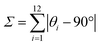Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rational experimental design and computational insights into novel heteroleptic mixed-ligand Ru(II) complexes possessing derivatized bipyridines and phendione for anti-cancer activities
Adewale Olufunsho
Adeloye
 *ab,
Damilare David
Babatunde
*ab,
Damilare David
Babatunde
 ac,
Kgaugelo Cornelius
Tapala
ac,
Kgaugelo Cornelius
Tapala
 a and
Nomampondo Penelope
Magwa
a
a and
Nomampondo Penelope
Magwa
a
aDepartment of Chemistry, College of Science, Engineering and Technology, University of South Africa, Florida Rooderpoort 1709, Johannesburg, South Africa
bDepartment of Analytical, Colloid Chemistry and Technology of Rare Elements, Al-Farabi Kazakh National University, 71 Al-Farabi Avenues, 050040 Almaty, Republic of Kazakhstan. E-mail: Adeloye.adewale@kaznu.edu.kz
cCentre for Materials Science, University of South Africa, Florida 1710, Johannesburg, South Africa
First published on 18th November 2025
Abstract
Several synthesized ruthenium(II) complexes, which are used as metallo-drug agents for cancer treatment, have fallen short of meeting expectations. In our continued efforts to search for potential anticancer drug agents for specific biological targets through rational design, this paper reports a facile, one-pot synthesis procedure, along with the photophysical and electro-redox properties, of three newly designed mixed-ligand ruthenium(II) complexes. These complexes contain triphenylphosphine (PPh3), 4-imidazole acrylic acid (mza), and either functionalized or unfunctionalized polypyridines, specifically 2,2′-bipyridine-4.4′-dicarboxylic acid (Hbpy), 1,10-phenanthroline-5,6-dione (ptd), and 2,2′-bipyridine (bpy). The complexes are formulated as follows: V2 = [RuCl2(PPh3)bpy(mza)·3H2O], W = [RuCl2(PPh3)Hbpy(mza)·H2O] and X = [RuCl2(PPh3)ptd(mza)·3H2O]. The complexes were characterized by elemental analysis, Fourier-Transform infrared spectroscopy (FT-IR), ultraviolet–visible spectroscopy (UV-vis), photoluminescence (PL), 1H, 13C, 31P nuclear magnetic resonance (NMR) spectroscopy, and mass spectrometry (MS). The UV-vis absorption spectra, showed a broad and intense metal-to-ligand charge transfer (MLCT) band with a strong emission intensity ratio near infra-red. The maximum absorbance wavelengths λmax were observed at 415–530 nm (ε ≈ 1.030 × 103 M−1 cm−1) and λem 730–640 nm respectively. The structure–activity relationships relate to a set of voltage-current data, with ligand-based electrochemical redox processes occuring at the Ru(III/II) redox range of +1.15 and –0.82 V. Density functional theory (DFT), molecular docking, and pharmacokinetic calculations confirm that complexes V2, W and X exhibit superior and strong binding interactions and biological inhibition trafficking involving ERα +, WT EGFR, and ALK receptors compared to standard drugs such as Crizotinib, Doxorubicin, Gemcitabine and Lorlatinib. However, the high molecular weights and poor lypophilicity of the complexes hinder their ability to cross the blood–brain barrier (BBB), thus limiting their utility for central nervous system (CNS) targets. This study primarily focuses on the synthesis, characterization, and theoretical evaluation of complexes V2, W and X for their potential anticancer properties, which is useful in the discovery of new materials for drug design. No complementary in-vitro wet laboratory tests for anticancer or other biological methods of analysis were conducted, as they were outside the scope of the present study.
Introduction
Cancer remains one of the deadliest diseases worldwide. It is a complex and multifaceted condition characterized by the uncontrolled growth and spread of abnormal cells. The pathogenesis of cancer can arise from various factors, including genetic mutations, environmental factors, infections, and genetic predisposition. Carcinomas, sarcomas, leukemias, lymphomas, and colorectal cancers are the most common types worldwide. Ongoing research efforts aim to find a solution for cancer and provide a better understanding of its spread. Current treatments, such as chemotherapy, surgery, immunotherapy, and radiation therapy are used to manage the disease.1,2 The refinement of current therapeutic and personalized approaches is ongoing.3In the treatment of cancer using chemotherapy, platinum-based drugs like cisplatin have been the primary agents for many years. However, due to their significant toxicity and debilitating side effects, ruthenium(II) complexes have shown great promise and have gained attention as potential anticancer drugs. They are being promoted as a viable alternative to platinum-based drugs because of their numerous advantages, including the ability to overcome drug resistance, stability in biological media, selectivity, lower toxicity, a wide range of biologically accessible oxidation states, and a distinct, clear mechanism of action.4–6 The oxidation states of most metal-based anticancer drugs play a crucial role in their bioactivity.7 Although no ruthenium complex has been commercialized for cancer treatment, certain derivatives such as NAMI-A, KP1019/NKP1339, and TLD1433 have advanced into various stages of clinical trials.8–10
Several complexes with Ru(II) have been reported, including those with homoleptic and/or heteroleptic ligands of mono-, bi-, or polydentate architectures.11–21 Of great importance is the use of ruthenium(II) polypyridine complexes as molecular targets of DNA base pairs in biomolecules, which are tailored towards different structures and binding modes following sequence-specific recognition.22–28 This recognition involves a combination of molecular interactions such as hydrogen bonding, van der Waals interactions, electrostatic and hydrophobic contacts.23,29,30 Ruthenium(II) complexes containing different ligand types, including triphenylphosphine, benzoic acid, imidazole, bipyridine, and phenanthroline derivatives, are abundant.
In this study, we prepared three ruthenium(II) heteroleptic complexes, each comprising a different combination of these ligands. This included a functionalized imidazole with an α,β-unsaturated carboxylic acid. The intrinsic properties of these individual ligand types are believed to enhance the anticancer properties of the resulting complexes. It is well-established that ruthenium(II) complexes containing triphenylphosphine often show increased delivery properties due to their high hydrophobicity and strong cytotoxicity.31,32
In particular, this work evaluates the photophysical, electro-redox, and pharmacokinetic properties of functionalized/non-functionalized polypyridyl ligands sandwiched by two common organic ligands: triphenylphosphine and functionalized imidazole-α,β-unsaturated carboxylic acid. Their potential anticancer properties are evaluated by density functional theory (DFT) and molecular docking techniques.
Results and discussion
General synthetic procedures for complexes V2, W and X
The synthetic reaction scheme for the general preparation of complexes V2, W, and X is shown in Scheme 1. Modifications to the general method reported by Goldman et al.,33 in terms of the solvent used and synthesis procedures were utilized in the reaction conditions due to ligand solubility. The reaction was carried out under reflux using absolute ethanol with the addition of a small volume of water to aid in the total dissolution of the reagents as well as in the coordination of the ligands to the ruthenium(II)dichloro-tris(triphenylphosphine), which served as the metal precursor. | ||
| Scheme 1 Synthetic reaction procedures for complexes: V2 = RuCl2(PPh3)bpy(mza)·3H2O; W = RuCl2(PPh3)Hbpy(mza)·H2O; and X = RuCl2(PPh3)ptd(mza)·3H2O. | ||
The ligands and metal precursor were added simultaneously in a one-pot reaction synthesis. All reagents and solvents used were of commercial grade with no further purification. All the complex products were obtained in solid form and in good percentage yield. Solubility tests carried out on the complexes showed that none of the complexes was soluble in dichloromethane, but they dissolved appreciably in DMSO and other protic solvents. Literature reports that RuCl2(PPh3)3 or similar complexes in solution can undergo intramolecular rearrangement or dissociate triphenylphosphine to some extent. This was observed in the reported complexes where two units of the tris(triphenylphosphine) dissociated to obtain the final products, as partially represented in eqn (1) below.
| RuCl2(PPh3)3 + bpy → RuCl2(bpy)(PPh3)2 + PPh3 | (1) |
| RuCl2(bpy)(PPh3)2 + mza → RuCl2(bpy)(mza)(PPh3) + PPh3 | (2) |
In fact, to obtain the six-coordinate Ru complex products, the dissociation of two triphenylphosphine units from the pentacoordinate precursor RuCl2(PPh3)3 during the reaction would have resulted from two effects. First, the chelate effect of the polypyridine brings about increased stability of a complex when a ligand binds with more than one atom. This is followed by the coordination of a nitrogen atom from the imidazole ring with the ruthenium center. Therefore, in the product structures, the bidentate nature of bipyridine and phenanthroline allows them to form a more stable chelate ring with the ruthenium center, favoring the displacement of a single monodentate PPh3 ligand.34–37
This observation was supported by the characteristic singlet peak of triphenylphosphine at approximately 25.48 ppm in the 31P NMR spectra for complexes V2 and W, although the peak was unresolved in complex X. It has been reported that for monodentate N-donor ligands, an upfield shift in 31P {1H} NMR is observed between 30–40 ppm, whereas complexes with bidentate N-donor ligands show their 31P {1H} resonances shifted upfield to approximately 20 ppm.38
All attempts to grow single crystals of complexes V2, W, and X by slow evaporation in different solvent systems were unsuccessful. The solvent combination used were n-hexane-toluene (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v), n-hexane-ethanol (1
4, v/v), n-hexane-ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, v/v), and toluene-ethanol (1
4, v/v), and toluene-ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The prepared samples were each kept in covered WHEATON liquid scintillation vials (20 mL capacity), but no crystal formation was obtained after a period of 2–3 months.39 It is believed that the hydrated complexes contain different stoichiometric ratios of water, which possibly makes it difficult to achieve the specific, repeating unit required for a well-ordered crystal.40 Factors such as steric hindrance of the bulky triphenylphosphine ligands, ligand arrangement disorder, strong or weak ligand interactions, and the thermodynamic properties of the complexes may also play significant roles.41 The proposed chemical structures of the complexes, as reported, are in very good agreement with the individual LS-MS, proton and carbon-13 NMR spectra data.
1, v/v). The prepared samples were each kept in covered WHEATON liquid scintillation vials (20 mL capacity), but no crystal formation was obtained after a period of 2–3 months.39 It is believed that the hydrated complexes contain different stoichiometric ratios of water, which possibly makes it difficult to achieve the specific, repeating unit required for a well-ordered crystal.40 Factors such as steric hindrance of the bulky triphenylphosphine ligands, ligand arrangement disorder, strong or weak ligand interactions, and the thermodynamic properties of the complexes may also play significant roles.41 The proposed chemical structures of the complexes, as reported, are in very good agreement with the individual LS-MS, proton and carbon-13 NMR spectra data.
FT-IR spectral studies of complexes V2, W and X
The FT-IR spectra for complexes V2, W, and X were recorded in the 4000–400 cm−1 region. These spectra were compared with the IR data of the free polypyridine ligands and 4-imidazoleacrylic acid. The IR spectra of the complexes also revealed all expected bands of interest, including the stretching vibrations of υ(O–H), υ(C–H), υ(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), υ(C
O), υ(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), υ(C
N), υ(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), υ(C–O), υ(Ru–Cl), and υ(Ru–P). The broad absorption bands at 3348 and 3406 cm−1 are assigned to the O–H stretching vibrations of the carboxylic acid group, as found for complexes W and X. However, this band was enhanced in complex V2 at 3417 cm−1. The vibration frequency bands in the 3066–3010 cm−1 region are assigned to the asymmetric carboxylate stretching bands, υasym(COO−).42 The bands at 2981–2754 cm−1 are assigned to the C–H aromatic stretching vibrations emanating from the ligands. The C
C), υ(C–O), υ(Ru–Cl), and υ(Ru–P). The broad absorption bands at 3348 and 3406 cm−1 are assigned to the O–H stretching vibrations of the carboxylic acid group, as found for complexes W and X. However, this band was enhanced in complex V2 at 3417 cm−1. The vibration frequency bands in the 3066–3010 cm−1 region are assigned to the asymmetric carboxylate stretching bands, υasym(COO−).42 The bands at 2981–2754 cm−1 are assigned to the C–H aromatic stretching vibrations emanating from the ligands. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching frequencies are assigned to bands at 1712 cm−1 for complex V2, 1658 cm−1 for complex W, and 1755 cm−1 for complex X. The aromatic carbon double bonds were assigned to bands at 1612 cm−1 for V2, 1658 cm−1 for W, and 1643 cm−1 for X. Two strong vibration bands were observed at 1589 and 1597 cm−1 in complexes W and X. These were possibly attributed to the influence of increased contributions from carboxylate or carbonyl carbon substitutions at the 4,4′-positions of the bipyridine ring and the 5,6-dione in the phenanthroline.43 The common C
O stretching frequencies are assigned to bands at 1712 cm−1 for complex V2, 1658 cm−1 for complex W, and 1755 cm−1 for complex X. The aromatic carbon double bonds were assigned to bands at 1612 cm−1 for V2, 1658 cm−1 for W, and 1643 cm−1 for X. Two strong vibration bands were observed at 1589 and 1597 cm−1 in complexes W and X. These were possibly attributed to the influence of increased contributions from carboxylate or carbonyl carbon substitutions at the 4,4′-positions of the bipyridine ring and the 5,6-dione in the phenanthroline.43 The common C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching frequencies are found in the 1566–1539 cm−1 region for all three complexes, while the C
N stretching frequencies are found in the 1566–1539 cm−1 region for all three complexes, while the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C aromatic band frequencies were found at 1481–1411 cm−1. The weak bands at 1369 and 1002 cm−1, common to all complexes, are tentatively assigned to the C–O stretching. In the fingerprint regions of the spectra, the literature has shown three intense characteristic vibrations for the PPh3 band group at 748, 698, 744, and 786 cm−1 for complexes V2, W, and X, respectively.44 The Ru–Cl and Ru–P band frequencies for all three complexes are found in the range of 420–698 cm−1.
C aromatic band frequencies were found at 1481–1411 cm−1. The weak bands at 1369 and 1002 cm−1, common to all complexes, are tentatively assigned to the C–O stretching. In the fingerprint regions of the spectra, the literature has shown three intense characteristic vibrations for the PPh3 band group at 748, 698, 744, and 786 cm−1 for complexes V2, W, and X, respectively.44 The Ru–Cl and Ru–P band frequencies for all three complexes are found in the range of 420–698 cm−1.
1H, 13C and 31P – NMR spectroscopic studies of Complexes V2, W and X
The 1H NMR spectrum of complexes V2, W, and X in deuterated DMSO exhibits a very complex pattern of resonance chemical shifts in the region of δ 7.78–6.26 ppm, which is indicative of the presence of mixed ligands comprising triphenylphosphine, 4-imidazoleacrylic acid, and polypyridine protons (see SI Sections S1, S4, and S7).It's obvious that there are multiple non-equivalent hydrogens, as the complexes are made of two different types of monovalent ligands: 4-imidazoleacrylic acid and triphenylphosphine. For complex V2, a total of 27 protons were accounted for in the spectrum at various chemical shifts. The high-intensity multiplet peaks between δ 7.41–7.39 ppm (integrating for 10 protons) and at δ 7.25–7.23 ppm (integrating for 5 protons) are unequivocally assigned to the aromatic protons in the triphenylphosphine ligand. It's obvious that the separation in these hydrogen nuclei may be indicative of the non-stereochemical molecular orientation of the phenyl rings of the coordinating triphenylphosphine moiety. The other peaks are assigned to the protons of the 4-imidazole ring and the unsubstituted bipyridine. For example, the eight protons on the unsubstituted bipyridine are assigned to the multiplet peaks. These peaks include a multiplet integrating for four protons at δ 7.64–7.60 ppm, another three-proton multiplet at δ 7.57–7.53 ppm, and a single remaining proton multiplet at δ 7.22 ppm. The two meta-coupled protons of the imidazole ring are found as singlet peaks at δ 7.78 and δ 7.49 ppm, respectively. Also observed in the spectrum is a doublet peak at δ 6.28 ppm, which integrates for one proton with a J-coupling constant of 12 Hz. This peak is attributed to the H-β of the α,β-unsaturated carbonyl of the imidazole moiety. The coupled H-α is likely one of the protons with a J-coupling constant of 12 Hz at δ 7.46 ppm. In the upfield region, a high-intensity singlet peak was observed at δ 3.36 ppm, which integrates for 71 exchangeable protons. These hydrogens clearly form water molecules within the chemical structure of complex V2, and relative to the total number of hydrogens in the aromatic region, they account for at least three molecules of water in the complex.
The 13C NMR spectrum showed the anticipated carbonyl carbon peak at δ 168.42 ppm which was assigned to the carboxylic acid group of the substituted 4-imidazoleacrylic acid. The other carbons in the structure were assigned as follows: δ 137.02 for C–2, δ 131.88 for C–4 (connected to acrylic side chain), δ 111.64 for C–5, δ 129.15 for the α-carbon, and δ 137.11 for the β-carbon. For the triphenylphosphine signals, the carbon directly linked to the phosphorus was assigned to the peak at δ 133.75, while the ortho-, meta- and para- carbons were assigned to the peaks at δ 133.60, 132.47 and 129.43 respectively. The unsubstituted bipyridine showed signals assigned to C–2/C–2′ at δ 138.12; C–3/C–3′ at δ 129.19; C–4/C–4′ at δ 131.95; C–5/C–5′ at δ 129.24; and C–6/C–6′ at δ 137.95. It is believed that the steric or shielding effects in the complex may have influenced the lowering of the chemical shifts in C–2/C–2′ and C–6/C–6′, which are ordinarily found in the regions of δ 155–160 ppm. The 31P NMR for complex V2 was found as a singlet peak at δ 25.46 ppm (see Fig. S1–S3).
In the 1H NMR of complex W, five major, distinct multiplet peaks, integrating to 25 protons, were observed in the aromatic region. These peaks are from the various mixed-ligand compositions of the complexes. A multiplet between δ 7.64 and 7.59 ppm integrated for six protons, while another at δ 7.56 and 7.52 ppm integrated for five protons. A broad peak at δ 7.39 ppm also integrated for five protons. Three more multiplets were found: one at δ 7.25–7.21 ppm and another at δ 7.11–7.07 ppm, both integrating for four protons each, and a third at δ 6.30–6.27 ppm, which integrated for one proton. A strong, broad peak at δ 3.43 ppm integrated for 25 protons, possibly indicating at least one stoichiometric value of water molecules in the complex, this is relative to the number of protons in the aromatic region.
The 13C spectrum showed peaks in the aromatic region at δ 169.36 and 165.67 ppm. These peaks were assigned to the carbonyl carbons of the 4-imidazoleacrylic acid and 2,2′-bipyridyl-4,4-dicarboxylic acid respectively. The peak at δ 159.21 and 159.17 ppm were assigned to the C–2/C–2′ and C–6/C–6′ of the 4.4′-disubstituted bipyridine moiety. The peak at δ 131.88 and δ 129.25 were assigned to the C–3/C–3′ and C–5/C–5′ signals while the C–4/C–4′ was assigned to the chemical shift at δ 131.95 ppm. The 13C peaks for the triphenylphosphine have the ipso-carbon at 137.11 ppm. The ortho-, meta-, and para-carbons were assigned to δ 133.75, 132.52 and 129.43 respectively. The C–2 carbon of the 4-imidazoleacrylic acid was assigned to the peak at δ 137.02 ppm, the C–4 has its peak at δ 133.60, and C–5 is at δ 129.15. The α-carbon and β-carbon were assigned to the peaks at δ 135.80 and 140.12 ppm respectively. The 31P NMR for complex W, also showed a singlet peak at δ 25.48 ppm (see Fig. S4–S6).
Complex multiplet peak patterns were observed in the aromatic region of the 1H NMR spectrum of X. The multiplet peaks were separated into eight major peaks at chemical shifts between δ 7.64–7.53 ppm, with all peaks integrating to a total of 25 protons. The complexity of the 1H spectrum is likely due to the more mixed ligand substitutions arising from the two dione substitutions at positions 5 and 6 of the 1,10-phenanthroline and 4-imidazoleacrylic acid ligands. Similarly, as was reported for complexes V2 and W, a broad singlet signal peak at δ 3.36 ppm integrating for 77 protons was attributed to the appreciable presence of water molecules in the complex. It could be inferred that the stoichiometry value for water molecules in complex X, relative to the total number of protons in the aromatic region is 3.
The 13C NMR spectrum for complex X shows the carbonyl carbon peak at δ 170.15, which, due to the symmetry of the 1,10-phendione ligand, was assigned to the carbon at position 6. The carbonyl peak at δ 166.91 was assigned to the carboxylic acid group of the 4-imidazoleacrylic acid. The other carbon peaks for the 1,10-phenadione include C–1 at δ 154.85, C–2 at δ 126.95, C–3 at δ 138.83, C–4 at δ 129.15, and C–5 at δ 148.78. The peaks for the triphenylphosphine ligand were observed for the ipso-carbon at 136.11 ppm. The ortho-, meta-, and para-carbons were assigned to δ 133.53, δ 132.72, and δ 129.25, respectively. The C–2 carbon of the 4-imidazoleacrylic acid was assigned to the peak at δ 131.95 ppm, the C–4 has its peak at δ 132.48, and C–5 is at δ 129.15. The α-carbon and β-carbon were assigned to the peaks at δ 131.88 and δ 132.50 ppm, respectively. The 31P NMR spectrum of complex X resonates at δ 25.46 ppm (see Fig. S7–S10).
Electronic absorption studies of complexes V2, W and X
The electronic absorption spectra of all the newly prepared complexes V2, W and X are shown in Fig. 1 and summarized in Table 1. The solid-line spectrum is for V2 (black), the dashed line is for W (red) and the dotted line is for X (blue). It has been established that at room temperature, the electronic absorption spectra of typical ruthenium(II) polypyridine complexes are often dominated by ligand-based spin-allowed π → π* and n → π* transitions in the UV region of the spectra and by 1MLCT absorption bands in the visible region.45,46 All the spectra for complexes V2, W and X showed absorption bands at the UV and visible regions. Prominent peak at λmax 228 nm common to all complexes but with different absorbances, is assigned to a spin-allowed ligand-centered π–π* transition, possibly originating from the 4-imidazoleacrylic acid and triphenylphosphine.| Complex | λ max/nm, (ε, M−1 cm−1) | λ em/nm | Quantum Yield (Φ) | Stokes shift (nm) | E 1/2 (V) (RuII/III) | E cathodic (V) | E anodic (V) | E 1/2 (V) Ligand Reductions |
|---|---|---|---|---|---|---|---|---|
| V2 | 492 (1615), 364 (3438), 283 (9563), 227 (6760) | 712 | 0.21 | 220 | +0.86 | +0.62 | –0.81 | –0.15 |
| W | 475 (1034), 336 (1603), 299 (5224), 272 (7889), 228 (9822) | 643 | 0.31 | 168 | +1.15 | +0.94 | –0.98 | –0.76 |
| X | 494 (4866), 378 (9205), 287 (6599), 272 (6863), 267 (6450), 228 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742) 742) |
641 | 0.66 | 147 | +1.08 | NA | –0.82 | –0.42 |
The second band was found in the region of 275–283 nm. Literature has shown that this region is assigned to the π–π* transition of phenanthroline and/or bipyridine containing ligands of typical ruthenium complexes.47 The small absorption peak at 299 nm which is conspicuously absent in V2 and X, is assumed to originate from the contribution of the bipyridine substituted carboxylic acid moieties present in complex W. This band has previously been assigned to intraligand π–π* transition in 2,2′ bipyridyl in which the position is not affected upon the introduction of phosphine ligands to the complex inner sphere.48 All complexes show a band ascribed to the ligand-to-ligand charge-transfer (LLCT) at λmax between 344–373 nm. The bands at 492 nm, 475 nm and 494 nm were adduced to the metal-to-ligand charge-transfer (MLCT) for complexes V2, W and X respectively.
The results are supported by previously reported data for heteroleptic complexes containing both mono- and bidentate ligands.49–51 The presence of the extra carboxylic acid in complex W enhanced the molar extinction coefficient, though with a blue-shift when compared to complexes V2 and X. This has been attributed to efficient π-delocalization as a result of additional electron-donating ability, which leads to a lowering of the π-orbital energy.52
Emission studies of complexes V2, W and X
Emission studies of metal complexes, particularly ruthenium(II) complexes, have provided a powerful tool for drug design, for investigating biochemical processes at the subcellular level, and as functional molecules for the rapid development of fluorescent probes. Complexes V2, W, and X are designed to possibly fit into such applications. As shown in Fig. 2, the emission spectra of complexes V2, W, and X were compared. Upon excitation at the 1LC and 1MLCT bands, (λexc. = 400 nm), all complexes display appreciable luminescence at room temperature. An emission wavelength maximum was found at 713 nm for V2, which is blue-shifted to lower intensity at 643 and 641 nm for complexes W and X, respectively. | ||
| Fig. 2 Excitation spectra at emission maxima for Complexes V2, W, and X (1 × 103 M) in DMSO; room temperature. (A) Full spectra, (B) Infra-red region of emission. | ||
It is well known that the presence of triphenylphosphine, a strong-field ligand, often affects the strong metal-to-ligand charge transfer (MLCT) transitions of polypyridines. It also influences the excited state and the energy ordering of its low-energy excited state, in particular, the orbital nature of its lowest excited state. This is likely due to the bulky nature of the ligand, which potentially influences the geometry of the complexes, thus leading to a shift in emission wavelength as well as emission intensity.53
It can be seen that the choice of ligands has a significant effect on the energy positions of the ligand center (LC), the metal center (MC), and the metal-to-ligand charge transfer (MLCT), which depends on the coordination of the donor/acceptor groups. It is obvious, when compared to V2, that the excess carboxylic acid group functionality at the 4,4′-positions of 2,2′-bipyridine in complex W, as well as the dione groups at the 5,6-positions of 1,10-phenanthroline in complex X, in combination with other ligands like 4-imidazole-acrylic acid and triphenylphosphine, significantly affect the energy ordering. This is such that the HOMO–LUMO energy gap and the singlet–triplet splitting energy are affected, thereby changing the overall energy splitting of the LC excited state, which has been found to depend on the intrinsic properties of each ligand.42
The quantum yield was found to depend on the intrinsic properties of each ligand.42 The calculated quantum yield of the complexes showed that complex X had the highest quantum yield at 0.66, followed by complex W at 0.31, and V2 at 0.21. The quantum yields seemed to increase following a pattern with a ratio of approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for complexes V2, W, and X, respectively. Ordinarily, it is well known that the dione functionality at positions 5 and 6 is able to introduce significant electronic perturbations into the ligand compared to unsubstituted 1,10-phenanthroline and/or bipyridine.54,55 This agrees with literature on the linear negative correlations between Stokes shift and emission quantum yield, which are a consequence of the reduced degrees of freedom of the intramolecular rotation, possibly due to the presence of the 1,10-phenanthroline-5,6-dione.
3 for complexes V2, W, and X, respectively. Ordinarily, it is well known that the dione functionality at positions 5 and 6 is able to introduce significant electronic perturbations into the ligand compared to unsubstituted 1,10-phenanthroline and/or bipyridine.54,55 This agrees with literature on the linear negative correlations between Stokes shift and emission quantum yield, which are a consequence of the reduced degrees of freedom of the intramolecular rotation, possibly due to the presence of the 1,10-phenanthroline-5,6-dione.
Accordingly, the intramolecular rotation restriction gives rise to an inverse relationship between their Stokes shift and quantum yield, whereby smaller Stokes shifts correspond to greater quantum yields, and vice versa56 (see Table 1). Although, in complex V2, the interaction between the metal d-orbital and the ligand π-systems may possibly be responsible for the significant contributions to the excited state, which leads to the observed intense emission and extension of the wavelength to the red region, better than W and X complexes.
Electrochemical studies of complexes V2, W and X
The voltammograms display the Ru(III)/Ru(II) couple at positive potentials and ligand-based reduction couples at negative potentials. A typical cyclic voltammogram and square-wave voltammogram are shown in Fig. 3 and Table 1 for comparison of the structural compositions of the complexes. It is a clear indication that reduction and oxidation processes are observed in complexes V2, W, and X, which have unfunctionalized or functionalized polypyridines. It appeared that complexes with 2,2′-bipyridyl-4,4′-dicarboxylic acid in coordination with 4-imidazoleacrylic acid, as shown in complex W (peak oxidation +1.15 V), had a greater influence on the redox processes compared to the corresponding unsubstituted 2,2′-bipyridine and 1,10-phenanthroline-5,6-dione. These latter complexes, V2 and X, had peak oxidation values of +0.86 V and +1.08 V, respectively. This was unequivocally assigned to the reversible one-electron oxidation process of the metal center, specifically the Ru(III)/Ru(II) wave couple.46 Often, the metal-centered (parent πM(t2g) in octahedral symmetry) orbital of polypyridine is ascribed to the formation of genuine Ru(III) complexes (low-spin 4d5 configuration) that are inert to ligand substitution, which relates to their oxidation.57| [Ru2+(LL3)]2+ ↔ [Ru3+(LL)3]3+ + e− |
Other redox waves include cathodic potentials at +0.62 and +0.94 V for V2 and W, respectively, neither of which is conspicuously shown for X. In addition, anodic potentials are found at −0.81, −0.98 V and −0.82 V for V2, W, and X. The half-potential for the ligand reductions was recorded at −0.15, −0.76 and −0.42 V for the complexes. The reduction corresponds to the reduction of the ligands that have the carboxyl acid and triphenylphosphine groups.58 In mixed-ligand complexes like these, electron transitions upon optical absorption would occur between the metal center and the ligand that is most easily reducible. The electron-withdrawing character of the carboxy group would shift the reduction potential of the ligand more positively relative to that of the unsubstituted bipyridine ligand. Thus, it was shown in this work that Ru(III)/Ru(II) redox couples are related to both donor/acceptor properties of Group 5 ligands. The effects of these properties are seen in terms of differential stabilization of Ru(III) over Ru(II) for the donor group properties, while the acceptor properties enhance the further stability of Ru(II).59 In general, cyclic voltammetry spectra of the complexes have shown that several reduction steps due to a strong ligand effect may be found in Ru(II) polypyridine-type complexes, which are most often caused by the presence of mixed-ligands or when the ligands are easily reduced.
Chemical reactivity descriptors and quantum chemical calculations
Density functional theory (DFT) was employed to investigate the electrostatic interactions, electronic properties, and chemical reactivity of compounds V2, W, and X. Key reactivity parameters are summarized in Table 2, and the optimized molecular orbitals are depicted in Fig. 4. Frontier molecular orbitals, particularly the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO), play a critical role in determining photochemical reactions, pharmaceutical activity, and biological interaction mechanisms. The energy gap (Eg) between HOMO and LUMO is a crucial indicator of a molecule's reactivity and stability. A smaller Eg facilitates electron transfer, enhancing reactivity, which is essential for effective drug-receptor interactions, leading to stabilizing effects and improved binding affinity.60 An optimal Eg ensures balanced binding, avoiding weak interactions associated with large gaps or excessive affinity that could result in toxicity due to very small or negative gaps.| EH (eV) | EL (eV) | E g (eV) | IP (eV) | EA (eV) | χ (eV) | μ (eV) | η (eV) | σ (eV) | ω (eV) | |
|---|---|---|---|---|---|---|---|---|---|---|
| V2 | −5.8401 | −3.3026 | 2.5375 | 5.8401 | 3.3026 | 4.5714 | −4.5714 | 1.2688 | 0.7881 | 8.2352 |
| W | −6.3748 | −3.3078 | 3.067 | 6.3748 | 3.3078 | 4.8413 | −4.8413 | 1.5335 | 0.6521 | 7.6421 |
| X | −5.7786 | −4.5329 | 1.2457 | 5.7786 | 4.5329 | 5.1558 | −5.1558 | 0.6229 | 1.6054 | 21.3375 |
 | ||
| Fig. 4 Frontier molecular orbital plots of V2, W, and X predicted by LANL2DZ basis set with DFT/B3LYP functional. | ||
For compound V2, the HOMO is primarily distributed over the 4-imidazole acrylic acid moiety, extending to the central ruthenium metal and one chlorine atom, with an energy of −5.8401 eV. The LUMO, on the other hand, is localized across the chlorine atoms and the central metal atom, with an energy of −3.3026 eV, resulting in an energy gap (ΔE) = 2.5375 eV (Fig. 4). This distribution indicates significant intramolecular charge transfer within the molecule. Similarly, for compound W, the HOMO is distributed over the 4-imidazole acrylic acid moiety, extending to the central ruthenium metal and both chlorine atoms, with an energy of −6.3748 eV. The LUMO is concentrated on the central metal atom bonded to both chlorine atoms, with slight spill-over onto one nitrogen atom of the 2,2′-bipyridine-4,4′-dicarboxylic acid, yielding an energy of −3.3078 eV and an energy gap (ΔE) = 3.067 eV (Fig. 4). In contrast, compound X exhibits a distinct electronic distribution. The HOMO is localized over the 4-imidazole acrylic acid moiety, with minor spillover onto the central ruthenium metal, and has energy of −5.7786 eV. The LUMO, however, is primarily localized on the 1,10-phenanthroline-5,6-dione moiety, with an energy of −4.5329 eV, resulting in a smaller energy gap (ΔE) of 1.2457 eV (Fig. 4). This smaller gap indicates higher reactivity and clearly defined electronegative and electron affinity sites, further confirming intramolecular charge transfer.
Additional reactivity descriptors, including chemical hardness (η) and softness (σ), are presented in Table 2. These parameters measure a molecule's resistance and susceptibility to charge transfer, respectively, and further elucidate its reactivity.61V2 (η = 1.2688 eV, σ = 0.7881 eV), W (η = 1.5335 eV, σ = 0.6521 eV), and X (η = 0.6229 eV, σ = 1.6054 eV) exhibit high softness, suggesting favorable interactions with biomolecules. The global electrophilicity index (ω), which quantifies a compound's tendency to attract or donate electrons, further supports their reactivity. High electrophilicity index values indicate that these molecules are likely to act as electrophiles in biological reactions,62 while lower values of electrophilicity index and chemical potential signify strong nucleophilic reactivity.63 Notably, a strong electrophile is characterized by ω > 1.5 eV.64 The electrophilicity index values for V2 (ω = 8.2352 eV), W (ω = 7.6421 eV), and X (ω = 21.3375 eV) are significantly high, underscoring their potential as promising drug candidates.
To investigate the coordination environment around the Ru(II) centre, such as ligand atoms (Cl, N, and P), and assess distortions from ideal octahedral geometry, we evaluated the angular distortion parameter Σ for three complexes (V2, W, and X) based on DFT-optimized geometries obtained using the B3LYP functional and LANL2DZ basis set in Gaussian 16. No symmetry constraints were imposed during optimization. In addition to electronic structural analysis, bond lengths (Å) and bond angles (°) around the central metal atom are provided in the Table 3.
| Property | Bond length (Å)/Bond angle (°) V2 | Bond length (Å)/Bond angle (°) W | Bond length (Å)/Bond angle (°) X |
|---|---|---|---|
| Ru–Cl39 (Cl1, atom 2) | 2.401 | 2.427 | 2.457 |
| Ru–Cl34 (Cl2, atom 4) | 2.457 | 2.413 | 2.448 |
| Ru–N (N13, atom 13) | 2.008 | 1.973 | 2.035 |
| Ru–N (N6, atom 6) | 2.145 | 2.247 | 2.160 |
| Ru–N (N7, atom 7) | 4.390 | 3.813 | 2.101 |
| Ru–P | 2.496 | 2.474 | 2.250 |
| Cl–Ru–Cl | 98.055 | 151.197 | 170.508 |
| N6–Ru–N | 89.267 (N13), 54.355 (N7) | 85.122 (N13), 55.549 (N7) | 170.956 (N13), 77.626 (N7) |
| N7–Ru–N13 | 60.672 | 121.431 | 93.538 |
| Cl–Ru–P | 91.056 (Cl1), 89.209 (Cl2) | 89.757 (Cl1), 87.240 (Cl2) | 88.475 (Cl1), 95.000 (Cl2) |
| Cl–Ru–N6 | 92.292 (Cl1), 85.627 (Cl2) | 84.997 (Cl1), 98.718 (Cl2) | 85.648 (Cl1), 84.985 (Cl2) |
| Cl–Ru–N13 | 92.074 (Cl1), 168.819 (Cl2) | 114.158 (Cl1), 94.639 (Cl2) | 96.106 (Cl1), 92.914 (Cl2) |
| Cl–Ru–N7 | 133.913 (Cl1), 108.456 (Cl2) | 104.417 (Cl1), 57.365 (Cl2) | 88.196 (Cl1), 88.301 (Cl2) |
| P–Ru–N | 174.195 (N6), 125.354 (N7), 95.352 (N13) | 174.042 (N6), 128.896 (N7), 94.482 (N13) | 102.194 (N6), 176.671 (N7), 86.741 (N13) |
| Σ (Cis-angle deviation) | 24.73° | 30.72° | 47.59° |
The Σ (Sigma) parameter, defined as the sum of absolute deviations of the 12 cis angles from 90°, provides a rapid and intuitive measure of angular distortion in pseudo-octahedral systems.65 This parameter is determined by the equation:
These findings are consistent with reported angular distortions in low-spin Ru(II) complexes bearing asymmetric or hydrogen-bonding ligands, where Σ values typically range from 20° to 50°, depending on ligand field strength and bite angles. Although high-spin states generally exhibit larger distortions,66,67 the present calculations were performed at the ground-state configuration, which for Ru(II) is typically low-spin, resulting in moderate but chemically reasonable deviations. Overall, the Σ parameter effectively captures angular strain and supports the structural integrity of the DFT-optimized geometries. These quantitative insights into ligand-induced distortion provide a valuable framework for comparing coordination environments and may inform the rational design of Ru-based catalysts, photochemical agents, or functional materials.
Molecular electrostatic potential (MEP)
The molecular electrostatic potential (MEP) surface analysis is a powerful tool for identifying reactive sites within a molecule by mapping the distribution of nuclear and electronic charges. This analysis highlights regions prone to nucleophilic and electrophilic attacks, providing insights into molecular reactivity. Using the B3LYP/LANL2DZ method, the MEP surfaces of compounds V2, W, and X, were examined. The electrostatic potentials are visualized using a red-green-blue colour gradient: blue regions indicate electrophilic sites (positive electrostatic potential), red regions correspond to nucleophilic sites (negative electrostatic potential), and green areas represent neutral zones (Fig. 5).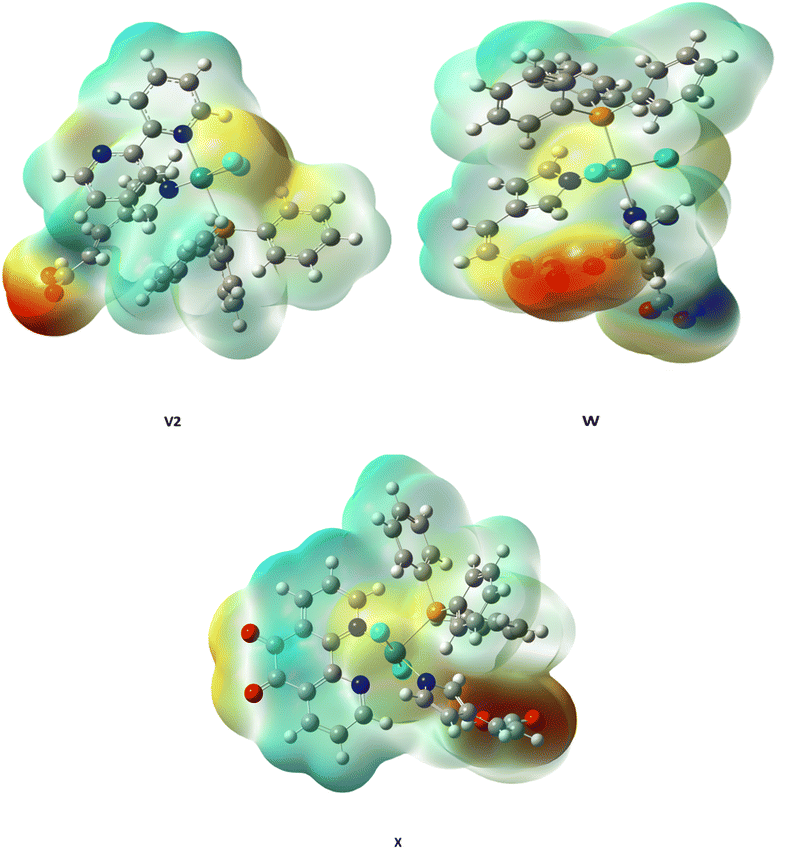 | ||
| Fig. 5 Three-dimensional plot of molecular electrostatic potential (MEP) for V2, W, and X molecules. | ||
For compound V2, the negative electrostatic potential regions are concentrated around the oxygen atom of the carboxylic group in the 4-imidazole acrylic acid moiety. Additionally, a yellow mapping on the chlorine atoms suggests a negative electrostatic potential, which may facilitate π–π stacking or hydrogen bond interactions with aromatic residues in receptor binding sites. This is supported by the molecular interactions observed between V2 and the ALK receptor (Fig. 6). The remaining surface of V2 exhibits a light blue, positive potential and a neutral electrostatic region. In the case of molecule W, nucleophilic centers (negative electrostatic potentials) are primarily located over one of the carboxylic groups in the 2,2′-bipyridine-4,4′-dicarboxylic acid moiety. The electron-rich regions, indicative of nucleophilic sites, are found near the carboxylic group of the 2,2′-bipyridine-4,4′-dicarboxylic acid moiety, adjacent to the carboxylic group of the 4-imidazole acrylic acid moiety. An intermediate potential, a less reactive region marked by yellow, is observed around the chlorine atom closest to the nitrogen atom on the imidazole attachment.
 | ||
| Fig. 6 3D and 2D representation of the molecular interaction between ALK (9GBE) and (A) V2; (B) W; (C) X. | ||
Compound X, however, displays a more scattered distribution of nucleophilic regions. These are predominantly localized on the carboxylic group of the 4-imidazole acrylic acid moiety, the chlorine atoms, and the carbonyl group of the 1,10-phenanthroline-5,6-dione moiety. The electrophilic sites, represented by light blue regions, correspond to electron-deficient areas. These patterns of electrostatic potential distribution reflect electron-rich π-interactions and suggest that the biological activity of these molecules is significantly influenced by variations in their atomic composition and electronic properties. Overall, the MEP analysis provides critical insights into the reactivity and potential binding interactions of V2, W, and X, underscoring the importance of electrostatic potential variations in determining their biological activity.
Molecular docking study
This study investigates the inhibition of biological trafficking involving ERα+, WT EGFR, and ALK receptors, focusing on the distinct binding interactions of compounds V2, W, and X. As shown in Table 4, compound W demonstrated the highest binding affinity to ERα+, with a MolDock score of −202.391. Its interaction with the ligand-binding domain of ERα+ is characterized by three conventional hydrogen bonds, one pi-donor hydrogen bond, one carbon–hydrogen bond, two Pi–Sigma bonds, one Pi–Pi T-shaped bond (with PHE 404), seven Pi–alkyl bonds, and one unfavorable bump with LEU 346. Notably, compound W forms a bifurcated hydrogen bond between ASP351 and PRO535, facilitating antagonistic docking of helix 12 (H12) in the activating function-2 (AF-2) cleft. This interaction is mediated by the carboxylic acid group on the 4-imidazole acrylic acid moiety, highlighting its role in stabilizing the binding.| ERα+ (8DU6) | WT EGFR (8PO4) | ALK (9GBE) | |
|---|---|---|---|
| Ligand | MolDock [GRID] score | MolDock [GRID] score | MolDock [GRID] score |
| V2 | −157.004 | −136.475 | −170.436 |
| W | −202.391 | −156.575 | −182.219 |
| X | −155.301 | −146.189 | −184.563 |
| Crizotinib | −138.783 | −121.169 | −124.025 |
| Doxorubicin | −121.179 | −94.0293 | −121.057 |
| Gemcitabine | −84.3502 | −95.4603 | −83.4191 |
| Lorlatinib | −115.028 | −114.656 | −148.465 |
Compounds V2 and X also exhibited strong binding affinities, outperforming standard reference molecules. However, V2 displayed the highest number of unfavorable bumps (5) and formed only two hydrogen bonds with active site residues. Unlike W and X, V2 lacked interactions with the catalytic residues ASP351 and VAL533, which are critical for the activity of selective estrogen receptor modulators (SERMs) and degraders (SERDs). The presence of hydrogen bonding with ASP351, a key feature of SERMs and SERDs, was observed in both W and X but not in V2. This interaction enhances the potential of W and X as anti-transcriptional agents.
Fig. S17 and S18 (see SI Section) illustrate the 2D and 3D molecular interactions and the distances of these interactions within the ERα+ active site. The figures also provide a detailed representation of the 2D interactions of the standard reference molecules within the same active site. These findings underscore the distinct binding mechanisms of V2, W, and X, highlighting their potential as inhibitors of biological trafficking and their varying efficacies in targeting ERα+ receptors.
For wild-type EGFR (PDB ID: 8PO4), a transmembrane receptor tyrosine kinase that regulates cell proliferation, survival, differentiation, and migration, compounds V2 (−156.575), W (−146.189), and X (−121.169) exhibited superior binding affinities compared to standard drugs. Although Crizotinib displayed a moderate binding energy (−121.169), it formed the highest number of hydrogen bonds (7), including a critical interaction with CYS 797. Compound V2 achieved the lowest binding energy, likely due to a pi–alkyl interaction with CYS 797 via its triphenylphosphine warhead. Notably, only Crizotinib and compound V2 interacted with key binding site residues (MET 766, MET 793, and PHE 856). The binding mode was illustrated in Fig. S19 and S20.
For ALK, a receptor tyrosine kinase often activated by gene fusion, mutations, or deletions in cancers such as NSCLC, neuroblastomas, gliomas, and inflammatory myofibroblastic tumors, compounds V2 (−170.436), W (−182.219), and X (−184.563) outperformed all standard drugs. Crizotinib, a first-generation ALK tyrosine kinase inhibitor (TKI), shows limited efficacy against acquired resistance mutations. Lorlatinib, a third-generation ALK TKI, is brain-penetrant and effective against many resistance mutations but is limited by the emergence of compound ALK mutations, particularly those involving ARG 1202, which confer resistance to all approved ALK TKIs.36
In this study, Lorlatinib and Crizotinib interacted with ARG 1202 via alkyl and pi–alkyl bonds, with binding scores of −148.465 and −124.025, respectively. Compound V2 formed a pi–alkyl interaction with ARG 1202 through its bipyridine moiety, while compound X demonstrated the highest binding affinity (−184.563) via hydrogen bonding and pi–alkyl interactions from its 1,10-phenanthroline-5,6-dione moiety. These interactions suggest that X is a potent inhibitor of ALK on coproteins, with minimal off-target effects on TRK in the nervous system. Most studied compounds interacted with key active site residues (LYS 1150, MET 1196, GLU 1197, LEU 1198, MET 1199, and ARG 1202). Detailed molecular interactions and distances are shown in Fig. 6 and S21. ERα+ (Breast cancer receptor), WT-EGFR, and ALK are all protein targets frequently dysregulated in cancer. WT-EGFR regulates cell growth and survival and is often overexpressed in cancers like non-small cell lung cancer (NSCLC). ALK drives tumorigenesis through gene rearrangements and is a common target in NSCLC and lymphoma.
Physicochemical and drug likeness properties
Optimizing lead compounds for drug development requires careful evaluation of physicochemical properties, lipophilicity, pharmacokinetics, and drug-likeness to ensure therapeutic efficacy. Here, we assess these parameters for V2, W, and X using SwissADME.69 Classical drug-likeness rules (Lipinski, Veber, Egan, Muegge) were examined with a focus on Lipinski, Verber, Lead-likeness, and pan-assay interference (PAINS) alerts. These rules are based on the values of some molecular properties, for example, according to Lipinski rule, poor absorption and permeation are more likely when the molecular weight (MW) > 500 g mol−1, lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) and hydrogen-bond donors (HBD) are more than five, and hydrogen-bond acceptors (HBA) are greater than ten. All compounds exhibited one violation (molecular weight > 500 g mol−1) of Lipinski's rule but otherwise complied (Table 5). However, compound W failed Verber's rule due to excessive rotatable bonds (RB > 10) or topological polar surface area (TPSA > 140 Å2). In drug discovery, “PAINS” (pan-assay interference compounds) are compounds that cause false-positive results in high-throughput screens, leading to the misidentification of potential drug candidates. PAINS alerts, which flag these compounds, are a common screening tool used to weed out such undesirable compounds in the early stages of drug discovery. None of the compounds triggered PAINS alerts, supporting their suitability as drug candidates.
P) and hydrogen-bond donors (HBD) are more than five, and hydrogen-bond acceptors (HBA) are greater than ten. All compounds exhibited one violation (molecular weight > 500 g mol−1) of Lipinski's rule but otherwise complied (Table 5). However, compound W failed Verber's rule due to excessive rotatable bonds (RB > 10) or topological polar surface area (TPSA > 140 Å2). In drug discovery, “PAINS” (pan-assay interference compounds) are compounds that cause false-positive results in high-throughput screens, leading to the misidentification of potential drug candidates. PAINS alerts, which flag these compounds, are a common screening tool used to weed out such undesirable compounds in the early stages of drug discovery. None of the compounds triggered PAINS alerts, supporting their suitability as drug candidates.
| Compounds | Molecular formula | M. W (g mol−1) | HA | AHA | RB | HBA | HBD | MR | TPSA (Å2) |
|---|---|---|---|---|---|---|---|---|---|
| Molecular weight (M. W), num. heavy atoms (HA), number of aromatic heavy atoms (AHA), number of rotatable bonds (RB), number of hydrogen bond acceptors (HBA), number of hydrogen bond donors (HBD), molar refractivity (MR), and topological polar surface area (TPSA). | |||||||||
| V2 | C34H29Cl2N4O2PRu·3H2O | 805.67 | 44 | 35 | 7 | 3 | 1 | 178.62 | 78.57 |
| W | C36H29Cl2N4O6PRu·H2O | 857.20 | 50 | 35 | 9 | 7 | 3 | 192.54 | 153.17 |
| X | C36H27Cl2N4O4PRu·3H2O | 857.21 | 48 | 37 | 7 | 5 | 1 | 191.92 | 112.71 |
The consensus lipophilicity partition Po/w ranged from 3.99 (V2) − 4.05 (X), indicating moderate to high lipophilicity (Table 6). All compounds were poorly soluble (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S < −9.75), with X showing the lowest solubility (log
S < −9.75), with X showing the lowest solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −11.85). This poor solubility correlates with low gastrointestinal (GI) absorption, necessitating alternative administration routes (e.g., intravenous) for systemic delivery. This is because low GI absorption means that a small fraction of the compound taken orally will enter the bloodstream, which is generally unfavorable for systemic cancer treatment, as it limits the sufficient concentration of the drug that reaches the tumor through oral administration, higher doses might be needed, potentially leading to increased side effects. The pharmacokinetic profiling is shown in Table 7.
S = −11.85). This poor solubility correlates with low gastrointestinal (GI) absorption, necessitating alternative administration routes (e.g., intravenous) for systemic delivery. This is because low GI absorption means that a small fraction of the compound taken orally will enter the bloodstream, which is generally unfavorable for systemic cancer treatment, as it limits the sufficient concentration of the drug that reaches the tumor through oral administration, higher doses might be needed, potentially leading to increased side effects. The pharmacokinetic profiling is shown in Table 7.
| Compounds | Consensus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w Po/w |
Water Solubility | Drug-likeness | ||||
|---|---|---|---|---|---|---|---|
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) S (ESOL) |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (Ali) S (Ali) |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (SILICOS-IT) S (SILICOS-IT) |
Lipinski | Verber | Lead likeness | ||
Solubility class (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S scale): Insoluble < −10 < Poorly < −6 < moderately < −4 < soluble < −2 < very < 0 < Highly. All compounds exhibited 0 alert for PAINS. S scale): Insoluble < −10 < Poorly < −6 < moderately < −4 < soluble < −2 < very < 0 < Highly. All compounds exhibited 0 alert for PAINS. |
|||||||
| V2 | 3.99 | −9.65 | −9.75 | −11.29 | Yes; 1 violation | Yes | No; 2 violations |
| W | 3.32 | −9.39 | −10.30 | −9.90 | Yes; 1 violation | No; 1 violation | No; 3 violations |
| X | 4.05 | −9.44 | −9.58 | −11.85 | Yes; 1 violation | Yes | No; 2 violations |
| Compounds | GI absorption | BBB permeant | P-gp substrate | CYP1A2 inhibitor | CYP2C19 inhibitor | CYP2C9 inhibitor | CYP2D6 inhibitor | CYP3A4 inhibitor | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp (skin permeation) cm s−1 Kp (skin permeation) cm s−1 |
|---|---|---|---|---|---|---|---|---|---|
| V2 | Low | No | No | No | No | No | No | No | −4.91 |
| W | Low | No | No | No | No | No | No | No | −6.12 |
| X | Low | No | No | Yes | No | No | No | No | −5.83 |
None of the compounds cross the blood–brain barrier (BBB), which limits their utility for CNS targets (such as brain tumors) but also reduces off-target neurotoxicity. Most compounds showed minimal cytochrome P450 (CYP) enzymes inhibition, which means they have a lower risk of having drug–drug interactions that can alter the metabolism of other drugs metabolized by the same drug. Additionally, low log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp (Skin permeation) values (−4.91 to −6.12 cm s−1) rule out transdermal delivery. In conclusion, while these ruthenium complexes exhibit promising drug-like properties and lack PAINS alerts, their high molecular weight, poor solubility, and low oral bioavailability pose challenges for conventional administration. Future work should explore formulation strategies (e.g., nanoparticulate delivery) or structural modifications to improve pharmacokinetics.
Kp (Skin permeation) values (−4.91 to −6.12 cm s−1) rule out transdermal delivery. In conclusion, while these ruthenium complexes exhibit promising drug-like properties and lack PAINS alerts, their high molecular weight, poor solubility, and low oral bioavailability pose challenges for conventional administration. Future work should explore formulation strategies (e.g., nanoparticulate delivery) or structural modifications to improve pharmacokinetics.
Experimental
General procedure for synthesis of complexes (V2, W and X)
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1612 (C
O), 1612 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1566 (C
C), 1566 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1481, 1435, 1369, 1265 (C–O), 1145, 1118, 1087 (C–N, aryl), 995, 748, 721, 690, (Ru–P), 528 (Ru–N), 420 (Ru–Cl). UV-Vis (ε, M−1 cm−1, nm, (DMSO)): λmax = 492 (1615), 364 (3438), 283 (9563), 227 (6760). 1H-NMR (400 MHz, DMSO-d6, δ): 7.78 (s, 1H), 7.64–7.60 (m, 4H), 7.57–7.53 (m, 3H), 7.49 (s, br, 1H), 7.46 (d, J = 12.0 Hz, 1H), 7.41–7.39 (m, 10H), 7.25–7.23 (m, 5H), 7.22 (m, 1H), 6.28 (d, J = 12.0 Hz, 1H). 13C NMR: 168.42, 138.12, 137.95, 137.11, 137.02, 133.75, 133.70, 133.60, 132.47, 131.95, 131.88, 129.43, 129.24, 129.19, 129.15, 115.29; 31P-NMR (162.00 MHz, DMSO-d6, δ): 25.46 (s, P). Anal. calcd for RuC34H29N4O2Cl2P·3H2O (%): C, 52.17; H, 4.52; N, 7.16,; found: C, 53.89; H, 4.99; N, 7.97. TOF-MS-ES +: m/z; found = 808.35 (Anal. calcd 805.67 for [M+Na]+. 3H2O).
N), 1481, 1435, 1369, 1265 (C–O), 1145, 1118, 1087 (C–N, aryl), 995, 748, 721, 690, (Ru–P), 528 (Ru–N), 420 (Ru–Cl). UV-Vis (ε, M−1 cm−1, nm, (DMSO)): λmax = 492 (1615), 364 (3438), 283 (9563), 227 (6760). 1H-NMR (400 MHz, DMSO-d6, δ): 7.78 (s, 1H), 7.64–7.60 (m, 4H), 7.57–7.53 (m, 3H), 7.49 (s, br, 1H), 7.46 (d, J = 12.0 Hz, 1H), 7.41–7.39 (m, 10H), 7.25–7.23 (m, 5H), 7.22 (m, 1H), 6.28 (d, J = 12.0 Hz, 1H). 13C NMR: 168.42, 138.12, 137.95, 137.11, 137.02, 133.75, 133.70, 133.60, 132.47, 131.95, 131.88, 129.43, 129.24, 129.19, 129.15, 115.29; 31P-NMR (162.00 MHz, DMSO-d6, δ): 25.46 (s, P). Anal. calcd for RuC34H29N4O2Cl2P·3H2O (%): C, 52.17; H, 4.52; N, 7.16,; found: C, 53.89; H, 4.99; N, 7.97. TOF-MS-ES +: m/z; found = 808.35 (Anal. calcd 805.67 for [M+Na]+. 3H2O).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v, 250 mL) to obtain a light brown solid, which was further purified by recrystallization in 30–40 mL of diethyl ether. The yield was 67%, with a melting point (mp) of 188–189 °C. FT-IR (cm−1): 3348, 3221 (O–H), 3010 (C–H, alkane), 1658 (C
3, v/v, 250 mL) to obtain a light brown solid, which was further purified by recrystallization in 30–40 mL of diethyl ether. The yield was 67%, with a melting point (mp) of 188–189 °C. FT-IR (cm−1): 3348, 3221 (O–H), 3010 (C–H, alkane), 1658 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1589 (C
O), 1589 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1539 (C
C), 1539 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1411, 1327, 1265, 1141, 1037 (C–N, aryl), 894, 813, 744, 698, 651, (Ru–P), 589 (Ru–N), 461 (Ru–Cl). UV-vis (ε, M−1 cm−1, nm, (DMSO)): λmax 475 (1034), 336 (1602), 299 (5224), 272 (7889), 228 (9822). 1H-NMR (400 MHz, DMSO d6, δ): 7.63–7.59 (m, br, 6H), 7.56–7.52 (m, br, 5H), 7.38 (s, br, 5H), 7.25–7.21 (m, br, 4H), 7.11–7.07 (m, br, 4H), 6.30–6.27 (m, 1H). 13C NMR: 169.36, 165.67, 159.21, 159.17, 140.12, 137.11, 137.02, 135.80, 133.75, 133.60, 132.71, 132.50, 132.48, 131.95, 131.88, 129.43, 129.25, 129.19, 129.15. 31P-NMR (162.00 MHz, DMSO-d6, δ): 25.48 (s, P). Anal. calcd for RuC36H29N4O6Cl2P·H2O (%): C, 52.96; H, 3.59; N, 6.86; found: C, 52.71; H, 3.76; N, 6.08. TOF-MS-ES +: m/z found = 857.20 (Anal. calcd. 857.66 for [M + Na]+·H2O).
N), 1411, 1327, 1265, 1141, 1037 (C–N, aryl), 894, 813, 744, 698, 651, (Ru–P), 589 (Ru–N), 461 (Ru–Cl). UV-vis (ε, M−1 cm−1, nm, (DMSO)): λmax 475 (1034), 336 (1602), 299 (5224), 272 (7889), 228 (9822). 1H-NMR (400 MHz, DMSO d6, δ): 7.63–7.59 (m, br, 6H), 7.56–7.52 (m, br, 5H), 7.38 (s, br, 5H), 7.25–7.21 (m, br, 4H), 7.11–7.07 (m, br, 4H), 6.30–6.27 (m, 1H). 13C NMR: 169.36, 165.67, 159.21, 159.17, 140.12, 137.11, 137.02, 135.80, 133.75, 133.60, 132.71, 132.50, 132.48, 131.95, 131.88, 129.43, 129.25, 129.19, 129.15. 31P-NMR (162.00 MHz, DMSO-d6, δ): 25.48 (s, P). Anal. calcd for RuC36H29N4O6Cl2P·H2O (%): C, 52.96; H, 3.59; N, 6.86; found: C, 52.71; H, 3.76; N, 6.08. TOF-MS-ES +: m/z found = 857.20 (Anal. calcd. 857.66 for [M + Na]+·H2O).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1755 (C
N), 1755 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1643 (C
O), 1643 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1597, 1566 (C
C), 1597, 1566 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1427, 1373, 1315, 1234, 1130, 1041, 1002 (C–N, aryl), 914, 786, 671, 601 (Ru–P), 516 (Ru–N), 420 (Ru–Cl). UV-Vis (ε, M−1 cm−1, nm, (DMSO)): λmax 494 (4866), 378 (9205), 287 (6599), 272 (6863), 267 (6450), 228 (12
N), 1427, 1373, 1315, 1234, 1130, 1041, 1002 (C–N, aryl), 914, 786, 671, 601 (Ru–P), 516 (Ru–N), 420 (Ru–Cl). UV-Vis (ε, M−1 cm−1, nm, (DMSO)): λmax 494 (4866), 378 (9205), 287 (6599), 272 (6863), 267 (6450), 228 (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742). 1H-NMR (400 MHz, DMSO-d6, δ): 7.64–7.63 (m, 3H), 7.62 (m, 5H), 7.60 (m, 4H), 7.59 (m, 3H), 7.56 (m, 4H), 7.55 (m, 2H), 7.54 (m, 2H), 7.53 (m, 2H). 13C NMR: 170.15, 166.91, 154.85, 148.78, 138.83, 136.11, 133.53, 132.72, 132.50, 132.48, 131.95, 131.88, 129.25, 129.15, 126.95. 31P-NMR (162.00 MHz, DMSO-d6, δ): Unresolved. Anal. calcd for RuC36H27N4O4Cl2P·3H2O (%): C, 51.69; H, 3.98; N, 6.70; found: C, 52.96; H, 4.89; N, 6.23. TOF-MS-ES +: m/z, found = 857.21 (Anal. calcd. 859.64 for [M+Na]+·3H2O).
742). 1H-NMR (400 MHz, DMSO-d6, δ): 7.64–7.63 (m, 3H), 7.62 (m, 5H), 7.60 (m, 4H), 7.59 (m, 3H), 7.56 (m, 4H), 7.55 (m, 2H), 7.54 (m, 2H), 7.53 (m, 2H). 13C NMR: 170.15, 166.91, 154.85, 148.78, 138.83, 136.11, 133.53, 132.72, 132.50, 132.48, 131.95, 131.88, 129.25, 129.15, 126.95. 31P-NMR (162.00 MHz, DMSO-d6, δ): Unresolved. Anal. calcd for RuC36H27N4O4Cl2P·3H2O (%): C, 51.69; H, 3.98; N, 6.70; found: C, 52.96; H, 4.89; N, 6.23. TOF-MS-ES +: m/z, found = 857.21 (Anal. calcd. 859.64 for [M+Na]+·3H2O).
Materials and methods
Experimental section
The starting materials used were RuCl2(PPh3)3, 4-imidazoleacrylic acid, 2,2′-bipyridine, 2,2′-bipyridyl-4,4′-dicarboxylic acid, and 1,10-phenanthroline-5,6-dione, which were purchased from Sigma-Aldrich, Alfa Aesar, and Ark Pharm. All complexes, V2, W, and X, were synthesized according to similar reported procedures, with the exception of changes in chemical reagents, reaction time, and purification solvents.71 All required solvents were ordered from Fischer Scientific. To confirm the purity of the synthesized complexes, data were collected using a Bruker Alpha FTIR spectrometer, which was equipped with an ATR platinum Diamond 1 reflectance accessory. Microanalyses (C, H, and N) were carried out with a (Vario Elementar), University of Witwaterstrand, South Africa. A 400 MHz Bruker NMR spectrometer equipped with an auto-sampler was used for the 1H, at 100 MHz for 13C, and at 162.00 MHz for 31P-NMR spectra. Chemical shift values were reported in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. Chemical shifts were also reported with respect to DMSO-d6 at δ C 40.98 ppm and DMSO-d6 at δ H 2.50 ppm. UV-vis absorption and emission data were recorded in a 1 cm path length quartz cell on a PerkinElmer Lambda 35 spectrophotometer and an LS-45 Fluorescence spectrometer, respectively. Luminescence quantum yields were calculated using eqn (1), where ΦD is the quantum yield, F and A are the gradients of luminescence versus absorbance, and η is a solvent refractive index. Subscripts D and std refer to donor and standard, respectively. Quinine sulphate in 0.1 M H2SO4 was employed as the standard (Φstd = 1.33; Astd = 0.050; Fstd = 1000); λex = 400 nm in DMSO = 1.479; room temperature. Melting points were determined by Stuart melting point apparatus analogue SMP 11 at a temperature range between 50–350 °C. Direct injections of the respective samples into a Waters Micromass LCT Premier MS instrument equipped with electrospray ionization (ESI) source and a time-of-flight (TOF) mass analyzer afford the mass spectra. Electrochemical experiments were conducted using an Autolab PGSTAT 302 N electrochemical workstation equipped with an Electrochemical Impedance Spectroscopy (EIS) module and the Autolab NOVA 1.7 software package. During these experiments, a three-electrode voltammetric cell including a glassy carbon working electrode, Ag|AgCl wire as a pseudo reference electrode and Pt counter electrode were used. The square-wave voltammetry settings include a step potential of 4 mV, a frequency of 25 Hz and amplitude of 20 mV. The cyclic and square wave voltammograms of the complexes V2, W, and X were examined in the potential range +1.5 to −1.5 V and at a scan rate 50 mV s−1 in acetonitrile solvent with 0.1 M tetrabutylammonium tetrafluoroborate as supporting electrolyte.Quantum chemical calculations
Density functional theory (DFT) calculations were conducted on the structures of V2, W, and X using Gaussian 16 software,72 on the Lengau cluster at the Center for High-Performance Computing (CHPC) in Cape Town, South Africa. Initial 3D molecular structures were constructed and minimized with molecular mechanics force field by Chem 3D software, incorporating synthesized inhibitors and the obtained standard drugs (Crizotinib, Doxorubicin, Gemcitabine, and Lorlatinib) from PubChem Chemical Database. The stable structures were saved in Mol2 format and fully optimized using the exchange–correlation B3LYP hybrid functional73 and ECP (effective core potential) LANL2DZ basis set was used to model the metal atoms74,75 in a vacuum. Meanwhile for the standard drugs to align with the molecular docking geometry, we optimized the molecules using PM6 basis set in semi-empirical method. Key quantum chemical properties, including frontier orbital energies (EHOMO and ELUMO), energy gap (Eg), ionization energy (IE), electron affinity (EA), electronegativity (χ), chemical hardness (η), chemical softness (σ), and global electrophilicity index (ω) were calculated.76 These parameters provide insight into molecular reactivity and potential interactions with biological systems. The optimized structures are saved in Mol2 format for further computational study.Molecular docking study
Molecular docking targeted three key cancer proteins: estrogen receptor alpha (ERα+) (PDB: 8DU6, 2.10 Å resolution), WT EGFR (PDB: 8PO4, 1.62 Å resolution) and the receptor tyrosine kinase anaplastic lymphoma kinase (ALK) (PDB: 9GBE, 1.58 Å resolution), retrieved from the RCSB Protein Data Bank.77–79 Protein preparation in Molegro Virtual Docker (MVD) involved repairing and optimizing the protein structure, while removing water molecules and cofactors from crystal structure files. Optimized V2, W, and X structures, alongside standard drugs, were docked against these proteins using the MolDock [GRID] algorithm, which integrates differential evolution with cavity prediction.80 MVD automatically identifies potential active sites (also referred to as cavities) by using its cavity detection algorithm. The cavity with the highest volume was selected for consideration. Ten independent docking runs generated single poses per run, ranked based on set of scoring functions, such as MolDock [GRID] and MolDock scores. However, we selected the MolDock [GRID] scores for ranking the inhibitor poses for the receptor targets. The highest-scoring poses were selected for each target receptor, and intermolecular interactions were analysed using Biovia Discovery Studio 2024 and ChimeraX.81 This methodology provided insights into binding affinities and structural interactions critical for evaluating inhibitory potential.ADME prediction
The SwissADME web tool (https://www.swissadme.ch/) evaluated physicochemical properties, pharmacokinetics, lipophilicity, and solubility of the compounds V2, W, and X. SMILES notations of optimized structures were submitted to predict drug-likeness and assess candidacy for further development. All computations were carried out using the computational cluster resources at the Centre for High Performance Computing (CHPC), Cape Town, South Africa.Conclusions
In conclusion, this study reports the facile synthesis of new mixed-ligand heteroleptic Ru(II) complexes. The optical, electrical, and molecular docking properties of compounds V2, W, and X demonstrate the influence of polypyridyl ligand substitutions, highlighting the superior quality of using combinations of bidentate and monodentate ligands in the buildup of potential bioactive anticancer agents. Although compound V2 shows the highest photoluminescent emission intensity and wavelength in the near-infrared region (>700 nm), it surprisingly gave the lowest quantum yield. The quantum yield ratio was found to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for compounds V2, W, and X, respectively. The biological investigations using DFT/molecular docking methods show that the HOMO in complexes V2 and W is primarily distributed over the 4-imidazole acrylic acid moiety, extending to the central ruthenium metal and one chlorine atom. The LUMO, in contrast, is localized across the chlorine atoms and the central metal atom with energy gaps (ΔE) of 2.5375 eV and 3.067 eV, respectively. In contrast, complex X has the HOMO localized over the 4-imidazole acrylic acid moiety, with a minor spill-over onto the central ruthenium metal. Its LUMO is primarily localized on the 1,10-phenanthroline-5,6-dione moiety and exhibits a smaller energy gap (ΔE) of 1.2457 eV. Complex V2 achieved the lowest binding energy compared to complex X, which had the highest binding affinity via hydrogen bonding and pi–alkyl interactions. This was attributed to the presence of 1,10-phenanthroline containing two carbonyl substitutions at positions 5 and 6. Overall, compounds V2, W, and X show superior anti-transcriptional activity compared to the standard drugs, which include crizotinib, doxorubicin, gemcitabine, and lorlatinib. Results of the physicochemical properties, lipophilicity, pharmacokinetics, and drug-likeness to ensure therapeutic efficacy show that the complexes have one violation or another. Specifically, their high molecular weights and/or poor lipophilicity hindered them from crossing the blood–brain barrier (BBB). This limited their utility for CNS targets (such as brain tumors) but reduced off-target neurotoxicity. In conclusion, this study provides new information on the complexes, revealing the strong anticancer potential of the individual complexes, which can be used as biological electrophiles. It is envisaged that the in vitro and in vivo biological activities of the reported complexes will be investigated in the future.
3 for compounds V2, W, and X, respectively. The biological investigations using DFT/molecular docking methods show that the HOMO in complexes V2 and W is primarily distributed over the 4-imidazole acrylic acid moiety, extending to the central ruthenium metal and one chlorine atom. The LUMO, in contrast, is localized across the chlorine atoms and the central metal atom with energy gaps (ΔE) of 2.5375 eV and 3.067 eV, respectively. In contrast, complex X has the HOMO localized over the 4-imidazole acrylic acid moiety, with a minor spill-over onto the central ruthenium metal. Its LUMO is primarily localized on the 1,10-phenanthroline-5,6-dione moiety and exhibits a smaller energy gap (ΔE) of 1.2457 eV. Complex V2 achieved the lowest binding energy compared to complex X, which had the highest binding affinity via hydrogen bonding and pi–alkyl interactions. This was attributed to the presence of 1,10-phenanthroline containing two carbonyl substitutions at positions 5 and 6. Overall, compounds V2, W, and X show superior anti-transcriptional activity compared to the standard drugs, which include crizotinib, doxorubicin, gemcitabine, and lorlatinib. Results of the physicochemical properties, lipophilicity, pharmacokinetics, and drug-likeness to ensure therapeutic efficacy show that the complexes have one violation or another. Specifically, their high molecular weights and/or poor lipophilicity hindered them from crossing the blood–brain barrier (BBB). This limited their utility for CNS targets (such as brain tumors) but reduced off-target neurotoxicity. In conclusion, this study provides new information on the complexes, revealing the strong anticancer potential of the individual complexes, which can be used as biological electrophiles. It is envisaged that the in vitro and in vivo biological activities of the reported complexes will be investigated in the future.
Author contributions
A. O. A. conceived and designed the research idea, carried out the synthesis of the mixed-ligand Ru complexes, performed UV-vis, PL, and electrochemical analyses, and interpreted IR and NMR data. T. K. C. was responsible for the IR and NMR acquisition and analysis. D.D.B carried out experiments on DFT, MEPs as well as the acquisition of data, analysis and interpretation of data. A. O. A. coordinated the writing of the manuscript. All authors read, edited and contributed to the manuscript and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that there are no conflicts to declare or any known competing financial interests of personal relationships that could have appeared to influence the work reported in this paper.Data availability
All authors declare here that all the data generated or analyzed during this study are included in the manuscript and its supplementary information (SI) files and link attached below: https://bit.ly/4c4Irsj. Supplementary information is available. See DOI: https://doi.org/10.1039/d5nj03704b.Acknowledgements
The authors sincerely acknowledge the Department of Chemistry, University of South Africa, for the use of analytical instrumentation. We also thank N.P. Magwa for allowing us to use the facilities at the Centre for Materials Science, University of South Africa, Florida, 1710, Johannesburg, South Africa.Notes and references
- J. Leng, L. Lei, S. F. Lei, Z. Zhu, A. Ocampo and F. Gany, Use of traditional Chinese herbal medicine concurrently with conventional Cancer treatment among Chinese Cancer patients, J Immigr Minor Health, 2020, 22, 1240 CrossRef PubMed.
- L. Paulus, M. Gallardo-Villagrán, C. Carrion, C. Ouk, F. Martin, B. Therrien, D. Y. Léger and B. Liagre, The effect of photosensitizer metalation incorporated into arene–ruthenium assemblies on prostate cancer, Int. J. Mol. Sci., 2023, 24, 13614 Search PubMed.
- J.-F. Beaulieu, Colorectal cancer research: Basic, preclinical, and clinical approaches, Cancers, 2020, 12, 416 CrossRef PubMed.
- H. Dragutan, V. Dragutan and A. Demonceau, Editorial of special issue ruthenium complex: the expanding chemistry of the ruthenium complexes, Molecules, 2015, 20, 17244 CrossRef.
- A. Levina, A. Mitra and P. Lay, Recent developments in ruthenium anticancer drugs, Metallomics, 2009, 1, 458 CrossRef CAS PubMed.
- Y. Qiong, W. Y. Shen, Q. Y. Yang, Z. F. Chen and H. Liang, Ru(III) complexes with pyrazolopyrimidines as anticancer agents: bioactivities and the underlying mechanisms, Dalton Trans., 2022, 51, 1333 RSC.
- S. Katheria, Ruthenium complexes as potential cancer cell growth inhibitors for targeted chemotherapy, ChemistrySelect, 2022, 7, e202201645 CrossRef CAS.
- K. Lin, Z.-Z. Zhao, H.-B. Bo, X.-J. Hao and J.-Q. Wang, Applications of ruthenium complex in tumor diagnosis and therapy, Front. Pharmacol., 2018, 9, 1323 CrossRef CAS PubMed.
- P. J. Dyson and G. Sava, Metal-based antitumour drugs in the post genomic era, Dalton Trans., 2006, 1929 RSC.
- E. S. Antonarakis and A. Emadi, Ruthenium-based chemotherapeutics: Are they ready for prime time?, Cancer Chemother. Pharmacol., 2010, 66, 1 CrossRef CAS.
- J. Honorato, K. M. Oliveira, C. M. Leite, L. Colina-Vegas, J. A. Nóbrega, E. E. Castellano, J. Ellena, R. S. Correa and A. A. Batista, “Half-sandwich”/RuII anticancer complexes containing triphenylphosphine and p-substituted benzoic acids, J. Brazil. Chem. Soc., 2020, 31, 2237 CAS.
- P. Srivastava, M. Shukla, G. Kaul, S. Chopra and A. K. Patra, Rationally designed curcumin based Ruthenium(II) antimicrobials effective against drug-resistant: Staphylococcus aureus, Dalton Trans., 2019, 48, 11822 Search PubMed.
- A. Catalano, A. Mariconda, M. S. Sinicropi, J. Ceramella, D. Iacopetta, C. Saturnino and P. Longo, Biological activities of ruthenium NHC complexes: An update, Antibiotics, 2023, 12, 365 CrossRef CAS PubMed.
- H. Hu, H. Zhang, R. Zhong, Y. Yang, C. Huang, J. Chen, L. Liang, Y. Chen and Y. Liu, Synthesis, RNA-sequence and evaluation of anticancer efficacy of ruthenium(II) polypyridyl complexes toward HepG2 cells, J. Inorg. Biochem., 2023, 244, 112230 CrossRef CAS PubMed.
- R. A. Khan, S. S. Alterary, I. I. Binsharfan, H. Alsaeedi, A. AlFawaz, M. S. Khan, M. H. Jaafar, Y. Shi, H. D. Arman and A. Alsalme, Piano-stool type (η6-p-cymene) ruthenium(II) thiazole-derived motifs complexes: Synthesis, crystal structures, DFT studies, molecular docking and in-vitro binding studies with HSA and cytotoxicity, Inorg. Chim. Acta, 2022, 537, 120925 CrossRef CAS.
- S. Parveen, Recent advances in anticancer ruthenium Schiff base complexes, Appl. Organometal. Chem., 2020, 34, e5687 CrossRef.
- G. H. Ribeiro, L. Colina-Vegas, J. C. T. Clavijo, J. Ellena, M. R. Cominetti and A. A. Batista, Ru(II)/N-N/PPh3 complexes as potential anticancer agents against MDA-MB-231 cancer cells (N-N = diimine or diamine), J. Inorg. Biochem., 2019, 193, 70 CrossRef.
- I. Warad, Synthesis, spectral and structural characterization of novel dichloro ruthenium (II)/triphenylphosphine/(3,4-diaminophenyl)(phenyl)methanone complex, J. Mater. Environ. Sci., 2013, 4, 822 Search PubMed.
- C. G. Hartinger, M. A. Jakupec, S. Zorbas-Seifried, M. Groessl, A. Egger, W. Berger, H. Zorbas, P. J. Dyson and B. K. Keppler, KP1019, a new redox-active anticancer agent–preclinical development and results of a clinical phase I study in tumor patients, Chem. Biodivers., 2008, 5, 2140 CrossRef.
- M. R. Gill, J. Garcia-Lara, S. J. Foster, C. S. G. Battaglia and J. A. Thomas, A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells, Nat. Chem., 2009, 1, 662 CrossRef.
- K. Lin, Z. Z. Zhao, H. B. Bo, X. I. Hao and J. Q. Wang, Applications of ruthenium complex in tumor diagnosis and therapy, Front. Pharmacol., 2018, 9, 1323 CrossRef PubMed.
- B. J. Pages, D. L. Ang, E. P. Wright and J. R. Aldrich-Wright, Metal complex interactions with DNA, Dalton Trans., 2015, 44, 3505 RSC.
- M. R. Gill and J. A. Thomas, Ruthenium(II) polypyridyl complexes and DNA--from structural probes to cellular imaging and therapeutics, Chem. Soc. Rev., 2012, 41, 3179 RSC.
- J. Karges, Encapsulation of Ru(II) polypyridine complexes for tumor-targeted anticancer therapy, BME Front., 2023, 4, 0024 CrossRef PubMed.
- A. Weiss, R. H. Berndsen, M. Dubois, C. Müller, R. Schibli, A. W. Griffioen, P. J. Dyson and P. Nowak-Sliwinsk, In vivo anti-tumor activity of the organometallic ruthenium(ii)-arene complex [Ru(η6-p-cymene)Cl2(pta)](RAPTA-C) in human ovarian and colorectal carcinomas, Chem. Sci., 2014, 5, 4742 RSC.
- D. R. Boer, L. Wu, P. Lincoln and M. Coll, Thread insertion of a Bis(dipyridophenazine) diruthenium complex into the DNA double helix by the extrusion of AT base pairs and cross-linking of DNA duplexes, Angew. Chem., Int. Ed., 2014, 53, 1949 CrossRef.
- J. G. Vos and J. M. Kelly, Ruthenium polypyridyl chemistry; from basic research to applications and back again, Dalton Trans., 2006, 4869 RSC.
- T. Biver, C. Cavazza, F. Secco and M. Venturini, The two modes of binding of Ru(phen)2dppz2+ to DNA: thermodynamic evidence and kinetic studies, J. Inorg. Biochem., 2007, 101, 461 CrossRef PubMed.
- G. S. Khan, A. Shah, R. Zia Ur and D. Barker, Chemistry of DNA minor groove binding agents, J. Photochem. Photobiol., B, 2012, 115, 105 CrossRef PubMed.
- J. K. Barton, A. Danishefsky and J. Goldberg, Tris(phenanthroline)- ruthenium(II): stereoselectivity in binding to DNA, J. Am. Chem. Soc., 1984, 106, 2172 CrossRef.
- B. N. Cunha, L. Colina-Vegas, A. M. Plutín, R. G. Silveira, J. Honorato, K. M. Oliveira, M. R. Cominetti, A. G. Ferreira, E. E. Castellano and A. A. Batista, Hydrolysis reaction promotes changes in coordination mode of Ru(II)/acylthiourea organometallic complexes with cytotoxicity against human lung tumor cell lines, J. Inorg. Biochem., 2018, 186, 147 CrossRef CAS.
- R. Saez, J. Lorenzo, M. J. Prieto, M. Font-Bardia, T. Calvet, N. Omeñaca, M. Vilaseca and V. Moreno, Influence of PPh3 moiety in the anticancer activity of new organometallic ruthenium complexes, J. Inorg. Biochem., 2014, 136, 1 CrossRef CAS PubMed.
- S. Malakar, B. M. Gordon, S. Mandal, T. J. Emge and A. S. Goldman, Ruthenium complexes of a triphosphorus-coordinating pincer ligand: Ru–P ligand-substituent exchange reactions driven by large variations of bond energies, Inorg. Chem., 2023, 62, 4525 CrossRef CAS PubMed.
- K. Akatsuka, R. Abe, T. Takase and D. Oyama, Coordination chemistry of Ru(II) complexes of an asymmetric bipyridine analogue: Synergistic effects of supporting ligand and coordination geometry on reactivities, Molecules, 2019, 19, 27 CrossRef.
- T. Munir, E. Aneggi, W. Baratta, L. Genesin, D. Zuccaccia and F. Trigatti, Rapid and efficient solid state mechanosynthesis of bipyridine metal complexes, Chem. – Eur. J., 2025, 31, 2202501214 CrossRef.
- N. Zacharopoulos, E. Kolovou, A. Peppas, K. Koukoulakis, E. Bakeas, G. Schnakenburg and A. I. Philippopoulos, Pyridyl based ruthenium(II) catalyst precursors and their dihydride analogues as the catalytically active species for the transfer hydrogenation of ketones, Polyhedron, 2018, 154, 27 CrossRef CAS.
- E. Arguello, A. Bolanos, F. Cuenu, M. Navarro, V. Herrera, A. Fuentes and R. A. Sánchez-Delgado, Synthesis, characterization and some catalytic properties of ruthenium complexes Ru(PPh3)2Cl2(L)2 [L = 4-But-py, 4-vinyl-py, 4-CN-py, 4-Me-py, 3-Me-py, L2 = 4,4′-bipy]. Kinetics of cyclohexene hydrogenation catalysed by Ru(PPh3)2Cl2(4-But-py)2, Polyhedron, 1996, 15, 909 CrossRef CAS.
- A. A. Batista, M. O. Santiago, C. L. Donnici, I. S. Moreira, P. C. Healy, S. J. Berners-Price and S. L. Queiroz, Electrochemical and spectroscopic studies on RuCl2(PPh3)2(N)2 and RuCl2(PPh3)2 (N–N) complexes (N = pyridine derivatives and N–N = phenanthroline or bipyridine derivatives). X-ray structure of RuCl2(PPh3)2(phen), Polyhedron, 2001, 20, 2123 CrossRef CAS.
- A. O. Adeloye, P. A. Ajibade, D. D. Babatunde and N. Magwa, Emissive luminescent, redox-active Ru(II) bischlorido- (triphenylphosphine) complexes containing functionalized imidazole tethered thiourea and dithiocarbamate: characterization and preliminary anticancer quantum chemical calculations, J. Mol. Struct., 2025, 1347, 143335 CrossRef CAS.
- F. Tian, H. Qu, A. Zimmermann, T. Munk, A. C. Jørgensen and J. Rantanen, Factors affecting crystallization of hydrates, J. Pharm. Pharmacol., 2010, 62, 1534 CrossRef CAS.
- A. Bartyzel, B. Cristóvão and R. Lyszczek, Crystal structure and thermal studies of coordination compounds, Crystals, 2020, 10, 1108 CrossRef CAS.
- A. O. Adeloye, Synthesis, photophysical and electrochemical properties of a mixed bipyridyl-phenanthrolyl ligand Ru(II) Heteroleptic complex having trans-2-Methyl-2-butenoic acid functionalities, Molecules, 2011, 16, 8353 CrossRef CAS.
- T. Fujihara, R. Okamura, T. Wada and K. Tanaka, Coordination ability of 1,10-phenanthroline-5,6-dione: syntheses and redox behavior of a Ru(ii) complex with an o-quinoid moiety and of bridged Ru(ii)–M(ii) complexes (M = Pd, Pt), Dalton Trans., 2003, 3221 RSC.
- N. W. Duffy, K. D. Dobson, K. C. Gordon, B. H. Robinson and A. J. McQuillan, In situ infrared spectroscopic analysis of the adsorption of ruthenium(II) bipyridyl dicarboxylic acid photosensitisers to TiO2 in aqueous solutions, Chem. Phys. Lett., 1997, 266, 451 CrossRef CAS.
- A. L. Rodriguez, G. Peron, C. Duprat, M. Vallier, E. Fouquet and F. Fages, The use of a monoorganotin derivative of pyrene in the palladium(0)-catalyzed synthesis of a new metal-cation complexing molecule displaying excited-state charge-transfer properties, Tetrahedron Lett., 1998, 39, 1179 CrossRef CAS.
- A. O. Adeloye and P. A. Ajibade, Towards the development of functionalized polypyridine ligands for Ru(II) complexes as photosensitizers in dye-sensitized solar cells (DSSCs), Molecules, 2014, 19, 12421 CrossRef.
- K. Kalyanasundaram and M. K. Nazeeruddin, Photophysics and photoredox reactions of ligand bridged binuclear polypyridyl complexes of ruthenium(II) and of their monomeric analogues, Inorg. Chem., 1990, 29, 1888 CrossRef CAS.
- A. Y. Ershov, S. V. Litke, A. S. Litke, T. V. Mezentseva, Y. M. Grigor'ev and G. N. Lyalin, Spectral properties of ruthenium(II) mixed-ligands complexes with 2,2’-bipyridyl and phosphines, Russian J. Gen. Chem., 2001, 71, 1329 CrossRef CAS.
- P. Sengupta, S. Ghosh and T. C. W. Mak, A new route for the synthesis of bis(pyridine dicarboxylato)bis (triphenyl phosphine) complexes of ruthenium(II) and X-ray structural characterization of the biologically active trans-[Ru(PPh3)2(L1H)2] (L1H2 = pyridine 2,3-dicarboxylic acid), Polyhedron, 2001, 20, 975 CrossRef CAS.
- E. Arguello, A. Bolaños, F. Cuenu, M. Navarro, V. Herrera, A. Fuentes and R. A. Sanchez-Delgado, Synthesis, characterization and some catalytic properties of ruthenium complexes Ru(PPh3)2Cl2(L)2 [L = 4-But-py, 4-vinyl-py, 4-CN-py, 4-Me-py, 3-Me-py, L2 = 4,4′-bipy]. Kinetics of cyclohexene hydrogenation catalysed by Ru(PPh3)2Cl2(4-But-py)2, Polyhedron, 1996, 15, 909 CrossRef CAS.
- A. Juris, V. Balzani, F. Barigelletti, S. Campagna, P. Belser and A. Von Zelewski, Ru(II) polypyridine complexes: photophysics, photochemistry, eletrochemistry, and chemiluminescence, Coord. Chem. Rev., 1988, 84, 85 CrossRef CAS.
- A. O. Adeloye, S. Koudjina, V. Kumar, K. C. Tapala, J. D. Gbenou and P. Chetti, One-pot synthesis, photophysical, electrochemical and molecular property of mixed-ligands ruthenium complex functionalities: Potential materials for dye-sensitized solar cells (DSSCs), Dyes Pigm., 2025, 235, 112626 CrossRef CAS.
- E. Biasin, D. R. Nascimento, B. I. Poulter, B. Abraham, K. Kunnus, A. T. Garcia-Esparza, S. H. Nowak, T. Kroll, R. W. Schoenlein, R. Alonso-Mori, M. Khalil, N. Govind and D. Sokaras, Revealing the bonding of solvated Ru complexes with valence-to-core resonant inelastic X-ray scattering, Chem. Sci., 2021, 12, 3713 RSC.
- S. S. Bhat, V. K. Revankar, R. V. Pinjari, V. Kumbar, K. Bhat and V. A. Kawade, Synthesis, characterization and biological properties of ruthenium(II) polypyridyl complexes containing 2(1H)-Quinolinone-3(1H-imidazo[4,5f][1,10]phenanthrolin-2-yl), Inorg. Chem. Commun., 2022, 138, 109260 CrossRef CAS.
- L. C.-C. Lee and K. K.-W. Lo, Shining new light on biological systems: Luminescent transition metal complexes for bioimaging and biosensing applications, Chem. Rev., 2024, 124, 8825 CrossRef CAS.
- J. J. Alcázar, L. García-Río, A. I. Robles, L. D. Villarroel, A. Fierroa, J. G. Santos and M. E. Aliaga, Linear relationship between emission quantum yield and Stokes shift in 3-styryl aza-coumarin based dyes in the presence of cyclodextrins, J. Mol. Liq., 2023, 381, 121790 CrossRef.
- A. Islam, H. Sugihara and H. Arakawa, Molecular design of ruthenium(II) polypyridyl photosensitizers for efficient nanocrystalline TiO2 solar cells, J. Photochem. Photobiol., A, 2003, 158, 131 CrossRef CAS.
- P.-H. Xie, Y.-J. Hou, B.-W. Zhang, Y. Cao, F. Wu and W.-J. Tian, J-C. Shen. Spectroscopic and electrochemical properties of ruthenium(II) polypyridyl complexes, J. Chem. Soc., Dalton Trans., 1999, 23, 4217 RSC.
- C. A. Tolman, Steric effects of phosphorus ligands in organometallic chemistry and homogeneous catalysis, Chem. Rev., 1977, 77, 313 CrossRef CAS.
- T. C. Jeyakumar, R. A. Timothy, O. C. Godfrey, R. Rajaram, E. C. Agwamba, O. E. Offiong and H. Louis, Insight into the crystal structure analysis, vibrational studies, reactivities (MESP, HOMO-LUMO, NBO), and the anticancer activities of ruthenium diazide [Ru(POP)(PPh3)(N3)2] complex by molecular docking approach, Inorg. Chem. Commun., 2023, 158, 111714 CrossRef CAS.
- O. E. Oyeneyin, A. Ibrahim, N. Ipinloju, A. J. Ademoyegun and N. D. Ojo, Insight into the corrosion inhibiting potential and anticancer activity of 1-(4-methoxyphenyl)-5-methyl-N’-(2-oxoindolin-3-ylidene)-1H-1,2,3-triazole-4-carbohydrazide via computational approaches, J. Biomol. Struct. Dyn., 2004, 42, 11149 CrossRef PubMed.
- A. Ramalingam, M. Kuppusamy, S. Sambandam, M. Medimagh, O. E. Oyeneyin, A. Shanmugasundaram, N. Issaoui and N. D. Ojo, Synthesis, spectroscopic, topological, Hirshfeld surface analysis, and anti-covid-19 molecular docking investigation of isopropyl 1-benzoyl-4-(benzoyloxy)-2,6-diphenyl-1,2,5,6-tetrahydropyridine-3-carboxylate, Heliyon, 2022, 8, e10831 CrossRef CAS PubMed.
- A. Mueller-Fahrnow and U. Egner, Ligand-binding domain of estrogen receptors, Curr. Opin. Biotechnol, 1999, 10, 550 CrossRef CAS PubMed.
- L. R. Domingo, M. Ríos-Gutiérrez and P. Pérez, Applications of the conceptual density functional theory indices to organic chemistry reactivity, Molecules, 2016, 21, 748 CrossRef PubMed.
- R. Ketkaew, Y. Tantirungrotechai, P. Harding, G. Chastanet, P. Guionneau, M. Marchivie and D. J. Harding, OctaDist: A tool for calculating distortion parameters in spin crossover and coordination complexes, Dalton. Trans., 2021, 50, 1086 RSC.
- I. Capel Berdiell, E. Michaels, O. Q. Munro and M. A. Halcrow, A Survey of the Angular Distortion Landscape in the Coordination Geometries of High-Spin Iron(II) 2,6-Bis(pyrazolyl)pyridine Complexes, Inorg. Chem., 2024, 63, 2732 CrossRef CAS PubMed.
- J. N. McPherson, R. W. Hogue, F. Sunday Akogun, L. Bondì, E. T. Luis, J. R. Price, A. L. Garden, S. Brooker and S. B. Colbran, Predictable Substituent Control of Co III/II Redox Potential and Spin Crossover in Bis(dipyridylpyrrolide)cobalt Complexes, Inorg. Chem., 2019, 58 Search PubMed.
- Y. Li, J. C. Huffman and A. H. Flood, Can terdentate 2,6-bis(1,2,3-triazol-4-yl)pyridines form stable coordination compounds?, Chem. Commun., 2007, 2692 RSC.
- A. Daina, O. Michielin and V. Zoete, SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules, Sci. Rep., 2017, 7, 1–13 CrossRef.
- D. S. Eggleston, K. A. Goldsby, D. J. Hodgson and T. J. Meyer, Structural variations induced by changes in oxidation state and their role in electron transfer crystal and molecular structures of cis-[Ru(bpy),Cl2].3.5H2O and cis-[Ru(bpy),C12]C1-2H2O, Inorg. Chem., 1985, 24, 4573 CrossRef CAS.
- B. P. Sullivan, D. J. Salmon and T. J. Meyer, Mixed phosphine 2,2'-bipyridine complexes of ruthenium, Inorg. Chem., 1978, 17, 3334 CrossRef CAS.
- M. J. Frisch, et al., Gaussian 16, Revision C.01, Gaussian Inc., Wallingford CT, vol. 572, 2016 Search PubMed.
- A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648 CrossRef CAS.
- P. J. Hay and W. R. Wadt, Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitale, J. Chem. Phys., 1985, 82, 299 CrossRef CAS.
- Y. Yang, M. N. Weaver and K. M. Merz, Assessment of the “6-31 + Gt; + LANL2DZ” mixed basis set coupled with density functional theory methods and the effective core potential: Prediction of heats of formation and ionization potentials for first-row-transition-metal complexes, J. Phys. Chem. A, 2009, 113, 9843 CrossRef CAS PubMed.
- I. A. Adejoro, D. D. Babatunde, Z. F. Okegbemi, A. O. Hikmat and C. A. G. Johnson, A D. Ikechukwu. Activities of analogues of carbamazepine and (R)– Lacosamide as potential inhibitors of epilepsy disease: Molecular docking, DFT, and pharmacokinetic study, Lett. Appl. NanoBioSci., 2024, 13, 1 Search PubMed.
- J. J. Lin, et al., NVL-655 Is a selective and brain-penetrant inhibitor of diverse ALK-mutant oncoproteins, including lorlatinib-resistant compound mutations, Cancer Discovery, 2024, 14, 2367–2386 CrossRef PubMed.
- G. R. Hancock, K. S. Young, D. J. Hosfield, C. Joiner, E. A. Sullivan, Y. Yildiz, M. Lainé, G. L. Greene and S. W. Fanning, Unconventional isoquinoline-based SERMs elicit fulvestrant-like transcriptional programs in ER+ breast cancer cells, Npj Breast Cancer, 2022, 8 Search PubMed.
- C. Thomson, P. Barton, E. Braybrooke, N. Colclough, Z. Dong, L. Evans, N. Floc’h, C. Guérot, D. Hargreaves, P. Khurana, S. Li, X. Li, A. Lister, W. McCoull, L. McWilliams, J. P. Orme, M. J. Packer, A. M. Swaih, R. A. Ward, P. Winlow and Y. Ye, Discovery and optimization of potent, efficacious and selective inhibitors targeting EGFR Exon20 insertion mutations, J. Med. Chem., 2024, 67, 8988 CrossRef CAS PubMed.
- R. Thomsen and M. H. Christensen, MolDock: A new technique for high-accuracy molecular docking, J. Med. Chem., 2006, 49, 3315 CrossRef CAS PubMed.
- E. C. Meng, T. D. Goddard, E. F. Pettersen, G. S. Couch, Z. J. Pearson, J. H. Morris and T. E. Ferrin, UCSF ChimeraX: Tools for structure building and analysis, Protein Sci., 2023, 32, e4792 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2025 |