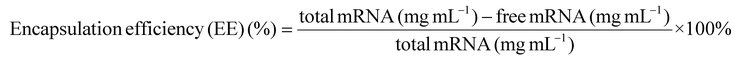Open Access Article
Open Access ArticlePEGylated lipid screening, composition optimization, and structure–activity relationship determination for lipid nanoparticle-mediated mRNA delivery†
Lingyun
Liu
 ,
Jae-Heon
Kim
,
Zhongyu
Li
,
Jae-Heon
Kim
,
Zhongyu
Li
 ,
Mengwei
Sun
,
Mengwei
Sun
 ,
Trent
Northen
,
Jackie
Tang
,
Trent
Northen
,
Jackie
Tang
 ,
Emma
Mcintosh
,
Shrirang
Karve
,
Emma
Mcintosh
,
Shrirang
Karve
 * and
Frank
DeRosa
* and
Frank
DeRosa
mRNA Center of Excellence, Sanofi, Waltham, MA 02451, USA. E-mail: Shri.Karve@Sanofi.com
First published on 19th March 2025
Abstract
Lipid nanoparticles (LNPs) have emerged as effective carriers for mRNA delivery in vaccine and therapeutic applications, attracting substantial attention since the COVID-19 pandemic. Continued efforts are crucial to optimize LNP composition for improved delivery efficacy and to elucidate the underlying mechanisms driving differences in protein expression. This study systematically screened PEGylated lipids for intramuscular mRNA delivery, followed by optimization of the formulation composition, physicochemical characterization, and investigation of the structure–activity relationship (SAR). Using a model ionizable lipid, we initially evaluated twenty-nine PEGylated lipids from four lipid families (glyceride, phosphoethanolamine (PE), cholesterol, and ceramide), each varying in linker chemistries, tail structures, or PEG molecular weights. 1,2-Dimyristoyl-rac-glycero-3-methoxypolyethylene glycol – 5000 (DMG-PEG5k) was identified as a promising candidate from this screening. Using a design of experiments (DoE) approach, we further optimized the formulation to increase in vivo transfection efficacy, achieving an increase in protein expression over the DMG-PEG2k benchmark. To explore the SAR of the DoE formulations, advanced physicochemical characterization was conducted including Laurdan assay, SAXS, Cryo-TEM, and QCM-D, alongside standard LNP analysis. Among the key factors examined, high mRNA encapsulation efficiency, LNP membrane integrity (especially under acidic conditions), and ordered internal structures were identified as the critical parameters for transfection efficiency. mRNA encapsulation efficiency increased with a lower PEG-lipid fraction. LNP membrane integrity, assessed by the generalized polarization (GP) ratio at pH 7.5 and 4.5 from the Laurdan assay, was strongly affected by the ionizable lipid ratio and, to a lesser extent, the cholesterol ratio. A lower GP7.5/GP4.5 ratio correlated with enhanced protein expression, primarily driven by a higher GP4.5 observed with lower ionizable lipid and higher cholesterol fractions. Overall, balancing the ratios of all LNP components is critical for maximizing LNP functionality. This study presents a systematic evaluation and characterization of LNPs with different PEG-lipid moieties, deepens SAR understanding, and provides valuable guidelines for rationally designing more effective next-generation LNPs.
Introduction
Lipid nanoparticles (LNPs), one of the most clinically advanced messenger ribonucleic acid (mRNA) delivery platforms, have gained tremendous attention over the past years.1–4 The FDA approval of Comirnaty® and Spikevax®, two commercial mRNA-LNP COVID-19 vaccines developed by Pfizer/BioNTech and Moderna, respectively, in response to the global SARS-CoV-2 pandemic, has truly underscored the significance of mRNA-LNP technologies in public health.1,2 The LNP platform not only protects the nucleic acid cargo from degradation and greatly enhances delivery efficacy, but also offers unique advantages over other platforms, including rapid developability, scalability due to the synthetic nature of its components, and safety as a nonviral delivery vehicle. Such advantages accelerate the development of mRNA-LNPs as next-generation nucleic acid-based drug modalities and open new avenues for developing novel vaccines and therapeutics. The wide-ranging applications include disease prevention, gene editing, cancer therapy, and rare disease treatment, all through the expression of functional proteins.5–10 As such, extensive efforts and contributions have been made by both industrial and academic research groups, from various perspectives, to improve mRNA-LNP drug products and elucidate the veiled mechanisms behind LNP-mediated mRNA delivery.11,12LNP formulations typically consist of four components: ionizable lipid, phospholipid, sterol, and PEGylated lipid (PEG-lipid). Each component plays a crucial role in maintaining nanoparticle integrity and enabling effective mRNA delivery.13,14 For instance, ionizable lipids are neutral at physiological pH, which improves tolerability for safe systemic delivery, and become protonated in acidic environments, which ensures mRNA encapsulation and facilitates endosomal escape by destabilizing the endosome membrane to release mRNA into the cytoplasm for protein translation.15 Phospholipids are used to form the lipid bilayer structure, facilitate membrane fusion, and aid in the bilayer disruption process during endosomal escape.16 Cholesterol enhances LNP integrity and membrane fluidity by intercalating between lipids. PEGylated lipids, located on the outer layer of nanoparticle surfaces, play a crucial role in preventing particle aggregation, controlling particle size, and affecting surface properties of LNPs. The ‘PEG layer’ through steric hindrance and hydration, not only reduces opsonization in the blood, yielding longer systemic circulation, but also provides particle stability in the vial.12,17
Although many research groups have focused on developing new ionizable lipids for LNPs to improve mRNA delivery,18,19 the PEG-lipid, despite being the least abundant component in LNPs, provides an optimization palette worth investigating. For example, Spikevax uses PEG 2000 with dimyristoyl glycerol (DMG-PEG2k) for the ionizable lipid SM-102, whereas Comirnaty uses PEG 2000 with ditetradecylacetamide (ALC-0159) for ionizable lipid ALC-0315. However, reports describing the impact of PEG-lipid features, such as PEG molecular weight, linker and lipid tail structures, and percentage, on intramuscular (IM) mRNA delivery are still rare. Many previous studies on PEG screening or formulation improvement have focused on delivering small interfering RNA (siRNA), antisense oligonucleotide (ASO), or mRNA intravenously (IV) or in vitro. For example, increasing the PEG-lipid molar ratio from 1.6 to 2.5% was shown to reduce particle size and increase ornithine transcarbamylase expression in the nonhuman primate liver by ∼8 folds when intravenously administering mRNA-LNPs.20 The impact of the alkyl chain length and molar percentage of PEG-lipids in LNPs was studied for the IV delivery of siRNA and hepatic gene silencing in mice.21 A library of ASO-LNP formulations, encompassing 18 PEG-lipids and 3 distinct lipid molar ratios, was assessed for neuronal gene silencing efficacy in vitro and analyzed for structure–activity relationship using small-angle X-ray scattering (SAXS).22,23 A nanoparticle tracking analysis method was developed to study the physicochemical behavior of the siRNA-containing LNPs made from five PEG-lipids with different lipid anchors upon contact with serum to predict the biological gene knockdown behavior in vitro.24
In this study, we conducted a comprehensive investigation into the impact of PEGylated lipids on LNP formulations and mRNA delivery efficacy when intramuscularly administering the mRNA-LNPs. A panel of 29 PEG-lipids from four distinct lipid families was screened for protein expression in mice. With one of the top identified PEG-lipid “hits”, we further optimized the formulations by systematically varying the molar percentages of the four lipid components, facilitated by a design of experiments (DoE) approach. Furthermore, the DoE formulations were comprehensively characterized to reveal structure–activity relationships. Investigating and gaining enhanced knowledge of how the composition and various structural features of LNPs affect mRNA delivery efficacy is crucial for designing more effective next-generation LNPs.
Materials and methods
Materials
All glyceride PEG-lipids, including 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol (DMG-PEG), 1,2-dipalmitoyl-rac-glycero-3-methylpolyoxyethylene (DPG-PEG), 1,2-distearoyl-rac-glycero-3-methylpolyoxyethylene (DSG-PEG), 1,2-dioleoyl-rac-glycerol-3-methoxypolyethylene glycol (DOG-PEG), with the PEG molecular weight of 2000 and 5000, were purchased from NOF Corporation (Tokyo). The phosphoethanolamine (PE) PEG-lipids, 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol) (DMPE-PEG) with the PEG molecular weight of 750, 1000, 2000, 3000, and 5000, were obtained from Avanti Polar Lipids (Alabaster, AL). 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine-methoxy(polyethylene glycol)-5000 (DLPE-PEG5k) and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-(polyethylene glycol) 5000-carboxyl acid (DMPE-PEG-COOH5k) were obtained from BiochemPEG (Watertown, MA). Other PE PEG-lipids, including 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol) (DPPE-PEG), 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol) (DSPE-PEG), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-methoxy(polyethylene glycol) (DOPE-PEG), with the PEG molecular weight of 2000 and 5000, were also obtained from BiochemPEG. The asymmetric ceramide PEG-lipids, N-octanoyl-sphingosine-1-succinyl[methoxy(polyethylene glycol)] (C8 PEG750, C8 PEG2k, C8 PEG5k) and N-palmitoyl-sphingosine-1-{succinyl[methoxy(polyethylene glycol)750]} (C16 PEG750) were purchased from Avanti. The cholesterol PEG lipids (chol-PEG), with PEG molecular weight of 600, 1000, 2000, and 5000, were obtained from BiochemPEG.The ionizable lipid OF-02 used in this study (Fig. 1) was provided internally by Sanofi. 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) was obtained from Avanti. Cholesterol was purchased from Sigma-Aldrich. All other chemicals and reagents were commercially available and used without further purification. Human erythropoietin (hEPO) encoding reporter mRNA was obtained internally.
Preparation of LNPs
The mRNA-encapsulating LNP formulations were prepared as described earlier.25 Briefly, an ethanolic mixture of PEG-lipid, ionizable lipid, cholesterol, and DOPE was vigorously mixed with an aqueous solution of mRNA in an acidic buffer, with a fixed nitrogen/phosphate (N/P) ratio and under controlled conditions, to yield a suspension of LNPs. After dialysis, ultrafiltration and diafiltration into a suitable diluent system, the suspension was filtered, diluted to final concentrations, and stored at −80 °C until use. For screening of PEGylated lipids, the molar composition was as follows: 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28.5
28.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 of PEG-lipid
30 of PEG-lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ionizable lipid
ionizable lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE. In cases where the PEG-lipid percentage was varied, the cholesterol percentage was adjusted to maintain a combined total of 30%. For composition optimization, JMP software was utilized to run a DoE that varied the fractions of all four lipids in the formulation, resulting in 20 different formulations for production.
DOPE. In cases where the PEG-lipid percentage was varied, the cholesterol percentage was adjusted to maintain a combined total of 30%. For composition optimization, JMP software was utilized to run a DoE that varied the fractions of all four lipids in the formulation, resulting in 20 different formulations for production.
Particle size and zeta potential
The particle size, polydispersity index (PDI), and zeta potential of the LNPs were characterized using a Dynamic Light Scattering (DLS) Zetasizer (Malvern, UK). The particle size was measured as the intensity-weighted mean hydrodynamic diameter. The zeta potential was determined in a potassium chloride (KCl) solution.mRNA encapsulation
Quant-iT RiboGreen RNA Assay Kit (invitrogen) was used to assess mRNA encapsulation efficiency following the manufacturer's instructions. The mRNA encapsulation efficiency (EE) was calculated using the following equation:Laurdan assay
Formulations were diluted to a theoretical lipid concentration of 3.12 mM with 10% trehalose solution. This solution was then added to a series of McIlvaine buffers with different pH values (pH 4.5, 5.5, 6.0, 6.5, 7.5) and gently inverted. After mixing, Laurdan dye (dissolved in DMSO at 0.2 mg mL−1) was added to the formulation in buffer solution, giving a final Laurdan concentration of 1.13 μM. The molar ratio of total lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Laurdan in the final test solution was 592
Laurdan in the final test solution was 592![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the volumetric fraction of DMSO was 0.2%. Solutions were allowed to sit at room temperature, protected from light, for three hours. Samples were then analyzed using a SpectraMax M5 multi-mode microplate reader (Molecular Devices, San Jose, CA). Laurdan fluorescence spectra with emission wavelength from 400 to 520 nm were recorded under a fluorescence excitation wavelength of 340 nm. Generalized polarization (GP) values were calculated using the following equation:
1 and the volumetric fraction of DMSO was 0.2%. Solutions were allowed to sit at room temperature, protected from light, for three hours. Samples were then analyzed using a SpectraMax M5 multi-mode microplate reader (Molecular Devices, San Jose, CA). Laurdan fluorescence spectra with emission wavelength from 400 to 520 nm were recorded under a fluorescence excitation wavelength of 340 nm. Generalized polarization (GP) values were calculated using the following equation:| GP = (I440 − I490)/(I440 + I490) |
TNS assay
The apparent acid dissociation constant (pKa) of each LNP formulation was evaluated using the 2-(p-toluidino)-6-naphthalene sulfonic acid (TNS) assay according to a standard protocol. Briefly, LNP formulations were added to a series of buffers with pH values ranging from 3.0 to 9.0. The TNS solution in DMSO was then incubated with LNPs in different pH buffers. The fluorescence intensity was recorded at excitation and emission wavelengths of 320 and 430 nm, respectively, using a spectrophotometer. The half-maximal inhibitory pH level (IC50) of the titration curve was reported as the apparent pKa of the LNPs.QCM-D characterization
The gold-coated quartz crystal microbalance with dissipation (QCM-D) sensors (QSX301, Biolin Scientific) with a fundamental resonance frequency of 4.95 MHz were initially cleaned in a solution of water, ammonium hydroxide (25%), and hydrogen peroxide (30%) (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v) for 5 minutes at 75 °C. Following thorough rinsing with water, the sensors were dried using nitrogen and subjected to UV/ozone cleaning for 10 minutes. The sensors were then installed in the flow module of QSense Pro (Q-Sense Inc., Gothenburg, Sweden). Shifts in resonance frequency (ΔF) and dissipation (ΔD) for odd overtones (n = 1, 3,…, 13) were recorded. All solutions were introduced using a peristaltic pump (IPC N-4, Ismatec). Experiments were performed at 37 °C.
1, v/v/v) for 5 minutes at 75 °C. Following thorough rinsing with water, the sensors were dried using nitrogen and subjected to UV/ozone cleaning for 10 minutes. The sensors were then installed in the flow module of QSense Pro (Q-Sense Inc., Gothenburg, Sweden). Shifts in resonance frequency (ΔF) and dissipation (ΔD) for odd overtones (n = 1, 3,…, 13) were recorded. All solutions were introduced using a peristaltic pump (IPC N-4, Ismatec). Experiments were performed at 37 °C.
Apolipoprotein E (ApoE) was deposited onto the sensors by injecting 0.1 mg mL−1 ApoE in PBS at 30 μL min−1 over the crystals. After 20 minutes of adsorption, the ApoE layers were rinsed with PBS at 40 μL min−1 until equilibrium was reached (approximately 10 minutes). Interactions between the LNP formulations and ApoE were investigated by flowing 50 μg mL−1 LNPs in PBS over the ApoE layers at a flow rate of 40 μL min−1 for 90 minutes. PBS buffer was continuously pumped over the surfaces afterwards.
PEG surface density
The surface density of PEG [Γ], defined as the number of PEG molecules per 100 nm2 of LNP surface, was determined following previous reports26,27 by the equation: | (1) |
The full surface mushroom coverage [Γ*], defined as the number of unconstrained PEG molecules per 100 nm2 of particle surface, was calculated using the following equations:
| ξ = 0.76m0.5 | (2) |
 | (3) |
Cryo-TEM
All mRNA-LNPs were buffer-exchanged into nuclease-free water (Millipore Sigma), vitrified by plunge-freezing, and imaged using brightfield cryogenic transmission electron microscopy (Cryo-TEM). TEM grids (Quantifoil GmbH) were glow-discharged for 25 seconds in rarefied room air using a Pelco EasiGlow (Ted Pella). Samples were applied to grids in the chamber of a Vitrobot Mk IV system (ThermoFisher) at 95% relative humidity and 22 °C, blotted, and then plunged into liquid ethane for vitrification. Each sample was vitrified on Quantifoil R2/1 200 mesh grids, both with and without a continuous carbon film of 2 nm nominal thickness. Imaging was performed either on a Talos Arctica transmission electron microscope (FEI Company) with a K2 direct electron detector (Gatan) controlled by SerialEM software at 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× and 130
000× and 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× nominal magnification, or on a Tundra transmission electron microscope (ThermoFisher) controlled by EPU software at 69
000× nominal magnification, or on a Tundra transmission electron microscope (ThermoFisher) controlled by EPU software at 69![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× and 110
000× and 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× nominal magnification. Images were collected from grids showing optimal ice thickness and low contamination. Regions of interest in the images displaying supramolecular structures were identified. Fast Fourier transform (FFT) and d-spacing determination were conducted using the Fiji distribution of ImageJ.
000× nominal magnification. Images were collected from grids showing optimal ice thickness and low contamination. Regions of interest in the images displaying supramolecular structures were identified. Fast Fourier transform (FFT) and d-spacing determination were conducted using the Fiji distribution of ImageJ.
SAXS
SAXS measurements were performed using a Xeuss 3.0 SAXS/WAXS system (Xenocs Inc, Holyoke, MA) equipped with a Genix 3D Cu microfocus source (Cu Kα λ = 1.5418 Å) and a movable Eiger 1M detector (Dectris). Samples were loaded into the BioCube sample environment using the robotic sample loader. The sample cell was maintained at 25 °C during measurements. Two-dimensional images were azimuthally averaged using XSACT. One-dimensional scattering profiles were then averaged and buffer-subtracted in XSACT. All samples were kept frozen and thawed to room temperature prior to measurement.In vivo protein expression
Animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC). The LNPs containing 0.1 μg of hEPO mRNA were intramuscularly injected into the right gastrocnemius muscle of female BALB/c mice (6–8 weeks old). Six hours post-injection, blood samples were collected from the mice, and hEPO levels in the sera were measured using enzyme-linked immunosorbent assay (ELISA), following the manufacturer's instructions.Correlation analysis
To explore the relationships between variables in our dataset, we conducted a correlation analysis using R's corrplot package and calculated Pearson correlation coefficients. The correlation matrix was visually represented using the corrplot function, which offers a variety of visualization methods, including circle and color-coded heatmaps, to effectively display the strength and direction of the correlations between variables.Results and discussion
We first formulated LNPs with a variety of PEG-lipids and evaluated hEPO expression in vivo. A total of 29 PEG-lipids were assessed from four lipid families – glyceride PEG, phosphoethanolamine (PE) PEG, cholesterol PEG, and ceramide PEG – each with different linker or tail structures and PEG molecular weights ranging from 600 to 5000. The structures of the ionizable lipid and all PEG-lipids screened in this work are illustrated in Fig. 1a. The process and timeline of the animal experiments, along with the overall design of the work described in this paper, are shown in Fig. 1b and c. The model ionizable lipid OF-02 features a cyclic lysine dipeptide core and four hydrophobic tails, each with 2 unsaturated bonds in its 18-carbon chain.28Eight glyceride PEG-lipids, with glyceride serving as the linker between the PEG head and two acyl lipid tails, were formulated, characterized, and evaluated for potency. In this study, all protein expression data was normalized to the levels from the DMG-PEG2k formulation, which exhibited an average 6 h-hEPO expression of approximately 30 ng mL−1. It was observed that as the acyl carbon chain length increased from 14 to 18, protein expression significantly decreased (Fig. 2a). Among the eight tested glyceride PEG-lipids, only the DMG-PEG5k formulation demonstrated comparable protein expression (slightly higher, ∼20%, than the DMG-PEG2k benchmark). Compared to PEG2k, PEG5k formulations showed smaller particle sizes (Fig. 2b). The PDI for all formulations was less than 0.2, with PEG5k LNPs showing a slight increase (Fig. 2c). The mRNA encapsulation efficiency (EE%) was close to or above 90% for all formulations, although PEG5k formulations exhibited slightly lower values (Fig. 2d). Decreased protein expression by DPG-, DSG-, and DOG-PEG formulations may be explained by different rates of PEG shedding from LNPs upon contact with complex media. PEG-lipids with shorter lipid tails shed faster than those with longer tails, promoting the formation of protein corona and enhancing cellular uptake.24,29 Additionally, PEG5k formulations with long acyl carbon chains (C16 to C18) exhibited decreased potency compared to their PEG2k counterparts, which could be attributed to reduced protein adsorption on higher molecular weight PEG on LNP surfaces. Notably, DMG-PEG, with its C14 tail, is the only lipid in this study where the PEG5k formulation demonstrated protein expression comparable to that of PEG2k. All other lipid tails tested, including the PE and ceramide PEG lipids discussed below, showed reduced potency of PEG5k formulations compared to their PEG2k counterparts.
Thirteen PE PEG-lipids being studied use glycero-3-phosphoethanolamine as the linker between the PEG head group and two lipid tails. As shown in Fig. 2f, LNPs made from DMPE-PEG2k, DMPE-PEG3k, and DPPE-PEG2k exhibited similar protein expression levels compared to the DMG-PEG2k control. DLPE (C12), DMPE (C14) with PEG750 and PEG1k, DPPE (C16) with PEG5k, DSPE (C18), and DOPE (C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) PEG-lipids all showed lower protein expression than the benchmark. A similar trend to that observed with the glyceride PEG-lipids was noted: PEG5k formulations generally demonstrated smaller particle sizes, slightly increased PDI, slightly lower EE%, and less protein expression compared to the respective PEG2k formulations (Fig. 2f–i). For the DMPE-PEG lipids, PEG750 and PEG1k formulations had much larger particle size of 196 and 161 nm, respectively, compared to the PEG2k to 5k formulations which had sizes around 100 nm. This difference in size may explain the low protein expression of the two formulations. Additionally, it was observed that as the molecular weight of PEG increased, the zeta potential of LNPs decreased (Fig. 2e and j). Notice that in most literature, the final buffer for LNPs is PBS (pH 7.4), typically resulting in zeta potentials near zero. In this work, a trehalose solution (pH ∼5) was used as the final buffer for LNPs, intended for cryostorage. Therefore, the LNPs exhibited higher zeta potentials.
1) PEG-lipids all showed lower protein expression than the benchmark. A similar trend to that observed with the glyceride PEG-lipids was noted: PEG5k formulations generally demonstrated smaller particle sizes, slightly increased PDI, slightly lower EE%, and less protein expression compared to the respective PEG2k formulations (Fig. 2f–i). For the DMPE-PEG lipids, PEG750 and PEG1k formulations had much larger particle size of 196 and 161 nm, respectively, compared to the PEG2k to 5k formulations which had sizes around 100 nm. This difference in size may explain the low protein expression of the two formulations. Additionally, it was observed that as the molecular weight of PEG increased, the zeta potential of LNPs decreased (Fig. 2e and j). Notice that in most literature, the final buffer for LNPs is PBS (pH 7.4), typically resulting in zeta potentials near zero. In this work, a trehalose solution (pH ∼5) was used as the final buffer for LNPs, intended for cryostorage. Therefore, the LNPs exhibited higher zeta potentials.
Since the DMG-PEG5k formulation showed promising protein expression, we decided to further investigate different molar ratios of DMPE-PEG5k from 1.5 to 5%. These formulations, however, did not show improved potency (Fig. S1†). Particle size across the samples remained at approximately 96 nm, and the zeta potential was consistently around 17.5 mV. Interestingly, as the PEG-lipid content increased from 1.5% to 5%, there was a noticeable increase in the PDI from 0.1 to 0.2. This may be attributed to the polarity of PEG, with which a high ratio of PEGylated lipid in the lipid mixture and higher PEG molecular weight could lead to heterogeneous mixing and phase separation. A similar phenomenon has been reported by others.30 As the mol% of DMPE-PEG5k increased, the EE% decreased (Fig. S1†). This reduction is likely due to the steric hindrance caused by excessive PEG molecules, which can limit the accessibility of ionizable lipids and interfere with lipid-mRNA interactions. When the mol% of DMPE-PEG5k reached 4% or higher, the EE% fell below 80%, and protein expression was significantly compromised.
Next, we investigated four ceramide PEG-lipids, with ceramide (8 or 16 acyl carbons) as the lipid anchor (Fig. 2k–n). LNPs formulated with two of these lipids (C8 PEG750, C16 PEG750) had diameters exceeding 200 nm and hence, not suitable for sterile filtration and further in vivo evaluation. The remaining two lipids (C8 PEG2k, C8 PEG5k) both exhibited much lower protein expression compared to the DMG-PEG2k control, although C8 PEG2k showed higher protein levels than C8 PEG5k.
Given cholesterol's role in membrane fluidity, we explored the cholesterol PEG-lipid (chol-PEG) family, where cholesterol serves as the anchor in the membrane. Notably, chol-PEG lipids have only one lipid tail. As depicted in Fig. 3a, nearly all chol-PEG lipids at the tested molar ratios exhibited lower expression than the DMG-PEG2k control, except for 5 mol% chol-PEG1k. The size of LNPs decreased as either the PEG molecular weight or PEG molar ratio increased (Fig. 3b). Further, chol-PEG LNPs were larger than those from other PEG-lipid families with the same PEG molecular weight. For instance, at 1.5 mol% of PEG2k and PEG5k, chol-PEG formulations had particle diameters around 200 nm and 150 nm, respectively, whereas glyceride, PE, and ceramide PEGs with equivalent PEG molecular weights resulted in particle diameters of less than 130 nm. Additionally, higher PEG molecular weights slightly increased PDI (Fig. 3c), consistent with observations from other PEG-lipid families (Fig. 2c and h). An increase in PEG molecular weight and molar ratio generally led to a reduction in mRNA encapsulation efficiency (Fig. 3d). When the PEG molecular weight was 2k or lower, greater protein expression was observed at higher PEG molar ratios, likely due to the larger particle sizes when both PEG molecular weight and mol% were low (Fig. 3b). In contrast, with a PEG molecular weight of 5k, maximal protein expression was seen at intermediate PEG molar ratios, probably attributed to the large particle size at low PEG molar ratios (Fig. 3b) and reduced EE% and protein binding at high PEG ratios (Fig. 3d). Lower PEG percentages in mRNA-LNPs are generally associated with higher protein expression.18 Interestingly, this study found that reduced chol-PEG content resulted in lower expression.
Among the 29 PEG-lipids and approximately 50 compositions screened, the DMG-PEG5k formulation was the only one that demonstrated improved biological potency compared to the DMG-PEG2k control. Given that PEG5k is more hydrophilic than PEG2k and provides additional ethylene glycol coverage even at lower molar ratios, which could impart distinct LNP properties, we decided to further investigate and optimize formulations with DMG-PEG5k, as well as explore their structure–activity relationship. It was observed that DMG-PEG5k did not fully dissolve in isopropanol (IPA) unless water was added whereas DMG-PEG2k exhibited good solubility in IPA (data not shown). For composition optimization, JMP software was used to conduct a DoE that varied the fractions of all four lipids in the formulation, resulting in 20 different formulations (Fig. 4a). The ranges of DMG-PEG5k, OF-02, cholesterol, and DOPE were set at 1–4, 20–60, 10–45, and 10–50 mol%, respectively.
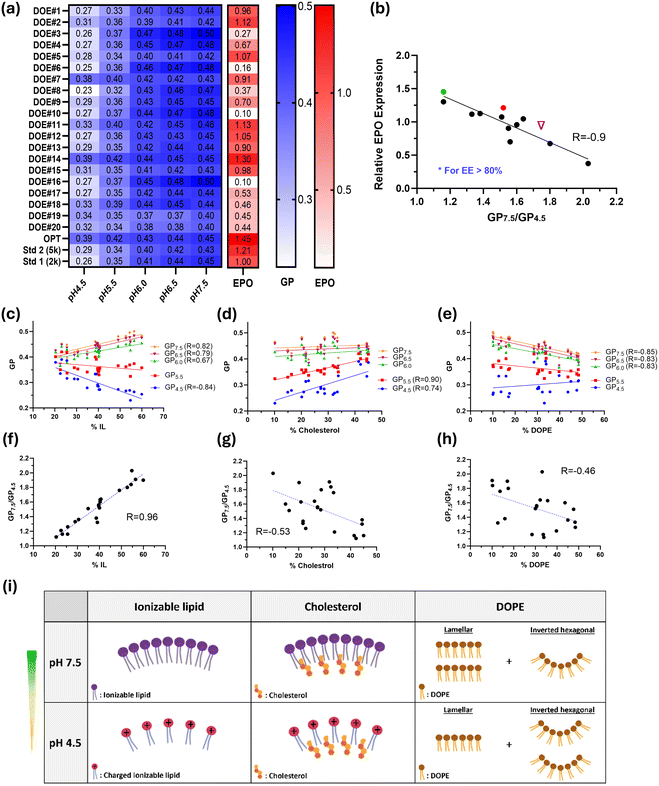
As shown in Fig. 4b, the twenty formulations with varying compositions yielded a broad range of protein expression, from 0.1 to 1.3 times that of the DMG-PEG2k control (Std1). These expression values were then fed back into JMP software, which predicted an optimal composition (OPT) that achieved a 1.5-fold increase in protein expression compared to Std1. The physicochemical properties of the 20 DoE formulations, along with those of the OPT and DMG-PEG5k standard (Std2) compositions, are detailed in Table 1. The hydrodynamic particle diameter, PDI, and EE% of these LNPs ranged from 90 to 128 nm, 0.05 to 0.17, and 64% to 95%, respectively.
| ID | DMG- PEG5k% | OF-02% | Chol% | DOPE% | Diameter (nm) | PDI | EE% | ZP (mV) | pKa | GP7.5/GP4.5 | [Γ] | [Γ]/[Γ*] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DOE 01 | 1.1 | 40.3 | 14.9 | 43.7 | 125 | 0.067 | 89 | 21 | 6.5 | 1.60 | 13.9 | 3.1 |
| DOE 02 | 1.0 | 29.0 | 21.7 | 48.3 | 119 | 0.111 | 91 | 20 | 6.2 | 1.33 | 13.8 | 3.1 |
| DOE 03 | 1.7 | 56.5 | 31.8 | 10.0 | 117 | 0.068 | 65 | 17 | 6.5 | 1.91 | 19.1 | 4.3 |
| DOE 04 | 1.1 | 52.7 | 28.7 | 17.5 | 125 | 0.049 | 88 | 19 | 6.5 | 1.80 | 14.3 | 3.2 |
| DOE 05 | 1.1 | 34.9 | 16.3 | 47.7 | 119 | 0.093 | 91 | 19 | 6.4 | 1.51 | 14.5 | 3.3 |
| DOE 06 | 2.9 | 59.9 | 20.2 | 17.0 | 100 | 0.083 | 70 | 15 | 6.6 | 1.90 | 25.1 | 5.7 |
| DOE 07 | 3.6 | 20.4 | 42.1 | 33.9 | 93 | 0.139 | 76 | 14 | 6.3 | 1.12 | 38.6 | 8.7 |
| DOE 08 | 2.0 | 54.8 | 10.1 | 33.1 | 110 | 0.077 | 86 | 18 | 6.7 | 2.03 | 19.5 | 4.4 |
| DOE 09 | 2.5 | 39.7 | 27.3 | 30.6 | 99 | 0.117 | 85 | 17 | 6.7 | 1.56 | 25.2 | 5.7 |
| DOE 10 | 3.8 | 49.1 | 33.4 | 13.7 | 98 | 0.119 | 66 | 16 | 6.6 | 1.76 | 34.3 | 7.8 |
| DOE 11 | 1.1 | 38.6 | 44.4 | 15.9 | 115 | 0.055 | 94 | 19 | 6.5 | 1.38 | 14.6 | 3.3 |
| DOE 12 | 1.0 | 40.7 | 26.1 | 32.2 | 128 | 0.107 | 95 | 20 | 6.7 | 1.64 | 14.0 | 3.2 |
| DOE 13 | 2.5 | 39.7 | 27.3 | 30.6 | 92 | 0.079 | 84 | 17 | 6.7 | 1.55 | 23.5 | 5.3 |
| DOE 14 | 1.8 | 23.1 | 41.4 | 33.7 | 93 | 0.113 | 90 | 19 | 6.2 | 1.16 | 20.8 | 4.7 |
| DOE 15 | 2.7 | 30.6 | 21.7 | 45.0 | 92 | 0.138 | 76 | 17 | 6.6 | 1.36 | 26.0 | 5.9 |
| DOE 16 | 3.0 | 54.4 | 32.4 | 10.2 | 102 | 0.084 | 64 | 17 | 6.6 | 1.84 | 28.1 | 6.4 |
| DOE 17 | 3.2 | 40.3 | 20.9 | 35.6 | 95 | 0.079 | 74 | 18 | 6.6 | 1.63 | 28.9 | 6.5 |
| DOE 18 | 3.8 | 39.1 | 44.4 | 12.6 | 97 | 0.084 | 73 | 15 | 6.6 | 1.32 | 38.0 | 8.6 |
| DOE 19 | 4.0 | 22.5 | 33.8 | 39.7 | 90 | 0.167 | 70 | 12 | 6.5 | 1.21 | 38.4 | 8.7 |
| DOE 20 | 3.2 | 25.6 | 22.7 | 48.5 | 96 | 0.161 | 71 | 16 | 6.5 | 1.26 | 32.3 | 7.3 |
| OPT | 1 | 25.5 | 45 | 28.5 | 120 | 0.111 | 91 | 19 | 6.0 | 1.16 | 15.5 | 3.5 |
| Std2 | 1.5 | 40 | 28.5 | 30 | 112 | 0.104 | 90 | 20 | 6.6 | 1.52 | 18.1 | 4.1 |
We next investigated how LNP composition and characteristics influence their biological potency. Protein expression showed a strong negative correlation with the fractions of ionizable lipid (Fig. 4c) and PEG-lipid (Fig. 4d), while correlations with DOPE% and cholesterol% were considerably weaker (Fig. S2a and b†). Importantly, there was a very strong positive correlation between protein expression and mRNA encapsulation efficiency, with an R value of 0.84 (Fig. 4e), whereas the correlation with particle size was weak, and no correlation was found with PDI (Fig. S2c and d†). Lower PEG ratios led to higher zeta potential (Fig. S2g†), which in turn increased EE% due to stronger electrostatic interactions with mRNA (Fig. S2h† and Fig. 4f). Since mRNA molecules encapsulated within LNPs are delivered into cells much more efficiently than free mRNA, high EE% at low PEG fractions and high zeta potential resulted in elevated protein expression (Fig. 4d, e and Fig. S2e†). Additionally, it was observed that as the ionizable lipid fraction increased, the pKa of LNPs also increased, which negatively impacted protein expression (Fig. S2f, i† and Fig. 4c).
To explore the structure–activity relationship, all DoE, Std1, Std2, and OPT formulations were thoroughly characterized using the Laurdan assay to gain a fundamental understanding of lipid packing. Laurdan is a fluorescent dye that penetrates lipid layers and is widely used to probe the packing and hydration of lipid membranes. Upon transition of lipid layers from a gel phase to a fluid phase, Laurdan spectra exhibit a red shift. Generalized polarization (GP), a normalized difference between Laurdan fluorescence intensities at 440 nm and 490 nm, is a commonly used parameter for analyzing Laurdan fluorescent spectra. Lower GP values indicate a more disordered or less tightly packed lipid bilayer.31
Laurdan spectra were measured at various pH values (7.5, 7.0, 6.5, 5.5, and 4.5) to simulate endosome maturation, which involves a decrease in pH following cellular uptake of LNPs. As shown in Fig. S3,† Laurdan spectra all shifted to higher wavelengths as the pH decreased, although the extent of this red shift varied significantly across different formulations. The GP values at each pH level along with the protein expression for all tested formulations are plotted in a heat map (Fig. 5a). GP values decreased with decreasing pH, indicating a more disordered lipid bilayer in acidic environments. Notably, the two formulations with the highest protein expression, OPT and DOE14, had the highest GP values at pH 4.5. The ratio of GP values at pH 7.5 to pH 4.5 (GP7.5/GP4.5) was then calculated for all formulations, and Laurdan spectra were categorized into five groups accordingly (Fig. S3†). In fact, a smaller GP7.5/GP4.5 ratio corresponds to a smaller red shift in Laurdan spectra, whereas a larger ratio indicates a greater shift. Interestingly, a general trend was observed where higher protein expression was associated with smaller GP7.5/GP4.5 ratio (Fig. S4a†), with a negative Pearson correlation coefficient of −0.6. Outliers with large deviations from the trendline were found to have poor encapsulation efficiency. When including only LNPs with EE > 80%, a strong negative correlation (R = −0.9) was observed between protein expression and GP7.5/GP4.5 (Fig. 5b). This finding explains why some formulations had high mRNA encapsulation but poor protein expression. For example, although DOE8 and DOE4 had high mRNA EE of 86% and 88%, respectively, their protein expression was only 37% and 67% of the Std1 benchmark. This arises from their high GP7.5/GP4.5 ratios of 2 and 1.8, which are primarily related to lower GP4.5, indicating poor membrane integrity at low pH. Conversely, DOE7, despite having the smallest GP7.5/GP4.5 ratio of 1.1 among all formulations, showed protein expression comparable to Std1, likely attributed to its reduced EE of 76%. Note that for the three standard and optimized formulations, GP7.5/GP4.5 follows the order OPT < Std2 < Std1, with EE values all above 90%, with protein expression exhibiting the opposite ranking, which agrees with the hypothesis. Overall, the Laurdan results suggest that both mRNA encapsulation efficiency and LNP membrane integrity, especially in acidic environments, are critical factors in determining protein expression.
We next delved deeper to gain insights into how each LNP component affects GP values at the molecular level. GP values at individual pHs and GP7.5/GP4.5 were plotted against the fraction of each lipid. The results show that GP correlates strongly with the ionizable lipid ratio at both high and low pHs, whereas for cholesterol, dependence is seen only at pH of 5.5 and below, and for DOPE, dependence is evident only at pH 6.0 and above (Fig. 5c–e). No correlation was found between GP values and PEG fraction (Fig. S4b†), which is expected since PEG-lipids are primarily located on the surface of the LNPs. Notably, a very strong correlation was observed between GP7.5/GP4.5 and the ionizable lipid ratio with an R value of 0.96 (Fig. 5f), whereas the correlations with cholesterol and DOPE ratios were weaker (Fig. 5g and 5h), and no correlation was found with the PEG ratio (Fig. S4c†). The acidic environment enhances electrostatic repulsion among protonated ionizable lipid molecules, leading to decreased lipid packing within LNPs (Fig. 5i) and increased membrane hydration, which in turn decreases GP4.5 and increases the GP7.5/GP4.5 ratio. The trend in GP value changes caused by cholesterol may be explained as follows: Incorporating cholesterol into the charged lipid bilayer promotes close packing of lipid molecules. At lower pHs (pH 4.5 and pH 5.5), ionizable lipids are protonated and repel each other. The addition of cholesterol brings these lipids closer together and decreases membrane fluidity, thereby enhancing GP and reducing GP7.5/GP4.5. DOPE lipid possesses an intrinsic negative curvature. Increased DOPE content in LNPs promotes the formation of inverted hexagonal structures and induces curvature stress,32,33 resulting in more voids between lipid tails (i.e., reduced lipid packing) and consequently lower GP values. Overall, the proportions of all LNP components must be balanced to maximize the biological potency of the LNPs. In our formulations, low ratios of ionizable and PEGylated lipids, combined with high ratios of cholesterol and DOPE, appear to be optimal. Consistent with our findings, previous research has shown that the lowest molar ratio of DOTAP to cholesterol in liposomes resulted in the highest mRNA transfection efficiency.11
When administered in vivo, LNP surfaces interact with various proteins, leading to the formation of a protein corona that affects the cellular uptake and biodistribution of the particles.34 The interactions between three mRNA-LNP formulations (Std1, Std2, and OPT) and Apolipoprotein E (ApoE) were studied using QCM-D. ApoE was chosen for this study due to its significant role in the clearance of LNPs and their targeting to the liver.35 QCM-D is a highly sensitive technique that monitors in situ changes in interfacial mass uptake and viscoelastic properties by detecting shifts in the frequency and dissipation of an oscillating quartz crystal sensor. Larger negative frequency shifts typically indicate a higher coupled mass. For simplicity, only the frequency and dissipation shift of the third overtone are presented here, though similar conclusions can be drawn from all measured overtones.
ApoE was first deposited onto the gold-coated sensor, followed by the introduction of LNPs over the pre-adsorbed ApoE layers for 90 minutes. The Std1, Std2, and OPT formulations exhibited irreversible adsorption on the protein layer, with frequency shifts of −470, −100, and −150 Hz, respectively (Fig. 6). The greater frequency shift observed for the Std1 formulation with 1.5% PEG2k indicates a higher degree of ApoE adsorption compared to Std2 formulated with 1.5% PEG5k and OPT formulated with 1% PEG5k. This suggests that LNPs formulated with lower molecular weight PEG-lipid exhibit stronger interactions with proteins than those with higher molecular weight PEG-lipid, which correlates well with the zeta potentials of Std1, Std2, and OPT, measured at 30, 20, and 19 mV, respectively (Table 1). Additionally, the PEG2k LNPs demonstrated a larger dissipation shift than the two PEG5k LNPs, indicating that the PEG2k LNPs formed a softer layer (Fig. 6).
 | ||
| Fig. 6 The frequency shift (a) and dissipation shift (b) of ApoE-LNP interactions measured by QCM-D. | ||
PEG surface density [Γ] denotes the number of PEG molecules per 100 nm2 of LNP surface, whereas [Γ*] represents the number of unconstrained PEG molecules within the same area. The ratio of [Γ]/[Γ*] reflects the packing density of PEG chains on the LNP surface and is used to assess the chain conformation. Typically, a ratio of <1 indicates a low PEG density with a mushroom conformation; a ratio between 1 and 2 indicates a brush conformation; and a ratio >2 suggests a dense brush conformation.12,17,36 In this study, the [Γ] values for the Std1, Std2, and OPT formulations are similar, at 19.3, 18.1, and 15.5, respectively (Table 1). The [Γ]/[Γ*] ratio for the Std1 formulation is 1.8, indicating a brush conformation, whereas the ratios for the Std2 and OPT formulations are 4.1 and 3.5 respectively, suggesting a dense brush conformation. Notice that the Comirnaty mRNA-LNP also exhibited a dense PEG brush-like conformation.12 The dense brush conformation results in a thicker hydrophilic surface barrier layer, leading to reduced nonspecific protein adsorption, hence smaller frequency shifts observed for the Std2 and OPT formulations. Although the OPT formulation showed significantly less protein adsorption than Std1, it achieved much higher protein expression, indicating that protein adsorption alone is not the sole determinant of biological potency. Additional critical factors including lipid packing, LNP structure, PEG shedding and others, all play a role in delivery efficacy.
Peaks and shifts in SAXS spectra provide valuable insights into the internal structures and phase transitions within LNPs. The SAXS profiles of OPT, Std2, and Std1 mRNA-LNP formulations were first characterized at a total mRNA concentration of 0.5 mg mL−1 in 10% trehalose (Fig. 7a). Two distinct Bragg peaks in the spectra – one at a scattering vector q1 ∼ 0.13 Å−1 and another at q2 ∼ 0.16 Å−1 – indicate the presence of two different internal structures (Fig. 7b). This was confirmed by Cryo-TEM images, which clearly revealed multilamellar and inverted hexagonal structures across all three formulations (Fig. 7c and Fig. S5†). The regions of interest were subjected to fast Fourier transform (FFT), yielding reciprocal lattices from which the repeating distances (i.e., d-spacings) of the structures were determined. The d-spacings for the multilamellar and inverted hexagonal structures of the OPT formulation were approximately 4.2 and 5.5 nm, respectively (Fig. 7c). Hence, peak 1 in the SAXS spectra at lower q corresponds to the inverted hexagonal structure, while peak 2 at higher q corresponds to the multilamellar phase, as the scattering vector is inversely proportional to the lattice spacing. Compared to Std1 and Std2, the OPT formulation exhibited a narrower and higher peak, yet a smaller peak area for the lamellar phase relative to the hexagonal peak (Fig. 7b). The lattice spacing d of the OPT formulation, calculated from q values of the SAXS spectra (d = 2πn/q), was 4.0 and 4.9 nm for the multilamellar and inverted hexagonal phases, respectively, consistent with the values derived from Cryo-TEM.
The pH-dependent phase behavior of the three formulations was subsequently evaluated using SAXS to investigate structural changes during endosomal maturation (Fig. 7d–g). The formulations were mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with citrate-phosphate buffers at pH levels of 7.5, 6.5, 5.5, or 4.5. At pH 7.5, the OPT formulation exhibited two strong peaks, whereas Std2 and Std1 showed weaker peaks, particularly for the multilamellar phase (Fig. 7g). As the pH decreased, the Bragg peaks of Std2 and Std1 became more pronounced, suggesting a transformation of their internal structures from a disordered state toward more ordered multilamellar and inverted hexagonal phases. This observation aligns with a previous study reporting the mesophase transition of LNPs from an amorphous state to lamellar and subsequently to inverted hexagonal phases upon acidification during endosomal maturation.37 Notably, the OPT formulation had the most pronounced inverted hexagonal peak at pH 4.5 compared to the other two formulations (Fig. 7d). The inverted hexagonal phase of LNPs is known to exhibit greater transfection efficiency compared to the lamellar structure.32 Although all three formulations showed significant laminar peaks at pH 4.5 (Fig. 7d), and the laminar phase could transition to the inverted hexagonal phase upon interaction with endosomal membrane-mimicking vesicles, the pre-programed inverted hexagonal phase facilitated a more efficient one-step delivery mechanism by bypassing this transition.32 Additionally, the SAXS peaks shifted to the left as the pH was lowered (Fig. S6†), which is consistent with an increase in d-spacing likely due to increased water content in the LNPs.
1 ratio with citrate-phosphate buffers at pH levels of 7.5, 6.5, 5.5, or 4.5. At pH 7.5, the OPT formulation exhibited two strong peaks, whereas Std2 and Std1 showed weaker peaks, particularly for the multilamellar phase (Fig. 7g). As the pH decreased, the Bragg peaks of Std2 and Std1 became more pronounced, suggesting a transformation of their internal structures from a disordered state toward more ordered multilamellar and inverted hexagonal phases. This observation aligns with a previous study reporting the mesophase transition of LNPs from an amorphous state to lamellar and subsequently to inverted hexagonal phases upon acidification during endosomal maturation.37 Notably, the OPT formulation had the most pronounced inverted hexagonal peak at pH 4.5 compared to the other two formulations (Fig. 7d). The inverted hexagonal phase of LNPs is known to exhibit greater transfection efficiency compared to the lamellar structure.32 Although all three formulations showed significant laminar peaks at pH 4.5 (Fig. 7d), and the laminar phase could transition to the inverted hexagonal phase upon interaction with endosomal membrane-mimicking vesicles, the pre-programed inverted hexagonal phase facilitated a more efficient one-step delivery mechanism by bypassing this transition.32 Additionally, the SAXS peaks shifted to the left as the pH was lowered (Fig. S6†), which is consistent with an increase in d-spacing likely due to increased water content in the LNPs.
We further examined the inter-relationships among protein expression, LNP composition, and physicochemical properties. Pearson correlation coefficients were used to assess the linear correlations between all pairs of variables, as shown in the matrix in Fig. 8a. These coefficients range from −1 to 1, where 1 indicates a perfect positive linear relationship, −1 indicates a perfect negative linear relationship, and 0 indicates no linear relationship. Red and blue circles represent positive and negative relationships, respectively. The hEPO expression was found to correlate most strongly with two LNP characteristics: mRNA encapsulation efficiency and GP7.5/GP4.5. While mRNA encapsulation largely depends on the PEG fraction, GP7.5/GP4.5, an index for lipid packing, is primarily impacted by the ratio of OF-02, followed by the ratios of cholesterol and DOPE.
A lower GP7.5/GP4.5 ratio resulted in higher protein expression, with the ratio being more strongly influenced by GP4.5 (Fig. 8a). Thus, a larger GP4.5 is desirable to enhance protein expression, underscoring the significance of membrane integrity at lower pH. Following endocytosis, LNPs encounter a highly acidic environment as endosomes mature. LNPs with poor lipid packing and compromised membrane integrity are more prone to suffer significant mRNA degradation under such acidic conditions. In contrast, LNPs with superior membrane integrity are more likely to fuse with endosome membrane, promoting endosomal escape and mRNA release, thereby improving transfection efficiency (Fig. 8b). Additionally, LNPs with more ordered internal structures may further enhance endosomal escape and exhibit even greater potency in such scenarios.
Conclusions
Screening a library of PEGylated lipids and optimizing the formulations using DoE resulted in the development of an optimal DMG-PEG5k mRNA-LNP formulation, which demonstrated improved transfection efficiency compared to the DMG-PEG2k benchmark. A series of advanced physicochemical characterizations were conducted to elucidate the structure–activity relationship. Among all the factors tested, mRNA encapsulation efficiency, lipid packing and membrane integrity (particularly at low pH), and the internal structure of the LNPs were identified as the three most critical factors influencing the transfection efficiency of mRNA-LNPs. High EE, enhanced membrane integrity as indicated by a low GP7.5/GP4.5 ratio, and more ordered internal phases, achieved through balanced ratios of lipid components, closely correlated with improved in vivo potency. With screening of novel PEG lipids and optimization of formulation compositions, this study offers a fundamental understanding of LNP structure–activity relationships for intramuscular route of administration and provides valuable guidelines and characterization techniques for the rational design, optimization, and application of mRNA-LNPs.Abbreviations
| LNP | Lipid nanoparticles |
| mRNA | Messenger ribonucleic acid |
| SAR | Structure–activity relationship |
| DOE | Design of Experiments |
| IM | Intramuscular |
| IV | Intravenous |
| SiRNA | Small interfering RNA |
| ASO | Antisense oligonucleotide |
| PEG | Polyethylene glycol |
| DMG-PEG2k | PEG 2000 with dimyristoyl glycerol |
| DMG-PEG5k | PEG 5000 with dimyristoyl glycerol |
| chol-PEG | Cholesterol PEG |
| DOPE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine |
| hEPO | Human erythropoietin |
| PE | Phosphoethanolamine |
| Cryo-TEM | Cryogenic transmission electron microscopy |
| SAXS | Small angle X-ray scattering |
| QCM-D | Quartz crystal microbalance with dissipation |
| GP | Generalized polarization |
| DLS | Dynamic light scattering |
| PDI | Polydispersity index |
| EE | mRNA encapsulation efficiency |
| pKa | Apparent acid dissociation constant |
| TNS | 2-(p-Toluidino)-6-naphthalene sulfonic acid |
| FFT | Fast Fourier transform |
| ELISA | Enzyme-linked immunosorbent assay |
| ApoE | Apolipoprotein E |
Associated content
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Lingyun Liu: conceptualization, methodology, investigation, writing – original draft, review & editing, visualization, supervision, project administration. Jae-Heon Kim: investigation, writing – original draft, visualization. Zhongyu Li: investigation, writing – original draft, review & editing, visualization. Mengwei Sun: investigation, writing – original draft, visualization. Trent Northen: methodology, visualization. Jackie Tang: visualization. Emma Mcintosh: investigation, writing – original draft, visualization. Frank DeRosa: writing – review & editing, project administration. Shrirang Karve: conceptualization, methodology, writing – review & editing, supervision, project administration.
Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
All authors are Sanofi employees and may hold shares and/or stock options in the company.Acknowledgements
The research included in the manuscript was funded by Sanofi. The authors would like to thank Barak Yahalom at Alpha Preclinical for managing all in vivo studies. We would also like to thank Dr Hui Wei and Prof. Jason Kaelber from the RCNF at Rutgers university for the Cryo-TEM images. We also thank Josue San Emeterio and Scott Barton at Xenocs for the SAXS spectra and analysis.References
- U. Sahin, A. Muik, E. Derhovanessian, I. Vogler, L. M. Kranz, M. Vormehr, A. Baum, K. Pascal, J. Quandt and D. Maurus, COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses, Nature, 2020, 586(7830), 594–599 CAS.
- L. R. Baden, H. M. El Sahly, B. Essink, K. Kotloff, S. Frey, R. Novak, D. Diemert, S. A. Spector, N. Rouphael and C. B. Creech, Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine, N. Engl. J. Med., 2021, 384(5), 403–416 CrossRef CAS.
- F. P. Polack, S. J. Thomas, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, J. L. Perez, G. Pérez Marc, E. D. Moreira and C. Zerbini, Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine, N. Engl. J. Med., 2020, 383(27), 2603–2615 CrossRef CAS.
- E. E. Walsh, R. W. Frenck Jr, A. R. Falsey, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, M. J. Mulligan and R. Bailey, Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates, N. Engl. J. Med., 2020, 383(25), 2439–2450 CrossRef CAS PubMed.
- W. Ho, M. Gao, F. Li, Z. Li, X. Q. Zhang and X. Xu, Next–generation vaccines: nanoparticle–mediated DNA and mRNA delivery, Adv. Healthcare Mater., 2021, 10(8), 2001812 CrossRef CAS.
- M. D. Buschmann, M. J. Carrasco, S. Alishetty, M. Paige, M. G. Alameh and D. Weissman, Nanomaterial delivery systems for mRNA vaccines, Vaccines, 2021, 9(1), 65 CAS.
- N. Chaudhary, D. Weissman and K. A. Whitehead, mRNA vaccines for infectious diseases: principles, delivery and clinical translation, Nat. Rev. Drug Discovery, 2021, 20(11), 817–838 CAS.
- J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana, M. L. Maitland, J. Seitzer, D. O'Connell, K. R. Walsh and K. Wood, CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis, N. Engl. J. Med., 2021, 385(6), 493–502 CAS.
- E. Kon, N. Ad-El, I. Hazan-Halevy, L. Stotsky-Oterin and D. Peer, Targeting cancer with mRNA–lipid nanoparticles: key considerations and future prospects, Nat. Rev. Clin. Oncol., 2023, 20(11), 739–754 CAS.
- W. Zhao, X. Hou, O. G. Vick and Y. Dong, RNA delivery biomaterials for the treatment of genetic and rare diseases, Biomaterials, 2019, 217, 119291 CAS.
- M. Sun, U. J. Dang, Y. Yuan, A. M. Psaras, O. Osipitan, T. A. Brooks, F. Lu and A. J. Di Pasqua, Optimization of DOTAP/chol cationic lipid nanoparticles for mRNA, pDNA, and oligonucleotide delivery, AAPS PharmSciTech, 2022, 23(5), 135 CAS.
- M. M. Wang, C. N. Wappelhorst, E. L. Jensen, Y.-C. T. Chi, J. C. Rouse and Q. Zou, Elucidation of lipid nanoparticle surface structure in mRNA vaccines, Sci. Rep., 2023, 13(1), 16744 CAS.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Lipid nanoparticles for mRNA delivery, Nat. Rev. Mater., 2021, 6(12), 1078–1094 CrossRef CAS PubMed.
- E. Samaridou, J. Heyes and P. Lutwyche, Lipid nanoparticles for nucleic acid delivery: Current perspectives, Adv. Drug Delivery Rev., 2020, 154, 37–63 CrossRef PubMed.
- S. C. Semple, A. Akinc, J. Chen, A. P. Sandhu, B. L. Mui, C. K. Cho, D. W. Sah, D. Stebbing, E. J. Crosley and E. Yaworski, Rational design of cationic lipids for siRNA delivery, Nat. Biotechnol., 2010, 28(2), 172–176 CAS.
- L. Miao, Y. Zhang and L. Huang, mRNA vaccine for cancer immunotherapy, Mol. Cancer, 2021, 20(1), 41 CAS.
- R. Tenchov, J. M. Sasso and Q. A. Zhou, PEGylated lipid nanoparticle formulations: immunological safety and efficiency perspective, Bioconjugate Chem., 2023, 34(6), 941–960 CAS.
- Z. Li, X.-Q. Zhang, W. Ho, F. Li, M. Gao, X. Bai and X. Xu, Enzyme-catalyzed one-step synthesis of ionizable cationic lipids for lipid nanoparticle-based mRNA COVID-19 vaccines, ACS Nano, 2022, 16(11), 18936–18950 CAS.
- F. Li, X.-Q. Zhang, W. Ho, M. Tang, Z. Li, L. Bu and X. Xu, mRNA lipid nanoparticle-mediated pyroptosis sensitizes immunologically cold tumors to checkpoint immunotherapy, Nat. Commun., 2023, 14(1), 4223 CAS.
- K. Lam, P. Schreiner, A. Leung, P. Stainton, S. Reid, E. Yaworski, P. Lutwyche and J. Heyes, Optimizing lipid nanoparticles for delivery in primates, Adv. Mater., 2023, 35(26), 2211420 CAS.
- B. L. Mui, Y. K. Tam, M. Jayaraman, S. M. Ansell, X. Du, Y. Y. C. Tam, P. J. Lin, S. Chen, J. K. Narayanannair and K. G. Rajeev, Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles, Mol. Ther.–Nucleic Acids, 2013, 2(12), e139 CAS.
- A. Sarode, Y. Fan, A. E. Byrnes, M. Hammel, G. L. Hura, Y. Fu, P. Kou, C. Hu, F. I. Hinz and J. Roberts, Predictive high-throughput screening of PEGylated lipids in oligonucleotide-loaded lipid nanoparticles for neuronal gene silencing, Nanoscale Adv., 2022, 4(9), 2107–2123 CAS.
- M. Hammel, Y. Fan, A. Sarode, A. E. Byrnes, N. Zang, P. Kou, K. Nagapudi, D. Leung, C. C. Hoogenraad and T. Chen, Correlating the structure and gene silencing activity of oligonucleotide-loaded lipid nanoparticles using small-angle X-ray scattering, ACS Nano, 2023, 17(12), 11454–11465 CAS.
- M. Berger, M. Degey, J. Leblond Chain, E. Maquoi, B. Evrard, A. Lechanteur and G. Piel, Effect of PEG anchor and serum on lipid nanoparticles: development of a nanoparticles tracking method, Pharmaceutics, 2023, 15(2), 597 CAS.
- F. DeRosa, B. Guild, S. Karve, L. Smith, K. Love, J. Dorkin, K. Kauffman, J. Zhang, B. Yahalom and D. Anderson, Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system, Gene Ther., 2016, 23(10), 699–707 CAS.
- Q. Xu, L. M. Ensign, N. J. Boylan, A. Schon, X. Gong, J.-C. Yang, N. W. Lamb, S. Cai, T. Yu and E. Freire, Impact of surface polyethylene glycol (PEG) density on biodegradable nanoparticle transport in mucus ex vivo and distribution in vivo, ACS Nano, 2015, 9(9), 9217–9227 CAS.
- Q. Xu, N. J. Boylan, S. Cai, B. Miao, H. Patel and J. Hanes, Scalable method to produce biodegradable nanoparticles that rapidly penetrate human mucus, J. Controlled Release, 2013, 170(2), 279–286 CAS.
- O. S. Fenton, K. J. Kauffman, R. L. McClellan, E. A. Appel, J. R. Dorkin, M. W. Tibbitt, M. W. Heartlein, F. DeRosa, R. Langer and D. G. Anderson, Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery, Adv. Mater., 2016, 28(15), 2939 CAS.
- S. C. Wilson, J. L. Baryza, A. J. Reynolds, K. Bowman, M. E. Keegan, S. M. Standley, N. P. Gardner, P. Parmar, V. O. Agir and S. Yadav, Real time measurement of PEG shedding from lipid nanoparticles in serum via NMR spectroscopy, Mol. Pharm., 2015, 12(2), 386–392 CAS.
- Y. Fan, C.-W. Yen, H.-C. Lin, W. Hou, A. Estevez, A. Sarode, A. Goyon, J. Bian, J. Lin and S. G. Koenig, Automated high-throughput preparation and characterization of oligonucleotide-loaded lipid nanoparticles, Int. J. Pharm., 2021, 599, 120392 CAS.
- K. Koitabashi, H. Nagumo, M. Nakao, T. Machida, K. Yoshida and K. Sakai-Kato, Acidic pH-induced changes in lipid nanoparticle membrane packing, Biochim. Biophys. Acta, Biomembr., 2021, 1863 (8), 183627 CrossRef.
- R. Pattipeiluhu, Y. Zeng, M. M. Hendrix, I. K. Voets, A. Kros and T. H. Sharp, Liquid crystalline inverted lipid phases encapsulating siRNA enhance lipid nanoparticle mediated transfection, Nat. Commun., 2024, 15(1), 1303 CAS.
- A. Szilágyi, E. Selstam and H.-E. Åkerlund, Laurdan fluorescence spectroscopy in the thylakoid bilayer: the effect of violaxanthin to zeaxanthin conversion on the galactolipid dominated lipid environment, Biochim. Biophys. Acta, Biomembr., 2008, 1778(1), 348–355 CrossRef PubMed.
- D. Chen, S. Ganesh, W. Wang and M. Amiji, Plasma protein adsorption and biological identity of systemically administered nanoparticles, Nanomedicine, 2017, 12(17), 2113–2135 CAS.
- Y. Suzuki and H. Ishihara, Structure, activity and uptake mechanism of siRNA-lipid nanoparticles with an asymmetric ionizable lipid, Int. J. Pharm., 2016, 510(1), 350–358 Search PubMed.
- L. Shi, J. Zhang, M. Zhao, S. Tang, X. Cheng, W. Zhang, W. Li, X. Liu, H. Peng and Q. Wang, Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery, Nanoscale, 2021, 13(24), 10748–10764 CAS.
- H. Yu, J. Iscaro, B. Dyett, Y. Zhang, S. Seibt, N. Martinez, J. White, C. J. Drummond, S. Bozinovski and J. Zhai, Inverse cubic and hexagonal mesophase evolution within ionizable lipid nanoparticles correlates with mRNA transfection in macrophages, J. Am. Chem. Soc., 2023, 145(45), 24765–24774 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: LNP characterization, correlations between protein expression and formulation parameters, overlaid Laurdan spectra, cryo-TEM images, SAXS diffractograms at different pH, formulation stability profile. See DOI: https://doi.org/10.1039/d5nr00433k |
| This journal is © The Royal Society of Chemistry 2025 |