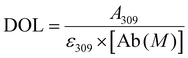Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The effects of buffer, pH, and temperature upon SPAAC reaction rates†
Toni A.
Pringle
 a and
James C.
Knight
a and
James C.
Knight
 *ab
*ab
aSchool of Natural and Environmental Sciences, Newcastle University, Newcastle Upon Tyne, UK. E-mail: james.knight2@newcastle.ac.uk
bNewcastle Centre for Cancer, Newcastle University, Newcastle upon Tyne, UK
First published on 28th January 2025
Abstract
This study investigates the effects of buffer type, pH, and temperature on the kinetics of strain-promoted alkyne–azide cycloaddition (SPAAC) reactions. Using 3-azido-L-alanine and 1-azido-1-deoxy-β-D-glucopyranoside as model azides and sulfo DBCO-amine as the alkyne, we examined reaction rates in a series of buffers, including PBS, HEPES, MES, borate buffer, and cell culture media (DMEM and RPMI), with pH values ranging from 5 to 10 and temperatures of 25 and 37 °C. Absorbance spectrophotometric data revealed that PBS (pH 7) exhibited among the lowest rate constants (0.32–0.85 M−1 s−1), whereas HEPES (pH 7) had the highest (0.55–1.22 M−1 s−1). Additionally, reactions in DMEM were faster than in RPMI (0.59–0.97 vs. 0.27–0.77 M−1 s−1). We observed that higher pH values generally increased reaction rates, except in HEPES buffer. Notably, 1-azido-1-deoxy-β-D-glucopyranoside reacted faster than 3-azido-L-alanine, highlighting the importance of considering the electron-donating capacity of azides in the optimisation of SPAAC reactions. Additional experiments with DBCO-modified antibodies (DBCO-trastuzumab and DBCO-PEG5-trastuzumab) corroborated the trends related to buffer and azide selection. The presence of a PEG linker notably enhanced reaction rates (0.18–0.37 M−1 s−1) by 31 ± 16%. This study offers useful insights into the factors affecting SPAAC kinetics, facilitating the development of optimised bioconjugation strategies.
Introduction
Bioorthogonal chemistry, a distinct subset of click chemistry, has become a key strategy for biomolecule conjugation, particularly in complex biological environments. The 2022 Nobel Prize in Chemistry was awarded to K. B. Sharpless, C. R. Bertozzi, and M. P. Meldal for their pioneering work in this field.1 Defined by Sharpless in 2001, click chemistry encompasses reactions that are fast, high yielding, and free from toxic solvents, resulting in easily purifiable products.2 Bioorthogonal reactions build on these principles, enabling highly specific, selective, and efficient modification of biomolecules, even in biological settings. These reactions typically involve small, reactive chemical groups that can be attached to biomolecules covalently with minimal structural or functional disruption, offering high utility in chemical biology and biotechnology.3,4The copper(I)-catalysed [3 + 2] alkyne–azide cycloaddition (CuAAC) reaction epitomises click chemistry, notable for its high second-order rate constants in the order of 10–100 M−1 s−1 and its widespread use in various systems.5,6 However, the application of this reaction in cellular assays and in vivo environments is hindered by the toxicity of copper ions.7 Despite copper being an essential metal in cells, the presence of free copper ions induces oxidative damage via ROS generation.8,9 While strategies have been developed to avoid Cu(I) toxicity using chelating agents, the development of copper-free click chemistry has become a popular alternative to overcome the challenges associated with CuAAC reactions, with wide-ranging utility in biomedical applications. Key examples include strain-promoted 1,3-dipolar cycloadditions, typically involving cyclooctyne derivatives and azides or nitrones, and inverse electron-demand Diels–Alder cycloadditions.10
Strain-promoted alkyne–azide cycloaddition (SPAAC) reactions involve the formation of a triazole-linked product, with reported second-order rate constants of 0.01–60 M−1 s−1.6,11 SPAAC reactions are driven by the relief of ring strain in the cycloalkyne, where the sp-hybridised carbon atoms have bond angles of ∼163°, rather than the ideal 180°.12
The first cyclooctyne utilised in SPAAC reactions, named OCT, was developed by Bertozzi et al. in 2004.13 Subsequently, an array of cyclooctyne derivatives have been developed with varying reactivity and stability. Notable enhancements in reactivity have been achieved by positioning electron-withdrawing groups near the alkyne (e.g. monofluorocyclooctyne and difluorocyclooctyne), and via the use of dibenzoannulated cyclooctynes, such as dibenzocyclooctyne (DBCO),14 with increased ring strain from sp2 hybridised carbon atoms adjacent the alkyne.15 DBCO-based reagents are commercially available and offer several advantages, including enhanced stability and higher reaction rates compared to other cyclooctynes, such as bicyclononyne (BCN).14 However, the presence of benzyl groups flanking the alkyne in DBCO can pose challenges due to steric hindrance and increased lipophilicity. The incorporation of hydrophilic groups around the benzyl rings or within the linker (e.g. PEG) has been used to reduce lipophilicity and improve solubility.16
SPAAC reactions involving DBCO are generally conducted in aqueous buffers to reflect biologically relevant conditions, although reported reaction rate constants are primarily determined in organic solvents such as dimethyl sulfoxide (DMSO), acetonitrile, or methanol. The use of organic solvents enhances solubility of reagents, but it has been observed that 1,3-dipolar cycloaddition reactions tend to increase in rate with increasing mole fraction of water in the solvent, which is likely due to the hydrogen-bonding effect that stabilises the transition state.17 Despite this, the impact of buffer type and pH on SPAAC reaction rates remains an understudied area. To the authors’ knowledge, only one study has explored this topic, which focused primarily on organic co-solvents, a single pH per buffer, and a single pair of click reagents, limiting the generalisability of the findings.18 Therefore, further research is necessary to gain a more comprehensive understanding of the effects of these various parameters on SPAAC reactions, which could inform their application across a broader range of reactions.
Results and discussion
Effect of buffer, pH, and temperature on the rate of SPAAC reactions
This study measured the rates of SPAAC reactions using biologically relevant alkynes and azides in multiple buffer types and pH values commonly used in bioconjugation protocols, at both 25 °C and 37 °C. The reaction kinetics were evaluated using three water-soluble DBCO reagents (including sulfo DBCO-amine [1] and two DBCO-conjugated trastuzumab derivatives, DBCO-Her [2] and DBCO-PEG5-Her [3]) (Fig. 1). Additionally, the reactivity of two commercially available azides, representing distinct biomolecular classes routinely applied in protein modification, were investigated: a sugar-based azide (1-azido-1-deoxy-β-D-glucopyranoside [4]) and an amino acid-based azide (3-azido-L-alanine hydrochloride [5]). | ||
| Fig. 1 Overview of SPAAC reactions between sulfo DBCO-amine (1), DBCO-trastuzumab (2), DBCO-PEG5-trastuzumab (3) and the azides 1-azido-1-deoxy-β-D-glucopyranoside (4) and 3-azido-L-alanine (5). | ||
For the determination of rate constants of SPAAC reactions, UV-Vis spectrophotometry was employed using pseudo first-order conditions, chosen over NMR due to its rapidity, simplicity, and ability to accommodate micromolar concentrations, particularly when combined with its efficient capability to monitor specific absorption peaks in modified biomolecules. The rate constant was determined using the Beer–Lambert law by measuring the decrease in the characteristic alkyne absorbance of DBCO after the addition of an excess of azide, which disrupts the chromophore during triazole formation.19
To monitor the SPAAC reaction rate, the optimal absorbance wavelength (λmax) of sulfo DBCO-amine (1) was first determined by acquisition of a UV-Vis spectrum. A λmax value of 308 nm was observed, consistent with the typical alkyne absorbance range of DBCO and other strained cyclooctyne species (Fig. 2).20–22 Notably, the λmax was found to remain constant for all buffers and pH values used in this study, affirming it as a reliable parameter.
The absorbance at 308 nm (A308) of the DBCO reagent was measured at a known concentration prior to the addition of azide. From this, the molar attenuation coefficient (ε308) was determined and then used with the A308 measurement at each time point to calculate the concentration of DBCO throughout the reaction. The resulting data was fitted to the integrated rate law, which established a linear relationship between the natural log of DBCO concentration and reaction time (Fig. 2B). To obtain the second-order rate constant (k2), the slope of the graph (kobs) was divided by the azide concentration (eqn (1)).
 | (1) |
Effect of buffer type and pH
The SPAAC reactions between DBCO reagents 1, 2 and 3 with azide reagents 4 and 5 were investigated in the following buffers: phosphate buffered saline (PBS), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), borate buffer, 2-ethanesulfonic acid (MES), and the cell culture media Roswell Park Memorial Institute (RPMI) and Dulbecco's Modified Eagle Medium (DMEM) (indicator free).Significant differences in the highest observed k2 values were observed based on the identity of the buffer, most prominently in the case of azide 4 at 37 °C (P < 0.0001, Table S1† and Fig. 3). SPAAC reactions in HEPES exhibited the highest reaction rate with a mean k2 of 1.22 ± 0.02 M−1 s−1 (pH 7, 37 °C, azide 4), which was significantly different from all other buffers (P < 0.0001) except for borate buffer (P = 0.701). The second highest reaction rate was observed in borate buffer, with a maximum k2 of 1.18 ± 0.01 M−1 s−1 (pH 10, 37 °C, azide 4). The reaction rates in DMEM, MES, PBS, and RPMI were lower, with highest observed k2 = 0.97 ± 0.01, 0.86 ± 0.02, 0.85 ± 0.03, and 0.77 ± 0.06 M−1 s−1, respectively (multiple comparisons P-values in ESI [Table S2†]).
These values are comparable to those obtained by Davis et al. in their similar investigation that focused primarily on the effect of organic co-solvents. Second-order rate constants in the range of 0.96–1.18 M−1 s−1 were reported for DBCO-PEG4-CO2H and N3-PEG3 in six different buffer types, including HEPES and MES.18 In contrast to our observations, the authors observed no significant differences in reaction rates among the buffers, but the highest rate was observed for HEPES at pH 7.5. Notably, HEPES has been reported as the most effective buffer for CuAAC reactions, though the reasons are still unclear.23,24 A possible explanation is that buffer composition influences the electrostatic stabilisation of the transition state via hydrogen bonding, enhancing reactivity.17 In addition, the ionic strengths of buffers, as well as the electrostatic interactions of zwitterionic buffers with charged molecules in solution, are also likely contributing factors.25,26 Computational modelling approaches have examined transition states in SPAAC reactions, though primarily focusing on cycloalkyne modifications. Other related investigations have suggested that certain disparities in reaction rates and regioselectivity can be attributed to ‘solvation effects’.27,28
Our observation that SPAAC reactions performed in PBS generally yield lower rate constants compared to other buffer types, except MES, is notable, as PBS is commonly used in SPAAC-based bioconjugation protocols. Based on the parameters used in this study, shorter reaction times and enhanced yields are likely to be achieved with the use of HEPES or borate buffer.
The optimal application of chemical reactions in in vitro cellular systems hinges on selecting a suitable cell culture medium that maintains pH, provides essential nutrients, and ensures biocompatibility. Therefore, the SPAAC reaction rates of DBCO 1 with azides 4 and 5 were compared in two widely used cell culture media, DMEM and RPMI, following the same experimental protocol. Statistical analysis revealed that the reaction rates for both azides were significantly higher in DMEM than in RPMI, with increases of 26% (P < 0.01) and 122% (P < 0.0001) for azides 4 and 5, respectively (Fig. S3†). Although the composition of cell culture media varies by brand and required additives, there are notable differences in the composition of DMEM and RPMI, such as the concentration of sodium bicarbonate buffer (3.7 g L−1 in DMEM vs. 2.0 g L−1 in RPMI), the presence of biotin, vitamin B12, and PABA in RPMI, and the absence of HEPES in the RPMI used in this study. Several factors may contribute to the observed variation in reaction rates, complicating the identification of specific media components responsible for these changes. Notably, the fastest reaction rate was observed in the media containing HEPES, which also yielded the fastest reaction rates overall among the tested buffers.
Given the paucity of literature on the influence of buffer pH on SPAAC reaction rates, this study systematically evaluated a range of pH values, within the effective buffering range for each buffer, aiming to provide insights into how pH modulation can optimise biomolecule conjugation efficiency. The results showed a discernible trend among the buffers studied (Fig. 4).
Overall, the SPAAC rate constants generally increased with rising pH, notably in borate and MES buffers. However, an anomalous trend was observed in HEPES buffer, where the reaction rate decreased as pH increased, and the reasons for this remain unclear. Additionally, elevating the temperature from 25 °C to 37 °C consistently enhanced the reaction rates across all buffers and pH levels, aligning with expected temperature effects on chemical reaction kinetics.
Effect of azide
Although there is limited literature on the effect of azide modification on SPAAC reaction rates, it is generally accepted that electron-rich azides lead to higher reaction rates, as SPAAC reactions proceed with HOMOazide–LUMOcyclooctyne interactions. Previous studies have shown that primary azides tend to have higher reaction rates than substituted azides. Interestingly, in all buffers, the secondary azide-containing compound 4 exhibited a higher reaction rate than the primary azide-containing compound 5 (Fig. S5–S9†). This may be due to azide 4 containing multiple electron-donating hydroxyl groups, while azide 5 contains an electron-withdrawing carboxyl group. This difference in electron-donating capacity suggests that azide 4 is more electron-rich than azide 5, likely accounting for its higher reaction rate. These findings highlight the importance of considering the electron-donating capacity of azides in the optimisation of SPAAC reactions, as even small structural modifications can significantly affect reaction kinetics.29Effect of buffer, pH, and temperature on the rate of SPAAC reactions with mAb-DBCO conjugates
An additional series of related experiments were performed using two DBCO-modified antibody conjugates based on the IgG1 trastuzumab (Herceptin, MW ∼ 150 kDa) to investigate the influence of these parameters in more complex biomolecular systems. These antibody conjugates were DBCO-Her (alkyne 2) and DBCO-PEG5-Her (alkyne 3), respectively. PEG linkers are known for their hydrophilicity and ability to extend from the antibody surface, creating a spacer that reduces steric hindrance between the antibody and the click partner. Therefore, the inclusion of alkyne 3 offered an additional opportunity to assess the potential impact of a PEG linker on SPAAC reaction rates, within the defined scope of this study.To modify lysine residues throughout the mAb, a 7-fold molar excess of DBCO-STP ester or DBCO-PEG5-NHS was used to achieve an optimal degree of labelling (DOL), ensuring effective DBCO group attachment without inducing mAb precipitation. Using 10% DMSO (v/v) as a co-solvent during DBCO-STP ester conjugation improved solubility and prevented mAb aggregation. DBCO-Her (2) and DBCO-PEG5-Her (3) were synthesised with an antibody recovery of 73.3 ± 11.9% (n = 8) and 78.8 ± 12.9% (n = 10), respectively, and with DOLDBCO of 6.41 ± 2.16 (n = 8) and 5.77 ± 0.90 (n = 10), respectively. The mAb concentration was adjusted to 0.016 ± 0.004 mM, optimal for monitoring the temporal reduction of A309 while minimising mAb-associated experimental costs. A λmax of 309 nm was determined for both 2 and 3 using UV-Vis spectrophotometry, and the pseudo first-order rate constant for each reaction was determined by monitoring the change in A309 over time (Fig. 5). Confirmation of DBCO-STP ester attachment to Herceptin was corroborated by MALDI-TOF mass spectrometry (Fig. S10†).
SPAAC reaction rates for DBCO-Her (2) and DBCO-PEG5-Her (3) were assessed in HEPES buffer (pH 7), reflecting the optimal conditions established by the earlier DBCO-STP ester model, and in PBS buffer (pH 7), due to its prevalent use in bioconjugation protocols, with the addition of azide 4 or azide 5.
The incorporation of a PEG linker in DBCO-PEG5-Her resulted in a faster rate constant under all conditions compared to DBCO-Her, with a mean increase of 31 ± 16% (n = 4). This effect was particularly pronounced in HEPES buffer with azide 4, showing a 53% increase in rate (P < 0.0001, Table 1 and Fig. S11†).
| Buffer | DBCO-Her (2) | DBCO-PEG5-Her (3) | ||
|---|---|---|---|---|
| Azide 4 | Azide 5 | Azide 4 | Azide 5 | |
| PBS | 0.17 ± 0.01 | 0.15 ± 0.00 | 0.23 ± 0.00 | 0.18 ± 0.01 |
| HEPES | 0.24 ± 0.01 | 0.18 ± 0.01 | 0.37 ± 0.00 | 0.22 ± 0.02 |
The faster SPAAC reaction rate of DBCO-PEG5-Her compared to DBCO-Her can be attributed to several factors. Firstly, the PEG linker increases the distance between the antibody and the DBCO group, minimising steric hindrance. This separation prevents the hydrophobic DBCO group from being buried within the antibody, thus avoiding isolation from the aqueous buffer and facilitating the approach of the azide molecule, thereby enhancing reaction efficiency.30 Additionally, PEG linkers improve solubility and dispersion of macromolecules.31,32 The hydrophilic nature of the PEG linker in DBCO-PEG5-Her increases the solubility of the DBCO-modified antibody, increasing the likelihood of successful collisions between reactive groups, thereby accelerating reaction kinetics.
Irrespective of the cycloalkyne or buffer utilised, azide 4 consistently exhibited a significantly higher rate constant than azide 5. Specifically, in PBS, the rate constants for azide 4 increased by 15% for DBCO-Her (P = 0.0231) and by 29% for DBCO-PEG5-Her (P = 0.0013) compared to azide 5. In HEPES, the rate constants for azide 4 increased by 36% for DBCO-Her (P = 0.0044) and by 69% for DBCO-PEG5-Her (P = 0.0001) compared to azide 5. Furthermore, reactions conducted in HEPES consistently exhibited a significantly higher rate constant than those performed in PBS, with an average rate increase of 37 ± 20% (P < 0.0179). The observed trends in the DBCO-modified antibodies, consistent with those in the simpler sulfo DBCO-amine model, highlight that the relationships between buffer conditions, azide physicochemical properties, and reaction kinetics also extend to more complex biomolecular systems.
Sulfo DBCO-amine exhibited significantly higher second-order rate constants compared to DBCO-modified antibodies (DBCO-Her and DBCO-PEG5-Her), showing average rate increases of 229 ± 73% and 151 ± 51%, respectively (P < 0.0001, Fig. S12†). The difference in reactivity between sulfo DBCO-amine and DBCO-Her conjugates is likely due to steric hindrance imposed by the antibody. The proximity of the DBCO group to the surrounding protein structure may impede its reaction with the azide, resulting in slower kinetics compared to the smaller, less sterically hindered sulfo DBCO-amine.
Conclusions
In this study, we systematically examined SPAAC reaction rates using biologically relevant alkynes and azides across various buffers, pH values, and temperatures commonly employed in bioconjugation protocols. Significant differences were observed in reaction rates depending on the buffer type. HEPES exhibited the highest reaction rates, followed by borate buffer, while PBS, despite its widespread use in bioconjugation protocols, exhibited slower kinetics. Additionally, higher reaction rates in DMEM compared to RPMI underscore the importance of media selection for optimal application of SPAAC in in vitro cellular experiments.The study also found that higher pH values (except for HEPES) and increased temperatures accelerated the reactions, thereby facilitating biomolecule conjugation. Notably, azide 4, which contains multiple electron-donating groups, exhibited higher reaction rates compared to azide 5 across all buffer types and pH values. This emphasises the importance of considering the electron-donating capacity of azides in SPAAC reactions, which can aid in designing more efficient biomolecule conjugation protocols. Further research into the interplay between azide physicochemical properties and reaction kinetics, particularly involving azide derivatives of other biomolecular classes, would be valuable.
For DBCO-modified antibodies, the effects of the studied parameters on reaction kinetics mirrored those observed with the smaller, less sterically hindered sulfo DBCO-amine. This suggests that these insights can be extended to larger, more complex biomolecules. The study also demonstrates that incorporating a PEG linker into the antibody conjugate enhances reaction rates, likely by reducing steric hindrance and increasing solubility. Further investigations on this topic, including an evaluation of a broader range of PEG linker lengths, as well as other linker types, are likely to provide additional useful insights to support the optimal application of SPAAC-based methodologies.
Experimental section
Reagents and methods
All reagents were used from the supplier without further purification. Sulfo DBCO-amine (Cat# 1227) was purchased from Click Chemistry Tools, 3-azido-L-alanine -L-HCl (Cal# CLK-AA003) was purchased from Jena Bioscience, 1-azido-1-deoxy-β-D-glucopyranoside (Cat# 514004) was purchased from Merck.Preparation of buffers
All buffer reagents were purchased from Fisher Scientific and used without further purification. Dulbecco's Modified Eagle Medium (DMEM, high glucose, no glutamine, no phenol red, Cat# 11594416) and Roswell Park Memorial Institute 1640 Medium (RPMI, no phenol red, Cat# 10363083) were used without altering the pH. Phosphate buffered saline (PBS, 1×, pH 7.2, Cat# 11530546), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, 1 M, Cat# 10397023), 2-ethanesulfonic acid (MES, 1 M, Cat# 475893) and borate buffer (1 M, Cat# 10522595) were made up through the addition of deionised water and their pH was adjusted by the addition of either HCl (1 M) or NaOH (6 M).Determining the rate constants of SPAAC reactions using UV-Vis spectrophotometry
The kinetics of the SPAAC reaction were monitored by measuring the absorbance of the alkyne (at 308–309 nm) over time using a UV-Vis spectrophotometer (Nanodrop OneC spectrophotometer). Focusing on pseudo first-order rate constants, no attempts were made to purify or analyse products. Rate constants were measured at either 25 or 37 °C. Prior to adding azide, an absorbance measurement at 308–309 nm was taken of the thermally equilibrated alkyne solution (0.1077 mM, 750 μL) in the appropriate buffer in a 10 mm quartz cuvette (Merck, Cat# Z803774). This initial absorbance was used to calculate the molar attenuation coefficient of the alkyne for each reaction using the Beer–Lambert law. Subsequently, a thermally equilibrated solution of azide (750 μL, 1 mM) was added to the cuvette, and the absorption was measured every 2.5 minutes for a total of 40 scans. The concentration of DBCO at each time point was determined using the Beer–Lambert law (eqn (2)). | (2) |
The pseudo first-order rate constant (kobs) was determined from the slope of the natural log of the DBCO concentration plotted against time. The second-order rate constant (k2) was calculated by dividing the pseudo first-order rate constant by the azide concentration (eqn (1)). Reactions were performed in triplicate for each reaction condition.
Preparation of Her-DBCO and Her-PEG5-DBCO
Approximately 5–10 mg of trastuzumab (Herceptin®, Her) was dissolved in 0.1 M NaHCO3 (pH 9, 500 μL), then passed through a pre-rinsed Amicon 30 kDa molecular weight cut-off 0.5 mL centrifugal filter (Merck, Cat# UFC503096) at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 8 min and washed three times (3 × 500 μL) with 0.1 M NaHCO3 (pH 9) to remove excipients. The antibody solution was adjusted to 5 mg mL−1 with 0.1 M NaHCO3 (pH 9) and the appropriate volume of DMSO was added to achieve 10% DMSO solution (v/v). A 7-fold molar excess of DBCO-STP ester (Click Chemistry Tools, Cat# 1259) or DBCO-PEG5-NHS ester (BroadPharm, Cat# BP-24055) was added from a 5 mM stock in DMSO to the antibody solution and the reaction mixture was incubated at 25 °C for 2 h with gentle shaking (450 rpm). DBCO-Her conjugates were isolated in PBS (450 μL, pH 7.2) or 1 M HEPES (450 μL, pH 7) using centrifugal filtration as previously described.
000g for 8 min and washed three times (3 × 500 μL) with 0.1 M NaHCO3 (pH 9) to remove excipients. The antibody solution was adjusted to 5 mg mL−1 with 0.1 M NaHCO3 (pH 9) and the appropriate volume of DMSO was added to achieve 10% DMSO solution (v/v). A 7-fold molar excess of DBCO-STP ester (Click Chemistry Tools, Cat# 1259) or DBCO-PEG5-NHS ester (BroadPharm, Cat# BP-24055) was added from a 5 mM stock in DMSO to the antibody solution and the reaction mixture was incubated at 25 °C for 2 h with gentle shaking (450 rpm). DBCO-Her conjugates were isolated in PBS (450 μL, pH 7.2) or 1 M HEPES (450 μL, pH 7) using centrifugal filtration as previously described.
The average number of DBCO moieties attached per antibody (DOLDBCO) was determined by measuring the absorbance of the conjugate at 280 nm (aromatic amino acid residues in mAb) and 309 nm (alkyne) using the Nanodrop OneC spectrophotometer.
CF = Correction factor (A280/A309), ε309 = molar attenuation coefficient at λ309, ε1% = 13.7 (percent molar attenuation coefficient for a 10 mg mL−1 IgG solution).
Statistical analysis
All statistical and regression analyses were performed using GraphPad Prism v9 (GraphPad Software, San Diego, CA, USA). A confidence interval of 95% (P < 0.05) was considered statistically significant. Statistical significance is indicated as follows: ns = not significant, * = P < 0.05, ** = P < 0.01, *** = P < 0.001, **** = P < 0.0001. Unpaired t-tests compared two groups, while one-way ANOVA followed by Tukey's post hoc test compared multiple groups. Simple linear regression analysis examined linear relationships, with the coefficient of determination (R2) assessing the goodness of fit. All data were obtained in at least triplicate and are reported as mean ± standard deviation.Author contributions
T. Pringle developed the methodology, performed the experiments and analysed the data. J. Knight conceived and designed the study; T. Pringle and J. Knight co-wrote the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful to Newcastle University for funding this research.References
- The Nobel Prize in Chemistry 2022, https://www.nobelprize.org/prizes/chemistry/2022/press-release/, (accessed 13/04/23).
- H. C. Kolb, M. G. Finn and K. B. Sharpless, Angew. Chem., Int. Ed., 2001, 40, 2004–2021 CrossRef CAS.
- E. M. Sletten and C. R. Bertozzi, Angew. Chem., Int. Ed., 2009, 48, 6974–6998 CrossRef CAS PubMed.
- T. Luu, K. Gristwood, J. C. Knight and M. Jörg, Bioconjugate Chem., 2024, 35, 715–731 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- B. L. Oliveira, Z. Guo and G. J. L. Bernardes, Chem. Soc. Rev., 2017, 46, 4895–4950 RSC.
- D. C. Kennedy, C. S. McKay, M. C. B. Legault, D. C. Danielson, J. A. Blake, A. F. Pegoraro, A. Stolow, Z. Mester and J. P. Pezacki, J. Am. Chem. Soc., 2011, 133, 17993–18001 CrossRef CAS PubMed.
- L. M. Gaetke and C. K. Chow, Toxicology, 2003, 189, 147–163 CrossRef CAS PubMed.
- S. Neumann, M. Biewend, S. Rana and W. H. Binder, Macromol. Rapid Commun., 2020, 41, 1900359 CrossRef CAS PubMed.
- L. Li and Z. Zhang, Molecules, 2016, 21, 1393–1414 CrossRef PubMed.
- E. Kim and H. Koo, Chem. Sci., 2019, 10, 7835–7851 RSC.
- J. Dommerholt, F. P. J. T. Rutjes and F. L. Van Delft, Top. Curr. Chem., 2016, 374, 16–35 CrossRef PubMed.
- N. J. Agard, J. A. Prescher and C. R. Bertozzi, J. Am. Chem. Soc., 2004, 126, 15046–15047 CrossRef CAS PubMed.
- J. Dommerholt, F. P. J. T. Rutjes and F. L. Van Delft, Cycloadditions in Bioorthogonal Chemistry, Springer International Publishing, 2016, pp. 57–76, DOI:10.1007/978-3-319-29686-9_4.
- C. G. Gordon, J. L. Mackey, J. C. Jewett, E. M. Sletten, K. N. Houk and C. R. Bertozzi, J. Am. Chem. Soc., 2012, 134, 9199–9208 CrossRef CAS PubMed.
- V. Terzic, G. Pousse, R. Méallet-Renault, P. Grellier and J. Dubois, J. Org. Chem., 2019, 84, 8542–8551 CrossRef CAS PubMed.
- R. N. Butler, W. J. Cunningham, A. G. Coyne and L. A. Burke, J. Am. Chem. Soc., 2004, 126, 11923–11929 CrossRef CAS PubMed.
- D. L. Davis, E. K. Price, S. O. Aderibigbe, M. X. H. Larkin, E. D. Barlow, R. Chen, L. C. Ford, Z. T. Gray, S. H. Gren, Y. Jin, K. S. Keddington, A. D. Kent, D. Kim, A. Lewis, R. S. Marrouche, M. K. O'Dair, D. R. Powell, M. L. H. C. Scadden, C. B. Session, J. Tao, J. Trieu, K. N. Whiteford, Z. Yuan, G. Yun, J. Zhu and J. M. Heemstra, J. Org. Chem., 2016, 81, 6816–6819 CrossRef CAS PubMed.
- S. Bijlsma, H. F. M. Boelens and A. K. Smilde, Appl. Spectrosc., 2001, 55, 77–83 CrossRef CAS.
- K. Yamagishi, K. Sawaki, A. Murata and S. Takeoka, Chem. Commun., 2015, 51, 7879–7882 RSC.
- P. Kanjilal, K. Singh, R. Das, J. Matte and S. Thayumanavan, Biomacromolecules, 2023, 24, 3638–3646 CrossRef CAS PubMed.
- V. Kardelis, R. C. Chadwick and A. Adronov, Angew. Chem., 2016, 128, 957–961 CrossRef.
- N. Darabedian and M. R. Pratt, in Methods in Enzymology, ed. A. K. Shukla, Academic Press, 2019, vol. 622, pp. 293–307 Search PubMed.
- C. G. Parker and M. R. Pratt, Cell, 2020, 180, 605–632 CrossRef CAS PubMed.
- E. Stellwagen, J. D. Prantner and N. C. Stellwagen, Anal. Biochem., 2008, 373, 407–409 CrossRef CAS PubMed.
- P. L. Breuer and M. I. Jeffrey, Hydrometallurgy, 2004, 72, 335–338 CrossRef CAS.
- K. Chenoweth, D. Chenoweth and W. A. Goddard III, Org. Biomol. Chem., 2009, 7, 5255–5258 RSC.
- F. Schoenebeck, D. H. Ess, G. O. Jones and K. N. Houk, J. Am. Chem. Soc., 2009, 131, 8121–8133 CrossRef CAS PubMed.
- J. A. Stewart, C. J. Wilson and B. M. Swarts, J. Carbohydr. Chem., 2014, 33, 408–419 CrossRef CAS.
- M. K. Rahim, R. Kota and J. B. Haun, Bioconjugate Chem., 2015, 26, 352–360 CrossRef CAS PubMed.
- A. Guillou, D. F. Earley, S. Klingler, E. Nisli, L. J. Nüesch, R. Fay and J. P. Holland, Bioconjugate Chem., 2021, 32, 1263–1275 CrossRef CAS PubMed.
- P. L. Turecek, M. J. Bossard, F. Schoetens and I. A. Ivens, J. Pharm. Sci., 2016, 105, 460–475 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ob01157k |
| This journal is © The Royal Society of Chemistry 2025 |