Open Access Article
Open Access ArticleTranscriptome profiling of serum exosomes by RNA-Seq reveals lipid metabolic changes as a potential biomarker for evaluation of roxadustat treatment of chronic kidney diseases
Ru
Zhou
ab,
YaXuan
Zhen
ab,
Hualin
Ma
ab,
Zhen
Wang
ab,
LiXia
Liu
ab,
Xinzhou
Zhang
*ab and
Baochun
Guo
 *ab
*ab
aThe Second Clinical Medical College, Jinan University (Shenzhen People’s Hospital), Shenzhen 518020, Guangdong, China. E-mail: echobbg@163.com; xin.zhou2@qq.com; Tel: +0755-25533018
bDepartment of Nephrology, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University, The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen 518020, Guangdong, China
First published on 17th March 2025
Abstract
The incidence of chronic kidney disease (CKD) is increasing globally; however, effective preventive and therapeutic strategies are currently limited. Roxadustat is being clinically used to treat renal anemia in CKD patients to reduce anemia-related complications and improve patients' life quality. Exosomes are small vesicles carrying important information that contribute to cell-to-cell communication and are present in various body fluids. However, little is known about the role of serum exosomes and their association with CKD after roxadustat treatment. Next-generation sequencing approaches have revealed that exosomes are enriched in noncoding RNAs and thus exhibit great potential as sensitive nucleic acid biomarkers in various human diseases. In this study, we aimed to identify the changed mRNAs–lncRNAs after roxadustat treatment as novel biomarkers for assessing the efficiency of the treatment. Through our study using RNA-seq data, we identified 957 mRNAs (626 upregulated and 331 downregulated after roxadustat treatment) and 914 lncRNAs (444 upregulated and 470 downregulated) derived from exosomes that were significantly changed, which was highly correlated to lipid metabolism. Our analysis through whole transcriptome profiling of exosome RNAs encompasses an identified differentially expressed mRNA–lncRNA regulatory axis in a larger patient cohort for the validation of suitable biomarkers for assessing CKD after roxadustat treatment.
1. Introduction
Chronic kidney disease (CKD) is a disease spread worldwide with life-threatening implications that occur under complex multifactorial conditions.1 Renal anemia is a common complication of CKD, especially in the late stage of non-dialysis CKD. It occurs because the kidneys cannot produce enough erythropoietin, a hormone that stimulates the production of red blood cells. Renal anemia can lead to fatigue, shortness of breath, and other symptoms and may increase the risk of cardiovascular disease and other serious complications.2 Therefore, various clinical practice guidelines require the monitoring, diagnosis and even treatment of anemia to improve the clinical outcomes and life quality of patients with CKD.3The primary reasons for renal anemia are insufficient production of erythropoietin (EPO), shortened lifespan of red blood cells, and disruptions in iron metabolism in the body. Traditional treatment methods, which include the administration of erythropoietin-stimulating agents (ESAs), iron supplements, and blood transfusions, have well-established foundations. However, these treatment outcomes are often compromised by various factors such as inflammation, iron absorption, production of EPO antibodies, and long-term safety concerns regarding drugs. To overcome these limitations, hypoxia-inducible factor-prolyl hydroxylase inhibitors (HIF-PHIs) were developed as an alternative strategy. This class of drugs, which includes roxadustat, was the first globally approved HIF-PHI with a completed phase III clinical trial for the treatment of renal anemia in dialysis and non-dialysis patients in China in 2018. By mimicking ketoglutarate, a substrate of prolyl hydroxylase (PH), roxadustat disrupts the role of PH in degrading HIFs. The stabilization of HIFs then improves anemia by increasing the expression of EPO, EPO-associated receptors, and proteins that facilitate iron absorption, leading to accelerated iron utilization and endogenous erythropoietin production.4,5 Multiple studies have demonstrated the effectiveness, safety, and cost-effectiveness of roxadustat.6–9 However, whether there are additional regulatory mechanisms of roxadustat for improving anemia in non-dialysis (ND-CKD) patients remains unclear. Therefore, further research into roxadustat's potential targets and related regulatory mechanisms will provide a more comprehensive theoretical basis for its clinical application.
Exosomes are extracellular vesicles secreted by cells, with diameter ranging from 30 to 150 nanometers. Almost all types of cells in the human body can produce exosomes, and they are present throughout the body in various bodily fluids such as blood, urine, cerebrospinal fluid, saliva, milk, semen, abdominal cavity fluid, and joint synovial fluid. Exosomes carry a variety of important proteins, lipids, DNA, and RNA, and their transfer can facilitate material transport and information exchange between cells.10,11 They contain a lipid bilayer membrane structure and carry typical transmembrane proteins and receptors, adhesion molecules, lipid raft-associated proteins, and immune regulatory molecules. Various proteins and nucleic acids can be found within the exosome lumen.11 Recent studies have demonstrated that exosomes obtained from different biological fluids and their contents (proteins, nucleic acids, glycoconjugates, and lipids) can serve as biomarkers for cancer diagnosis, prognosis, and therapeutic response.12 Moreover, the discovery of exosomes as therapeutic delivery vehicles has attracted much attention in antineoplastic drug delivery; thus, they are regarded as a potential therapeutic target in translational medicine.
In the cellular environment, the repertoire of RNA extends beyond protein-coding messenger RNAs (mRNAs) to include non-coding RNAs (ncRNAs) such as microRNAs (miRNAs), circular noncoding RNAs (circRNAs) and long noncoding RNAs (lncRNAs).13,14 These diverse RNA species serve as central players in the genesis and progression of various pathologies, including cancer and viral diseases.15,16 MiRNAs, spanning approximately 17–25 nucleotides in length, exert influence on gene expression by binding specifically to the 3′ untranslated regions (UTR) of target mRNAs, thus inhibiting gene expression. The expression profiling of RNAs in CKD has been characterized previously,17 and interactions between different RNA species such as miRNA–mRNA, lncRNA–mRNA, and miRNA–lncRNA have been investigated extensively using CKD clinical samples.18 Research has demonstrated that treatment with roxadustat alters the expression levels of specific microRNAs and long non-coding RNAs in patients, potentially impacting cellular metabolism and function.19 This suggests that roxadustat's therapeutic effect extends beyond the direct promotion of erythropoiesis; it may also influence the overall physiological state of CKD patients through complex molecular mechanisms. Currently, the complex molecular mechanisms involved require further elucidation. A comprehensive analysis of regulatory networks derived from transcriptome profiles could improve our understanding of CKD patients’ physiological changes post-roxadustat treatment, particularly regarding the extracellular vesicle environment and treatment efficacy evaluation. In this study, we aim to elucidate the co-regulatory functional RNA profile network in ND-CKD patients undergoing roxadustat treatment. This will be based on transcriptome sequencing of the expression profiles of non-coding RNAs and mRNAs with varying expression levels, which will help us better understand the comprehensive therapeutic effects of roxadustat.
2. Materials and methods
2.1. Clinical serum specimen collection
Clinical specimen of human blood samples (5 mL for each sample) were collected from 13 ND-CKD anemia patients diagnosed in our hospital before and one month20 after roxadustat treatment. This included the experimental group (after roxadustat treatment, n = 13) and the control group (before roxadustat treatment, n = 13). The samples were centrifuged at 1000g (centrifuge 5427 R, Eppendorf) at 4 °C for 10 min, and the supernatant was transferred to new tubes and stored at −80 °C before use. Detailed sample information, including age, gender, and clinical information, is listed in Table 1. The patients were notified, and signed informed consent forms were obtained before sample use. All the sample treatments and procedures were strictly performed under the guidance of the guidelines for the collection and application of human-related specimens of our hospital and approved by the institutional review boards of the Second Clinical Medical College (Shenzhen People's Hospital) of Jinan University (KY-LL-2021681-01).| Roxadustat (n = 13) | |
|---|---|
| The average (±standard deviation) values are reported in Table 1. | |
| Age (years) | 54 ± 13.80 |
| Male sex, n (%) | 7(53.85) |
| Comorbidities, n (%) | |
| Type 2 diabetes | 5(38.46) |
| Coronary artery disease | 2(15.38) |
| Congestive heart failure | 3(23.08) |
| Cerebrovascular events | 2(15.38) |
| Hypertension | 10(76.92) |
| eGFR (mL min−1 per 1.73 m2) | 12.30 ± 9.53 |
| Serum creatinine (μmol L−1) | 531.85 ± 253.24 |
| Albumin (g L−1) | 38.24 ± 5.72 |
| Glutamic pyruvic transaminase (U L−1) | 16.08 ± 12.37 |
| Glutamic oxaloacetic transaminase (U L−1) | 15.31 ± 7.04 |
| Hemoglobin (g L−1) | 79.54 ± 11.52 |
| Erythrocytic count | 3.02 ± 1.45 |
| HCT | 25 ± 3.91 |
| Low-density lipoprotein (mmol L−1) | 2.26 ± 0.61 |
| Triglycerides (mmol L−1) | 2.09 ± 1.24 |
| Total cholesterol (mmol L−1) | 4.35 ± 0.88 |
| hsCRP (mg L−1) | 7.05 ± 6.22 |
| SBP (mmHg) | 138.54 ± 19.06 |
| DBP (mmHg) | 84.08 ± 8.63 |
| Uric acid (mmol L−1) | 504.08 ± 94.46 |
| Serum potassium (mmol L−1) | 4.32 ± 0.44 |
| Blood calcium (mmol L−1) | 2.21 ± 0.24 |
| Blood phosphorus (mmol L−1) | 1.49 ± 0.43 |
| Carbon-dioxide combining power | 22.67 ± 3.68 |
2.2. Exosome isolation from serum
Exosomes were isolated from CKD patient serum samples using differential centrifugation and two commercial kits following the manufacturer's instructions (ExoQuick Serum Exosome Precipitation Solution (System Biosciences, Palo Alto, CA, USA) and exoEasy kit (Qiagen GmbH, Hilden, Germany)). Briefly, the above collected serum samples were centrifuged to remove cellular debris. Then, the supernatants were filtered through a 0.2 μm pore size filter to exclude particles > 200 nm in diameter. For exosome isolation by differential ultracentrifugation, the filtered supernatant was collected into ultracentrifuge tubes and centrifuged in an Optima MAX-XP ultracentrifuge with an MLA-130 rotor at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 1 h at 4 °C. The pellets were washed with PBS, ultracentrifuged and resuspended in PBS. For exosome isolation by ExoQuick and exoEasy, manufacturers’ instructions were followed. Exosome preparations were conserved at
000g for 1 h at 4 °C. The pellets were washed with PBS, ultracentrifuged and resuspended in PBS. For exosome isolation by ExoQuick and exoEasy, manufacturers’ instructions were followed. Exosome preparations were conserved at![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) −80 °C for later use.
−80 °C for later use.
2.3. Transmission electron microscopy (TEM)
The above collected samples were placed onto freshly glow-discharged 300 mesh carbon formvar grids for 10 min. The solution was wicked away with filter paper, and 5 μL 2% glutaraldehyde was placed on the grid for 3 min. The grid was wicked dry, rinsed with water and stained with 2% uranyl acetate for 1 min. Each grid was then wicked and air-dried for 10 min before examining at 80 kV on a JEOL 100CX transmission electron microscope at the Electron Microscopy Core Facility. Images were obtained via a chamberscope film camera.2.4. Western blotting analysis
The collected exosome was boiled in the 6 times volume of sample loading buffer at 99 °C for 10 min, and the boiled samples were subjected to estimation of protein levels following the protocol supplied by cell signaling technology. Briefly, equal amounts of proteins were resolved by 6–15% SDS-PAGE and transferred to nitrocellulose or PVDF membranes. After blocking with 5% non-fat milk or bovine serum albumin, the blots were then probed with primary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500–1
500–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). The bound antibodies were detected with an HRP-conjugated secondary antibody (1
000). The bound antibodies were detected with an HRP-conjugated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000–1
2000–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and visualized using a Tanon 5200 imaging system using medium-sensitive ECL substrates (4A-Biotech).
000) and visualized using a Tanon 5200 imaging system using medium-sensitive ECL substrates (4A-Biotech).
2.5. RNA library construction and sequencing
For RNA samples preparation, total RNA was extracted from the above collected exosomes in different groups. To remove rRNAs, the Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI, USA) was used following the manufacturer's instructions. Subsequently, six sequencing libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA). The resulting libraries were sequenced on the Illumina Hiseq 2500 platform with paired-end reads generated. For small RNAs sequencing, the RNA per sample was used for library construction. The NEBNext Ultra Small RNA Sample Library Prep Kit for Illumina (NEB, USA) was employed according to the manufacturer's guidelines to generate sequencing libraries. The quality of the constructed libraries was assessed using an Agilent Bioanalyzer 2100 system. Sequencing was performed on an Illumina Hiseq X Ten platform with single-end reads generated.2.6. Data processing
The above sequencing collected data were further analyzed using bioinformatics programs. The samples were subjected to differential gene expression analysis (transcript expression analysis, circRNA expression analysis, and lncRNA expression analysis) after multiplexing and sequencing. For quality assurance, sequences were trimmed using Trimmomatic 0.32, followed by data mapping to the musculus genome sequence (version GRCm38.72). To determine the expression levels of each gene, gene hit counts and reads were calculated within the CLC bio software environment (CLC Genomics Workbench 7.0.2, CLC genomics Server). Using bigwig files (converted from bam files), mapped reads were visualized on the UCSC browser, and differentially expressed mRNAs were identified (cutoff of p < 0.05).2.7. Functional enrichment analysis
The above identified DEGs were subjected to functional analysis using gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) analyses. These gene functional enrichment analyses were conducted using the cluster profiler package in R. The threshold for GO and KEGG enrichment analysis was P![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) <
< ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.05. The GO plot package in R was used to visualize the GO and KEGG results.
0.05. The GO plot package in R was used to visualize the GO and KEGG results.
2.8. Statistical analysis
All data were organized after random collection and expressed as mean ± SEM. Statistical analysis was performed using the relevant tests including two-tailed, unpaired Student's t-test, and one-way/two-way ANOVA accordingly. The post-hoc Tukey method was employed for pairwise comparisons between means in case ANOVA showed a significant difference. Values of P < 0.05 were considered statistically significant. GraphPad Prism 8.0 software was used for this analysis.3. Results
3.1. Enrichment and validation of exosome from CKD serum
A workflow for exosome enrichment, validation, RNA extraction, and RNA-Seq library generation is illustrated in Fig. 1A. Serum samples from 13 clinically diagnosed CKD patients were collected before and after roxadustat treatment, and these samples were further subjected to the optimized methods for exosome purification and exosomal RNA extraction. Exosomes were purified from equal volumes of cell-free serum using ultracentrifugation and the ExoQuick reagent, and two approaches were then used to validate each approach. First, TEM was used to identify vesicles in the size range of 30–100 nm. Exosomes were present in all preparations generated using both ultracentrifugation and ExoQuick protocols (Fig. 1B). Second, we performed western blotting for exosome markers analysis, including CD63, Calnexin, HSP70 and TSG1 using specific antibodies, on five representative samples to text the purity and density of exosome isolated (Fig. 1C). Immunoblotting results showed that all these markers were expressed in isolated exosomes from the serum samples, suggesting that exosomes were indeed isolated from all tissues.3.2. Overview of the differentially expressed mRNAs and lncRNAs after roxadustat treatment based on transcriptional profiling
After validation of the exosome isolated from the serum of CKD patients before and after roxadustat treatment, we further subjected the exosome to RNA extraction and RNA-sequencing analyses to investigate differently expressed genes in the collected samples. After data normalization and processing (with a fold change > 2.0 and P < 0.05), we found that 957 mRNAs (626 upregulated and 331 downregulated after roxadustat treatment) (Fig. 2A and B), 914 lncRNAs (444 upregulated and 470 downregulated) (Fig. 2C and D), were significantly differently expressed. The DEGs were presented in the heatmap diagram and volcano plot.3.3. GO and KEGG pathway analyses of differentially expressed mRNAs in roxadustat-treated CKD patient
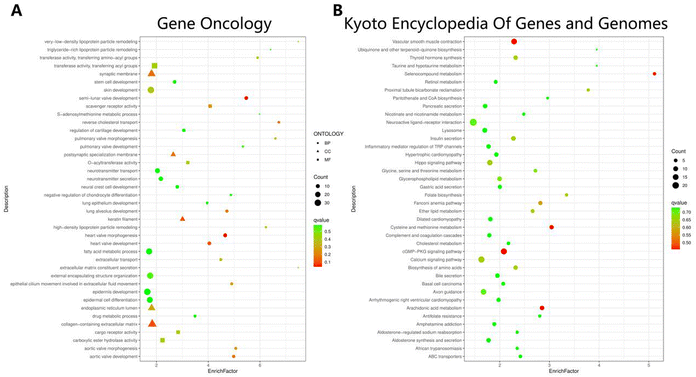
We next subjected these differentially expressed mRNA for KEGG and GO analysis for further insight into the functional networks. As shown in Fig. 3A by GO analysis, our analysis revealed an enrichment of up- and down-regulated DEGs in regulation of metabolic changes, including lipoprotein particle remodeling, reverse cholesterol transport, S-adenosylmethionine metabolic process, as well as amino-acyl-transferase activity.KEGG pathway analysis also revealed that these DEGs were considerably enriched for vascular smooth muscle contraction, cGMP-PKG signaling pathway, as well as multiple types of metabolic pathways, including cysteine and methionine metabolism, selenocompound metabolism, pantothenate and CoA biosynthesis, and cholesterol metabolism (Fig. 3B). Together, all these data suggest that roxadustat treatment has a significant effect on metabolic changes, especially lipid-associated metabolism in CKD patients.
3.4. Signaling transduction network of DEGs after roxadustat treatment of CKD patients
Based on the differently expressed genes and above-analyzed enrichment pathways, we further structured a global signal transduction network based on the STRING database to pinpoint the most crucial genes and mRNAs (Fig. 4). From the network, we concluded that the ENSG00000134594, ENSG00000168158, ENSG00000204019 and ENSG00000159182 genes were the ones at the center that linked to others and had the greatest degree value, suggesting that these genes are crucial for evaluation of roxadustat treatment efficiency. Furthermore, several other genes with high degree values in the network were also found to perform crucial roles, including ENSG00000212899, ENSG00000262814, ENSG00000130748 ENSG00000183024, ENSG00000204366 and ENSG00000158477. By functional classification, we also observed that these DEGs were highly enriched for the pathways of cGMP-PKG-regulated inflammation process and fatty acids metabolic process, which is in line with the KEGG analysis. All these observations together suggest that lipid-associated metabolic process is a potential evaluation biomarker for roxadustat treatment of CKD patients.3.5. LncRNA target pathway network
We further constructed a lncRNA target pathway network to seek the detailed regulation network on roxadustat treatment of CKD patients and found that many lncRNAs, including ENST00000458443, NONHSAT176305.1, NONHSAT156031.1, NONHSAT215690.1, NONHSAT145315.2, NONHSAT191411.1 and NONHSAT212423.1, were highly linked to other nodes in the regulatory network (Fig. 5), suggesting that these lncRNAs played crucial roles in roxadustat treatment on CKD patients. By pathway enrichment analysis, we found that these significantly differently expressed lncRNAs were involved in a wide variety of signaling pathways, such as inflammation regulation and fatty acids metabolic process, which is consistently with the pathway enrichment identified by mRNA profiling. Taken together, these data further suggest that lipid-associated metabolic process is a potential evaluation biomarker for roxadustat treatment of CKD patients. | ||
| Fig. 5 Analysis of lncRNA – target pathways as a network. Circles stand for each differently expressed lncRNAs, while the red boxes denote enriched pathways involved in these DEG-lncRNAs. | ||
3.6. Validation of mRNA–lncRNA regulatory network in response to roxadustat treatment
Based on the differently expressed mRNA–lncRNA identified in our study, we next focused our analysis on lipid metabolic process since this axis is significantly changed by roxadustat treatment, including ENSG00000103089 (FA2H)-NONHSAT174050.1, ENSG00000100336 (APOL)-ENST00000567376, and ENSG00000158201 (ABHD3)-NONHSAT177422.1, which may be characterized as significant nodes of the lipid (fatty acids) metabolic network (Fig. 6A). To further confirm that these identified lipids metabolic regulatory network are indeed important for assessing the roxadustat treatment, we further validated these targets by real-time PCR experimentally to examine the expression levels in five pairs of serum samples collected before and after roxadustat treatment clinically. As shown in Fig. 6B and D, the ENSG00000103089 (FA2H)-NONHSAT174050.1 axis was upregulated, while ENSG00000100336 (APOL)-ENST00000567376 and ENSG00000158201 (ABHD3)-NONHSAT177422.1 axis were downregulated after roxadustat treatment, which was consistent with our RNA-Seq results. Taken together, all these observations suggest that lipid metabolic regulatory network is a potential target for the evaluation of the roxadustat treatment efficiency.4. Discussion
As one of the first-class HIF–PHIs drugs approved by the FDA, roxadustat is widely used for anemia treatment in patients with CKD in clinical. Even though the detailed mechanisms and targets of roxadustat are well-identified, the potential evaluation and assessment of treatment efficiency of roxadustat on CKD patients remain poorly understood.6–9 Herein, we compared the differences in whole transcriptome profiling in ND-CKD patients before and after roxadustat treatment for the first time to clarify the potential assessment of roxadustat treatment efficiency, which may provide scientific evidence for the discovery of a new assessment biomarker for ND-CKD patients.There has been a long-term bias toward studying protein encoding genes. Non-coding RNAs, such as lnRNA and circRNA, have been shown to significantly regulate physiological function and disease in recent years, such as cancer21,22 and CKD.23,24 Despite not encoding proteins or just encoding certain short peptides, lncRNAs are still considered as functional regulators in disease development. Currently, mRNA, lncRNA, and circRNA are the most frequently considered biomarkers for diseases diagnosis and prognosis.22,23 However, the interplay between these different RNAs and their impact on CKD recurrence after roxadustat treatment and the regulatory network involved in CKD pathogenesis is less understood. Even previous studies have summarized the roles of non-coding RNAs in kidney diseases and perspectives in kidney associated-diseases’ therapy the whole picture on the interplay between RNAs and this kind of diseases is related, especially CKD.25–27 Our present study by whole transcriptional profiling analysis identified numerous changed mRNAs and lncRNAs after roxadustat treatment on the exosome of CKD patients. It is interesting to note that some of the mRNAs and lncRNAs previously identified to be associated with CKD progression, such as circulating miRNAs (miR-223 and miR-126), were involved in CKD pathogenesis.28 By RNA-Seq and novel computational algorithm analysis, another study identified 30 differentially expressed ncRNA (including 16 miRNAs, 2 tRFs, 3 mt-tRNAs and 9 lincRNAs) in the early stages of CKD compared to healthy controls.29 Nevertheless, our study highlighted the importance of RNAs and lncRNAs on CKD after roxadustat treatment, which may provide more information and evidences for assessing the recurrence of CKD after clinical therapies.
Currently, the biomarkers used for monitoring CKD patients' therapeutic effects and prognosis are limited in clinical practice. Many studies have confirmed that exosomes have great potential as future biomarkers, which is of great help for the early intervention and personalized treatment of patients.30 In this study, we identified a series of mRNA–lncRNA regulatory axis in exosome samples of CKD patients after roxadustat treatment, which may serve as potential biomarkers for evaluating the recurrence efficiency of roxadustat on CKD patients in clinic. By whole transcriptome analysis, we uncovered the lipid metabolic regulatory network was dramatically changed after roxadustat treatment. Our identified DEG RNAs are highly correlated with lipid metabolic process, which was highlighted in numerous previous studies.29,31 One study demonstrated that roxadustat treatment significantly reduced total cholesterol (TC), low-density lipoprotein cholesterol (LDL-C), and triglyceride levels in CKD patients, while simultaneously improving hemoglobin levels.32 Additionally, multi-omics analyses have revealed that roxadustat alters metabolic pathways related to cholesterol metabolism, suggesting that it may help mitigate the lipid abnormalities commonly observed in CKD.33 Furthermore, roxadustat has been shown to influence various signaling pathways, including the Ras signaling pathway, which is implicated in both erythropoiesis and lipid metabolism. This interaction underscores the potential of roxadustat to act as a dual modulator, improving both red blood cell production and lipid profiles.34 In addition, studies have shown that lipotoxicity plays a key role in the pathogenesis of CKD, manifested by the excessive accumulation of lipids, leading to cellular dysfunction, oxidative stress, and increased inflammatory response, all of which are important factors in the progression of CKD.35 In the context of CKD, researchers have found that dysregulation of lipid metabolism not only affects the kidneys’ structure and function but can also damage renal tubules and glomerular cells by activating oxidative stress and inflammatory signaling pathways.36,37 Based on our observations as well as previous discoveries, we thus undoubtedly hypothesize that roxadustat has potential for improving the progression of CKD by reversing the status of lipid metabolic status on CKD patients, thus achieving the treatment efficiency on clinical. Our present study illustrated the mechanism of roxadustat on treatment of CKD by improving the lipid metabolic process, which may serve as a potential biomarker for evaluation of the treatment efficiency by targeting the mRNA–lncRNA axis.
Together, we have shown that the potential mRNA–lncRNA regulatory network-mediated lipid metabolism coordinately manifested after roxadustat treatment in ND-CKD patients and our analysis by whole transcriptome profiling of exosome RNAs encompassing identified differentially expressed mRNA–lncRNA regulatory axis. Identifying specific mRNA–lncRNA interactions within exosomes could provide insight into the pathophysiological changes associated with CKD and the therapeutic response to roxadustat. These exosomal RNAs are not only reflective of the cellular state but also exhibit potential as non-invasive biomarkers. Our findings suggest that further validating these biomarkers across a larger patient cohort is essential to establish their clinical utility.
Author contributions
Data curation: ZW, HM, and LL; formal analysis: RZ, YZ, ZW, HM, and LL; methodology: XZ and BG; software: RZ, YZ, ZW, HM, and LL; supervision: XZ and BG; visualization: ZW; writing – original draft: RZ and YZ; writing – review & editing: XZ and BG.Ethics approval and consent to participate
This study protocol was reviewed and approved by Ethics Committee of Shenzhen People's Hospital, approval number LL-KY-2023161-02. All subjects had signed the informed consent.Data availability
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author. All original data supporting this study are publicly available in the NCBI repository under BioProject ID: [PRJNA1118673] (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1118673).Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
This work was supported by the Medical Research Foundation in Guangdong Province (no. B2023174), the Shenzhen Fund for Guangdong Provincial High-Level Clinical Key Specialties (no. SZGSP001) and the Shenzhen Key Laboratory of Kidney Diseases (ZDSYS201504301616234).References
- V. Jha, et al., Chronic kidney disease: global dimension and perspectives, Lancet, 2013, 382(9888), 260–272 CrossRef PubMed.
- R. N. Foley, et al., The impact of anemia on cardiomyopathy, morbidity, and and mortality in end-stage renal disease, Am. J. Kidney Dis., 1996, 28(1), 53–61 CrossRef CAS PubMed.
- S. Padhi, et al., Management of anaemia in chronic kidney disease: summary of updated NICE guidance, BMJ, 2015, 350, h2258 CrossRef PubMed.
- D. Groenendaal-van de Meent, et al., Effect of Kidney Function and Dialysis on the Pharmacokinetics and Pharmacodynamics of Roxadustat, an Oral Hypoxia-Inducible Factor Prolyl Hydroxylase Inhibitor, Eur. J. Drug Metab. Pharmacokinet., 2021, 46(1), 141–153 CrossRef CAS PubMed.
- C. Ogawa, et al., A Hypoxia-Inducible Factor Stabilizer Improves Hematopoiesis and Iron Metabolism Early after Administration to Treat Anemia in Hemodialysis Patients, Int. J. Mol. Sci., 2020, 21, 19 Search PubMed.
- Q. Zheng, et al., Efficacy and safety of HIF prolyl-hydroxylase inhibitor vs epoetin and darbepoetin for anemia in chronic kidney disease patients not undergoing dialysis: A network meta-analysis, Pharmacol. Res., 2020, 159, 105020 CrossRef CAS PubMed.
- L. Jia, et al., Effectiveness of hypoxia-inducible factor prolyl hydroxylase inhibitor roxadustat on renal anemia in non-dialysis-dependent chronic kidney disease: a systematic review and meta-analysis, Ann. Transl. Med., 2019, 7(23), 720 CrossRef CAS PubMed.
- D. Groenendaal-van de Meent, et al., Effect of Multiple Doses of Omeprazole on the Pharmacokinetics, Safety, and Tolerability of Roxadustat in Healthy Subjects, Eur. J. Drug Metab. Pharmacokinet., 2018, 43(6), 685–692 CrossRef CAS PubMed.
- Z. Hu, et al., The efficacy and economic evaluation of roxadustat treatment for anemia in patients with kidney disea se not receiving dialysis, Expert Rev. Pharmacoeconomics Outcomes Res., 2020, 20(4), 411–418 CrossRef PubMed.
- M. N. Huda, et al., Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications, ACS Biomater. Sci. Eng., 2021, 7(6), 2106–2149 CrossRef CAS PubMed.
- L. Xu, L. F. Wu and F. Y. Den, Exosome: An Emerging Source of Biomarkers for Human Diseases, Curr. Mol. Med., 2019, 19(6), 387–394 CrossRef CAS PubMed.
- X. Wang, et al., Exosomes and cancer-Diagnostic and prognostic biomarkers and therapeutic vehicle, Oncogenesis, 2022, 11, 1 CrossRef PubMed.
- M. Soutschek and G. Schratt, Non-coding RNA in the wiring and remodeling of neural circuits, Neuron, 2023, 111(14), 2140–2154 CrossRef CAS PubMed.
- S. Guil and M. Esteller, RNA-RNA interactions in gene regulation: the coding and noncoding players, Trends Biochem. Sci., 2015, 40(5), 248–256 CrossRef CAS PubMed.
- N. Sarfaraz, S. Somarowthu and M. J. Bouchard, The interplay of long noncoding RNAs and hepatitis B virus, J. Med. Virol., 2023, 95, 1 CrossRef PubMed.
- F. J. Slack and A. M. Chinnaiyan, The Role of Non-coding RNAs in Oncology, Cell, 2019, 179(5), 1033–1055 CrossRef CAS PubMed.
- M. Rudnicki, et al., Renal microRNA- and RNA-profiles in progressive chronic kidney disease, Eur. J. Clin. Invest., 2016, 46(3), 213–226 CrossRef CAS PubMed.
- D. D. Motshwari, et al., MicroRNAs Associated with Chronic Kidney Disease in the General Population and High-Risk Subgroups-A Systematic Review, Int. J. Mol. Sci., 2023, 24, 2 Search PubMed.
- N. Qu, et al., Roxadustat Attenuates the Disruption of Epithelial Tight Junction in Caco2 Cells and a Rat Model of CKD Through MicroRNA-223, Front. Med., 2022, 9, 850966 CrossRef PubMed.
- N. Chen, et al., Roxadustat Treatment for Anemia in Patients Undergoing Long-Term Dialysis, N. Engl. J. Med., 2019, 381(11), 1011–1022 CrossRef CAS PubMed.
- X. Guo and H. Z. Piao, Research Progress of circRNAs in Glioblastoma, Front. cell dev. biol., 2021, 9, 791892 CrossRef PubMed.
- Y. Li, et al., Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers, Nat. Commun., 2020, 11(1), 1000 CrossRef CAS PubMed.
- M. S. Hu, et al., Long Non-Coding RNAs in the Pathogenesis of Diabetic Kidney Disease, Front. cell dev. biol., 2022, 10, 845371 CrossRef PubMed.
- M. Ignarski, R. Islam and R. U. Müller, Long Non-Coding RNAs in Kidney Disease, Int. J. Mol. Sci., 2019, 20(13) CAS.
- J. A. Moreno, et al., Non-Coding RNAs in Kidney Diseases: The Long and Short of Them, Int. J. Mol. Sci., 2021, 22(11) CAS.
- F. Chen, et al., Total Glucosides of Paeony Alleviate Cell Apoptosis and Inflammation by Targeting the Long Noncoding RNA XIST/MicroRNA-124-3p/ITGB1 Axis in Renal Ischemia/Reperfusion Injury, Mediators Inflamm, 2020, 2020, 8869511 Search PubMed.
- Y. Yang, et al., Analysis of circulating lncRNA expression profiles in patients with diabetes mellitus and diabetic nephropathy: Differential expression profile of circulating lncRNA, Clin. Nephrol., 2019, 92(1), 25–35 CrossRef CAS PubMed.
- O. Fourdinier, et al., Serum levels of miR-126 and miR-223 and outcomes in chronic kidney disease patients, Sci. Rep., 2019, 9(1), 4477 CrossRef PubMed.
- R. Khurana, et al., Identification of urinary exosomal noncoding RNAs as novel biomarkers in chronic kidney disease, RNA, 2017, 23(2), 142–152 CrossRef CAS PubMed.
- Y. Zheng, et al., Extracellular vesicles in chronic kidney disease: diagnostic and therapeutic roles, Front. Pharmacol., 2024, 15, 1371874 CrossRef CAS PubMed.
- H. Noels, et al., Lipoproteins and fatty acids in chronic kidney disease: molecular and metabolic alterations, Nat. Rev. Nephrol., 2021, 17(8), 528–542 CrossRef CAS PubMed.
- K. Hirai, et al., Effects of roxadustat on anemia, iron metabolism, and lipid metabolism in patients with non-dialysis chronic kidney disease, Front. Med., 2023, 10, 1071342 CrossRef PubMed.
- X. You, et al., Integrated proteomic and metabolomic profiling of urine of renal anemia patients uncovers the molecular mechanisms of roxadustat, Mol. Omics, 2023, 19(6), 473–483 RSC.
- R. Provenzano, et al., Oral Hypoxia–Inducible Factor Prolyl Hydroxylase Inhibitor Roxadustat (FG-4592) for the Treatment of Anemia in Patients with CKD, Clin. J. Am. Soc. Nephrol., 2016, 11(6), 982–991 CrossRef CAS PubMed.
- E. Giardini, et al., The dual role of lipids in chronic kidney disease: Pathogenic culprits and therapeutic allies, Atherosclerosis, 2024, 398, 118615 CrossRef CAS PubMed.
- Y.-J. Kim, et al., Impact of Ring Finger Protein 20 and Its Downstream Regulation on Renal Tubular Injury in a Unilateral Nephrectomy Mouse Model Fed a High-Fat Diet, Nutrients, 2023, 15(23), 4959 CrossRef CAS PubMed.
- X.-s Jiang, et al., Activation of the Nrf2/ARE signaling pathway protects against palmitic acid-induced renal tubular epithelial cell injury by ameliorating mitochondrial reactive oxygen species-mediated mitochondrial dysfunction, Front. Med., 2022, 9, 939149 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2025 |