Open Access Article
Open Access ArticleLuminescent insulin–Au(III) conjugate retains insulin biological properties in human microglia
Dusica
Maysinger
 *a,
Issan
Zhang
*a,
Issan
Zhang
 a,
Hao
Yuan
a,
Hao
Yuan
 b,
Vlasta
Bonačić-Koutecký
b,
Vlasta
Bonačić-Koutecký
 cd,
Željka
Sanader Maršić
cd,
Željka
Sanader Maršić
 ce and
Rodolphe
Antoine
ce and
Rodolphe
Antoine
 *b
*b
aDepartment of Pharmacology and Therapeutics, McGill University, 3655 Promenade Sir-William-Osler, H3G 1Y6 Montreal, Canada. E-mail: dusica.maysinger@mcgill.ca
bInstitut Lumière Matière, CNRS UMR 5306, Université Claude Bernard Lyon 1, Univ. Lyon, 69622 Villeurbanne Cedex, France. E-mail: rodolphe.antoine@univ-lyon1.fr
cCenter of Excellence for Science and Technology, Integration of Mediterranean Region (STIM), Faculty of Science, University of Split, Ruđera Boškovića 33, 21000 Split, Croatia
dChemistry Department, Humboldt University of Berlin, Brook-Taylor-Strasse 2, 12489 Berlin, Germany
eFaculty of Science, University of Split, Rudera Boskovica 33, 21000 Split, Croatia
First published on 2nd July 2025
Abstract
The aims of this study were to establish if insulin conjugated with auric gold Au(III) can be visualized in living cells expressing insulin receptors, and if key steps in insulin signaling are undisturbed. Au(III) is coordinated with three tyrosines of insulin and is strongly stabilized by coordination with these tyrosines into a metal-ion coordinated insulin template. We combined theoretical and experimental approaches, including computational analyses, immunocytochemistry and functional assays to analyze the cellular effects of this construct. Results in this study show that the insulin–Au(III) conjugate does not impair several steps in insulin signaling and downstream metabolic functions in lysosomes and lipid droplets. Namely, the signaling kinases protein kinase B (Akt1) and mitogen-activated protein kinases (ERK1/2) are phosphorylated to a comparable extent as with unmodified insulin. Immunocytochemistry also shows a comparable decrease in nuclear transcription factor EB (TFEB), an essential regulator of lysosomal biogenesis. Fluorescence co-labelling of lysosomes with LysoTracker and lipid droplets with BODIPY 493/503 shows normal organellar morphology and slightly enhanced lipid droplet content. The luminescent properties of insulin–Au(III) could serve to combine biochemical and imaging studies in human cells beyond microglia.
Introduction
The brain was once considered to be insensitive to insulin, but more recent research has indicated otherwise.1,2 Insulin concentrations, abundance of insulin receptors and signaling triggered by insulin–insulin receptor interaction is disturbed in several neurological disorders.1,3,4 Microglia are important players in the modulation of insulin in the brain. Microglia respond to stressful stimuli, trophic factors, metabolic and hormonal disturbances.5–7 Because microglia express insulin receptors and necessary molecules to signal from the cell surface to the nucleus, they detect and respond to abnormalities in insulin abundance.8 Hyperinsulinemia exerts series of adverse effects on neural cells and can profoundly affects microglia capacity to phagocytose biological aggregates (e.g. amyloid beta in Alzheimer's disease, disintegrated or malfunctional organelles), thus contributing to the severity and progression of neurological disorders.9,10 Impaired phagocytosis is associated with impaired lysosomal functions, which is in turn dependent on lysosomal pH. Microglia, being the main central nervous system phagocytes, lose their phagocytic abilities with aging and hyperinsulinemia.11 Lysosomal biogenesis regulated by the master transcription factor EB (TFEB) is thus linked to insulin signaling, and lysosomes have become therapeutic targets in clinical and basic research due to their fundamental roles under physiological and disease conditions.12–14Liu et al. developed an insulin-protected Au nanocluster which emits red luminescence while maintaining good bioactivity and biocompatibility, an attractive tool for bioimaging.15 Such gold nanoclusters were luminescent when excited by two-photon excitation at 800 nm, or in the visible region at 400 nm. The authors show that myoblasts can internalize the nanoclusters. Mouse brain homogenate quenches the fluorescence at 670 nm, but it can be inhibited by racecadotril and thiorphran.15 Although quite appealing as fluorescent nanomaterials, these insulin constructs with gold nanoclusters contained an undefined number of gold atoms in different oxidation states (approximately 24% Au(I) and 76% Au(0)). The intact insulin entity provides an optimized structure to foster the formation of Au nanoclusters; the counterpart being that a weak interaction between insulin and Au nanoclusters may cause the breakdown of unprotected gold clusters. This might have deleterious consequences regarding the stability of these constructs, even if insulin-stabilized Au nanoclusters have shown biocompatibility when tested in mouse myoblasts. As an alternative and for more robust luminescent probes, the insulin–Au(III) conjugate was recently synthesized and characterized to possess attractive luminescence properties. The single gold atom is not predicted to interfere with the biological activity of insulin, but has not been tested in brain cells, insulin pathways and metabolic organelles.16 The Au(III) is coordinated and strongly stabilized with three tyrosines of insulin. This study aims to establish if insulin strongly coordinated with Au(III) can be detected in human cells expressing insulin receptors, and if key steps in insulin signaling are undisturbed.
We here focus on human microglia and the effects of the gold–insulin construct on several biomarkers. Insulin binding to its receptors leads to the phosphorylation of kinases Akt1 and ERK1/2, then to anabolic effects (stimulation of lipid droplets biogenesis and decrease in lysosomal biogenesis).17 Lipid droplets are abundant in adipocytes and relevant to metabolic syndrome disorders, yet their multiple roles in brain cells are only beginning to be elucidated.18–20 We combined theoretical and experimental approaches, including computational analyses, immunocytochemistry and functional assays to reveal effects of the insulin–Au(III) construct on insulin signaling in microglia. Computational analysis showing the arrangement of insulin with its receptor suggests that favorable properties of insulin–Au(III) render this nanostructure suitable for rapid, sensitive and inexpensive assays for insulin and insulin receptors in biological specimens under physiological and pathological conditions.
Materials and methods
Insulin–Au(III) synthesis and characterization
The insulin–Au(III) conjugate was synthesized using a one pot “green” synthesis method. The reaction was carried out at physiological temperature (37 °C) and elevated pH (∼11.5) as reported by Bain et al.16 Insulin (100 mg) was dissolved in 5 mL ultrapure water and stirred. After dissolving well, 5 mL of 5 mM Au3+ solution was added. After stirring for 5 min, 400 μL of 1 M NaOH solution was added to adjust the pH to ∼11.5. The reaction was kept at 37 °C for 4 h. The insulin–Au(III) conjugate was purified by ultrafiltration with membrane tubes (3 kDa cut off). The prepared sample was centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C), exchanged water 5 times to remove extra salts, then lyophilized to powder for further applications. Tetrachloroauric acid trihydrate (HAuCl4·3H2O), sodium hydroxide (NaOH) were purchased from Sigma-Aldrich. Recombinant human insulin (C257H383N65O77S6, molecular weight 5807.65 g mol−1) was purchased from Life Technologies Corporation. Ultrapure (Milli-Q) water with a resistivity of 18.2 MΩ was used for all experiments.
000 rpm, 4 °C), exchanged water 5 times to remove extra salts, then lyophilized to powder for further applications. Tetrachloroauric acid trihydrate (HAuCl4·3H2O), sodium hydroxide (NaOH) were purchased from Sigma-Aldrich. Recombinant human insulin (C257H383N65O77S6, molecular weight 5807.65 g mol−1) was purchased from Life Technologies Corporation. Ultrapure (Milli-Q) water with a resistivity of 18.2 MΩ was used for all experiments.
UV-vis absorption spectra were recorded using an AvaSpec2048FT spectrophotometer, with a continuous spectrum of halogen lamp coupled with an AvaLightDH-S deuterium lamp. Photoluminescence spectra were recorded with a Horiba Jobin Yvon Fluoromax4 spectrophotometer, and the data were collected and plotted with the FluorEssence software.
In silico modelling
Gromacs version 2020.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 was used for all-atom molecular dynamics (MD) simulation of insulin in explicit solvent with parameters previously described.16 Such simulations were analyzed in the terms of distances between tyrosine residues. All combinations of distances between residues have been calculated using a Gromacs built-in tool (gmx distance) for each time frame during the dynamics. Then, distances for each time frame were averaged and the time frame with the shortest average distance was selected as a representation of the structure possible with Au coordination. Chimera22 was used for visual representation as well as for alignment of insulin and insulin MD structures.
21 was used for all-atom molecular dynamics (MD) simulation of insulin in explicit solvent with parameters previously described.16 Such simulations were analyzed in the terms of distances between tyrosine residues. All combinations of distances between residues have been calculated using a Gromacs built-in tool (gmx distance) for each time frame during the dynamics. Then, distances for each time frame were averaged and the time frame with the shortest average distance was selected as a representation of the structure possible with Au coordination. Chimera22 was used for visual representation as well as for alignment of insulin and insulin MD structures.
Cell culture and treatments
The HMC3 human microglia were originally obtained from the American Type Culture Collection. Cells were used within 25 passages, and maintained in Dulbecco's Modified Eagle's Medium (DMEM, Thermo Fisher Scientific) supplemented with 5% (v/v) fetal bovine serum (Wisent) and 1% (v/v) penicillin–streptomycin (Thermo Fisher Scientific). Cells were kept in incubation at 37 °C, 5% CO2 and 95% relative humidity.Insulin–Au(III) stocks (300 μM) were prepared fresh in ultrapure water before each experiment. Cells were treated with the equivalence of 0.3 nM insulin, or 0.3 nM insulin–Au(III), or 0.3 nM Au for the HAuCl4 negative control. Cells treated in DMEM without serum were washed twice with phosphate-buffered saline (PBS, Wisent) before treatment.
Immunocytochemistry
Cells were seeded on 12 mm diameter glass coverslips (Assistent) at 7000 cells per coverslip. Cells were kept in culture overnight before treatment. Cells were deprived from serum for 24 h before treatment. At the end of the treatment time, cells were washed twice with PBS, then fixed with 4% (w/v) paraformaldehyde (BDH) in PBS for 10 min. Permeabilization was achieved with 0.1% (v/v) Triton X-100 (Millipore-Sigma) in PBS for 10 min. Non-specific binding was reduced by blocking with 10% (v/v) donkey serum in PBS for 1 h. Cells were incubated with primary antibodies diluted in 10% donkey serum overnight at 4 °C in a humidified container. The primary antibodies used were: rabbit anti-phosphorylated Akt1 (Ser473), 1/500, Chemicon, AB3132; mouse anti-phosphorylated ERK1/2 (Thr202/Tyr204), 1/500, Cell Signaling, #9106; rabbit anti-TFEB, 1/500, Proteintech, 13372-1-AP. The next day, cells were washed thrice with PBS for 5 min. Cells were then incubated with secondary antibodies (goat anti-rabbit Alexa Fluor 647, 1/500, Thermo Fisher Scientific; goat anti-mouse Alexa Fluor 647, 1/500, Thermo Fisher Scientific) diluted in 10% donkey serum and Hoechst 33342 (10 μM, Millipore-Sigma) for 1 h at room temperature, protected from light. Secondary antibodies were washed off thrice with PBS for 5 min, then coverslips were mounted onto microscope slides (Fisher Scientific) using Aqua-Poly/Mount (Polysciences) and left to dry for at least 90 min before imaging.Cells were imaged using a fluorescence microscope (Leica DMI4000 B) at 63× magnification, using the following filters: DAPI-1160A (Semrock, ex: 387 nm, em: 447 nm) and CY5-404A (Semrock, ex: 628 nm, em: 692 nm). Fluorescence signals were quantified using the software Fiji (version 2.16.0).23
Lipid droplet and lysosome labelling
Cells were seeded on glass coverslips as for immunocytochemistry. At the end of the treatment time, cells were incubated in phenol-free DMEM (Thermo Fisher Scientific) with BODIPY 403/503 (10 μM, Thermo Fisher Scientific), LysoTracker Deep Red (50 nM, Thermo Fisher Scientific) and Hoechst 33342 (10 μM) for 20 min at 37 °C. Cells were washed in phenol-free DMEM before live imaging with a fluorescence microscope (Leica DMI4000 B) at 63×, with the filters GFP-3035B (Semrock, ex: 472 nm, em: 520 nm), CY5-404A and DAPI-1160A.2-NBD-glucose imaging
Cells were seeded on glass coverslips as for immunocytochemistry, and cultured for 24 h. Cells were deprived from serum for 24 h, then incubated for 1 h with insulin, insulin–Au(III) or HAuCl4 in medium without glucose and phenol red (Hanks buffered salt solution, Thermo Fisher Scientific). Fluorescent glucose (2-NBD-glucose, Abcam) was added at the end of treatment time for a final concentration of 100 μg mL−1. After 10 min, cells were washed twice with PBS, then fixed in 4% (w/v) paraformaldehyde in PBS for 10 min. Cell nuclei were fluorescently labelled with Hoechst 33342 (10 μM, 10 min). Cells were washed thrice with PBS, then mounted on microscope slides with Aqua-Poly/Mount mounting medium. Cells were imaged using a fluorescence microscope (Leica DMI4000 B) at 63×, with the filters GFP-3035B.Phagocytosis
Cells were seeded on glass coverslips as for immunocytochemistry and kept for 24 h. Cells were deprived from serum for 24 h, then incubated for 4 h with insulin, insulin–Au(III) or HAuCl4, in the presence of fluorescent FluoSpheres polystyrene microbeads (Invitrogen, ex: 580 nm, em: 605 nm) at 1 × 106 beads per mL. Cells were then washed twice with PBS and fixed in 4% (w/v) paraformaldehyde for 10 min. Nuclei were labelled with Hoechst 33342 (10 μM, 10 min) and F-actin was labelled with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). Coverslips were mounted on microscope slides with Aqua-Poly/Mount, then cells were imaged using a fluorescence microscope (Leica DMI4000 B) at 20×, with the filters DAPI-1160A, GFP-3035B, and TRITC-A (Semrock, ex: 542 nm, em: 620 nm).Statistics
Statistical analyses were performed with one-way ANOVA followed by Dunnett's post hoc test. A Student's t-test was applied to compare the means of two groups. p values lower than 0.05 were considered significant. Experiments were performed independently at least three times.Results and discussion
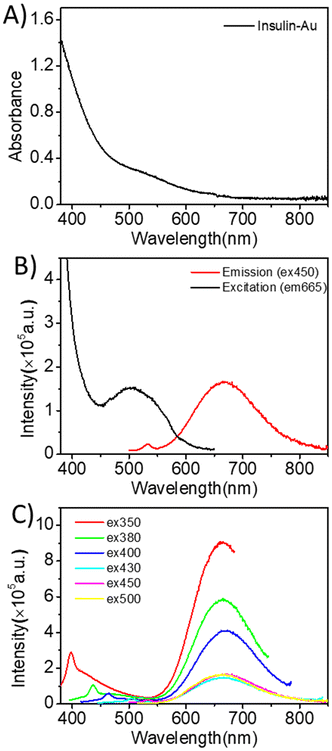
Results from this study are obtained using insulin–Au(III) conjugate in human microglia expressing insulin receptors. The biological investigations comparing insulin–Au(III) to insulin alone support the notion that conjugating one auric gold to insulin does not adversely change the key steps in insulin signaling and effects on organelles. The relevance of the pathways investigated in this study to the biological role of insulin in cells and tissues was thoroughly reviewed.4 Other literature highlighting the role of insulin in neurological disorders, where microglia and insulin impact brain metabolism and pathologies are represented by Yang, Doust and others.1,2,10,11The physicochemical characterization of insulin–Au(III) is summarized in Table 1 and optical spectra are shown in Fig. 1. The interaction of one Au(III) atom with one insulin was demonstrated by electrospray mass spectrometry. The structure of insulin contains the A and B chains, linked together by disulfide bonds.24 The secondary structure comprises of 47% alpha helices and 4% beta sheets in chain A, and 46% alpha helices and 3% beta sheets in chain B.25 The presence of Au(III) only affects the local alpha-helix environment of the coordination site, where 3 of 4 tyrosine residues are in helix motifs.
| NAME | Composition | Size: hydrodynamic diameter (nm) | Luminescence properties |
|---|---|---|---|
| Insulin–Au(III) | Insulin–3H·Au(III): 3 deprotonated phenolic groups in tyrosine residues serve as anchors for stabilizing Au(III) ions. | 7.4, Size from fluorescence anisotropy26 | Photoemission peak at 667 nm. |
| Formula: C257H380N65O77S6Au | 7, Size from DLS16 | Luminescence quantum yield (QYs) 5.40% | |
| Molecular weight: 6001.62 g mol−1 | Luminescence lifetime components: 237 ns and 1723 ns (ref. 16) | ||
| ESI-mass spectrometry16 |
The strong red emission and high fluorescence quantum yield (QY = 5.4%)16 of insulin–Au(III) are attractive for targeted bioimaging. The absorption (Fig. 1A) and emission spectra (Fig. 1B and C) show an absorption within the visible region and a strong emission peak at ∼665 nm regardless of the excitation wavelength (350 nm, 380 nm, 400 nm, 420 nm, 450 nm and 500 nm). However, the intensity of the emission was remarkably different, with excitation at 350 nm yielding a maximal emission (red line). Such a remarkable emission was more difficult to detect in low concentrations of insulin–Au(III) corresponding to physiological post-prandial insulin levels (0.3 nM). We have previously shown that gold nanoclusters can be detected in glia and neuron-like differentiated SH-SY5Y cells,27 and studies by Liu et al. showed internalized insulin–gold nanoclusters at supraphysiological concentration (250 mg L−1) in C2C12 myoblasts.15 Liu's studies also show that fluorescence of the insulin gold nanoclusters is strongly quenched in rodent brain homogenate, suggesting that abundance of insulin degrading enzymes (IDE) affects detection.28,29
In order to better understand the structural implications of Au(III) and insulin interactions with its receptor, we provide computational and theoretical data from molecular dynamic simulations (Fig. 2). First, we analyzed the structure of the insulin receptor and its interaction with insulin. Insulin is a hormone consisting of chain A (21 amino acids) and chain B (30 amino acids) linked by disulfide bridges.24,30 The insulin receptor is a homodimer, in which each monomer consists of 3 different chains (chain A with 211 amino acids, chain B with 214 amino acids and chain C with 593 amino acids), as shown in Fig. 2A.31 Interactions involve the insulin chain B (Ser 9, Glu13, Tyr 16 and Glu 21) and the insulin receptor chain C (Phe 39, Lys 40, Arg 65 and Glu97) through the formation of a salt bridge between Glu13, Arg65 and nonbonded contacts (Fig. 2B, left panel).
Next, we assessed differences that Au(III) could induce into the structure of insulin, and how it would influence its interaction with the insulin receptor. The position of the Au atom within insulin was previously described,16 and tyrosine residues were identified as the coordinating residues for Au. At the atomic level, molecular dynamics simulations of pure insulin were performed to show structural changes in insulin. We were interested in the distance between four tyrosine residues that are candidates for the coordination of the auric gold. Distances between those four tyrosine residues have been analyzed, and the structure with the shortest average distance was selected as representative of the effect of Au: its coordination brings tyrosine residues closer. The right panel of Fig. 2B shows the selected MD simulation of insulin in overlap with the insulin receptor. The overall structure of insulin was well conserved in the MD simulations. Further analysis shows that insulin Tyr16 interacts with the insulin receptor, leaving Tyr14, Tyr19 and Tyr26 to coordinate around Au.
Given that the addition of the gold atom is unlikely to disrupt the biological activity of insulin, we asked whether insulin–Au(III) can engage the same signaling pathways as insulin. One of the first kinases activated at the insulin receptor is Akt1, which becomes phosphorylated within the same time frame and to a comparable extent with insulin–Au(III) as with insulin (Fig. 3A and B). The tyrosine phosphorylation of some substrates activates phosphatidylinositol-3-kinase (PI3K), which produces polyphosphoinositides that interact with protein kinases, leading to activation of the kinase Akt1.4 Phosphorylation of Akt1 (Phospho-Akt1) is a reliable readout of Akt1 activation by insulin.4,32 The second major signaling cascade downstream of the insulin receptor involves the phosphorylation of ERK1/2, which translocate to the nucleus as a transcription factor for cell growth and survival.33 The response to insulin can differ depending on the brain tissue; in the mediobasal hypothalamus, insulin preferentially activates Akt, whereas ERK1/2 was mainly activated in the dorsal vagal complex.34 This partitioned response is likely tightly regulated to prevent dysregulated signaling. Hyperinsulinemia and chronic ERK1/2 activation are both detrimental to cell function.35 Insulin–Au(III) exerted comparable effects as insulin on the activation of ERK1/2, which increased in the nucleus following its phosphorylation (Fig. 3C and D).36 This demonstrates that insulin–Au(III) usage in human cells does not cause aberrances in the canonical effects of insulin, and is effective in the concentration range (∼0.3 nM) relevant to post-prandial physiology.
In response to insulin, cells adjust their metabolic processes towards anabolic pathways. Among the cellular processes controlled by insulin are activities of metabolic enzymes, transcriptional factors, and degradation of insulin itself.4 Disrupting the expression of the insulin receptor in the mouse brain results in significant changes in fat metabolism and body weight.37 Downstream of Akt1 activation are the mammalian target of rapamycin complexes (mTORC), which stimulate protein synthesis at the expense of protein breakdown.33 Thus, mTORC inhibits TFEB by phosphorylation at Ser211, retaining it in the cytosol and preventing its translocation to the nucleus to regulate lysosomal biogenesis.13,38 We show that insulin–Au(III) exerts comparable effects to that of insulin in decreasing nuclear TFEB in microglia cells (Fig. 4A and B). TFEB plays multiple roles in cells and has been considered a therapeutic target in several pathologies.13 TFEB together with TFE3 play critical roles in lysosomal biogenesis, cellular adaptation to stress,39,40 as well as whole body energy metabolism.41,42 TFEB modulation by mTORC1 forms a feedback loop that coordinates the balance between lysosomal catabolism and anabolism to adapt to different metabolic conditions.43
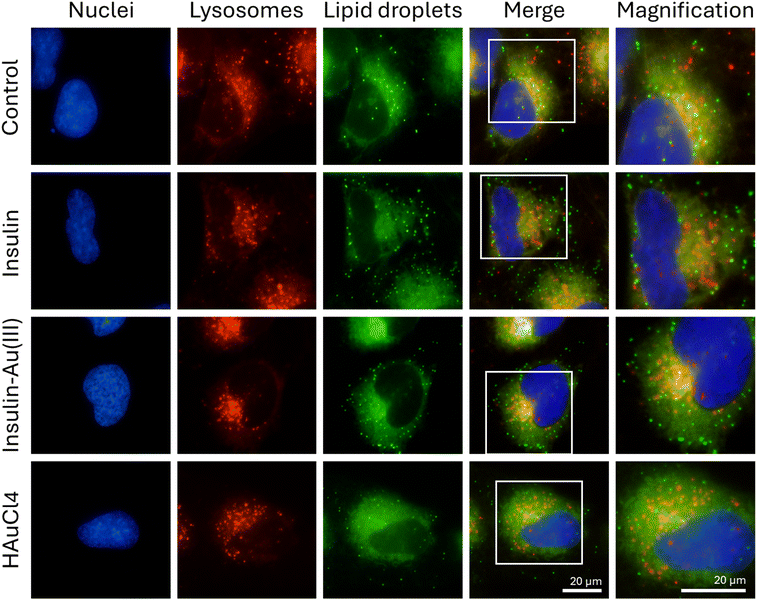
Lysosomes act as storage and degradation sites not only for proteins, but also lipids. In lipophagy, the lysosome-associated membrane protein 2 (LAMP2) interact with perilipins 2 and 3 on the surface of lipid droplets. Lipid droplets thus engulfed and degraded by lysosomes provide fatty acids to mitochondrial β-oxidation.44 Conversely, activation of anabolic pathways and inhibition of the lysosomal master regulator TFEB result in lipogenesis.45 Our results suggest that lipid droplet abundance is slightly increased following treatment with either insulin or insulin–Au(III) (Fig. 4C and D), as they interact closely with lysosomes in microglia cells (Fig. 5) in the perinuclear area. This physiological fluctuation in lipid droplet numbers resulting from a metabolic shift is unlike the significant increase observed in stressed microglia in inflammation. Lipid droplets assist mitochondria during autophagy,46 sequester lipophilic agents, and serve as signaling hubs for lipid metabolism and catabolism.47–49 The modulation of lipid droplets is therefore within the scope of therapeutic avenues considered for neurological disorders. For example, some gold nanoclusters were effective in reducing excessive oxidative stress and keeping lipid droplets within the physiological range.27,50 Many studies, including ours, have shown that bacterial lipopolysaccharide can increase lipid droplets concurrently with levels of inflammatory cytokines and reactive oxygen species.51,52 Lipid droplets also accumulate in microglia under dysfunctional and pro-inflammatory states of the aging brain.53 Nevertheless, lipid droplets are crucial in supporting normal cell functions, particularly under restricted nutrient supply. The functional outcomes of insulin–Au(III) on microglia were verified in Fig. 6. Insulin and insulin–Au(III) increased the uptake of fluorescently-labelled glucose to a comparable extent within the time frame of its physiological effectiveness (Fig. 6A and B). Microglial functionality was also assessed by the phagocytosis of fluorescent beads (Fig. 6C and D). Microglia phagocytosis is a crucial function that fulfills synaptic health as well as defense and clean up roles in the brain.54 In many neurodegenerative diseases, functionally impaired microglia result in the aberrant accumulation of neurotoxic plaques, tangles, etc.55 Insulin and insulin–Au(III) slightly increased microglial phagocytosis within a few hours, showing that these cells are overall sensitive to insulin at several levels: cell signaling, metabolic adaptation, and functionality.
The present study focuses on microglia and employs a novel nanostructure where one single atom of gold is conjugated with insulin molecule. This unique nano-construct is distinct from gold nanoclusters with multiple gold atoms, or the even bigger gold nanoparticles. The uniqueness is associated with its extremely long lifetime, not yet exploited for bioimaging, or for the development of assays to determine insulin receptor abundance and insulin-receptor interactions. The advantage of a single gold atom bound to insulin is that it does not significantly change the size of insulin, does not interfere with insulin biological activity, and provides an imaging tool for live cells in potentially different brain regions, as well as peripheral tissues. Insulin receptors are present in the brain, and insulin signalling specifically modulates brain function with diverse metabolic or cognitive outcomes, affecting memory, olfactory perception, emotional regulation, eating behaviour, and peripheral metabolism. For example, the hippocampus is known for its prominent role in learning and memory, and hippocampal insulin receptors regulate structural and functional plasticity to enhance cognition. Hippocampal insulin resistance might occur independently of peripheral insulin resistance. Insulin signaling in microglia plays a key role in microglial cellular metabolism and neuroinflammation; reduced insulin signaling in microglia alters mood and social behavior, and accelerates Alzheimer's pathogenesis.56 Improving central insulin action could represent a therapeutic option for people at an increased risk of developing metabolic and cognitive diseases.57 Taken together, these results suggest that the modification of insulin with Au(III) does not markedly change the biological activity of the protein at key signaling steps in human microglia. It is an interesting demonstration of “tagging” a protein relevant for tracing experiments, valuable for further investigations in human cells and human cerebral organoids.
Conclusion
Investigations by numerous laboratories and clinical studies indicate that insulin controls a complex series of intertwining pathways that instruct cells on how and when to store energy. While extensive progress has been made, numerous gaps remain in our understanding of the precise molecular events of insulin action in different tissues. We only started to appreciate the modification of proteins at the nanoscale level, and how the complex insulin structure arranges around a single auric gold. It is clear that the single Au(III) ion within insulin–Au(III) does not cause major molecular perturbation, but is stabilized with the protein. Insulin–Au(III) exerts comparable effects as that of insulin in human microglia, which is desirable to avoid disrupting glucose imbalance and proteasomal homeostasis. Lipid droplets, lysosomes and their transcription factors are essential for cellular function, or negative effects ensue if they are too abundant or reduced. Insulin–Au(III) could have several advantages for studies and medical uses of insulin, such as stability, solubility, and further mechanistic investigations in models of neurological disorders using human brain organoids. The insulin–Au(III) conjugate can be excited in the visible spectral region and has a long excited state lifetime, which are desirable properties for cell imaging and therapeutic purposes. Moreover, large two-photon absorption cross sections renders insulin–Au(III) conjugate a suitable candidate for multiphoton excited luminescence (670 nm) for living cells and organoids.Author contributions
D. M., I. Z., Z. S. M. conceptualized the studies and wrote the manuscript drafts and revisions. H. Y. performed synthesis and characterization of insulin–Au(III). I. Z. performed all experiments in cells. Z. S. M. carried out modeling, analyzed and prepared the results in publishable format. V. B. K. and R. A. participated in the initial experimental plan and provided feedback on the text.Conflicts of interest
There are no conflicts of interest to declare.Data availability
All quantified data in the manuscript show individual data points within the main text, and publically available softwares are used.Acknowledgements
DM acknowledges funding from NSERC (RGPIN 2020-07011). ZSM and VBK were supported by the project STIM-REI (contract number: KK.01.1.1.01.0003), funded by the European Union through the European Regional Development Fund—the Operational Programme Competitiveness and Cohesion 2014–2020 (KK.01.1.1.01). HY is grateful for PhD fellowships donated by the China Scholarship Council (CSC). RA and HY acknowledge the Shanghai Science and Technology Innovation Program (22520712500) for support. The Agence Nationale de la Recherche (project nanoGOLD, ANR-22-CE29-0022) is acknowledged for support.References
- J. L. Milstein and H. A. Ferris, The brain as an insulin-sensitive metabolic organ, Mol. Metab., 2021, 52, 101234 CrossRef CAS PubMed.
- L. A. Post, J. A. Kulas, J. L. Milstein, S. V. L. Sebastian, S. Hosseini-Barkooie and M. E. Stevenson, et al., Inceptor as a regulator of brain insulin sensitivity, Sci. Rep., 2023, 13(1), 11582 CrossRef CAS PubMed.
- A. Kleinridders, W. Cai, L. Cappellucci, A. Ghazarian, W. R. Collins and S. G. Vienberg, et al., Insulin resistance in brain alters dopamine turnover and causes behavioral disorders, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(11), 3463–3468 CrossRef CAS PubMed.
- A. R. Saltiel, Insulin signaling in health and disease, J. Clin. Invest., 2021, 131(1), e142241 CrossRef CAS PubMed.
- E. Broberg, J. English, D. M. Clarke, M. J. Shin, B. T. Bikman and P. R. Reynolds, et al., Differential Regulation of PKM2, AMPK, and mTOR in Response to Insulin and Dietary Management, Cells, 2025, 14(6), 416 CrossRef CAS PubMed.
- E. Młynarska, W. Czarnik, N. Dzieża, W. Jędraszak, G. Majchrowicz and F. Prusinowski, et al., Type 2 Diabetes Mellitus: New Pathogenetic Mechanisms, Treatment and the Most Important Complications, Int. J. Mol. Sci., 2025, 26(3), 1094 CrossRef PubMed.
- M. Soták, M. Clark, B. E. Suur and E. Börgeson, Inflammation and resolution in obesity, Nat. Rev. Endocrinol., 2025, 21(1), 45–61 CrossRef PubMed.
- C. D. Rae, J. A. Baur, K. Borges, G. Dienel, C. M. Díaz-García and S. R. Douglass, et al., Brain energy metabolism: A roadmap for future research, J. Neurochem., 2024, 168(5), 910–954 CrossRef CAS PubMed.
- C. Green, V. Zaman, K. Blumenstock, N. L. Banik and A. Haque, Dysregulation of Metabolic Peptides in the Gut–Brain Axis Promotes Hyperinsulinemia, Obesity, and Neurodegeneration, Biomedicines, 2025, 13(1), 132 CrossRef CAS PubMed.
- X. Yang, Y. Xu, W. Gao, L. Wang, X. Zhao and G. Liu, et al., Hyperinsulinemia-induced microglial mitochondrial dynamic and metabolic alterations lead to neuroinflammation in vivo and in vitro, Front. Neurosci., 2022, 16 Search PubMed , [cited 2025 Mar 29]. Available from: https://www.frontiersin.org/journals/neuroscience/articles/10.3389/fnins.2022.1036872/full.
- Y. V. Doust, N. Sumargo, J. M. Ziebell and D. Premilovac, Insulin Resistance in the Brain: Evidence Supporting a Role for Inflammation, Reactive Microglia, and the Impact of Biological Sex, Neuroendocrinology, 2022, 112(11), 1027–1038 CrossRef CAS PubMed.
- M. Cao, X. Luo, K. Wu and X. He, Targeting lysosomes in human disease: from basic research to clinical applications, Signal Transduction Targeted Ther., 2021, 6(1), 1–28 CrossRef PubMed.
- H. Chen, S. Gong, H. Zhang, Y. Chen, Y. Liu and J. Hao, et al., From the regulatory mechanism of TFEB to its therapeutic implications, Cell Death Discovery, 2024, 10, 84 CrossRef CAS PubMed.
- X. Yan, L. Yang, X. Fu, X. Luo, C. Wang and Q. P. Xie, et al., Transcription factor EB, a promising therapeutic target in cardiovascular disease, PeerJ, 2024, 12, e18209 CrossRef PubMed.
- C. L. Liu, H. T. Wu, Y. H. Hsiao, C. W. Lai, C. W. Shih and Y. K. Peng, et al., Insulin-directed synthesis of fluorescent gold nanoclusters: preservation of insulin bioactivity and versatility in cell imaging, Angew. Chem., Int. Ed., 2011, 50(31), 7056–7060 CrossRef CAS PubMed.
- D. Bain, H. Yuan, A. Pniakowska, A. Hajda, C. Bouanchaud and F. Chirot, et al., One- and two-photon brightness of proteins interacting with gold. A closer look at gold–insulin conjugates, Nanoscale, 2024, 16(31), 14953–14958 RSC.
- L. Zhang, Y. Zhou, Z. Yang, L. Jiang, X. Yan and W. Zhu, et al., Lipid droplets in central nervous system and functional profiles of brain cells containing lipid droplets in various diseases, J. Neuroinflammation, 2025, 22(1), 7 CrossRef PubMed.
- R. Verma, P. Sharma, V. Sharma and T. G. Singh, Modulating lipid droplet dynamics in neurodegeneration: an emerging area of molecular pharmacology, Mol. Biol. Rep., 2025, 52(1), 277 CrossRef CAS PubMed.
- M. A. Welte, Expanding roles for lipid droplets, Curr. Biol., 2015, 25(11), R470–R481 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, C. Zhuang, J. Qi, R. C. Zhao and J. Wang, Lipid droplets in the nervous system: involvement in cell metabolic homeostasis, Neural Regener. Res., 2025, 20(3), 740–750 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith and B. Hess, et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch and J. H. Morris, et al., UCSF ChimeraX: Meeting modern challenges in visualization and analysis, Protein Sci., 2018, 27(1), 14–25 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair and T. Pietzsch, et al., Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9(7), 676–682 CrossRef CAS PubMed.
- S. G. Chang, K. D. Choi, S. H. Jang and H. C. Shin, Role of Disulfide Bonds in the Structure and Activity of Human Insulin, Mol. Cells, 2003, 16(3), 323–330 CrossRef CAS PubMed.
- S. Delbeck and H. M. Heise, Quality Assurance of Commercial Insulin Formulations: Novel Assay Using Infrared Spectroscopy, J. Diabetes Sci. Technol., 2021, 15(4), 865–873 CrossRef CAS PubMed.
- A. Soleilhac, F. Bertorelle and R. Antoine, Sizing protein-templated gold nanoclusters by time resolved fluorescence anisotropy decay measurements, Spectrochim. Acta, Part A, 2018, 193, 283–288 CrossRef CAS PubMed.
- I. Zhang, D. Maysinger, M. Beus, A. Mravak, Z. Yu and M. P. Bakulić, et al., Gold nanoclusters Au25AcCys18 normalize intracellular ROS without increasing cytoplasmic alarmin acHMGB1 abundance in human microglia and neurons, Nanoscale, 2025, 17(2), 1092–1104 RSC.
- M. A. Bessard, A. Moser, E. Waeckel-Énée, V. Lindo, A. Gdoura and S. You, et al., Insulin-degrading enzyme regulates insulin-directed cellular autoimmunity in murine type 1 diabetes, Front. Immunol., 2024, 15 Search PubMed , [cited 2025 Mar 30]. Available from: https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2024.1474453/full.
- M. Corraliza-Gomez, T. Bermejo, J. Lilue, N. Rodriguez-Iglesias, J. Valero and I. Cozar-Castellano, et al., Insulin-degrading enzyme (IDE) as a modulator of microglial phenotypes in the context of Alzheimer's disease and brain aging, J. Neuroinflammation, 2023, 20(1), 233 CrossRef CAS PubMed.
- V. I. Timofeev, R. N. Chuprov-Netochin, V. R. Samigina, V. V. Bezuglov, K. A. Miroshnikov and I. P. Kuranova, X-ray investigation of gene-engineered human insulin crystallized from a solution containing polysialic acid, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2010, 66(Pt 3), 259–263 CrossRef CAS PubMed.
- J. G. Menting, J. Whittaker, M. B. Margetts, L. J. Whittaker, G. K. W. Kong and B. J. Smith, et al., How insulin engages its primary binding site on the insulin receptor, Nature, 2013, 493(7431), 241–245 CrossRef CAS PubMed.
- Y. B. Kim, S. E. Nikoulina, T. P. Ciaraldi, R. R. Henry and B. B. Kahn, Normal insulin-dependent activation of Akt/protein kinase B, with diminished activation of phosphoinositide 3-kinase, in muscle in type 2 diabetes, J. Clin. Invest., 1999, 104(6), 733–741 CrossRef CAS PubMed.
- P. De Meyts, The Insulin Receptor and Its Signal Transduction Network, ed. K. R. Feingold, S. F. Ahmed, B. Anawalt, M. R. Blackman, A. Boyce, G. Chrousos, et al., MDText.com, Inc., South Dartmouth (MA), 2000, [cited 2025 Apr 1]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK378978/ Search PubMed.
- B. M. Filippi, C. S. Yang, C. Tang and T. K. T. Lam, Insulin Activates Erk1/2 Signaling in the Dorsal Vagal Complex to Inhibit Glucose Production, Cell Metab., 2012, 16(4), 500–510 CrossRef CAS PubMed.
- N. Rachdaoui, L. Polo-Parada and F. Ismail-Beigi, Prolonged Exposure to Insulin Inactivates Akt and Erk1/2 and Increases Pancreatic Islet and INS1E β-Cell Apoptosis, J. Endocr. Soc., 2018, 3(1), 69–90 CrossRef PubMed.
- R. Roskoski, ERK1/2 MAP kinases: Structure, function, and regulation, Pharmacol. Res., 2012, 66(2), 105–143 CrossRef CAS PubMed.
- J. C. Brüning, D. Gautam, D. J. Burks, J. Gillette, M. Schubert and P. C. Orban, et al., Role of Brain Insulin Receptor in Control of Body Weight and Reproduction, Science, 2000, 289(5487), 2122–2125 CrossRef PubMed.
- J. A. Martina, Y. Chen, M. Gucek and R. Puertollano, MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB, Autophagy, 2012, 8(6), 903–914 CrossRef CAS PubMed.
- S. J. Jeong, J. Stitham, T. D. Evans, X. Zhang, A. Rodriguez-Velez and Y. S. Yeh, et al., Trehalose causes low-grade lysosomal stress to activate TFEB and the autophagy-lysosome biogenesis response, Autophagy, 2021, 17(11), 3740–3752 CrossRef CAS PubMed.
- N. Raben and R. Puertollano, TFEB and TFE3: Linking Lysosomes to Cellular Adaptation to Stress, Annu. Rev. Cell Dev. Biol., 2016, 32(1), 255–278 CrossRef CAS PubMed.
- N. Pastore, A. Vainshtein, T. J. Klisch, A. Armani, T. Huynh and N. J. Herz, et al., TFE3 regulates whole-body energy metabolism in cooperation with TFEB, EMBO Mol. Med., 2017, 9(5), 605–621 CrossRef CAS PubMed.
- C. Settembre, R. De Cegli, G. Mansueto, P. K. Saha, F. Vetrini and O. Visvikis, et al., TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop, Nat. Cell Biol., 2013, 15(6), 647–658 CrossRef CAS PubMed.
- R. E. Lawrence and R. Zoncu, The lysosome as a cellular centre for signalling, metabolism and quality control, Nat. Cell Biol., 2019, 21(2), 133–142 CrossRef CAS PubMed.
- S. Zhang, X. Peng, S. Yang, X. Li, M. Huang and S. Wei, et al., The regulation, function, and role of lipophagy, a form of selective autophagy, in metabolic disorders, Cell Death Dis., 2022, 13(2), 1–11 Search PubMed.
- S. Kersten, Mechanisms of nutritional and hormonal regulation of lipogenesis, EMBO Rep., 2001, 2(4), 282–286 CrossRef CAS PubMed.
- T. Klecker, R. J. Braun and B. Westermann, Lipid Droplets Guard Mitochondria during Autophagy, Dev. Cell, 2017, 42(1), 1–2 CrossRef CAS PubMed.
- C. Klose, M. A. Surma and K. Simons, Organellar lipidomics–background and perspectives, Curr. Opin. Cell Biol., 2013, 25(4), 406–413 CrossRef CAS PubMed.
- P. Prakash, C. E. Randolph, K. A. Walker and G. Chopra, Lipids: Emerging Players of Microglial Biology, Glia, 2025, 73(3), 657–677 CrossRef CAS PubMed.
- M. E. Traetta, H. A. Vecchiarelli and M. È. Tremblay, Fundamental Neurochemistry Review: Lipids across microglial states, J. Neurochem., 2025, 169(1), e16259 CrossRef CAS PubMed.
- D. Z. Wei, D. Li, D. M. Zheng, Z. M. An, X. J. Xing and D. W. Jiang, et al., Curcumin Conjugated Gold Nanoclusters as Perspective Therapeutics for Diabetic Cardiomyopathy, Front. Chem., 2021, 9 Search PubMed , [cited 2025 Apr 1]. Available from: https://www.frontiersin.org/journals/chemistry/articles/10.3389/fchem.2021.763892/full.
- M. Lalancette-Hébert, A. Moquin, A. O. Choi, J. Kriz and D. Maysinger, Lipopolysaccharide-QD Micelles Induce Marked Induction of TLR2 and Lipid Droplet Accumulation in Olfactory Bulb Microglia, Mol. Pharm., 2010, 7(4), 1183–1194 CrossRef PubMed.
- M. E. Tremblay, I. Zhang, K. Bisht, J. C. Savage, C. Lecours and M. Parent, et al., Remodeling of lipid bodies by docosahexaenoic acid in activated microglial cells, J. Neuroinflammation, 2016, 13(1), 116 CrossRef PubMed.
- J. Marschallinger, T. Iram, M. Zardeneta, S. E. Lee, B. Lehallier and M. S. Haney, et al., Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain, Nat. Neurosci., 2020, 23(2), 194–208 CrossRef CAS PubMed.
- C. Arcuri, C. Mecca, R. Bianchi, I. Giambanco and R. Donato, The Pathophysiological Role of Microglia in Dynamic Surveillance, Phagocytosis and Structural Remodeling of the Developing CNS, Front. Mol. Neurosci., 2017, 10, 191 CrossRef PubMed.
- S. Nizami, H. Hall-Roberts, S. Warrier, S. A. Cowley and E. Di Daniel, Microglial inflammation and phagocytosis in Alzheimer's disease: Potential therapeutic targets, Br. J. Pharmacol., 2019, 176(18), 3515–3532 CrossRef CAS PubMed.
- W. Chen, X. Liu, V. R. Munoz and C. R. Kahn, Loss of Insulin Signaling in Microglia Impairs Cellular Uptake of Aβ and Neuroinflammatory Response Exacerbating Alzheimer-like Neuropathology [Internet]. bioRxiv, 2024, preprint [cited 2025 Jun 9]. p. 2024.08.22.609112. Available from: https://www.biorxiv.org/content/10.1101/2024.08.22.609112v1.
- R. Agrawal, C. M. Reno, S. Sharma, C. Christensen, Y. Huang and S. J. Fisher, Insulin action in the brain regulates both central and peripheral functions, Am. J. Physiol.: Endocrinol. Metab., 2021, 321(1), E156–E163 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |