all three. pSer341 and pSer356 are more exposed, and in contrast to pSer352, do not have significant interactions with their surroundings.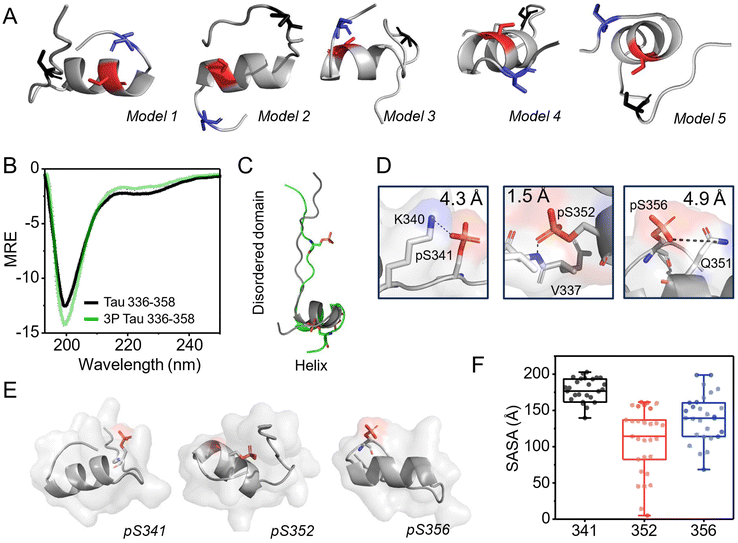 | ||
| Fig. 6 (A) The five most probable models calculated using PEP-FOLD347 show that Tau R4 has an IDR tail, a helical domain, and a disordered region. In all models Ser341 (black) is located in the disordered region, Ser352 (red) in the helical domain and Ser356 (blue) in the IDR tail. (B) CD spectra of 0P (black) and 3P (green) indicate the same secondary structures, comprising of helical and disordered domains. (C) models of 0P (black) and 3P (green) as calculated by PEP-FOLD3 and PEPstrMOD.48 Phosphate groups are shown in orange. (D) Zoom in on the chemical environment and interactions of each phosphorylation site (Ser341 and 356 shown in model 1, Ser352 in model 2). (E) The position of each phosphorylation site in the Tau peptide, as shown by Pymol for model 1. (F) SASA of each phosphorylation site. Boxplots represent the median value and interquartile (25–75% percentiles) of SASA. Each dot represents one of the 29 models that were examined. | ||
Using the Pymol software we calculated the solvent-accessible surface area (SASA)51 for pSer341, pSer352 and pSer356 in the most probable 29 models of the peptide. The phosphate groups were added by PyTM.49 to the models that were generated by the predictor. The average SASA of pSer352 was 108 ± 45 Å2 while pSer341 had an average SASA of 169 ± 36 Å2. pSer356 had an average SASA of 137 ± 36 Å2.
Discussion
In the current study, we combined phosphorylated peptide libraries with an ensemble of biophysical methods to elucidate the specific effects of phosphorylation patterns on the different association processes of Tau R4. We found that phosphorylation at specific sites of the β-helix motif dictate whether the peptides can assemble to form oligomers at all, and which type of oligomerization process they undergo or induce (condensation or amyloid aggregation). Our results highlight the importance of Ser352 as a major phosphorylation site for inducing Tau oligomerization. This result could be obtained only with the help of our synthetic methods enabling the synthesis of phosphorylated peptide libraries.The role of the phosphorylation pattern in switching between condensation and aggregation of Tau R4
Our results show that the Tau R4 peptide (0P) does not form any type of high-order oligomers by itself and does not induce oligomerization of Tau-RD. Phosphorylation of one or more serine residues within peptides derived from this R4 domain results in significant changes in oligomerization processes. The tri-phosphorylated peptide 3P formed condensates or aggregates, depending on the time and ionic strength, and induced aggregation of the Tau-RD. This indicates that phosphorylation controls both the condensation and aggregation of this domain and the specific phosphorylation is critical for the decision which of the processes will take place. These phenomena are unique, as recombinant non-phosphorylated Tau monomers do not tend to aggregate even within days.52The contributions of each phosphorylation within these patterns are not necessarily additive. Phosphorylation of Ser352 activated condensation of R4 as shown by the ability of the 1Pb peptide to undergo condensation, while phosphorylation of Ser341 activated aggregation of R4 as shown by the aggregation of the 1Pa peptide. Phosphorylation of Ser356 prevented any oligomerization of R4 as reflected by the inability of 1Pc to form condensates or aggregates. The effects of combinations of two phosphorylations were synergistic. 2Pa, which contained phosphorylations at residues Ser341 and Ser352, generated condensates with similar size and propensity to 1Pb containing a single phosphorylation at Ser352. We conclude that Ser352 is more dominant in promoting condensation than Ser341. Combining phosphorylation of Ser356 with phosphorylation of Ser341 (2Pb) or with phosphorylation of Ser352 (2Pc) resulted in no condensates, like the effect of Ser356 phosphorylation alone (1Pc) (Fig. 7C). We conclude that Ser356 phosphorylation prevents condensate formation even in the presence of the oligomerization–activating phosphorylation at Ser341 or Ser352. However, when the 3 phosphorylations were combined (3P), we observed condensate formation despite the phosphorylation at position 356. This shows that a combination of both Ser341 and Ser352 phosphorylations overcame the inhibitory effect of phosphorylating Ser 356. Using a peptide library with all possible combinations of phosphorylations makes it possible to elucidate the role of each phosphorylation in controlling the oligomerization processes of Tau. Moreover, it enables demonstrating the synergism between the different phosphorylation sites. The library thus provides a detailed mechanistic insight about how the combinations of the phosphorylations contribute to the oligomerization processes.
Our results revealed that phosphorylation of Ser352 is highly important for Tau aggregation. Phosphorylated Ser352 can be found in a type of Tau filament, named PHF, which was isolated from brains of AD patients.53 However, it could not be phosphorylated in vitro by kinases combination including PKA, GSK, PKA + GSK, P12, P20, PKA + GSK3β and PKA + SAPK4.21,53–55 This suggests that phosphorylation of this site in vitro is challenging, explaining why this site was not studied extensively in previous research. Working at the peptide level, using our advanced phosphorylated peptide synthesis methods, allowed us to pinpoint Ser352 as crucial site for Tau aggregation.
The effect of the phosphorylated Tau R4 peptides on the aggregation of Tau-RD protein
Studying biological systems in their native forms is ideal, but creating a relevant library of phosphoproteins is currently not feasible. To overcome this challenge, we combined synthetic phosphopeptide libraries with recombinant Tau-RD to investigate the effect of phosphorylation on aggregation at the protein level. We focused on the Tau-RD domain because it plays a critical role in amyloid formation in the brains of Alzheimer's disease (AD) and chronic traumatic encephalopathy (CTE) patients. While inducers such as heparin and poly-RNA have been widely used to study Tau aggregation and condensation by simulating certain aspects of the physiological environment,56 they can introduce non-native interactions, potentially distorting the interpretation of aggregation mechanisms. Our study shows that using 100 μM of peptides with specific phosphorylation patterns alone can induce Tau aggregation and condensation without the need for exogenous inducers. This approach provides native cellular conditions, ensuring that the observed effects are driven solely by the intrinsic properties of the phosphorylated peptides, and most importantly, in the specific phosphorylation pattern.The micro-environment of the phosphorylation sites is responsible for controlling Tau R4 oligomerization
The computational part represents a monomeric, non-aggregated peptide, which is why its structure does not align with the beta-sheet structure found in full-length Tau amyloids (Fig. 6). This difference in structure highlights the effect of the microenvironment on peptide conformation, that is crucial for understanding aggregation processes. SASA calculations of the 29 models of Tau R4 β-helix motif show that pSer352 is significantly less exposed to the solvent compared with pSer341 and pSer356. In the calculated conformations, pSer352 is buried and forms hydrogen bonds with the backbone of Val337 (e.g.Fig. 6D model 2). A closer examination of model 1 shows that pSer341 and pSer356 are located at the peripheral parts of the peptide with the closest interactions being at distances larger than 4 Å (Fig. 6D). This suggests that these phosphate groups do not bind any backbone or side chain residues within the peptide (Fig. 6E). Ser341 and Ser356 still differ in their micro-environments. Ser341 is in the IDR of Tau R4, while Ser356 is located within the IDR tail, adjacent to the structured domain of the peptide. The residues surrounding Ser341 are the charged Lys340 and Glu342, while the residues surrounding Ser356 are the small Gly355 and the hydrophobic Leu357.The microenvironments described above for each phosphorylation site explain the observed effects of the different phosphorylations on the oligomerization of the Tau R4 peptide. Phosphorylation of Ser352 resulted in phosphoserine which is less accessible to water and salt. Thus, the peptides including this phosphorylation, 1Pb, 2Pa, 2Pc and 3P, are less soluble and more available to form interactions with additional peptides or proteins (such as Tau-RD) resulting in condensation or aggregation. Phosphorylation of Ser356, as in 1Pc, did not show any type of oligomerization. This can be explained by a possible interaction of the phosphate group with water molecules, which leads to better solubility of the entire peptide. Our suggested model correlates with the experimental findings regarding the activating and inhibiting phosphorylation.
High salt concentration inhibited condensate formation by 1Pa, containing phosphorylation only at Ser341, but not by peptides containing phosphorylation at position 352. This correlates well with our model and can be explained by the higher exposure of Ser 341 to the environment and thus to the salt, which at high concentrations can inhibit the molecular interaction leading to condensation. The micro-environment of the peptides can also explain why 1Pa formed smaller condensates than 1Pb. The same principles also explain why 1Pa condensates are smaller and the peptide is concentrated at the boundaries of the condensate. The buried pSer352 was not reactive enough to initiate aggregation on its own (it initiated condensation) but it did induce Tau-RD aggregation. pS356 on the other hand is too exposed to allow participation of the peptide in aggregation events with or without Tau-RD.
In conclusion, by using phosphopeptide libraries, our study pinpoints the essential role of specific phosphorylation sites in driving Tau's self-assembly and the decision when condensation or aggregation will take place, further advancing the understanding of its role in neurodegenerative diseases.
Methods
Peptide synthesis, labeling and purification
The peptides were synthesized on a rink amid resin with a 0.48 mmol gr−1 loading after 30 min of swelling in DMF. All peptides were synthesized using the Stirring-based accelerated peptide synthesis approach.24 A reactor containing a sintered glass filter, and a heating jacket connected to a circulating 90 °C water bath. The reactor was equipped with an overhead 5-fin turbine PTFE impeller. Solvents and reagents were inserted directly by a feeding line and drained by vacuum filtration. Each cycle started with Fmoc deprotection followed by a 5 mL DMF wash, continued with 1 min coupling and a second wash. Fmoc-Ser/(PO3Bzl)–OH and its sequential residue were double coupled. 3 mL of coupling mixture containing 3 equiv. of protected amino acid, 2.9 equiv. of HATU, and 8 equiv. of DIEA was added. Fmoc deprotection was done by adding 3 mL of a 0.5% DBU solution in DMF for 10 s. Peptides were cleaved using a freshly prepared TFA cocktail, and were purified by Waters preparative HPLC, using a reverse-phase C18 column with a TDW/ACN gradient. The column was equilibrated with 5% ACN for 5 minutes, followed by a linear increase from 5% to 60% ACN over 30 minutes. After elution, the column was washed with 95% ACN for 12 minutes to remove residual hydrophobic components. Finally, the ACN concentration was reduced to 5% over 10 minutes to re-equilibrate the column for subsequent runs. All peptides were characterized by electron spray ionization mass spectrometry (ESI-MS) (ESI Fig. 1†). Peptide purity was tested by dissolving 80 μL of peptide in water injecting into analytical HPLC. Peptides were fluorescein labeled For FRAP and fluorescence microscopy experiments, with 5(6)-carboxy fluorescein at their N′ termini. Fractions containing the desired phosphorylated peptides were lyophilized and the peptides were used without further purifications.Turbidity assay
150 μL 0P and 3P (with 5% FL-peptide each) were tested at peptide concentrations of 50, 300 and 1000 μM. The solutions contained 25 mM phosphate buffer at pH of 7.4 and 10% PEG 400 (buffer A) with NaCl concentrations of 0, 150, 300 and 1000 mM. Triplicates were measured at λ = 350 in BioTek Synergy H1 plate reader.Fluorescence microscopy
150 μL 0P and 3P (with 5% FL-peptide each) at peptide concentrations of 50, 300 and 1000 μM were also studied using fluorescence microscopy. 10 μL of 50 μM peptide from each sample were applied onto a glass slip and visualized under Zeiss Axio scope A1 microscope with a green-fluorescent filter. Images were taken with an AxioCam ICc 3 camera, using Zen software.Fluorescence recovery after photobleaching (FRAP) experiments
FRAP experiments were performed using an FV-1200 confocal microscope (Olympus, Japan) equipped with a sim scanner which enables bleaching while scanning. All experiments were done using a 60×/1.42 oil objective. Imaging was done using the 488 nm laser line and emission was collected 500–540 nm. 5% of FL-3P and 95% of 3P were dissolved in 100 μL buffer A to final concentration of 25 mM phosphate buffer and 150 mM NaCl and 50 μM peptide. The measurement started by adding 10% (V/V) PEG-400 and samples were bleached after 15 and 45 min to evaluate the maturation of condensates. A specific region in the sample was exposed to 100% laser intensity for 120 ms. The interval between image scans was 2 s. Photobleaching correction and recovery were calculated using the OriginPro 2022 software. Final FRAP recovery curves are the average from measuring three different condensates.CD of 0P and 3P
120 μM of 0P and 3P, in a 25 mM phosphate buffer at pH of 7.4 and 150 mM NaCl CD spectrum were determined using JASCO J-810 spectropolarimeter and Spectra manager software. The temperature was 25 °C and controlled by Peltier thermostat, in a 0.1 cm quartz cuvette for far-UV CD spectrometry, ranged from 190 to 260 nm.Aggregation assay
To examine the aggregation properties of the peptides, 100 μM of peptides were incubated with and without 10 μM Tau-RD and 20 mM phosphate buffer at pH of 7.4 and 150 mM NaCl. 150 μL of the final solutions were incubated at a temperature of 37 °C for 10 days. The solution was then dried on silicon wafers and examined using SEM Apero 2S LoVac. Each experiment was repeated between three to nine times.Conflicts of interest
The authors declare no conflict of interest.Data availability
The data supporting this article have been included as part of the ESI.†Acknowledgements
MH is supported by the European Innovation Council (EIC) under the European Union's Horizon Europe research and innovation program (No. 101046369) and by the Israel Science Foundation grant no. 1805/22. SGDR was supported by grants of the Campaign Team Huntington and Alzheimer Nederland (No. WE.03-2019-03). He is PIs of the gravitation consortium FLOW, funded by the Nederlands Minister of Education, Culture and Science. AF thanks the Minerva Center for Bio-Hybrid complex systems and the Saerree K. and Louis P. Fiedler Chair in Chemistry. This study was supported by a visiting professorship in HUJI (to SR). We thank Dr Ofrah Faust and Prof Rina Rosenzweig from the Department of Chemical and Structural Biology, Weizmann Institute Israel for generously contributing the Tau-RD protein.References
- Q. Zhong, X. Xiao, Y. Qiu, Z. Xu, C. Chen, B. Chong, X. Zhao, S. Hai, S. Li, Z. An and L. Dai, MedComm, 2023, 4, e261 CrossRef CAS PubMed.
- F. Ardito, M. Giuliani, D. Perrone, G. Troiano and L. Lo Muzio, Int. J. Mol. Med., 2017, 40, 271–280 CrossRef CAS PubMed.
- S. G. Bressler, A. Mitrany, A. Wenger, I. Nathke and A. Friedler, Int. J. Mol. Sci., 2023, 24, 6478 CrossRef CAS PubMed.
- T. Pawson and J. D. Scott, Trends Biochem. Sci., 2005, 30, 286–290 CrossRef CAS PubMed.
- M. Samarasimhareddy, G. Mayer, M. Hurevich and A. Friedler, Org. Biomol. Chem., 2020, 18, 3405–3422 RSC.
- M. Mann, S.-E. Ong, M. Grønborg, H. Steen, O. N. Jensen and A. Pandey, Trends Biotechnol., 2002, 20, 261–268 CrossRef CAS PubMed.
- T. Garfagnini, L. Ferrari, M. B. Koopman, F. A. Dekker, S. Halters, E. Van Kappel, G. Mayer, S. Bressler, M. M. Maurice, S. G. D. Rudiger and A. Friedler, Chem. – Eur. J., 2024, 30, e202400080 CrossRef CAS PubMed.
- B. Wang, L. Zhang, T. Dai, Z. Qin, H. Lu, L. Zhang and F. Zhou, Signal Transduction Targeted Ther., 2021, 6, 290 CrossRef PubMed.
- T. P. Lopez-Palacios and J. L. Andersen, Trends Cell Biol., 2023, 33, 649–666 CrossRef CAS PubMed.
- X. Tong, R. Tang, J. Xu, W. Wang, Y. Zhao, X. Yu and S. Shi, Signal Transduction Targeted Ther., 2022, 7, 211 CrossRef PubMed.
- A. Aguzzi and T. O'Connor, Nat. Rev. Drug Discovery, 2010, 9, 237–248 CrossRef CAS PubMed.
- M. Samarasimhareddy, D. Mayer, N. Metanis, D. Veprintsev, M. Hurevich and A. Friedler, Org. Biomol. Chem., 2019, 17, 9284–9290 RSC.
- A. Kozelekova, A. Naplavova, T. Brom, N. Gasparik, J. Simek, J. Houser and J. Hritz, Front. Chem., 2022, 10, 835733 CrossRef CAS PubMed.
- P. E. Thorsness and D. E. Koshland, J. Biol. Chem., 1987, 262, 10422–10425 CrossRef CAS PubMed.
- N. E. Mammeri, A. J. Dregni, P. Duan and M. Hong, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2316175121 CrossRef PubMed.
- S. M. Pearlman, Z. Serber and J. E. Ferrell Jr., Cell, 2011, 147, 934–946 CrossRef CAS PubMed.
- N. M. Otto, W. G. McDowell, D. M. Dickey and L. R. Potter, Mol. Pharmacol., 2017, 92, 67–74 CrossRef CAS PubMed.
- S. A. Johnson and T. Hunter, Nat. Methods, 2005, 2, 17–25 CrossRef CAS PubMed.
- K. E. Paleologou, A. W. Schmid, C. C. Rospigliosi, H.-Y. Kim, G. R. Lamberto, R. A. Fredenburg, P. T. Lansbury Jr., C. O. Fernandez, D. Eliezer, M. Zweckstetter and H. A. Lashuel, J. Biol. Chem., 2008, 283, 16895–16905 CrossRef CAS PubMed.
- G. Tesei, T. K. Schulze, R. Crehuet and K. Lindorff-Larsen, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2111696118 CrossRef CAS PubMed.
- J. X. Meng, Y. Zhang, D. Saman, A. M. Haider, S. De, J. C. Sang, K. Brown, K. Jiang, J. Humphrey, L. Julian, E. Hidari, S. F. Lee, G. Balmus, R. A. Floto, C. E. Bryant, J. L. P. Benesch, Y. Ye and D. Klenerman, Nat. Commun., 2022, 13, 2692 CrossRef CAS PubMed.
- D. Ellmer, M. Brehs, M. Haj-Yahya, H. A. Lashuel and C. F. Becker, Angew. Chem., 2019, 131, 1630–1634 CrossRef.
- Y. Li, J. Heng, D. Sun, B. Zhang, X. Zhang, Y. Zheng, W.-W. Shi, T.-Y. Wang, J.-Y. Li and X. Sun, J. Am. Chem. Soc., 2021, 143, 17566–17576 CrossRef CAS PubMed.
- D. Grunhaus, E. R. Molina, R. Cohen, T. Stein, A. Friedler and M. Hurevich, Org. Process Res. Dev., 2022, 26, 2492–2497 CrossRef CAS PubMed.
- J. N. Naoum, I. Alshanski, G. Mayer, P. Strauss and M. Hurevich, Org. Process Res. Dev., 2022, 26, 129–136 CrossRef CAS.
- D. Ben Abba Amiel and M. Hurevich, Eur. J. Org. Chem., 2022, 202200623 CrossRef.
- P. Strauss, F. Nuti, M. Quagliata, A. M. Papini and M. Hurevich, Org. Biomol. Chem., 2023, 21, 1674–1679 RSC.
- M. Samarasimhareddy, G. Mayer, M. Hurevich and A. Friedler, Org. Biomol. Chem., 2020, 18, 3405–3422 RSC.
- M. Samarasimhareddy, D. Mayer, N. Metanis, D. Veprintsev, M. Hurevich and A. Friedler, Org. Biomol. Chem., 2019, 17, 9284–9290 Search PubMed.
- R. Rajmohan and P. H. Reddy, J. Alzheimer's Dis., 2017, 57, 975–999 CAS.
- D. Eisenberg and M. Jucker, Cell, 2012, 148, 1188–1203 CrossRef CAS PubMed.
- T. P. Knowles, M. Vendruscolo and C. M. Dobson, Nat. Rev. Mol. Cell Biol., 2014, 15, 384–396 CrossRef CAS PubMed.
- S. Boyko and W. K. Surewicz, Trends Cell Biol., 2022, 32, 611–623 CrossRef CAS PubMed.
- S. Ambadipudi, J. Biernat, D. Riedel, E. Mandelkow and M. Zweckstetter, Nat. Commun., 2017, 8, 275 CrossRef PubMed.
- N. M. Kanaan, C. Hamel, T. Grabinski and B. Combs, Nat. Commun., 2020, 11, 2809 CrossRef CAS PubMed.
- S. Wegmann, B. Eftekharzadeh, K. Tepper, K. M. Zoltowska, R. E. Bennett, S. Dujardin, P. R. Laskowski, D. MacKenzie, T. Kamath, C. Commins, C. Vanderburg, A. D. Roe, Z. Fan, A. M. Molliex, A. Hernandez-Vega, D. Muller, A. A. Hyman, E. Mandelkow, J. P. Taylor and B. T. Hyman, EMBO J., 2018, 37, e98049 CrossRef PubMed.
- D. Ellmer, M. Brehs, M. Haj-Yahya, H. A. Lashuel and C. F. W. Becker, Angew. Chem., Int. Ed., 2019, 58, 1616–1620 CrossRef CAS PubMed.
- A. W. P. Fitzpatrick, B. Falcon, S. He, A. G. Murzin, G. Murshudov, H. J. Garringer, R. A. Crowther, B. Ghetti, M. Goedert and S. H. W. Scheres, Nature, 2017, 547, 185–190 CrossRef CAS PubMed.
- B. Falcon, J. Zivanov, W. Zhang, A. G. Murzin, H. J. Garringer, R. Vidal, R. A. Crowther, K. L. Newell, B. Ghetti, M. Goedert and S. H. W. Scheres, Nature, 2019, 568, 420–423 CrossRef CAS PubMed.
- M. Von Bergen, P. Friedhoff, J. Biernat, J. Heberle, E. M. Mandelkow and E. Mandelkow, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 5129–5134 CrossRef CAS PubMed.
- S. Lovestam, D. Li, J. L. Wagstaff, A. Kotecha, D. Kimanius, S. H. McLaughlin, A. G. Murzin, S. M. V. Freund, M. Goedert and S. H. W. Scheres, Nature, 2024, 625, 119–125 CrossRef CAS PubMed.
- M. Haj-Yahya, P. Gopinath, K. Rajasekhar, H. Mirbaha, M. I. Diamond and H. A. Lashuel, Angew. Chem., Int. Ed., 2020, 59, 4059–4067 CrossRef CAS PubMed.
- D. N. Edun, M. R. Flanagan and A. L. Serrano, Chem. Sci., 2020, 12, 2474–2479 RSC.
- M. Abbas, W. P. Lipinski, K. K. Nakashima, W. T. S. Huck and E. Spruijt, Nat. Chem., 2021, 13, 1046–1054 CrossRef CAS PubMed.
- D. Grunhaus, A. Friedler and M. Hurevich, Eur. J. Org. Chem., 2021, 2021, 3737–3742 Search PubMed.
- S. Wegmann, B. Eftekharzadeh, K. Tepper, K. M. Zoltowska, R. E. Bennett, S. Dujardin, P. R. Laskowski, D. MacKenzie, T. Kamath, C. Commins, C. Vanderburg, A. D. Roe, Z. Fan, A. M. Molliex, A. Hernandez-Vega, D. Muller, A. A. Hyman, E. Mandelkow, J. P. Taylor and B. T. Hyman, EMBO J., 2018, 37, e98049–e98049 CrossRef PubMed.
- A. Lamiable, P. Thevenet, J. Rey, M. Vavrusa, P. Derreumaux and P. Tuffery, Nucleic Acids Res., 2016, 44, W449–W454 CrossRef CAS PubMed.
- S. Singh, H. Singh, A. Tuknait, K. Chaudhary, B. Singh, S. Kumaran and G. P. Raghava, Biol. Direct, 2015, 10, 73 CrossRef PubMed.
- A. Warnecke, T. Sandalova, A. Achour and R. A. Harris, BMC Bioinf., 2014, 15, 370 CrossRef PubMed.
- The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC, New York, NY, USA, 2015 Search PubMed.
- S. Mitternacht, F1000Research, 2016, 5, 189 Search PubMed.
- W. Zhang, B. Falcon, A. G. Murzin, J. Fan, R. A. Crowther, M. Goedert and S. H. Scheres, eLife, 2019, 8, 43584 CrossRef CAS PubMed.
- C. X. Gong, F. Liu, I. Grundke-Iqbal and K. Iqbal, J. Neural Transm., 2005, 112, 813–838 CrossRef CAS PubMed.
- K. Tepper, J. Biernat, S. Kumar, S. Wegmann, T. Timm, S. Hubschmann, L. Redecke, E. M. Mandelkow, D. J. Muller and E. Mandelkow, J. Biol. Chem., 2014, 289, 34389–34407 CrossRef CAS PubMed.
- D. P. Hanger, H. L. Byers, S. Wray, K. Y. Leung, M. J. Saxton, A. Seereeram, C. H. Reynolds, M. A. Ward and B. H. Anderton, J. Biol. Chem., 2007, 282, 23645–23654 CrossRef CAS PubMed.
- P. Duan, A. J. Dregni, N. E. Mammeri and M. Hong, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2310067120 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5ob00885a |
| ‡ These authors equally contributed to the paper |
| This journal is © The Royal Society of Chemistry 2025 |