Open Access Article
Open Access ArticleDiscovery, biosynthesis, and bioactivities of peptidic natural products from marine sponges and sponge-associated bacteria
Weimao
Zhong†
 a,
Zhenjian
Lin†
b,
Eric W.
Schmidt
a,
Zhenjian
Lin†
b,
Eric W.
Schmidt
 *b and
Vinayak
Agarwal
*b and
Vinayak
Agarwal
 *ac
*ac
aSchool of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, GA 30332, USA. E-mail: vagarwal@gatech.edu
bDepartment of Medicinal Chemistry, University of Utah, Salt Lake City, UT 84112, USA. E-mail: ews1@utah.edu
cSchool of Biological Sciences, Georgia Institute of Technology, Atlanta, GA 30332, USA
First published on 12th September 2025
Abstract
Covering 2010 to 2025
Sponges are benthic, sessile invertebrate metazoans that are some of the most prolific sources of natural products in the marine environment. Sponge-derived natural products are often endowed with favorable pharmaceutical bioactivities, and paired with their structural complexity, have long served as title compounds for chemical syntheses. Sponges are holobionts, in that the sponge host is associated with symbiotic and commensal microbiome. Natural products isolated from sponges can be produced by the sponge host, or the associated microbiome. Recent genomic studies have shed light on the sponge eukaryotic host as the true producer of several classes of sponge-derived peptidic natural products. In this review spanning years 2010–2025, we describe peptidic natural products isolated from the sponge hosts and the associated microbiome, detail their biosynthetic processes where known, and offer forward looking insights into future innovation in discovery and biosynthesis of peptidic natural products from marine sponges.
1. Introduction
Sponges (phylum Porifera) are among the most ancient marine invertebrates. The inventory of sponge species in the ocean continues to expand, with nearly 9000 species described to date.1 Sponges are multicellular sessile organisms living across diverse marine ecological habitats from tropics to poles, oceans and freshwater, intertidal zones to deep seas.2,3 Sponges live by heterotrophic filter-feeding, a process that involves pumping several liters of water per gram of sponge biomass per day.4 In addition, carnivorous sponges use specialized appendages to capture prey.5,6Sponges are holobionts consisting of sponge hosts, symbiotic bacteria, and commensal bacteria, as well as other microbes.7 Tightly associated symbiotic bacteria are embedded in the sponge host physiology and the holobiont metabolism with endosymbionts residing within the sponge tissue. Consequently, symbiotic bacteria can be challenging to cultivate (often called “uncultivated”, or less accurately, “uncultivable”) under traditional laboratory culturing conditions. Photosynthetic endosymbionts contribute to the nutrient supply to the sponge holobiont.8 On the other hand, commensal bacteria that are loosely associated with the sponge host are usually amenable to cultivation.
Secondary metabolites—colloquially referred to as natural products—are cornerstones for pharmaceutical discovery.9,10 Among natural products, peptidic natural products (pNPs) are a particularly promising group of bioactive compounds that target a wide range of pathologies including infectious diseases, cancers, diabetes, HIV, chronic pain, among others.11 For the purposes of this review, pNPs are divided according to their biosynthetic origins: ribosomally synthesized and post-translationally modified peptides (RiPPs), and non-ribosomally synthesized peptides (NRPs) (Fig. 1). For the purposes of this review, dipeptide alkaloids are not treated as canonical pNPs (vide infra).
 | ||
| Fig. 1 Chemical classes of pNPs discussed in this review and sites of their biosynthesis within the sponge holobiont. | ||
The abovementioned organismal complexity of the sponge holobiont has prompted genetic investigations aimed at deciphering which component of the holobiont—the sponge host, or the symbiotic microbiome—is the true producer of the pNPs that are detected in sponge extracts. As many pNPs isolated from sponges share structure similarities with those originating from bacteria, it was proposed that the symbiotic microbiome would be the source of sponge-derived pNPs.12 This hypothesis has indeed borne out (vide infra). In addition, the sponge commensal microbiome is now appreciated as a repository possessing numerous biosynthetic gene clusters (BGCs) that encode cryptic pNPs that are not detected in the sponge extracts but can be accessed using microbiology and synthetic biology workflows.13–16
In light of the demonstrated proclivity of the sponge microbiome to furnish pNPs, a question endured if the sponge host itself could also produce pNPs?17 Certainly, the production of bioactive pNPs—such as venoms and toxins—is widespread in Eukarya, and sponges have been shown to produce other classes of natural products.18–20 In recent years, the sponge hosts have themselves shown to be producers of pNPs.21,22 These studies provide new avenues to understand the entire sponge holobionts as pNP producers and posit new hypotheses regarding the differential ecological roles of the host-derived and the microbiome-derived pNPs in sponge ecology.23
With the motivation to juxtapose the sponge holobiont architecture against the biosynthetic routes of sponge-derived pNPs, this review is structured such as to provide a description of the three classes of pNPs according to the site of their biosynthesis within the sponge holobiont—the eukaryotic sponge host itself, the symbiotic microbiome, or the commensal microbiome. Biosynthetic origins for several sponge-derived pNPs have not been deciphered yet; these molecules are discussed separately.
Among pNPs, RiPP biosynthesis starts with the ribosomal synthesis of a precursor peptide. Typically, RiPP precursor peptides are divided into an N-terminal leader and a C-terminal core regions. The leader region is recognized by and binds to the RiPP biosynthetic enzymes which enables post-translational modification to be affected upon the C-terminal core. Proteolytic removal of the unmodified leader region furnishes the mature RiPP.24,25 As described in later sections, the microbes are a well validated source of RiPPs, both from the symbiotic and the commensal microbiomes (Fig. 1). The sponge host has been shown to be a producer of RiPPs as well.
The biosynthesis of NRPs is catalyzed by large megadalton non-ribosomal peptide synthetases (NRPSs) that construct pNPs in an assembly line fashion.26–29 Upon ATP-dependent adenylation of the amino acyl carboxylates by the NRPS adenylation domain (A domain), the amino acids are thioesterified to the phosphopantetheinyl arm of thiolation domain (T domain; also referred to peptidyl carrier protein). The NRPS condensation domain (C domain) then catalyzes the peptide bond formation between the downstream T domain-tethered amino acid building block, and the upstream T-domain tethered peptide chain and transfers the growing peptide chain onto the downstream T domain phosphopantetheine arm for the next round of chain elongation by the NRPS A–T–C module. A thioesterase domain (TE domain) terminates chain extension by offloading the mature NRP.30,31 These are typical rules; a vast array of other chemical diversity-amplifying modifications have been reported in NRPS assembly lines.32 The symbiotic and commensal microbiomes of marine sponges are sites for biosynthesis of NRPs and various examples are described below. To date, the sponge host has not been found to synthesize NRPs (Fig. 1).
Numerous sponge-derived pNPs cannot be classified among RiPPs and NRPs as their biosynthetic origins are unknown. Principal among these are dipeptide alkaloids that belong to the pyrrole aminoimidazole alkaloid (PIA) and the bromotyrosine alkaloid (BTA) chemical classes (Fig. 1).33–39 The PIAs and BTAs possess a dipeptidic structure with conserved proline-derived brominated pyrroles and brominated tyrosine building blocks, respectively. PIAs and BTAs have attracted attention due to their structural complexity that has provided attractive targets for chemical syntheses, as well as due to their chemical diversity and geographical ubiquity.40–42 The biosynthetic routes for sponge-derived PIA and BTA alkaloids have not been deciphered, and hence, the site of their biosynthesis with the sponge holobiont is not known (Fig. 1). In addition to ATP-grasp enzymes and cyclodipeptide synthetases,43–47 among other ribosome-independent peptide bond forming modalities,48–50 numerous enzymatic routes can be envisaged for the assembly of sponge-derived dipeptide alkaloids. As PIAs and BTAs do not fall under the purview of canonical pNPs, this review does not cover into their discovery, biosynthesis, and progress in deciphering their possible biosynthetic origins.51–55 Instead, the reader is directed to excellent review and primary literature articles referenced above.
In this review, we cover the literature published between the years 2010 and 2025, illustrating representative pNPs discovered from marine sponges. We describe the structures of these pNPs, their biological activities, and the biosynthetic routes where known. Finally, we provide a brief description of the chemical and bioinformatic routes that guide pNP discovery and present prospects for pNP discovery from marine sponges and sponge-associated bacteria. The readers are also directed toward other excellent reviews focusing on comprehensive inventory of natural products from sponges and associated microorganisms.56–59
2. pNPs with known biosynthetic origins
2.1. pNPs encoded by the sponge host
PRMPs are present in low abundance in sponge extracts, making it challenging to determine their structures using traditional natural product isolation and spectroscopic/crystallographic methods without sacrificing a large amount of sponge tissue biomass. Guided by the propensity of a peptide chain to fragment at the N-terminus of a Pro residue, a mass spectrometry-based sequencing protocol to determine PRMP sequences using sub-gram amounts of dry sponge biomass was developed.63 Supplemented by biomimetic synthesis of head-to-tail macrocyclic peptides, the analytical workflow reliably differentiated between Leu/Ile residues and rapidly recovered the two-dimensional PRMP topologies; note that the cis/trans isomerization around the prolyl amide bond could not be discerned using this workflow. In addition to known PRMP sequences, numerous additional PRMPs were detected in Stylissa and Axinella sponge extracts allowing us to posit that the chemical space explored by PRMPs could still be transformatively expanded by analytical innovation despite decades of isolation-based studies by the natural products community;63 similar inferences were made for other classes of sponge-derived natural products as well.64–66
2.1.1.1 Biosynthesis of PRMPs. Querying the microbiome architectures led to the identification that phylogenetically disperse low microbial abundance (LMA) sponges as well as high microbial abundance (HMA) sponges harbored PRMPs.63,67–69 To further query the molecular basis for these pNP biosynthesis, Schmidt and coworkers sequenced and assembled draft transcriptomes and draft genomes of Axinella and Stylissa sponge specimens that were prolific sources of PRMPs. These investigations led to the identification of PRMP-encoding genes in the sponge nuclear genomes and established PRMPs as RiPPs and not NRPs (Fig. 2A).22 Multiple PRMP-encoding cassettes were detected to be embedded within a single open reading frame (ORF), with each cassette flanked by conserved amino acid sequences at both the N- and the C-termini, reminiscent of other multi-cassette RiPP precursor peptide architectures (Fig. 2B).70–73 This study also revealed that a vast diversity of PRMPs were encoded within each sponge genome and that the detection and isolation of PRMPs from sponge specimens are likely limited by analytical and compound purification workflows (Fig. 2C).
 | ||
| Fig. 2 Sponge genome encodes the precursors for the biosynthesis of PRMPs. (A) General architecture of sponge PRMP precursor genes in the genome. (B) Proposed biosynthetic route of PRMPs. (C) The core peptides found in the intact precursor peptides from genome assembly. Each different color represents a single precursor peptide. The numbers on the x-axis indicate the counts for each core peptide in the corresponding precursor peptide. Figure based on ref. 22. | ||
The finding that the PRMPs are derived from linear precursor peptides indicates that the head-to-tail macrocyclization is a post-translational modification likely catalyzed by a peptidase similar to plant and microbial peptidases that furnish macrocyclic products.74–80 The peptidase catalyzed macrocyclization to furnish RiPPs (such as PRMPs) is akin to macrocyclic NRP offloading catalyzed by the NRPS TE domains using an intramolecular nucleophile; both reactions proceed via an acyl intermediate generated using a serine side chain hydroxyl or a cysteine side chain thiol and nucleophilic resolution of the acyl intermediate to install the macrocyclizing amide bond (Fig. 2B).30,31 The plausible PRMP-furnishing peptidases in marine sponge proteomes have not been reported. In the section below, we highlight some of the recent description of PRMP structures and bioactivities.
2.1.1.2 Recent advances in PRMP discovery. Marine sponges of the genus Stylissa sponges are well validated to be prolific sources of PRMPs.81 Prior to 2010, Köck had described the PRMPs stylissamides A–D from Stylissa caribica.82 In 2010, the stylissamide family of PRMPs was extended by Köck to include congeners E (1) and F (2) from S. caribica collected in the Bahamas (Fig. 3).83 The planar structures of 1–2 were established spectroscopically and using mass spectrometric fragmentation of the peptide chain. The configurations of the prolyl peptide bonds were established by empirical rules correlating with the differences in the 13C chemical shifts of their β and γ carbons, and all amino acids were determined to be L-amino acids. No bioactivity was reported. The stylissamide family was further extended in 2014 by Molinski with the description of stylissamides G (3) and H (4) from S. caribica collected in the Bahamas (Fig. 3).84 Molecule 4 showed mild cytotoxicity against HCT-116 with EC50 of 5.7 μM, while no bioactivity was assigned to 3. Interestingly, the planar structure of 4 was identical to that of a previously described PRMP euryjanicin A (vide infra) with all amino acids possessing L-configuration.85 The authors had noted conformational equilibria in solution and the structure of euryjanicin A was established using crystallography. It is thus noteworthy that 4 exhibited a cisPro–transPro configuration, while euryjanicin A demonstrated a cisPro–cisPro configuration (Fig. 3). Konno and co-workers have synthesized 4, revisiting its structure using NMR and X-ray diffraction methods.86 They demonstrated that, in solution, synthetic 4 has a cisPro–transPro configuration, but adopts the cisPro–cisPro configuration crystallographically. Therefore, the authors concluded that 4 and euryjanicin A could be identical.
The discoveries of stylissamides I (5), L (6), and X (7) by Gonoi, Mangoni, and Kobayashi have further extended the stylissamide family of PRMPs described from the Stylissa genus; noteworthy is the geographical spread of the stylissamides across the Pacific and the Caribbean (Fig. 3).87–89 Molecule 5 exhibited antifungal activity against Aspergillus niger with IC50 of 4 μg mL−1 while 7 exhibited EGF-induced migration of cultured HeLa cells; no bioactivity has been ascribed to 6.
From the sponge Stylissa massa, another family of PRMPs termed the stylissatins have been reported with the congeners A (8), B (9), C (10), and D (11) by Kigoshi and Lin described with all amino acids in the L-configuration (Fig. 3).90,91 In line with the conformational flexibility of 4 described above, 10 and 11 were isolated as inseparable tautomers, with the three proline residues in either the cis–trans–cis or trans–cis–trans configurations.91 Molecule 11 is the aspartyl methyl ester of 10. Molecule 8 inhibited the production of nitric oxide in LPS-stimulated murine macrophage cells, an activity not previously reported for PRMPs while 9 exhibited moderate inhibitory effects against a panel of human tumor cell lines.90,91 The chemical synthesis of 8 and derivatives has been achieved.92 The planar structures of stylissatins E–G (cyclo(FVPELWP), cyclo(FPWVPLTP), and cyclo(WLPLTPLP), respectively) were discerned using mass spectrometry based fragmentation.63
Yet another family of PRMPs—heptapeptides called carteritins—have been described by Tsukamoto from Stylissa carteri with the structures of congeners A (12) and B (13) deciphered using the usual spectroscopic, fragmentation, and chemical degradation and derivatization procedures (Fig. 3).93 Molecule 12 was cytotoxic against mammalian HeLa, HCT-116, and RAW264 cell lines with IC50 from 0.7 to 1.5 μM. Three new phakellistatin congeners 20–22 (14–16) were isolated by Shin from a tropical sponge Stylissa flabelliformis.94 The same sponge specimen also yielded 12–13, in addition to several other previously reported PRMPs.94 The heptapeptide 14 was isolated as an inseparable mixture of sulfoxide epimers. The decapeptides 15 and 16 were likewise epimers at the methionine side chain sulfoxide (Fig. 3). Molecules 15 and 16 were found to be significantly cytotoxic against several cell lines IC50 ranging from 0.01 to 0.4 μM while 14 was inactive.
Congeners of the family of PRMPs, the phakellistatins, have been isolated from Phakellia sponges.95 There is no underlying chemical distinction between the different PRMP families, and a PRMP with sequence and topology identical to that of phakellistatin 18 was detected in an Axinella sp. sponge specimen, reinforcing a potential lack of sponge species specificity in the PRMPs.63 Proceeding from the description of phakellistatin 1 from Indo-Pacific sponges Phakellia costata and Stylotella aurantium,96 numerous phakellistatin congeners have now been reported.97–106 Of these phakellistatins 15–18 (17–20, Fig. 4) from Phakellia fusca were reported by Lin in 2010. Structures of 17–20 were established using NMR and peptide fragmentation.106 The absolute configurations of their amino acid residues were determined to be L. In DMSO-d6, 18 had two sets of NMR signals in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, corresponding to isomerization around the prolyl amide bond. The Xaa-Pro amide bonds in 17 and 19 were all trans; molecule 20 contains all cis prolyl amide bonds. Molecule 17 demonstrated cytotoxicity against cancer cell line P388 with IC50 of 8.5 μM, while 18 against P388 and BEL-7402 with IC50 of 5.4 and 14.3 μM, respectively. Several phakellistatins feature successive Pro–Pro residues, leading to a considerable synthetic challenge.107
1 ratio, corresponding to isomerization around the prolyl amide bond. The Xaa-Pro amide bonds in 17 and 19 were all trans; molecule 20 contains all cis prolyl amide bonds. Molecule 17 demonstrated cytotoxicity against cancer cell line P388 with IC50 of 8.5 μM, while 18 against P388 and BEL-7402 with IC50 of 5.4 and 14.3 μM, respectively. Several phakellistatins feature successive Pro–Pro residues, leading to a considerable synthetic challenge.107
 | ||
| Fig. 4 Structures of phakellistatins 17–20, fuscasins 21–24, and phakefustatins 25–27. Additional PTMs beyond cyclization are highlighted in red. | ||
Another family of PRMPs, the fuscasins, have been described from P. fusca. The heptapeptides fuscasins A–D (21–24) were isolated by Lin from Phakellia fusca collected in the South China Sea (Fig. 4).108 Even though PRMPs are RiPPs, posttranslational modifications (PTMs) beyond the head-to-tail macrocyclization are few and far between. Based on the key HMBC correlation of Phe-Hα with the γ carbonyl carbon of Asp, a pyrrolidine-2,5-dione fragment formed between the Asp side chain carboxyl and the Phe nitrogen was assigned to 21.108 Another PTM was observed for the PRMPs phakefustatins A–C (25–27) also isolated by Lin from P. fusca wherein a tryptophan side chain was modified to yield a kynurenine residue.109 Molecule 26 was isolated as a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of cis/trans-Pro2 isomers while 25 selectively exhibited cytotoxicity against cancer cell lines.
1 mixture of cis/trans-Pro2 isomers while 25 selectively exhibited cytotoxicity against cancer cell lines.
Proceeding from the isolation of congeners A–D, heptapeptides euryjanicins E–G (28–30) were described by Rodriguez in 2013 from the sponge Prosuberites laughlini collected off the west coast of Puerto Rico (Fig. 5).85,110,111 As described above for euryjanicin A for which the structure was established using X-ray crystallography,85 multiple conformations for 29 and 30 were observed in solution that were only resolved in pyridine-d5.111 Euryjanicins were inactive in a tumor cell line panel. Four heptapeptides reniochalistatins A–D (31–34) and octapeptide reniochalistatin E (35) were isolated by Lin from Reniochalina stalagmitis (Fig. 5).112 Only 35 showed moderate activity against tumor cell lines myeloma RPMI-8226 and gastric MGC-803 with IC50 of 4.9 and 9.7 μM. As before and consistent with their biosynthetic origins, all amino acids were determined to possess the L-configuration.
2.1.2.1 Biosynthesis of CBPs. There is a rich body of literature describing the discovery of CBPs from marine sponges, and their chemical syntheses. Just as for PRMPs, marine sponge-derived CBPs also comprise of proteinogenic L-amino acids with minimal PTMs other than cyclization, in this instance by disulfide bond formation (PTMs observed in sponge-derived CBPs are discussed below). These factors indicated a ribosomal origin of sponge-derived CBPs, akin to other eukaryotic CBPs.19,121 Indeed, by transcriptomic sequencing of the marine sponge Geodia barretti, Cárdenas and coworkers recovered precursor peptide sequences originating from the sponge, and not the microbiome, that encoded CBPs belong to the barrettide family (vide infra).21 In addition to correlating the precursor peptide sequences to previously described chemical structures of barrettide congeners A (36) and B (37),122 Cardenas described the discovery of an additional barrettide congener—barrettide C (38)—by correlating the barrettide sequences unveiled by their transcriptomics data to the mass spectrometry-based profiling of Geodia barretti extract (Fig. 6).21 The architecture of the barrettide precursor sequences was in line with that of canonical RiPP precursor peptides wherein an N-terminal signal peptide is followed by a leader sequence then followed by the cysteine-rich core sequence.21 Peptidases that would conceivably remove the leader sequences from the barrettide cores are not yet known.
2.1.2.2 Recent advances in CBP discovery from marine sponges. As mentioned above, CBPs 36 and 37 were reported by Göransson from G. barretti collected in the Swedish Kosterfjord.122 Only a single amino acid differentiates the two CBPs. Two disulfide bonds—between the side chain thiols of Cys5/Cys23 and Cys7/Cys18—rigidify the barrettide structures. The presence of the disulfide linkages was established by chemical reduction of the pNP and alkylation of the reduced thiols. The topology of the disulfide bond formation and the conformation of the prolyl amide bonds were discerned spectroscopically. The structure of 36 was confirmed by chemical synthesis of the linear peptide and oxidative installation of the disulfide bonds. The authors also established the three-dimensional solution structure of 36 revealing two antiparallel β–sheets each containing five amino acid residues forming a β-hairpin structure, which were braced by two disulfide bonds arranged in a ladder-like mode (Fig. 6). Molecules 36 and 37 did not demonstrate antimicrobial activity but exhibited antifouling activity against barnacle larvae at 0.6 and 6.0 μM, respectively. Guided by the sponge transcriptome mining, the CBP 38 was detected in G. barretti extracts by mass spectrometry and the structure was established using a synthetic linear peptide that was oxidatively folded.21 The three-dimensional structure of 38 was similar to that of 36 (Fig. 6). Yet another congener, barrettide D, was also detected to be present in sponge extract and the CBP topology was inferred based on sequence similarity with 36–38. Additional barrettide sequences were detected in the sponge transcriptomes which would correlate to congeners not yet detected and isolated from G. barretti. All barrettides, known and bioinformatically predicted, contain two disulfide bonds.
The knottins—CBPs containing three or more disulfide bonds—belonging to the family asteropsins have been described by Jung from the Asteropus sp. sponge specimens.123–126 To date, the two-dimensional structures of asteropsins A–F (39–44) have been established. In addition, the three-dimensional structures of all asteropsins except for 42 have been determined using solution NMR spectroscopy, and for 39, also using X-ray crystallography (Fig. 7). Containing more than thirty all L-amino acids, most asteropsins congeners possess an N-terminal pyroglutamate residue and a highly conserved tertiary structure. As would be expected of knottins, all asteropsins exhibited remarkable stability against proteases (trypsin, chymotrypsin, pepsin, elastase) in the gastrointestinal tract and blood plasma.123–126 Molecule 44 was non-hemolytic to human and fish red blood cells.126 Molecule 39 enhanced veratridine-induced Ca2+ influx in murine cerebrocortical neurons, and its heterologous production in bacterial expression systems has been acheived.123,127 A solitary N-terminal pyroglutamate residue containing knottin recifin A (45) has been described from an Axinella sp. sponge that targeted the tyrosyl-DNA phosphodiesterase 1 in an FRET assay with IC50 of 190 nM.128
Theoretically, for a peptide containing four (barrettides) or six (asteropsins) cysteine residues, multiple different oxidative outcomes are possible. However, in the sponge extract, only a single topological isomer is detected. The originally proposed connectivity of three disulfide bonds in the sponge-derived CBPs neopetrosiamides A and B was revised by comparison to chemically synthesized topological isomers.129,130 The topological fidelity of disulfide bond formation in barrettides and asteropsins, and CBPs in general should be considered in context of their three-dimensional structures. Because only one of several theoretically possible outcomes is realized, topology of disulfide bond formation is likely chaperoned by an enzyme active site. Various enzymes catalyze disulfide bond formation. For example, protein disulfide isomerase is a large protein family that can catalyze disulfide formation and isomerization. Peroxiredoxins and glutathione peroxidases are another two widespread protein families that can install disulfide bonds. Quinone and flavin cofactor employing enzymes also oxidize thiols and catalyze disulfide bond formation.131,132
It is conceivable that the chaperoned oxidative formation of the first disulfide bond biases the conformation of the intermediate in a manner that the formation of the subsequent disulfide bonds can proceed in an unguided manner. In such a scenario, an enzyme would be needed to construct only the first disulfide bond with high fidelity.
This plausible scenario was computationally explored for the construction of halichondamide A (46), an 11-residue peptide containing all L-amino acids with two disulfide bonds that was discovered in the extracts of the sponge Halichondria bowerbanki (Fig. 7).133 The structure of 46 was established using spectroscopy, chemical derivatization, and degradation studies. Molecular dynamics simulations revealed that if disulfide bond between Cys3 and Cys10 residues would be installed first (conceivably by a biosynthetic enzyme), then the second disulfide bond between Cys2 and Cys9 residues became highly favorable and could be installed in a non-catalytic manner. The parsimonious biosynthetic scheme presented here is in line with the conservation of disulfide ring topologies of CBPs families such as barrettides and asteropsins described above; a singular dedicated biosynthetic enzyme could construct the entire family of CBP congeners. Molecule 46 was mildly cytotoxic against a panel of cancer cell lines.
The complication of an oxidative cascade is not required for the biosynthesis of CBPs that contain only a single disulfide bond; several such pNPs have been described from marine sponges. Proceeding from the description of the founding member of the gombamide family of CBPs—the heptapeptide gombamide A (47) from sponge Clathria gombawuiensis by Shin—additional congeners gombamides B–D (48–50) were discovered by Proksch from sponge Clathria basilana (Fig. 8).134,135 The gombamides all possess a single disulfide bond, an N-terminal pyroglutamate residue and two contiguous proline residues in the macrocycle. While 47 terminates in a p-hydroxystyrylamide unit which could conceivably arise from the oxidative decarboxylation of a C-terminal tyrosine, 48 and 49 contain E- or Z-2-phenylethen-1-amine (PEA) residues, respectively, at this position analogously derived from a C-terminal phenylalanine. The oxidative decarboxylation of C-terminal amino acids in bacterial RiPP biosynthesis has been reported previously.136–138 The chemical synthesis of 47 has been realized.139,140 Divergent reports for the bioactivity of 47 are available; while the isolated natural product demonstrated moderate cytotoxicity and inhibition of the Na+/K+-ATPase, these effects were not recovered by the synthetic molecule. Similarly, biological activities of several natural PRMPs have not been replicated following their total synthesis.95,134,140
Another family of proline-rich peptides containing a single disulfide linkage among a constellation of all L-amino acids are the chujamides with congeners A (51) and B (52) isolated from sponge Suberites waedoensis by Shin (Fig. 8).141 Both 51 and 52 demonstrated moderate cytotoxicity against A549 and K562 with IC50 from 10.1 to 55.6 μM with 51 inhibiting Na+/K+-ATPase with IC50 of 17.2 μM, reminiscent of 47.
Oxidative decarboxylation of the C-terminal carboxylate is also observed in the sponge-derived single disulfide bridge containing CBPs belonging to the microcionamides family. Proceeding from the description of microcionamides A and B in 2004 that were isolated from the sponge Clathria abietina, congeners C (53) and D (54) were isolated by Ireland, along with the gombamides 48–50, from Clathria basilana collected in Indonesia (Fig. 8).135,142 Unlike gombamides, the microcionamides do not possess the N-terminal pyroglutamate or any proline residues. However, the oxidative decarboxylation of the C-terminal residue is conserved. Molecules 53 and 54 demonstrated antibacterial activity against Staphylococcus aureus and Enterococcus faecium with minimal inhibiting concentration between 6.2 and 12 μM, and induced apoptotic cell death in Jurkat J16 and blocked autophagy in starvation conditions.135
In the section above, the biosynthetic routes and recent new molecular discoveries regarding PRMP and CBP pNPs have been presented. Both classes of molecules are produced by marine sponges and possess rather restrictive set of modifications to the peptide chain with macrocyclization being the principal and conserved chemical transformation to the ribosomally synthesized peptide. A much more expanded set of modifications are displayed by pNPs that are synthesized by the symbiotic microbiome associated with the sponge host. These are discussed in the ensuing section.
2.2. pNPs from sponge symbiotic bacteria
Sponges are a rich repository of pNPs that are produced by bacterial symbionts that maintain a close metabolic and physiological relationship with the sponge host.143 The BGCs encoding production of these pNPs have been recovered by metagenomic sequencing of the entire sponge symbiont followed by bioinformatically enriching the microbial fraction of the reads and mining the assembled metagenome(s) as guided by the chemical logic of pNP biosynthesis. Another route has been the physical enrichment of bacterial symbionts as guided by their distinguishing physiological traits followed by metagenomic sequencing or single cell sequencing of the enriched bacterial fractions followed by retrobiosynthesis-guided genome mining. Both directions have been successfully applied for discovery of pNP encoding BGCs from sponge bacterial symbionts.8,17,144,145 Examples will be discussed below.Polytheonamides represent the most complex pNPs isolated from T. swinhoei with two- and three-dimensional structures established and chemical synthesis realized.149,159–161 Polytheonamides were originally proposed to be NRPs due to the presence of many nonproteinogenic and D-amino acid residues. Instead, Piel and co-workers investigated the metagenome of T. swinhoei, describing a poy BGC demonstrating the biosynthesis of polytheonamides via a RiPP pathway.162 The poy locus contains seven genes that install 48 post-translational modifications, including C- and N-methylation, hydroxylation, dehydration, and amino acid epimerization distributed across 49 residues (Fig. 9).163–166 The enzymatic activities discovered as a part of unraveling the polytheonamides biosynthetic pathway served as leads for genome mining which further enriched the tapestry of RiPP pNPs by prioritizing the exploration of BGCs using synthetic biology workflows.15,167,168
 | ||
| Fig. 9 (A) The poy BGC with the functions of the Poy enzymes listed. (B) Structures of polytheonamides A–B (epimers around the sulfoxide) with the chemical modifications catalyzed by the Poy enzymes annotated. (C) Tertiary structure of an NHLP possessing RiPP precursor peptide (PDB: 8TB1) colored according to the secondary structural elements.169 The region separating the leader from the core is colored in black. | ||
The architecture of the polytheonamide precursor peptide—PoyA—was intriguing. The PoyA leader peptide was uncharacteristically long as compared to other canonical RiPP leader sequences.162,170 Due to its sequence similarity to the sequence of the nitrile hydratase alpha subunits, the PoyAleader belonged to the family of RiPP leader peptides called the nitrile hydratase-like leader peptides (NHLPs) that had been detected bioinformatically.171 The polytheonamides were the first RiPPs that were functionally connected to NHLP precursor peptides. As mentioned above, PoyA-like RiPP precursor peptides were subsequently mined in genetic database leading to the access to cryptic RiPPs.15,167,172,173 The structure of an NHLP has since been determined leading to the appreciation that these leader peptides are akin to proteins rather than peptides wherein they possess rigid tertiary structures leading to new protein/protein interaction modalities underlying RiPP biosynthesis.169,174 Microbiomes of geographically and phylogenetically disperse sponges contain an exceptionally high abundance of a cryptic RiPP-encoding BGC with the precursor peptide adopting the NHLP architecture—the ecological and organismal implications of the conservation of this BGC in sponge microbiomes are as yet not clear.13
In contrast to the polytheonamides that are RiPPs, two families of NRPS-derived NRPs—the theonellamides and the theopalauamides—have been isolated from Theonella sp. sponges. Despite the different names, both families refer to highly similar glycopeptides bridged by a histidinoalanine residue (Fig. 10A).14,146–148,175–177 The founding members for these classes of pNPs, theonellamides A–F and theopalauamide were described prior to 2010.146–148 Since 2010, theonellamides G–K (55–59) and 5-cis-Apoa-theopalauamide (60; Apoa: 3-amino-4-hydroxy-6-methyl-8-phenyl-5E,7E-octadienoic acid) have been described from Theonella swinhoei sponges from different locations by different groups with the structure of 56 established by interpretation of mass spectrometry fragmentation spectra.14,175–177 The absolute configurations of the sugar moieties were determined by Tanaka's method, involving mild acid hydrolysis and derivatization.178 Some of these NRPs possess cytotoxic and antifungal activities, possibly because they bind membrane sterols. In fungi, this induces glucan overproduction and activates Rho1-mediated 1,3-β-D-glucan synthesis.179,180
Using single cell sequencing, Piel and co-workers have described the hybrid non-ribosomal peptide synthetase/polyketide synthase (NRPS/PKS) encoding tna BGC from the Theonella symbiont Candidatus Entotheonella serta that is responsible for the production of theonellamides (Fig. 10B).14 The NRPS-PKS architecture was rationalized to fit the assembly line biosynthetic pathway furnishing the theonellamide aglycone with a candidate glycosyl transferase enzyme identified to be encoded within the tna BGC. Crucially, as rationalized from the adenylation domain specificities encoded within the tna BGC, the histidinoalanine ring was proposed to be constructed using histidine and serine amino acid side chains. This transformation would require dehydration of the serine side chain to yield an α/β unsaturated dehydroalanine residue and a stereospecific Michael-type addition of the histidine side chain imidazole nitrogen to the β-carbon of the dehydroalanine residue reminiscent of the lanthionine ring formation in lanthipeptides. Other enzymatic transformations have been realized for the formation of macrocyclic pNPs containing histidine side chain cross links.181 As for theonellamides, a candidate enzyme with similarity to kinases and tetratricopeptide repeat domains was identified in the tna BGC and proposed to be involved in the formation of the histidinoalanine cross link; this proposal remains to be experimentally verified.14
In addition to polytheonamides, theonellamides, and theopalauamides, Theonella sponges have been a rich repository of other pNPs, most of which are NRPS-derived NRPs or peptide/polyketide hybrids. These include the keramamides,150,182–184 cyclotheonamides,152 nazumamide A,153 and konbamides (Fig. 11). Putative BGCs—ker BGC, cth BGC, naz BGC, and the kon BGC, respectively—that were collinear with the assembly line biosynthetic logic of these NRPs were detected in Entotheonella symbionts as well.157 The reader is directed to some excellent review articles on this topic that detail the isolation, chemical syntheses, and bioactivities of these marine natural products.185–187 The natural product biosynthetic potential of Entotheonella symbionts extends beyond pNPs to include structurally elaborate polyketide natural products. While a discussion of polyketide natural products is outside the scope of this review, the reader is directed to the primary literature ref. 157, 188 and 189.
In the sponge Discodermia calyx, a Ca. Entotheonella sp. symbiont was shown to be the producer of the polyketide natural product calyculin A diphosphate.194 The previously described natural product, calyculin A, was furnished upon dephosphorylation of calyculin A diphosphate in response to sponge tissue disruption.195 Following the identification of the calyculin A diphosphate encoding BGC, Wakimoto, Abe and coworkers detected the presence of kasumigamide (64) encoding kas BGC in the same D. calyx symbiont Ca. Entotheonella sp. which directed the isolation and structure elucidation of 64 from marine sponges; 64 had previously been described from marine cyanobacterium Microcystis aeruginosa NIES-87 (Fig. 13A and B).196–198 The makas BGC for the production of 64 in M. aeruginosa NIES-87 was subsequently identified.196,199
 | ||
| Fig. 13 (A) Structure of 64. (B) The assembly line biosynthesis of 64 as predicted by the organization of the kas BGC. (C) Structures of 65 and 66 and similarity to keramamide F. | ||
Another intriguing case of Ca. Entotheonella sp.-derived pNPs in D. calyx sponges are the N-formylated macrolactams calyxamides A and B (65–66) isolated by Abe that demonstrated cytotoxicity against the murine leukemia cell line P388 with IC50 of 3.9 and 0.9 μM, respectively.200 The structures as well as absolute configurations were determined by spectroscopic data and chemical degradation experiments. Structurally, 65 and 66 are a pair of diastereomers differing only in the configuration of the isoleucine residue (Fig. 13C). The calyxamides share several structural features with the keramamides, and as mentioned previously, the ker BGC encoding production of the keramamides has been detected in Ca. Entotheonella symbionts in Theonella sponges.157 Indeed, microscopic analysis and 16S rRNA encoding gene amplification established the presence of Ca. Entotheonella symbionts in the calyxamide harboring D. calyx sponge specimens.200 It is thus likely that a ker-like BGC remains to be identified in the D. calyx Enthotheonella symbionts. Prior to the studies describing the isolation of pNPs 64–66 from D. calyx, the presence of Entotheonella symbionts in the sponge Discodermia dissoluta had been noted.201
There are numerous other families of pNPs isolated from Theonella, Discodermia, and other sponge species for which the biosynthetic routes have not been identified to date. These molecules will be highlighted in Section 3. It is likely that metagenomic workflows and retrobiosynthesis-guided genome mining workflows will connect these natural products to the corresponding BGCs in sponge holobionts.
2.3. pNPs encoded by commensal bacteria
In contrast to symbiotic bacteria that are tightly interwoven into the metabolism and physiology of their sponge hosts and are thusly onerous to culture in the laboratory, loosely associated commensal bacteria are amenable for culturing. Recent reports have established that sponge-derived commensal bacteria are likewise sources of novel pNPs. Some of the recent work in this space will be highlighted below.Aquimarins A–H (67–74) are peptide/polyketide hybrid natural products isolated by Piel from a sponge-derived bacterium Aquimarina sp. Aq135.202,203 Of these, 67–70 and 73–74 are dodecapeptides with a diketide 3-hydroxy-4-methyl pentanoic acid moiety at the N-terminus and a primary amine substituting the C-terminal carboxylate with some congeners possessing chlorinated aliphatic amino acid side chains. Molecules 71 and 72 are truncated analogs. The structures of the molecules were established using spectroscopy, fragmentation, and degradation experiments (Fig. 14A).204 While the molecules were identified guided by their activity against Mycobacterium tuberculosis, they were found to possess antibiotic activity against a range of human pathogens such as methicillin-resistant Staphylococcus aureus with MIC ranging from 2 to 8 μM. Genome sequencing and mining of the aquimarin producer strain led to the identification of the hybrid NRPS/PKS encoding aqm BGC. The Aqm assembly line consisted of a bimodular PKS followed by twelve NRPS modules containing three epimerase (E) domains, which was in conformity with the presence of three D-amino acids in the peptide chain. Aliphatic chlorination was experimentally demonstrated to be catalyzed by the FeII/α-ketoglutarate-dependent halogenase AqmA.204 The chemical syntheses of 69 and 70, and various derivatives was realized.202 Structure activity relationship studies revealed that the primary amine at the peptide C-terminus was critical for activity; the primary amine was conceivably installed by the reductive offloading of the peptide chain from the NRPS assembly line by the terminal thioester reductase domain of the AqmG NRPS followed by reduction by the NAD-dependent aminotransferase AqmH (Fig. 14B).30
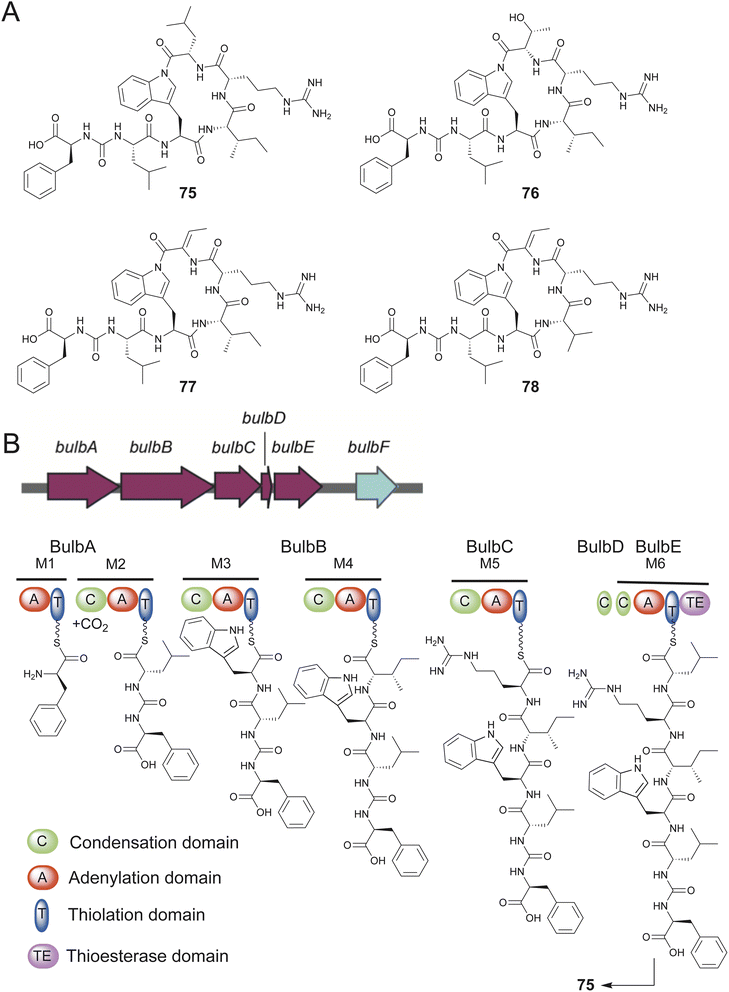
Obligate marine Proteobacteria of the genus Microbulbifer are commonly cultured from sponges and other marine invertebrates and are best known for their proclivity to degrade polysaccharides.205,206 Concomitant with Igarashi, Agarwal reported the discovery of ureido hexapeptidic bulbiferamides A–D (75–78) from sponge- and coral-derived Microbulbifer strains.207,208 The natural product structures were established using standard spectroscopy and degradation experiments with all amino acids being in the L-configuration. Genome mining led to the identification of the bulb BGC encoding a hexamodular NRPS consistent with the bulbiferamide structures (Fig. 15). The presence of the bulb BGC was widespread among Microbulbifer strains.207 In light of previous studies, the ureido bond in bulbiferamides was postulated to the installed by the C domain of the initiating NRPS module.209
An intriguing structural feature among bulbiferamides was the presence of the macrocyclizing amide bond generated using the tryptophan side chain indole nitrogen. While lysine and ornithine side chain primary amines are commonly observed to participate in macrocyclizing amide bond formation in cyanobacterial ureidopeptide natural products, the bulbiferamides were the first such examples of bacterial natural products possessing a macrocyclizing indolylamide.210–212 The terminal TE domain of the BulbE NRPS was experimentally characterized to install the indolylamide bond, making it a unique biocatalytic transformation that potentially complements the synthetically onerous installation of late-stage indolylamides owing to the poor nucleophilicity of the indolic nitrogen.213,214 The enzymatic installation of macrocyclizing indolylamides in RiPPs has recently been reported.215
The mass spectrometric fragmentation pattern associated with the ureido bond was observed for a different set of pNPs from the marine sponge-derived bacterium Microbulbifer sp. MKSA007; this strain also produced bulbiferamides. Isolation of these pNPs followed by spectroscopic and degradation experiments established the structures of heptapeptidic pseudobulbiferamides A–E (79–83) which likewise bear a ureido bond at the N-terminus (Fig. 16).216 The pseudobulbiferamides were highly similar to the heptapeptidic pseudovibriamides A congeners (84–89) that were isolated from the bacterium Pseudovibrio brasiliensis strain Ab134 that was cultivated from the marine sponge Arenosclera brasiliensis.217,218 In addition to pseudovibriamides A congeners, the P. brasiliensis strain Ab134 also produced further elaborated hybrid peptide/polyketide pseudovibriamides B congeners 90–95; pseudovibriamides B analogs were not detected in Microbulbifer sp. MKSA007 extracts. Genome sequencing enabled identification of the ppp BGC in P. brasiliensis strain Ab134 encoding production of pseudovibriamides and the mbp BGC in Microbulbifer sp. MKSA007 encoding production of pseudobulbiferamides.216,217 Curiously, the architecture of the ppp and the mbp BGCs was highly similar, and both BGCs were positioned on plasmids raising the possibility that the genetic potential for making these ureidopeptidic pNPs might be broadly distributed among marine invertebrate associated commensal microbiomes. The pseudovibriamides were also shown to be fundamentally important in modulating the flagellar motility and biofilm formation in Pseudovibrio bacteria.219
Other pNPs sourced from sponge-derived bacteria include the N-terminal pyroglutamate containing proline-rich linear peptides 96 and 97 isolated from Microbacterium sp. V1 which was cultured from the sponge Petrosia ficiformis (Fig. 17).220 The macrocyclic hexapeptide nocardiotide A (98) containing all L-amino acids was isolated from Nocardiopsis sp. UR67 which was cultivated from the sponge Callyspongia sp. and demonstrated cytotoxicity against a panel of human cancer cell lines of MM. 1S multiple myeloma, HeLa cervix carcinoma, and murine CT26 colon carcinoma with IC50 of 8, 11, and 12 μM mL−1, respectively.221 The macrocyclic heptadepsipeptide xanthostatin B (99) containing a D-valine residue was isolated by Zhang from Streptomyces sp. SCSIO 40046 and demonstrated α-glucosidase inhibitory activity.222
3. pNPs with as yet unknown biosynthetic origins
In the sections below, macrocyclic and linear pNPs for which biosynthetic genes, enzymes, and transformations have not been identified are described. While sponge-derived PIAs and BTAs are dipeptidic in nature, they defy characterization as either macrocyclic or linear pNPs; they resemble and can be thought of as alkaloids, rather than canonical pNPs. The reader is directed to some excellent reviews that describe the discovery, synthesis, and pharmacology of PIAs and BTAs.33–36,38,40 While the biosynthesis basis for these classes of sponge-derived natural products are as yet unknown, recent metabolomic investigations which build upon prior labeled precursor feeding studies point toward the identities of the core amino acid building blocks.3.1. Macrocyclic pNPs
Several instances in which the bacterial Entotheonella sp. symbionts were responsible for synthesizing pNPs derived from Theonella sponges have been described in the previous section. It is thus instructive to note that several other classes of pNPs have been described from Theonella sponges that have yet to be connected to their biosynthetic origins and the corresponding BGCs identified. These are likely targets for future genomic investigations.Theonellapeptolides are a group of cyclic tridecadepsipeptides characterized by the presence of N-methyl amino acids, D-amino acids, and β-amino acids mainly isolated from T. swinhoei with more than 15 congeners reported to date (Fig. 18).223,224 Theonellapeptolides show various biological activities, such as cytotoxicity and inhibition of Na+ and K+ ion transport. Recently, pNPs sulfinyltheonellapeptolide (100) and theonellapeptolide If (101) from T. swinhoei were reported with structures determined using spectroscopy, mass spectrometry, and chemical degradation experiments (Fig. 18).225 These compounds significantly reduced proliferation rate of HepG2 cells to 30% and 50% at the dose of 10 μM. A further three analogues theonellapeptolides IIb (102), IIa (103), and IIc (104) were isolated from T. swinhoei.226 As is characteristic of theonellapeptolides, these recently reported congeners contained multiple amino acids with non-polar aliphatic side chains.
Two recent reports by Carmeli and Carroll described the isolation and structures of the pNPs cyclotheonellazoles A–C (105–107) and cyclotheonellazoles D–I (108–113) from Theonella sponges with the chemical synthesis of 105 now realized (Fig. 19).227–229 The cyclotheonellazoles structures resemble the pNP oriamide described from Theonella sponges and the calyxamides described from Discodermia sponges, and are likely to be biosynthesized by a Entotheonella-derived NRPS assembly line.200,230 Structural correspondence between cyclotheonellazoles and calyxamides extends to jamaicensamide A (114) isolated by Molinski from sponge Plakina jamaicensis.231 Molecules 105–107 inhibited chymotrypsin with IC50 of 0.62, 2.8, and 2.3 nM, and elastase with IC50 of 0.034, 0.10, and 0.099 nM. Molecules 108–113 displayed inhibition of elastase with IC50 values ranging from 16.0 to 61.8 nM. The cyclotheonellazoles were found to be potent protease inhibitors, including the SARS-CoV-2 3-chymotrypsin-like protease. The recurrent theme of pNPs being shared among Theonella and Discodermia sponges was also realized for the pNP cyclolithistide A (115). Molecule 115 was originally isolated by Crews from T. swinhoei and its structure was revised by Abe upon isolation from D. japonica with the configuration of the chloroisoleucine residue in the antifungal cyclodepsipeptide determined by synthesis of chemical standards and comparisons thereof.232–234 The presence of a chlorinated pyrrole residue was observed in gunungamide A (116), a macrocyclic depsipeptide isolated from Discodermia sp.235 Solomonamide A (117) and solomonamide B (118) are cyclic tetrapeptides isolated from T. swinhoei (Fig. 19).236 The solomonamide structures were revised upon chemical syntheses which also furnished derivatives to explore their anti-inflammatory activities.237–243
 | ||
| Fig. 19 Cyclotheonellazoles 105–113, jamaicensamide A 114, cyclolithistide A 115, gunungamide A 116, and solomonamides 117 and 118. | ||
Progressing from the description of neamphamide A, the depsipeptides neamphamides B (119), C (120), and D (121) were described from sponge Neamphius huxleyi by Kobayashi and Quinn (Fig. 20).244–247 The neamphamides were reported to possess myriad bioactivities which include protective activity against viral infection, anti-mycobacterial activity, and cytotoxicity against human cell lines. Another class of macrocyclic depsipeptides, stellatolides A–G (122–128) were isolated from the sponge Ecionemia acervus by Cuevas (Fig. 20).248 The chemical synthesis of 122 was realized, and most stellatolides were found to be cytotoxic to A549, HT-29, and MDA-MB-231 human tumor cell lines with GI50 from 0.08 to 2.7 μM except for molecules 127 and 128 that were found to be much less potent.248 Among other structural features, the neamphamides and the stellatolides both possess the β-methoxy tyrosine residue in the peptide macrocycle. The cytotoxic stellatolide H (129) was described by Matsunaga from Discodermia sp. sponge.249 The cyclodepsipeptides pipecolidepsins A (130) and B (131) were described from Homophymia lamellose with the chemical synthesis of 130 realized.250,251 Molecules 130 and 131 displayed cytotoxic against a panel of human tumor cell lines with GI50 from 0.01 to 1.12 μM.251 The depsipeptide structures point toward a hybrid NRPS/PKS-involving biosynthetic route that could be encoded within a bacterial symbiont. The biosynthesis of non-proteinogenic amino acid building blocks constituting these depsipeptides, such as the pipecolic residue in 130 and 131, are well described in bacterial natural product biosynthetic schema.252,253 The cyclic sulfated depsipeptide mutremdamide A (132) was isolated by Bewley from Theonella sp. sponges (Fig. 20).254
Lipodiscamides A–C (133–135) are cyclic lipodepsipeptides bearing a dilactone macrocycle described isolated by Abe from the sponge Discodermia kiiensis (Fig. 21).255 The peptidic macrocycle of lipodiscamides constitutes various non-proteinogenic amino acids such as a dehydronorvaline, ureidoalanine, and citrulline. The citrulline side chain was found to be sulfated in the sulfolipodiscamides A–C (136–138) that were isolated from the same sponge.256 Molecules 133–135 demonstrated cytotoxicity against mammalian HeLa cell line with IC50 of 18, 26, and 46 μM, and against P388 with IC50 of 23, 20, and 31 μM, respectively, while molecules 136–138 exhibited cytotoxicity against P388 with IC50 of 15, 30, and 29 μM, respectively.
Progressing from Faulkner's description of aciculitin congeners A–C from sponge Aciculites orientalis, aciculitin D (139) was described by Matsunaga from the deep-sea sponge Poecillastra sp. and found to possess cytotoxic bioactivity against HeLa and HCT-116 cancer cells with IC50 of 4.5 and 1.4 μM, respectively.257,258 It is tantalizing to propose that the aciculitin peptidic macrocycle to which the glycosylated lipid moiety is appended to arises from a NRPS assembly line. However, a ribosomal route to the peptidic macrocycle can also be envisaged with a bi-radical coupling between the histidine and epimerized tyrosine residue side chains, as have been observed in other RiPP biosynthetic schemes, leading to the formation of the coupled biaryl bridge (Fig. 22).181,259–261 Differences in configurational assignments between aciculitins A–C and 139 suggest that the amino acid configurations should be reinvestigated.
Predicting a plausible biosynthetic route is difficult for the callyaerins A–M (140–152), cyclic peptides isolated by Proksch from sponge Callyspongia aerizusa (Fig. 23).262–264 The chemical synthesis of 140 has been described by Brimble.265 While all amino acids in callyaerins are proteinogenic amino acids or derivatives therein that are well represented among RiPPs, an assembly line NRPS-mediated construction cannot be ruled out. The biosynthesis of the macrocyclizing aminoacrylamide moiety is an enzymological novelty. The callyaerins demonstrate antitubercular activity by targeting a membrane protein that causes dysregulation of various primary biological processes in the pathogen. Specifically, molecules 140 and 141 exhibited the most potent activity with MIC90 of 2 and 5 μM.266
Natural product chemical scaffolds are often shared among marine bacteria and marine sponges with the biosynthesis of several of these sponge-derived natural products attributed to the symbiotic bacteria residing within the sponge holobiont community.12 As mentioned previously, marine cyanobacteria are prolific producers of ureidopeptidic molecules that are macrocyclized by the terminal primary amine of lysine or ornithine side chains.210–212 From marine sponge Siliquariaspongia mirabilis, Bewley described isolation, structure determination, and chemical synthesis of the macrocyclic ureidopeptide namalide (153) (Fig. 24).267 Molecule 153 demonstrated inhibition to carboxypeptidase A with IC50 of 250 nM. Subsequently, namalides B (154) and C (155) were isolated from the freshwater cyanobacterium Sphaerospermopsis torques-reginae.268 The namalides were also detected to be produced by cyanobacterium Nostoc sp. CENA543 along with the macrocyclic ureidopeptides of the anabaenopeptin family.210 These observations perhaps point toward a cyanobacterial symbiont being the possible primary producer of 153 in the sponge S. mirabilis. Numerous pNPs isolated from marine sponges have been connected to symbiotic cyanobacterial producers using genomics as well as microscopy and mass spectrometry-based techniques.64,269 Alternatively, akin to konbamide biosynthesis, an Entotheonella symbiont could encode production of namalides.157,270 These cross-organismal biosynthesis has been observed for swinholides that were isolated from Theonella sponges and from marine cyanobacteria, and BGCs for their production have now been detected in terrestrial cyanobacteria as well.271–273 Along the same lines, homophymamide A (156) was isolated by Matsunaga from the Homophymia sp. marine sponge and its chemical synthesis realized (Fig. 24).274
3.2. Linear pNPs
Amide bond N-methylation can alter the structure and physiochemical properties of peptides, rendering them resistant to proteolysis and improving upon their bioavailability. Several highly N-methylated linear peptides (HNMLPs) have been reported from marine sponges with as yet unknown biosynthetic origins. Note that iterative N-methylation in the context of fungal RiPPs is well established.275–277Numerous HNMLPs from marine cyanobacteria have been described. Recently, the linear octapeptide pembamide (157) containing all L-amino acids was described by Fernández from the sponge Cribrochalina sp. which demonstrated cytotoxicity against cancer cell lines of MDA-MB-231, HT-29, and A549 with GI50 of 3.35, 3.80, and 2.46 μM, respectively.278 From the Antartic sponge Inflatella coelosphaeroides, Baker and co-workers described the isolation and structure of friomaramide A (158) which demonstrated inhibition of liver-stage Plasmodium falciparum parasite development.279 A second friomaramide congener, friomaramide B (159), was also described by Baker from I. coelosphaeroides along with six new HNLMPs termed shagamides A–F (160–165) which inhibited different activities against three blood-stage P. falciparum strains, in which molecules 159, 160, and 162 are the most potent ones with IC50 values ranging from 1.07 to 8.65 μM.280 The friomaramides possess a C-terminal tryptenamine moiety that is absent in shagamides (Fig. 25).
The linear tridecapeptides yaku'amides A (166) and B (167) were isolated from a deep-sea sponge Ceratopsion sp. by Matsunaga and coworkers.281 Their structures were initially elucidated by analysis of NMR spectroscopic data and chemical degradation, with complete assignment achieved by total synthesis by Inoue and coworkers.282,283 Among the several non-proteinogenic amino acids comprising the yaku'amides are residues bearing α,β-unsaturated as well as β-hydroxylated side chains (Fig. 26). Molecules 166 and 167 exhibited cell-growth inhibitory against P388 cancer cell line. Molecule 167 was further tested against a wide panel of human cancer lines and the molecular target was identified to be the mitochondrial FoF1-ATP synthase.284 Recent efforts have been aimed at streamlining synthetic schemes and furnishing derivatives for biological evaluation.285–287
The linear heptapeptide bearing all L-amino acids, subarmigeride A (168), was isolated from Callyspongia subarmigera.288 Several analogs of 168 were detected to be present in the sponge extract and a cyanobacterial symbiont was proposed to be the producer of the pNP, though that remains to be validated using genomic and biochemical investigations.
4. Future directions in sponge pNP discovery
The already extensive inventory of natural products sourced from marine sponges continues to expand.289 Among these, pNPs have always attracted attention due to their structural complexity and biological activities (vide supra). Sponges are also among the most abundant sources of cultivated marine bacteria described in natural products manuscripts, leading to the discovery of many pNPs. This amazing, continued abundance promises many future discoveries. Perhaps more strikingly, metagenomic methods indicate that this is just the beginning. Ongoing research continues to uncover the vast microbial dark matter present in sponge metagenomes, revealing a largely untapped biosynthetic potential.290 For example, Loureiro et al. performed metagenomic analysis on four sponge specimens spanning three species and identified 5082 BGCs, with NRPS and RiPP-related BGCs accounting for approximately 30% of the total.291 And these are just from bacterial symbionts; relatively little is known about the biosynthetic potential encoded in host sponge genomes, or about the metabolic potential of non-bacterial sponge symbionts such as dinoflagellates (as speculated for halichondrin biosynthesis), archaea, and many other branches of life.292,293 Limited data from sponge genomes and transcriptomes also reveals a high potential for undiscovered chemical novelty, with a single sponge potentially encoding 22 cyclic PRMPs, including several new compounds that were then detected by mass spectrometry.22 In addition, the same sponge had more than 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 transcripts encoding secretion signals, reinforcing how much completely unknown chemical diversity is out there.
000 transcripts encoding secretion signals, reinforcing how much completely unknown chemical diversity is out there.
Despite the wealth of potential new compounds, the field faces at least three significant limiting factors: first, as found throughout natural products research, the newly described pNPs largely fall into known structural groups previously identified in sponges. In short, structural novelty is much harder to find than in the recent past.294 Second, although thousands of new biosynthetic gene clusters (BGCs) have been discovered in sponge metagenomes, the majority of corresponding molecules have not been accessed in the sponge metabolomes or by synthetic biology approaches.295 It has remained quite hard (with no published examples despite significant effort) to express NRPS biosynthetic pathways from the uncultivated microbial pool in sponges. Third, it is still common that few of the sponge-derived pNPs are sourced in the significant amounts needed to fully validate their biological activities or to take the needed preclinical steps to advance therapeutic candidates to application. Below, we describe some important considerations and future developments to enhance discovery and development of sponge pNPs.
As a cautionary note, it is important to mention that it has been popular to infer the origin of metabolites based upon cellular localization in host or symbiont, or by structural homology. However, in the modern era, it is important to take an agnostic approach that considers that either, both, or even multiple symbionts might be responsible for making the same compound.17 Such an approach simultaneously employs the chemical, metagenomic, and transcriptomic methods described below.
4.1. Chemical approaches
Traditional approaches, and innovative modifications to those approaches remain the primary tools for discovery. In the initial decades of the field, new compounds were identified from easily accessible sponges found in the shallow water, by snorkeling, or SCUBA. By the late 1980s or early 1990s, the rate of rediscovery was rapidly increasing. Collection of sponges began to move to difficult-to-reach locales, deep waters (often via dredging or submarine collection), or harsher environments such as polar regions.296,297 An increased emphasis was placed on cryptic biodiversity (of both host sponges and microbial symbionts), as biodiversity and chemical diversity are often coupled.298Sponge extracts have traditionally been investigated via bioassay-guided fractionation, chemistry-directed methods, or a combination of the two.299 Most of the papers cited in this review used these approaches, demonstrating their continued importance to the field. Reinvestigation of sponges using unique bioassays has been a powerful method of discovery. A limitation to that approach can be that sponges are often chemically defended by major, abundant compounds that can mask the bioactivities of less abundant metabolites, specially as it related to antibiosis bioactivities. Pre-fractionation has been applied to mitigate the problem. This approach is particularly a noteworthy feature of the National Cancer Institute compound library, which has thousands of sponge extracts and fraction libraries, in addition to efforts at other institutes.300–302
Chemistry-directed methods have evolved significantly over the years, with the most recent trend focusing on widespread adoption of mass spectrometry-based metabolomic mining. These methods have continued to increase in importance especially for pNPs that demonstrate highly ordered fragmentation patterns.303 For instance, the unique fragmentation patterns of the prolyl amide bond and the ureidopeptide bond guided the discovery of novel PRMPs in sponges and bulbiferamides and pseudobulbiferamides in commensal bacteria, respectively.22,63,207,216
4.2. Genomic approaches
Recently, Nguyen et al. analyzed 13 sponge metagenomes and identified a well-conserved and widely distributed RiPP BGC in marine sponge microbiomes.13,16 This BGC encodes a broad-spectrum regiospecific peptidyl halogenase, an enzyme responsible for site-specific halogenation of peptide substrates which provided a valuable biotechnological tool for peptide diversification.304,305 The study found that this BGC was present in all queried sponges with high microbial diversity, emphasizing its ecological and functional importance. By applying genome mining and metagenomic sequencing, they established a systematic approach to uncovering cryptic BGCs and discovering new peptide compounds and enzymes from marine sponge metagenomes. This is particularly significant because the integration of metagenomic mining with synthetic biology enables the expression and engineering of BGCs in heterologous hosts. This powerful combination opens new avenues for the discovery and production of new-to-nature pNPs with potential therapeutic value.
In some cases, these types of methods can also be applied to NRPS-derived pNPs and not just to RiPPs. Although not from sponges, Brady and co-workers pioneered the application of direct synthesis of predicted NRPS-derived peptides found in genomes.306 In sponges, there is the unique opportunity to directly query whether those compounds are present, using mass spectrometry in tandem with (bio)synthetic standards.
The above is an example of an ideal case. It is still not trivial to identify sponge-derived pNP transcripts without prior knowledge of the sequence from proteomics or chemical approaches. Often, the high complexity of sponge transcriptomes necessitates additional informatics methods. For example, in the case of discovering PRMP genes from sponges, 79 possible precursor peptides were discovered using BLAST. To determine which peptide(s) was likely to be the correct one, unique characteristics of bioactive peptides were queried.22 Usually, in such peptides, the sequences encoding the pNPs (core peptides) are hypervariable and not sequence conserved, but parts of the precursor peptide that are proteolyzed and discarded (recognition sequences and signal peptides) are highly conserved and often highly repetitive. Such sequences are discoverable using specialized tools such as MEME-ChiP, which was applied to discover the PRMP transcripts.307 Subsequently, many new and known PRMP genes were found. The resulting products are readily amenable to chemical synthesis.
Lack of gene co-localization is another challenge for host-derived pNPs. Although animals often contain true animal-derived BGCs,308 peptides such as the PRMP precursors are so far found distributed across the sponge chromosomes and not co-localized with genes encoding modifying enzymes.22 In better-studied systems, such as cone snails, transcriptomic discovery is aided by a good understanding of prepropeptide regions. Some types of host-derived bioactive peptides are also recognizable because of their döppelganger nature, in which their sequences converge with important hormones found in their prey organisms.116,309 These and other transcriptomic analysis tools provide virtually untapped opportunities for discovery and synthesis of novel host-derived peptides. It should also be noted that sponge genomes encode many NRPSs,310 as well as other host-encoded BGCs, which are relatively unexplored.
5. Conclusion and prospects
All components of the sponge holobiont are prolific sources for pNP discovery. Though widely proposed associated microorganisms are the real origin of many pNPs isolated from sponges, the animal hosts indeed can produce pNPs themselves. However, biosynthesis study of sponge animals is still in its infancy with the genetic potential in sponge genomes needs to be unlocked and complemented with molecular and chemical tool development. Furthermore, with principals well established from decades of investigations from microbial systems, currently, most biosynthetic investigations for sponge-derived pNPs are focused on NRPs and RiPPs while other chemical classes of sponge-derived pNPs, such as the PIAs and BTAs have received lesser attention. It is foreseeable that with rapid evolvements in (meta)genome sequencing, bioinformatics, molecular biology, and synthetic biology, biosynthesis of all types of pNPs and alkaloids from the three components of the sponge holobiont will garner considerable attention and grow significantly. As modern genome mining-based, metabolomics-guided, and traditional bioassay-guided strategies are increasingly more accessible and continue to be refined, it is undoubtable that the sponge holobionts would continue to be cornucopias of pNP discovery. With more chemical entities in hand, not only their biological activities but also their ecological roles will need to be evaluated. We believe mining sponge derived pNPs and alkaloids and investigating their biosynthesis will provide perspective drug leads, new enzymatic and synthetic biology tools, and development in ecological protection.96. Conflicts of interest
The authors declare no conflicts of interest.7. Data availability
No primary research results, software or code have been included and no new data were generated or analyzed as part of this review.8. Acknowledgements
The authors acknowledge funding support from the National Institutes of Health (R35GM142882 to V. A. and R35GM148283 to E. W. S.) and the National Science Foundation (CHE-2238650 to V. A.).9. References
- R. W. M. Van Soest, N. Boury-Esnault, J. Vacelet, M. Dohrmann, D. Erpenbeck, N. J. De Voogd, N. Santodomingo, B. Vanhoorne, M. Kelly and J. N. A. Hooper, Global Diversity of Sponges (Porifera), PLoS One, 2012, 7, e35105 CrossRef CAS PubMed.
- N. S. Webster and T. Thomas, The sponge hologenome, mBio, 2016, 7, 2 CrossRef.
- T. R. Thomas, D. P. Kavlekar and P. A. LokaBharathi, Marine drugs from sponge-microbe association–a review, Mar. Drugs, 2010, 8, 1417–1468 CrossRef CAS.
- J. J. Bell, The functional roles of marine sponges, Estuarine, Coastal Shelf Sci., 2008, 79, 341–353 CrossRef.
- J. T. Hestetun, G. Tompkins-Macdonald and H. T. Rapp, A review of carnivorous sponges (Porifera: Cladorhizidae) from the Boreal North Atlantic and Arctic, Zool. J. Linn. Soc., 2017, 181, 1–69 CrossRef.
- M. Ekins and N. G. Wilson, New carnivorous sponges (Porifera: Cladorhizidae) from Western Australia, collected by a Remotely Operated Vehicle (ROV), Sci. Rep., 2024, 14, 22173 CrossRef CAS PubMed.
- L. Pita, L. Rix, B. M. Slaby, A. Franke and U. Hentschel, The sponge holobiont in a changing ocean: from microbes to ecosystems, Microbiome, 2018, 6, 46 CrossRef CAS.
- V. J. Paul, C. J. Freeman and V. Agarwal, Chemical Ecology of Marine Sponges: New Opportunities through “-Omics”, Integr. Comp. Biol., 2019, 59, 765–776 CrossRef CAS PubMed.
- B. S. Moore and D. J. Newman, The Extraordinary Benefit of Nature’s Chemistry to Health, Society, and the Economy, J. Nat. Prod., 2025, 88, 1541–1548 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS.
- M. Muttenthaler, G. F. King, D. J. Adams and P. F. Alewood, Trends in peptide drug discovery, Nat. Rev. Drug Discovery, 2021, 20, 309–325 CrossRef CAS.
- E. P. McCauley, I. C. Piña, A. D. Thompson, K. Bashir, M. Weinberg, S. L. Kurz and P. Crews, Highlights of marine natural products having parallel scaffolds found from marine-derived bacteria, sponges, and tunicates, J. Antibiot., 2020, 73, 504–525 CrossRef CAS PubMed.
- N. A. Nguyen, Z. Lin, I. Mohanty, N. Garg, E. W. Schmidt and V. Agarwal, An obligate peptidyl brominase underlies the discovery of highly distributed biosynthetic gene clusters in marine sponge microbiomes, J. Am. Chem. Soc., 2021, 143, 10221–10231 CrossRef CAS.
- T. Mori, J. K. B. Cahn, M. C. Wilson, R. A. Meoded, V. Wiebach, A. F. C. Martinez, E. J. N. Helfrich, A. Albersmeier, D. Wibberg, S. Dätwyler, R. Keren, A. Lavy, C. Rückert, M. Ilan, J. Kalinowski, S. Matsunaga, H. Takeyama and J. Piel, Single-bacterial genomics validates rich and varied specialized metabolism of uncultivated Entotheonella sponge symbionts, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 1718–1723 CrossRef CAS.
- M. Rust, E. J. N. Helfrich, M. F. Freeman, P. Nanudorn, C. M. Field, C. Rückert, T. Kündig, M. J. Page, V. L. Webb, J. Kalinowski, S. Sunagawa and J. Piel, A multiproducer microbiome generates chemical diversity in the marine sponge Mycale hentscheli, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 9508 CrossRef CAS.
- V. Nowak Vincent, P. Hou and G. Owen Jeremy, Microbial communities associated with marine sponges from diverse geographic locations harbor biosynthetic novelty, Appl. Environ. Microbiol., 2024, 90, e00726–24 Search PubMed.
- M. Morita and E. W. Schmidt, Parallel lives of symbionts and hosts: chemical mutualism in marine animals, Nat. Prod. Rep., 2018, 35, 357–378 RSC.
- S. S. Pineda, Y. K. Y. Chin, E. A. B. Undheim, S. Senff, M. Mobli, C. Dauly, P. Escoubas, G. M. Nicholson, Q. Kaas, S. Guo, V. Herzig, J. S. Mattick and G. F. King, Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11399–11408 CrossRef CAS.
- N. Y. Shaikh and K. Sunagar, The deep-rooted origin of disulfide-rich spider venom toxins, eLife, 2023, 12, e83761 CrossRef CAS PubMed.
- K. Wilson, T. de Rond, I. Burkhardt, T. S. Steele, R. J. B. Schäfer, S. Podell, E. E. Allen and B. S. Moore, Terpene biosynthesis in marine sponge animals, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2220934120 CrossRef CAS.
- K. Steffen, Q. Laborde, S. Gunasekera, C. D. Payne, K. J. Rosengren, A. Riesgo, U. Göransson and P. Cárdenas, Barrettides: a peptide family specifically produced by the deep-sea sponge Geodia barretti, J. Nat. Prod., 2021, 84, 3138–3146 CrossRef CAS.
- Z. Lin, V. Agarwal, Y. Cong, S. A. Pomponi and E. W. Schmidt, Short macrocyclic peptides in sponge genomes, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2314383121 CrossRef CAS.
- V. J. Paul, M. P. Puglisi and R. Ritson-Williams, Marine chemical ecology, Nat. Prod. Rep., 2006, 23, 153–180 RSC.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K. D. Entian, M. A. Fischbach, J. S. Garavelli, U. Goransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Muller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. Reaney, S. Rebuffat, R. P. Ross, H. G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Sussmuth, J. R. Tagg, G. L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature, Nat. Prod. Rep., 2013, 30, 108–160 RSC.
- G. A. Hudson and D. A. Mitchell, RiPP antibiotics: biosynthesis and engineering potential, Curr. Opin. Microbiol., 2018, 45, 61–69 CrossRef CAS.
- M. A. Fischbach and C. T. Walsh, Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms, Chem. Rev., 2006, 106, 3468–3496 CrossRef CAS PubMed.
- C. T. Walsh, Insights into the chemical logic and enzymatic machinery of NRPS assembly lines, Nat. Prod. Rep., 2016, 33, 127–135 RSC.
- E. A. Felnagle, E. E. Jackson, Y. A. Chan, A. M. Podevels, A. D. Berti, M. D. McMahon and M. G. Thomas, Nonribosomal Peptide Synthetases Involved in the Production of Medically Relevant Natural Products, Mol. Pharm., 2008, 5, 191–211 CrossRef CAS.
- H. D. Mootz, D. Schwarzer and M. A. Marahiel, Ways of Assembling Complex Natural Products on Modular Nonribosomal Peptide Synthetases, ChemBioChem, 2002, 3, 490–504 CrossRef CAS.
- R. F. Little and C. Hertweck, Chain release mechanisms in polyketide and non-ribosomal peptide biosynthesis, Nat. Prod. Rep., 2022, 39, 163–205 RSC.
- M. E. Horsman, T. P. A. Hari and C. N. Boddy, Polyketide synthase and non-ribosomal peptide synthetase thioesterase selectivity: logic gate or a victim of fate?, Nat. Prod. Rep., 2016, 33, 183–202 RSC.
- S. Nelson and E. I. Parkinson, Synthetic-bioinformatic natural product-inspired peptides, Nat. Prod. Rep., 2025, 42, 50–66 RSC.
- J. Peng, J. Li and M. T. Hamann, The marine bromotyrosine derivatives, Alkaloids, 2005, 61, 59–262 CAS.
- B. Forte, B. Malgesini, C. Piutti, F. Quartieri, A. Scolaro and G. Papeo, A submarine journey: the pyrrole-imidazole alkaloids, Mar. Drugs, 2009, 7, 705–753 CrossRef CAS PubMed.
- A. Al-Mourabit, M. A. Zancanella, S. Tilvi and D. Romo, Biosynthesis, asymmetric synthesis, and pharmacology, including cellular targets, of the pyrrole-2-aminoimidazole marine alkaloids, Nat. Prod. Rep., 2011, 28, 1229–1260 RSC.
- X. Wang, Z. Ma, X. Wang, S. De, Y. Ma and C. Chen, Dimeric pyrrole–imidazole alkaloids: synthetic approaches and biosynthetic hypotheses, Chem. Commun., 2014, 50, 8628–8639 RSC.
- K. S. Singh, and M. S. Majik, Chapter 10 - Pyrrole-Derived Alkaloids of Marine Sponges and Their Biological Properties, in Studies in Natural Products Chemistry, ed. R. Atta ur, Elsevier, 2019, vol. 62, pp. , pp. 377–409 Search PubMed.
- H. Niemann, A. Marmann, W. Lin and P. Proksch, Sponge Derived Bromotyrosines: Structural Diversity through Natural Combinatorial Chemistry, Nat. Prod. Commun., 2015, 10, 1934578X1501000143 CrossRef CAS.
- T. Lindel, Chapter Three – Chemistry and Biology of the Pyrrole–Imidazole Alkaloids, in The Alkaloids: Chemistry and Biology, ed. H.-J. Knölker, Academic Press, 2017, vol. 77, pp. 117–219 Search PubMed.
- M. Kock, A. Grube, I. B. Seiple and P. S. Baran, The pursuit of palau'amine, Angew. Chem., Int. Ed., 2007, 46, 6586–6594 CrossRef PubMed.
- Z. Ma, X. Wang, R. A. Rodriguez, C. E. Moore, S. Gao, X. Tan, Y. Ma, A. L. Rheingold, P. S. Baran and C. Chen, Asymmetric syntheses of sceptrin and massadine and evidence for biosynthetic enantiodivergence, Science, 2014, 346, 219–224 CrossRef CAS PubMed.
- P. S. Baran, Natural Product Total Synthesis: As Exciting as Ever and Here To Stay, J. Am. Chem. Soc., 2018, 140, 4751–4755 CrossRef CAS.
- N. Canu, M. Moutiez, P. Belin and M. Gondry, Cyclodipeptide synthases: a promising biotechnological tool for the synthesis of diverse 2,5-diketopiperazines, Nat. Prod. Rep., 2020, 37, 312–321 RSC.
- L. Sauguet, M. Moutiez, Y. Li, P. Belin, J. Seguin, M.-H. Le Du, R. Thai, C. Masson, M. Fonvielle, J.-L. Pernodet, J.-B. Charbonnier and M. Gondry, Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis, Nucleic Acids Res., 2011, 39, 4475–4489 CrossRef CAS.
- Y. Ogasawara and T. Dairi, Biosynthesis of Oligopeptides Using ATP-Grasp Enzymes, Chem.–Eur. J., 2017, 23, 10714–10724 CrossRef CAS.
- M. V. Fawaz, M. E. Topper and S. M. Firestine, The ATP-grasp enzymes, Bioorg. Chem., 2011, 39, 185–191 CrossRef CAS PubMed.
- J. J. Cui, Y. Zhang and K.-S. Ju, Phosphonoalamides Reveal the Biosynthetic Origin of Phosphonoalanine Natural Products and a Convergent Pathway for Their Diversification, Angew. Chem., Int. Ed., 2024, 63, e202405052 CrossRef CAS.
- L. M. Iyer, S. Abhiman, A. Maxwell Burroughs and L. Aravind, Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins, Mol. BioSyst., 2009, 5, 1636–1660 RSC.
- Y. Yu and W. A. van der Donk, PEARL-Catalyzed Peptide Bond Formation after Chain Reversal by Ureido-Forming Condensation Domains, ACS Cent. Sci., 2024, 10, 1242–1250 CrossRef CAS PubMed.
- C. Maruyama and Y. Hamano, tRNA-dependent amide bond–forming enzymes in peptide natural product biosynthesis, Curr. Opin. Chem. Biol., 2020, 59, 164–171 CrossRef CAS.
- P. Andrade, R. Willoughby, S. A. Pomponi and R. G. Kerr, Biosynthetic studies of the alkaloid, stevensine, in a cell culture of the marine sponge Teichaxinella morchella, Tetrahedron Lett., 1999, 40, 4775–4778 CrossRef CAS.
- T. Lindel, M. Hochgürtel, M. Assmann and M. Köck, Synthesis of the marine natural product Nα-(4-bromopyrrolyl-2-carbonyl)-l-homoarginine, a putative biogenetic precursor of the pyrrole–imidazole alkaloids, J. Nat. Prod., 2000, 63, 1566–1569 CrossRef.
- I. Mohanty, S. G. Moore, D. Yi, J. S. Biggs, D. A. Gaul, N. Garg and V. Agarwal, Precursor-guided mining of marine sponge metabolomes lends insight into biosynthesis of pyrrole–imidazole alkaloids, ACS Chem. Biol., 2020, 15, 2185–2194 CrossRef.
- I. Mohanty, S. G. Moore, J. S. Biggs, C. J. Freeman, D. A. Gaul, N. Garg and V. Agarwal, Stereochemical assignment and absolute abundance of nonproteinogenic amino acid homoarginine in marine sponges, ACS Omega, 2021, 6, 33200–33205 CrossRef.
- I. Mohanty, S. Tapadar, S. G. Moore, J. S. Biggs, C. J. Freeman, D. A. Gaul, N. Garg and V. Agarwal, Presence of bromotyrosine alkaloids in marine sponges is independent of metabolomic and microbiome architectures, mSystems, 2021, 6, e01387 CrossRef PubMed.
- M. M. Cheng, X. L. Tang, Y. T. Sun, D. Y. Song, Y. J. Cheng, H. Liu, P. L. Li and G. Q. Li, Biological and Chemical Diversity of Marine Sponge-Derived Microorganisms over the Last Two Decades from 1998 to 2017, Molecules, 2020, 25, 853 CrossRef.
- P. Li, H. Lu, Y. Zhang, X. Zhang, L. Liu, M. Wang and L. Liu, The natural products discovered in marine sponge-associated microorganisms: structures, activities, and mining strategy, Front. Mar. Sci., 2023, 10, 1191858 CrossRef.
- B.-N. Han, L.-L. Hong, B.-B. Gu, Y.-T. Sun, J. Wang, J.-T. Liu, and H.-W. Lin, Natural Products from Sponges. Symbiotic Microbiomes of Coral Reefs Sponges and Corals, 2019, pp. 329–463 Search PubMed.
- P. B. Svendsen, L. Alfastsen, L. Gram, N. N. S. E. Henriksen, M. Bentzon-Tilia and S.-D. Zhang, Roles of Marine Microbial Natural Products, Annu. Rev. Microbiol., 2025, 79 DOI:10.1146/annurev-micro-040824-022431.
- W. Y. Fang, R. Dahiya, H. L. Qin, R. Mourya and S. Maharaj, Natural Proline-Rich Cyclopolypeptides from Marine Organisms: Chemistry, Synthetic Methodologies and Biological Status, Mar. Drugs, 2016, 14, 194 CrossRef PubMed.
- R. Sakai, K. L. Rinehart, V. Kishore, B. Kundu, G. Faircloth, J. B. Gloer, J. R. Carney, M. Namikoshi, F. Sun, R. G. Hughes, D. G. Grávalos, T. G. de Quesada, G. R. Wilson and R. M. Heid, Structure–Activity Relationships of the Didemnins, J. Med. Chem., 1996, 39, 2819–2834 CrossRef PubMed.
- D. L. Herald, G. L. Cascarano, G. R. Pettit and J. K. Srirangam, Crystal Conformation of the Cyclic Decapeptide Phakellistatin 8: Comparison with Antamanide, J. Am. Chem. Soc., 1997, 119, 6962–6973 CrossRef.
- I. Mohanty, N. A. Nguyen, S. G. Moore, J. S. Biggs, D. A. Gaul, N. Garg and V. Agarwal, Enzymatic synthesis assisted discovery of proline-rich macrocyclic peptides in marine sponges, Chembiochem, 2021, 22, 2614–2618 Search PubMed.
- T. P. Cantrell, C. J. Freeman, V. J. Paul, V. Agarwal and N. Garg, Mass spectrometry-based integration and expansion of the chemical diversity harbored within a marine sponge, J. Am. Soc. Mass Spectrom., 2019, 30, 1373–1384 CrossRef.
- I. Mohanty, S. Podell, J. S. Biggs, N. Garg, E. E. Allen and V. Agarwal, Multi-Omic Profiling of Melophlus Sponges Reveals Diverse Metabolomic and Microbiome Architectures that Are Non-overlapping with Ecological Neighbors, Mar. Drugs, 2020, 18, 124 CrossRef PubMed.
- S. Khushi, A. A. Salim and R. J. Capon, Case Studies in Molecular Network-Guided Marine Biodiscovery, Mar. Drugs, 2023, 21, 413 CrossRef PubMed.
- K. Bayer, J. Kamke and U. Hentschel, Quantification of bacterial and archaeal symbionts in high and low microbial abundance sponges using real-time PCR, FEMS Microbiol. Ecol., 2014, 89, 679–690 Search PubMed.
- V. Gloeckner, M. Wehrl, L. Moitinho-Silva, C. Gernert, P. Schupp, J. R. Pawlik, N. L. Lindquist, D. Erpenbeck, G. Wörheide and U. Hentschel, The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species, Biol. Bull., 2014, 227, 78–88 Search PubMed.
- L. Moitinho-Silva, G. Steinert, S. Nielsen, C. C. P. Hardoim, Y.-C. Wu, G. P. McCormack, S. López-Legentil, R. Marchant, N. Webster, T. Thomas and U. Hentschel, Predicting the HMA-LMA status in marine sponges by machine learning, Front. Microbiol., 2017, 8, 752 CrossRef PubMed.
- M. S. Donia, J. Ravel and E. W. Schmidt, A global assembly line for cyanobactins, Nat. Chem. Biol., 2008, 4, 341–343 CrossRef.
- Y. Zhang, K. Li, G. Yang, J. L. McBride, S. D. Bruner and Y. Ding, A distributive peptide cyclase processes multiple microviridin core peptides within a single polypeptide substrate, Nat. Commun., 2018, 9, 1780 CrossRef PubMed.
- O. M. Manley, T. J. Shriver, T. Xu, I. A. Melendrez, P. Palacios, S. A. Robson, Y. Guo, N. L. Kelleher, J. J. Ziarek and A. C. Rosenzweig, A multi-iron enzyme installs copper-binding oxazolone/thioamide pairs on a nontypeable Haemophilus influenzae virulence factor, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2408092121 CrossRef.
- L. Leprevost, S. Jünger, G. Lippens, C. Guillaume, G. Sicoli, L. Oliveira, E. Falcone, E. de Santis, A. Rivera-Millot, G. Billon, F. Stellato, C. Henry, R. Antoine, S. Zirah, S. Dubiley, Y. Li and F. Jacob-Dubuisson, A widespread family of ribosomal peptide metallophores involved in bacterial adaptation to metal stress, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2408304121 CrossRef PubMed.
- J. A. McIntosh, C. R. Robertson, V. Agarwal, S. K. Nair, G. W. Bulaj and E. W. Schmidt, Circular logic: nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst, J. Am. Chem. Soc., 2010, 132, 15499–15501 CrossRef PubMed.
- V. Agarwal, E. Pierce, J. McIntosh, E. W. Schmidt and S. K. Nair, Structures of cyanobactin maturation enzymes define a family of transamidating proteases, Chem. Biol., 2012, 19, 1411–1422 CrossRef PubMed.
- J. Koehnke, A. Bent, W. E. Houssen, D. Zollman, F. Morawitz, S. Shirran, J. Vendome, A. F. Nneoyiegbe, L. Trembleau, C. H. Botting, M. C. M. Smith, M. Jaspars and J. H. Naismith, The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain, Nat. Struct. Mol. Biol., 2012, 19, 767–772 CrossRef.
- C. M. Czekster, H. Ludewig, S. A. McMahon and J. H. Naismith, Characterization of a dual function macrocyclase enables design and use of efficient macrocyclization substrates, Nat. Commun., 2017, 8, 1045 CrossRef PubMed.
- H. Ludewig, C. M. Czekster, E. Oueis, E. S. Munday, M. Arshad, S. A. Synowsky, A. F. Bent and J. H. Naismith, Characterization of the fast and promiscuous macrocyclase from plant PCY1 enables the use of simple substrates, ACS Chem. Biol., 2018, 13, 801–811 CrossRef.
- J. R. Chekan, P. Estrada, P. S. Covello and S. K. Nair, Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6551 CrossRef PubMed.
- W. Gu, S.-H. Dong, S. Sarkar, S. K. Nair, and E. W. Schmidt, Chapter Four – The biochemistry and structural biology of cyanobactin pathways: enabling combinatorial biosynthesis, in Methods in Enzymology, ed. B. S. Moore, Academic Press, 2018, vol. 604, pp. 113–163 Search PubMed.
- R. Mohammed, J. Peng, M. Kelly and M. T. Hamann, Cyclic Heptapeptides from the Jamaican Sponge Stylissa caribica, J. Nat. Prod., 2006, 69, 1739–1744 CrossRef.
- G. Schmidt, A. Grube and M. Köck, Stylissamides A–D – new proline-containing cyclic heptapeptides from the marine sponge Stylissa caribica, Eur. J. Org Chem., 2007, 2007, 4103–4110 CrossRef.
- C. Cychon and M. Köck, Stylissamides E and F, cyclic heptapeptides from the Caribbean sponge Stylissa caribica, J. Nat. Prod., 2010, 73, 738–742 CrossRef PubMed.
- X. Wang, B. I. Morinaka and T. F. Molinski, Structures and solution conformational dynamics of stylissamides G and H from the Bahamian sponge Stylissa caribica, J. Nat. Prod., 2014, 77, 625–630 CrossRef.
- J. Vicente, B. Vera, A. D. Rodríguez, I. Rodríguez-Escudero, R. G. Raptis and A. Euryjanicin, a new cycloheptapeptide from the Caribbean marine sponge Prosuberites laughlini, Tetrahedron Lett., 2009, 50, 4571–4574 CrossRef PubMed.
- Y. Li, S. Osanai, K. Nakajima, F. Xu, H. Sano, H. Sato, H. Katagiri and H. Konno, Synthesis and Chemical Structure of the Cyclic Heptapeptide Stylissamide H, J. Nat. Prod., 2025, 88, 593–598 CrossRef.
- J. i. Kobayashi, T. Kubota, K. Nakamura, S.-i. Kurimoto, K. Sakai, J. Fromont, T. Gonoi and I. Stylissamide, a New Cyclic Heptapeptide from an Okinawan Marine Sponge Stylissa sp, Heterocycles, 2017, 95, 799 CrossRef.
- S. Scarpato, R. Teta, G. Della Sala, J. R. Pawlik, V. Costantino and A. Mangoni, New Tricks with an Old Sponge: Feature-Based Molecular Networking Led to Fast Identification of New Stylissamide L from Stylissa caribica, Mar. Drugs, 2020, 18, 443 CrossRef PubMed.
- M. Arai, Y. Yamano, M. Fujita, A. Setiawan, M. Kobayashi and X. Stylissamide, a new proline-rich cyclic octapeptide as an inhibitor of cell migration, from an Indonesian marine sponge of Stylissa sp, Bioorg. Med. Chem. Lett., 2012, 22, 1818–1821 CrossRef PubMed.
- M. Kita, B. Gise, A. Kawamura, H. Kigoshi and A. Stylissatin, a cyclic peptide that inhibits nitric oxide production from the marine sponge Stylissa massa, Tetrahedron Lett., 2013, 54, 6826–6828 CrossRef.
- J. Sun, W. Cheng, N. J. de Voogd, P. Proksch and W. Lin, Stylissatins B–D, cycloheptapeptides from the marine sponge Stylissa massa, Tetrahedron Lett., 2016, 57, 4288–4292 CrossRef.
- T. Akindele, B. Gise, T. Sunaba, M. Kita and H. Kigoshi, Total Synthesis of Stylissatin A, A Cyclic Peptide That Inhibits Nitric Oxide Production, Bull. Chem. Soc. Jpn., 2015, 88, 600–609 CrossRef.
- A. H. Afifi, A. H. El-Desoky, H. Kato, R. E. P. Mangindaan, N. J. de Voogd, N. M. Ammar, M. S. Hifnawy and S. Tsukamoto, Carteritins A and B, cyclic heptapeptides from the marine sponge Stylissa carteri, Tetrahedron Lett., 2016, 57, 1285–1288 CrossRef.
- O.-S. Kwon, C.-K. Kim, W. S. Byun, J. Oh, Y.-J. Lee, H.-S. Lee, C. J. Sim, D.-C. Oh, S. K. Lee, K.-B. Oh and J. Shin, Cyclopeptides from the sponge Stylissa flabelliformis, J. Nat. Prod., 2018, 81, 1426–1434 CrossRef PubMed.
- A. Meli, C. Tedesco, G. Della Sala, R. Schettini, F. Albericio, F. De Riccardis and I. Izzo, Phakellistatins: An Underwater Unsolved Puzzle, Mar. Drugs, 2017, 15, 78 CrossRef PubMed.
- G. R. Pettit, Z. Cichacz, J. Barkoczy, A.-C. Dorsaz, D. L. Herald, M. D. Williams, D. L. Doubek, J. M. Schmidt, L. P. Tackett, D. C. Brune, R. L. Cerny, J. N. A. Hooper and G. J. Bakus, Isolation and Structure of the Marine Sponge Cell Growth Inhibitory Cyclic Peptide Phakellistatin 1, J. Nat. Prod., 1993, 56, 260–267 CrossRef PubMed.
- G. R. Pettit, R. Tan, M. D. Williams, L. Tackett, J. M. Schmidt, R. L. Cerny and J. N. A. Hooper, Isolation and structure of phakellistatin 2 from the eastern indian ocean marine sponge Phakellia carteri, Bioorg. Med. Chem. Lett., 1993, 3, 2869–2874 CrossRef.
- G. R. Pettit, R. Tan, D. L. Herald, M. D. Williams and R. L. Cerny, Antineoplastic Agents. 277. Isolation and Structure of Phakellistatin 3 and Isophakellistatin 3 from a Republic of Comoros Marine Sponge, J. Org. Chem., 1994, 59, 1593–1595 CrossRef.
- G. R. Pettit, J.-p. Xu, Z. A. Cichacz, M. D. Williams, A.-C. Dorsaz, D. C. Brune, M. R. Boyd and R. L. Cerny, Antineoplastic agents 315. Isolation and structure of the marine sponge cancer cell growth inhibitor phakellistatin 5, Bioorg. Med. Chem. Lett., 1994, 4, 2091–2096 CrossRef.
- G. R. Pettit, J.-P. Xu, Z. A. Cichacz, M. D. Williams, J.-C. Chapuis and R. L. Cerny, Antineoplastic agents 323. Isolation and structure of phakellistatin 6 from a Chuuk archipelago marine sponge, Bioorg. Med. Chem. Lett., 1994, 4, 2677–2682 CrossRef.
- G. R. Pettit, J.-p. Xu, A.-C. Dorsaz, M. D. Williams, M. R. Boyd and R. L. Cerny, Isolation and structure of the human cancer cell growth inhibitory cyclic decapeptides phakellistatins 7, 8 and 91,2, Bioorg. Med. Chem. Lett., 1995, 5, 1339–1344 CrossRef.
- G. R. Pettit, R. Tan, Y. Ichihara, M. D. Williams, D. L. Doubek, L. P. Tackett, J. M. Schmidt, R. L. Cerny, M. R. Boyd and J. N. A. Hooper, Antineoplastic Agents, 325. Isolation and Structure of the Human Cancer Cell Growth Inhibitory Cyclic Octapeptides Phakellistatin 10 and 11 from Phakellia sp, J. Nat. Prod., 1995, 58, 961–965 CrossRef.
- G. R. Pettit and R. Tan, Antineoplastic Agents 390. Isolation and Structure of Phakellistatin 12 from a Chuuk Archipelago Marine Sponge, Bioorg. Med. Chem. Lett., 2003, 13, 685–688 CrossRef PubMed.
- W.-L. Li, Y.-H. Yi, H.-M. Wu, Q.-Z. Xu, H.-F. Tang, D.-Z. Zhou, H.-W. Lin and Z.-H. Wang, Isolation and Structure of the Cytotoxic Cycloheptapeptide Phakellistatin 13, J. Nat. Prod., 2003, 66, 146–148 CrossRef CAS.
- G. R. Pettit and R. Tan, Isolation and Structure of Phakellistatin 14 from the Western Pacific Marine Sponge Phakellia sp.,1, J. Nat. Prod., 2005, 68, 60–63 CrossRef CAS PubMed.
- H.-J. Zhang, Y.-H. Yi, G.-J. Yang, M.-Y. Hu, G.-D. Cao, F. Yang and H.-W. Lin, Proline-Containing Cyclopeptides from the Marine Sponge Phakellia fusca, J. Nat. Prod., 2010, 73, 650–655 CrossRef.
- M. Pelay-Gimeno, A. Meli, J. Tulla-Puche and F. Albericio, Rescuing Biological Activity from Synthetic Phakellistatin 19, J. Med. Chem., 2013, 56, 9780–9788 CrossRef PubMed.
- Y. Wu, L. Liu, H.-F. Chen, W.-H. Jiao, F. Sun, L.-Y. Liu, H.-R. Zhu, S.-P. Wang, H.-W. Lin and A. -D. Fuscasins, Cycloheptapeptides from the Marine Sponge Phakellia fusca, J. Nat. Prod., 2019, 82, 970–979 CrossRef PubMed.
- Y. Wu, H. Liao, L.-Y. Liu, F. Sun, H.-F. Chen, W.-H. Jiao, H.-R. Zhu, F. Yang, G. Huang, D.-Q. Zeng, M. Zhou, S.-P. Wang, H.-W. Lin and A. -C. Phakefustatins, Kynurenine-Bearing Cycloheptapeptides as RXRα Modulators from the Marine Sponge Phakellia fusca, Org. Lett., 2020, 22, 6703–6708 CrossRef PubMed.
- B. Vera, J. Vicente and A. D. Rodríguez, Isolation and Structural Elucidation of Euryjanicins B–D, Proline-Containing Cycloheptapeptides from the Caribbean Marine Sponge Prosuberites laughlini, J. Nat. Prod., 2009, 72, 1555–1562 CrossRef PubMed.
- E. Avilés, A. D. Rodríguez and E. -G. Euryjanicins, poly-phenylalanine, and poly-proline cyclic heptapeptides from the Caribbean sponge Prosuberites laughlini, Tetrahedron, 2013, 69, 10797–10804 CrossRef PubMed.
- K.-X. Zhan, W.-H. Jiao, F. Yang, J. Li, S.-P. Wang, Y.-S. Li, B.-N. Han and H.-W. Lin, Reniochalistatins A–E, Cyclic Peptides from the Marine Sponge Reniochalina stalagmitis, J. Nat. Prod., 2014, 77, 2678–2684 CrossRef PubMed.
- S. J. Moore, C. L. Leung and J. R. Cochran, Knottins: disulfide-bonded therapeutic and diagnostic peptides, Drug Discovery Today: Technol., 2012, 9, e3–e11 CrossRef PubMed.
- J. R. Kintzing and J. R. Cochran, Engineered knottin peptides as diagnostics, therapeutics, and drug delivery vehicles, Curr. Opin. Chem. Biol., 2016, 34, 143–150 CrossRef PubMed.
- B. Molesini, D. Treggiari, A. Dalbeni, P. Minuz and T. Pandolfini, Plant cystine-knot peptides: pharmacological perspectives, Br. J. Clin. Pharmacol., 2017, 83, 63–70 CrossRef PubMed.
- T. L. Koch, S. D. Robinson, P. F. Salcedo, K. Chase, J. Biggs, A. E. Fedosov, M. Yandell, B. M. Olivera and H. Safavi-Hemami, Prey Shifts Drive Venom Evolution in Cone Snails, Mol. Biol. Evol., 2024, 41, msae120 CrossRef PubMed.
- G. P. Miljanich, Ziconotide: Neuronal Calcium Channel Blocker for Treating Severe Chronic Pain, Curr. Med. Chem., 2004, 11, 3029–3040 CrossRef PubMed.
- Z. Miao, G. Ren, H. Liu, R. H. Kimura, L. Jiang, J. R. Cochran, S. S. Gambhir and Z. Cheng, An Engineered Knottin Peptide Labeled with 18F for PET Imaging of Integrin Expression, Bioconjugate Chem., 2009, 20, 2342–2347 CrossRef PubMed.
- S. J. Moore, M. G. Hayden Gephart, J. M. Bergen, Y. S. Su, H. Rayburn, M. P. Scott and J. R. Cochran, Engineered knottin peptide enables noninvasive optical imaging of intracranial medulloblastoma, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14598–14603 CrossRef CAS PubMed.
- S. W. B. Yu and S. S. C. Rao, Advances in the management of constipation-predominant irritable bowel syndrome: the role of linaclotide, Ther. Adv. Gastroenterol., 2014, 7, 193–205 CrossRef CAS.
- A.-H. Jin, M. Muttenthaler, S. Dutertre, S. W. A. Himaya, Q. Kaas, D. J. Craik, R. J. Lewis and P. F. Alewood, Conotoxins: chemistry and biology, Chem. Rev., 2019, 119, 11510–11549 CrossRef CAS.
- B. B. Carstens, K. J. Rosengren, S. Gunasekera, S. Schempp, L. Bohlin, M. Dahlström, R. J. Clark and U. Göransson, Isolation, characterization, and synthesis of the barrettides: disulfide-containing peptides from the marine sponge Geodia barretti, J. Nat. Prod., 2015, 78, 1886–1893 CrossRef CAS PubMed.
- H. Li, J. J. Bowling, F. R. Fronczek, J. Hong, S. V. Jabba, T. F. Murray, N.-C. Ha, M. T. Hamann, J. H. Jung and A. Asteropsin, An unusual cystine-crosslinked peptide from porifera enhances neuronal Ca2+ influx, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 2591–2599 CrossRef PubMed.
- H. Li, J. J. Bowling, M. Su, J. Hong, B.-J. Lee, M. T. Hamann, J. H. Jung and B. -D. Asteropsins, sponge-derived knottins with potential utility as a novel scaffold for oral peptide drugs, Biochim. Biophys. Acta, Gen. Subj., 2014, 1840, 977–984 CrossRef PubMed.
- H. Li, M. Su, M. T. Hamann, J. J. Bowling, H. S. Kim and J. H. Jung, Solution structure of a sponge-derived cystine knot peptide and its notable stability, J. Nat. Prod., 2014, 77, 304–310 CrossRef PubMed.
- M. Su, H. Li, H. Wang, E. L. Kim, H. S. Kim, E.-H. Kim, J. Lee and J. H. Jung, Stable and biocompatible cystine knot peptides from the marine sponge Asteropus sp, Bioorg. Med. Chem., 2016, 24, 2979–2987 CrossRef PubMed.
- Z. Chang, M. Lu, Y. Ma, D.-G. Kwag, S.-H. Kim, J.-M. Park, B.-H. Nam, Y.-O. Kim, C.-M. An, H. Li, J. H. Jung and J.-S. Park, Production of disulfide bond-rich peptides by fusion expression using small transmembrane proteins of Escherichia coli, Amino Acids, 2015, 47, 579–587 CrossRef.
- L. R. H. Krumpe, B. A. P. Wilson, C. Marchand, S. N. Sunassee, A. Bermingham, W. Wang, E. Price, T. Guszczynski, J. A. Kelley, K. R. Gustafson, Y. Pommier, K. J. Rosengren, C. I. Schroeder, B. R. O'Keefe and A. Recifin, Initial Example of the Tyr-Lock Peptide Structural Family, Is a Selective Allosteric Inhibitor of Tyrosyl-DNA Phosphodiesterase I, J. Am. Chem. Soc., 2020, 142, 21178–21188 CrossRef.
- D. E. Williams, P. Austin, A. R. Diaz-Marrero, R. V. Soest, T. Matainaho, C. D. Roskelley, M. Roberge and R. J. Andersen, Neopetrosiamides, peptides from the marine sponge Neopetrosia sp. that inhibit amoeboid invasion by human tumor cells, Org. Lett., 2005, 7, 4173–4176 CrossRef PubMed.
- H. Liu, M. A. Boudreau, J. Zheng, R. M. Whittal, P. Austin, C. D. Roskelley, M. Roberge, R. J. Andersen and J. C. Vederas, Chemical synthesis and biological activity of the neopetrosiamides and their analogues: revision of disulfide bond connectivity, J. Am. Chem. Soc., 2010, 132, 1486–1487 CrossRef PubMed.
- D. Fass and C. Thorpe, Chemistry and enzymology of disulfide cross-linking in proteins, Chem. Rev., 2018, 118, 1169–1198 CrossRef PubMed.
- A. Kam, S. Loo, Y. Qiu, C.-F. Liu and J. P. Tam, Ultrafast Biomimetic Oxidative Folding of Cysteine-rich Peptides and Microproteins in Organic Solvents, Angew. Chem., Int. Ed., 2024, 63, e202317789 CrossRef PubMed.
- W. Zhong, J. O. Olugbami, P. Rathakrishnan, I. Mohanty, S. G. Moore, N. Garg, A. K. Oyelere, T. L. Turner, A. C. McShan and V. Agarwal, Discovery and Folding Dynamics of a Fused Bicyclic Cysteine Knot Undecapeptide from the Marine Sponge Halichondria bowerbanki, J. Org. Chem., 2024, 89, 12748–12752 CrossRef.
- J.-K. Woo, J.-e. Jeon, C.-K. Kim, C. J. Sim, D.-C. Oh, K.-B. Oh, J. Shin and A. Gombamide, a cyclic thiopeptide from the sponge Clathria gombawuiensis, J. Nat. Prod., 2013, 76, 1380–1383 CrossRef CAS.
- A. Mokhlesi, F. Stuhldreier, K. W. Wex, A. Berscheid, R. Hartmann, N. Rehberg, P. Sureechatchaiyan, C. Chaidir, M. U. Kassack, R. Kalscheuer, H. Brötz-Oesterhelt, S. Wesselborg, B. Stork, G. Daletos and P. Proksch, Cyclic Cystine-Bridged Peptides from the Marine Sponge Clathria basilana Induce Apoptosis in Tumor Cells and Depolarize the Bacterial Cytoplasmic Membrane, J. Nat. Prod., 2017, 80, 2941–2952 CrossRef CAS.
- T. Kupke, C. Kempter, G. Jung and F. Götz, Oxidative Decarboxylation of Peptides Catalyzed by Flavoprotein EpiD: Determination Of Substrate Specificity Using Peptide Libraries And Neutral Loss Mass Spectrometry*, J. Biol. Chem., 1995, 270, 11282–11289 CrossRef CAS PubMed.
- N. A. Bruender and V. Bandarian, The Radical S-Adenosyl-l-methionine Enzyme MftC Catalyzes an Oxidative Decarboxylation of the C-Terminus of the MftA Peptide, Biochemistry, 2016, 55, 2813–2816 CrossRef CAS PubMed.
- M. A. Ortega, D. P. Cogan, S. Mukherjee, N. Garg, B. Li, G. N. Thibodeaux, S. I. Maffioli, S. Donadio, M. Sosio, J. Escano, L. Smith, S. K. Nair and W. A. van der Donk, Two flavoenzymes catalyze the post-translational generation of 5-chlorotryptophan and 2-aminovinyl-cysteine during NAI-107 biosynthesis, ACS Chem. Biol., 2017, 12, 548–557 CrossRef CAS.
- P. M. Garcia-Barrantes and C. W. Lindsley, Total Synthesis of Gombamide A, Org. Lett., 2016, 18, 3810–3813 CrossRef CAS PubMed.
- M. R. Vippila, S. Nikhar, A. P. Gracia and G. D. Cuny, Divergent Approach for the Synthesis of Gombamide A and Derivatives, Org. Lett., 2016, 18, 4726–4729 CrossRef CAS PubMed.
- J. Song, J.-e. Jeon, T. H. Won, C. J. Sim, D.-C. Oh, K.-B. Oh and J. Shin, New Cyclic Cystine Bridged Peptides from the Sponge Suberites waedoensis, Mar. Drugs, 2014, 12, 2760–2770 CrossRef CAS PubMed.
- R. A. Davis, G. C. Mangalindan, Z. P. Bojo, R. R. Antemano, N. O. Rodriguez, G. P. Concepcion, S. C. Samson, D. de Guzman, L. J. Cruz, D. Tasdemir, M. K. Harper, X. Feng, G. T. Carter and C. M. Ireland, Microcionamides A and B, Bioactive Peptides from the Philippine Sponge Clathria (Thalysias) abietina, J. Org. Chem., 2004, 69, 4170–4176 CrossRef CAS.
- U. Hentschel, J. Piel, S. M. Degnan and M. W. Taylor, Genomic insights into the marine sponge microbiome, Nat. Rev. Microbiol., 2012, 10, 641–654 CrossRef CAS.
- C. Gurgui, and J. Piel, Metagenomic Approaches to Identify and Isolate Bioactive Natural Products from Microbiota of Marine Sponges, in Metagenomics: Methods and Protocols, ed. W. R. Streit, and R. Daniel, Humana Press, Totowa, NJ, 2010, pp. 247–264 Search PubMed.
- J. K. B. Cahn and J. Piel, Opening up the single-cell toolbox for microbial natural products research, Angew. Chem., Int. Ed., 2021, 60, 18412–18428 CrossRef CAS PubMed.
- S. Matsunaga, N. Fusetani, K. Hashimoto, M. Walchli and F. Theonellamide, A novel antifungal bicyclic peptide from a marine sponge Theonella sp, J. Am. Chem. Soc., 1989, 111, 2582–2588 CrossRef CAS.
- S. Matsunaga, N. Fusetani and A.-E. Theonellamides, cytotoxic bicyclic peptides, from a marine sponge Theonella sp, J. Org. Chem., 1995, 60, 1177–1181 CrossRef CAS.
- E. W. Schmidt, C. A. Bewley and D. J. Faulkner, Theopalauamide, a bicyclic glycopeptide from filamentous bacterial symbionts of the lithistid sponge Theonella swinhoei from Palau and Mozambique, J. Org. Chem., 1998, 63, 1254–1258 CrossRef CAS.
- T. Hamada, S. Matsunaga, G. Yano and N. Fusetani, Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei, J. Am. Chem. Soc., 2005, 127, 110–118 CrossRef CAS.
- J. i. Kobayashi, M. Sato, M. Ishibashi, H. Shigemori, T. Nakamura, Y. Ohizumi and A. Keramamide, a novel peptide from the Okinawan marine sponge Theonella sp, J. Chem. Soc., Perkin Trans. 1, 1991, 2609–2611 RSC.
- N. Fusetani, T. Sugawara, S. Matsunaga and H. Hirota, Orbiculamide A: a novel cytotoxic cyclic peptide from a marine sponge Theonella sp, J. Am. Chem. Soc., 1991, 113, 7811–7812 CrossRef CAS.
- N. Fusetani, S. Matsunaga, H. Matsumoto and Y. Takebayashi, Bioactive marine metabolites. 33. Cyclotheonamides, potent thrombin inhibitors, from a marine sponge Theonella sp, J. Am. Chem. Soc., 1990, 112, 7053–7054 CrossRef CAS.
- N. Fusetani, Y. Nakao, S. Matsunaga and A. Nazumamide, a thrombin-inhibitory Tetrapeptide, from a marine sponge, Theonella sp, Tetrahedron Lett., 1991, 32, 7073–7074 CrossRef CAS.
- C. A. Bewley, N. D. Holland and D. J. Faulkner, Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts, Experientia, 1996, 52, 716–722 CrossRef CAS.
- C. A. Bewley and D. J. Faulkner, Lithistid Sponges: Star Performers or Hosts to the Stars, Angew. Chem., Int. Ed., 1998, 37, 2162–2178 CrossRef PubMed.
- E. W. Schmidt, A. Y. Obraztsova, S. K. Davidson, D. J. Faulkner and M. G. Haygood, Identification of the antifungal peptide-containing symbiont of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis”, Mar. Biol., 2000, 136, 969–977 CrossRef CAS.
- M. C. Wilson, T. Mori, C. Rückert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. E. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. N. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crüsemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, An environmental bacterial taxon with a large and distinct metabolic repertoire, Nature, 2014, 506, 58–62 CrossRef CAS PubMed.
- A. Bhushan, E. E. Peters, and J. Piel, Entotheonella Bacteria as Source of Sponge-Derived Natural Products: Opportunities for Biotechnological Production, in Blue Biotechnology: From Gene to Bioactive Product, ed. W. E. G. Müller, H. C. Schröder, and X. Wang, Springer International Publishing, Cham, 2017, pp. 291–314 Search PubMed.
- T. Hamada, T. Sugawara, S. Matsunaga and N. Fusetani, Polytheonamides, unprecedented highly cytotoxic polypeptides from the marine sponge Theonella swinhoei 2. Structure elucidation, Tetrahedron Lett., 1994, 35, 609–612 CrossRef CAS.
- T. Hamada, S. Matsunaga, M. Fujiwara, K. Fujita, H. Hirota, R. Schmucki, P. Güntert and N. Fusetani, Solution structure of polytheonamide B, a highly cytotoxic nonribosomal polypeptide from marine sponge, J. Am. Chem. Soc., 2010, 132, 12941–12945 CrossRef CAS.
- M. Inoue, N. Shinohara, S. Tanabe, T. Takahashi, K. Okura, H. Itoh, Y. Mizoguchi, M. Iida, N. Lee and S. Matsuoka, Total synthesis of the large non-ribosomal peptide polytheonamide B, Nat. Chem., 2010, 2, 280–285 CrossRef CAS PubMed.
- M. F. Freeman, C. Gurgui, M. J. Helf, B. I. Morinaka, A. R. Uria, N. J. Oldham, H.-G. Sahl, S. Matsunaga and J. Piel, Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides, Science, 2012, 338, 387 CrossRef CAS.
- M. F. Freeman, A. L. Vagstad and J. Piel, Polytheonamide biosynthesis showcasing the metabolic potential of sponge-associated uncultivated 'Entotheonella' bacteria, Curr. Opin. Chem. Biol., 2015, 31, 8–14 CrossRef.
- A. Parent, A. Benjdia, A. Guillot, X. Kubiak, C. Balty, B. Lefranc, J. Leprince and O. Berteau, Mechanistic Investigations of PoyD, a Radical S-Adenosyl-l-methionine Enzyme Catalyzing Iterative and Directional Epimerizations in Polytheonamide A Biosynthesis, J. Am. Chem. Soc., 2018, 140, 2469–2477 CrossRef CAS.
- M. J. Helf, M. F. Freeman and J. Piel, Investigations into PoyH, a promiscuous protease from polytheonamide biosynthesis, J. Ind. Microbiol. Biotechnol., 2019, 46, 551–563 CrossRef CAS.
- M. F. Freeman, M. J. Helf, A. Bhushan, B. I. Morinaka and J. Piel, Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium, Nat. Chem., 2017, 9, 387–395 CrossRef CAS PubMed.
- A. Bhushan, P. J. Egli, E. E. Peters, M. F. Freeman and J. Piel, Genome mining- and synthetic biology-enabled production of hypermodified peptides, Nat. Chem., 2019, 11, 931–939 CrossRef CAS PubMed.
- N. M. Bösch, M. Borsa, U. Greczmiel, B. I. Morinaka, M. Gugger, A. Oxenius, A. L. Vagstad and J. Piel, Landornamides: antiviral ornithine-containing ribosomal peptides discovered through genome mining, Angew. Chem., Int. Ed., 2020, 59, 11763–11768 CrossRef PubMed.
- N. A. Nguyen, F. N. U. Vidya, N. H. Yennawar, H. Wu, A. C. McShan and V. Agarwal, Disordered regions in proteusin peptides guide post-translational modification by a flavin-dependent RiPP brominase, Nat. Commun., 2024, 15, 1265 CrossRef CAS.
- M. Montalbán-López, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud, Y. Goto, M. J. Grande Burgos, C. Hill, S. Kim, J. Koehnke, J. A. Latham, A. J. Link, B. Martínez, S. K. Nair, Y. Nicolet, S. Rebuffat, H.-G. Sahl, D. Sareen, E. W. Schmidt, L. Schmitt, K. Severinov, R. D. Süssmuth, A. W. Truman, H. Wang, J.-K. Weng, G. P. van Wezel, Q. Zhang, J. Zhong, J. Piel, D. A. Mitchell, O. P. Kuipers and W. A. van der Donk, New developments in RiPP discovery, enzymology and engineering, Nat. Prod. Rep., 2021, 38, 130–239 RSC.
- D. H. Haft, M. K. Basu and D. A. Mitchell, Expansion of ribosomally produced natural products: a nitrile hydratase- and Nif11-related precursor family, BMC Biol., 2010, 8, 70 CrossRef PubMed.
- N. A. Nguyen, Y. Cong, R. C. Hurrell, N. Arias, N. Garg, A. W. Puri, E. W. Schmidt and V. Agarwal, A Silent Biosynthetic Gene Cluster from a Methanotrophic Bacterium Potentiates Discovery of a Substrate Promiscuous Proteusin Cyclodehydratase, ACS Chem. Biol., 2022, 17, 1577–1585 CrossRef CAS PubMed.
- K. Delawská, J. Hájek, K. Voráčová, M. Kuzma, J. Mareš, K. Vicková, A. Kádek, D. Tučková, F. Gallob, P. Divoká, M. Moos, S. Opekar, L. Koch, K. Saurav, D. Sedlák, P. Novák, P. Urajová, J. Dean, R. Gažák, T. J. H. Niedermeyer, Z. Kameník, P. Šimek, A. Villunger and P. Hrouzek, Discovery of nostatin A, an azole-containing proteusin with prominent cytostatic and pro-apoptotic activity, Org. Biomol. Chem., 2025, 23, 449–460 RSC.
- M. A. Wakeel, E. A. Corbin, A. C. McShan and V. Agarwal, Extending the Peptide/Protein Interaction Paradigm to a Protein/Protein Engagement Model in RiPP Biosynthesis, ACS Chem. Biol., 2025 DOI:10.1021/acschembio.5c00411.
- D. T. A. Youssef, L. A. Shaala, G. A. Mohamed, J. M. Badr, F. H. Bamanie, S. R. M. Ibrahim and G. Theonellamide, a Potent Antifungal and Cytotoxic Bicyclic Glycopeptide from the Red Sea Marine Sponge Theonella swinhoei, Mar. Drugs , 2014, 12, 1911–1923 CrossRef PubMed.
- O. Hasin, S. Shoham, Y. Kashman, M. Ilan and S. Carmeli, Theonellamides J and K and 5-cis-Apoa-theopalauamide, Bicyclic Glycopeptides of the Red Sea Sponge Theonella swinhoei, Mar. Drugs, 2022, 20, 31 CrossRef CAS.
- K. Fukuhara, K. Takada, R. Watanabe, T. Suzuki, S. Okada and S. Matsunaga, Colony-wise Analysis of a Theonella swinhoei Marine Sponge with a Yellow Interior Permitted the Isolation of Theonellamide I, J. Nat. Prod., 2018, 81, 2595–2599 CrossRef CAS.
- T. Tanaka, T. Nakashima, T. Ueda, K. Tomii and I. Kouno, Facile Discrimination of Aldose Enantiomers by Reversed-Phase HPLC, Chem. Pharm. Bull., 2007, 55, 899–901 Search PubMed.
- S. Nishimura, K. Ishii, K. Iwamoto, Y. Arita, S. Matsunaga, Y. Ohno-Iwashita, S. B. Sato, H. Kakeya, T. Kobayashi and M. Yoshida, Visualization of Sterol-Rich Membrane Domains with Fluorescently-Labeled Theonellamides, PLoS One, 2013, 8, e83716 CrossRef.
- S. Nishimura, Y. Arita, M. Honda, K. Iwamoto, A. Matsuyama, A. Shirai, H. Kawasaki, H. Kakeya, T. Kobayashi, S. Matsunaga and M. Yoshida, Marine antifungal theonellamides target 3β-hydroxysterol to activate Rho1 signaling, Nat. Chem. Biol., 2010, 6, 519–526 CrossRef CAS PubMed.
- S. K. Kandy, M. A. Pasquale and J. R. Chekan, Aromatic side-chain crosslinking in RiPP biosynthesis, Nat. Chem. Biol., 2025, 21, 168–181 CrossRef CAS.
- F. Itagaki, H. Shigemori, M. Ishibashi, T. Nakamura, T. Sasaki, J. Kobayashi and F. Keramamide, a new thiazole-containing peptide from the Okinawan marine sponge Theonella sp, J. Org. Chem., 1992, 57, 5540–5542 CrossRef CAS.
- J. Kobayashi, F. Itagaki, H. Shigemori, M. Ishibashi, K. Takahashi, M. Ogura, S. Nagasawa, T. Nakamura and H. Hirota, Keramamides B .apprx. D, novel peptides from the Okinawan marine sponge Theonella sp, J. Am. Chem. Soc., 1991, 113, 7812–7813 CrossRef CAS.
- H. Uemoto, Y. Yahiro, H. Shigemori, M. Tsuda, T. Takao, Y. Shimonishi and J. i. Kobayashi, Keramamides K and L, new cyclic peptides containing unusual tryptophan residue from Theonella sponge, Tetrahedron, 1998, 54, 6719–6724 CrossRef CAS.
- T. F. Molinski, Cyclic azole-homologated peptides from Marine sponges, Org. Biomol. Chem., 2018, 16, 21–29 RSC.
- P. L. Winder, S. A. Pomponi and A. E. Wright, Natural Products from the Lithistida: A Review of the Literature since 2000, Mar. Drugs, 2011, 9, 2643–2682 CrossRef CAS.
- T. Wakimoto, Biosynthesis of Bioactive Natural Products Derived from Theonellidae Family Marine Sponges, Chem. Pharm. Bull., 2023, 71, 1–8 CrossRef CAS PubMed.
- J. Piel, D. Hui, G. Wen, D. Butzke, M. Platzer, N. Fusetani and S. Matsunaga, Antitumor polyketide biosynthesis by an uncultivated bacterial symbiont of the marine sponge Theonella swinhoei, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16222–16227 CrossRef CAS.
- S. Reiter, J. K. B. Cahn, V. Wiebach, R. Ueoka and J. Piel, Characterization of an Orphan Type III Polyketide Synthase Conserved in Uncultivated “Entotheonella” Sponge Symbionts, ChemBioChem, 2020, 21, 564–571 CrossRef CAS PubMed.
- H. Kim, J. Ahn, J. Kim and H.-S. Kang, Metagenomic insights and biosynthetic potential of Candidatus Entotheonella symbiont associated with Halichondria marine sponges, Microbiol. Spectrum, 2024, 13, e0235524 CrossRef.
- H.-y. Li, S. Matsunaga, N. Fusetani and A.-C. Halicylindramides, antifungal and cytotoxic depsipeptides from the marine sponge Halichondria cylindrata, J. Med. Chem., 1995, 38, 338–343 Search PubMed.
- H.-y. Li, S. Matsunaga and N. Fusetani, Halicylindramides D and E, Antifungal Peptides from the Marine Sponge Halichondria cylindrata, J. Nat. Prod., 1996, 59, 163–166 CrossRef CAS PubMed.
- D. Hahn, H. Kim, I. Yang, J. Chin, H. Hwang, D. H. Won, B. Lee, S.-J. Nam, M. Ekins, H. Choi and H. Kang, The Halicylindramides, Farnesoid X Receptor Antagonizing Depsipeptides from a Petrosia sp. Marine Sponge Collected in Korea, J. Nat. Prod., 2016, 79, 499–506 CrossRef CAS.
- T. Wakimoto, Y. Egami, Y. Nakashima, Y. Wakimoto, T. Mori, T. Awakawa, T. Ito, H. Kenmoku, Y. Asakawa, J. Piel and I. Abe, Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont, Nat. Chem. Biol., 2014, 10, 648–655 Search PubMed.
- Y. Kato, N. Fusetani, S. Matsunaga, K. Hashimoto, S. Fujita and T. Furuya, Bioactive marine metabolites. Part 16. Calyculin A. A novel antitumor metabolite from the marine sponge Discodermia calyx, J. Am. Chem. Soc., 1986, 108, 2780–2781 CrossRef CAS.
- Y. Nakashima, Y. Egami, M. Kimura, T. Wakimoto and I. Abe, Metagenomic Analysis of the Sponge Discodermia Reveals the Production of the Cyanobacterial Natural Product Kasumigamide by ‘Entotheonella’, PLoS One, 2016, 11, e0164468 Search PubMed.
- K. Ishida and M. Murakami, Kasumigamide, an Antialgal Peptide from the Cyanobacterium Microcystis aeruginosa, J. Org. Chem., 2000, 65, 5898–5900 CrossRef CAS.
- T. Kuranaga, K. Matsuda, M. Takaoka, C. Tachikawa, A. Sano, K. Itoh, A. Enomoto, K. Fujita, I. Abe and T. Wakimoto, Total Synthesis and Structural Revision of Kasumigamide, and Identification of a New Analogue, ChemBioChem, 2020, 21, 3329–3332 Search PubMed.
- H. Yamaguchi, S. Suzuki and M. Kawachi, Draft Genome Sequence of Microcystis aeruginosa NIES-87, a Bloom-Forming Cyanobacterium from Lake Kasumigaura, Japan, Genome Announc., 2018, 6, 8 Search PubMed.
- M. Kimura, T. Wakimoto, Y. Egami, K. C. Tan, Y. Ise and I. Abe, Calyxamides A and B, Cytotoxic Cyclic Peptides from the Marine Sponge Discodermia calyx, J. Nat. Prod., 2012, 75, 290–294 Search PubMed.
- W. M. Brück, S. H. Sennett, S. A. Pomponi, P. Willenz and P. J. McCarthy, Identification of the bacterial symbiont Entotheonella sp. in the mesohyl of the marine sponge Discodermia sp, ISME J., 2008, 2, 335–339 Search PubMed.
- C. L. Dieterich, S. I. Probst, R. Ueoka, I. Sandu, D. Schäfle, M. D. Molin, H. A. Minas, R. Costa, A. Oxenius, P. Sander and J. Piel, Aquimarins, Peptide Antibiotics with Amino-Modified C-Termini from a Sponge-Derived Aquimarina sp. Bacterium, Angew. Chem., Int. Ed., 2022, 61, e202115802 CrossRef CAS.
- A. I. S. Esteves, C. C. P. Hardoim, J. R. Xavier, J. M. S. Gonçalves and R. Costa, Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic, FEMS Microbiol. Ecol., 2013, 85, 519–536 Search PubMed.
- V. Agarwal, Z. D. Miles, J. M. Winter, A. S. Eustaquio, A. A. El Gamal and B. S. Moore, Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse, Chem. Rev., 2017, 117, 5619–5674 Search PubMed.
- W. Zhong and V. Agarwal, Polymer degrading marine Microbulbifer bacteria: an un(der)utilized source of chemical and biocatalytic novelty, Beilstein J. Org. Chem., 2024, 20, 1635–1651 Search PubMed.
- J. M. Deutsch, M. O. Green, P. Akavaram, A. C. Davis, S. S. Diskalkar, I. A. Du Plessis, H. A. Fallon, E. M. Grason, E. G. Kauf, Z. M. Kim, J. R. Miller, A. L. Neal, T. Riera, S.-E. Stroeva, J. Tran, V. Tran, A. V. Coronado, V. V. Coronado, B. T. Wall, C. M. Yang, I. Mohanty, N. H. Abrahamse, C. J. Freeman, C. G. Easson, C. L. Fiore, A. E. Onstine, N. Djeddar, S. Biliya, A. V. Bryksin, N. Garg and V. Agarwal, Limited metabolomic overlap between commensal bacteria and marine sponge holobionts revealed by large scale culturing and mass spectrometry-based metabolomics: an undergraduate laboratory pedagogical effort at Georgia Tech, Mar. Drugs, 2023, 21, 53 Search PubMed.
- W. Zhong, J. M. Deutsch, D. Yi, N. H. Abrahamse, I. Mohanty, S. G. Moore, A. C. McShan, N. Garg and V. Agarwal, Discovery and biosynthesis of ureidopeptide natural products macrocyclized via indole N-acylation in marine Microbulbifer spp. bacteria, ChemBioChem, 2023, 24, e202300190 Search PubMed.
- S. Lu, Z. Zhang, A. R. Sharma, J. Nakajima-Shimada, E. Harunari, N. Oku, A. Trianto and Y. Igarashi, Bulbiferamide, an antitrypanosomal hexapeptide cyclized via an N-acylindole linkage from a marine obligate Microbulbifer, J. Nat. Prod., 2023, 86, 1081–1086 Search PubMed.
- H. J. Imker, C. T. Walsh and W. M. Wuest, SylC catalyzes ureido-bond formation during biosynthesis of the proteasome inhibitor syringolin A, J. Am. Chem. Soc., 2009, 131, 18263–18265 Search PubMed.
- T. K. Shishido, J. Jokela, D. P. Fewer, M. Wahlsten, M. F. Fiore and K. Sivonen, Simultaneous production of anabaenopeptins and namalides by the cyanobacterium Nostoc sp. CENA543, ACS Chem. Biol., 2017, 12, 2746–2755 Search PubMed.
- S. T. Lima, D. O. Alvarenga, A. Etchegaray, D. P. Fewer, J. Jokela, A. M. Varani, M. Sanz, F. A. Dörr, E. Pinto, K. Sivonen and M. F. Fiore, Genetic organization of anabaenopeptin and spumigin biosynthetic gene clusters in the cyanobacterium Sphaerospermopsis torques-reginae ITEP-024, ACS Chem. Biol., 2017, 12, 769–778 Search PubMed.
- M. Baunach, A. Guljamow, M. Miguel-Gordo and E. Dittmann, Harnessing the potential: advances in cyanobacterial natural product research and biotechnology, Nat. Prod. Rep., 2024, 41, 347–369 Search PubMed.
- W. Zhong, Z. L. Budimir, L. O. Johnson, E. I. Parkinson and V. Agarwal, Activity and Biocatalytic Potential of an Indolylamide Generating Thioesterase, Org. Lett., 2024, 26, 9378–9382 Search PubMed.
- H. Maruyama, Y. Yamada, Y. Igarashi, K. Matsuda and T. Wakimoto, Enzymatic peptide macrocyclization via indole-N-acylation, Chem. Sci., 2025, 16, 3872–3877 Search PubMed.
- A. Rajput, K. S. Butler, D. A. Springer and J. R. Chekan, Indolylamide Macrocyclization by a Streptococcus pneumoniae ThiF-like Enzyme Family Member, Org. Lett., 2025, 27, 5765–5770 Search PubMed.
- W. Zhong, N. Aiosa, J. M. Deutsch, N. Garg and V. Agarwal, Pseudobulbiferamides: plasmid-encoded ureidopeptide natural products with biosynthetic gene clusters shared among marine bacteria of different genera, J. Nat. Prod., 2023, 86, 2414–2420 Search PubMed.
- L. P. Ióca, Y. Dai, S. Kunakom, J. Diaz-Espinosa, A. Krunic, C. M. Crnkovic, J. Orjala, L. M. Sanchez, A. G. Ferreira, R. G. S. Berlinck and A. S. Eustáquio, A family of nonribosomal peptides modulate collective behavior in Pseudovibrio bacteria isolated from marine sponges, Angew. Chem., Int. Ed., 2021, 60, 15891–15898 Search PubMed.
- C. P. J. Rua, A. E. Trindade-Silva, L. R. Appolinario, T. M. Venas, G. D. Garcia, L. S. Carvalho, A. Lima, R. Kruger, R. C. Pereira, R. G. S. Berlinck, R. A. B. Valle, C. C. Thompson and F. Thompson, Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis, PeerJ, 2014, 2, e419 Search PubMed.
- Y. Dai, V. Lourenzon, P. Ióca Laura, D. Al-Smadi, L. Arnold, I. McIntire, G. S. Berlinck Roberto and S. Eustáquio Alessandra, Pseudovibriamides from Pseudovibrio marine sponge bacteria promote flagellar motility via transcriptional modulation, mBio, 2024, 16, e0311524 Search PubMed.
- G. A. Vitale, S. Scarpato, A. Mangoni, M. V. D'Auria, G. Della Sala and D. de Pascale, Enhanced Molecular Networking Shows Microbacterium sp. V1 as a Factory of Antioxidant Proline-Rich Peptides, Mar. Drugs, 2023, 21, 256 Search PubMed.
- A. H. Ibrahim, E. Z. Attia, D. Hajjar, M. A. Anany, S. Y. Desoukey, M. A. Fouad, M. S. Kamel, H. Wajant, T. A. M. Gulder and U. R. Abdelmohsen, New Cytotoxic Cyclic Peptide from the Marine Sponge-Associated Nocardiopsis sp. UR67, Mar. Drugs, 2018, 16, 290 Search PubMed.
- S. Chen, Z. Qingbo, Z. Xinya, J. Xiaodong, Z. Haibo, Z. Yiguang, Z. Changsheng and L. Zhang, A new xanthostatin analogue from the marine sponge-associated actinomycete Streptomyces sp. SCSIO 40064, Nat. Prod. Res., 2022, 36, 3529–3537 Search PubMed.
- I. Kitagawa, N. K. Lee, M. Kobayashi and H. Shibuya, Structure Of Theonellapeptolide Ie, A New Tridecapeptide Lactone From An Okinawan Marine Sponge, Theonella Sp. (Theonellidae), Chem. Pharm. Bull., 1987, 35, 2129–2132 CrossRef CAS PubMed.
- M. Kobayashi, N. K. Lee, H. Shibuya, T. Momose and I. Kitagawa, Marine Natural Products. XXVI. Biologically Active Tridecapeptide Lactones from the Okinawan Marine Sponge Theonella swinhoei(Theonellidae).(2).Structures of Theonellapeptolides Ia, Ib, Ic, and Ie, Chem. Pharm. Bull., 1991, 39, 1177–1184 Search PubMed.
- A. Sinisi, B. Calcinai, C. Cerrano, H. A. Dien, A. Zampella, C. D'Amore, B. Renga, S. Fiorucci and O. Taglialatela-Scafati, New tridecapeptides of the theonellapeptolide family from the Indonesian sponge Theonella swinhoei, Beilstein J. Org. Chem., 2013, 9, 1643–1651 Search PubMed.
- J. R. Haedar, A. R. Uria, S. Lallo, D. F. Dibwe and T. Wakimoto, New Theonellapeptolides from Indonesian Marine Sponge Theonella swinhoei as Anti-Austerity Agents, Mar. Drugs, 2022, 20, 661 Search PubMed.
- M. Issac, M. Aknin, A. Gauvin-Bialecki, N. De Voogd, A. Ledoux, M. Frederich, Y. Kashman and S. Carmeli, Cyclotheonellazoles A–C, Potent Protease Inhibitors from the Marine Sponge Theonella aff. swinhoei, J. Nat. Prod., 2017, 80, 1110–1116 Search PubMed.
- D. C. Holland, W. A. Schroder, M. J. Calcott, E. Kaemmerer, V. M. Avery, M. G. Ekins, A. R. Carroll and D.-I. Cyclotheonellazoles, Potent Elastase Inhibitory Thiazole-Containing Cyclic Peptides from Theonella sp. (2131), J. Nat. Prod., 2023, 86, 2216–2227 Search PubMed.
- B. Long, L. Pu, Z. Liu, X. Zeng and Z. Wu, Formal synthesis of cyclotheonellazole A, Org. Biomol. Chem., 2023, 21, 3531–3536 Search PubMed.
- L. Chill, Y. Kashman and M. Schleyer, Oriamide, a new cytotoxic cyclic peptide containing a novel amino acid from the marine sponge Theonella sp, Tetrahedron, 1997, 53, 16147–16152 CrossRef CAS.
- M. T. Jamison and T. F. Molinski, Jamaicensamide A, a Peptide Containing β-Amino-α-keto and Thiazole-Homologated η-Amino Acid Residues from the Sponge Plakina jamaicensis, J. Nat. Prod., 2016, 79, 2243–2249 CrossRef CAS.
- D. P. Clark, J. Carroll, S. Naylor and P. Crews, An Antifungal Cyclodepsipeptide, Cyclolithistide A, from the Sponge Theonella swinhoei, J. Org. Chem., 1998, 63, 8757–8764 CrossRef CAS.
- H. Tajima, T. Wakimoto, K. Takada, Y. Ise and I. Abe, Revised Structure of Cyclolithistide A, a Cyclic Depsipeptide from the Marine Sponge Discodermia japonica, J. Nat. Prod., 2014, 77, 154–158 CrossRef CAS.
- T. Tian, Y. Ogura, Y. Ise, H. Takikawa, S. Okada, S. Matsunaga and G.-L. Tang, Absolute Configuration of 4-Chloroisoleucine in Cyclolithistide A: An Antifungal Peptide Lactone Discovered in Marine Lithistid Sponges, J. Org. Chem., 2024, 89, 9135–9138 CrossRef CAS.
- G. Tarazona, R. Fernández, P. G. Cruz, M. Pérez, J. Rodríguez, C. Jiménez and C. Cuevas, Combining JBCA and Marfey's methodology to determine the absolute configuration of threonines: the case of gunungamide A, a new cyclic depsipeptide containing chloropyrrole from the sponge Discodermia sp, Org. Chem. Front., 2019, 6, 15–21 RSC.
- C. Festa, S. De Marino, V. Sepe, M. V. D'Auria, G. Bifulco, C. Débitus, M. Bucci, V. Vellecco and A. Zampella, Solomonamides A and B, New Anti-inflammatory Peptides from Theonella swinhoei, Org. Lett., 2011, 13, 1532–1535 CAS.
- N. Vasudevan, K. Kashinath and D. S. Reddy, Total Synthesis of Deoxy-solomonamide B by Mimicking Biogenesis, Org. Lett., 2014, 16, 6148–6151 CrossRef CAS.
- N. Kavitha and S. Chandrasekhar, Scalable synthesis of the unusual amino acid segment (ADMOA unit) of marine anti-inflammatory peptide: solomonamide A, Org. Biomol. Chem., 2015, 13, 6242–6248 RSC.
- K. Kashinath, G. R. Jachak, P. R. Athawale, U. K. Marelli, R. G. Gonnade and D. S. Reddy, Total Synthesis of the Marine Natural Product Solomonamide B Necessitates Stereochemical Revision, Org. Lett., 2016, 18, 3178–3181 CrossRef CAS.
- I. Cheng-Sánchez, C. García-Ruíz and F. Sarabia, An olefin metathesis approach towards the solomonamides, Tetrahedron Lett., 2016, 57, 3392–3395 CrossRef.
- G. R. Jachak, P. R. Athawale, H. Agarwal, M. K. Barthwal, G. Lauro, G. Bifulco and D. S. Reddy, Total synthesis of the potent anti-inflammatory natural product solomonamide A along with structural revision and biological activity evaluation, Org. Biomol. Chem., 2018, 16, 9138–9142 RSC.
- P. Carrillo, B. Martínez-Poveda, I. Cheng-Sánchez, J. Guerra, C. Tobia, J. M. López-Romero, F. Sarabia, M. Á. Medina and A. R. Quesada, Exploring the Antiangiogenic Potential of Solomonamide A Bioactive Precursors: In Vitro and In Vivo Evidences of the Inhibitory Activity of Solo F-OH During Angiogenesis, Mar. Drugs, 2019, 17, 228 CrossRef CAS PubMed.
- G. R. Jachak, K. Kashinath, N. Vasudevan, P. R. Athawale, R. Choudhury, S. S. Dange, H. Agarwal, M. K. Barthwal and D. S. Reddy, Comprehensive Study on Solomonamides: Total Synthesis, Stereochemical Revision, and SAR Studies toward Identification of Simplified Lead, J. Org. Chem., 2023, 88, 17088–17133 CrossRef CAS PubMed.
- N. Oku, K. R. Gustafson, L. K. Cartner, J. A. Wilson, N. Shigematsu, S. Hess, L. K. Pannell, M. R. Boyd and J. B. McMahon, Neamphamide A, a New HIV-Inhibitory Depsipeptide from the Papua New Guinea Marine Sponge Neamphius huxleyi, J. Nat. Prod., 2004, 67, 1407–1411 CrossRef CAS PubMed.
- N. Oku, R. Krishnamoorthy, A. G. Benson, R. L. Ferguson, M. A. Lipton, L. R. Phillips, K. R. Gustafson and J. B. McMahon, Complete Stereochemistry of Neamphamide A and Absolute Configuration of the β-Methoxytyrosine Residue in Papuamide B, J. Org. Chem., 2005, 70, 6842–6847 CrossRef CAS.
- Y. Yamano, M. Arai, M. Kobayashi and B. Neamphamide, new cyclic depsipeptide, as an anti-dormant mycobacterial substance from a Japanese marine sponge of Neamphius sp, Bioorg. Med. Chem. Lett., 2012, 22, 4877–4881 CrossRef CAS.
- T. D. Tran, N. B. Pham, G. Fechner, D. Zencak, H. T. Vu, J. N. A. Hooper and R. J. Quinn, Cytotoxic Cyclic Depsipeptides from the Australian Marine Sponge Neamphius huxleyi, J. Nat. Prod., 2012, 75, 2200–2208 CrossRef CAS.
- M. J. Martín, R. Rodríguez-Acebes, Y. García-Ramos, V. Martínez, C. Murcia, I. Digón, I. Marco, M. Pelay-Gimeno, R. Fernández, F. Reyes, A. M. Francesch, S. Munt, J. Tulla-Puche, F. Albericio and C. Cuevas, Stellatolides, a New Cyclodepsipeptide Family from the Sponge Ecionemia acervus: Isolation, Solid-Phase Total Synthesis, and Full Structural Assignment of Stellatolide A, J. Am. Chem. Soc., 2014, 136, 6754–6762 CrossRef PubMed.
- S. Nakamukai, K. Takada, K. Furihata, Y. Ise, S. Okada, Y. Morii, N. Yamawaki, T. Takatani, O. Arakawa, K. R. Gustafson, S. Matsunaga and H. Stellatolide, a cytotoxic peptide lactone from a deep-sea sponge Discodermia sp, Tetrahedron Lett., 2018, 59, 2532–2536 CrossRef CAS.
- M. Pelay-Gimeno, Y. García-Ramos, M. Jesús Martin, J. Spengler, J. M. Molina-Guijarro, S. Munt, A. M. Francesch, C. Cuevas, J. Tulla-Puche and F. Albericio, The first total synthesis of the cyclodepsipeptide pipecolidepsin A, Nat. Commun., 2013, 4, 2352 CrossRef PubMed.
- L. Coello, F. Reyes, M. J. Martín, C. Cuevas and R. Fernández, Isolation and Structures of Pipecolidepsins A and B, Cytotoxic Cyclic Depsipeptides from the Madagascan Sponge Homophymia lamellosa, J. Nat. Prod., 2014, 77, 298–303 CrossRef CAS.
- G. J. Gatto, M. T. Boyne, N. L. Kelleher and C. T. Walsh, Biosynthesis of Pipecolic Acid by RapL, a Lysine Cyclodeaminase Encoded in the Rapamycin Gene Cluster, J. Am. Chem. Soc., 2006, 128, 3838–3847 CrossRef CAS.
- J. B. Hedges and K. S. Ryan, Biosynthetic Pathways to Nonproteinogenic α-Amino Acids, Chem. Rev., 2020, 120, 3161–3209 CrossRef CAS.
- A. Plaza, G. Bifulco, M. Masullo, J. R. Lloyd, J. L. Keffer, P. L. Colin, J. N. A. Hooper, L. J. Bell and C. A. Bewley, Mutremdamide A and Koshikamides C–H, Peptide Inhibitors of HIV-1 Entry from Different Theonella Species, J. Org. Chem., 2010, 75, 4344–4355 CrossRef CAS PubMed.
- K. C. Tan, T. Wakimoto and I. Abe, Lipodiscamides A–C, New Cytotoxic Lipopeptides from Discodermia kiiensis, Org. Lett., 2014, 16, 3256–3259 CrossRef CAS PubMed.
- K. C. Tan, T. Wakimoto and I. Abe, Sulfoureido Lipopeptides from the Marine Sponge Discodermia kiiensis, J. Nat. Prod., 2016, 79, 2418–2422 CrossRef CAS.
- C. A. Bewley, H. He, D. H. Williams and D. J. Faulkner, Aciculitins A–C: Cytotoxic and Antifungal Cyclic Peptides from the Lithistid Sponge Aciculites orientalis, J. Am. Chem. Soc., 1996, 118, 4314–4321 CrossRef CAS.
- K. Sugawara, D. Kanki, R. Watanabe, R. Matsushima, Y. Ise, H. Yokose, Y. Morii, N. Yamawaki, A. Ninomiya, S. Okada, S. Matsunaga and D. Aciculitin, a cytotoxic heterodetic cyclic peptide from a Poecillastra sp. marine sponge, Tetrahedron, 2022, 119, 132859 CrossRef CAS.
- L. Padva, J. Gullick, L. J. Coe, M. H. Hansen, J. J. De Voss, M. Crüsemann and M. J. Cryle, The Biarylitides: Understanding the Structure and Biosynthesis of a Fascinating Class of Cytochrome P450 Modified RiPP Natural Products, ChemBioChem, 2025, 26, e202400916 CrossRef CAS PubMed.
- S. Guo, S. Wang, S. Ma, Z. Deng, W. Ding and Q. Zhang, Radical SAM-dependent ether crosslink in daropeptide biosynthesis, Nat. Commun., 2022, 13, 2361 CrossRef CAS PubMed.
- K. A. Clark and M. R. Seyedsayamdost, Bioinformatic Atlas of Radical SAM Enzyme-Modified RiPP Natural Products Reveals an Isoleucine–Tryptophan Crosslink, J. Am. Chem. Soc., 2022, 144, 17876–17888 CrossRef CAS PubMed.
- S. R. M. Ibrahim, C. C. Min, F. Teuscher, R. Ebel, C. Kakoschke, W. Lin, V. Wray, R. Edrada-Ebel and P. Proksch, Callyaerins A–F and H, new cytotoxic cyclic peptides from the Indonesian marine sponge Callyspongia aerizusa, Bioorg. Med. Chem., 2010, 18, 4947–4956 CrossRef CAS.
- S. R. M. Ibrahim, R. Edrada-Ebel, G. A. Mohamed, D. T. A. Youssef, V. Wray, P. Proksch and G. Callyaerin, a new cytotoxic cyclic peptide from the marine sponge Callyspongia aerizusa, ARKIVOC, 2008, 2008, 164–171 Search PubMed.
- G. Daletos, R. Kalscheuer, H. Koliwer-Brandl, R. Hartmann, N. J. de Voogd, V. Wray, W. Lin and P. Proksch, Callyaerins from the Marine Sponge Callyspongia aerizusa: Cyclic Peptides with Antitubercular Activity, J. Nat. Prod., 2015, 78, 1910–1925 CrossRef CAS.
- S. Zhang, L. M. De Leon Rodriguez, I. K. H. Leung, G. M. Cook, P. W. R. Harris and M. A. Brimble, Total Synthesis and Conformational Study of Callyaerin A: Anti-Tubercular Cyclic Peptide Bearing a Rare Rigidifying (Z)-2,3- Diaminoacrylamide Moiety, Angew. Chem., Int. Ed., 2018, 57, 3631–3635 CrossRef CAS.
- D. Podlesainski, E. T. Adeniyi, Y. Gröner, F. Schulz, V. Krisilia, N. Rehberg, T. Richter, D. Sehr, H. Xie, V. E. Simons, A.-L. Kiffe-Delf, F. Kaschani, T. R. Ioerger, M. Kaiser and R. Kalscheuer, The anti-tubercular callyaerins target the Mycobacterium tuberculosis-specific non-essential membrane protein Rv2113, Cell Chem. Biol., 2024, 31, 1755–1771e73 CrossRef PubMed.
- P. Cheruku, A. Plaza, G. Lauro, J. Keffer, J. R. Lloyd, G. Bifulco and C. A. Bewley, Discovery and Synthesis of Namalide Reveals a New Anabaenopeptin Scaffold and Peptidase Inhibitor, J. Med. Chem., 2012, 55, 735–742 CrossRef PubMed.
- M. Sanz, R. K. Salinas and E. Pinto, Namalides B and C and Spumigins K–N from the Cultured Freshwater Cyanobacterium Sphaerospermopsis torques-reginae, J. Nat. Prod., 2017, 80, 2492–2501 CrossRef.
- T. L. Simmons, R. C. Coates, B. R. Clark, N. Engene, D. Gonzalez, E. Esquenazi, P. C. Dorrestein and W. H. Gerwick, Biosynthetic origin of natural products isolated from marine microorganism-invertebrate assemblages, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4587–4594 CrossRef.
- J. i. Kobayashi, M. Sato, T. Murayama, M. Ishibashi, M. R. Wälchi, M. Kanai, J. Shoji and Y. Ohizumi, Konbamide, a novel peptide with calmoduiin antagonistic activity from the Okinawan marine sponge Theonella sp, J. Chem. Soc. Chem. Commun., 1991, 1050–1052 RSC.
- E. H. Andrianasolo, H. Gross, D. Goeger, M. Musafija-Girt, K. McPhail, R. M. Leal, S. L. Mooberry and W. H. Gerwick, Isolation of Swinholide A and Related Glycosylated Derivatives from Two Field Collections of Marine Cyanobacteria, Org. Lett., 2005, 7, 1375–1378 CrossRef PubMed.
- R. Ueoka, A. R. Uria, S. Reiter, T. Mori, P. Karbaum, E. E. Peters, E. J. N. Helfrich, B. I. Morinaka, M. Gugger, H. Takeyama, S. Matsunaga and J. Piel, Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms, Nat. Chem. Biol., 2015, 11, 705–712 CrossRef PubMed.
- A. Humisto, J. Jokela, L. Liu, M. Wahlsten, H. Wang, P. Permi, P. Machado João, A. Antunes, P. Fewer David and K. Sivonen, The Swinholide Biosynthesis Gene Cluster from a Terrestrial Cyanobacterium, Nostoc sp. Strain UHCC 0450, Appl. Environ. Microbiol., 2018, 84, e0232117 CrossRef PubMed.
- D. Kanki, S. Nakamukai, Y. Ogura, H. Takikawa, Y. Ise, Y. Morii, N. Yamawaki, T. Takatani, O. Arakawa, S. Okada and S. Matsunaga, Homophymamide A, heterodetic cyclic tetrapeptide from a Homophymia sp. marine sponge: a cautionary note on configurational assignment of peptides that contain a ureido linkage, J. Nat. Prod., 2021, 84, 1848–1853 CrossRef PubMed.
- N. S. van der Velden, N. Kälin, M. J. Helf, J. Piel, M. F. Freeman and M. Künzler, Autocatalytic backbone N-methylation in a family of ribosomal peptide natural products, Nat. Chem. Biol., 2017, 13, 833–835 CrossRef PubMed.
- M. R. Quijano, C. Zach, F. S. Miller, A. R. Lee, A. S. Imani, M. Künzler and M. F. Freeman, Distinct Autocatalytic α-N-Methylating Precursors Expand the Borosin RiPP Family of Peptide Natural Products, J. Am. Chem. Soc., 2019, 141, 9637–9644 CrossRef PubMed.
- A. R. Lee, R. S. Carter, A. S. Imani, S. R. Dommaraju, G. A. Hudson, D. A. Mitchell and M. F. Freeman, Discovery of Borosin Catalytic Strategies and Function through Bioinformatic Profiling, ACS Chem. Biol., 2024, 19, 1116–1124 CrossRef.
- C. Urda, M. Pérez, J. Rodríguez, C. Jiménez, C. Cuevas and R. Fernández, Pembamide, a N-methylated linear peptide from a sponge Cribrochalina sp, Tetrahedron Lett., 2016, 57, 3239–3242 Search PubMed.
- M. A. Knestrick, N. G. Wilson, A. Roth, J. H. Adams and B. J. Baker, Friomaramide, a Highly Modified Linear Hexapeptide from an Antarctic Sponge, Inhibits Plasmodium falciparum Liver-Stage Development, J. Nat. Prod., 2019, 82, 2354–2358 CrossRef PubMed.
- J. Bracegirdle, D. Casandra, J. R. Rocca, J. H. Adams and B. J. Baker, Highly N-Methylated Peptides from the Antarctic Sponge Inflatella coelosphaeroides Are Active against Plasmodium falciparum, J. Nat. Prod., 2022, 85, 2454–2460 CrossRef PubMed.
- R. Ueoka, Y. Ise, S. Ohtsuka, S. Okada, T. Yamori and S. Matsunaga, Yaku'amides A and B, Cytotoxic Linear Peptides Rich in Dehydroamino Acids from the Marine Sponge Ceratopsion sp, J. Am. Chem. Soc., 2010, 132, 17692–17694 CrossRef PubMed.
- T. Kuranaga, Y. Sesoko, K. Sakata, N. Maeda, A. Hayata and M. Inoue, Total Synthesis and Complete Structural Assignment of Yaku'amide A, J. Am. Chem. Soc., 2013, 135, 5467–5474 CrossRef PubMed.
- H. Mutoh, Y. Sesoko, T. Kuranaga, H. Itoh and M. Inoue, The total synthesis and functional evaluation of fourteen stereoisomers of yaku'amide B. The importance of stereochemistry for hydrophobicity and cytotoxicity, Org. Biomol. Chem., 2016, 14, 4199–4204 RSC.
- K. Kitamura, H. Itoh, K. Sakurai, S. Dan and M. Inoue, Target Identification of Yaku'amide B and Its Two Distinct Activities against Mitochondrial FoF1-ATP Synthase, J. Am. Chem. Soc., 2018, 140, 12189–12199 CrossRef.
- K. Kamiya, K. Miura, H. Itoh and M. Inoue, Divergent Solid-Phase Synthesis and Biological Evaluation of Yaku'amide B and Its Seven E/Z Isomers, Chem.–Eur. J., 2021, 27, 1088–1093 CrossRef CAS PubMed.
- Y. Cai, Z. Ma, J. Jiang, C. C. L. Lo, S. Luo, A. Jalan, J. M. Cardon, A. Ramos, D. A. Moyá, D. Joaquin and S. L. Castle, Convergent Total Synthesis of Yaku'amide A, Angew. Chem., Int. Ed., 2021, 60, 5162–5167 CrossRef CAS PubMed.
- C. C. L. Lo, D. Joaquin, D. A. Moyá, A. Ramos, D. W. Kastner, S. M. White, B. L. Christensen, J. G. Naglich, W. J. Degnen and S. L. Castle, Synthesis and evaluation of potent yaku'amide A analogs, Chem. Sci., 2022, 13, 1899–1905 RSC.
- A. Castaldi, R. Teta, G. Esposito, M. A. Beniddir, N. J. De Voogd, S. Duperron, V. Costantino and M.-L. Bourguet-Kondracki, Computational Metabolomics Tools Reveal Subarmigerides, Unprecedented Linear Peptides from the Marine Sponge Holobiont Callyspongia subarmigera, Mar. Drugs, 2022, 20, 673 CrossRef CAS.
- A. R. Carroll, B. R. Copp, T. Grkovic, R. A. Keyzers and M. R. Prinsep, Marine natural products, Nat. Prod. Rep., 2025, 42, 257–297 RSC.
- J.-Y. Jiao, L. Liu, Z.-S. Hua, B.-Z. Fang, E.-M. Zhou, N. Salam, B. P. Hedlund and W.-J. Li, Microbial dark matter coming to light: challenges and opportunities, Natl. Sci. Rev., 2021, 8, nwaa280 CrossRef CAS PubMed.
- C. Loureiro, A. Galani, A. Gavriilidou, M. Chaib de Mares, J. van der Oost, H. Medema Marnix and D. Sipkema, Comparative Metagenomic Analysis of Biosynthetic Diversity across Sponge Microbiomes Highlights Metabolic Novelty, Conservation, and Diversification, mSystems, 2022, 7, e0035722 CrossRef.
- R. M. Van Wagoner, M. Satake and J. L. C. Wright, Polyketide biosynthesis in dinoflagellates: what makes it different?, Nat. Prod. Rep., 2014, 31, 1101–1137 RSC.
- C. M. Preston, K. Y. Wu, T. F. Molinski and E. F. DeLong, A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 6241–6246 CrossRef.
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Retrospective analysis of natural products provides insights for future discovery trends, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5601–5606 CrossRef.
- S. Leopold-Messer, C. Chepkirui, M. F. J. Mabesoone, J. Meyer, L. Paoli, S. Sunagawa, A. R. Uria, T. Wakimoto and J. Piel, Animal-associated marine Acidobacteria with a rich natural-product repertoire, Chem, 2023, 9, 3696–3713 Search PubMed.
- S. Abbas, M. Kelly, J. Bowling, J. Sims, A. Waters and M. Hamann, Advancement into the Arctic Region for Bioactive Sponge Secondary Metabolites, Mar. Drugs, 2011, 9, 2423–2437 CrossRef PubMed.
- D. Skropeta and L. Wei, Recent advances in deep-sea natural products, Nat. Prod. Rep., 2014, 31, 999–1025 RSC.
- D. G. I. Kingston, Modern Natural Products Drug Discovery and Its Relevance to Biodiversity Conservation, J. Nat. Prod., 2011, 74, 496–511 CrossRef.
- D. Newman, Screening and identification of novel biologically active natural compounds, F1000Research, 2017, 6, 783 Search PubMed.
- C. C. Thornburg, J. R. Britt, J. R. Evans, R. K. Akee, J. A. Whitt, S. K. Trinh, M. J. Harris, J. R. Thompson, T. L. Ewing, S. M. Shipley, P. G. Grothaus, D. J. Newman, J. P. Schneider, T. Grkovic and B. R. O'Keefe, NCI Program for Natural Product Discovery: A Publicly-Accessible Library of Natural Product Fractions for High-Throughput Screening, ACS Chem. Biol., 2018, 13, 2484–2497 CrossRef.
- L. Martínez-Fructuoso, S. J. R. Arends, V. F. Freire, J. R. Evans, S. DeVries, B. D. Peyser, R. K. Akee, C. C. Thornburg, R. Kumar, S. Ensel, G. M. Morgan, G. D. McConachie, N. Veeder, L. R. Duncan, T. Grkovic and B. R. O'Keefe, Screen for New Antimicrobial Natural Products from the NCI Program for Natural Product Discovery Prefractionated Extract Library, ACS Infect. Dis., 2023, 9, 1245–1256 CrossRef.
- T. Grkovic, R. K. Akee, C. C. Thornburg, S. K. Trinh, J. R. Britt, M. J. Harris, J. R. Evans, U. Kang, S. Ensel, C. J. Henrich, K. R. Gustafson, J. P. Schneider and B. R. O'Keefe, National Cancer Institute (NCI) Program for Natural Products Discovery: Rapid Isolation and Identification of Biologically Active Natural Products from the NCI Prefractionated Library, ACS Chem. Biol., 2020, 15, 1104–1114 CrossRef.
- H. Mohimani, A. Gurevich, A. Mikheenko, N. Garg, L. F. Nothias, A. Ninomiya, K. Takada, P. C. Dorrestein and P. A. Pevzner, Dereplication of peptidic natural products through database search of mass spectra, Nat. Chem. Biol., 2017, 13, 30–37 CrossRef.
- N. Saha, F. N. U. Vidya, R. Xie and V. Agarwal, Halogenase-Assisted Alkyne/Aryl Bromide Sonogashira Coupling for Ribosomally Synthesized Peptides, J. Am. Chem. Soc., 2024, 146, 30009–30013 CrossRef CAS PubMed.
- N. A. Nguyen and V. Agarwal, A leader-guided substrate tolerant RiPP brominase allows Suzuki–Miyaura cross-coupling reactions for peptides and proteins, Biochemistry, 2023, 62, 1838–1843 CrossRef CAS.
- X. Vila-Farres, J. Chu, D. Inoyama, M. A. Ternei, C. Lemetre, L. J. Cohen, W. Cho, B. V. B. Reddy, H. A. Zebroski, J. S. Freundlich, D. S. Perlin and S. F. Brady, Antimicrobials Inspired by Nonribosomal Peptide Synthetase Gene Clusters, J. Am. Chem. Soc., 2017, 139, 1404–1407 CrossRef CAS.
- W. Ma, W. S. Noble and T. L. Bailey, Motif-based analysis of large nucleotide data sets using MEME-ChIP, Nat. Protoc., 2014, 9, 1428–1450 CrossRef CAS.
- N. E. Grayson, P. D. Scesa, M. L. Moore, J.-B. Ledoux, J. Gomez-Garrido, T. Alioto, T. P. Michael, I. Burkhardt, E. W. Schmidt and B. S. Moore, A widespread metabolic gene cluster family in metazoans, Nat. Chem. Biol., 2025 DOI:10.21203/rs.3.rs-4859447/v1.
- T. L. Koch, J. P. Torres, R. P. Baskin, P. F. Salcedo, K. Chase, B. M. Olivera and H. Safavi-Hemami, A toxin-based approach to neuropeptide and peptide hormone discovery, Front. Mol. Neurosci., 2023, 16, 1176662 CrossRef CAS PubMed.
- K. Steffen, E. Proux-Wéra, L. Soler, A. Churcher, J. Sundh and P. Cárdenas, Whole genome sequence of the deep-sea sponge Geodia barretti (Metazoa, Porifera, Demospongiae), G3:Genes, Genomes, Genet., 2023, 13, jkad192 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |