Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Advances in the discovery and study of Trichoderma natural products for biological control applications
Sophie
Jin
 a and
Fabrizio
Alberti
a and
Fabrizio
Alberti
 *b
*b
aLeibniz Institute for Natural Product Research and Infection Biology – Hans Knöll Institute (HKI), Beutenbergstraße 11a, 07745 Jena, Germany. E-mail: sophie.jin@leibniz-hki.de; Tel: +49 3641 532-1740
bSchool of Life Sciences, University of Warwick, Coventry CV4 7AL, UK. E-mail: F.Alberti@warwick.ac.uk; Tel: +44 (0) 24 765 23516
First published on 6th June 2025
Abstract
Covering: up to 2025
Reducing the prevalence of phytopathogens and their impact on crops is essential to reach sustainable agriculture goals. Synthetic pesticides have been commonly used to control crop disease but are now strongly linked to disease resistance, environmental pollution, depletion of soil biodiversity, and bioaccumulation, leading to adverse effects on human health. As a alternative, the prolific Trichoderma genus has been studied for its biocontrol properties, as well as its ability to promote plant growth and increase nutrient uptake. This is done through various mechanisms, one of which is the production of bioactive natural products with high chemical diversity. These include terpenoids, alkaloids, non-ribosomal peptides, polyketides and RiPPs. One of the most studied examples is 6-pentyl-2H-pyran-2-one, a volatile organic polyketide, which induces systemic acquired resistance, morphogenesis, and natural product biosynthesis in plants. Methods for culturing Trichoderma spp., isolating and characterising unique bioactive metabolites are discussed here, with an emphasis on dereplication strategies using metabolomics to optimise discovery. In addition, the role of genome mining for the study of natural product biosynthesis in Trichoderma, and more generally, filamentous fungi is discussed. Examples of bioinformatics tools available to date are listed here with applications in Trichoderma and other ascomycetes. New advances in genome engineering in Trichoderma are also detailed, providing insights into available strategies for the validation of biosynthetic gene clusters identified using genome mining. Finally, the use of a combination of omics approaches, namely metabologenomics, is presented as a growing field for natural product discovery in fungi.
1 Introduction
Reducing the prevalence of phytopathogens and their impact on crops is essential to reach sustainable agriculture. Synthetic pesticides have been commonly used to control crop disease but are not sustainable. Indeed, they are linked to disease resistance, environmental pollution, depletion of soil biodiversity, and bioaccumulation linked to adverse effects on human health.1,2 Many chemical pesticides are now banned in the EU due to their concerning effects on environmental and human health.3 Instead, biocontrol agents (BCA) or biopesticides are getting increasing traction for their environmentally friendly characteristics. Examples of BCA include Streptomyces griseoviridis strain K61 (Mycostop®),4Pythium oligandrum strain M1/ATCC 38472 (Polyversum®, commercialised by De Sangosse), and Trichoderma asperellum strain TV1 (Xedavir®, commercialised by Xeda Italia S.r.l).Trichoderma is a well-known genus of filamentous ascomycetes from the Hypocreaceae family and is widely distributed around the world, colonising a variety of ecological habitats such as soil, decaying wood, living plants (within the rizhosphere or as endophytes) and marine ecosystems.5,6 To date, the genus contains over 400 species spanning over several clades,7 of which the Harzianum clade includes most of the biocontrol agents used in agriculture.8Trichoderma spp. display biocontrol properties against plant pathogens through various mechanisms such as antibiosis, competition, or mycoparasitism.9–12 Aside from biotic stresses, Trichoderma spp. can also protect plants from abiotic stresses such as drought or salt stress via transcriptional activation of defence responses resulting in inhibition of seed germination and plant development, as well as stomatal regulation.13–15 In addition to biocontrol potential, Trichoderma spp. can also enhance plant development by improving nutrient uptake, photosynthesis and overall plant growth.16–18
2 Trichoderma spp. as a solution for crop management
One of the most used and researched genus of BCA is Trichoderma and a list of commercially available Trichoderma-based formulations can be found in the recent review by Martinez et al.53 and Woo et al.54 Species such as T. asperellum, T. atroviride, T. gamsii, T. hamatum, T. polysporum, T. virens, T. viride and T. harzianum are commercialised in Europe, the US, Canada, Australia, New Zealand, South Africa, Vietnam and India as biological control agents. One example of product available in Europe is ASPERELLO® T34 Biocontrol (Trichoderma asperellum T34), a fungicide sold as a powder to be mixed with the substrate before transplantation.54 Another strain which is available in Europe and the US is T. harzianum Rifai T-22 (T-22™ HC, BioWorks®), a formulated preventative fungicide which can be applied to seeds, soil and other propagative parts, protecting the plants from root pathogens. Other products utilising T. harzianum include AkTRIvator® (Canna International BV, Breda, the Netherlands) Trichosan® (Vitalin Pflanzengesundheit GmbH, Ober-Ramstadt, Germany) and Promot® WP (JH Biotech Inc., Ventura, California).Many Trichoderma species exhibit an antagonistic effect on plant pathogens through antibiosis, competition or mycoparasitism11 and can thus improve productivity,10,55 and this review will focus mainly on antibiosis as a mode of action for plant disease suppression. Antibiosis is a common phenomenon seen in Trichoderma spp. and can be defined as the interaction between microorganisms through the production of specialised metabolites and resulting in toxicity or growth inhibition for one of the microorganism.56 Many recent studies have explored the potential of Trichoderma spp. as biocontrol agents against pathogenic fungi and bacteria of plants34,35,38,43,45,50,51 and the subject has been extensively reviewed.54,57–59 Examples of species of Trichoderma which have shown antagonism against plant pathogens and their mechanisms are detailed in Table 1. An exhaustive list of interactions between Trichoderma spp. and plants can be found in the review by Sood et al.59 Reports of Trichoderma spp. as an insect pests BCA have also been reviewed by Poveda et al.60
| Species | Mode of action | Pathogens | Reference |
|---|---|---|---|
| Trichoderma viride | Specialised metabolites and volatile organic compounds (VOCs) | Fusarium oxysporum, Pythium aphanidermatum, Rhizoctonia solani, Sclerotium rolfsii, Candida albicans, Pythium ultimum, Nigrospora oryzae | 19–21 |
| Trichoderma atroviride | Cell wall degrading enzymes, antibiosis (polyketides against R. solani), VOCs | Verticillium dahlia, Rhizoctonia solani, Botrytis cinerea, Phytophthora capsica, Plasmopara viticola, Nigrospora oryzae | 21–25 |
| Trichoderma harzianum | Cell wall degrading enzymes, VOCs, specialised metabolites (gliotoxin, peptaibols, antrhaquinones, methyl dihydrojasmonate) | Sclerotinia sclerotiorum, Rhizoctonia solani, Plasmopara viticola, Pythium aphanidermatum, Fusarium oxysporum | 19 and 25–29 |
| Trichoderma koningii | Parasitism and specialised metabolites: trichokonins | Sclerotinia sclerotiorum | 30 and 31 |
| Trichoderma pseudokoningii | Specialised metabolites: peptaibols and trichokonins | Fusarium oxysporum | 32 |
| Trichoderma koningiopsis | Specialised metabolites: trichodermin, azetidine, 2-phenylethanol, and ethyl hexadecanoate, polyketides, VOCs | Pyricularia oryzae, Aspergillus fumigatus, Botrytis cinera, Colletotrichum gloeosporioides, Fusarium oxysporum, Verticillium dahliae | 33–37 |
| Trichoderma asperellum | VOCs, chitinases | Fusarium incarnatum, Plasmopara viticola, Nigrospora oryzae | 21, 25 and 38–40 |
| Trichoderma virens | Gliotoxin and trichodermamides | Rhizoctonia solani | 23 and 41 |
| Trichoderma longibrachiatum | Parasitism through cell wall degrading enzymes against nematodes, peptaibol production | Magnaporthiopsis maydis, Meloidogyne incognita, Heterodera avenae, Pseudomonas syringae | 42–44 |
| Trichoderma reesei | Cell wall degrading enzymes and secretion of phenols, and antifungal compounds (peptaibols, anthocyanins, β-caryophyllens) | Rhizoctonia solani, Fusarium oxysporum | 23 and 45 |
| Trichoderma lignorum | Specialised metabolites: gliotoxin | Rhizoctonia solani, Sclerotinia americana | 46 |
| Trichoderma brevicompactum | Cell wall degrading enzymes, secretion of indole acetic acid, trichodermin | Fusarium oxysporum | 47 and 48 |
| Trichoderma carraovejensis | Uncharacterised antagonism | Phaeoacremonium minimum, Phaeomoniella chlamydospora, Diplodia seriata | 49 |
| Trichoderma spp. | Spore adhesion and niche exclusion, stimulating gene expression involved in plant-disease resistance, (±)-trichodermatrione A production | Phaeoacremonium minimum, Fusarium oxysporum, Xanthomonas oryzae | 50–52 |
3 Specialised metabolism of Trichoderma spp.
Trichoderma species are talented producers of specialised metabolites. With over 200 isolated compounds reported recently, the potential of Trichoderma spp. still remains untapped.61,62 Not only are those compounds chemically diverse, but they also exhibit a broad spectrum of bioactivity, with a wide range of applications. The vast majority of these specialised metabolites are synthesised by biosynthetic gene clusters (BGC). Genes in a BGC encode for specific steps in the biosynthesis pathway of metabolites and are co-localised in the genome, which is thought to be due to evolutionary pressure (mainly coinheritance and coregulation).63 In fungi, a typical BGC contains genes encoding one or more core enzymes which catalyse the synthesis of the backbone of the final product, several accessory enzymes (hydrolases, epimerases, oxidoreductases, and methyltransferases amongst others) which further modify the backbone, regulatory proteins, transport-related proteins and in some cases, proteins related to resistance mechanisms.643.1 Chemical diversity of natural products in Trichoderma spp.
Reviews of specialised metabolites from the Trichoderma genus have been published in 2016,65 2021![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 62 and most recently in 2023.66 The most recent review on specialised metabolites from Trichoderma spp. focused on marine strains and listed the isolation of 445 specialised metabolites over the past 30 years, some of which presented new carbon skeletons.67 As this subject has been extensively investigated, this section will focus on the most relevant natural product classes with a discussion on some of their corresponding examples. Structures of each example can be found in Fig. 1.
62 and most recently in 2023.66 The most recent review on specialised metabolites from Trichoderma spp. focused on marine strains and listed the isolation of 445 specialised metabolites over the past 30 years, some of which presented new carbon skeletons.67 As this subject has been extensively investigated, this section will focus on the most relevant natural product classes with a discussion on some of their corresponding examples. Structures of each example can be found in Fig. 1.
In the class of terpenoids, trichothecenes and their derivatives are mycotoxins which have been reported in many Trichoderma species like T. brevicompactum, T. harzianum, T. viride, T. longibrachiatum, T. atroviride, T. erinaceum, T. citrinoviride76 and more broadly in species of Fusarium, Cryptomela, Spicellum, Myrothecium, Stachybotyrs, Cephalosporium and Trichothecium.77 Trichothecenes are sesquiterpene epoxides with a tricyclic 12,13-epoxytrichothec-9-ene (EPT) core and differ from the substitution found on the EPT, classifying them into 4 types (A, B, C, D).78 While the epoxide is essential for bioactivity, the nature of the activity can vary based on the substitutions found on the EPT.79,80 For example, trichodermin (2) inhibits protein synthesis whereas harzianum A (3) acts as a powerful herbicide.81–84 The biosynthetic gene cluster (TRI) for both compounds has been established in Fusarium species85 and orthologs were found in Trichoderma.86 Briefly, the gene cluster for trichothecene biosynthesis in Fusarium species is composed of 15 genes, spread over 3 chromosomes, and is initiated by the trichodiene synthase Tri5, which catalyses the cyclisation of farnesyl pyrophosphate to trichodiene. Tri1, Tri3, Tri4, Tri8, Tri11, and Tri10 catalyse the other steps of the biosynthesis.87–89
A comprehensive list of other terpenoids produced by Trichoderma can be found in the reviews by Zhang et al.,62 and most recently by Bai et al.90 and Guo et al.66
 | ||
| Fig. 2 Examples of alkaloids and peptides from Trichoderma. Gliotoxin (14); gliovirin (15); alamethicin (16); harzianin A (17); trichokonin V (18); trichogin GA IV (19); pentadecaibin (20). | ||
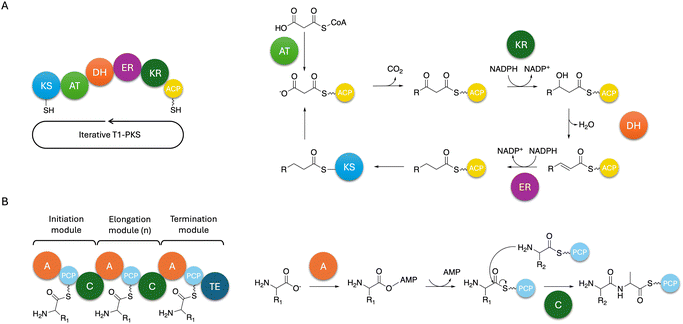
NRPSs are multi-domain enzymes with a multi-modular architecture described in Fig. 3. NRPSs are composed of an adenylation (A), a condensation (C), a peptidyl carrier protein (PCP) and a thioesterase (TE) domain organised in modules where each module usually contains one instance of those domains. Despite this modular organisation, it is still challenging to predict the structure of these peptides due to module skipping.94 Indeed, it was reported that Tex2, an NRPS from T. virens, produces two classes of peptaibols with either 11 or 14 residues.94
In Trichoderma, peptide specialised metabolites are predominantly peptaibols. These peptides range from 5–20 amino acids and are characterised by their high content of 2-aminoisobutyric acid, their acetylated (peptaibols) or acylated (peptaibiotics) N-terminus and their C-terminal amino alcohol.95 Some examples of peptaibols include alamethicin (16), harzianins (17), trichokonins (18), and lipopeptaibols like trichogin GA IV (19) as shown in Fig. 2. Their antimicrobial activity has been linked to their helical structure and amphipathic nature. Due to those properties, they can form ion channels in the lipid membranes causing permeabilisation of the cells.96,97 Over 440 peptaibol sequences have been reported and databases of peptaibols and peptaibiotics containing details of their biological source, activity, 3D structure and accompanying bibliographical data can be accessed or downloaded for research purposes.97–100
Another class of peptides include ribosomally synthesised and post-translationally modified peptides (RiPP) but scarce research is found on that class of specialised metabolites in Trichoderma. What is known is through genome mining using tools like RIPPMiner,101 RRE-Finder102 or RiPPER103 and rely heavily on known BGC structures of RiPPs like ustiloxins.104 This limits the number of identifiable RiPPs as novel classes of RiPPs can be missed. Recently, a combined genomic and transcriptomics approach along with careful manual curation enabled the discovery of a series of potential new RiPP gene clusters in Trichoderma, missed by other traditional methods like antiSMASH.105,106
In fungi, polyketides are classified in two categories: aromatic and aliphatic compounds, due to the domain structure of fungal PKSs.111 These structures range from non-reducing (no reductive steps) to highly reducing (varied levels of reduction) based on their domain composition. Most known fungal PKSs fall into the type I iterative PKSs and resemble mammalian fatty acid synthases112 but type III PKSs also exist.113,114 In type I iterative PKSs, the same module is used over cycles of elongation to produce the final product as opposed to bacterial type I PKSs. Most recently, a review on highly reducing fungal PKSs (hr-PKSs) was published by Cox110 detailing the catalytic activities of each domain involved in hr-PKS biosynthesis as well as their stereoselectivity.
Examples of polyketides from Trichoderma are shown in Fig. 4 and include sorbicillinoids (sorbicillin 21), anthraquinones (emodin 22), cyclopentones (trichoderone 23), naphthopyrones (hypochromin A 24) and koninginins (koninginin A 25).67 Sorbicillinoids have been isolated in many species of Trichoderma and are responsible for the yellow pigmentation observed in cultures. They hold a wide range of bioactivity including antioxidant115 and antimicrobial.116 The gene cluster for their biosynthesis has been described in T. reesei and contains 8 genes, two of them being transcription regulators.117,118 Briefly, Sor1 forms the polyketide chain, which is then further elongated and methylated by Sor2. The released aldehyde undergoes spontaneous cylcisation to form sorbicillin. The rest of the tailoring enzymes like Sor3 (FAD-dependent monooxygenase) and Sor4 (FAD/flavin mononucleotide-containing dehydrogenase) are responsible for the formation of the key intermediate sorbicillinol.119 Emodin (22) corresponds to a type of anthraquinone from Trichoderma with antifungal, antibacterial, and antioxidative properties which have been linked to the antagonistic activity against phytopathogens.120–122 Koninginins were found in multiple species of Trichoderma, most notably in T. koningii where the first koninginin (koninginin A 25) was isolated,123 which displayed cytotxic activity and plant growth regulating properties.123,124
3.2 Traditional methods for natural product discovery and examples in Trichoderma spp.
For the longest time, bioactive natural products were discovered in cultivable microorganisms using activity-guided methods, for which penicillin is a prime example.125 However, due to the complexity of the environments harbouring those microorganisms, the number of specialised metabolites which can be isolated from microbial cultures is limited. One traditional approach to promote the isolation of new natural products is the “One Strain Many Compounds” (OSMAC) technique, a concept that has been first introduced by Schiewe et al.,126 25 years ago.However, the traditional OSMAC approach has limitations due to low ability to reproduce the natural habitats of isolates within laboratory settings. This method has then included other techniques such as co-cultures and epigenetic modifications to circumvent those limitations.
In the first instance, co-cultivation of Trichoderma spp. with either bacteria or fungi has proven to induce production of metabolites as demonstrated by the discovery of two new sesquiterpenes microsphaeropsisin B/C (5) along with two new methyllasiodiplodins (6) from co-cultures of Trichoderma sp. strain 307 and Acenitobacter jonhsonii strain B2.135 Similarly, co-cultures of T. atroviride SG343 and B. subtilis 22 were found to inhibit the growth of Fusarium graminearum, when monocultures failed to show any antifungal activity.136 Similar findings were observed in co-cultures of B. amyloliquefaciens ACCC11060 and T. asperellum GDFS1009, where increase antimicrobial production was detected compared to monocultures.137 Fungal–fungal co-cultures have also displayed their potential for natural product discovery in Trichoderma spp. Indeed, co-cultivation of T. harzianum M10 with Talaromyces pinophilus F36CF was found to induce production of harziaphilic acid (7),138 whereas co-cultivation of Chaunopycnis sp. (CMB-MF028) and T. hamatum (CMB-MF030) activated production of chaunopyran A (26), identified as a broad-spectrum antifungal.139 More recently, efforts have been directed at optimisation of Trichoderma consortia for plant growth promotion as exemplified in studies on cucumber plants.140,141 In one study, researchers found that simultenaous inoculation of Trichoderma strains promoted better growth of cucumber seedlings and fermentation of 96 h to 120 h yielded the best production of growth promoting metabolites.140 Another study focused on optimising the combination of Trichoderma strains and found that co-cultures with T. asperellum GDSF1009, T. asperelloides Z4-1, T. harzianum 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 569, and T. asperellum 10
569, and T. asperellum 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 264 yielded the best results for both seed germination and antagonistic activity against F. oxysporum.141
264 yielded the best results for both seed germination and antagonistic activity against F. oxysporum.141
The second one relies on epigenetic modification as its role in regulating gene expression involved in secondary metabolism is well-known and was proven for the first time in Aspergillus.142–144 Indeed it was shown that deletion of a histone deacetylase (HDAC) resulted in overexpression of three biosynthetic gene clusters involved in the synthesis of penicillin, sterigmatocystin and terraquinone A.142 However, examples of the use of chemical epigenetic modifiers in Trichoderma spp. for natural product discovery remain scarce.145,146 In T. harzianum XS-20090075, the use of 10 μM sodium butyrate, a known histone deacetylase inhibitor, induced expression of genes involved in terpenoid biosynthesis and led to the isolation of harzianolic acid A (8), harzianone E (9), and 3,7,11-trihydroxy-cycloneran (10).146 In T. atroviride, application of the histone deacetylase inhibitor TSA1 induced specialised metabolism, with expression of genes related to peptaibol and terpene biosynthesis and linked to increased inhibitory activity against R. solani.145
Another method is ribosome engineering, which relies on the appearance of random mutations in RNA polymerase or ribosomes when organisms are exposed to antibiotics which target ribosomes. This approach has been proven to increase yield and even enable the production of new specialised metabolites. It has been used successfully in Actinomycetes with the activation of cryptic BGCs in Streptomyces species, amongst others.147–150 However, in fungi, examples of this approach have only been reported in Penicillium and Aspergillus species. In Penicillium purpurogenum G59, gentamycin induced the production of janthinone, fructigenine A, aspterric acid methyl ester and citrinin, while neomycin induced the production of curvularin, citrinin, penicitrinone A, erythro-23-O-methylneocyclocitrinol and 22E-7α-methoxy-5α, 6α-epoxyergosta-8(14),22-dien-3β-ol in selected resistant mutants.151,152 In Aspergillus versicolor ZBY-3, neomycin-resistant mutants were shown to produce six peptides with antitumor activity, absent in the wild type extracts.153 Most recently, a study in Actinomadura sp. used random mutagenesis methods including ribosome engineering with streptomycin to increase the yield of pentostatin, an antitumor drug, by close to 34%.154
4 The role of omics in the study of specialised metabolites biosynthesis in Trichoderma spp.
Along with analytical chemistry techniques, the advent of next generation sequencing has enabled the use of genome mining as a tool for dereplication. Using genome sequences to predict BGCs and prioritise candidates with unknown products has recently been the driving force for natural product discovery.1694.1 Sequencing efforts towards Trichoderma genomes
Efforts in isolation and identification of Trichoderma species are continuously reported on the International Commission on Trichoderma Taxonomy (ICTT) platform.170 Guidelines for molecular taxonomy of Trichoderma were established in 2020 by Cai et al.,170 and an inventory of over 450 unique species was drawn out. Methods for identification now focus on DNA-based techniques like barcoding.170,171 In 2008, the Joint Genome Institute (JGI) published the first full genome sequence for Trichoderma reesei.172 This major achievement paved the way for many other species of Trichoderma to follow.173,174 Now, over 150 genomes of Trichoderma can be found on NCBI, and 9 reference genomes are available as RefSeq annotations.175 These annotations include T. reesei, T. atroviride, T. asperellum, T. breve, T. harzianum, T. aggressivum, T. virens, T. citrinoviride, and T. gamsii. More efforts by the JGI community are underway to increase the number of available Trichoderma genomes.173 In a comparative genomic study on 12 species of Trichoderma, Kubicek et al.108 found that the number of PKS BGCs are similar to the numbers in Aspergillus spp. but that Aspergillus NRPS and terpenoid BGCs are significantly outnumbered by the ones in Trichoderma. However, the relationship between those BGCs and their metabolites remains elusive.4.2 Advances in genome mining tools and applications in Trichoderma spp.
The number of tools for automated BGC prediction and analysis based on genomic data has been increasing ever since the first iteration of antiSMASH in 2011,176 now reaching its 7th version.177 This pipeline uses a rule-based approach to find BGCs in bacteria, fungi and plant genomes based on available information on BGCs in public databases like MiBIG.178 AntiSMASH can now identify up to 81 cluster types with its latest update, which also includes improvements in substrate specificity for PKS and NRPS genes, prediction of RiPP clusters, and prediction of transcription factor binding sites. Following on this work, other tools were developed like PRISM, MIDDAS-M and DeepBGC for BGC detection;179–181 BiG-SCAPE/CORASON, Big-SLICE, MultiGeneBlast, and EvoMining for BGC clustering and phylogenetic analyses.182–185 The amount of interest towards gene cluster families (GCF) based research led to the generation of BiG-FAM, a dedicated database created in 2021.185 Recent reviews on bioinformatics tools for BGC mining are numerous106,169,186–191 with the most recent one by Cano-Prieto et al.192Tools specific for fungal natural products mining like TOUCAN,193 FunOrder,194 CO-OCCUR,195 CLOCI196 and FunGeneClusterS197 have also been reported. The first software, alike to DeepBGC, uses supervised learning to predict BGCs but uses amino acid sequences to do so. Compared to fungiSMASH, DeepBGC, TOUCAN provided better performance in BGC prediction and identification of core enzymes in both A. niger and A. nidulans.193 FunOrder performs co-evolution analysis to identify essential genes in BGCs and prioritises them for further studies. Using CO-OCCUR, Gluck et al.195 found over 3000 putative BGC and 719 unique GCFs within the Dothideomycetes fungal lineage using co-occurrence frequency of gene pairs. Of those, known BGCs and their respective compounds include aflatoxin-like dothistromin,198 dimethylcoprogen,199 alternapyrone,200 and chaetoglobosins.201 Similarly to FunOrder, CLOCI identified gene clusters based on co-evolution, outperforming antiSMASH.196 Lastly, FunGeneClusterS uses a combination of genomic and transcriptomic data to predict BGC based on co-regulation patterns.197 Using transcriptomics to guide natural product discovery has been done in A. niger. Indeed, based on 283 transcriptomes, Kwon et al.202 generated co-expression networks and identified six transcription factors which regulate specialised metabolism in A. niger. The metabolome of the strains overexpressing those transcription factors displayed over 140 more metabolites compared to the control strain, some of which were associated with known gene clusters such as the alkylcitrates BGC.202,203
In Trichoderma, genome mining has uncovered new specialised metabolites with great chemical diversity including polyketides, terpenes, non-ribosomal peptides, and RiPPs. Indeed, Yan et al.204 found a novel class of hybrid polyketides with a terpene-like structure and a D-glucose esterified core in Trichoderma afroharzianum T-22, namely treconorin (28). In the same fungus, genome mining of PKS-NRPS hybrid clusters combined with heterologous expression in A. nidulans yielded six new tetronate SMs, trihazones A–F (29).205 In T. viride, genome mining enabled the isolation of a novel 5/6 bicyclic sesquiterpene (Fig. 1, 11) and its esterified derivative.206 In the class of non-ribosomal peptides, genome sequencing of Trichoderma spp. MMS1255 led to the discovery of a set of 5 unique 15-residue peptaibols, subsequently named pentadecaibins I–V (20).207 Finally, using whole-genome sequencing and transcriptomics, Vignolle et al.106 developed a new approach for mining RiPP clusters which revealed over 600 potential RiPP BGCs across 4 Trichoderma genomes. This work still requires molecular validation to confirm the accuracy of the framework.
4.3 Innovations in resistance-guided genome mining in fungi
Another approach to mine for bioactive natural products is to exploit the presence of resistance genes.208,209 One of the first examples of antibiotic discovery using self-resistance-based genome mining was the discovery of a set of thiotetronic acid derivatives from Salinispora bacteria.210 Research on this topic has historically been applied to bacteria but growing interest has been shown to fungi recently. One of the first pioneering works in this field was done in Penicillium brevicompactum. By looking for a BGC containing a IMPDH homolog (target of mycophenolic acid), a 25-kb BGC was identified and linked to the biosynthesis of mycophenolic acid.211 Work in A. nidulans followed, where the presence of a proteasome inhibitor (inpE) in a cryptic BGC was hypothesised to be involved in self-resistance. Using serial promoter exchanges to activate the cluster, the elucidation of fellutamide B biosynthesis212 was achieved. These findings illustrated the potential of resistance-guided genome mining in fungi. Following this study, the same approach was applied to T. afroharzianum to elucidate the biosynthesis of harzianic acid, where a acetohydroxyacid synthase homolog was found within the BGC and proven to be a target of harzianic acid.213 Similarly, the BGC for restricticin in A. nomius was identified based on co-localisation of its target, a lanosterol 14α-demethylase (CYP51) paralog, displaying unique mutations and less susceptible to inhibition.214Subsequently, more automated methods were used to facilitate discovery. Currently, 3 frameworks exist for computer-aided discovery using resistance genes in fungi. In 2019, the FRIGG pipeline was developed based on 50 Aspergillus genomes and was able to predict over 70 unique clusters containing putative resistance genes. It was further validated with the correct identification of the previously characterised fellutamide B cluster.215 Later on, Jenkinson et al.216 developed a Python script to query the MycoCosm genome database (https://mycocosm.JGI.doe.gov/mycocosm/home) for SMs targeting either the proteasome β6 subunit or the HMG-CoA reductase. This resulted in the identification of putatively novel inhibitors of HMG-CoA reductase in Aspergillus genomes.216 However, both aforementioned tools required bioinformatics skills for correct usage. FunARTS (“fungal bioactive compound resistant target seeker”) was then created by Yılmaz et al.,217 and made publicly available at https://funarts.ziemertlab.com as a user-friendly web tool. This workflow built on their bacterial version ARTS and extended their search to fungal genomes.218,219 FunARTS uses Hidden Markov Models to identify core genes of a BGC and co-localised known resistance genes to prioritise novel BGCs. It can then be linked to BiG-SCAPE184 to perform gene cluster network analysis.
4.4 Recent progress in machine learning for BGC prediction
Novel approaches using machine learning for BGC prediction have now seen the light with ClusterFinder220 or DeepBGC181 as prime examples. Specifically for fungi, a model using reinforcement learning was built and tested on A. niger and A. nidulans. The model, relying on protein domains and functional annotation, outperformed other software like fungiSMASH, TOUCAN and DeepBGC in cluster prediction.221 Other machine learning programs also focus on prediction of bioactivity. In antiSMASH, the core structure of polyketides or non-ribosomal peptides can be roughly predicted thanks to the integration of NRPSPredictor2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 222 which uses support vector machines, and methods from Minowa et al.223 and Starcevic et al.224 However, the accuracy of prediction remains subpar and based on bacterial-generated data. More recently, Walker et al.225 built a machine learning program to predict SM antibacterial, antifungal and cytotoxic activities directly from gene sequences. They achieved accuracy of up to 80% using the PFAM, MiBIG and Resistance Gene Identifier databases. As the model is trained prior to predictions, it can be applied to any well-curated database, including fungal ones. Finally, a fungi-specific platform for bioactivity prediction was created by Riedling et al.,226 but its performance is still lacking accuracy, as trained models were only able to reach scores of up to 68%. Bigger databases with well-curated data are crucial to advance the field of machine learning-based discovery of natural products, one of the main points of the extensive review by Mullowney et al.227 on the use of artificial intelligence in natural product discovery. Careful usage of those tools remains the golden rule and thorough reflection is needed when it comes to choosing algorithms. Towards the curation of better repositories for fungal genomes, Robey et al.228 constructed an atlas of 1037 fungal genomes with their respective biosynthetic content, paving the way for improved genome mining strategies.
222 which uses support vector machines, and methods from Minowa et al.223 and Starcevic et al.224 However, the accuracy of prediction remains subpar and based on bacterial-generated data. More recently, Walker et al.225 built a machine learning program to predict SM antibacterial, antifungal and cytotoxic activities directly from gene sequences. They achieved accuracy of up to 80% using the PFAM, MiBIG and Resistance Gene Identifier databases. As the model is trained prior to predictions, it can be applied to any well-curated database, including fungal ones. Finally, a fungi-specific platform for bioactivity prediction was created by Riedling et al.,226 but its performance is still lacking accuracy, as trained models were only able to reach scores of up to 68%. Bigger databases with well-curated data are crucial to advance the field of machine learning-based discovery of natural products, one of the main points of the extensive review by Mullowney et al.227 on the use of artificial intelligence in natural product discovery. Careful usage of those tools remains the golden rule and thorough reflection is needed when it comes to choosing algorithms. Towards the curation of better repositories for fungal genomes, Robey et al.228 constructed an atlas of 1037 fungal genomes with their respective biosynthetic content, paving the way for improved genome mining strategies.
4.5 Metabologenomics: a framework for natural product discovery
Metabologenomics is a concept that was first introduced in 2016 by the Kelleher lab.229 When dealing with large datasets housing hundreds if not thousands or different strains, the use of pattern-based genome mining coupled with molecular networking has proven to be game-changing. One significant study on this topic described the investigation of Salinispora genomes and enabled the characterisation of retimycin A, a quinomycin-like depsipeptide.230 Most recently, the NegMDF strategy was created to standardise this approach for the discovery of novel polyketides in bacteria.231 In this study, the authors employ a BGC-guided mass defect filtering (MDF) approach in parallel with negative mode MS scans to screen for novelty and use targeted MS/MS and NMR for validation. The MDF approach showed great advantages for the detection and prediction of bacterial PKSs as it does not rely on MS/MS fragmentation to infer product ions, offering limited bias towards abundant ions. Using this method, Liu et al.231 were able to identify novel polyketides in Streptomyces cattleya NRRL 8057, namely cattleyatetronates. Additional studies on bacteria and reviews on the subject attest of the rising interest in this approach.162,229,232–239 A community resource was created in 2021, the Paired Omics Data Platform, to offer scientists a database which facilitates the study of natural product biosynthesis based on both metabolomics and genomics data, with over 4800 genome-metabolome links with attached metadata.240 Recently, it was applied in 110 ascomycetes and was able to link more than 200 specialised metabolites to gene cluster families, within which the biosynthesis of pestalamides was uncovered.241 This workflow was then linked to bioactivity to further improve prioritisation of SM discovery, leading to the isolation of three novel stemphones, 19-acetylstemphones G, B and E.242 However, most studies using this approach focus on bacterial natural products, leaving fungal metabolites under-represented in this area of natural product research. More interest needs to be given to this highly promising approach for discovery.5 Genetic manipulation in Trichoderma for the study of natural product biosynthesis
After prioritising BGCs using all aforementioned methods, experimental validation is necessary to link the novel natural products to the BGCs. This can be done using various strategies for fungal organisms, depending on their cultivability, genetic tractability and genetic engineering tools available for the fungal host. A decision tree to establish a workflow was designed in the review by Kjærbølling et al.2155.1 Engineering in the host organism
If the fungal strain is cultivable and genetically tractable, investigation using the native host can be used and transformation protocols have been established for Trichoderma spp.243–245 Strategies to activate silent clusters through cultivation have already been detailed previously. Using genetic engineering approaches like transcription factor (TF) overexpression and promoter replacement or heterologous expression can also trigger and/or increase expression.In order to manipulate TFs effectively, the use of promoter engineering is needed. Reports of promoter engineering in actinomycetes and other bacteria are now abundant,264 but examples of implementations in Trichoderma are scarcer, with most studies focusing on T. reesei and overproduction of cellulases.265–268 In Trichoderma, a list of promoters and their use for overproduction of cellulase-encoding genes was established by Adnan et al.267 Most recently, a study in Aspergillus nidulans uncovered 93 promoter sequences from 454 TFs from transcriptome data and tested them for relative transcriptional strength using a single cell flow cytometry-based quantification method. From those, two strong promoters were chosen and introduced into A. fumigatus to drive transcription of the NRPS gene Afpes1. This led to the isolation and identification of fumiganins A and B.269 This work establishes a more comprehensive set of promoter sequences and applies them to other fungal species for natural product discovery. However, the use of promoter exchange is commonly labour intensive and requires a robust method for homologous recombination in the host.
Another approach involves the use of CRISPR/Cas technologies (Clustered Regularly Interspaced Short Palindormic Repeats).The clustered regularly interspaced short palindromic repeats and associated Cas9 nuclease (CRISPR-Cas9) have revolutionised the field of genetic engineering for its versatility and preciseness.270 In this system, the endonuclease Cas9 is guided by a single guide RNA (sgRNA) to a target locus in the genome and performs a double-strand break. The break can then be repaired via the Non-Homologous End Joining pathway which introduces mutations (insertions, deletions), or homologous recombination if provided with donor DNA. This is now well-established in model organisms such as Escherichia coli,271Saccharomyces cerevisiae,272 and Streptomyces spp.273 CRISPR-Cas9 systems have also been developed for filamentous fungi, including Aspergillus spp.,274–276Fusarium spp.277–279 and Trichoderma spp.280 A recent extensive review on the different tools available for CRISPR-mediated editing in filamentous fungi has been published by Woodcraft et al.,281 but specific studies of applications in Trichoderma spp. are scarce, with only two recent examples. Indeed, in a study by Wang et al.,282 researchers were able to activate the silent ilicicolin H (30) BGC in T. reesei using a new quinic acid-inducible Cas9. In the same species, Fang et al.283 were able to transform the 32.7 kb sorbicillinoids BGC, split into 10 fragments, into the clr2 locus using CRISPR/Cas9 and in a single transformation step. However, no production of sorbicillinoids was observed due to the presence of three point mutations in the biosynthetic genes. Nevertheless, this method called simultaneous in vivo assembly and targeted genome integration of multiple DNA fragments (SATIMD), opens new avenues for the use of CRISPR/Cas9 in Trichoderma for the heterologous expression of BGCs from Trichoderma spp.283
5.2 Heterologous expression in model organisms
When the fungal host is not genetically tractable or cultivable, researchers will rely on heterologous expression. The choice of host is crucial and highly dependent on the nature of the cryptic BGC. Escherichia coli is a powerful host due to its fast growth, simple cultivation and easy and highly efficient transformation. It is generally used to express a single gene from the cluster of interest and coupled with in vitro experiments to study enzymatic structure and function.285 This was done in T. atroviride and led to the isolation of a new sesquiterpene alcohol trichobrasinelol (12). The sesquiterpene cyclase was cloned into E. coli BL21 and the expressed protein was purified and characterised for substrate specificity.286 However, the use of E. coli as a host for fungal BGCs usually shows limitations due to the presence of introns, codon bias, the lack of post-translational modifications, the potential toxicity of the products, and the availability of the precursors.285Saccharomyces cerevisiae is one of the preferred hosts for the heterologous expression of fungal BGCs due to its low specialised metabolism background, its fast growth for a eukaryotic organism, its well-curated genetic engineering tools, its high homologous recombination rates, and its ability to correctly synthesise and fold fungal proteins, being a fungal organism.287–289 In the case of Trichoderma spp., S. cerevisiae has been used as heterologous host to reconstitute the biosynthetic pathway of several trichothecenes.86,88,290 Indeed, the final product trichodermol (13) was successfully biosynthesised heterologously in yeast using a codon optimised trichodiene synthase gene, coupled with a multicopy integration plasmid targeting the repetitive chromosomal rDNA.290 More recently, a heterologous expression platform (HEx) was developed to enable expression of fungal BGCs into S. cerevisiae.291 Using this platform, 41 BGCs from various ascomycetes, and basidiomycetes were integrated into yeast and of those, 54% produced specialised metabolites novel to yeast, including a PKS BGC from T. virens. This strategy enables high-throughput expression of a multitude of BGCs from various origins.
However, for Trichoderma natural products, the use of filamentous fungi remains the most used, with A. oryzae,292A. niger (ATNT system),293A. nidulans294 and more recently T. reesei295–297 as heterologous hosts. Examples of studies using A. oryzae or A. nidulans as host have been described previously and can be found in the review by Shenouda et al.,298 with the ilicicolin H BGC as a recent case. In A. niger, a heterologous expression strain with altered NHEJ and altered pigmentation, has now been optimised for expression of long biosynthetic genes (over 20 kb) from ascomycetes, basidiomycetes and early diverging fungi.299 However, the main advantages of using T. reesei as a heterologous host for investigating Trichoderma BGCs is its phylogenetic closeness with other Trichoderma species and its ability to grow on cellulosic biomass, enabling the valorisation of waste material. This is the rationale behind the work by Shenouda et al.,296 where T. reesei was grown on peels from various fruits, used coffee grounds or barley straw to produce several specialised metabolites like pretenellin A (Fig. 5, 31) in a newly engineered strain, designed for SM production. This strain was provided a cleaner SM background by knocking out its sorbicillin BGC, and transformed with a vector containing the pretenellin A PKS-NRPS megasynthase with its trans-acting ER under the control of 2 constitutive promoters.296
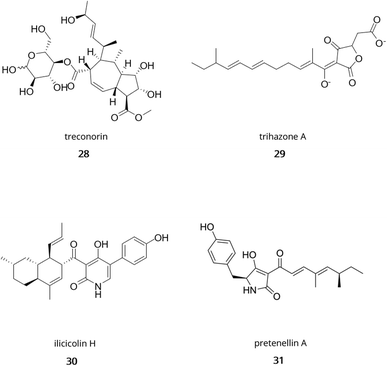 | ||
| Fig. 5 Examples of hybrid molecules from Trichoderma. Treconorin (28); trihazone A (29); ilicicolin H (30); pretenellin A (31). | ||
However, heterologous expression can still prove challenging. This is exemplified by Hang et al.300 and their studies on the PKS Tv6-931 from T. virens. Their first attempt at heterologous expression of the PKS, as well as the entire BGC, failed to produce a new compound in S. cerevisiae and A. nidulans. They were only able to isolate new tetraketide products when a proper offloading substrate was added to the reaction. This shows that availability of substrates can impose limitations in various heterologous hosts.
6 Conclusions
To conclude, a combination of culture-based approaches and molecular approaches should be used to explore the biosynthetic potential of Trichoderma species, guided by genome mining strategies. The advances in this field and the increasingly available tools for genome engineering in Trichoderma and other filamentous fungi are tremendously helping discovery rates. Alongside, the development of tools for metabolomics are becoming indispensable for dereplication and prioritisation of compounds to prevent re-discovery of known SMs. Finally, an integrated platform combining omics strategies is now proving highly efficient in selecting cryptic BGCs to focus efforts on the discovery of new bioactive metabolites (Fig. 6). | ||
| Fig. 6 Strategies for natural product discovery in fungi. The OSMAC (“One Strain Many Compounds”) approach which includes modulating cultivation conditions or using molecular approaches like epigenetic modifications or ribosome engineering. Dereplication is essential to ensure novelty and is exemplified by the discovery of scopularide H from Scopulariopsis sp. CMB-F115 using GNPS.284 If the product can be detected, a combination of genomics, transcriptomics and metabolomics can be used to isolate the target SM, identify its structure, and link it to a gene cluster using several genome mining tools. This is the concept behind metabologenomics where the use of pattern-based genome mining coupled with molecular networking can provide insight into which clusters to prioritise for BGC-SM discovery. Validation of the link can then be done via a variety of different genetic engineering strategies such as TF or promoter engineering, heterologous expression or CRISPR/Cas technologies. This was exemplified by the discovery of pestalamide B along with its BGC from the native host Aspergillus brasiliensis and heterologously expressed in A. nidulans.241 | ||
7 Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.8 Conflicts of interest
There are no conflicts to declare.9 Acknowledgements
F. A. was supported by a UKRI Future Leaders Fellowship award (MR/V022334/1). S. J. was supported by a BBSRC Midlands Integrative Biosciences Training Partnership (BB/T00746X/1) scholarship. The authors would also like to acknowledge support from BBSRC through an International Partnering Award (BB/X018369/1).10 Notes and references
- L. Rani, K. Thapa, N. Kanojia, N. Sharma, S. Singh, A. S. Grewal, A. L. Srivastav and J. Kaushal, J. Cleaner Prod., 2021, 283, 124657 CrossRef CAS.
- V. M. Pathak, V. K. Verma, B. S. Rawat, B. Kaur, N. Babu, A. Sharma, S. Dewali, M. Yadav, R. Kumari, S. Singh, A. Mohapatra, V. Pandey, N. Rana and J. M. Cunill, Front. Microbiol., 2022, 13, 962619 CrossRef PubMed.
- N. Donley, Environ. Health, 2019, 18, 44 CrossRef PubMed.
- M.-L. Lahdenperä, E. Simon and J. Uoti, Developments in Agricultural and Managed Forest Ecology, Elsevier, 1991, vol. 23, pp. 258–263 Search PubMed.
- M. Xue, R. Wang, C. Zhang, W. Wang, F. Zhang, D. Chen, S. Ren, Z. Manman, J. Hou and T. Liu, BioMed Res. Int., 2021, 2021, 7913950 CrossRef PubMed.
- I. Gal-Hemed, L. Atanasova, M. Komon-Zelazowska, I. S. Druzhinina, A. Viterbo and O. Yarden, Appl. Environ. Microbiol., 2011, 77, 5100–5109 CrossRef CAS PubMed.
- N. Wijayawardene, Mycosphere, 2020, 11, 1060–1456 CrossRef.
- P. Chaverri, F. Branco-Rocha, W. Jaklitsch, R. Gazis, T. Degenkolb and G. J. Samuels, Mycologia, 2015, 107, 558–590 CrossRef CAS.
- I. S. Druzhinina, V. Seidl-Seiboth, A. Herrera-Estrella, B. A. Horwitz, C. M. Kenerley, E. Monte, P. K. Mukherjee, S. Zeilinger, I. V. Grigoriev and C. P. Kubicek, Nat. Rev. Microbiol., 2011, 9, 749–759 CrossRef CAS PubMed.
- R. Hermosa, A. Viterbo, I. Chet and E. Monte, Microbiology, 2012, 158, 17–25 CrossRef CAS.
- A. Viterbo and B. A. Horwitz, Cellular and Molecular Biology of Filamentous Fungi, ASM Press, Washington, DC, USA, 2014, pp. 676–693 Search PubMed.
- A. K. Singh, H. K. Rana and A. K. Pandey, Recent Advancement in White Biotechnology Through Fungi: Volume 2: Perspective for Value-Added Products and Environments, Springer International Publishing, Cham, 2019, pp. 229–248 Search PubMed.
- N. W. Zaidi, M. H. Dar, S. Singh and U. S. Singh, Biotechnology and Biology of Trichoderma, Elsevier, Amsterdam, 2014, pp. 515–525 Search PubMed.
- S. A. Mona, A. Hashem, E. F. Abd Allah, A. A. Alqarawi, D. W. K. Soliman, S. Wirth and D. Egamberdieva, J. Integr. Agric., 2017, 16, 1751–1757 CrossRef CAS.
- C. Yu, X. Jiang, H. Xu and G. Ding, J. Fungi, 2023, 9, 694 CrossRef CAS.
- W. Asghar and R. Kataoka, Arch. Microbiol., 2021, 203, 4281–4291 CrossRef CAS PubMed.
- R. Abdenaceur, B.-t. Farida, D. Mourad, H. Rima, O. Zahia and S.-H. Fatma, Int. Microbiol., 2022, 25, 817–829 CrossRef CAS PubMed.
- R. A. A. Khan, S. Najeeb, J. Chen, R. Wang, J. Zhang, J. Hou and T. Liu, Physiol. Plant., 2023, 175, e14133 CrossRef.
- R. P. John, R. Tyagi, D. Prévost, S. K. Brar, S. Pouleur and R. Surampalli, Crop Prot., 2010, 29, 1452–1459 CrossRef.
- N. E. Awad, H. A. Kassem, M. A. Hamed, A. M. El-Feky, M. A. A. Elnaggar, K. Mahmoud and M. A. Ali, Mycology, 2018, 9, 70–80 CrossRef CAS PubMed.
- J. Han, Z. Lu, H. Zhang, S. Ji, B. Liu, N. Kong, Y. Yang, B. Xing and Z. Liu, Physiol. Plant., 2024, 176, e14556 CrossRef CAS PubMed.
- B. Reithner, E. Ibarra-Laclette, R. L. Mach and A. Herrera-Estrella, Appl. Environ. Microbiol., 2011, 77, 4361–4370 CrossRef CAS.
- L. Atanasova, S. L. Crom, S. Gruber, F. Coulpier, V. Seidl-Seiboth, C. P. Kubicek and I. S. Druzhinina, BMC Genom., 2013, 14, 121 CrossRef CAS.
- M. E. Morán-Diez, I. Carrero-Carrón, M. B. Rubio, R. M. Jiménez-Díaz, E. Monte and R. Hermosa, Front. Microbiol., 2019, 10, 1120 CrossRef.
- V. Lazazzara, B. Vicelli, C. Bueschl, A. Parich, I. Pertot, R. Schuhmacher and M. Perazzolli, Physiol. Plant., 2021, 172, 1950–1965 CrossRef CAS.
- S. Vázquez-Garcidueñas, C. A. Leal-Morales and A. Herrera-Estrella, Appl. Environ. Microbiol., 1998, 64, 1442–1446 CrossRef PubMed.
- R. F. Troian, A. S. Steindorff, M. H. S. Ramada, W. Arruda and C. J. Ulhoa, Biotechnol. Lett., 2014, 36, 2095–2101 CrossRef CAS PubMed.
- G. Hernández, A. Ponce de la Cal, Y. Louis, Y. Baró Robaina, Y. Coll, I. Spengler and Y. Mirabal-Gallardo, J. Fungi, 2024, 10, 547 CrossRef.
- I. Verma, S. K. Soni and P. C. Singh, Plant Physiol. Biochem., 2024, 215, 108953 CrossRef CAS.
- P. Trutmann and P. Keane, Soil Biol. Biochem., 1990, 22, 43–50 CrossRef.
- S. Xiao-Yan, S. Qing-Tao, X. Shu-Tao, C. Xiu-Lan, S. Cai-Yun and Z. Yu-Zhong, FEMS Microbiol. Lett., 2006, 260, 119–125 CrossRef.
- M. Shi, L. Chen, X.-W. Wang, T. Zhang, P.-B. Zhao, X.-Y. Song, C.-Y. Sun, X.-L. Chen, B.-C. Zhou and Y.-Z. Zhang, Microbiology, 2012, 158, 166–175 CrossRef CAS PubMed.
- K. Liu, Y.-B. Yang, J.-L. Chen, C.-P. Miao, Q. Wang, H. Zhou, Y.-W. Chen, Y.-Q. Li, Z.-T. Ding and L.-X. Zhao, Nat. Prod. Bioprospect., 2016, 6, 49–55 CrossRef CAS.
- S. Leylaie and D. Zafari, Front. Microbiol., 2018, 9, 1484 CrossRef PubMed.
- O.-U. Ruangwong, C. Pornsuriya, K. Pitija and A. Sunpapao, J. Fungi, 2021, 7, 276 CrossRef CAS.
- J. You, G. Li, C. Li, L. Zhu, H. Yang, R. Song and W. Gu, J. Fungi, 2022, 8, 131 CrossRef CAS.
- W.-L. Kong, H. Ni, W.-Y. Wang and X.-Q. Wu, Front. Microbiol., 2022, 13, 1–13 CAS.
- W. Intana, S. Kheawleng and A. Sunpapao, J. Fungi, 2021, 7, 46 CrossRef CAS.
- C. Shanmugaraj, D. Kamil, R. Parimalan, P. K. Singh, P. R. Shashank, M. A. Iquebal, Z. Hussain, A. Das, R. Gogoi and K. Nishmitha, 3 Biotech, 2024, 14, 210 CrossRef CAS PubMed.
- F. Berini, A. Montali, R. Liguori, G. Venturini, M. Bonelli, L. Shaltiel-Harpaz, M. Reguzzoni, M. Siti, F. Marinelli, M. Casartelli and G. Tettamanti, Pest Manage. Sci., 2024, 80, 3401–3411 CrossRef CAS.
- E. Garo, C. M. Starks, P. R. Jensen, W. Fenical, E. Lobkovsky and J. Clardy, J. Nat. Prod., 2003, 66, 423–426 CrossRef CAS.
- S. Zhang, Y. Gan and B. Xu, Appl. Soil Ecol., 2015, 94, 21–29 CrossRef.
- O. Degani and S. Dor, J. Fungi, 2021, 7, 315 CrossRef CAS.
- N. Luo, Y. Jiao, J. Ling, Z. Li, W. Zhang, J. Zhao, Y. Li, Z. Mao, H. Li and B. Xie, J. Agric. Food Chem., 2024, 72, 20763–20774 CrossRef CAS PubMed.
- T. Damodaran, S. Rajan, M. Muthukumar, R. Gopal, K. Yadav, S. Kumar, I. Ahmad, N. Kumari, V. K. Mishra and S. K. Jha, Front. Microbiol., 2020, 11, 595845 CrossRef PubMed.
- R. Weindling and O. H. Emerson, Phytopathology, 1936, 26, 1068–1070 CAS.
- L. Tian, X. Zhu, Y. Guo, Q. Zhou, L. Wang and W. Li, Front. Microbiol., 2024, 15, 1454670 CrossRef PubMed.
- X. Yao, J. Xie, Y. Qi, B. Wang, W. Fang, G. Tao and X. Jiang, Shengwu Gongcheng Xuebao, 2024, 40, 211–225 CAS.
- L. Zanfaño, G. Carro-Huerga, Á. Rodríguez-González, S. Mayo-Prieto, R. E. Cardoza, S. Gutiérrez and P. A. Casquero, Front. Plant Sci., 2024, 15, 1–15 Search PubMed.
- G. Carro-Huerga, S. Compant, M. Gorfer, R. E. Cardoza, M. Schmoll, S. Gutiérrez and P. A. Casquero, Front. Plant Sci., 2020, 11, 1170 CrossRef PubMed.
- J. Chen, L. Zhou, I. U. Din, Y. Arafat, Q. Li, J. Wang, T. Wu, L. Wu, H. Wu, X. Qin, G. R. Pokhrel, S. Lin and W. Lin, Front. Microbiol., 2021, 12, 579920 CrossRef PubMed.
- W. Yan, W. Ji, C. Ping, T. Zhang, Y. Li, B. Wang, T. Chen, B. He and Y. Ye, Phytochemistry, 2022, 196, 1–13 CrossRef PubMed.
- Y. Martinez, J. Ribera, F. W. M. R. Schwarze and K. D. France, Appl. Microbiol. Biotechnol., 2023, 107, 55–95 Search PubMed.
- S. L. Woo, M. Ruocco, F. Vinale, M. Nigro, R. Marra, N. Lombardi, A. Pascale, S. Lanzuise, G. Manganiello and M. Lorito, Open Mycol. J., 2014, 8, 71–126 CrossRef.
- G. E. Harman, C. R. Howell, A. Viterbo, I. Chet and M. Lorito, Nat. Rev. Microbiol., 2004, 2, 43–56 CrossRef CAS.
- B. A. Bailey, H. Bae, M. D. Strem, J. Crozier, S. E. Thomas, G. J. Samuels, B. T. Vinyard and K. A. Holmes, Biol. Control, 2008, 46, 24–35 CrossRef.
- M. Verma, S. K. Brar, R. D. Tyagi, R. Y. Surampalli and J. R. Valéro, Biochem. Eng. J., 2007, 37, 1–20 CrossRef.
- Savita and A. Sharma, Biofertilizers for Sustainable Agriculture and Environment, Springer International Publishing, Cham, 2019, pp. 395–411 Search PubMed.
- M. Sood, D. Kapoor, V. Kumar, M. S. Sheteiwy, M. Ramakrishnan, M. Landi, F. Araniti and A. Sharma, Plants, 2020, 9, 762 CrossRef CAS PubMed.
- J. Poveda, Biol. Control, 2021, 159, 104634 CrossRef CAS.
- V. K. Gupta, M. Schmoll, A. Herrera-Estrella, R. S. Upadhyay, I. Druzhinina and M. G. Tuohy, Biotechnology and Biology of Trichoderma, Elsevier, Amsterdam, 2014, pp. 125–137 Search PubMed.
- J.-L. Zhang, W.-L. Tang, Q.-R. Huang, Y.-Z. Li, M.-L. Wei, L.-L. Jiang, C. Liu, X. Yu, H.-W. Zhu, G.-Z. Chen and X.-X. Zhang, Front. Microbiol., 2021, 12, 723828 CrossRef PubMed.
- M. A. Fischbach, C. T. Walsh and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4601–4608 CrossRef CAS PubMed.
- N. P. Keller, Nat. Rev. Microbiol., 2019, 17, 167–180 CrossRef CAS.
- S. Zeilinger, S. Gruber, R. Bansal and P. K. Mukherjee, Fungal Biol. Rev., 2016, 30, 74–90 CrossRef.
- Q. Guo, L. Shi, X. Wang, D. Li, Z. Yin, J. Zhang, G. Ding and L. Chen, J. Agric. Food Chem., 2023, 71, 13612–13632 CrossRef CAS PubMed.
- Y.-P. Song and N.-Y. Ji, Nat. Prod. Bioprospect., 2024, 14, 14 CrossRef CAS PubMed.
- A. Herrmann, The Chemistry and Biology of Volatiles, John Wiley & Sons, 2011 Search PubMed.
- J. F. Jiménez-Bremont, E. González-Pérez, M. A. Ortega-Amaro, S. Madrigal-Ortiz, A. Duque-Ortiz and A. Mendoza-Mendoza, Sci. Hortic., 2024, 325, 112656 CrossRef.
- R. Scarselletti and J. L. Faull, Mycol. Res., 1994, 98, 1207–1209 CrossRef CAS.
- C. Mutawila, F. Vinale, F. Halleen, M. Lorito and L. Mostert, Plant Pathol., 2016, 65, 104–113 CrossRef CAS.
- Y. Wu, X. Li, L. Dong, T. Liu, Z. Tang, R. Lin, J. Norvienyeku and M. Xing, J. Fungi, 2023, 9, 863 CrossRef CAS PubMed.
- M. Xing, J. Zhao, J. Zhang, Y. Wu, R. A. A. Khan, X. Li, R. Wang, T. Li and T. Liu, J. Agric. Food Chem., 2023, 71, 19488–19500 CrossRef CAS PubMed.
- A. Garnica-Vergara, S. Barrera-Ortiz, E. Muñoz-Parra, J. Raya-González, A. Méndez-Bravo, L. Macías-Rodríguez, L. F. Ruiz-Herrera and J. López-Bucio, New Phytol., 2016, 209, 1496–1512 CrossRef CAS PubMed.
- A. Mendoza-Mendoza, E. U. Esquivel-Naranjo, S. Soth, H. Whelan, H. Alizadeh, J. F. Echaide-Aquino, D. Kandula and J. G. Hampton, Front. Plant Sci., 2024, 15, 1420068 CrossRef PubMed.
- K. F. Nielsen, T. Gräfenhan, D. Zafari and U. Thrane, J. Agric. Food Chem., 2005, 53, 8190–8196 CrossRef CAS PubMed.
- R. H. Proctor, S. P. McCormick, H.-S. Kim, R. E. Cardoza, A. M. Stanley, L. Lindo, A. Kelly, D. W. Brown, T. Lee, M. M. Vaughan, N. J. Alexander, M. Busman and S. Gutiérrez, PLoS Pathog., 2018, 14, e1006946 CrossRef PubMed.
- S. P. McCormick, A. M. Stanley, N. A. Stover and N. J. Alexander, Toxins, 2011, 3, 802 CrossRef CAS PubMed.
- E. Cundliffe, M. Cannon and J. Davies, Proc. Natl. Acad. Sci. U. S. A., 1974, 71, 30–34 CrossRef CAS.
- W. Wang, Y. Zhu, N. Abraham, X.-Z. Li, M. Kimber and T. Zhou, Int. J. Mol. Sci., 2021, 22, 1604 CrossRef CAS PubMed.
- L. Carrasco, M. Barbacid and D. Vazquez, Biochim. Biophys. Acta, 1973, 312, 368–376 CrossRef CAS PubMed.
- A. E. Desjardins, T. M. Hohn and S. P. McCormick, Microbiol. Rev., 1993, 57, 595–604 CrossRef CAS PubMed.
- Q. Wu, V. Dohnal, K. Kuca and Z. Yuan, Curr. Drug Metab., 2013, 14, 641–660 CrossRef CAS PubMed.
- M. Yin, O. E. Fasoyin, C. Wang, Q. Yue, Y. Zhang, B. Dun, Y. Xu and L. Zhang, AMB Express, 2020, 10, 118 CrossRef CAS.
- D. W. Brown, R. B. Dyer, S. P. McCormick, D. F. Kendra and R. D. Plattner, Fungal Genet. Biol., 2004, 41, 454–462 CrossRef CAS PubMed.
- R. E. Cardoza, M. G. Malmierca, M. R. Hermosa, N. J. Alexander, S. P. McCormick, R. H. Proctor, A. M. Tijerino, A. Rumbero, E. Monte and S. Gutiérrez, Appl. Environ. Microbiol., 2011, 77, 4867–4877 CrossRef CAS PubMed.
- M. Kimura, T. Tokai, K. O'Donnell, T. J. Ward, M. Fujimura, H. Hamamoto, T. Shibata and I. Yamaguchi, FEBS Lett., 2003, 539, 105–110 CrossRef CAS PubMed.
- T. Tokai, H. Koshino, N. Takahashi-Ando, M. Sato, M. Fujimura and M. Kimura, Biochem. Biophys. Res. Commun., 2007, 353, 412–417 CrossRef CAS PubMed.
- Y. Chen, H. C. Kistler and Z. Ma, Annu. Rev. Phytopathol., 2019, 57, 15–39 CrossRef CAS PubMed.
- B. Bai, C. Liu, C. Zhang, X. He, H. Wang, W. Peng and C. Zheng, J. Adv. Res., 2023, 49, 81–102 CrossRef CAS PubMed.
- A. M. A. El-Bondkly, Biotechnology and Biology of Trichoderma, Elsevier, Amsterdam, 2014, pp. 377–392 Search PubMed.
- D. M. Gardiner and B. J. Howlett, FEMS Microbiol. Lett., 2005, 248, 241–248 CrossRef CAS PubMed.
- C. J. Balibar and C. T. Walsh, Biochemistry, 2006, 45, 15029–15038 CrossRef CAS PubMed.
- P. K. Mukherjee, A. Wiest, N. Ruiz, A. Keightley, M. E. Moran-Diez, K. McCluskey, Y. F. Pouchus and C. M. Kenerley, J. Biol. Chem., 2011, 286, 4544–4554 CrossRef CAS PubMed.
- J. F. d. S. Daniel and E. R. Filho, Nat. Prod. Rep., 2007, 24, 1128–1141 RSC.
- J. K. Chugh and B. A. Wallace, Biochem. Soc. Trans., 2001, 29, 565–570 CrossRef CAS PubMed.
- C. d. l. Fuente-Núñez, L. Whitmore and B. A. Wallace, Handbook of Biologically Active Peptides, Academic Press, Boston, 2nd edn, 2013, pp. 150–156 Search PubMed.
- L. Whitmore and B. A. Wallace, Nucleic Acids Res., 2004, 32, D593 CrossRef CAS PubMed.
- N. Stoppacher, N. K. N. Neumann, L. Burgstaller, S. Zeilinger, T. Degenkolb, H. Brückner and R. Schuhmacher, Chem. Biodiversity, 2013, 10, 734–743 CrossRef CAS.
- N. K. N. Neumann, N. Stoppacher, S. Zeilinger, T. Degenkolb, H. Brückner and R. Schuhmacher, Chem. Biodiversity, 2015, 12, 743–751 CrossRef CAS.
- P. Agrawal, S. Khater, M. Gupta, N. Sain and D. Mohanty, Nucleic Acids Res., 2017, 45, W80–W88 CrossRef CAS PubMed.
- A. M. Kloosterman, K. E. Shelton, G. P. van Wezel, M. H. Medema and D. A. Mitchell, mSystems, 2020, 5, e00267–20 CrossRef CAS PubMed.
- A. D. Moffat, J. Santos-Aberturas, G. Chandra and A. W. Truman, Methods Mol. Biol., 2021, 2296, 227–247 CrossRef CAS PubMed.
- R. E. Ford, G. D. Foster and A. M. Bailey, Fungal Biol. Biotechnol., 2022, 9, 12 CrossRef CAS PubMed.
- K. Blin, S. Shaw, K. Steinke, R. Villebro, N. Ziemert, S. Y. Lee, M. H. Medema and T. Weber, Nucleic Acids Res., 2019, 47, W81–W87 CrossRef CAS PubMed.
- G. A. Vignolle, R. L. Mach, A. R. Mach-Aigner and C. Derntl, BMC Genom., 2020, 21, 258 CrossRef CAS PubMed.
- Y. A. Chan, A. M. Podevels, B. M. Kevany and M. G. Thomas, Nat. Prod. Rep., 2009, 26, 90 RSC.
- C. P. Kubicek, A. S. Steindorff, K. Chenthamara, G. Manganiello, B. Henrissat, J. Zhang, F. Cai, A. G. Kopchinskiy, E. M. Kubicek, A. Kuo, R. Baroncelli, S. Sarrocco, E. F. Noronha, G. Vannacci, Q. Shen, I. V. Grigoriev and I. S. Druzhinina, BMC Genom., 2019, 20, 485 CrossRef PubMed.
- J. Wang, R. Zhang, X. Chen, X. Sun, Y. Yan, X. Shen and Q. Yuan, Microb. Cell Fact., 2020, 19, 110 CrossRef CAS PubMed.
- R. J. Cox, Nat. Prod. Rep., 2023, 40, 9–27 RSC.
- R. J. Cox, E. Skellam and K. Williams, Physiology and Genetics: Selected Basic and Applied Aspects, Springer International Publishing, Cham, 2018, pp. 385–412 Search PubMed.
- J. M. Crawford and C. A. Townsend, Nat. Rev. Microbiol., 2010, 8, 879 CrossRef CAS.
- M. B. Austin and J. P. Noel, Nat. Prod. Rep., 2003, 20, 79–110 RSC.
- Y. Seshime, P. R. Juvvadi, I. Fujii and K. Kitamoto, Biochem. Biophys. Res. Commun., 2005, 331, 253–260 CrossRef CAS PubMed.
- N. Abe and A. Hirota, Chem. Commun., 2002, 662–663 RSC.
- Z. Yang, Y. Qiao, J. Li, F.-G. Wu and F. Lin, Langmuir, 2020, 36, 13227–13235 CrossRef CAS PubMed.
- C. Derntl, A. Rassinger, E. Srebotnik, R. L. Mach and A. R. Mach-Aigner, Appl. Environ. Microbiol., 2016, 82, 6247–6257 CrossRef CAS.
- I. S. Druzhinina, E. M. Kubicek and C. P. Kubicek, BMC Evol. Biol., 2016, 16, 269 CrossRef PubMed.
- C. Derntl, F. Guzmán-Chávez, T. M. Mello-de Sousa, H.-J. Busse, A. J. M. Driessen, R. L. Mach and A. R. Mach-Aigner, Front. Microbiol., 2017, 8, 1–12 Search PubMed.
- D. M. X. Donnelly and M. H. Sheridan, Phytochemistry, 1986, 25, 2303–2304 CrossRef CAS.
- Y.-R. Lin, C.-T. Lo, S.-Y. Liu and K.-C. Peng, J. Agric. Food Chem., 2012, 60, 2123–2128 CrossRef CAS.
- X. Zhan, R. Wang, M. Zhang, Y. Li, T. Sun, J. Chen, J. Li and T. Liu, Pest Manage. Sci., 2024, 80, 1039–1052 CrossRef CAS.
- H. G. Cutler, D. S. Himmelsbach, R. F. Arrendale, P. D. Cole and R. H. Cox, Agric. Biol. Chem., 1989, 53, 2605–2611 CAS.
- G. d. C. Ramos, I. N. d. F. Ramos, L. A. Watanabe, L. A. W. Castro, A. J. G. de Moraes, G. R. dos Santos, J. E. d. S. Siqueira, A. S. Khayat, A. M. d. R. Marinho and P. S. B. Marinho, Molecules, 2024, 29, 5278 CrossRef CAS.
- A. Fleming, Br. J. Exp. Pathol., 1929, 10, 226 CAS.
- H.-J. Schiewe and A. Zeeck, J. Antibiot., 1999, 52, 635–642 CrossRef CAS.
- G. Bills, G. Platas, A. Fillola, M. Jiménez, J. Collado, F. Vicente, J. Martín, A. González, J. Bur-Zimmermann, J. Tormo and F. Peláez, J. Appl. Microbiol., 2008, 104, 1644–1658 CrossRef CAS PubMed.
- W. Guo, J. Peng, T. Zhu, Q. Gu, R. A. Keyzers and D. Li, J. Nat. Prod., 2013, 76, 2106–2112 CrossRef CAS PubMed.
- R. T. Hewage, T. Aree, C. Mahidol, S. Ruchirawat and P. Kittakoop, Phytochemistry, 2014, 108, 87–94 CrossRef CAS PubMed.
- A. Lutfia, E. Munir, Y. Yurnaliza, M. Basyuni and H. Oku, Curr. Res. Microb. Sci., 2024, 7, 100288 CAS.
- F. C. Özkaya, W. Ebrahim, M. El-Neketi, T. Tansel Tanrıkul, R. Kalscheuer, W. E. G. Müller, Z. Guo, K. Zou, Z. Liu and P. Proksch, Fitoterapia, 2018, 131, 9–14 CrossRef PubMed.
- A. Staropoli, G. Iacomino, P. De Cicco, S. L. Woo, L. Di Costanzo and F. Vinale, Chem. Biol. Technol. Agric., 2023, 10, 28 CrossRef CAS.
- R. Pan, X. Bai, J. Chen, H. Zhang and H. Wang, Front. Microbiol., 2019, 10, 1–20 CrossRef CAS PubMed.
- C. Pinedo-Rivilla, J. Aleu and R. Durán-Patrón, Mar. Drugs, 2022, 20, 84 CrossRef CAS PubMed.
- L. Zhang, S. I. Niaz, D. Khan, Z. Wang, Y. Zhu, H. Zhou, Y. Lin, J. Li and L. Liu, Mar. Drugs, 2017, 15, 35 CrossRef PubMed.
- T. Li, J. Tang, V. Karuppiah, Y. Li, N. Xu and J. Chen, Biol. Control, 2020, 140, 104122 CrossRef CAS.
- Q. Wu, M. Ni, K. Dou, J. Tang, J. Ren, C. Yu and J. Chen, Microb. Cell Factories, 2018, 17, 155 CrossRef PubMed.
- F. Vinale, R. Nicoletti, F. Borrelli, A. Mangoni, O. A. Parisi, R. Marra, N. Lombardi, F. Lacatena, L. Grauso, S. Finizio, M. Lorito and S. L. Woo, Sci. Rep., 2017, 7, 14330 CrossRef CAS PubMed.
- Z. Shang, A. A. Salim and R. J. Capon, J. Nat. Prod., 2017, 80, 1167–1172 CrossRef CAS PubMed.
- H. Liu, D. Hao, Y. Li, X. Wang and J. Chen, Front. Microbiol., 2022, 13, 1–18 Search PubMed.
- D. Hao, B. Lang, Y. Wang, X. Wang, T. Liu and J. Chen, Microb. Cell Factories, 2022, 21, 234 CrossRef CAS PubMed.
- J. W. Bok, Y.-M. Chiang, E. Szewczyk, Y. Reyes-Dominguez, A. D. Davidson, J. F. Sanchez, H.-C. Lo, K. Watanabe, J. Strauss, B. R. Oakley, C. C. C. Wang and N. P. Keller, Nat. Chem. Biol., 2009, 5, 462–464 CrossRef CAS PubMed.
- J. Collemare and M. F. Seidl, FEMS Microbiol. Rev., 2019, 43, 591–607 CrossRef CAS PubMed.
- B. T. Pfannenstiel and N. P. Keller, Biotechnol. Adv., 2019, 37, 107345 CrossRef CAS PubMed.
- E. Y. Gómez-Rodríguez, E. E. Uresti-Rivera, O. A. Patrón-Soberano, M. A. Islas-Osuna, A. Flores-Martínez, L. Riego-Ruiz, M. T. Rosales-Saavedra and S. Casas-Flores, PLoS One, 2018, 13, e0193872 CrossRef PubMed.
- T. Shi, C.-L. Shao, Y. Liu, D.-L. Zhao, F. Cao, X.-M. Fu, J.-Y. Yu, J.-S. Wu, Z.-K. Zhang and C.-Y. Wang, Front. Microbiol., 2020, 11, 1–12 CrossRef PubMed.
- Y. Tanaka, K. Kasahara, Y. Hirose, K. Murakami, R. Kugimiya and K. Ochi, J. Bacteriol., 2013, 195, 2959–2970 CrossRef CAS PubMed.
- R. H. Baltz, J. Antibiot., 2014, 67, 619–624 CrossRef CAS PubMed.
- X. Shentu, N. Liu, G. Tang, Y. Tanaka, K. Ochi, J. Xu and X. Yu, J. Antibiot., 2016, 69, 406–410 CrossRef CAS.
- S. Zhu, Y. Duan and Y. Huang, Antibiotics, 2019, 8, 133 CrossRef CAS PubMed.
- Y.-J. Chai, C.-B. Cui, C.-W. Li, C.-J. Wu, C.-K. Tian and W. Hua, Mar. Drugs, 2012, 10, 559–582 CrossRef CAS PubMed.
- C.-J. Wu, L. Yi, C.-B. Cui, C.-W. Li, N. Wang and X. Han, Mar. Drugs, 2015, 13, 2465–2487 CrossRef CAS PubMed.
- Y. Dong, C.-B. Cui, C.-W. Li, W. Hua, C.-J. Wu, T.-J. Zhu and Q.-Q. Gu, Mar. Drugs, 2014, 12, 4326–4352 CrossRef CAS PubMed.
- H. Zhang, D. Zhang, R. Liu, T. Lou, R. Tan and S. Wang, Fermentation, 2023, 9, 398 CrossRef CAS.
- CHEMnetBASE, https://dnp.chemnetbase.com/chemical/ChemicalSearch.xhtml?dswid=-6137.
- J. A. van Santen, G. Jacob, A. L. Singh, V. Aniebok, M. J. Balunas, D. Bunsko, F. C. Neto, L. Castaño-Espriu, C. Chang, T. N. Clark, J. L. Cleary Little, D. A. Delgadillo, P. C. Dorrestein, K. R. Duncan, J. M. Egan, M. M. Galey, F. J. Haeckl, A. Hua, A. H. Hughes, D. Iskakova, A. Khadilkar, J.-H. Lee, S. Lee, N. LeGrow, D. Y. Liu, J. M. Macho, C. S. McCaughey, M. H. Medema, R. P. Neupane, T. J. O'Donnell, J. S. Paula, L. M. Sanchez, A. F. Shaikh, S. Soldatou, B. R. Terlouw, T. A. Tran, M. Valentine, J. J. J. van der Hooft, D. A. Vo, M. Wang, D. Wilson, K. E. Zink and R. G. Linington, ACS Cent. Sci., 2019, 5, 1824–1833 CrossRef CAS PubMed.
- C. Guijas, J. R. Montenegro-Burke, X. Domingo-Almenara, A. Palermo, B. Warth, G. Hermann, G. Koellensperger, T. Huan, W. Uritboonthai, A. E. Aisporna, D. W. Wolan, M. E. Spilker, H. P. Benton and G. Siuzdak, Anal. Chem., 2018, 90, 3156 CrossRef CAS.
- M. Sorokina, P. Merseburger, K. Rajan, M. A. Yirik and C. Steinbeck, J. Cheminf., 2021, 13, 2 Search PubMed.
- M. Sorokina and C. Steinbeck, J. Cheminf., 2020, 12, 20 CAS.
- A. F. Tawfike, C. Viegelmann and R. Edrada-Ebel, Methods Mol. Biol., 2013, 1055, 227–244 CrossRef CAS PubMed.
- K. F. Nielsen and T. O. Larsen, Front. Microbiol., 2015, 6, 1–15 Search PubMed.
- T. Hautbergue, E. L. Jamin, L. Debrauwer, O. Puel and I. P. Oswald, Nat. Prod. Rep., 2018, 35, 147–173 RSC.
- H. Mohimani, A. Gurevich, A. Shlemov, A. Mikheenko, A. Korobeynikov, L. Cao, E. Shcherbin, L.-F. Nothias, P. C. Dorrestein and P. A. Pevzner, Nat. Commun., 2018, 9, 4035 CrossRef PubMed.
- G. Li, T. Jian, X. Liu, Q. Lv, G. Zhang and J. Ling, Molecules, 2022, 27, 7365 CrossRef CAS.
- S. P. Gaudêncio, E. Bayram, L. Lukić Bilela, M. Cueto, A. R. Díaz-Marrero, B. Z. Haznedaroglu, C. Jimenez, M. Mandalakis, F. Pereira, F. Reyes and D. Tasdemir, Mar. Drugs, 2023, 21, 308 CrossRef.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. P. Boya, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- K. Dührkop, M. Fleischauer, M. Ludwig, A. A. Aksenov, A. V. Melnik, M. Meusel, P. C. Dorrestein, J. Rousu and S. Böcker, Nat. Methods, 2019, 16, 299–302 CrossRef PubMed.
- L. Flores-Bocanegra, Z. Y. Al Subeh, J. M. Egan, T. El-Elimat, H. A. Raja, J. E. Burdette, C. J. Pearce, R. G. Linington and N. H. Oberlies, J. Nat. Prod., 2022, 85, 614–624 CrossRef CAS PubMed.
- E. Kenshole, M. Herisse, M. Michael and S. J. Pidot, Curr. Opin. Chem. Biol., 2021, 60, 47–54 CrossRef CAS PubMed.
- F. Cai and I. S. Druzhinina, Fungal Diversity, 2021, 107, 1–69 CrossRef CAS.
- I. S. Druzhinina, A. G. Kopchinskiy, M. Komoń, J. Bissett, G. Szakacs and C. P. Kubicek, Fungal Genet. Biol., 2005, 42, 813–828 CrossRef CAS PubMed.
- D. Martinez, R. M. Berka, B. Henrissat, M. Saloheimo, M. Arvas, S. E. Baker, J. Chapman, O. Chertkov, P. M. Coutinho, D. Cullen, E. G. J. Danchin, I. V. Grigoriev, P. Harris, M. Jackson, C. P. Kubicek, C. S. Han, I. Ho, L. F. Larrondo, A. L. de Leon, J. K. Magnuson, S. Merino, M. Misra, B. Nelson, N. Putnam, B. Robbertse, A. A. Salamov, M. Schmoll, A. Terry, N. Thayer, A. Westerholm-Parvinen, C. L. Schoch, J. Yao, R. Barabote, M. A. Nelson, C. Detter, D. Bruce, C. R. Kuske, G. Xie, P. Richardson, D. S. Rokhsar, S. M. Lucas, E. M. Rubin, N. Dunn-Coleman, M. Ward and T. S. Brettin, Nat. Biotechnol., 2008, 26, 553–560 CrossRef CAS PubMed.
- M. Schalamun and M. Schmoll, Front. Fungal Biol., 2022, 3, 1–22 Search PubMed.
- A.-A. Tamizi, N. Mat-Amin, J. A. Weaver, R. T. Olumakaiye, M. A. Akbar, S. Jin, H. Bunawan and F. Alberti, J. Fungi, 2022, 8, 246 CrossRef CAS PubMed.
- N. A. O'Leary, M. W. Wright, J. R. Brister, S. Ciufo, D. Haddad, R. McVeigh, B. Rajput, B. Robbertse, B. Smith-White, D. Ako-Adjei, A. Astashyn, A. Badretdin, Y. Bao, O. Blinkova, V. Brover, V. Chetvernin, J. Choi, E. Cox, O. Ermolaeva, C. M. Farrell, T. Goldfarb, T. Gupta, D. Haft, E. Hatcher, W. Hlavina, V. S. Joardar, V. K. Kodali, W. Li, D. Maglott, P. Masterson, K. M. McGarvey, M. R. Murphy, K. O'Neill, S. Pujar, S. H. Rangwala, D. Rausch, L. D. Riddick, C. Schoch, A. Shkeda, S. S. Storz, H. Sun, F. Thibaud-Nissen, I. Tolstoy, R. E. Tully, A. R. Vatsan, C. Wallin, D. Webb, W. Wu, M. J. Landrum, A. Kimchi, T. Tatusova, M. DiCuccio, P. Kitts, T. D. Murphy and K. D. Pruitt, Nucleic Acids Res., 2016, 44, D733–D745 CrossRef PubMed.
- M. H. Medema, K. Blin, P. Cimermancic, V. de Jager, P. Zakrzewski, M. A. Fischbach, T. Weber, E. Takano and R. Breitling, Nucleic Acids Res., 2011, 39, W339–W346 CrossRef CAS PubMed.
- K. Blin, S. Shaw, H. E. Augustijn, Z. L. Reitz, F. Biermann, M. Alanjary, A. Fetter, B. R. Terlouw, W. W. Metcalf, E. J. N. Helfrich, G. P. van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2023, 51, W46–W50 CrossRef CAS PubMed.
- B. R. Terlouw, K. Blin, J. C. Navarro-Muñoz, N. E. Avalon, M. G. Chevrette, S. Egbert, S. Lee, D. Meijer, M. J. J. Recchia, Z. L. Reitz, J. A. van Santen, N. Selem-Mojica, T. Tørring, L. Zaroubi, M. Alanjary, G. Aleti, C. Aguilar, S. A. A. Al-Salihi, H. E. Augustijn, J. A. Avelar-Rivas, L. A. Avitia-Domínguez, F. Barona-Gómez, J. Bernaldo-Agüero, V. A. Bielinski, F. Biermann, T. J. Booth, V. J. Carrion Bravo, R. Castelo-Branco, F. O. Chagas, P. Cruz-Morales, C. Du, K. R. Duncan, A. Gavriilidou, D. Gayrard, K. Gutiérrez-García, K. Haslinger, E. J. N. Helfrich, J. J. J. van der Hooft, A. P. Jati, E. Kalkreuter, N. Kalyvas, K. B. Kang, S. Kautsar, W. Kim, A. M. Kunjapur, Y.-X. Li, G.-M. Lin, C. Loureiro, J. J. R. Louwen, N. L. L. Louwen, G. Lund, J. Parra, B. Philmus, B. Pourmohsenin, L. J. U. Pronk, A. Rego, D. A. B. Rex, S. Robinson, L. R. Rosas-Becerra, E. T. Roxborough, M. A. Schorn, D. J. Scobie, K. S. Singh, N. Sokolova, X. Tang, D. Udwary, A. Vigneshwari, K. Vind, S. P. J. M. Vromans, V. Waschulin, S. E. Williams, J. M. Winter, T. E. Witte, H. Xie, D. Yang, J. Yu, M. Zdouc, Z. Zhong, J. Collemare, R. G. Linington, T. Weber and M. H. Medema, Nucleic Acids Res., 2023, 51, D603–D610 CrossRef CAS PubMed.
- M. Umemura, H. Koike, N. Nagano, T. Ishii, J. Kawano, N. Yamane, I. Kozone, K. Horimoto, K. Shin-ya, K. Asai, J. Yu, J. W. Bennett and M. Machida, PLoS One, 2013, 8, e84028 CrossRef PubMed.
- M. A. Skinnider, C. A. Dejong, P. N. Rees, C. W. Johnston, H. Li, A. L. H. Webster, M. A. Wyatt and N. A. Magarvey, Nucleic Acids Res., 2015, 43, 9645–9662 CAS.
- G. D. Hannigan, D. Prihoda, A. Palicka, J. Soukup, O. Klempir, L. Rampula, J. Durcak, M. Wurst, J. Kotowski, D. Chang, R. Wang, G. Piizzi, G. Temesi, D. J. Hazuda, C. H. Woelk and D. A. Bitton, Nucleic Acids Res., 2019, 47, e110 CrossRef CAS PubMed.
- M. H. Medema, E. Takano and R. Breitling, Mol. Biol. Evol., 2013, 30, 1218 CrossRef CAS PubMed.
- N. Sélem-Mojica, C. Aguilar, K. Gutiérrez-García, C. E. Martínez-Guerrero and F. Barona-Gómez, Microb. Genomics, 2019, 5, e000260 Search PubMed.
- J. C. Navarro-Muñoz, N. Selem-Mojica, M. W. Mullowney, S. A. Kautsar, J. H. Tryon, E. I. Parkinson, E. L. C. De Los Santos, M. Yeong, P. Cruz-Morales, S. Abubucker, A. Roeters, W. Lokhorst, A. Fernandez-Guerra, L. T. D. Cappelini, A. W. Goering, R. J. Thomson, W. W. Metcalf, N. L. Kelleher, F. Barona-Gomez and M. H. Medema, Nat. Chem. Biol., 2020, 16, 60–68 CrossRef PubMed.
- S. A. Kautsar, K. Blin, S. Shaw, T. Weber and M. H. Medema, Nucleic Acids Res., 2021, 49, D490–D497 CrossRef CAS PubMed.
- L. Foulston, Curr. Opin. Microbiol., 2019, 51, 1–8 CrossRef CAS PubMed.
- L. Albarano, R. Esposito, N. Ruocco and M. Costantini, Mar. Drugs, 2020, 18, 199 CrossRef CAS PubMed.
- L. Chu, J. Huang, M. Muhammad, Z. Deng and J. Gao, Crit. Rev. Biotechnol., 2020, 40, 571–589 CrossRef CAS PubMed.
- K. D. Bauman, K. S. Butler, B. S. Moore and J. R. Chekan, Nat. Prod. Rep., 2021, 38, 2100 RSC.
- E. Pereira-Flores, M. Medema, P. L. Buttigieg, P. Meinicke, F. O. Glöckner and A. Fernández-Guerra, Mining metagenomes for natural product biosynthetic gene clusters: unlocking new potential with ultrafast techniques, 2021, https://www.biorxiv.org/content/10.1101/2021.01.20.427441v1.
- S. C. Kessler and Y.-H. Chooi, Nat. Prod. Rep., 2022, 39, 222–230 RSC.
- C. Cano-Prieto, A. Undabarrena, A. C. d. Carvalho, J. D. Keasling and P. Cruz-Morales, Annu. Rev. Biochem., 2024, 93, 411–445 CrossRef CAS PubMed.
- H. Almeida, S. Palys, A. Tsang and A. B. Diallo, NAR: Genomics Bioinf., 2020, 2, lqaa098 CrossRef PubMed.
- G. A. Vignolle, D. Schaffer, L. Zehetner, R. L. Mach, A. R. Mach-Aigner and C. Derntl, PLoS Comput. Biol., 2021, 17, e1009372 CrossRef CAS PubMed.
- E. Gluck-Thaler, S. Haridas, M. Binder, I. V. Grigoriev, P. W. Crous, J. W. Spatafora, K. Bushley and J. C. Slot, Mol. Biol. Evol., 2020, 37, 2838–2856 CrossRef CAS PubMed.
- Z. Konkel, L. Kubatko and J. C. Slot, Nucleic Acids Res., 2024, 52, e75 CrossRef CAS PubMed.
- T. C. Vesth, J. Brandl and M. R. Andersen, Synth. Syst. Biotechnol., 2016, 1, 122–129 CrossRef PubMed.
- R. E. Bradshaw, J. C. Slot, G. G. Moore, P. Chettri, P. J. G. M. de Wit, K. C. Ehrlich, A. R. D. Ganley, M. A. Olson, A. Rokas, I. Carbone and M. P. Cox, New Phytol., 2013, 198, 525–535 CrossRef CAS PubMed.
- S. Oide, W. Moeder, S. Krasnoff, D. Gibson, H. Haas, K. Yoshioka and B. G. Turgeon, Plant Cell, 2006, 18, 2836–2853 CrossRef CAS PubMed.
- I. Fujii, N. Yoshida, S. Shimomaki, H. Oikawa and Y. Ebizuka, Chem. Biol., 2005, 12, 1301–1309 CrossRef CAS PubMed.
- M. Xue, Q. Zhang, J.-M. Gao, H. Li, J.-M. Tian and G. Pescitelli, Chirality, 2012, 24, 668–674 CrossRef CAS PubMed.
- M. J. Kwon, C. Steiniger, T. C. Cairns, J. H. Wisecaver, A. L. Lind, C. Pohl, C. Regner, A. Rokas and V. Meyer, Microbiol. Spectr., 2021, 9, e0089821 CrossRef PubMed.
- S. Palys, T. T. M. Pham and A. Tsang, Front. Microbiol., 2020, 11, 1–13 CrossRef PubMed.
- C. Yan, W. Han, Q. Zhou, K. Niwa, M. J. Tang, J. E. Burch, Y. Zhang, D. A. Delgadillo, Z. Sun, Z. Wu, S. E. Jacobsen, H. Nelson, K. N. Houk and Y. Tang, J. Am. Chem. Soc., 2023, 145, 25080–25085 CrossRef CAS PubMed.
- Y. Zhu, J. Wang, P. Mou, Y. Yan, M. Chen and Y. Tang, Org. Biomol. Chem., 2021, 19, 1985–1990 RSC.
- X. Sun, Y.-S. Cai, Y. Yuan, G. Bian, Z. Ye, Z. Deng and T. Liu, Beilstein J. Org. Chem., 2019, 15, 2052 CrossRef CAS PubMed.
- A.-I. van Bohemen, N. Ruiz, A. Zalouk-Vergnoux, A. Michaud, T. Robiou du Pont, I. Druzhinina, L. Atanasova, S. Prado, B. Bodo, L. Meslet-Cladiere, B. Cochereau, F. Bastide, C. Maslard, M. Marchi, T. Guillemette and Y. F. Pouchus, J. Nat. Prod., 2021, 84, 1271–1282 CrossRef CAS PubMed.
- Y. Yan, N. Liu and Y. Tang, Nat. Prod. Rep., 2020, 37, 879–892 RSC.
- C. Hobson, A. N. Chan and G. D. Wright, Chem. Rev., 2021, 121, 3464–3494 CrossRef CAS PubMed.
- X. Tang, J. Li, N. Millán-Aguiñaga, J. J. Zhang, E. C. O'Neill, J. A. Ugalde, P. R. Jensen, S. M. Mantovani and B. S. Moore, ACS Chem. Biol., 2015, 10, 2841–2849 CrossRef CAS PubMed.
- T. B. Regueira, K. R. Kildegaard, B. G. Hansen, U. H. Mortensen, C. Hertweck and J. Nielsen, Appl. Environ. Microbiol., 2011, 77, 3035–3043 CrossRef CAS PubMed.
- H.-H. Yeh, M. Ahuja, Y.-M. Chiang, C. E. Oakley, S. Moore, O. Yoon, H. Hajovsky, J.-W. Bok, N. P. Keller, C. C. C. Wang and B. R. Oakley, ACS Chem. Biol., 2016, 11, 2275–2284 CrossRef CAS PubMed.
- L. Xie, X. Zang, W. Cheng, Z. Zhang, J. Zhou, M. Chen and Y. Tang, J. Am. Chem. Soc., 2021, 143, 9575–9584 CrossRef CAS PubMed.
- N. Liu, E. D. Abramyan, W. Cheng, B. Perlatti, C. J. B. Harvey, G. F. Bills and Y. Tang, J. Am. Chem. Soc., 2021, 143, 6043–6047 CrossRef CAS PubMed.
- I. Kjærbølling, T. Vesth and M. R. Andersen, mSystems, 2019, 4, e00085 CrossRef PubMed.
- C. B. Jenkinson, A. R. Podgorny, C. Zhong and B. R. Oakley, J. Ind. Microbiol. Biotechnol., 2023, 50, kuad045 CrossRef CAS PubMed.
- T. M. Yılmaz, M. D. Mungan, A. Berasategui and N. Ziemert, Nucleic Acids Res., 2023, 51, W191 CrossRef PubMed.
- M. Alanjary, B. Kronmiller, M. Adamek, K. Blin, T. Weber, D. Huson, B. Philmus and N. Ziemert, Nucleic Acids Res., 2017, 45, W42 CrossRef CAS PubMed.
- M. D. Mungan, M. Alanjary, K. Blin, T. Weber, M. H. Medema and N. Ziemert, Nucleic Acids Res., 2020, 48, W546–W552 CrossRef CAS PubMed.
- A. S. Anker, U. Friis-Jensen, F. L. Johansen, S. J. L. Billinge and K. M. Jensen, Acta Crystallogr., Sect. A:Found. Adv., 2024, 80, 213 CrossRef CAS PubMed.
- H. Almeida, A. Tsang and A. B. Diallo, Bioinformatics, 2022, 38, 3984–3991 CrossRef CAS PubMed.
- M. Röttig, M. H. Medema, K. Blin, T. Weber, C. Rausch and O. Kohlbacher, Nucleic Acids Res., 2011, 39, W362 CrossRef PubMed.
- Y. Minowa, M. Araki and M. Kanehisa, J. Mol. Biol., 2007, 368, 1500–1517 CrossRef CAS PubMed.
- A. Starcevic, J. Zucko, J. Simunkovic, P. F. Long, J. Cullum and D. Hranueli, Nucleic Acids Res., 2008, 36, 6882 CrossRef CAS PubMed.
- A. S. Walker and J. Clardy, J. Chem. Inf. Model., 2021, 61, 2560–2571 CrossRef CAS PubMed.
- O. Riedling, A. S. Walker and A. Rokas, Microbiol. Spectrum, 2024, 12, e03400–e03423 Search PubMed.
- M. W. Mullowney, K. R. Duncan, S. S. Elsayed, N. Garg, J. J. J. Van Der Hooft, N. I. Martin, D. Meijer, B. R. Terlouw, F. Biermann, K. Blin, J. Durairaj, M. Gorostiola González, E. J. N. Helfrich, F. Huber, S. Leopold-Messer, K. Rajan, T. De Rond, J. A. Van Santen, M. Sorokina, M. J. Balunas, M. A. Beniddir, D. A. Van Bergeijk, L. M. Carroll, C. M. Clark, D.-A. Clevert, C. A. Dejong, C. Du, S. Ferrinho, F. Grisoni, A. Hofstetter, W. Jespers, O. V. Kalinina, S. A. Kautsar, H. Kim, T. F. Leao, J. Masschelein, E. R. Rees, R. Reher, D. Reker, P. Schwaller, M. Segler, M. A. Skinnider, A. S. Walker, E. L. Willighagen, B. Zdrazil, N. Ziemert, R. J. M. Goss, P. Guyomard, A. Volkamer, W. H. Gerwick, H. U. Kim, R. Müller, G. P. Van Wezel, G. J. P. Van Westen, A. K. H. Hirsch, R. G. Linington, S. L. Robinson and M. H. Medema, Nat. Rev. Drug Discov., 2023, 22, 895–916 CrossRef CAS PubMed.
- M. T. Robey, L. K. Caesar, M. T. Drott, N. P. Keller and N. L. Kelleher, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020230118 CrossRef CAS PubMed.
- A. W. Goering, R. A. McClure, J. R. Doroghazi, J. C. Albright, N. A. Haverland, Y. Zhang, K.-S. Ju, R. J. Thomson, W. W. Metcalf and N. L. Kelleher, ACS Cent. Sci., 2016, 2, 99–108 CrossRef CAS PubMed.
- K. R. Duncan, M. Crüsemann, A. Lechner, A. Sarkar, J. Li, N. Ziemert, M. Wang, N. Bandeira, B. S. Moore, P. C. Dorrestein and P. R. Jensen, Chem. Biol., 2015, 22, 460–471 CrossRef CAS PubMed.
- R.-Z. Liu, Z. Zhang, M. Li and L. Zhang, Chem. Sci., 2025, 16, 1696–1706 RSC.
- J. R. Doroghazi, J. C. Albright, A. W. Goering, K.-S. Ju, R. R. Haines, K. A. Tchalukov, D. P. Labeda, N. L. Kelleher and W. W. Metcalf, Nat. Chem. Biol., 2014, 10, 963–968 CrossRef CAS PubMed.
- J. J. J. v. d. Hooft, H. Mohimani, A. Bauermeister, P. C. Dorrestein, K. R. Duncan and M. H. Medema, Chem. Soc. Rev., 2020, 49, 3297–3314 RSC.
- S. Soldatou, G. H. Eldjárn, A. Ramsay, J. J. J. v. d. Hooft, A. H. Hughes, S. Rogers and K. R. Duncan, Mar. Drugs, 2021, 19, 103 CrossRef PubMed.
- Y. Qi, K. K. Nepal and J. A. V. Blodgett, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2103515118 CrossRef CAS PubMed.
- L. K. Caesar, R. Montaser, N. P. Keller and N. L. Kelleher, Nat. Prod. Rep., 2021, 38, 2041 RSC.
- N. M. Maimone, M. C. P. Junior, L. F. P. de Oliveira, D. Rojas-Villalta, S. P. de Lira, L. Barrientos and K. Núñez-Montero, Front. Microbiol., 2023, 14, 1–13 Search PubMed.
- G. A. Vitale, G. G. January, E. Oppong-Danquah, G. Della Sala, F. Palma Esposito, D. Tasdemir and D. de Pascale, PNAS Nexus, 2023, 2, pgad221 CrossRef PubMed.
- C. E. Yancey, L. Hart, S. Hefferan, O. G. Mohamed, S. A. Newmister, A. Tripathi, D. H. Sherman and G. J. Dick, mSystems, 2024, 9, e0033424 CrossRef PubMed.
- M. A. Schorn, S. Verhoeven, L. Ridder, F. Huber, D. D. Acharya, A. A. Aksenov, G. Aleti, J. A. Moghaddam, A. T. Aron, S. Aziz, A. Bauermeister, K. D. Bauman, M. Baunach, C. Beemelmanns, J. M. Beman, M. V. Berlanga-Clavero, A. A. Blacutt, H. B. Bode, A. Boullie, A. Brejnrod, T. S. Bugni, A. Calteau, L. Cao, V. J. Carrión, R. Castelo-Branco, S. Chanana, A. B. Chase, M. G. Chevrette, L. V. Costa-Lotufo, J. M. Crawford, C. R. Currie, B. Cuypers, T. Dang, T. de Rond, A. M. Demko, E. Dittmann, C. Du, C. Drozd, J.-C. Dujardin, R. J. Dutton, A. Edlund, D. P. Fewer, N. Garg, J. M. Gauglitz, E. C. Gentry, L. Gerwick, E. Glukhov, H. Gross, M. Gugger, D. G. Guillén Matus, E. J. N. Helfrich, B.-F. Hempel, J.-S. Hur, M. Iorio, P. R. Jensen, K. B. Kang, L. Kaysser, N. L. Kelleher, C. S. Kim, K. H. Kim, I. Koester, G. M. König, T. Leao, S. R. Lee, Y.-Y. Lee, X. Li, J. C. Little, K. N. Maloney, D. Männle, C. H. Martin, A. C. McAvoy, W. W. Metcalf, H. Mohimani, C. Molina-Santiago, B. S. Moore, M. W. Mullowney, M. Muskat, L.-F. Nothias, E. C. O'Neill, E. I. Parkinson, D. Petras, J. Piel, E. C. Pierce, K. Pires, R. Reher, D. Romero, M. C. Roper, M. Rust, H. Saad, C. Saenz, L. M. Sanchez, S. J. Sørensen, M. Sosio, R. D. Süssmuth, D. Sweeney, K. Tahlan, R. J. Thomson, N. J. Tobias, A. E. Trindade-Silva, G. P. van Wezel, M. Wang, K. C. Weldon, F. Zhang, N. Ziemert, K. R. Duncan, M. Crüsemann, S. Rogers, P. C. Dorrestein, M. H. Medema and J. J. J. van der Hooft, Nat. Chem. Biol., 2021, 17, 363–368 CrossRef CAS PubMed.
- L. K. Caesar, F. A. Butun, M. T. Robey, N. J. Ayon, R. Gupta, D. Dainko, J. W. Bok, G. Nickles, R. J. Stankey, D. Johnson, D. Mead, K. B. Cank, C. E. Earp, H. A. Raja, N. H. Oberlies, N. P. Keller and N. L. Kelleher, Nat. Chem. Biol., 2023, 19, 846 CrossRef CAS PubMed.
- N. J. Ayon, C. E. Earp, R. Gupta, F. A. Butun, A. E. Clements, A. G. Lee, D. Dainko, M. T. Robey, M. Khin, L. Mardiana, A. Longcake, M. Rangel-Grimaldo, M. J. Hall, M. R. Probert, J. E. Burdette, N. P. Keller, H. A. Raja, N. H. Oberlies, N. L. Kelleher and L. K. Caesar, Metabolomics, 2024, 20, 90 CrossRef CAS PubMed.
- M. G. Malmierca, R. E. Cardoza and S. Gutiérrez, Genetic Transformation Systems in Fungi, Springer International Publishing, Cham, 2015, vol. 1, pp. 41–48 Search PubMed.
- F. Cai, C. P. Kubicek and I. S. Druzhinina, Biofuels and Biodiesel, Springer US, New York, NY, 2021, pp. 171–185 Search PubMed.
- S. Jin and F. Alberti, STAR Protoc., 2025, 6, 103668 CrossRef CAS PubMed.
- J. W. Bok and N. P. Keller, Eukaryotic Cell, 2004, 3, 527–535 CrossRef CAS PubMed.
- M. Schalamun, S. Beier, W. Hinterdobler, N. Wanko, J. Schinnerl, L. Brecker, D. E. Engl and M. Schmoll, Sci. Rep., 2023, 13, 1912 CrossRef CAS PubMed.
- J. Yang, J.-X. Li, F. Zhang and X.-Q. Zhao, Eng. Microbiol., 2023, 3, 100065 CrossRef CAS PubMed.
- P. K. Mukherjee and C. M. Kenerley, Appl. Environ. Microbiol., 2010, 76, 2345 CrossRef CAS PubMed.
- R. Karimi Aghcheh, Z. Németh, L. Atanasova, E. Fekete, M. Paholcsek, E. Sándor, B. Aquino, I. S. Druzhinina, L. Karaffa and C. P. Kubicek, PLoS One, 2014, 9, e112799 CrossRef PubMed.
- V. Karuppiah, Y. Li, J. Sun, M. Vallikkannu and J. Chen, Biol. Control, 2020, 151, 104374 CrossRef CAS.
- V. Karuppiah, L. Zhixiang, H. Liu, M. Vallikkannu and J. Chen, Microb. Cell Factories, 2021, 20, 57 CrossRef CAS PubMed.
- Z. Ding, X. Wang, F.-D. Kong, H.-M. Huang, Y.-N. Zhao, M. Liu, Z.-P. Wang and J. Han, Front. Microbiol., 2020, 11, 1–8 CrossRef PubMed.
- J.-C. Shi, W.-L. Shi, Y.-R. Zhou, X.-L. Chen, Y.-Z. Zhang, X. Zhang, W.-X. Zhang and X.-Y. Song, Front. Microbiol., 2020, 11, 1–13 CrossRef PubMed.
- R. K. Aghcheh, I. S. Druzhinina and C. P. Kubicek, PLoS One, 2013, 8, e67144 CrossRef CAS PubMed.
- S. Bergmann, J. Schümann, K. Scherlach, C. Lange, A. A. Brakhage and C. Hertweck, Nat. Chem. Biol., 2007, 3, 213–217 CrossRef CAS PubMed.
- S. Bergmann, A. N. Funk, K. Scherlach, V. Schroeckh, E. Shelest, U. Horn, C. Hertweck and A. A. Brakhage, Appl. Environ. Microbiol., 2010, 76, 8143–8149 CrossRef CAS PubMed.
- S. Janevska and B. Tudzynski, Appl. Microbiol. Biotechnol., 2018, 102, 615–630 CrossRef CAS PubMed.
- C. Derntl, R. L. Mach and A. R. Mach-Aigner, Biotechnol. Biofuels, 2019, 12, 231 CrossRef PubMed.
- F. Wang, R. Zhang, L. Han, W. Guo, Z. Du, K. Niu, Y. Liu, C. Jia and X. Fang, Biotechnol. Biofuels, 2019, 12, 244 CrossRef PubMed.
- L. Han, Y. Tan, W. Ma, K. Niu, S. Hou, W. Guo, Y. Liu and X. Fang, Front. Bioeng. Biotechnol., 2020, 8, 1–9 CrossRef PubMed.
- Q.-S. Meng, F. Zhang, W. Wang, C.-G. Liu, X.-Q. Zhao and F.-W. Bai, Front. Bioeng. Biotechnol., 2020, 8, 1–9 CrossRef PubMed.
- M. Chen, Q. Liu, S.-S. Gao, A. E. Young, S. E. Jacobsen and Y. Tang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 5499–5504 CrossRef CAS PubMed.
- H. Ren, B. Wang and H. Zhao, Curr. Opin. Biotechnol., 2017, 48, 21–27 CrossRef CAS PubMed.
- G. Zou, S. Shi, Y. Jiang, J. van den Brink, R. P. de Vries, L. Chen, J. Zhang, L. Ma, C. Wang and Z. Zhou, Microb. Cell Fact., 2012, 11, 21 CrossRef CAS PubMed.
- E. Fitz, F. Wanka and B. Seiboth, Front. Bioeng. Biotechnol., 2018, 6, 1–15 CrossRef PubMed.
- M. Adnan, X. Ma, S. Olsson, J. Wang and G. Liu, Microbiol. Res., 2022, 259, 127011 CrossRef CAS PubMed.
- M. Adnan and G. Liu, Synthetic Promoters: Methods and Protocols, Springer US, New York, NY, 2024, pp. 47–68 Search PubMed.
- P.-L. Wei, J. Fan, J. Yu, Z. Ma, X. Guo, N. P. Keller, E. Li, C. Lou and W.-B. Yin, Sci. China Life Sci., 2023, 66, 848–860 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- W. Jiang, D. Bikard, D. Cox, F. Zhang and L. A. Marraffini, Nat. Biotechnol., 2013, 31, 233–239 CrossRef CAS PubMed.
- J. E. DiCarlo, J. E. Norville, P. Mali, X. Rios, J. Aach and G. M. Church, Nucleic Acids Res., 2013, 41, 4336–4343 CrossRef CAS PubMed.
- R. E. Cobb, Y. Wang and H. Zhao, ACS Synth. Biol., 2015, 4, 723–728 CrossRef CAS PubMed.
- K. K. Fuller, S. Chen, J. J. Loros and J. C. Dunlap, Eukaryotic Cell, 2015, 14, 1073–1080 CrossRef CAS PubMed.
- T. Katayama, Y. Tanaka, T. Okabe, H. Nakamura, W. Fujii, K. Kitamoto and J.-I. Maruyama, Biotechnol. Lett., 2016, 38, 637–642 CrossRef CAS PubMed.
- S.-Y. Shih, U. H. Mortensen, F.-R. Chang and H. Tsai, Synth. Biol., 2022, 7, ysac031 CrossRef PubMed.
- D. M. Gardiner and K. Kazan, Fungal Biol., 2018, 122, 131–137 CrossRef CAS PubMed.
- Q. Wang, P. A. Cobine and J. J. Coleman, Fungal Genet. Biol., 2018, 117, 21–29 CrossRef CAS PubMed.
- M. Ferrara, M. Haidukowski, A. F. Logrieco, J. F. Leslie and G. Mulè, Sci. Rep., 2019, 9, 19836 CrossRef CAS PubMed.
- Y. Wang, H. Chen, L. Ma, M. Gong, Y. Wu, D. Bao and G. Zou, Microb. Biotechnol., 2022, 15, 2521–2532 CrossRef CAS PubMed.
- C. Woodcraft, Y.-H. Chooi and I. Roux, Nat. Prod. Rep., 2023, 40, 158–173 RSC.
- L. Wang, J. Liu, J. Tang, Y. Dang, L. Sun, B. Liu, H. Li, X. He, Q. Shuai, Z. Peng, T. Huang, Y. Sun, Y. Feng and J. Xie, Int. J. Biol. Macromol., 2024, 279, 135339 CrossRef PubMed.
- Y. Fang, X. Meng, L. Liu, Z. Li, K. Jia and W. Liu, ACS Synth. Biol., 2025, 14, 575–584 CrossRef CAS PubMed.
- X. Wang, K. Subko, S. Kildgaard, J. C. Frisvad and T. O. Larsen, Front. Fungal Biol., 2021, 2, 1–13 CrossRef PubMed.
- F. Alberti, G. D. Foster and A. M. Bailey, Appl. Microbiol. Biotechnol., 2017, 101, 493–500 CrossRef CAS PubMed.
- K. Murai, L. Lauterbach, K. Teramoto, Z. Quan, L. Barra, T. Yamamoto, K. Nonaka, K. Shiomi, M. Nishiyama, T. Kuzuyama and J. S. Dickschat, Angew. Chem., Int. Ed., 2019, 58, 15046–15050 CrossRef CAS PubMed.
- M. S. Siddiqui, K. Thodey, I. Trenchard and C. D. Smolke, FEMS Yeast Res., 2012, 12, 144–170 CrossRef CAS PubMed.
- K. Ishiuchi, T. Nakazawa, T. Ookuma, S. Sugimoto, M. Sato, Y. Tsunematsu, N. Ishikawa, H. Noguchi, K. Hotta, H. Moriya and K. Watanabe, ChemBioChem, 2012, 13, 846–854 CrossRef CAS PubMed.
- C. Bond, Y. Tang and L. Li, Fungal Genet. Biol., 2016, 89, 52–61 CrossRef CAS PubMed.
- J. Liu, Y. Zhai, Y. Zhang, S. Zhu, G. Liu and Y. Che, Front. Microbiol., 2018, 9, year Search PubMed.
- C. J. B. Harvey, M. Tang, U. Schlecht, J. Horecka, C. R. Fischer, H.-C. Lin, J. Li, B. Naughton, J. Cherry, M. Miranda, Y. F. Li, A. M. Chu, J. R. Hennessy, G. A. Vandova, D. Inglis, R. S. Aiyar, L. M. Steinmetz, R. W. Davis, M. H. Medema, E. Sattely, C. Khosla, R. P. S. Onge, Y. Tang and M. E. Hillenmeyer, Sci. Adv., 2018, 4, eaar5459 CrossRef PubMed.
- C. M. Lazarus, K. Williams and A. M. Bailey, Nat. Prod. Rep., 2014, 31, 1339–1347 RSC.
- E. Geib and M. Brock, Fungal Biol. Biotechnol., 2017, 4, 13 CrossRef PubMed.
- Y.-M. Chiang, C. E. Oakley, M. Ahuja, R. Entwistle, A. Schultz, S.-L. Chang, C. T. Sung, C. C. C. Wang and B. R. Oakley, J. Am. Chem. Soc., 2013, 135, 7720–7731 CrossRef CAS PubMed.
- I. Tomico-Cuenca, R. L. Mach, A. R. Mach-Aigner and C. Derntl, Fungal Biol. Biotechnol., 2021, 8, 11 CrossRef CAS PubMed.
- M. L. Shenouda, M. Ambilika, E. Skellam and R. J. Cox, J. Fungi, 2022, 8, 355 CrossRef CAS PubMed.
- H. Mathis, D. Naquin, A. Margeot and F. Bidard, Appl. Microbiol. Biotechnol., 2024, 108, 470 CrossRef CAS PubMed.
- M. L. Shenouda and R. J. Cox, RSC Adv., 2021, 11, 3622–3635 RSC.
- L. Kirchgaessner, J. M. Wurlitzer, P. S. Seibold, M. Rakhmanov and M. Gressler, Fungal Biol. Biotechnol., 2023, 10, 1–16 CrossRef PubMed.
- L. Hang, M.-C. Tang, C. J. B. Harvey, C. G. Page, J. Li, Y.-S. Hung, N. Liu, M. E. Hillenmeyer and Y. Tang, Angew. Chem., 2017, 129, 9684–9688 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |