Light-activatable photochemically targeting chimeras (PHOTACs) enable the optical control of targeted protein degradation of HDAC6†
Silas L.
Wurnig‡
a,
Maria
Hanl‡
a,
Thomas M.
Geiger
b,
Shiyang
Zhai
a,
Ina
Dressel
b,
Dominika E.
Pieńkowska
b,
Radosław P.
Nowak
b and
Finn K.
Hansen
 *a
*a
aDepartment of Pharmaceutical and Cell Biological Chemistry, Pharmaceutical Institute, University of Bonn, An der Immenburg 4, 53121 Bonn, Germany. E-mail: finn.hansen@uni-bonn.de
bInstitute of Structural Biology, Medical Faculty, University of Bonn, Venusberg Campus 1, 53127 Bonn, Germany
First published on 13th March 2025
Abstract
Proteolysis targeting chimeras (PROTACs) are heterobifunctional modalities that induce protein degradation via a catalytic mode of action. Photochemically targeting chimeras (PHOTACs) are a subset of PROTACs designed for light-activated protein degradation, thereby offering precise spatiotemporal control. In this study, we report the design, solid-phase synthesis, and characterization of the first PHOTACs targeting histone deacetylase 6 (HDAC6). We achieved this by incorporating an azobenzene photoswitch into our previously developed HDAC6-selective PROTAC A6. Among the synthesized compounds, PHOTAC 12 demonstrated no HDAC6 degradation in the absence of light but showed significant degradation upon activation to its cis-state with 390 nm light irradiation. Notably, we find that PHOTAC 12 in the cis-state shows significantly improved ternary complex formation compared to the trans-state correlating with its degradation efficacy. Overall, PHOTAC 12 is a promising lead compound for the development of light-activatable HDAC6 degraders.
Introduction
Proteolysis targeting chimeras (PROTACs) have become a leading technology in targeted protein degradation.1 These molecules consist of three moieties: a protein of interest (POI) ligand and an E3 ligase ligand tethered by a linker. PROTACs induce proximity between the POI and the E3 ligase, thereby resulting in efficient ubiquitin transfer and polyubiquitination of the target protein.2 This ubiquitination then serves as recognition for the proteasome and the polyubiquitinated protein is subsequently degraded by the proteasome.3 Therefore, PROTACs act as catalysts and can participate in successive ternary complex formations. This approach offers various advantages over traditional occupancy-driven therapeutic modalities. While occupancy-driven approaches need extensive optimization of the ligand to ensure adequate physiological effects, PROTACs often do not need high affinity towards their POI due to the catalytic mode of action.4 Additionally, the POI ligand moiety of PROTACs does not necessarily have to bind to an active site of a protein. The POI can be targeted at any pocket addressable by a small molecule ligand, thereby reducing the competition with endogenous ligands, which usually necessitates high affinity of the ligands to the target protein.5 With the first PROTACs entering late-stage clinical trials, there is growing interest in the exploration of new methodologies to expand the toolkit for targeted protein degradation. To broaden the scope of targeted protein degradation, innovative approaches such as molecular glues, lysosome targeting chimeras (LYTACs), autophagy targeting chimeras (AUTACs), and antibody targeted chimeras (AbTACs) have emerged as promising strategies.6–9While these novel methods have expanded the range of proteins that can be targeted, they have limitations in terms of spatial and temporal control of their activity. Once administered, these degraders typically act continuously, degrading target proteins regardless of their physiological context or location, which can lead to unwanted toxicity.10 Despite their versatility, the lack of precise modulation of protein function remains a challenge for therapeutic applications.
One approach to achieve localized drug effects is through light control. The use of light to precisely modulate biological pathways has gained attention in recent years.11,12 Optical control methods include, among others, photocaged compounds and synthetic photoswitches, which use photochemical isomerization and thermal relaxation processes to alter compound activity, thereby providing precise temporal and spatial control over their effects.13–18
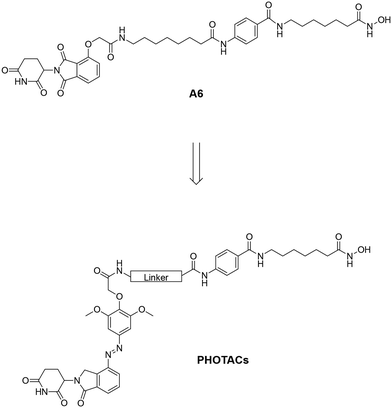
In 2020, Trauner and co-workers introduced the first photochemically targeting chimeras (PHOTACs).10 These molecules are light-activatable PROTACs and consist of a POI ligand, a photoswitch, and an E3 ligase ligand. In their inactive form, PHOTACs prevent ternary complex formation by introducing a steric clash between the binary complex and the second protein (Fig. 1A).10 When irradiated with light of a specific wavelength, the internal photoswitch changes its conformation, enabling the PHOTAC to form a productive ternary complex (Fig. 1A and B). This leads to polyubiquitination and subsequent proteasomal degradation of the POI. The active isomer gradually reverts to the inactive form through thermal relaxation, losing its activity, and can also be rapidly inactivated photochemically.18 This approach could potentially minimize adverse effects, as PHOTACs can be selectively activated by light at the desired location. Their inactivation is also less dependent on clearance and metabolism. Thus, PHOTACs provide a level of spatiotemporal and local control that is unmatched by other targeted protein degradation methods. In this study, we synthesized and characterized a set of three PHOTACs for targeted HDAC6 degradation by incorporating an azobenzene photoswitch, using our previously published HDAC6 PROTAC A6 as a template (Fig. 2).19
 | ||
| Fig. 2 Design of the PHOTACs based on the existing HDAC6 degrader A6.19 | ||
Results and discussion
The thalidomide moiety of A6 was replaced by a photoswitchable lenalidomide-based cereblon (CRBN)-recruiter to obtain the desired PHOTACs. The synthesis of the photoswitch (outlined in Scheme 1A) was performed by dissolving lenalidomide (1) in hydrochloric acid followed by the addition of sodium nitrite to obtain the aryldiazonium salt 2. This intermediate was then exposed to a solution of 2,6-dimethoxyphenol to obtain the diazo compound 3. In the next step, compound 3 was treated with tert-butyl bromoacetate to obtain the photoswitchable CRBN ligand 4via a nucleophilic substitution reaction.The synthesis of the HDAC ligand was performed on a 2-chlorotrityl chloride (2-CTC) resin. Firstly, the commercially available 2-CTC resin (5) was treated with N-hydroxyphthalimide to obtain the protected immobilized hydroxylamine 6. In the next step, the phthalimide protecting group was cleaved by treatment with hydrazine hydrate and the subsequent HATU-mediated amide coupling with Fmoc-7-aminoheptanoic acid gave the intermediate 7. Subsequent Fmoc-deprotection by treatment with a piperidine solution and HATU-mediated amide coupling with Fmoc-4-aminobenzoic acid yielded the resin-bound key intermediate 8 (Scheme 1B). From this point, the diversification of linker length was introduced in the last steps (Scheme 1C). To this end, the Fmoc-protecting group of 8 was cleaved and the subsequent HATU-mediated amide coupling with either Fmoc-8-aminooctanoic acid or Fmoc-4-aminobutanoic acid yielded the PHOTAC precursors 9 and 10. In the last step of the synthesis, the photoswitchable CRBN ligand 4 and the intermediates 8, 9, and 10 were subjected to an HATU-mediated amide coupling to obtain the final PHOTACs 11–13. First, the CRBN ligand 4 was converted into the corresponding carboxylic acid by treatment with trifluoroacetic acid (TFA). The resin-bound HDAC6 ligands 8, 9 and 10 were then Fmoc-deprotected followed by an amide coupling with the photoswitchable CRBN ligand. After the successful amide coupling reactions, the products were cleaved from the resin and purified by preparative HPLC to yield the PHOTACs 11–13 in > 95% purity. The positive control A6 was synthesized according to our previously published procedure.19
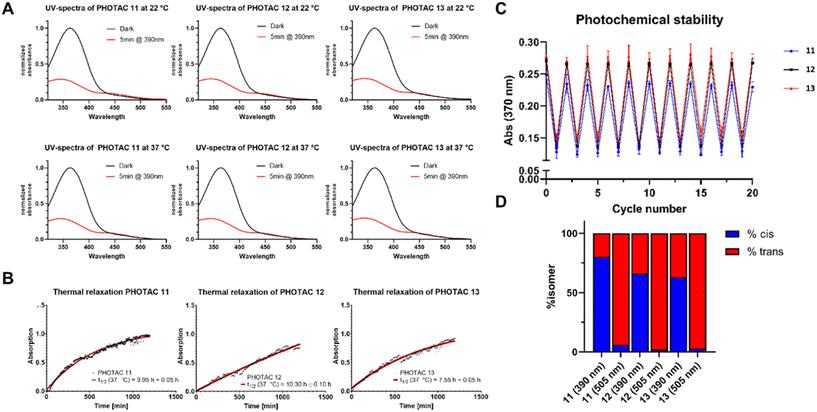
The most important step in the photochemical evaluation of the synthesized PHOTACs was to confirm their ability to switch from the trans- to the cis-state. In order to investigate this, the UV spectra of the respective PHOTACs 11–13 were measured in the dark. After illumination with light at a wavelength of 390 nm, the UV spectra were measured again, and the two distinct spectra were overlaid (Fig. 3A). This experiment was performed at a room temperature of 22 °C and at the physiological temperature of 37 °C. All synthesized PHOTACs showed the same overlaid spectra regardless of the temperature, confirming their intrinsic ability to switch from the trans- to the cis-state independently of temperature. This ensures the possibility for a cellular use of the synthesized PHOTACs.
To investigate whether the photoactivated PHOTACs show a half-life long enough to induce degradation of the target protein, the half-lifes of PHOTACs 11–13 were measured photometrically. To this end, the absorbance at 370 nm was measured prior to illumination for normalization. After irradiation with the appropriate wavelength for 5 minutes the absorbance at 370 nm was measured over a time period of 20 hours at a temperature of 37 °C to calculate the respective half-life times of the cis-isomers at physiological temperatures. The results are shown in Fig. 3B. PHOTAC 12 showed the longest half-life with 10.30 h, while the PHOTACs 11 and 13 demonstrated shorter half-life times of 3.95 h and 7.55 h, respectively. The photochemical stability of the synthesized PHOTACs was then tested by rapidly switching them between the cis- and the trans-state and measuring the absorbance at 370 nm. All three PHOTACs showed no change in absorbance at both states over the course of 20 cycles ensuring the stability of the photoswitch (Fig. 3C). Lastly the amount of cis- and trans-isomers at the photostationary states (PSSs) were measured photometrically. All three PHOTACs showed a photostationary state containing 63–80% of the cis-isomer after irradiation at 390 nm and could be switched back to >94% of trans-isomer following irradiation at 505 nm (Fig. 3D).
In the next step, we evaluated the ability of PHOTACs 11–13 to degrade HDAC6 using PROTAC A6 as positive control. To this end, we treated MM.1S cells with 1 μM of 11–13 or A6 for 6 hours. The MM.1S cell line was selected due to its well-established use in PROTAC characterization and our prior findings demonstrating A6’s high degradation activity in these cells.20,21 The experiments were conducted in two treatment groups. Both treatment groups were first treated with light of 505 nm to ensure full conversion into the trans-state. Next, one group was kept in the dark, and the second group was treated with light of 390 nm to switch the PHOTACs into the cis-state (see ESI† for experimental details; the results are summarized in Fig. 4). The positive control A6 achieved significant HDAC6 degradation after treatment with 390 and 505 nm. PHOTAC 11 induced HDAC6 degradation after irradiation with both wavelengths, likely due to its relatively long and flexible linker. The linker-less PHOTAC 13 failed to degrade HDAC6 in both treatment groups. In contrast, PHOTAC 12 with a relatively short γ-aminobutyric acid linker was unable to degrade HDAC6 after irradiation with 505 nm (<10% Dmax) but showed a pronounced and significant degradation of HDAC6 with a Dmax of ∼50% at 1 μM concentration upon irradiation with light of 390 nm. These results indicate that the cis-state of PHOTAC 12 is an active degrader, while the trans-state is nearly inactive (<10% Dmax).
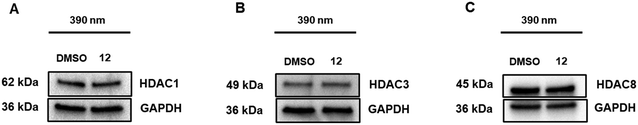
Subsequently, we analyzed the HDAC degradation selectivity profile of 12 after activation to its cis-state by irradiation with light of 390 nm. Consistent with its parent PROTAC A6,19 activated PHOTAC 12 selectively degraded HDAC6 while causing minimal degradation of HDAC1, HDAC3, and HDAC8 (Fig. 5). Similarly, when preirradiated with 505 nm to ensure the trans-form, PHOTAC 12 did not induce degradation of HDAC1, HDAC3, or HDAC8 (Fig. S1, ESI†).
After confirming that 12 can be activated by treatment with light of 390 nm into an active and selective HDAC6 degrader, we analyzed the concentration dependency of 12-mediated HDAC6 degradation. To this end, we treated MM.1S cells with varying concentrations of 12 (0.123, 0.37, 1.11, 3.33, and 10 μM) and determined the resulting HDAC6 protein levels by immunoblot analysis. The maximum degradation was observed at a concentration of 0.37 μM and the HDAC6 protein levels remained stable up to the highest tested concentration (see Fig. S2, ESI†).
To further investigate the mode of action and confirm the expected degradation mechanism, we conducted co-treatment experiments of activated 12 with the HDAC inhibitor vorinostat (SAHA, 1 μM), neddylation inhibitor MLN4924 (1 μM), CRBN ligand lenalidomide (5 μM), and the proteasome inhibitors MG132 (1 μM), and bortezomib (0.5 μM). In all co-treatment conditions, the HDAC6 levels increased (see Fig. 6), indicating that HDAC6 degradation requires the binding of 12 to both HDAC6 and CRBN and is neddylation- and proteasome-dependent.
To explore the differential activities of the cis- and trans-state of PHOTAC 12, we monitored ternary complex formation through TR-FRET between purified CRBN-DDB1ΔB and HDAC6. After irradiation with 390 nm light (cis-state), PHOTAC 12 led to approximately 2-fold increase in ternary complex between CRBN and HDAC6 compared to 505 nm condition (trans-state) (Fig. 7A), which explains the reduced degradation in a cellular environment. Our prior experiments showed that 390 nm irradiation results in ∼65% transition to cis-state (Fig. 3D), while 505 nm light results in near 95% transition to trans-state indicating that the pure cis-state would result in even higher ternary complex formation. To gain deeper insights on a molecular level, we performed modeling studies utilizing an established protocol for ternary complex modeling.22,23 To this end, crystal structures of HDAC6 (PDB ID: 5EEI)24 and CRBN (PDB ID: 4CI2)25 were selected as templates. The results (Fig. 7B) demonstrate that both cis- and trans-states of PHOTAC 12 are capable of forming distinct ternary complexes with HDAC6 and CRBN. Specifically, the HDAC warhead in both states of PHOTAC 12 engages in metal chelation with Zn2+ within the HDAC6 pocket. In addition, the CRBN warhead of PHOTAC 12 in both states forms hydrogen bonds with N353, H380, and W382 within the CRBN binding pocket, which are crucial for CRBN recruitment. These interactions provide the basis for ternary complex formation and corroborate the findings from immunoblot analysis and TR-FRET assay.
Conclusions
In conclusion, in this study we successfully designed, synthesized, and characterized the first photochemically targeting chimeras (PHOTACs) for the targeted degradation of histone deacetylase 6 (HDAC6). By incorporating an azobenzene photoswitch into our previously developed HDAC6-selective PROTAC A6, we created PHOTAC 12, which exhibited significant HDAC6 degradation only upon activation to its cis-state by irradiation with light of 390 nm. In vitro ternary complex dimerization assay indicated that the cis-isomer of PHOTAC 12 results in significant increase in ternary complex formed, consistent with potent degradation of HDAC6. These findings establish PHOTAC 12 as a promising lead for the development of light-activatable HDAC6 degraders, highlighting the potential of PHOTACs for the precise spatiotemporal control in targeted protein degradation.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. Z. is funded by China Scholarship Council (grant no. 202106150022). The new NMR console for the 500 MHz NMR spectrometer used in this research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under project number 507275896. M. H. and F. K. H. acknowledge partial funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)–GRK2873 (494832089). R. P. N. is a member of the excellence cluster ImmunoSensation2 funded by the Deutsche Forschungsgemeinschaft (DFG) under Germany's Excellence Strategy (EXC2151–390873048). We would like to thank Christoph Kick for crafting the Cell Disco LED panels. Experimental support by Svenja Henze is gratefully acknowledged.References
- M. Békés, D. R. Langley and C. M. Crews, PROTAC targeted protein degraders: the past is prologue, Nat. Rev. Drug Discovery, 2022, 21(3), 181–200, DOI:10.1038/s41573-021-00371-6.
- Z. Liu, M. Hu, Y. Yang, C. Du, H. Zhou, C. Liu, Y. Chen, L. Fan, H. Ma, Y. Gong and Y. Xie, An overview of PROTACs: a promising drug discovery paradigm, Mol. Biomed., 2022, 3(1), 46, DOI:10.1186/s43556-022-00112-0.
- G. L. Grice and J. A. Nathan, The recognition of ubiquitinated proteins by the proteasome, Cell. Mol. Life Sci., 2016, 73(18), 3497–3506, DOI:10.1007/s00018-016-2255-5.
- H. Graham, The mechanism of action and clinical value of PROTACs: A graphical review, Cell. Signalling, 2022, 99, 110446, DOI:10.1016/j.cellsig.2022.110446.
- V. Haridas, S. Dutta, A. Munjal and S. Singh, Inhibitors to degraders: Changing paradigm in drug discovery, iScience, 2024, 27(5), 109574, DOI:10.1016/j.isci.2024.109574.
- A. D. Cotton, D. P. Nguyen, J. A. Gramespacher, I. B. Seiple and J. A. Wells, Development of Antibody-Based PROTACs for the Degradation of the Cell-Surface Immune Checkpoint Protein PD-L1, J. Am. Chem. Soc., 2021, 143(2), 593–598, DOI:10.1021/jacs.0c10008.
- J. M. Sasso, R. Tenchov, D. Wang, L. S. Johnson, X. Wang and Q. A. Zhou, Molecular Glues: The Adhesive Connecting Targeted Protein Degradation to the Clinic, Biochemistry, 2023, 62(3), 601–623, DOI:10.1021/acs.biochem.2c00245.
- Q. Zheng, J. Guo, R. Ma and W. Pu, Lysosome-targeting Chimera (LYTAC): A Silver Bullet for Targeted Degradation of Oncogenic Membrane Proteins, MedComm: Oncol., 2024, 3(1), 2–5, DOI:10.1002/mog2.64.
- L. Zhao, J. Zhao, K. Zhong, A. Tong and D. Jia, Targeted protein degradation: mechanisms, strategies and application, Signal Transduction Targeted Ther., 2022, 7(1), 113, DOI:10.1038/s41392-022-00966-4.
- M. Reynders, B. S. Matsuura, M. Bérouti, D. Simoneschi, A. Marzio, M. Pagano and D. Trauner, PHOTACs enable optical control of protein degradation, Sci. Adv., 2020, 6(8), eaay5064, DOI:10.1126/sciadv.aay5064.
- L. Z. Fan and M. Z. Lin, Optical control of biological processes by light-switchable proteins, Wiley Interdiscip. Rev.:Dev. Biol., 2015, 4(5), 545–554, DOI:10.1002/wdev.188.
- N. Ankenbruck, T. Courtney, Y. Naro and A. Deiters, Optochemical Control of Biological Processes in Cells and Animals, Angew. Chem., Int. Ed., 2018, 57(11), 2768–2798, DOI:10.1002/anie.201700171.
- P. Pfaff, K. T. G. Samarasinghe, C. M. Crews and E. M. Carreira, Reversible Spatiotemporal Control of Induced Protein Degradation by Bistable PhotoPROTACs, ACS Cent. Sci., 2019, 5(10), 1682–1690, DOI:10.1021/acscentsci.9b00713.
- F. B. Kraft, M. Hanl, F. Feller, L. Schäker-Hübner and F. K. Hansen, Photocaged Histone Deacetylase Inhibitors as Prodrugs in Targeted Cancer Therapy, Pharmaceuticals, 2023, 16(3), 356, DOI:10.3390/ph16030356.
- R. Chen, Z. Wang, L. Liu and Z. Pan, Discovery of novel photocaged ERK1/2 inhibitors as light-controlled anticancer agents, Chem. Commun., 2022, 58(31), 4901–4904, 10.1039/D2CC00456A.
- Q. Zhang, C. S. Kounde, M. Mondal, J. L. Greenfield, J. R. Baker, S. Kotelnikov, M. Ignatov, C. P. Tinworth, L. Zhang, D. Conole, E. de, D. Kozakov, A. McCluskey, J. D. Harling, M. J. Fuchter and E. W. Tate, Light-mediated multi-target protein degradation using arylazopyrazole photoswitchable PROTACs (AP-PROTACs), Chem. Commun., 2022, 58(78), 10933–10936, 10.1039/D2CC03092F.
- J. Cheng, J. Zhang, S. He, M. Li, G. Dong and C. Sheng, Photoswitchable PROTACs for Reversible and Spatiotemporal Regulation of NAMPT and NAD, Angew. Chem., 2024, 63(12), e202315997, DOI:10.1002/anie.202315997.
- T. Ko, C. Jou, A. B. Grau-Perales, M. Reynders, A. A. Fenton and D. Trauner, Photoactivated Protein Degrader for Optical Control of Synaptic Function, ACS Chem. Neurosci., 2023, 14(19), 3704–3713, DOI:10.1021/acschemneuro.3c00390.
- L. Sinatra, J. Yang, J. Schliehe-Diecks, N. Dienstbier, M. Vogt, P. Gebing, L. M. Bachmann, M. Sönnichsen, T. Lenz, K. Stühler, A. Schöler, A. Borkhardt, S. Bhatia and F. K. Hansen, Solid-Phase Synthesis of Cereblon-Recruiting Selective Histone Deacetylase 6 Degraders (HDAC6 PROTACs) with Antileukemic Activity, J. Med. Chem., 2022, 65(24), 16860–16878, DOI:10.1021/acs.jmedchem.2c01659.
- F. Feller, I. Honin, M. Miranda, H. Weber, S. Henze, M. Hanl and F. K. Hansen, Development of the First-in-Class FEM1B-Recruiting Histone Deacetylase Degraders, J. Med. Chem., 2025, 68(2), 1824–1843, DOI:10.1021/acs.jmedchem.4c02569.
- C. Steinebach, A. Bricelj, A. Murgai, I. Sosič, L. Bischof, Y. L. D. Ng, C. Heim, S. Maiwald, M. Proj, R. Voget, F. Feller, J. Košmrlj, V. Sapozhnikova, A. Schmidt, M. R. Zuleeg, P. Lemnitzer, P. Mertins, F. K. Hansen, M. Gütschow, J. Krönke and M. D. Hartmann, Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs, J. Med. Chem., 2023, 66(21), 14513–14543, DOI:10.1021/acs.jmedchem.3c00851.
- M. L. Drummond and C. I. Williams, In Silico Modeling of PROTAC-Mediated Ternary Complexes: Validation and Application, J. Chem. Inf. Model., 2019, 59(4), 1634–1644, DOI:10.1021/acs.jcim.8b00872.
- M. L. Drummond, A. Henry, H. Li and C. I. Williams, Improved Accuracy for Modeling PROTAC-Mediated Ternary Complex Formation and Targeted Protein Degradation via New In Silico Methodologies, J. Chem. Inf. Model., 2020, 60(10), 5234–5254, DOI:10.1021/acs.jcim.0c00897.
- Y. Hai and D. W. Christianson, Histone deacetylase 6 structure and molecular basis of catalysis and inhibition, Nat. Chem. Biol., 2016, 12(9), 741–747, DOI:10.1038/nchembio.2134.
- E. S. Fischer, K. Böhm, J. R. Lydeard, H. Yang, M. B. Stadler, S. Cavadini, J. Nagel, F. Serluca, V. Acker, G. M. Lingaraju, R. B. Tichkule, M. Schebesta, W. C. Forrester, M. Schirle, U. Hassiepen, J. Ottl, M. Hild, R. E. J. Beckwith, J. W. Harper, J. L. Jenkins and N. H. Thomä, Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide, Nature, 2014, 512(7512), 49–53, DOI:10.1038/nature13527.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplemental figures, experimental procedures, 1H and 13C NMR data, and HPLC chromatograms. See DOI: https://doi.org/10.1039/d4md00972j |
| ‡ S. L. W. and M. H. contributed equally and share the first authorship. |
| This journal is © The Royal Society of Chemistry 2025 |