 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Exploring medium and long arm extensions of 1,2,4-triazole derivatives as Candida albicans 14α-demethylase (CYP51) inhibitors†
Marwa
Alsulaimany‡
a,
Faizah A.
Binjubair§
a,
Esra
Tatar
b,
Diane E.
Kelly
c,
Steven L.
Kelly
c,
Andrew G.
Warrilow
c,
Mikhail V.
Keniya¶
d,
Brian C.
Monk
d,
Josie E.
Parker
*e and
Claire
Simons
 *a
*a
aSchool of Pharmacy and Pharmaceutical Sciences, Cardiff University, King Edward VII Avenue, Cardiff, CF10 3NB, UK. E-mail: simonsc@cardiff.ac.uk
bDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, Marmara University, 34668 Istanbul, Turkey
cCentre for Cytochrome P450 Biodiversity, Institute of Life Science, Swansea University Medical School, Swansea University, Swansea, SA2 8PP, UK
dFaculty of Dentistry, Sir John Walsh Research Institute, University of Otago, Dunedin 9016, New Zealand
eSchool of Biosciences, Cardiff University, Museum Avenue, Cardiff, CF10 3AX, UK. E-mail: parkerj21@cardiff.ac.uk
First published on 12th March 2025
Abstract
Fungal infections have been described as a silent crisis affecting more than one billion people each year. At least 150 million of these cases involve severe and life threatening invasive fungal infections, accounting for approximately 1.7 million deaths annually. 1,2,4-Trizoles such as fluconazole and posaconazole are widely used antifungal agents, but azole resistance is an increasing problem requiring further study. 1,2,4-Triazole derivatives with medium and long arm extensions designed to bind within the Candida albicans CYP51 (CaCYP51) access channel were synthesised to study their inhibition of CaCYP51 (IC50, MIC) and binding affinity (Kd). A long arm extension using the amide linker was found to be most effective (e.g.13), giving an antifungal profile vs. wild-type and resistant model fungal strains comparable with posaconazole.
Introduction
Candida albicans (C. albicans), the predominant cause of candidiasis, is a commensal that resides in different parts of the human body including the gastrointestinal tract, genitourinary tract, oropharynx, and the skin.1 Under certain conditions C. albicans can cause infections that range from superficial to invasive, life-threatening, infections such as candidemia.1,2 Worldwide C. albicans is one of the most common healthcare associated bloodstream infections in intensive care units with a mortality of 30–50% for candidemia.3According to the Centre for Disease Control and prevention (CDC), many patients with severe COVID-19 symptoms in 2020 developed invasive candidemia from C. albicans as well as resistance to antifungal treatment owing to immune suppression caused by the virus.4–6 This emphasises the need for improved antifungal treatments as the current range of effective antifungal agents is limited and susceptible to drug resistance.
Ergosterol is a fungal-specific and major sterol component of the fungal cell membrane that affects its physiochemical characteristics including fluidity. A key cytochrome P450 (CYP) enzyme involved in the synthesis of ergosterol is sterol 14α-demethylase (CYP51, Erg11).7 The azole antifungals are the largest class of drugs that act by inhibiting CYP51.8 They are type II inhibitors that bind directly with the haem iron. The incidence of drug resistance to the available azole antifungals in yeast and other fungi is increasing and is attributed to the long treatments and prophylactic use in the clinic, and widespread use of agricultural azole fungicides to protect crops.9,10
Fluconazole (FLZ, Fig. 1) is one of the most commonly used antifungal agents and significant target-based FLZ resistance has been found in clinical isolates of C. albicans.10–14 FLZ resistance has been attributed to single and double mutations within CYP51, with common single amino acid mutations involving Y132F and K143R located within the haem active site, and a G450E mutation within the electron transfer area that interacts with the NADPH-cytochrome P450 reductase redox partner. Y132 forms a key H-bonding interaction with azole antifungals via the tertiary hydroxy group of triazoles such as FLZ and voriconazole (VCZ, Fig. 1). Double mutations such as C. albicans CYP51 (CaCYP51) Y132H/K143R and Y132F/F145L further reduce the binding affinity of FLZ, increase the IC50 value,10 and probably account for increased MIC values found in C. albicans strains containing these mutations.11–14 Conformational changes induced by such mutations are also likely to affect the binding between the azole nitrogen and the haem iron as well as the haem propionates with Y118 and Y132, e.g. by affecting H-bonding interactions of haem and by distorting the ionic interaction with K143.9,10,15–18
Azole antifungals, especially the triazoles, are often a first-choice treatment due to their high oral bioavailability, low toxicity, selectivity, tolerability by patient and the broad spectrum of activity. There are some limitations for the use of these azoles. For example, while VCZ (Fig. 1) has a broad spectrum of activity against most Candida species and fungal pathogens compared with FLZ, it has more adverse effects and drug–drug interactions than FLZ.19 Itraconazole (ITZ, Fig. 1) and posaconazole (PCZ, Fig. 1) have broad spectrum activities against candidiasis, but their low bioavailability has limited their clinical use.20
Recent studies on azole antifungals21 have focussed on azole-hybrid structures e.g. FLZ-COX inhibitor hybrids,22 to explore dual activity and tetrazole derivatives e.g. VT-1161 (oteseconazole),23 to improve CYP selectivity, althoughVT-1161 is susceptible to azole resistance.24 The increased incidence of resistant clinical isolates highlights the importance of developing new antifungals with activity against such strains, with azoles the focus of the research presented here. Our goal was to develop mid-sized and extended arm azoles targeting the CaCYP51 enzyme by using FLZ as a pharmacophore and applying the promising results obtained in our previous published study17 to enhance activity, in particular against resistant fungal strains, and binding affinity. The following design was proposed: one triazole moiety of FLZ was retained as the haem-binding group and the 2,4-difluorobenzene ring was replaced with the more lipophilic 4-chlorobenzene and 2,4-dichlorobenzene to enhance uptake across the lipophilic fungal cell wall and still fit in the small hydrophobic binding site of CaCYP51. The second triazole ring was replaced with either a medium or long arm extension (Fig. 2). The medium extensions contain an amide linked to either acetophenone or a 2-chlorothiazole ring identified through computational modelling to increase binding interactions. The long arm extensions contain a benzamide attached to a second benzene ring through either a thiourea, urea, amide or sulfonamide linker. The rationale for adding thiourea, urea, amide or sulfonamide linkers was to add H-bond acceptor and/or donor capability, which might form additional binding interactions with amino acid residues in the CaCYP51 access channel binding site, to compensate for the loss of the key H-bonding interaction between the azole tertiary hydroxy group and common active site mutations, such as Y132H/F (C. albicans) or Y140H/F (S. cerevisiae models) in resistant strains.
 | ||
| Fig. 2 Rationale for modifications to generate azole antifungals with medium and long arm extensions. | ||
Results and discussion
Chemistry
The mid-sized acetyl (6a–b) and thiazole (7a–b) derivatives were obtained by a 5-step synthesis (Scheme 1) starting with introduction of the triazole ring by reaction of acetophenone (1) with 1,2,4-triazole and activated K2CO3 to give the 1-(substituted phenyl)-2-(1H-1,2,4-triazol-1-yl)ethan-1-one (2) derivatives.25 Corey-Chaykovsky epoxidation reaction of the ethanone derivative (2) using TMSOI and 20% aqueous NaOH with heating at 60 °C for 6 h (ref. 26) gave the crude epoxides (3), which were directly converted to the azides (4) on treatment with NaN3 and NH4Cl in good yields (4a 60% and 4b 63%) over two steps from 2. Azide reduction to the free amines (5) was performed using a Staudinger reaction,27 with initial reaction of azides (4) with triphenylphosphine in THF at room temperature for 1 h followed by the addition of H2O and heating at 60 °C for 4 h. The mid-sized derivatives (6 and 7) were prepared by reaction of the amines (5) with either 3-acetylbenzoic acid or 2-chlorothiazole-4-carboxylic acid using 1,1′-carbonyldiimidazole (CDI) as the amide coupling reagent (Scheme 1). The mid-sized amides (6a–b and 7a–b) were obtained in yields ranging from 45–68% (Table 1) and structure and purity confirmed by 1H/13C NMR and HPLC respectively.| Cmpda | Yield (%) | MP (°C) | Amide/linker 1H NMR (δ)b | Amide/linker 13C NMR (δ)b |
|---|---|---|---|---|
| a a R1 = Cl, R2 = H; b R1 = Cl, R2 = Cl. b DMSO-d6. | ||||
| 6a | 52 | Semisolid | NH 8.55 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 167.1 O 167.1 |
| 6b | 45 | 160–162 | NH 8.72 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 167.5 O 167.5 |
| 7a | 68 | Semisolid | NH 8.10 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 160.1 O 160.1 |
| 7b | 68 | Semisolid | NH 8.26 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 160.4 O 160.4 |
| 11a | 68 | 146–148 | 2× NH 10.05, 10.00 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S 180.0 S 180.0 |
| 11b | 56 | 138–140 | 2× NH 10.06, 10.03 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S 179.9 S 179.9 |
| 12a | 63 | 226–228 | 2× NH 8.99, 8.90 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 152.63 O 152.63 |
| 12b | 52 | 216–218 | 2× NH 9.06, 8.97 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 152.66 O 152.66 |
| 13a | 80 | 222–224 | NH 10.50 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 165.13 O 165.13 |
| 13b | 76 | 190–192 | NH 10.51 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 165.14 O 165.14 |
| 14a | 77 | 236–238 | NH 10.73 | — |
| 14b | 83 | 228–230 | NH 10.76 | — |
The nitro intermediates (9) for the preparation of the extended derivative, were prepared by reaction of 4-nitrobenzoyl chloride (8) with the free amines (5) at room temperature overnight, (Scheme 2). The nitro derivatives (9) were reduced to the amines (10) using 10% palladium on carbon in dry MeOH and H2 balloon for 3 h at room temperature. The synthesis of the final compounds (11–14) with the different linkers was achieved by adding 4-chlorophenyl derivatives to 4-amino-N-(2-(substituted phenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (12) in dry pyridine, and the reaction was left at room temperature overnight (Scheme 2). The extended compounds were obtained in good yields as white solids (Table 1), although lower yields were obtained for compounds 11b and 12b owing to more difficult purifications. Purity was determined either by elemental analysis or HPLC, while structure characterisation was performed by 1H/13C NMR.
 | ||
| Scheme 2 Reagents and conditions: (i) sat. aq. NaHCO3, CH2Cl2, r.t. o/n, 55–74% (ii) H2, Pd/C, EtOH, r.t., 3 h, 71–100% (iii) pyridine, r.t., o/n, 52–83% [a R1 = Cl, R2 = H; b R1 = Cl, R2 = Cl]. | ||
In vitro biological assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P data as the more lipophilic compounds should more readily penetrate the fungal membrane resulting in improved uptake or may relate to optimal fill of the small hydrophobic pocket leading to improved binding.
P data as the more lipophilic compounds should more readily penetrate the fungal membrane resulting in improved uptake or may relate to optimal fill of the small hydrophobic pocket leading to improved binding.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values for mid-sized and extended compounds
P values for mid-sized and extended compounds
| Cmpd | MIC (μg mL−1) | IC50 (μM) | K d (nM) | clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pa Pa |
|
|---|---|---|---|---|---|
| CA14 | SC5314 | ||||
a clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P by Crippen's fragmentation.29 P by Crippen's fragmentation.29
|
|||||
| 6a | 0.25 | 0.25 | 1.30 | 1445 ± 266 | 1.85 |
| 6b | <0.03 | 0.06 | 0.49 | 229 ± 45 | 2.46 |
| 7a | 0.25 | 0.25 | 0.77 | 635 ± 181 | 1.95 |
| 7b | <0.03 | <0.03 | 0.41 | 106 ± 15 | 2.55 |
| 11a | 4 | 4 | — | — | 3.13 |
| 11b | 0.25 | 0.5 | 0.67 | 125 ± 41.0 | 3.73 |
| 12a | 1 | 2 | — | — | 3.86 |
| 12b | 0.125 | 0.125 | 0.47 | 137 ± 14 | 4.46 |
| 13a | <0.03 | <0.03 | 0.56 | 46 ± 8 | 3.54 |
| 13b | <0.03 | <0.03 | 0.48 | 166 ± 39 | 4.15 |
| 14a | 1 | 1 | — | — | 3.45 |
| 14b | 0.25 | 0.25 | 0.78 | 94 ± 24 | 4.01 |
| FLZ | 0.125 | 0.125 | 0.31 | 31 ± 8 | 0.86 |
The dichloro with the thiourea, urea and sulfonamide linker of the extended derivatives (11b, 12b and 14b) were more effective at inhibiting C. albicans growth, for example: the dichloro thiourea 11b showed MIC 0.25 μg mL−1 compared with the monochloro 11a MIC 4 μg mL−1. Even though the clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P of the amide inhibitors (13a, clog
P of the amide inhibitors (13a, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.53 and 13b, clog
P 3.53 and 13b, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 4.15) was more lipophilic than thiourea and sulfonamide (11a, clog
P 4.15) was more lipophilic than thiourea and sulfonamide (11a, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.13, 11b, clog
P 3.13, 11b, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.73, 15a, clog
P 3.73, 15a, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.45 and 15b, clog
P 3.45 and 15b, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 4.01) and less than urea (12a, clog
P 4.01) and less than urea (12a, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.86 and 12b, clog
P 3.86 and 12b, clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 4.46), the mono and dichloro derivatives of the amide linker (13a and 13b) were the most effective over the three linkers as well as the standard FLZ with MIC <0.03 μg mL−1 (Table 2). This could be owing to the higher polarity of the thiourea and urea functional group compared with the amide, which may lead to a reduced uptake across the lipophilic fungal membrane, although other factors such as efflux and binding affinity may also play a role.
P 4.46), the mono and dichloro derivatives of the amide linker (13a and 13b) were the most effective over the three linkers as well as the standard FLZ with MIC <0.03 μg mL−1 (Table 2). This could be owing to the higher polarity of the thiourea and urea functional group compared with the amide, which may lead to a reduced uptake across the lipophilic fungal membrane, although other factors such as efflux and binding affinity may also play a role.
The binding affinity (Kd) values for FLZ and the novel inhibitors were determined using saturation curves for CaCYP51 that measured the change in the absorbance (ΔA peak-trough) plotted against antifungal concentration (Fig. 3, S1 and S2†). For the mid-sized series, specifically the dichloro derivatives, the binding affinity for the thiazole (7b) compound was acceptable (Kd = 106 ± 15 nM) compared with fluconazole (Kd = 31 ± 8 nM), which showed tight binding with the haem iron. The binding affinity for the chloro and dichloro of the acetyl derivatives (6a–b) as well as the chloro of the thiazole (7a) derivative indicated weaker binding affinities (Table 2). For the extended series, the monochloro of the amide derivative (13a) showed the tightest binding (Kd = 46 ± 8 nM) with CaCYP51 haem Fe3+ compared with the mid-sized series as well as FLZ. Good binding affinity for the dichloro derivative of the thiourea (11b), urea (12b) and sulfonamide (14b) was observed (11b, Kd = 125 ± 41 nM, 12b, Kd = 137 ± 14 nM and 14b, Kd = 94 ± 24 nM) (Table 2).
| Compound | 7b | 13a | 13b | FLZ | PCZ |
|---|---|---|---|---|---|
| Strain | MIC (μg mL−1) | ||||
| Y2411 – azole sensitive control strain, Y2300 ScCYP51 overexpressed, Y2301 ScCYP51 Y140F mutant, Y2513 ScCYP51 Y140H mutant, Y525 MFS efflux pump CaMDR1a overexpressed, Y570 ABC efflux pump CaCDR1B overexpressed. FLZ fluconazole, PCZ posaconazole. | |||||
| S. cerevisiae models | |||||
| Y2411 parent | 0.125 | 0.25 | <0.03–0.06 | 4 | <0.03 |
| Y2300 ScCYP51 | 0.5–1.0 | 1–2 | 0.25 | 16 | 0.06 |
| Y2301 Y140F | 4 | 1–2 | 0.25 | 32 | 0.06 |
| Y2513 Y140H | 2–4 | 1 | 0.125 | 32 | <0.03 |
| C. albicans models | |||||
| Y525 CaMDR1a | 2 | 0.125 | <0.03–0.06 | 32 | <0.03 |
| Y570 CaCDR1B | >16 | >16 | >16 | >128 | 8 |
| H. sapiens model | |||||
| Y2760 HsCYP51 | >16 | >16 | 4–8 | >128 | 8 |
The most promising thiazole derivative 7b, and the extended amide derives 13a and 13b were all active against the susceptible strain Y2411 and S. cerevisiae CYP51 overexpressing strain Y2300. The extended amide derivatives (13a/b) retained good activity against the azole-resistant strains Y2301 and Y2513, expressing ScCYP51 Y140F and Y140H respectively and with 13b the most effective. Against the C. albicans efflux pump models, 13a and 13b retained activity against strain Y525, which overexpresses the C. albicans MFS transporter MDR1a. However inhibitory activity was significantly reduced against strain Y570, which overexpresses the C. albicans ABC transporter CDR1B, consistent with both FLZ and PCZ. Good selectivity was observed for ScCYP51 compared with HsCYP51 (Y2760 strain). All three derivatives were more effective than FLZ and 13b displayed a profile comparable with PCZ (Table 3) at both 24 and 48 h. Micafungin, a β-(1–3)-D-glucan synthase inhibitor, served as a negative control with MIC values of 0.125 μg mL−1 for all strains.
Computational studies
Protein–ligand complexes were prepared through docking of the R- and S-enantiomers of ligands 6a/b, 7a/b, 11a/b, 12a/b, 13a/b and 14a/b using Molecular Operating Environment (MOE) software33 and the crystal structure of CaCYP51 co-crystallised with PCZ (pdb 5FSA).34 The protein–ligand complexes were subjected to 200 ns molecular dynamics simulations using the Desmond programme of Schrödinger.35,36 All complexes equilibrated, although fluctuations were observed in RMSD for some compounds due to changes in binding interactions over the 200 ns MD simulations (Fig. S3 and S4†). The ligands should be positioned with the triazole above the haem Fe3+ and perpendicular to the haem with a distance <3 Å for optimal binding. For nine of the ligands (6a, 6b, 7b, 11a, 12a, 12b, 13a, 13b and 14b) only the S-enantiomer was positioned optimally, for 11b only the R-enantiomer was optimally positioned and for 7a and 14a both enantiomers were optimally positioned for binding with the haem (Fig. S5†).The ligands were positioned in contact with the haem, and within the small hydrophobic pocket and the access channel (Fig. 4). The small hydrophobic pocket accommodates the chlorobenzene (e.g.S-7b) or dichlorobenzene (e.g.S-13a) ring and the haem accommodates the triazole and amide linker (Fig. 4). The medium extended compounds (6 and 7) form π–π stacking interactions with Tyr118, Tyr132 and Phe380 and H-bond interactions with Tyr132, Ser378 and Met508. The long extensions of compounds 11–14 form additional binding interactions along the access channel including Tyr64, Leu121, Phe233, Thr229, Ser506 and Ser507 (ligand interactions for each compound is provided in detail in Fig. S5 and S6†).
 | ||
| Fig. 4 Positioning of exemplar compounds S-7b (orange) and S-13a (cyan) in the CaCYP51 protein (for clarity not all amino acids are shown). | ||
Conclusions
In general, the dichlorobenzene derivatives (b) displayed better inhibitory activity (MIC, IC50) and binding affinity (Kd) compared with the monochlorobenzene derivatives (a). Among the medium arm extensions, the benzene acetyl derivative (6b) and the thiazole derivative (7b) had MIC and IC50 values comparable with FLZ (Table 2), however 7b had improved binding affinity (Kd = 106 ± 15 nM) (Table 2). Comparison of the extended arm derivatives demonstrated the optimal linker with respect to antifungal activity was the amide (13), then urea (12), thiourea (11) and lastly sulfonamide (14), which was the least effective. The amide linker compounds (13a and 13b) were highly active against C. albicans strains CA14 and SC5314 with MIC <0.03 μg mL−1 and gave potent IC50 values (13a IC50 = 0.56 μM, 13b IC50 = 0.48 μM), but only compound 13a showed very tight binding to CaCYP51 (13aKd = 46 ± 8 nM) while a good binding affinity was noted for 13b (Kd = 166 ± 39 nM,) compared with fluconazole (MIC = 0.125 μg mL−1, IC50 = 0.31 μM and Kd = 31 ± 8 nM) (Table 2).Evaluation (MIC) against S. cerevisiae models indicated the amide of the three lead compounds 7b, 13a and 13b all outperformed FLZ, with 13b optimal as it retained activity against mutant ScCYP51 Y140F/H mutant strains and the strain overexpressing C. albicans efflux pump MDR1a with a similar profile to PCZ. Computational studies suggest the S-enantiomers are generally preferred for optimal binding and positioning within the CaCYP51 active site, with the extended arm derivatives forming additional binding interactions in the access channel (Fig. 4 and S5†).
In summary (Fig. 5), the dichlorobenzene ring was optimal for binding within the small hydrophobic pocket and, in combination with an extended arm (e.g. as seen in 13b) performed better in resistant strains than FLZ owing to additional binding opportunities along the access channel to stabilise ligand–protein interactions.
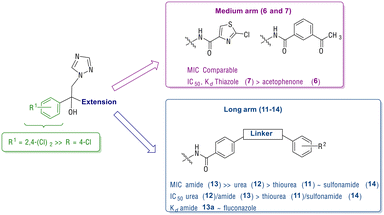 | ||
| Fig. 5 Summary of antifungal (MIC), enzyme inhibition (IC50) and binding affinity (Kd) of the medium (6 and 7) and long arm (11–14) extended compounds. | ||
Consideration of the physicochemical properties of the most promising medium (7b) and extended (13a/b) arm derivatives compared with reference antifungal agents (Table 4), showed that the thiazole derivative 7b is comparable with FLZ, while the extended derivatives (13a/b) are intermediate between FLZ and PCZ. Compounds 13a and 13b are just over the ideal molecular weight resulting in one violation of Lipinski's Ro5, however this is still a significant improvement compared with PCZ with three violations. Improved physicochemical properties would make these compounds more suitable for oral administration, a major limitation of ITZ and PCZ, which combined with the addition of linkers and long arms capable of H-bonding interactions enables antifungal activity against resistant strains, in particular common mutations in the CYP51 active site.
| Cmpd | MW | clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
n ON | n OHNH | n rotb | n viol |
|---|---|---|---|---|---|---|
| MW = Molecular weight; nON = H-bond acceptor; nOHNH = H-bond donor; nrotb = number of rotable bonds; nviol = number of Lipinsky violations. | ||||||
| 7b | 399.81 | 2.55 | 7 | 2 | 6 | 0 |
| 13a | 510.38 | 3.54 | 8 | 3 | 8 | 1 |
| 13b | 544.83 | 4.15 | 8 | 3 | 8 | 1 |
| FLZ | 306.27 | 0.86 | 7 | 1 | 5 | 0 |
| PCZ | 700.77 | 5.74 | 12 | 1 | 12 | 3 |
Experimental
General
All chemicals, reagents and solvents were purchased from Sigma-Aldrich, Alfa Aesar, VWR, Acros and Fluka. Solvents were dried over molecular sieves (4 Å) prior to use. For column chromatography, a glass column was slurry packed in the appropriate eluent with silica gel (Fluka Kieselgel 60). TLC was performed on pre-coated silica plates (ALUGRAM® SIL G/UV254) with visualisation via UV light (254 nm). Melting points were determined using an electrothermal instrument (Gallenkamp melting point apparatus) and were uncorrected. 1H and 13C NMR spectra were recorded on a Bruker Advance DP500 spectrometer operating at 500 MHz and 125 MHz, respectively. Chemical shifts are given in parts per million (ppm) relative to the internal standard tetramethylsilane (Me4Si). HPLC-HRMS or HPLC was performed at the Department of Pharmacy and Pharmacology, University of Bath, Bath, UK (method A) or at Cardiff University (method B). Method A: was performed on a Zorbax Eclipse Plus C18 Rapid Resolution 2.1 × 50 mm, 1.8 μm particle size using a 7.5 minute gradient method 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 v/v water
95 v/v water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol with 0.1% formic acid as additive; method B: was performed on a Shimadzu LC-2030C Plus C18 Rapid Resolution 250 × 4.6 mm, 5 μm particle size using a 7–10 min gradient method 5
methanol with 0.1% formic acid as additive; method B: was performed on a Shimadzu LC-2030C Plus C18 Rapid Resolution 250 × 4.6 mm, 5 μm particle size using a 7–10 min gradient method 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95 v/v water: methanol. Details for the preparation of the intermediate amines (5 and 12) are provided in the ESI.†
95 v/v water: methanol. Details for the preparation of the intermediate amines (5 and 12) are provided in the ESI.†
Chemistry
3-Acetyl-N-(2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (6a, R1 = Cl, R2 = H). Prepared from 1-amino-2-(4-chlorophenyl)-3-(1H-1,2,4-triazol-1-yl)propan-2-ol (5a) (0.15 g, 0.59 mmol) and 3-acetyl benzoic acid (0.14 g, 0.88 mmol). Purified by gradient column chromatography eluting with 3.5% MeOH in CH2Cl2. Product obtained as a semisolid, yield 0.12 g (52%). TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf 0.4. 1H NMR (DMSO-d6): δ 8.55 (t, J = 5.7 Hz, 1H, NH), 8.27 (d, J = 7.2 Hz, 1H, Ar), 8.26 (s, 1H, triaz), 8.09 (d, J = 7.8 Hz, 1H, Ar), 7.98 (d, J = 8.3 Hz, 1H, Ar), 7.83 (s, 1H, triaz), 7.60 (d, J = 7.8 Hz, 1H, Ar), 7.45 (d, J = 8.8 Hz, 2H, Ar), 7.32 (d, J = 8.8 Hz, 2H, Ar), 6.07 (s, 1H, OH, ex), 4.61 (q, J = 14.4 Hz, 2H, C
0.5 v/v), Rf 0.4. 1H NMR (DMSO-d6): δ 8.55 (t, J = 5.7 Hz, 1H, NH), 8.27 (d, J = 7.2 Hz, 1H, Ar), 8.26 (s, 1H, triaz), 8.09 (d, J = 7.8 Hz, 1H, Ar), 7.98 (d, J = 8.3 Hz, 1H, Ar), 7.83 (s, 1H, triaz), 7.60 (d, J = 7.8 Hz, 1H, Ar), 7.45 (d, J = 8.8 Hz, 2H, Ar), 7.32 (d, J = 8.8 Hz, 2H, Ar), 6.07 (s, 1H, OH, ex), 4.61 (q, J = 14.4 Hz, 2H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 3.90 (dd, J = 6.8, 14.0 Hz, 1H, C
-triaz), 3.90 (dd, J = 6.8, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–NH), 3.65 (dd, J = 5.3, 14.0 Hz, 1H, C
2–NH), 3.65 (dd, J = 5.3, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2–NH), 2.61 (s, 3H, C
2–NH), 2.61 (s, 3H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ). 13C NMR (DMSO-d6): δ 197.99 (C, CH3C
). 13C NMR (DMSO-d6): δ 197.99 (C, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.08 (C, C
O), 167.08 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, amide), 150.01 (CH, triaz), 145.45 (CH, triaz), 141.40 (C, Ar), 137.19 (C, Ar), 134.92 (C, Ar), 132.31 (CH, Ar), 132.17 (C, C–Cl), 131.41 (CH, Ar), 129.29 (CH, Ar), 128.37 (2× CH, Ar), 128.09 (2× CH, Ar), 127.29 (2× CH, Ar), 76.28 (
O, amide), 150.01 (CH, triaz), 145.45 (CH, triaz), 141.40 (C, Ar), 137.19 (C, Ar), 134.92 (C, Ar), 132.31 (CH, Ar), 132.17 (C, C–Cl), 131.41 (CH, Ar), 129.29 (CH, Ar), 128.37 (2× CH, Ar), 128.09 (2× CH, Ar), 127.29 (2× CH, Ar), 76.28 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 57.01 (
–OH), 57.01 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 48.24 (
H2-triaz), 48.24 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH), 27.30 (
H2–NH), 27.30 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3). HPLC (method A): 100%, RT = 4.37 min. HRMS (ESI, m/z). Calcd for C20H19ClN4O3 [M + H]+, 399.1224; found, 399.1225.
H3). HPLC (method A): 100%, RT = 4.37 min. HRMS (ESI, m/z). Calcd for C20H19ClN4O3 [M + H]+, 399.1224; found, 399.1225.
3-Acetyl-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1yl)propyl)benzamide (6b, R1 = Cl, R2 = Cl). Prepared from 1-amino-2-(2,4-dichlorophenyl)-3-(1H-1,2,4-triazol-1-yl)propan-2-ol (5b) (0.15 g, 0.52 mmol) and 3-acetyl benzoic acid (0.13 g, 0.78 mmol). Purified by gradient column chromatography eluting with 3.5% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.1 g (45%). M.p.: 160–162 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.45. 1H NMR (DMSO-d6): δ 8.72 (t, J = 6.0 Hz, 1H, NH), 8.34 (s, 1H, triaz), 8.30 (s, 1H, Ar), 8.10 (d, J = 7.8 Hz, 1H, Ar), 8.01 (d, J = 8.3 Hz, 1H, Ar), 7.74 (s, 1H, triaz), 7.59 (m, 3H, Ar), 7.29 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.35 (s, 1H, OH, ex), 5.10 (d, J = 14.4 Hz, 1H, C
0.5 v/v), Rf = 0.45. 1H NMR (DMSO-d6): δ 8.72 (t, J = 6.0 Hz, 1H, NH), 8.34 (s, 1H, triaz), 8.30 (s, 1H, Ar), 8.10 (d, J = 7.8 Hz, 1H, Ar), 8.01 (d, J = 8.3 Hz, 1H, Ar), 7.74 (s, 1H, triaz), 7.59 (m, 3H, Ar), 7.29 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.35 (s, 1H, OH, ex), 5.10 (d, J = 14.4 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.70 (d, J = 14.4 Hz, 1H, C
-triaz), 4.70 (d, J = 14.4 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.08 (dd, J = 5.7, 14.0 Hz, 1H, C
-triaz), 4.08 (dd, J = 5.7, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -NH), 3.97 (dd, J = 6.4, 14.0 Hz, 1H, C
-NH), 3.97 (dd, J = 6.4, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 2.61 (s, 3H, C
–NH), 2.61 (s, 3H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[3 with combining low line]](https://www.rsc.org/images/entities/char_0033_0332.gif) ). 13C NMR (DMSO-d6): δ 198.00 (C, CH3C
). 13C NMR (DMSO-d6): δ 198.00 (C, CH3C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 167.58 (C, C
O), 167.58 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, amide), 151.02 (CH, triaz), 145.56 (CH, triaz), 138.11 (C, Ar), 137.20 (C, Ar), 134.71 (C, Ar), 133.29 (C, C–Cl), 132.40 (CH, Ar), 131.51 (CH, Ar), 131.50 (C, C–Cl), 130.43 (CH, Ar), 129.30 (2× CH, Ar), 127.39 (2× CH, Ar), 76.90 (
O, amide), 151.02 (CH, triaz), 145.56 (CH, triaz), 138.11 (C, Ar), 137.20 (C, Ar), 134.71 (C, Ar), 133.29 (C, C–Cl), 132.40 (CH, Ar), 131.51 (CH, Ar), 131.50 (C, C–Cl), 130.43 (CH, Ar), 129.30 (2× CH, Ar), 127.39 (2× CH, Ar), 76.90 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 54.09 (
–OH), 54.09 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 45.95 (
H2-triaz), 45.95 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH), 27.32 (
H2–NH), 27.32 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3). HPLC (method A): 100%, RT = 4.49 min. HRMS (ESI, m/z), calcd for C20H18Cl2N4O3 [M + H]+, 433.0834; found, 433.0832.
H3). HPLC (method A): 100%, RT = 4.49 min. HRMS (ESI, m/z), calcd for C20H18Cl2N4O3 [M + H]+, 433.0834; found, 433.0832.
2-Chloro-N-(2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)thiazole-4-carboxamide (7a, R1 = Cl, R2 = H). Prepared from 1-amino-2-(2,4-dichlorophenyl)-3-(1H-1,2,4-triazol-1-yl)propan-2-ol (5a) (0.16 g, 0.63 mmol) and 2-chlorothiazole-4-carboxylic acid (0.15 g, 0.95 mmol). Purified by gradient column chromatography eluting with 3.5% MeOH in CH2Cl2. Product obtained as a semisolid, yield 0.17 g (68%). TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.45. 1H NMR (DMSO-d6): δ 8.23 (s, 1H, thiazole), 8.22 (s, 1H, triaz), 8.10 (t, J = 5.2 Hz, 1H, NH), 7.84 (s, 1H, triaz), 7.41 (d, J = 8.8 Hz, 2H, Ar), 7.32 (d, J = 8.8 Hz, 2H, Ar), 6.12 (s, 1H, OH, ex), 4.53 (s, 2H, C
0.5 v/v), Rf = 0.45. 1H NMR (DMSO-d6): δ 8.23 (s, 1H, thiazole), 8.22 (s, 1H, triaz), 8.10 (t, J = 5.2 Hz, 1H, NH), 7.84 (s, 1H, triaz), 7.41 (d, J = 8.8 Hz, 2H, Ar), 7.32 (d, J = 8.8 Hz, 2H, Ar), 6.12 (s, 1H, OH, ex), 4.53 (s, 2H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 3.96 (dd, J = 7.3, 14.1 Hz, 1H, C
-triaz), 3.96 (dd, J = 7.3, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.58 (dd, J = 5.1, 14.1 Hz, 1H, C
–NH), 3.58 (dd, J = 5.1, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH). 13C NMR (DMSO-d6): δ 160.06 (C, C
–NH). 13C NMR (DMSO-d6): δ 160.06 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, amide), 151.44 (C, C–Cl), 151.13 (CH, triaz), 147.72 (C, Ar), 145.45 (CH, triaz), 141.28 (C, thiazole), 132.28 (C, Ar), 128.30 (2× CH, Ar), 128.19 (2× CH, Ar), 128.03 (CH, thiazole), 75.72 (
O, amide), 151.44 (C, C–Cl), 151.13 (CH, triaz), 147.72 (C, Ar), 145.45 (CH, triaz), 141.28 (C, thiazole), 132.28 (C, Ar), 128.30 (2× CH, Ar), 128.19 (2× CH, Ar), 128.03 (CH, thiazole), 75.72 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 57.26 (
–OH), 57.26 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 47.22 (
H2-triaz), 47.22 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-NH). HPLC (method A): 100%, RT = 4.52 min. HRMS (ESI, m/z). Calcd for C15H13Cl2N5O2S [M + H]+, 398.0245; found, 398.0242.
H2-NH). HPLC (method A): 100%, RT = 4.52 min. HRMS (ESI, m/z). Calcd for C15H13Cl2N5O2S [M + H]+, 398.0245; found, 398.0242.
2-Chloro-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)thiazole-4-carboxamide (7b, R1 = Cl, R2 = Cl). Prepared from 1-amino-2-(2,4-dichlorophenyl)-3-(1H-1,2,4-triazol-1-yl)propan-2-ol (5b) (0.15 g, 0.52 mmol) and 2-chlorothiazole-4-carboxylic acid (0.13 g, 0.78 mmol). Purified by gradient column chromatography eluting with 2.5% MeOH in CH2Cl2. Product obtained as a pale yellow wax, yield 0.15 g (68%). TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.55. 1H NMR (DMSO-d6): δ 8.32 (s, 1H, thiazole), 8.26 (t, J = 5.9 Hz, 1H, NH), 8.25 (s, 1H, triaz), 7.72 (s, 1H, triaz), 7.54 (d, J = 7.7 Hz, 2H, Ar), 7.29 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.33 (s, 1H, OH, ex), 5.03 (d, J = 14.4 Hz, 1H, C
0.5 v/v), Rf = 0.55. 1H NMR (DMSO-d6): δ 8.32 (s, 1H, thiazole), 8.26 (t, J = 5.9 Hz, 1H, NH), 8.25 (s, 1H, triaz), 7.72 (s, 1H, triaz), 7.54 (d, J = 7.7 Hz, 2H, Ar), 7.29 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.33 (s, 1H, OH, ex), 5.03 (d, J = 14.4 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.64 (d, J = 14.4 Hz, 1H, C
-triaz), 4.64 (d, J = 14.4 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.05 (dd, J = 6.7, 14.0 Hz, 1H, C
-triaz), 4.05 (dd, J = 6.7, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.99 (dd, J = 5.8, 14.0 Hz, 1H, C
–NH), 3.99 (dd, J = 5.8, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH). 13C NMR (DMSO-d6): δ 160.43 (C, C
–NH). 13C NMR (DMSO-d6): δ 160.43 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, amide), 151.45 (C, C–Cl), 151.03 (CH, triaz), 147.67 (C, Ar), 145.58 (CH, triaz), 137.98 (C, thiazole), 133.35 (C, C–Cl), 132.07 (C, C–Cl), 131.49 (CH, Ar), 130.47 (CH, Ar), 128.18 (CH, thiazole), 127.30 (CH, Ar), 76.27 (
O, amide), 151.45 (C, C–Cl), 151.03 (CH, triaz), 147.67 (C, Ar), 145.58 (CH, triaz), 137.98 (C, thiazole), 133.35 (C, C–Cl), 132.07 (C, C–Cl), 131.49 (CH, Ar), 130.47 (CH, Ar), 128.18 (CH, thiazole), 127.30 (CH, Ar), 76.27 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 54.14 (
–OH), 54.14 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 45.06 (
H2-triaz), 45.06 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH). HPLC (method A): 100%, RT = 4.63 min. HRMS (ESI, m/z). Calcd for C15H12Cl3N5O2S [M + H]+, 431.9855; found, 433.9826.
H2–NH). HPLC (method A): 100%, RT = 4.63 min. HRMS (ESI, m/z). Calcd for C15H12Cl3N5O2S [M + H]+, 431.9855; found, 433.9826.
N-(2-(4-Chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)-4-(3-(4-chlorophenyl)thioureido)benzamide (11a, R1 = Cl, R2 = H, X = S). Prepared from 4-amino-N-(2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10a) (0.16 g, 043 mmol) and 4-chlorophenyl isothiocyanate (0.11 g, 0.64 mmol). Purified by gradient column chromatography eluting with 2.5% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.15 g (68%). M.p.: 146–148 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.23. 1H NMR (DMSO-d6): δ 10.05 (s, 1H, NH–thiourea), 10.00 (s, 1H, NH–thiourea), 8.22 (t, J = 5.9 Hz, 1H, NH–CH2), 8.20 (s, 1H, triaz), 7.84 (s, 1H, triaz), 7.70 (d, J = 8.9 Hz, 2H, Ar), 7.55 (dd, J = 8.8, 21.2 Hz, 4H, Ar), 7.43 (d, J = 7.1 Hz, 2H, Ar), 7.39 (d, J = 9.0 Hz, 2H, Ar), 7.29 (m, 2H, Ar), 6.05 (s, 1H, OH, ex), 5.74 (dd, J = 14.3, 20.3 Hz, 2H, C
0.5 v/v), Rf = 0.23. 1H NMR (DMSO-d6): δ 10.05 (s, 1H, NH–thiourea), 10.00 (s, 1H, NH–thiourea), 8.22 (t, J = 5.9 Hz, 1H, NH–CH2), 8.20 (s, 1H, triaz), 7.84 (s, 1H, triaz), 7.70 (d, J = 8.9 Hz, 2H, Ar), 7.55 (dd, J = 8.8, 21.2 Hz, 4H, Ar), 7.43 (d, J = 7.1 Hz, 2H, Ar), 7.39 (d, J = 9.0 Hz, 2H, Ar), 7.29 (m, 2H, Ar), 6.05 (s, 1H, OH, ex), 5.74 (dd, J = 14.3, 20.3 Hz, 2H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 3.86 (dd, J = 6.7, 14.0 Hz, 1H, C
-triaz), 3.86 (dd, J = 6.7, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.63 (dd, J = 5.2, 14.1 Hz, 1H, C
–NH), 3.63 (dd, J = 5.2, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH). 13C NMR (DMSO-d6): δ 179.97 (C, C
–NH). 13C NMR (DMSO-d6): δ 179.97 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 167.33 (C, C
S), 167.33 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 150.96 (CH, triaz), 145.38 (CH, triaz), 142.70 (C, Ar), 142.45 (C, Ar), 141.48 (C, Ar), 138.75 (C, C–Cl), 132.14 (C, C–Cl), 129.85 (C, Ar), 129.84 (2× CH, Ar), 128.23 (2× CH, Ar), 128.17 (2× CH, Ar), 126.24 (2× CH, Ar), 125.74 (2× CH, Ar), 122.74 (2× CH, Ar), 76.48 (C,
O), 150.96 (CH, triaz), 145.38 (CH, triaz), 142.70 (C, Ar), 142.45 (C, Ar), 141.48 (C, Ar), 138.75 (C, C–Cl), 132.14 (C, C–Cl), 129.85 (C, Ar), 129.84 (2× CH, Ar), 128.23 (2× CH, Ar), 128.17 (2× CH, Ar), 126.24 (2× CH, Ar), 125.74 (2× CH, Ar), 122.74 (2× CH, Ar), 76.48 (C, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 57.40 (
–OH), 57.40 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 48.35 (
H2-triaz), 48.35 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-NH). HPLC (method B): 99%, RT = 4.78 min. HRMS (ESI, m/z). Calcd for C25H22Cl2N6O2S [M + H]+, 541.0980; found, 541.0971.
H2-NH). HPLC (method B): 99%, RT = 4.78 min. HRMS (ESI, m/z). Calcd for C25H22Cl2N6O2S [M + H]+, 541.0980; found, 541.0971.
4-(3-(4-Chlorophenyl)thioureido)-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (11b, R1 = Cl, R2 = Cl, X = S). Prepared from 4-amino-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl) benzamide (10b) (0.17 g, 0.41 mmol) and 4-chlorophenyl isothiocyanate (0.1 g, 0.62 mmol). Purified by gradient column chromatography eluting with 3.5% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.13 g (56%). M.p.: 138–140 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.32. 1H NMR (DMSO-d6): δ 10.06 (s, 1H, NH–thiourea), 10.03 (s, 1H, NH–thiourea), 8.47 (t, J = 5.9 Hz, 1H, NH-CH2), 8.34 (s, 1H, triaz), 7.74 (s, 1H, triaz), 7.73 (d, J = 8.8 Hz, 2H, Ar), 7.57 (m, 4H, Ar), 7.51 (d, J = 8.9 Hz, 2H, Ar), 7.39 (d, J = 8.9 Hz, 2H, Ar), 7.29 (dd, J = 2.3, 8.7 Hz, 1H, Ar), 6.46 (s, 1H, OH-ex), 5.05 (d, J = 14.4 Hz, 1H, C
0.5 v/v), Rf = 0.32. 1H NMR (DMSO-d6): δ 10.06 (s, 1H, NH–thiourea), 10.03 (s, 1H, NH–thiourea), 8.47 (t, J = 5.9 Hz, 1H, NH-CH2), 8.34 (s, 1H, triaz), 7.74 (s, 1H, triaz), 7.73 (d, J = 8.8 Hz, 2H, Ar), 7.57 (m, 4H, Ar), 7.51 (d, J = 8.9 Hz, 2H, Ar), 7.39 (d, J = 8.9 Hz, 2H, Ar), 7.29 (dd, J = 2.3, 8.7 Hz, 1H, Ar), 6.46 (s, 1H, OH-ex), 5.05 (d, J = 14.4 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.68 (d, J = 14.4 Hz, 1H, C
-triaz), 4.68 (d, J = 14.4 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.02 (dd, J = 5.7, 14.2 Hz, 1H, C
-triaz), 4.02 (dd, J = 5.7, 14.2 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.97 (dd, J = 6.5, 14.2 Hz, 1H, C
–NH), 3.97 (dd, J = 6.5, 14.2 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -NH). 13C NMR (DMSO-d6): δ 179.90 (C, C
-NH). 13C NMR (DMSO-d6): δ 179.90 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 168.02 (C, C
S), 168.02 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.01 (CH, triaz), 145.59 (CH, triaz), 142.89 (C, Ar), 138.76 (C, Ar), 138.19 (C, C–Cl), 133.26 (C, C–Cl), 132.17 (C, C–Cl), 132.04 (C, C–Cl), 131.54 (CH, Ar), 130.41 (CH, Ar), 128.81 (2× CH, Ar), 128.32 (2× CH, Ar), 127.27 (CH, Ar), 125.72 (2× CH, Ar), 122.70 (2× CH, Ar), 76.99 (C,
O), 151.01 (CH, triaz), 145.59 (CH, triaz), 142.89 (C, Ar), 138.76 (C, Ar), 138.19 (C, C–Cl), 133.26 (C, C–Cl), 132.17 (C, C–Cl), 132.04 (C, C–Cl), 131.54 (CH, Ar), 130.41 (CH, Ar), 128.81 (2× CH, Ar), 128.32 (2× CH, Ar), 127.27 (CH, Ar), 125.72 (2× CH, Ar), 122.70 (2× CH, Ar), 76.99 (C, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 54.18 (
–OH), 54.18 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 46.05 (
H2-triaz), 46.05 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH). Anal. calcd for C25H21Cl3N6O2S (575.89): C 52.13%, H 3.68%, N 14.59%. Found: C 52.29%, H 3.82%, N 14.71%.
H2–NH). Anal. calcd for C25H21Cl3N6O2S (575.89): C 52.13%, H 3.68%, N 14.59%. Found: C 52.29%, H 3.82%, N 14.71%.
N-(2-(4-Chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)-4-(3-(4-chlorophenyl)ureido)benzamide (12a, R1 = Cl, R2 = H, X = O). Prepared from 4-amino-N-(2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10a) (0.16 g, 0.43 mmol) and 4-chlorophenyl isocyanate (0.1 g, 0.64 mmol). Purified by gradient column chromatography eluting with 6% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.14 g (63%). M.p.: 226–228 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.35. 1H NMR (DMSO-d6): δ 8.99 (s, 1H, NH–urea), 8.90 (s, 1H, NH–urea), 8.25 (s, 1H, triaz), 8.22 (t, J = 5.7 Hz, 1H, NH–CH2), 7.83 (s, 1H, triaz), 7.54 (dd, J = 3.2, 8.9 Hz, 2H, Ar), 7.48 (m, 4H, Ar), 7.43 (d, J = 8.8 Hz, 2H, Ar), 7.31 (m, 4H, Ar), 6.16 (s, 1H, OH-ex), 4.60 (dd, J = 14.3, 21.5 Hz, 2H, C
0.5 v/v), Rf = 0.35. 1H NMR (DMSO-d6): δ 8.99 (s, 1H, NH–urea), 8.90 (s, 1H, NH–urea), 8.25 (s, 1H, triaz), 8.22 (t, J = 5.7 Hz, 1H, NH–CH2), 7.83 (s, 1H, triaz), 7.54 (dd, J = 3.2, 8.9 Hz, 2H, Ar), 7.48 (m, 4H, Ar), 7.43 (d, J = 8.8 Hz, 2H, Ar), 7.31 (m, 4H, Ar), 6.16 (s, 1H, OH-ex), 4.60 (dd, J = 14.3, 21.5 Hz, 2H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 3.85 (dd, J = 6.7, 14.1 Hz, 1H, C
-triaz), 3.85 (dd, J = 6.7, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.63 (dd, J = 5.2, 14.0 Hz, 1H, C
–NH), 3.63 (dd, J = 5.2, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH). 13C NMR (DMSO-d6): δ 167.51 (C, C
–NH). 13C NMR (DMSO-d6): δ 167.51 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 152.63 (C, C
O), 152.63 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.00 (CH, triaz), 145.45 (CH, triaz), 143.01 (C, Ar), 142.50 (C, Ar), 141.52 (C, Ar), 138.88 (C, C–Cl), 132.12 (C, C–Cl), 129.10 (C, Ar), 128.80 (2× CH, Ar), 128.38 (2× CH, Ar), 128.09 (2× CH, Ar), 120.34 (2× CH, Ar), 117.66 (2× CH, Ar), 76.34 (C,
O), 151.00 (CH, triaz), 145.45 (CH, triaz), 143.01 (C, Ar), 142.50 (C, Ar), 141.52 (C, Ar), 138.88 (C, C–Cl), 132.12 (C, C–Cl), 129.10 (C, Ar), 128.80 (2× CH, Ar), 128.38 (2× CH, Ar), 128.09 (2× CH, Ar), 120.34 (2× CH, Ar), 117.66 (2× CH, Ar), 76.34 (C, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 57.14 (
–OH), 57.14 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 48.24 (
H2-triaz), 48.24 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH). Anal. calcd. for C25H22Cl2N6O3, (525.39): C 57.15%, H 4.22%, N 15.99%. Found: C 57.53%, H 4.28%, N 15.94%.
H2–NH). Anal. calcd. for C25H22Cl2N6O3, (525.39): C 57.15%, H 4.22%, N 15.99%. Found: C 57.53%, H 4.28%, N 15.94%.
4-(3-(4-Chlorophenyl)ureido)-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (12b, R1 = Cl, R2 = Cl, X = O). Prepared from 4-amino-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10b) (0.16 g, 0.43 mmol) and 4-chlorophenyl isocyanate (0.1 g, 0.64 mmol). Purified by gradient column chromatography eluting with 6% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.11 g (52%). M.p.: 216–218 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.37. 1H NMR (DMSO-d6): δ 9.06 (s, 1H, NH–urea), 8.97 (s, 1H, NH–urea), 8.42 (t, J = 6.0 Hz, 1H, NH–CH2), 8.34 (s, 1H, triaz), 7.72 (d, J = 11.0 Hz, 2H, Ar), 7.70 (s, 1H, triaz), 7.57 (dd, J = 8.6, 11.0 Hz, 2H, Ar), 7.49 (m, 4H, Ar), 7.33 (d, J = 8.9 Hz, 2H, Ar), 7.28 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.53 (s, 1H, OH, ex), 5.05 (d, J = 14.3 Hz, 1H, C
0.5 v/v), Rf = 0.37. 1H NMR (DMSO-d6): δ 9.06 (s, 1H, NH–urea), 8.97 (s, 1H, NH–urea), 8.42 (t, J = 6.0 Hz, 1H, NH–CH2), 8.34 (s, 1H, triaz), 7.72 (d, J = 11.0 Hz, 2H, Ar), 7.70 (s, 1H, triaz), 7.57 (dd, J = 8.6, 11.0 Hz, 2H, Ar), 7.49 (m, 4H, Ar), 7.33 (d, J = 8.9 Hz, 2H, Ar), 7.28 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.53 (s, 1H, OH, ex), 5.05 (d, J = 14.3 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.67 (d, J = 14.3 Hz, 1H, C
-triaz), 4.67 (d, J = 14.3 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.02 (dd, J = 5.5, 14.1 Hz, 1H, C
-triaz), 4.02 (dd, J = 5.5, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.95 (dd, J = 6.3, 14.1 Hz, 1H, C
–NH), 3.95 (dd, J = 6.3, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH). 13C NMR (DMSO-d6): δ 168.21 (C, C
–NH). 13C NMR (DMSO-d6): δ 168.21 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 152.66 (C, C
O), 152.66 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.00 (CH, triaz), 145.45 (CH, triaz), 143.18 (C, Ar), 138.90 (C, Ar), 138.25 (C, C–Cl), 133.25 (C, C–Cl), 132.02 (C, C–Cl), 131.55 (CH, Ar), 131.24 (CH, Ar), 130.04 (CH, Ar), 129.10 (2× CH, Ar), 128.93 (CH, Ar), 127.27 (CH, Ar), 120.35 (2× CH, Ar), 117.64 (2× CH, Ar), 77.03 (C,
O), 151.00 (CH, triaz), 145.45 (CH, triaz), 143.18 (C, Ar), 138.90 (C, Ar), 138.25 (C, C–Cl), 133.25 (C, C–Cl), 132.02 (C, C–Cl), 131.55 (CH, Ar), 131.24 (CH, Ar), 130.04 (CH, Ar), 129.10 (2× CH, Ar), 128.93 (CH, Ar), 127.27 (CH, Ar), 120.35 (2× CH, Ar), 117.64 (2× CH, Ar), 77.03 (C, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 54.19 (
–OH), 54.19 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 46.12 (
H2-triaz), 46.12 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH). Anal. calcd. For C25H21Cl3N6O3 (559.83): C 53.64%, H 3.78%, N 15.00%. Found: C 53.84%, H 3.68%, N 14.63%.
H2–NH). Anal. calcd. For C25H21Cl3N6O3 (559.83): C 53.64%, H 3.78%, N 15.00%. Found: C 53.84%, H 3.68%, N 14.63%.
4-Chloro-N-(4-((2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl) carbamoyl) phenyl)benzamide (13a, R1 = Cl, R2 = H, Y = C(O)). Prepared from 4-amino-N-(2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10a) (0.15 g, 0.42 mmol) and 4-chlorobenzoyl chloride (0.1 g, 0.64 mmol). Purified by gradient column chromatography eluting with 6% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.16 g (80%). M.p.: 222–224 °C. TLC (CH2Cl2-MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.33. 1H NMR (DMSO-d6): δ 10.50 (s, 1H, NH-amide), 8.28 (t, J = 5.7 Hz, 1H, NH-CH2), 8.26 (s, 1H, triaz), 7.99 (d, J = 8.8 Hz, 2H, Ar), 7.84 (s, 1H, triaz), 7.83 (dd, J = 3.8, 8.9 Hz, 2H, Ar), 7.76 (d, J = 8.9 Hz, 2H, Ar), 7.62 (d, J = 8.8 Hz, 2H, Ar), 7.44 (d, J = 8.8 Hz, 2H, Ar), 7.30 (m, 2H, Ar), 6.14 (s, 1H, OH-ex), 4.59 (dd, J = 14.4, 21.9 Hz, 2H, C
0.5 v/v), Rf = 0.33. 1H NMR (DMSO-d6): δ 10.50 (s, 1H, NH-amide), 8.28 (t, J = 5.7 Hz, 1H, NH-CH2), 8.26 (s, 1H, triaz), 7.99 (d, J = 8.8 Hz, 2H, Ar), 7.84 (s, 1H, triaz), 7.83 (dd, J = 3.8, 8.9 Hz, 2H, Ar), 7.76 (d, J = 8.9 Hz, 2H, Ar), 7.62 (d, J = 8.8 Hz, 2H, Ar), 7.44 (d, J = 8.8 Hz, 2H, Ar), 7.30 (m, 2H, Ar), 6.14 (s, 1H, OH-ex), 4.59 (dd, J = 14.4, 21.9 Hz, 2H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 3.86 (dd, J = 6.8, 14.1 Hz, 1H, C
-triaz), 3.86 (dd, J = 6.8, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.64 (dd, J = 5.3, 14.1 Hz, 1H, C
–NH), 3.64 (dd, J = 5.3, 14.1 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -NH). 13C NMR (DMSO-d6): δ 167.38 (C, C
-NH). 13C NMR (DMSO-d6): δ 167.38 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 165.13 (C, C
O), 165.13 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.00 (CH, triaz), 145.45 (CH, triaz), 142.26 (C, Ar), 141.49 (C, Ar), 137.10 (C, C–Cl), 133.76 (C, Ar), 132.14 (C, C–Cl), 130.17 (2× CH, Ar), 129.34 (C, Ar), 128.97 (2× CH, Ar), 128.53 (2× CH, Ar), 128.09 (2× CH, Ar), 126.25 (2× CH, Ar), 119.91 (2× CH, Ar), 76.33 (C,
O), 151.00 (CH, triaz), 145.45 (CH, triaz), 142.26 (C, Ar), 141.49 (C, Ar), 137.10 (C, C–Cl), 133.76 (C, Ar), 132.14 (C, C–Cl), 130.17 (2× CH, Ar), 129.34 (C, Ar), 128.97 (2× CH, Ar), 128.53 (2× CH, Ar), 128.09 (2× CH, Ar), 126.25 (2× CH, Ar), 119.91 (2× CH, Ar), 76.33 (C, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 57.10 (
–OH), 57.10 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 48.24 (
H2-triaz), 48.24 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH). Anal. calcd. for C25H21Cl2N5O3 (510.38): C 58.83%, H 4.15%, N 13.72%. Found: C 58.78%, H 4.07%, N 13.68%.
H2–NH). Anal. calcd. for C25H21Cl2N5O3 (510.38): C 58.83%, H 4.15%, N 13.72%. Found: C 58.78%, H 4.07%, N 13.68%.
4-Chloro-N-(4-((2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl) carbamoyl)phenyl)benzamide (13b, R1 = Cl, R2 = Cl, Y = C(O)). Prepared from 4-amino-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10b) (0.16 g, 0.39 mmol) and 4-chlorobenzoyl chloride (0.16 g, 0.6 mmol). Purified by gradient column chromatography eluting with 6% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.16 g (76%). M.p.: 190–192 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.32. 1H NMR (DMSO-d6): δ 10.51 (s, 1H, NH–amide), 8.49 (t, J = 5.9 Hz, 1H, NH–CH2), 8.35 (s, 1H, triaz), 7.99 (d, J = 8.6 Hz, 2H, Ar), 7.84 (d, J = 8.8 Hz, 2H, Ar), 7.79 (d, J = 8.8 Hz, 2H, Ar), 7.74 (s, 1H, triaz), 7.62 (d, J = 8.6 Hz, 2H, Ar), 7.57 (dd, J = 8.6, 13.4 Hz, 2H, Ar), 7.29 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.50 (s, 1H, OH, ex), 5.07 (d, J = 14.3 Hz, 1H, C
0.5 v/v), Rf = 0.32. 1H NMR (DMSO-d6): δ 10.51 (s, 1H, NH–amide), 8.49 (t, J = 5.9 Hz, 1H, NH–CH2), 8.35 (s, 1H, triaz), 7.99 (d, J = 8.6 Hz, 2H, Ar), 7.84 (d, J = 8.8 Hz, 2H, Ar), 7.79 (d, J = 8.8 Hz, 2H, Ar), 7.74 (s, 1H, triaz), 7.62 (d, J = 8.6 Hz, 2H, Ar), 7.57 (dd, J = 8.6, 13.4 Hz, 2H, Ar), 7.29 (dd, J = 2.2, 8.6 Hz, 1H, Ar), 6.50 (s, 1H, OH, ex), 5.07 (d, J = 14.3 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.68 (d, J = 14.3 Hz, 1H, C
-triaz), 4.68 (d, J = 14.3 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) -triaz), 4.04 (dd, J = 5.5, 14.0 Hz, 1H, C
-triaz), 4.04 (dd, J = 5.5, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH), 3.97 (dd, J = 6.5, 14.0 Hz, 1H, C
–NH), 3.97 (dd, J = 6.5, 14.0 Hz, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif)
![[2 with combining low line]](https://www.rsc.org/images/entities/char_0032_0332.gif) –NH). 13C NMR (DMSO-d6): δ 168.02 (C, C
–NH). 13C NMR (DMSO-d6): δ 168.02 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 165.14 (C, C
O), 165.14 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.00 (CH, triaz), 145.58 (CH, triaz), 142.38 (C, Ar), 138.21 (C, Ar), 137.11 (C, C–Cl), 133.75 (C, C–Cl), 133.26 (C, C–Cl), 132.04 (C, Ar), 131.55 (CH, Ar), 130.41 (CH, Ar), 130.17 (2× CH, Ar), 129.06 (C, Ar), 128.98 (2× CH, Ar), 128.65 (2× CH, Ar), 127.27 (CH, Ar), 119.89 (2× CH, Ar), 76.99 (C,
O), 151.00 (CH, triaz), 145.58 (CH, triaz), 142.38 (C, Ar), 138.21 (C, Ar), 137.11 (C, C–Cl), 133.75 (C, C–Cl), 133.26 (C, C–Cl), 132.04 (C, Ar), 131.55 (CH, Ar), 130.41 (CH, Ar), 130.17 (2× CH, Ar), 129.06 (C, Ar), 128.98 (2× CH, Ar), 128.65 (2× CH, Ar), 127.27 (CH, Ar), 119.89 (2× CH, Ar), 76.99 (C, ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) –OH), 54.16 (
–OH), 54.16 (![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2-triaz), 46.68(
H2-triaz), 46.68(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2–NH). Anal. calcd. for C25H20Cl3N5O3 (544.82): C 54.39%, H 3.65%, N 12.68%. Found: C 54.05%, H 3.80%, N 12.31%.
H2–NH). Anal. calcd. for C25H20Cl3N5O3 (544.82): C 54.39%, H 3.65%, N 12.68%. Found: C 54.05%, H 3.80%, N 12.31%.
N-(2-(4-Chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)-4-((4-chlorophenyl) sulfonamido)benzamide (14a R1 = Cl, R2 = H, Y = S(O2)). Prepared from 4-amino-N-(2-(4-chlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10a) (0.30 g, 0.82 mmoL) and 4-chlorobenzenesulfonyl chloride (0.26 g, 1.23 mmol). Purified by gradient column chromatography eluting with 3.5% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.35 g (77%). M.p. 236–238 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.45. 1H NMR (DMSO-d6): δ 10.73 (br.s, 1H, NHSO2), 8.23 (s, 1H, triazole), 8.21 (t, J = 6.0 Hz, 1H, NH), 7.81 (s, 1H, triazole), 7.79 (d, J = 8.8 Hz, 2H, Ar), 7.63 (m, 4H, Ar), 7.41 (d, J = 8.7 Hz, 2H, Ar), 7.28 (m, 2H, Ar), 7.16 (m, 2H, Ar), 6.04 (s, 1H, OH), 4.56 (dd, J = 14.4, 25.0 Hz, 2H, CH2-triazole), 3.79 (dd, J = 6.6, 14.0 Hz, 1H, NHCHaHb), 3.59 (dd, J = 5.3, 14.0 Hz, 1H, NHCHaHb). 13C NMR (DMSO-d6): δ 167.19 (C, C
0.5 v/v), Rf = 0.45. 1H NMR (DMSO-d6): δ 10.73 (br.s, 1H, NHSO2), 8.23 (s, 1H, triazole), 8.21 (t, J = 6.0 Hz, 1H, NH), 7.81 (s, 1H, triazole), 7.79 (d, J = 8.8 Hz, 2H, Ar), 7.63 (m, 4H, Ar), 7.41 (d, J = 8.7 Hz, 2H, Ar), 7.28 (m, 2H, Ar), 7.16 (m, 2H, Ar), 6.04 (s, 1H, OH), 4.56 (dd, J = 14.4, 25.0 Hz, 2H, CH2-triazole), 3.79 (dd, J = 6.6, 14.0 Hz, 1H, NHCHaHb), 3.59 (dd, J = 5.3, 14.0 Hz, 1H, NHCHaHb). 13C NMR (DMSO-d6): δ 167.19 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.00 (CH, triazole), 145.44 (CH, triazole), 141.45 (C, Ar), 140.85 (C, Ar), 138.65 (C, Ar), 138.48 (C, C–Cl), 132.15 (C, C–Cl), 130.03 (2× CH, Ar), 129.76 (C, Ar), 129.15 (2× CH, Ar), 129.04 (2× CH, Ar), 128.35 (2× CH, Ar), 128.06 (2× CH, Ar), 119.09 (2× CH, Ar), 76.25 (C, C–OH), 57.04 (CH2-triazole), 48.18 (CH2–NH). HPLC (method B): 100%, RT = 4.77 min. HRMS (ESI), m/z. Calcd for C24H21Cl2N5O4S [M + H]+, 546.0769; found, 546.0769.
O), 151.00 (CH, triazole), 145.44 (CH, triazole), 141.45 (C, Ar), 140.85 (C, Ar), 138.65 (C, Ar), 138.48 (C, C–Cl), 132.15 (C, C–Cl), 130.03 (2× CH, Ar), 129.76 (C, Ar), 129.15 (2× CH, Ar), 129.04 (2× CH, Ar), 128.35 (2× CH, Ar), 128.06 (2× CH, Ar), 119.09 (2× CH, Ar), 76.25 (C, C–OH), 57.04 (CH2-triazole), 48.18 (CH2–NH). HPLC (method B): 100%, RT = 4.77 min. HRMS (ESI), m/z. Calcd for C24H21Cl2N5O4S [M + H]+, 546.0769; found, 546.0769.
4-((4-Chlorophenyl)sulfonamido)-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (14b, R1 = Cl, R2 = Cl, Y = S(O2)). Prepared from 4-amino-N-(2-(2,4-dichlorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)benzamide (10b) (0.17 g, 0.42 mmoL) and 4-chlorobenzenesulfonyl chloride (0.13 g, 0.63 mmoL). Purified by gradient column chromatography eluting with 3.5% MeOH in CH2Cl2. Product obtained as a white solid, yield 0.2 g (83%). M.p. 228–230 °C. TLC (CH2Cl2–MeOH 9.5
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 v/v), Rf = 0.46. 1H NMR (DMSO-d6): δ 10.76 (br.s, 1H, NHSO2), 8.40 (t, J = 6.0 Hz, 1H, NH), 8.32 (s, 1H, triazole), 7.79 (d, J = 8.8 Hz, 2H, Ar), 7.72 (s, 1H, triazole), 7.60 (dd, J = 4.8, 8.9 Hz, 4H, Ar), 7.54 (dd, J = 3.2, 5.5 Hz, 2H, Ar), 7.27 (dd, J = 2.2, 8.7 Hz, 1H, Ar), 713 (d, J = 8.8 Hz, 2H, Ar), 6.36 (s, 1H, OH), 5.04 (d, J = 14.4 Hz, 1H, CHaHb-triazole), 4.63 (d, J = 14.4 Hz, 1H, CHaHb-triazole), 4.01 (dd, J = 5.7, 14.1 Hz, 1H, CHaHbNH), 3.89 (dd, J = 6.3, 14.0 Hz, 1H, CHaHbNH). 13C NMR (DMSO-d6): δ 167.78 (C, C
0.5 v/v), Rf = 0.46. 1H NMR (DMSO-d6): δ 10.76 (br.s, 1H, NHSO2), 8.40 (t, J = 6.0 Hz, 1H, NH), 8.32 (s, 1H, triazole), 7.79 (d, J = 8.8 Hz, 2H, Ar), 7.72 (s, 1H, triazole), 7.60 (dd, J = 4.8, 8.9 Hz, 4H, Ar), 7.54 (dd, J = 3.2, 5.5 Hz, 2H, Ar), 7.27 (dd, J = 2.2, 8.7 Hz, 1H, Ar), 713 (d, J = 8.8 Hz, 2H, Ar), 6.36 (s, 1H, OH), 5.04 (d, J = 14.4 Hz, 1H, CHaHb-triazole), 4.63 (d, J = 14.4 Hz, 1H, CHaHb-triazole), 4.01 (dd, J = 5.7, 14.1 Hz, 1H, CHaHbNH), 3.89 (dd, J = 6.3, 14.0 Hz, 1H, CHaHbNH). 13C NMR (DMSO-d6): δ 167.78 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 151.00 (CH, triazole), 145.56 (CH, triazole), 141.01 (C, Ar), 138.65 (C, Ar), 138.48 (C, Ar), 138.15 (C, C–Cl), 133.26 (C, C–Cl), 132.01 (C, C–Cl), 131.50 (CH, Ar), 130.41 (CH, Ar), 130.03 (2× CH, Ar), 129.44 (C, Ar), 129.26 (2× CH, Ar), 129.08 (2× CH, Ar), 127.27 (CH, Ar), 119.02 (2× CH, Ar), 76.91 (C, C–OH), 45.09 (CH2-triazole), 45.96 (CH2–NH). HPLC (method B): 99.7%, RT = 4.76 min. HRMS (ESI), m/z. calcd for C24H20Cl3N5O4S [M + H]+, 580.038; found, 580.0304.
O), 151.00 (CH, triazole), 145.56 (CH, triazole), 141.01 (C, Ar), 138.65 (C, Ar), 138.48 (C, Ar), 138.15 (C, C–Cl), 133.26 (C, C–Cl), 132.01 (C, C–Cl), 131.50 (CH, Ar), 130.41 (CH, Ar), 130.03 (2× CH, Ar), 129.44 (C, Ar), 129.26 (2× CH, Ar), 129.08 (2× CH, Ar), 127.27 (CH, Ar), 119.02 (2× CH, Ar), 76.91 (C, C–OH), 45.09 (CH2-triazole), 45.96 (CH2–NH). HPLC (method B): 99.7%, RT = 4.76 min. HRMS (ESI), m/z. calcd for C24H20Cl3N5O4S [M + H]+, 580.038; found, 580.0304.
CaCYP51 susceptibility testing
Antifungal MIC values were obtained according to the CLSI microdilution method for yeasts.28C. albicans strains CA14 and SC5314 were tested in triplicate. Cultures were diluted to 2.5 × 103 cells per mL in RPMI 1640 (Sigma), buffered with 0.165 M MOPS, pH 7.0. Antifungal compounds were dissolved in DMSO and added at a final concentration of 1% v/v DMSO. Plates were incubated at 37 °C and read at 48 h.CaCYP51 IC50
C. albicans CYP51 was overexpressed in E. coli and purified and CaCYP51 reconstitutions assays performed.10,37 The final reaction volume of 500 μL, contained; 1 μM C. albicans CYP51, 2 μM Homo sapiens cytochrome P450 reductase (CPR) (UniProtKB accession number P16435), 60 μM lanosterol, 50 μM dilaurylphosphatidylcholine, 4% w/v 2-hydroypropyl-cyclodextrin, 0.4 mg mL−1 isocitrate dehydrogenase, 25 mM trisodium isocitrate, 50 mM NaCl, 5 mM MgCl2, and 40 mM MOPS (morpholinepropanesulfonic acid, pH 7.2). Antifungal compounds were added in 2.5 μL DMSO. The reaction was started by the addition of 4 mM NADPH-tetrasodium salt. Samples were shaken for 15 min at 37 °C and sterol metabolites extracted with ethyl acetate and derivatised with BSTFA.Binding affinity (Kd)
The ligand–CaCYP51 complex was determined by non-linear regression (Levenberg–Marquardt algorithm) using a rearrangement of the Morrison equation for tight ligand binding,38 while weak ligand binding was calculated by the Michaelis–Menten equation. Curve fitting for novel azole saturation curves was performed using the computer program ProFit 6.1.12 (QuantumSoft, Zurich, Switzerland) for Mac OSX. Kd values were determined for each of the three replicate titrations per azole compound and then mean Kd values and standard deviations calculated.Preparation of S. cerevisiae strain Y2760
The HsCYP51 ORF (NP_000777.1) was codon harmonized (ch) using DNA 2.0 for expression in S. cerevisiae and synthesized by ATUM (Newark, Ca, USA). The 3′ end of the ORF was extended to encode EQKI of the ScErg11 C-terminus, followed by a GGR linker and a 6xHis tag. Strain Y2514 was prepared by transforming host strain Y1857 with a transformation cassette including the above construct together with the ScPGK transcription terminator and URA3 selection marker located downstream, bordered by upstream and downstream parts of ScPDR5 gene identical to sequences in ScErg11 over-expressing strain Y941. Strain Y2514 constitutively overexpressed HsCYP51ch-6xHis and URA3 from the ScPDR5 locus.Strain Y2757 (Y2514, PDR15::HsCPRch-6xHis-LoxP-HIS1-LoxP) was prepared by co-expressing the cognate reductase HsCRP in strain Y2514. Codon harmonisation and synthesis of human NADPH-cytochrome P450 reductase isoform 2 (HsCPRch, NP_001369584.1) ORF was performed by ATUM. The gene was 3′ extended with a sequence encoding GGR-6xHIS. A transformation cassette was prepared by recombinant PCR to include the HsCPRch ORF with 657 nucleotides the ScPDR5 promoter upstream, and the ScPGK terminator followed by the LoxP flanked ScHIS1 selection marker downstream. ScPDR15 specific arms bordering the construct enabled the cassette to be integrated between the nucleotide 441 of the 5′ UTR and nucleotide 1 of the 3′ UTR in PDR15.
Strain Y2758 (Y2757 ΔHIS1) was made using the following steps. Strain Y2757 was transformed with the pSH69 pGAL1-cre plasmid (Euroscarf, Germany) via selection on hygromycin (75 μg mL−1) containing YPD plates. A single hygromycin positive colony was inoculated in 1% yeast extract, 2% peptone and 2% of galactose broth and incubated for 16 hours at 30 °C with shaking. The diluted culture was then plated on YPD agar and replica-plated on YPD + hygromycin and SD-HIS dropout agar plates. Deletion of the HIS selection marker downstream of HsCPRch-6xHis in the PDR15 locus was confirmed in double His and hygromycin negative isolates by PCR and DNA sequence analysis.
The endogenous ScERG11 ORF in strain Y2758 was removed by transforming with a LoxP- ScHIS1 containing cassette with arms complementing from +387 nucleotides in the 5′UTR and −60 nucleotides in the 3′UTR of the ScERG11 locus, resulting in strain Y2760 (ADΔΔ PDR5::HsCYP51ch-6×HIS-URA3; PDR15::HsCPRch6×HIS-LoxP, ΔERG11-LoxP-HIS1-LoxP).
Antifungal susceptibility testing of recombinant S. cerevisiae strains expressing CYP51
Minimum inhibitory concentration (MIC) were determined according to recommendations outlined in the Clinical and Laboratory Standards Institute (CLSI) document M27-Ed4E39 with modifications. Sabouraud dextrose (SD) medium (pH 6.8) was used instead of RPMI-1640 for inoculum and growth of the S. cerevisiae strains and cultures were incubated at 30 °C. Assays were performed in round-bottomed 96-well plates (Corning, NY, US) and MICs were measured as the lowest concentrations of each antifungal agent that resulted in 80% inhibition of growth as compared with a drug-free, positive control. Stock solutions of each agent were prepared in DMSO and the final concentration of DMSO was 1% (v/v) in SD. The final concentration of fluconazole ranged from 0.25 to 128 μg mL−1. The final concentration of all other compounds ranged from 0.03–16 μg mL−1. Three biological repeats were performed.Computational studies
CaCYP51 protein–ligand complexes of all final compounds (R- and S-enantiomers) were obtained with Molecular Operating Environment (MOE) software33 using the crystal structure of wild-type CaCYP51 (PDB 5FSA).34 Minimisations were performed with the MMFF94 force field (ligands) and partial charges were automatically calculated. The charge of the haem iron at physiological pH was set to 3+ (geometry d2sp3) through the atom manager in MOE. Results were refined using the MMFF94 force field and scored by applying the London ΔG scoring function. The output database dock file was created with different poses for each ligand and arranged according to the final S-score function, which is the score of the last stage that was not set to zero. Molecular dynamics (MD) simulations were performed on CaCYP51-ligand complexes, which were optimised with the protein preparation wizard in Maestro (Schrödinger release 2020-1),35 by assigning bond orders, adding hydrogens, and correcting incorrect bond types. A default quick relaxation protocol was used to minimise the MD systems with the Desmond programme.36 The orthorhombic water box allowed for a 10 Å buffer region between protein atoms and box sides. Overlapping water molecules were deleted, and the systems were neutralised with Na+ ions and a salt concentration of 0.15 M. Force-field parameter assignment used the OPLS_2005 force field, for a 200 ns molecular dynamic run in the NPT ensemble (T = 300 K) at a constant pressure of 1 bar. Energy and trajectory atomic coordinate data were recorded at 1.2 ns intervals.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
MA and FAB performed the chemical synthesis and ET performed computational studies supervised by CS. AGW performed the binding affinity studies supervised by DEK and SLK. JEP performed the MIC and IC50 assays and BCM and MVK contributed to the S. cerevisiae model assays. All authors contributed to manuscript preparation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Thanks to Taibah University (M. A.) and King Saud University (F. A. B.) and the Royal Embassy of Saudi Arabia for full PhD scholarships. We are grateful to the European Union for support through the European Regional Development Fund (ERDF) as part of the Welsh Government funded BEACON project (Swansea University). Recombinant S. cerevisiae strains were prepared with the support of the Health Research Council of New Zealand. Molecular dynamics simulations were undertaken using the supercomputing facilities at Cardiff University operated by Advanced Research Computing at Cardiff (ARCCA) on behalf of the Cardiff Supercomputing Facility and the HPC Wales and Supercomputing Wales (SCW) projects. We acknowledge support of the latter, which is part-funded by the European Regional Development Fund (ERDF) via the Welsh Government.Notes and references
- F. L. Mayer, D. Wilson and B. Hube, Virulence, 2013, 4, 119–128 CrossRef PubMed.
- A. Mohamed, T. R. Rogers and A. F. Talento, J. Fungi, 2020, 6, 1–14 Search PubMed.
- F. Bongomin, S. Gago, R. Oladele and D. Denning, J. Fungi, 2017, 3, 1–29 CrossRef.
- Fungal diseases and COVID-19 | CDC, https://www.cdc.gov/fungal/covid-fungal.html.
- M. Nucci, G. Barreiros, L. F. Guimarães, V. A. S. Deriquehem, A. C. Castiñeiras and S. A. Nouér, Mycoses, 2021, 64, 152–156 CrossRef CAS.
- B. Posteraro, R. Torelli, A. Vella, P. M. Leone, G. De Angelis, E. De Carolis, G. Ventura, M. Sanguinetti and M. Fantoni, J. Fungi, 2020, 6, 1–11 Search PubMed.
- Q.-Z. Lv, L. Yan and Y.-Y. Jiang, Virulence, 2016, 7, 649–659 CAS.
- L. E. Cowen, D. Sanglard, S. J. Howard, P. D. Rogers and D. S. Perlin, Cold Spring Harbor Perspect. Med., 2015, 5, a019752 CrossRef PubMed.
- J. E. Parker, A. G. Warrilow, C. L. Price, J. G. L. Mullins, D. E. Kelly and S. L. Kelly, J. Chem. Biol., 2014, 7, 143–161 CrossRef PubMed.
- A. G. Warrilow, A. T. Nishimoto, J. E. Parker, C. L. Price, S. A. Flowers, D. E. Kelly, P. D. Rogers and S. L. Kelly, Antimicrob. Agents Chemother., 2019, 63, 1–16 CrossRef PubMed.
- P. Marichel, L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. S. Ramaekers and F. C. Odds, Microbiology, 1999, 145, 2701–2713 CrossRef PubMed.
- F. Morio, C. Loge, B. Besse, C. Hennequin and P. Le Pape, Diagn. Microbiol. Infect. Dis., 2010, 66, 373–384 CrossRef CAS PubMed.
- S. Perea, J. L. López-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillán, M. Martínez, D. Calabrese, D. Sanglard and T. F. Patterson, Antimicrob. Agents Chemother., 2001, 45, 2676–2684 CrossRef CAS PubMed.
- D. Sanglard, F. Ischer, L. Koymans and J. Bille, Antimicrob. Agents Chemother., 1998, 42, 241–253 CrossRef CAS PubMed.
- S. A. Flowers, B. Colón, S. G. Whaley, M. A. Schuler and P. D. Rogers, Antimicrob. Agents Chemother., 2015, 59, 450–460 Search PubMed.
- A. A. Sagatova, M. V. Keniya, R. K. Wilson, M. Sabherwal, J. D. A. Tyndall and B. C. Monk, Sci. Rep., 2016, 6, 26213 CAS.
- F. A. Binjubair, J. E. Parker, A. G. Warrilow, K. Puri, P. J. Braidley, E. Tatar, S. L. Kelly, D. E. Kelly and C. Simons, ChemMedChem, 2020, 15, 1294–1309 CAS.
- A. G. S. Warrilow, J. G. L. Mullins, C. M. Hull, J. E. Parker, D. C. Lamb, D. E. Kelly and S. L. Kelly, Antimicrob. Agents Chemother., 2012, 56, 2099–2107 CrossRef CAS PubMed.
- R. Ben-Ami, J. Fungi, 2018, 4, 97 CAS.
- T. J. Gintjee, M. A. Donnelley and G. R. Thompson, J. Fungi, 2020, 6, 28 CrossRef CAS PubMed.
- A. Tiwari, S. Thakur, R. Verma, R. Kumar, N. Khurana, C. Kaur and G. Singh, Curr. Top. Med. Chem., 2024, 24, 2288–2313 CrossRef PubMed.
- R. Elias, P. Basu and M. Fridman, Fluconazole-COX inhibitor hybrids: as dual-acting class of antifungal azoles, J. Med. Chem., 2022, 65, 2361–2373 CrossRef CAS PubMed.
- S. M. Hoy, Drugs, 2022, 82, 1017–1023 CrossRef CAS PubMed.
- B. C. Monk, M. V. Keniya, M. Sabherwal, R. K. Wilson, D. O. Graham, H. F. Hassan, D. Chen and J. D. A. Tyndall, Antimicrob. Agents Chemother., 2018, 63, e02114-18 Search PubMed.
- K. Sekimata, S.-Y. Han, K. Yoneyama, Y. Takeuchi, S. Yoshida and T. A. Asami, J. Agric. Food Chem., 2002, 50, 3486–3490 CAS.
- Y. Wang, G. L. V. Damu, J.-S. Lv, R.-X. Geng, D.-C. Yang and C.-H. Zhou, Bioorg. Med. Chem. Lett., 2012, 22, 5363–5366 CAS.
- V. S. Pore, S. G. Agalave, P. Singh, P. K. Shukla, V. Kumar and M. I. Siddiqi, Org. Biomol. Chem., 2015, 13, 6551–6561 RSC.
- Clinical and Laboratory Standards Institute, CLSI document M27-S4, 2012, Wayne, PA Search PubMed.
- A. K. Ghose and G. M. Crippen, J. Chem. Inf. Comput. Sci., 1987, 27, 21–35 CAS.
- A. G. S. Warrilow, C. M. Martel, J. E. Parker, N. Melo, D. C. Lamb, W. D. Nes, D. E. Kelly and S. L. Kelly, Antimicrob. Agents Chemother., 2010, 54, 4235–4245 CAS.
- E. Lamping, B. C. Monk, K. Niimi, A. R. Holmes, S. Tsao, K. Tanabe, M. Niimi, Y. Uehara and R. D. Cannon, Eukaryotic Cell, 2007, 6, 1150–1165 CAS.
- A. R. Holmes, S. Tsao, S. W. Ong, E. Lamping, K. Niimi, B. C. Monk, M. Niimi, A. Kaneko, B. R. Holland, J. Schmid and R. D. Cannon, Mol. Microbiol., 2006, 62, 170–186 CAS.
- Molecular Operating Environment (MOE) CCI, 2022, https://www.chemcomp.com/Products.htm Search PubMed.
- T. Y. Hargrove, L. Friggeri, Z. Wawrzak, A. Qi, W. J. Hoekstra, R. J. Schotzinger, J. D. York, F. P. Guengerich and G. I. Lepesheva, J. Biol. Chem., 2017, 292, 672–6743 Search PubMed.
- Schrödinger Release 2020–1: Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2020, https://www.schrodinger.com/products/desmond Search PubMed.
- K. J. Bowers, E. Chow, H. Xu, R. O. Dror, M. P. Eastwood, B. A. Gregersen, J. L. Klepeis, I. Kolossvary, M. A. Moraes, F. D. Sacerdoti, K. K. Salmon, Y. Shan and D. E. Shaw, Proceedings of the ACM/IEEE Conference on Supercomputing (SC06), Tampa, Florida, 2006, DOI:10.1145/1188455.
- G. I. Lepesheva, R. D. Ott, T. Y. Hargrove, Y. Y. Kleshchenko, I. Schuster, W. D. Nes, G. C. Hill, F. Villalta and M. R. Waterman, Chem. Biol., 2007, 14, 1283–1293 CAS.
- J. D. Lutz, V. Dixit, C. K. Yeung, L. J. Dickmann, A. Zelter, J. A. Thatcher, W. L. Nelson and N. Isoherranen, Biochem. Pharmacol., 2009, 77, 258–268 CAS.
- Clinical and Laboratory Standards Institute, Reference method for broth dilution antifungal susceptibility testing of yeasts; fourth informational supplement, CLSI document M27-Ed4E, 2017, Wayne, PA Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental methods, analytical and computational data. See DOI: https://doi.org/10.1039/d4md00863d |
| ‡ Present address: College of Pharmacy, Department of Pharmacognosy & Pharmaceutical Chemistry, Taibah University, Medina, 42353, Saudi Arabia. |
| § Present address: College of Pharmacy, Department of Pharmaceutical Chemistry, King Saud University, P.O. Box 2457, Riyadh 11451, Saudi Arabia. |
| ¶ Present address: Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ 07110, USA. |
| This journal is © The Royal Society of Chemistry 2025 |



