Open Access Article
Open Access ArticleSesterterpenoids: sources, structural diversity, biological activity, and data management†
Valeria
Iobbi‡
 a,
Valentina
Parisi‡
a,
Valentina
Parisi‡
 bg,
Mauro
Giacomini
bg,
Mauro
Giacomini
 c,
Francesco
De Riccardis
c,
Francesco
De Riccardis
 d,
Paola
Brun
d,
Paola
Brun
 e,
Laura
Núñez-Pons
e,
Laura
Núñez-Pons
 fg,
Giuliana
Drava
a,
Paolo
Giordani
a,
Maria Chiara
Monti
fg,
Giuliana
Drava
a,
Paolo
Giordani
a,
Maria Chiara
Monti
 h,
Roberto
Poggi
h,
Roberto
Poggi
 i,
Ylenia
Murgia
c,
Nunziatina
De Tommasi
i,
Ylenia
Murgia
c,
Nunziatina
De Tommasi
 *bg and
Angela
Bisio
*bg and
Angela
Bisio
 *a
*a
aDepartment of Pharmacy, University of Genova, Viale Cembrano 4, 16148 Genova, Italy. E-mail: angela.bisio@unige.it
bDepartment of Pharmacy, Via Giovanni Paolo II, 132, 84084 Fisciano, Salerno, Italy. E-mail: detommasi@unisa.it
cDepartment of Informatics, Bioengineering, Robotics and System Science, University of Genova, Via all’Opera Pia 13, 16146 Genova, Italy
dDepartment of Chemistry and Biology “A. Zambelli”, Via Giovanni Paolo II, 132, 84084 Fisciano, Salerno, Italy
eDepartment of Molecular Medicine, Section of Microbiology, University of Padova, Via A. Gabelli, 63, 35121 Padova, Italy
fDepartment of Integrative Marine Ecology (EMI), Stazione Zoologica Anton Dohrn, Villa Comunale, 80121 Napoli, Italy
gNBFC, National Biodiversity Future Center, 90133 Palermo, Italy
hDepartment of Pharmacy, University of Napoli “Federico II”, Via T. De Amicis 95, 80131 Napoli, Italy
iMuseo Civico di Storia Naturale Giacomo Doria, Via Brigata Liguria 9, 16121 Genova, Italy
First published on 20th January 2025
Abstract
Reviewing the literature published up to October 2024.
Sesterterpenoids are one of the most chemically diverse and biologically promising subgroup of terpenoids, the largest family of secondary metabolites. The present review article summarizes more than seven decades of studies on isolation and characterization of more than 1600 structurally novel sesterterpenoids, supplemented by biological, pharmacological, ecological, and geographic distribution data. All the information have been implemented in eight tables available on the web and a relational database https://sesterterpenoids.unige.net/. The interface has two sections, one open to the public for reading only and the other, protected by an authentication mechanism, for timely updating of published results.
1 Introduction
Despite originating from a single biosynthetic pentaprenyl linear precursor, sesterterpenoids1–9 epitomize the astounding strive of nature towards molecular diversity and complexity. The incredibly vast chemical space covered by sesterterpenoids embodies a myriad of forms, skeletal architectures, and substitution patterns. To date, over 1600 structures have been reported, with tens of unique hetero- and carbocyclic ring systems.Since the isolation of the first members, in the late fifties and early sixties,8 it has been clear that sesterterpenoids were widespread in several phyletic groups, including marine sponges, nudibranchs, bacteria, lichens, fungi, higher plants and insects (Fig. 1).7,10–12 Although only a few macrocategories of sesterterpenoids are known for some taxa, most phyla can synthesize a considerable variety of compounds, from the simplest, such as the acyclic linear (AI), to the most complex, such as the hexacarbocyclic (HC). In general, several evolutionary mechanisms have been described that can lead to biosynthetic diversity and cause biosynthetic pathways to converge or diverge within or between different groups of organisms. These include gene duplication (and gene loss), horizontal and endosymbiotic gene transfer, and gene fusion.13 With regard to sesterterpenoids, the picture of possible co-evolution of biosynthetic pathways is still unclear, and this is certainly a gap worth exploring.
Among the natural sources, the Porifera phylum (sponges) is the most prolific.14–17 The original producers of most natural products, including sesterterpenoids, in sponges, but also in other pluricellular holobionts, are often suspected to be their associated microbes, with larger metabolic capabilities.18–23 This widely approved hypothesis has been at the moment scarcely proved.14,16,17 Nonetheless, technological advances in omics and biological spatial approaches, single cell analyses, and cultivation procedures may soon provide suitable tools to demonstrate a symbiotic implication in the synthesis or biotransformation of secondary metabolites.24 In the intricate interactions between microbes and their hosts and within microbial communities, sesterterpenoids act as chemical defence and communication systems to enhance beneficial associations,25 and thus they can be considered allelochemicals mediating species interactions.14,16,17 Marine sponges, algae, and terrestrial plants produce sesterterpenoids to disrupt microbial membranes or ignite reactive species inside bacterial and fungal predators, whereas, in some plant–microbe interactions, sesterterpenoids produced by the plant can attract beneficial microbes that promote plant health and growth.26 Certain spongivore molluscs are known to specifically feed on sesterterpenoid rich prey, in order to bioaccumulate or biotransform them for their own defence.27 In other instances, molluscs and cnidarians seem to produce these bioactive molecules de novo.14,16,17,27 In microbial communities, microbes produce sesterterpenoids to inhibit the growth of competing species, thereby securing their niche or influence quorum sensing, a mechanism that bacteria use to coordinate their behaviour and eventually control biofilm formation, virulence, and metabolism.28
In other groups of organisms, such as insects and plants, the function of sesterterpenoids has been less well investigated. Future studies should ascertain the involvement of these compounds in mediating the relationships between organism and their environment.
The driving force behind sesterterpenoids research, besides the purely structural, and synthetic studies, has been the wide range of biological and pharmacological properties they often exhibit (i.e.: anticancer, antimicrobial, anti-inflammatory, antifeedant, and antiviral activities). Sesterterpenoids also play a key role in the modulation of neurodegenerative processes, they have been studied for the treatment of type-II diabetes, hypercholesterolemia and obesity and as potential immunosuppressive agents.
With their rich oxidation patterns and three-dimensional complexity, these pentaprenyl terpenoids constitute a vast chemical library that can be easily morphed in new chemical entities (via ingenious semisynthetic or synthetic approaches). However, no synthetic study is reported in this review (unless for structural/stereochemical confirmation of the isolated secondary metabolites).
This review considers the extensive literature on sesterterpenoids to identify the state of the art on the subject and to highlight gaps in knowledge that need to be addressed in future studies. Specifically, the review is structured as follows: Section 2 presents a database that collects and organises the available information on the over 1600 sesterterpenoids. Sections 3 and 4 review the main compounds found in the different organism groups, while Section 5 outlines the biological and pharmacological properties already tested for these compounds.
2 Information infrastructure
2.1 Conceptual model
Since the 1970s, the Entity-Relationship (ER) diagram has played a fundamental role in the initial phases of data modelling projects. In this project, the conceptual model based on ER diagrams played a crucial role in defining the common design of the project's information support. The diagrams' graphic nature facilitated effective collaboration among the project's experts, enabling the proper exchange of individual collaborators' skills.The diagrams depict compounds, organisms, and bibliographic resources as the primary entities, with production, biological activity, and corresponding bibliography descriptions as the primary relationships connecting them.
2.2 Logical model
To ensure a reliable and efficient database, we translated the ER diagram into a logical scheme using the well-known relational model. This allowed us to transfer the agreed-upon knowledge organization of all project participants and experts. The logical scheme was implemented in a relational database management system (DBMS). The Microsoft SQL Server 2022 DBMS29 was chosen and mounted on a server running the Windows Server 2022 operating system.30 The server is hosted within the IT structure of Genoa University, which is backed up daily. In this project, specific measures were applied in addition to the standard rules of the relational model. Among these measures, we included an identification code for the compounds specific to this project. This is necessary because different bibliographic sources do not refer to a common nomenclature standard. Additionally, translating the graphic peculiarities (such as the use of italics, superscripts, and subscripts) typical of the nomenclature rules of both compounds and organisms into HTML is necessary. Unicode coding was used to name the compounds to ensure the letters of the Greek alphabet are essential for their correct naming.However, when it comes to organisms, scientific names that follow well-established taxonomic rules are used in the literature. These names have been stored in the database as they appear in the bibliographic resources used for this project. It is important to note that the taxonomy of many organisms can rapidly change, rendering some nomenclatures obsolete and introducing new names that are recognized internationally. All names listed in international standard nomenclators are stored in the database. For each organism, all taxonomic levels are stored in the database using a normalized relational structure. This allows for quick provision of descriptive statistics of the database contents.
2.3 Data presentation
Although the DBMS allows for adequate and efficient management of data storage, excessive normalization results in a high number of tables connected by numerical indexes. This can make table management difficult for non-experts and irrelevant to ordinary people. To address this issue, a web interface was developed, divided into two sections. The initial section is publicly accessible and read-only and is structured for easy navigation among the data collected for this research. It can be explored based on the three major entities listed above: compounds, organisms, and bibliographic resources. The second section is restricted to authorized personnel and requires first-level authentication. It enables regular maintenance of the database content. The need to feed the database during its development suggested implementing the possibility of feeding data stored in batch mode through a set of Ms Excel files. The format of these files is based on the results of the conceptual analysis explained above.2.4 External connections
The working group frequently updates the database, but the taxonomies of the organisms involved in the research can frequently change, which can quickly render the stored data obsolete. To address this issue, automatic query mechanisms have been planned for the main global databases in the sector, often providing access via web services. The system enables interested users to access updated data on a particular organism by requesting the same interface used in this project to query the relevant services. The updated data is then presented on a page that is appropriately formatted for the purpose. This allows non-expert users to access the most recent data on the subject. The sources of the updates are clearly indicated on the page.3 Sesterterpenoids isolated from marine and terrestrial organisms
The current review categorises sesterterpenoids into ten subgroups (Table S1, ESI,† Sections 1.1–1.10), based on their structural features and increasing molecular complexity, ranging from linear to hexacarbocyclic backbones. All previous reviews on the subject in Natural Product Reports have followed, directly or indirectly, this subdivision criterion.1–3,6,31,32 However, due to the large number of linear sesterterpenoids reported, a further partition has been implemented based on the growing number of heterocyclic nuclei present in the terpenoid backbone (Table S1, ESI,† Sections 1.1–1.4). Accordingly, Section 1.1 of Table S1† presents the structures of the simplest linear acyclic (AI) sesterterpenoids decorated with various functional groups (i.e.: AI-11 from Oryza sativa,33Fig. 2); in Section 1.2 linear sesterterpenoids incorporating a single heterocyclic nucleus are collected and labelled as linear monoheterocyclic (MH) sesterterpenoids (i.e.: granuloside, MH-52 from Charcotia granulosa, Fig. 2);34 Section 1.3 includes the structures of linear diheterocyclic (DH) sesterterpenoids (i.e.: hippolide A, DH-80 from Hippospongia lachne, Fig. 2);35 Section 1.4 reports the structures of linear triheterocyclic (TH) sesterterpenoids (i.e.: ircinialactam F, TH-24 from Ircinia oros, Fig. 2)36 and their possible dimeric counterparts. From Sections 1.5 to 1.10 a variety of structurally diverse and morphologically complex sesterterpenoids including carbocyclic moieties have been reported. In particular, Section 1.5 reports monocarbocyclic (MC) sesterterpenoids (i.e.: manoalide, MC-13, from Luffariella variabilis, Fig. 3);37 Section 1.6 includes dicarbocyclic (DC) sesterterpenoids (i.e.: terpestacin, DC-104, from Arthrinium sp.);38 Section 1.7 reports tricarbocyclic (TrC) sesterterpenoids (i.e.: ophiobolin A, TrC-2 from Ophiobolus miyabeanus);39 Section 1.8 comprehends tetracarbocyclic (TeC) sesterterpenoids (i.e.: bipolarolide A, TeC-1 from Bipolaris sp.);40 Section 1.9, pentacarbocyclic (PC) sesterterpenoids [i.e.: phyllofenone D, PC-13 from Carteriospongia (syn. of Phyllospongia foliascens)];41 and Section 1.10, hexacarbocyclic (HC) sesterterpenoids (i.e.: niduterpenoid A, HC-1 from Aspergillus nidulans).42 For Table S1 (ESI†) consultation, it is important to consider the following relevant information: (1) the sesterterpenoids included in each of the ten sections have been listed without any specific structural, biogenetic, phylogenetic, or chronological order. The only criterion followed is that they belong to the structural class indicated by the denomination of the section; (2) where a revision or a new stereochemical assignment has been published, the correct structures and configurations are given; (3) carbocycles are counted as single independent units, even if they have a different biogenetic origin (i.e.: see phenyl rings in sesterterpenoids MC-85,43TrC-112,44HC-3 (ref. 45)); (4) dimeric structures are always reported in the section of the corresponding monomeric counterparts (i.e.: sulawesin C, TH-25, from Psammocinia sp. in Section 1.4,46 and molliorin-B, TeC-362, from Cacospongia mollior in Section 1.8, Fig. 2);47 (5) heterocyclic and carbocyclic rings are counted as single independent units even when they are present as bridged units (two rings share more than two atoms: i.e.: DH-99,48DC-205,49TrC-177,50PC-9 (ref. 51)); (6) Table S1 (ESI)† includes the sesterterpenoids isolated from natural sources and by biosynthetic experiments and reported since the sixties of the last century. In some cases, meroterpenoid derivatives have been included when the pentaprenyl co-substrate is easily recognisable in the structure. Unfortunately, in most of the cases, meroterpenoids are difficult to be classified and lack of proper biogenetic studies. This hampers proper skeleton recognition and structural sorting. | ||
| Fig. 2 Representative structures of formally linear sesterterpenoids incorporating no heterocycles (AI-11), or incorporating one (MH-52), two (DH-80), and three heterocyclic rings (TH-24), as reported in Sections 1.1–1.4, respectively, of Table S1.† Sulawesin C (TH-25) and molliorin-B (TeC-362) represent rare dimeric sesterterpenoids. | ||
 | ||
| Fig. 3 Representative structures of sesterterpenoids incorporating one (MC-13), two (DC-104), three (TrC-2), four (TeC-1), five (PC-13), and six carbocyclic rings (HC-1), as reported in Sections 1.5–1.10, respectively, of Table S1.† | ||
4 Distribution of sesterterpenoids
4.1 Sesterterpenoids in animals
Most soft-bodied and sessile marine organisms, lacking physical or other mechanical means of protection or locomotion for escape, have evolved chemical defences for survival. Often these defensive compounds are noxious to potential predators, toxic, or have some type of bioactivity that directly or indirectly interferes with the behaviour or biology of co-occurring competing species.16,17 Among these allelochemicals, terpenoids are the most abundant and important compounds of marine origin. In this large family, the sesterterpenoids form a relatively narrow group of molecules, found mainly in sponges, and to lesser extent in molluscs and cnidarians, as well as in a reduced group of terrestrial producers, as soft scales insects (Fig. 4a).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 taxon names), distributed in four classes – Calcarea, Hexactinellida, Homoscleromorpha, and Demospongiae.54 Sponges have simple bodied anatomy, consisting of a diploblastic cellular organization with an intraepithelial mesenchyme called mesophyl, composed by collagen, amoeboid cells, and skeletal elements. Their body plan lacks true tissues and consists of a system of branched canals and choanocyte chambers, that produce a water flow for feeding, respiration, excretion, and reproduction. The majority of sponges are heterotrophic filter feeders, with some exceptions that are partially (mixotrophic) or completely (phototropic, chemotrophic) dependent on photosynthetic symbionts trophic exchange, and a few carnivorous species that feed on small invertebrates.18,52
000 taxon names), distributed in four classes – Calcarea, Hexactinellida, Homoscleromorpha, and Demospongiae.54 Sponges have simple bodied anatomy, consisting of a diploblastic cellular organization with an intraepithelial mesenchyme called mesophyl, composed by collagen, amoeboid cells, and skeletal elements. Their body plan lacks true tissues and consists of a system of branched canals and choanocyte chambers, that produce a water flow for feeding, respiration, excretion, and reproduction. The majority of sponges are heterotrophic filter feeders, with some exceptions that are partially (mixotrophic) or completely (phototropic, chemotrophic) dependent on photosynthetic symbionts trophic exchange, and a few carnivorous species that feed on small invertebrates.18,52
Sponges form intimate associations with a wide range of microorganisms, including bacteria, archaea, fungi, protists, and viruses, which are essential for their health and survival. The meta-organismal systems formed by sponge hosts and their microbiota should be considered as the minimal functional biological units.19,21 Microbial symbionts provide their hosts with nutrients, process waste products, and appear to be involved in nutrient recycling processes.22,23 Moreover, sponge microbiomes may enhance growth and competitive ability within benthic communities, through the exchange of certain bioactive molecules and precursors. In certain sponges, the accumulation of microbial-derived natural products has been shown to provide various chemical defence strategies, such as deterrence of predatory fish from feeding, anti-fouling to prevent overgrowth and suffocation, or growth inhibition against competing or pathogenic microbes.19 Much of the repertoire of secondary metabolites in eukaryotic organisms, particularly sponges, is thought to be derived from associated microorganisms.55,56 These compounds include mostly terpenes, sterols, cyclic peptides, unusual nucleosides, alkaloids, fatty acids, peroxides, and amino acid derivatives. Many of these products show promising therapeutic potential due to their anti-inflammatory, anticancer, antimicrobial, anti-atherosclerotic and antiherpetic properties (as seen in previous reviews of the series).10,15,57,58 However, apart from their biotechnological applicability, the presence of these molecules in the sponge host should primarily respond to ecological means in the first place, which in most cases have yet to be revealed. Although there is much evidence suggesting the symbiotic production of many sponge secondary metabolites, few studies have empirically demonstrated the microbial synthesis of these compounds, including the sesterterpenoid family.19,55,56 Recently, the discovery of type I terpene synthases in the sponge holobiomes may call into question the absolute production of secondary metabolites by microbiome associates, implying the involvement of the animal host in several terpenoid biosynthetic pathways.59 In the following lines, we will illustrate some examples of sesterterpenoids occurrence in Porifera (Table S2, ESI†). Tropical and temperate shallow-water Porifera provide most of the known bioactive molecules. The reason is probably due to the intense allelochemical interactions in the highly biodiverse tropical ecosystems (as seen in previous review of the series).10,15,17 The order Dictyoceratida represents the most prolific taxon of known secondary metabolites, while the scalarane tetracarbocyclic sesterterpenoids form by far the broadest structural group of the terpene family60 (and this review). Scalaranes are mainly found in sponges, but also in nudibranchs, largely due to trophic transfer (see below). In both organisms, they exert a deterrent effect against generalist consumers, serving as an efficient antipredator or multipurpose defence mechanism.17 These products further exhibit a wide array of pharmacological activities (anti-cancer, anti-inflammatory and antimicrobial being the most frequently described).61 Scalarin (TeC-156, Fig. 5) was the pioneering compound giving name to this large group of sesterterpeneoids. It was originally elucidated from the Mediterranean sponge Scalarispongia scalaris.62 Since the first discovery, TeC-156 has been recurrently found in Dictyoceratida sponges from diverse geographic locations (e.g., Scalarispongia sp. from Korea, Hyrtios erectus from South China, Ircinia sp. and Dysidea sp. from China and New Zealand, Spongia (Spongia) matamata from Palau, Spongia (Spongia) tubulifera from Mexico, Spongia (Spongia) virgultosa from Spain, Hyattella intestinalis from Mexico).63–67 Subsequently, many other scalarane-type compounds have been unveiled. The widespread species Hyrtios erectus seems to be the most productive in tetracarbocyclic sesterterpenoids, revealing scalaradial (TeC-75), heteronemin (TeC-326) and numerous derivatives, together with hyrtial (TeC-48) products, hyrtiosins (TeC-305–TeC-309) or salmahyrtisols (TeC-123, TeC-373, TeC-342, and TeC-504), in specimens from various regions, including China, Japan, Egypt, Saudi Arabia, the Maldives, New Guinea, New Caledonia, and Tonga.68–85 Congeneric Hyrtios species from Fiji, Thailand, Paracel Islands, and New Caledonia contained scalaranes and other molecules such as thorectolide (DH-76), erectusolide (DH-79), sesterstamide (TeC-345).86–89 At the same time, monocarbocyclic thorectidaeolides (MC-5–MC-8), acantholide A (MC-12) and luffariellolide (MC-70) were documented from H. communis from Palau.90 Similarly, Indopacific Phyllospongia foliascens collected in several areas (e.g., South China, Japan, Indonesia, New Guinea, Australia, and India) revealed an extensive array of scalarane-related products in addition to other metabolites.91–94 These included rare scalarane-derived pentacarbocyclic sesterterpenoids with an additional cyclobutene, like carteriofenones A–D (TeC-37–TeC-40) and E–K (TeC-247, PC-11, PC-20–PC-23, and TeC-258),95 and other sesterterpenes like foliaspongin (TeC-33) and derivatives as dehydrofoliaspongin (TeC-412), phyllofoliaspongin (TeC-413),96–98 phyllactones F–G (TeC-217–TeC-218) and phyllofolactones A–D and M (TeC-246, TeC-206–TeC-208).99–102 Phyllofolactones and scalaranes were additionally found in congeneric sponges Phyllospongia sp., P. lamellosa, P. vermicularis.103–107 Phyllactones were found in P. papyracea from specimens coming from Egypt, Madagascar, Indonesia, New Guinea and China.108–110 Indian Hyattella cribriformis and a Korean Hyattella sp. were found to produce scalaranes, together with H. intestinalis from Australia and Mexico, which reported in addition to the repertoire of TeC-156 and TeC-326 relative metabolites, also norscalarals hyatolides (TrC-171, TrC-172 and TeC-299–TeC-301), mooloolabenes (TeC-66–TeC-70 and TeC-248–TeC-257), furoscalarol (TeC-329), and hyatelactam (TeC-344).66,111–114 Sponges Hyattella sp. from Indonesia were found to possess hyattellactones (TeC-239 and TeC-240) and phyllofolactones (TeC-246, TeC-205–TeC-214, TeC-302, TeC-303, TeC-492).115Lendenfeldia sponges represent another taxon with an extended presence of scalaranes, homoscalaranes and related molecules, including furodendins, homoscalarates and homoscalaralactone from L. chondrodes from Palau, Australian L. dendyi and L. frondosa from the Salomon Islands and New Guinea, as well as sesterterpenes like lendenfeldaranes, felixins (TeC-100) or furanolipids, in Lendenfeldia sp. from Madagascar.116–123 The genus Spongia has afforded another notable repertoire of scalaranes, including the original TeC-156, TeC-75 and derivatives, as well as numerous other tetracyclic sesterterpenoids and different sesterterpene type molecules like scalalactams, furospongins, hyrtiosal (TrC-130), igernellin (MC-71), hipposulfates, ircinins, cometins, petrosaspongiolides, and plenty of others. These compounds were obtained from diverse species and locations such as Spongia (Spongia) agaricina, S. (S.) nintens and S. (S.) officinalis from Mediterranean, S. (S.) hispida and S. (S.) matamata from Papua New Guinea, S. (S.) oceanica from Hawaii, and from a number of unidentified Spongia sp. from the USA, Australia, Borneo, Japan, and Korea.63,64,124–140 Other genera of sponge typically containing scalarane-related products include Smenospongia specimens from Korea,141,142 or the only species in the Collospongia genus, C. auris from Australia, containing TeC-326 and terpene derivatives.143Strepsichordaia lendenfeldi from Australia revealed scalarane derivatives,144 whereas Indonesian Strepsichordaia aliena yielded honulactones, phyllofenone C (TeC-287) and phyllofolactones.145 Older investigations frequently described scalarane compound series in sponges whose terminology has changed after taxonomic revisions. TeC-75 and related scalardysins and scalarherbacins were described from Lamellodysidea herbacea from Gulf of Suez,146 and TeC-326, scalarolide (TeC-173), scalarafuran (TeC-331), furospinosulin-1 (MH-18), idiadione (MH-28) among others in Leiosella idia from USA.147TeC-75 and scalarane-type molliorins were isolated from Mediterranean Cacospongia mollior,47,148–155 and Pacific Cacospongia sp. unveiled cacolic acid (MH-53) and several cacolides, which are mainly linear sesterterpene compounds.156 Scalarane-type including TeC-326 and manoalide-type product (MC-27) were recovered from Thai Brachiaster sp. sponges.157 Tetracyclic scalarane-types and pentacyclic sesterterpenoids are commonly found in Dysidea genus. Scalarane compounds have been reported in Dysidea sp. from China,67 and in D. gumminae from Thailand, in addition to similan A, hyrtiolide, TeC-173 and scalafuran.158 Chinese D. granulosa produced various tetracyclic and pentacyclic sesterterpenoids.159 Bilosespenes were elucidated from Eritrean D. cinerea160 and dysidiolide (DC-22, Fig. 5) from D. etheria from USA,161 while halisulfates 1, 3, and 5 (TrC-115, DC-38, and DC-40), dysideapalaunic acid (DC-23) and coscinoquinol (TrC-112) were found in Dysidea sp. from Palau and Micronesia.162,163Psammocinia sp. sponges have been reported to produce variable suites of sesterterpenoids. For example, some specimens from Korea yielded scalarane-type products,164,165 or psammocinins A1, A2, and B (DH-30, DH-31, and DH-69) and variabilin (DH-16).166 Meanwhile, Australian relatives were found to contain bicarbocyclic sesterterpenoids such as ircinianin (DC-159), ircinianin sulfate (DC-179, Fig. 5), ircinianin sulfate lactam (DC-177) and derivatives, as well as isopalinurin (DH-4),167 and linear sesterterpenes ircinins 1–2 (TH-8 and TH-9) and sulawesins A–C (MC-9, MC-10, and TH-25) were also isolated from Psammocinia sp. in Indonesia.46 Hippospongide A (TeC-385), hippospongide B (TeC-172) and other scalaranes were isolated from Taiwanese Hippospongia sp.168 Other unidentified Hippospongia provided furanoterpene hippospongins (TH-26–TH-31) from Australian specimens,169 or TrC-115 and DC-40 from Micronesia collections.170 Linear and bicyclic hippolides A–J (DH-80, DH-81, MH-11–MH-16, DC-216, DC-217) and monocarbocyclic manoalides (MC-16 and MC-17) were isolated from H. lachne collected in South China.35,48,171
Disparate tetracarbocyclic sesterterpenoids toxistylides A–B were identified in Mediterranean Clathria (Clathria) toxistyla,172 and in C. (C.) gombawuiensis from Korea, including ansellone C (TrC-206, Fig. 5), gombaspiroketals A–C (DC-195–DC-197) and phorone B (TeC-389).173 Related phorones A (TeC-386) and C (TeC-481) were also identified in Korean Phorbas sp. Anvilones A (TeC-390) and B (TeC-391), phorbadione (TrC-199), ansellones C–K (TrC-207–TrC-211 and TrC-323–TrC-326), and monocyclic phorbaketals A–C and L–M (MC-119–MC-121 and MC-130–MC-132) were purified from congenerics from British Columbia.174–181 The antarctic collections of P. areolatus, in its place, revealed suberitenones (TeC-23–TeC-25, TeC-30) and suberitane derivatives, including isosuberitenone B (TeC-394), 19-epi-suberitenone (TeC-25), and isoxaspirosuberitenone (TeC-395).182 Furthermore, monocyclic spirocyclic MC-119–MC-129 and phorbin A (MC-4) were also obtained from Korean Monanchora sp. sponges.183 A number of norsesterterpene peroxides have been reported in genus Diacarnus. Tasnemoxides A–C (MC-40–MC-42), muqubilin (MC-36–MC-37) and derivatives were found in D. erythraeanus from the Red Sea.184,185 Aikupikoxide A (MH-30) and sigmosceptrellin B (DC-17) were also identified in the same species. Sigmosceptrellins A–C (DC-60, DC-17, and DC-63) and diacarnoxides A–D (MC-49–MC-52) were respectively identified in D. laevis and D. levii both from Papua New Guinea.186 Diacarperoxides and MC-36 were recovered in Indonesian D. megaspinorhabdosa.187 Additionally, muqubilin relative products were also found in D. spinipoculum from Solomon Islands.188 Other cyclic norsesterterpenes, known as mycaperoxides A–B (DC-15 and DC-16), were retrieved from Thai Mycale sp.189,190 Tricarbocyclic sesterterpenoids called coscinolactams A–G (TrC-200, TrC-154, TrC-201–TrC-205), including suvanine (TrC-143) derivatives, were isolated from Coscinoderma sp. from Micronesia,191–193 and from C. mathewsi coming from Solomon Islands, along with coscinalactone (TeC-402) and coscinafuran (TeC-403).194–196
The genus Ircinia probably comprises one of the most diversified in sesterterpene series, reporting a vast range of molecules including cyclic and linear furanosesterterpenoids, scalaranes, 24-homoscalaranes, tetronic acid related compounds, cheilanthane sesterterpenoids or C22-trinorsesterterpenoids, among others. Felixin scalaranes (TeC-100, TeC-101, TeC-165–TeC-169) were described from I. felix in Taiwan, ircinins (TH-8–TH-11) were detected in I. oros,36,197 and a numerous group of irciformonins (DH-10, DH-13, TH-19–TH-21) and ircinialactams (DH-96–DH-98, TH-24, TH-32) were isolated from Ircinia sp. from Taiwan and Australia.198,199 Moreover, strobilinins, felixinins and variabilins are frequently found in several congeneric species and regions: e.g., I. oros and I. variabilis from Mediterranean, I. campana from Colombia, I. strobilina from Brazil, and Ircinia sp. from several Pacific areas. DC-159 and wistarin (DC-160) were isolated from the Australian species I. wistarii.198,200–205 Chinese collections of Dactylospongia elegans provided γ-oxygenated butenolides, the dactylospenes A–E (MH-57, DC-224–DC-227).206Jaspis sponges from New Guinea (Jaspis cf. johnstoni), China (Jaspis sp.), and Japan (J. stellifera) yielded jaspic acid (TrC-110), jaspolide F (TrC-135), and jaspiferals C–F (TrC-131–TrC-134), respectively.207–209 Acantholides C–E (MH-10, MC-1 and MC-2) were recorded from Pacific Acanthodendrilla sp.,210 while agelisamines A and B (DC-29 and DC-30) and aplysinoplides A–C (MC-30, MC-33, and MC-34) were obtained from Agelas mauritiana; and Aplysinopsis elegans and Aplysinopsis sp. respectively all from Japanese collections.211–213 New Caledonian Petrosaspongia nigra were found to possess numerous petrosaspongiolides, A–L (TrC-212–TrC-221, Fig. 5 and TrC-222), M–P (TrC-178–TrC-180), Q and R (TrC-182 and TrC-183),214 and Petrosaspongia sp. from Fiji revealed several petrosaspongiolactams.215 Aurorals 1–4 (TrC-255–TrC-258) and globostelletins C–G (TrC-264–TrC-267) were isolated from Rhabdastrella globostellata from New Caledonia and South China, respectively.216 Rhopaloic acids A–C (MH-7, MH-1–MH-3) were found in Rhopaloeides sp. from Japan.217,218
Luffariella representatives produce specific bicyclic, monocyclic, and acyclic sesterterpenoids, including luffarins A–O (DC-43–DC-57), P (MC-65), Q (MH-8), R (DH-58), S (DH-106), T and U (DH-59) and DH-60) from Australian L. geometrica,219 luffariolides A–G (MC-77–MC-83, MC-70, luffalides A–F (MC-57–MC-62) or luffolide (TrC-280) from Luffariella sp. from several Pacific collections (e.g., Australia, Micronesia, Japan, Taiwan, Palau). Additionally, Luffariella sp. produces a variable suite of product series from L. variabilis according to the collection site. For example, specimens from Australia produce luffariellin A (MC-53), 25-acetoxyluffariellin (MC-55), 25-acetoxyluffariellin B (MC-69), and manoalide-type products (MC-13–MC-15, MC-28), while Palau sponges produce luffariellin B (MC-56), luffalactone (DC-162), several manoalides (MC-13–MC-16, MC-19, MC-21, MC-28) in Malaysian samples, or oshimalides A and B (MC-146 and MC-147) from deep sea populations.37,220–229Sarcotragus sp. sponges are similarly sources of norsesterterpenoids, linear sesterterpenes and other secondary metabolites. Collections from Korea have revealed a notable number of bioactive compounds, including a list of sarcotins A–C (DH-36–DH-38), D and E (TH-6 and TH-7), F (DH-74), G and H (TH-22–TH-22), I and J (MH-46 and MH-47), M (DH-40), O and P (MH-20 and MH-21), sarcotrines (DH-87–DH-91), sarcotragins A–C (MC-85, MH-42, and MH-54).43,230–234 Unidentified congenerics from New Zealand or Australia have yielded ircinialactams (DH-96, MH-32–MH-34, MH-55–MH-56) ircinialactone A (DH-62), DH-16 or MH-18.235,236 Antarctic Suberites sp. demonstrated to possess a series of suberitenones (TeC-23, TeC-24, TeC-27, and TeC-28, Fig. 5)237,238 and S. caminatus also from Southern Polar waters contained suberitenones derivatives (TeC-26 and TeC-30), together with caminatal (TrC-250).239 Pacific Thorectandra sp. from Palau unveiled the presence of thorectandrols (DC-8, DC-9, DC-11–DC-13), palauolol (DC-2) and DH-58.240 Thorectolide monoacetate (DH-77) was reported in Australian T. excavatus.241 Among the few linear sesterterpene products known in marine habitats, a group of the so-called balibalosides and derivatives (MH-4–MH-6, and MH-9) were described in the Mediterranean Oscarella balibaloi from France.242
Nudibranchs have a diverse diet, consuming various food items, including Chlorophyta, Ochrophyta, Rhodophyta (green, brown, and red algae, respectively), Porifera, Cnidaria, Bryozoa, Chordata (tunicates), and other Mollusca. However, their feeding behaviour is highly specialized, and almost each species predates on one or few prey.246 Such pattern is correlated with the ecological competence and defensive tactics of these animals, due to their ability to “steal” functional structures (cleptoplasty) or chemical products (cleptochemistry) from their prey. For example, they can incorporate chloroplasts or zooxanthellae from algae to obtain energy and camouflage,249,250 or nematocysts from cnidarian prey to use as protective weapons.251 Cleptochemistry is the process of sequestrating natural products derived from food items, and then the usage for self-defence. Some nudibranchs can biotransform the dietary metabolites into less toxic compounds to allow bioaccumulation, or to more effective noxious products to deter predators. While a few species can uptake simple precursors and de novo biosynthesize defensive molecules, most rely on sequestering compounds from their diet.27,252,253 For efficient energetic and protective purposes, the resulting bioactive compounds are often translocated to susceptible anatomical sites, glands, or exposed parts, or they may be exhausted within mucous secretions.27,245,254,255 Moreover, in synchrony with allelochemistry warning (aposematic) colorations frequently advise the presence of chemical defences or toxicity towards putative predators in exposed habitats. Consumers learn the connection between bright colorations and bad taste.27,245,256
It has been hypothesized that the reduction of the shell in the evolution of Opisthobranchia was facilitated by modifications in the foraging habits towards food items containing secondary metabolites, which would have been sequestered and used as chemical defences. This also suggests that the protection based on dietary allelochemicals would have preceded de novo synthesized defensive mechanisms.27,252 Dietary sesterterpenoids are common in dorids, particularly in family Chromodorididae. Most of these compounds are bioactive tetracyclic sesterterpenoid products, which are assimilated from sponge prey.27,257 The scalarane compounds of dietary acquisition, specifically TeC-75 (a potent anti-inflammatory metabolite) and other related bioactive molecules present in demosponge prey are of particular relevance.258 Once trophically incorporated, these metabolites are usually further bio transformed into derivatives by in chromodorid slugs as part of detoxification processes. They are frequently allocated towards the mantle border and dermal formation-like structures for the purpose of antipredation protection. Glossodoris sedna from Costa Rica besides reporting sponge scalaranes,259 it was previously described to possess sednolide (TeC-365).260 Similarly, luffariellin C (MC-63) and D (MC-64) were detected along scalarane derivatives (TeC-164, TeC-281, and TeC-432) in C. funerea from Palau.261TeC-326 is a recurrent scalarane-type sesterterpene trophically transferred from sponges (e.g. Heteronema erecta, several Spongia sp.) to nudibranch slugs [e.g. Glossodoris (syn. of Doriprismatica) atromarginata, atromarginata].262 This compound with antimycobacterial properties263 was allocated in the viscera of the Australian slugs G. hikuerensis and G. vespa, in contrast with TeC-75, 12-deacetyl-12-oxoscalaradial (TeC-46, Fig. 6), and 12-deacetoxy-12-oxo-deoxoscalarin (TeC-141), which were detected in the mantle.264 Ten norscalaranes mooloolabenes D–O (TeC-69, TeC-70, TeC-248–TeC-257), along with scalaranes (TeC-141 and TeC-433) were isolated from Australian D. atromarginata from diverse locations, indicating varied sponge diet and/or diversified enzymatic detoxification mechanisms in these slugs.265 Other dietary tetracyclic sesterterpenoids found in sea slugs include ansellone A (TrC-192, Fig. 6) from Cadlina luteomarginata from Canada and its prey Phorbas sp.,177 as well as MH-28 and luteone (TrC-232).147,266 Hamiltonin E (TrC-142) was isolated from South African Chromodoris hamiltoni.267 Inorolides A, B and C (TeC-368, TeC-369, and TrC-281) were found in Japanese Chromodoris inornata,268 along with TeC-156 or scalaradial derivatives (TeC-428–TeC-431). Variabilin derivatives (DH-22Fig. 6, DH-49, DH-107, and DH-109) were obtained from the South African Hypselodoris capensis and its demosponge prey Fasciospongia sp.269 Finally, a unique example of a non-dietary linear sesterterpenoid is granuloside (MH-52), isolated from the Antarctic cladobranch Charcotia granulosa.34,270
In the Mediterranean, the most significant bioconstructing zooxanthellate scleractinid coral is the madreporaria Cladocora caespitosa,273 which was found to contain bioactive sesterterpenes called cladocoranes A and B (DC-6, Fig. 6, and DC-7). These products revealed biologcal properties in the treatment of various diseases, including cancer, together with potential antitubercular and antibacterial activities inhibiting the growth of Gram-positive strains.276
4.2 Sesterterpenoids in microrganisms
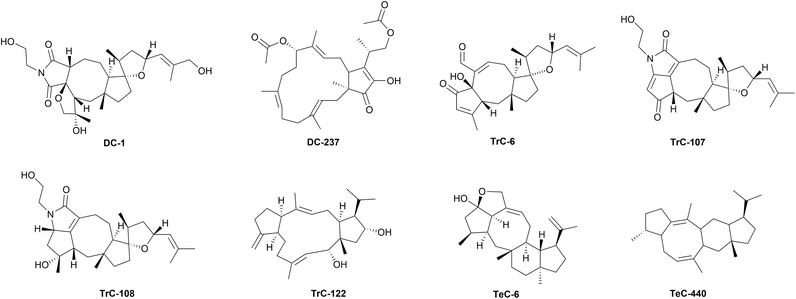
Fungi and bacteria represent significant sources of natural sesterterpenoids (Table S6, ESI,† and Fig. 4b). In the microbiological world, sesterterpenoids are metabolites endowed with different physiological effects. They drive interactions (i.e., symbiosis, competition, or proliferation) with neighbouring commensal or invader organisms, and some are endowed with anti-inflammatory and anticancer activities.297 This section will examine the significance of sesterterpenoids in the ecological setting of microbes.Bipolaris species are known to produce ophiobolins. 6-epi-Ophiobolin A (TrC-3), 3-anhydroophiobolin A (TrC-4),324 and 3-anhydro-6-epi-ophiobolin A (TrC-5),304 as well ophiobolin F (TrC-17),279,325 25-hydroxyophiobolin I (TrC-34)304 has been firstly isolated from B. maydis, TrC-6,326 ophiobolin B (TrC-8),327 ophiobolin I (TrC-32),312,328 6-epi-ophiobolin I (TrC-33),329 ophiobolin J (TrC-35),329 and 8-deoxyophiobolin J (TrC-37)329 have been isolated from B. oryzae. Bipolarolides, ophiobolin derived sesterterpenoids have been isolated from Bipolaris sp. (TJ403-B1).40 Bipolarolides A and B (TeC-1 and TeC-2) are characterized by a multicyclic caged oxapentacyclo[9.3.0.0 (ref. 1 and 6).0 (ref. 5 and 9).1 (ref. 8 and 12)]pentadecane-bridged system. Bipolarolides C and D (TeC-3 and TeC-4) show a 5/5/5/5-fused core skeleton, and bipolarolide C also contains a C-3–C-14 oxygen bridge to construct the caged architecture. Bipolarolides E–G (DC-1, TrC-107, and TrC-108, Fig. 7) are highly modified pentacyclic oxaspiro[4.4]nonane-containing sesterterpene-alkaloid hybrids.40 From the same strain, growing on fermented rice medium, others ophiobolin-type metabolites, bipolaricins A–I (TrC-87–TrC-95),330 and bipolarins A–H (TrC-79–TrC-86), tetracyclic ophiobolin-type sesterterpenes characterized by an oxaspiro[4.4]nonane moiety, together with ophiobotriol (TrC-78) have been characterized.331 Ophiobolin-type sesterterpenoids maydispenoid A (TrC-306) with a decahydro-3-oxacycloocta[cd]pentalene moiety, and maydispenoid B (and TrC-307), have been isolated from B. maydis collected from Anoectochilus roxburghii (Wall.) Lindl leaves.332
Ophiobolins are also commonly found in the genus Aspergillus.326,331,333,334 A large number of compounds have been first obtained from the crude extracts of the liquid and solid cultures of the mangrove fungus A. ustus, namely ophiobolins G and H (TrC-18 and TrC-28),335 ophiobolin K (TrC-38),336 ophiobolin O (TrC-49), ophiobolin P (TrC-52) ophiobolins U–Z (TrC-57, TrC-60–TrC-64),337,338 21-epi-ophiobolin O (TrC-51),337 21-dehydroophiobolin U (TrC-59),337 21-epi-ophiobolin Z (TrC-65),337 21-deoxyophiobolin K (TrC-42),337 6-epi-ophiobolin K (TrC-39),336 (6α)-18,19,21,21-O-tetrahydro-18,19-dihydroxyophiobolin G (TrC-24),338 (6α)-21-deoxyophiobolin G (TrC-26),338 (6α)-16,17-dihydro-21-deoxyophiobolin G (TrC-27),338 (5α,6α)-ophiobolin H (TrC-29),339 (5α,6α)-5-O-methylophiobolin H (TrC-30),339 5-O-methylophiobolin H (TrC-31),339 and (6α)-21,21-O-dihydroophiobolin G (TrC-25).339 The marine fungus A. flocculosus afforded 14,15-dehydro-6-epi-ophiobolin K (TrC-41), 14,15-dehydro-ophiobolin K (TrC-40),340 14,15-dehydro-6-epi-ophiobolin G (TrC-23),340 14,15-dehydroophiobolin G (TrC-22),340 and 14,15-dehydro-(Z)-14-ophiobolin G (TrC-21),340 and ophiobolin N (TrC-47).340 6-epi-Ophiobolin N (TrC-48) has been obtained from A. insuetus.341
Other ophiobolins and congeners have been isolated firstly from other fungi. Ophiobolin C (zizanin A) (TrC-13) has been obtained from Drechslera zizaniae (as Helminthosporium zizaniae),320 ophiobolin D (cephalonic acid) (TrC-15) from Cephalosporium caerulens,342–344 ophiobolin E (TrC-16) and 8-epi-ophiobolin J (TrC-36) from D. gigantea,318,319 and 6-epi-ophiobolin G (TrC-19) from Emericella variecolor obtained from a marine sediment.345,346Cochliobolus heterostrophus yielded ophiobolin L (TrC-43),315 ophiobolin M (TrC-45),347 6-epi-ophiobolin M (TrC-46),347 the degradation products 3-anhydro-6-epi-Δ10(14)-ophiobolin B and the dimer di-3-anhydro-6-epi-ophiobolin B (TrC-10 and TrC-11).316
Variculanol (TrC-122, Fig. 7), a sesterterpenoid having a 5/12/5 ring system has been isolated from A. variecolor.348 This compound is probably produced from geranylfarnesylpyrophosphate after a requisite folding and cyclizations followed by a 1,5-hydride shift to the carbocation.348 Spectanoids A–G (TrC-311–TrC-317), showing the same ring system, have been later isolated from A. spectabilis.349 This unusual 5/12/5 ring system is also present in nitiol (TrC-184), isolated from Gentianella nitida.350 Spectanoid H (TeC-461), with a 5/8/6/5 ring system, and a different biosynthetic pathway compared to other spectanoids, has been isolated from A. spectabilis.349 Terretonin (TrC-160), a meroterpenoid with a heavily oxidized 25-carbon skeleton, produced from polyketide and terpenoid precursors,351 was firstly isolated from A. terreus in 1979.352 Further congeners terretonins A–D (TrC-161–TrC-164),353 terretonin G (TrC-168),354 and terretonins E and F (TrC-165 and TrC-166)355 have been subsequently isolated from the same species, form another Aspergillus sp. strain OPMF00272, and from A. insuetus, respectively. Terretonins H and I (TrC-169 and TrC-170)356 and 1,2-dihydroterretonin F (TrC-167)338 have been isolated from A. ustus. Halorosellinic acid (TrC-96) and 17-dehydroxyhalorosellinic acid (TrC-333) have been isolated from the culture broth of the marine fungus Halorosellinia oceanica.357,358 Clavaphyllene (DC-230), a byciclic hydrocarbon sesterterpenoid structurally distinct from the ophiobolanes, has been isolated from A. clavatus.359 Another byciclic sesterterpenoid, terpestacin (DC-104), was firstly isolated from the marine fungal strain Arthrinium sp.38 Oxidative derivatives of terpestacin have been isolated later from the same fungus, 21-hydroxyterpestacin (DC-107), terpestacin B (DC-109).360 11-epi-Terpestacin (siccanol) (DC-105)361 and 11-epi-terpestacin glycoside (DC-106)362 have been isolated from D. siccans and B. sorokiniana, respectively. Bipolarenic acid (DC-210) has been obtained from a marine isolated of the fungus Lophiostoma bipolare (BCC25910).363 Variecolin (TeC-5), a tetracyclic sesterterpenoid with a 5/8/6/5 ring system, was firstly isolated from A. variecolor.364 This compound was later isolated from other fungi, Emericella purpurea,365E. aurantiobrunnea366,367 and Phoma sp.368TeC-5 congeners, variecolol (TeC-6, Fig. 7) and variecolactone (TeC-11), were isolated from the mycelium of E. purpurea,369 and emericolins A–D (TeC-7–TeC-10) have been isolated from E. aurantiobrunnea,366 as well as variecoacetals A and B (TeC-12 and TeC-13).367 The asperanes are hydroxylated 7/6/6/5 tetracyclic sesterterpenoids featuring with a hydroxylated skeleton isolated from Aspergillus fungi. The first asperane-type sesterterpenoid aspergilloxide (TeC-22) was discovered in 2002, from Aspergillus sp.370 Later, six other asperane, asperunguisin A–F (TeC-396–TeC-399 and TeC-353), were found in A. unguis, which inhabits the lichen Xanthoria sp.371 Aspergstressin (DC-253) hybrid polyketide sesterterpenoid, was discovered from A. sp. WU 243.372 Niduterpenoids A and B (HC-1 and HC-2), characterized by a highly congested hexacyclic 5/5/5/5/3/5 carbon skeleton, have been isolated from A. nidulans.42 The involvement of a cyclopropane in the ring system makes this skeleton uncommon. A hypothetical biosynthetic pathway from GFPP has been proposed, including a series of cyclization and Wagner–Meerwein hydride and alkyl shift reactions, to the formation of an intermediate with a hexacyclic 5/5/5/5/3/5 ring system, which after oxidation reactions forms HC-1 and HC-2.42 Several species of the diverged fungal classes Dothideomycetes (Bipolaris, Alternaria, Aplosporella, Pyrenophora) and Sordariomycetes (Arthrinium) are now known to possess highly conserved gene clusters that are mandatory for the sesterterpenoids biosynthesis.373 Co-evolution with host plants, however, has forced pathogenic fungi to acquire novel gene functions and pathways mining terpene scaffolds with new biological potential to overcome competitors or acquire novel physiological functions.374
The fungal terpene biosynthesis has been studied from the last decades of twentieth century. Sesterterpene synthases in fungi are all chimeric proteins consisting of a prenyltransferase (PT) domain at the C-terminus and a terpene cyclase (TC) domain at the N-terminus.297,375 Genome mining and heterologous expression of fungal bifunctional sesterterpene synthases have led to the discovery of new sesterpenoids.376–380 The first sesterterpene synthase, Aspergillus clavatus ophiobolin synthase (AcOS), responsible for the biosynthesis of TrC-17 was identified from the genome of A. clavatus in 2013. Two catalytically independent domains (prenyltransferase/terpene cyclase), homologous to those of diterpene synthase, fusicoccadiene synthase, were identified.381 A single transformant with the ACLA_76850 gene from A. clavatus produced TrC-17 and three minor sesterterpene hydrocarbons, namely DC-230, 3,20-anhydroophiobolin F (ophiobolane 2) (TrC-297), and ophiobola-1,7,18-triene (ophiobolane 1) (TrC-298).297,359 Heterologous expression of bifunctional sesterfisherol synthase gene (NfSS) and cytochrome P450 monooxygenase (NfP450) from Neosartorya fischeri in A. oryzae system afforded the nitidasane sesterterpenoid sesterfisherol (TeC-20), containing a characteristic 5/6/8/5 tetracyclic ring system. TeC-20 is next modified by cytochrome P450 monooxygenase (NfP450) to sesterfisheric acid (TeC-21).382 An unified biogenesis for sesterterpenes branching from bicyclic (5/15), tricyclic (5/12/5), and tetracyclic (5/6/8/5) cation intermediates, distinct from that of separate class of sesterterpenes including ophiobolins, has been proposed.382
The bifunctional sesterterpene synthase Stl-SS in the genome of E. variecolor has been identified as responsible for the biosynthesis of the tricyclic sesterterpenoid stellata-2,6,19-triene (TrC-299)378 with a 11/6/5 fused ring system. Investigation of the Stl-SS gene revealed a gene encoding a cytochrome P450 monooxygenase located next to the Stl-SS gene, that catalyzes three successive oxidation reactions on the C-20 methyl group to generate the carboxylic acid stellatic acid (TrC-121),383 previously identified in A. stellatus.378
The sesterterpene synthase EvQS, obtained from E. variecolor NBRC 32302, heterologously expressed in A. oryzae NSAR1, afforded quiannulatene (PC-1), further oxidized to quiannulatic acid (PC-2) by the cytochrome P450 Qnn-P450.375 Genome mining of bifunctional terpene synthase PbTS1 (BtcAPb) against two phytopathogens, Phoma betae and Colletotrichum orbiculare resulted in the production of betaestacins I–IV, Va–c, and VI (TrC-224–TrC-231).384 Functional expression of a terpene synthase (EvAS) from E. variecolor NBRC 32302 in A. oryzae led to the production of astellifadiene (TeC-16), showing a 6/8/6/5 ring system.377 Heterologous expression of four clade-A bifunctional terpene synthases (BFTSs), BmTS1, BmTS2, and BmTS3 from B. maydis ATCC48331 and PbTS1 from Phoma betae PS-13 giving di/sesterterpenes with unique polycyclic carbon skeletons such as sesterfisherol, enabled the isolation of the sesterterpene 5/12/5 tricyclic hydrocarbons Bm1 (TrC-308) and betaestacin I (Pb1, TrC-224), the 5/6/8/5-tetracyclic hydrocarbon Bm2 (TeC-505), and of the sesterterpene 5/15 bicyclic alcohol Bm3 (DC-235).385
Based on the initial carbocation formation strategy, the cyclization mechanisms of terpene synthase have been classified into two types, namely A and B. Type A cyclization (C1-IV-V) is initiated between the C1-C15/C14-C18 of geranylfarnesyl diphosphate (GFPP) to yield a 5/15 ring system. Type B cyclization (C1-III-IV) is initiated between the C1-C11/C10-C14 of GFPP/geranylgeranyl diphosphate (GGPP) to yield a 5/11 ring system.386 Cyclization-based classification reflects the phylogenetic relationships among bifunctional terpene synthases. The known enzymes catalysing types A and B cyclization have been classified into two clades, namely clade I, catalysing type A (C1-IV-V) cyclization, and containing terpene synthase genes from five lineages, and clade II, catalysing type B (C1-III-IV) cyclization and containing terpene synthase genes from seven lineages of fungi.386 NfSS is a clade A enzyme, while PaFS and AcOS belong to clade B.382 Anyway, more than 90% of the chimeric terpene synthase genes is still functionally unknown.386 Systematic search of sequenced fungal genomes among diverse taxa revealed that chimeric terpene synthase genes were restricted to Dikarya subkingdom.386 Phylogenetic analysis led to the discovery of a sub-clade involved in the biosynthesis of a 5–15 trans-fused bicyclic sesterterpene, namely preterpestacin I (DC-245). A bifunctional terpene synthase, preterpestacin I synthase (BmTS3), from B. maydis, that catalyses a chain elongation and a cyclization to afford preterpestacin I, was identified. Oxidative modifications from DC-245 to DC-104 are catalysed by enzymes encoded by genes adjacent to BmTS3 (renamed as tpcA), two cytochrome P450 genes (tpcB and tpcC) and a single flavin-dependent oxidase gene (tpcD). The total biosynthesis of DC-104 was then obtained by artificial reconstitution of the biosynthetic machinery in A. oryzae. Heterologous expression in A. oryzae was applied to characterize the function of the putative modification enzyme genes tpcBCD, and this led to the isolation of two biosynthetic intermediates, preterpestacin II and preterpestacin III, and the natural product DC-104.387
Recently, a genomic organization analysis revealed a unique glycosyltransferase gene cluster in the graminaceous pathogen B. sorokiniana. This has resulted in the identification of two new metabolites, sestersorokinicin A (DC-237, Fig. 7) and sestersorokiniside A (DC-238), featuring glucosyl moieties that can enhance the pathogenic effects of bacterial lipopolysaccharide.373
In 2022, Yan and colleagues elegantly demonstrated that the oxidase OblCAu of A. ustus catalyzes dehydrogenation at the C16 and C17 sites of TrC-17 and TrC-13 which are intermediates of TrC-38. Subsequently, TrC-38 is transported and stored in a space between the cell wall and membrane. Feedback mechanisms regulate the production of TrC-38 and its precursors, as their excessive accumulation is closely related to cell toxicity.388
Genome analysis of Penicillium brasilianum NBRC 6234 and Penicillium verruculosum TPU1311 revealed the presence of two bifunctional StTPS genes with prenyl transferase (PT) and terpene synthase (TPS) domains, P. brasilianum sesterbrasiliatriene synthase (PbSS) and P. verruculosum preasperterpenoid A synthase (PvPS), that were heterologously expressed in A. oryzae NSAR1 affording sesterbrasiliatriene (Trc-293) and preasperterpenoid A (PC-79).379 Phylogenetic analysis of the TC domain of protein JNUA3651 from Talaromyces wortmannii ATCC 26942 with those derived from known sesterterpene synthases revealed that its closest neighbor was PvPS, suggesting that JNUA3651 is likely to play the same role as PvPS to synthesize PC-79. Stepwise reconstitution of this gene cluster in A. oryzae NSAR1 revealed that the terpene synthase AstC encodes a sesterterpene cyclase to synthesize PC-79. The P450 enzyme AstB oxidizes PC-79 to give PC-31 along with the minor product asperterpenoid B (PC-64). Subsequently another P450 enzyme AstA oxidizes PC-31 to asperterpenoid C (PC-65).389 Probable pathways catalyzed by AstB were then proposed, the oxidation order of C-19 and C-21 was revealed by quantum chemistry calculations and HPLC-MS analysis, and the intermediates, the three new asperterpenoids D–F (PC-66–PC-68) were obtained.390 Finally, other ten new asperterpenoids, namely asperterpenoids G–P (PC-69–PC-78), featuring a 5/7/3/6/5 skeleton, were obtained from two A. oryzae transformants with heterologous expression of a terpene cyclase gene AstC with one or two P450 genes AstB and AstA, by using a molecular networking approach.391 Heterologous expression of chimeric enzymes PTTS010 (ZbSS), isolated from Zymoseptoria brevis, and Colletotrichum orbiculare sesterorbiculene synthase (CoSS) in Saccharomyces cerevisiae afforded sesterorbiculene (DC-249) and the 5/8/6/5 tetracyclic sesterevisene (TeC-440, Fig. 7).386
In fungi, the in vitro production of ophiobolins changes with the conditions of the culture. For example, B. maydis produces TrC-2, 3-anhydroophiobolin A (TrC-4), ophiobolin B (TrC-8), and ophiobolin L (TrC-43) in liquid broth, whereas it generates ophiobolin M (TrC-45), 6-epi-ophiobolin M (TrC-46), TrC-13, 6-epi-ophiobolin C (TrC-14), TrC-38, and 6-epi-ophiobolin K (TrC-39), when grown in agar media. Ophiobola-7,19-dien-25-oic acid (14,18(R)-epoxy-3,5-dihydroxy-γ-lactone) (TrC-66) is produced by Cochliobolus miyabeanus under modified fermentation conditions.392 More importantly, adding specific substrates, such as methionine, to the culture media of Bipolaris spp. increases the production of TrC-2 precursor.315,347 Generally, sesterterpenoid production increases with the culture time of fermentation in liquid broth. Production of sclareol peaks in old-stage cultures of Fusarium spp., Rhizopus spp., and Aspergillus spp. cultured for more than six days in standard media, indicating that fungal conidia are mainly involved in sesterterpenoids biosynthesis.393,394
Given the broad spectrum of biological activities of ophiobolins, against nematodes, fungi, and bacteria, studies have been done to predict the metabolic pathway of these compounds. Transformations of TrC-3 with Polyangium cellulosum produced TrC-32 and ophiobolin A lactone (TrC-7), while Pseudomonas aeruginosa produced ophiobolin B lactone (TrC-12). Resting-cell preparations of Penicillium patulum afforded TrC-3, and 6-epi-ophiobolin L (TrC-44).315
The in vitro standardized production of sesterterpenoids yielded the possibility of better understanding their biological role in the ecology of fungi. Most of the research has been focused on the phytotoxic properties of ophiobolins produced by Bipolaris spp. and Alternaria spp. as they have been implicated in significant plant disease epidemics (the Bengal rice famine in India, 1943 and the spotted leaf blight).304,395 When applied to plants, ophiobolins cause detrimental effects, such as growth inhibition of roots and coleoptiles, reduced seed germination, and decreased photosynthesis. The effects of ophiobolins depend on proton extrusion alterations and membrane permeability changes. Indeed, TrC-2 alters the permeability of the plasma membrane to potassium and impairs transport processes resulting in the leakage of electrolytes and glucose from roots and the impaired synthesis of the primary cell wall in plants.396 The phytotoxic effects of ophiobolins require concentrations of approximately 100 μM, which are typically only reached during epidemics. However, during endophytic relationships, fungi produce ophiobolins at concentrations that allow them to obtain ions and sugars from the host without causing significant plant damage.397
In Streptomyces spp., cosmopolitan soil bacteria providing valuable secondary metabolites, especially antibiotics,399 a synthase with sesqui-, di-, and sesterterpene synthase activity, coupled with the cyclase StsC, catalyses the formation of a new dicarbocyclic sesterterpenoid, somaliensene A (DC-239, Fig. 8), and of one monocarbocyclic (−)-somaliensene B (MC-149, Fig. 8) from geranylfarnesyl pyrophosphate.400
Yang and colleagues reported that StsC is a membrane-bound sesterterpene cyclase belonging to the UbiA superfamily of proteins in bacteria. DC-239 and MC-149 were obtained by expressing the corresponding gene in an engineered Escherichia coli strain. Among the 990 homologues of the UbiA family proteins reported in nature, 28 homologues have been identified in the Streptomyces genus However, the sts operon in Streptomyces flanks specific repressors that make StsC and the related products unstable, at least in vitro.401 Sequence analysis revealed that the Bacillus clausii genome contains a promiscuous terpene synthase homologue (Bcl-TS) (a sesterterpene/triterpene synthase), which potentially catalyses the conversion of linear C35 isoprenoid into monocyclic isoprenoid. However, the cyclization step does not result in the sesterterpenoids production, as revealed by GC-MS analysis of the culture media.402 Nevertheless, the authors were able to purify functional Bcl-TS protein that successfully converted GFPP into the linear sesterterpenoid β-geranylfarnesene, by introducing the Bcl-TS gene of B. clausii in stable transfected Escherichia coli.402 Recent reports suggest that culture-related factors, such as the media's oxygenation or pH, dramatically change cyclization and the profile production of sesterterpenoids in bacteria.403 Therefore, enzyme activities select the substrates and restrict the range of products, making the in vitro bacterial cultures unsuitable tools for the research of sesterterpenoids. In nature, bacteria grow in structured communities, forming microcolonies that bound to surfaces to form biofilms. Biofilm formation is a dynamic process initiated by the early colonizers, which determine the three-dimensional expansion of the bacterial communities.404 The colonizers elaborate the outer polymer matrix to capture nutrients and new microbes, leading to a complex, multi-species biofilm that represents a protected environment allowing cells to survive in hostile situations. In the biofilm, bacteria communicate through small diffusible molecules to regulate gene expression, control the production of secondary metabolites, organize the community's structure and relationships, and colonize new surfaces by dispersing bacteria.405Streptomyces albus forms a compact biofilm in which cells are embedded in an extracellular protein matrix composed of a network of fimbriae.406 The biofilm allows S. albus to maintain its metabolic functions, including the catalysis of regio- and stereo-selective hydroxylation.407 The co-cultivation of S. albus with Bacillus amyloliquefaciens leads to an increase in the formation of fimbriae and biofilm stability. This suggests that secondary metabolites extend the half-life of bacteria in the biofilm.406,408
Improved yeast-based promoter engineering platform (mCRISTAR),409 has been used to activate the previously uncharacterized Class II cyclase-containing gene cluster, called atolypene (ato) gene cluster, cloned from the genome of the cultured actinomycete Amycolatopsis tolypomycina. Heterologous expression of ato gene cluster into S. albus led to the characterization of atolypenes A and B (TrC-144 and TrC-145, Fig. 8).410
Two clade III promiscuous terpene synthases (Fusarium graminearum mangicdiene synthase, FgMS and F. graminearum GJ1012 synthase, FgGS) from an endophytic fungus, F. graminearum, showed to produce variable terpenoids in vivo by converting precursor polyisoprenoid diphosphates of different lengths (C10, C15, C20, C25). Six Escherichia coli variants, obtained by combining two terpene synthases and three PTs, afforded 50 different terpenoids, including the 11–6 bicyclic variecoltetraene (DC-234), the 5/5/6/5 tetracyclic mangicdiene (TeC-482), and (2E)-α-cericerene (MC-114). Further exploitation of mutants F65L and F159G afforded the 5/8/6/6 tetracyclic sesterterpene Tec-509, Fig. 8.411 Recently, Gu et al. reported the discovery of the sesterterpene synthases Streptomyces violens sesterviolene synthase (SvSS) from S. violens, that converted GFPP into a sesterterpene hydrocarbon, sesterviolene A (TeC-464), and a few trace compounds. Enzyme engineering through site-directed mutagenesis gave access to a high-yielding enzyme variant that provided six additional minor products sesterviolenes B–G (TrC-331, TeC-466, TeC-467, DC-250–DC-252) and the main product TeC-464.412 The guanidine-containing scalarane scytoscalarol (TeC-366) has been isolated from the cyanobacterium Scytonema sp. (Scytonemataceae).413 Other guanidine-bearing compounds are cybastacines A and B (TeC-359 and TeC-360, Fig. 8), isolated from another cyanobacterium, Nostoc sp. (Nostocaceae).414 Only compounds with simpler structures, such as the acyclic sesterterpenoids (AI, Table S1, ESI†), are known in benthic diatoms. C25 highly branched isoprenoid alkenes (haslenes) are ubiquitous in marine sediments.415 These polyunsaturated sesterterpene oils have been isolated from several species of Haslea (Naviculaceae), i.e. H. ostrearia (AI-14–AI-21, AI-23 and AI-24), H. crucigera, H. pseudostrearia (AI-18) and H. saltstonica (AI-16).416,417 Similar compounds (AI-12 and AI-13 rhizenes) have been isolated from the marine diatom Rhizosolenia setigera (Rhizosoleniaceae).418 A promising source of sesterterpenoid synthesis could be that of lichenised Ascomycota species. As these compounds have been shown to mediate trophic or defensive interactions in many fungi, it can be expected that a wide variety of sesterterpenoids will be present in lichen species that have to cope with biotic relationships between mycobionts, algal or cyanobacterial phototrophs and a major component of the microbiota. To date, however, knowledge in this regard is relatively scarce. From Gypsoplaca macrophylla (Gypsoplacaceae), gypmacrophin A (PC-34, Fig. 8), showing a skeleton similar to asperterpenoids isolated from Aspergillus and other fungal species, has been isolated.379,391 Retigeranic acid A (PC-7) has been purified from Lobaria isidiosa var. subisidiosa, L. retigera, and L. subretigera (Peltigeraceae), while the epimer retigeranic acid B (PC-56) has been described only from Lobaria isidiosa var. subisidiosa.419–421 Retigeran-11-ol (PC-32) and 4-hydroxyretigeran-11-ol (PC-33) have been isolated from Leprocaulon microscopicum (Leprocaulaceae).422
4.3 Sesterterpenoids in plants
With over 350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 known species, plants (Table S7, ESI†), are a megadiverse kingdom of organisms.449 They were among the first organisms to colonise terrestrial environments, and have acted as habitat builders, significantly modifying the chemical composition of the Earth's atmosphere and the chemical and physical properties of the soil. Plant species have developed a complex array of primary and secondary metabolic pathways in response to environmental pressures. These pathways facilitate to the synthesis of metabolites that are biologically active within the plant and potentially useful for regulating the physiology of other organisms. Plants have been a significant source of food and medicine since ancient times, owing to their properties. In a constant balance between an adaptive response to the pressures of the environment and a modifying effect on it, plant species have differentiated in their genome a very rich complexity of primary and secondary metabolic pathways capable of leading to the synthesis of different compounds. These compounds can be functionally active both for the plant healthiness and potentially for regulation of the physiology of other organisms. Regarding sesterterpenoids, vascular plants (Tracheophyta) are one of the richest phyla with 219 known compounds, following Porifera and Ascomycota (Fig. 1). The diversity of compound classes (6 out of the 10 considered in this review) is remarkable, and the most represented class being dicarbocycles (DCs) with 114 known compounds (Fig. 1). TrCs (Table S1, ESI,† and Fig. 4d) are the class of sesterterpenoids most shared between different plant families, even those that are phylogenetically distant from each other. In some families, this is the predominant or even exclusive class of sesterterpenoids, such as Pteridaceae.450,451 Cheilanthane sesterterpenoids have been isolated from Cheilanthes farinosa, cheilarinosin (TrC-102, Fig. 9)452 and cheilanthatriol (TrC-138),453Aleuritopteris khunii (TrC-236 and TrC-237),450A. agetae (TrC-300–TrC-302).451 17-Oxo-18,19-bisnorcheilanth-13(24)-en-6α-ol, a 19,20-bisnorcheilanthane (C23 sesterpenoid) (TrC-303) and 13,17-dioxo-18,19,24-trisnorcheilanth-6α-ol, a 19,20,21-trisnorcheilanthane (C22 sesterpenoid) (TrC-304), have been also isolated from A. agetae.451 Ancepsone A (TrC-320), isolated from A. anceps,454 and (17Z)-13,19-epoxycheilanth-17-en-6α-ol (Tec-506, Fig. 9), from A. mexicana,455 showed a 13,19-epoxycheilanthane skeleton. 18-epi-Scalar-16-ene-6α,19-diol (Tec-507) and 16α,19-epi-dioxy-18-episcalar-17(25)-en-6α-ol (Tec-508) have been also isolated from A. mexicana. Other cheilantanes, cristasesterterpenoic acid and cristasesterterpinol glucoside (TrC-276 and TrC-277), have been obtained from Caesalpinia crista (Fabaceae).456 Involudispirones A and B (TrC-278 and TrC-279, Fig. 9), containing a 1,2-dioxadispiro[5.2.5.2]hexadecane ring system have been isolated from Stahlianthus involucratus (Zingiberaceae).457 It has been speculated that this spiro system could be formed by an enzyme-catalyzed Diels–Alder addition between the derivative of cadalenequinone, a major constituent in S. involucratus, and myrcene.457,458 Similarly, it has been hypothesized that heliocide H2 (TrC-157), isolated together with heliocides B1 (TrC-155) and H1, H3 and H4 (TrC-156, TrC-158, and TrC-159) from Gossypium hirsutum (Malvaceae),459–462 could be biosynthesized by a Diels–Alder addition between the sesquiterpenoid gossypolone and myrcene.461 A Diels–Alder reaction has been used to synthesize TrC-155 and TrC-159 from hemigossypolone and trans-β-ocimene.462 Linderasesterterpenoids A and B (TrC-319 and TrC-319) with an unusual 7-cyclohexyldecahydroazulene carbon skeleton have been isolated from the roots of Lindera glauca (Lauraceae).463 2′-Isopicrasin A (TrC-288)464 and picrasin A (TrC-289),465 simaroubolides having the C25 simarolidane skeleton which consists of three carbocycles and two lactone rings, and is closely related to simarolide, have been isolated from Picrasma quassioides (Simaroubaceae).465 Vulgarosides 1–4 (TrC-245–TrC-248), sesterterpene esters whose base aglycon skeleton is quite similar to TrC-280 (except for the stereochemistry at C-13 and C-14), have been obtained from Cydonia vulgaris (Rosaceae). The distribution of TrCs in vascular plants suggests a more ancestral inheritance of the genes involved in the synthesis of these compounds. While a detailed discussion of the phylogeny of sesterterpenoids in plants is not a primary objective of this review, it is worth noting that there are several gaps in our knowledge regarding this topic. Based on current knowledge, Lamiaceae is the largest source of compounds among the 22 vascular plant families for which sesterterpenoids have been described, with DC and MC being the most important. Coleifolides A and B (MH-50 and MH-51), characterized by a β-methyl-α,β-unsaturated-γ-lactone and structurally similar to manoalide derivatives, have been isolated from Scutellaria coleifolia.466 Leucosceptrane sesterterpenoids (leucosceptroids), colquhounoids and eurysoloids have been isolated from the Lamiaceae Leucosceptrum canum, Colquhounia coccinea var. mollis and Eurysolen gracilis. From the trichome exudates and the leaves of L. canum bicarbocyclic sesterterpenoids have been isolated, i.e. leucosceptroids A and B, E–Q (DC-168 and DC-169),467 leucosceptroids E–N (DC-185–DC-192, DC-138, and DC-229) showing an α,β-unsaturated γ-lactone moiety,468 leucosceptroids P and Q (DC-198–DC-199),469 leucosesterlactone (DC-164),470 17α-hydroxyleucosceptrine (DC-166),471 leucosceptrine (DC-211).472 Leucosceptroid O (DC-193), possessing a spiro α,β-unsaturated γ-lactone moiety, has been isolated from the flowers of the same species.473 Additionally, tricarbocyclic compounds with antipodal cyclopentenones leucosceptroids C–D (TrC-190 and TrC-191),474 leucosesterterpenone
(TrC-188, Fig. 9),470 and 14β-methylleucosesterterpenone (TrC-189),471 have been found. Leucosceptroid degradation products, i.e. the C20 norleucosceptroids A–C (DC-172–DC-174), and the C21 norleucosceptroids D–H (DC-200–DC-204).469,475 have been isolated from L. canum of Nepalese and Chinese origin, respectively. 1α-Hydroxyleucosceptrine (DC-212) and 8α-hydroxyleucosceptrine (DC-213) were produced microbial transformation of DC-211 by Rhizopus stolonifer.394 Colquhounoids A–D (DC-182–DC-184 and DC-246), and 14-epi-colquhounoid D (DC-247) have been isolated from Colquhounia coccinea var. mollis.476,477 Eurysoloids A and B (TeC-441 and TeC-442), characterized by a pentacyclic 5/6/5/10/5 scaffold with an unusual macrocyclic ether system have been isolated by Eurysolen gracilis.478 The genus Salvia has received much attention within Lamiaceae. Sesterterpenoids have been found in both cultivated plants e.g. S. tingitana479 and wild species, such as in the case of S. dominica from subarid regions of Jordan.480,481 Prenyllabdane-type sesterterpenoids25 have been isolated from several Salvia species, as S. syriaca (DC-59, Fig. 9),482S. hypoleuca (DC-64, DC-67, DC-68, and DC-70),483,484S. palaestina (DC-73–DC-75),485S. dominica (DC-81–DC-91, DC-94–DC-96, DC-101 and DC-102),481S. tingitana (DC-120, DC-127, DC-129–DC-132, DC-146, DC-147),479S. yosgadensis (DC-134), and S. mirzayanii (DC-121).486 Prenyllabdane sesterterpenoids with a lactone, ester or acetal functionality between C6 and C23 have been found in various species, as S. hypoleuca (DC-65, DC-66, DC-69, and TrC-223),483,484S. tingitana (DC-127, DC-143–DC-145),479S. sahendica (DC-71 and DC-72),487S. dominica (DC-76–DC-80, DC-92–DC93, DC-101 and DC-102),481S. lachnocalyx (DC-97–DC-98).488 Other metabolites with a tetrahydropyran ring similar to manoyloxide-type diterpenoids between C-8 and C-13 were obtained from S. mirzayanii (DC-122–DC-126)489 as well as other hydroxymanoyloxide derivatives (DC-140–DC-142) from S. limbata,490 (14E)-methylmanoyloxide-14,16,18-trien-19,16-oxide-23-carboxylate (DC-139) from S. tingitana,479 lachnocalyxolides C and D (DC-99–DC-100) from S. lachnocalyx,488 yosgadensolides A and B (DC-134 and DC-135) along with their epimers from S. yosgadensis,491 salviaethiopisolide derivatives (DC-219–DC-220)492 and others hydroxymanoyloxides (DC-221–DC-223)493 from S. aethiopis. A norprenyllabdane-type sesterterpenoid, (13E)-4α,6α,8α-trihydroxylabd-13(14),17(18)-dien-16,19-olide (DC-132), was isolated from S. tingitana.479 Compounds arising from degradation of C-19 and C-20, 19,20-dinorsesterterpenoids were yosgadensenol (DC-133) and 13-epi-yosgadensenol (DC-42) from S. yosgadensis,494 and 6-dehydroxyyosgadensenol (DC-137) and 6-dehydroxy-13-epi-yosgadensenol (DC-136) from S. limbata.490 The sesterpene γ-lactone genepolide (DC-167, Fig. 9), isolated from Artemisia umbelliformis (Asteraceae) has been described as a formal Diels–Alder adduct of the exomethylene-γ-lactone germacranolide costunolide and the diene myrcene.495 Another intermolecular Diels–Alder reaction between thujanone, a thujane-type monoterpene one of main constituents in the essential oil of A. argyi, and a guaianolide, has been hypothesized for the biosynthesis of isoartemisolide (PC-26), isolated from the same species.496 Raoulic acid (DC-218), isolated from Raoulia australis, probably biosynthesized by diprenylation of a sesquiterpenoid precursor of the germacrene type, common in other Asteraceae species.497 Dibritannilactone A (TeC-45), isolated from Inula britannica, may derive from a monoterpene and a sesquiterpenoid.25,61 The trinorsesterterpene glycoside 3-[6-(4,8-dimethyl-nona-1,3,7-trienyl)-4-hydroxy2,6-dimethyl-cyclohex-1-enyl]-3-hydroxypropionic acid 1 glucoside (MC-99) has been obtained from the fern Woodwardia virginica (Aspleniaceae).498 The scalarane corallocarpscalarolide (TeC-410) has been isolated from the roots of Corallocarpus epigaeus (Cucurbitaceae).499 Biyoulactones D and E (DC-170 and DC-171), tricyclic meroterpenoids possessing an octahydroindene ring, a γ-butyrolactone ring, and a β-diketone moiety, have been isolated from Hypericum chinense (Hypericaceae).500 From the fruit of Phellodendron chinense var. glabriusculum (Rutaceae), phellogine (TeC-32), whose structure is similar to tirucalla-7-ene derivatives in the same species, has been obtained.501 Goniocarpic acid (DC-214) has been described within the constituents of the leaves of Serjania goniocarpa (Sapindaceae). Nevertheless, the classification of DC-214 as a sesterterpenoid has been questioned as it cannot be derived from GFDP.25 Among Monocotiledonous species, Aletris farinosa (Nartheciaceae), afforded two sclarane (TeC-226 and TeC-227) and four cheilantane sesterterpenoids (TrC-274, TrC-275, TrC-290, and TrC-291) have been studied.502,503 The scalarane sesterterpenoid perisomalien A (TeC-34)504 and the monocyclic n-non-2′-en-1′-yl-13(15,19,19-trimethyl-cyclohex-14,16-dienyl)-2,6,10-trimethyl-tetradec-6-ol-13-on-1-oate (hemidesmu sesterterpenoid ester) (MC-98), have been isolated from Periploca somaliensis and Hemidesmus indicus, respectively.505 The potential of sesterterpenoids in plants is still largely untapped, as evidenced by the case of Gentianaceae.25 Nitidasin (TeC-352)506 was the first gentianellane-type25 sesterterpenoid isolated from the Andean species Gentianella nitida. From the same species, nitiol was isolated (TrC-184).350 It has been speculated that the seco-gentianellane alborosin (TrC-109) from G. alborosea, could be derived from the cyclization of GFPP through TeC-352 as a key intermediate, and after the final oxidative cleavage of the C11/C12 bond.507 Later, other gentianelloids have been isolated from the Chinese G. turkestanorum, namely the C-6 epimeric sesterterpenoids gentianelloids A and B (TrC-286 and TrC-287), two compounds containing an unusual 10,11-seco-gentianellane skeleton,508 and 18-epi-nitidasin (PC-80), gentianelloids C–F (TeC-447, TeC-449, TeC-451, TeC-453), 18-epi-gentianelloids C–F (TeC-448, TeC-450, TeC-452, TeC-454), and 18-epi-alborosin (TeC-455).509 Another species belonging to Gentianaceae, Swertia bimaculata, afforded aspterpenacid C (PC-40, Fig. 9)510 with the 5/3/7/6/5 pentacyclic skeleton similar to aspterpenacids A and B (PC-38 and PC-39) mangrove endophytic fungus Aspergillus terreus, isolated from Kandelia obovata (Rhizophoraceae).435 Their biosynthetic pathway could be derived from GFPP with a series of cyclizations, rearrangements, redox and acetylation reactions.435 Bolivianine (PC-53) and isobolivianine (PC-53), two sesterterpenoids with an unusual skeleton, have been isolated from the bark of Hedyosmum angustifolium.511 The authors hypothesized their biosynthesis starting from onoseriolide, a sesquiterpene lactone already isolated from other Chloranthaceae. Another species, H. brasiliense, afforded hedyosulide (TrC-20), probably biogenetically related to the α-exo-methylene-γ-lactone precursor hedyosumin A and myrcene.512 In the cormophytes, acyclic sesterterpenoids (AI, Table S1, ESI†) have been obtained from Poaceae, Euphorbiaceae, and Solanaceae. 2,6,10,14-Tetramethyl-18-butanecarboxymethylene-henecos-12-en-17β-ol (AI-11) has been isolated from Oryza sativa,33 and (2Z,6Z,10E,14E)-geranylfarnesol (AI-8) from Triticum aestivum.513 Other linear sesterterpenoids, 2E-3,7,11,15,19-pentamethyleicos-2-en-1-ol (AI-22) (2Z,6E,10E,14E)-geranylfarnesol (AI-9), have been isolated from the aerial parts of Croton hieronymi (Euphorbiaceae).514 3,7,11,15,19-Pentamethyl-2-cis-6-trans-eicosadien-1-ol (AI-4) has been obtained from the unsaponifiable matter of the leaves of Solanum tuberosum (Solanaceae).515
000 known species, plants (Table S7, ESI†), are a megadiverse kingdom of organisms.449 They were among the first organisms to colonise terrestrial environments, and have acted as habitat builders, significantly modifying the chemical composition of the Earth's atmosphere and the chemical and physical properties of the soil. Plant species have developed a complex array of primary and secondary metabolic pathways in response to environmental pressures. These pathways facilitate to the synthesis of metabolites that are biologically active within the plant and potentially useful for regulating the physiology of other organisms. Plants have been a significant source of food and medicine since ancient times, owing to their properties. In a constant balance between an adaptive response to the pressures of the environment and a modifying effect on it, plant species have differentiated in their genome a very rich complexity of primary and secondary metabolic pathways capable of leading to the synthesis of different compounds. These compounds can be functionally active both for the plant healthiness and potentially for regulation of the physiology of other organisms. Regarding sesterterpenoids, vascular plants (Tracheophyta) are one of the richest phyla with 219 known compounds, following Porifera and Ascomycota (Fig. 1). The diversity of compound classes (6 out of the 10 considered in this review) is remarkable, and the most represented class being dicarbocycles (DCs) with 114 known compounds (Fig. 1). TrCs (Table S1, ESI,† and Fig. 4d) are the class of sesterterpenoids most shared between different plant families, even those that are phylogenetically distant from each other. In some families, this is the predominant or even exclusive class of sesterterpenoids, such as Pteridaceae.450,451 Cheilanthane sesterterpenoids have been isolated from Cheilanthes farinosa, cheilarinosin (TrC-102, Fig. 9)452 and cheilanthatriol (TrC-138),453Aleuritopteris khunii (TrC-236 and TrC-237),450A. agetae (TrC-300–TrC-302).451 17-Oxo-18,19-bisnorcheilanth-13(24)-en-6α-ol, a 19,20-bisnorcheilanthane (C23 sesterpenoid) (TrC-303) and 13,17-dioxo-18,19,24-trisnorcheilanth-6α-ol, a 19,20,21-trisnorcheilanthane (C22 sesterpenoid) (TrC-304), have been also isolated from A. agetae.451 Ancepsone A (TrC-320), isolated from A. anceps,454 and (17Z)-13,19-epoxycheilanth-17-en-6α-ol (Tec-506, Fig. 9), from A. mexicana,455 showed a 13,19-epoxycheilanthane skeleton. 18-epi-Scalar-16-ene-6α,19-diol (Tec-507) and 16α,19-epi-dioxy-18-episcalar-17(25)-en-6α-ol (Tec-508) have been also isolated from A. mexicana. Other cheilantanes, cristasesterterpenoic acid and cristasesterterpinol glucoside (TrC-276 and TrC-277), have been obtained from Caesalpinia crista (Fabaceae).456 Involudispirones A and B (TrC-278 and TrC-279, Fig. 9), containing a 1,2-dioxadispiro[5.2.5.2]hexadecane ring system have been isolated from Stahlianthus involucratus (Zingiberaceae).457 It has been speculated that this spiro system could be formed by an enzyme-catalyzed Diels–Alder addition between the derivative of cadalenequinone, a major constituent in S. involucratus, and myrcene.457,458 Similarly, it has been hypothesized that heliocide H2 (TrC-157), isolated together with heliocides B1 (TrC-155) and H1, H3 and H4 (TrC-156, TrC-158, and TrC-159) from Gossypium hirsutum (Malvaceae),459–462 could be biosynthesized by a Diels–Alder addition between the sesquiterpenoid gossypolone and myrcene.461 A Diels–Alder reaction has been used to synthesize TrC-155 and TrC-159 from hemigossypolone and trans-β-ocimene.462 Linderasesterterpenoids A and B (TrC-319 and TrC-319) with an unusual 7-cyclohexyldecahydroazulene carbon skeleton have been isolated from the roots of Lindera glauca (Lauraceae).463 2′-Isopicrasin A (TrC-288)464 and picrasin A (TrC-289),465 simaroubolides having the C25 simarolidane skeleton which consists of three carbocycles and two lactone rings, and is closely related to simarolide, have been isolated from Picrasma quassioides (Simaroubaceae).465 Vulgarosides 1–4 (TrC-245–TrC-248), sesterterpene esters whose base aglycon skeleton is quite similar to TrC-280 (except for the stereochemistry at C-13 and C-14), have been obtained from Cydonia vulgaris (Rosaceae). The distribution of TrCs in vascular plants suggests a more ancestral inheritance of the genes involved in the synthesis of these compounds. While a detailed discussion of the phylogeny of sesterterpenoids in plants is not a primary objective of this review, it is worth noting that there are several gaps in our knowledge regarding this topic. Based on current knowledge, Lamiaceae is the largest source of compounds among the 22 vascular plant families for which sesterterpenoids have been described, with DC and MC being the most important. Coleifolides A and B (MH-50 and MH-51), characterized by a β-methyl-α,β-unsaturated-γ-lactone and structurally similar to manoalide derivatives, have been isolated from Scutellaria coleifolia.466 Leucosceptrane sesterterpenoids (leucosceptroids), colquhounoids and eurysoloids have been isolated from the Lamiaceae Leucosceptrum canum, Colquhounia coccinea var. mollis and Eurysolen gracilis. From the trichome exudates and the leaves of L. canum bicarbocyclic sesterterpenoids have been isolated, i.e. leucosceptroids A and B, E–Q (DC-168 and DC-169),467 leucosceptroids E–N (DC-185–DC-192, DC-138, and DC-229) showing an α,β-unsaturated γ-lactone moiety,468 leucosceptroids P and Q (DC-198–DC-199),469 leucosesterlactone (DC-164),470 17α-hydroxyleucosceptrine (DC-166),471 leucosceptrine (DC-211).472 Leucosceptroid O (DC-193), possessing a spiro α,β-unsaturated γ-lactone moiety, has been isolated from the flowers of the same species.473 Additionally, tricarbocyclic compounds with antipodal cyclopentenones leucosceptroids C–D (TrC-190 and TrC-191),474 leucosesterterpenone
(TrC-188, Fig. 9),470 and 14β-methylleucosesterterpenone (TrC-189),471 have been found. Leucosceptroid degradation products, i.e. the C20 norleucosceptroids A–C (DC-172–DC-174), and the C21 norleucosceptroids D–H (DC-200–DC-204).469,475 have been isolated from L. canum of Nepalese and Chinese origin, respectively. 1α-Hydroxyleucosceptrine (DC-212) and 8α-hydroxyleucosceptrine (DC-213) were produced microbial transformation of DC-211 by Rhizopus stolonifer.394 Colquhounoids A–D (DC-182–DC-184 and DC-246), and 14-epi-colquhounoid D (DC-247) have been isolated from Colquhounia coccinea var. mollis.476,477 Eurysoloids A and B (TeC-441 and TeC-442), characterized by a pentacyclic 5/6/5/10/5 scaffold with an unusual macrocyclic ether system have been isolated by Eurysolen gracilis.478 The genus Salvia has received much attention within Lamiaceae. Sesterterpenoids have been found in both cultivated plants e.g. S. tingitana479 and wild species, such as in the case of S. dominica from subarid regions of Jordan.480,481 Prenyllabdane-type sesterterpenoids25 have been isolated from several Salvia species, as S. syriaca (DC-59, Fig. 9),482S. hypoleuca (DC-64, DC-67, DC-68, and DC-70),483,484S. palaestina (DC-73–DC-75),485S. dominica (DC-81–DC-91, DC-94–DC-96, DC-101 and DC-102),481S. tingitana (DC-120, DC-127, DC-129–DC-132, DC-146, DC-147),479S. yosgadensis (DC-134), and S. mirzayanii (DC-121).486 Prenyllabdane sesterterpenoids with a lactone, ester or acetal functionality between C6 and C23 have been found in various species, as S. hypoleuca (DC-65, DC-66, DC-69, and TrC-223),483,484S. tingitana (DC-127, DC-143–DC-145),479S. sahendica (DC-71 and DC-72),487S. dominica (DC-76–DC-80, DC-92–DC93, DC-101 and DC-102),481S. lachnocalyx (DC-97–DC-98).488 Other metabolites with a tetrahydropyran ring similar to manoyloxide-type diterpenoids between C-8 and C-13 were obtained from S. mirzayanii (DC-122–DC-126)489 as well as other hydroxymanoyloxide derivatives (DC-140–DC-142) from S. limbata,490 (14E)-methylmanoyloxide-14,16,18-trien-19,16-oxide-23-carboxylate (DC-139) from S. tingitana,479 lachnocalyxolides C and D (DC-99–DC-100) from S. lachnocalyx,488 yosgadensolides A and B (DC-134 and DC-135) along with their epimers from S. yosgadensis,491 salviaethiopisolide derivatives (DC-219–DC-220)492 and others hydroxymanoyloxides (DC-221–DC-223)493 from S. aethiopis. A norprenyllabdane-type sesterterpenoid, (13E)-4α,6α,8α-trihydroxylabd-13(14),17(18)-dien-16,19-olide (DC-132), was isolated from S. tingitana.479 Compounds arising from degradation of C-19 and C-20, 19,20-dinorsesterterpenoids were yosgadensenol (DC-133) and 13-epi-yosgadensenol (DC-42) from S. yosgadensis,494 and 6-dehydroxyyosgadensenol (DC-137) and 6-dehydroxy-13-epi-yosgadensenol (DC-136) from S. limbata.490 The sesterpene γ-lactone genepolide (DC-167, Fig. 9), isolated from Artemisia umbelliformis (Asteraceae) has been described as a formal Diels–Alder adduct of the exomethylene-γ-lactone germacranolide costunolide and the diene myrcene.495 Another intermolecular Diels–Alder reaction between thujanone, a thujane-type monoterpene one of main constituents in the essential oil of A. argyi, and a guaianolide, has been hypothesized for the biosynthesis of isoartemisolide (PC-26), isolated from the same species.496 Raoulic acid (DC-218), isolated from Raoulia australis, probably biosynthesized by diprenylation of a sesquiterpenoid precursor of the germacrene type, common in other Asteraceae species.497 Dibritannilactone A (TeC-45), isolated from Inula britannica, may derive from a monoterpene and a sesquiterpenoid.25,61 The trinorsesterterpene glycoside 3-[6-(4,8-dimethyl-nona-1,3,7-trienyl)-4-hydroxy2,6-dimethyl-cyclohex-1-enyl]-3-hydroxypropionic acid 1 glucoside (MC-99) has been obtained from the fern Woodwardia virginica (Aspleniaceae).498 The scalarane corallocarpscalarolide (TeC-410) has been isolated from the roots of Corallocarpus epigaeus (Cucurbitaceae).499 Biyoulactones D and E (DC-170 and DC-171), tricyclic meroterpenoids possessing an octahydroindene ring, a γ-butyrolactone ring, and a β-diketone moiety, have been isolated from Hypericum chinense (Hypericaceae).500 From the fruit of Phellodendron chinense var. glabriusculum (Rutaceae), phellogine (TeC-32), whose structure is similar to tirucalla-7-ene derivatives in the same species, has been obtained.501 Goniocarpic acid (DC-214) has been described within the constituents of the leaves of Serjania goniocarpa (Sapindaceae). Nevertheless, the classification of DC-214 as a sesterterpenoid has been questioned as it cannot be derived from GFDP.25 Among Monocotiledonous species, Aletris farinosa (Nartheciaceae), afforded two sclarane (TeC-226 and TeC-227) and four cheilantane sesterterpenoids (TrC-274, TrC-275, TrC-290, and TrC-291) have been studied.502,503 The scalarane sesterterpenoid perisomalien A (TeC-34)504 and the monocyclic n-non-2′-en-1′-yl-13(15,19,19-trimethyl-cyclohex-14,16-dienyl)-2,6,10-trimethyl-tetradec-6-ol-13-on-1-oate (hemidesmu sesterterpenoid ester) (MC-98), have been isolated from Periploca somaliensis and Hemidesmus indicus, respectively.505 The potential of sesterterpenoids in plants is still largely untapped, as evidenced by the case of Gentianaceae.25 Nitidasin (TeC-352)506 was the first gentianellane-type25 sesterterpenoid isolated from the Andean species Gentianella nitida. From the same species, nitiol was isolated (TrC-184).350 It has been speculated that the seco-gentianellane alborosin (TrC-109) from G. alborosea, could be derived from the cyclization of GFPP through TeC-352 as a key intermediate, and after the final oxidative cleavage of the C11/C12 bond.507 Later, other gentianelloids have been isolated from the Chinese G. turkestanorum, namely the C-6 epimeric sesterterpenoids gentianelloids A and B (TrC-286 and TrC-287), two compounds containing an unusual 10,11-seco-gentianellane skeleton,508 and 18-epi-nitidasin (PC-80), gentianelloids C–F (TeC-447, TeC-449, TeC-451, TeC-453), 18-epi-gentianelloids C–F (TeC-448, TeC-450, TeC-452, TeC-454), and 18-epi-alborosin (TeC-455).509 Another species belonging to Gentianaceae, Swertia bimaculata, afforded aspterpenacid C (PC-40, Fig. 9)510 with the 5/3/7/6/5 pentacyclic skeleton similar to aspterpenacids A and B (PC-38 and PC-39) mangrove endophytic fungus Aspergillus terreus, isolated from Kandelia obovata (Rhizophoraceae).435 Their biosynthetic pathway could be derived from GFPP with a series of cyclizations, rearrangements, redox and acetylation reactions.435 Bolivianine (PC-53) and isobolivianine (PC-53), two sesterterpenoids with an unusual skeleton, have been isolated from the bark of Hedyosmum angustifolium.511 The authors hypothesized their biosynthesis starting from onoseriolide, a sesquiterpene lactone already isolated from other Chloranthaceae. Another species, H. brasiliense, afforded hedyosulide (TrC-20), probably biogenetically related to the α-exo-methylene-γ-lactone precursor hedyosumin A and myrcene.512 In the cormophytes, acyclic sesterterpenoids (AI, Table S1, ESI†) have been obtained from Poaceae, Euphorbiaceae, and Solanaceae. 2,6,10,14-Tetramethyl-18-butanecarboxymethylene-henecos-12-en-17β-ol (AI-11) has been isolated from Oryza sativa,33 and (2Z,6Z,10E,14E)-geranylfarnesol (AI-8) from Triticum aestivum.513 Other linear sesterterpenoids, 2E-3,7,11,15,19-pentamethyleicos-2-en-1-ol (AI-22) (2Z,6E,10E,14E)-geranylfarnesol (AI-9), have been isolated from the aerial parts of Croton hieronymi (Euphorbiaceae).514 3,7,11,15,19-Pentamethyl-2-cis-6-trans-eicosadien-1-ol (AI-4) has been obtained from the unsaponifiable matter of the leaves of Solanum tuberosum (Solanaceae).515
Among terrestrial organisms, vascular plants are therefore the most important source of sesterterpenoids.25 It is interesting to examine the geographical distribution of the source species of sesterterpenoids on a global scale. Fig. 10 shows the most noteworthy areas for raw materials collection to be exploited for a potential use in the pharmaceutical field. The spatial distribution patterns of the compounds also reveal regions where it may be beneficial to increase eco-metabolomic research on sesterterpenoids. Overall, China, India and, more generally, the Asian continent are the areas with the greatest wealth of sesterterpenoids rich vascular plants even if Australia (for MC, DC and TrC), the Andean regions of South America (for AI) and the Mediterranean areas (for DC) are also notable. It should be noted that this distribution partially overlaps with the world's floristic hotspots. However, some areas with high specific richness and high rates of endemism, such as Macaronesia, Amazonia, or the Alpine regions, appear surprisingly underrepresented and may still contain a considerable wealth of sesterterpenoid diversity.
Thanks to the investigation of new biosynthetic chemistry, undescribed plant sesterterpenoids have been defined. By genome mining and gene clusters analysis, it appears that many genes and gene clusters do not direct the production of known metabolites of the organism in which they are found and that novel gene clusters provide the biosynthesis of known natural products.516 Moreover, the induced expression of silent genes can lead to the isolation of new metabolites.382 Terpenoid diversity in plants is determined by short-chain prenyltransferases (SC-PTs) and terpene synthases (TPSs). Among the SC-PTs, geranyl-farnesyl pyrophosphate synthases (GFPPS) produce the sesterterpene base scaffold GFPP.28,381,517 Sesterterpene synthases (sester-TPSs), a clade of the TPS-a subfamily, are a branch of class I TPSs, firstly identified in Brassicaceae,28 and widely distributed in the plant kingdom. Sester-TPSs catalyse diphosphate abstraction, cyclization of C1–C15 and C14–C18, producing a 5/15 bicyclic carbocation, which then undergoes structural modifications to yield diverse polycyclic sesterterpene scaffolds.516 Many sesterterpenoids contain a cyclopentane moiety, generated via type A (C1-IV-V) (between the C1 cation, the C14–C15 double bond (IV), and the C18–C19 double bond (V)) or type B (C1-III-IV) (between the C1 cation, the C10–C11 double bond (III), and the C14–C15 double bond (IV)) early-stage cyclization mechanisms of GFPP in the catalytic pocket of plant sesterTPSs.28,51,518 Type A cyclization is mainly represented in plants. Examples are (−)-caprudiene A (TeC-435, Fig. 9), produced by transient expression of individual STS with a GFPPS gene by “agro-infiltration”, with Agrobacterium tumefaciens infiltrated into the undersides of leaves of Nicotiana benthamiana,519 and (−)-variculatriene A (TrC-117).51,519 Type-B Sester-TPSs, considered as possible progenitors of Type-A Sester-TPSs, have been discovered in Brassicaceae. AtTPS06, identified in Arabidopsis thaliana, is the first gene found in plants which produces the E/Z isomer of flocerene (TrC-292) via type-B cyclization mechanism.28,517,520 In the genome of A. thaliana, AtsesterTPS1, responsible for the biosynthesis of the new tricyclic sesterterpene (+)-thalianatriene (TrC-120) with the unprecedented 11–6–5 fused ring system, and AtsesterTPS2, which produced (−)-retigeranin B (PC-4), with the characteristic 5/5/5/6/5 fused ring system, were identified.521 Transient expression of TPSs from A. thaliana, Capsella rubella, and Brassica oleracea in Nicotiana benthamiana afforded sesterterpenoids with various scaffolds, as (−)-caprutriene (TrC-116), (−)-variculatriene A (TrC-117), (−)-aleurodiscalene A (TeC-17), (−)-ent-quiannulatene (PC-3), (+)-boleracene (PC-6), and (+)-astellatene (PC-9).51 Engineered Nicotiana benthamiana expressing LcTPS2, characterized from Leucosceptrum canum, produced two 18- and four 14-membered ring sesterterpenoids including (S,2E,6E,10E,14E)-3,7,11,15-tetramethyl-18-(1-hydroxyisopropyl)cyclooctadecatetraene (MC-141), (S,2E,6E,10E,14E)-3,7,11,15-tetramethyl-18-(1-methylethenyl)cyclooctadecatetraene (MC-142), (S,2E,15Z)-a-cericerene (MC-143), (S,2E)-cericerene (MC-114), (S,2E)-α-cericerene and (14S,15R,2E)-15-hydroxy-α-cericerene (MC-145).522 Engineered Escherichia coli expressing CcTPS1, obtained from Colquhounia coccinea var. mollis, produced (+)-α-geranylbisabolene (MC-150) and (+)-somaliensene B (MC-151), characterized by an alotane skeleton, not previously isolated from any plants.523Trans-prenyltransferase (PT) and N-terminal terpene synthase (TPS) gene pairs from Arabidopsis thaliana have been reported to synthesize sesterterpenoids. AtsesterTPS2 produced the pentacyclic sesterterpenoid (−)-retigeranin B (PC-4) with the characteristic 5/5/5/6/5 fused ring system. AtsesterTPS1 produced the tricyclic (+)-thalianatriene (TrC-120).521
5 Biological and pharmacological properties of sesterterpenoids
Over the years, sesterterpenoids have been investigated for their action on a wide range of biological activities of pharmacological importance including suppression of cancer cell growth, anti-inflammatory properties, mainly counteracting PLA2 activity, and modulation of neurodegenerative processes (Fig. 11). Moreover, they were explored for their key functions such as antimicrobial effects against fungi, Gram + and − bacteria, viruses such as HIV or as anti-malarial and anti-tuberculosis agents. Sesterterpenoids have been also inspected as ichthyo- and phyto-toxins, as nematocidals, and antifeedants from an ecological point of view. Few of them were tested for the involvement in the treatment of metabolic diseases such as type-II diabetes, hypercholesterolemia and obesity and as immunosuppressive agents. Here, we briefly discuss those sesterterpenoids whose mechanism of action has been thoroughly investigated, focusing on their target enzymes. In parallel, a more comprehensive list of sesterterpenoids biological activity can be found in the annexed Table S8, ESI,† and at https://sesterterpenoids.unige.net/. Selected sesterpenoids structures were numbered to help reading.5.1 Sesterterpenoids with anti-cancer activity
The majority of sesterterpenoids were probed for their cytotoxicity, predominantly against cancer cells. Principally, ophiobolins were screened versus multiple cancer cell lines: among the approximately 80 identified ophiobolins, about 20 were reported for their cytotoxic capability. Mainly, TrC-2 (Fig. 3) displayed growing inhibition against 25 human cancer cells: it principally counteracts apoptosis-resistant glioblastoma cells by inducing a non-apoptotic cell death via reaction with primary amines prompting pyrrolylation of lysine residues on its intracellular target protein.524 Intrigued by this property, Fujiwara studied the behaviour of TrC-2 in the L1210 cell line in 2000, showing a concentration-dependent cytotoxicity.525 This compound provoked paraptosis-like cell death in human glioblastoma cells by altering the cytoskeleton dynamic pathway.526 Furthermore, TrC-2 selectively inhibited the correct development of solid and haematological cancer cells with a very low IC50; it also prompted apoptosis in MDA-MB-231 cells through the inhibition of PI3K/mTOR, Ras/Raf/ERK and CDK/RB pathways313 and it has been discovered to significantly repress the mammosphere formation.527 This super active compound has also a role in fighting multi-drug resistant cancer cells, as it acts against HL60 cells that are resilient to combined chemotherapy, ovarian carcinoma cells that are resistant to cisplatin, small lung carcinoma cell line GLC4 that is resistant to adriamycin, and the others.526 Ophiobolins not only were extensively studied for their in vitro cell inhibition, but also reduced tumour development in in vivo animal models. For instance, TrC-2 was inoculated in a mouse model of glioblastoma528 and of melanoma.526 Later on, TeC-326, heteronemin acetate (TeC-349), hyrtiosin E (TeC-309), 12-deacetoxy-scalarin 19-acetate (TeC-176) and TrC-130 were tested against a large panel of cells such as HuCCA-1, HeLa, MDA-MB-231, MCF7, HT-29, and H69AR, KB, human colon adenocarcinoma (DLD-1 and HCT-116), hormone-dependent breast cancer (T-47D) and human chronic myelogenous leukaemia (K562) cell lines and TeC-326, Fig. 12, was the most potent on all cell lines.88,529 On examining the mechanism of TeC-326 action, it was found to induce apoptosis in leukaemia Molt4 cells by acting on the oxidative stress pathway, on mitochondrial dysfunction and on talin expression. More in detail, it was able to upregulate both talin and phosphorylated talin expression and, additionally, to interfere with actin microfilament formation inducing morphological alteration.530TeC-326 was found to inhibit the proliferation of the HCC cell lines HA22T and HA59T and to induce apoptosis via the caspase pathway. TeC-326 treatment also induced the formation of reactive oxygen species (ROS), which is associated with TeC-326-induced cell death, and to triggers ROS removal by mitochondrial SOD2 instead of cytosolic SOD1. The mitogen-activated protein kinase (MAPK) signalling pathway was linked to ROS-induced cell death, and TeC-326 reduced the expression of ERK, a MAPK that is associated with cell proliferation. Inhibitors of JNK and p38, which are MAPKs associated with apoptosis, restored TeC-326-induced cell death. In addition, TeC-326 treatment reduced the expression of GPX4, a protein that inhibits ferroptosis, which is a novel form of non-apoptotic programmed cell death. Treatment with a ferroptosis inhibitor also restored TeC-326-induced cell death.531TeC-326, 12-epi-heteronemin acetate (TeC-328), TeC-331 and TrC-130 were found to be toxic also against human epidermoid carcinoma KB cells532 and TeC-326 is also reported to inhibit the proteasome, promoting apoptotic cell death.533 A very deep analysis has been carried out by Lai et al. 2016 on two scalarane sesterterpenoids, 12β-(3′β-hydroxybutanoyloxy)-20,24-dimethyl-24-oxo-scalara-16-en-25-al (TeC-105) and 12β-(3′β-hydroxypentanoyloxy)-20,24-dimethyl-24-oxo-scalara-16-en-25-al (TeC-320) on various cancer cells. In leukaemia Molt 4 cells, TeC-105, Fig. 12, triggered mitochondrial membrane potential disruption, induced ROS production, calcium release and ER stress and inactivates topoisomerase IIα, determining apoptosis. Moreover, this molecule is able to target the molecular chaperone Hsp90 modulating its client proteins expression.534 In 2009, sesterterpene lactones isolated from Salvia dominica were discovered to interact with tubulin-tyrosine ligase (TTL), an enzyme involved in tubulin tyrosination in cancer cells, by chemical proteomics and surface plasmon resonance assays. An inhibition of this enzyme has also been assessed by 8α,15(S),23α-trihydroxy-23,6 α-epoxy-labd-13(14),17-dien-16(S),19-olide (DC-78), 8α,15(S)-dihydroxy-23α-O-ethyl-23,6α-epoxy-labd-13(14),17-dien-16(S),19-olide (DC-79), 8α-hydroxy-23α-O-ethyl-23,6α-epoxy-labd-13(14),17-dien-16(R),19-olide (DC-80), 6α,8α,15(S),23-tetrahydroxy-labd-13(14), 17-dien-16(S),19-olide (DC-81), 6α,8α,15(S)-trihydroxy-23-carbossi-labd-13(14),17-dien-16(S),19-olide (DC-83), 6α,8α-dihydroxy-23-carbossi-labd-13(14),17-dien-16,19-olide (DC-84), 6α,8α,15(S)-trihydroxy-23-oxo-labd-13(14),17-dien-16(S),19-olide (DC-85), 6α,8α-dihydroxy-23-oxo-labd-13(14),17-dien-16(R),19-olide (DC-86), 6α,15(S),23-trihydroxy-labd-8(22),13(14),17-trien-16(S),19-olide (DC-87), 6α,15(S)-dihydroxy-23-oxo-labd-8(22),13(14),17-trien-16(S),19-olide (DC-88), 6α,8α,23-trihydroxy-labd-13(14),15,17-trien-16,19-olide (DC-89), 6α,8α-dihydroxy-23-carbossi-labd-13(14),15,17-trien-16,19-olide (DC-90), 6α,8α-dihydroxy-23-oxo-13(14),15,17-trien-16,19-olide (DC-91), 8α-hydroxylabd-13(14),15,17-trien-6R,23-16,19-diolide (DC-92) and 8α-23-dihydroxy-23,6R-epoxy-labd-13(14),15,17-trien-16,19-olide (DC-93).481 Also kohamaic acid A (DC-4, Fig. 12), besides other bioactivities, has been described to prevent the growth of human cancer cell (promyelocytic leukaemia cell line, HL-60) and its derivatives by a binding to four mammalian polymerase Beta residues (Leu11, Lys35, His51 and Thr79).535 Starting from 2010, Prof. Monti's group published some results on TrC-178's, new targets obtained by chemical proteomics. In brief, this molecule was able to interact with the proteasome inhibiting its function and thus becoming a candidate as an anticancer drug. It also altered the autophagy pathway.536–538DH-80, Fig. 2, and DH-81, Fig. 12, presented an anti-proliferative effect against A549, HeLa, and HCT-116 cells with IC50 values in the micromolar range through the inhibition of protein-tyrosine phosphatase 1B.35 Differently, MC-9, MC-10, and TH-25 targeted a deubiquitinating enzyme named USP7 blocking its function with IC50 values in the range of 2.7–4.6 μM and are being considered as new anticancer lead compounds.46 Also the suvanines, sulfate-containing sesterterpenoids,539 differently decorated on their scaffold, including TrC-143, Fig. 12, suvanine N,N-dimethyl-1,3-dimethylherbipoline salt (TeC-387), suvanine N,N-dimethylguanidium salt (TeC-388), and TrC-200–TrC-205, revealed moderate cytotoxicity against the K562 and A549 cell lines.192 More in detail, using a chemical proteomics approach, Cassiano et al.540 revealed a direct interaction of suvanine and the heat shock protein 60, a key chaperonin involved in several tumoral and inflammatory diseases.5.2 Sesterterpenoids with anti-inflammatory activity
Sesterterpenoids have an exceptional potential as anti-inflammatory compounds. In the latter part of 80s, many compounds were isolated from the marine sponge Luffariella sp. with a consistent anti-inflammatory activity: the most well-known is manoalide (MC-13, Fig. 3),541–543 which significantly lowers chemically-induced inflammation in vivo and irreversibly blocks the in vitro hydrolysis of phosphotidylcholine by phospholipase A2 (PLA2).544,545 PLA2 is a key step at the beginning of the inflammatory cascade releasing arachidonic acid from membranes. MC-13 is a non-specific inhibitor of phospholipases that modulates the activity of phospholipase C, human sPLA2 (IC50 = 1.7 μM), cPLA2 (IC50 = 10 μM) and snake venom sPLA2 (IC50 = 0.03 μM).543 This latter activity has been confirmed by Dal Piaz et al.546 in deciphering the mechanism of MC-13 along with TrC-178, Fig. 12, binding to bee venom PLA2: the γ-hydroxybutenolide ring with its masked aldehyde is able to covalently bind the N-terminus of PLA2 giving rise to a Schiff base. The same inhibition mechanism has also been demonstrated for different petrosaspongiolides,547–549 21-hydroxy-petrosaspongiolide K (TeC-65, Fig. 13), and 21-hydroxy-petrosaspongiolide P (TrC-181).133 Prof. Gomez-Paloma group also extended this work to human synovial PLA2 and demonstrated that TrC-178 was able to covalently modify a certain lysine on the protein's surface, reducing the adhesion of this protein on the membrane and inhibiting its function.550MC-13 was registered by Allergan pharmaceuticals and reached phase II clinical trials as a topical antipsoriatic. However, because of formulation hitches, it stopped in that phase. Several congeners of MC-13 have been extracted from different organisms. For example, three sesterterpenes fitting in the same family, i.e. (4E,6E)-dehydromanoalide (MC-68), deoxysecomanoalide (MC-29) and deoxymanoalide (MC-22), showed good inactivation degree on the snake venom PLA2 at 0.2 to 0.5 μM even if lower than MC-13.551 Noteworthy, these metabolites are reduced forms of MC-13, and thus their PLA2 inhibitory activity is much weaker.552MC-70 is a powerful antagonist of topical phorbol myristate acetate promoted inflammation in the mouse ear. It inhibited in vitro hydrolysis of phosphatidyl choline by bee venom PLA2, too (IC50 = 0.23 μM).553 The determined inhibition achievable by MC-70 is only 80% as compared to MC-13 because its binding was partially reversible (around 30%). Indeed, a detailed kinetic analysis of the MC-70 reaction with PLA2 confirmed also a non-competitive type of inhibition.220 Furthermore, the luffariellins (mainly MC-53, Fig. 13 and MC-56, Fig. 13) from the sponge Luffariella variabilis were also considered anti-inflammatory sesterterpenes by the research group of Kernan in 1987.223 They are powerful antagonists of topical phorbol myristate acetate-induced inflammation in the mouse ear and also bee venom PLA2 was significantly downregulated by MC-53 (IC50 = 0.056 μM) and MC-56 (IC50 = 0.060 μM). Barbara Potts confirmed in 1992 that even DC-162 displayed approximately 50% inhibition of oedema in the mouse ear assay.554 Other PLA2-modulating sesterterpenes from marine origin included cacospongionolides B (DC-19, Fig. 13) and E (DC-20) (IC50 = 300 nM against human and bee venom sPLA2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 555,556 and DH-16 (IC50 = 6.9 μM against human sPLA2 and cPLA2
555,556 and DH-16 (IC50 = 6.9 μM against human sPLA2 and cPLA2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 557 De Marino et al.133 in 2000 proposed that spongidines A–D (TrC-259–TrC-262) inhibited hsPLA2 as well. Interestingly, studies on diastereoisomeric mixture of DC-6 and DC-7, both bearing a hydroxybutenolide moiety, demonstrated that this group itself is not sufficient for PLA2 inhibitory activity but that non-covalent interactions between the counterparts also play key roles in terms of compound potency.558,559TeC-75, a di-aldehyde containing sesterterpene, is considered a good anti-inflammatory compound,560,561 mainly inactivating PLA2s.562 In 2000, Prof. Cimino's group isolated many scalarane and homoscalarane compounds from the nudibranchs Glossodoris sedna and G. dalli and tested them against mammalian cytosolic PLA2. The di-aldehyde-bearing compounds exhibited a consistent inhibition of the enzyme although it occurred at high concentration.259 Next, Monti et al. assessed the role of di-aldehyde moiety of TeC-75 and of 12-epi-scalaradial (TeC-78) on secretory PLA2.537,563,564 Also DH-76 and its monoacetate analogue, isolated from the sponge Hyrtios sp., were tested on cobra venom PLA2, showing good results in terms of inactivation.565 In 2009, Prof. Zampella isolated TrC-200, Fig. 13 and TrC-154, Fig. 13, two sesterterpenoids bearing nitrogen atoms together with suvanine (TrC-143), the latter exerting anti-inflammatory activity by the inhibition of different secretory PLA2 such as groups IIA, IA (Naja naja venom), IB (porcine pancreatic enzyme) and III (bee venom enzyme).195
557 De Marino et al.133 in 2000 proposed that spongidines A–D (TrC-259–TrC-262) inhibited hsPLA2 as well. Interestingly, studies on diastereoisomeric mixture of DC-6 and DC-7, both bearing a hydroxybutenolide moiety, demonstrated that this group itself is not sufficient for PLA2 inhibitory activity but that non-covalent interactions between the counterparts also play key roles in terms of compound potency.558,559TeC-75, a di-aldehyde containing sesterterpene, is considered a good anti-inflammatory compound,560,561 mainly inactivating PLA2s.562 In 2000, Prof. Cimino's group isolated many scalarane and homoscalarane compounds from the nudibranchs Glossodoris sedna and G. dalli and tested them against mammalian cytosolic PLA2. The di-aldehyde-bearing compounds exhibited a consistent inhibition of the enzyme although it occurred at high concentration.259 Next, Monti et al. assessed the role of di-aldehyde moiety of TeC-75 and of 12-epi-scalaradial (TeC-78) on secretory PLA2.537,563,564 Also DH-76 and its monoacetate analogue, isolated from the sponge Hyrtios sp., were tested on cobra venom PLA2, showing good results in terms of inactivation.565 In 2009, Prof. Zampella isolated TrC-200, Fig. 13 and TrC-154, Fig. 13, two sesterterpenoids bearing nitrogen atoms together with suvanine (TrC-143), the latter exerting anti-inflammatory activity by the inhibition of different secretory PLA2 such as groups IIA, IA (Naja naja venom), IB (porcine pancreatic enzyme) and III (bee venom enzyme).195
5.3 Sesterterpenoids with anti-microbial activity
Hyrtiosin B (TeC-306, Fig. 14) from Hyrtios erecta was found to modulate the protein isocitrate lyase from Candida albicans.566 This compound was a good inhibitor of this enzyme which plays a crucial role in fungal development. Furospongin-4 (MH-44, Fig. 14) and isofurospongin-4 (MH-25, Fig. 14) showed weak influence on Staphylococcus aureus at 100 μg mL−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 137 while PC-31 exhibited strong influence against Mycobacterium tuberculosis protein tyrosine phosphatase B.433 Several sesterterpenoids have been described to have bacteriostatic and bactericidal effects against tested pathogens. The antibiotic profile of MC-13 was investigated early, in 1980,567 alongside (6E)-neomanoalide (MC-16), and (6Z)-neomanoalide (MC-17) which were found to be effective selectively against Gram-positive bacteria.37 The proposed mechanism regards the modulation of cell–cell communication by MC-13 and by manoalide 25-monoacetate (MC-14).568 In the field of HIV inhibitors, phyllolactones were verified for their effect in downregulating HIV-1 envelope-mediated fusion.104 Additionally, TrC-130 inactivates HIV-1 integrase by binding to DNA as reported by surface plasmon resonance and docking analysis.569 Using a model of HIV-1 infection, TrC-192 and TrC-211, alotaketals C and D (MC-117 and MC-118) and TeC-390 were found to promote HIV proviral gene expression as well as being at least as potent as MC-117 and MC-118.179PC-40 inhibits HIV-1 replication.510 Also PC-31 and PC-64 had potent inhibition against Mycobacterium tuberculosis protein tyrosine phosphatase B with IC50 values of 3–6 μM.389
137 while PC-31 exhibited strong influence against Mycobacterium tuberculosis protein tyrosine phosphatase B.433 Several sesterterpenoids have been described to have bacteriostatic and bactericidal effects against tested pathogens. The antibiotic profile of MC-13 was investigated early, in 1980,567 alongside (6E)-neomanoalide (MC-16), and (6Z)-neomanoalide (MC-17) which were found to be effective selectively against Gram-positive bacteria.37 The proposed mechanism regards the modulation of cell–cell communication by MC-13 and by manoalide 25-monoacetate (MC-14).568 In the field of HIV inhibitors, phyllolactones were verified for their effect in downregulating HIV-1 envelope-mediated fusion.104 Additionally, TrC-130 inactivates HIV-1 integrase by binding to DNA as reported by surface plasmon resonance and docking analysis.569 Using a model of HIV-1 infection, TrC-192 and TrC-211, alotaketals C and D (MC-117 and MC-118) and TeC-390 were found to promote HIV proviral gene expression as well as being at least as potent as MC-117 and MC-118.179PC-40 inhibits HIV-1 replication.510 Also PC-31 and PC-64 had potent inhibition against Mycobacterium tuberculosis protein tyrosine phosphatase B with IC50 values of 3–6 μM.389
 | ||
| Fig. 14 Representative structures of sesterterpenoids with anti-micro, anti-neurodegenerative and anti-feedant activities. | ||
5.4 Sesterterpenoids with anti-neurodegenerative activity
Few sesterterpenoids have been studied for their ability to counteract neurodegeneration. In 2014, Prof. Monti's group identified TeC-326, Fig. 12, a marine sesterterpenoids isolated from Hyrtios sponge,570 as an interactor of trans-activation response DNA-binding protein of 43 kDa (TDP-43) by using chemical proteomics. This metabolite can bind the protein and can alter its tendency to create insoluble aggregates in brain cells. After this, a small library of natural scalarane derivatives such as petrosaspongiolactams A–C (TeC-126–TeC-128) has been investigated for their ability to modulate the activity of TDP-43 giving the identification of protein aggregates inhibitors.87 In parallel, TrC-6 from Bipolaris oryzae has been discovered to strongly induce the autophagic degradation pathways of insoluble aggregates involved in neurodegenerative diseases.571 In this case, the targeted protein was alpha-synuclein in PC12 cells. Also, erinacine S (TeC-392), isolated from the ethanol extract of the mycelia of Hericium erinaceus, downregulates the formation of Aβ insoluble precipitates in the brains of transgenic mice after an oral administration as reported by Chen et al.572 Following this, in 2019, Hu et al. deeper investigated the mechanism of TeC-392, examining its capability in reducing amyloid plaque formation and improving neurogenesis in rats. Furthermore, this study showed that TeC-392 can penetrate the blood–brain barrier.573DC-164 and DC-211 showed their anti-neurodegenerative profile by acting against prolylendopeptidase470,472 as well as MC-36 was tested against Alzheimer showing a good profile as PPARα/γ-RXRα agonist and RARα positive allosteric modulator.574TeC-97 and TeC-98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 434 played as acetylcholinesterase inhibitors in rats whereas the compound YW3699 inhibited the glycosylphosphatidylinositol (GPI) synthesis.575 Finally, palinurin (DH-3) acted by an allosteric regulation on GSK-3.576
434 played as acetylcholinesterase inhibitors in rats whereas the compound YW3699 inhibited the glycosylphosphatidylinositol (GPI) synthesis.575 Finally, palinurin (DH-3) acted by an allosteric regulation on GSK-3.576
5.5 Sesterterpenoids with phytotoxicity
In 1984, TrC-2, Fig. 3, and its congeners were deeply analysed as photosynthesis inhibitors. Furthermore, their biological profile was acutely evaluated using spinach leaf slices and chlorella. TrC-2 resulted in well counteracting photosynthesis causing brown spots in rice plants.577 Later on, other ophiobolins were investigated by a mitochondrial electron transport assay designed to identify malate oxidation inhibitors: TrC-2 and TrC-5 were found to inhibit oxidation in maize mitochondria.324 Moreover, using the so-called leaf spot assay, their influence has been measured against several plants such as sorghum, maize, bentgrass, sicklepod and morning glory based on the necrosis superficial amplitude: TrC-2 and TrC-3 were more phytotoxic than their anhydro derivatives.314 Additionally, in 1991, Prof. Ballio deeply studied the mode of action of these sesterterpenoids. He postulated that the probable role of TrC-2 in the development of brown spot of rice is due to its involvement in a precise membrane transport process related to K+ permeability. Indeed, he demonstrated that in vitroTrC-2 irreversibly inhibits the calmodulins of bovine brain, maize and spinach by a covalent bond of the toxin to lysine 63, identifying this enzyme as ophiobolins target: the Ca2+-calmodulin complex is accountable for the regulation of many cellular functions.578 In 2006, ophiobolins were also investigated as phytotoxins by Prof. Evidente's group: in particular, TrC-2 instigated the appearance of large necrosis areas, even at low concentrations on grass weeds.318 Also, ophiobolins TrC-8, TrC-16 and TrC-35 were toxic to various weed species by the leaf-puncture bioassay.3195.6 Sesterterpenoids with nematocidal and anti-feedant activity
In plants, also TrC-38, Fig. 14, and TrC-39 were tested as nematocidal agents and they were toxic against the free-living worm Caenorhabditis elegans.348 In 1996, Tsipouras deepened the understanding of the mechanism of TrC-45 and its congeners.347 Indeed, all of these compounds exerted their nematocidal property by altering the ivermectin binding to nematode membranes. The same sesterterpenoids along with TrC-13 were also effective in the C. elegans motility assays and they were toxic in the low μM range.347 Some defensive sesterterpenoids with unusual cyclopentenone rings were extracted and purified from the leaves of L. canum: they were leucosceptroids DC-168, Fig. 14, TrC-190 and TrC-191.467 The potent antifeedant activity recommended them as self-protective molecules.474 The related DC-172–DC-174 showed the same bioactivity as well as DC-187 and DC-192 and, finally, in 2014 also DC-200–DC-202.468 Moreover, it was discovered that leucosceptrane sesterterpenoid degradation products could also be involved in the plant defence mechanisms against insects.469 In 2013, in a very innovative paper, Li group studied DC-182, DC-183 and DC-184 from leaves through a laser-microdissection coupled to mass spectrometry analysis and X-ray diffraction and planned a defensive role for these sesterterpenoids since they were closely related to the above-described leucosceptroid-class. All three compounds disincentivised the beet armyworm and the cotton boll worm.477 Finally, TrC-20 has been indicated as trypanosomicidal: mainly, its anti-protozoal activity was exerted against trypomastigote and amastigote forms of Trypanosoma cruzi.5125.7 Sesterterpenoids with immunosuppressive activity
In 2007, irregularasulfate (DC-26), hipposulfate A (TrC-113) and halisulfate-7 (DC-31) were discovered to be moderate inhibitors of the catalytic subunits of the mammalian Ser/Thr protein phosphatases calcineurin and could be used as immunosuppressive molecules.579 Also TrC-286 and TrC-287 were considered immunosuppressive by a different mechanism of action inhibiting the proliferation, activation, and cytokine IFN-γ production on T cells.580 Also TeC-5, TeC-6, TeC-11, TeC-12, and TeC-13, studied by Fujimoto et al., were immunosuppressive, acting on the human chemokine receptor CCR5.367 It's intriguing that more and more sesterterpenoids have been revealed to have immunosuppressive properties after the Covid-19 epidemic. Initially, the activity of leucosceptrane-type sesterterpenoids was assessed by preventing the release of cytokines in LPS-induced macrophages RAW264.7 and 3-H-2,17α-dihydroxy-leucosceptroid N (DC-270) demonstrated immunosuppressive activity against TNF-α and IL-6, with IC50 values of 13.39 and 19.34 μM, respectively.581 Then, a deep analysis of leucosceptroid N (DC-192), 5,13-dehydro-leucosceptroid A (DC-306), and 5α,16α-epoxy-leucosceptroid R (DC-291) showed their block on T cells and macrophages activations thought the inhibition of AKT-mTOR, JNK, p38 MAPK, and ERK pathway. Furthermore, DC-291 and DC-306 caused T cell G0/G1 cell arrest, whereas DC-192 markedly reduced IL-6 and TNF-α levels in peripheral blood serum and lessened the multiorgan damage in mice with LPS-induced sepsis, at 25 mg per kg dose. Additionally, there was a dose-dependent suppression of T cell proliferation and IFN-γ release when activated by anti-CD3/CD28.582 The TNF-α secretion was also effectively inhibited by leucosceptrodine (MC-153) and nor-leucosceptroid L (DC-264), with IC50 values of 11.21 μM and 12.19 μM, respectively.583 Lastly, few ophiobolin-like substances were studied in this area, including maydispenoids and undobolins: maydispenoids A (TrC-306) and B (TrC-307) demonstrated proliferation inhibitory activity on differently stimulated murine splenocytes332 and undobolin F (TrC-362) established significant inactivation against ConA-induced T lymphocyte proliferation with an IC50 value of 2.3 μM.5845.8 Miscellaneous
In 2006, Nam and coworkers isolated several sesterterpenoids from the sponge Spongia sp. including TeC-109 and TeC-110 which have a structure similar to guggulsterone and have been tested for their ability to modulate the transactivation of the farnesoid-X-receptor (FXR). These scalaranes inhibited FXR and were not cytotoxic for cells opening the way to treat hypercholesterolemia and, more in general, metabolic diseases.135 An anti-obesity profile has been reported for DC-169, based on a different mechanism of action: this metabolite acts reducing fat amassing through destroying unsaturated fatty acid biosynthesis. More deeply, it downregulates the expression of two stearoyl-CoA desaturase (SCD) genes fat-6 and fat-7, and a fatty acid elongase gene elo-2 in wild-type C. elegans.580 Inhibition of protein tyrosine phosphatase 1B has also been explored by a few sesteterpenoids as, for instance, hyattellactones A and B (TeC-239 and TeC-240), in an interesting paper115 since this protein can be considered a therapeutic target for the treatment of type-II diabetes and obesity. The same inactivation has been reported for phyllofolactones characterized by an alpha-beta-unsaturated lactone.115 In 2011, phorbasones were discovered to induce osteoblast differentiation, specifically phorbasone A (TrC-195) revealed a stimulatory effect on the calcium deposition activity in C3H10T1/2 cells.174MC-119, also, significantly excites osteoblast differentiation by activating the ERK pathway.5856 Conclusions
Within the terpenoids family, which is the biggest group of secondary metabolites, sesterterpenoids represent an incredible chemically varied and pharmacologically relevant subgroup, widespread in more than a few phyletic groups, including marine sponges, nudibranchs, bacteria, lichens, fungi, higher plants and insects. Over seven decades of research on their isolation and characterization, around 1600 structurally unique sesterterpenoids are summarized in the current review paper, which is supported by information on biological, pharmacological, ecological, and geographic distribution. It is highly probable that in the next years novel understudied taxa groups will bring up new interesting members of these pentaprenyl terpenes. Moreover, it is expected that the synthetic origin of some of these compounds will be disentangled, unveiling interconnecting mechanisms of symbiotic or trophic exchanges, bioaccumulation, biotransformation, and de novo synthesis. By regulating microbial metabolism, sesterterpenoids have facilitated the co-speciation of bacteria with fungi and the interaction of fungi with marine and terrestrial plants, thus generating complex interactions essential for microbial survival. Recently, there has been a growing recognition of the role of specific sesterterpenoids in establishing the composition of leaf and root microbiota in which the dynamic complementation of enzymatic pathways controls plant health.51 From a biological point of view, microorganisms as well as humans use them as antimicrobial, antiviral, antifeedants, and to combat neurodegeneration, hypercholesterolemia, diabetes, and obesity, in addition to their regal role as anti-cancer and anti-inflammatory (ophiobolin and manoalide derivatives, about everything). Recently, tests for the inhibition of T cell proliferation and the release of IFN-γ, TNF-α, and IL-6 have been conducted in an effort to counterbalance overactive immune system components. This could potentially be a future avenue for the repurposing of these poly-pharmacological metabolites. Furthermore, it should be emphasized that genome mining and heterologous expression approaches of sesterterpene synthases, increasingly in recent years, have led to the identification of new sesterpenoids, drawing a path that will be surely implemented in the future for the discovery of new molecular scaffolds. In the present review, we reported selected examples of naturally occurring marine and terrestrial sesterterpenoids within their structures, ecological, geographical and biological details. But without a question, the most important component of this enormous undertaking is the exhaustive list of known metabolites, which is included in the annexed Tables, and at the friendly consultable https://sesterterpenoids.unige.net/. The authors chose to publish the results of their work using an architecture formed by the integration between a relational database and a web-based interface. This architecture allows for rapid updating of research contents which will be implemented through two channels. The first is the direct intervention of the authors who will be able, after their authentication on the site, to manually insert new evidence relating to the subject matter present in new publications. The second concerns the interaction with public databases, selected by the authors of this work, for their evident authority, which will provide automatic access to updated in-depth content on the description of the individual organisms involved in the research. Pipelines based on Natural Language Processing to obtain automatic insertion of new evidence detected from scientific articles will be applied.7 Author contributions
Conceptualization: A. B., M. G., and N. De T.; data curation: V. I., A. B., N. De T., V. P., and F. De R.; formal analysis: A. B., N. De T. and M. G.; funding acquisition: A. B. and F. De R.; investigation: V. I., M. G., F. De R., P. B., L. N.-P., G. D., P. G., M. C. M., R. P., Y. M., V. P., A. B.; methodology: A. B., M. G. and N. De T.; project administration: A. B., and N. De T.; resources: A. B. and N. De T.; software: M. G.; supervision: A. B., M. G., and N. De T.; validation: A. B., M. G., F. De R., and N. De T.; writing – original draft: M. G., F. De R., P. B., L. N.-P., G. D., P. G., M. C. M., R. P., and A. B.; writing – review & editing: V. P., M. C. M, A. B., G. D., P. G., and N. De T.8 Conflicts of interest
There are no conflicts to declare.9 Acknowledgements
FDR: Financial support from the University of Salerno (FARB), from PRIN 2020: “Natural Products-Assisted Organic Synthesis” (2020AEX4TA). “Graphical Abstract was Created in BioRender. Núñez-Pons, L. (2024) BioRender.com/j70k119”. NDT and VP: Finacial support by National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4 - Call for tender No. 3138 of December 16, 2021, rectified by Decree n.3175 of December 18, 2021 of Italian Ministry of University and Research funded by the European Union – NextGenerationEU; Award Number: Project code CN_00000033, Concession Decree No. 1034 of June 17, 2022 adopted by the Italian Ministry of University and Research, CUP: D43C22001260001, Project title “National Biodiversity Future Center - NBFC”.10 References
- J. R. Hanson, Nat. Prod. Rep., 1986, 3, 123–132 RSC.
- J. R. Hanson, Nat. Prod. Rep., 1992, 9, 481–489 RSC.
- J. R. Hanson, Nat. Prod. Rep., 1996, 13, 529–535 RSC.
- T. Mitsuhashi and I. Abe, in Progress in the Chemistry of Organic Natural Products, ed. A. D. Kinghorn, H. Falk, S. Gibbons, J. Kobayashi, Y. Asakawa and J. K. Liu, Springer Nature, Cham, Switzerland, 2020, vol. 111, pp. 1–79 Search PubMed.
- S. K. Talaprata, in Chemistry of Plant Natural Products, ed. S. K. Talapatra and B. Talapatra, Springer-Verlag, Berlin Heidelberg, 2015, pp. 511–515, DOI:10.1007/978-3-642-45410-3_9.
- L. Wang, B. Yang, X.-P. Lin, X.-F. Zhou and Y. Liu, Nat. Prod. Rep., 2013, 30, 455–473 RSC.
- P. Crews and S. Naylor, in Fortschritte der Chemie organischer Naturstoffe/Progress in the Chemistry of Organic Natural Products, ed. W. Herz, H. Grisebach, G. W. Kirby and C. Tamm, Springer, Vienna, 1985, vol. 48, pp. 204–269 Search PubMed.
- G. A. Cordell, Phytochemistry, 1974, 13, 2343–2364 CrossRef CAS.
- J. R. Hanson, Terpenoids and Steroids, London, UK, 1974, vol. 4 Search PubMed.
- S. S. Ebada, W. Lin and P. Proksch, Mar. Drugs, 2010, 8, 313–346 CrossRef CAS PubMed.
- R. Ebel, Mar. Drugs, 2010, 8, 2340–2368 CrossRef CAS PubMed.
- P. Máximo and A. Lourenço, Curr. Org. Chem., 2018, 22, 2381–2393 CrossRef.
- A. J. Michael, Biochem. J., 2017, 474, 2277–2299 CrossRef CAS PubMed.
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2019, 36, 122–173 RSC.
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2023, 40, 275–325 RSC.
- L. Nunez-Pons and C. Avila, Nat. Prod. Rep., 2015, 32, 1114–1130 RSC.
- V. J. Paul, K. E. Arthur, R. Ritson-Williams, C. Ross and K. Sharp, Biol. Bull., 2007, 213, 226–251 CrossRef CAS PubMed.
- M. Maldonado and P. R. Bergquist, in Atlas of Marine Invertebrate Larvae, A. Press, 2002, ch. 2, pp. 21–50 Search PubMed.
- M. W. Taylor, R. Radax, D. Steger and M. Wagner, Microbiol. Mol. Biol. Rev., 2007, 71, 295–347 CrossRef CAS PubMed.
- N. S. Webster and L. L. Blackall, ISME J., 2009, 3, 1–3 CrossRef CAS PubMed.
- N. S. Webster and M. W. Taylor, Environ. Microbiol., 2012, 14, 335–346 CrossRef CAS PubMed.
- M. Hudspith, L. Rix, M. Achlatis, J. Bougoure, P. Guagliardo, P. L. Clode, N. S. Webster, G. Muyzer, M. Pernice and J. M. De Goeij, Microbiome, 2021, 9, 44 CrossRef CAS PubMed.
- L. Pita, L. Rix, B. M. Slaby, A. Franke and U. Hentschel, Microbiome, 2018, 6, 46 CrossRef CAS PubMed.
- J. M. Deutsch, M. O. Green, P. Akavaram, A. C. Davis, S. S. Diskalkar, I. A. Du Plessis, H. A. Fallon, E. M. Grason, E. G. Kauf, Z. M. Kim, J. R. Miller, A. L. Neal, T. Riera, S.-E. Stroeva, J. Tran, V. Tran, A. V. Coronado, V. V. Coronado, B. T. Wall, C. m. Yang, I. Mohanty, N. H. Abrahamse, C. J. Freeman, C. G. Easson, C. L. Fiore, A. E. Onstine, N. Djeddar, S. Biliya, A. V. Bryksin, N. Garg and V. Agarwal, Mar. Drugs, 2023, 21, 53 CrossRef CAS PubMed.
- K. Guo, Y. Liu and S.-H. Li, Nat. Prod. Rep., 2021, 38, 2293–2314 RSC.
- F. Bibi, M. Faheem, E. I. Azhar, M. Yasir, S. A. Alvi, M. A. Kamal, I. Ullah and M. I. Naseer, Curr. Drug Metab., 2017, 18, 11–15 CrossRef CAS PubMed.
- C. Avila, L. Núñez-Pons and J. Moles, in Chemical ecology: the ecological impacts of marine natural products, ed. M. P. Puglisi and M. A. Becerro, CRC Press, Boca Raton, FL, USA, 1st edn, 2018, pp. 71–163 Search PubMed.
- Q. Chen, J. Li, Y. Ma, W. Yuan, P. Zhang and G. Wang, Plant Commun., 2021, 2, 100184 CrossRef CAS PubMed.
- Microsoft Corporation, Microsoft SQL Server, https://www.microsoft.com/en-us/sql-server/sql-server-2022.
- Microsoft Corporation, Windows Server, 2022, https://learn.microsoft.com/en-us/windows-server/get-started/whats-new-in-windows-server-2022.
- K. Li and K. R. Gustafson, Nat. Prod. Rep., 2021, 38, 1251–1281 RSC.
- Y. Liu, L. Wang, J. H. Jung and S. Zhang, Nat. Prod. Rep., 2007, 24, 1401–1429 RSC.
- Y. S. Yang, I. M. Chung, S. H. Kim and A. Ahmad, Asian J. Chem., 2014, 26, 7792–7794 CrossRef.
- A. Cutignano, J. Moles, C. Avila and A. Fontana, J. Nat. Prod., 2015, 78, 1761–1764 CrossRef CAS PubMed.
- S. J. Piao, H. J. Zhang, H. Y. Lu, F. Yang, W. H. Jiao, Y. H. Yi, W. S. Chen and H. W. Lin, J. Nat. Prod., 2011, 74, 1248–1254 CrossRef CAS PubMed.
- G. Chianese, J. Silber, P. Luciano, C. Merten, D. Erpenbeck, B. Topaloglu, M. Kaiser and D. Tasdemir, J. Nat. Prod., 2017, 80, 2566–2571 CrossRef CAS PubMed.
- E. D. De Silva and P. J. Scheuer, Tetrahedron Lett., 1981, 22, 3147–3150 CrossRef CAS.
- S. Iimura, M. Oka, Y. Narita, M. Konishi, H. Kakisawa, Q. Gao and T. Oki, Tetrahedron Lett., 1993, 34, 493–496 CrossRef CAS.
- S. Nozoe, M. Morisaki, K. Tsuda, Y. Iitaka, N. Takahashi, S. Tamura, K. Ishibashi and M. Shirasaka, J. Am. Chem. Soc., 1965, 87, 4968–4970 CrossRef CAS PubMed.
- M. Liu, W. Sun, L. Shen, Y. He, J. Liu, J. Wang, Z. Hu and Y. Zhang, Angew. Chem., Int. Ed., 2019, 58, 12091–12095 CrossRef CAS PubMed.
- H. J. Zhang, H. F. Tang, Y. H. Yi and H. W. Lin, Helv. Chim. Acta, 2009, 92, 762–767 CrossRef CAS.
- Q. Li, C. Chen, M. Wei, C. Dai, L. Cheng, J. Tao, X. N. Li, J. Wang, W. Sun, H. Zhu and Y. Zhang, Org. Lett., 2019, 21, 2290–2293 CrossRef CAS PubMed.
- J. Shin, J. R. Rho, Y. Seo, H. S. Lee, K. W. Choa and C. J. Simb, Tetrahedron Lett., 2001, 42, 3005–3007 CrossRef CAS.
- G. Alea, A. Carroll and B. Bowden, Aust. J. Chem., 1994, 47, 191–194 CrossRef CAS.
- G. Cimino, P. De Luca, S. De Stefano and L. Minale, Tetrahedron, 1975, 31, 271–275 CrossRef CAS.
- A. H. Afifi, I. Kagiyama, A. H. El-Desoky, H. Kato, R. E. P. Mangindaan, N. J. de Voogd, N. M. Ammar, M. S. Hifnawy and S. Tsukamoto, J. Nat. Prod., 2017, 80, 2045–2050 CrossRef CAS PubMed.
- F. Cafieri, L. De Napoli, E. Fattorusso and C. Santacroce, Experientia, 1977, 33, 994–995 CrossRef CAS PubMed.
- S. J. Piao, W. H. Jiao, F. Yang, Y. H. Yi, Y. T. Di, B. N. Han and H. W. Lin, Mar. Drugs, 2014, 12, 4096–4109 CrossRef PubMed.
- Y. M. Xu, P. Espinosa-Artiles, M. X. Liu, A. E. Arnold and A. A. Gunatilaka, J. Nat. Prod., 2013, 76, 2330–2336 CrossRef CAS PubMed.
- E. Fattorusso, V. Lanzotti, S. Magno, L. Mayol and M. Pansini, J. Org. Chem., 1992, 57, 6921–6924 CrossRef CAS.
- A. C. Huang, S. A. Kautsar, Y. J. Hong, M. H. Medema, A. D. Bond, D. J. Tantillo and A. Osbourn, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E6005–E6014 CrossRef CAS PubMed.
- J. J. Bell, Estuar. Coast Shelf Sci., 2008, 79, 341–353 CrossRef.
- N. J. de Voogd, B. Alvarez, N. Boury-Esnault, P. Cárdenas, M.-C. Díaz, M. Dohrmann, R. Downey, C. Goodwin, E. Hajdu, J. N. A. Hooper, M. Kelly, M. Klautau, S. C. Lim, R. Manconi, C. Morrow, U. Pinheiro, A. B. Pisera, P. Ríos, K. Rützler, C. Schönberg, T. Turner, J. Vacelet, R. W. M. van Soest and J. Xavier, The World Porifera Database, https://www.marinespecies.org/porifera/, (accessed February 20th 2024).
- R. W. M. Van Soest, N. Boury-Esnault, J. Vacelet, M. Dohrmann, D. Erpenbeck, N. J. De Voogd, N. Santodomingo, B. Vanhoorne, M. Kelly and J. N. A. Hooper, PLoS One, 2012, 7, e35105 CrossRef CAS PubMed.
- U. Hentschel, J. Piel, S. M. Degnan and M. W. Taylor, Nat. Rev. Microbiol., 2012, 10, 641–654 CrossRef CAS PubMed.
- S. Zhang, W. Song, L.-F. Nothias, S. P. Couvillion, N. Webster and T. Thomas, Microbiome, 2022, 10, 22 CrossRef CAS PubMed.
- D. Varijakzhan, J.-Y. Loh, W.-S. Yap, K. Yusoff, R. Seboussi, S.-H. E. Lim, K.-S. Lai and C.-M. Chong, Mar. Drugs, 2021, 19, 246 CrossRef CAS PubMed.
- T. Hughes, A. Robinson, S. Rogers and V. Tillett, MarinLit – Dedicated to marine natural products research, https://marinlit.rsc.org, (accessed February 20th 2024).
- K. Wilson, T. De Rond, I. Burkhardt, T. S. Steele, R. J. B. Schäfer, S. Podell, E. E. Allen and B. S. Moore, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2220934120 CrossRef CAS PubMed.
- E. R. Abdelaleem, M. N. Samy, S. Y. Desoukey, M. Liu, R. J. Quinn and U. R. Abdelmohsen, RSC Adv., 2020, 10, 34959–34976 RSC.
- X.-F. Zhang, J. Ren, X.-R. Cheng, H.-Z. Jin and W.-D. Zhang, RSC Adv., 2015, 5, 1979–1982 RSC.
- E. Fattorusso, S. Magno, C. Santacroce and D. Sica, Tetrahedron, 1972, 28, 5993–5997 CrossRef CAS.
- Q. Lu and D. J. Faulkner, J. Nat. Prod., 1997, 60, 195–198 CrossRef CAS PubMed.
- G. Cimino, S. De Stefano, L. Minale and E. Trivellone, J. Chem. Soc., Perkin Trans. 1, 1977,(13), 1587–1593 RSC.
- R. C. Cambie, C. E. F. Rickard, P. S. Rutledge and X. S. Yang, Acta Crystallogr., 1999, C55, 112–114 CAS.
- C. J. Hernández-Guerrero, E. Zubía, M. J. Ortega and J. L. Carballo, Tetrahedron, 2006, 62, 5392–5400 CrossRef.
- X. Yang, Z. Shao and X. Zhang, Z. Naturforsch., 2010, 65b, 625–627 CrossRef.
- P. Crews, P. Bescansa and G. J. Bakus, Experientia, 1985, 41, 690–691 CrossRef CAS PubMed.
- P. Crews and P. Bescansa, J. Nat. Prod., 1986, 49, 1041–1052 CrossRef CAS PubMed.
- Y. Kashman and A. Rudi, Tetrahedron, 1977, 33, 2997–2998 CrossRef CAS.
- R. Kazlauskas, P. T. Murphy, R. J. Quinn and R. J. Wells, Tetrahedron Lett., 1976, 17, 2631–2634 CrossRef.
- G. R. Pettit, Z. A. Cichacz, R. Tan, M. S. Hoard, N. Melody and R. K. Pettit, J. Nat. Prod., 1998, 61, 13–16 CrossRef CAS PubMed.
- N. Tsuchiya, A. Sato, T. Hata, N. Sato, K. Sasagawa and T. Kobayashi, J. Nat. Prod., 1998, 61, 468–473 CrossRef CAS PubMed.
- H. Miyaoka, S. Nishijima, H. Mitome and Y. Yamada, J. Nat. Prod., 2000, 63, 1369–1372 CrossRef CAS PubMed.
- D. T. A. Youssef, R. K. Yamaki, M. Kelly and P. J. Scheuer, J. Nat. Prod., 2002, 65, 2–6 CrossRef CAS PubMed.
- D. T. A. Youssef, L. A. Shaala and S. Emara, J. Nat. Prod., 2005, 68, 1782–1784 CrossRef CAS PubMed.
- Y. Qiu, Z. Deng, Y. Pei, H. Fu, J. Li, P. Proksch and W. Lin, J. Nat. Prod., 2004, 67, 921–924 CrossRef CAS PubMed.
- Z. G. Yu, K. S. Bi and Y. W. Guo, Helv. Chim. Acta, 2005, 88, 1004–1009 CrossRef CAS.
- A. R. Diaz-Marrero, T. Matainaho, R. van Soest, M. Roberge and R. J. Andersen, Nat. Prod. Rep., 2008, 22, 1304–1309 CrossRef CAS PubMed.
- L. Yin, H. Li, X. Chen and Y. Qiu, Rec. Nat. Prod., 2014, 8, 417–421 Search PubMed.
- S. S. Elhady, A. M. El-Halawany, A. M. Alahdal, H. A. Hassanean and S. A. Ahmed, Molecules, 2016, 21(82), 81–89 Search PubMed.
- A. M. Alahdal, H. Z. Asfour, S. A. Ahmed, A. O. Noor, A. M. Al-Abd, M. A. Elfaky and S. S. Elhady, Molecules, 2018, 23, 1–11 CrossRef PubMed.
- P. Francis and K. Chakraborty, Med. Chem. Res., 2021, 30, 886–896 CrossRef CAS.
- M.-Z. Su, Q. Zhang, L.-G. Yao, B. Wu and Y.-W. Guo, J. Asian Nat. Prod. Res., 2022, 1–7, DOI:10.1080/10286020.2022.2150614.
- H. N. K. Tran, M. J. Kim and Y.-J. Lee, Mar. Drugs, 2022, 20, 604 CrossRef CAS PubMed.
- S. Jaisamut, S. Thengyai, S. Yuenyongsawad, C. Karalai, A. Plubrukarn and K. Suwanborirux, Pure Appl. Chem., 2009, 81, 1019–1026 CrossRef CAS.
- C. Festa, C. Cassiano, M. V. D'Auria, C. Debitus, M. C. Monti and S. De Marino, Org. Biomol. Chem., 2014, 12, 8646–8655 RSC.
- F. Yang, J.-H. Gan, X.-Y. Liu and H.-W. Lin, Nat. Prod. Commun., 2014, 9, 1934578X1400900608 CrossRef.
- W. Kaweetripob, C. Mahidol, P. Tuntiwachwuttikul, S. Ruchirawat and H. Prawat, Mar. Drugs, 2018, 16(474), 471–415 Search PubMed.
- J. Li, L. Du, M. Kelly, Y. D. Zhou and D. G. Nagle, J. Nat. Prod., 2013, 76, 1492–1497 CrossRef CAS PubMed.
- J. C. Braekman and D. Daloze, Pure Appl. Chem., 1986, 58, 357–364 CrossRef CAS.
- J. C. Braekman, D. Daloze, M. Kaisin and B. Moussiaux, Tetrahedron, 1985, 41, 4603–4614 CrossRef CAS.
- R. J. Quinn and D. J. Tucker, Aust. J. Chem., 1989, 42, 751–755 CrossRef CAS.
- D. E. Williams, I. Hollander, L. Feldberg, E. Frommer, R. Mallon, A. Tahir, R. van Soest and R. J. Andersen, J. Nat. Prod., 2009, 72, 1106–1109 CrossRef CAS PubMed.
- F. Cao, Z. H. Wu, C. L. Shao, S. Pang, X. Y. Liang, N. J. de Voogd and C. Y. Wang, Org. Biomol. Chem., 2015, 13, 4016–4024 RSC.
- H. Kikuchi, Y. Tsukitani, I. Shimizu, M. Kobayashi and I. Kitagawa, Chem. Pharm. Bull., 1981, 29, 1492–1494 CrossRef CAS.
- H. Kikuchi, Y. Tsukitani, I. Shimizu, M. Kobayashi and I. Kitagawa, Chem. Pharm. Bull., 1983, 31, 552–556 CrossRef CAS.
- I. Kitagawa, M. Kobayashi, N.-K. Lee, Y. Oyama and Y. Kyogoku, Chem. Pharm. Bull., 1989, 37, 2078–2082 CrossRef CAS.
- X. Fu, L. Zeng, J. Su, M. Païs and P. Potier, J. Nat. Prod., 1993, 56, 1985–1988 CrossRef CAS.
- X. Fu, L. Zeng, J. Su and F. J. Schmitz, J. Nat. Prod., 1999, 62, 644–646 CrossRef CAS PubMed.
- L. Zeng, X. Fu, J. Su, E. O. Pordesimo, S. C. Traeger and F. J. Schmitz, J. Nat. Prod., 1991, 54, 421–427 CrossRef CAS.
- H. J. Zhang, Y. H. Yi, F. Yang, W. S. Chen and H. W. Lin, Molecules, 2010, 15, 834–841 CrossRef CAS PubMed.
- M. Jaspars, E. Jackson, E. Lobkovsky, J. Clardy, M. C. Diaz and P. Crews, J. Nat. Prod., 1997, 60, 556–561 CrossRef CAS PubMed.
- L. C. Chang, S. Otero-Quintero, G. M. Nicholas and C. A. Bewely, Tetrahedron, 2001, 57, 5731–5738 CrossRef CAS.
- M. C. Roy, J. Tanaka, N. de Voogd and T. Higa, J. Nat. Prod., 2002, 65, 1838–1842 CrossRef CAS PubMed.
- M. H. A. Hassan, M. E. Rateb, M. Hetta, T. A. Abdelaziz, M. A. Sleim, M. Jaspars and R. Mohammed, Tetrahedron, 2015, 71, 577–583 CrossRef CAS.
- S. Hayes, A. C. Taki, K. Y. Lum, J. J. Byrne, J. M. White, M. G. Ekins, R. B. Gasser and R. A. Davis, J. Nat. Prod., 2022, 85, 1723–1729 CrossRef CAS PubMed.
- L. P. Ponomarenko, A. I. Kalinovsky and V. A. Stonik, J. Nat. Prod., 2004, 67, 1507–1510 CrossRef CAS PubMed.
- H. J. Li, T. Amagata, K. Tenney and P. Crews, J. Nat. Prod., 2007, 70, 802–807 CrossRef CAS PubMed.
- H. Zhang, P. Crews, K. Tenney and F. A. Valeriote, Med. Chem., 2017, 13, 295–300 CrossRef CAS PubMed.
- P. Karuso, R. C. Cambie, B. F. Bowden and P. R. Bergquist, J. Nat. Prod., 1989, 52, 289–293 CrossRef CAS.
- M. J. Somerville, J. N. A. Hooper and M. J. Garson, J. Nat. Prod., 2006, 69, 1587–1590 CrossRef CAS PubMed.
- M. M. Kumar, N. Krishna, P. Muralidhar, B. S. Sastry and D. V. Rao, Indian J. Chem., 2008, 47B, 325–328 CAS.
- J. E. Jeon, J. Bae, K. J. Lee, K. B. Oh and J. Shin, J. Nat. Prod., 2011, 74, 847–851 CrossRef CAS PubMed.
- D. B. Abdjul, H. Yamazaki, O. Takahashi, R. Kirikoshi, R. E. Mangindaan and M. Namikoshi, Bioorg. Med. Chem. Lett., 2015, 25, 904–907 CrossRef CAS PubMed.
- R. Kazlauskas, P. T. Murphy and R. J. Wells, Aust. J. Chem., 1982, 35, 51–59 CrossRef CAS.
- K. A. Alvi and P. Crews, J. Nat. Prod., 1992, 55, 859–865 CrossRef CAS PubMed.
- Y. Sera, K. Adachi and Y. Shizuri, J. Nat. Prod., 1999, 62, 152–154 CrossRef CAS PubMed.
- C. C. Stessman, R. Ebel, A. J. Corvino and P. Crews, J. Nat. Prod., 2002, 65, 1183–1186 CrossRef CAS PubMed.
- L. Chill, A. Rudi, M. Aknin, S. Loya, A. Hizi and Y. Kashman, Tetrahedron, 2004, 60, 10619–10626 CrossRef CAS.
- J. Dai, Y. Liu, Y.-D. Zhou and D. G. Nagle, J. Nat. Prod., 2007, 70, 1824–1826 CrossRef CAS PubMed.
- Y. Liu, R. Liu, S. C. Mao, J. B. Morgan, M. B. Jekabsons, Y. D. Zhou and D. G. Nagle, J. Nat. Prod., 2008, 71, 1854–1860 CrossRef CAS PubMed.
- B. R. Peng, K. H. Lai, Y. Y. Chen, J. H. Su, Y. M. Huang, Y. H. Chen, M. C. Lu, S. S. Yu, C. Y. Duh and P. J. Sung, Mar. Drugs, 2020, 18, 76–86 CrossRef CAS PubMed.
- G. Cimino, S. De Stefano and L. Minale, Tetrahedron, 1972, 28, 5983–5991 CrossRef CAS.
- G. Cimino, S. De Stefano, L. Minale and E. Fattorusso, Tetrahedron, 1972, 28, 267–273 CrossRef CAS.
- G. Cimino, S. De Stefano and A. Di Luccia, Experientia, 1979, 35, 1277–1278 CrossRef CAS.
- B. Terem and P. J. Scheuer, Tetrahedron, 1986, 42, 4409–4412 CrossRef CAS.
- A. De Giulio, S. De Rosa, G. Di Vincenzo and N. Zavodnik, J. Nat. Prod., 1989, 52, 1258–1262 CrossRef CAS.
- R. Davis and R. J. Capon, Aust. J. Chem., 1993, 46, 1295–1299 CrossRef CAS.
- H. He, P. Kulanthaivel and B. J. Baker, Tetrahedron Lett., 1994, 35, 7189–7192 CrossRef CAS.
- P. A. Searle and T. F. Molinski, Tetrahedron, 1994, 50(33), 9893–9990 CrossRef CAS.
- A. Rueda, E. Zubía, M. J. Ortega, J. L. Carballo and J. Salvá, J. Nat. Prod., 1998, 61, 258–261 CrossRef CAS PubMed.
- S. De Marino, M. Iorizzi, F. Zollo, C. Debitus, J.-L. Menou, L. F. Ospina, M. J. Alcaraz and M. Payá, J. Nat. Prod., 2000, 63, 322–326 CrossRef CAS PubMed.
- S. Tsukamoto, S. Miura, R. W. M. van Soest and T. Ohta, J. Nat. Prod., 2003, 66, 438–440 CrossRef CAS PubMed.
- S. J. Nam, H. Ko, M. K. Ju, H. Hwang, J. Chin, J. Ham, B. Lee, J. Lee, D. H. Won, H. Choi, J. Ko, K. Shin, T. Oh, S. Kim, J. R. Rho and H. Kang, J. Nat. Prod., 2007, 70, 1691–1695 CrossRef CAS PubMed.
- S. J. Nam, H. Ko, M. Shin, J. Ham, J. Chin, Y. Kim, H. Kim, K. Shin, H. Choi and H. Kang, Bioorg. Med. Chem. Lett., 2006, 16, 5398–5402 CrossRef CAS PubMed.
- E. Manzo, M. L. Ciavatta, G. Villani, M. Varcamonti, S. M. Sayem, R. van Soest and M. Gavagnin, J. Nat. Prod., 2011, 74, 1241–1247 CrossRef CAS PubMed.
- C.-S. Phan, T. Kamada, T. Hamada and C. S. Vairappan, Rec. Nat. Prod., 2018, 12, 643–647 CrossRef CAS.
- I. Yang, J. Lee, J. Lee, D. Hahn, J. Chin, D. H. Won, J. Ko, H. Choi, A. Hong, S. J. Nam and H. Kang, Molecules, 2018, 23(3187), 3181–3110 Search PubMed.
- C. Hertzer, S. Kehraus, N. Böhringer, F. Kaligis, R. Bara, D. Erpenbeck, G. Wörheide, T. F. Schäberle, H. Wägele and G. M. König, Beilstein J. Org. Chem., 2020, 16, 1596–1605 CrossRef CAS PubMed.
- J. R. Rho, H. S. Lee, H. J. Shin, J. W. Ahn, J. Y. Kim, C. J. Sim and J. Shin, J. Nat. Prod., 2004, 67, 1748–1751 CrossRef CAS PubMed.
- J. Song, W. Jeong, N. Wang, H.-S. Lee, C. J. Sim, K.-B. Oh and J. Shin, J. Nat. Prod., 2008, 71, 1866–1871 CrossRef CAS PubMed.
- P. R. Bergquist, R. C. Cambie and M. R. Kernan, Biochem. Syst. Ecol., 1990, 18, 349–357 CrossRef CAS.
- B. F. Bowden, J. C. Coll, H. Li, R. C. Cambie, M. R. Kernan and P. R. Bergquist, J. Nat. Prod., 1992, 55, 1234–1240 CrossRef CAS PubMed.
- J. I. Jiménez, W. Y. Yoshida, P. J. Scheuer, E. Lobkovsky, J. Clardy and M. Kelly, J. Org. Chem., 2000, 65, 6837–6840 CrossRef PubMed.
- Y. Kashman and M. Zviely, Tetrahedron Lett., 1979, 20, 3879–3882 CrossRef.
- R. P. Walker, J. E. Thompson and D. J. Faulkner, J. Org. Chem., 1980, 45, 4976–4979 CrossRef CAS.
- G. Cimino, S. De Stefano and L. Minale, Experientia, 1974, 30, 846–847 Search PubMed.
- G. Cimino, A. Fontana, F. Giménez, A. Marin, E. Mollo, E. Trivellone and E. Zubía, Experientia, 1993, 49, 582–586 CrossRef CAS.
- F. Cafieri, L. De Napoli, E. Fattorusso, C. Santacroce and D. Sica, Tetrahedron Lett., 1977, 18, 477–480 CrossRef.
- F. Cafieri, L. De Napoli, E. Fattorusso, C. Santacroce and D. Sica, Gazz. Chim. Ital., 1977, 107, 71–74 CAS.
- F. Cafieri, L. De Napoli, A. Iengo and C. Santacroce, Experientia, 1978, 34, 300–301 CrossRef CAS.
- F. Cafieri, L. De Napoli, A. Iengo and C. Santacroce, Experientia, 1979, 35, 157–158 CrossRef CAS.
- M. Puliti, C. A. Mattia and L. Mazzarella, Acta Crystallogr., 1995, C51, 1703–1707 Search PubMed.
- S. De Rosa, S. De Stefano and N. Zavodnik, J. Org. Chem., 1988, 53, 5020–5023 CrossRef CAS.
- S. Khushi, L. Nahar, A. A. Salim and R. J. Capon, Mar. Drugs, 2018, 16, 456 CrossRef CAS PubMed.
- S. Wonganuchitmeta, S. Yuenyongsawad, N. Keawpradub and A. Plubrukarn, J. Nat. Prod., 2004, 67, 1767–1770 CrossRef CAS PubMed.
- C. Mahidol, H. Prawat, S. Sangpetsiripan and S. Ruchirawat, J. Nat. Prod., 2009, 72, 1870–1874 CrossRef CAS PubMed.
- Q. Wang, Y. Sun, L. Yang, X. Luo, N. J. de Voogd, X. Tang, P. Li and G. Li, J. Nat. Prod., 2020, 83, 516–523 CrossRef CAS PubMed.
- A. Rudi, T. Yosief, M. Schleyer and Y. Kashman, Org. Lett., 1999, 1, 471–472 CrossRef CAS PubMed.
- S. P. Gunasekera, P. J. McCarthy, M. Kelly-Borges, E. Lobkovsky and J. Clardy, J. Am. Chem. Soc., 1996, 118, 8759–8760 CrossRef CAS.
- H. Hagiwara and H. Uda, J. Chem. Soc., Chem. Commun., 1988, 815–817, 10.1039/C39880000815.
- D. Lee, J. Shin, K. M. Yoon, T. I. Kim, S. H. Lee, H. S. Lee and K. B. Oh, Bioorg. Med. Chem. Lett., 2008, 18, 5377–5380 CrossRef CAS PubMed.
- L. Murray, H. Hamit, J. Hooper, L. Hobbs and R. Capon, Aust. J. Chem., 1995, 48, 1899–1902 CrossRef CAS.
- D. Hahn, D. H. Won, B. Mun, H. Kim, C. Han, W. Wang, T. Chun, S. Park, D. Yoon, H. Choi, S. J. Nam, M. Ekins, J. Chin and H. Kang, Bioorg. Med. Chem. Lett., 2013, 23, 2336–2339 CrossRef CAS PubMed.
- K. Choi, J. Hong, C.-O. Lee, D.-k. Kim, C. J. Sim, K. S. Im and J. H. Jung, J. Nat. Prod., 2004, 67, 1186–1189 CrossRef CAS PubMed.
- W. Balansa, R. Islam, F. Fontaine, A. M. Piggott, H. Zhang, X. Xiao, T. I. Webb, D. F. Gilbert, J. W. Lynch and R. J. Capon, Org. Biomol. Chem., 2013, 11, 4695–4701 RSC.
- Y. C. Chang, S. W. Tseng, L. L. Liu, Y. Chou, Y. S. Ho, M. C. Lu and J. H. Su, Mar. Drugs, 2012, 10, 987–997 CrossRef CAS PubMed.
- S. Rochfort, D. Atkin, L. Hobbs and R. Capon, J. Nat. Prod., 1996, 59, 1024–1028 CrossRef CAS.
- H. S. Lee, T. H. Lee, S. H. Yang, H. J. Shin, J. Shin and K. B. Oh, Bioorg. Med. Chem. Lett., 2007, 17, 2483–2486 CrossRef CAS PubMed.
- W.-H. Jiao, L.-L. Hong, J.-B. Sun, S.-J. Piao, G.-D. Chen, H. Deng, S.-P. Wang, F. Yang and H.-W. Lin, Eur. J. Org Chem., 2017, 2017, 3421–3426 CrossRef CAS.
- G. Cimino, S. De Stefano, L. Minale and R. Riccio, Tetrahedron Lett., 1979, 38, 3619–3622 CrossRef.
- J. K. Woo, C. K. Kim, S. H. Kim, H. Kim, D. C. Oh, K. B. Oh and J. Shin, Org. Lett., 2014, 16, 2826–2829 CrossRef CAS PubMed.
- J.-R. Rho, B. S. Hwang, S. Joung, M. R. Byun, J.-H. Hong and H.-Y. Lee, Org. Lett., 2011, 13, 884–887 CrossRef CAS PubMed.
- J. R. Rho, B. S. Hwang, C. J. Sim, S. Joung, H. Y. Lee and H. J. Kim, Org. Lett., 2009, 11, 5590–5593 CrossRef CAS PubMed.
- J. Daoust, M. Chen, M. Wang, D. E. Williams, M. A. Garcia Chavez, Y. A. Wang, C. E. Merchant, A. Fontana, T. J. Kieffer and R. J. Andersen, J. Org. Chem., 2013, 78, 8267–8273 CrossRef CAS PubMed.
- J. Daoust, A. Fontana, C. E. Merchant, N. J. De Voogd, B. O. Patrick, T. J. Kieffer and R. J. Andersen, Org. Lett., 2010, 12, 3208–3211 CrossRef CAS PubMed.
- W. Wang, Y. Lee, T. G. Lee, B. Mun, A. G. Giri, J. Lee, H. Kim, D. Hahn, I. Yang, J. Chin, H. Choi, S.-J. Nam and H. Kang, Org. Lett., 2012, 14, 4486–4489 CrossRef CAS PubMed.
- M. Wang, I. Tietjen, M. Chen, D. E. Williams, J. Daoust, M. A. Brockman and R. J. Andersen, J. Org. Chem., 2016, 81, 11324–11334 CrossRef CAS PubMed.
- M. Wang, A. Sciorillo, S. Read, D. N. Divsalar, K. Gyampoh, G. Zu, Z. Yuan, K. Mounzer, D. E. Williams, L. J. Montaner, N. de Voogd, I. Tietjen and R. J. Andersen, J. Nat. Prod., 2022, 85, 1274–1281 CrossRef CAS PubMed.
- Y. Lee, W. Wang, H. Kim, A. G. Giri, D. H. Won, D. Hahn, K. R. Baek, J. Lee, I. Yang, H. Choi, S. J. Nam and H. Kang, Bioorg. Med. Chem. Lett., 2014, 24, 4095–4098 CrossRef CAS PubMed.
- H. Solanki, C. Angulo-Preckler, K. Calabro, N. Kaur, P. Lasserre, B. Cautain, M. de la Cruz, F. Reyes, C. Avila and O. P. Thomas, Tetrahedron Lett., 2018, 59, 3353–3356 CrossRef CAS.
- W. Wang, B. Mun, Y. Lee, M. Venkat Reddy, Y. Park, J. Lee, H. Kim, D. Hahn, J. Chin, M. Ekins, S. J. Nam and H. Kang, J. Nat. Prod., 2013, 76, 170–177 CrossRef CAS PubMed.
- D. T. A. Youssef, W. Y. Yoshida, M. Kelly and P. J. Scheuer, J. Nat. Prod., 2001, 64, 1332–1335 CrossRef CAS PubMed.
- D. T. A. Youssef, J. Nat. Prod., 2004, 67, 112–114 CrossRef CAS PubMed.
- J. Dai, Y. Liu, Y.-D. Zhou and D. G. Nagle, J. Nat. Prod., 2007, 70, 130–133 CrossRef CAS PubMed.
- S. R. M. Ibrahim, R. Ebel, V. Wray, W. E. G. Müller, R. A. Edrada-Ebel and P. Proksch, J. Nat. Prod., 2008, 71, 1358–1364 CrossRef CAS PubMed.
- S. Sperry, F. A. Valeriote, T. H. Corbett and P. Crews, J. Nat. Prod., 1998, 61, 241–247 CrossRef CAS PubMed.
- P. Phuwapraisirisan, S. Matsunaga, N. Fusetani, N. Chaitanawisuti, S. Kritsanapuntu and P. Menasveta, J. Nat. Prod., 2003, 66, 289–291 CrossRef CAS PubMed.
- J. Tanaka, T. Higa, K. Suwanborirux, U. Kokpol, G. Bernardinelli and C. W. Jefford, J. Org. Chem., 1993, 58, 2999–3002 CrossRef CAS.
- J. Bae, J. E. Jeon, Y. J. Lee, H. S. Lee, C. J. Sim, K. B. Oh and J. Shin, J. Nat. Prod., 2011, 74, 1805–1811 CrossRef CAS PubMed.
- C. K. Kim, I. H. Song, H. Y. Park, Y. J. Lee, H. S. Lee, C. J. Sim, D. C. Oh, K. B. Oh and J. Shin, J. Nat. Prod., 2014, 77, 1396–1403 CrossRef CAS PubMed.
- J.-W. Lee, H.-S. Lee, J. Shin, J. S. Kang, J. Yun, H. J. Shin, J. S. Lee and Y.-J. Lee, Arch. Pharmacal Res., 2015, 38, 1005–1010 CrossRef CAS PubMed.
- J. Kimura, E. Ishizuka, Y. Nakao, W. Y. Yoshida, P. J. Scheuer and M. Kelly-Borges, J. Nat. Prod., 1998, 61, 248–250 CrossRef CAS PubMed.
- S. De Marino, C. Festa, M. V. D'Auria, M.-L. Bourguet-Kondracki, S. Petek, C. Debitus, R. M. Andrés, M. C. Terencio, M. Payá and A. Zampella, Tetrahedron, 2009, 65, 2905–2909 CrossRef CAS.
- F. S. Di Leva, C. Festa, C. D'Amore, S. De Marino, B. Renga, M. V. D'Auria, E. Novellino, V. Limongelli, A. Zampella and S. Fiorucci, J. Med. Chem., 2013, 56, 4701–4717 CrossRef CAS PubMed.
- G. Cimino, S. De Stefano and L. Minale, Tetrahedron, 1972, 28, 331–341 Search PubMed.
- W. Balansa, R. Islam, F. Fontaine, A. M. Piggott, H. Zhang, T. I. Webb, D. F. Gilbert, J. W. Lynch and R. J. Capon, Bioorg. Med. Chem., 2010, 18, 2912–2919 CrossRef CAS PubMed.
- Y.-C. Shen, K.-L. Lo, Y.-C. Lin, A. T. Khalil, Y.-H. Kuo and P.-S. Shih, Tetrahedron Lett., 2006, 47, 4007–4010 CrossRef CAS.
- D. J. Faulkner, Tetrahedron Lett., 1973, 39, 3821–3822 CrossRef.
- I. Rothberg and P. Shubiak, Tetrahedron Lett., 1975, 16, 769–772 CrossRef.
- A. Martinez, C. Duque, N. Sato and Y. Fujimoto, Chem. Pharm. Bull., 1997, 45, 181–184 CrossRef CAS.
- S. De Rosa, A. Milone, A. De Giulio, A. Crispino and C. Iodice, Nat. Prod. Lett., 1996, 8, 245–251 CrossRef CAS.
- U. Höller, G. M. König and A. D. Wright, J. Nat. Prod., 1997, 60, 832–835 CrossRef.
- A. Fontana, I. Fakhr, E. Mollo and G. Cimino, Tetrahedron: Asymmetry, 1999, 10, 3869–3872 CrossRef CAS.
- H. B. Yu, B. B. Gu, A. Iwasaki, W. L. Jiang, A. Ecker, S. P. Wang, F. Yang and H. W. Lin, Mar. Drugs, 2020, 18(10), 491 CrossRef CAS PubMed.
- L. M. Murray, A. Johnson, M. C. Diaz and P. Crews, J. Org. Chem., 1997, 62, 5638–5641 CrossRef CAS.
- S. Tang, Y. Pei, H. Fu, Z. Deng, J. Li, P. Proksch and W. Lin, Chem. Pharm. Bull., 2006, 54, 4–8 CrossRef CAS PubMed.
- J. i. Kobayashi, K. Yuasa, T. Kobayashi, T. Sasaki and M. Tsuda, Tetrahedron, 1996, 52, 5745–5750 CrossRef CAS.
- E. Elkhayat, R. Edrada, R. Ebel, V. Wray, R. van Soest, S. Wiryowidagdo, M. H. Mohamed, W. E. G. Müller and P. Proksch, J. Nat. Prod., 2004, 67, 1809–1817 CrossRef CAS PubMed.
- M. Ohba, K. Iizuka, H. Ishibashi and T. Fujii, Tetrahedron, 1997, 53, 16977–16986 CrossRef CAS.
- R. Ueoka, Y. Nakao, S. Fujii, R. W. M. van Soest and S. Matsunaga, J. Nat. Prod., 2008, 71, 1089–1091 CrossRef CAS PubMed.
- P. Crews, C. Jiménez and M. O'Neil-Johnson, Tetrahedron, 1991, 47, 3585–3600 CrossRef CAS.
- L. Gomez Paloma, A. Randazzo, L. Minale, C. Debitus and C. Roussakis, Tetrahedron, 1997, 53, 10451–10458 CrossRef CAS.
- A. Randazzo, C. Debitus, L. Minale, P. García Pastor, M. J. Alcaraz, M. Payá and L. Gomez-Paloma, J. Nat. Prod., 1998, 61, 571–575 CrossRef CAS PubMed.
- J. Li, B. Xu, J. Cui, Z. Deng, N. J. de Voogd, P. Proksch and W. Lin, Bioorg. Med. Chem., 2010, 18, 4639–4647 CrossRef CAS PubMed.
- S. Ohta, M. Uno, M. Yoshimura, Y. Hiraga and S. lkegami, Tetrahedron Lett., 1996, 37, 2265–2266 CrossRef CAS.
- M. Yanai, S. Ohta, E. Ohta and S. Ikegami, Tetrahedron, 1998, 54, 15607–15612 CrossRef CAS.
- M. S. Butler and R. J. Capon, Aust. J. Chem., 1992, 45, 1705–1743 CrossRef CAS.
- K. F. Albizati, T. Holman, D. J. Faulkner, K. B. Glaser and R. S. Jacobs, Experientia, 1987, 43, 949–950 CrossRef CAS.
- G. M. Köenig, A. D. Wright and O. Sticher, J. Nat. Prod., 1992, 55, 174–178 CrossRef PubMed.
- P. Ettinger-Epstein, C. A. Motti, R. de Nys, A. D. Wright, C. N. Battershill and D. M. Tapiolas, J. Nat. Prod., 2007, 70, 648–651 CrossRef CAS PubMed.
- M. R. Kernan, D. J. Faulkner and R. S. Jacobs, J. Org. Chem., 1987, 52, 3081–3083 CrossRef CAS.
- M. R. Kernan, D. J. Faulkner, L. Parkanyi, J. Clardy, M. S. de Carvalho and R. S. Jacobs, Experientia, 1989, 45, 388–390 CrossRef CAS PubMed.
- J. i. Kobayashi, C.-m. Zeng, M. Ishibashi and T. Sasaki, J. Nat. Prod., 1993, 56, 436–439 CrossRef CAS PubMed.
- M. Tsuda, T. Endo, Y. Mikami, J. Fromont and J. Kobayashi, J. Nat. Prod., 2002, 65, 1507–1508 CrossRef CAS PubMed.
- T.-R. Su, M.-C. Lu, Y.-J. Wu and J.-H. Su, Bull. Chem. Soc. Jpn., 2015, 88, 176–182 CrossRef.
- P. Ahmadi, M. Higashi, N. J. Voogd and J. Tanaka, Mar. Drugs, 2017, 15, 1–8 CrossRef PubMed.
- D. Kanki, K. Imai, Y. Ise, S. Okada and S. Matsunaga, J. Nat. Prod., 2021, 84, 1676–1680 CrossRef CAS PubMed.
- Y. Liu, B. H. Bae, N. Alam, J. Hong, C. J. Sim, C.-O. Lee, K. S. Im and J. H. Jung, J. Nat. Prod., 2001, 64, 1301–1304 CrossRef CAS PubMed.
- Y. Liu, J. Hong, C. O. Lee, K. S. Im, N. D. Kim, J. S. Choi and J. H. Jung, J. Nat. Prod., 2002, 65, 1307–1314 CrossRef CAS PubMed.
- Y. Liu, T. A. Mansoor, J. Hong, C. O. Lee, C. J. Sim, K. S. Im, N. D. Kim and J. H. Jung, J. Nat. Prod., 2003, 66, 1451–1456 CrossRef CAS PubMed.
- J.-K. Woo, J. E. Jeon, B. Kim, C. Sim, D.-C. Oh, K.-B. Oh and J. Shin, Nat. Prod. Sci., 2015, 21, 237 CrossRef CAS.
- W. He, X. Lin, T. Xu, J. H. Jung, H. Yin, B. Yang and Y. Liu, Chem. Nat. Compd., 2012, 48, 208–210 CrossRef CAS.
- C. J. Barrow, J. W. Blunt, M. H. G. Munro and N. B. Perry, J. Nat. Prod., 1988, 51, 275–281 CrossRef CAS.
- P. Prasad, A. Zhang, A. A. Salim and R. J. Capon, Fitoterapia, 2018, 126, 83–89 CrossRef CAS PubMed.
- J. Shin, Y. Seo, J.-R. Rho, E. Baek, B.-M. Kwon, T.-S. Jeong and S.-H. Bok, J. Org. Chem., 1995, 60, 7582–7588 CrossRef CAS.
- H. S. Lee, J. W. Ahn, Y. H. Lee, J. R. Rho and J. Shin, J. Nat. Prod., 2004, 67, 672–674 CrossRef CAS PubMed.
- A. R. Díaz-Marrero, I. Brito, M. Cueto, A. San-Martín and J. Darias, Tetrahedron Lett., 2004, 45, 4707–4710 CrossRef.
- R. D. Charan, T. C. McKee and M. R. Boyd, J. Nat. Prod., 2001, 64, 661–663 CrossRef CAS PubMed.
- R. C. Cambie, P. A. Craw, P. R. Bergquist and P. Karuso, J. Nat. Prod., 1988, 51, 331–334 CrossRef.
- C. Audoin, D. Bonhomme, J. Ivanisevic, M. de la Cruz, B. Cautain, M. C. Monteiro, F. Reyes, L. Rios, T. Perez and O. P. Thomas, Mar. Drugs, 2013, 11, 1477–1489 CrossRef CAS PubMed.
- W. F. Ponder and D. R. Lindberg, Zool. J. Linn. Soc., 1997, 119, 83–265 CrossRef.
- H. Wägele, A. Klussmann-Kolb, E. Verbeek and M. Schrödl, Org. Divers. Evol., 2014, 14, 133–149 CrossRef.
- H. Wägele and A. Klussmann-Kolb, Front. Zool., 2005, 2, 1–18 CrossRef PubMed.
- K. Göbbeler and A. Klussmann-Kolb, Thalassas, 2011, 27, 121–153 Search PubMed.
- M. Medina, S. Lal, Y. Vallès, T. L. Takaoka, B. A. Dayrat, J. L. Boore and T. Gosliner, Mar. Genomics, 2011, 4, 51–59 CrossRef PubMed.
- C. Avila, L. Núñez-Pons and J. Moles, in Chemical Ecology, ed. M. P. Puglisi and M. A. Becerro, CRC Press, 1st edn, 2018, pp. 71–163 Search PubMed.
- H. Wagele, Org. Divers. Evol., 2004, 4, 175–188 CrossRef.
- B. Burghardt, J. Evertsen, G. Johnsen and H. Wägele, Symbiosis, 2005, 38, 227–250 Search PubMed.
- P. G. Greenwood and R. N. Mariscal, Tissue Cell, 1984, 16, 719–730 CrossRef CAS PubMed.
- G. Cimino and M. T. Ghiselin, Chemoecology, 1999, 9, 187–207 CrossRef CAS.
- C. Avila, Oceanogr. Mar. Biol. Annu. Rev., 1995, 33, 487–559 Search PubMed.
- C. Avila and V. Paul, Mar. Ecol. Prog. Ser., 1997, 150, 171–180 CrossRef CAS.
- A. Fontana, F. Giménez, A. Marin, E. Mollo and G. Cimino, Experientia, 1994, 50, 510–516 CrossRef CAS.
- T. M. Gosliner, Boll. Malacol., 2001, 37, 163–170 Search PubMed.
- C. Avila, Mar. Drugs, 2020, 18, 162 CrossRef CAS.
- R. S. Jacobs, M. A. Bober, I. Pinto, A. B. Williams, P. B. Jacobson and M. S. De Carvalho, in Pharmaceutical and Bioactive Natural Products, ed. D. H. Attaway and O. R. Zaborsky, Springer US, Boston, MA, 1993, pp. 77–99 Search PubMed.
- A. Fontana, E. Mollo, J. Ortea, M. Gavagnin and G. Cimino, J. Nat. Prod., 2000, 63, 527–530 CrossRef CAS PubMed.
- J. E. Hochlowski, D. J. Faulkner, L. S. Bass and J. Clardy, J. Org. Chem., 1983, 48, 1738–1740 CrossRef CAS.
- M. R. Kernan, E. B. Barrabee and D. John Faulkner, Comp. Biochem. Physiol., B, 1988, 89, 275–278 CrossRef.
- A. Fontana, P. Cavaliere, N. Ungur, L. D'Souza, P. S. Parameswaram and G. Cimino, J. Nat. Prod., 1999, 62, 1367–1370 CrossRef CAS PubMed.
- K. A. El Sayed, P. Bartyzel, X. Shen, T. L. Perry, J. K. Zjawiony and M. T. Hamann, Tetrahedron, 2000, 56, 949–953 CrossRef CAS.
- A. E. Winters, A. M. White, A. S. Dewi, I. W. Mudianta, N. G. Wilson, L. C. Forster, M. J. Garson and K. L. Cheney, J. Chem. Ecol., 2018, 44, 384–396 CrossRef CAS PubMed.
- K. W. L. Yong, I. W. Mudianta, K. L. Cheney, E. Mollo, J. T. Blanchfield and M. J. Garson, J. Nat. Prod., 2015, 78, 421–430 CrossRef CAS PubMed.
- J. Hellou, R. J. Andersen, S. Rafii, E. Arnold and J. Clardy, Tetrahedron Lett., 1981, 22, 4173–4176 CrossRef CAS.
- J. Pika and D. John Faulkner, Tetrahedron, 1995, 51, 8189–8198 CrossRef CAS.
- T. Miyamoto, K. Sakamoto, H. Amano, Y. Arakawa, Y. Nagarekawa, T. Komori, R. Higuchi and T. Sasaki, Tetrahedron, 1999, 55, 9133–9142 CrossRef CAS.
- K. McPhail, M. T. Davies-Coleman and P. Coetzee, J. Nat. Prod., 1998, 61, 961–964 CrossRef CAS PubMed.
- J. Moles, H. Wägele, A. Cutignano, A. Fontana and C. Avila, Mar. Biol., 2016, 163, 54 CrossRef.
- J. E. N. Veron, Coral in Space and Time The Biogeography and Evolution of the Scleractinia, Cornell University Press, Ithaca, NY, 1995 Search PubMed.
- S. K. Davy, D. Allemand and V. M. Weis, Microbiol. Mol. Biol. Rev., 2012, 76, 229–261 CrossRef CAS PubMed.
- E. Ballesteros, Oceanogr. Mar. Biol. Annu. Rev., 2006, 44, 123–195 Search PubMed.
- R. G. Waller, in Cold-Water Corals and Ecosystems, ed. A. Freiwald and J. M. Roberts, Springer Berlin Heidelberg, Berlin, Heidelberg, 2005, pp. 691–700 Search PubMed.
- Z. Dubinsky and N. Stambler, Coral reefs: an ecosystem in transition, Springer Netherlands, Dordrecht, 2011 Search PubMed.
- A. Fontana, M. L. Ciavatta and G. Cimino, J. Org. Chem., 1998, 63, 2845–2849 CrossRef CAS.
- T. Noda, C. Kitamura, S. Takahashi, K. Takagi, T. Kashio and M. Tanaka, Appl. Entomol. Zool., 1982, 17, 350–357 CrossRef.
- T. Ríos and S. Pérez C, J. Chem. Soc. D, 1969, 214–215, 10.1039/C29690000214.
- S. Nozoe, M. Morisaki, K. Fukushlma and S. Okuda, Tetrahedron Lett., 1968, 42, 4457–4458 CrossRef.
- L. Quijano, J. S. Calderón and T. Ríos, Chem. Lett., 1979, 1387–1388 CrossRef CAS.
- R. Kazlauskas, P. T. Murphy, R. J. Quinn and R. J. Wells, Tetrahedron Lett., 1976, 30, 2635–2636 CrossRef.
- R. Veloz, L. Quijano, J. S. Calderon and T. Rios, J. Chem. Soc., Chem. Commun., 1975, 191–192 RSC.
- F. Miyamoto, H. Naoki, T. Takemoto and Y. Naya, Tetrahedron, 1979, 35, 1913–1917 CrossRef CAS.
- J. K. Pawlak, M. S. Tempesta, T. Iwashita, K. Nakanishi and Y. Naya, Chem. Lett., 1983, 1069–1072 CrossRef CAS.
- M. Toki, T. Ooi and T. Kusumi, J. Nat. Prod., 1999, 62, 1504–1509 CrossRef CAS.
- T. Rios and F. Colunga, Chem. Ind., 1965, 26, 1184–1185 Search PubMed.
- Y. Iitaka, I. Watanabe, I. T. Harrison and S. Harrison, J. Am. Chem. Soc., 1968, 90, 1092–1093 CrossRef CAS.
- T. Rios and L. Quijano, Tetrahedron Lett., 1969, 17, 1317–1318 CrossRef PubMed.
- T. Rios and F. Gomez, Tetrahedron Lett., 1969, 34, 2929–2930 CrossRef PubMed.
- J. S. Calderon, L. Quijano and T. Rios, Chem. Ind., 1978, 15, 584–585 Search PubMed.
- J. S. Calderon, L. Quijano and T. Rios, Experientia, 1978, 34, 421–422 CrossRef CAS.
- R. K. Boeckman, D. M. Blum, E. V. Arnold and J. Clardy, Tetrahedron Lett., 1979, 20, 4609–4612 CrossRef.
- R. K. Boeckman, D. M. Blum and S. D. Arthur, J. Am. Chem. Soc., 1979, 101, 5060–5062 CrossRef CAS.
- G. Brochere-Ferreol and J. Polonsky, Bull. Soc. Chim. Fr., 1960, 963–967 CAS.
- R. Scartazzini, Doctoral thesis, ETH, 1966, DOI:10.3929/ethz-a-000088868.
- Y. Naya, K. Yoshihara, T. Iwashita, H. Komura, K. Nakanishi and Y. Hata, J. Am. Chem. Soc., 1981, 103, 7009–7011 CrossRef CAS.
- P. Zhang, J. Qi, Y. Duan, J. M. Gao and C. Liu, J. Fungi, 2022, 8, 1080 CrossRef CAS PubMed.
- R. Lucking, M. C. Aime, B. Robbertse, A. N. Miller, T. Aoki, H. A. Ariyawansa, G. Cardinali, P. W. Crous, I. S. Druzhinina, D. M. Geiser, D. L. Hawksworth, K. D. Hyde, L. Irinyi, R. Jeewon, P. R. Johnston, P. M. Kirk, E. Malosso, T. W. May, W. Meyer, H. R. Nilsson, M. Opik, V. Robert, M. Stadler, M. Thines, D. Vu, A. M. Yurkov, N. Zhang and C. L. Schoch, Nat. Microbiol., 2021, 6, 540–548 CrossRef PubMed.
- T. A. Richards, G. Leonard and J. G. Wideman, Microbiol. Spectr., 2017, 5 DOI:10.1128/microbiolspec.FUNK-0044-2017.
- D. L. Hawksworth and R. Lucking, Microbiol. Spectr., 2017, 5 DOI:10.1128/microbiolspec.funk-0052-2016.
- Q. Dai, F. L. Zhang and T. Feng, J. Fungi, 2021, 7, 1026 CrossRef CAS PubMed.
- W. Tian, Z. Deng and K. Hong, Mar. Drugs, 2017, 15(229), 221 Search PubMed.
- K. Ishibashi, J. Agric. J. Agric. Chem. Soc. Jpn., 1962, 36, 226–228 CAS.
- F. Sugawara, G. Strobel, R. N. Strange, J. N. Siedow, G. D. Van Duyne and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 3081–3085 CrossRef CAS PubMed.
- M. Orsenigo, Phytopathol. Z., 1957, 29, 189–196 Search PubMed.
- M. Orsenigo and D. Pavan, Ann. Facul. Agric., 1958, 6, 19–54 Search PubMed.
- M. Nakamura, K. Ishibashi and J. Agric, J. Agric. Chem. Soc. Jpn., 1958, 32, 739–744 CAS.
- L. Canonica, A. Fiecchi, M. G. Kienle and A. Scala, Tetrahedron Lett., 1966, 7, 1211–1218 CrossRef.
- L. Radics, M. Kajtar-Peredy, S. Nozoe and H. Kobayashi, Tetrahedron Lett., 1975, 49, 4415–4418 CrossRef.
- J. Z. Xiao, M. Tsuda, N. Doke and S. Nishimura, Phytopathology, 1991, 81, 58–64 CrossRef CAS.
- P. Phuwapraisirisan, K. Sawang, P. Siripong and S. Tip-pyang, Tetrahedron Lett., 2007, 48, 5193–5195 CrossRef CAS.
- C.-H. Yun, F. Sugawara and G. A. Strobel, Plant Sci., 1988, 54, 237–243 CrossRef CAS.
- D. R. Bhatia, P. Dhar, V. Mutalik, S. K. Deshmukh, S. A. Verekar, D. C. Desai, R. Kshirsagar, P. Thiagarajan and V. Agarwal, Nat. Prod. Rep., 2016, 30, 1455–1458 CrossRef CAS PubMed.
- L. M. Pena-Rodriguez and W. S. Chilton, J. Nat. Prod., 1989, 52, 1170–1172 CrossRef CAS.
- E. Li, A. M. Clark, D. P. Rotella and C. D. Hufford, J. Nat. Prod., 1995, 58, 74–81 CrossRef CAS PubMed.
- X. Shen, S. B. Krasnoff, S.-W. Lu, C. D. Dunbar, J. O'Neal, B. G. Turgeon, O. C. Yoder, D. M. Gibson and M. T. Hamann, J. Nat. Prod., 1999, 62, 895–897 CrossRef CAS PubMed.
- H. Ohkawa and T. Tamura, Agric. Biol. Chem., 1966, 30, 285–291 CAS.
- A. Evidente, A. Andolfi, A. Cimmino, M. Vurro, M. Fracchiolla and R. Charudattan|, J. Agric. Food Chem., 2006, 54, 1779–1783 CrossRef CAS PubMed.
- A. Evidente, A. Andolfi, A. Cimmino, M. Vurro, M. Fracchiolla, R. Charudattan and A. Motta, Phytochemistry, 2006, 67, 2281–2287 CrossRef CAS PubMed.
- K. Ishibashi, J. Antibiot., 1962, 15, 88–92 CAS.
- S. Nozoe, K. Hirai and K. Tsuda, Tetrahedron Lett., 1966, 7, 2211–2216 CrossRef.
- K. Ishibashi, J. Agric. Chem. Soc. Jpn., 1961, 35, 257–262 CAS.
- J.-M. Kim, S.-B. Hyeon, A. Isogai and A. Suzuki, Agric. Biol. Chem., 2014, 48, 803–805 Search PubMed.
- M. W. Canales and G. R. Gray, Phytochemistry, 1988, 27, 1653–1663 CrossRef CAS.
- S. Nozoe and M. Morisaki, Chem. Commun., 1969, 1319–1320 RSC.
- Q. X. Wang, J. L. Yang, Q. Y. Qi, L. Bao, X. L. Yang, M. M. Liu, P. Huang, L. X. Zhang, J. L. Chen, L. Cai and H. W. Liu, Bioorg. Med. Chem. Lett., 2013, 23, 3547–3550 CrossRef CAS PubMed.
- L. Canonica, A. Friecchi, K. U. Galli and A. Scala, Tetrahedron Lett., 1966, 7, 1329–1333 CrossRef.
- J. W. Ahn, M. K. Lee, S. U. Choi, C. O. Lee and B. S. Kim, J. Microbiol. Biotechnol., 1998, 8, 406–408 CAS.
- F. Sugawara, N. Takahashi, G. Strobel, C. H. Yun, G. George, Y. Fu and J. Clardy, J. Org. Chem., 1988, 53, 2170–2172 CrossRef CAS.
- M. Liu, W. Sun, L. Shen, X. Hao, W. H. Al Anbari, S. Lin, H. Li, W. Gao, J. Wang, Z. Hu and Y. Zhang, J. Nat. Prod., 2019, 82, 2897–2906 CrossRef CAS PubMed.
- M.-T. Liu, Y. He, L. Shen, Z.-X. Hu and Y.-H. Zhang, Chin. J. Nat. Med., 2019, 17, 935–944 CAS.
- X. Duan, X. Tan, L. Gu, J. Liu, X. Hao, L. Tao, H. Feng, Y. Cao, Z. Shi, Y. Duan, M. Deng, G. Chen, C. Qi and Y. Zhang, Bioorg. Chem., 2020, 99, 103816 CrossRef CAS PubMed.
- R. Cai, H. Jiang, Y. Mo, H. Guo, C. Li, Y. Long, Z. Zang and Z. She, J. Nat. Prod., 2019, 82, 2268–2278 CrossRef CAS PubMed.
- Q. X. Wang, L. Bao, X. L. Yang, D. L. Liu, H. Guo, H. Q. Dai, F. H. Song, L. X. Zhang, L. D. Guo, S. J. Li and H. W. Liu, Fitoterapia, 2013, 90, 220–227 CrossRef CAS PubMed.
- H. G. Cutler, F. G. Crumley, R. H. Cox, J. P. Springer, R. F. Arrendale, R. J. Cole and P. D. Cole, J. Agric. Food Chem., 1984, 32, 778–782 CrossRef CAS.
- S. B. Singh, J. L. Smith, G. S. Sabnis, A. W. Dombrowski, J. M. Schaeffer, M. A. Goetz and G. F. Bills, Tetrahedron, 1991, 47, 6931–6938 CrossRef CAS.
- T. Zhu, Z. Lu, J. Fan, L. Wang, G. Zhu, Y. Wang, X. Li, K. Hong, P. Piyachaturawat, A. Chairoungdua and W. Zhu, J. Nat. Prod., 2018, 81, 2–9 CrossRef CAS PubMed.
- X.-H. Liu, F.-P. Miao, M.-F. Qiao, R. H. Cichewicz and N.-Y. Ji, RSC Adv., 2013, 3, 588–595 RSC.
- H.-B. Liu, R. Edrada-Ebel, R. Ebel, Y. Wang, B. Schulz, S. Draeger, W. E. G. Müller, V. Wray, W.-H. Lin and P. Proksch, Helv. Chim. Acta, 2011, 94, 623–631 CrossRef CAS.
- B. K. Choi, P. T. H. Trinh, H. S. Lee, B. W. Choi, J. S. Kang, N. T. D. Ngoc, T. T. T. Van and H. J. Shin, Mar. Drugs, 2019, 17(346), 341–349 Search PubMed.
- T. T. Bladt, C. Dürr, P. B. Knudsen, S. Kildgaard, J. C. Frisvad, C. H. Gotfredsen, M. Seiffert and T. O. Larsen, Molecules, 2013, 18, 14629–14650 CrossRef CAS PubMed.
- A. Itai, S. Nozoe, K. Tsuda, S. Okuda and Y. Iitaka, Tetrahedron Lett., 1967, 42, 4111–4112 CrossRef CAS PubMed.
- S. Nozoe, A. Itai, K. Tsuda and S. Okuda, Tetrahedron Lett., 1967, 8, 4113–4117 CrossRef PubMed.
- A. Itai, S. Nozoe, S. Okuda and Y. Iitaka, Acta Crystallogr. Sect. B Struct. Sci., 1969, 25, 872–881 CrossRef CAS.
- H. Wei, T. Itoh, M. Kinoshita, Y. Nakai, M. Kurotaki and M. Kobayashi, Tetrahedron, 2004, 60, 6015–6019 CrossRef CAS.
- M. Arai, H. Niikawa and M. Kobayashi, J. Nat. Med., 2013, 67, 271–275 CrossRef CAS PubMed.
- A. Tsipouras, A. A. Adefarati, J. S. Tkacz, E. G. Frazier, S. P. Rohrer, E. Birzin, A. Rosegay, D. L. Zink, M. A. Goetz, S. B. Singh and J. M. Schaeffer, Bioorg. Med. Chem., 1996, 4, 531–536 CrossRef CAS PubMed.
- S. B. Singh, R. A. Reamer, D. Zink, D. Schmatz, A. Dombrowski and M. A. Goetz, J. Org. Chem., 1991, 56, 5618–5622 CrossRef CAS.
- M. Wei, P. Zhou, L. Huang, J. Yin, Q. Li, C. Dai, J. Wang, L. Gu, Q. Tong, H. Zhu and Y. Zhang, Phytochemistry, 2021, 191, 112910 CrossRef CAS PubMed.
- N. Kawahara, M. Nozawa, A. Kurata, T. Hakamatsuka, S. Sekita and M. Satake, Chem. Pharm. Bull., 1999, 47, 1344–1345 CrossRef CAS PubMed.
- C.-J. Guo, B. P. Knox, Y.-M. Chiang, H.-C. Lo, J. F. Sanchez, K.-H. Lee, B. R. Oakley, K. S. Bruno and C. C. C. Wang, Org. Lett., 2012, 14, 5684–5687 CrossRef CAS PubMed.
- J. P. Springer, J. W. Dorner, R. J. Cole and R. H. Cox, J. Org. Chem., 1979, 44, 4852–4854 CrossRef CAS.
- G. Y. Li, B. G. Li, T. Yang, J. H. Yin, H. Y. Qi, G. Y. Liu and G. L. Zhang, J. Nat. Prod., 2005, 68, 1243–1246 CrossRef CAS PubMed.
- T. Fukuda, Y. Kurihara, A. Kanamoto and H. Tomoda, J. Antibiot., 2014, 67, 593–595 CrossRef CAS PubMed.
- M. P. López-Gresa, N. Cabedo, M. C. González-Mas, M. L. Ciavatta, C. Avila and J. Primo, J. Nat. Prod., 2009, 72, 1348–1351 CrossRef PubMed.
- G. K. Oleinikova, V. A. Denisenko, D. V. Berdyshev, M. A. Pushilin, N. N. Kirichuk, N. I. Menzorova, A. S. Kuzmich, E. A. Yurchenko, O. I. Zhuravleva and S. S. Afiyatullov, Phytochem. Lett., 2016, 17, 135–139 CrossRef CAS.
- M. Chinworrungsee, P. Kittakoop, M. Isaka, A. Rungrod, M. Tanticharoen and Y. Thebtaranonth, Bioorg. Med. Chem. Lett., 2001, 11, 1965–1969 CrossRef CAS PubMed.
- M. Chinworrungsee, P. Kittakoop, M. Isaka, R. Chanphen, M. Tanticharoen and Y. Thebtaranonth, J. Chem. Soc., Perkin Trans. 1, 2002, 2473–2476, 10.1039/B207887M.
- R. Chiba, A. Minami, K. Gomi and H. Oikawa, Org. Lett., 2013, 15, 594–597 CrossRef CAS PubMed.
- B. Ye, W. Ding, P.-M. Wang and J. Xu, Chem. Nat. Compd., 2019, 55, 281–284 CrossRef CAS.
- C. H. Lim, Agric. Chem. Biotechnol., 1996, 39, 241–244 CAS.
- C.-H. Lim and Y. Nihashi, J. Appl. Bio Chem., 2018, 61, 79–82 CrossRef.
- C. Intaraudom, S. Nitthithanasilp, P. Rachtawee, T. Boonruangprapa, S. Prabpai, P. Kongsaeree and P. Pittayakhajonwut, Phytochemistry, 2015, 120, 19–27 CrossRef CAS PubMed.
- O. D. Hensens, D. Zink, J. M. Williamson, V. J. Lotti, R. S. L. Chang and M. A. Goetz, J. Org. Chem., 1991, 56, 3399–3403 CrossRef CAS.
- K.-i. Kawai, K. Nozawa and S. Nakajima, J. Chem. Soc., Perkin Trans. 1, 1994, 1673–1674, 10.1039/P19940001673.
- K. Yoganathan, C. Rossant, R. P. Glover, S. Cao, J. J. Vittal, S. Ng, Y. Huang, A. D. Buss and M. S. Butler, J. Nat. Prod., 2004, 67, 1681–1684 CrossRef CAS PubMed.
- H. Fujimoto, E. Nakamura, E. Okuyama and M. Ishibashi, Chem. Pharm. Bull., 2000, 48, 1436–1441 CrossRef CAS PubMed.
- Y. Tezuka, A. Takahashi, M. Maruyama, T. Tamamura, S. Kutsuma, H. Naganawa and T. Takeuchi, Novel antibiotics, AB5362-A, B, and C, their manufacture, their use as fungicides, and Phoma species, Japan Pat., JP 10045662, 1998 Search PubMed.
- H. Takahashi, T. Hosoe, K. Nozawa and K.-i. Kawai, J. Nat. Prod., 1999, 62, 1712–1713 CrossRef CAS.
- M. Cueto, P. R. Jensen and W. Fenical, Org. Lett., 2002, 4, 1583–1585 CrossRef CAS PubMed.
- Y.-L. Li, Y. Gao, C.-Y. Liu, C.-J. Sun, Z.-T. Zhao and H.-X. Lou, J. Nat. Prod., 2019, 82, 1527–1534 CrossRef CAS PubMed.
- C. Ding, X. Wu, B. N. Auckloo, C.-T. A. Chen, Y. Ye, K. Wang and B. Wu, Molecules, 2016, 21, 105 CrossRef PubMed.
- L. Jiang, G. Zhu, J. Han, C. Hou, X. Zhang, Z. Wang, W. Yuan, K. Lv, Z. Cong, X. Wang, X. Chen, L. Karthik, H. Yang, X. Wang, G. Tan, G. Liu, L. Zhao, X. Xia, X. Liu, S. Gao, L. Ma, M. Liu, B. Ren, H. Dai, R. J. Quinn, T. Hsiang, J. Zhang, L. Zhang and X. Liu, Appl. Microbiol. Biotechnol., 2021, 105, 5407–5417 CrossRef CAS PubMed.
- S. Kusari, C. Hertweck and M. Spiteller, Chem. Biol., 2012, 19, 792–798 CrossRef CAS PubMed.
- M. Okada, Y. Matsuda, T. Mitsuhashi, S. Hoshino, T. Mori, K. Nakagawa, Z. Quan, B. Qin, H. Zhang, F. Hayashi, H. Kawaide and I. Abe, J. Am. Chem. Soc., 2016, 138, 10011–10018 CrossRef CAS PubMed.
- K. D. Bauman, K. S. Butler, B. S. Moore and J. R. Chekan, Nat. Prod. Rep., 2021, 38, 2100–2129 RSC.
- Y. Matsuda, T. Mitsuhashi, S. Lee, M. Hoshino, T. Mori, M. Okada, H. Zhang, F. Hayashi, M. Fujita and I. Abe, Angew. Chem., Int. Ed., 2016, 55, 5785–5788 CrossRef CAS PubMed.
- Y. Matsuda, T. Mitsuhashi, Z. Quan and I. Abe, Org. Lett., 2015, 17, 4644–4647 CrossRef CAS PubMed.
- T. Mitsuhashi, J. Rinkel, M. Okada, I. Abe and J. S. Dickschat, Chem. Eur J., 2017, 23, 10053–10057 CrossRef CAS PubMed.
- O. Mosunova, J. C. Navarro-Muñoz and J. Collemare, in Encyclopedia of Mycology, ed. Ó. Zaragoza and A. Casadevall, Elsevier, Oxford, 2021, vol. 2, pp. 458–476 Search PubMed.
- D. W. Christianson, Chem. Rev., 2017, 117, 11570–11648 CrossRef CAS PubMed.
- Y. Ye, A. Minami, A. Mandi, C. Liu, T. Taniguchi, T. Kuzuyama, K. Monde, K. Gomi and H. Oikawa, J. Am. Chem. Soc., 2015, 137, 11846–11853 CrossRef CAS PubMed.
- I. H. Qureshi, S. A. Husain, R. Noorani, N. Murtaza, Y. Iitaka, S. Iwasaki and S. Okuda, Tetrahedron Lett., 1980, 21, 1961–1962 CrossRef CAS.
- L. Gao, K. Narita, T. Ozaki, N. Kumakura, P. Gan, A. Minami, C. Liu, X. Lei, K. Shirasu and H. Oikawa, Tetrahedron Lett., 2018, 59, 1136–1139 CrossRef CAS.
- K. Narita, H. Sato, A. Minami, K. Kudo, L. Gao, C. Liu, T. Ozaki, M. Kodama, X. Lei, T. Taniguchi, K. Monde, M. Yamazaki, M. Uchiyama and H. Oikawa, Org. Lett., 2017, 19, 6696–6699 CrossRef CAS PubMed.
- R. Chen, Q. Jia, X. Mu, B. Hu, X. Sun, Z. Deng, F. Chen, G. Bian and T. Liu, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2023247118 CrossRef CAS PubMed.
- K. Narita, A. Minami, T. Ozaki, C. Liu, M. Kodama and H. Oikawa, J. Org. Chem., 2018, 83, 7042–7048 CrossRef CAS PubMed.
- J. Yan, J. Pang, J. Liang, W. Yu, X. Liao, A. Aobulikasimu, X. Yi, Y. Yin, Z. Deng and K. Hong, Int. J. Mol. Sci., 2022, 23(3), 1903 CrossRef CAS PubMed.
- J. H. Huang, J. M. Lv, Q. Z. Wang, J. Zou, Y. J. Lu, Q. L. Wang, D. N. Chen, X. S. Yao, H. Gao and D. Hu, Org. Biomol. Chem., 2019, 17, 248–251 RSC.
- H.-Y. Huang, J.-H. Huang, Y.-H. Wang, D. Hu, Y.-J. Lu, Z.-G. She, G.-D. Chen, X.-S. Yao and H. Gao, Front. Chem., 2021, 9, 785431 CrossRef CAS PubMed.
- W. Yang, T. Chen, Y. Chen, Q. Tan, Y. Ou, G. Li, B. Wang, D. Hu, H. Yao and Z. She, J. Org. Chem., 2022, 87, 16807–16819 CrossRef CAS PubMed.
- C. Rossi and L. Tuttobello, Tetrahedron Lett., 1978, 19, 307–308 CrossRef.
- C. Heddergott, A. M. Calvo and J. P. Latge, Eukaryot. Cell, 2014, 13, 1014–1025 CrossRef CAS PubMed.
- M. I. Choudhary, R. Ranjit, R. Atta Ur, K. P. Devkota, S. G. Musharraf and T. M. Shrestha, Phytochemistry, 2006, 67, 439–443 CrossRef CAS PubMed.
- A. C. Stierle, J. H. Cardellina and G. A. Strobel, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 8008–8011 CrossRef CAS PubMed.
- L. Gianani, S. Cocucci, D. Pardi and G. Randazzo, Planta, 1979, 146, 271–274 CrossRef CAS PubMed.
- T. K. Au, W. S. Chick and P. C. Leung, Life Sci., 2000, 67, 733–742 CrossRef CAS PubMed.
- Y. Yamada, T. Kuzuyama, M. Komatsu, K. Shin-Ya, S. Omura, D. E. Cane and H. Ikeda, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 857–862 CrossRef CAS PubMed.
- O. Genilloud, Nat. Prod. Rep., 2017, 34, 1203–1232 RSC.
- Y. Yang, Y. Zhang, S. Zhang, Q. Chen, K. Ma, L. Bao, Y. Tao, W. Yin, G. Wang and H. Liu, J. Nat. Prod., 2018, 81, 1089–1092 CrossRef CAS PubMed.
- J. Rinkel, L. Lauterbach and J. S. Dickschat, Angew Chem. Int. Ed. Engl., 2017, 56, 16385–16389 CrossRef CAS PubMed.
- T. Sato, H. Yamaga, S. Kashima, Y. Murata, T. Shinada, C. Nakano and T. Hoshino, ChemBioChem, 2013, 14, 822–825 CrossRef CAS PubMed.
- F. Lopez-Gallego, S. A. Agger, D. Abate-Pella, M. D. Distefano and C. Schmidt-Dannert, ChemBioChem, 2010, 11, 1093–1106 CrossRef CAS PubMed.
- A. J. Paula, G. Hwang and H. Koo, Nat. Commun., 2020, 11, 1354 CrossRef CAS PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- M. Winn, E. Casey, O. Habimana and C. D. Murphy, FEMS Microbiol. Lett., 2014, 352, 157–164 CrossRef CAS PubMed.
- S. Schulz, M. Girhard and V. B. Urlacher, ChemCatChem, 2012, 4, 1889–1895 CrossRef CAS.
- K. G. Domínguez-González, J. J. Robledo-Medrano, J. J. Valdez-Alarcón, O. Hernández-Cristobal, H. E. Martínez-Flores, J. F. Cerna-Cortés, M. G. Garnica-Romo and R. Cortés-Martínez, Appl. Sci., 2022, 12, 11425 CrossRef.
- H.-S. Kang, Z. Charlop-Powers and S. F. Brady, ACS Synth. Biol., 2016, 5, 1002–1010 CrossRef CAS PubMed.
- S. H. Kim, W. Lu, M. K. Ahmadi, D. Montiel, M. A. Ternei and S. F. Brady, ACS Synth. Biol., 2019, 8, 109–118 CrossRef CAS PubMed.
- G. Bian, Y. Han, A. Hou, Y. Yuan, X. Liu, Z. Deng and T. Liu, Metab. Eng., 2017, 42, 1–8 CrossRef CAS PubMed.
- B. Gu, B. Goldfuss and J. S. Dickschat, Angew. Chem., Int. Ed., 2023, 62, e202215688 CrossRef CAS PubMed.
- S. Mo, A. Krunic, S. D. Pegan, S. G. Franzblau and J. Orjala, J. Nat. Prod., 2009, 72, 2043–2045 CrossRef CAS PubMed.
- A. H. Cabanillas, V. Tena Perez, S. Maderuelo Corral, D. F. Rosero Valencia, A. Martel Quintana, M. Ortega Domenech and A. Rumbero Sanchez, J. Nat. Prod., 2018, 81, 410–413 CrossRef CAS PubMed.
- S. J. Rowland and J. N. Robson, Mar. Environ. Res., 1990, 30, 191–216 CrossRef CAS.
- W. G. Allard, S. T. Belt, G. Massé, R. Naumann, J. M. Robert and S. Rowland, Phytochemistry, 2001, 56, 795–800 CrossRef CAS PubMed.
- S. J. Rowland, S. T. Belt, E. J. Wraige, G. Massé, C. Roussakis and J. M. Robert, Phytochemistry, 2001, 56, 597–602 CrossRef CAS PubMed.
- S. T. Belt, G. Massé, W. G. Allard, J.-M. Robert and S. J. Rowland, Tetrahedron Lett., 2003, 44, 9103–9106 CrossRef CAS.
- M. Kaneda, Y. Iitaka and S. Shibata, Acta Crystallogr., 1974, B30, 358–364 CrossRef.
- P. S. Rao, K. G. Sarma and T. R. Seshadi, Curr. Sci., 1966, 6, 147–148 Search PubMed.
- H. Sugawara, A. Kasuya, Y. Iitaka and S. Shibata, Chem. Pharm. Bull., 1991, 39, 3051–3054 CrossRef CAS.
- M. Millot, M. Martin-de-Lassalle, M. Chollet-Krugler, Y. Champavier, L. Mambu, J.-A. Chulia and M.-A. Lacaille-Dubois, Helv. Chim. Acta, 2016, 99, 169–173 CrossRef CAS.
- D. Li, M. Yang, R. Mu, S. Luo, Y. Chen, W. Li, A. Wang, K. Guo, Y. Liu and S. Li, Chin. Chem. Lett., 2023, 34, 107469 CrossRef CAS.
- D.-L. Guo, M. Zhao, S.-J. Xiao, B. Xia, B. Wan, Y.-C. Gu, L.-S. Ding and Y. Zhou, Phytochem. Lett., 2015, 14, 260–264 CrossRef CAS.
- Y. Gao, F. Stuhldreier, L. Schmitt, S. Wesselborg, L. Wang, W. E. G. Müller, R. Kalscheuer, Z. Guo, K. Zou, Z. Liu and P. Proksch, Fitoterapia, 2020, 146, 104652 CrossRef CAS PubMed.
- D. Zhang, S. Fukuzawa, M. Satake, X. Li, T. Kuranaga, A. Niitsu, A. Niitsu, K. Yoshizawa and K. Tachibana, Nat. Prod. Commun., 2012, 7, 1411–1414 CAS.
- M. K. Renner, P. R. Jensen and W. Fenical, J. Org. Chem., 2000, 65, 4843–4852 CrossRef CAS PubMed.
- G. Randazzo, V. Fogliano, A. Ritieni, L. Mannina, E. Rossi, A. Scarallo and A. L. Segre, Tetrahedron, 1993, 49, 10883–10896 CrossRef CAS.
- A. Ritieni, V. Fogliano, G. Randazzo, A. Scarallo, A. Logrieco, A. Moretti, L. Manndina and A. Bottalico, Nat. Toxins, 1995, 3, 17–20 CrossRef CAS PubMed.
- A. Santini, A. Ritieni, V. Fogliano, G. Randazzo, L. Mannina, A. Logrieco and E. Benedetti, J. Nat. Prod., 1996, 59, 109–112 CrossRef CAS PubMed.
- D. Liu, X.-M. Li, C.-S. Li and B.-G. Wang, Helv. Chim. Acta, 2013, 96, 437–444 CrossRef CAS.
- N. Hoque, C. M. Hasan, M. S. Rana, A. Varsha, M. H. Sohrab and K. M. Rahman, Molecules, 2018, 23(3288), 3281–3288 Search PubMed.
- X. Huang, H. Huang, H. Li, X. Sun, H. Huang, Y. Lu, Y. Lin, Y. Long and Z. She, Org. Lett., 2013, 15, 721–723 CrossRef CAS PubMed.
- Z. e. Xiao, H. Huang, C. Shao, X. Xia, L. Ma, X. Huang, Y. Lu, Y. Lin, Y. Long and Z. She, Org. Lett., 2013, 15, 2522–2525 CrossRef CAS PubMed.
- Z. Liu, Y. Chen, S. Chen, Y. Liu, Y. Lu, D. Chen, Y. Lin, X. Huang and Z. She, Org. Lett., 2016, 18, 1406–1409 CrossRef CAS PubMed.
- T. Shiina, K. Nakagawa, Y. Fujisaki, T. Ozaki, C. Liu, T. Toyomasu, M. Hashimoto, H. Koshino, A. Minami, H. Kawaide and H. Oikawa, Biosci., Biotechnol., Biochem., 2019, 83, 192–201 CrossRef CAS PubMed.
- T. Roncal, S. Cordobes, O. Sterner and U. Ugalde, Eukaryot. Cell, 2002, 1, 823–829 CrossRef CAS PubMed.
- S. Kusari and M. Spiteller, Nat. Prod. Rep., 2011, 28, 1203–1207 RSC.
- C. A. Young, S. Felitti, K. Shields, G. Spangenberg, R. D. Johnson, G. T. Bryan, S. Saikia and B. Scott, Fungal Genet. Biol., 2006, 43, 679–693 CrossRef CAS PubMed.
- L. Keller and M. G. Surette, Nat. Rev. Microbiol., 2006, 4, 249–258 CrossRef CAS PubMed.
- V. Schroeckh, K. Scherlach, H. W. Nutzmann, E. Shelest, W. Schmidt-Heck, J. Schuemann, K. Martin, C. Hertweck and A. A. Brakhage, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14558–14563 CrossRef CAS PubMed.
- T. T. Shen, X. H. Mo, L. P. Zhu, L. L. Tan, F. Y. Du, Q. W. Wang, Y. M. Zhou, X. J. Yuan, B. Qiao and S. Yanga, Appl. Environ. Microbiol., 2019, 85, e00293–e00219 CrossRef PubMed.
- S. Bertrand, O. Schumpp, N. Bohni, M. Monod, K. Gindro and J.-L. Wolfender, J. Nat. Prod., 2013, 76, 1157–1165 CrossRef CAS PubMed.
- R. S. T. Kamdem, H. Wang, P. Wafo, W. Ebrahim, F. C. Özkaya, G. Makhloufi, C. Janiak, P. Sureechatchaiyan, M. U. Kassack, W. Lin, Z. Liu and P. Proksch, Fitoterapia, 2018, 124, 132–136 CrossRef CAS PubMed.
- S. Bertrand, N. Bohni, S. Schnee, O. Schumpp, K. Gindro and J.-L. Wolfender, Biotechnol. Adv., 2014, 32, 1180–1204 CrossRef CAS PubMed.
- E. Cohen, L. Koch, K. M. Thu, Y. Rahamim, Y. Aluma, M. Ilan, O. Yarden and S. Carmeli, Bioorg. Med. Chem., 2011, 19, 6587–6593 CrossRef CAS PubMed.
- J. P. Engelberts, S. J. Robbins, J. M. de Goeij, M. Aranda, S. C. Bell and N. S. Webster, ISME J., 2020, 14, 1100–1110 CrossRef CAS PubMed.
- R. W. Thacker and C. J. Freeman, Adv. Mar. Biol., 2012, 62, 57–111 Search PubMed.
- WFO, World flora online, http://www.worldfloraonline.org, (accessed April 03 2024).
- R. Kamaya and H. Ageta, Chem. Pharm. Bull., 1990, 38, 342–346 CrossRef CAS.
- R. Kamaya, K. Masuda, H. Ageta and H.-C. Chang, Chem. Pharm. Bull., 1996, 44, 695–698 CrossRef CAS.
- R. T. Iyer, K. N. N. Ayengar and S. Rangaswami, Indian J. Chem., 1972, 10, 482–484 CAS.
- H. Khan, A. Zaman, G. L. Chetty, A. S. Gupta and S. Dev, Tetrahedron Lett., 1971, 12, 4443–4446 CrossRef.
- T. Jiang, X. Dai, T. Gao, L. Wang, F. Yang, Y. Zhang, N. Wang, G. Huang and J. Cao, Tetrahedron Lett., 2022, 100, 153869 CrossRef CAS.
- R. Kamaya, K. Masuda, K. Suzuki, H. Ageta and H.-Y. Hsu, Chem. Pharm. Bull., 1996, 44, 690–694 CrossRef CAS.
- K. Zaman, D. Chetia and M. Ali, Asian J. Chem., 2017, 29, 485–488 CrossRef CAS.
- Q.-M. Li, J.-G. Luo, H.-J. Zhao, W.-Y. Yu, X.-B. Wang, M.-H. Yang, J. Luo, H.-B. Sun, Y.-J. Chen, Q.-L. Guo and L.-Y. Kong, Asian J. Org. Chem., 2015, 4, 1366–1369 CrossRef CAS.
- Q.-M. Li, J.-G. Luo, Y.-M. Zhang, Z.-R. Li, X.-B. Wang, M.-H. Yang, J. Luo, H.-B. Sun, Y.-J. Chen and L.-Y. Kong, Chem.–Eur. J., 2015, 21, 13206–13209 CrossRef CAS PubMed.
- D. H. O'Brien and R. D. Stipanovic, J. Org. Chem., 1978, 43, 1105–1111 CrossRef.
- R. D. Stipanovic, A. A. Bell and M. J. Lukefahr, Phytochemistry, 1978, 17, 151–152 CrossRef CAS.
- R. D. Stipanovic, A. A. Bell, D. H. O'Brien and M. J. Lukefahr, Tetrahedron Lett., 1977, 18, 567–570 CrossRef.
- R. D. Stipanovic, A. A. Bell, D. H. O'Brien and M. J. Lukefahr, J. Agric. Food Chem., 1978, 26, 115–118 CrossRef CAS.
- Z. Chen, X. Chen, Y. Tang, Y. Zhou, H. Deng, J. He, Y. Liu, Z. Zhao and H. Cui, Org. Lett., 2022, 24, 3717–3720 CrossRef CAS PubMed.
- W. H. Jiao, H. Gao, F. Zhao, F. He, G. X. Zhou and X. S. Yao, Chem. Biodivers., 2011, 8, 1163–1169 CrossRef CAS PubMed.
- H. Hikino, T. Ohta and T. Takemoto, Phytochemistry, 1975, 14, 2473–2481 CrossRef CAS.
- S. Kurimoto, J. X. Pu, H. D. Sun, Y. Takaishi and Y. Kashiwada, Chem. Biodivers., 2015, 12, 1200–1207 CrossRef CAS PubMed.
- S.-H. Luo, Q. Luo, X.-M. Niu, M.-J. Xie, X. Zhao, B. Schneider, J. Gershenzon and S.-H. Li, Angew. Chem., Int. Ed., 2010, 49, 4471–4475 CrossRef CAS PubMed.
- S.-H. Luo, J. Hua, X.-M. Niu, Y. Liu, C.-H. Li, Y.-Y. Zhou, S.-X. Jing, X. Zhao and S.-H. Li, Phytochemistry, 2013, 86, 29–35 CrossRef CAS PubMed.
- S. H. Luo, C. L. Hugelshofer, J. Hua, S. X. Jing, C. H. Li, Y. Liu, X. N. Li, X. Zhao, T. Magauer and S. H. Li, Org. Lett., 2014, 16, 6416–6419 CrossRef CAS PubMed.
- M. I. Choudhary, R. Ranjit, A.-u. Rahman, S. Hussain, K. P. Devkota, T. M. Shrestha and M. Parvez, Org. Lett., 2004, 6, 4139–4142 CrossRef CAS PubMed.
- M. I. Choudhary, R. R. Atta-ur-Rahman, K. P. Devkotaa and T. M. Shrestha, Z. Naturforsch., 2007, 62b, 587–592 CrossRef.
- M. I. Choudhary, R. Ranjit, A.-u. Rahman, T. M. Shrestha, A. Yasin and M. Parvez, J. Org. Chem., 2004, 69, 2906–2909 CrossRef CAS PubMed.
- S.-H. Luo, J. Hua, C.-H. Li, Y. Liu, X.-N. Li, X. Zhao and S.-H. Li, Tetrahedron Lett., 2013, 54, 235–237 CrossRef CAS.
- S.-H. Luo, L.-H. Weng, M.-J. Xie, X.-N. Li, J. Hua, X. Zhao and S.-H. Li, Org. Lett., 2011, 13, 1864–1867 CrossRef CAS PubMed.
- S.-H. Luo, J. Hua, C.-H. Li, S.-X. Jing, Y. Liu, X.-N. Li, X. Zhao and S.-H. Li, Org. Lett., 2012, 14, 5768–5771 CrossRef CAS PubMed.
- S.-X. Jing, R. Fu, C.-H. Li, T.-T. Zhou, Y.-C. Liu, Y. Liu, S.-H. Luo, X.-N. Li, F. Zeng and S.-H. Li, J. Org. Chem., 2021, 86, 11169–11176 CrossRef CAS PubMed.
- C.-H. Li, S.-X. Jing, S.-H. Luo, W. Shi, J. Hua, Y. Liu, X.-N. Li, B. Schneider, J. Gershenzon and S.-H. Li, Org. Lett., 2013, 15, 1694–1697 CrossRef CAS PubMed.
- L.-L. Teng, R.-F. Mu, Y.-C. Liu, C.-J. Xiao, D.-S. Li, J.-X. Gao, K. Guo, X.-N. Li, Y. Liu, F. Zeng and S.-H. Li, Org. Lett., 2021, 23, 2232–2237 CrossRef CAS PubMed.
- A. Bisio, A. M. Schito, F. Pedrelli, O. Danton, J. K. Reinhardt, G. Poli, T. Tuccinardi, T. Burgi, F. De Riccardis, M. Giacomini, D. Calzia, I. Panfoli, G. C. Schito, M. Hamburger and N. De Tommasi, J. Nat. Prod., 2020, 83, 1027–1042 CrossRef CAS PubMed.
- M. R. Hasan, H. I. Al-Jaber, M. A. Al-Qudah and M. H. Abu Zarga, Phytochem. Lett., 2016, 16, 12–17 CrossRef CAS.
- F. Dal Piaz, A. Vassallo, L. Lepore, A. Tosco, A. Bader and N. De Tommasi, J. Med. Chem., 2009, 52, 3814–3828 CrossRef CAS PubMed.
- A. Rustaiyan and A. Sadjadi, Phytochemistry, 1987, 26, 3078–3079 CrossRef CAS.
- A. Rustaiyan, A. Niknejad, L. Nazarians, J. Jakupovic and F. Bohlmann, Phytochemistry, 1982, 21, 1812–1813 CrossRef CAS.
- A. Rustaiyan and S. Koussari, Phytochemistry, 1988, 27, 1767–1769 CrossRef CAS.
- G. Cioffi, A. Bader, A. Malafronte, F. Dal Piaz and N. De Tommasi, Phytochemistry, 2008, 69, 1005–1012 CrossRef CAS PubMed.
- F. M. Moghaddam, R. Amiri, M. Alam, M. B. Hossain and D. van der Helm, J. Nat. Prod., 1998, 61, 279–281 CrossRef CAS PubMed.
- F. M. Moghaddam, M. M. Farimani, M. Seirafi, S. Taheri, H. R. Khavasi, J. Sendker, P. Proksch, V. Wray and R. Edrada, J. Nat. Prod., 2010, 73, 1601–1605 CrossRef CAS PubMed.
- M. Moridi Farimani and Z. Mazarei, Fitoterapia, 2014, 98, 234–240 CrossRef CAS PubMed.
- S. N. Ebrahimi, M. M. Farimani, F. Mirzania, M. A. Soltanipoor, M. De Mieri and M. Hamburger, J. Nat. Prod., 2014, 77, 848–854 CrossRef CAS PubMed.
- A. Ulubelen, G. Topcu, U. Sönmez, C. Eriş and U. Özgen, Phytochemistry, 1996, 43, 431–434 CrossRef CAS.
- G. Topcu, A. Ulubelen, T. C. M. Tam and T.-C. Chun, Phytochemistry, 1996, 42, 1089–1092 CrossRef CAS.
- M. S. Gonzalez, J. M. San Segundo, M. C. Grande, M. Medarde and I. S. Bellido, Tetrahedron, 1989, 45, 3575–3582 CrossRef CAS.
- A. Malafronte, F. Dal Piaz, G. Cioffi, A. Braca, A. Leone and N. De Tommasi, Nat. Prod. Commun., 2008, 3, 877–880 CrossRef CAS.
- G. Topcu, A. Ulubelen, T. C.-M. Tam and C.-T. Che, J. Nat. Prod., 1996, 59, 113–116 CrossRef CAS.
- G. Appendino, O. Taglialatela-Scafati, A. Romano, F. Pollastro, C. Avonto and P. Rubiolo, J. Nat. Prod., 2009, 72, 340–344 CrossRef CAS PubMed.
- S. Wang, J. Li, J. Sun, K. W. Zeng, J. R. Cui, Y. Jiang and P. F. Tu, Fitoterapia, 2013, 85, 169–175 CrossRef CAS PubMed.
- S. J. Bloor, Tetrahedron Lett., 1993, 34, 5617–5620 CrossRef CAS.
- L. O. Hanus, T. Rezanka and V. M. Dembitsky, Phytochemistry, 2003, 63, 869–875 CrossRef CAS PubMed.
- M. Ali and J. Gupta, J. Med. Aromat. Plant Sci., 1996, 18, 791–794 CAS.
- N. Tanaka, S. Abe and J. i. Kobayashi, Tetrahedron Lett., 2012, 53, 1507–1510 CrossRef CAS.
- C. Yan, Y. Wang and X. J. Hao, Chin. Chem. Lett., 2008, 19, 937–939 CrossRef CAS.
- V. L. Challinor, S. Chap, R. P. Lehmann, P. V. Bernhardt and J. J. De Voss, J. Nat. Prod., 2013, 76, 485–488 CrossRef CAS PubMed.
- V. L. Challinor, R. C. Johnston, P. V. Bernhardt, R. P. Lehmann, E. H. Krenske and J. J. De Voss, Chem. Sci., 2015, 6, 5740–5745 RSC.
- K. A. Jabal, H. M. Abdallah, G. A. Mohamed, I. A. Shehata, M. Y. Alfaifi, S. E. I. Elbehairi, A. A. Koshak and S. R. M. Ibrahim, Nat. Prod. Res., 2019, 1–6, DOI:10.1080/14786419.2019.1577842.
- S. K. Roy, M. Sanjrani, M. Sharma and R. Ramachandram, Indian J. Chem., Sect. B:Org. Chem. Incl. Med. Chem., 2002, 41, 2390–2394 Search PubMed.
- N. Kawahara, M. Nozawa, D. Flores, P. Bonilla, S. Sekita, M. Satake and K. I. Kawai, Chem. Pharm. Bull., 1997, 45, 1717–1719 CrossRef CAS.
- N. Kawahara, M. Nozawa, D. Flores, P. Bonilla, S. Sekita and M. Satake, Phytochemistry, 2000, 53, 881–884 CrossRef CAS PubMed.
- K. Guo, X. Liu, T. T. Zhou, Y. C. Liu, Y. Liu, Q. M. Shi, X. N. Li and S. H. Li, J. Org. Chem., 2020, 85, 5511–5515 CrossRef CAS PubMed.
- K. Guo, Y.-C. Liu, Y. Liu, H. Zhang, W.-Y. Li, Q.-M. Shi, X.-N. Li, F. Zeng and S.-H. Li, Phytochemistry, 2021, 187, 112780 CrossRef CAS PubMed.
- M. Dong, L. Q. Quan, W. F. Dai, S. L. Yan, C. H. Chen, X. Q. Chen and R. T. Li, Planta Med., 2017, 83, 1368–1373 CrossRef CAS PubMed.
- L. Acebey, M. Sauvain, S. Beck, C. Moulis, A. Gimenez and V. Jullian, Org. Lett., 2007, 9, 4693–4696 CrossRef CAS PubMed.
- C. Murakami, R. S. A. Cabral, K. S. Gomes, T. A. Costa-Silva, M. Amaral, M. Romanelli, A. G. Tempone, J. H. G. Lago, V. da S. Bolzani, P. R. H. Moreno and M. C. M. Young, Phytochem. Lett., 2019, 33, 6–11 CrossRef CAS.
- T. Akihisa, K. Koike, Y. Kimura, N. Sashida, T. Matsumoto, M. Ukiya and T. Nikaido, Lipids, 1999, 34, 1151–1157 CrossRef CAS PubMed.
- C. A. N. Catalán, C. S. de Heluani, C. Kotowicz, T. E. Gedris and W. Herz, Phytochemistry, 2003, 64, 625–629 CrossRef PubMed.
- M. Toyoda, M. Asahina and H. Fukawa, Tetrahedron Lett., 1969, 55, 4879–4882 CrossRef.
- C. Corre and G. L. Challis, Nat. Prod. Rep., 2009, 26, 977–986 RSC.
- C. Wang, Q. Chen, D. Fan, J. Li, G. Wang and P. Zhang, Mol. Plant, 2016, 9, 195–204 CrossRef CAS PubMed.
- A. Minami, T. Ozaki, C. Liu and H. Oikawa, Nat. Prod. Rep., 2018, 35, 1330–1346 RSC.
- A. C. Huang, Y. J. Hong, A. D. Bond, D. J. Tantillo and A. Osbourn, Angew. Chem., Int. Ed., 2018, 57, 1291–1295 CrossRef CAS PubMed.
- Q. Chen, J. Li, Z. Liu, T. Mitsuhashi, Y. Zhang, H. Liu, Y. Ma, J. He, T. Shinada, T. Sato, Y. Wang, H. Liu, I. Abe, P. Zhang and G. Wang, Plant Commun., 2020, 1, 100051 CrossRef PubMed.
- J. Shao, Q.-W. Chen, H.-J. Lv, J. He, Z.-F. Liu, Y.-N. Lu, H.-L. Liu, G.-D. Wang and Y. Wang, Org. Lett., 2017, 19, 1816–1819 CrossRef CAS PubMed.
- Y.-G. Chen, D.-S. Li, Y. Ling, Y.-C. Liu, Z.-L. Zuo, L.-S. Gan, S.-H. Luo, J. Hua, D.-Y. Chen, F. Xu, M. Li, K. Guo, Y. Liu, J. Gershenzon and S.-H. Li, Angew. Chem., Int. Ed., 2021, 60, 25468–25476 CrossRef CAS PubMed.
- D.-S. Li, J. Hua, S.-H. Luo, Y.-C. Liu, Y.-G. Chen, Y. Ling, K. Guo, Y. Liu and S.-H. Li, Plant Commun., 2021, 2, 100233 CrossRef CAS PubMed.
- M. Masi, R. Dasari, A. Evidente, V. Mathieu and A. Kornienko, Bioorg. Med. Chem. Lett., 2019, 29, 859–869 CrossRef CAS PubMed.
- H. Fujiwara, K. Matsunaga, H. Kumagai, M. Ishizuka and Y. Ohizumi, Pharm. Pharmacol. Commun., 2000, 6, 427–431 CrossRef CAS.
- M. Bury, A. Girault, V. Megalizzi, S. Spiegl-Kreinecker, V. Mathieu, W. Berger, A. Evidente, A. Kornienko, P. Gailly, C. Vandier and R. Kiss, Cell Death Dis., 2013, 4, e561 CrossRef CAS PubMed.
- A. K. Najumudeen, A. Jaiswal, B. Lectez, C. Oetken-Lindholm, C. Guzman, E. Siljamaki, I. M. Posada, E. Lacey, T. Aittokallio and D. Abankwa, Oncogene, 2016, 35, 5248–5262 CrossRef CAS PubMed.
- R. Dasari, M. Masi, R. Lisy, M. Ferderin, L. R. English, A. Cimmino, V. Mathieu, A. J. Brenner, J. G. Kuhn, S. T. Whitten, A. Evidente, R. Kiss and A. Kornienko, Bioorg. Med. Chem. Lett., 2015, 25, 4544–4548 CrossRef CAS PubMed.
- S. S. Elhady, A. M. Al-Abd, A. M. El-Halawany, A. M. Alahdal, H. A. Hassanean and S. A. Ahmed, Mar. Drugs, 2016, 14(130), 131–114 Search PubMed.
- Y. C. Chen, M. C. Lu, M. El-Shazly, K. H. Lai, T. Y. Wu, Y. M. Hsu, Y. L. Lee and Y. C. Liu, Mar. Drugs, 2018, 16(212), 211–222 CAS.
- W. T. Chang, Y. D. Bow, P. J. Fu, C. Y. Li, C. Y. Wu, Y. H. Chang, Y. N. Teng, R. N. Li, M. C. Lu, Y. C. Liu and C. C. Chiu, Oxid. Med. Cell. Longev., 2021, 2021, 7689045 CrossRef PubMed.
- Y. Doi, H. Shigemori, M. Ishibashi, F. Mizobe, A. Kawashima, S. Nakaike and J. Kobayashi, Chem. Pharm. Bull., 1993, 41, 2190–2191 CrossRef CAS PubMed.
- M. Schumacher, C. Cerella, S. Eifes, S. Chateauvieux, F. Morceau, M. Jaspars, M. Dicato and M. Diederich, Biochem. Pharmacol., 2010, 79, 610–622 CrossRef CAS PubMed.
- K. H. Lai, Y. C. Liu, J. H. Su, M. El-Shazly, C. F. Wu, Y. C. Du, Y. M. Hsu, J. C. Yang, M. K. Weng, C. H. Chou, G. Y. Chen, Y. C. Chen and M. C. Lu, Sci. Rep., 2016, 6(36170), 36171–36114 Search PubMed.
- Y. Mizushina, D. Manita, T. Takeuchi, F. Sugawara, Y. Kumamoto-Yonezawa, Y. Matsui, M. Takemura, M. Sasaki, H. Yoshida and H. Takikawa, Molecules, 2008, 14, 102–121 CrossRef PubMed.
- L. Margarucci, M. C. Monti, A. Tosco, R. Riccio and A. Casapullo, Angew Chem. Int. Ed. Engl., 2010, 49, 3960–3963 CrossRef CAS PubMed.
- L. Margarucci, A. Tosco, R. De Simone, R. Riccio, M. C. Monti and A. Casapullo, ChemBioChem, 2012, 13, 982–986 CrossRef CAS PubMed.
- M. C. Monti, L. Margarucci, R. Riccio, L. Bonfili, M. Mozzicafreddo, A. M. Eleuteri and A. Casapullo, Biochim. Biophys. Acta, 2014, 1844, 713–721 CrossRef CAS PubMed.
- L. V. Manes, P. Crews, M. R. Kernan, D. J. Faulkner, F. R. Fronczek and R. D. Gandour, J. Org. Chem., 1988, 53, 570–575 CrossRef CAS.
- C. Cassiano, M. C. Monti, C. Festa, A. Zampella, R. Riccio and A. Casapullo, ChemBioChem, 2012, 13, 1953–1958 CrossRef CAS PubMed.
- J. C. de Freitas, L. A. Blankemeier and R. S. Jacobs, Experientia, 1984, 40, 864–865 CrossRef CAS PubMed.
- C. F. Bennett, S. Mong, M. A. Clarke, L. I. Kruse and S. T. Crooke, Biochem. Pharmacol., 1987, 36, 733–740 CrossRef CAS PubMed.
- K. B. Glaser and R. S. Jacobs, Biochem. Pharmacol., 1986, 35, 449–453 CrossRef CAS PubMed.
- K. B. Glaser and R. S. Jacobs, Biochem. Pharmacol., 1987, 36, 2079–2086 CrossRef CAS PubMed.
- K. B. Glaser, T. S. Vedvick and R. S. Jacobs, Biochem. Pharmacol., 1988, 37, 3639–3646 CrossRef CAS PubMed.
- F. Dal Piaz, A. Casapullo, A. Randazzo, R. Riccio, P. Pucci, G. Marino and L. Gomez-Paloma, ChemBioChem, 2002, 3, 664–671 CrossRef CAS PubMed.
- M. C. Monti, A. Casapullo, R. Riccio and L. Gomez-Paloma, Bioorg. Med. Chem. Lett., 2004, 12, 1467–1474 CrossRef CAS PubMed.
- M. C. Monti, A. Casapullo, R. Riccio and L. Gomez-Paloma, FEBS Lett., 2004, 578, 269–274 CrossRef CAS PubMed.
- M. C. Monti, R. Riccio and A. Casapullo, Bioorg. Chem., 2009, 37, 6–10 CrossRef CAS PubMed.
- M. C. Monti, A. Casapullo, C. N. Cavasotto, A. Tosco, F. Dal Piaz, A. Ziemys, L. Margarucci and R. Riccio, Chemistry, 2009, 15, 1155–1163 CrossRef CAS PubMed.
- M. H. Uddin, M. Otsuka, T. Muroi, A. Ono, N. Hanif, S. Matsuda, T. Higa and J. Tanaka, Chem. Pharm. Bull., 2009, 57, 885–887 CrossRef CAS PubMed.
- F. Folmer, M. Jaspars, M. Schumacher, M. Dicato and M. Diederich, Biochem. Pharmacol., 2010, 80, 1793–1800 CrossRef CAS PubMed.
- K. B. Glaser, M. L. Sung, D. A. Hartman, Y. W. Lock, J. Bauer, T. Walter and R. P. Carlson, Skin Pharmacol., 1995, 8, 300–308 CrossRef CAS PubMed.
- B. C. M. Potts, R. J. Capon and D. J. Faulkner, J. Org. Chem., 1992, 57, 2965–2967 CrossRef CAS.
- P. Garcia Pastor, S. De Rosa, A. De Giulio, M. Paya and M. J. Alcaraz, Br. J. Pharmacol., 1999, 126, 301–311 CrossRef CAS PubMed.
- R. P. Murelli, A. K. Cheung and M. L. Snapper, J. Org. Chem., 2007, 72, 1545–1552 CrossRef CAS PubMed.
- V. Escrig, A. Ubeda, M. L. Ferrandiz, J. Darias, J. M. Sanchez, M. J. Alcaraz and M. Paya, J. Pharmacol. Exp. Ther., 1997, 282, 123–131 CrossRef CAS PubMed.
- H. Miyaoka, M. Yamanishi and H. Mitome, Chem. Pharm. Bull., 2006, 54, 268–270 CrossRef CAS PubMed.
- M. C. Monti, M. G. Chini, L. Margarucci, R. Riccio, G. Bifulco and A. Casapullo, ChemBioChem, 2011, 12, 2686–2691 CrossRef CAS PubMed.
- M. S. de Carvalho and R. S. Jacobs, Biochem. Pharmacol., 1991, 42, 1621–1626 CrossRef CAS PubMed.
- J. D. Winkler, C. M. Sung, W. C. Hubbard and F. H. Chilton, Biochem. Pharmacol., 1992, 44, 2055–2066 CrossRef CAS PubMed.
- M. S. Barnette, J. Rush, L. A. Marshall, J. J. Foley, D. B. Schmidt and H. M. Sarau, Biochem. Pharmacol., 1994, 47, 1661–1667 CrossRef CAS PubMed.
- M. C. Monti, A. Casapullo, C. N. Cavasotto, A. Napolitano and R. Riccio, ChemBioChem, 2007, 8, 1585–1591 CrossRef CAS PubMed.
- M. C. Monti, A. Casapullo, R. Riccio and L. Gomez-Paloma, Rapid Commun. Mass Spectrom., 2005, 19, 303–308 CrossRef CAS PubMed.
- M. L. Bourguet-Kondracki, C. Debitus and M. Guyot, J. Chem. Res., 1996,(S), 192–193 CAS.
- H. S. Lee, K. M. Yoon, Y. R. Han, K. J. Lee, S. C. Chung, T. I. Kim, S. H. Lee, J. Shin and K. B. Oh, Bioorg. Med. Chem. Lett., 2009, 19, 1051–1053 CrossRef CAS PubMed.
- E. Dillp de Sllva and P. J. Scheuer, Tetrahedron Lett., 1980, 21, 1611–1614 CrossRef.
- M. E. Skindersoe, P. Ettinger-Epstein, T. B. Rasmussen, T. Bjarnsholt, R. de Nys and M. Givskov, Mar. Biotechnol., 2008, 10, 56–63 CrossRef CAS PubMed.
- L. Du, L. Shen, Z. Yu, J. Chen, Y. Guo, Y. Tang, X. Shen and H. Jiang, ChemMedChem, 2008, 3, 173–180 CrossRef CAS PubMed.
- C. Cassiano, R. Esposito, A. Tosco, A. Zampella, M. V. D'Auria, R. Riccio, A. Casapullo and M. C. Monti, Chem. Commun., 2014, 50, 406–408 RSC.
- D. Xue, Q. Wang, Z. Chen, L. Cai, L. Bao, Q. Qi, L. Liu, X. Wang, H. Jin, J. Wang, H. Wu, H. Liu and Q. Chen, Bioorg. Med. Chem. Lett., 2015, 25, 1464–1470 CrossRef CAS PubMed.
- C. C. Chen, T. T. Tzeng, C. C. Chen, C. L. Ni, L. Y. Lee, W. P. Chen, Y. J. Shiao and C. C. Shen, J. Nat. Prod., 2016, 79, 438–441 CrossRef CAS PubMed.
- J. H. Hu, I. C. Li, T. W. Lin, W. P. Chen, L. Y. Lee, C. C. Chen and C. F. Kuo, Molecules, 2019, 24(1624), 1621–1613 Search PubMed.
- E. D’Aniello, F. A. Iannotti, L. G. Falkenberg, A. Martella, A. Gentile, F. De Maio, M. L. Ciavatta, M. Gavagnin, J. S. Waxman, V. Di Marzo, P. Amodeo and R. M. Vitale, Mar. Drugs, 2019, 17, 110 CrossRef PubMed.
- Y. Wang, M. Dreyfuss, M. Ponelle, L. LukasOberer and H. Riezman, Tetrahedron, 1998, 54, 6415–6426 CrossRef CAS.
- A. Bidon-Chanal, A. Fuertes, D. Alonso, D. I. Perez, A. Martinez, F. J. Luque and M. Medina, Eur. J. Med. Chem., 2013, 60, 479–489 CrossRef CAS PubMed.
- J.-M. Kim, S.-B. Hyeon, A. Isogai and A. Suzuki, Agric. Biol. Chem., 1984, 48, 803–805 CAS.
- A. Ballio, Experientia, 1991, 47(8), 783–790 CrossRef CAS.
- G. Carr, M. Raszek, R. Van Soest, T. Matainaho, M. Shopik, C. F. B. Holmes and R. J. Andersen, J. Nat. Prod., 2007, 70, 1812–1815 CrossRef CAS PubMed.
- Y. Ling, L.-L. Teng, J. Hua, D.-S. Li, S.-H. Luo, Y.-C. Liu, Y. Liu and S.-H. Li, Chin. J. Nat. Med., 2019, 17, 892–899 CAS.
- K. Guo, L. Feng, H. Zhang, T.-T. Zhou, Y.-Y. Chen, C.-L. Tan, Y.-C. Liu, Y. Liu and S.-H. Li, Fitoterapia, 2024, 178, 106158 CrossRef CAS PubMed.
- K. Guo, T.-T. Zhou, S.-H. Luo, Y.-C. Liu, Y. Liu and S.-H. Li, J. Med. Chem., 2024, 67, 513–528 CrossRef CAS PubMed.
- M. Li, L. Feng, H. Zhang, Y.-C. Liu, T.-T. Zhou, Y. Zheng, K. Guo, Y. Liu and S.-H. Li, Org. Biomol. Chem., 2024, 22, 3019–3024 RSC.
- Y. Zheng, Q. Li, M. Gu, H. Liao, Y. Liang, F. Liu, X.-N. Li, W. Sun, C. Chen, Y. Zhang and H. Zhu, J. Nat. Prod., 2024, 87, 1965–1974 CrossRef CAS PubMed.
- M. R. Byun, A. R. Kim, J. H. Hwang, M. K. Sung, Y. K. Lee, B. S. Hwang, J. R. Rho, E. S. Hwang and J. H. Hong, FEBS Lett., 2012, 586, 1086–1092 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4np00041b |
| ‡ These authors share the first authorship. |
| This journal is © The Royal Society of Chemistry 2025 |