Open Access Article
Open Access ArticleMagnetic nanosheets: from iron oxide nanocubes to polydopamine embedded 2D clusters and their multi-purpose properties†
Giacomo
Mandriota
 a,
Sahitya Kumar
Avugadda
a,
Ehsan
Sadeghi
a,
Sahitya Kumar
Avugadda
a,
Ehsan
Sadeghi
 ab,
Niccolò
Silvestri
ab,
Niccolò
Silvestri
 a,
Roberto
Marotta
a,
Helena
Gavilán
a,
Ulf
Olsson
a,
Roberto
Marotta
a,
Helena
Gavilán
a,
Ulf
Olsson
 c,
Cinzia
Giannini
d,
Yu Hsin
Tsai
c,
Cinzia
Giannini
d,
Yu Hsin
Tsai
 e,
Anna Cristina S.
Samia
e and
Teresa
Pellegrino
e,
Anna Cristina S.
Samia
e and
Teresa
Pellegrino
 a
a
aItalian Institute of Technology, via Morego 30, 16163, Genoa, Italy. E-mail: teresa.pellegrino@iit.it
bChemical and Chemical Industry Department, via Dodecaneso, 31, Genoa, 16146, Italy
cPhysical Chemistry, Lund University, Box 124, Lund SE-22100, Sweden
dInstitute of Crystallography, National Research Council, via Amendola 122/O, 70126, Bari, Italy
eDepartment of Chemistry, Case Western Reserve University, 10900 Euclid Avenue, Cleveland, Ohio 44106, USA
First published on 24th March 2025
Abstract
We here develop stable bidimensional magnetic nanoclusters (2D-MNCs) of iron oxide nanocubes (IONCs) arranged in thin nanosheets of closed-packed nanocubes. The assembly occurs by means of a two-step approach: in the first one, the ionic surfactant, sodium dodecyl sulfate (SDS), acts as a transient water transfer agent and as 2D clustering agent to induce formation of a monolayer of nanocubes arranged in thin nanosheets. Next, the addition of dopamine followed by solution basification, induces the in situ polymerization of dopamine with a tunable shell tickness depending on the dopamine amount, which helps to compact the clusters and ensures the long term water stability of the clusters. TEM, cryo-EM, and SAXS techniques helped to reveal structural features of the 2D-clusters. The pH-dependent degradation properties of polydopamine, enable to disassemble the clusters in acidic tumour microenviroment leading to a four-fold increase in the magnetic particle imaging signal and a concomitant increase of the magnetic heat losses of these clusters, makes them appealing in magnetic hyperthermia, while the shortening of T2 relaxation time suggests their use as contrast in magnetic resonance imaging. Finally, with crystal violet dye, used as drug molecule, the feasibility to release payloads pre-encapsulated with the polydopamine polymer shell has been also shown.
New conceptsWe reported the successful preparation, via anionic surfactant interaction, of nanosheet clusters (2D-MNCs@PDO) composed of closely packed iron oxide nanocubes (IONCs), with control bidimensional structures enwrapped in a polydopamine (PDO) shell. The PDO coating besides ensuring water stability to the structure with or without exposure to magnetic fields can disassemble in acidic tumor microenvironments, making these clusters suitable for drug delivery. Structural characterization techniques such as SAXS, cryo-EM, and TEM techniques confirmed the bidimensional lamellar organization of the clusters. Magneto heating performance of 2D-MNCs@PDO, showed higher heating performance with respect to clinical used iron oxide nanoparticles. Remarkable, the heating performance as well as the magnetic particle imaging signal showing a pH-dependent enhancement increase, together with their r2 relaxation rates higher than the individual nanocubes provide proofs of these multipurpose nanoplatform tools. |
Introduction
In recent years, the assembly of iron oxide nanoparticles (IONPs) is exploited as a means to re-design magnetic properties1,2 resulting in nanostructures with applications in nanoelectronics,3 sensors,4 food,5 cosmetics,6 drug delivery, and controlled drug release.7–9 Magnetic nanoclusters (MNCs) are also proposed as a mean to redisperse hydrophobic IONPs into aqueous media.10–12 In the liquid–liquid interface method11,13,14 interactions between IONPs and nanoparticles/polymer induce self-organization into ordered meso-/microscopic structures.15–18 Alternatively, assembly is driven by polyelectrolytes19–21 in layer-by-layer (lbl) approach, or by employing biomolecules which act as linkers22–24 Indeed, in this latter approach different molecules surface as proteins,25 nucleic acids,26 polymers27 or surfactants,28 which can complement with specific molecular counterparts are introduced on the IONPs and their self-assembly drives their organization into chains,29–32 sheets,33,34 vesicles,30,35 or more complex 3D architectures.36–39Cation surfactants, like cetyltrimethylammonium bromide (CTAB), stabilize IONPs and induce clustering into water stable centrosymmetric spheres in oil-in-water emulsion.40 However, CTAB, it is not considered biocompatible and may represent a problem for biological exploitation. Alternatively, biocompatible polymers, like glycidyl methacrylate (GMA) and ethylglycol dimethacrylate (EGDMA), can stabilize IONP clusters and enhance their functionalization in water, as demonstrated by Paquet et al.27 Among these polymers polydopamine (PDO) is a bio-adhesive polymer inspired by mussels, useful for clustering and forming thin layers on various surfaces.41 PDO is also used for cancer therapy to construct multifunctional nanomaterials with excellent therapeutic performance shown both in vitro and in vivo.42–44 Moreover, PDO can form a shell via oxidation and self-polymerization under alkaline conditions, simplifying and reducing the cost of clustering processes.45–51
MNCs play a role in both diagnosing and monitoring tumors, acting as contrast agents for magnetic resonance imaging (MRI), as a tracer in magnetic particle imaging (MPI) as heat mediators for cancer therapy by magnetic hyperthermia (MHT) treatment52–54 and as drug-delivery platform for targeted chemotherapy.55–57
The magnetic heating efficiency of IONPs under the application of AMFs is evaluated through the Specific absorption rate (SAR) parameter, which is influenced by factors like size, shape, and composition of the nanoparticles, as well as AMF parameters.58–60 For clinical use, SAR values must remain within certain limits.61,62 Recent studies showed that iron oxide nanocubes (IONCs), with unique shape anisotropy and lower spin counting surface disorders, offer higher SAR values than spherical nanoparticles.63–69 Efforts to control the aggregation of IONCs in polymeric beads have shown promising results, though SAR values for clusters decrease due to demagnetization effects and changes in hydrodynamic size.62,70–72
One-dimensional (1D) chain-like assemblies73,74 and bidimensional arrangements (2D), are clusters types with better heating performance than centrosymmetrical once. We reported,75 the synthesis of 2D-MNCs coated with polyhydroxyalkanoate (PHA), a bio-resorbable bacteria-derived polymer, which possess higher heating efficiency than spherical beads. Moreover, some MRI studies on nanocubes and nanocube clusters have also suggested their good T2-weight contrast for MRI imaging with diagnostic performances comparable to those of spherical shaped IONPs.76 In the last decade, few studies have also focused on the MPI signal generated by IONC and IONC clusters since, by this technique and differently by MRI, in MPI magnetic nanoparticles can be quantified within a short image acquisition times.77,78 This technique provides detailed maps of IONP locations and combines well with high-resolution imaging methods.79–82 Due to uniaxial magnetic dipolar coupling, MPI measurements of dimers and trimers show higher signals than 3D-clusters or commercial tracers.83 Therefore, for MHT, MRI and MPI applications high and precise clustering control is becoming central to reveal the relationship between the conformational structure of the clusters and their relative properties.
In this frame, here we report a versatile, direct and quick one-pot method to control the assembling of IONCs in close-packed 2D arrangement (2D-MNCs). This clustering process is based on an oil-in-water emulsion obtained by mixing a chloroform solution of well dispersed IONCs with a SDS aqueous solution. Both the choice of oil dispersant and the SDS concentration are parameters that can be tuned to control the 2D arrangement of IONCs when using iron oxide nanocubes at different cube edge (12, 17, 22 and 24 nm).
Following the formation of the 2D-MNCs in SDS, the MNCs are coated by PDO polymer (2D-MNCs@PDO) via in situ polymerization to provide a shell that promotes higher water stability of the pre-formed bi-dimensional clusters. This study, by using a combination of different material characterization techniques (TEM, cryo-EM, SAXS), aims at correlating the structural features of 2D-MNCs with their unique magnetic properties characterized via static and dynamic magnetic measurements including SQUID-magnetization curves, MHT calorimetric, MRI relaxation measurements, and MPI signal analysis.
Experimental section
Materials
All chemicals were obtained from commercial suppliers and except otherwise stated, used without further purification. Triethylamine (TEA), toluene (99%), chloroform (CHCl3, 99%), sodium dodecyl sulfate (SDS), dopamine hydrochloride (DOPA, >98%), oleic acid (OA, ≥99%) were purchased from Sigma-Aldrich. 1-Octanol (anhydrous, ≥99%; Sigma-Aldrich) hexadecylamine (98%; Sigma-Aldrich), Iron pentacarbonyl (>99.99%; Sigma-Aldrich), Benzaldehyde (≥99%, ReagentPlus; Sigma-Aldrich, didodecylamine (>97%, Sigma-Aldrich), Ultrapure water (18.2 MΩ × cm at 25 °C filtered with filter pore size 0.22 μM) obtained using a Milli-Q (MQ) water system was used throughout all experiments. Gallol-poly(ethylene glycol) (PEG) carboxylic-terminated ligand (GA-PEG-OH) was prepared accordingly to our previously reported protocol.71,84Synthesis of IONCs
The synthesis of IONCs of 12–24 nm was conducted by a solvothermal-based process reported by some of us with minor modifications.85,86 Briefly, oleic acid and an alkyl amine are dissolved in 1-octanol at 60 °C for 30 min and magnetic stirring of 800 rpm, using a three-neck round-bottom flask (with a capacity of 25 mL) and a heating mantle and a hot plate stirrer. This solution is allowed to cool down to room temperature. Then, iron pentacarbonyl is added, along with benzaldehyde molecule, which acts as the key shape-directing agent. This solution is mixed vigorously (1100 rpm) for 30 min. The obtained mixture is transferred to a Teflon-lined autoclave for solvothermal crystallization at 200 °C for certain time. The autoclave volume is 25 mL except for sample IONCs-22, which is 100 mL. The precise amounts and experimental conditions used for each sample are summarized in Table 1.| Solvent 1-Octanol | Surfactant Oleic acid | Alkyl amine | Iron precursor Iron pentacarbonyl | Aldehyde Benzaldehyde | Temperature | Time | Sample name |
|---|---|---|---|---|---|---|---|
| Volume (mL) | Volume (mL) | Mass (g) | Volume (mL) | Volume (mL) | (°C) | (h) | |
| Hexadecylamine | 2 | 200 | 4 | IONCs-12 | |||
| 7 | 0.6 | 0.2 | 2 | ||||
| Hexadecylamine | 1.2 | 200 | 4 | IONCs-17 | |||
| 7.8 | 0.6 | 0.2 | 2 | ||||
| Hexadecylamine | 2 | 220 | 6 | IONCs-22 | |||
| 8 | 0.6 | 0.2 | 1 | ||||
| Didodecylamine | 2 | 200 | 4 | IONCs-24 | |||
| 8 | 0.6 | 0.294 | 2 |
After solvothermal crystallization, the autoclave is removed from the oven, and it is allowed to cool down to room temperature. The content of the autoclave is recovered with chloroform and transferred to two falcon tubes. The sample is washed once with acetone (4500 rpm, 20 min). The supernatant is discarded, and the pellet sample is redispersed in approximately 20 mL of chloroform.
2D Clustering of IONCs (2D-MNCs)
In a typical synthesis, 2D-MNCs were first obtained by an oil-phase evaporation-microemulsion which induces self-assembly strategy. 90 μL of IONCs-12 nm in cube edge (64 mM, 10 gFe L−1) were dispersed in 155 μL of Chloroform and mixed with 4 mL of sodium dodecyl sulfate (SDS, 1.8 mg mL−1) in a 40 mL glass vial. The solution was left to sonicate for 1 h at 80 °C in a hot water bath sonicator thus enabling to slowly evaporate the organic solvent. After sonication, to narrow the size distribution of the nanoclusters, 2D-MNCs were transferred to a 50 mL falcon tube and two centrifugation steps were performed: the first centrifugation was done at 700 rpm for 30 min Sigma 3-16PK centrifuge with a swing-out rotor (no. 11180); the supernatant was collected and transferred in a new 50 mL falcon tube, while the pellet containing big MNCs (size > 400 nm) was discarded. The collected supernatant was again centrifuged at 3600 rpm for 30 min on the same centrifuge, the supernatant was discarded and the 2D-MNCs fraction (D < 150 nm) was collected and redispersed in 200 μL of Milli-Q water. A typical concentration of the obtained solution is of 2 mg mL−1 in Iron.In situ polymerization of PDO on 2D-MNCs (2D-MNCs@PDO)
To coat the 2D-MNCs with PDO polymer (2D-MNCs@PDO) a one-step approach was adapted from a published protocol.87 The aqueous solution of 2D-MNCs12 nm (200 μL, 3 mgFe mL−1) was mixed with 300 μL of a DOPA solution (67 mM in 10 mM Tris–HCl, pH 7.4). The pH was adjusted to 8.5 with NaOH 1 M and the mixture was continuously shaken on a orbital shaker at RT for 3 h. The polymerization of DOPA monomers immediately occurred and a change in color from pale brown to dark black was observed, which is consistent to the observation by other group.88 Next, the excess of unbounded PDO was removed by magnetic separation of the 2D-MNCs@PDO samples (using a permanent magnet of 0.3 T) leaving the 8 mL vial solution on the magnet for 2 h. The solution was discarded, and the pellet of 2D-MNCs@PDO collected on the magnet was re-suspended in 500 μL of Milli-Q water. This washing step was repeated 4 times.Stability test of 2D-MNCs12@PDO in PBS at different pH
The stability test of 2D-MNCs12@PDO was assessed at RT in both acidic (pH 5) and alkaline (pH 9) environments. To this aim, 1 mL of 2D-MNCs12@PDO solution was centrifugated to remove the MilliQ-water and redispersed in 1 mL of Phosphate Buffer Solution (PBS) solution, to simulate physiological conditions. The solution was divided into two different aliquots, and the pH was adjusted using 0.4 M hydrochloric acid (HCl) and 1 M sodium hydroxide (NaOH), respectively. The pH was measured with a pH meter. Samples were shaken at RT, with aliquots taken at four different time points: 0, 3, 12, and 24 h. The stability test on 2DMNCs12@PDO was performed to evaluate the behaviour of the PDO coating under different pH conditions through TEM characterization.Encapsulation of crystal violet (CV) in 2D-MNCs
For CV loading, 200 μL of 2D-MNCs (3 mgFe mL−1, 17 ± 1 nm nanocubes size), 300 μL DOPA in buffer 7.4 (33 mM) and 300 μL CV in water (2 mg mL−1) were mixed and the mixture was left on an orbital shaker (1000 rpm) for 30 min. Then, the pH was adjusted to 8.5 by addition of 10 μL NaOH 1 M and the mixture was maintained under shaking at room temperature for 24 h. The clusters were then magnetically collected by exposing the 4-mL vial to a 0.3 T magnet for 24 h and the pellet resuspended in 200 μL Milli-Q water. The supernatant solution, containing the excess of CV dye, which was not loaded into the clusters, was used to measure the characteristic emission peak of CV dye by using a Varian Cary Eclipse fluorescence spectrophotometer equipped with a xenon lamp source. The supernatant samples were transferred to 1 mL quartz cuvette. The Photoluminescence (PL) spectra intensity of the supernatant with a maximum at 628 nm when excited at 590 nm, was compared with the initial CV dye solution on a calibration curve reporting the PL (628 nm) signal intensity versus dye concentration. The encapsulation efficiency was calculated as the percentage of the difference between the initial concentration of the CV solution and that of the supernatant (after cluster separation) normalized to the concentration of the initial CV solution.pH release experiment on CV-loaded PDO coated 2D clusters
To 500 μL of CV-loaded 2D clusters at 1.2 mgFe mL−1 (containing an encapsulated amount of 0.48 mg of CV), 50 μL of 0.36 M HCl solution were added such that the pH reached 5 and the sample was shaken on an orbital shaker (1000 rpm) at RT for 1, 3 or 6 h. Four aliquots were prepared and the 2D-MNCs17@PDO cluster sample at time 0 and the supernatants taken at 1, 3, and 6 h after exposure at pH 5 having magnetically separated the clusters (as explained in the previous paragraph), were transferred into a 1-mL cuvette and diluted to 1 mL with Milli-Q water to then read their emission peak intensity at 628 nm thus extracting the corresponding dye concentration on the dye calibration curve.Water transfer of individual IONCs
The water transfer of single IONCs, with edge lengths of 12 ± 1 nm (IONCs12 nm, Fig. S1, ESI†), was done following a ligand exchange protocol previously reported by us with minor modifications.71,84 Briefly, in a 40-mL glass vial, 1 mL of IONCs12 nm in chloroform (1 gFe L−1) was added to a 8.65 mL chloroform solution of Gallol modified PEG (GA-PEG, 0.05 M), and corresponding to 150 ligands per nm2 of IONCs surface. Next, 0.865 mL of trimethylamine (TEA) was added to the mixture, which was vigorously shaken overnight at room temperature (RT). Later, the mixture was transferred to a separation funnel, which contained 100 mL of water and 40 mL of toluene; an emulsion was created by shaking the funnel. After leaving the solution undisturbed for 1 hour, the lower aqueous phase containing the water transferred nanocubes, was carefully collected and the phase extraction process was repeated until all the nanoparticles fraction was extracted in the water phase (no more brown colour in the organic phase). Residues of organic solvent were removed by bubbling nitrogen inside the aqueous solution, followed by evaporation of organic solvent with rotavapor, under reduced pressure conditions at 40 °C for 1 hour. The excess of free GA-PEG ligands was removed by five centrifugation washings, using centrifuge filter (Amicon filters, with molecular weight cut-off-MWCO- of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da, 1500 rpm Sigma 3-16PK centrifuge with a swing-out rotor (no. 11180). The PEG-IONCs12 nm were concentrated to 2 mL and sonicated for 30 min at 65 °C. The same protocol was also applied to IONCs of edge length 22 ± 4 nm following a very similar approach.
000 Da, 1500 rpm Sigma 3-16PK centrifuge with a swing-out rotor (no. 11180). The PEG-IONCs12 nm were concentrated to 2 mL and sonicated for 30 min at 65 °C. The same protocol was also applied to IONCs of edge length 22 ± 4 nm following a very similar approach.
Magnetic hyperthermia measurements
The calorimetric measurements were performed using a commercially available magnetic nano-heating device (DM 100 series, nanoScale Biomagnetics Corp), by comparing the SAR values of the 2D-MNCs12@PDO and 2D-MNCs22@PDO with the respective single IONCs in water. For the measure, 150 μL of each sample at 4 gFe L−1, after sonication, were exposed to the AMF at two different frequencies (110 and 182 kHz) and magnetic field amplitudes of 12, 16, 20, and 24 kA m−1. The temperature increase was monitored with an optic fiber thermosensor (LumaSense). To calculate and compare the heating efficiency of these samples, in terms of SAR, the initial slope of the heating curve T vs. t curve was considered. The SAR can be estimated from the following eqn (1): | (1) |
SAR signal evolution of PDO-coated 2D clusters at pH 5
We investigated the trend of SAR variations in 2D-MNCs12@PDO clusters at pH 5, over a period of 7 hours. For this experiment, a calorimetric set up (DM 100 series, nanoScale Biomagnetics Corp) was used at frequency (f) of 300 kHz and a magnetic field amplitude (H) of 24 kA m−1. For the sample preparation, 150 μL of 2D-MNCs12@PDO dispersed in water (4 mgFe mL−1) were acidified at pH 5 at RT by addition of 10 μL of a 0.36 M HCl solution. The SAR values of the 2D-MNCs12@PDO were recorded as previously described, at different time points: 0, 1, 2, 3, 4, 5, 6 and 7 h. The temperature increase was monitored with an optic fiber thermosensor (LumaSense). To calculate and compare the heating efficiency of 2D-clusters, in terms of SAR, the initial slope of the heating curve T vs. t curve was considered and eqn (1) was applied.Magnetic relaxation measurements
Longitudinal (T1) and transverse (T2) proton relaxation times of the samples (2D-MNCs12@PDO, 2D-MNCs22@PDO, PEG-IONCs12 nm and PEG-IONCs22 nm) were measured using commercially available Minispec spectrometers (Bruker, Germany) of three different magnetic fields; mq 20 (0.5 T), mq 40 (1 T), and mq 60 (1.5 T). For the measurements, calibration curves were prepared by serial dilutions of each sample at an iron concentration in the range between 1 mM and 0.015 mM (i.e. 8 dilution points, 500 μL of volume each), in quartz NMR tubes. Before the analysis, the samples were kept at 40 °C for 20 min. Firstly, the T1 and T2 proton relaxation times of samples were collected; the T1 relaxation times were derived from the saturation-recovery sequence, with 16 data points and 3 acquisitions for each measurement, and the T2 relaxation time was obtained from a Carr-Purcell Meiboom Gill (CPMG) spin-echo pulse sequence (100 data points, 3 acquisitions). The data provided is an average of three independent measures, which are reproducible with a standard deviation of less than 5%. Here, the r1 and r2 values were estimated by deriving the slope of a linear fit obtained by plotting the reciprocal of relaxation times (1/T1 and 1/T2) as a function of iron concentration.76Magnetic particle imaging
MPI analyses were performed in a custom made x-space magnetic particle relaxometer.69 Representative samples of 2D-MNCs and single IONCs in water (500 μL each) were measured at a fixed iron concentration of 1.0 mgFe mL−1; the samples were sonicated prior to perform each Magnetic Particle Spectroscopy (MPS) relaxometry measurement. All MPS studies were conducted under an AMF field intensity of 16 kA m−1 and frequency of 16.8 kHz. The MPS signals were normalized to the MPS signal of a commercial MPI tracer, VivoTraxTM (Resorvist® licensed for distribution in USA through Magnetic Insight for pre-clinical MPI studies, 500 μL, 1.0 mgFe mL−1).MPI signal evolution of PDO-coated 2D clusters at pH 5
2D-MNCs17@PDO clusters relaxometry was performed on a MOMENTUM™ system (Magnetic Insight Inc.) using the Relax mode. For the preparation of the sample, four aliquots were prepared each made of 20 μL of a 0.36 M HCl solution and 20 μL of PDM-coated 2D clusters at 4 mgFe mL−1 such that the pH of the solution was set at 5 and the clusters concentration was fixed at 2 mgFe mL−1. After shaking the Eppendorf at RT, one aliquot was measured immediately while the other three were left at RT for 1, 3 or 6 hours before performing the relaxation measurements on the MPS relaxometry of the MOMENTUM™ system.Small angle X-ray scattering (SAXS) characterization
SAXS measurements were performed on 2D-MNCs12 nm, 2D-MNCs12@PDO, 2D-MNCs24 nm, and 2D-MNCs24@PDO. Experiments were performed on a GANESHA pin-hole SAXS instrument (SAXSLAB ApS, Skovlunde, Denmark) equipped with a two-dimensional 300k Pilatus detector (Dectris Ltd, Baden, Switzerland) and a Genix 3D X-ray source (Xenocs SA, Sassenage, France). The source had a wavelength of 1.54 Å. Two different sample-to-detector distances were used, giving an effective q-range of 0.004–0.15 Å−1. Here q is the scattering vector given by q = 4π/λ sin{θ/2} where λ is the X-ray wavelength and θ is the scattering angle. Measurements were performed under vacuum at ambient temperature (≈21 °C). Samples were injected into 1.5 mm disposable quartz capillaries that were sealed with glue. The isotropic two-dimensional (2D) scattering pattern was radially averaged using the SAXSGui software, to obtain the scattered intensity, (q).Elemental analysis characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 HCl
1 HCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3), subsequently diluted to 25 mL using Milli-Q water and finally, before analysis, the solutions were filtered through 0.45 μM PTFE filter.
HNO3), subsequently diluted to 25 mL using Milli-Q water and finally, before analysis, the solutions were filtered through 0.45 μM PTFE filter.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 corresponding to a magnified pixel size of 3.6 Å. Tilt series assembling and tomograms generation was performed by both WBP and SIRT algoritms using IMOD IMOD 4.9.89 3D reconstructions have been performed using Amira™ Software (Thermo Fisher Scientific).
000 corresponding to a magnified pixel size of 3.6 Å. Tilt series assembling and tomograms generation was performed by both WBP and SIRT algoritms using IMOD IMOD 4.9.89 3D reconstructions have been performed using Amira™ Software (Thermo Fisher Scientific).
Results and discussion
Clustering 2D-MNCs
For the 2D-MNCs protocol in our one-pot approach, in a water bath set at 80 °C under sonication, the emulsion was achieved by dispersing surfactant-coated IONCs in chloroform with an aqueous solution of sodium dodecyl sulfate (SDS), used as micellization agent. Indeed, during the microemulsion process, the hydrophobic chains of SDS, intercalated with the hydrocarbon tails of OA at the surface of the IONCs, while the extended polar heads of SDS offer the water solubility of the emulsion in which IONC are entrapped.27,90 Such high temperature of the bath enables the slow evaporation of CHCl3 solvent thus inducing a gradual change of the media polarity. These changes drive the interaction between the flat surface of the nanocubes, and those of nanocubes and SDS surfactant molecules leading to a 2D ordering of nanocubes as observed under TEM (see 2D-MNCs in Fig. 1).28 Indeed, the formation of 2D-MNCs starting from nanocubes of 12 nm and 22 nm after SDS intercalation/micellization was confirmed by respective DLS profiles in which monomodal peaks were recorded, with average size distribution by intensity of 146 ± 62 nm and 106 ± 36 nm, and with polydispersity indexes (PDI's) of 0.150 and 0.167 respectively (Fig. S2, ESI†). The clusters, at this stage, showed surface ζ-potential values in the range between −41 ± 8 mV and −54 ± 10 mV, likely attributed to the presence of SDS (Fig. S2B, ESI†). To explain the 2D ordering of nanocubes, we refer to phase diagrams from literature on ternary systems of chloroform, water, and SDS surfactant.91–94 Our mixture was near the water vertex, with small amounts of chloroform and SDS, indicating a predominantly aqueous region where chloroform is dispersed as droplets stabilized by SDS. At higher SDS concentrations and a larger organic-to-aqueous phase ratio, the system enters a lamellar region, leading to the self-assembly of ordered structures like bilayers or 2D clusters. Moreover, the geometry of the SDS molecule plays an important role in nanoparticle assembly due to its self-assembly behavior in aqueous solutions, which is governed by the packing parameter ‘P’ defined as the ratio between the volume of the surfactant tail and the product of the length of the surfactant tail and the surface area of the hydrophilic headgroup. SDS can form different structures in water, such as micelles, vesicles, or bilayers, depending on its P value.95 Du et al.96 demonstrated as SDS exhibits a low P value (0.30–0.44) at the liquid–liquid interface, favoring spherical micelle formation. However, interaction with solid surfaces (the nanocube surface here) increases the P value (0.53–0.70), enabling vesicle formation. A P value approaching 1 on glass particles suggests the formation of bilayer structures. These considerations can provide a reasonable explanation for the nanoparticle 2D assemblies observed by us.The effects of the nanoparticles dispersing solvent as well as the amount of free OA into the nanoparticle solution were studied to correlate them to the final arrangement of the clusters. In fact, the particle–particle interaction can be adjusted by changing the particles surrounding,97 likewise, changing the solvent98 in which the IONCs are dispersed or by tuning the optimal surfactant amount in the IONCs dispersion. A first experiment was performed by replacing CHCl3 with toluene on 12 nm IONCs by simply drying CHCl3 under a nitrogen flow and redispersing the IONCs powder in toluene. After microemulsion of the toluene sample with SDS water solution the resulting clusters had a three-dimensional centrosymmetric ordering (3D-MNCs12 nm, Fig. S3A, ESI†) rather than 2D order obtained on the sample from CHCl3 on the same batch of nanocubes (Fig. S3B, ESI†). This was also the case when changing the nanocubes size (Fig. S3C and D, ESI†). A reasonable explanation for this change can be found in a previous study by Park et al.,99 which highlighted how the nature of the organic phase in terms of polarity and different boiling point of the solvent, affected the morphology of the colloidal interfacial assembly of nanoparticles: by using a mixture composed of a chloroform solution of iron oxide nanoparticles and an aqueous solution of hexadecyltrimethylammonium bromide (CTAB) well-defined nanoparticle assemblies with a 2D arrangement were achieved while with toluene solution of the same nanoparticles’ solid assemblies with small cavities and a 3D arrangement were obtained. They speculated as the higher solubility of chloroform in water (0.82% w/w versus 0.052% w/w of toluene at 25 °C) and the fast evaporation of CHCl3 (68 °C versus 110 °C) determined the miscible properties and the different morphology.
Moreover, in the attempt to reproduce these 2D structures from batch to batch of nanocubes, we noticed that the OA amount in the nanocubes solution was crucial to obtain the 2D-MNCs, likely because these molecules at the surface of the nanoparticles are also involved in the direct interdigitating interactions with the anionic SDS surfactant molecules.100–103 The assembling experiments, by tuning the OA percentages at the surface of the nanocubes (from 4 to 76 wt%), were performed on IONCs12 nm. The TGA analysis was used to evaluate the amount of OA covering IONCs affecting the arrangement of the final magnetic clusters (see materials and methods and Fig. S4, ESI†). As observed, the surfactant amount dictated the particles ordering in the cluster (Fig. S4, ESI†) with the best 2D orderings obtained when the OA's content was close to 50–60 wt% (Fig. S4C, ESI†). At this optimal range, OA likely forms a balanced structure, where a double layer of OA is partially adsorbed and partially bonded to the nanoparticle surface, promoting both steric stabilization and uniform assembly behavior. This double layer OA coating is supported by the two TGA peaks with a first weight loss in the temperature range from 150 to 280 °C corresponding to the physisorbed OA layer (the predominant fraction) and a second weight loss from 280 to 400 °C (the less abundant fraction) likely due to the chemisorbed OA layer (Fig. S4, ESI†).104 Instead, deformed shaped clusters were observed at 24 and 76 wt% (Fig. S4B and D, ESI†) with the prevalence of 3D-MNCs at 4 wt% (Fig. S4A, ESI†).
These data suggest that, when the OA amount is rather low it maybe not enough to entirely cover the IONCs surface and properly interdigitate with SDS and it likely leads to a high-density assembling, like 3D-MNCs, in which hydrophobic nanocubes interactions become more predominant with each others.105 Conversely, the optimal tuning of free OA dispersed in the IONCs could lead to a gradual increase of the interparticle interactions originating from the van der Waals interactions due to the intercalation of the hydrocarbon chain,106,107 thus, reducing the surface tension with the SDS surfactant during the clustering process.108–110
To note that other synthesis parameters including SDS concentration, sonication temperature, and solvent evaporation time, were also considered to tune the structural parameters of the clusters. For SDS concentrations below or above the SDS value of 1.8 mg mL−1 adopted in the optimal protocol to form 2D clusters, MNCs showed both 2D and 3D arrangements (Fig. S5A, ESI† for 1.1 mg mL−1 and Fig. S5B, ESI† for 2.6 mg mL−1). These observations suggested that the assembly process is highly sensitive to small variations in SDS concentration. For the same reason sonication temperature was set at 80 °C in the optimal 2D clustering protocol because in the attempt to reduce the temperature to 25 °C double populations of 3D and 2D-MNCs were found (Fig. S6, ESI†).
The solvent evaporation time played also a key role in guiding the self-assembly of IONCs over the final arrangement. The influence of this parameter was analyzed by monitoring the TEM structural evolution of the MNCs by taking different aliquots of samples at specific time points during the sonication process, ranging from 5 to 50 minutes (Fig. S7, ESI†). TEM images revealed a clear progression from an initial disorganized state at 5 and 20 minutes (Fig. S7, ESI†) to increasingly ordered 2D arrangements at 30, 40, 50 minutes, choosing 40 minutes as the ideal evaporation time.
Scaling up the synthesis of magnetic assemblies has always been a challenge, because on bulk production, often the assembly suffers from either low yields or poor reproducibility, while still controlling the desired morphology.71,75 Here, by starting from IONCs-12 nm and IONCs-22 nm simply by increasing the reaction volumes by 20 folds (with respect to small scale) and maintaining the water solution – to – CHCl3 phase ratio equal to 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, the 2D assemblies morphology was maintained without no remarkable difference in size distribution (see Fig. S8, ESI†).
1 v/v, the 2D assemblies morphology was maintained without no remarkable difference in size distribution (see Fig. S8, ESI†).
PDO coating on 2D-MNCs
The 2D-MNCs transferred in water by SDS were unstable and irreversibly aggregates if exposed to permanent magnets or AMFs. To overcome this issue, 2D-MNCs were additionally coated with PDO polymer layer, to increase the stability of the nanosystems in water for further exposure to magnetic fields.The self polymerization of DOPA molecules on 2D-MNCs was obtained via a simple method already reported in literature.111 As schematized in Fig. 2(A), a mixture of 200 μL of 2D-MNCs (3 mgFe mL−1) and 300 μL 300 μL of DOPA solution (67 mM in 10 mM Tris–HCl, pH 7.4) were mixed and the pH of the solution was adjusted to 8.5 by addition of NaOH 1 M. The mixture was continuously shaken at room temperature for 3 h at 1000 rpm to promote the polymerization of DOPA. Indeed, the interaction of the DOPA with the surface of the nanoclusters was achieved through the ionic interaction between the amino groups (–NH2) of dopamine and the sulfonic groups (–SO3−) of the SDS.112 After 3 h of polymerization, the solution colour changes from orange to dark black, which indicates the formation of PDO on the surface of the 2D-MNCs.
From the analysis of TEM images (Fig. 2(B)) the 2D structure of 2D-MNC12 nm (named as 2DMNCs12@PDO) was unaltered by the coating procedure and the thickness of PDO shell was around 3 nm in these clusters. The TEM size distribution analysis indicated that the 2D-MNCs12@PDO have an average edge-length of 132 ± 46 nm (inset Fig. 2(B)), which is in agreement with the hydrodynamic size of the same sample measured by DLS (Fig. S2, ESI†). The ζ-potential in water of 2D-MNCs12@PDO was measured at −21 ± 8 mV, due to the multi catechol –OH groups of the PDO shell. Indeed, the electrostatic repulsive forces generated by the negative charge on the surface of 2D-MNC12 nm@PDO ensured the colloidal stability in water of these assemblies without undesired aggregation.
The parameters used to coat 2D-MNCs12 with PDO were adapted also for 2D-MNCs22 nm obtaining 2D-MNCs22@PDO (Fig. 2(C)). Interestingly, the same 3 nm thickness of the PDO shell was obtained while the average edge-length was 112 ± 33 nm and the arrangement of the IONCs was also kept (inset in Fig. 2(C)). To tune the thickness of the PDO shell on 2D-MNCs12 nm, the DOPA amount was adjusted using three different solution concentrations: 20 mM, 33 mM, and 65 mM respectively. As the DOPA concentration increased, the thickness of the PDO shell deposited on the 2D-MNC12 nm was gradually growing from 3 nm to 20 nm (Fig. 2(D)). Finally, in order to simulate physiological conditions, the 2D-MNCs12@PDO was redispersed in PBS at pH 5 and pH 9 at four different time points, 0, 3, 12 and 24 h at RT. Interestingly, after 3 hours of shaking in PBS at pH 5 (Fig. 3(A)), the PDO shell started to destabilize as seen by the different polymer edge on the clusters but it took 24 hours to observe the detachment of some of the nanocubes from the core of the cluster, confirming the pH degradation behaviour of PDO reported by another group.113 This is due to the protonation of the hydroxyl groups of catechol in an acidic condition.114 To note that if 2D-MNCs17@DPO with thinner PDO shell were exposed to the acidic solution, the degradation at pH 5 is much quicker and already at 1 hour, a different shadow of the polymer shell was visible while the nanocube's release was observed already at 6 hours Fig. 3(B)). Instead, when the 2D-MNCs12@PDO were incubated at pH 9, no degradation of the PDO shell was observed within 24 h, because catechol group of PDO cannot be deprotonated in an alkaline environment (Fig. S9, ESI†).115
Crystal violet loading and release on 2D-MNCs17@PDO
With the aim to quantify the effect of the polymer degradation, CV dye was used as a probe. For the encapsulation, during PDO shell formation on the clusters, it was enough to add to the aqueous solution of 2D-MNCs, first CV then DOPA and finally adjusting the pH to 8.5 with NaOH solution to initiate the polymerization. The amount of CV trapped in the PDO shell, was measured at 628 nm by PL intensity difference between the initial CV stock solution and the PL signal of CV left in the supernatant after magnetic separation of the CV-loaded clusters (Fig. S10, ESI† for the calibration curve). The amount of loaded CV associated to the 2D clusters corresponds to 80% of the initial CV (meaning 0.48 mg of CV per 0.6 mgFe of the 2D clusters). To note also that the TEM structure of the CV-loaded 2D-clusters is fully preserved like that of 2D clusters with no CV encapsulated in the shell (Fig. 3(C1)). On the supernatant we could also follow the release of the CV dye at pH 5. Again, after having magnetically separated the clusters left at pH 5 for 1, 3 and 6 hours, the PL intensity of the supernatant solutions at 628 nm (when excited at 590 nm) was measured: there was a constant increase of the PL signal intensity, indicating a progressive release of the CV dye (Fig. 3(D)). The estimated dye release at 6 hours, read on the calibration curve, corresponds to about 70% of that loaded CV amount. The release was also confirmed by the visible change in color of the 2D clusters solutions at 0 with respect to the solution of the supernatant at 3 and 6 h (Fig. 3(D) insets) and additionally confirmed by the TEM images of the progressive disassembling of the CV-loaded 2D clusters overtime (Fig. 3(C)). As a final consideration, CV can be considered as a small drug model, and these results suggest the use of the PDO shell as a pH-delivery system for small cargo molecules.Cryo-TEM characterization and cryo-electron tomography
To investigate, in-depth, the assembly arrangement we performed cryo-electron microscopy-(cryo-EM) followed by cryo-electron tomography (CET), an approach in which the IONPs assemblies were three-dimensionally imaged in their native fully hydrated state, thus, providing a full 3D view of the clusters and potentially minimizing the artefacts arising from the 2D projection obtained by conventional TEM imaging (Fig. 4(B)–(D) and (F)–(I)). CET performed on vitrified 2D-MNCs12@PDO and 2D-MNCs22@PDO allowed the reconstruction of their 3D spatial distribution consisting, for both samples in ordered monolayers composed of nanocubes (Fig. 4(C), (D) and (G)–(I)) respectively. This data further demonstrated that the proposed clustering procedure allowed us to obtain a two-dimensional arrangement of the IONCs used as building blocks, complementing the information that was shown by conventional TEM.Small-angle X-ray scattering
The structural features of the 2D-MNCs12@PDO were investigated more in detail, through small-angle X-ray scattering (SAXS). Precisely, SAXS data were analyzed with a paracrystal lamellar Model116 model which describes the scattering from a stack of repeating lamellae, treated as a paracrystal. The polydispersity of lamellae periodicity is described by a Gaussian function. Moreover, a further evaluation was performed by testing 2D-MNCs24@PDO composed of IONCs with edge size of 24 ± 4 nm (IONCs24 nm, Fig. S11A, ESI†). From the fitted curves, (Fig. 5), a two-dimensional planar organization was displayed for all the samples tested. In addition, all of them showed that the ordering and the spacing values of the IONCs were reasonable and in agreement with the TEM characterization (see Fig. S11, ESI†). By fitting the experimental data for the 2D-MNCs12 nm and 2D-MNCs12@PDO samples, a 5 times periodicity was obtained, with an A + B repetition, where A refers to the intra-nanocube space and B refers to the nanocube size. These data demonstrated that the interparticle distance in the 2D-MNCs12@PDO sample was 2.4 nm (Fig. 5(B and C)). On the other hand, analyzing the 2D-MNCs24 nm and 2D-MNCs24@PDO, 2 times periodicity was found, a lower value compared to the result obtained for 2D-MNCs12@PDO. Finally, the intra-cube space was 2.4 nm in agreement with the TEM characterization (Fig. 5(E and F)).SQUID measurements
The magnetic properties of the 2D-MNCs12@PDO and the 2D-MNCs22@PDO were studied in comparison with their respective single IONCs (12 nm and 22 nm) after water transferring the nanocubes samples through a ligand exchange protocol, using a gallol-PEG derivate as hydrophilic polymer and with a protocol previously reported84 and here named PEG-IONCs12 nm and PEG-IONCs22 nm, respectively (Fig. 6 and Fig. S12, ESI†). The magnetic response to the external magnetic field (sweeping between −70 and +70 kOe) was measured at 298 K (Fig. 6) and 5 K (Fig. S12, ESI†) for all samples. The saturation magnetization (Ms) of the 2D-MNCs22@PDO sample was slightly higher (120 emu g−1) than that of the PEG-IONCs22 nm (about 100 emu g−1), while 2D-MNCs12@PDO (Fig. 6(A)) showed a similar Ms profile (about 100 emu g−1) compared to PEG-IONCs12 nm (100 emu g−1) as seen in Fig. 6(A)). This effect can be ascribed to the packing and to the morphology of the clusters: when the IONCs12 nm and IONCs22 nm are close-packed and highly ordered within the clusters, their magnetic dipolar coupling could influence the Ms.117,118 Therefore, we counted the average number of IONCs per cluster for both samples and we found that the 2D-MNCs12@PDO sample was composed of ca. 30–100 IONCs12 nm (see Fig. S8D, ESI†). This may account for higher interparticle interaction for 2D-MNCs12@PDO, which are reflected in the decreasing of the Ms values (Fig. 6(a) and Fig. S12, ESI†). Instead, in 2D-MNCs22@PDO clusters composed of approximately 4–9 of 22 nm nanocubes (see Fig. S8H, ESI†), the interaction between these nanocubes may be beneficial as it can lead to a more favorable alignment of the individual magnetic moments, promoting a parallel alignment that results in a greater overall magnetic moment compared to that of a single nanocube (Fig. 6(C) and Fig. S12, ESI†).104 Further assessment of the magnetic properties of the 2D-MNCs@PDO was obtained by measuring the magnetization curves as a function of temperature taken in zero-field cooling (ZFC) and field cooling (FC) modes. In this regard, as shown in Fig. 6(B), in agreement with the smaller size of the IONCs used as building blocks, the blocking temperature (Tb) estimated from the maximum of ZFC curves, increased from 325 K for PEG-IONCs12 nm to 350 K for 2D-MNCs12@PDO. This shift in Tb towards higher value provides also an indication of interparticle dipole interactions occurring only in the clusters and not on individual nanocubes in water. At the same time, the assembling did not alter the superparamagnetic behavior of the IONCs packed.119,120Hyperthermia measurements
The magnetic heating efficiencies of the clusters were measured by using a calorimeter and evaluating the SAR values in aqueous media for 2D-MNCs12@PDO and 2D-MNCs22@PDO (4 mgFe mL−1), in comparison to single IONCs, still in water (PEG-IONC12 nm and PEG-IONCs22 nm). The SAR values were measured at different AMF amplitudes (from 12 to 24 kA m−1) and at two different frequencies (110 kHz and 182 kHz) whose H × f products were below the biological safety limit (<5 × 109 A ms−1).77 The SAR values of 2D clusters were always lower than their respective single IONCs at any frequency and field conditions (Fig. 7(A) and (B)). The SAR values of 2D-MNCs12@PDO and 2D-MNCs22@PDO clusters were 64 ± 4 W gFe−1 and 144 ± 13 W gFe−1, respectively (f: 110 kHz, H: 24 kA m−1). These results were in agreement with a recent work of our group in which the SAR performances of 2D clusters coated with a bacterial extracted biopolymer were investigated.75 It was interesting to note, that for the clusters, the absolute SAR values doubled by increasing the frequency (see Fig. 7(B)). In fact, by exposing 2D-MNCs12@PDO and 2D-MNCs22@PDO at a higher frequency (182 kHz) at the same field strength (24 kA m−1), the SAR increased up to 164 ± 8 W gFe−1 and 242 ± 11 W gFe−1, respectively.To evaluate the heating performances of the produced 2D-MNCs when enclosed in viscous media which can resemble the biological tumor or cell environment, the SAR of both 2D-MNCs12@PDO and 2D-MNCs22@PDO was measured in four different aqueous solution containing an increasing percentage in volume of glycerol (25%, 50%, 75% and 81%) (Fig. 7(C)–(F)).
At a frequency of 110 kHz and at a field strength of 24 kA m−1 the heating losses for 2DMNCs12@PDO was less than 25%, while for 2D-MNCs22@PDO the losses reached 40% (Fig. 7(C) and (E)). Moreover, by exposing 2D-MNCs12@PDO and 2D-MNCs22@PDO at a higher frequency (182 kHz) at the same field strength (24 kA m−1), the percentage losses were in the range of 30% and 45% respectively, thus showing slight heat losses as the frequency increases (Fig. 7(D) and (F)). Therefore, the heating capability of 2D-clusters exposed to viscous media is also frequency dependent.121 The best SAR values were reached for a 25% mixture of glycerol, in the 2DMNCs12@PDO the value was 131 ± 6 W gFe−1 (f: 182 KHz, H: 24 kA m−1) and in the 2D-MNCs22@PDO the maximum value reached 223 ± 12 W gFe−1 (f: 182 KHz; H: 24 kA m−1). In the case of 2DMNCs12@PDO the differences in SAR values between 25, 50% and 75% of the glycerol mixture (f: 110 KHz, H: 12 kA m−1, 16 kA m−1 and 20 kA m−1) were negligible as also expected for the 12 nm nanocubes used for the assemblies. This is consistent with the fact that the Brown relaxation becomes negligible with respect to Néel relaxation for clusters made of this nanocube size.122
Relaxivity measurements
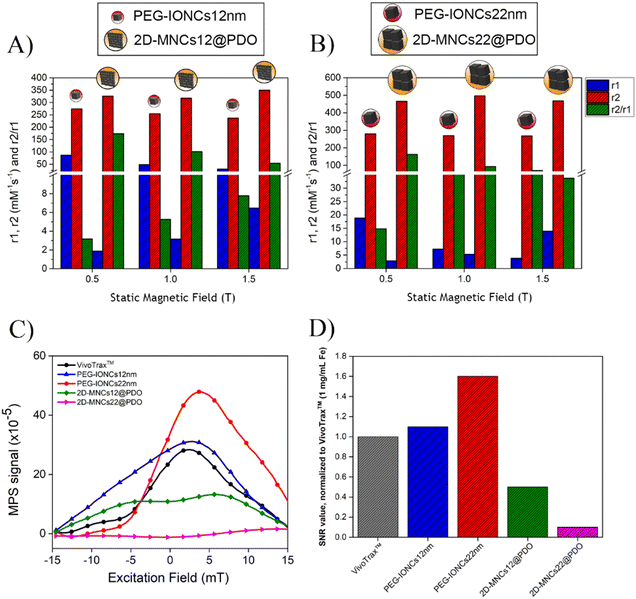
Next, water proton relaxation signals for the 2D-MNCs12@PDO and 2D-MNCs22@PDO were measured at three different static magnetic fields (0.5 T, 1 T, and 1.5 T) by making a comparison with their respective single-coated PEG-IONCs12 nm and PEG-IONCs22 nm nanocubes. The r1 and r2 relaxivities for all samples indicate that 2D-MNCs12@PDO and 2D-MNCs22@PDO clusters exhibited higher absolute r2 values than their respective single-coated nanocubes for all field conditions (Fig. 8(A), (B) and Table 2 Fig. S13, ESI†). For instance, at 1.5 T (Table 2), the r2 values of 2D-MNCs12@PDO (325 mM−1 s−1) and 2D-MNCs22@PDO (426 mM−1 s−1) are higher than the corresponding single coated nanocubes (236 mM−1 s−1 for PEG-IONCs12 nm and 268 mM−1 s−1 for PEG-IONCs22 nm, respectively). The enhancement of r2 in 2D-ordered IONCs may be likely attributed to their multimagnetic core features, hydrodynamic size (DH), and spatial confinement. Here, the magnetic exchange coupling between the cubes plays a crucial role in reducing the spin–spin relaxation time (T2). Magnetic exchange coupling refers to the interaction between the magnetic moments of adjacent nanocubes within the 2D clusters. This interaction can create strong local magnetic field gradients and enhance the degree of magnetic inhomogeneity within the system. As water protons diffuse through these magnetic field gradients, their transverse magnetization dephases faster, shortening T2 relaxation time. This phenomenon is observed in other multicomponent nanostructures, where magnetic coupling enhances spin-dephasing and shortens the spin–spin relaxation time (T2).123–126 For example, Paquet et al.127 showed that clustering iron oxide nanoparticles increased magnetic field gradients, amplifying T2 relaxivity. Similarly, Lartigue et al.128 described an enhanced relaxation effect due to cooperative dipole interactions. These insights align with our observations for 2D-MNCs12@PDO and 2D-MNCs22@PDO clusters, where the magnetic coupling effect contributes significantly to their superior r2 relaxivity compared to single-coated PEG-IONCs. Furthermore, the spatial arrangement within the clusters enhances the exchange of magnetic energy, further supporting the observed relaxivity trends. Moreover, since our clusters have an increased DH size, which is known to enhance the r2 relaxivity within an optimal size range, this parameter may also have played a role. Pöselt, Elmar, et al.,129 had for instance, self-assembled SPIO of different sizes into various hydrodynamic cluster sizes using an amphiphilic polymer and demonstrated an increase in r2 rates with DH up to a critical size threshold which was correlated to the motional average regime and static de-phase regime, and echo-limiting regime.| 0.5 T | 1 T | 1.5 T | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Relaxivity (mM−1 s−1) | Relaxivity (mM−1 s−1) | Relaxivity (mM−1 s−1) | |||||||
| Sample | r 1 | r 2 | r 2/r1 | r 1 | r 2 | r 2/r1 | r 1 | r 2 | r 2/r1 |
| PEG-IONCs12 nm | 86.63 | 274 | 3.16 | 48.4 | 254 | 5.24 | 30.4 | 236 | 7.76 |
| PEG-IONCs22 nm | 18.82 | 279 | 14.82 | 7.22 | 270 | 37.4 | 3.82 | 268 | 70.2 |
| 2D-MNCs12@PDO | 6.47 | 349 | 54.94 | 3.15 | 317 | 100 | 1.88 | 325 | 172 |
| 2D-MNCs22@PDO | 2.87 | 465 | 162 | 5.34 | 497 | 93.1 | 13.9 | 468 | 33.7 |
Likewise to this work, our 2D clusters with a larger hydrodynamic size than single nanoparticles were assumed to be in the motional average regime or at the static de-phasing regime thus showing higher r2 rates (or shortened T2 time).130 In addition, the IONCs in such 2D-confinement, as a whole entity, offer a higher magnetic diffusion surface for water molecules, in comparison to the smaller structures, because of the broader planar surface, thus the faster dephasing. Overall, the r2 values of our magnetic clusters are higher than many other iron oxide-based commercial products,131 because of the cubic and magnetically anisotropic feature of nanoparticles used.12 For the MNCs12@PDO, the relatively lower r2 relaxivity can be due to the smaller nanocube size (12 nm) and their lower saturation magnetization. On the other hand, the absolute r1 values of 2D-MNCs@PDO clusters were lower than those of the PEG-IONCs. The poor r1 values measured for the clusters are likely related to the presence of the extra hydrophobic interface (OA) between the outer PDO layer and inner magnetic cluster that prevents the free access of water molecules. Instead, for single IONCs, the hydrophilic PEG molecules are directly bound to the cube surface thus enabling a direct water retention close to the surface of the iron atoms, resulting in higher r1 rate. In addition, at lower magnetic fields (Fig. 8(B)) the differences in r1 between MNCs22@PDO and IONCs22 were less significant because dipole–dipole interactions and local magnetic field inhomogeneities were weaker, and relaxation is mainly influenced by factors like Brownian motion and intrinsic spin.120 In contrast, at 1.5 T, the strong magnetic field amplified local magnetic field inhomogeneities in magnetic clusters, enhancing dipole–dipole interactions and accelerating proton relaxation, resulting in a higher r1 for MNCs22@PDO. The collective magnetic effects and larger effective magnetic moment of clusters further increased their impact on proton relaxation. Additionally, the spatial arrangement of cubes in 2D clusters created anisotropic effects that also contributed to higher r1.132,133 To better describe a magnetic material, either as a T1 (positive) or T2 (negative) weighted MR imaging agent, it is been said theoretically, that, when the r2/r1 ratio was higher than 2, the agents are considered T2 contrast agents for MRI.134 Accordingly, all our magnetic clusters have r2/r1 ratios much larger than 2. For instance, 2D-MNC12@PDO have a r2/r1 ratio range between 54.94 to 172 depending on the magnetic fields where they were measured, while the 2D-MNCs22@PDO ranged between 162 to 33.7. The higher r2/r1 ratios for our clusters are consistent with those found by group's previous reports.71,135 From these observations, it can be deduced that 2D-MNCs12@PDO and 2D-MNCs22@PDO are excellent T2 or negative contrast enhancers for MR imaging.
Despite the several studies on magnetic clusters, in relation to MH and MRI,12,62,75,104,136 only few studies have been reported on the MPI signals of magnetic clusters.83 For this reason, MPI signal studies were performed on aqueous dispersions of 2D-MNCs12@PDO, 2D-MNCs22@PDO, PEG-IONCs12 nm and PEG-IONCs22 nm (all at 500 μL and 1 mgFe mL−1), using a custom made x-space magnetic particle relaxometer137 available at Case Western Reserve University, that operated at a fixed sinusoidal magnetic excitation field of 16.8 kHz and 20 mT (Fig. 8(C) and (D)). 2D-MNCs@PDO cluster samples exhibited relatively lower MPI signals than their single particle counterparts (Fig. 8(C)). For instance, the point spread function (PSF) of 2D-MNCs12@PDO (Fig. 8(C)), was 50% less intense and much broader (FWHM = 20.2 mT, which indicates the low resolution), of that of corresponding isolated PEG-IONCs12 nm similar values were also recorded for the 2D-MNCs22@PDO.
Moreover, it should be noted that even, the Vivo Trax™ used as a standard for comparative MPI signal measurements, shows higher MPI signal than our 2D clusters but lower values than that of single nanocubes. The reduced MPI response of the 2D assembly could be correlated to their unique spatial arrangement: the interparticle magnetic dipolar interactions of the nanocubes that stabilize them in the 2D confined architecture may promote reduced overall magnetization response of the cluster assembly to the external field at the low frequency and field intensity used in MPI measurements: in contrast from magnetic field conditions used in the MH measurements, the low frequency and field amplitude may be not enough to oscillate the structure under this time varying field.83
MPI and SAR signal evolution of PDO-coated 2D clusters at acidic environment
Provided that the degradation of PDO shell in acidic environment results in important structural differences of the clusters before and after exposure to pH 5, which, in turn, can affect the magnetic properties of the clusters, we have designed an in-test tube experiment to follow the evolution of MPI signal and SAR values upon continuous exposure of the clusters to pH 5.For the MPI signal, on four aliquots of the same PDO-coated 2D-MNCs (2 gFe L−1) sample, the pH was adjusted to pH 5 and the MPI signal amplitude was recorded at 0, 1, 3, and 6 h time points. A gradual increment of the MPI signal amplitude was observed over a period of 6 hours with a signal intensity that was almost 4 times higher than the value recorded when starting the experiment (Fig. 9(A)). As expected, this change is likely a direct consequence of the degradation of the PDO coating, leading to the disaggregation of the nanocubes previously packed in the 2D-sheets and to the changing of the surrounding environment of the nanocubes which are no longer blocked in the PDO shell thus gaining freedom to switch their magnetic moments in MPI.
It is remarkable to observe a very similar signal trend when considering the evolution over the same time window of the SAR values at pH 5 (Fig. 9(B)). In the first three hours, the SAR performances of 2D clusters showed a decrease of the signal with a minimum value of 214 ± 5 W gFe−1 at 1 h with respect to SAR value measured at the begging of the experiment (369 ± 29 W gFe−1 at t = 0). In the next four hours the SAR value gradually increase to reach a value as high as 481 ± 17 W gFe−1 after 7 h overcoming the SAR signal of the initial clusters. The initial low SAR values may be due to a first destabilization effect of the 2D clusters in an acid environment, however, with the time, upon exposure to the acidic environment with the degradation of the PDO shell and the release of single nanocubes, the freed nanocubes enhanced their ability to absorb magneto energy, thus explaining the observed SAR increase. To note that these data are supported by the structural change in morphology and destabilization of the PDO shell observed under TEM (Fig. 3) and, also, confirmed by the change in hydrodynamic size of the clusters in acidic environment measured by dynamic light scattering (Fig. S14, ESI†). Altogether, these data suggest the possibility to use MPI signal of the cluster as a tracer signal to gain information on the status of the disassembling of the clusters thus enable to plan the magnetic hyperthermia therapy only when the heat generated by the disassembled clusters will be maximized.
Conclusions
In this work, we have succeeded to prepare nanosheet clusters (2D-MNCs@PDO) with unique structural and magnetic properties, composed of close-packed IONCs, with control on the 2D structure and to their stability in water thanks to the PDO shell. The assembling requires a simple microemulsion process in hot water bath sonicator of an organic CHCl3 solution of magnetic nanocubes with a aqueous SDS surfactant solution. We found that the water: CHCl3 couple choice is crucial to obtain these specific 2D-clusters. The use of PDO shell, obtained through a self-polymerization process of DOPA in alkaline environment, is required to increase stability under magnetic field but also to introduce a protective shell.SAXS and cryo-EM techniques are used to confirm the 2D ordered configuration. Noteworthy, these techniques highlight the formation of the 2D-MNCs, in which the IONCs are assembled in a two-dimensional lamellar organization. Moreover, the IONCs order, the IONCs number per cluster and the interparticle spacing values are reasonable and in agreement to those measured by TEM characterization.
The final shell thickness of the PDO polymer is controlled by tuning the initial amount of DA while the kinetic of degradation is also adjustable by the thickness of the shell: it is more rapid when the PDO shell is thinner (around 7 nm) and it requires longer time when the PDO shell is ticker (ca. 20 nm). Finally, as here shown with CV dye, the PDO shell provides also a versatile framework for encapsulating small antitumoral drug molecules and, eventually, the release occurs under acidic pH. Although 2D-MNCs@PDO exhibit lower hyperthermia performance than their respective individually dispersed IONCs, the SAR values of the 2D-MNCs12@PDO are better than those of iron oxide nanosphere used in clinic.70 A remarkable aspect of the 2D-MNCs@PDO is their enhanced magnetic properties as a function of the pH. Under acidic environment, as that of tumor microenvironment, the PDO shell of the 2D clusters can be destabilized enabling the disassembly of the clusters. This has a direct effect on the MPI signals which gradually increase overtime. In concomitance, a SAR value enhancement is recorded in acidic pH solution, which is attributed to the release of free nanocubes upon disassembling of the clusters.
Moreover, the 2D-MNCs@PDO allows for higher r2 relaxation rates compared to their respective IONCs due to a faster dephasing of the hydrogen protons and therefore, to a shortening of T2. Having established such a theranostic platform, future work will aim at applying such systems as drug delivery tools, magnetic hyperthermia agent, MPI tracer and MRI agents first in vitro and then in vivo on tumor cells.
Author contributions
G. M. and T. P. designed the research. H. G. and E. S. synthesized and characterized the IONCs. G. M. performed water transfer of IONCs, the 2D clustering of IONCs, in situ polymerization of PDO on 2D-clusters, DLS, TEM and magnetic hyperthermia measurements. S. K. A. performed the magnetic relaxation measurements. E. S. performed the CV loading and release experiments. E. S. and N. S performed the MPI study at acidic pH. G. M. performed the SAR study at acidic pH. A. C. S. S. and Y. H. T. performed MPI. C. G. and U. O. performed SAXS characterization. R. M. performed cryo-TEM and cryo-electron tomography analyses. N. S. performed SQUID characterization. G. M., H. G., N. S., S. K. A. and T. P. analyzed and discussed the results. G. M. and T. P. wrote the first draft, and the remaining authors contributed to revise the manuscript.Data availability
The data supporting this article ‘Magnetic Nanosheets: from iron oxide nanocubes to polydopamine embedded 2D clusters and their multi-purpose properties’ by the authors ‘Giacomo Mandriota, Sahitya Kumar Avugadda, Ehsan Sadeghi, Niccolò Silvestri, Roberto Marotta, Helena Gavilán, Ulf Olsson, Cinzia Giannini, Yu Hsin Tsai4, Anna Cristina S. Samia and Teresa Pellegrino’ have been included as part of the ESI.†Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was partially funded by the European Research Council (Consolidator grant GIULIa, contract no. ID N. 101044020) and the AIRC project (contract no. AIRC IG-29323). The authors acknowledge Peter Holmqvist for performing the SAXS data collection and Giammarino Pugliese for having performed TGA measurement.References
- M. R. Jones, N. C. Seeman and C. A. Mirkin, Science, 2015, 347, 1260901 CrossRef PubMed.
- M. A. Boles, M. Engel and D. V. Talapin, Chem. Rev., 2016, 116, 11220–11289 CrossRef CAS PubMed.
- B. A. Patil and R. D. Kokate, Procedia Manuf., 2018, 20, 147–153 CrossRef.
- L. Gloag, M. Mehdipour, D. Chen, R. D. Tilley and J. J. Gooding, Adv. Mater., 2019, 31, 1904385 CrossRef CAS PubMed.
- V. A. Bragina, S. L. Znoyko, A. V. Orlov, A. V. Pushkarev, M. P. Nikitin and P. I. Nikitin, Anal. Chem., 2019, 91, 9852–9857 CrossRef CAS PubMed.
- P. Miralles, I. van Gemert, A. Chisvert and A. Salvador, J. Chromatogr. A, 2019, 1604, 460465 CrossRef CAS PubMed.
- B. T. Mai, S. Fernandes, P. B. Balakrishnan and T. Pellegrino, Acc. Chem. Res., 2018, 51, 999–1013 CrossRef CAS PubMed.
- V. F. Cardoso, A. Francesko, C. Ribeiro, M. Bañobre-López, P. Martins and S. Lanceros-Mendez, Adv. Healthcare Mater., 2018, 7, 1700845 CrossRef PubMed.
- J. F. Liu, B. Jang, D. Issadore and A. Tsourkas, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2019, 11, e1571 Search PubMed.
- L. Geng and Z. Luo, J. Phys. Chem. Lett., 2024, 15, 1856–1865 CAS.
- S. K. Ghosh and A. Böker, Macromol. Chem. Phys., 2019, 220, 1900196 Search PubMed.
- S. K. Avugadda, N. Soni, E. M. Rodrigues, S. Persano and T. Pellegrino, ACS Appl. Mater. Interfaces, 2024, 16, 6743–6755 CAS.
- T. Raczka, A. Wolf, J. Reichstein, C. Stauch, B. Schug, S. Müssig and K. Mandel, J. Magn. Magn. Mater., 2024, 598, 172042 CAS.
- Y. Lin, H. Skaff, T. Emrick, A. D. Dinsmore and T. P. Russell, Science, 2003, 299, 226–229 CAS.
- S. A. Majetich, T. Wen and R. A. Booth, ACS Nano, 2011, 5, 6081–6084 CAS.
- Z. Nie, A. Petukhova and E. Kumacheva, Nat. Nanotechnol., 2010, 5, 15–25 CAS.
- T. Wang, J. Zhuang, J. Lynch, O. Chen, Z. Wang, X. Wang, D. LaMontagne, H. Wu, Z. Wang and Y. C. Cao, Science, 2012, 338, 358 CAS.
- K. Deng, Z. Luo, L. Tan and Z. Quan, Chem. Soc. Rev., 2020, 49, 6002–6038 CAS.
- Z. Ferjaoui, S. Nahle, C. S. Chang, J. Ghanbaja, O. Joubert, R. Schneider, L. Ferrari, E. Gaffet and H. Alem, ACS Omega, 2020, 5, 4770–4777 CAS.
- N. A. Kotov, I. Dekany and J. H. Fendler, J. Phys. Chem., 1995, 99, 13065–13069 CAS.
- R. P. Pothukuchi, V. K. Prajapat and M. Radhakrishna, Langmuir, 2021, 37, 12007–12015 CAS.
- L. E. Low, H. P. Lim, Y. S. Ong, S. P. Siva, C. S. Sia, B.-H. Goh, E. S. Chan and B. T. Tey, J. Controlled Release, 2022, 345, 231–274 CrossRef CAS PubMed.
- W. E. Ford, O. Harnack, A. Yasuda and J. M. Wessels, Adv. Mater., 2001, 13, 1793–1797 CrossRef CAS.
- G. Lee, J. Song, H. Han, D. Kwon, J. Park, S. Jeon, S. Jeong and S. Kim, Bioconjugate Chem., 2021, 32, 1052–1057 CrossRef CAS PubMed.
- E. Maltas and M. Ozmen, Mater. Sci. Eng., C, 2015, 54, 43–49 CrossRef CAS PubMed.
- B. Liu and J. Liu, Nanoscale, 2015, 7, 13831–13835 RSC.
- C. Paquet, L. Pagé, A. Kell and B. Simard, Langmuir, 2010, 26, 5388–5396 CrossRef CAS PubMed.
- C.-H. Wu, J. Cook, S. Emelianov and K. Sokolov, Adv. Funct. Mater., 2014, 24, 6862–6871 CrossRef CAS.
- G. A. DeVries, M. Brunnbauer, Y. Hu, A. M. Jackson, B. Long, B. T. Neltner, O. Uzun, B. H. Wunsch and F. Stellacci, Science, 2007, 315, 358 CrossRef CAS PubMed.
- S. Park, J. H. Lim, S. W. Chung and C. A. Mirkin, Science, 2004, 303, 348–351 CrossRef CAS PubMed.
- Y. Kang, K. J. Erickson and T. A. Taton, J. Am. Chem. Soc., 2005, 127, 13800–13801 CrossRef CAS PubMed.
- K. K. Caswell, J. N. Wilson, U. H. F. Bunz and C. J. Murphy, J. Am. Chem. Soc., 2003, 125, 13914–13915 CrossRef CAS PubMed.
- Z. Tang, Z. Zhang, Y. Wang, S. C. Glotzer and N. A. Kotov, Science, 2006, 314, 274–278 CrossRef CAS PubMed.
- N. Zhao, K. Liu, J. Greener, Z. Nie and E. Kumacheva, Nano Lett., 2009, 9, 3077–3081 CrossRef CAS PubMed.
- M. S. Nikolic, C. Olsson, A. Salcher, A. Kornowski, A. Rank, R. Schubert, A. Frömsdorf, H. Weller and S. Förster, Angew. Chem., Int. Ed., 2009, 48, 2752–2754 CrossRef CAS PubMed.
- A. M. Kalsin, M. Fialkowski, M. Paszewski, S. K. Smoukov, K. J. M. Bishop and B. A. Grzybowski, Science, 2006, 312, 420 CrossRef CAS PubMed.
- D. Nykypanchuk, M. M. Maye, D. van der Lelie and O. Gang, Nature, 2008, 451, 549–552 CrossRef CAS PubMed.
- S. Y. Park, A. K. R. Lytton-Jean, B. Lee, S. Weigand, G. C. Schatz and C. A. Mirkin, Nature, 2008, 451, 553–556 CrossRef CAS PubMed.
- C. R. Iacovella and S. C. Glotzer, Nano Lett., 2009, 9, 1206–1211 CrossRef CAS PubMed.
- P. H. Qiu, C. Jensen, N. Charity, R. Towner and C. B. Mao, J. Am. Chem. Soc., 2010, 132, 17724–17732 CrossRef CAS PubMed.
- M. E. Yu, J. Y. Hwang and T. J. Deming, J. Am. Chem. Soc., 1999, 121, 5825–5826 CrossRef CAS.
- M. L. Alfieri, T. Weil, D. Y. W. Ng and V. Ball, Adv. Colloid Interface Sci., 2022, 305, 102689 CrossRef CAS PubMed.
- H. Hemmatpour, O. De Luca, D. Crestani, M. C. A. Stuart, A. Lasorsa, P. C. A. van der Wel, K. Loos, T. Giousis, V. Haddadi-Asl and P. Rudolf, Nat. Commun., 2023, 14, 664 CAS.
- M. E. Lynge, P. Schattling and B. Stadler, Nanomedicine, 2015, 10, 2725–2742 CrossRef CAS PubMed.
- S. S. Ruppel and J. Liang, Langmuir, 2022, 38, 5020–5029 CAS.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CAS.
- Y. Liu, C. K. K. Choi, H. Hong, Y. Xiao, M. L. Kwok, H. Liu, X. Y. Tian and C. H. J. Choi, ACS Nano, 2021, 15, 13871–13890 CAS.
- M. Battaglini, M. Emanet, A. Carmignani and G. Ciofani, Nano Today, 2024, 55, 102151 CAS.
- G. Qi, S. Wang, Q. Yin, Z. Zhang, X. Wen and L. Hao, ACS Appl. Nano Mater., 2023, 6, 23184–23195 CAS.
- M. Witkowska, E. Golusińska-Kardach, W. Golusiński and E. Florek, Int. J. Mol. Sci., 2023, 24, 4890 CrossRef CAS PubMed.
- Q. Wei, F. L. Zhang, J. Li, B. J. Li and C. S. Zhao, Polym. Chem., 2010, 1, 1430–1433 RSC.
- D. Ortega and Q. A. Pankhurst, Nanoscience: Volume 1: Nanostructures through Chemistry, The Royal Society of Chemistry, 2013, vol. 1, pp. 60–88 Search PubMed.
- A. E. Deatsch and B. A. Evans, J. Magn. Magn. Mater., 2014, 354, 163–172 CrossRef CAS.
- I. M. Obaidat, B. Issa and Y. Haik, Nanomaterials, 2015, 5, 63–89 CrossRef PubMed.
- D. Yoo, J.-H. Lee, T.-H. Shin and J. Cheon, Acc. Chem. Res., 2011, 44, 863–874 CAS.
- M. Bañobre-López, A. Teijeiro and J. Rivas, Rep. Pract. Oncol. Radiother., 2013, 18, 397–400 Search PubMed.
- E. Duguet, S. Vasseur, S. Mornet and J.-M. Devoisselle, Nanomedicine, 2006, 1, 157–168 CAS.
- J. van der Zee, Ann. Oncol., 2002, 13, 1173–1184 CrossRef CAS PubMed.
- L. F. Fajardo, Cancer Res., 1984, 44, 4826s CAS.
- J. Carrey, B. Mehdaoui and M. Respaud, J. Appl. Phys., 2011, 109, 083921 CrossRef.
- I. Andreu, E. Natividad, L. Solozábal and O. Roubeau, ACS Nano, 2015, 9, 1408–1419 CAS.
- H. Gavilán, S. K. Avugadda, T. Fernández-Cabada, N. Soni, M. Cassani, B. T. Mai, R. Chantrell and T. Pellegrino, Chem. Soc. Rev., 2021, 50, 11614–11667 RSC.
- P. Guardia, R. Di Corato, L. Lartigue, C. Wilhelm, A. Espinosa, M. Garcia-Hernandez, F. Gazeau, L. Manna and T. Pellegrino, ACS Nano, 2012, 6, 3080–3091 Search PubMed.
- R. Das, J. Alonso, Z. Nemati Porshokouh, V. Kalappattil, D. Torres, M.-H. Phan, E. Garaio, J. Á. García, J. L. Sanchez Llamazares and H. Srikanth, J. Phys. Chem. C, 2016, 120, 10086–10093 CAS.
- C. Martinez-Boubeta, K. Simeonidis, A. Makridis, M. Angelakeris, O. Iglesias, P. Guardia, A. Cabot, L. Yedra, S. Estradé, F. Peiró, Z. Saghi, P. A. Midgley, I. Conde-Leborán, D. Serantes and D. Baldomir, Sci. Rep., 2013, 3, 1652 Search PubMed.
- Z. Nemati, J. Alonso, L. M. Martinez, H. Khurshid, E. Garaio, J. A. Garcia, M. H. Phan and H. Srikanth, J. Phys. Chem. C, 2016, 120, 8370–8379 CAS.
- Z. Nemati, J. Alonso, I. Rodrigo, R. Das, E. Garaio, J. Á. García, I. Orue, M.-H. Phan and H. Srikanth, J. Phys. Chem. C, 2018, 122, 2367–2381 CrossRef CAS.
- S.-h Noh, W. Na, J.-t Jang, J.-H. Lee, E. J. Lee, S. H. Moon, Y. Lim, J.-S. Shin and J. Cheon, Nano Lett., 2012, 12, 3716–3721 CrossRef CAS PubMed.
- L. M. Bauer, S. F. Situ, M. A. Griswold and A. C. S. Samia, Nanoscale, 2016, 8, 12162–12169 RSC.
- B. Thiesen and A. Jordan, Int. J. Hyperthermia, 2008, 24, 467–474 CAS.
- M. E. Materia, P. Guardia, A. Sathya, M. Pernia Leal, R. Marotta, R. Di Corato and T. Pellegrino, Langmuir, 2015, 31, 808–816 CAS.
- B. Mehdaoui, R. P. Tan, A. Meffre, J. Carrey, S. Lachaize, B. Chaudret and M. Respaud, Phys. Rev. B: Condens. Matter Mater. Phys., 2013, 87, 174419 Search PubMed.
- D. Serantes, D. Baldomir, M. Pereiro, B. Hernando, V. M. Prida, J. L. Sánchez Llamazares, A. Zhukov, M. Ilyn and J. González, J. Phys. D: Appl. Phys., 2009, 42, 215003 CrossRef.
- A. N. Generalova, V. A. Oleinikov and E. V. Khaydukov, Adv. Colloid Interface Sci., 2021, 297, 102543 CAS.
- S. K. Avugadda, M. E. Materia, R. Nigmatullin, D. Cabrera, R. Marotta, T. F. Cabada, E. Marcello, S. Nitti, E. J. Artés-Ibañez, P. Basnett, C. Wilhelm, F. J. Teran, I. Roy and T. Pellegrino, Chem. Mater., 2019, 31, 5450–5463 CAS.
- N. C. Bigall, C. Wilhelm, M.-L. Beoutis, M. García-Hernandez, A. A. Khan, C. Giannini, A. Sánchez-Ferrer, R. Mezzenga, M. E. Materia, M. A. Garcia, F. Gazeau, A. M. Bittner, L. Manna and T. Pellegrino, Chem. Mater., 2013, 25, 1055–1062 CAS.
- B. Gleich and J. Weizenecker, Nature, 2005, 435, 1214–1217 CAS.
- P. W. Goodwill and S. M. Conolly, IEEE Trans. Med. Imaging, 2010, 29, 1851–1859 Search PubMed.
- T.-H. Shin, Y. Choi, S. Kim and J. Cheon, Chem. Soc. Rev., 2015, 44, 4501–4516 RSC.
- J. Rahmer, A. Antonelli, C. Sfara, B. Tiemann, B. Gleich, M. Magnani, J. Weizenecker and J. Borgert, Phys. Med. Biol., 2013, 58, 3965–3977 CrossRef CAS PubMed.
- S. Harvell-Smith, L. D. Tung and N. T. K. Thanh, Nanoscale, 2022, 14, 3658–3697 RSC.
- R. M. Ferguson, A. P. Khandhar and K. M. Krishnan, J. Appl. Phys., 2012, 111, 07B318 CrossRef PubMed.
- S. K. Avugadda, S. Wickramasinghe, D. Niculaes, M. Ju, A. Lak, N. Silvestri, S. Nitti, I. Roy, A. C. S. Samia and T. Pellegrino, Nanomaterials, 2020, 11, 62 Search PubMed.
- A. Quarta, A. Curcio, H. Kakwere and T. Pellegrino, Nanoscale, 2012, 4, 3319–3334 CAS.
- T. Pellegrino, H. Gavilán Rubio, B. T. Mai and R. Cingolani, Eur. Pat., EP3962863A1, 2022 Search PubMed.
- H. Gavilán, G. M. R. Rizzo, N. Silvestri, B. T. Mai and T. Pellegrino, Nat. Protoc., 2023, 18, 783–809 Search PubMed.
- N. Liao, M. Wu, F. Pan, J. Lin, Z. Li, D. Zhang, Y. Wang, Y. Zheng, J. Peng, X. Liu and J. Liu, Sci. Rep., 2016, 6, 18746 CAS.
- R. Batul, T. Tamanna, A. Khaliq and A. Yu, Biomater. Sci., 2017, 5, 1204–1229 CAS.
- J. R. Kremer, D. N. Mastronarde and J. R. McIntosh, J. Struct. Biol., 1996, 116, 71–76 CAS.
- Z. Lu and Y. Yin, Chem. Soc. Rev., 2012, 41, 6874–6887 CAS.
- R. G. Alany, I. G. Tucker, N. M. Davies and T. Rades, Drug Dev. Ind. Pharm., 2001, 27, 31–38 Search PubMed.
- A. V. Sineva, D. S. Ermolat’ev and A. V. Pertsov, Colloid J., 2007, 69, 89–94 CAS.
- M. Choudhary and S. M. Kamil, ACS Omega, 2020, 5, 22891–22900 CAS.
- N. Akter, S. Radiman, F. Mohamed and M. Reza Imam Hasan, Mini-Rev. Med. Chem., 2013, 13, 1327–1339 CAS.
- A. Rahman and J. Eastoe, Soft Matter, 2022, 18, 9133–9152 RSC.
- N. Du, R. Song, H. Zhang, J. Sun, S. Yuan, R. Zhang and W. Hou, Colloids Surf., A, 2016, 509, 195–202 CrossRef CAS.
- T. K. Sau and C. J. Murphy, Langmuir, 2005, 21, 2923–2929 CAS.
- L. Hu, C. Wang, R. M. Kennedy, L. D. Marks and K. R. Poeppelmeier, Inorg. Chem., 2015, 54, 740–745 CrossRef CAS PubMed.
- J.-e Park, D. R. Hickey, S. Jun, S. Kang, X. Hu, X.-J. Chen and S.-J. Park, Adv. Funct. Mater., 2016, 26, 7791–7798 CAS.
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418 CAS.
- C. B. Murray, C. R. Kagan and M. G. Bawendi, Annu. Rev. Mater. Sci., 2000, 30, 545–610 CAS.
- M. M. Maye, I. I. S. Lim, J. Luo, Z. Rab, D. Rabinovich, T. Liu and C.-J. Zhong, J. Am. Chem. Soc., 2005, 127, 1519–1529 CAS.
- C. P. Collier, T. Vossmeyer and J. R. Heath, Annu. Rev. Phys. Chem., 1998, 49, 371–404 CAS.
- D. Niculaes, A. Lak, G. C. Anyfantis, S. Marras, O. Laslett, S. K. Avugadda, M. Cassani, D. Serantes, O. Hovorka, R. Chantrell and T. Pellegrino, ACS Nano, 2017, 11, 12121–12133 CrossRef CAS PubMed.
- B. P. Pichon, A. Demortière, M. Pauly, K. Mougin, A. Derory and S. Bégin-Colin, J. Phys. Chem. C, 2010, 114, 9041–9048 CAS.
- P. N. Njoki, I. I. S. Lim, D. Mott, H.-Y. Park, B. Khan, S. Mishra, R. Sujakumar, J. Luo and C.-J. Zhong, J. Phys. Chem. C, 2007, 111, 14664–14669 CAS.
- S. I. Lim and C.-J. Zhong, Acc. Chem. Res., 2009, 42, 798–808 CAS.
- J. Winkelmann, Ber. Bunsenges. Phys. Chem., 1997, 101, 641–642 Search PubMed.
- N. B. Bowden, A. Terfort, J. D. Carbeck and G. M. Whitesides, Science, 1997, 276, 233–235 CAS.
- T. Wang, X. Wang, D. LaMontagne, Z. Wang, Z. Wang and Y. C. Cao, J. Am. Chem. Soc., 2012, 134, 18225–18228 CAS.
- Q. Wei, F. Zhang, J. Li, B. Li and C. Zhao, Polym. Chem., 2010, 1, 1430–1433 CAS.
- G. Mandriota, R. Di Corato, M. Benedetti, F. De Castro, F. P. Fanizzi and R. Rinaldi, ACS Appl. Mater. Interfaces, 2019, 11, 1864–1875 CAS.
- A. M. Maurelli, V. De Leo, V. Daniello, C. D. Calvano, F. Ciriaco, F. Milano, C. Ingrosso, T. R. I. Cataldi, S. Di Gioia, M. Conese, A. Agostiano and L. Catucci, Mater. Today Chem., 2024, 37, 101994 CAS.
- X. Zheng, J. Zhang, J. Wang, X. Qi, J. M. Rosenholm and K. Cai, J. Phys. Chem. C, 2015, 119, 24512–24521 CAS.
- Q. Liu, B. Yu, W. Ye and F. Zhou, Macromol. Biosci., 2011, 11, 1227–1234 CAS.
- M. Bergström, J. S. Pedersen, P. Schurtenberger and S. U. Egelhaaf, J. Phys. Chem. B, 1999, 103, 9888–9897 Search PubMed.
- R. D. Zysler, D. Fiorani and A. M. Testa, J. Magn. Magn. Mater., 2001, 224, 5–11 Search PubMed.
- E. Tronc, D. Fiorani, M. Noguès, A. M. Testa, F. Lucari, F. D’Orazio, J. M. Grenèche, W. Wernsdorfer, N. Galvez, C. Chanéac, D. Mailly and J. P. Jolivet, J. Magn. Magn. Mater., 2003, 262, 6–14 Search PubMed.
- R. Fu, Y. Yan, C. Roberts, Z. Liu and Y. Chen, Sci. Rep., 2018, 8, 4704 Search PubMed.
- S. Ota and Y. Takemura, J. Phys. Chem. C, 2019, 123, 28859–28866 CAS.
- A. Rousseau, M. Tellier, L. Marin, M. Garrow, C. Madelaine, N. Hallali and J. Carrey, J. Magn. Magn. Mater., 2021, 518, 167403 CAS.
- D. Cabrera, A. Lak, T. Yoshida, M. E. Materia, D. Ortega, F. Ludwig, P. Guardia, A. Sathya, T. Pellegrino and F. J. Teran, Nanoscale, 2017, 9, 5094–5101 Search PubMed.
- Y. X. Wang, S. M. Hussain and G. P. Krestin, Eur. Radiol., 2001, 11, 2319–2331 CAS.
- T. Ahmad, H. Bae, I. Rhee, Y. Chang, J. Lee and S. Hong, Curr. Appl. Phys., 2012, 12, 969–974 Search PubMed.
- M. Talelli, C. J. F. Rijcken, T. Lammers, P. R. Seevinck, G. Storm, C. F. van Nostrum and W. E. Hennink, Langmuir, 2009, 25, 2060–2067 CAS.
- D. Cheng, G. Hong, W. Wang, R. Yuan, H. Ai, J. Shen, B. Liang, J. Gao and X. Shuai, J. Mater. Chem., 2011, 21, 4796–4804 RSC.
- C. Paquet, H. W. de Haan, D. M. Leek, H.-Y. Lin, B. Xiang, G. Tian, A. Kell and B. Simard, ACS Nano, 2011, 5, 3104–3112 Search PubMed.
- L. Lartigue, P. Hugounenq, D. Alloyeau, S. P. Clarke, M. Lévy, J.-C. Bacri, R. Bazzi, D. F. Brougham, C. Wilhelm and F. Gazeau, ACS Nano, 2012, 6, 10935–10949 CAS.
- E. Pöselt, H. Kloust, U. Tromsdorf, M. Janschel, C. Hahn, C. Maßlo and H. Weller, ACS Nano, 2012, 6, 1619–1624 Search PubMed.
- S. Fu, Z. Cai and H. Ai, Adv. Healthcare Mater., 2021, 10, 2001091 CAS.
- M. Rohrer, H. Bauer, J. Mintorovitch, M. Requardt and H.-J. Weinmann, Invest. Radiol., 2005, 40, 715–724 Search PubMed.
- Y. Zhang, J. Cheng and W. Liu, Sensors, 2019, 19, 3396 Search PubMed.
- P. Ilg and M. Kröger, Sci. Rep., 2023, 13, 16523 Search PubMed.
- Q. L. Vuong, J.-F. Berret, J. Fresnais, Y. Gossuin and O. Sandre, Adv. Healthcare Mater., 2012, 1, 502–512 CAS.
- S. K. Avugadda, S. Wickramasinghe, D. Niculaes, M. Ju, A. Lak, N. Silvestri, S. Nitti, I. Roy, A. C. S. Samia and T. Pellegrino, Nanomaterials, 2021, 11, 62 Search PubMed.
- Z. Cai, C. Wu, L. Yang, D. Wang and H. Ai, ACS Biomater. Sci. Eng., 2020, 6, 2533–2542 CAS.
- M. Ju, M. Navarreto-Lugo, S. Wickramasinghe, N. B. Milbrandt, A. McWhorter and A. C. S. Samia, Nanoscale, 2019, 11, 18582–18594 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: TEM characterization of the IONCs and PEG-IONCs and the obtained nanosystems, TGA analysis, DLS analysis, normalized magnetization curves measured at 5 K, SAR evolution vs. glycerol content, relaxivities and MPS measurements. See DOI: https://doi.org/10.1039/d4nh00566j |
| This journal is © The Royal Society of Chemistry 2025 |