Open Access Article
Open Access ArticleModulating peptide co-assembly via macromolecular crowding: Recipes for co-assembled structures
Xin Y.
Dong
 *a,
Madisen
Domayer
b,
Gregory A.
Hudalla
*a,
Madisen
Domayer
b,
Gregory A.
Hudalla
 b and
Carol K.
Hall
b and
Carol K.
Hall
 a
a
aDepartment of Chemical and Biomolecular Engineering, North Carolina State University, USA. E-mail: hall@ncsu.edu
bDepartment of Biomedical Engineering, University of Florida, USA
First published on 13th June 2025
Abstract
Peptide-based biomaterials are commonly found in applications such as tissue engineering, wound healing, and drug delivery. Control over the size and morphology of the peptide supramolecular structure remains a challenge. One way to influence peptide assembly is through macromolecular crowding. Here we use discontinuous molecular dynamics simulation combined with the PRIME20 force field to investigate the effect of hydrophobic crowders on the architecture of co-assembled peptide aggregates. The peptide system used in this work is a mixture of oppositely-charged synthetic peptides: “CATCH(6K+)” (KQKFKFKFKQK) and “CATCH(6E−)” (EQEFEFEFEQE). The systems explored contained a mixture of 50 CATCH(6K+) and 50 CATCH (6E−) peptides at peptide concentrations of 5 mM and 20 mM, and crowders with diameters of 10, 20, 40 and 80 Å. Crowders were modeled as spheres with either hard-sphere or square-well/square-shoulder interactions. At low concentrations where CATCH co-assembly typically does not occur, the crowders were effective chaperones to trigger co-assembly. Small hard-sphere crowders promoted formation of multilayer fibrils. Large highly hydrophobic crowders promoted the formation of monolayer β-sheet structures and suppressed the formation of fibril structures. Overall, the simulations demonstrate that the crowder size and crowder–sidechain interaction strength govern the supramolecular architecture of peptide co-assemblies.
Introduction
Peptide self-assembly is a process in which peptides spontaneously come together to form organized structures. It is a hallmark of amyloidogenic and neurodegenerative diseases, e.g. the self-assembly of Aβ in Alzheimer's, α-synuclein in Parkinson's, and amylin in diabetes. The self-assembling properties of peptides can be leveraged to develop supramolecular biomaterials such as hydrogels, vesicles, lipid bilayers, and tubular structures.1 Peptide-based biomaterials are a particularly attractive choice for biological applications such as biosensing, drug delivery, and tissue engineering due to their mechanical stability, biocompatibility, and biodegradability.2–4 As peptide-based structures rely solely on physical cross-linking, as opposed to chemical modifications, there is low risk of introducing harmful agents upon administration.5 Any artifacts from peptide degradation are naturally-occurring amino acids that can be metabolized by the cells. An added benefit of peptide biomaterials is that they can be functionalized by conjugating the self-assembling peptide to a molecule with the desired functionality.6The bulk properties of biomaterials govern their performance for biomedical applications. For example, network alignment, pore size, and stiffness can impact cell behavior, which can be consequential in tissue engineering or regenerative medicine as cells are responsive to the stiffness of their environment.7 In drug delivery, the rigidity of the nanostructure can determine how well it can transport across biological hydrogels and cellular barriers.8 In addition, the size, shape, and charge of the carrier material directly impact how it interacts with its target.9 For supramolecular peptide-based biomaterials it is advantageous to understand how assembly conditions affect fibril structure, which underlies bulk material properties.
Peptide-based hydrogels can be constructed from short and long-chain peptides, β-structures (sheets, hairpins, or turns), α-helical peptides, and cyclic peptides,10–12 with advances in peptide synthesis broadening the options for peptides sequences. Previous research conducted on the effect of amino acid sequence on peptide assembly provides general guidelines for designing peptide-based supramolecular structures with predictable architectural features.13–17 Self-assembling peptides are commonly amphiphiles with alternating hydrophobic and hydrophilic residues. Altering the peptide composition impacts the secondary structure formed, the interactions between peptides, the fibril persistence length, the mesh size of the fibril network, and in turn the bulk properties of the resulting fibril structure.18,19 Increasing the strength of hydrophobic interactions can lead to stiffer fibril networks.20 Additionally, incomplete burial of hydrophobic resides may lead to displaced β-sheet layers and inhibit fibril growth in the linear direction.21 The net charge of a polypeptide can influence mechanical properties. Previous work by Zhao and coworkers on the RATEA16 peptide demonstrated that balancing attractive and repulsive electrostatic forces facilitates self-assembly and the formation of a self-supporting gel.22 Inter-backbone hydrogen bonds, which can be modulated by side-chain interactions, also contribute to fibril rigidity.23 These interactions can alter the twist in the self-assembled β-sheets, which is related to the strength of the gel. Greater gel stiffness has been previously associated with β-sheets with less twisting and disorder.16–18,24 The chain length and structure of the peptide can also influence the fibril architecture. Using a series of truncated peptides, Pogostin and coworkers illustrated the impact of sequence length on secondary structure formation, fibril morphology, and ultimately material properties (i.e. hydrogel stiffness).25
The morphology of a peptide fibril can be modulated by transforming its environment through crowding rather than by altering its primary structure through sequence mutations or truncations. Macromolecular crowding is the phenomenon in which a high concentration of macromolecules added to a solution of peptides impacts the interactions and transport properties of the peptides.26 The concept of macromolecular crowding was first introduced by Minton in 1981 and has since been extensively researched and reviewed.27–31 Motivation for research on macromolecular crowding in biological systems comes in part from the observation that cells themselves are crowded.32 Understanding of how crowders impact peptide assembly has advanced our understanding of peptide aggregation. For example, within the Aβ system, the introduction of metal nanoparticles (NPs) has been shown to both accelerate and inhibit fibrillization.33–36 In the field of biomaterials, previous work has demonstrated that macromolecular crowding can be used to alter hydrogel stiffness. Ranamukhaarachchi and coworkers used 8 kDa PEG crowders to fine-tune fibril architecture and the degree of confinement in collagen hydrogels.37 They showed that increasing the amount of PEG added during collagen assembly increased the tightness and durability of collagen fiber networks. Dewavrin and coworkers found that the introduction of Ficoll crowders to collagen gels increased their resistance to mechanical stress.38 Experiments by Hirota and coworkers revealed that PEG crowders converted a linear cytochrome c trimer to a smaller cyclic trimer.39 Restuccia and coworkers showed that crowded conditions led to the alignment of glycosylated peptide fibrils.40 Collectively, these examples demonstrate that the impact of crowding on peptide assembly is complex, as trends in one system cannot always be generalized to another. Inspired by the afore-mentioned previous work, we hypothesize that macromolecular crowding can be used to influence the size and morphology of supramolecular peptide nanostructures.
The objective of this work is to determine the effect of crowder size and crowder–sidechain interaction strength on peptide co-assembly—a process in which two distinct peptides spontaneously associate to form a supramolecular structure. Our peptide system is a mixture of oppositely-charged synthetic peptides, referred to as CATCH (Co-Assembly Tags based on Charge complementarity).41 Although various CATCH pairs have been reported, here we limit our study to the “CATCH(6K+)” (KQKFKFKFKQK) and “CATCH(6E−)” (EQEFEFEFEQE). CATCH peptides of the same charge resist self-assembly due to strong electrostatic repulsion. The CATCH(6K+/6E−) system has been experimentally and computationally shown to selectively co-assemble into β-sheet nanofibers with an alternating “ABAB” motif of positively-charged and negatively-charged peptide strands above a critical concentration in the low 0.1 mM range.15–17,42 At mM concentration, CATCH(6K+/6E−) nanofibers can undergo a sol–gel transition to form a physically crosslinked hydrogel. Here we used discontinuous molecular dynamics simulations to understand the effect of hydrophobic crowders on CATCH(6K+/6E−) co-assembly in dilute and concentrated conditions.
In this work, we apply discontinuous molecular dynamics simulation with the intermediate-resolution PRIME20 force field to study the effect of crowders on peptide co-assembly. We consider two conditions: (1) a low peptide concentration where co-assembly typically does not take place, and (2) a high peptide concentration where co-assembly typically does take place. Crowders are modeled as spheres for simplicity. A factorial design was carried out to systematically investigate the impact of varying crowder size (diameter = 10, 20, 40, 80 Å) and crowder–sidechain interaction strength (0, ½, 1, 2, 5ε) on peptide co-assembly at low (Cpeptide = 5 mM) and high (Cpeptide = 20 mM) peptide concentrations. All simulations involving crowders were performed at a crowder volume fraction φcrowder = 0.20.
Highlights of our results include the following: At a low peptide concentration where aggregation is typically not observed, the addition of any size crowder resulted in co-assembly—save the addition of small extremely-hydrophobic crowders. Small hard-sphere crowders were the most effective for guiding peptide co-assembly into fibrils containing two or more β-sheet layers. The presence of crowders can alter the relative orientation of neighboring β-strands in a β-sheet structure. Adding crowders into the CATCH(6K+/6E−) mixture increased the instances of mismatched nearest neighbors (instances of AA or BB pattern rather than AB) and “flipped” nearest neighbors in a β-sheet—instances where the hydrophobic residues that lie on one side of a β-strand are flipped along the backbone relative to the hydrophobic residues of the neighboring β-strand. Introduction of small hard-sphere, weakly-hydrophobic, or hydrophobic crowders increased fibril thickness. Large highly-hydrophobic crowders promoted and stabilized the formation of monolayer β-sheet structures and prevented formation of cylindrins and multilayer fibril structures. In summary, simulations show that crowder size and interaction strength influence the organization of peptides. Our work provides a comprehensive approach to systematically evaluate the effect of crowders on peptide assembly; the method presented can be extended to other peptide sequences and crowder–sidechain interactions.
Results and discussion
In this work we considered peptide concentrations of 5 and 20 mM, crowder–sidechain interaction strengths of 0, ½, 1, 2, and 5ε, and crowder diameters of 10, 20, 40 and 80 Å, at a constant crowder volume fraction of φcrowder = 0.20. The crowder volume fraction is defined as φcrowder = NVCR/V (where N is the number of crowders, VCR is the volume occupied by a single crowder, and V is the volume of the simulation box). This was the highest crowder volume fraction that allowed us to consider a wide range of crowder diameters and a timescale of ∼10 μs without demanding excessive CPU time. The unit for crowder–sidechain interaction strength ε is based on the phenylalanine sidechain in PRIME20 (see Methods). For example, 0ε represents a hard-sphere interaction and 1ε represents a hydrophobic interaction equivalent to a phenylalanine sidechain group. The interaction energy parameters are provided in Table 1 in Methods. The hydrophobic crowders have attractive interactions with phenylalanine (F) sidechains, and repulsive interactions with glutamine (Q), glutamic acid (E), and aspartic acid (D) sidechains—consistent with how a phenylalanine sidechain is modeled in the PRIME20 force field. A visual of each crowding condition is shown in Fig. 1. The peptide concentration throughout this paper refers to the number of peptides per unit volume and is defined as: Cpeptide = Npeptide/(NAV), where NA is Avogadro's number and V is the volume of the simulation box. The addition of crowders increases the excluded volume (volume inaccessible to peptides due to the presence of crowders and other peptides) and effectively, the peptide concentration. This increased peptide concentration is referred to as the “effective peptide concentration” and is defined as: Ceff = Npeptide/[V(1 − ϕ)]. At a crowder volume fraction of φcrowder = 0.20, the effective peptide concentrations for 5 and 20 mM systems become 6.3 mM and 25.3 mM, respectively.| Residue | Interaction strength ε and range λ between crowder and K, E, and F sidechains | ||||
|---|---|---|---|---|---|
| ½ε | 1ε | 2ε | 5ε | λ | |
| Q | 0.0075 | 0.015 | 0.03 | 0.075 | 1.517 |
| K | 0.0075 | 0.015 | 0.03 | 0.075 | 1.676 |
| E | 0.0075 | 0.015 | 0.03 | 0.075 | 2.195 |
| F | −0.1025 | −0.205 | −0.41 | −1.025 | 1.767 |
Aggregation in the absence of crowders
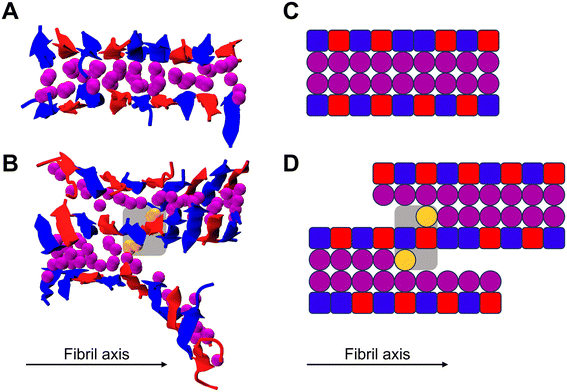
Simulations of CATCH(6K+/6E−) peptides in the absence of crowders were performed to establish a baseline for peptide co-assembly. Systems of equimolar CATCH(6K+) and CATCH(6E−) mixtures containing a total of 100 peptides were simulated for 300 billion collisions (∼10 μs), at a reduced temperature of T* = 0.20 (∼342 K) and concentrations of 5 and 20 mM. This simulation temperature is the fibrilization temperature, above which systems of CATCH(6K+/6E−) peptides at Cpeptide = 20 mM are unlikely to form fibril structures. The final simulation snapshots are shown in Fig. 2. At a peptide concentration of 5 mM, nearly all of the CATCH(6K+/6E−) peptides remained in a random coil conformation throughout the simulation. The final simulation snapshot in this case revealed the presence of a cylindrin-like structure. Cylindrins are β-sheet structures connected by backbone hydrogen bonds that have wrapped around to form a β-barrel-like structure with a hydrophobic core—typically composed of six to eight peptides. The structure formed at Cpeptide = 5 mM agrees with previous observations of oligomer formation at lower concentrations in the CATCH(4K+/6E−) system.43 At a peptide concentration of 20 mM, the CATCH(6K+/6E−) peptides organized into a long antiparallel β-sheet bilayer with an alternating motif of CATCH(6K+) and CATCH(6E−) peptides. This β-sheet structure is consistent with previous experimental and simulation-based observations of CATCH(6K+/6E−) structures.15–17 | ||
| Fig. 2 (Left) Final simulation snapshots of CATCH(6K+/6E−) mixtures at Cpeptide = 5 mM and (right) 20 mM, at T* = 0.20. CATCH(6K+) is shown in blue. CATCH(6E−) is shown in red. | ||
Fibrils are the largest ordered structures that form during CATCH(6K+/6E−) co-assembly in DMD simulations. In experiments, CATCH(6K+/6E−) fibrils extend to form nanofibers that can further crosslink into hydrogels.15,16 The fibrils formed at Cpeptide = 20 mM contained a maximum of two β-sheets stacked upon one another, with the hydrophobic phenylalanine sidechains pointing inwards (between the sheets) and the charged lysine and glutamic acid sidechains pointing outwards. The β-sheets within the fibril were antiparallel and exhibited the expected ABAB pattern between strands of positively-charged and negatively-charged peptides. For a third β-sheet to laminate or form upon the two existing β-sheets, the negatively-charged residues on the CATCH(6E−) peptides on one β-sheet would need to align with the positively-charged residues on the CATCH(6K+) peptides on the other β-sheet, and vice versa. We speculate that we have not captured this lamination event in DMD/PRIME20 simulations because at a reduced temperature of T* = 0.20, which is relatively high, it is unlikely that this alignment of negatively and positively-charged residues will occur.
Effect of crowding on peptide co-assembly kinetics
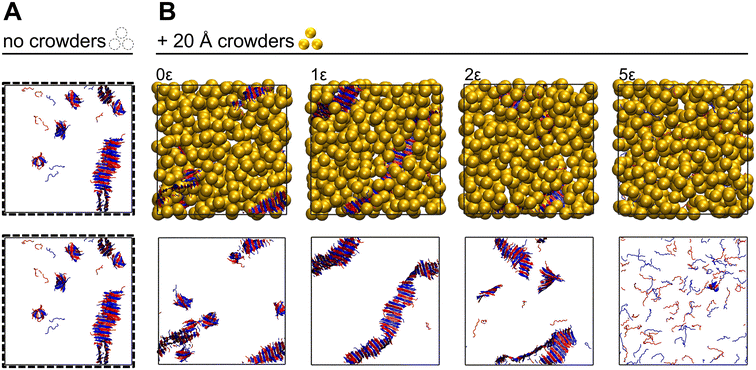
In this section we summarize the impact of crowding on peptide co-assembly kinetics at three crowder–sidechain interaction strengths—hard-sphere (0ε), moderately hydrophobic (2ε), and strongly hydrophobic (5ε)—and detail how the behavior at each interaction strength changed with crowder size. The impact of crowding on peptide co-assembly was characterized by calculating the number of peptides in assembled structures and the number of sidechain–sidechain interactions over the course of a simulation. An “assembled” structure is defined to be a group of peptides that share either four hydrogen bonding interactions or four sidechain–sidechain interactions. The effect of the crowder–sidechain interaction strength and the crowder size on assembly kinetics was more pronounced for Cpeptide = 20 mM than for 5 mM and is discussed in detail below. The effect of crowders on peptide assembly fluctuated at Cpeptide = 5 mM due to the sparseness of CATCH(6K+/6E−) peptides in a low concentration system and the timescale explored in our simulations (∼10 μs).Hard-sphere (0ε) crowders increased the rate of co-assembly for CATCH(6K+/6E−) peptides and stabilized co-assemblies. Interestingly, the rate of assembly for Cpeptide = 5 mM in the presence of 10 Å hard-sphere crowders was higher than in a noncrowded Cpeptide = 20 mM system, even though the effective concentration (Ceff = 6.3 mM) was much lower. This result demonstrated that a crowded system with hard-spheres is more effective than a non-crowded high concentration system in driving peptide assembly. For both Cpeptide = 5 mM and 20 mM, the rate of peptide co-assembly increased with decreasing crowder diameter (Fig. 3 shows results for Cpeptide = 20 mM). Decreasing the crowder diameter increased the depletion forces between peptides, in agreement with theory and with previous computational and experimental observations.44–46 In a hard-sphere crowded environment, CATCH(6K+/6E−) peptides are incentivized to assemble as the peptide aggregates occupy less space and experience a greater number of favorable interactions than in the random coil conformation. The increase in peptide co-assembly in the presence of inert crowders demonstrates that hard-sphere repulsions are entropically stabilizing.30,31 Our results are consistent with previous simulations carried out by Latshaw et al. in which the addition of hard-sphere crowders increased the rate of aggregation of Aβ(16–22).44 Experimentally, excluded volume effects by inert polymers have been reported to increase the rate of amyloid aggregation and stabilize compact conformations.47–49
Strongly-hydrophobic 2ε crowders of diameter 40 and 80 Å were effective in initiating CATCH(6K+/6E−) co-assembly, but later obstructed the formation of sidechain–sidechain interactions between peptides (Fig. 4). In the presence of strongly-hydrophobic 2ε crowders, the phenylalanine sidechains on CATCH(6K+/6E−) peptides favored interactions with the crowders over interactions with other phenylalanine sidechains; this led to a reduction in the maximum number of sidechain–sidechain interactions that formed compared to systems with hard-sphere crowders or no crowders. The rate of formation of sidechain–sidechain interactions increased with crowder size. At the start of the simulation, crowders with 40 and 80 Å diameters promoted sidechain–sidechain interactions by recruiting peptides to their surfaces and increasing the local concentration. In other words, the surface of the large strongly-hydrophobic crowders with 40 and 80 Å diameters acted as a template for CATCH(6K+/6E−) peptide co-assembly. However, since CATCH(6K+/6E−) phenylalanine sites were occupied by the crowders, the maximum number of sidechain–sidechain interactions that formed is reduced compared to the case with no crowders (dashed line Fig. 4). This explains why the β-sheet structures form in the presence of strongly-hydrophobic 2ε crowders of diameter 40 and 80 Å, as will be discussed in later sections.
Extremely-hydrophobic (5ε) crowders with diameters of 10 and 20 Å suppressed peptide co-assembly (Fig. 5). For both Cpeptide = 5 (not shown) and 20 mM, the 10 and 20 Å diameter crowders with 5ε interaction strength inhibited co-assembly as the CATCH(6K+/6E−) peptides could not escape the strong interaction with the crowder. At a fixed crowder volume fraction, as the crowder diameter decreased, the number of crowders needed to maintain the same crowder volume fraction increased; smaller crowders created a more homogenous and well-dispersed void space and increased the likelihood of a crowder–sidechain interaction compared to systems of large crowders. As a result, CATCH(6K+/6E−) peptides in the presence of small extremely-hydrophobic crowders were restrained in movement due to their interaction with the crowder and remained relatively dispersed in the system in random coil conformations.
The inhibition of higher order peptide co-assemblies by small interacting crowders is substantiated by previous studies on macromolecular crowding in protein systems. Simulations by Latshaw showed that highly-hydrophobic crowders led to disordered Aβ(16–22) oligomers.50 Similarly, nanoparticles have been explored as a potential strategy for inhibiting fibrillation of amyloidogenic proteins such as Aβ and amylin.51–55 Gao et al. used L-glutathione (GSH) coated AuNPs as their crowder model; the GSH has free carboxyl and amino groups that have non-specific electrostatic and hydrogen bonding interactions with Aβ. They showed that larger AuNPs accelerated Aβ40 fibrillation, while smaller AuNPs suppressed Aβ40 fibrillation, and Au nanoclusters completely inhibited Aβ40 fibrillation.34 The inhibition mechanism typically stems from the binding of monomeric species to the nanoparticle, which prevents fibrillation by reducing the amount of monomer in solution and by blocking the sites on the monomers that initiate assembly.51,53 Generally, previous studies have shown that attractive crowder–protein interactions can destabilize protein–protein interactions.56–58 At a high attractive crowder–protein interaction strength, there is a large enthalpic barrier for dissociating crowder–protein interactions.59 These findings align with the observations made from simulations in this work.
It helps to summarize the full range of interaction strengths explored: Hard-sphere (0ε) and weakly-hydrophobic (½ε) crowders were more effective in driving co-assembly as the crowder diameter decreased. In contrast, moderately (1ε) to extremely (5ε) hydrophobic crowders were more effective in driving co-assembly as the crowder diameter increased. Extremely-hydrophobic (5ε) crowders of 10 and 20 Å diameters suppressed peptide co-assembly.
Effect of crowders on fibril structure
The assembled CATCH(6K+/6E−) structures that were observed in the presence of crowders were classified as either oligomers or fibrils. As mentioned earlier, an “assembled structure” here is a cluster of CATCH(6K+/6E−) peptides in which each of the peptides share at least four hydrogen bonding interactions or share at least four sidechain–sidechain interactions with another peptide in the cluster. An oligomer is defined as an assembled structure that contains less than or equal to eight peptides or is a monolayer β-sheet structure. A fibril is an assembled structure that contains greater than eight peptides and has more than one β-sheet layer.CATCH(6K+/6E−) fibril formation was promoted by the addition of small (10 Å) crowders at Cpeptide = 5 and 20 mM (Fig. 6). At a low peptide concentration (Cpeptide = 5 mM), fibril formation typically does not occur in the absence of crowders. However, the addition of small hard-sphere (0ε) or weakly-hydrophobic (½ε) crowders led to the formation of fibrils that contained two β-sheet layers. The addition of small hydrophobic (1ε) and strongly-hydrophobic (2ε) crowders led to the formation of single-layer β-sheet structures. The addition of small extremely-hydrophobic (5ε) crowders prevented any assembly. As the crowder–peptide interaction strength increased, the level of organization in the assembled structures decreased. At a high peptide concentration (Cpeptide = 20 mM), where fibril formation typically does occur in the absence of crowders, these fibrils contained a maximum of two β-sheet layers. However, the addition of small 0, ½, 1ε, and 2ε crowders at Cpeptide = 20 mM led to the formation of fibrils that contained greater than two β-sheet layers. At Cpeptide = 5 and 20 mM, addition of small hard-sphere crowders was more effective at promoting fibril formation than the addition of larger and more hydrophobic crowders. To summarize, the addition of small 10 Å crowders led to the formation of: (1) fibril structures in low concentration systems where assembly typically does not occur, and (2) thicker fibril structures in high concentration systems where assembly typically does occur.
The addition of small 0ε to 2ε crowders led to CATCH(6K+/6E−) fibril structures with instances of “flipped” neighboring β-strands within a β-sheet. Typically, in non-crowded systems, the hydrophobic residues on adjacent β-strands point in the same direction (Fig. 7A and C). The term “flipped” is used here to describe neighboring β-strands in which the hydrophobic residues on one β-strand are facing the opposite direction relative to the direction faced by the hydrophobic residues on the adjacent β-strand (Fig. 7B and D). To the best of our knowledge, this study is the first to suggest the presence of “flipped” peptide pairs. This structural heterogeneity has not been verified experimentally due to the buried placement of these peptides within the fibril structure, the difference between the peptide concentrations in simulations and experiments, and the rare occurrence of “flipped” peptides in simulations. It is possible that these “flipped” peptides may be an artifact of the simulation resulting from truncated electrostatic interactions.60–63 The presence of crowders also increased the number of mismatches between neighboring CATCH(6K+/6E−) peptides in a β-sheet; by mismatch here we mean that nearest neighbor CATCH peptides are of the same charge. Previous simulations on CATCH systems, including CATCH(6K+/6E−) have predicted instances of mismatched peptides and have been confirmed experimentally via ssNMR measurements.43 Given the previous experimental verification of mismatched peptide pairs in the CATCH system and the previous work demonstrating the impact of macromolecular crowding on protein structure,35,64–70 we consider “flipped” peptide pairs to be conceivable under crowded conditions; in our opinion this observation provides relevant insight into the impact of crowding on secondary structure formation.
Flipped peptide pairs facilitated the formation of multilayer fibrils containing greater than two β-sheet layers (Fig. 6). This misalignment between peptides stems from the association between two organized structures (e.g. two cylindrin-like structures, two β-sheet structures, or a cylindrin-like structure and a β-sheet). A β-strand from each structure interacts with another β-strand through backbone hydrogen bonds. In this instance, if the hydrophobic sidechains on the two interacting β-strands are facing opposite directions, then the peptide pair is considered “flipped”. Otherwise, if the hydrophobic sidechains are facing the same direction, then it is considered to be a typical growth event.17 That says that not every association between two organized structures leads to “flipped” peptides. We surmise that in the presence of crowders, it is difficult for CATCH β-strands that are flipped to reorient their backbones to make the hydrophobic sidechains are face in the same direction. When the association is between a cylindrin-like structure and a bilayer, the cylindrin may unravel into a β-sheet structure. If the interacting β-strands are flipped, this leads to the formation of a β-sheet bilayer with a hydrophobic core, but with a segment of hydrophobic residues facing the implicit solvent on the end of a β-sheet. The same resulting structure forms when a β-sheet structure associates with a bilayer and the interacting β-strands are flipped. In either case, CATCH(6K+/6E−) peptides can then add on to the exposed hydrophobic face forming a third layer that elongates further. Fig. 8 illustrates the formation of a flipped peptide pair starting from two separate assemblies. Overall, we observe that the fibrils formed in the presence of hydrophobic crowders tend to be more contorted than ones formed in the absence of crowders. This is expected as CATCH(6K+/6E−) fibrils must maneuver around the crowders themselves and new peptide–peptide interactions must form around already-existing crowder–peptides interactions.
The influence of crowders on the secondary structure of polypeptides has been explored experimentally. Yeung and coworkers showed that the crowded environment of a reversed micelle led to the formation of antiparallel β-sheet Aβ structures, as opposed to the expected parallel arrangement.70 This antiparallel orientation of Aβ was also observed by Gladytz and coworkers in molecular dynamics simulations employing gold nanoparticles as crowders.35 In mixtures of an aspherical protein and Ficoll 70, Homouz et al. observed via circular dichroism increased helical content with increased crowding.65 Similarly, simulations by Parray et al. found that ethylene glycol crowders resulted in a small increase in α-helical content in cytochrome c proteins.69 The atypical secondary structures observed in our simulations and in work by others are likely due to the restrictive void spaces produced by the presence of crowders.
Effect of crowding on β-sheet structures
CATCH(6K+/6E−) peptides formed monolayer β-sheets in the presence of 2ε and 5ε hydrophobic crowders with diameters of 20, 40, and 80 Å structures at Cpeptide = 5 and 20 mM. Fig. 9 shows the impact of increasing the crowder–sidechain interaction strength on CATCH(6K+/6E−) co-assembly for 80 Å crowders at Cpeptide = 20 mM. When the crowder–sidechain interaction is greater than 1ε, it is more energetically favorable for the phenylalanine residues on CATCH(6K+/6E−) to interact with the crowder than with the phenylalanine residues on another CATCH peptide. The hydrophobic crowders continually recruited the CATCH peptides to their surfaces; there the peptides assembled into β-sheet structures or added onto existing β-sheet structures. The strong crowder–sidechain interaction prevented β-sheets from delaminating and assembling into multilayer fibril structures and led to a system of dispersed β-sheets. The crowder-CATCH(6K+/6E−) β-sheet complex maximizes the number of hydrophobic interactions as it places CATCH(6K+/6E−) phenylalanine sidechains in contact with the hydrophobic crowder and other phenylalanine sidechains. Increasing the peptide–crowder interaction strength from 2ε to 5ε led to similar monolayer β-sheet structures. Our results align with previous simulations performed by Latshaw that showed that increasing the crowder–peptide interaction strength for the hydrophobic crowders resulted in formation of only β-sheets in systems of Aβ(16–22).50 Radic and coworkers observed a similar trend in DMD simulations of coarse-grained Aβ peptides and nanoparticles with explicit surface atoms.36 They found that strong NP–peptide attractions led all of the Aβ peptides to be bound to the NP surface and reduced peptide mobility. Additionally, proteins have been shown to form a monolayer corona around nanoparticles, a potential method for ensuring monodispersity in nanoparticle-based medicines.71The formation of dispersed crowder-β-sheet complexes in the presence of large hydrophobic crowders is likely a consequence of the compartments formed by the crowders and is chaperoned by the crowder–sidechain interactions. Hydrophobic crowders have both attractive and repulsive interactions with CATCH(6K+/6E−) sidechains. Scaling up the crowder–sidechain interaction, ε, strengthened both types of interactions. The duality of attractive and repulsive crowder–sidechain interactions incentivized hydrophobic interactions between CATCH(6K+/6E−) phenylalanine residues and hydrophobic crowders, leading to crowder-β-sheet complexes with a hydrophobic center and charged CATCH(6K+/6E−) residues facing outwards. When a repulsive interaction occurs between a crowder and a charged residue, the crowder appears to have a larger volume (in comparison to a hard-sphere crowder) as the charged residue cannot easily enter the region defined by the square-shoulder potential. The augmented excluded volume generated by the repulsive interactions between the hydrophobic crowders and the charged residues further stabilized the crowder-β-sheet complexes. Simulations results are supported by experimental work from Munishkina et al., who found that fibrillation of oligomeric species was slowed down or inhibited by polymeric crowders.72 Similarly, Mittal and Singh observed a decrease in bovine carbonic anhydrase aggregation with the addition of both Ficoll 70 and Dextran 70.73 They suggest that the reduction in aggregation may be due to the reduced mobility of aggregation-prone intermediates and the decrease in the population of aggregation-prone partially folded species (a consequence of increased native state stability). The latter suggestion is in agreement with work by Benton et al., who showed that the addition of Ficoll 70 increased the enthalpy of denaturation and stabilized monomer intermediates.74 Additionally, theoretical work by Kim and Mittal demonstrated that repulsive crowder–protein interactions reduced the change in binding free energy between protein structures, stabilizing protein association.59 Our simulations and previous work suggest that excluded volume effects produced by crowders can stabilize oligomeric species and hinder fibrillation.
The CATCH(6K+/6E−) peptides assembled the fastest in the presence of 80 Å extremely-hydrophobic (5ε) crowders. This is likely because the flatter surface of the 80 Å crowders “straightens out” the CATCH(6K+/6E−) peptides. In a simulation study of hydrophobic crowders, Latshaw calculated the minimum diameter, 24.4 Å, of a sphere that would allow for a 3-peptide β-sheet to form along the surface.44 As the crowder diameter increased, the angle between consecutive peptides in a β-sheet laying on the crowder surface decreased, facilitating β-sheet nucleation on a crowder surface. Latshaw also determined that crowders with diameters below 24.4 Å have too small a curvature and hinder hydrogen bond formation. This is best exemplified in our simulation containing CATCH(6K+/6E−) peptide and extremely-hydrophobic 10 Å crowders; in this system the small 5ε crowders inhibited the formation of any organized structure, as discussed earlier.
CATCH(6K+/6E−) β-sheet structures spanned the surfaces of many 20 and 40 Å crowders—allowing an ordered assembly to interact with multiple crowders at a time. CATCH(6K+/6E−) peptides formed into β-sheet structures longer than the length of the simulation box in the presence of 1ε hydrophobic crowders of 20 Å diameter (Fig. 10). In contrast, in the presence of 80 Å hydrophobic crowders, (Fig. 9), CATCH(6K+/6E−) cannot easily span across multiple crowders, likely due to the large gaps created in between the crowders. In the latter case, the CATCH(6K+/6E−) formed individual β-sheet structures that wrapped around the large crowders. We surmise that ∼20 Å is the optimal crowder diameter for facilitating the formation of long β-sheet structures. To summarize this section, CATCH(6K+/6E−) peptides transitioned from forming thick fibrils to monolayer β-sheet structures as the size and the hydrophobicity of the crowders increased.
Methods
Peptide model
The simulation method used in this work is discontinuous molecular dynamics, a fast alternative to conventional molecular dynamics (DMD).75 DMD is an event-driven approach that approximates a continuous intermolecular potential with step and shoulder potentials. The PRIME20 force field was used for the peptide model. PRIME20 was developed by the Hall group to study the formation of ordered structures in peptides.76 In the PRIME20 force field, each amino acid is represented using a four-bead-per-residue model: three for the backbone (NH, Cα, and CO) and one for the sidechain group (R). Parameters for all 20 essential amino acids are available in the force field; each amino acid has unique geometric and energetic parameters. The combination of DMD and the PRIME20 force field allows molecular simulation of systems at long timescales that are not accessible using all-atom simulations. DMD/PRIME20 has been used to study multiple amyloidogenic peptides: Aβ, tau, and prion protein peptides.77–79 Greater detail on the force field can be found in earlier work.76,80Crowder model
Crowder diameters of 10, 20, 40, and 80 Å were chosen as they roughly correspond to the hydrodynamic radius of PEG 300, 1000, 4000, and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively.81,82 The mass of each crowder was based on a PEG density of 1.125 and calculated from the volume of the crowder. The crowder volume fraction used was φcrowder = 0.20. The crowder volume fraction is defined as φcrowder = NVCR/V, where N is the number of crowders, VCR is the volume occupied by a single crowder, and V is the volume of the simulation box.
000, respectively.81,82 The mass of each crowder was based on a PEG density of 1.125 and calculated from the volume of the crowder. The crowder volume fraction used was φcrowder = 0.20. The crowder volume fraction is defined as φcrowder = NVCR/V, where N is the number of crowders, VCR is the volume occupied by a single crowder, and V is the volume of the simulation box.
Crowder–sidechain interactions are modeled as either hard-sphere interactions or hydrophobic (square-well and square-shoulder) interactions. For simplicity, crowder–crowder interactions are modeled only as hard-sphere interactions. Multiple sets of crowder–sidechain interaction strengths were explored. The variable ε describes the magnitude of the interaction strength between a crowder and the peptide sidechain groups. The crowder–sidechain interaction strength, ε, is based on the set of interaction energies between a phenylalanine sidechain and the 20 amino acid sidechain groups in the PRIME20 force field. When the crowder–sidechain interaction strength is 1ε, the interaction energies between a crowder and the amino acid sidechain groups are equivalent to the interaction energies between a phenylalanine sidechain and the amino acid sidechain groups. When the crowder–sidechain interaction strength is 2ε, the interaction energies between a crowder and the amino acid sidechain groups are twice the interaction energies between a phenylalanine sidechain and the amino acid sidechain groups, and so forth. The interaction energies for the phenylalanine sidechain group were chosen to model the crowder–sidechain interactions because it is a hydrophobic residue. In addition, phenylalanine is the only hydrophobic residue present in the CATCH(6K+/6E−) system and plays a major role in driving co-assembly. We investigate how the crowder size and crowder–sidechain interaction strength affect competition between the crowders and CATCH(6K+/6E−) peptides for hydrophobic interactions when the crowder–sidechain interaction is weaker than, equivalent to, and stronger than sidechain–sidechain interactions. The crowder–sidechain interaction strengths that were explored are: 0, ½, 1, 2, and 5ε; these are referred to as hard-sphere, weakly-, moderately-, strongly- and extremely-hydrophobic interactions. A summary of the interaction energies used in this work is given in Table 1, where positive values denote a repulsive square-shoulder interaction, and negative values denote an attractive square-well interaction. The interaction energies in Table 1 are scaled based on the hydrogen bonding energy, εHB = 12.00 kJ mol−1. To relate the crowder–sidechain interaction energy to real units, simply multiple the value by εHB. The interaction range is defined as, λ = (wsquare-well − σ)/2, where wsquare-well is the width of the square-well and σ is the average diameter for the two interacting coarse-grained sites. The interaction range, λ, is kept constant regardless of interaction strength.
Simulation procedure
Initial configurations for each simulation were generated using PACKMOL.83 Peptides and crowders were randomly placed in the simulation box. The reduced temperature is defined as T* = KT/εHB, where εHB is the hydrogen bonding well depth. The reduced temperature can be related to real temperature in Kelvin by the following approximation: T[K] = 2288.467T* − 115.79.84 The concentration is defined as Cpeptide = Npeptide/(NAV), where Npeptide is the number of peptides, NA is Avogadro's number, and V is the volume of the simulation box. The initial system temperature for each simulation was set to T* = 0.50 to transform the peptides from extended to random coil structures. The temperature was then gradually reduced to T* = 0.20. At Cpeptide = 20 mM, CATCH(6K+/6E−) does not form fibrils above T* = 0.20. In addition to Cpeptide = 20 mM, systems of Cpeptide = 5 mM were also considered. At Cpeptide = 5 mM and T* = 0.20, CATCH(6K+/6E−) does not typically assemble. Investigating peptide concentrations of both 5 and 20 mM reveals the effect of crowders on nonideal and ideal co-assembly conditions, respectively. Simulation snapshots were rendered using Visual Molecular Dynamics.85,86Peptide classification
To determine whether a peptide was a part of an oligomer, β-sheet, or fibril, we constructed graphs containing nodes (peptides) and edges (interactions) to model pairwise interactions between peptides. For each simulation snapshot, two graphs were constructed using the NetworkX python package:87 (1) based only on hydrogen bonding interactions and (2) based on hydrogen bonding interactions and sidechain–sidechain interactions. When considering only hydrogen bonding interactions, an edge is drawn between two nodes (peptides) when there are at least four hydrogen bonding interactions between the two peptides. β-Sheets are defined as the resulting connected components of the hydrogen bonding graph. When considering both hydrogen bonding interactions and sidechain–sidechain interactions, an edge is drawn between two nodes when there are either at least four hydrogen bonding interactions or at least four sidechain–sidechain interactions. Considering both types of interactions allows us to identify formations that contain more than one single β-sheet layer. From the hydrogen bonding/sidechain–sidechain interaction graph, an oligomer is defined as a connected component containing eight or less peptides or containing only one β-sheet layer. A fibril is defined as a connected component containing more than eight peptides and at least two β-sheet layers. Peptides that do not fall into the oligomer or the fibril classification are classified as a free peptide. Data from peptide classification were fitted to a sigmoid curve to reduce noise and increase readability. The R2 value from curve fitting was 0.85.Conclusion
In this work we used DMD/PRIME20 simulations to investigate the effect of hard-sphere and hydrophobic crowders on CATCH(6K+/6E−) co-assembly. We focused on how the different contributions of attractive, repulsive, and hard-sphere interactions affect co-assembly kinetics and the morphology of the co-assembled aggregates. In addition, we explored how these contributions change with crowder size in systems of low (Cpeptide = 5 mM) and high (Cpeptide = 20 mM) peptide concentrations. Hard-sphere and weakly-hydrophobic crowders were more effective in promoting co-assembly as the crowder diameter decreased. The small hard-sphere (0ε) crowders were the most effective for triggering co-assembly for CATCH systems at low concentrations where fibrillation typically does not occur, and for accelerating the rate of co-assembly for CATCH systems at high concentrations where fibrillation is expected. In comparison, hydrophobic crowders with an interaction strength of 1ε or greater were more effective in promoting co-assembly as the crowder diameter increased. Small extremely-hydrophobic (5ε) crowders inhibited co-assembly and the formation of organized structures. Future experimental validation for this computational study may involve engineering nanoparticles in a range of sizes with various coatings to examine different crowder–peptide interaction strengths. Mixtures of nanoparticles and CATCH peptides may be assessed for assembly and fibrillation using biophysical techniques such as Thioflavin T assays, circular dichroism, and transmission electron microscopy. In systems that form a gel, mechanical properties may be estimated with oscillatory rheology.The results from DMD/PRIME20 simulations provide new insights for directing peptide co-assembly to produce specific nanostructures. Thicker fibril structures were produced in the presence of small hard-sphere, weakly-hydrophobic, and moderately-hydrophobic crowders. In these cases, the addition of small crowders led to instances of mismatched and “flipped” neighboring β-strands in a β-sheet. The “flipped” nearest neighbors facilitated the formation of thicker fibril structures with multiple β-sheet layers. Formation of monolayer β-sheet structures was accelerated by large highly-hydrophobic crowders. In contrast the small highly-hydrophobic crowders constrained the CATCH(6K+/6E−) peptides to a random coil conformation. Overall, we showed that crowders can be used to modulate the morphology of co-assembled structures by influencing peptide–peptide interactions and that simulations can be used to efficiently investigate a range of crowder sizes and crowder–sidechain interaction strengths.
Macromolecular crowding has broad impacts in medicine and biotechnology. It has been well-established that living cells are a crowded environment. Broadening our understanding of macromolecular crowding provides insight into amyloidogenic diseases as changes in crowding may contribute to accelerated fibrillation of amyloidogenic peptides.49 In addition, crowding can also stabilize smaller oligomeric amyloid species that may be more toxic than their fibrillar counterparts.88 We foresee that the connection between macromolecular crowding and amyloidosis will likely become of interest with the rise of micro- and nano plastics contamination in our drinking water and food supply.89,90 In pharmaceuticals, polyethylene glycol (PEG) crowders are commonly used in drug formulation as a solubilizer, stabilizer, or as a carrier. Although PEG has been commonly considered an “inert” molecule, studies have now shown that interactions exist between PEG and proteins.91–95 In biomaterials, crowders have been used to modulate peptide fibril architecture and in turn, hydrogel properties. The findings from research on macromolecular crowding in different disciplines—whether it be for drug formulation, developing biomaterials, or developing therapeutics against amyloidogenic diseases—can be used to inform one another. Understanding macromolecular crowding effects on peptide aggregation is important for both biotechnology and human health.
There are a great number of crowder candidates and biological systems to consider. The effect of macromolecular crowding on a peptide's propensity to aggregate and a peptide's assembled morphology depends on the crowder's size, shape, and interactions.96–98 Studying crowded biological systems becomes even more complex with polymeric crowders (e.g. PEG) as they can crosslink to form networks and meshes.99 In addition, the conclusions drawn from the behavior of one peptide system cannot necessarily be generalized to another peptide system, as the crowder–sidechain interactions and the bottlenecks for assembly may change from system to system.100 For example, a study by Breydo et al. showed that PEG accelerated fibril formation in α-synuclein and insulin, but UCON (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 copolymer of ethylene glycol and propylene glycol) directed aggregation of both proteins towards oligomeric structures, inhibiting fibril formation.101 Ma et al. showed Ficoll 70 accelerates fibril formation in human prion protein (PrP) and tau segments, but the same crowder inhibits fibril formation in rabbit PrP and hen lysozyme.102 Screening through libraries of peptide sequences to determine their behavior in crowded environments can be expensive and time consuming. However, given prior knowledge of the crowder–peptide interactions, simulations can be used to help predict the type of structures that may or may not form in new crowded systems. In the absence of prior knowledge, simulations can be used as a first pass to elucidate the interactions between crowders and biomolecules. The number of potential crowding agents is sizable, but one that can be tackled through a systemic exploration of conditions via experimental and computational means. We hope that our findings may stimulate future experimental investigations on the use of crowders (e.g. engineered nanoparticles) for governing peptide co-assembly.
1 copolymer of ethylene glycol and propylene glycol) directed aggregation of both proteins towards oligomeric structures, inhibiting fibril formation.101 Ma et al. showed Ficoll 70 accelerates fibril formation in human prion protein (PrP) and tau segments, but the same crowder inhibits fibril formation in rabbit PrP and hen lysozyme.102 Screening through libraries of peptide sequences to determine their behavior in crowded environments can be expensive and time consuming. However, given prior knowledge of the crowder–peptide interactions, simulations can be used to help predict the type of structures that may or may not form in new crowded systems. In the absence of prior knowledge, simulations can be used as a first pass to elucidate the interactions between crowders and biomolecules. The number of potential crowding agents is sizable, but one that can be tackled through a systemic exploration of conditions via experimental and computational means. We hope that our findings may stimulate future experimental investigations on the use of crowders (e.g. engineered nanoparticles) for governing peptide co-assembly.
Author contributions
Conceptualization: Xin Y. Dong, Madisen Domayer, Gregory A. Hudalla, Carol K. Hall; data curation: Xin Y. Dong; formal analysis: Xin Y. Dong; funding acquisition: Gregory A. Hudalla, Carol K. Hall; investigation: Xin Y. Dong, Gregory A. Hudalla, Carol K. Hall; methodology: Xin Y. Dong; project administration: Gregory A. Hudalla, Carol K. Hall; resources: Gregory A. Hudalla, Carol K. Hall; software: Xin Y. Dong; supervision: Gregory A. Hudalla, Carol K. Hall; validation: Xin Y. Dong; visualization: Xin Y. Dong; writing – original draft: Xin Y. Dong; writing – review & editing: Xin Y. Dong, Gregory A. Hudalla, Carol K. Hall.Data availability
DMD/PRIME20 code is publicly available at https://github.com/CarolHall-NCSU-CBE/Serial-DMD-PRIME20. Data generated from DMD simulations and the analysis scripts used are available at https://doi.org/10.5061/dryad.6wwpzgn70.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation by grant number CBET 1743432 to CKH and GAH.References
- T. P. J. Knowles and R. Mezzenga, Adv. Mater., 2016, 28, 6546–6561 CrossRef CAS PubMed
.
- R. Binaymotlagh, L. Chronopoulou, F. H. Haghighi, I. Fratoddi and C. Palocci, Materials, 2022, 15, 5871 CrossRef CAS PubMed
.
- W. Y. Seow and C. A. E. Hauser, Mater. Today, 2014, 17, 381–388 CrossRef CAS
.
- S. Koutsopoulos, J. Biomed. Mater. Res., Part A, 2016, 104, 1002–1016 CrossRef CAS PubMed
.
- N. Bakhtiary, B. Ghalandari, F. Ghorbani, S. N. Varma and C. Liu, Polymers, 2023, 15, 1068 CrossRef CAS PubMed
.
- R. Liu and G. A. Hudalla, Molecules, 2019, 24, 1450 CrossRef CAS PubMed
.
- A. L. Sieminski, A. S. Was, G. Kim, H. Gong and R. D. Kamm, Cell Biochem. Biophys., 2007, 49, 73–83 CrossRef CAS PubMed
.
- M. Yu, L. Xu, F. Tian, Q. Su, N. Zheng, Y. Yang, J. Wang, A. Wang, C. Zhu, S. Guo, X. Zhang, Y. Gan, X. Shi and H. Gao, Nat. Commun., 2018, 9, 2607 CrossRef PubMed
.
- J. Di, X. Gao, Y. Du, H. Zhang, J. Gao and A. Zheng, Asian J. Pharm. Sci., 2021, 16, 444–458 Search PubMed
.
- A. Mishra, Y. Loo, R. Deng, Y. J. Chuah, H. T. Hee, J. Y. Ying and C. A. E. Hauser, Nano Today, 2011, 6, 232–239 CrossRef CAS
.
- E. F. Banwell, E. S. Abelardo, D. J. Adams, M. A. Birchall, A. Corrigan, A. M. Donald, M. Kirkland, L. C. Serpell, M. F. Butler and D. N. Woolfson, Nat. Mater., 2009, 8, 596–600 CrossRef CAS PubMed
.
- M. R. Ghadiri, J. R. Granja, R. A. Milligan, D. E. McRee and N. Khazanovich, Nature, 1993, 366, 324–327 CrossRef CAS PubMed
.
- G. Yoon, M. Lee, J. I. Kim, S. Na and K. Eom, PLoS One, 2014, 9, e88502 CrossRef PubMed
.
- J. K. Leman, P. Szczerbiak, P. D. Renfrew, V. Gligorijevic, D. Berenberg, T. Vatanen, B. C. Taylor, C. Chandler, S. Janssen, A. Pataki, N. Carriero, I. Fisk, R. J. Xavier, R. Knight, R. Bonneau and T. Kosciolek, Nat. Commun., 2023, 14, 2351 CrossRef PubMed
.
- D. T. Seroski, X. Dong, K. M. Wong, R. Liu, Q. Shao, A. K. Paravastu, C. K. Hall and G. A. Hudalla, Commun. Chem., 2020, 3, 1–11 CrossRef PubMed
.
- R. Liu, X. Dong, D. T. Seroski, B. S. Morales, K. M. Wong, A. S. Robang, L. Melgar, T. E. Angelini, A. K. Paravastu, C. K. Hall and G. Hudalla, Angew. Chem., Int. Ed., 2023, 62, e202314531 CrossRef CAS PubMed
.
- X. Y. Dong, R. Liu, D. T. Seroski, G. A. Hudalla and C. K. Hall, PLoS Comput. Biol., 2023, 19, e1011685 CrossRef CAS PubMed
.
- A. Scelsi, B. Bochicchio, A. Smith, V. L. Workman, L. A. Castillo Diaz, A. Saiani and A. Pepe, J. Biomed. Mater. Res., Part A, 2019, 107, 535–544 CrossRef CAS PubMed
.
- J. Gao, C. Tang, M. A. Elsawy, A. M. Smith, A. F. Miller and A. Saiani, Biomacromolecules, 2017, 18, 826–834 CrossRef CAS PubMed
.
- S. A. Semerdzhiev, S. Lindhoud, A. Stefanovic, V. Subramaniam, P. van der Schoot and M. M. A. E. Claessens, Phys. Rev. Lett., 2018, 120, 208102 CrossRef CAS PubMed
.
- T. Yucel, C. M. Micklitsch, J. P. Schneider and D. J. Pochan, Macromolecules, 2008, 41, 5763–5772 CrossRef CAS PubMed
.
- Y. Zhao, H. Yokoi, M. Tanaka, T. Kinoshita and T. Tan, Biomacromolecules, 2008, 9, 1511–1518 CrossRef CAS PubMed
.
- T. P. Knowles, A. W. Fitzpatrick, S. Meehan, H. R. Mott, M. Vendruscolo, C. M. Dobson and M. E. Welland, Science, 2007, 318, 1900–1903 CrossRef CAS PubMed
.
- S. Boothroyd, A. F. Miller and A. Saiani, Faraday Discuss., 2014, 166, 195–207 RSC
.
- B. H. Pogostin, K. Godbe, M. Dubackic, I. Angstman, W. Fox, N. Giovino, M. Lagator, A. Payson, M. LaBarca, B. Frohm, K. Bernfur, S. Linse, C. H. Londergan, U. Olsson, L. Gentile and K. S. Åkerfeldt, ACS Nano, 2024, 18, 31109–31122 CrossRef CAS PubMed
.
- A. P. Minton, Curr. Biol., 2006, 16, R269–R271 CrossRef CAS PubMed
.
- A. P. Minton, Biophys. J., 1980, 32, 77–79 CrossRef CAS PubMed
.
- G. Rivas and A. P. Minton, Trends Biochem. Sci., 2016, 41, 970–981 CrossRef CAS PubMed
.
- H.-X. Zhou, G. Rivas and A. P. Minton, Annu. Rev. Biophys., 2008, 37, 375–397 CrossRef CAS PubMed
.
- S. L. Speer, C. J. Stewart, L. Sapir, D. Harries and G. J. Pielak, Annu. Rev. Biophys., 2022, 51, 267–300 CrossRef CAS PubMed
.
- C. Alfano, Y. Fichou, K. Huber, M. Weiss, E. Spruijt, S. Ebbinghaus, G. De Luca, M. A. Morando, V. Vetri, P. A. Temussi and A. Pastore, Chem. Rev., 2024, 124(6), 3186–3219 CrossRef CAS PubMed
.
- S. B. Zimmerman and S. O. Trach, J. Mol. Biol., 1991, 222, 599–620 CrossRef CAS PubMed
.
- J. Dong, J. E. Shokes, R. A. Scott and D. G. Lynn, J. Am. Chem. Soc., 2006, 128, 3540–3542 CrossRef CAS PubMed
.
- G. Gao, M. Zhang, D. Gong, R. Chen, X. Hu and T. Sun, Nanoscale, 2017, 9, 4107–4113 RSC
.
- A. Gladytz, B. Abel and H. J. Risselada, Angew. Chem., Int. Ed., 2016, 55, 11242–11246 CrossRef CAS PubMed
.
- S. Radic, T. P. Davis, P. C. Ke and F. Ding, RSC Adv., 2015, 5, 105489–105498 RSC
.
- S. K. Ranamukhaarachchi, R. N. Modi, A. Han, D. O. Velez, A. Kumar, A. J. Engler and S. I. Fraley, Biomater. Sci., 2019, 7, 618–633 RSC
.
- J.-Y. Dewavrin, N. Hamzavi, V. P. W. Shim and M. Raghunath, Acta Biomater., 2014, 10, 4351–4359 CrossRef CAS PubMed
.
- S. Hirota, Y. Hattori, S. Nagao, M. Taketa, H. Komori, H. Kamikubo, Z. Wang, I. Takahashi, S. Negi, Y. Sugiura, M. Kataoka and Y. Higuchi, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 12854–12859 CrossRef CAS PubMed
.
- A. Restuccia, D. T. Seroski, K. L. Kelley, C. S. O'Bryan, J. J. Kurian, K. R. Knox, S. A. Farhadi, T. E. Angelini and G. A. Hudalla, Commun. Chem., 2019, 2, 1–12 CrossRef CAS
.
- D. T. Seroski, A. Restuccia, A. D. Sorrentino, K. R. Knox, S. J. Hagen and G. A. Hudalla, Cell. Mol. Bioeng., 2016, 9, 335–350 CrossRef CAS
.
- K. M. Wong, A. S. Robang, A. H. Lint, Y. Wang, X. Dong, X. Xiao, D. T. Seroski, R. Liu, Q. Shao, G. A. Hudalla, C. K. Hall and A. K. Paravastu, J. Phys. Chem. B, 2021, 125, 13599–13609 CrossRef CAS PubMed
.
- Q. Shao, K. M. Wong, D. T. Seroski, Y. Wang, R. Liu, A. K. Paravastu, G. A. Hudalla and C. K. Hall, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 4710–4717 CrossRef CAS PubMed
.
- D. C. Latshaw, M. Cheon and C. K. Hall, J. Phys. Chem. B, 2014, 118, 13513–13526 CrossRef CAS PubMed
.
- D. Marenduzzo, K. Finan and P. R. Cook, J. Cell Biol., 2006, 175, 681–686 CrossRef CAS PubMed
.
- S. Asakura and F. Oosawa, J. Polym. Sci., 1958, 33, 183–192 CrossRef CAS
.
- D. M. Hatters, A. P. Minton and G. J. Howlett, J. Biol. Chem., 2002, 277, 7824–7830 CrossRef CAS PubMed
.
- T. E. R. Werner, I. Horvath and P. Wittung-Stafshede, QRB Discov., 2021, 2, e2 CrossRef PubMed
.
- C. F. Lee, S. Bird, M. Shaw, L. Jean and D. J. Vaux, J. Biol. Chem., 2012, 287, 38006–38019 CrossRef CAS PubMed
.
- D. C. Latshaw and C. K. Hall, Biophys. J., 2015, 109, 124–134 CrossRef CAS PubMed
.
- C. Cabaleiro-Lago, F. Quinlan-Pluck, I. Lynch, S. Lindman, A. M. Minogue, E. Thulin, D. M. Walsh, K. A. Dawson and S. Linse, J. Am. Chem. Soc., 2008, 130, 15437–15443 CrossRef CAS PubMed
.
- C. Cabaleiro-Lago, O. Szczepankiewicz and S. Linse, Langmuir, 2012, 28, 1852–1857 CrossRef CAS PubMed
.
- S. K. Sonawane, A. Ahmad and S. Chinnathambi, ACS Omega, 2019, 4, 12833–12840 CrossRef CAS PubMed
.
- P. Ghosh and P. De, ACS Appl. Bio Mater., 2020, 3, 6598–6625 CrossRef CAS PubMed
.
- K. Debnath, A. K. Sarkar, N. R. Jana and N. R. Jana, Acc. Mater. Res., 2022, 3, 54–66 CrossRef CAS
.
- J. F. Douglas, J. Dudowicz and K. F. Freed, Phys. Rev. Lett., 2009, 103, 135701 CrossRef PubMed
.
- J. Rosen, Y. C. Kim and J. Mittal, J. Phys. Chem. B, 2011, 115, 2683–2689 CrossRef CAS PubMed
.
- M. Jiao, H.-T. Li, J. Chen, A. P. Minton and Y. Liang, Biophys. J., 2010, 99, 914–923 CrossRef CAS PubMed
.
- Y. C. Kim and J. Mittal, Phys. Rev. Lett., 2013, 110, 208102 CrossRef PubMed
.
- P. Mark and L. Nilsson, J. Comput. Chem., 2002, 23, 1211–1219 CrossRef CAS PubMed
.
- H. Schreiber and O. Steinhauser, Chem. Phys., 1992, 168, 75–89 CrossRef CAS
.
- R. J. Loncharich and B. R. Brooks, Proteins:Struct., Funct., Bioinf., 1989, 6, 32–45 CrossRef CAS PubMed
.
- S. Izadi, R. Anandakrishnan and A. V. Onufriev, J. Chem. Theory Comput., 2016, 12, 5946–5959 CrossRef CAS PubMed
.
- A. P. Minton, Curr. Opin. Struct. Biol., 2000, 10, 34–39 CrossRef CAS PubMed
.
- D. Homouz, L. Stagg, P. Wittung-Stafshede and M. S. Cheung, Biophys. J., 2009, 96, 671–680 CrossRef CAS PubMed
.
- I. Horvath, R. Kumar and P. Wittung-Stafshede, Biophys. J., 2021, 120, 3374–3381 CrossRef CAS PubMed
.
- A. Dhar, A. Samiotakis, S. Ebbinghaus, L. Nienhaus, D. Homouz, M. Gruebele and M. S. Cheung, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 17586–17591 CrossRef CAS PubMed
.
- R. J. Ellis, Trends Biochem. Sci., 2001, 26, 597–604 CrossRef CAS PubMed
.
- Z. A. Parray, A. A. T. Naqvi, I. A. Ahanger, M. Shahid, F. Ahmad, M. I. Hassan and A. Islam, Polymers, 2022, 14, 4808 CrossRef CAS PubMed
.
- P. S.-W. Yeung and P. H. Axelsen, J. Am. Chem. Soc., 2012, 134, 6061–6063 CrossRef CAS PubMed
.
- S. Dominguez-Medina, J. Blankenburg, J. Olson, C. F. Landes and S. Link, ACS Sustainable Chem. Eng., 2013, 1, 833–842 CrossRef CAS PubMed
.
- L. A. Munishkina, A. Ahmad, A. L. Fink and V. N. Uversky, Biochemistry, 2008, 47, 8993–9006 CrossRef CAS PubMed
.
- S. Mittal and L. R. Singh, J. Biochem., 2014, 156, 273–282 CrossRef CAS PubMed
.
- L. A. Benton, A. E. Smith, G. B. Young and G. J. Pielak, Biochemistry, 2012, 51, 9773–9775 CrossRef CAS PubMed
.
- B. J. Alder and T. E. Wainwright, J. Chem. Phys., 1959, 31, 459–466 CrossRef CAS
.
- M. Cheon, I. Chang and C. K. Hall, Proteins:Struct., Funct., Bioinf., 2010, 78, 2950–2960 CrossRef CAS PubMed
.
- S. J. Bunce, Y. Wang, K. L. Stewart, A. E. Ashcroft, S. E. Radford, C. K. Hall and A. J. Wilson, Sci. Adv., 2019, 5, eaav8216 CrossRef PubMed
.
- M. Cheon, I. Chang and C. K. Hall, Protein Sci., 2012, 21, 1514–1527 CrossRef CAS PubMed
.
- Y. Wang and C. K. Hall, Protein Sci., 2018, 27, 1304–1313 CrossRef CAS PubMed
.
- A. V. Smith and C. K. Hall, Proteins:Struct., Funct., Bioinf., 2001, 44, 344–360 CrossRef CAS PubMed
.
- X. Dong, A. Al-Jumaily and I. C. Escobar, Membranes, 2018, 8, 23 CrossRef PubMed
.
-
O. V. Krasilnikov, in Structure and Dynamics of Confined Polymers, ed. J. J. Kasianowicz, M. S. Z. Kellermayer and D. W. Deamer, Springer Netherlands, Dordrecht, 2002, pp. 97–115 Search PubMed
.
- L. Martínez, R. Andrade, E. G. Birgin and J. M. Martínez, J. Comput. Chem., 2009, 30, 2157–2164 CrossRef PubMed
.
- Y. Wang, Q. Shao and C. K. Hall, J. Biol. Chem., 2016, 291, 22093–22105 CrossRef CAS PubMed
.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed
.
-
J. Stone, Master's Thesis, Computer Science Department, University of Missouri-Rolla, 1998
.
-
A. Hagberg, P. J. Swart and D. A. Schult, in Exploring network structure, dynamics, and function using NetworkX, Los Alamos National Laboratory (LANL), Los Alamos, NM (United States), 2008 Search PubMed
.
- I. Benilova, E. Karran and B. De Strooper, Nat. Neurosci., 2012, 15, 349–357 CrossRef CAS PubMed
.
- J. Windheim, L. Colombo, N. C. Battajni, L. Russo, A. Cagnotto, L. Diomede, P. Bigini, E. Vismara, F. Fiumara, S. Gabbrielli, A. Gautieri, G. Mazzuoli-Weber, M. Salmona and L. Colnaghi, Int. J. Mol. Sci., 2022, 23, 10329 CrossRef CAS PubMed
.
- K. Ziani, C.-B. Ioniţă-Mîndrican, M. Mititelu, S. M. Neacşu, C. Negrei, E. Moroşan, D. Drăgănescu and O.-T. Preda, Nutrients, 2023, 15, 617 CrossRef CAS PubMed
.
- Z. A. Parray, M. I. Hassan, F. Ahmad and A. Islam, Polym. Test., 2020, 82, 106316 CrossRef CAS
.
- Z. A. Parray, F. Ahmad, M. F. Alajmi, A. Hussain, M. I. Hassan and A. Islam, Sci. Rep., 2021, 11, 6475 CrossRef CAS PubMed
.
- I. A. Shkel, D. B. Knowles and M. T. Record Jr., Biopolymers, 2015, 103, 517–527 CrossRef CAS PubMed
.
- D. B. Knowles, I. A. Shkel, N. M. Phan, M. Sternke, E. Lingeman, X. Cheng, L. Cheng, K. O'Connor and M. T. Record, Biochemistry, 2015, 54, 3528–3542 CrossRef CAS PubMed
.
- J. Wu, C. Zhao, W. Lin, R. Hu, Q. Wang, H. Chen, L. Li, S. Chen and J. Zheng, J. Mater. Chem. B, 2014, 2, 2983–2992 RSC
.
- L. A. Munishkina, E. M. Cooper, V. N. Uversky and A. L. Fink, J. Mol. Recognit., 2004, 17, 456–464 CrossRef CAS PubMed
.
- T. Skóra, F. Vaghefikia, J. Fitter and S. Kondrat, J. Phys. Chem. B, 2020, 124, 7537–7543 CrossRef PubMed
.
- F. C. Zegarra, D. Homouz, A. G. Gasic, L. Babel, M. Kovermann, P. Wittung-Stafshede and M. S. Cheung, J. Phys. Chem. B, 2019, 123, 3607–3617 CrossRef CAS PubMed
.
-
M. Rubinstein and R. H. Colby, Polymer Physics, Oxford University Press, Oxford, New York, 2003 Search PubMed
.
- A. H. Gorensek-Benitez, B. Kirk and J. K. Myers, Biomolecules, 2022, 12, 950 CrossRef CAS PubMed
.
- L. Breydo, A. E. Sales, T. Frege, M. C. Howell, B. Y. Zaslavsky and V. N. Uversky, Biochemistry, 2015, 54, 2957–2966 CrossRef CAS PubMed
.
- Q. Ma, J.-B. Fan, Z. Zhou, B.-R. Zhou, S.-R. Meng, J.-Y. Hu, J. Chen and Y. Liang, PLoS One, 2012, 7, e36288 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2025 |