 Open Access Article
Open Access ArticleComparative study of the therapeutic potential of C24, C32, B12N12, and B16N16 nanocages as drug delivery carriers for delivering an erlotinib derivative: DFT and QTAIM investigations†
Khourshid
Mehdizadeh
 a,
Sourour
PourFalatoon
b,
Milad
Nouraliei
c,
Majid
Farsadrooh
a,
Sourour
PourFalatoon
b,
Milad
Nouraliei
c,
Majid
Farsadrooh
 *d,
Hanseung
Kim
e,
Marzieh
Ramezani Farani
*d,
Hanseung
Kim
e,
Marzieh
Ramezani Farani
 *e and
Yun Suk
Huh
*e and
Yun Suk
Huh
 *e
*e
aDepartment of Chemistry, Roudsar and Amlash Branch, Islamic Azad University, Roudsar, Iran
bDepartment of Chemistry, Science and Research Branch, Islamic Azad University, Tehran, Iran
cDepartment of Chemistry, Islamic Azad University, Central Tehran Branch, Tehran, Iran
dRenewable Energies Research Laboratory, Department of Chemistry, Faculty of Science, University of Sistan and Baluchestan, P.O. Box 98135 674, Zahedan, Iran. E-mail: farsadroohmajid@gmail.com; Tel: +989111369044
eNanoBio High-Tech Materials Research Center, Department of Biological Sciences and Bioengineering, Inha University, Incheon 22212, Republic of Korea. E-mail: farani.marzi@inha.ac.kr; yunsuk.huh@inha.ac.kr; Tel: +82-10-9609-1367 Tel: +82-10-9633-6377
First published on 14th March 2025
Abstract
The use of nanostructures as drug delivery vehicles for a wide range of anticancer medications to lessen their severe side effects by delivering them to the targeted tumor cell location is presently a broadly studied innovative biomedical application of different nanostructures. To investigate the capability of C24 and C32, B12N12, and B16N16 nanocages as nanocarriers for delivering the methyl erlotinib molecule, we conducted density functional theory (DFT) computations using the M06-2X/6-311G(d,p) and M06-2X/6-31G(d) levels of theory. The calculation of the adsorption energy of methyl erlotinib on the nanocages was performed in aqueous and gaseous phases. The adsorption energy values associated with the interaction between the nanocages and methyl erlotinib were negative, indicating that this interaction was exothermic in nature. The adsorption energy values in the aqueous state were higher than those in the gaseous state, suggesting a stronger interaction in the aqueous state, with the exception of the C32 nanocage. Analyses of the density of states (DOS) and projected density of states (PDOS) were performed in order to examine the effect of methyl erlotinib adsorption on the electronic characteristics of selected nanocages. The findings indicated that the B12N12 nanocage following methyl erlotinib molecule adsorption came nearer to the Fermi level than the other nanocages examined. Calculations based on the Quantum Theory of Atoms in Molecules (QTAIM) indicated that methyl erlotinib had a weak interaction with all selected nanocages. According to the values of the adsorption energy derived from both methodologies, the interaction between methyl erlotinib and the B12N12 nanocage was determined to be more robust than the interaction between methyl erlotinib and the C24 nanocage, while the interaction between methyl erlotinib and the B16N16 nanocage was also stronger than that with the C32 nanocage. Notable variations in the ΔEg values were detected for methyl erlotinib@B12N12 and methyl erlotinib@B16N16 across all methods, suggesting that the conductivity of these two nanostructures improved more significantly following the adsorption of methyl erlotinib than that of other nanostructures. Consequently, the B12N12 and B16N16 nanocages can function as nanosensors for methyl erlotinib.
1. Introduction
Cancer is a crucial global health issue marked by the uncontrolled proliferation and spread of abnormal cells, and it remains a primary cause of death worldwide.1 Lung cancer is the most common type of cancer globally and the leading cause of cancer-related deaths. Lung cancer can be categorized into two main types based on its histopathological characteristics: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), with NSCLC accounting for approximately 80% of all clinical lung cancer cases.2 A significant proportion of NSCLC patients harbor mutations in the epidermal growth factor receptor (EGFR), with estimates indicating that around 40%–60% of these individuals are of Asian origin.3 EGFR tyrosine kinase inhibitors, including gefitinib, afatinib, and erlotinib, have shown significant efficacy in treating NSCLC patients with EGFR-activating mutations, which have led to remarkable treatment responses. Erlotinib, a quinazoline derivative chemically known as N-(3-ethynylphenyl)-6,7-bis(2-methoxyethoxy)-4-quinazolinamine, reversibly and selectively inhibits the EGFR tyrosine kinase, thereby blocking downstream signaling pathways involved in angiogenesis, metastasis, and cell proliferation.4–6 Approved by the U.S. FDA in 2004 for the treatment of pancreatic cancer and metastatic NSCLC,5 erlotinib binds reversibly to the adenosine triphosphate (ATP) binding site within the tyrosine kinase domain of the EGFR, inhibiting auto-phosphorylation and promoting apoptosis, suppressing angiogenesis, and preventing excessive cell proliferation.7,8 However, poor water solubility of erlotinib and extensive metabolism, primarily by CYP3A4 enzymes in the intestines and liver, result in reduced and inconsistent bioavailability (59%).9 This necessitates higher doses to achieve effective tumor growth inhibition, which in turn is associated with common side effects such as anemia, Stevens–Johnson syndrome, neutropenia, mucositis, headaches, diarrhea, and skin rash.9–11 Therefore, developing a delivery system that enhances efficacy at reduced doses is crucial for mitigating side effects and preventing or delaying acquired resistance.Nanoscale drug delivery systems have emerged as a promising approach for addressing the challenges associated with chemotherapy. These systems offer several advantages, including reduced toxicity, lower effective doses, and specific targeting of tumor sites. These systems can bypass physiological barriers, evade drug resistance mechanisms, and release drugs precisely at the tumor site, often triggered by specific stimuli. The enhanced permeability and retention (EPR) effect, first identified in 1986, allows drug-loaded carriers to accumulate at tumor sites due to the significantly higher vascular permeability in tumors compared to normal tissues. Nanoscale drug delivery systems offer several advantages, including reduced toxicity to healthy cells, lower effective doses, and the ability to specifically target tumor sites.
Among the various nanostructures explored for drug delivery, fullerenes, carbon nanotubes,12,13 and boron nitride fullerenes have garnered significant attention due to their unique properties. Fullerenes, a distinctive group of carbon allotropes, exhibit attractive photochemical, electrochemical, and physical characteristics, making them promising candidates for nanoscale drug carriers.14,15 Their significant apolar nature facilitates drug absorption, while their low toxicity and biocompatibility enhance metabolic efficacy and make them suitable for medical applications.16 Additionally, non-covalent interactions, such as π–π stacking, between fullerenes and the aromatic rings of drug molecules further enhance their drug delivery potential.17 The small size of fullerenes provides significant benefits, including modified pharmacokinetic properties and enhanced payload capacity, compared to conventional drug delivery systems.18 In summary, fullerenes are utilized in drug delivery due to their ability to (i) enable effective drug incorporation and release, (ii) enhance formulation stability and shelf life, (iii) exhibit biocompatibility, (iv) facilitate biodistribution and targeting, and (v) offer functional versatility.19 Similarly, boron nitride (BN) fullerenes, which share isoelectronic similarities with carbon fullerenes, have gained attention for their chemical stability and unique properties. The ionic bonds between positively charged boron atoms and negatively charged nitrogen atoms contribute to their stability,20 while the sp3 hybridization of nitrogen atoms with boron atoms results in locally pyramidal geometries.21 These structural features enable BN fullerenes to interact with other molecules through physical or chemical binding, making them suitable for drug delivery applications. Recent theoretical studies have demonstrated the responsiveness of BN nanocages to biomolecules, highlighting their potential as biochemical sensors.22
The primary aim of this research is to examine the adsorption behavior of methyl erlotinib molecules onto the C24, C32, B12N12, and B16N16 nanocages as potential drug delivery carriers, as well as the anticancer properties of their complexes, using density functional theory (DFT) and molecular docking simulations. The quantum theory of atoms in molecules (QTAIM) was employed to analyze the interactions between methyl erlotinib and the nanocages. Additionally, to understand the impact of nanocage volume on drug adsorption, C24 and C32 nanocages were selected for comparative analysis.
2. Computational methods
C24, B12N12, C32, and B16N16 fullerenes were selected as nanocarriers to investigate their ability to deliver the methyl erlotinib drug. GAMESS-US software was used to perform DFT calculations utilizing the M06-2X approach with the basis sets of 6-311G(d,p) and 6-31G(d).23–25 The objective was to conduct natural bond orbital (NBO) analysis,26 as well as to obtain the outcomes of the density of states (DOS) and the levels of molecular electrostatic potential.27,28 Furthermore, we sought to identify the characteristics of the interaction between the medication and the nanostructures by employing calculations derived from the QTAIM. DFT calculations were conducted based on the ground state of the compounds. Additionally, the compounds were assigned a charge of 0 and a spin of 1 (singlet). Software tools Gauss View 5.0 and Nanotube Modeler were employed to simulate nanotubes and interact with their structures. Using GaussSum v3.0 software,29 we plotted density of states (DOS) diagrams. The following equation was employed to determine the energy of adsorption (Eads) of methyl erlotinib molecules at the selected nanocages:21,30| Eads = (Ecomplex) − (Enanostructure + Edrug) | (1) |
3. Results and discussion
3.1. Adsorption of methyl erlotinib onto the C24 nanocage
The optimum C24 nanocage geometric shape is composed of eight hexagonal rings and six quadrilateral rings. In the tetragonal rings, the C–C bonds were 1.48 Å long, whereas in the hexagonal rings, the C–C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds were 1.48 Å and 1.37 Å long, respectively. The C24 nanocage studied consisted of 24 carbon atoms. The methyl erlotinib molecule was initially positioned in multiple locations on the outer surface of the C24 nanocage, each with distinct orientations. Fig. 2a depicts the most stable and best position of methyl erlotinib on the C24 nanocage surface. Analysis based on natural bond orbital (NBO) techniques suggested that positive charge transfer (QT) occurs in the C24 fullerene. The findings indicate that charge is transferred from methyl erlotinib to the C24 nanocage. Following the adsorption process, the energy gap (Eg) of the C24 nanocage was diminished. This reduction in the energy gap (Eg) suggests that the conductivity of the C24 nanocage has improved.
C bonds were 1.48 Å and 1.37 Å long, respectively. The C24 nanocage studied consisted of 24 carbon atoms. The methyl erlotinib molecule was initially positioned in multiple locations on the outer surface of the C24 nanocage, each with distinct orientations. Fig. 2a depicts the most stable and best position of methyl erlotinib on the C24 nanocage surface. Analysis based on natural bond orbital (NBO) techniques suggested that positive charge transfer (QT) occurs in the C24 fullerene. The findings indicate that charge is transferred from methyl erlotinib to the C24 nanocage. Following the adsorption process, the energy gap (Eg) of the C24 nanocage was diminished. This reduction in the energy gap (Eg) suggests that the conductivity of the C24 nanocage has improved.
Fig. 1 displays the optimum structures for the methyl erlotinib molecule and the C24, C32, B12N12, and B16N16 nanocages. The methyl erlotinib molecule was initially positioned at multiple locations on the outer surface of the C24 nanocage, exploring different orientations. The most stable and optimal orientation of methyl erlotinib on the surface of C24 is depicted in Fig. 2a. Table 1 presents the calculated values for adsorption energy (Eads), energy gap (Eg), and the variation in the energy gap of the nanostructures before and after the adsorption of methyl erlotinib (ΔEg). The analysis conducted using NBO reveals a positive transferred charge (QT), indicating that charge transfer occurs from methyl erlotinib to the C24 atom. The energy gap value (Eg) in C24 diminished following adsorption, suggesting alterations in its conductivity compared to the state prior to adsorption. The calculated values of Eads for both the gaseous and aqueous states reveal that methyl erlotinib was chemically adsorbed onto the surface of C24. The negative adsorption energy values of methyl erlotinib in both aqueous and gaseous states signify that its adsorption on the C24 nanocage is exothermic. The obtained adsorption energy for methyl erlotinib on C24 indicates that C24 could serve as a suitable nanocarrier for delivering the methyl erlotinib molecule.
 | ||
| Fig. 1 Optimized structures of (a) the methyl erlotinib molecule, (b) C24 nanocage, (c) B12N12 nanocage, (d) C32 nanocage and (e) B16N16 nanocage. | ||
 | ||
| Fig. 2 Optimized structures of (a) the methyl erlotinib@C24 nanocage, (b) methyl erlotinib@B12N12 nanocage, (c) methyl erlotinib@C32 nanocage, and (d) methyl erlotinib@B16N16 nanocage. | ||
| Nanostructures | HOMO (eV) | LUMO (eV) | E ads (eV) in gas | E ads (eV) in water | BSSE (eV) | E g (eV) | ΔEg (eV) | Q T (e) |
|---|---|---|---|---|---|---|---|---|
| C24 nanocage | −7.133 | −2.653 | — | — | — | 4.480 | — | — |
| B12N12 nanocage | −9.278 | +0.160 | — | — | — | 9.439 | — | — |
| C32 nanocage | −6.890 | −3.259 | — | — | — | 3.631 | — | — |
| B16N16 nanocage | −9.313 | −0.209 | — | — | — | 9.104 | — | — |
| Methyl erlotinib@C24 nanocage | −6.823 | −2.669 | −0.084 | −0.1063 | 0.002 | 4.153 | −0.326 | +0.003 |
| Methyl erlotinib@B12N12 nanocage | −7.029 | −0.819 | −1.144 | −1.2269 | 0.008 | 6.210 | −3.229 | −0.003 |
| Methyl erlotinib@C32 nanocage | −6.801 | −3.180 | −0.150 | −0.136 | 0.0018 | 3.620 | −0.010 | +0.006 |
| Methyl erlotinib@B16N16 nanocage | −7.035 | −0.827 | −1.112 | −1.419 | 0.0034 | 6.208 | −2.896 | −0.003 |
3.2. Methyl erlotinib adsorption onto the B12N12 nanocage
The B12N12 nanocage being examined consisted of 12 nitrogen atoms and 12 boron atoms. The optimal structure of the B12N12 nanostructure is composed of eight hexagonal rings and six quadrilateral rings. The B–N bond lengths were found to be 1.480 Å and 1.430 Å in the hexagonal rings and 1.480 Å in the tetragonal rings. The molecule of methyl erlotinib was initially positioned in different locations on the outer surface of the B12N12 nanocage utilizing varying orientations. The optimal and most stable placement of methyl erlotinib on the surface of the B12N12 nanocage is depicted in Fig. 2b. There was a 2.2490 eV increase in the HOMO energy level following interaction with methyl erlotinib. This suggests that methyl erlotinib likely acts as an electron acceptor, while the B12N12 nanocage functions as an electron donor. On the other hand, the nitrogen atoms (atomic numbers 16, 24, 17, and 24) and boron atom (atomic number 10) in the B12N12 nanostructure interact with the oxygen atom (atomic number 35) and the hydrogen atoms (atomic numbers 66, 64, 69, and 59) of methyl erlotinib. In the B12N12 nanocage, electron transfer is much easier than in the C24 nanocage. As a result, the B12N12 nanocage has considerably higher adsorption energy than the C24 nanocage. The charge transfer (QT) obtained from the NBO analysis was found to be negative (Table 1), suggesting that methyl erlotinib acts as an electron acceptor within the methyl erlotinib@B12N12 configuration. The energy gap (Eg) showed a notable reduction following the adsorption process. The evaluation of Eads values in both aqueous and gaseous states indicates that methyl erlotinib has a stronger affinity for B12N12 compared to C24, B16N16, and C32. In both the aqueous and gaseous phases, the adsorption energies of methyl erlotinib were negative, signifying that the binding process of methyl erlotinib to B12N12 is exothermic. The values of the adsorption energy of methyl erlotinib on B12N12 indicate that its binding occurs through chemisorption, resulting in a significant enhancement of B12N12's conductivity. Thus, B12N12 can be used in drug delivery as a recognition system for methyl erlotinib molecules.3.3. Methyl erlotinib adsorption onto the C32 nanocage
There are four different kinds of carbon–atom bonds in the optimal geometric shape of the C32 nanocage. In hexagonal structures, the lengths of C–C bonds were found to be 1.48 Å, 1.38 Å, and 1.45 Å, while in octagonal configurations, the C–C bond lengths were found to be 1.52 Å and 1.38 Å. The C32 nanocage studied consisted of 32 carbon atoms. The methyl erlotinib molecule was initially positioned in multiple locations on the outer surface of the C32 nanocage, employing various orientations. The most stable and optimal binding site of methyl erlotinib on the C32 nanocage is illustrated in Fig. 2c. The analysis using NBO indicated that the charge transfer (QT) in the C32 nanocage was positive. This suggests that there is a transfer of charge from methyl erlotinib to the C32 nanocage. The adsorption process led to a reduction in the energy gap value (Eg) of the C32 nanocage. The reduction in Eg suggests that the conductivity of the C32 nanocage has increased.3.4. Methyl erlotinib adsorption onto the B16N16 nanocage
The B16N16 nanocage being examined consisted of 16 nitrogen atoms and 16 boron atoms. In the optimized geometric structure of the B16N16 nanocage, five distinct kinds of B–N bonds with varying lengths can be identified. The lengths of B–N bonds in the hexagonal structure were found to be 1.46 Å, 1.43 Å, and 1.50 Å, while in the octagonal configurations, the lengths of B–N bonds were found to be 1.48 Å and 1.43 Å. The methyl erlotinib molecule was initially situated in multiple positions on the outer surface of B16N16, with varying orientations. The most stable and optimal binding site of methyl erlotinib on the surface of B16N16 is illustrated in Fig. 2d. There was a 2.2781 eV increase in the HOMO energy level following interaction with methyl erlotinib. This suggests that methyl erlotinib likely acts as an electron acceptor, while the B16N16 nanocage functions as an electron donor. On the other hand, in the B16N16 nanocage, the boron atom (atomic number 5) and the nitrogen atoms (atomic numbers 26, 27, and 31) interact with the oxygen atom (atomic number 43) and the hydrogen atoms (atomic numbers 67, 72, 74, and 77) of methyl erlotinib. In the B16N16 nanocage, electron transfer is much easier than in the C32 nanocage. As a result, the B16N16 nanocage has considerably higher adsorption energy than the C32 nanocage. The charge transfer (QT) obtained from the NBO analysis was found to be negative (Table 1), suggesting that methyl erlotinib acts as an electron acceptor within the methyl erlotinib@B16N16 configuration. The energy gap (Eg) showed a notable reduction following the adsorption process. The evaluation of Eads values in both aqueous and gaseous states indicates that methyl erlotinib has a stronger affinity for B16N16 than C24 and C32. In both the aqueous and gaseous phases, the adsorption energies of methyl erlotinib were negative, signifying that the binding process of methyl erlotinib to B16N16 is exothermic. The values of adsorption energy of methyl erlotinib on B16N16 indicate that its binding occurs through chemisorption, resulting in a significant enhancement of B16N16's conductivity. Based on the analysis, the B16N16 nanocage could serve as an effective recognition platform for targeting and delivering methyl erlotinib, a drug molecule, to the desired locations.3.5. Projected density of states (PDOS) and density of states (DOS)
To investigate how the surface adsorption of the methyl erlotinib molecule affects the electronic properties of nanocages, the DOS was analyzed. Moreover, the primary factor characterizing the interaction between the methyl erlotinib molecule and selected nanocages is the projected density of states (PDOS) spectra, which are associated with the atoms involved in the active sites. The results of PDOS are presented in Fig. S1.† The DOS diagrams of the C24, methyl erlotinib@C24, B12N12, methyl erlotinib@B12N12, C32, methyl erlotinib@C32, B16N16 and methyl erlotinib@B16N16 nanostructures are depicted in Fig. 3(a–h). Following the interaction between the nanocages and methyl erlotinib, the findings indicated that the LUMO and HOMO energy levels of the B12N12 and B16N16 nanocages approached the Fermi energy level (represented by the yellow line) more closely than those of the other nanostructures. Under optimal conditions, the adsorption of methyl erlotinib onto the C24, B12N12, C32, and B16N16 nanocages resulted in ΔEg values of −0.3269 eV, −3.2296 eV, −0.0106 eV, and −2.8966 eV, respectively. The DOS diagrams illustrate that the adsorption of the methyl erlotinib molecules onto the B12N12 nanostructure results in a modification of the energy gap. According to the DOS analysis presented in Table 1, the valence and conduction levels of molecular orbitals in the methyl erlotinib@B12N12 nanocage and the methyl erlotinib@B16N16 nanocage were found to be closer to lower and higher energy levels, respectively. As a result, the energy gap values of the B12N12 and B16N16 nanocages decreased more significantly than those of the C24 and C32 nanocages. Additionally, compared to other nanostructures, the valence level of the B12N12 nanocage in the methyl erlotinib@B12N12 nanocage was closer to the Fermi level. As a result of the interaction, the B12N12 nanocage's conductivity increased more significantly in comparison with the other nanostructures. The following equation can be used to determine the electrical conductivity (σ) of molecular complexes at a specific temperature:12 | (2) |
In this case, electrical conductivity is denoted by σ. Temperature and the Boltzmann constant are denoted by K and T. According to eqn (2), a lower value of Eg at a given hypothetical temperature results in an increased electrical conductivity. The adsorption of methyl erlotinib onto B16N16 and B12N12 results in a significant reduction in Eg. Since conductivity is directly related to the reduction in the Eg value, it can be inferred that a decrease in the Eg value leads to an increase in conductivity. The electrical properties of the B12N12 and B16N16 nanocages become quite pronounced following the adsorption of the methyl erlotinib molecule. Consequently, the B12N12 and B16N16 nanocages serve not only as potential drug delivery systems for methyl erlotinib but may also function as detectors or nanosensors for this compound.
3.6. Analysis of the surfaces of molecular electrostatic potential (MEP)
The distribution of charge was analyzed through MEP analysis to understand how the methyl erlotinib molecule interacts with various nanostructures, including the C24, B12N12, C32, and B16N16 nanocages. The MEP derived from the distribution of molecular charge was characterized using the following equation:21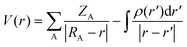 | (3) |
The charge of nucleus A, denoted as ZA, is positioned at RA. The sign of V(r) relies on which factor, the nucleus or electrons, has a predominant effect at a particular point. According to the MEP analysis, atoms with a positive charge appeared in blue, while those with a negative charge appeared in red (see Fig. 4).35 It is anticipated that when methyl erlotinib interacts with the C24 and C32 nanocages, there will be a transfer of charge from methyl erlotinib to both the C24 (see Fig. 4a) and C32 nanocages (see Fig. 4c). On the other hand, when methyl erlotinib interacts with B12N12 and B16N16, charge is transferred from B12N12 (Fig. 4b) and B16N16 (Fig. 4d) to methyl erlotinib. The interaction between methyl erlotinib and the nanostructures will be weak due to the transfer of charge.
3.7. FMO analysis
The energy surfaces of frontier molecular orbitals, namely the HOMO and the LUMO, indicate how effectively a molecule can accept and donate electrons. The energy difference between the surfaces of the HOMO and the LUMO serves as a valuable indicator for assessing a molecule's biological activity, kinetic stability, and chemical reactivity. Fig. 5 depicts the surfaces of the HOMO and the LUMO of methyl erlotinib when interacting with the C24, B12N12, C32, and B16N16 nanocages. The LUMO and HOMO orbitals are mostly focused on the methyl erlotinib medication in the methyl erlotinib@B12N12 and methyl erlotinib@B16N16 nanocages. In contrast, in the methyl erlotinib@C32 nanocage, the LUMO and HOMO orbitals are localized on the C32 nanocage itself. The HOMO orbital of the methyl erlotinib@C24 nanocage is positioned on the methyl erlotinib medication, while the LUMO orbital is found on the C24 nanocage component, implying electron transfer from methyl erlotinib to the C24 nanocage. Compounds possessing a small energy gap typically demonstrate lower stability and high chemical reactivity, while the opposite is true for compounds with a larger energy gap. The calculated Eg values for the methyl erlotinib@C24 nanocage, the methyl erlotinib@B12N12 nanocage, the methyl erlotinib@C32 nanocage and the methyl erlotinib@B16N16 nanocage are found to be 4.153, 6.210, 3.620 and 6.208 eV, respectively. The findings suggest that the methyl erlotinib@C32 nanocage system exhibits superior chemical reactivity in comparison with the other systems. The following is the order of the systems’ stability:methyl erlotinib@B12N12 nanocage > methyl erlotinib@B16N16 nanocage > methyl erlotinib@C24 nanocage > methyl erlotinib@C32 nanocage and the chemical reactivity for the systems is as follows:
methyl erlotinib@C32 nanocage > methyl erlotinib@C24 nanocage > methyl erlotinib@B16N16 nanocage > methyl erlotinib@B12N12 nanocage.
3.8. Analysis of the quantum theory of atoms in molecules (QTAIM)
Electrons are scattered within the gravitational field generated by the nuclei, as explained by the quantum theory of atoms in molecules (QTAIM). The charge density ρ(r) indicates that the nucleus functions as an attractor within a cloud of negative charge. In this theory, the BCP is recognized as a collection of specific points that can be identified. Certain characteristics they possess can be utilized to identify intermolecular interactions. During crucial moments in these interactions, the distribution of electron density plays a significant role in determining the type of bond formed. The maximal charge density is exhibited by the critical point (CP). At the BCP, the value of ρ(r) reflects both the degree and strength of the bond. In this context, VC indicates the potential electron energy density, while KC stands for the kinetic electron energy density, and HC signifies the electron energy density at the BCP. Given that VC is negative and KC is positive, Wirral's theorem indicates that the potential energy at the bond's Cp reduces significantly, leading to a negative value for ∇2ρ(r). Based on eqn (4):| HC = KC + VC | (4) |
The  ratio can be employed to assess the nature of intermolecular bonding. A value greater than 1 signifies the presence of non-covalent bonds, whereas a value ranging from 0.5 to 1 indicates the existence of partial covalent hydrogen bonds. The outcomes presented in Table 2 indicate that all examined nanocages including methyl erlotinib@C24, methyl erlotinib@B12N12, methyl erlotinib@C32, and methyl erlotinib@B16N16 exhibit a ratio higher than 1, implying their noncovalent nature. In this study, it is assumed that all interactions are weak because HC is greater than zero. The intermolecular interactions are described by the vital parameter of the electron density ρ(r). In the examined nanostructures, the electron density (ρ(r)) varies considerably, spanning from 0.00239 to 0.09658 atomic units. This wide range suggests the occurrence of diverse intermolecular forces with varying intensities. As a result, the strongest interaction is related to B10–O35 (ρ(r) = 0.09658 a.u.), whereas the weakest interaction is associated with C72–H45 (ρ(r) = 0.00239 a.u.). Molecular graphs of the optimized complexes, including the methyl erlotinib@C24 nanocage, the methyl erlotinib@B12N12 nanocage, the methyl erlotinib@C32 nanocage and the methyl erlotinib@B16N16 nanocage, are displayed in Fig. 6. The values of Eads for the methyl erlotinib@C24 nanocage, methyl erlotinib@B12N12 nanocage, methyl erlotinib@C32 nanocage and methyl erlotinib@B16N16 nanocage complexes indicate a weak interaction between the nanocages and the inhibitors. The AIM outcomes for the methyl erlotinib@C24 nanocage, methyl erlotinib@B12N12 nanocage, methyl erlotinib@C32 nanocage, and methyl erlotinib@B16N16 nanocage complexes (see Table 2) align with the interaction trends predicted using the Eads and MEP. This confirms that the C24, B12N12, C32, and B16N16 nanocages are effective candidates for drug delivery of the methyl erlotinib molecule.
ratio can be employed to assess the nature of intermolecular bonding. A value greater than 1 signifies the presence of non-covalent bonds, whereas a value ranging from 0.5 to 1 indicates the existence of partial covalent hydrogen bonds. The outcomes presented in Table 2 indicate that all examined nanocages including methyl erlotinib@C24, methyl erlotinib@B12N12, methyl erlotinib@C32, and methyl erlotinib@B16N16 exhibit a ratio higher than 1, implying their noncovalent nature. In this study, it is assumed that all interactions are weak because HC is greater than zero. The intermolecular interactions are described by the vital parameter of the electron density ρ(r). In the examined nanostructures, the electron density (ρ(r)) varies considerably, spanning from 0.00239 to 0.09658 atomic units. This wide range suggests the occurrence of diverse intermolecular forces with varying intensities. As a result, the strongest interaction is related to B10–O35 (ρ(r) = 0.09658 a.u.), whereas the weakest interaction is associated with C72–H45 (ρ(r) = 0.00239 a.u.). Molecular graphs of the optimized complexes, including the methyl erlotinib@C24 nanocage, the methyl erlotinib@B12N12 nanocage, the methyl erlotinib@C32 nanocage and the methyl erlotinib@B16N16 nanocage, are displayed in Fig. 6. The values of Eads for the methyl erlotinib@C24 nanocage, methyl erlotinib@B12N12 nanocage, methyl erlotinib@C32 nanocage and methyl erlotinib@B16N16 nanocage complexes indicate a weak interaction between the nanocages and the inhibitors. The AIM outcomes for the methyl erlotinib@C24 nanocage, methyl erlotinib@B12N12 nanocage, methyl erlotinib@C32 nanocage, and methyl erlotinib@B16N16 nanocage complexes (see Table 2) align with the interaction trends predicted using the Eads and MEP. This confirms that the C24, B12N12, C32, and B16N16 nanocages are effective candidates for drug delivery of the methyl erlotinib molecule.
| Complex | BCP | ρ(r) | ∇2ρ(r) | K C | V C | H C | |−KC/VC| |
|---|---|---|---|---|---|---|---|
| Methyl erlotinib@C24 | C22–H69 | 0.00281 | −0.00202 | 0.00152 | −0.00050 | 0.00102 | 3.04 |
| C1–H58 | 0.00293 | −0.00231 | 0.00181 | −0.00050 | 0.00131 | 3.62 | |
| C4–H55 | 0.00394 | −0.00297 | 0.00233 | −0.00064 | 0.00169 | 3.64 | |
| C3–O26 | 0.00361 | −0.00343 | 0.00271 | −0.00071 | 0.0020 | 3.82 | |
| Methyl erlotinib@B12N12 | N16–H66 | 0.01563 | −0.01390 | 0.01240 | −0.00150 | 0.01090 | 8.26 |
| B10–O35 | 0.09658 | −0.08727 | 0.14352 | 0.05625 | 0.19977 | 2.55 | |
| N24–H64 | 0.00727 | −0.00575 | 0.00492 | −0.00082 | 0.00410 | 6.00 | |
| N24–H59 | 0.00499 | −0.00406 | 0.00312 | −0.00094 | 0.00218 | 3.32 | |
| N17–H69 | 0.01432 | −0.01368 | 0.01179 | −0.00189 | 0.00990 | 6.24 | |
| Methyl erlotinib@C32 | C72–H45 | 0.00239 | −0.00201 | 0.00156 | −0.00045 | 0.00111 | 3.46 |
| C59–H35 | 0.00297 | −0.00230 | 0.00174 | −0.00057 | 0.00117 | 3.05 | |
| C59–O11 | 0.00441 | −0.00419 | 0.00338 | −0.00081 | 0.00257 | 4.17 | |
| C75–O11 | 0.00635 | −0.00553 | 0.00468 | −0.00084 | 0.00384 | 5.57 | |
| C56–H42 | 0.00299 | −0.00255 | 0.00198 | −0.00057 | 0.00141 | 3.47 | |
| Methyl erlotinib@B16N16 | N27–H74 | 0.01576 | −0.01367 | 0.01231 | −0.00136 | 0.01095 | 9.05 |
| N31–H72 | 0.00698 | −0.00552 | 0.00471 | −0.00081 | 0.00390 | 5.81 | |
| B5–O43 | 0.09509 | −0.08612 | 0.14140 | 0.05528 | 0.19668 | 2.56 | |
| N26–H77 | 0.01477 | −0.01389 | 0.01208 | −0.00181 | 0.01027 | 6.67 | |
| N27–H67 | 0.00425 | −0.00350 | 0.00265 | −0.00085 | 0.00180 | 3.12 |
3.9. Comparison of calculations from the 6-31G(d) basis set with the larger 6-311G(d,p) basis set
Table 3 displays computations utilizing the 6-311G(d,p) basis set, indicating that the process of methyl erlotinib adsorption across all nanocages is exothermic. Furthermore, the aqueous phase had higher adsorption energy (Eads) values than that of the gas phase (with the exception of the C32 nanocage). The Eads order is from the greatest to the least, related to the B12N12, B16N16, C32 and C24 nanocages, respectively, which aligns closely with the results obtained using the 6-31G(d) basis set. The calculations using the 6-311G(d,p) basis set revealed that both the LUMO and HOMO energy levels were lower compared to those obtained with the 6-31G(d) basis set and the values of energy gap (Eg) were also found to be smaller than those from the 6-31G(d) basis set.| Nanostructures | HOMO (eV) | LUMO (eV) | E ads (eV) in gas | E ads (eV) in water | BSSE (eV) | E g (eV) | ΔEg (eV) |
|---|---|---|---|---|---|---|---|
| C24 nanocage | −7.471 | −3.040 | — | — | — | 4.431 | — |
| B12N12 nanocage | −9.401 | −0.051 | — | — | — | 9.349 | — |
| C32 nanocage | −7.224 | −3.641 | — | — | — | 3.582 | — |
| B16N16 nanocage | −9.436 | −0.397 | — | — | — | 9.042 | — |
| Methyl erlotinib@C24 nanocage | −7.031 | −3.043 | −0.084 | −0.110 | 0.001 | 3.988 | −0.443 |
| Methyl erlotinib@B12N12 nanocage | −7.230 | −1.049 | −1.107 | −1.179 | 0.008 | 6.181 | −3.167 |
| Methyl erlotinib@C32 nanocage | −7.043 | −3.551 | −0.159 | −0.141 | 0.001 | 3.492 | −0.090 |
| Methyl erlotinib@B16N16 nanocage | −7.236 | −1.058 | −1.078 | −1.135 | 0.006 | 6.178 | −2.863 |
3.10. Molecular docking (MD) simulations
Table 4 provides a list of models from this research that feature erlotinib derivatives as ligands. Molecular dynamics simulations were used to examine the interactions between each ligand structure and each TKI enzyme target separately. Values of binding energy (Eb) indicating efficacious interactions between the ligands and their targets are presented in Table 5. Furthermore, Fig. 7 provides a comprehensive overview of the names of amino acids, as well as the various kinds of non-hydrogen and hydrogen bonds present in the complex systems. Eventually, an examination of the stability of the obtained complexes reveals that E4 (4-methyl erlotinib), with a binding energy (Eb) of −5.0 kcal mol−1, shows potential for effective interactions with TKI enzyme targets.| No. | Chemspider ID | Name | R1 | R2 | R3 | R4 |
|---|---|---|---|---|---|---|
| E | 154044 | Erlotinib | CH3 | CH3 | CH | H |
| E1 | 8493030 | Desmethyl erlotinib | CH3 | OH | CH | H |
| E2 | 13174069 | 2-[4-(3-Ethynylanilino)-7-(2-hydroxyethoxy)quinazolin-6-yl]oxyethanol | OH | OH | CH | H |
| E3 | 17349184 | N-(3-Ethylphenyl)-6,7-bis(2-methoxyethoxy)-4-quinazolinamine | OH | OH | CH | H |
| E4 | 30646730 | 4-Methyl erlotinib | CH3 | CH3 | CH | CH3 |
| E5 | 5142935 | N-(3-Ethylphenyl)-6,7-bis(2-methoxyethoxy)-4-quinazolinamine | CH3 | CH3 | C2H5 | H |
| No. | E b (kcal mol−1) | Interacting amino acids | |
|---|---|---|---|
| Hydrogen bonds | Non-hydrogen bonds | ||
| E | −4.98 | LYS. A:320 & GLN. A:170 | ARG.A:79 & GLY.A:314 & GLU.A:315 & GLU. A:76 & GLU.A:239 & GLU.A:258 & SER. A:167 & VAL. A:318 & LEU. A:80 & HOH.C:O & LYS.A:168 & MET.A:260 |
| E1 | −4.60 | LYS. A:320 & GLU. A:239 | ARG.A:79 & GLY.A:314 & GLU.A:315 & GLU. A:76 & GLU.A:258 & SER. A:167 & VAL. A:318 & LEU. A:80 & HOH.C:O & LYS.A:240 & MET.A:260 & GLN.A:170& HIS.A:238 |
| E2 | −4.89 | LYS. A:320 & GLU. A:239 | ARG.A:79 & GLY.A:314 & GLU. A:76 & GLU.A:258 & SER. A:167 & VAL. A:318 & LEU. A:80 & HOH.C:O & MET.A:260 & THR.A:166 & ILE.A:72 & GLN.A:170 |
| E3 | −3.83 | — | GLY.A:314 & GLU. A:76 & GLU.A:239 & GLU.A:315 & SER. A:167 & VAL. A:318 & LEU. A:80 & HOH.C:O & MET.A:260 & THR.A:166 & ILE.A:72 & LYS.A:320 & HIS.A:238 |
| E4 | −5.00 | LYS.A:320 | PRO.A:169 & GLY.A:314 & GLU. A:76 & GLU.A:239 & GLU.A:315 & GLU.A:258 & SER. A:167 & VAL. A:318 & LEU. A:80 & HOH.C:O & MET.A:260 & LYS.A:168 & HIS.A:238 & GLN.A:170 |
| E5 | −4.48 | LYS.A:320 | PRO.A:169 & GLU. A:76 & GLU.A:239 & GLU.A:258 & SER. A:167 & VAL. A:318 & LEU. A:80 & HOH.C:O & MET.A:260 & ARG.A:79 & HIS.A:238 & GLN.A:170& THR.A:166 & ILE.A:72 |
4. Conclusions
This study aimed to perform DFT calculations to explore how methyl erlotinib molecules interact with various nanostructures, including C24, B12N12, C32, and B16N16 nanocages. The findings demonstrated that all four nanocages were capable of interacting with methyl erlotinib, as evidenced by negative adsorption energy (Eads) values in both aqueous and gaseous states. These negative Eads values confirm that the adsorption of methyl erlotinib onto the nanocages is an exothermic process, indicating favorable interactions. Among the nanocages, B12N12 exhibited the strongest interaction with methyl erlotinib, followed by B16N16, C32, and C24, as reflected by their respective adsorption energies. Notably, the adsorption of methyl erlotinib onto B12N12 and B16N16 led to significant reductions in their energy gaps (ΔEg), with values of −3.229 eV and −2.896 eV, respectively. This reduction in energy gaps suggests an enhancement in the conductivity of these nanostructures, making them promising candidates for applications in nanosensors.The analysis of charge transfer, supported by the MEP results, revealed distinct mechanisms for the interaction of methyl erlotinib with the nanocages. For the C24 and C32 nanocages, charge was transferred from methyl erlotinib to the nanocages, whereas for the B12N12 and B16N16 nanocages, charge was transferred from the nanocages to methyl erlotinib. This difference in charge transfer behavior highlights the unique electronic properties of each nanocage and their varying interactions with methyl erlotinib. Despite these differences, QTAIM analysis confirmed that the interactions between methyl erlotinib and all four nanocages are weak and non-covalent, with electron density (ρ(r)) values ranging from 0.0023 to 0.0965 a.u. These weak interactions are advantageous for drug delivery systems, as they allow for the controlled release of methyl erlotinib at target sites. Frontier molecular orbital (FMO) analysis revealed that the chemical reactivity and stability of the methyl erlotinib–nanocage systems varied depending on the type of nanocage. Among the systems studied, B12N12 exhibited the highest stability, while C32 showed the highest chemical reactivity. These findings further support the potential of the B12N12 and B16N16 nanocages for applications requiring stable and efficient interactions with methyl erlotinib, such as drug delivery and sensing platforms.
In conclusion, the results of this study demonstrate that all four nanocages, C24, B12N12, C32, and B16N16, can effectively function as drug delivery carriers for methyl erlotinib. However, the B12N12 and B16N16 nanocages stand out due to their superior adsorption energies, significant reductions in energy gaps, and enhanced conductivity, making them particularly suitable for applications as nanosensors. The findings underscore the importance of selecting nanocages based on their specific properties, such as conductivity, interaction strength, and charge transfer behavior, for targeted drug delivery and sensing applications. This study provides valuable insights into the design and optimization of nanocage-based systems for biomedical applications, paving the way for future research in this field.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data, took part in drafting the article for important intellectual content, and agreed to be accountable for all aspects of the work.Data availability
Data will be made available in a repository on acceptance.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korean government (MSIT) (RS-2023-00240052). This work was also supported by the Korea Environmental Industry & Technology Institute (KEITI), funded by the Ministry of Environment (MOE) of the Republic of Korea (No. 2022002980004).References
- W. M. Eldehna, M. A. El Hassab, M. F. Abo-Ashour, T. Al-Warhi, M. M. Elaasser, N. A. Safwat, H. Suliman, M. F. Ahmed, S. T. Al-Rashood and H. A. Abdel-Aziz, Development of isatin-thiazolo [3, 2-a] benzimidazole hybrids as novel CDK2 inhibitors with potent in vitro apoptotic anti-proliferative activity: Synthesis, biological and molecular dynamics investigations, Bioorg. Chem., 2021, 110, 104748 CrossRef CAS PubMed.
- F. Li, H. Mei, X. Xie, H. Zhang, J. Liu, T. Lv, H. Nie, Y. Gao and L. Jia, Aptamer-conjugated chitosan-anchored liposomal complexes for targeted delivery of erlotinib to EGFR-mutated lung cancer cells, AAPS J., 2017, 19, 814–826 CrossRef CAS PubMed.
- D. Wang, J. Zhou, W. Fang, C. Huang, Z. Chen, M. Fan, M.-R. Zhang, Z. Xiao, K. Hu and L. Luo, A multifunctional nanotheranostic agent potentiates erlotinib to EGFR wild-type non-small cell lung cancer, Bioact. Mater., 2022, 13, 312–323, DOI:10.1016/j.bioactmat.2021.10.046.
- P. Pandey, K. Dua and H. Dureja, Erlotinib loaded chitosan nanoparticles: Formulation, physicochemical characterization and cytotoxic potential, Int. J. Biol. Macromol., 2019, 139, 1304–1316 CrossRef CAS PubMed.
- S. Jain, K. Dongare, B. Nallamothu, C. P. Dora, V. Kushwah, S. S. Katiyar and R. Sharma, Enhanced stability and oral bioavailability of erlotinib by solid self nano emulsifying drug delivery systems, Int. J. Pharm., 2022, 622, 121852 CrossRef CAS PubMed.
- K. M. Yang, I. C. Shin, J. W. Park, K.-S. Kim, D. K. Kim, K. Park and K. Kim, Nanoparticulation improves bioavailability of Erlotinib, Drug Dev. Ind. Pharm., 2017, 43(9), 1557–1565 CrossRef CAS PubMed.
- H. Zhong and J. Phillip Bowen, Recent, advances in small molecule inhibitors of VEGFR and EGFR signaling pathways, Curr. Top. Med. Chem., 2011, 11(12), 1571–1590 CrossRef CAS PubMed.
- H. Xu, C. He, Y. Liu, J. Jiang and T. Ma, Novel therapeutic modalities and drug delivery–erlotinib liposomes modified with galactosylated lipid: In vitro and in vivo investigations, Artif. Cells, Nanomed., Biotechnol., 2018, 46(8), 1902–1907 CAS.
- R. Rampaka, K. Ommi and N. Chella, Role of solid lipid nanoparticles as drug delivery vehicles on the pharmacokinetic variability of Erlotinib HCl, J. Drug Delivery Sci. Technol., 2021, 66, 102886, DOI:10.1016/j.jddst.2021.102886.
- D. H. Truong, V. K. H. Le, T. T. Pham, A. H. Dao, T. P. D. Pham and T. H. Tran, Delivery of erlotinib for enhanced cancer treatment: An update review on particulate systems, J. Drug Delivery Sci. Technol., 2020, 55, 101348, DOI:10.1016/j.jddst.2019.101348.
- C. P. Dora, V. Kushwah, S. S. Katiyar, P. Kumar, V. Pillay, S. Suresh and S. Jain, Improved oral bioavailability and therapeutic efficacy of erlotinib through molecular complexation with phospholipid, Int. J. Pharm., 2017, 534(1), 1–13, DOI:10.1016/j.ijpharm.2017.09.071.
- S. Taghavi, M. Taghavi, M. Ghaemy, M. Farsadrooh and H. Javadian, Green and selective synthesis of sulfonated poly(pyrimidine-amides) in ionic liquid and their nanocomposites based on carboxylated MWCNTs: Investigation on photophysical, solubility, thermal, and removal of ions behaviors, Colloids Surf., A, 2021, 631, 127759, DOI:10.1016/j.colsurfa.2021.127759.
- S. H. Khoshravesh, Z. Azizi, H. Javadian, M. Farsadrooh, N. Hashemifard, M. Soltani and M. Taghavi, Synthesis of resorcinol-functionalized multi-walled carbon nanotubes as a nanoadsorbent for the solid-phase extraction and determination of diclofenac in human plasma and aqueous samples, Colloid Interface Sci. Commun., 2022, 46, 100555, DOI:10.1016/j.colcom.2021.100555.
- Q. Tan, Y. Chu, M. Bie, Z. Wang and X. Xu, Preparation and Investigation of Amphiphilic Block Copolymers/Fullerene Nanocomposites as Nanocarriers for Hydrophobic Drug, Materials, 2017, 10(2), 192 CrossRef PubMed.
- N. Jafari and S. Zeinali, Highly Rapid and Sensitive Formaldehyde Detection at Room Temperature Using a ZIF-8/MWCNT Nanocomposite, ACS Omega, 2020, 5(9), 4395–4402, DOI:10.1021/acsomega.9b03124.
- C. Parlak, Ö. Alver and Ö. Bağlayan, Quantum mechanical simulation of Molnupiravir drug interaction with Si-doped C60 fullerene, Comput. Theor. Chem., 2021, 1202, 113336, DOI:10.1016/j.comptc.2021.113336.
- E. B. Kalika, K. P. Katin, A. I. Kochaev, S. Kaya, M. Elik and M. M. Maslov, Fluorinated carbon and boron nitride fullerenes for drug Delivery: Computational study of structure and adsorption, J. Mol. Liq., 2022, 353, 118773, DOI:10.1016/j.molliq.2022.118773.
- S. V. Prylutska, L. M. Skivka, G. V. Didenko, Y. I. Prylutskyy, M. P. Evstigneev, G. P. Potebnya, R. R. Panchuk, R. S. Stoika, U. Ritter and P. Scharff, Complex of C60 fullerene with doxorubicin as a promising agent in antitumor therapy, Nanoscale Res. Lett., 2015, 10, 1–7 CrossRef CAS PubMed.
- C. Misra, N. Thotakura, R. Kumar, B. Singh, G. Sharma, O. P. Katare and K. Raza, Improved cellular uptake, enhanced efficacy and promising pharmacokinetic profile of docetaxel employing glycine-tethered C60-fullerenes, Mater. Sci. Eng., C, 2017, 76, 501–508, DOI:10.1016/j.msec.2017.03.073.
- H. Xu, X. Tu, G. Fan, Q. Wang, X. Wang and X. Chu, Adsorption properties study of boron nitride fullerene for the application as smart drug delivery agent of anti-cancer drug hydroxyurea by density functional theory, J. Mol. Liq., 2020, 318, 114315, DOI:10.1016/j.molliq.2020.114315.
- K. Mehdizadeh, F. Toiserkani, M. J. Khodabakhshi, N. Hajali and M. Farsadrooh, Investigation of drug delivery capability of single-walled carbon and boron-nitride nanotubes, boron-nitride (B16N16), and C32 fullerenes as nanocarriers of captopril drug; DFT study, Diamond Relat. Mater., 2024, 146, 111195, DOI:10.1016/j.diamond.2024.111195.
- M. T. Baei, M. R. Taghartapeh, E. T. Lemeski and A. Soltani, A computational study of adenine, uracil, and cytosine adsorption upon AlN and BN nano-cages, Phys. B, 2014, 444, 6–13 CrossRef CAS.
- J. Gong, J. W. Lam and B. Z. Tang, Benchmark and parameter tuning of hybrid functionals for fast calculation of excitation energies of AIEgens, Phys. Chem. Chem. Phys., 2020, 22(32), 18035–18039 RSC.
- D. Jacquemin, E. A. Perpete, I. Ciofini, C. Adamo, R. Valero, Y. Zhao and D. G. Truhlar, On the performances of the M06 family of density functionals for electronic excitation energies, J. Chem. Theory Comput., 2010, 6(7), 2071–2085 CrossRef CAS PubMed.
- M. Walker, A. J. Harvey, A. Sen and C. E. Dessent, Performance of M06, M06-2X, and M06-HF density functionals for conformationally flexible anionic clusters: M06 functionals perform better than B3LYP for a model system with dispersion and ionic hydrogen-bonding interactions, J. Phys. Chem. A, 2013, 117(47), 12590–12600 CrossRef CAS PubMed.
- M. Nouraliei, H. Javadian, K. Mehdizadeh, N. Sheibanian, A. S. Douk, F. Mohammadzadeh and N. Osouleddini, Fullerene carbon nanostructures for the delivery of phenelzine derivatives as new drugs to inhibit monoamine oxidase enzyme: Molecular docking interactions and density functional theory calculations, Colloids Surf., A, 2023, 657, 130599, DOI:10.1016/j.colsurfa.2022.130599.
- Y. Wang, P. Verma, X. Jin, D. G. Truhlar and X. He, Revised M06 density functional for main-group and transition-metal chemistry, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(41), 10257–10262 CrossRef CAS PubMed.
- B. C. Yeo, D. Kim, C. Kim and S. S. Han, Pattern learning electronic density of states, Sci. Rep., 2019, 9(1), 5879 CrossRef PubMed.
- N. M. O'boyle, A. L. Tenderholt and K. M. Langner, Cclib: a library for package–independent computational chemistry algorithms, J. Comput. Chem., 2008, 29(5), 839–845 CrossRef PubMed.
- S. Soleimani Gorgani, M. Nouraliei and S. Soleimani Gorgani, Heterogeneous C16Zn8O8 nanocluster as a selective CO/NO nanosensor: computational investigation, Int. J. Environ. Sci. Technol., 2016, 13, 1573–1580 CrossRef CAS.
- K. F. Biegler, J. Schnbohm and D. Bayles, A program to analyze and visualize atoms in molecules, 2001 Search PubMed.
- J. Pilme, E. Renault, F. Bassal, M. Amaouch, G. Montavon and N. Galland, QTAIM analysis in the context of quasirelativistic quantum calculations, J. Chem. Theory Comput., 2014, 10(11), 4830–4841 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility, J. Comput. Chem., 2009, 30(16), 2785–2791 CrossRef CAS PubMed.
- S. Kim, P. A. Thiessen, E. E. Bolton, J. Chen, G. Fu, A. Gindulyte, L. Han, J. He, S. He and B. A. Shoemaker, PubChem substance and compound databases, Nucleic Acids Res., 2016, 44(D1), D1202–D1213 CrossRef CAS PubMed.
- S. Zinatloo-Ajabshir, S. Rakhshani, Z. Mehrabadi, M. Farsadrooh, M. Feizi-Dehnayebi, S. Rakhshani, M. Dušek, V. Eigner, S. Rtimi and T. M. Aminabhavi, Novel rod-like [Cu(phen)2(OAc)]·PF6 complex for high-performance visible-light-driven photocatalytic degradation of hazardous organic dyes: DFT approach, Hirshfeld and fingerprint plot analysis, J. Environ. Manage., 2024, 350, 119545, DOI:10.1016/j.jenvman.2023.119545.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr05393a |
| This journal is © The Royal Society of Chemistry 2025 |






