DOI:
10.1039/D5MA00164A
(Paper)
Mater. Adv., 2025,
6, 5701-5712
Fabrication and catalytic efficiency of functionalized bacterial cellulose nanofibers for the reduction of 4-nitrophenol and methylene blue
Received
20th February 2025
, Accepted 27th June 2025
First published on 8th July 2025
Abstract
Bacterial cellulose nanofibers (BCNFs) produced from freeze-dried bacterial cellulose (BC) exhibit unique properties such as low cost, flexibility, environmental friendliness, and structural stability. In this study, a novel catalyst, BCNF-CPTMS-Gu-Cu(II), was successfully fabricated by modifying BCNFs with 3-(chloropropyl)-trimethoxysilane (CPTMS) and guanidine hydrochloride (Gu) to create ligand-functionalized BCNFs, followed by the stabilization of copper(II) nanoparticles on guanidine groups. This synthesis process maintained the original morphology of BCNFs. The BCNF-CPTMS-Gu-Cu(II) catalyst demonstrated excellent catalytic activity for the rapid reduction of 4-nitrophenol (4-NP) and methylene blue (MB) in the presence of sodium borohydride (NaBH4). For the reduction of 4-NP to 4-aminophenol (4-AP), the catalytic activity of BCNF-CPTMS-Gu-Cu(II) was optimized by increasing NaBH4 concentration and reaction temperature. It was observed that higher temperatures positively affected the reaction rate. Similarly, for the reduction of MB to leuco-methylene blue (LMB), the catalytic efficiency was enhanced by increasing the NaBH4 concentration and optimizing the pH, with the best catalytic performance achieved at pH 6. The reduced products, 4-AP and LMB, are highly soluble in water and have potential utility in various applications. This study introduces an effective and low-cost catalyst based on bacterial cellulose nanofibers, with promising applications in industrial processes.
1. Introduction
Today, the increasing contamination of water sources substantially threatens ecosystems and human health.1,2 Concurrently, the wastewater industry is transforming owing to the emergence of pollutants such as pharmaceuticals, antibiotics, pesticides, heavy metals, toxic dyes, and organic compounds.3–5 Recent reports highlight the remediation of toxic dyes and organic compounds in water.6–10 4-NP is an organic phenolic compound known to be released in the aquatic environment through various industries and chemical processing.11–14 Toxicological tests have indicated the presence of 4-NP in the effluents of commercial coloring industries, which can cause nephrotoxic impacts, serious sensitivities, dermatitis, shaking, and cerebral pains, and it is just as harmful to the kidneys, liver, and nervous system.15–17 However, some harmful dye pollutants include azo dyes such as MB, which can release carcinogenic aromatic amines.18–20 Hence, the development of methods such as designing conversion catalysts and adsorbents as well as photocatalytic degradation is a valuable strategy for converting dangerous materials into harmless compounds in wastewater.10,21–26 Catalytic conversion is the most preferred method to reduce wastewater pollution and can convert harmful materials to less harmful compounds with high efficiency.16,27,28 Therefore, finding more green and sustainable materials with multifunctional applications is imperative.29,30 Nowadays, the utilization of natural resources as engineering materials has aroused great interest owing to their intrinsic advantages, including environmental friendliness and inexpensiveness.31,32 Cellulose nanofibers (CNFs) represent a remarkable and emerging class of nature-derived nanomaterials distinguished by their extraordinary mechanical properties.33 CNFs are web-like structures made of flexible, entangled fibril strands with diameters of 10–50 nm and lengths of a few micrometers, which are influenced by the cellulose source and extraction methods.33 Bacterial cellulose (BC) is one of the most promising polysaccharides with a microbial origin and a highly hydrated natural polymer cellulose pellicle derived from the extracellular metabolic products of certain bacteria.34 It exhibits a superb 3D microfibril structure with notable advantages, including high purity, excellent mechanical properties (flexibility and tensile strength), high porosity, and significant water absorption capability.25,35–40 In addition, BC possesses hydroxyl functional groups, which facilitate the formation of coordination and hydrogen bonds with other metal nanoparticles that are used as catalysts in catalytic hydrogenation, catalytic hydrodesulfurization, etc.41 It has been reported as an excellent and strong support for catalysts in the article published by Liu et al., who successfully prepared AuNPs/AOBC from amidoxime-surface-functionalized BC and an aqueous HAuCl4 solution and used it to catalyze the reduction of 4-NP.42 Because of their favorable properties, transition metals such as Cu43 and Ni13,44 have recently gained attention owing to their high catalytic activity and excellent selectivity on catalyst surfaces.45 Aditya, A. Pal, and T. Pal reviewed the NaBH4-mediated reduction of nitroarenes, particularly from 4-nitrophenol to 4-aminophenol, as a standard test for nanoparticle catalysts. They examined various metal and metal oxide nanocatalysts, discussed synthesis methods, and analyzed catalytic performance. Their review highlighted real-time UV-visible monitoring and kinetic models such as Langmuir–Hinshelwood and Eley–Rideal, confirming the reliability and widespread use of the reaction in catalyst evaluation.46 Two agarose (AG) materials, one coated with Ni nanoparticles and the other with Cu nanoparticles, were synthesized using polyurethane by Ali et al. Moreover, 20 mg of either catalyst was able to reduce 0.13 mM of 4-NP and 0.08 mM of MB in the presence of 0.2 M NaBH4 within more than 5 minutes.47 Copper oxide nanoparticles synthesized derived from apple peels and copper sulfate via microwave methods by Rajamohan et al. catalyzed the reduction of methylene blue with 80% efficiency within 6 min.48 Ni@luffa was produced from luffa fibers and NiCl2 by Eisa et al. and was shown to reduce the concentration of 0.08 mM 4-NP using 12 mM NaBH4 within 160 s.49 These mentioned catalysts have some problems such as being harmful to the environment and requiring a long time to reduce 4-NP and MB. Because NaBH4 is toxic and carcinogenic for humans, its use is limited. One of the problems of the mentioned studies is that low concentrations of 4-NP and MB require a large quantity of the catalyst and a high concentration of NaBH4 and greater time to reduce.
In this research article, we designed a novel catalytic material based on bacterial cellulose nanofibers (BCNFs). It was obtained by first freeze-drying bacterial cellulose (BC) and subsequently freezing it with liquid nitrogen to enhance its stability, flexibility, and lightweight properties. The BCNF was chosen as the support material owing to its environmental friendliness. Fig. 1 shows that the BCNF maintains its structural integrity after folding and rolling, demonstrating its lightweight, flexible, and superior mechanical properties. A sample of this material was prepared as a low-cost, highly active catalyst for the reduction of 4-NP and MB. The catalyst, which was functionalized with guanidine hydrochloride (Gu) and copper acetate, efficiently converted 4-NP and MB to 4-AP and LMB, respectively, in a short period with high efficiency, as confirmed by UV-visible spectroscopy. One of the main advantages of this catalyst is its easy separation from the reaction solution, making it an ideal candidate for applications requiring recyclability. The composite properties of the BCNF were characterized by various analytical techniques, including X-ray diffraction (XRD), field-emission scanning electron microscopy (FE-SEM), Fourier transform infrared (FT-IR) spectroscopy, energy-dispersive X-ray spectroscopy (EDS), inductively coupled plasma (ICP) spectroscopy, and zeta potential (ζ) measurements. The study also demonstrated that altering the pH of the reaction medium could significantly enhance catalyst activity.
 |
| | Fig. 1 Images of the flexible and lightweight features of the BCNF. | |
2. Experimental
2.1. Materials
Kombucha, tea, and sugar were procured from a local confectionery in Tehran Province, Iran. 4-Nitrophenol (4-NP) (99%, Sigma-Aldrich), methylene blue (MB) (99%, Sigma-Aldrich), sodium borohydride (NaBH4) (>98%, Sigma-Aldrich), sodium hydroxide (NaOH) (96%, Sigma-Aldrich), 3-(chloropropyl)-trimethoxysilane (CPTMS) (≥97%, Sigma-Aldrich), toluene distilled for 48 h before using (99.5%, Sigma-Aldrich), guanidine hydrochloride (Gu) (≥99.5%, Sigma-Aldrich), sodium carbonate (Na2CO3) (99.999%, Sigma-Aldrich), potassium iodide (KI) (99.8–99.9%, Sigma-Aldrich), ethanol for washing (70% solution, Sigma-Aldrich), methanol (CH3OH) (99.85%, Sigma-Aldrich), and a salt of copper(II) acetate hydrate [Cu(OAc)2·H2O] (+99%, Sigma-Aldrich) were used.
2.2. Preparation of BC
To grow bacterial cellulose (BC), bacteria from tea and sugar are required. For this process, 10 g of Iranian tea was brewed in a teapot, and the strained tea was added to 3000 mL of boiled water in a 5000-mL beaker. Then, 180 g of sugar was dissolved in this solution and stirred for 1 hour. Once the solution reached ambient temperature, 0.2 g of a Kombucha culture was added. Then, the beaker was covered with a clean cloth and placed in a dark, clean, and dry location. After 30 days, 10 g of bacterial cellulose (BC) was formed and ready for use.50
2.3. Preparation of the BCNF
The BCNF is an excellent support for catalysts and possesses amazing stability and flexibility. Ten grams of grown bacterial cellulose were cut into small pieces and washed with deionized water several times. Then, the pieces of BC along with 1000 mL of a NaOH solution (0.25 mol L−1) were placed in a 2000-mL Erlenmeyer flask and stirred using a shaker for 4 h. The BC washed with NaOH was soaked with distilled water several times to reach a neutral pH.31 The washed BC, after freezing in liquid nitrogen at −96 °C, was put into a freezer for 24 h and then freeze-dried at −70 °C for 48 h to prepare 3 g of the BCNF.
2.4. Preparation of BCNF-CPTMS
The bond between the hydroxyl functional group of the BCNF and guanidine ligand is a non-covalent bond of the hydrogen bond type; this bond is not suitable for catalytic reactions. Therefore, a linker such as CPTMS was used to establish strong covalent bonds between BCNF and CPTMS as well as CPTMS and guanidine. In a 50-mL round-bottom flask, 0.5 g of the BCNF was added to 20 mL of dry toluene. Then, 2.5 mL of 3-(chloropropyl)-trimethoxysilane (CPTMS), drawn using a 3-mL syringe, was added dropwise to the reaction solution and refluxed at 110 °C for 24 h in a nitrogen atmosphere. Finally, the as-prepared sample of the BCNF was washed with dry toluene twice and dried at 80 °C for 12 h.
2.5. Preparation of BCNF-CPTMS-Gu
A ligand such as guanidine was used to coordinate the copper metal. In a 50-mL round-bottom flask, salts including 0.75 g of guanidine hydrochloride, 0.17 g of Na2CO3, and 0.2 g of KI in 15 mL of a water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH3OH solvent (1
CH3OH solvent (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, volume ratio) were mixed. Then, 0.5 g of BCNF-CPTMS was dispersed in the as-prepared solution and stirred at room temperature in a nitrogen atmosphere for 24 h. Finally, the semi-finished product was washed with water and EtOH twice and dried at 80 °C for 12 h.
1, volume ratio) were mixed. Then, 0.5 g of BCNF-CPTMS was dispersed in the as-prepared solution and stirred at room temperature in a nitrogen atmosphere for 24 h. Finally, the semi-finished product was washed with water and EtOH twice and dried at 80 °C for 12 h.
2.6. Preparation of BCNF-CPTMS-Gu-Cu(II)
A metal such as copper was used to carry out the catalytic reaction. First, 0.5 g of BCNF-CPTMS-Gu was dissolved in a 50-mL round-bottom flask with 10 mL of distilled water. Then, 0.5 g of Cu(OAc)2·H2O was added and stirred at room temperature for 24 h. Finally, the product was washed with water and EtOH three times and dried at 80 °C for 12 h (Fig. 2).
 |
| | Fig. 2 Synthesis of the BCNF-CPTMS-Gu-Cu(II) catalyst. | |
2.7. Catalytic activity of BCNF-CPTMS-Gu-Cu(II)
The reduction of 4-NP and MB by NaBH4 was used to evaluate the catalytic activity of BCNF-CPTMS-Gu-Cu(II). In a 50-mL round-bottom flask, 10 mL of a freshly prepared NaBH4 solution (0.1 M) was added to 10 mL of a 4-NP solution (0.7 mM), which turned the light yellow color of the 4-NP solution into a deep yellow, indicating the formation of 4-nitrophenolate. Under stirring at room temperature (25 °C), 0.01 g of the BCNF-CPTMS-Gu-Cu(II) catalyst was added to the 4-nitrophenolate solution until it became colorless. The progress of the reaction was monitored by recording the UV-visible spectrum at 80-second intervals. The absorption peak of 4-NP appeared at 317 nm, and after the addition of NaBH4, a new peak corresponding to 4-nitrophenolate was observed at 400 nm. The appearance of a peak at 296 nm indicated the reduction of 4-NP. The rate of catalytic activity was affected by the concentrations of NaBH4 and 4-NP as well as the temperature of the reaction system. In another experiment, in a 50-mL round-bottom flask, 20 mL of an aqueous solution adjusted to pH = 6 was prepared by mixing 10 mL of a freshly prepared NaBH4 solution (0.05 M) and 10 mL of an MB solution (2.2 mM). Then, 0.01 g of the BCNF-CPTMS-Gu-Cu(II) catalyst was added to this blue solution and stirred at room temperature (25 °C) until it became colorless. The reduction process was monitored by recording the UV-visible spectrum during the 46-second reaction. The appearance and disappearance of the absorption peak at 663 nm indicated the presence of MB and its reduced form (i.e., LMB), respectively. The effects of NaBH4 and MB concentrations as well as the pH of the reaction system were also examined for their influence on catalytic activity.
2.8. Characterization and measurements
Changes in the reduction of organic pollutants were measured using a UV-visible spectrophotometer (UV-2550, Shimadzu Company). The pH of various solutions was adjusted using a Sat 401 automatic pH meter. FT-IR, FE-SEM, XRD, EDS, and ICP analyses confirmed the structural and chemical properties of BCNF-CPTMS-Gu-Cu(II). The FT-IR spectrum (Spectrum RX I, PerkinElmer) was used to identify the functional groups in the material. Field-emission scanning electron microscopy (FE-SEM) images obtained using a ZEISS Sigma 300 microscope revealed the particle size and surface morphology of BCNF-CPTMS-Gu-Cu(II). X-ray diffraction (XRD) analysis (D8 Advance) was used to determine the structural and chemical properties of the synthesized materials, and energy-dispersive X-ray spectroscopy (EDS) was performed using a MIRA3 system. The amount of coordinated copper was determined by inductively coupled plasma (ICP) mass spectrometry.
3. Results and discussion
3.1. Fourier transform infrared (FT-IR) spectroscopy
FT-IR spectra were studied to characterize the functional groups of the BCNF, BCNF-CPTMS, and BCNF-CPTMS-Gu. The pure bacterial cellulose nanofiber, having a hydroxyl functional group (O–H), has a broad absorption in the region of 3200–3650 cm−1, which indicates the stretching vibration of the O–H bond,51,52 and the peak at 1654 cm−1 is related to the bending vibrations of this bond.52,53 The absorption bands caused by the symmetric and asymmetric stretching vibrations of C–H can be seen at 2920 cm−1 and those for CH2 at 2852 cm−1.54 A band is observed at 1118 cm−1, which is related to the ether functional group C–O–C of bacterial cellulose.54 The bacterial cellulose nanofiber substrate, owing to its strong gas absorption properties, absorbs CO2 gas, and the absorption band related to the asymmetric stretching of this gas can be seen at 2370 cm−1.55 To confirm the synthesis of BCNF-CPTMS, the spectral signals related to the bonds present in the CPTMS structure should be visible. The absorption band near 800 cm−1 indicates the stretching vibration of the C–Cl bond, which is observed at 798 cm−1 for this composite.56 The 1032 cm−1 and 1170 cm−1 peaks of the stretching vibrations of Si–O–Si and Si–O–C bonds, respectively, were visible.56 The presence of the broad absorption band from 3100 cm−1 to 3600 cm−1 signifies the overlapping bands of symmetric and asymmetric stretching vibrations of the O–H and H–N–H bonds of the BCNF-CPTMS-Gu composite at the same wavelength.57 The presence of an absorption spectral peak at 1114 cm−1 from the stretching vibrations of the aliphatic C–N bond related to the molecular structure of guanidine indicates that bacterial cellulose nanofibers are functionalized.58 In addition, for more certainty, the wagging-bending vibration of guanidine N–C–N–H bonds is observed at 618 cm−1.57 The 1638 cm−1 peak also shows the scissor-bending vibration of H–N–C bonds (Fig. 3).57
 |
| | Fig. 3 FT-IR spectra of each step in the synthesis of BCNF-CPTMS-Gu. | |
3.2. Crystal characterization
X-ray diffraction (XRD) analysis was used to investigate the crystallographic nature of synthetic nanoparticles. Bacterial cellulose nanofibers and their composites are semi-crystalline polymers, and this crystallinity mainly results from the alignment and accumulation of linear chains within the polymer structure. Several sharp peaks in the X-ray diffraction pattern indicate the presence of crystalline regions, reflecting a high degree of crystallinity. The observed peaks at 22.5°, 16.59° and 14.3° correspond to the (200), (110), and (1![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 0) planes of bacterial cellulose nanofibers, respectively.42 The decrease in the intensity of the peaks of the synthesized composite indicates the reduced crystal structure of bacterial cellulose, indicating the successful formation of the nanofiber composite. In addition, the broad peak at 9.9° shows that the guanidine component is amorphous (Fig. 4).59
0) planes of bacterial cellulose nanofibers, respectively.42 The decrease in the intensity of the peaks of the synthesized composite indicates the reduced crystal structure of bacterial cellulose, indicating the successful formation of the nanofiber composite. In addition, the broad peak at 9.9° shows that the guanidine component is amorphous (Fig. 4).59
 |
| | Fig. 4 XRD patterns of the BCNF and BCNF-CPTMS-Gu-Cu(II). | |
3.3. Elemental composition
Energy-dispersive X-ray spectroscopy (EDS) is an analytical technique used for the structural analysis and determination of the chemical composition of a sample. EDS was employed to confirm the elemental composition and successful formation of the BCNF-CPTMS-Gu-Cu(II) composite. The presence of silicon (Si) and chlorine (Cl) elements indicates that the CPTMS linker is successfully attached to bacterial cellulose. The detection of nitrogen (N) confirms the functionalization of the substrate with guanidine. Finally, the presence of copper (Cu) confirms the coordination of copper ions by guanidine (Fig. 5).
 |
| | Fig. 5 EDS analysis results of BCNF-CPTMS-Gu-Cu(II). | |
3.4. FE-SEM micrographs
Scanning electron microscopy (SEM) is one of the most effective techniques for studying the surface morphology of synthesized materials. Field-emission SEM (FE-SEM) was used to investigate the morphology of the bacterial cellulose nanofiber (BCNF) composite. Fig. 6a shows the surface of pure BCNFs, with fiber diameters measuring less than 100 nm. Fig. 6b presents the cross-sectional morphology of BCNFs, where the layered structure—an important feature contributing to the mechanical stability of the BCNF network—is evident. Fig. 6c and d display the morphology of the BCNF-CPTMS-Gu-Cu(II) composite, confirming that the fibrous structure of BCNFs is preserved after functionalization. These images also demonstrate the successful synthesis of the BCNF-based composite. Overall, the SEM images in Fig. 6 highlight the structural integrity and robustness of BCNFs, supporting their suitability as an effective substrate for catalytic applications.
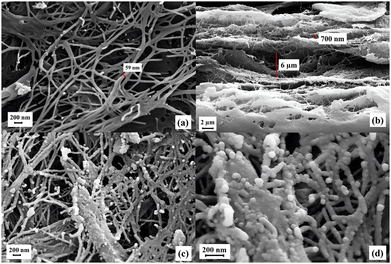 |
| | Fig. 6 FE-SEM images of the (a) surface and (b) cross section of BCNFs and (c) and (d) BCNF-CPTMS-Gu-Cu(II). | |
3.5. Inductively coupled plasma (ICP) mass spectrometry
Elements such as carbon (C), hydrogen (H), oxygen (O), nitrogen (N), and halogens cannot be detected by inductively coupled plasma (ICP) methods. Therefore, the only element in the bacterial cellulose nanofiber (BCNF) composite measured using ICP was copper (Cu). To quantify the copper content, the catalyst substrate was decomposed using a strong acid solvent. Then, the remaining copper was measured by inductively coupled plasma mass spectrometry (ICP-MS), which indicates that the composite contains 7.1% copper by weight.
3.6. Zeta potential (ζ)
Because the catalytic activity of BCNF-CPTMS-Gu-Cu(II) is dependent on pH, its behavior at different pH results in changes in the absorption and reduction processes, and using zeta potential measurements of the catalyst system, these processes between the adsorbent and adsorbate can be comprehended. It can be seen in Fig. 7 that these measurements were carried out under different conditions (pH = 3–11) on BCNF-CPTMS-Gu-Cu(II). Due to the pH-sensitive nature, pH changes influence the protonation and deprotonation of heteroatoms (N–H and O–H) existing on the catalyst surface; hence, the catalyst surface exhibits a wide range of negative, positive, and neutral charges with changing pH values. At a pH value of <6 (acidic), the catalyst shows the maximum zeta potential at pH 3, with positive charges present on the catalyst surface because the N–H group is protonated.31 As the pH increases, its zeta potential gradually decreases, reaching pH 6, where positive charges remain on the surface of the catalyst. In a highly acidic environment, the N–H group is in a protonated state, leading to the expansion of the powerful intermolecular hydrogen bond. By exceeding the pH beyond 6, the zeta potential decreases owing to the presence of O–H on the surface of the catalyst causing deprotonation and accepting negative charges.31
| Protonated state: –NH + H+ → –NH2+ |
| Deprotonated state: –OH + OH− → –O− + H2O |
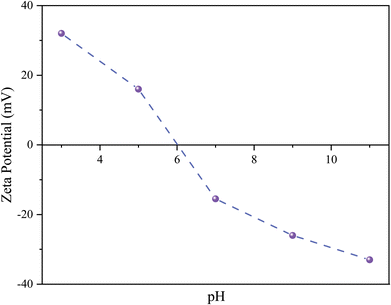 |
| | Fig. 7 The zeta potential as a function of pH for BCNF-CPTMS-Gu-Cu(II). | |
3.7. BCNF-CPTMS-Gu-Cu(II) for the catalytic reduction of 4-NP and MB
The reduction of 4-nitrophenol (4-NP) by NaBH4 was significantly accelerated upon the addition of the BCNF-CPTMS-Gu-Cu(II) catalyst. The influence of the temperature, NaBH4 and 4-NP concentrations, and catalyst amount on the reaction efficiency and rate was examined, as summarized in Table 1. Initially, the light-yellow 4-NP solution (0.7 mM) turns deep yellow after adding a fresh NaBH4 solution (0.1 M), forming 4-nitrophenolate ions. Upon the addition of 0.01 g of the catalyst at room temperature, the solution becomes colorless within 80 seconds, indicating complete reduction. Throughout the reaction, aliquots were withdrawn using an insulin syringe at 15-second intervals. After equal dilution, samples were placed in a UV-visible spectrophotometer cell, and their absorbance was measured. As shown in Fig. 8a, the initial absorbance peak of 4-NP appears at 317 nm. After the addition of NaBH4, a new peak corresponding to 4-nitrophenolate emerges at 400 nm, which gradually decreases over time with catalyst addition. The appearance of a peak at 296 nm confirms the formation of the reduced product. Table 1 further demonstrates the effect of the temperature: the reduction of a more-concentrated 4-NP solution (2.1 mM) at room temperature requires 296 seconds to reach 40% efficiency. However, when the reaction is conducted at 65 °C, 95% reduction is achieved within just 67 seconds. This increase in the reaction rate with the temperature is consistent with the Arrhenius equation (eqn (1)), which predicts that higher temperatures enhance the reaction kinetics and overall efficiency.60| |  | (1) |
In the Arrhenius equation, T represents the reaction temperature, R is the universal gas constant, Ea is the activation energy of the reaction, and A is the Arrhenius constant. As shown in Fig. 8b, the plot of ln(At/A0) versus time was used to analyze the reduction of 4-NP at concentrations of 0.35 mM, 0.47 mM, and 0.7 mM, each reduced using a 0.1 M NaBH4 solution. A0 and At are the initial and final absorption intensities of 4-nitrophenolate at a wavelength of 400 nm, respectively. The kapp is a reaction rate constant obtained from eqn (2).61| |  | (2) |
In eqn (2), t represents the reaction time. A higher value of kapp indicates a faster reaction rate. In this study, it was observed that at lower concentrations of 4-NP, the reduction reaction occurred more rapidly and within a shorter time.
Table 1 Optimization of the conditions for the reduction reaction of 4-NP
|
|
Mass of the catalyst (g) |
Concentration of NaBH4 solution (M) |
Concentration of 4-NP solution (mM) |
Temperature (°C) |
Time (s) |
Yield (%) |
| Reaction conditions: 4-NP (10 mL, 0.7 mM), catalyst (0.01 g), NaBH4 (10 mL, 0.1 M) and room temperature. |
| 1 |
0.01 |
0.1 |
0.35 |
25 |
25 |
95 |
| 2 |
0.01 |
0.1 |
0.47 |
25 |
45 |
95 |
|
3
|
0.01
|
0.1
|
0.7
|
25
|
80
|
95
|
| 4 |
0.01 |
0.1 |
0.875 |
25 |
165 |
90 |
| 5 |
0.01 |
0.1 |
1.05 |
25 |
290 |
90 |
| 6 |
0.01 |
0.1 |
1.4 |
25 |
296 |
60 |
| 7 |
0.01 |
0.1 |
2.1 |
25 |
296 |
40 |
| 8 |
0.01 |
0.05 |
0.7 |
25 |
460 |
88 |
| 9 |
0.01 |
0.15 |
0.7 |
25 |
46 |
95 |
| 10 |
0.01 |
0.1 |
0.7 |
45 |
40 |
95 |
| 11 |
0.01 |
0.1 |
0.7 |
55 |
26 |
95 |
| 12 |
0.01 |
0.1 |
2.1 |
45 |
180 |
57 |
| 13 |
0.01 |
0.1 |
2.1 |
55 |
98 |
83 |
| 14 |
0.01 |
0.1 |
2.1 |
65 |
67 |
95 |
| 15 |
0.005 |
0.1 |
0.7 |
25 |
280 |
90 |
| 16 |
0.02 |
0.1 |
0.7 |
25 |
10 |
95 |
| 17 |
0.01 |
0.15 |
0.7 |
25 |
46 |
95 |
| 18 |
0.01 |
0.05 |
0.7 |
25 |
460 |
88 |
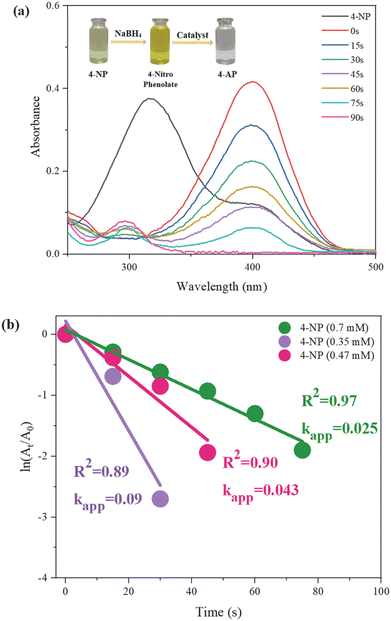 |
| | Fig. 8 (a) Time-dependent UV-visible spectra of the reduction of 4-NP (0.7 mM) and (b) plot of ln(At/A0) versus time for the reduction of different 4-NP concentrations using NaBH4 (0.1 M). The mass of BCNF-CPTMS-Gu-Cu(II) was 0.01 g. | |
A blue solution of methylene blue (MB) was reduced by a freshly prepared NaBH4 solution, resulting in a colorless solution. The catalyst played a key role in accelerating the reaction. The concentrations of MB and NaBH4, the amount of the catalyst, and the pH of the solution were optimized. According to Table 2, the reduction of a 2.2 mM MB solution using 0.05 M fresh NaBH4 and 0.01 g of the catalyst at pH = 6 and room temperature is completed within 46 seconds. The reduction process was monitored using UV-vis spectroscopy. Initially, an absorption peak is observed at 663 nm, which gradually decreases in intensity and disappears after 46 seconds, indicating the successful reduction of MB (Fig. 9a). In Fig. 9b, the plot of ln(At/A0) for the reduction of MB (2.2 mM) under optimized conditions is shown.
Table 2 Optimization of the conditions for the reduction reaction of MB
|
|
Mass of the catalyst (g) |
Concentration of NaBH4 solution (M) |
Concentration of MB solution (mM) |
Temperature (°C) |
pH |
Time (s) |
Yield (%) |
| Reaction conditions: MB (10 mL, 2.2 mM), catalyst (0.01 g), NaBH4 (10 mL, 0.05 M), pH = 6 and room temperature. |
| 1 |
0.01 |
0.05 |
0.94 |
25 |
6 |
9 |
95 |
| 2 |
0.01 |
0.05 |
1.1 |
25 |
6 |
17 |
95 |
| 3 |
0.01 |
0.05 |
1.26 |
25 |
6 |
23 |
95 |
| 4 |
0.01 |
0.05 |
1.41 |
25 |
6 |
28 |
95 |
| 5 |
0.01 |
0.05 |
1.57 |
25 |
6 |
34 |
95 |
| 6 |
0.01 |
0.05 |
1.9 |
25 |
6 |
39 |
95 |
|
7
|
0.01
|
0.05
|
2.2
|
25
|
6
|
46
|
95
|
| 8 |
0.01 |
0.05 |
2.5 |
25 |
6 |
77 |
76 |
| 9 |
0.01 |
0.025 |
2.2 |
25 |
6 |
100 |
95 |
| 10 |
0.01 |
0.10 |
2.2 |
25 |
6 |
30 |
95 |
| 11 |
0.005 |
0.05 |
2.2 |
25 |
6 |
298 |
88 |
| 12 |
0.02 |
0.05 |
2.2 |
25 |
6 |
4 |
95 |
| 13 |
0.01 |
0.05 |
2.2 |
25 |
3 |
113 |
84 |
| 14 |
0.01 |
0.05 |
2.2 |
25 |
4 |
110 |
89 |
| 15 |
0.01 |
0.05 |
2.2 |
25 |
5 |
76 |
95 |
| 16 |
0.01 |
0.05 |
2.2 |
25 |
7 |
60 |
95 |
| 17 |
0.01 |
0.05 |
2.2 |
25 |
8 |
180 |
88 |
| 18 |
0.01 |
0.05 |
2.2 |
25 |
9 |
169 |
88 |
| 19 |
0.01 |
0.05 |
2.2 |
25 |
10 |
121 |
80 |
| 20 |
0.01 |
0.05 |
2.2 |
25 |
11 |
115 |
78 |
 |
| | Fig. 9 (a) Time-dependent UV-visible spectra and (b) plot of ln(At/A0) versus time for the reduction of MB (2.2 mM) using NaBH4 (0.1 M). The mass of BCNF-CPTMS-Gu-Cu(II) was 0.01 g. | |
3.8. Mechanism of the reduction reaction on the catalyst BCNF-CPTMS-Gu-Cu(II)
The catalytic reduction process is governed by a redox cycle, in which Cu0 (E° ≈ +0.34 V)62 plays a central role as an electron mediator. Although Cu0 is initially responsible for accepting electrons from BH4− (E° ≈ −1.24 V)63 and transferring them to the target molecules, such as methylene blue (MB) or 4-nitrophenol (4-NP), it may undergo partial oxidation to Cu2+, possibly forming CuO under the reaction conditions. The standard reduction potentials (E°) indicate that MB (E° ≈ +0.011 V)64 can be readily reduced by Cu0, while 4-NP (E° ≈ −0.76 V)65 cannot. Therefore, in the presence of NaBH4, the BH4− ions act as the primary electron source, and Cu0 merely facilitates electron transfer. During this process, Cu0 is transiently oxidized and then regenerated by BH4−, completing a redox cycle; hence, the color of the catalyst changes from light blue to black, and after conversion to Cu2+, the catalyst color changes back to blue. This dynamic transition between Cu0 and Cu2+/CuO is crucial for sustaining the catalytic activity. The mechanism is schematically illustrated in Fig. 10a and b, and this redox interaction is now discussed in detail to clarify the electron transfer pathway. The catalyst recovery diagram confirms this oxidation–reduction cycle (Fig. 12a and b). NaBH4 and water act as the reducing agent and hydrogen source, respectively.66 Initially, the reaction between BH4− and water releases hydrogen anions (H−), which are adsorbed onto the catalyst surface. In the presence of NaBH4, neither 4-NP nor MB is efficiently reduced without a catalyst. The catalyst acts as a redox mediator in this reduction reaction. After dispersing the negative charge of BH4−, the copper species in the catalyst facilitate hydrogen anion (H−) transfer.67,68 Copper ions (Cu2+) initially accept the hydrogen anion (H−) and are reduced to metallic copper nanoparticles (Cu0). Then, these Cu0 species transfer the hydrogen anion (H−) to 4-NP and MB molecules and are oxidized back to Cu2+. Due to the resonance structures in the molecular frameworks of 4-NP and MB, atoms such as oxygen, nitrogen (in the nitro group) and sulfur (in the MB molecule) act as hydrogen anion (H−) acceptors. 4-Nitrophenol (4-NP) is initially reduced to 4-nitrophenolate in the presence of NaBH4. Subsequently, the oxygen and nitrogen atoms in the nitro group accept a hydride ion (H−) from the catalyst surface, leading to its partial reduction. First, one of the oxygen atoms in the nitro group receives a hydride (H−) from the catalyst surface. Then, the double bond shifts to the nitrogen atom. In the next step, the lone pair on one of the hydroxyl groups forms a double bond between oxygen and nitrogen, while the other hydroxyl group, after receiving a hydrogen atom, leaves as a water molecule. Subsequently, the nitrogen atom of the nitro group receives another hydride from the catalyst surface, and its double bond shifts to the oxygen atom. The oxygen atom, after accepting two hydrogen atoms, is released as a water molecule.69 Finally, 4-nitrophenolate is reduced to 4-aminophenol. Similarly, the sulfur atom in MB accepts hydrogen, and the molecule is reduced to leuco-methylene blue (LMB). Fig. 10a and b illustrate the proposed reduction mechanisms of 4-NP and MB over the BCNF-CPTMS-Gu-Cu(II) catalyst, respectively.
 |
| | Fig. 10 Proposed mechanism for the reduction reactions of (a) 4-NP and (b) MB performed using the novel BCNF-CPTMS-Gu-Cu(II) catalyst. | |
3.9. Effective ambient pH on the catalyst BCNF-CPTMS-Gu-Cu(II) in the reduction reaction of MB
The highest rate of the reduction reaction at pH = 6 is due to the absence of charges on the surface of the catalyst. As mentioned in the Zeta potential section, in an acidic environment (pH < 6), the protonation of the NH group in the composite leads to a positive charge on the surface of the catalyst. Consequently, the cationic dye methylene blue is repelled, resulting in a decrease in the reaction rate and efficiency compared to those under the conditions at pH = 6.31 In a basic environment (pH > 6), the deprotonation of OH groups on the substrate leads to a negative charge on the surface of the catalyst, which repels BH4− ions from the NaBH4 solution. Under more basic reaction conditions, similar to acidic environments, the rate of the reaction and its efficiency decrease compared to those under the conditions at pH = 6.70Fig. 11 shows the reduction of methylene blue at different pH values.
 |
| | Fig. 11 Effect of ambient pH on the BCNF-CPTMS-Gu-Cu(II) catalyst in the MB reduction reaction. | |
 |
| | Fig. 12 Recyclability of BCNF-CPTMS-Gu-Cu(II) in the reduction reactions of (a) 4-NP and (b) MB. (c) FT-IR analyses of fresh and used BCNF-CPTMS-Gu-Cu(II). | |
3.10. Catalyst recyclability after 4-NP and MB reduction
Catalyst recyclability is known as one of the most influential parameters in any reduction reaction system and directly affects the final cost of the process. Therefore, the performance of BCNF-CPTMS-Gu-Cu(II) in catalytic cycles was investigated. After the easy separation of composite bacterial cellulose nanofiber pieces following each cycle, the catalyst is washed several times with water and ethanol and retains >60% of its efficiency after five cycles. The main reason for the decrease in the reaction efficiency and rate after five cycles is the inactivation of copper ions following its transformation into copper nanoparticles. Another reason is that the catalyst BCNF-CPTMS-Gu-Cu(II) is a solid piece, and after recovery, it is removed with forceps, maintained, and used in the same amount for the next reaction; hence, a slightly lower yield is observed after five cycles. Fig. 12a and b show catalyst recyclability after 4-NP and MB reduction, respectively. To further ensure the stability of the functional groups of the catalyst after five cycles, FT-IR analysis was used. Fig. 12c exhibits that all peaks related to the guanidine functional group and CPTMS linker are preserved after five cycles; hence, this catalyst is stable.
3.11. Comparison of the BCNF-CPTMS-Gu-Cu(II) catalyst with previous reported catalysts for the reduction reaction
According to the structure of the BCNF, it possesses excellent specific surface area properties, and the activity of the copper metal accelerates the reduction reaction. Table 3a and b demonstrate that this catalyst provides an optimal reaction time and high efficiency for the reduction of 4-NP and MB, respectively. Additionally, it offers significant advantages, such as environmental friendliness and easy separation from the reaction solution, compared to other catalysts.
Table 3 (a) Comparison of the BCNF-CPTMS-Gu-Cu(II) catalyst with previously reported catalysts on the 4-NP reduction reaction. (b) Comparison of the BCNF-CPTMS-Gu-Cu(II) catalyst with previously reported catalysts on the MB reduction reaction
| (a) |
|
|
Catalyst |
Concentration of 4-NP solution (mM) |
Mass of catalyst (g) |
Time (s) |
k
app (s−1) |
Ref. |
| 1 |
Fe3O4/CS-Me@Pd |
0.007 |
0.012 |
300 |
0.0165 |
51
|
| 2 |
CuBDC.DMF |
0.25 |
0.05 |
960 |
0.0151 |
52
|
| 3 |
Sulfurized graphene (SG) |
0.001 |
0.001 |
3600 |
0.00048 |
71
|
| 4 |
Ag/TP |
1 |
0.01 |
360 |
0.0052 |
72
|
| 5 |
Cu-AG-sponge |
0.13 |
0.02 |
660 |
0.0054 |
47
|
| 6 |
BCNF-CPTMS-Gu-Cu(II) |
0.7 |
0.01 |
80 |
0.025 |
This work |
| (b) |
|
|
Catalyst |
Concentration of MB solution (mM) |
Mass of catalyst (g) |
Time (s) |
k
app (s−1) |
Ref. |
| 1 |
CuO@CS |
0.125 |
0.025 |
480 |
0.0027 |
55
|
| 2 |
AgNC |
0.045 |
0.01 |
720 |
0.0026 |
56
|
| 3 |
Ag/SiO2 |
0.125 |
0.01 |
180 |
0.0354 |
57
|
| 4 |
Cu-AG-sponge |
0.08 |
0.02 |
300 |
0.0096 |
47
|
| 5 |
BCNF-CPTMS-Gu-Cu(II) |
2.2 |
0.01 |
46 |
0.062 |
This work |
4. Conclusions
In this study, the catalyst BCNF-CPTMS-Gu-Cu(II) was synthesized by coordinating copper onto functionalized bacterial cellulose nanofibers (BCNFs) modified with 3-(chloropropyl)-trimethoxysilane (CPTMS) and guanidine. This catalyst was characterized and utilized for the reduction reactions of 4-nitrophenol (4-NP) and methylene blue (MB), as identified using a UV-vis spectrophotometer. Using just 0.01 g of the catalyst, the reduction of 4-NP and MB was achieved with excellent efficiency (95%) within 80 and 46 seconds, respectively.
The optimization of reaction conditions revealed that the reduction of 4-NP is fastest at an ambient temperature of 65 °C. During pH optimization, it was determined that the surface charge of the BCNF-CPTMS-Gu-Cu(II) catalyst is neutral at pH = 6. At this pH, the reduction rate of MB is the highest because, in the absence of charges on the catalyst surface, the cationic dye MB can be more easily reduced by NaBH4.
This catalyst offers several advantages, including good performance, high efficiency (95%), novelty of applications, reusability, easy separation, low cost, and environmental friendliness.
Data availability
No primary research results, software or code has been included and no new data were generated or analysed as part of this study.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors gratefully acknowledge the support of the Research Council of the Iran University of Science and Technology, Tehran, IRAN.
References
- R. Karimi, F. Yousefi, M. Ghaedi, A. Ghoorchian, K. Dashtian and G. Yasin, Chem. Eng. J., 2024, 497, 154892 CrossRef CAS.
- M. Batool and M. N. Zafar, Mater. Adv., 2025, 6(1), 224–240 RSC.
- A. Nasri, B. Jaleh, Z. Nezafat, M. Nasrollahzadeh, S. Azizian, H. W. Jang and M. Shokouhimehr, Ceram. Int., 2021, 47, 3565–3572 CrossRef CAS.
- J. Zhu, X. Zhang, Z. Qin, L. Zhang, Y. Ye, M. Cao, L. Gao and T. Jiao, Colloids Surf., A, 2021, 611, 125889 CrossRef CAS.
- A. Zahid, H. H. Nawaz, A. Siddique, B. Ahmed, S. Razzaque, X. Liu, H. Razzaq and M. Umar, Mater. Adv., 2024, 5, 9471–9487 RSC.
- A. B. Hamou, M. Enneiymy, S. Farsad, A. Amjlef, A. Chaoui, N. Nouj, A. Majdoub, A. Jada, M. Ez-zahery and N. El Alem, Mater. Adv., 2024, 5, 1960–1976 RSC.
- S. Kundu and M. K. Naskar, Mater. Adv., 2021, 2, 3600–3612 RSC.
- W. Lu, C. Duan, Y. Zhang, K. Gao, L. Dai, M. Shen, W. Wang, J. Wang and Y. Ni, Carbohydr. Polym., 2021, 258, 117676 CrossRef CAS PubMed.
- F. Ali, S. B. Khan, T. Kamal, K. A. Alamry, E. M. Bakhsh, A. M. Asiri and T. R. Sobahi, Carbohydr. Polym., 2018, 192, 217–230 CrossRef CAS PubMed.
- P. Sirajudheen, S. Vigneshwaran, N. Thomas, M. Selvaraj, K. Venkatesan and C. M. Park, Carbohydr. Polym., 2024, 335, 122071 CrossRef CAS PubMed.
- M. S. Mustafa, N. N. Mohammad, F. H. Radha, K. F. Kayani, H. O. Ghareeb and S. J. Mohammed, RSC Adv., 2024, 14, 16045–16055 RSC.
- G. Rathee, H. Chugh, S. Kohli, R. K. Gaur and R. Chandra, Mater. Adv., 2023, 4, 1656–1663 RSC.
- F. Eshrati, H. Ghafuri, P. Hanifehnejad and H. Dogari, Mater. Adv., 2025, 6(1), 278–297 RSC.
- T. Ma, W. Yang, S. Liu, H. Zhang and F. Liang, Catalysts, 2017, 7, 38 CrossRef.
- D. Sun, C. Song, X. Zhang, D. Chen, Y. Ma and W. Yang, Mater. Adv., 2023, 4, 1135–1145 RSC.
- G. Anjum, A. Kumar, G. Ramanathan and J. N. Moorthy, Mater. Adv., 2024, 5(23), 9279–9291 RSC.
- W. Shen, Y. Qu, X. Pei, S. Li, S. You, J. Wang, Z. Zhang and J. Zhou, J. Hazard. Mater., 2017, 321, 299–306 CrossRef CAS PubMed.
- S. Ahmad, X. Liu, J. Tang and S. Zhang, Chem. Eng. J., 2022, 431, 133187 CrossRef CAS.
- Z. Li, D. Liu, W. Huang, X. Wei and W. Huang, Sci. Total Environ., 2020, 721, 137764 CrossRef CAS PubMed.
- N. Aramesh and B. Yadollahi, Mater. Adv., 2024, 5(14), 5781–5793 RSC.
- Z. Sun, Y. Zhang, S. Guo, J. Shi, C. Shi, K. Qu, H. Qi, Z. Huang, V. Murugadoss and M. Huang, Adv. Compos. Hybrid Mater., 2022, 5, 1566–1581 CrossRef CAS.
- M. Cao, Y. Shen, Z. Yan, Q. Wei, T. Jiao, Y. Shen, Y. Han, Y. Wang, S. Wang and Y. Xia, Chem. Eng. J., 2021, 405, 126647 CrossRef CAS.
- K. Selvaraj, P. Palanisamy, M. Manikandan, P. B. Managutti, P. Sangeetha, S. Mohamed, R. Pamanji, J. Selvin, S. Nasiri and S. Kment, Mater. Adv., 2023, 4, 6259–6270 RSC.
- T. Ran, C. Ji, Q. Zhang, S. Wang, Y. Zhang, W. Niu, T. Wei and Y. Shi, Carbohydr. Polym., 2024, 122425 CrossRef CAS PubMed.
- S. D. Gallegos-Cerda, J. D. Hernández-Varela, J. J. C. Pérez, C. A. Huerta-Aguilar, L. G. Victoriano, B. Arredondo-Tamayo and O. R. Hernández, Carbohydr. Polym., 2024, 324, 121476 CrossRef CAS PubMed.
- S. Gazi and R. Ananthakrishnan, Appl. Catal., B, 2011, 105, 317–325 CrossRef CAS.
- D. Patra, S. R. Nalluri, H. R. Tan, M. S. Saifullah, R. Ganesan and B. Gopalan, Nanoscale Adv., 2020, 2, 5384–5395 RSC.
- S. Iniyan, A. Vijayaprabhakaran, C. Sebastian and M. Kathiresan, Mater. Adv., 2024, 5, 7006–7015 RSC.
- E. S. Beach, Z. Cui and P. T. Anastas, Energy Environ. Sci., 2009, 2, 1038–1049 RSC.
- H. Dogari, N. Salimi-Turkamani, H. Ghafuri and R. Peymanfar, Mater. Adv., 2025, 6, 992–1005 RSC.
- E. Mosaffa, D. Patel, N. A. Ramsheh, R. I. Patel, A. Banerjee and H. Ghafuri, Int. J. Biol. Macromol., 2024, 254, 127794 CrossRef CAS PubMed.
- J. M. Dorresteijn, R. Conradi, L. D. Mandemaker, K. Schnabl, V. Cirriez, A. Welle, D. Curulla-Ferré, F. Meirer, E. T. Vogt and B. M. Weckhuysen, Mater. Adv., 2024, 6(1), 201–213 RSC.
- H. Haridevan, D. A. Evans, D. J. Martin and P. K. Annamalai, Mater. Adv., 2024, 5, 1540–1551 RSC.
- N. F. Lima, G. M. Maciel, N. P. Lima, I. D. A. A. Fernandes and C. W. I. Haminiuk, Int. J. Biol. Macromol., 2024, 133396 CrossRef CAS PubMed.
- V. Gayathri, N. P. Lobo, V. L. Vikash, N. R. Kamini and D. Samanta, ACS Biomater. Sci. Eng., 2023, 9, 625–641 CrossRef CAS PubMed.
- H. Sai, R. Fu, L. Xing, J. Xiang, Z. Li, F. Li and T. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 7373–7381 CrossRef CAS PubMed.
- K. Kamiński, M. Jarosz, J. Grudzień, J. Pawlik, F. Zastawnik, P. Pandyra and A. M. Kołodziejczyk, Cellulose, 2020, 27, 5353–5365 CrossRef.
- D. Li, E.-J. Li, L. Li, B. Li, S. Jia, Y. Xie and C. Zhong, Int. J. Biol. Macromol., 2024, 264, 130344 CrossRef CAS PubMed.
- H. El-Saied, A. H. Basta and R. H. Gobran, Polym.-Plast. Technol. Eng., 2004, 43, 797–820 CrossRef CAS.
- M. Fernandes, M. Gama, F. Dourado and A. P. Souto, Microb. Biotechnol., 2019, 12, 650–661 CrossRef CAS PubMed.
- M.-C. Li, X. Liu, K. Lv, J. Sun, C. Dai, B. Liao, C. Liu, C. Mei, Q. Wu and M. Hubbe, Prog. Mater. Sci., 2023, 101187 CrossRef CAS.
- M. Chen, H. Kang, Y. Gong, J. Guo, H. Zhang and R. Liu, ACS Appl. Mater. Interfaces, 2015, 7, 21717–21726 CrossRef CAS PubMed.
- G. Y. Duan, X. Q. Li, G. R. Ding, L. J. Han, B. H. Xu and S. J. Zhang, Angew. Chem., 2022, 134, e202110657 CrossRef.
- P. Yan, J. Mensah, A. Adesina, E. Kennedy and M. Stockenhuber, Appl. Catal., B, 2020, 267, 118690 CrossRef CAS.
- Z. Lu, T. Li, S. R. Mudshinge, B. Xu and G. B. Hammond, Chem. Rev., 2021, 121, 8452–8477 CrossRef CAS PubMed.
- T. Aditya, A. Pal and T. Pal, Chem. Commun., 2015, 51, 9410–9431 RSC.
- T. Kamal, A. M. Asiri and N. Ali, Spectrochim. Acta, Part A, 2021, 261, 120019 CrossRef CAS PubMed.
- R. Rajamohan and Y. R. Lee, J. Mol. Struct., 2023, 1276, 134803 CrossRef CAS.
- A. M. Abozied, A. Abouelsayed, B. Anis, M. F. Zayed, W. H. Eisa and A. B. Abdelrazzak, Mater. Today Sustainability, 2024, 27, 100814 CrossRef.
- D. Morales, R. Gutiérrez-Pensado, F. I. Bravo and B. Muguerza, LWT, 2023, 189, 115482 CrossRef CAS.
- M. Åkerholm, B. Hinterstoisser and L. Salmén, Carbohydr. Res., 2004, 339, 569–578 CrossRef PubMed.
- T. Kondo, Cellulose, 1997, 4, 281–292 CrossRef CAS.
- V. Kovalenko, R. Mukhamadeeva, L. Maklakova and N. Gustova, J. Struct. Chem., 1994, 34, 540–547 CrossRef.
- M. Makarem, C. M. Lee, K. Kafle, S. Huang, I. Chae, H. Yang, J. D. Kubicki and S. H. Kim, Cellulose, 2019, 26, 35–79 CrossRef CAS.
- G. Zhang and X. Wu, Opt. Lasers Eng., 2004, 42, 219–231 CrossRef.
- H. FaniMoghadam, M. G. Dekamin and N. Rostami, Res. Chem. Intermed., 2022, 48, 3061–3089 CrossRef CAS.
- M. Drozd, Mater. Sci. Eng.: B, 2007, 136, 20–28 CrossRef CAS.
- M. Ara, H. Ghafuri and N. Ghanbari, Colloid Interface Sci. Commun., 2023, 53, 100704 CrossRef CAS.
- J. B. Habinshuti, J. P. Munganyinka, A. R. Adetunji, B. Mishra, H. Tanvar, J. Mukiza, G. Ofori-Sarpong and A. P. Onwualu, Results Eng., 2022, 14, 100415 CrossRef CAS.
- T. Kanti Das, S. Ganguly, S. Remanan and N. C. Das, ChemistrySelect, 2019, 4, 3665–3671 CrossRef CAS.
- Y. Ban, T. Asakura and Y. Morita, J. Radioanal. Nucl. Chem., 2009, 279, 423–429 CrossRef CAS.
-
A. J. Bard, L. R. Faulkner and H. S. White, Electrochemical methods: fundamentals and applications, John Wiley & Sons, 2022 Search PubMed.
- S. Colominas, J. McLafferty and D. Macdonald, Electrochim. Acta, 2009, 54, 3575–3579 CrossRef CAS.
-
H.-E. Jacob, Methods in microbiology, Elsevier, 1970, vol. 2, pp. 91–123 Search PubMed.
- L.-Z. Huang, H. C. B. Hansen and M. J. Bjerrum, J. Hazard. Mater., 2016, 306, 175–183 CrossRef CAS PubMed.
- N. R. Jana, T. K. Sau and T. Pal, J. Phys. Chem. B, 1999, 103, 115–121 CrossRef CAS.
- D. Pal, T. Pal and A. Pal, Catal. Commun., 2023, 181, 106730 CrossRef CAS.
- N. Pradhan, A. Pal and T. Pal, Colloids Surf., A, 2002, 196, 247–257 CrossRef CAS.
- A. Gholami, Z. Nasri and H. Ghafuri, J. Environ. Chem. Eng., 2025, 117237 CrossRef CAS.
- J.-M. Song, C.-C. Liu, S.-S. Zhang, H.-L. Niu, C.-J. Mao, S.-Y. Zhang and Y.-H. Shen, Sep. Purif. Technol., 2014, 124, 148–154 CrossRef CAS.
- Z. Wang, R. Su, D. Wang, J. Shi, J.-X. Wang, Y. Pu and J.-F. Chen, Ind. Eng. Chem. Res., 2017, 56, 13610–13617 CrossRef CAS.
- M. Ismail, M. Khan, S. B. Khan, K. Akhtar, M. A. Khan and A. M. Asiri, J. Mol. Liq., 2018, 268, 87–101 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ,
Mohamadmahdi
Moghadari
,
Mohamadmahdi
Moghadari
 ,
Hossein
Ghafuri
,
Hossein
Ghafuri
 * and
Haniyeh
Dogari
* and
Haniyeh
Dogari
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH3OH solvent (1
CH3OH solvent (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, volume ratio) were mixed. Then, 0.5 g of BCNF-CPTMS was dispersed in the as-prepared solution and stirred at room temperature in a nitrogen atmosphere for 24 h. Finally, the semi-finished product was washed with water and EtOH twice and dried at 80 °C for 12 h.
1, volume ratio) were mixed. Then, 0.5 g of BCNF-CPTMS was dispersed in the as-prepared solution and stirred at room temperature in a nitrogen atmosphere for 24 h. Finally, the semi-finished product was washed with water and EtOH twice and dried at 80 °C for 12 h.
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 0) planes of bacterial cellulose nanofibers, respectively.42 The decrease in the intensity of the peaks of the synthesized composite indicates the reduced crystal structure of bacterial cellulose, indicating the successful formation of the nanofiber composite. In addition, the broad peak at 9.9° shows that the guanidine component is amorphous (Fig. 4).59
0) planes of bacterial cellulose nanofibers, respectively.42 The decrease in the intensity of the peaks of the synthesized composite indicates the reduced crystal structure of bacterial cellulose, indicating the successful formation of the nanofiber composite. In addition, the broad peak at 9.9° shows that the guanidine component is amorphous (Fig. 4).59














