Open Access Article
Open Access ArticleEmerging SERS and TERS MoS2 platforms for the characterization of plasma-derived extracellular vesicles†
Lorena
Veliz
 a,
Cédric
Lambin
a,
Cédric
Lambin
 a,
Tyler T.
Cooper
a,
Tyler T.
Cooper
 bc,
W. Michael
McCarvell
a,
Gilles A.
Lajoie
b,
Lynne-Marie
Postovit
c and
François
Lagugné-Labarthet
bc,
W. Michael
McCarvell
a,
Gilles A.
Lajoie
b,
Lynne-Marie
Postovit
c and
François
Lagugné-Labarthet
 *a
*a
aDepartment of Chemistry, Western University (The University of Western Ontario), 1151 Richmond Street, London, ON N6A 5B7, Canada. E-mail: flagugne@uwo.ca
bDepartment of Biochemistry, Western University, 1151 Richmond Street, London, ON N6A 5B7, Canada
cDepartment of Biomedical and Molecular Sciences, Queen's University, 99 University Ave, Kingston, ON K7L 3N6, Canada
First published on 17th March 2025
Abstract
Extracellular vesicles (EVs) play a crucial role in intercellular communication processes. In addition, their biomolecular cargoes such as lipids, proteins, and nucleic acids are useful for identifying potential biomarkers related to different stages of cancer disease. However, the small size and heterogenicity of tumor-related EVs represent a major challenge in properly identifying the content of EVs' cargoes with common characterization protocols. To address these issues, surface-enhanced Raman spectroscopy (SERS) and tip-enhanced Raman spectroscopy (TERS) are powerful alternatives to assign the vibrational fingerprints to the biomolecules contained in cancer EVs, providing high specificity and spatial resolution. Transition metal dichalcogenides are particularly interesting as SERS and TERS substrates due to the high sensitivity of their 2D surface through coulombic and van der Waals interactions when in contact with an analyte or small object such as the charged membranes of EVs. These interactions induce subtle changes in the work function of the flakes which can be measured through drastic changes of optical processes. We investigate the use of MoS2 flakes synthesized by atmospheric pressure chemical vapor deposition as a potential label-free SERS and TERS platform for the identification of plasma EVs. To exemplify this technology, we isolated plasma EV samples from donors with early-stage [FIGO (I/II)] with high-grade serous carcinoma (HGSC) by size exclusion chromatography (SEC). Both surface- and tip-enhanced measurements were conducted individually, enabling the identification of a series of markers from ovarian cancer donors, highlighting the complementarity of SERS and TERS measurements.
1. Introduction
Ovarian cancer is recognized as one of the most lethal gynecological cancers leading to the death of thousands of women around the world.1 High-grade serous carcinoma (HGSC) is the most common type of ovarian cancer and tends to proliferate at an aggressive rate masked by nonspecific symptoms; thus, the diagnosis procedures often occur when HGSC has metastasized outside the ovaries.1,2 As for any cancer, a common diagnosis focuses on the genetic and pathological characterization of tumor biopsy in an effort to determine the histology subtype and course of therapeutic intervention. Although necessary, these methods are invasive and subject patients to painful recovery time. For this reason, the direct analysis of body fluids to detect prospective biomarkers is an attractive alternative due to its ease of acquisition, low cost of processing and high specificity needed for diagnosis.3Extracellular vesicles (EVs) isolated from blood plasma offer a source of lipid-enclosed vesicles that contain diverse lipids, nucleic acids and proteomic cargoes responsible for long-distance communication to the sites of metastasis.4,5 The principal challenge with EVs derived from biofluids is the heterogeneity and large dynamic range of proteomic content from multiple non-tumor sources, complicating biomarker discovery. Knowing the proteomic content in EVs is one of the major concerns because it allows us to understand the role played by some specific proteins and the discovery of new biomarkers to determine the progression of a disease such as ovarian cancer.
Classical methods for analyzing proteins, such as mass spectrometry, immunoblotting and ELISA techniques, while being powerful, are often limited by high cost, a long sample preparation time and low specificity.6 Regardless, our recent study demonstrated the utility of MS for profiling EV proteomes and identifying biomarkers for early-stage HGSC. Importantly, we also provided foundational evidence that Raman spectral characterization of plasma EVs, combined with machine learning, can effectively distinguish early-stage HGSC from benign gynecological conditions. Improving analytical Raman spectroscopy of biofluid EVs is essential for advancing biomarker discovery and enhancing early diagnostic capabilities for HGSC.
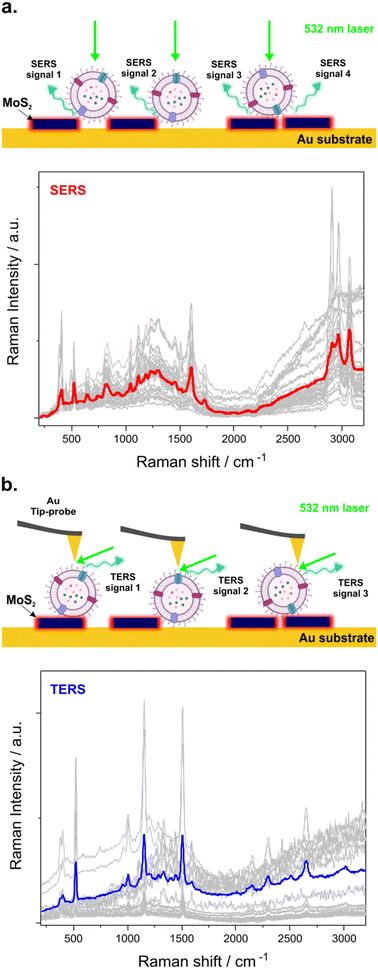
A recent approach for advancing Raman analysis has been dedicated to the combination of novel nanostructure fabrication and vibrational spectroscopy methods such as surface-enhanced Raman spectroscopy (SERS).7–9 SERS is a label-free non-destructive technique that allows the collection of the chemical profile of EVs based on the interaction between excitation light, the sample and a material – generally a metal – that exhibits local surface enhancement.9–11 The information contained in a vibrational SERS spectrum produces a unique fingerprint for each biomolecule contained in the EV.7,9,10,12 Similarly, tip-enhanced Raman spectroscopy (TERS) allows the amplification of the Raman signal in the vicinity of a metal-coated atomic force microscope (AFM) tip that provides a sub-20 nm nanoscale spatial resolution which can be used to map a single isolated EV.13 In TERS, the tip, usually coated with gold (Au) and/or silver (Ag), acts as a nanoantenna to generate a strong electromagnetic field confined at its apex through a plasmonic effect.13,14 The plasmonic contribution refers to the excitation and confinement of the localized surface plasmon in the vicinity of the tip apex when illuminated with resonant incident light. The spatial resolution of TERS enables the possibility to map nano and microscale materials and biomaterials with spatial resolution typically better than 20 nm.15–18 The spatial resolution of TERS experiments depends on many factors including the tip activity, the scattering Raman cross-section of the molecules to be probed and the substrate used for the measurements.19 Under ambient conditions, spatial resolution of a few nm for biological objects such as isolated DNA chains onto an ultra-flat substrate has been reported.20–22 On the other hand, specific SERS from transition-metal dichalcogenides is a plasmon-free chemical enhancement due mainly to charge transfer between the substrate and the analyte that finds its origin from the multiple densities of states of the TMD near the Fermi level.23,24 Subsequently, due to the different origins of the enhancements in SERS and TERS measurements, the intensities of the enhanced Raman modes may differ.
In this work, we investigate SERS and TERS measurements of EVs deposited onto transition metal dichalcogenide (TMD) flakes of MoS2. TMDs have recently been investigated for their SERS activity due to stability, ease of synthesis, biocompatibility and tunable bandgap.25 Intriguingly, despite the semiconducting character, these 2D materials exhibit SERS activity when combined with other materials, such as graphene or metallic nanoparticles, with better enhancement than plasmonic nanostructures. It was shown for MoS2 and MoSe2 that the metallic phase (1T) provides a larger enhancement compared to the semi-conductive phase (2H), highlighting the role of charge transfer in the SERS activity of TMD materials. This charge transfer can be further amplified by altering the electronic structure of TMDs through defect engineering, doping and fabrication processes, leading to optimized 2D SERS substrates that are competitive with noble metal substrates.26–28
Herein, MoS2 flakes with different sizes and shapes were fabricated by chemical vapor deposition, transferred onto an Au surface and used as SERS and TERS substrates for the characterization of plasma EVs isolated from patients with early-stage HGSC. Our label-free new platform has demonstrated the capability to identify the biochemical profile of EVs as well as provide the correlation between the Raman signals and the surface of the EVs absorbed onto the MoS2 flakes at the nanoscale level. Herein, we report the first comparative investigation of SERS and TERS measurements of isolated plasma EVs deposited over 2D MoS2 flakes, highlighting the complementarity of both approaches and their potential as biosensing platforms.
2. Experimental
2.1 Fabrication of MoS2 flakes by CVD
MoS2 flakes were fabricated following the protocol reported by Lambin et al.28 Briefly, the flakes were grown in a silicon (Si) substrate coated with a 280 nm layer of silicon dioxide (SiO2) by atmospheric pressure chemical vapor deposition (CVD). The substrate, with dimensions of 2 × 2 cm2, was previously cleaned with acetone, isopropanol and ultrapure water and placed above the molybdenum trioxide powder (MoO3). The precursors (sulfur and MoO3) were placed in two different alumina boats in a one-zone furnace using a quartz tube (d = 2.5 cm) with a separation of 21 cm between them. To control the melting of sulfur, the ramping rate was initially set at 12 °C min−1 until the temperature reached 550 °C. After this step, the ramp temperature was changed to 6 °C min−1 to reach 850 °C. The furnace was held at this temperature for 15 min and finally, it was let to cool down naturally at room temperature.2.2 Transfer of MoS2 flakes onto a gold substrate
A 50 mg mL−1 solution of PMMA (120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1) in CHCl3 was prepared and used as a transfer layer. About 5–10 drops of the solution were placed in the MoS2 flakes in a Si/SiO2 substrate and spin-coated at 2000 rpm for 80 s. The substrate with the thin PMMA film was then baked at 150 °C for 10 min to evaporate the residual solvent. A 1 M solution of NaOH was prepared to perform the etching process. The baked substrate was placed in a small container with NaOH and let float at 80 °C for 30 min–2 hours depending on the sample size. The PMMA film containing the nanostructures was placed in ultrapure water for 30 min to eliminate the etching solution. Then, the film was slowly and carefully placed in the target substrate (80 nm gold film) and baked for 20 min at 150 °C. Finally, the PMMA film was redissolved in CHCl3 and the new gold substrate containing the MoS2 flakes was let dry overnight.
000 g mol−1) in CHCl3 was prepared and used as a transfer layer. About 5–10 drops of the solution were placed in the MoS2 flakes in a Si/SiO2 substrate and spin-coated at 2000 rpm for 80 s. The substrate with the thin PMMA film was then baked at 150 °C for 10 min to evaporate the residual solvent. A 1 M solution of NaOH was prepared to perform the etching process. The baked substrate was placed in a small container with NaOH and let float at 80 °C for 30 min–2 hours depending on the sample size. The PMMA film containing the nanostructures was placed in ultrapure water for 30 min to eliminate the etching solution. Then, the film was slowly and carefully placed in the target substrate (80 nm gold film) and baked for 20 min at 150 °C. Finally, the PMMA film was redissolved in CHCl3 and the new gold substrate containing the MoS2 flakes was let dry overnight.
2.3 Isolation of HGSC blood plasma by SEC
The plasma samples were collected from women patients with early-stage (I/II) HGSC and obtained from the Banque Cancer de l'ovaire, Centre de recherche du CHUM (CRCHUM), in Montréal, Québec, Canada. The samples were obtained from individual patients (2009–2020) before any treatment such as chemotherapy or radiotherapy and were stored at −80 °C for less than 15 days before the isolation procedure. SEC of HGSC plasma (pooled donors) was performed using an LV70 column following the instructions established by the manufacturer. Briefly, three washes of PBS buffer were added to the column followed by ∼200 μL of blood plasma. After 5 min, 2 mL of PBS buffer was loaded onto the column and let pass until no more eluent was observed. Finally, 1.2 μL of PBS buffer containing 2.5 mM trehalose was added to the column to collect the fraction enriched with EVs (1.2 mL). The aliquots were stored at −80 °C until further use.2.4 AFM characterization of EVs
EV aliquots were analyzed by atomic force microscopy (AFM) to determine their height and size. Isolated EVs were diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in ultrapure water, deposited onto a freshly cleaved mica substrate and let dry overnight in a biosafety cabinet. Preliminary AFM measurements were performed in tapping mode following the protocol reported in previous work7,9 using a Bioscope Catalyst atomic force microscope (Bruker).
20 in ultrapure water, deposited onto a freshly cleaved mica substrate and let dry overnight in a biosafety cabinet. Preliminary AFM measurements were performed in tapping mode following the protocol reported in previous work7,9 using a Bioscope Catalyst atomic force microscope (Bruker).
2.5 Proteomic analysis of plasma EVs by gas-phase fractionation and data-independent acquisition
The MS/MS spectra were obtained using the protocol previously reported by our group.29 Briefly, proteins from SEC-isolated plasma EVs were isolated by single-pot protein precipitation (SP3) using 50% ethanol and proteolysis (50 mM ammonium bicarbonate, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 TrypLysC, 18 hours, 37 °C) and analyzed on a Thermo Eclipse using gas-phase fractionation operating in data-independent acquisition (GPF-DIA). Spectral libraries were initially searched in DIA-NN against human proteome prior to quantitative analysis using Skyline software. The software parameters were set to a precursor mass tolerance of 20 ppm and a fragment ion mass tolerance of 10 ppm. The search parameters allowed for up to 2 missed cleavages and a maximum of 2 post-translational modifications per peptide. Missing value imputation, transformations, and proteomic data visualizations were performed using in-house Python code.
50 TrypLysC, 18 hours, 37 °C) and analyzed on a Thermo Eclipse using gas-phase fractionation operating in data-independent acquisition (GPF-DIA). Spectral libraries were initially searched in DIA-NN against human proteome prior to quantitative analysis using Skyline software. The software parameters were set to a precursor mass tolerance of 20 ppm and a fragment ion mass tolerance of 10 ppm. The search parameters allowed for up to 2 missed cleavages and a maximum of 2 post-translational modifications per peptide. Missing value imputation, transformations, and proteomic data visualizations were performed using in-house Python code.
2.6 Characterization of MoS2 flakes
Before the deposition of EVs over the platform, the MoS2 flakes in the gold substrate were characterized using a LEO 1530 (Zeiss) scanning electron microscope with a 3.0 kV EHT voltage and a 30.0 μm aperture. The optical images and SERS Raman spectra were obtained with a confocal microscope (HORIBA, Xplora) using a white light source with a 100× objective (N.A. 0.9). A laser with an excitation wavelength of 532 nm and a 600 or 1200 groove per mm grating were selected to perform all the experiments in conjunction with a slit width of 100 μm and a pinhole of 300 μm. Finally, to determine the thickness of the flakes, AFM measurements were performed using the Bioscope Catalyst (Bruker) in tapping mode.2.7 Sample preparation and TERS measurements
The MoS2 flakes were transferred onto a silicon substrate coated with a thin film of gold and served as the TERS platform. The Au film beneath the MoS2 flakes was used to work in gap mode TERS. The gap mode implies that the object of interest, here the EVs/MoS2, are located between the TERS tip and the Au substrate. This geometry yields higher confinement of excitation electric fields which subsequently yield to an enhancement of Raman scattering. The EVs were deposited over the platform with an aliquot of enriched EVs. A dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 of the EVs was prepared and deposited over the platform of the substrate. Then the sample was placed in a Petri dish surrounded by wet Kimwipes to slow down the evaporation and ensure a homogeneous deposition. After 48 hours, the substrate was rinsed and dried gently with nitrogen gas. The TERS experiments were carried out using the same confocal microscope (HORIBA Scientific, Xplora) equipped with an atomic force microscope (HORIBA Scientific, AIST-NT OmegaScope). The TERS tips (VTESPA-300, Bruker) were coated with a 5 nm layer of titanium as an adhesion layer and an 80 nm layer of gold by electron beam evaporation. The tip radius varies between 12.5 and 25 nm with a force constant of 42 N m−1 and a length of 150 μm. A 532 nm laser was focused on the tip apex of the cantilever with side illumination (tilted by 60 °C) using a 100× objective (0.70 NA) and a 600 gr mm−1 grating. First, a topographic image is obtained in the tapping mode, then a TERS Raman map is obtained in point-by-point mode. The increase in the Raman signal as a function of the tip–sample distance is shown in Fig. S1.† The tip-in spectrum is obtained when the tip is in direct contact with the surface. In such measurements, the signal combines the contribution of both the near- and far-fields. Accordingly, to obtain the far-field, the tip was lifted slightly away from the surface, this separation of a few nanometers produced a drastic drop in the signal-to-noise ratio.
10 of the EVs was prepared and deposited over the platform of the substrate. Then the sample was placed in a Petri dish surrounded by wet Kimwipes to slow down the evaporation and ensure a homogeneous deposition. After 48 hours, the substrate was rinsed and dried gently with nitrogen gas. The TERS experiments were carried out using the same confocal microscope (HORIBA Scientific, Xplora) equipped with an atomic force microscope (HORIBA Scientific, AIST-NT OmegaScope). The TERS tips (VTESPA-300, Bruker) were coated with a 5 nm layer of titanium as an adhesion layer and an 80 nm layer of gold by electron beam evaporation. The tip radius varies between 12.5 and 25 nm with a force constant of 42 N m−1 and a length of 150 μm. A 532 nm laser was focused on the tip apex of the cantilever with side illumination (tilted by 60 °C) using a 100× objective (0.70 NA) and a 600 gr mm−1 grating. First, a topographic image is obtained in the tapping mode, then a TERS Raman map is obtained in point-by-point mode. The increase in the Raman signal as a function of the tip–sample distance is shown in Fig. S1.† The tip-in spectrum is obtained when the tip is in direct contact with the surface. In such measurements, the signal combines the contribution of both the near- and far-fields. Accordingly, to obtain the far-field, the tip was lifted slightly away from the surface, this separation of a few nanometers produced a drastic drop in the signal-to-noise ratio.
3. Results
MoS2 flakes were synthesized by atmospheric pressure CVD. Fig. 1a depicts the representation of the one-zone furnace and the reaction between the two precursors (MoO3 and S). The protocol optimized by Lambin et al.28 allows careful and precise control of the temperature during the melting process, producing a thin layer of MoS2 with a variety of flakes in terms of shape and size. Due to the weak van der Waals forces between the TMD flakes and the Si wafer, the MoS2 flakes were transferred to suitable substrates for SERS and TERS measurements. As shown in the diagram in Fig. 1b, the flakes were transferred from the Si/SiO2 substrate to an 80 nm gold substrate via a PMMA-assisted method. The gold surface is of particular interest for gap mode TERS measurements, enabling better confinement of the electromagnetic field and resulting in higher enhancement. The optical images of some flakes before and after the transferring process are shown in Fig. 1c and d; different geometries of flakes from triangular to dendritic structures were obtained. The typical thickness of the flakes was around 1.8 nm as determined by AFM (Fig. 1e), which is indicative of a double-layer MoS2 flake.3.1 Characterization of MoS2 flakes by SEM and Raman spectroscopy
The transferred MoS2 flakes were characterized by different techniques before the functionalization with EVs. A bare flake with a surface of about 35 × 35 μm2 was selected to ensure a large, clean and homogeneous surface to perform SERS and TERS experiments, as shown in the SEM image of Fig. 2a for a flake transferred onto a gold-coated substrate. Confocal point scanning maps were collected over the flake for both the Raman modes of MoS2 and photoluminescence (PL). The confocal maps consist of 25 × 25 points with a spacing of 1.5 μm, an acquisition time of 2 s, and an irradiance of 2.8 × 104 W cm−2 at 532 nm excitation using a 100× objective. Fig. 2b shows the optical image of the chosen flake, while Fig. 2c and d show the corresponding maps for the E2g (383 cm−1) and A1g (409 cm−1) Raman modes (blue map in Fig. 2e) characteristic of MoS2 and the photoluminescence at 3500–5000 cm−1 (red-Fig. 2f). The separation between the E2g and A1g modes is 26 cm−1, indicative of ultrathin layers, confirming the AFM measurements. The Raman and PL maps reveal a uniform flake surface in correspondence with the SEM and optical images.3.2 Preliminary characterization of EVs: AFM and proteomics
A preliminary characterization of the extracted EVs was performed to determine the dimensions and size distribution. Fig. 3a shows the AFM image, demonstrating the existence of EV-like particles with a flattened spherical shape due to the restructuring of the EVs over the surface under ambient conditions. The statistical analysis in Fig. 3b indicates a diameter distribution of 129 ± 38 nm, while the height distribution is 50 ± 12 nm, in agreement with our previous characterization of size by nano particle tracking analysis.7,29 To further validate the enrichment of EVs from plasma using SEC, the proteomic content of these EV samples was analyzed using GPF-DIA. Three different donor pools of HGSC-EVs were analyzed by MS to identify a total of 1427 unique proteins, of which 837 were suitable for label-free quantification (Fig. 3c). Fig. 3d shows an enrichment of EV-specific and HGSC-relevant biomarkers. For example, we detected the EV marker CD9 and the current HGSC biomarker MUC16. Importantly, our proteomic analysis was able to uncover prospective early-stage biomarkers, such as MUC1, despite a persistent presence of common plasma proteins.29 Despite significant depletion of common plasma protein using SEC, we still detected significant levels of albumin, tissue factor (TF), fibrinogen (FGG), and complement factors (e.g. CFHR4) that can be explained by the formation of a proteomic corona often observed in EVs from complex biofluids. While additional EV purity could be obtained with combinatorial techniques to SEC, such as ultracentrifugation or ultrafiltration, we opted to maintain a simplified EV workflow to retain the high yield for downstream analysis and technological development.3.3 Characterization of EVs using SERS
Here, different substrates such as an Au thin layer (control), MoS2@SiO2 and MoS2@Au were evaluated for SERS measurements to investigate the content of EVs. The MoS2@Au platform produced the highest-quality spectrum under low-intensity laser power and short acquisition time conditions, highlighting the interest of a hybrid TMD/Au heterostructure for SERS measurements. A comparison between the three substrates is provided in Fig. S2.†As discussed in previous sections, SERS is a novel approach for characterizing EVs because it provides access to the vibrational signature of the EV content, revealing the presence of specific chemical groups that belong to the proteins, lipids and nucleic acid cargoes encapsulated inside the vesicles. Raman measurements were performed after depositing 10 μL of EV solution over the MoS2@Au platform and letting it dry before the experiments. First, several SEM images were taken in the selected area of analysis, allowing the identification of an EV-like particle at the top of the MoS2 flakes, as shown in Fig. 4a. Then, several Raman maps were obtained in the same area to record the EV signals; the overlay of the map and the optical image is shown in Fig. 4b. The SERS maps consisted of 15 × 15 points with a spacing of 0.8 μm, an acquisition time of 4 s, an irradiance of 2.8 × 104 W cm−2 using a 100× objective, an excitation wavelength of 532 nm and a 600 groove per mm grating and collected over the 300–5000 cm−1 spectral range (Fig. 4c). The SERS spectra are indicative of the presence of amino acids and proteins such as the C–C twisting (651 cm−1) and out-of-plane ring breathing (824 cm−1) of tyrosine, the symmetric breathing (752 cm−1) and ring breathing (1192 cm−1) of tryptophan, and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending mode for phenylalanine at 1606 cm−1. In this case, vibrational modes associated with amino acids could be related to the presence of cancer; however, the SERS signals obtained are still weak in intensity, challenging the interpretation of the complete spectra. In addition, the detected Raman modes indicate the presence of carbohydrates; the vibrational mode at 931 cm−1 and the glucose signal at 1118 cm−1 can be assigned to the C–C and C–O stretching vibrational modes, respectively. Other Raman modes were assigned to lipids such as the CH2 deformation at 1297 cm−1, the C
C bending mode for phenylalanine at 1606 cm−1. In this case, vibrational modes associated with amino acids could be related to the presence of cancer; however, the SERS signals obtained are still weak in intensity, challenging the interpretation of the complete spectra. In addition, the detected Raman modes indicate the presence of carbohydrates; the vibrational mode at 931 cm−1 and the glucose signal at 1118 cm−1 can be assigned to the C–C and C–O stretching vibrational modes, respectively. Other Raman modes were assigned to lipids such as the CH2 deformation at 1297 cm−1, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester stretching mode at 1735 cm−1, and the CH3 stretching bands at 2960 cm−1 and 3070 cm−1. The importance of carbohydrates lies in the alteration of the glycemic index in a patient; it was reported that high amounts of these biomolecules increase the risk of generating illnesses such as cancer.30 However, even when a correlation between a high carbohydrate-based diet and cancer risk can be defined, there are still not enough studies to establish a tendency regarding the significant presence of carbohydrate modes and cancer stages. Finally, a few characteristic signals that can be associated with tumors were also identified such as the formalin peaks appearing in fixed tumor tissues at 1039 cm−1, the asymmetric PO2− from the phosphodiester group at 1240 cm−1, which suggests a possible increase in the nucleic acids in malignant tissue, and the structural protein (C–H bending) mode of tumors at 1455 cm−1.31 A summary of the Raman peaks identified in this sample and their assignment is provided in Table 1, following the values reported in the literature.
O ester stretching mode at 1735 cm−1, and the CH3 stretching bands at 2960 cm−1 and 3070 cm−1. The importance of carbohydrates lies in the alteration of the glycemic index in a patient; it was reported that high amounts of these biomolecules increase the risk of generating illnesses such as cancer.30 However, even when a correlation between a high carbohydrate-based diet and cancer risk can be defined, there are still not enough studies to establish a tendency regarding the significant presence of carbohydrate modes and cancer stages. Finally, a few characteristic signals that can be associated with tumors were also identified such as the formalin peaks appearing in fixed tumor tissues at 1039 cm−1, the asymmetric PO2− from the phosphodiester group at 1240 cm−1, which suggests a possible increase in the nucleic acids in malignant tissue, and the structural protein (C–H bending) mode of tumors at 1455 cm−1.31 A summary of the Raman peaks identified in this sample and their assignment is provided in Table 1, following the values reported in the literature.
| Position (cm−1) | Possible assignment of Raman peaks | Ref. |
|---|---|---|
| 651 | C–C twisting mode of tyrosine | 32 |
| 752 | Symmetric breathing of tryptophan | 32 |
| 824 | Out-of-plane ring breathing of tyrosine | 31 and 32 |
| 931 | Carbohydrate peak | 31 |
| 1039 | Formalin peaks appear in the fixed tumor tissue | 31 |
| 1118 | Glucose or C–C stretching (breast lipid) | 33 |
| 1192 | Tryptophan ring breathing | 32 |
| 1240 | Asymmetric PO2− from the phosphodiester of nucleic acids | 31 |
| 1297 | CH2 deformation in lipids | 31 and 34 |
| 1455 | Structural protein mode from tumors | 31 |
| 1606 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending of phenylalanine C bending of phenylalanine |
35 |
| 1736 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O ester (lipids) stretching O ester (lipids) stretching |
36 |
| 2960 | Out-of-plane chain end antisymmetric CH3 stretching band | 31 and 37 |
| 3070 | CH3 stretching band | 34 |
3.4 Characterization of EVs using TERS
TERS has been used to characterize biological materials such as proteins, lipids, nucleic acids, and EVs.34,37 In this work, the TERS experiments were performed similarly to the SERS experiments after depositing 10 μL of HGSC-EV solution in the MoS2@Au/Si platform and letting it dry (Fig. 5). A shorter exposure time (t = 0.3 s per spectrum) during the acquisition of the maps and a lower intensity (I = 0.89 mW) at the samples were selected to minimize thermal effects on the sample. Topography over a surface of 15 × 15 μm2 and TERS maps over a surface of 7.12 × 7.87 μm2 are presented in Fig. 5a. The edges of a MoS2 flake and the EVs deposited at the top of the substrate are observed clearly in the AFM image. Selected Raman modes (1154 cm−1 and 1507 cm−1) reveal the location of the EVs in Fig. 5b and c, while PL in Fig. 5d provides contrast for the MoS2 flake. These maps enable the correlation of the specific spectral features of the EVs over the MoS2 flakes. TERS spectra obtained after several measurements taken at different spots over the vesicle are reported in Fig. 5e. Two major Raman modes were observed in the TERS spectra, the first one reveals the C–C and C–N stretching modes of proteins at 1154 cm−1 and can be related to its presence all around the EV membrane. The conformation of the EVs is normally based on the phospholipid bilayer to protect the internal cargo; however, EV proteins also play a crucial role. For instance, they maintain their functionality and conformation for long periods, allowing the transfer of unaltered information from one cell to another. The proteome of EVs provides better and more precise details regarding the parental cells, the microenvironment and even the progression of the disease.38 In addition, the second intense signal at 1507 cm−1 related to the vibration of the N–H bond in nucleic acids is shown and supports the previous literature on DNA and RNA species within EVs. This Raman mode contains information about cytosine; this nitrogen base is well recognized for its role in the demethylation reaction which is an important part of the DNA transcription process.39,40 The control of gene expression plays a substantial role in the regulation of cell proliferation; thus a small change in the gene sequence or the overexpression of the enzyme that catalyzes the reaction will produce an alteration in the DNA strand, leading to catastrophic biological events such as cancer development. The intensity of this peak can be associated with the deregulation of cytosine content and work as a possible marker peak for early ovarian cancer stages.40 In order to compare the SERS and TERS measurements collected over an EV, the average spectra collected from a series of 30 spectra are reported in Fig. 6. Differential EV fingerprints were obtained for TERS compared to SERS, thus supporting that the enhancement of the signals depends on the substrate and the effect of the coated tip. Comparison between SERS and TERS signal intensities are provided in Table S1.†Besides the two major Raman modes discussed previously (1154 and 1507 cm−1), several Raman signals were also detected in the TERS spectra such as the vibrational modes of carotenoids and cholesterol at 959 cm−1 and 1395 which were reported in the literature and are strongly related to the presence of tumoral cells in cervical and breast cancer.32 Also, Raman modes from amino acids such as phenylalanine (1006 cm−1) and signals from tyrosine and tryptophan as well as amide III (1207 cm−1) were also observed.31,41 In speculation with our proteomic analysis, the presence of differential proteins in a malignant tissue could be related to inflammation derived from the proliferation of the malignant ovarian cancer cells.42,43 Although we did not directly compare HGSC cohorts in this study, we speculate the intensity and stability of some of these signals can be potentially used to differentiate between the presence and absence of cancer in further prognosis assays. Likewise, vibrational modes for the glycine backbone and proline chains were also identified at 1283 cm−1; the presence of these signals can be connected to the activity of serine hydroxymethyl transferase (SHMT1) that produces glycine as part of nucleotide biosynthesis. Overexpression of this catalytic enzyme was reported in patients with HGSC, enabling tumor growth and cell propagation.44 A signal from collagen and CH3–CH2 wagging modes for nucleic acids is noticed at 1331 cm−1 and 1448 cm−1. The importance of these signals lies in the role of collagen in creating an adequate microenvironment for the tumor, influencing the cancer cell activity and intensifying the progression of the illness.43 In addition, the peak at 1331 cm−1 can be potentially useful for detecting slight differences in the amount of DNA (nucleic acids), suggesting the evolution of tumorigenesis. Collectively, these results demonstrate the potential of TERS to provide comprehensive Raman fingerprints of biofluid EVs which may be used for diagnostic applications.
Interestingly, extra signals are observed in the region of 2000–3000 cm−1 for the TERS spectra compared to the SERS spectra (Fig. 6a and b).
It is worth mentioning that this region is usually known as a silent region due to the scarcity of vibrational modes derived from biomolecules. The first clear signal at around 2150 cm−1 is attributed to the possible presence of the thiocyanate group (C–N stretch). This ion is not produced naturally by the body; however, there are some vegetables and foods recognized as natural sources of thiocyanate such as brassica vegetables, milk or cheese. The presence of thiocyanate was reported in saliva samples for oral cancer diagnosis, yet it can also be found in different fluids such as human serum or urine. Indeed, the presence of this ion can be associated with diseases, such as cancer. In the work presented by Fălămaş et al.,45 all cancer samples presented this characteristic peak, highlighting the importance of tracking this signal and considering it a possible biomarker. Moreover, signals at 2300 and 2650 cm−1 can be related to the vibrational stretching modes of CN, NH and OH groups, which are important components of a wide variety of proteins and carbohydrates in human body fluids. Once again, these peaks were not observed in the SERS experiments. Lastly, contributions of intense lipid signals and acyl-related vibrational modes at around 2800–3100 cm−1 were observed in the SERS spectra. Lipids also play a key role in the development and progression of cancer, serving as an energy source for tumoral cells. Thus, an alteration in the lipid content can suggest an uncontrollable growth of the cells from early stages to metastasis.46 However, as shown in the TERS spectra (Fig. 6b), the lipid signals are lower in intensity. A summary of the TERS signals observed is shown in Table 2.
| Position (cm−1) | Possible assignment of Raman peaks | Ref. |
|---|---|---|
| 959 | Carotenoids (absent in normal tissue), cholesterol | 32 |
| 1006 | Phenylalanine, protein assignment. Carotenoids | 11 |
| 1154 | C–C (and C–N) stretching of proteins | 11 and 31 |
| 1207 | Hydroxyproline, tyrosine or tryptophan. Amide III (protein) | 31 and 32 |
| 1283 | Amide III and CH2 wagging vibrations from the glycine backbone and proline chains | 31 |
| 1331 | CH3–CH2 wagging mode in nucleic acids. Twisting and wagging modes in collagen. | 32 |
| 1395 | CH rocking, carotenoids. | 31 and 35 |
| 1448 | CH3CH2 deformation and CH2 deformation, collagen | 32 |
| 1507 | N–H bending, cytosine | 31 |
| 2150 | C–N stretch | 45 |
| 2300–3000 | Stretching vibration of CH, NH and OH groups | 34 and 37 |
| 3005 | Contribution from acyl chains | 34 and 37 |
The presence of the new modes observed in TERS spectra and the difference in lipid peak intensity are related to the configuration of the TERS system. The presence of the Au-coated AFM tip and its subsequent localized Raman enhancement can be affected by several parameters such as the illumination angle of the AFM probe with respect to the surface. The in- and out-of-plane vibrations may be enhanced differently since the z-component of the field along the tip axis is mostly enhanced in the TERS configuration. As a result, not only the MoS2 modes but also the EV fingerprints are affected in TERS by producing the presence of new signals or the absence of some of the modes detected previously in the SERS spectra due to different selection rules. Additionally, recent work highlights the challenges of obtaining homogeneous EV fingerprints due to the variety of binding phenomena between the sample and the substrate (illustrated in Fig. 6) and the complex composition of the EVs with high concentrations of biological compounds at the surface of the phospholipid membrane.47 Moreover, EVs are normally larger than the possible hotspots generated in the substrate. Then, the position of the EV over the substrate has a strong and direct effect on the signal intensity because not all cargoes can be in proximity to an effective hotspot, resulting in a low amplification of the signal and the collection of different spectra even in the same sample.
As discussed before, small fluctuations or alterations in the spectral pattern can also be observed depending on the deposition strategies, capturing methods, and the distribution of the EV around the sample. Thus, further efforts and methodologies must be applied to ensure good reproducibility and sensitivity in the analysis of EV fingerprints.
In summary, meanwhile SERS provides an averaged analysis in the EV population, TERS makes possible single EV analysis opening the characterization and mapping the distribution of biomolecules inserted into the EV surface. TERS is suited for the study of proteins, nucleic acids and lipids within a complex biological object with a good signal/noise ratio for the analysis of critical Raman modes such as the ones observed at 1154 and 1507 cm−1 (proteins and nucleic acids, respectively). Furthermore, TERS was used for the identification of new signals from distinct domains such as the ones observed at around 2300 and 2650 cm−1 (OH, CH and NH stretching vibrations) associated with carbohydrates and proteins present in the EV lipid membrane.
4. Conclusions
In this work, SERS and TERS platforms using MoS2 flakes deposited over a gold substrate were used for the characterization of HGSC-EVs derived from plasma fluid. The isolation of the samples was achieved using SEC to enrich our EV samples; additionally, the size and proteomic content were determined by AFM and MS. To our knowledge, this is the first time that a comparative study between two spectroscopy techniques such as SERS and TERS is performed in a complex EV sample. The new MoS2@Au substrate has shown excellent capabilities as SERS and TERS platforms, enhancing the Raman signal in the HGSC-EV sample and providing a complete fingerprint with characteristic signals from nucleic acids, proteins, and lipids. Of interest is that TERS allowed the complete characterization of the sample, distinguishing between the presence of raw protein cargoes and the potential HGSC-EVs, highlighting the correlation between the topography and the Raman spectra. Moreover, TERS spectra permit the identification of three new vibrational modes (at 2150, 2300 and 2650 cm−1) that were not present in SERS measurements, which can be used for diagnosis purposes and work as powerful differentiation Raman modes in machine learning procedures.Author contributions
The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript. L. Veliz and C. Lambin have contributed equally to this manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research (L. V., C. L., W. M. and F. L.-L.) was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada through a Discovery Grant (DGRGPIN-2020-06676). T. T. C. and L. M. P wish to acknowledge the Cancer Research Society (#945817). The authors would gratefully thank the Nanofabrication Facility at the University of Western Ontario (Western University) for their assistance in the fabrication of TERS tips and the SiO2/Si substrates. In addition, the authors would also like to thank the Banque Cancer de l'ovaire, Centre de recherche du CHUM (CRCHUM), in Montreal, Québec, Canada for providing the plasma samples for HGSC.References
- A. M. Karst and R. Drapkin, J. Oncol., 2010, 2010, 1–13 Search PubMed.
- K. Levanon, C. Crum and R. Drapkin, J. Clin. Oncol., 2008, 26, 5284–5293 Search PubMed.
- D. R. Nebgen, K. H. Lu and R. C. Bast, Curr. Oncol. Rep., 2019, 21, 1–9 CAS.
- P. E. Geyer, L. M. Holdt, D. Teupser and M. Mann, Mol. Syst. Biol., 2017, 13, 477 Search PubMed.
- E. Serrano-Pertierra, M. Oliveira-Rodríguez, M. Rivas, P. Oliva, J. Villafani, A. Navarro, M. Blanco-López and E. Cernuda-Morollón, Bioengineering, 2019, 6, 1–13 Search PubMed.
- X. Tan, K. C. Day, X. Li, L. J. Broses, W. Xue, W. Wu, W. Y. Wang, T.-W. Lo, E. Purcell, S. Wang, Y.-L. Sun, M. K. Khaing Oo, B. M. Baker, S. Nagrath, M. L. Day and X. Fan, Biosens. Bioelectron.:X, 2021, 8, 100066 CAS.
- L. Veliz, T. T. Cooper, I. Grenier-Pleau, S. A. Abraham, J. Gomes, S. H. Pasternak, B. Dauber, L. M. Postovit, G. A. Lajoie and F. Lagugné-Labarthet, ACS Sens., 2024, 9, 272–282 CAS.
- M. Erzina, A. Trelin, O. Guselnikova, A. Skvortsova, K. Strnadova, V. Svorcik and O. Lyutakov, Sens. Actuators, B, 2022, 367, 132057 CrossRef CAS.
- N. M. Ćulum, T. T. Cooper, G. A. Lajoie, T. Dayarathna, S. H. Pasternak, J. Liu, Y. Fu, L. M. Postovit and F. Lagugne-Labarthet, Analyst, 2021, 146, 7194–7206 RSC.
- N. M. Ćulum, T. T. Cooper, G. I. Bell, D. A. Hess and F. Lagugné-Labarthet, Anal. Bioanal. Chem., 2021, 413, 5013–5024 CrossRef PubMed.
- G. Shetty, C. Kendall, N. Shepherd, N. Stone and H. Barr, Br. J. Cancer, 2006, 94, 1460–1464 CrossRef CAS PubMed.
- J. Penders, A. Nagelkerke, E. M. Cunnane, S. V. Pedersen, I. J. Pence, R. C. Coombes and M. M. Stevens, ACS Nano, 2021, 15, 18192–18205 CrossRef CAS PubMed.
- Z. Zhang, S. Sheng, R. Wang and M. Sun, Anal. Chem., 2016, 88, 9328–9346 CrossRef CAS PubMed.
- N. N. Kurus, I. A. Milekhin, N. A. Nebogatikova, I. V. Antonova, E. E. Rodyakina, A. G. Milekhin, A. V. Latyshev and D. R. T. Zahn, J. Phys. Chem. C, 2023, 127, 5013–5020 CrossRef CAS.
- N. Kazemi-Zanjani, H. Chen, H. A. Goldberg, G. K. Hunter, B. Grohe and F. Lagugné-Labarthet, J. Am. Chem. Soc., 2012, 134, 17076–17082 CrossRef CAS PubMed.
- Y. Pandey, A. Ingold, N. Kumar and R. Zenobi, Nanoscale, 2024, 16, 10578–10583 RSC.
- F. Pashaee, F. Sharifi, G. Fanchini and F. Lagugné-Labarthet, Phys. Chem. Chem. Phys., 2015, 17, 21315–21322 RSC.
- L. Xiao and Z. D. Schultz, Anal. Chem., 2018, 90, 440–458 CrossRef CAS PubMed.
- C. Höppener, J. Aizpurua, H. Chen, S. Gräfe, A. Jorio, S. Kupfer, Z. Zhang and V. Deckert, Nat. Rev. Methods Primers, 2024, 4, 47 CrossRef.
- E. Bailo and V. Deckert, Angew. Chem., Int. Ed., 2008, 47, 1658–1661 CAS.
- F. Pashaee, M. Tabatabaei, F. A. Caetano, S. S. G. Ferguson and F. Lagugné-Labarthet, Analyst, 2016, 141, 3251–3258 CAS.
- Z. He, Z. Han, M. Kizer, R. J. Linhardt, X. Wang, A. M. Sinyukov, J. Wang, V. Deckert, A. V. Sokolov, J. Hu and M. O. Scully, J. Am. Chem. Soc., 2019, 141, 753–757 CAS.
- J. P. Fraser, P. Postnikov, E. Miliutina, Z. Kolska, R. Valiev, V. Švorčík, O. Lyutakov, A. Y. Ganin and O. Guselnikova, ACS Appl. Mater. Interfaces, 2020, 12, 47774–47783 CrossRef CAS PubMed.
- X. Song, Y. Wang, F. Zhao, Q. Li, H. Q. Ta, M. H. Rümmeli, C. G. Tully, Z. Li, W.-J. Yin, L. Yang, K.-B. Lee, J. Yang, I. Bozkurt, S. Liu, W. Zhang and M. Chhowalla, ACS Nano, 2019, 13, 8312–8319 CAS.
- S. A. Ghopry, S. M. Sadeghi, C. L. Berrie and J. Z. Wu, Biosensors, 2021, 11, 1–14 Search PubMed.
- M. Chen, D. Liu, X. Du, K. H. Lo, S. Wang, B. Zhou and H. Pan, TrAC, Trends Anal. Chem., 2020, 130, 115983 Search PubMed.
- M. Jalali, C. del Real Mata, L. Montermini, O. Jeanne, I. I. Hosseini, Z. Gu, C. Spinelli, Y. Lu, N. Tawil, M. C. Guiot, Z. He, S. Wachsmann-Hogiu, R. Zhou, K. Petrecca, W. W. Reisner, J. Rak and S. Mahshid, ACS Nano, 2023, 17, 12052–12071 Search PubMed.
- C. Lambin, M. O. Avilés, J. Jelken and F. Lagugné-Labarthet, J. Phys. Chem. C, 2023, 127, 24281–24290 CAS.
- T. T. Cooper, D. Z. Dieters-Castator, J. Liu, G. M. Siegers, D. Pink, L. Veliz, J. D. Lewis, F. Lagugné-Labarthet, Y. Fu, H. Steed, G. A. Lajoie and L.-M. Postovit, J. Ovarian Res., 2024, 17, 149 CAS.
- S. Sieri, C. Agnoli, V. Pala, S. Grioni, F. Brighenti, N. Pellegrini, G. Masala, D. Palli, A. Mattiello, S. Panico, F. Ricceri, F. Fasanelli, G. Frasca, R. Tumino and V. Krogh, Sci. Rep., 2017, 7, 9757 CAS.
- A. C. S. Talari, Z. Movasaghi, S. Rehman and I. U. Rehman, Appl. Spectrosc. Rev., 2015, 50, 46–111 CAS.
- N. Stone, C. Kendall, J. Smith, P. Crow and H. Barr, Faraday Discuss., 2004, 126, 141–157 Search PubMed.
- E. Wiercigroch, E. Szafraniec, K. Czamara, M. Z. Pacia, K. Majzner, K. Kochan, A. Kaczor, M. Baranska and K. Malek, Spectrochim. Acta, Part A, 2017, 185, 317–335 CAS.
- T. Stepanenko, K. Sofińska, N. Wilkosz, J. Dybas, E. Wiercigroch, K. Bulat, E. Szczesny-Malysiak, K. Skirlińska-Nosek, S. Seweryn, J. Chwiej, E. Lipiec and K. M. Marzec, Analyst, 2024, 149, 778–788 Search PubMed.
- N. Huang, M. Short, J. Zhao, H. Wang, H. Lui, M. Korbelik and H. Zeng, Opt. Express, 2011, 19, 22892–22909 CAS.
- H. Nawaz, F. Bonnier, A. D. Meade, F. M. Lyng and H. J. Byrne, Analyst, 2011, 136, 2450–2463 CAS.
- L. Buccini, A. Proietti, G. La Penna, C. Mancini, F. Mura, S. Tacconi, L. Dini, M. Rossi and D. Passeri, Nanoscale, 2024, 16, 8132–8142 CAS.
- J. Tian, G. Casella, Y. Zhang, A. Rostami and X. Li, Int. J. Biol. Sci., 2020, 16, 620–632 CAS.
- S. K. Patra, A. Patra, F. Rizzi, T. C. Ghosh and S. Bettuzzi, Cancer Metastasis Rev., 2008, 27, 315–334 CAS.
- K. L. Terry, I. De Vivo, L. Titus-Ernstoff, M. C. Shih and D. W. Cramer, Cancer Res., 2005, 65, 5974–5981 CrossRef CAS PubMed.
- G. Neurauter, A. V. Grahmann, M. Klieber, A. Zeimet, M. Ledochowski, B. Sperner-Unterweger and D. Fuchs, Cancer Lett., 2008, 272, 141–147 CrossRef CAS PubMed.
- A. Watanabe, T. Higashi, T. Sakata and H. Nagashima, Cancer, 1984, 54, 1875–1882 CrossRef CAS PubMed.
- S. Xu, H. Xu, W. Wang, S. Li, H. Li, T. Li, W. Zhang, X. Yu and L. Liu, J. Transl. Med., 2019, 17, 309 Search PubMed.
- R. Murali, V. Balasubramaniam, S. Srinivas, S. Sundaram, G. Venkatraman, S. Warrier, A. Dharmarajan and R. K. Gandhirajan, Metabolites, 2023, 13, 1–39 CrossRef PubMed.
- A. Fălămaş, H. Rotaru and M. Hedeşiu, Laser Med. Sci., 2020, 35, 1393–1401 CrossRef PubMed.
- S. Vasseur and F. Guillaumond, Oncogenesis, 2022, 11, 1–15 CrossRef PubMed.
- H. Shin, D. Seo and Y. Choi, Molecules, 2020, 25, 5209 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional experimental details and tables of Raman signals. See DOI: https://doi.org/10.1039/d4nr04926h |
| This journal is © The Royal Society of Chemistry 2025 |