Open Access Article
Open Access ArticleMachine learning-guided fabrication of carbon dot–pepsin nano-conjugates for enhanced bioimaging, synergistic drug delivery, and visible light-induced photosensitization†
Subhrajeet
Banerjee
 a,
Saptarshi
Mandal‡
a,
Saptarshi
Mandal‡
 a,
Sneha
Singh‡
b,
Abhik
Sen
*b and
Prolay
Das
a,
Sneha
Singh‡
b,
Abhik
Sen
*b and
Prolay
Das
 *a
*a
aDepartment of Chemistry, Indian Institute of Technology Patna, Bihta, 801103, India. E-mail: prolay@iitp.ac.in
bDepartment of Molecular Biology, Indian Council of Medical Research-Rajendra Memorial Research Institute of Medical Sciences, Agam Kuan, Patna, 800007, India. E-mail: abhik.sen@icmr.gov.in
First published on 23rd May 2025
Abstract
Carbon dots (CDs) are emerging as next-generation bioimaging agents due to their strong fluorescence, photobleaching resistance, and biocompatibility. However, their small size often limits efficient cell internalization, leading to unspecified cellular targets and high endosomal escape. Herein, we showcase the potential of pepsin, a simple proteolytic enzyme, to address the limitations of CDs through its covalent conjugation and subsequent creation of CD–pepsin nanoparticles (NPs). Interestingly, repeated failures in achieving CD–pepsin NPs prompted us to explore machine learning (ML), which, in a novel approach, successfully predicted the most favourable conditions for their formation. These nanoparticles unlocked two critical functionalities beyond enhanced imaging: visible light-induced reactive oxygen species (ROS) generation and sustained drug delivery, highlighting their potential for chemo-photodynamic combination therapy. The photophysical properties of the green-emissive CDs, synthesized from 3-hydroxy-2-naphthoic acid, were significantly enhanced through covalent conjugation with pepsin. This enhancement was evident in the efficient confocal imaging of human macrophage cells (THP1 cells) using both the CD–pepsin covalent conjugate and CD–pepsin NPs. Beyond enhancing the imaging potential of CDs by stabilizing their surface states through covalent conjugation with pepsin, this modification also significantly increased ROS generation, surpassing conventional photosensitizers like protoporphyrin (PpIX). Irradiation of the cells with a simple 23 W white LED induced increased oxidative stress, leading to reduced cell viability. Simultaneously, the CD–pepsin NPs functioned as a drug delivery vehicle, enabling the sustained release of the model drug, doxorubicin, over 4–5 days. These combined functionalities—bioimaging, photosensitization, and drug delivery—highlight CD–pepsin NPs as a versatile and effective multipurpose platform, overcoming key limitations of pristine CDs for advanced theranostics.
1. Introduction
A high fluorescence quantum yield of a molecule indicates efficient radiative transitions, typically accompanied by suppressed internal conversion and other non-radiative pathways.1 Interestingly, carbon dots (CDs) are a rare nanoscale material exhibiting intense fluorescence while maintaining exceptional energy transfer and electron transfer proficiency.2–4 When harnessed effectively, these unique photophysical properties of CDs can become a valuable asset for advancing biomedical research.5–7 Specifically, their potential for fluorescence tracking and bioimaging, combined with simultaneous photosensitization, is contextually highlighted. Not all CDs can fulfill this dual role, and predicting their suitability remains a significant challenge. The selection of substrates, reaction conditions, and purity are critical factors in fine-tuning the photophysical properties of CDs.8–11 Beyond influencing their photophysics, these factors also play a key role in equipping CDs with chemical connectors, such as surface functional groups, further enhancing their scope and purpose.12 Using appropriate conjugation chemistry, these functional groups allow CDs to covalently attach to biomolecules such as proteins, DNA, RNA, and lipids, enabling the formation of functional nanoparticles (NPs) through various methods.13–15 However, due to the diverse physical and chemical properties of biomolecules, predicting the optimal conditions for formulating biomolecule-based NPs remains challenging, with failed attempts often going unnoticed. Herein, we report the successful conjugation of a CD with the digestive enzyme pepsin, resulting in a CD–pepsin conjugate that was subsequently formulated into stable CD–pepsin NPs. This was achieved through artificial intelligence (AI) intervention to predict the optimal fabrication conditions, enabling simultaneous enhancement of bioimaging, model drug delivery, and photodynamic sensitization.CDs, a highly promising yet inexpensive class of carbon-based nanomaterials, possess exceptional biocompatibility, photostability, tuneable fluorescence, and distinctive surface chemistry, making them suitable for a wide range of biomedical applications.16,17 The bioimaging potential of CDs is well documented in the literature, but pristine CDs often face cell internalization issues mostly due to non-compatible surface charge and impurities.18 To overcome this, we hypothesized the conjugation of CDs with pepsin, leading to conjugates and ultimately to NPs that enhance the bioavailability of CDs within cells, facilitating improved cellular uptake for bioimaging applications. Moreover, the choice of pepsin is driven by the abundance of aromatic amino acids present in it, which reportedly play a critical role in band gap reduction, thereby enhancing the ROS generation capability of chemical entities.19,20
When excited by visible light, CDs generate ROS through energy or electron transfer processes that convert triplet oxygen molecules into singlet oxygen or other reactive species to induce oxidative stress, leading to cell damage or apoptosis, making them highly effective against cancer cells and pathogens.21–25 However, the relatively modest ROS production and lack of endocytosis of unmodified CDs can limit their effectiveness in therapeutic settings. To address this limitation, functional modifications with biomolecules like proteins have been explored to enhance ROS generation by improving energy transfer and providing more targeted interactions with oxygen molecules in the microenvironment, ultimately enhancing the specificity and efficacy of photodynamic therapy (PDT) applications.14 Encouragingly, pepsin itself significantly influences cellular oxidative stress26,27 and this effect could be further enhanced when conjugated with CDs, which have the potential to generate ROS. Pepsin, known for its ability to break down other proteins in the stomach's highly acidic environment,28,29 offers notable stability, specificity, and biocompatibility. This prompted us to utilize pepsin for bioconjugation with CDs, aiming to synergistically enhance their theranostics potential for bioimaging and photosensitization. To our knowledge, this effect has never been demonstrated with CDs conjugated to pepsin—a proteolytic enzyme primarily known for its role in digestion rather than therapeutic applications.
Protein-based nano-formulations hold promise for drug delivery, tissue engineering, and cancer therapy.30 However, variations in primary and secondary structures, polarity, solubility, charge, and other interdependent factors make a universal solution elusive, necessitating rigorous optimization. In this context, the limited number of cationic residues in pepsin hindered NP formation. To overcome this, we leveraged AI, as human intuition often struggles to recognize patterns in highly correlated and nonlinear factors. Using a machine learning (ML) model, we systematically predicted the most favourable conditions for NP formation, optimized reaction parameters, and significantly improved yield, effectively tackling complex formulation challenges in nanomedicine development. This is evidenced by the successful synthesis of CD–pepsin NPs, derived from 3-hydroxy-2-naphthoic acid-based green fluorescent CD and pepsin. Remarkably, the CD–pepsin NPs demonstrated enhanced ROS generation under irradiation from a simple 23 W white LED, surpassing the effectiveness of traditional photosensitizers like protoporphyrin IX (PpIX). Additionally, the nano-assembly displayed sustained release of the infused model drug payload (doxorubicin), thereby flexing its therapeutic prowess. This nano-assembly offers structural stability, preserves the photophysical properties of the CD–pepsin conjugate, and ensures consistent ROS generation and fluorescence under irradiation. It highlights the potential of enzyme-based nanomaterials to enhance CD functionality, marking a significant step forward in theranostics by seamlessly integrating imaging and therapeutic capabilities within a single nanostructure (Scheme 1).
2. Experimental section
2.1. Materials and reagents
3-Hydroxy-2-naphthoic acid, porcine pepsin, 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC·HCl), N-hydroxysuccinimide (NHS), sodium dodecylsulphate (SDS), acylamide, N,N’-methylenebisacrylamide, ammonium persulfate (≥98%), tetramethylethylenediamine (TEMED), sodium hydroxide, hydrochloric acid, magnesium chloride (MgCl2), acrylamide for electrophoresis, N,N-bisacrylamidedimethylformamide for electrophoresis, acetonitrile (HPLC grade), methanol, n-hexane, chloroform, dimethylformamide (DMF), protoporphyrin IX (PpIX), ethyl acetate, Coomassie brilliant blue, sodium azide, p-benzoquinone, terephthalic acid (TA), TiO2 (anatase), dihydrorhodamine-123 (DHR-123), RPMI 1640, FBS, L-glutamine, penicillin–streptomycin, paraformaldehyde, ProLong Gold Antifade reagent, poly-lysine, and 4′,6-diamidino-2-phenylindole (DAPI) were procured from commercial sources (Sigma, CDH, TCI, and SRL, India) and were used without further purification. Potassium bromide, a dialysis membrane (molecular weight cut off 2 kDa), doxorubicin hydrochloride (DOX·HCl), 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), and HPLC grade solvents were purchased from Sigma. Carbon-coated copper TEM grids (300 mesh) were obtained from Ted Pella Inc. A white LED (23 W) was purchased from Phillips. DI water was used as the solvent unless specified.2.2. Instrumentation
Transmission electron microscopy (TEM) images were captured on a JEM 2100 Plus multi-purpose electron microscope (JEOL). The samples were prepared by drop-casting dilute water dispersions of CD on copper TEM grids and then air-dried. Field emission scanning electron microscopy (FE-SEM) images were obtained on a Gemini SEM 500 microscope (ZEISS). Powder XRD (pXRD) analysis was performed at room temperature on an Empyrean, Malvern Panalytical, using Cu-Kα (λ = 1.54 Å) irradiation at a 2θ angle of 5°–60°. Fourier transform infrared (FTIR) spectra were obtained from a PerkinElmer Spectrum 400. The samples were dried in a vacuum concentrator (Eppendorf) and then pelletized with KBr in a hydraulic press. X-ray photoelectron (XPS) studies were performed using a Kratos Axis Ultra spectrometer with an incident monochromatic X-ray source of Al-Kα (KE = 1.4866 keV). A reverse-phase high-performance liquid chromatography (RP-HPLC) system (controller: CBM-20Alite, pump: LC-20AD, PDA detector: SPD-M20A; Shimadzu, Kyoto, Japan) with an Enable C18G column (250 × 4.6 mm) was used for obtaining HPLC chromatograms. UV-Vis absorption spectra and steady-state fluorescence spectra were recorded on a UV-2550 spectrophotometer (Shimadzu, Japan) and a Fluoromax-4P spectrofluorometer (Horiba Jobin Yvon), respectively, using a quartz cuvette with a path length of 1 cm. Time-resolved fluorescence emission decays were recorded using a laser of wavelength 375 nm on a time-resolved fluorescence spectrophotometer (LifeSpec-II, Edinburgh Instruments, UK), working based on the picosecond time-correlated single photon counting (TCSPC) technique. Circular dichroism measurements were performed with a JASCO 1500 circular dichroism spectrophotometer equipped with a Peltier based temperature control system. The data were acquired between wavelengths of 190 and 320 nm at a scan rate of 100 nm min−1 using a micro-cuvette with a path length of 1 mm. Digital images were captured on a Canon EOS 600D. The hydrodynamic radius (size) was determined by dynamic light scattering (DLS) analysis using water as a diluent on a Zetasizer ultra-particle analyzer (Malvern Panalytical). Zeta potential measurements were also carried out on the same instrument by electrophoretic light scattering.2.3. Synthesis of CDs
The CDs were synthesized by dissolving 1 mmol of 3-hydroxy-2-naphthoic acid in 5 mL of DMF. The solution was then heated to 220 °C for 10 h in a Teflon-lined hydrothermal autoclave. After cooling to room temperature, the mixture was centrifuged to remove large carbonized agglomerates, and the supernatant was collected. The supernatant was then filtered through a 0.22 μm syringe filter and subjected to dialysis against water for 48 h using a membrane (2 kDa MWCO) to eliminate any remaining by-products and impurities.2.4. Synthesis of the CD–pepsin conjugate
The CDs were conjugated with pepsin following carbodiimide chemistry31 using an earlier reported procedure with a slight modification. The CD solution (1.25 μL, 50 μg) was added to 800 μL of water, followed by the addition of EDC·HCl (100 μL, 52 μmol) and NHS solution (100 μL, 86 μmol). The pH of the solution was adjusted to around 7. The reaction mixture was kept for 1 h with occasional stirring to activate carboxyl groups on the surface of the CDs. Subsequently, 30 mg of pepsin was added to the carboxyl-activated CD solution, and pH was maintained at 8.5 with dilute NaOH solution. The reaction mixture was kept overnight at 4 °C for conjugation. To remove excess EDC, NHS, CD, and byproducts, the solution was taken in a mini-dialysis unit (MWCO 2 kDa) and dialyzed overnight against water with occasional changing of water. The dialyzed solution was collected and stored at 4 °C for further use.2.5. ML modelling for CD–pepsin nanoparticle synthesis
75 examples of CD–pepsin NP formation were created through practical experimentation. The data consist of different conditions used for the NP formation and results in terms of yield. The data were cleaned, and the NP formation yield was categorized into 5 classes (yield <20 as 1, <40 as 2, <60 as 3, <80 as 4, and <100 as 5). ML modelling was performed using the Orange software.32 The feed data consisted of 8 features and one target in terms of yield class. The features are categorical: salt and desolvation agent, and numeric: pepsin%, salt concentration (mM), pH, desolvation agent volume in mL, desolvation agent dipole moment, and desolvation agent dielectric constant. The data set was split into 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 percentages for training and testing the models. After training the models, the impact of the features on the successfully trained model output was visualized using the Explain Model widget.
20 percentages for training and testing the models. After training the models, the impact of the features on the successfully trained model output was visualized using the Explain Model widget.
2.6. Fabrication of nanoparticles by CD–pepsin and DOX loading
CD–pepsin NPs were prepared by the desolvation method with a few modifications. CD–pepsin aqueous solution with 3% protein content in 20 mM MgCl2 was prepared, and the pH was adjusted accordingly with dilute NaOH solution. A desolvating agent, 1.5 mL of methanol/chloroform mixture (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), was added dropwise to 1 mL of the CD–pepsin conjugate solution under magnetic stirring and kept overnight at room temperature. The NPs, thus formed, were harvested by ultracentrifugation at 20
1), was added dropwise to 1 mL of the CD–pepsin conjugate solution under magnetic stirring and kept overnight at room temperature. The NPs, thus formed, were harvested by ultracentrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min. The supernatant was also collected to calculate the percentage yield and entrapment efficiency of CD–pepsin NPs. For the encapsulation of the model drug, 20 μg of DOX was dispersed in the organic solvent by ultrasonication for 5 min, and then the dispersion was added dropwise to the CD–pepsin solution. The rest of the procedure was the same as that used for CD–pepsin NP preparation.
000 rpm for 30 min. The supernatant was also collected to calculate the percentage yield and entrapment efficiency of CD–pepsin NPs. For the encapsulation of the model drug, 20 μg of DOX was dispersed in the organic solvent by ultrasonication for 5 min, and then the dispersion was added dropwise to the CD–pepsin solution. The rest of the procedure was the same as that used for CD–pepsin NP preparation.
2.7. HPLC study and gel electrophoresis
A reverse phase high-performance liquid chromatography (RP-HPLC) system (controller: CBM-20Alite, pump: LC-20AD, PDA detector: SPD-M20A; Shimadzu, Kyoto, Japan) with an enable C18G column (250 × 4.6 mm) was used for obtaining HPLC chromatograms. 10 mM sodium phosphate buffer of pH 7.4 and acetonitrile (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15) was used as the mobile phase at a flow rate of 0.2 mL min−1 for HPLC analysis of CD, pepsin and CD–pepsin. The injection volume was 5 μL and the final chromatogram was extracted at a wavelength of 278 nm.
15) was used as the mobile phase at a flow rate of 0.2 mL min−1 for HPLC analysis of CD, pepsin and CD–pepsin. The injection volume was 5 μL and the final chromatogram was extracted at a wavelength of 278 nm.
12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was employed for analysing the conjugation between CD and pepsin in the BioRad gel electrophoresis system. The gel was run at 120 V for 2 h and then stained with Coomassie blue following the standard protocol. The stained gel was imaged using a Canon digital camera.
2.8 Yield (%) and entrapment efficiency (%) of the nanoparticles
The CD–pepsin yield was calculated using the following equation: | (1) |
Nanoparticle yield was calculated using the following equation:
 | (2) |
Entrapment efficiency was calculated using the following equation:
 | (3) |
The amount of CD–pepsin and drug (DOX) was calculated using a steady-state fluorescence spectrophotometer.
2.9 ROS generation study
DHR 123 (5 μL, 1 nM) solution was added to respective sample solutions and separately irradiated with a 23 W white LED. DHR 123 was oxidized to rhodamine 123 by ROS species generated by the samples, and the fluorescence of rhodamine 123 was measured using steady-state fluorescence spectroscopy at different intervals of time. For this, the excitation wavelength was kept fixed at 488 nm, and fluorescence emission was taken between 500 and 600 nm. As a standard PS, PpIX solution (5 μL, 1 mM) was used to perform the same experiment under similar conditions with the DHR 123 solution. Furthermore, to elucidate the mechanism of ROS generation by CD–pepsin NPs under visible light, 1 mM solutions of p-benzoquinone (p-BZQ), sodium azide (NaN3) and methanol (MeOH) were used as scavengers for superoxide (O2˙−), singlet oxygen (1O2) and hydroxy radical (˙OH), respectively. A decrease in ROS-mediated rhodamine 123 fluorescence upon the addition of a specific scavenger indicates the presence of the corresponding radical species.2.10. In vitro drug release profile
DOX-loaded CD–pepsin NPs (0.5 mL), equivalent to 14.1 μg of DOX load, were taken in a mini-dialysis unit placed in a small beaker containing 3 mL of phosphate buffer (pH 7.4). The whole system was set on a shaker at 37 °C, and the volume of the release medium was maintained at 3 mL. The amount of DOX released at a series of time intervals was measured by monitoring the characteristic emission maxima peak of DOX at 590 nm upon excitation by 480 nm wavelength light and recorded on a steady-state fluorescence spectrophotometer.2.11. Human macrophage (THP1 cell) culture, CD–pepsin treatment and uptake
THP1 cells were cultured in a cell culture flask containing RPMI 1640 medium (Gibco) supplemented with 10% FBS (Gibco), 2 mM L-glutamine, and 200 U mL−1 penicillin–streptomycin (Gibco) in a CO2 incubator at 37 °C. To evaluate the cellular uptake of the drug, the cultured THP1 cells were seeded at the density of 1 × 105 cells in four sets of microcentrifuge tubes. Each microcentrifuge tube containing the THP1 cells was treated with a concentration of 25 g ml−1 CDs, pepsin, CD–pepsin conjugate, and CD–pepsin NPs, respectively, and incubated at 37 °C with 5% CO2 for 4 h. Following drug treatment, the cells were washed thrice with 1× PBS and fixed with 4% paraformaldehyde for 4 min at rt. The cells were again washed thrice with 1× PBS before treatment with ProLong Gold Antifade (Invitrogen) containing 4′,6-diamidino-2-phenylindole (DAPI; stain for the nucleus).2.12. Measurement of cellular oxidative stress
THP1 cells were cultured and treated with 25 μg ml−1 CDs, pepsin, CD–pepsin conjugate, and CD–pepsin NPs, respectively, as mentioned above. The incubated cells were irradiated with or without a 23 W white LED for 100 min. After the treatment, the cells were incubated at 37 °C with 5% CO2 for 3 h. Following treatments, the cells were washed three times with 1× PBS, and 100 μl of 20 μM H2DCFDA was added to each microcentrifuge tube and incubated in the dark for 45 min at 37 °C. The cells were centrifuged at 350g for 5 min, the H2DCFDA solution was aspirated, and finally, the cells were washed with 1× PBS. Next, the THP-1 cells were seeded on poly-lysine (0.001%)-coated coverslips 20 × 20 mm in ProLong Gold Antifade (Invitrogen) containing 4′,6-diamidino-2-phenylindole (DAPI; stain for the nucleus). A baseline was established using CD-treated cells alone at 488 nm excitation to account for any background fluorescence from the CDs. The ROS per cell was calculated by normalizing the total H2DCFDA channel intensity against the number of DAPI-positive cells.2.13. Confocal microscopy
The mounted slides were examined at 405 excitation and 522 to 565 nm emissions for CDs and CD conjugate internalization assay and at 405 excitation and 465 nm emissions for DAPI using a confocal microscope (LSM 880, Zeiss) following methods described earlier.33,34 The H2DCFDA intensity was measured at 488 excitation and 530 nm emission.3. Results and discussion
3.1. Synthesis and characterization of CDs
3-Hydroxy-2-naphthoic acid (BON acid), known for its intramolecular proton transfer capabilities, was used as the sole precursor for synthesizing green-emitting CDs. Its ability to undergo excited-state intramolecular proton transfer (ESIPT) endows it with unique excited-state properties, making it highly significant in photophysical studies.35 A solvothermal method was employed using DMF as the solvent at 220 °C for 10 h. Following careful synthesis, extraction, and purification, the CDs were obtained with a formation yield of 15 wt% (based on 3-hydroxy-2-naphthoic acid). The product was thoroughly characterized using various analytical techniques to investigate its surface functional groups and morphological features. Following careful synthesis, extraction, and purification, the CDs were thoroughly characterized using various analytical techniques to investigate their surface functional groups and morphological features. The quantum yield (QY) of CDs is determined as 14.02% using rhodamine 6G as the standard.FTIR spectroscopy was used to investigate the surface functionality of CDs (Fig. 1A). A broad peak in the 3000–3400 cm−1 range is attributed to N–H and O–H stretching vibrations from amine and hydroxyl groups. Peaks at 1632 cm−1 and 1592 cm−1 were identified as stretching vibrations from C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C groups, respectively. Additional evidence for the presence of hydroxyl groups was confirmed through peaks at 1384 cm−1 and 1128 cm−1, corresponding to C–O–H in-plane bending and C–OH stretching, respectively. X-ray photoelectron spectroscopy (XPS) was employed to investigate the elemental composition and surface states of the CD. The survey spectra showed characteristic peaks at C 1s (285 eV), O 1s (532 eV), and N 1s (399 eV), with atomic compositions of 81.92%, 7.72%, and 10.37%, respectively (Fig. 1B). The nitrogen content can likely be traced back to the solvent, DMF, employed in the synthesis. Detailed analysis of the C 1s spectrum revealed peaks associated with C
C groups, respectively. Additional evidence for the presence of hydroxyl groups was confirmed through peaks at 1384 cm−1 and 1128 cm−1, corresponding to C–O–H in-plane bending and C–OH stretching, respectively. X-ray photoelectron spectroscopy (XPS) was employed to investigate the elemental composition and surface states of the CD. The survey spectra showed characteristic peaks at C 1s (285 eV), O 1s (532 eV), and N 1s (399 eV), with atomic compositions of 81.92%, 7.72%, and 10.37%, respectively (Fig. 1B). The nitrogen content can likely be traced back to the solvent, DMF, employed in the synthesis. Detailed analysis of the C 1s spectrum revealed peaks associated with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C (284.2 eV), C–H (285 eV), C
C/C–C (284.2 eV), C–H (285 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (287.5 eV), and C(O)OR (290.9 eV) bonds (Fig. 1C). The O 1s peak was deconvoluted into C–OH/C–O–C (530.7 eV), C
O (287.5 eV), and C(O)OR (290.9 eV) bonds (Fig. 1C). The O 1s peak was deconvoluted into C–OH/C–O–C (530.7 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (531.4 eV), and C–O (533.3 eV) signals (Fig. 1D), while the N 1s spectrum showed contributions from pyridinic N (398.7 eV), N–H (399.2 eV) and graphitic nitrogen (401.5 eV) (Fig. S1†).
O (531.4 eV), and C–O (533.3 eV) signals (Fig. 1D), while the N 1s spectrum showed contributions from pyridinic N (398.7 eV), N–H (399.2 eV) and graphitic nitrogen (401.5 eV) (Fig. S1†).
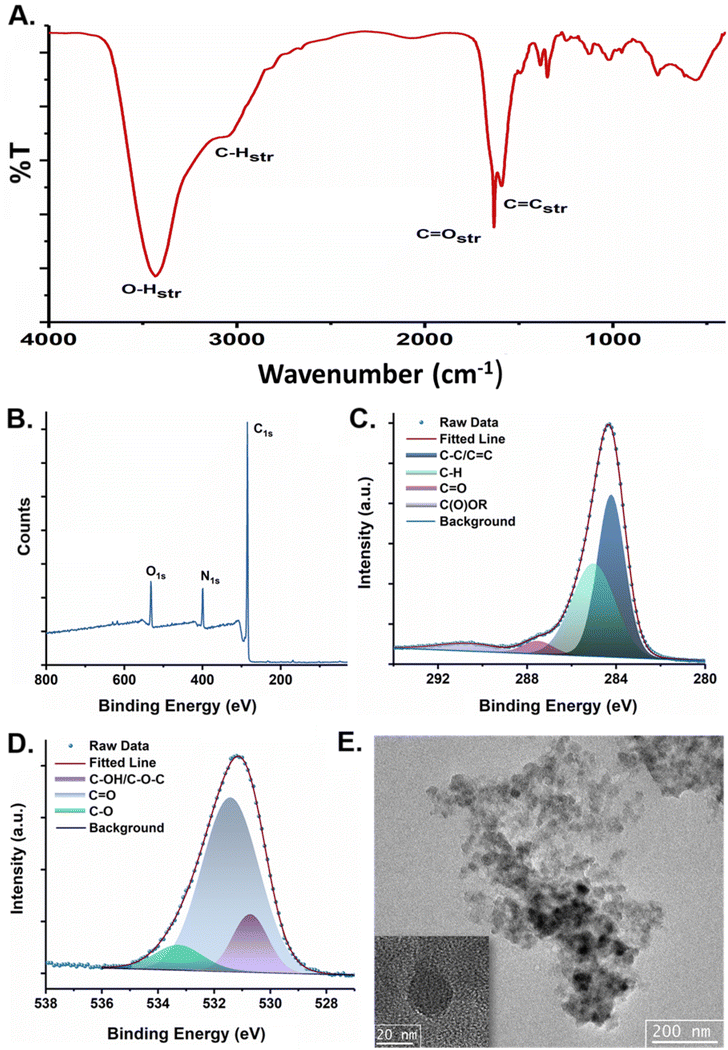 | ||
| Fig. 1 (A) FTIR spectra of CD and (B) XPS survey spectrum of CD. (C) Deconvoluted XPS spectrum for C 1s of CD. (D) Deconvoluted XPS spectrum for O 1s of CD and (E) TEM image of CD. | ||
Powder X-ray diffraction (PXRD) analysis was done to gain insights into the phase structural aspects of the CDs (Fig. S2†). Prominent diffraction peaks at approximately 28.01° and 41.15° were observed, corresponding to the (002) and (100) planes of the graphitic framework, respectively. The broadening and reduced intensity of these peaks indicates partial graphitization with considerable disorder, and the faint peak at 41.15° further suggests the substantial presence of amorphous carbon within the structure. These observations confirm that the synthesized CDs have a multilayered and heterogeneous structure with discrete crystallinity consistent with disordered carbon.36 Morphological analysis through TEM images revealed a spherical shape with an average particle size (diameter) of approximately 27 nm (Fig. 1E and S3A†). The selected area electron diffraction (SAED) pattern obtained from TEM confirmed the predominantly amorphous nature with limited crystalline regions corroborating the finding of PXRD (Fig. S3B†). Overall, the CDs have retained the key functional groups such as –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, –COOH, and –OH from their molecular precursors embedded into a quasi-crystalline core with prominent amorphicity.
O, –COOH, and –OH from their molecular precursors embedded into a quasi-crystalline core with prominent amorphicity.
3.2. Synthesis and characterization of the CD–pepsin conjugate
The CD–pepsin covalent conjugate was synthesized following carbodiimide chemistry with EDC/NHS as activating agents for the carboxyl group on the CD surface, allowing them to react with the amine groups of pepsin. To enhance coupling efficiency, the reaction pH was adjusted to 8.5 after adding pepsin, promoting the deprotonation of amine groups. The structural alterations in pepsin induced by CD conjugation were initially assessed using circular dichroism spectroscopy, a widely recognized biophysical technique frequently used to investigate the secondary structures of proteins in solution. Native pepsin exhibits a characteristic negative band at 210.5 nm and a positive band at 193 nm, corresponding to its predominant β-strand secondary structure. Upon conjugation with CD, these spectral features shift significantly, with a strong negative ellipticity observed at 198.5 nm. This shift indicates notable alterations in the secondary structure of pepsin, specifically a change in its β-sheet content37 (Fig. 2A). The efficiency of conjugation was quantified by calculating the yield of the CD–pepsin conjugate (∼94%) using eqn (1), based on the CD content in the dialysate obtained using a steady state spectrophotometer.Further insights into the successful conjugation of CDs with pepsin were obtained from changes in surface functionalities observed through FTIR spectroscopy. As evident from Fig. 2B, the covalent attachment of CDs to pepsin resulted in a marked reduction in the intensity of the amide II (1550 cm−1) band, while the amide B band (around 3100 cm−1) became nearly indistinguishable. Additionally, a substantial change in the amide A peak at 3500 cm−1 and a slight blue shift in the amide I band (1650 cm−1) indicated considerable modifications in pepsin's secondary structure.38 The fusion of C–O stretching bands at 1172 and 1033 cm−1 further corroborated the successful coupling between CD and pepsin. The conjugation was also confirmed by 12% SDS-PAGE analysis, where the CD–pepsin conjugate exhibited reduced mobility compared to native pepsin, reflecting an increase in molecular weight due to the covalent attachment of CDs (Fig. 2C). Zeta potential measurements show a significant shift from −4.08 mV for innate CDs to −9.78 mV after conjugation with pepsin, approaching the native zeta potential of pepsin at basic pH (−10.77 mV) (Fig. S4–S6†). The isoelectric point of pepsin typically ranges between 2 and 3, meaning at highly alkaline pH, pepsin carries a strong negative charge which induces conformational changes, leading to alterations in its secondary structure. The unfolded pepsin forms a corona around the CDs, thereby influencing the conjugate's electrostatic properties. This is evident from the intermediate zeta potential value of the conjugate, suggesting a partial surface coverage of pepsin over CD, with some charges likely shielded due to steric effects or charge neutralisation at basic pH. These results highlight the effective modifications of CDs post conjugation, substantiating successful conjugation, with pepsin dominating the surface charge of the conjugate. Finally, RP-HPLC analysis, optimized with an in-house developed eluent system, showed distinct retention times for CDs, native pepsin, and the CD–pepsin conjugate (Fig. 2D). The absence of free CDs in the dialyzed conjugate confirmed the completeness of the conjugation process. These comprehensive findings from circular dichroism, FTIR, SDS-PAGE, and RP-HPLC analyses convincingly proved the conjugation of CDs with pepsin, wherein the secondary structure of pepsin was observed to alter.
3.3. Photophysics of CDs and the CD–pepsin conjugate
The UV–Vis absorption spectrum of the CD–pepsin conjugate displayed a distinct peak at 274 nm, corresponding to the π–π* transition from the aromatic amino acids such as tryptophan (5 residues), tyrosine (15 residues), and phenylalanine (14 residues) in pepsin.39 This characteristic absorption peak indicates that the aromatic residues of pepsin remain structurally intact upon conjugation with the CDs. The conjugation process likely induces subtle structural adjustments, leading to the observed spectral feature, which is distinct from the inherent absorption peaks of the CDs at 283 nm and 352 nm. These findings confirm the successful integration of pepsin with CDs while preserving its native aromatic characteristics, underscoring the compatibility and structural stability of the hybrid system. Additionally, the CD–pepsin conjugate exhibited an extended absorption spectrum that reaches up to 500 nm, effectively covering a decent portion of the visible range, which enhances its potential as a photosensitizer for visible light-driven therapeutic applications (Fig. 3A).Fig. 3B depicts the steady-state fluorescence spectra of the CDs under different excitation wavelengths from 340 to 480 nm, revealing excitation-independent emission with the maximum emission at 530 nm under 360 nm excitation. This is attributed to surface states that are enriched with oxygen-containing functional groups, including carbonyl, hydroxyl, and carboxyl groups. However, conjugation with pepsin caused a decrease in fluorescence intensity accompanied by a redshift of about 10 nm in the emission maximum, ascribed to a decrease in the band gap due to the amine modifications on the surface of CDs40 (Fig. 3C). Time-resolved fluorescence spectroscopy demonstrated a notable increase in the average fluorescence lifetime, from 1.78 ns in the CDs to 3.88 ns in the CD–pepsin conjugate (Fig. 3D). This increase in lifetime indicates a prolonged stay in the excited state, enhancing the probability of intersystem crossing (ISC) and likely promoting triplet exciton formation, which in turn boosts ROS generation.19
3.4. Synthesis of ML-optimized CD–pepsin nanoparticles
Pepsin has been widely utilized as a stabilizer and template in nanomaterials; however, it has rarely been crafted into nanoparticles themselves.20 This study, to our knowledge, pioneers the use of pepsin—a protease enzyme—as a nanoparticle through a desolvation method, establishing it as both a potent generator of photoactive ROS and a highly efficient drug transport vehicle. However, the limited number of charged amino acids, particularly cationic residues in pepsin, appeared to hinder NP formation. Since nanoparticle synthesis via the desolvation method involves multiple interdependent parameters—including pepsin percentage, solvent selection, salt type and concentration, and pH—manually optimizing these factors proved challenging. To address this complexity, we employed ML models to predict the most favourable conditions for NP formation.A total of 10 ML models were employed to make models for the CD–pepsin NP formation, out of which the neural network, kNN, SVM, and stochastic gradient descent functioned well (Table S1†). The Explain Model widget further unveiled these models with SHAP (Shapley Additive exPlanations)41 summary plots (Fig. 4A and Fig. S7–S9†). SHAP values transform the “black box” nature of many machine learning models into a transparent and comprehensible one. Each point on a plot is a SHAP value for a specific feature and a data instance in the dataset. Higher SHAP values, as indicated by greater deviation from the center of the graph, signify a greater impact on the prediction for the selected class. Positive SHAP values (right of the center) contribute towards the prediction for the selected class, while negative SHAP values (left of the center) act against classification in that class. The colors represent the value of each feature, with red indicating higher and blue denoting lower values. The color range is determined based on an entire dataset for each feature. The features in the plot are ordered according to their importance to the prediction. Herein, we focused on identifying the top parameters that are substantially impacting the class 1 output (yield value <20%). We anticipated that the factors that are negatively affecting the class 1 output could be the critical factors to practically achieve a higher yield. We observed that a higher percentage of pepsin negatively impacts the Class 1 output, suggesting that an even higher percentage may be required for greater yield. Additionally, we identified desolvation agents and their various factors as critical components in the process. Among the desolvation agents, MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHCl3 (4
CHCl3 (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was found to negatively affect the model output, which is further supported by the negative impact of the major proportion of higher dielectric constant of the desolvation agents. Although mixed results were obtained for the volume of desolvation agents in different models, the negative impact of lower values on the neural network model output indicates that a lower desolvation agent volume could be more beneficial for a higher yield. All four models’ outputs were negatively impacted by higher salt concentrations. Among the various salts, only MgCl2 was found to negatively impact the model outputs. Hence, a high concentration of MgCl2 could favor efficient NP formation. The negative impact of both the low and high pH values and the positive impact of only high pH values (pH range 6.5 to 12) on the model outputs specify the importance of keeping the pH value only slightly alkaline for a better yield. Following these ML-directed pieces of information, we further synthesized pepsin CD-NP, and the higher yields of the NPs in the range of 62–78% justified the validity and importance of the ML models built herein (Table S2†). Briefly, the desolvating agent facilitated protein aggregation, and as the pH increased, more functional groups on pepsin were exposed, encouraging cross-linking interactions at the expense of structural integrity and proteolytic activity. Nevertheless, adding CDs to the system introduced further functional groups, with nanoparticle stability achieved through a network of electrostatic interactions and hydrogen bonding.
1) was found to negatively affect the model output, which is further supported by the negative impact of the major proportion of higher dielectric constant of the desolvation agents. Although mixed results were obtained for the volume of desolvation agents in different models, the negative impact of lower values on the neural network model output indicates that a lower desolvation agent volume could be more beneficial for a higher yield. All four models’ outputs were negatively impacted by higher salt concentrations. Among the various salts, only MgCl2 was found to negatively impact the model outputs. Hence, a high concentration of MgCl2 could favor efficient NP formation. The negative impact of both the low and high pH values and the positive impact of only high pH values (pH range 6.5 to 12) on the model outputs specify the importance of keeping the pH value only slightly alkaline for a better yield. Following these ML-directed pieces of information, we further synthesized pepsin CD-NP, and the higher yields of the NPs in the range of 62–78% justified the validity and importance of the ML models built herein (Table S2†). Briefly, the desolvating agent facilitated protein aggregation, and as the pH increased, more functional groups on pepsin were exposed, encouraging cross-linking interactions at the expense of structural integrity and proteolytic activity. Nevertheless, adding CDs to the system introduced further functional groups, with nanoparticle stability achieved through a network of electrostatic interactions and hydrogen bonding.
The loading of DOX, a well-known anticancer drug, into the CD–pepsin NPs resulted in an impressive drug-loading capacity of 74%. The DLS measurements showed a slight increase in the hydrodynamic diameter after drug loading, from 303 ± 10 nm for unloaded particles to approximately 472.9 ± 10 nm for the loaded form (Fig. S10†). At neutral pH, the zeta potential of CD–pepsin NPs measured −25.18 mV, typical of protein-based nanoparticles (below −15 mV), indicating strong electrostatic repulsion resulting in high nanoparticle stability,42 and this high negative value persisted post-drug loading, highlighting the stability and resistance of the drug-loaded CD–pepsin NPs to flocculation (Fig. S11†). Morphological investigation of the nanoparticles using FESEM revealed a uniform, blunt square shape with a smooth surface and an average diameter of 200–220 nm, distinct from the naturally disordered structure typical of globular proteins (Fig. 4B and S12†). While diffusion may be slower for larger particles, an important advantage of our system is its protein-based composition, which enhances cellular uptake. Unlike rigid inorganic nanoparticles, protein-based nanoparticles can interact favorably with cellular receptors, facilitating efficient internalization through endocytosis, even at slightly larger sizes. Overall, this study presents a novel approach for utilizing pepsin in the nanoparticle form, creating a multifunctional platform with both photoactive properties and efficient drug delivery potential, which may broaden the application of enzymes in nanomedicine.
3.5. Photoactivated ROS generation study
To evaluate the photodynamic potential of our synthesized materials, we aimed to measure ROS generation in vitro using DHR-123, a universal molecular probe. DHR 123 was introduced to aqueous solutions of CDs, CD–pepsin conjugate, and CD–pepsin NPs, followed by visible light irradiation from a 23 W white LED source for 100 minutes (distance 15 cm). As controls, a physical mixture of pepsin and CD, along with the well-known photosensitizer PpIX, was tested. ROS generation was monitored by tracking the change in fluorescence intensity, corresponding to the conversion of nonfluorescent DHR 123 to fluorescent rhodamine 123. To enable quantitative comparison, the fluorescence intensities at each time point were normalized to the initial zero-time intensity, allowing for a detailed assessment of the photoactive capabilities of these materials for potential photodynamic applications. Our findings revealed that while CDs alone could generate ROS, the CD–pepsin conjugate and CD–pepsin NPs produced significantly higher ROS levels, surpassing even those generated by the well-established photosensitizer PpIX (Fig. 4C). This result is compelling as it underscores that a simple enzyme, pepsin, can transform into a highly effective photosensitizer through CD conjugation. The reason for the augmented ROS generation is proposed here as a synergistic effect arising from the conjugation process. The aromatic amino acids in pepsin (tryptophan, tyrosine, and phenylalanine) are known to donate electrons under basic pH conditions. During the conjugation process, the increased pH likely promotes electron transfer from these residues to the photogenerated holes of the CD, thereby lowering its band gap and enhancing its photoactivity.19 Furthermore, the structural framework of pepsin plays a pivotal role in promoting efficient charge transfer43 and sustained interaction with molecular oxygen during light irradiation. This combination of electron donation and structural support is proposed to significantly amplify ROS generation, positioning the CD–pepsin NPs as a highly promising candidate for photodynamic therapy applications.To probe the nature of the generated ROS, we conducted control experiments using selective quenchers: p-benzoquinone (superoxide), methanol (hydroxyl radicals), and sodium azide (singlet oxygen). Significant suppression of DHR fluorescence was observed only with methanol, indicating that hydroxyl radicals (˙OH) are the dominant ROS in our system (Fig. S13†). This was further validated using a terephthalic acid (TA) probe, which selectively reacts with ˙OH to form fluorescent 2-hydroxyterephthalic acid (HTA). Upon visible light irradiation with CD–pepsin NPs, a time-dependent increase in blue fluorescence (∼425 nm) was recorded, confirming efficient ˙OH generation (Fig. S14†). Furthermore, to quantify ˙OH generation, the relative quantum yield was estimated using TiO2 (QY = 4%) as the reference. Based on HTA fluorescence intensity under identical conditions, CD–pepsin NPs exhibited a hydroxyl radical quantum yield of 3.3%, highlighting its excellent photoactivity and suitability for efficient PDT.
3.6. In vitro drug release profile
To investigate the in vitro drug release kinetics of DOX from CD–pepsin NPs, the cumulative drug release (%) was plotted against time (h). The release profile exhibited zero-order kinetics, suggesting that the rate of DOX release was constant and independent of its concentration within the NPs. Over a period of 40 h, approximately 70% of the drug payload was released gradually, demonstrating a diffusion-driven mechanism (Fig. 4D). Unlike conventional drug carriers, where drug release often occurs via complete degradation of the carrier, CD–pepsin NPs facilitated controlled release through a structured collapse of their framework. This zero-order release pattern is indicative of advanced drug delivery systems, wherein the gradual disintegration of the carrier is a hallmark. The release mechanism appears to rely on reversible and dynamic non-covalent interactions, particularly hydrogen bonding between the DOX and the CD–pepsin conjugate. Remarkably, the CD–pepsin NPs did not exhibit the initial burst release commonly associated with many protein-based nanoparticles, enabling a sustained release profile that promotes improved pharmacokinetics. This steady release allows for the maintenance of therapeutic levels over an extended period, making the CD–pepsin NPs a promising carrier for effective drug delivery applications and combined therapeutic strategies. DOX was found to have a higher loading efficiency (76%) compared to ciprofloxacin (54%), just taken as a hydrophobic model drug that aligns with the nature of our CD–pepsin nanoparticles (NPs) and the desolvation method used that creates a hydrophilic protein matrix that strongly interacts with DOX through hydrogen bonding and electrostatic interactions, leading to higher encapsulation. Interestingly, FE-SEM images (Fig. S15†) show progressive morphological changes in the nanoparticles during the drug release period that reveal a clear transition of the CD–pepsin NPs to a deformed and collapsed structure, indicating a gradual structural relaxation and breakdown of the NP framework over time.3.7. Cellular uptake of CD–pepsin for imaging and ROS-mediated oxidative stress
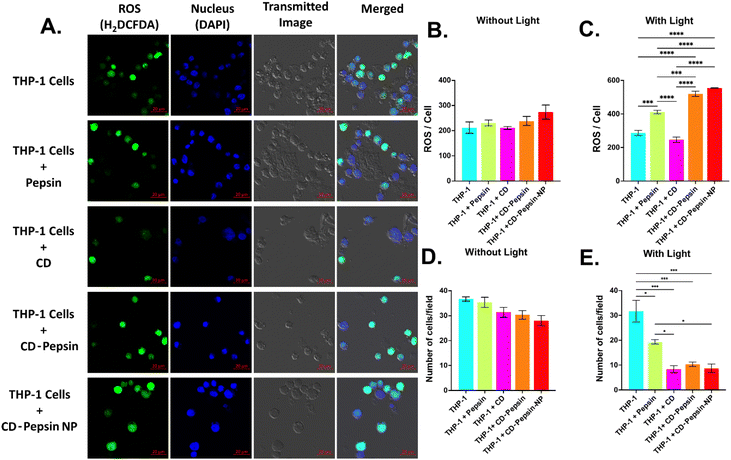
The uptake of CD, CD–pepsin, and CD–pepsin NPs by THP1 cells was measured using confocal microscopy. Image analysis showed that CD–pepsin and CD–pepsin NPs accumulated in higher amounts compared to free CDs in the cells (Fig. 5). These data indicate that the loading of pepsin in CDs through covalent conjugation increases the bioavailability of CDs.Furthermore, upon the treatment of cells with 23 W LED light, ROS levels were found to increase in pepsin, CD–pepsin, and CD–pepsin NP treated cells (F(4,10) = 116.7, P < 0.0001) compared to the dark, unirradiated control (Fig. 6A–C). The total number of THP-1 cells per microscopic field was also assessed in the presence and absence of light. It was observed that the number of cells/field decreased in CDs, CD–pepsin, and CD–pepsin NP-treated groups in the presence of light (F(4,10) = 19.35, P < 0.0001) compared to the control (Fig. 6A, D and E). Thus, it is evident that the CD–pepsin conjugate induces more oxidative stress than pristine CDs as well as CD–pepsin NPs. While the effect of aromatic amino acids of pepsin in augmenting ROS generation is evident in the CD–pepsin conjugate through band gap reduction, increased structural rigidity in the CD–pepsin NPs somewhat negates that effect, slightly compromising ROS generation. However, as noted earlier, the quantum of ROS generation from CD–pepsin NPs is higher than the PpIX control under the given irradiation conditions with a 23 W LED lamp. Given the importance of a practical formulation and colloidal stability for storage, this slight decrease in ROS-mediated cellular oxidative stress of CD–pepsin NPs is highly acceptable.
Summarily, CDs derived from controlled pyrolysis of 3-hydroxy-2-naphthoic acid were well characterized for surface functionality and morphology. The –COOH groups of CDs were made to conjugate with –NH2 groups of pepsin using specific amide formation chemistry under mild conditions. The resultant CD–pepsin conjugate was further transformed into CD–pepsin NPs through desolvation. It is important to note that CD contains multiple –COOH groups, while pepsin has multiple –NH2 groups. This enables one CD molecule to conjugate with multiple pepsin molecules and vice versa during the conjugation process. Although all possibilities are theoretically acceptable, as the goal is to enhance photophysics through bioimaging and increase ROS generation, the evidence suggests that one CD is most likely conjugated with one pepsin molecule. This is supported by the sharp peak of the CD–pepsin conjugate in the HPLC spectrum and the close Rf values of pepsin and CD–pepsin in SDS-PAGE. While enhanced imaging was observed for CD–pepsin NPs confirming its better bioavailability, ROS generation was possible using a non-UV low wattage source (23 W white LED) to induce cellular oxidative stress. Moreover, the CD–pepsin NPs were able to carry drug cargo only to be sustainably released without bursting. Thus, we are able to demonstrate multiple potentials of a simple conjugation strategy of a carbonaceous nanoparticle with a protein, both of which are inexpensive yet strong contenders for combination photo-theranostics.
4. Conclusions
CDs can be prepared from any plant or animal source, may seem like attractive multifaceted nanoparticles, and are consequently reported over-enthusiastically. However, with little-known chemical constituents and impurities in the substrates, the resulting CDs have limited use. Nevertheless, one common use does not escape the scrutiny, which is the inherent fluorescence applicable to bioimaging. We believe that pure chemicals as substrates have the edge that could be engineered to produce CDs with predictable properties. The choice of substrates is perhaps the most important aspect of synthesizing CDs when tuning surface properties and photophysics of CDs are concerned. Herein, the precursor chosen for the synthesis of CDs, 3-hydroxy-naphthoic acid, was selected for two primary reasons: first, it provides suitable binding sites by harboring the carboxyl moieties that facilitate protein attachment, enabling functional conjugation with pepsin, and second, it possesses inherent photoactive properties due to its origins in the dye and pigment industry. While CDs were easily conjugated to pepsin, transforming the CD–pepsin covalent conjugate into a stable nanoformulation proved challenging. In a novel approach, we used ML to optimize the conditions for NP formation, where the summary plots using SHAP values proved invaluable. This study suggests that relevant machine-learning tools can help predict optimal conditions for formulating biomolecule-based nanoparticles, potentially reducing time, cost, and labour. It is noteworthy that the ML modelling performed here is an example of how AI could be used for optimizing protein-based nanoparticle formation but is not to be generalized. The same model may not be used for other protein–CD conjugates, and making a generalizable ML model needs a large amount of data (in millions) with numerous protein–CD conjugates. The CDs’ photophysical properties, including their fluorescence quantum yield, were significantly enhanced while leveraging pepsin's intrinsic oxidative stress-inducing capability. Most importantly, this improvement increased cellular permeability, making them more promising for imaging studies. Although near-infrared (NIR) fluorescence is advantageous for deep tissue imaging, green-emitting nanomaterials remain widely utilized in bioimaging, particularly for cellular imaging, intracellular tracking, and confocal microscopy, due to their high contrast, photostability, and compatibility with the standard clinical imaging system. While we acknowledge the advantages of NIR fluorescence, our approach represents a trade-off between optimizing ROS generation and maintaining strong bioimaging capability. These unique photophysical properties make CDs versatile platforms for both therapy and diagnostics, seamlessly combining bioimaging with ROS-mediated photodynamic action using a simple LED source. This approach is especially effective in targeting cancer cells and microbial pathogens, as demonstrated with THP-1 cells that originate from human monocytic leukemia and are widely used for nanoparticle uptake studies, particularly in bioimaging and drug delivery research. Additionally, the CD–pepsin NPs exhibited excellent structural stability and sustained drug release capabilities, emphasizing their potential for multifunctional theranostic applications. This work not only demonstrates the use of pepsin to enhance the theranostic potential of CDs but also underscores the broader impact of AI-guided protein-functionalized nanomaterials in advancing chemo-photodynamic combination therapy alongside bioimaging.Data availability
Data will be made available on request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the Indian Institute of Technology (IIT) Patna and ICMR-Rajendra Memorial Research Institute of Medical Sciences for infrastructural and financial assistance. S. B. thanks IIT Patna for the Institute Research Fellowship. SB & PD thank SAIF IIT Patna for the instrumentation facility.References
- J. R. Lakowicz and J. R. Lakowicz, Princ. Fluoresc. Spectrosc, 1999, pp. k1–23 Search PubMed.
- J. Liu, R. Li and B. Yang, ACS Cent. Sci., 2020, 6, 2179–2195 CrossRef CAS PubMed.
- Y. Zhang, Q. Jia, F. Nan, J. Wang, K. Liang, J. Li, X. Xue, H. Ren, W. Liu, J. Ge and P. Wang, Biomaterials, 2023, 293, 121953 CrossRef CAS PubMed.
- N. A. Pechnikova, K. Domvri, K. Porpodis, M. S. Istomina, A. V. Iaremenko and A. V. Yaremenko, Aggregate, 2024, e707 Search PubMed.
- M. Behi, L. Gholami, S. Naficy, S. Palomba and F. Dehghani, Nanoscale Adv., 2022, 4, 353–376 RSC.
- W. Su, H. Wu, H. Xu, Y. Zhang, Y. Li, X. Li and L. Fan, Mater. Chem. Front., 2020, 4, 821–836 RSC.
- S. Ghorai, K. Sen and P. S. Dash, ACS Appl. Nano Mater., 2024, 7, 19803–19820 CrossRef CAS.
- A. N. Duman and A. S. Jalilov, Mater. Adv., 2024, 5, 7097–7112 RSC.
- H. Guo, Y. Lu, Z. Lei, H. Bao, M. Zhang, Z. Wang, C. Guan, B. Tang, Z. Liu and L. Wang, Nat. Commun., 2024, 15, 4843 CrossRef CAS PubMed.
- R. D. Senanayake, X. Yao, C. E. Froehlich, M. S. Cahill, T. R. Sheldon, M. McIntire, C. L. Haynes and R. Hernandez, J. Chem. Inf. Model., 2022, 62, 5918–5928 CrossRef CAS PubMed.
- C. Xing, G. Chen, X. Zhu, J. An, J. Bao, X. Wang, X. Zhou, X. Du and X. Xu, Nano Res., 2024, 17, 1984–1989 CrossRef CAS.
- X. Xu, Y. Li, G. Hu, L. Mo, M. Zheng, B. Lei, X. Zhang, C. Hu, J. Zhuang and Y. Liu, J. Mater. Chem. C, 2020, 8, 16282–16294 RSC.
- Y. Park, J. Yoo, B. Lim, W. Kwon and S.-W. Rhee, J. Mater. Chem. A, 2016, 4, 11582–11603 RSC.
- S. Mandal, S. R. Prasad, D. Mandal and P. Das, ACS Appl. Mater. Interfaces, 2019, 11, 33273–33284 CrossRef CAS PubMed.
- S. Nayak and P. Das, ACS Omega, 2021, 6, 21425–21435 CrossRef CAS PubMed.
- P. Zhu, S. Wang, Y. Zhang, Y. Li, Y. Liu, W. Li, Y. Wang, X. Yan and D. Luo, ACS Appl. Bio Mater., 2022, 5, 2031–2045 CrossRef CAS PubMed.
- S. Mandal, Next Res., 2024, 1, 100051 CrossRef.
- Q. Mao, Y. Meng, Y. Feng, H. Li and T. Ma, Inorg. Chem. Front., 2024, 11, 713–734 RSC.
- Y. Zhang, C. Yang, D. Yang, Z. Shao, Y. Hu, J. Chen, L. Yuwen, L. Weng, Z. Luo and L. Wang, Phys. Chem. Chem. Phys., 2018, 20, 17262–17267 RSC.
- H. Kawasaki, K. Hamaguchi, I. Osaka and R. Arakawa, Adv. Funct. Mater., 2011, 21, 3508–3515 CrossRef CAS.
- Q. Li, X. Shen and D. Xing, Dyes Pigm., 2023, 208, 110784 CrossRef.
- R. Nie, J. Zhang, Q. Jia, Y. Li, W. Tao, G. Qin, X. Liu, Y. Tao, Y. Zhang and P. Li, ACS Nano, 2024, 18, 22055–22070 CrossRef CAS PubMed.
- M. Chowdhury, S. Sarkar and P. K. Das, ACS Appl. Bio Mater., 2019, 2, 4953–4965 CrossRef CAS PubMed.
- Y. Nosaka and A. Y. Nosaka, Chem. Rev., 2017, 117, 11302–11336 CrossRef CAS PubMed.
- R. L. Ferreira, W. M. Jr, L. E. A. Souza, H. M. C. Navarro, L. R. de Mello, V. R. Mastelaro, T. O. Sales, C. D. Barbosa, A. S. Ribeiro and E. R. da Silva, ACS Appl. Bio Mater., 2023, 6, 4345–4357 CrossRef CAS PubMed.
- Y. Li, G. Xu, B. Zhou, Y. Tang, X. Liu, Y. Wu, Y. Wang, J. Kong, T. Xu, C. He, S. Zhu, X. Wang and J. Zhang, Eur. Arch. Otorhinolaryngol., 2022, 279, 2743–2752 CrossRef PubMed.
- J.-J. Tan, Y.-F. Dai, F. Wang, Z.-H. Lv, L.-J. Huang, L.-Y. Peng and X.-P. Li, Cytokine, 2024, 178, 156568 CrossRef CAS PubMed.
- Y.-Q. Wang and H.-M. Zhang, J. Agric. Food Chem., 2014, 62, 11303–11311 CrossRef CAS PubMed.
- T. R. Hollands and J. S. Fruton, Proc. Natl. Acad. Sci. U. S. A., 1969, 62, 1116–1120 CrossRef CAS PubMed.
- W. Lohcharoenkal, L. Wang, Y. C. Chen and Y. Rojanasakul, Biomed Res. Int., 2014, 2014, 180549 Search PubMed.
- F. Song and W. C. W. Chan, Nanotechnology, 2011, 22, 494006 CrossRef PubMed.
- J. Demšar, T. Curk, A. Erjavec, Č. Gorup, T. Hočevar, M. Milutinovič, M. Možina, M. Polajnar, M. Toplak and A. Starič, J. Mach. Learn. Res., 2013, 14, 2349–2353 Search PubMed.
- A. Sen, T. J. Nelson and D. L. Alkon, J. Neurosci., 2015, 35, 7538–7551 CrossRef CAS PubMed.
- R. Shankar, S. Nayak, S. Singh, A. Sen, N. Kumar, R. Bhushan, M. Aggarwal and P. Das, ACS Appl. Bio Mater., 2023, 6, 1556–1565 CrossRef CAS PubMed.
- P. S. Sardar, S. Samanta, S. S. Maity, S. Dasgupta and S. Ghosh, J. Phys. Chem. B, 2008, 112, 3451–3461 CrossRef CAS PubMed.
- K. J. Mintz, M. Bartoli, M. Rovere, Y. Zhou, S. D. Hettiarachchi, S. Paudyal, J. Chen, J. B. Domena, P. Y. Liyanage and R. Sampson, Carbon, 2021, 173, 433–447 CrossRef CAS.
- H. H. Ahn, K. S. Kim, J. H. Lee, M. S. Lee, I. B. Song, M. H. Cho, Y. N. Shin, M. S. Kim, G. Khang and H. B. Lee, Int. J. Biol. Macromol., 2007, 41, 590–596 CrossRef CAS PubMed.
- Y. Wang, Y. Sun, J. Yang, L. Dai, N. Ji, L. Xiong and Q. Sun, J. Agric. Food Chem., 2020, 68, 10174–10183 CrossRef CAS PubMed.
- J. Tang, P. Sepulveda, J. Marciniszyn Jr, K. C. S. Chen, W. Y. Huang, N. Tao, D. Liu and J. P. Lanier, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 3437–3439 CrossRef CAS PubMed.
- S. Zhu, J. Zhang, S. Tang, C. Qiao, L. Wang, H. Wang, X. Liu, B. Li, Y. Li and W. Yu, Adv. Funct. Mater., 2012, 22, 4732–4740 CrossRef CAS.
- S. Lundberg and S. Lee, arXiv, 2017, preprint, DOI:10.48550/arXiv.1705.07874.
- A. L. Martínez-López, C. Pangua, C. Reboredo, R. Campión, J. Morales-Gracia and J. M. Irache, Int. J. Pharm., 2020, 581, 119289 CrossRef PubMed.
- A. Cellini, M. K. Shankar, A. Nimmrich, L. A. Hunt, L. Monrroy, J. Mutisya, A. Furrer, E. V. Beale, M. Carrillo and T. N. Malla, Nat. Chem., 2024, 16, 624–632 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5nr00752f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |