DOI:
10.1039/D5MA00144G
(Paper)
Mater. Adv., 2025,
6, 3533-3545
In vitro scratch assay, cytotoxicity, DNA/BSA binding properties, and antimicrobial activity of green synthesized TiO2 nanoparticles via neem flowers
Received
15th February 2025
, Accepted 11th April 2025
First published on 14th April 2025
Abstract
Titanium dioxide nanoparticles (TiO2 NPs) were prepared using an extract from Azadirachta indica (Neem) flowers. Their properties were examined using UV-visible and infrared spectroscopy, while their crystallinity was assessed through pXRD analysis. Their shape, size, and morphology were further studied using FESEM and TEM techniques. XRD analysis confirmed the anatase phase of the nanoparticles. TEM results indicated particle sizes in the range of 20 to 30 nm. The EDAX spectrum confirms the presence of both titanium and oxygen in the sample. The TiO2 NPs demonstrated significant biological activities, including antioxidant, antibacterial, antifungal, and cytotoxic properties. Their antioxidant activity exhibited effective IC50 values, while antimicrobial studies indicated strong activity against pathogens. Additionally, the nanoparticles showed significant binding affinity toward DNA and BSA, highlighting their potential for biomedical applications. Cytotoxicity tests on normal HEK-293 cell lines revealed minimal toxicity, suggesting suitability for further clinical research. In vitro scratch assays were carried out with L929 fibroblast cells which show significant cell migration and proliferation. The present study concludes that TiO2 NPs have potential for diverse applications, including antioxidant, antimicrobial, and bioengineering uses.
Introduction
Advancements in nanotechnology have heralded a transformative era in materials science, enabling the synthesis of nanoparticles with tailored physicochemical properties and diverse applications. Among various nanoparticles, titanium dioxide (TiO2) has garnered significant attention due to its remarkable properties such as high surface area, photocatalytic activity, non-toxicity, and chemical stability.1–3 TiO2 nanoparticles (NPs) have been extensively explored for applications ranging from photocatalysis and environmental remediation to biomedical uses, including antimicrobial agents, drug delivery systems, and cancer therapies.4–8 However, the synthesis method profoundly influences the properties and effectiveness of TiO2 NPs. In recent years, green synthesis has emerged as a sustainable and eco-friendly alternative to conventional chemical and physical methods for nanoparticle production. The green synthesis approach leverages biological materials such as plant extracts, microorganisms, and enzymes as reducing and stabilizing agents. This methodology offers significant advantages, including reduced use of hazardous chemicals, lower energy consumption, and the generation of biocompatible and environmentally benign nanoparticles.9–11 Plant-based green synthesis, in particular, has gained traction due to the rich reservoir of bioactive compounds in plants that facilitate the rapid and efficient synthesis of nanoparticles. These bioactive compounds, including phenolics, flavonoids, alkaloids, and terpenoids, serve as natural reducing agents, imparting additional functionalities to the synthesized nanoparticles. Green-synthesized titanium dioxide nanoparticles (TiO2 NPs) have garnered significant attention due to their sustainable production methods and broad applicability across various fields. These nanoparticles are extensively used in cosmetics, pharmaceuticals, biosensors, cancer therapy, drug delivery, and genetic engineering due to their unique physicochemical and biological properties. One of the key advantages of TiO2 NPs is their multifunctionality, exhibiting remarkable antibacterial, antibiofilm, antifungal, antileishmanial, antioxidant, and larvicidal properties. Their potent anticancer activity is particularly noteworthy, as they can induce cytotoxic effects in tumor cells, contributing to the suppression of cancerous growth. Additionally, their photocatalytic properties make them valuable in environmental remediation and biomedical applications, enhancing their efficacy in targeted drug delivery and cancer therapy. Due to their biocompatibility and eco-friendly synthesis, TiO2 NPs have emerged as a promising alternative to conventional nanoparticles, offering safer and more efficient solutions in both medical and industrial domains.12–15 The neem tree (Azadirachta indica), a highly versatile medicinal plant, has been widely researched for its health benefits. Different parts of the tree, including its leaves, bark, seeds, and flowers, are abundant in bioactive compounds known for their antioxidant, antimicrobial, anticancer, and anti-inflammatory effects.16–24 Neem flowers, in particular, contain a plethora of secondary metabolites, including flavonoids, terpenoids, and phenolic compounds, making them an excellent candidate for green nanoparticle synthesis.25–29 Utilizing neem flower extracts for the synthesis of TiO2 nanoparticles presents a dual advantage: the production of functionalized nanoparticles with enhanced biological activities and the valorization of plant-based resources. This study focuses on the synthesis of TiO2 nanoparticles using neem flower extract as a reducing and stabilizing agent. The green synthesis process is explored for its simplicity, cost-effectiveness, and ability to produce biocompatible nanoparticles. The resulting TiO2 NPs are characterized to evaluate their structural, morphological, and physicochemical properties.
Materials and methods
Chemicals
All the metal salts and solvents were purchased from Merck India and were of analytical grade. The experiment utilized double distilled water at all stages.
Preparation of neem flower extract
Flowers from a single neem tree were collected in Vellore during February and March 2020. After harvesting, the flowers were dried in the shade and stored in an airtight container for future use. The extraction process and phytochemical analysis of the neem flower extract were carried out using the methods described in our earlier studies.30
Synthesis of TiO2 nanoparticles
TiO2 nanoparticles (TiO2 NPs) were synthesized using an aqueous extract of neem flowers, following a slightly modified version of a standard procedure.31 In summary, 10 mg of concentrated neem flower extract was dissolved in 20 mL of water and gradually added dropwise to a solution containing TTIP (titanium tetraisopropoxide, 0.750 μL in 100 mL of water) under magnetic stirring at room temperature overnight. The solution was stirred until a reddish-brown color developed and then incubated at room temperature for two hours. The mixture was centrifuged at 7000 rpm for 15 minutes, and the resulting pellets were washed thoroughly with water and ethanol to remove impurities and residual organic material. The cleaned pellets were dried at 100 °C for two hours, ground into a fine powder using a mortar and pestle, and calcined at 500 °C for two hours. The final powder was stored in an airtight container at 4 °C for future analysis.
Characterization
The synthesized TiO2 nanoparticles (NPs) were thoroughly characterized using a range of analytical techniques, including FTIR, UV-vis spectroscopy, XRD, BET analysis, photoluminescence (PL), zeta potential analysis, TEM, and FE-SEM. FTIR analysis was performed with a SHIMADZU IR Affinity spectrometer over a wavelength range of 4000–400 cm−1, with samples prepared using anhydrous KBr pellets. Elemental composition was determined using an Elementar Vario EL III instrument. Optical properties were studied with a JASCO V-670 UV-vis spectrophotometer, while XRD patterns were recorded using a BRUKER D8 Advance diffractometer, and the crystalline size and structure were calculated using the Scherrer equation (D = Kλ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ).
θ).
The morphological and structural features were analyzed through field emission scanning electron microscopy (FE-SEM) and energy-dispersive X-ray spectroscopy (EDX) using an FEI Quanta 250 FEG system. High-resolution transmission electron microscopy (TEM) was conducted with a JEOL JEM-2100 Plus microscope. The stability and size distribution of the TiO2 nanoparticles were assessed through zeta potential and dynamic light scattering (DLS) analyses using a Horiba SZ-100. This comprehensive characterization approach provided in-depth insights into the physicochemical properties of the synthesized nanoparticles.
Anti-oxidant activity
The antioxidant potential of TiO2 NPs was evaluated using different methods.32,33
DPPH radical scavenging activity.
The antioxidant capacity of TiO2 nanoparticles (NPs) was evaluated using a DPPH assay. Different concentrations of TiO2 NPs (20–100 μg mL−1) prepared in methanol were combined with a 1 mM DPPH solution and allowed to incubate at rt for 30 minutes. The abs. of the samples and the control were then recorded at 517 nm. The percentage of inhibition was calculated as follows:
| % Inhibition = [(Abscontrol − Abssample)/Abscontrol] × 100. |
Reducing power assay.
TiO2 nanoparticles (NPs) were tested at concentrations ranging from 20 μg mL−1 to 100 μg mL−1. The samples were combined with 0.2 M phosphate buffer and 1% potassium ferricyanide, with ascorbic acid serving as a positive control. The reducing power was evaluated by measuring the absorbance of the samples at 700 nm.
Phospho-molybdenum assay.
TiO2 nanoparticles (NPs) were tested at concentrations ranging from 20 to 100 μg mL−1. Each test tube contained the TiO2 NPs, 3 mL of dd water, and 1 mL of molybdate reagent. The samples and ascorbic acid abs. were measured at 695 nm.
Antimicrobial activity
The antibacterial properties of TiO2 nanoparticles (NPs) were investigated using the standard disc diffusion method.34,35 The antimicrobial activity of the samples was evaluated based on a previously reported protocol, with dimethyl sulfoxide (DMSO) employed as a negative control. Various concentrations of TiO2 NPs (25, 50, 75, and 100 μg mL−1) were tested against Staphylococcus aureus (ATCC 25923), Pseudomonas aeruginosa (ATCC 27853), Candida tropicalis (ATCC 10231), and Candida glabrata (MTCC 3019). Standard antibiotics, including ampicillin for bacterial strains and ketoconazole for fungal strains, were used as positive controls. All experiments were conducted in triplicate to ensure reliability, with the inoculated plates incubated at 37 °C for 24 hours. The antimicrobial efficacy was determined by measuring the diameter of the inhibition zones around the discs, recorded in millimeters (mm).
DNA binding study
The DNA binding ability of the extracts was investigated following established protocols.36,37 The sample was prepared in a Tris–HCl/NaCl buffer, and the concentration of calf thymus DNA (CT-DNA) was determined from its absorbance spectrum. Titration was performed by gradually increasing the CT-DNA concentration with absorbance measurements taken after each addition. A plot of [DNA]/(εa − εf) vs. [DNA] was constructed, and the binding constant (Kb) was determined from the linear fit of the plot using the equation:| |  | (1) |
BSA binding study
Serum albumin proteins play a crucial role in drug transport and metabolism within the bloodstream. The structural similarity between human serum albumin (HSA) and BSA makes BSA a useful model for quenching studies. In binding studies, BSA was used to assess the drug–protein binding capacity.36,37 Tris–HCl/NaCl buffer was used to prepare the BSA solution (2 × 106 M) and the aqueous samples were added to this solution. The fluorescence intensity of BSA was measured, and a decrease in fluorescence indicated the interaction between BSA and the compounds. The quenching constant (KBSA) was calculated using the Stern–Volmer equation. Additionally, the number of binding sites (n) was determined using the Scatchard equation (eqn (2)).| | | I0/I = 1 + KBSA[Q] = 1 + kqτ0[Q] | (2) |
| | log(I0 − I/I) = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K + n K + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q] | (3) |
Cytotoxicity
Monolayer cell cultures were trypsinized, and the cell concentration was adjusted to 1.0 × 105 cells per mL in a medium supplemented with 10% FBS. A 100 μL aliquot of this cell suspension (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) was seeded into a 96-well microtiter plate. After 24 hours of incubation, allowing a partial monolayer to form, the supernatant was carefully removed, and the wells were gently rinsed with fresh medium. Subsequently, 100 μL of various concentrations of the test drugs were added to the wells. The plates were then incubated at 37 °C with 5% CO2 for 24 hours. Following this incubation, the test solutions were aspirated, and 100 μL of MTT solution (5 mg/10 mL in PBS) was added to each well. The plates were incubated for another 4 hours at 37 °C in a 5% CO2 environment. After incubation, the supernatant was removed, and 100 μL of DMSO was added to each well to dissolve the formazan crystals. The plates were gently shaken, and absorbance was measured at 590 nm using a microplate reader.38 The percentage of cell growth inhibition and the IC50 value were calculated from the dose–response curves.
000 cells per well) was seeded into a 96-well microtiter plate. After 24 hours of incubation, allowing a partial monolayer to form, the supernatant was carefully removed, and the wells were gently rinsed with fresh medium. Subsequently, 100 μL of various concentrations of the test drugs were added to the wells. The plates were then incubated at 37 °C with 5% CO2 for 24 hours. Following this incubation, the test solutions were aspirated, and 100 μL of MTT solution (5 mg/10 mL in PBS) was added to each well. The plates were incubated for another 4 hours at 37 °C in a 5% CO2 environment. After incubation, the supernatant was removed, and 100 μL of DMSO was added to each well to dissolve the formazan crystals. The plates were gently shaken, and absorbance was measured at 590 nm using a microplate reader.38 The percentage of cell growth inhibition and the IC50 value were calculated from the dose–response curves.
| % inhibition = 100 − (OD of sample/OD of control) × 100 |
In vitro wound scratch assay
Cells were detached from the tissue culture dish following standard passaging procedures. A 6-well culture plate was prepared by adding 1–2 mL of pre-warmed culture medium to each well. Cells were then seeded in 24-well plates at a density of 4000 cells per well and incubated at 37 °C with 5% CO2 for 24 hours to allow attachment. After incubation, cells were treated with test samples at their IC50 concentrations (μM) and further incubated for the required duration. Once the cells reached confluence, typically after 18–24 hours, a linear scratch was made in the monolayer using a sterile cell scraper or pipette tip. The monolayer was then gently washed with phosphate-buffered saline (PBS) to remove detached cells, and fresh culture medium was added to support the cell viability and migration. Immediately after the scratch, cells were visualized using a fluorescence microscope (Olympus CKX-53, Japan), and the percentage of dead cells was quantified in at least three randomly selected microscopic fields.
Results and discussion
UV vis analysis and bandgap calculations
The UV-vis absorption spectrum of the biosynthesized TiO2 nanoparticles (TiO2 NPs) is shown in Fig. 1a, revealing a distinct absorption peak at 315 nm. This observation aligns with previously reported studies.31,39 The excitonic absorption of the biogenically synthesized TiO2 NPs exhibits a notable blue shift at 325 nm, reflecting size-dependent optical properties. The sharp and well-defined absorption peak indicates the formation of monodispersed nanoparticles, likely due to surface plasmon resonance. The band-gap energy of the TiO2 NPs was calculated as depicted in Fig. 1b. The band-gap energy was determined by extrapolating the linear portion of the Tauc plot, which represents the variation of (αhν)2 as a function of photon energy (hν). The band-gap energy values were derived using the Tauc equation:
 |
| | Fig. 1 (A) UV spectrum of the TiO2 NPs. (B) Tauc plot for bandgap energy calculations. (C) FTIR spectrum of TiO2 NPs. (D) XRD spectrum of TiO2 NPs. | |
In this equation, α denotes the absorption coefficient, h represents Planck's constant, ν signifies the frequency of the incident light, Eg corresponds to the band-gap energy, and A is a proportionality constant. The biosynthesized TiO2 nanoparticles exhibited a band-gap energy of 3.13 eV, highlighting their potential for use in semiconductors, optics, and biomedical applications. The high band-gap energy of the TiO2 NPs contributes to their excellent catalytic activity, UV filtering properties, wound healing capabilities, and anti-inflammatory effects.40,41 Consistent with previous research, the band gap energy of TiO2 nanoparticles produced through the chemical method has been reported as 3.1 eV. In the present study, the band gap energy determined using our approach was 3.13 eV, yielding a relative error of 0.9%. This minor discrepancy suggests strong agreement between the two values, which may arise from variations in synthesis parameters, nanoparticle size, or measurement precision.42
FTIR and pXRD analysis
The FTIR spectrum of the aqueous neem flower extract and the green-synthesized TiO2 nanoparticles (TiO2 NPs) is shown in Fig. 1c. A distinct peak at 3445 cm−1 corresponds to O–H stretching vibrations, indicating the presence of hydroxyl groups associated with phenolics and flavonoids in the extract. Peaks at 2932 cm−1 and 2851 cm−1 represent C–H stretching vibrations, characteristic of aliphatic groups in phytochemicals. The band at 1640 cm−1 is attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations, suggesting the presence of aldehyde or ketone groups, while the peak at 1380 cm−1 corresponds to C–H bending vibrations, confirming the involvement of organic compounds. Aromatic stretching vibrations, indicative of aromatic compounds derived from the extract, are also observed. These peaks are present in both the extract and the TiO2 NPs, confirming that the phytochemicals in the extract play a crucial role in nanoparticle formation. Additionally, the peak at 530 cm−1 is associated with metal–oxygen (Zn–O) stretching vibrations, verifying the formation of TiO2. These functional groups emphasize the role of the plant extract as both a reducing and capping agent in the synthesis of TiO2 NPs. X-ray diffraction (p-XRD) analysis was conducted to evaluate the crystallinity and phase purity of the synthesized TiO2 nanoparticles (TiO2 NPs), as depicted in Fig. 1d. The broadening of the diffraction peaks confirms the nanoscale size of the particles. The prominent peaks, along with their corresponding Miller indices, confirm the crystalline structure of the TiO2 NPs. The observed reflections are consistent with the hexagonal wurtzite phase of TiO2, matching the standard reference pattern (JCPDS card no. 00-021-1272). Using the Debye–Scherrer equation, the average crystalline size of the nanoparticles was estimated to be around 23 nm. In previous studies, TiO2 nanoparticles synthesized using lemon peel extract were reported to have an average crystalline size of 60 nm.43 However, in our study, the average crystalline size was determined to be 23 nm. These findings suggest that neem flower extract serves as an effective green fuel for the synthesis of TiO2 nanoparticles.
O stretching vibrations, suggesting the presence of aldehyde or ketone groups, while the peak at 1380 cm−1 corresponds to C–H bending vibrations, confirming the involvement of organic compounds. Aromatic stretching vibrations, indicative of aromatic compounds derived from the extract, are also observed. These peaks are present in both the extract and the TiO2 NPs, confirming that the phytochemicals in the extract play a crucial role in nanoparticle formation. Additionally, the peak at 530 cm−1 is associated with metal–oxygen (Zn–O) stretching vibrations, verifying the formation of TiO2. These functional groups emphasize the role of the plant extract as both a reducing and capping agent in the synthesis of TiO2 NPs. X-ray diffraction (p-XRD) analysis was conducted to evaluate the crystallinity and phase purity of the synthesized TiO2 nanoparticles (TiO2 NPs), as depicted in Fig. 1d. The broadening of the diffraction peaks confirms the nanoscale size of the particles. The prominent peaks, along with their corresponding Miller indices, confirm the crystalline structure of the TiO2 NPs. The observed reflections are consistent with the hexagonal wurtzite phase of TiO2, matching the standard reference pattern (JCPDS card no. 00-021-1272). Using the Debye–Scherrer equation, the average crystalline size of the nanoparticles was estimated to be around 23 nm. In previous studies, TiO2 nanoparticles synthesized using lemon peel extract were reported to have an average crystalline size of 60 nm.43 However, in our study, the average crystalline size was determined to be 23 nm. These findings suggest that neem flower extract serves as an effective green fuel for the synthesis of TiO2 nanoparticles.
FESEM and EDX
The FESEM images presented in Fig. 2a–c at varying magnifications demonstrate that the synthesized particles exhibit distorted spherical morphologies. This deformation is likely attributed to the presence of pores within the structural framework of the TiO2 nanoparticles. Elemental analysis of the nanoparticles was performed using EDX spectroscopy, as shown in Fig. 2d. The EDX spectra revealed prominent peaks corresponding to titanium and oxygen, confirming the presence of TiO2. The quantitative analysis yielded elemental compositions of 41.50% titanium and 58.50% oxygen, which are consistent with previously reported findings.
 |
| | Fig. 2 (A)–(C) FESEM images of the TiO2 NPs of different magnifications and (D) EDAX spectrum of the TiO2 NPs. | |
TEM, particle size analysis, and zeta potential analysis
Transmission electron microscopy (TEM) images of the G-TiO2 nanoparticles reveal an average particle size in the range of 20–30 nm (Fig. 3a–c). HRTEM analysis further confirms the crystalline nature of the TiO2 NPs, showing well-aligned lattice fringes. The calculated interplanar spacing (d-spacing) is 0.263 nm (Fig. 3c), aligning with the structure of TiO2. The selected area electron diffraction (SAED) pattern shows concentric rings, confirming the polycrystalline nature of the synthesized nanoparticles (Fig. 3d). As per the previous studies, TiO2 nanoparticles (NPs) synthesized using lemon peel extract have been reported to have particle sizes ranging from 80 to 110 nm.43 In contrast, the TiO2 NPs synthesized in our study exhibited a much smaller particle size of 20–30 nm. These findings highlight the effectiveness of neem flower extract as a green resource for producing TiO2 nanoparticles with reduced particle size.
 |
| | Fig. 3 (A)–(C) TEM images of the TiO2 NPs. (D) SAED pattern. (E) Histogram of particle size analysis. (F) zeta potential distribution analysis of TiO2. | |
Particle size analysis (DLS) was performed to determine the size distribution of the TiO2 nanoparticles (TiO2 NPs) in a dispersed medium. The analysis revealed an average particle size of 26 nm (Fig. 3e). The polydispersity index (PDI) was determined using dynamic light scattering (DLS) analysis to assess the size heterogeneity of the NPs in the sample. The measured PDI value was 0.118 ± 0.03, indicating high particle uniformity. Reports suggest that a PDI below 0.3 is suitable for drug delivery applications. These results highlight the potential of the synthesized TiO2 nanoparticles for various applications. Zeta potential, a key parameter for assessing the stability and functionality of nanoparticles, was measured for the synthesized TiO2 NPs. The zeta potential value was determined to be −20.4 mV (Fig. 3f). This relatively low zeta potential suggests limited electrostatic repulsion between particles, indicating a tendency for agglomeration in the dispersed medium.
TGA
The thermal stability of titanium dioxide nanoparticles (TiO2 NPs) is essential for their use in electronics, optics, and catalysis. TGA analysis shows that the sample undergoes decomposition as the temperature increases, with complete degradation occurring at 1180 °C (Fig. 4a). The first weight loss of nearly 1.2%, at 35–150 °C, is attributed to the removal of moisture and volatile components that act as capping agents around the nanoparticles. A second degradation of approximately 1.5%, between 190 and 450 °C, corresponds to the breakdown of organic molecules, such as phenolics, flavonoids, precursors, and other biomolecules involved in the biosynthesis of TiO2 NPs. The final degradation of ∼0.03% between 500 and 1000 °C may be due to the phase transformation of anatase to rutile TiO2. In total 2.73% of mass loss was observed, which is aligned with previous reports suggesting that the quality and thermostability synthesised TiO2 NPs.44 The residue left after decomposition was 97.27% of the initial residue, indicating that TiO2 NPs maintain excellent thermal stability, with minimal mass loss even at higher temperatures. This stability makes them suitable for a wide range of industrial, technological, and scientific applications.
 |
| | Fig. 4 (A) TGA analysis of the TiO2 NPs. (B)–(D) Antioxidant activity of the TiO2 NPs. | |
Antioxidant activity
The antioxidant activity of TiO2 nanoparticles (TiO2 NPs) was assessed using three analytical methods: the DPPH assay, ferric-reducing power assay, and the phosphomolybdenum method, with ascorbic acid employed as the standard. The results, depicted in Fig. 4(b–d), revealed that all three assays exhibited inhibition rates exceeding 75%, indicating notable antioxidant efficacy. A positive correlation was observed between the concentration of TiO2 NPs and the percentage of inhibition, demonstrating a concentration-dependent enhancement of antioxidant activity. The IC50 values were determined to be 63.24, 54.11, and 58.25 μg mL−1 for the DPPH, ferric-reducing power, and phosphomolybdenum assays, respectively. These findings confirm the strong free-radical scavenging capability of TiO2 NPs, underscoring their potential as effective natural antioxidants. These results were compared with earlier reports45 and determined their significant importance as a potential antioxidant agent. Furthermore, there is a non-linear correlation between the antioxidant activity and band gap energy of NPs. In molecules like flavonoids, the highest band gap results in the lowest reactivity and vice versa.46,47 Similarly, TiO2 NPs with a moderate band gap energy (3.13 eV) have significant antioxidant activity due to their ability to participate in the electron transfer mechanism. The antioxidant activity of the nanoparticles is another focus area, as oxidative stress is implicated in numerous diseases, including cancer, cardiovascular disorders, and neurodegenerative conditions. Earlier studies reported on TiO2 nanoparticles synthesized using plum and kiwi peel extracts exhibited DPPH inhibition of 69% and 59%, respectively.48 In contrast, TiO2 NPs synthesized using neem flower extract showed over 75% inhibition. These findings confirm the strong antioxidant potential of neem flower-mediated TiO2 nanoparticles. By scavenging free radicals, antioxidant nanoparticles have the potential to mitigate oxidative damage and improve health outcomes.
Anti-bacterial activity
The antibacterial activity of nanoparticles is crucial for addressing global challenges such as antibiotic resistance, food safety, healthcare-associated infections, and environmental contamination. Their versatility and effectiveness make them a valuable tool for developing innovative solutions across multiple disciplines. The antibacterial activity of the synthesized TiO2 nanoparticles (TiO2 NPs) was evaluated against both Gram-positive and Gram-negative bacterial pathogens, specifically Staphylococcus aureus (ATCC 25923) and Pseudomonas aeruginosa (ATCC 27853) (Fig. 5a and b). The agar well diffusion method was employed to assess the antibacterial efficacy of the TiO2 NPs. The results demonstrated a concentration-dependent antibacterial effect against the tested microorganisms. The ZOI was measured to be 22 ± 0.20 mm for S. aureus and 20 ± 0.50 mm for P. aeruginosa (Fig. 5a and b). Interestingly, the TiO2 NPs exhibited greater efficiency against Gram-negative bacteria. This enhanced activity may be attributed to the porous structure of the Gram-negative cell wall, which facilitates the penetration of TiO2 NPs, a feature not observed in the thicker peptidoglycan layer of Gram-positive bacteria. In this study, TiO2 nanoparticles exhibited a MIC value of 100 μg mL−1 against the tested bacterial strain(s), indicating a significant antibacterial effect. The results suggest that these nanoparticles have potential as an effective antimicrobial agent for various biomedical and environmental applications. The current findings were compared with previous studies on the antibacterial activity of S. aureus and P. aeruginosa using TiO2 nanoparticles synthesized from plum, kiwi, and peach peel extracts. These extracts exhibited a zone of inhibition (ZOI) ranging from 15 to 18 mm.48,49 However, TiO2 NPs synthesized using neem flower extract demonstrated a ZOI exceeding 20 mm. This indicates that our material possesses strong antibacterial properties against pathogens and has potential as an effective antibacterial agent.
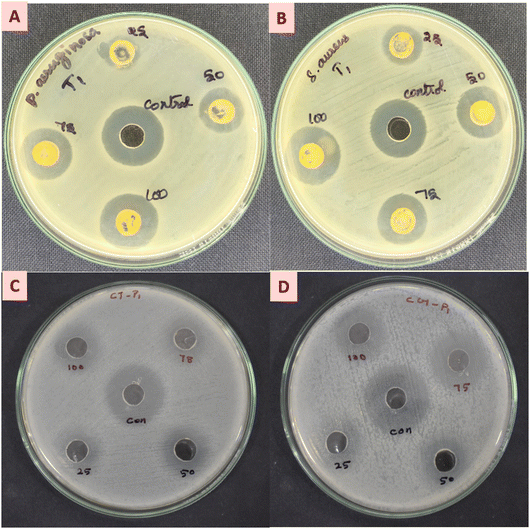 |
| | Fig. 5 Antimicrobial activity of the TiO2 NPs. (A) and (B) Antibacterial activity against (A) P. aeruginosa and (B) S. aureus. antifungal activity against (C) C. tropicalis and (D) C. glabrata. | |
Anti-fungal activity
The antifungal activity of titanium dioxide nanoparticles (TiO2 NPs) plays a crucial role in developing advanced therapeutic strategies for combating fungal infections, particularly in the face of increasing resistance to conventional antifungal agents. The unique properties of TiO2 NPs enable them to target various fungal species, disrupt biofilm formation, and enhance the effectiveness of traditional treatments, making them an important tool in modern medicine. In this study, the antifungal potential of TiO2 NPs was evaluated against Candida tropicalis (ATCC 10231) and Candida glabrata (MTCC 3019), with ketoconazole used as a control. The zone of inhibition (ZOI) observed was 23 ± 0.10 mm and 22 ± 0.20 mm at a concentration of 100 μg mL−1 (Fig. 5c and d) (Table 1). These findings are consistent with previous reports, highlighting the promising potential of TiO2 NPs as effective antifungal agents.31
Table 1 % Of wound closure on L929 cells at different time intervals
| S. no. |
Time (h) |
% Of wound closure |
| 01 |
0 |
0 |
| 02 |
12 |
62.36 |
| 03 |
24 |
75.14 |
| 04 |
48 |
92.15 |
DNA binding activity
The UV-visible titration method was employed to evaluate the binding efficacy of titanium dioxide nanoparticles with calf thymus DNA, aiming to assess their potential for cancer cell targeting and DNA interaction. DNA, as a primary pharmacological target for anticancer therapies, plays a key role in determining the binding characteristics of these agents. The results revealed a hyperchromic shift in absorbance intensity with increasing DNA concentration, indicating both covalent and noncovalent binding modes. These binding interactions include electrostatic interactions and groove or intercalative binding. The intrinsic binding constant was calculated to be 2.658 × 106 M−1, suggesting a strong affinity between TiO2 NPs and DNA exhibiting the base stacking interactions indicating groove binding (Fig. 6a and b).50 Molecules that bind to DNA can influence processes like replication, transcription, and repair, and may protect DNA from oxidative damage. Since TiO2 NPs exhibit good antioxidant activities and also good DNA binding properties, they are potential candidates for therapeutic applications via protecting DNA in diseases linked to oxidative stress i.e., cancer and neurodegenerative disorders.51–53
 |
| | Fig. 6 DNA binding assay of the TiO2 NPs. (A) DNA binding via UV titration, (B) linear plot, (C) EtBr quenching assay, (D) Stern–Volmer plot, (E) Scatchard plot and (F) viscosity analysis. | |
Competitive EtBr quenching assay
A competitive EtBr assay was conducted to analyze the interaction between TiO2 nanoparticles (NPs) and DNA, utilizing fluorescence quenching to determine the binding mode. Ethidium bromide (EtBr), a fluorescent compound, emits light at 630 nm in its free state, with its emission intensity increasing upon binding to DNA. The assay assessed the competitive binding of molecules to DNA through fluorescence spectroscopy by gradually increasing the concentration of TiO2 NPs. As the concentration of TiO2 rose, it displaced EtBr from DNA and attached to the DNA base pairs, suggesting a groove-binding mechanism (Fig. 6c). The gradual reduction in fluorescence intensity of the EtBr–DNA complex in the presence of TiO2 NPs indicated that the nanoparticles primarily followed a groove-binding mode rather than intercalation. This conclusion was further supported by the higher binding constant (Kb – 0.986 × 105 M−1) and the lower Stern–Volmer constant (KSV – 0.3321 × 106 M−1) and the apparent binding constant (Kapp – 1.221 × 106 M−1) (Fig. 6d and e).
BSA binding activity (UV titration method)
UV-vis spectral analysis of BSA (bovine serum albumin) binding studies provides crucial information about interactions between BSA and a ligand. Typically, BSA alone exhibits a prominent absorption peak around 278–280 nm, attributed to aromatic amino acids like tryptophan, tyrosine, and phenylalanine. Any shift or intensity variation in this peak upon ligand addition suggests structural alterations due to binding. As shown in Fig. 7a, free BSA exhibits an absorption peak near 280 nm. Upon the addition of TiO2 (10 μL), the absorption shifts to a lower wavelength with increased intensity, indicating a hyperchromic shift. This shift suggests that ligand binding leads to BSA unfolding, making aromatic residues more exposed to the solvent. Furthermore, as the TiO2 concentration increases from 10 to 80 μL, the hyperchromic effect becomes more pronounced. These spectral changes suggest the formation of a stable ground-state complex between BSA and the ligand. The observed fluorescence quenching confirms that the primary quenching mechanism is static quenching, resulting from BSA–ligand complex formation at the ground-state level.54,55
 |
| | Fig. 7 BSA binding assay of the TiO2 NPs. (A) BSA binding by the UV method, (B) BSA binding by FL, (C) Stern–Volmer plot and (D) Scatchard plot. | |
BSA binding activity (by fluorescence analysis)
The fluorescence titration method was employed to evaluate the binding efficacy of titanium dioxide nanoparticles (TiO2 NPs) with BSA, a structural analog of HSA. In both static and dynamic modes, metal complexes bind to proteins, leading to quenching of the intrinsic fluorescence of aromatic residues such as phenylalanine, tyrosine, and tryptophan. In this study, BSA exhibited fluorescence at 345 nm, which significantly decreased as the concentration of TiO2 NPs increased (Fig. 7b). The quenching constants (Kq) and binding constants (KBSA) were determined using the Stern–Volmer equation, eqn (2). The values for Kq and KBSA were calculated to be 3.32 × 1012 M−1 and 0.332 × 105 M−1, respectively. The binding constant (K) was found to be 1.050 × 106 M−1. Furthermore, the number of binding sites and the binding constants were determined using the Scatchard equation, eqn (3) (Fig. 7c and d). These results strongly suggest that TiO2 NPs exhibit a high binding affinity toward serum albumin, supporting their potential application in cancer treatment due to their robust binding nature.
Viscosity studies
Viscosity measurements serve as a valuable tool for investigating the interaction between TiO2 nanoparticles (NPs) and DNA, offering insights into the binding mechanism. An increase in viscosity typically indicates intercalation, whereas minimal changes suggest groove binding or electrostatic interactions, and a decrease implies DNA condensation or aggregation. In this study, minimal or no change in viscosity was observed (Fig. 6f), indicating that TiO2 NPs interact strongly with DNA through groove binding. These findings are consistent with results from prior UV-vis titration experiments.
In vitro cytotoxicity
Cytotoxicity assessment is a crucial step in determining the biocompatibility and therapeutic potential of nanoparticles. The cytotoxic effects of the synthesized TiO2 NPs are evaluated against cancer cell lines to explore their potential as anticancer agents. The mechanism of cytotoxicity, including the induction of reactive oxygen species (ROS) and apoptosis, is also investigated to provide insights into their mode of action. To evaluate the cytotoxicity and cell viability of the nanoparticle formulation, the MTT assay was conducted using cisplatin as the positive control and DMSO as the solvent control. Various concentrations of NF–TiO2 NPs, ranging from 10 μg mL−1 to 100 μg mL−1, were tested against normal HEK-294 cells (Fig. 8a–c). The results demonstrated that cell viability remained above 80% even at the highest concentration of 100 μg mL−1, indicating that NF–TiO2 NPs exhibit low toxicity within the tested concentration range (Fig. 8d). Compared to previous studies, commercially available TiO2 nanoparticles (NPs) have been reported to exhibit only 50% cell viability in HEK-293 cell lines at a concentration of 100 μg mL−1.56 In contrast, our green-synthesized TiO2 NPs demonstrated significantly higher cell viability at 80%. Similarly, in another study, TiO2 NPs synthesized using Tulbhagia violacea extract showed only 20% cell viability at the same concentration.57 These findings suggest that neem flower-mediated TiO2 NPs exhibit lower toxicity and better biocompatibility with normal HEK-293 cells, resulting in higher cell viability. These findings indicate the low toxicity of metal oxide nanoparticles in non-cancerous cell lines, further supporting their potential for safe clinical application.
 |
| | Fig. 8 (A)–(C) Treatment of the TiO2 NPs with HEK-293 cells. (D) Cell viability of the TiO2 NPs. | |
In vitro wound scratch assay
An in vitro wound scratch assay was performed to assess the impact of titanium dioxide (TiO2) nanoparticles on L929 fibroblast cell lines. Cell migration was tracked at various time points after the scratch was introduced. The findings revealed a progressive, time-dependent enhancement in wound closure, reaching 62% at 12 hours and peaking at 92% by 48 hours (Table 1 and Fig. 9). These results highlight the notable wound-healing properties of green-synthesized TiO2 nanoparticles in L929 cells, underscoring their potential as a promising therapeutic option for skin-related ailments.
 |
| | Fig. 9 (A) Normal and (B) fluorescence images of the in vitro wound scratch assay of the TiO2 NPs on L929 cells at 0, 12, 24, and 48 h. | |
Conclusions
This study underscores the potential of neem flower-mediated green synthesis as an eco-friendly method for producing multifunctional TiO2 nanoparticles. Key attributes such as binding properties, antioxidant activity, cytotoxicity, and antimicrobial effectiveness are explored due to their significance in biomedical and environmental applications. The binding interactions of TiO2 nanoparticles synthesized with neem flower extract are analyzed to understand their compatibility with biomolecules and other substances, which is crucial for applications like drug delivery, biosensing, and catalysis. The cytotoxic potential of these nanoparticles is assessed against cancer cell lines to evaluate their suitability as anticancer agents. Additionally, their antimicrobial activity is tested against various pathogenic bacteria and fungi, demonstrating their efficacy in combating microbial infections. The wound healing activity of synthesized NPs shows good to excellent activity. Leveraging the bioactive compounds in neem flowers enhances the biological performance of the synthesized nanoparticles, positioning them as promising tools for medical and environmental advancements. This research contributes to the field of green nanotechnology by highlighting the value of plant-based resources in nanoparticle synthesis for sustainable development.
Author contributions
Palanivelmurugan Mohanasundaram: conceptualization, formal analysis, investigation, methodology, and writing – original draft. Mary Saral A: funding acquisition, project administration, resources, supervision, validation, visualization, and writing – review & editing.
Data availability
The data will be available upon request from the corresponding author.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
The authors are grateful to VIT Management for providing laboratory facilities for carrying out the research.
Notes and references
- C. S. Santos, B. Gabriel, M. Blanchy, O. Menes, D. García, M. Blanco and V. Neto, Mater. Today Proc., 2015, 2(1), 456–465, DOI:10.1016/j.matpr.2015.04.056.
- M. A. Irshad, R. Nawaz, M. Z. Rehman, M. Adrees, M. Rizwan, S. Ali and S. Tasleem, Ecotoxicol. Environ. Saf., 2021, 212, 111978, DOI:10.1016/j.ecoenv.2021.111978.
- G. Nabi, W. Raza and M. Tahir, J. Inorg. Organomet. Polym. Mater., 2019, 1–5, DOI:10.1007/s10904-019-01248-3.
- M. A. Mousa, A. E. Nemr, E. A. Gomaa, S. M. Eldafrawy and E. T. Helmy, Egypt. J. Aquat. Biol. Fish, 2018, 22, 149–162, DOI:10.21608/ejabf.2018.8343.
- A. A. Kashale, K. P. Gattu, K. Ghule, V. H. Ingole, S. Dhanayat, R. Sharma and A. V. Ghule, Compos. Part B: Eng., 2016, 99, 297–304, DOI:10.1016/j.compositesb.2016.06.015.
- S. M. Hunagund, V. R. Desai, J. S. Kadadevarmath, D. A. Barretto, S. Vootla and A. H. Sidarai, RSC Adv., 2016, 6(99), 97438–97444, 10.1039/C6RA22163G.
- S. Marimuthu, A. A. Rahuman, C. Jayaseelan, A. V. Kirthi, T. Santhoshkumar, K. Velayutham and K. V. B. Rao, Asian Pac. J. Trop. Med., 2016, 9, 682–688, DOI:10.1016/S1995-7645(13)60118-2.
- S. Kalyanasundaram and M. J. Prakash, Int. Lett. Chem., Phys. Astron., 2015, 50, 80–95, DOI:10.56431/p-0c3605.
- S. Syamsol Bahri, Z. Harun, S. Khadijah Hubadillah, W. Norhayati Wan Salleh, N. Rosman, N. Hasliza Kamaruddin and H. Basri, IOP Conf. Ser.: Mater. Sci. Eng., 2021, 1142(1), 012005, DOI:10.1088/1757-899x/1142/1/012005.
- S. M. Roopan, A. Bharathi, A. Prabhakaran, A. A. Rahuman, K. Velayutham, G. Rajakumar and G. Madhumitha, Spectrochim. Acta, Part A, 2012, 98, 86–90, DOI:10.1016/j.saa.2012.08.055.
- J. Rajkumari, C. Magdalane, B. Siddhardha, J. Madhavan, G. Ramalingam and N. A. Al-Dhabi, K. Kaviyarasu, J. Photochem. Photobiol., B, 2019, 201, 111667, DOI:10.1016/j.jphotobiol.2019.111667.
- S. N. Nandhini, N. Sisubalan, A. Vijayan, C. Karthikeyan, M. Gnanaraj, D. Gideon and R. Sadiku, Heliyon., 2023, 9(2), e13128, DOI:10.1016/j.heliyon.2023.e13128.
- M. Z. Ahmad, A. S. Alasiri, J. Ahmad, A. A. Alqahtani, M. M. Abdullah, B. A. Abdel-Wahab and S. A. Alzahrani, Molecules, 2022, 27(22), 7712, DOI:10.3390/molecules27227712.
- P. Singh Jassal, D. Kaur, R. Prasad and J. Singh, J. Agric. Food Res., 2022, 10, 100361, DOI:10.1016/j.jafr.2022.100361.
- Z. Chen, S. Han, S. Zhou, H. Feng, Y. Liu and G. Jia, NanoImpact., 2020, 18, 100224, DOI:10.1016/j.impact.2020.100224.
- T. M. Braga, L. Rocha and T. Y. Chung, Molecules, 2021, 26(2), 252, DOI:10.3390/molecules26020252.
- V. Singh, M. Roy, N. Garg, A. Kumar, S. Arora and D. S. Malik, Recent Adv. Anti-Infect. Drug Discovery, 2021, 16(2), 94–121, DOI:10.2174/2772434416666210604105251.
- M. Iman, M. Taheri and Z. Bahari, J. Complementary Integr. Med., 2021, 19(2), 203–211, DOI:10.1515/jcim-2021-0009.
- P. Bairagi, N. Tiwari, V. Singh and R. Padiyar, World J. Pharm. Pharm. Sci., 2023, 12(9), 2467–2476, DOI:10.20959/wjpps20239-25783.
- S. M. PatiL, P. S. Shirahatti and R. Ramu, J. Pharm. Pharmacol., 2022, 74(5), 681–710, DOI:10.1093/jpp/rgab098.
- K. W. Sarika, S. B. Siddhi, K. W. Sakshi and S. K. Anuja, World J. Pharm. Pharm. Sci., 2022, 11(12), 829–842, DOI:10.20959/wjpps202212-23753.
- S. Adusei and S. Azupio, J. Chem., 2022, 6778554, DOI:10.1155/2022/6778554.
- M. Saman, K. Hafsa, Y. Ramit, A. Mohd, A. Nadeem, D. Mohd and B. Preeti, World J. Pharm. Pharm. Sci., 2022, 11(10), 325–351, DOI:10.20959/wjpr202210-24862.
- S. Vivek, M. D. Daneyal Khurshid and K. A. Bhupendra, Int. J. Pharm. Biol. Sci., 2020, 10(3), 172–180, DOI:10.21276/ijpbs.2020.10.3.1.
- B. M. Olabinri, E. A. Adepoju, A. A. Zainab and A. A. Ahmed, J. Pharmacogn. Phytother., 2014, 6(2), 17–23, DOI:10.5897/JPP2013.0308.
- B. S. Siddiqui, S. T. Ali, M. Rasheed and M. N. Kardar, Helv. Chim. Acta, 2003, 86(8), 2787–2796, DOI:10.1002/hlca.200390229.
- B. S. Siddiqui, S. T. Ali, M. T. Rajput, T. Gulzar, M. Rasheed and R. Mehmood, Nat. Prod. Res., 2009, 23(3), 271–283, DOI:10.1080/14786410802006082.
- B. S. Siddiqui, S. Tariq Ali and S. Kashif Ali, Nat. Prod. Res., 2006, 20(3), 241–245, DOI:10.1080/14786410500059318.
- K. K. D. Somathilaka Ranaweera, K. D. Prasanna Priyantha Gunathilake and G. Janarny, Croat. J. Food Sci. Technol., 2023, 15(1), 37–47, DOI:10.17508/cjfst.2023.15.1.05.
- P. Mohanasundaram and A. Mary Saral, ECS Trans., 2022, 107(1), 15021–15031, DOI:10.1149/10701.15021ecst.
- S. Jeyaraj and A. Mary Saral, ChemistrySelect, 2024, 9(21), e202401603, DOI:10.1002/slct.202401603.
- S. Jeyaraj and A. Mary Saral, Results Chem., 2024, 11, 101747, DOI:10.1016/j.rechem.2024.101747.
- P. Mohanasundaram and A. Mary Saral, Chem. Biodiversity, 2023, 20(3), e202201049, DOI:10.1002/cbdv.202201049.
- F. D. Gonelimali, J. Lin, W. Miao, J. Xuan, F. Charles, M. Chen and S. R. Hatab, Front. Microbiol., 2018, 9, 1639, DOI:10.3389/fmicb.2018.01639.
- M. Balouiri, M. Sadiki and S. K. Ibnsouda, J. Pharm. Anal., 2016, 6(2), 71–79, DOI:10.1016/j.jpha.2015.11.005.
- S. V. Salimath and M. Pathak, Inorg. Chem. Commun., 2024, 162, 112226, DOI:10.1016/j.inoche.2024.112226.
- S. V. Salimath and M. Pathak, ChemistrySelect, 2024, 9(7), e202304565, DOI:10.1002/slct.202304565.
- T. Mosmann, J. Immunol. Methods, 1983, 65(1–2), 55–63, DOI:10.1016/0022-1759(83)90303-4.
- W. Ahmad, A. Singh, K. K. Jaiswal and P. Gupta, J. Inorg. Organomet. Polym. Mater., 2021, 31(2), 614–623, DOI:10.1007/s10904-020-01703-6.
- W. Ahmad, A. Singh, V. S. Mishrwan, S. Joshi and A. Rawat, Asian J. Chem., 2023, 35(8), 1770–1774, DOI:10.14233/ajchem.2023.27287.
- S. Sagadevan, S. Imteyaz, B. Murugan, J. Anita Lett, N. Sridewi, G. Weldegebrieal and W. C. Oh, Green Process Synth., 2022, 11(1), 44–63, DOI:10.1515/gps-2022-0005.
- H. Kamil, N. Hiba and A. Ali, J. Opt., 2024, 54(3), 2698–2703, DOI:10.1007/s12596-023-01393-6.
- G. Nabi, M. Bilal Tahir, K. Riaz, T. Iqbal, M. Rafique, S. Hussain, W. Raza, I. Aslam and M. Rizwan, Int. J. Environ. Anal. Chem., 2020, 102(2), 434–442, DOI:10.1080/03067319.2020.1722816.
- G. Behailu Yitagesu, D. Tsegaye Leku and G. Adam Workneh, ACS Omega, 2023, 8, 43999–44012, DOI:10.1021/acsomega.3c06142.
- B. Blessymol, P. Yasotha, V. Kalaiselvi and S. Gopi, Results Chem., 2024, 7, 101291, DOI:10.1016/j.rechem.2023.101291.
- W. Cai, Y. Chen, L. Xie, H. Zhang and C. Hou, Eur. Food Res. Technol., 2014, 238(1), 121–128, DOI:10.1007/s00217-013-2091-x.
- L. Sens, A. S. De Oliveira, A. Mascarello, I. M. C. Brighente, R. A. Yunes and R. J. Nunes, J. Braz. Chem. Soc., 2018, 29(2), 343–352, DOI:10.21577/0103-5053.20170146.
- N. Ajmal, K. Saraswat, Md Afroz Bakht, Y. Riadi, M. Jawed Ahsan and Md Noushad, Green Chem. Lett. Rev., 2019, 12(3), 244–254, DOI:10.1080/17518253.2019.1629641.
- R. Saini and P. Kumar, Inorg. Chem. Commun., 2023, 156, 111221, DOI:10.1016/j.inoche.2023.111221.
- S. Patel, P. Patel, S. B. Undre, S. R. Pandya, M. Singh and S. Bakshi, J. Mol. Liq., 2016, 213, 304–311, DOI:10.1016/j.molliq.2015.11.002.
- J. Kruk, B. H. Aboul-Enein, E. Duchnik and M. Marchlewicz, J. Physiol. Sci., 2022, 72, 19, DOI:10.1186/s12576-022-00845-1.
- M. Ibrahim, H. U. Nabi, N. Muhammad, M. Ikram, M. Khan, M. Ibrahim and A. Alshammari, Molecules, 2023, 28(15), 5926, DOI:10.3390/molecules28155926.
- S. Patel, P. Patel and S. R. Bakshi, Cytotechnology, 2017, 69(2), 245–263, DOI:10.1007/s10616-016-0054-3.
- G. Zhang, X. Chen, J. Guo and J. Wang, J. Mol. Struct., 2009, 921(1–3), 346–351, DOI:10.1016/j.molstruc.2009.01.036.
- K. B. Hiremath, M. Shivashankar and N. Chandrasekaran, ChemistrySelect, 2023, 8(25), e202301772, DOI:10.1002/slct.202301772.
- M. Khater, G. Kulkarni and S. Khater, ES Mater. Manuf., 2024, 26, 1266, DOI:10.30919/esmm1266.
- Y. Mbenga, J. O. Adeyemi, D. M. N. Mthiyane, M. Singh and D. C. Onwudiwe, Results Chem., 2023, 6, 101007, DOI:10.1016/j.rechem.2023.101007.
|
| This journal is © The Royal Society of Chemistry 2025 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *
*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ).
θ).

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K + n
K + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q]
log[Q]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) was seeded into a 96-well microtiter plate. After 24 hours of incubation, allowing a partial monolayer to form, the supernatant was carefully removed, and the wells were gently rinsed with fresh medium. Subsequently, 100 μL of various concentrations of the test drugs were added to the wells. The plates were then incubated at 37 °C with 5% CO2 for 24 hours. Following this incubation, the test solutions were aspirated, and 100 μL of MTT solution (5 mg/10 mL in PBS) was added to each well. The plates were incubated for another 4 hours at 37 °C in a 5% CO2 environment. After incubation, the supernatant was removed, and 100 μL of DMSO was added to each well to dissolve the formazan crystals. The plates were gently shaken, and absorbance was measured at 590 nm using a microplate reader.38 The percentage of cell growth inhibition and the IC50 value were calculated from the dose–response curves.
000 cells per well) was seeded into a 96-well microtiter plate. After 24 hours of incubation, allowing a partial monolayer to form, the supernatant was carefully removed, and the wells were gently rinsed with fresh medium. Subsequently, 100 μL of various concentrations of the test drugs were added to the wells. The plates were then incubated at 37 °C with 5% CO2 for 24 hours. Following this incubation, the test solutions were aspirated, and 100 μL of MTT solution (5 mg/10 mL in PBS) was added to each well. The plates were incubated for another 4 hours at 37 °C in a 5% CO2 environment. After incubation, the supernatant was removed, and 100 μL of DMSO was added to each well to dissolve the formazan crystals. The plates were gently shaken, and absorbance was measured at 590 nm using a microplate reader.38 The percentage of cell growth inhibition and the IC50 value were calculated from the dose–response curves.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations, suggesting the presence of aldehyde or ketone groups, while the peak at 1380 cm−1 corresponds to C–H bending vibrations, confirming the involvement of organic compounds. Aromatic stretching vibrations, indicative of aromatic compounds derived from the extract, are also observed. These peaks are present in both the extract and the TiO2 NPs, confirming that the phytochemicals in the extract play a crucial role in nanoparticle formation. Additionally, the peak at 530 cm−1 is associated with metal–oxygen (Zn–O) stretching vibrations, verifying the formation of TiO2. These functional groups emphasize the role of the plant extract as both a reducing and capping agent in the synthesis of TiO2 NPs. X-ray diffraction (p-XRD) analysis was conducted to evaluate the crystallinity and phase purity of the synthesized TiO2 nanoparticles (TiO2 NPs), as depicted in Fig. 1d. The broadening of the diffraction peaks confirms the nanoscale size of the particles. The prominent peaks, along with their corresponding Miller indices, confirm the crystalline structure of the TiO2 NPs. The observed reflections are consistent with the hexagonal wurtzite phase of TiO2, matching the standard reference pattern (JCPDS card no. 00-021-1272). Using the Debye–Scherrer equation, the average crystalline size of the nanoparticles was estimated to be around 23 nm. In previous studies, TiO2 nanoparticles synthesized using lemon peel extract were reported to have an average crystalline size of 60 nm.43 However, in our study, the average crystalline size was determined to be 23 nm. These findings suggest that neem flower extract serves as an effective green fuel for the synthesis of TiO2 nanoparticles.
O stretching vibrations, suggesting the presence of aldehyde or ketone groups, while the peak at 1380 cm−1 corresponds to C–H bending vibrations, confirming the involvement of organic compounds. Aromatic stretching vibrations, indicative of aromatic compounds derived from the extract, are also observed. These peaks are present in both the extract and the TiO2 NPs, confirming that the phytochemicals in the extract play a crucial role in nanoparticle formation. Additionally, the peak at 530 cm−1 is associated with metal–oxygen (Zn–O) stretching vibrations, verifying the formation of TiO2. These functional groups emphasize the role of the plant extract as both a reducing and capping agent in the synthesis of TiO2 NPs. X-ray diffraction (p-XRD) analysis was conducted to evaluate the crystallinity and phase purity of the synthesized TiO2 nanoparticles (TiO2 NPs), as depicted in Fig. 1d. The broadening of the diffraction peaks confirms the nanoscale size of the particles. The prominent peaks, along with their corresponding Miller indices, confirm the crystalline structure of the TiO2 NPs. The observed reflections are consistent with the hexagonal wurtzite phase of TiO2, matching the standard reference pattern (JCPDS card no. 00-021-1272). Using the Debye–Scherrer equation, the average crystalline size of the nanoparticles was estimated to be around 23 nm. In previous studies, TiO2 nanoparticles synthesized using lemon peel extract were reported to have an average crystalline size of 60 nm.43 However, in our study, the average crystalline size was determined to be 23 nm. These findings suggest that neem flower extract serves as an effective green fuel for the synthesis of TiO2 nanoparticles.








