Open Access Article
Open Access ArticleTapeTech microfluidic connectors: adhesive tape-enabled solution for organ-on-a-chip system integration†
Terry
Ching
 abc,
Abraham C. I.
van Steen
abc,
Abraham C. I.
van Steen
 abc,
Delaney
Gray-Scherr
abc,
Delaney
Gray-Scherr
 abc,
Jessica L.
Teo
abc,
Anish
Vasan
ab,
Joshua
Jeon
ab,
Jessica
Shah
abc,
Jessica L.
Teo
abc,
Anish
Vasan
ab,
Joshua
Jeon
ab,
Jessica
Shah
 d,
Aayush
Patel
ab,
Amy E.
Stoddard
abcd,
Jennifer L.
Bays
abc,
Jeroen
Eyckmans
d,
Aayush
Patel
ab,
Amy E.
Stoddard
abcd,
Jennifer L.
Bays
abc,
Jeroen
Eyckmans
 abc and
Christopher S.
Chen
abc and
Christopher S.
Chen
 *abc
*abc
aBiological Design Center, Boston University, Boston, MA 02215, USA. E-mail: chencs@bu.edu
bDepartment of Biomedical Engineering, Boston University, Boston, MA 02215, USA
cWyss Institute for Biologically Inspired Engineering, Harvard University, Boston, MA 02115, USA
dHarvard-MIT Division of Health Sciences and Technology, Institute for Medical Engineering and Science, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
First published on 5th February 2025
Abstract
A longstanding challenge in microfluidics has been the efficient delivery of fluids from macro-scale pumping systems into microfluidic devices, known as the “world-to-chip” problem. Thus far, the entire industry has accepted the use of imperfect, rigid tubing and connectors as the ecosystem within which to operate, which, while functional, are often cumbersome, labor-intensive, prone to errors, and ill-suited for high-throughput experimentation. In this paper, we introduce TapeTech microfluidics, a flexible and scalable solution designed to address the persistent “world-to-chip” problem in microfluidics, particularly in organ-on-a-chip (OoC) applications. TapeTech offers a streamlined alternative, utilizing adhesive tape and thin-film polymers to create adaptable, integrated multi-channel ribbon connectors that simplify fluidic integration with pumps and reservoirs. Key features of TapeTech include reduced pressure surges, easy priming, rapid setup, easy multiplexing, and broad compatibility with existing devices and components, which are essential for maintaining stable fluid dynamics and protecting sensitive cell cultures. Furthermore, TapeTech is designed to flex around the lids of Petri dishes, enhancing sterility and transportability by enabling easy transfer between incubators, biosafety cabinets (BSCs), and microscopes. The rapid design-to-prototype iteration enabled by TapeTech allows users to quickly develop connectors for a wide range of microfluidic devices. Importantly, we showcase the utility of TapeTech in OoC cultures requiring fluid flow. We also highlight other utilities, such as real-time microscopy and a well-plate medium exchanger. The accessibility of this technology should enable more laboratories to simplify design and setup of microfluidic experiments, and increase technology adoption.
Tribute to George WhitesidesThe authors dedicate this article to George M. Whitesides, who pioneered the field of microfluidics amongst countless others. Christopher Chen thanks Dr. Whitesides for mentoring him during his formative years, and for fostering a cauldron of creativity, rigor, and exceptionalism that has had a lasting impact on the community.Christopher Chen |
1. Introduction
In 1990, Andreas Manz published the seminal paper “Microchemical Total Analysis Systems”, which proposed an approach to integrate and automate the entire spectrum of chemical analysis within microfluidic devices.1 These devices employed microfabrication techniques akin to those used in semiconductor manufacturing and showed initial promise, but widespread adoption was limited due to high production costs.2 The landscape of microfluidic research experienced a pivotal shift in 1998 and 2000 following the publication of papers by George Whitesides and Steve Quake, which introduced “PDMS microfluidics” and “soft lithography”.3,4 These methods simplified the microfabrication process, reduced costs and production time, and broadened access to microfluidic technology across research laboratories.5 Over the years, Whitesides and his colleagues have advanced the democratization of microfluidics through the concept of “frugal microfluidics”, which aims to make advanced research tools more affordable and accessible to a broader scientific community.6–11While PDMS microfluidics and soft lithography have largely addressed issues of device accessibility and cost, the field of microfluidics continues to confront the “world-to-chip” problem.12–18 This challenge involves the efficient integration of macro-scale fluid delivery systems, such as pumps, with micro-scale devices, a bottleneck that remains a critical area of ongoing research and development within the microfluidic community.19–23 The current suite of solutions addressing this problem presents numerous limitations and challenges. Existing options for connecting tubing, pumps and reservoirs to devices are cumbersome and require the manual labor of physically joining many pieces of tubing, connector fittings, and devices. This often leads to tangled tubing and mistaken connections, making such approaches not ideal for scaling to complex or high(er) throughout applications.24–27 Additionally, pressure buildup during the connection of existing connectors can compromise sensitive biological experiments. Sudden surges can create damaging fluid shear forces, disrupting cell cultures or delicate structures within OoC devices.28,29
Many existing world-to-chip solutions offered by manufacturers are proprietary, expensive, and limited in accessibility, making them unsuitable for most laboratories.14,16 These bespoke systems are designed to work exclusively within specific product suites, hindering the broader adaptability and scalability of microfluidic systems, particularly for labs with limited resources. As a result, the complexity and cost of current solutions hinder widespread adoption of microfluidics in research and clinical settings. Addressing these issues is critical for advancing the field and enabling broader application of microfluidic technologies in various disciplines.
In this paper, we introduce TapeTech microfluidics, a flexible, scalable solution to address the world-to-chip problem. Drawing inspiration from the impact of flexible printed circuits (FPCs) and ribbon connectors in the consumer electronics industry, TapeTech leverages adhesive tape and thin-film polymers to create a modular, adaptable system that simplifies fluidic connections. Designed in the context of organ-on-chip (OoC) applications, TapeTech overcomes key challenges such as minimizing pressure buildup, facilitating easy priming, and enabling rapid setup without disrupting fluid flow across cell cultures. By streamlining the integration of OoCs with pumps, TapeTech enhances the potential for high(er)-throughput experimentation, making it an ideal solution for scaling complex biological studies.
Our approach emphasizes accessibility, cost-effectiveness, and ease of production using readily available materials and basic laboratory tools like cutting plotters (<USD $400). The modularity of TapeTech allows users to customize configurations based on their experimental requirements, providing performance on par with commercially available systems. We further demonstrate the versatility of TapeTech in dynamic fluidic flow applications for OoC devices and its seamless integration into existing workflows, including real-time live-cell imaging and automation of fluid handling setups, offering a practical and scalable solution for the microfluidics community.
2. Results and discussion
2.1. Experimental design: key considerations for OoC devices connectivity
We designed TapeTech microfluidic systems to simplify the challenge of building fluidic connections between various components such as pumps, devices, valves, bubble traps, and more. While cutting tubing and manually connecting each fluidic path is feasible for very simple setups, the associated time and effort can become burdensome for progressively more complex circuits. For example, some setups involve dozens of connections wherein a single misconnection or entanglement of tubing would lead to system failure (Fig. 1A). To address this challenge, we take inspiration from the ribbon connector used in the electronic industry to replace individual wire connections and proposed that an integrated array of microfluidic channels could be used to avoid the challenges of manually connecting individual tubing and connectors (Fig. 1B).In addition to simplifying the implementation of wiring diagrams, another challenge is the connectors themselves. Multiple types of connectors between tubing and devices have been developed (Fig. 1C), yet these often require time-consuming and labor-intensive manual one-connector-at-a-time assembly, especially in setups with multiple channels. Furthermore, the lack of cross-compatibility between connectors means that devices from different systems may not interface seamlessly, leading to limitations in experimental flexibility. Additionally, many traditional connectors have a large vertical profile, which can prevent properly fitting systems within enclosed environments such as Petri dishes, making it difficult to maintain sterility and fit into standard lab equipment.
Our solution, TapeTech, utilizes xurography combined with adhesive tape to create flexible, ribbon-like microfluidic connectors that reduce clutter and simplify fluidic circuits. This ribbon layout simplifies the implementation of complex fluidic circuits by organizing all fluidic paths into a single, cohesive assembly (Fig. 1B and D). The layout can enable simultaneous sealing to all connection points to a device in a single step. The low-profile design fits within Petri dishes, maintaining sterility and minimizing vertical height (Fig. 1D), while integrated flat connectors provide seamless compatibility with devices and pumps. This compact solution accelerates setup time, is easily modifiable to ensure compatibility with various devices, and is designed for easy transport between incubators, microscopes, and biosafety cabinets.
2.2. Demonstrating OoC connections with TapeTech
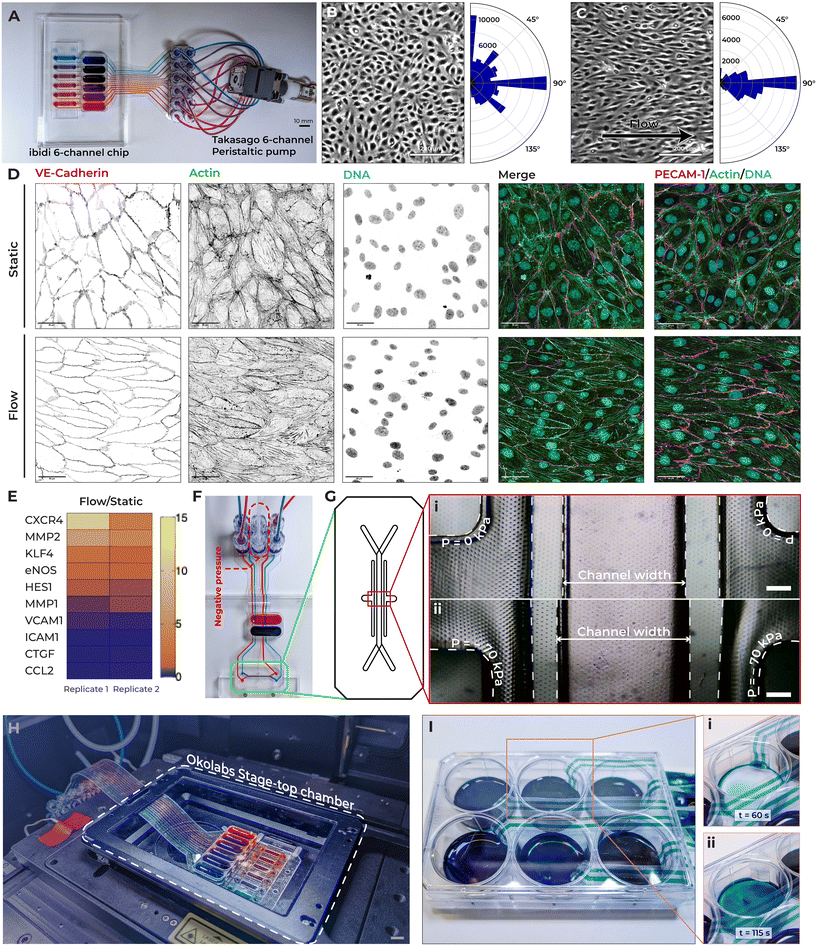
To test the TapeTech concept, we focused on an initial use case with an ibidi 6-channel device designed for parallel flow assays. Traditionally, establishing flow into an OoC device relies on rigid tubing and connectors to interface with pumps, reservoirs, and bubble traps. While these are effective in delivering culture medium to and from fluidic chips, challenges arise when connecting multiple channels. For example, connecting an ibidi 6-channel chamber slide to a pump, reservoir, and bubble trap results in a setup that is not only labor intensive but also bulky (Fig. 2A). The system requires a minimum of 24 separate pieces of silicone or rigid tubing, and 36 connector fittings, all of which need to be manually tightened and assembled (Fig. 2B). This dense network of tubing and fittings often overlaps and interlaces, leading to increased clutter. The number of components creates a cumbersome setup that takes up considerable space both within and outside the incubator, making the system difficult to transport. This bulkiness further complicates essential tasks such as transferring the setup between the incubator and microscope or BSC for medium changes or imaging. Moreover, the total volume of fittings, tubing, and the chip prevents the system from fitting within standard sterile enclosures, increasing the risk of contamination as components remain exposed during transfers.To address these challenges, we introduce TapeTech microfluidics. Much like how flexible printed circuits (FPCs) complement traditional wiring in electronics, TapeTech microfluidics offers a streamlined alternative to rigid tubing for interconnecting fluidic components. By adopting the FPC form factor, TapeTech assemblies can be quickly designed and fabricated to interface seamlessly with both existing and custom 3D-printed chips.
We designed and fabricated an integrated assembly that streamlines connecting a pump to the same ibidi 6-channel chamber slide as above (Fig. 2C, Video S1†). With TapeTech, the number of connector fittings was reduced, with only 12 fittings needed (Fig. 2D), versus the 36 required in the traditional setup (Fig. 2A). With this assembly, the reservoir is integrated into the TapeTech assembly, simplifying the connection process and reducing overall complexity (Fig. 2C–E). The reservoir itself is designed to serve as a passive bubble trap, allowing any bubbles that rise to the surface to be diverted away from the fluid flowing into the cell chamber (Fig. 2F).
The open-format reservoir, which facilitates easy medium changes and drug delivery, is enabled by the slender and flexible structure of TapeTech. This design allows the connectors to conform around the lid of a Petri dish, ensuring that both the chip and reservoirs remain enclosed within the dish, maintaining sterility throughout handling and operation. The streamlined design reduces the risk of incorrect connections and creates a more compact system, making it easier to transport into and out of incubators or BSCs for imaging or medium changes.
To further demonstrate the versatility and ease of integrating TapeTech connectors across various use cases, we selected two representative format of OoC devices: a PDMS-based emulate lung chip and an acrylate-based 3D-printed fluidic chip (Fig. 2G and H, Video S2†). In each case, custom-designed TapeTech connectors tailored to the specific form factors and material properties of the chips seamlessly interfaced with pumps and reservoirs. This adaptability highlights how the TapeTech system can easily be customized for different chip materials and geometries, ensuring reliable connections and minimizing the complexity associated with multi-channel setups.
2.3. Benefits of TapeTech connectors
A key advantage of TapeTech connectors is their ability to reduce the risk of harmful pressure surges during connections. Traditional plug-based or tolerance-based connectors often cause a pressure surge when plugs are inserted into inlets or outlets (Fig. 3A and B). For example, when a plug is inserted into a device inlet, we observed a sudden spike in flow rate to approximately 50 μL min−1, displacing around 30 μL of fluid (Fig. 3E and F). Similarly, inserting a rigid tube into a PDMS device inlet (tolerance fit) caused a flow rate spike of about 40 μL min−1, displacing approximately 4 μL of fluid. These abrupt changes in flow rate caused by pressure fluctuations can be detrimental to sensitive components, particularly adherent cells in microfluidic channels, due to the increased shear stress.30 In more delicate OoC systems, where soft extracellular matrices (ECMs) were employed,31 these sudden pressure surges can be damaging. For example, in a microfluidic device with a fibrin gel cast into a chamber to form a hollow channel (using an acupuncture needle as a mold), we observed a 23% expansion in the width of the channel when using plug-based connectors due to the abrupt pressure surge (Fig. 3K, Video S3†). Similarly, using tolerance-based fittings caused a 5% expansion in the fibrin gel, both due to the pressure surges (Fig. 3L).To mitigate pressure surges in traditional tubing, techniques have been employed such as pre-compressing the downstream silicone tubing before inserting the plug, then gradually releasing it during insertion, can help balance the pressure. Users can also insert the plug slowly to prevent sudden surges. While these methods can reduce shear stress, their effectiveness is highly dependent on the skill and precision of the user. Additionally, improper priming during plug insertion can result in bubble formation, which can obstruct flow and harm cells.32 To minimize this risk, overfilling the inlet so that a visible droplet remains at the end of the plug before insertion can help prevent bubbles. However, this approach can inadvertently introduce excess fluid, leading to additional pressure surges.
TapeTech connectors avoid such drastic spikes in fluidic flow. Our method involves a two-step process to adhere our connectors to the devices (Fig. 3C, Video S1†). First, we adhere our connectors to the devices. Our connector features a vent hole on top of 1 mm in diameter, allowing air to escape while preventing fluid overflow due to surface tension maintaining the fluid in place. Once the fluidic space between the Tapetech ribbon and the device is completely filled with fluid, we then place a second piece of tape over the vent hole to seal the system. The flowrate measured during the application of TapeTech connectors was negligible, and the volume of fluid displaced by the system was less than 1 μL (Fig. 3I and J). Additionally, the width change of the fibrin gel channel was minimal (Fig. 3M).
2.4. Design guidelines and durability of adhesive connection
To ensure the robustness and leak-proof performance of the double-sided tape, we subjected our connectors to high-pressure conditions and assessed their ability to maintain a sealed environment and prevent leakage. Leakage occurs when adhesive failure happens. As shown in Fig. 4A, pressure in the fluidic channel exerts a force on the opening, described by the equation P = F/A, where pressure (P) generates force (F) over the area (A = πd2/4). If the force generated by the pressure exceeds the adhesive strength of the tape, adhesion failure occurs, resulting in leakage.Our goal was to determine the optimal design parameters for maintaining a leak-proof seal. We investigated four key factors: 1) tape width (w), 2) hole diameter (d), 3) substrate material (m), and 4) the impact of extended duration inside the incubator (in+, in−) on adhesive integrity. At baseline, we set w = 2 mm, d = 2 mm, the substrate was polystyrene (PS), and no incubation was applied. For each test (n = 6), the samples were subjected to a five-step pressure ramp profile, consisting of 40 kPa for 60 s, 80 kPa for 60 s, 120 kPa for 60 s, 160 kPa for 60 s, and 200 kPa for 60 s. At the conclusion of the experiment, the results were visualized in Fig. 4B–E as heatmaps, which depict the survival of samples (n) under progressively increasing pressure levels across various conditions or substrates.
When the tape width (w) was varied between 1 mm and 3 mm, the survival outcomes were reduced for samples with w = 1 mm (Fig. 4B). This suggests that wider tape provides greater surface contact, thereby distributing the applied pressure more effectively and enhancing adhesive strength. When the hole diameter (d) was varied, the survival outcomes were uniform across all test conditions (d1, d2, d3), with all samples (n = 6) successfully withstanding the entire five-step pressure ramp profile, from 40 kPa to 200 kPa (Fig. 4C). These results indicate that hole diameter does not have a significant impact on survivability.
Based on these findings, a tape width greater than 2 mm is necessary to ensure robust survivability under pressure, as narrower tapes are less effective at distributing stress. On the other hand, hole diameter variations between 1 mm and 3 mm were found to have minimal influence on survivability.
We conducted tests on a range of substrate materials commonly used in OoC systems, including polystyrene (PS), polypropylene (PP), 3D-printed acrylate, polymethyl methacrylate (PMMA), and polydimethylsiloxane (PDMS). For our tests, we used 3M double-coated tape (9495LE), designed to adhere effectively to low surface energy materials. For PDMS devices, we used a tape (Nitto, 5302A) specifically engineered to bond with silicone rubber. The tape adhered to PS, PP, 3DP, and PMMA demonstrated perfect survivability (n = 6) across all pressure levels tested (Fig. 4D and S1†). Notably, even the 3D-printed substrate, which has inherent surface roughness due to the printing process (produced using a Formlabs 3D printer), exhibited strong adhesion. The pressure-sensitive adhesive in the double-coated tape was able to effectively fill in surface irregularities, demonstrating its ability to maintain a robust seal even on textured materials.
In contrast, PDMS exhibited a decline in performance under higher pressures. Specifically, at 200 kPa, no PDMS samples (n = 0) survived, and at 160 kPa, only two samples (n = 2) remained intact. The reduced adhesion performance on PDMS can be attributed to its low surface energy, which makes it inherently resistant to bonding. This behavior is consistent with expectations, as silicone-based materials like PDMS resist forming strong adhesive bonds due to their unique surface chemistry. Despite the limitations of PDMS, the level of bonding achieved was still adequate for most OoC applications, as these devices typically operate under pressures well below 20 kPa—an order of magnitude lower than the pressures tested in this study.33–37 This indicates that, while PDMS does not form as strong a bond compared to other substrates, the tape still provides a sufficient seal suitable for standard OoC conditions. Additionally, our devices rely on negative pressure flow (Fig. 2E); the tape is not subjected to the positive pressures that typically lead to adhesive failure. Overall, our study demonstrated that the tape can effectively seal a variety of materials which are frequently used in microfluidic manufacturing. This versatility underscores the potential utility across diverse microfluidic workflows, making it a reliable option for ensuring leak-proof connections in OoC devices.
Additionally, we evaluated the performance of the tape under two environmental conditions to assess its suitability for OoC applications, which require controlled environments to ensure cell viability. Our goal was to determine whether extended exposure to incubator conditions would affect adhesive strength. First, we tested the tape immediately after adhesion at room temperature. Second, we conditioned tape samples in an incubator at 37 °C with 85–95% humidity for four days to simulate typical OoC operating conditions and assess the long-term durability of the adhesive. The tape in both conditions demonstrated perfect survivability (n = 6) across all pressure levels tested (Fig. 4E). This suggests that the adhesive bond was not affected by humidity and temperature over time, environments like those found in incubators. This result is particularly important for OoC applications, where devices are typically housed in incubators for extended periods.38 These findings indicate that the tape not only performs well across a variety of substrates but also maintains its performance under real-world incubator conditions, making it highly suitable for long-term use in OoC systems.
To further confirm that the adhesive tape provides a reliable, leak-proof seal and prevents fluid leakage at the tape-device interface, we conducted a five-day perfusion culture test at a flow rate of approximately 50 μL min−1 using a PS microfluidic chip. A pressure not exceeding 0.61 kPa is required to achieve a flow rate of 50 μL min−1 through the TapeTech ribbon channels, which measure 1 mm in width, 170 μm in height, and approximately 300 mm in length, as calculated using the Elveflow online microfluidic calculator. A phosphate-buffered saline (PBS) solution mixed with green fluorescein dye was circulated through the system, allowing us to visually track any potential leakage. Fluorescent intensity values from microscopy images captured at the interface between the tape and chip show no observable leakage after five days of continuous perfusion in an incubator (Fig. 4F). Importantly, statistical analysis of the fluorescence signals at the tape region showed no significant differences in mean gray values between day 0 and day 5, confirming that the tape maintained a robust, leak-proof seal throughout the experiment (Fig. 4G). We note that, based on empirical observations, the adhesive strength of the tape diminishes after detachment; therefore, we do not recommend reusing it after detachment. The TapeTech ribbon assembly is inexpensive (∼$0.63 per ribbon; Fig. 2B, Table S2†) and best employed for single-use applications.
2.5. Fabrication and suggested workflow for cell-based applications
The fabrication of TapeTech ribbon begins with designing the cut files in computer-aided design (CAD) software. Only two-dimensional (2D) files are required, and the cutting plotter precisely cuts polyethylene terephthalate (PET) films (100 μm) and double-sided tapes (170 μm) according to the CAD design (Fig. 5A). A through-hole was also cut over the cell culture chamber to prevent potential autofluorescence from the tape during microscopy, ensuring clear imaging of the region of interest. The tape was cut with the release liner still on. To ensure accurate alignment during assembly, two additional alignment holes were included in the cut files, which fit into a jig. Once the films and tape were cut, the layers were carefully placed on the jig, and a scraper tool was used to ensure conformal contact between the layers, promoting strong adhesion (Fig. 5B). Additional components, such as the PMMA reservoirs and outlets, were attached to the assembly using double-sided tape (Fig. 5C). After assembly, the TapeTech ribbon were baked at 80 °C for a minimal of 2 hours to assist in the bonding. All TapeTech ribbon assemblies followed this method, with adjustments to the CAD design file as needed for customization (Fig. S2–S8†).To demonstrate the process, we showcase the workflow of connecting the TapeTech ribbon to an ibidi 6-channel chip. The procedure begins with a chip pre-seeded with cells, as per the protocol suggested by OoC chip manufacturers (Fig. 5D). This standard protocol usually involves coating the channels, seeding the cells, and allowing several hours for the cells to settle and adhere to the channel surface before connecting the chip to flow systems. A laser-cut jig with alignment pins was placed around the chip to assist in aligning the TapeTech assembly (Fig. 5E; Video S1†) The release liner on the adhesive tape was then removed from the TapeTech ribbon assembly, and the assembly was aligned with the pins on the jig (Fig. 5F). Gentle pressure was applied using a scraper tool to ensure the adhesive formed a conformal contact with the chip (Fig. 5G).
Next, a 200 μL multipipette was used to prime the channels (Fig. 5H). Fluid was slowly dispensed until it visibly reached the inlet of the cell chamber. The TapeTech assembly features a small 1 mm vent hole directly over the inlets and outlets of the cell chamber to allow air to escape while preventing fluid overflow, as surface tension keeps the fluid in place. Once the fluid reaches the cell chamber inlets, hydrostatic pressure naturally fills the inlet and outlet. When the fluid had completely filled the inlet and outlet spaces, a second piece of tape was applied over the vent holes to seal the system. For easier application and providing greater tolerance for alignment errors, a larger strip of tape, ibiSeal (7 × 56 mm), that covers all six aligned vents was employed (Fig. 5I). The reservoirs were then filled with fluid using the multipipette (Fig. 5J). Finally, the pump was activated to begin perfusion culture, ensuring that the flow direction followed the orientation illustrated in Fig. 2E.
The fabrication of the TapeTech ribbon assembly is highly customizable for a variety of chip designs. While we specifically demonstrated its application with the ibidi 6-channel chip, other assemblies can be quickly designed and fabricated, as shown in Fig. 2G and H. It is important to note, however, that the materials used in TapeTech (i.e., PET film and acrylic-based adhesive) may limit its suitability for certain microfluidic applications. For instance, it may not be appropriate for environments involving harsh solvents, corrosive chemicals, or high-temperature operations exceeding 100 °C.39 Tape-based microfluidics, however, has been successfully tested in applications such as polymerase chain reaction (PCR) chips, which require temperatures up to 95 °C.40 Nonetheless, we believe that TapeTech is ideally suited for OoC applications, where the use of corrosive chemicals and extreme temperatures is not typically required.
2.6. Demonstrating TapeTech in cell-based applications requiring flow
To demonstrate the practicability of TapeTech for cell-based applications, we integrated a TapeTech ribbon assembly to apply fluid shear stress on human umbilical vein endothelial cells (HUVECs) monolayer that were seeded in a fluidic flow chamber slide. In response to fluid shear stress, endothelial cells align to the direction of flow, rearrange their cytoskeleton,41 reduce inflammatory status,42 mature their glycocalyx maturation,43 and improve barrier function.44 Disruptions to the cells ability to respond to fluid shear stress is a hallmark of, and is often the underlying cause of, many vascular pathologies.45 In this demonstration, we used the TapeTech ribbon assembly to connect a multi-channel chip to a peristaltic pump (Fig. 6A). Specifically, we employed an multi-channel chip with channel dimensions of 100 μm (height) by 2000 μm (width), similar to the commercially available 6-channel fluidic chips from ibidi GmbH. A Takasago 6-channel peristaltic pump facilitated fluid flow through the chip.HUVECs were seeded into the fluidic chambers and subjected to a 2-day perfusion culture, starting 6 hours after seeding. A flow rate of 167 μL min−1, corresponding to approximately 6 dynes cm−2 of shear stress, was applied to the chambers. We did not observe any signs of cytotoxicity, consistent with previous reports demonstrating that tape-based microfluidic systems are compatible with cell culture.39,46,47 Additionally, we conducted a live/dead assay to further verify this, as shown in Fig. S9.† In the static control condition, HUVECs exhibited the classic cobblestone-like morphology (Fig. 6B). However, under flow conditions, the cells developed an elongated phenotype and aligned in the direction of fluid flow (Fig. 6C). Quantification of cell alignment across multiple channels revealed that HUVECs under flow predominantly aligned at a 90° angle to the flow, while cells in static conditions displayed a more randomized alignment pattern (Fig. 6B and C).
In addition to alignment, flow conditions led to the reorganization of the actin cytoskeleton and the adherens junction protein VE-cadherin along the flow direction (Fig. 6D). Moreover, the surface protein PECAM-1, a surface molecule trapped at junctions via homophilic interactions with the PECAM-1 on adjacent cells,48 clearly exhibits the overlapping fish scale phenotype characteristic of in vivo endothelial cells (Fig. 6E).49 We further investigated the effects of laminar shear stress on gene expression in HUVECs by conducting quantitative polymerase chain reaction (qPCR) analyses. Previous studies have shown that endothelial cells exposed to laminar shear stress activate numerous mechanosensitive genes while suppressing proinflammatory genes.44,50–52 Comparison of endothelial cells cultured under flow conditions using TapeTech versus those grown statically show consistent upregulation of mechanosensitive genes such as KLF4 and eNOS, and downregulation of inflammatory genes such as ICAM1 and CTGF (Fig. 6E, Table S1†). Together, these findings demonstrate that the laminar shear stress generated by the TapeTech system effectively induces phenotypical and transcriptional changes in endothelial cells that align with prior research.
These demonstrations highlight the utility of the TapeTech system in seamlessly connecting OoC devices to existing pumps, enabling researchers to investigate biological questions involving fluidic flow. Its flexibility and adaptability in integrating with a variety of OoC platforms open new avenues for studying complex cellular behaviors under tightly controlled fluidic conditions. The ability to connect multiple channels easily and replicate physiological mechanical forces, such as shear stress, presents exciting opportunities for tissue engineering and regenerative medicine, where understanding the influence of mechanical cues on cell differentiation and tissue formation is essential. It is important to note that in the context of cell-based applications, where proteins and growth factors are critical for influencing cell responses, the double-sided tape may absorb these molecules (Fig. S10†). This is a well-documented phenomenon in the microfluidic field and various surface passivation methods can be applied to mitigate this issue and reduce molecular absorption, ensuring more accurate biological interactions.53–55 Finally, the scalability of TapeTech for multi-channel setups positions it as a valuable tool for expanding into multi-organ models or complex co-culture systems, offering deeper insights into systemic biological interactions and advancing the field of personalized medicine.
2.7. Other cell-based applications
To further demonstrate the versatility of TapeTech in advancing biological research, we applied the system in three distinct applications: (1) applying negative pressure to stretch the porous membrane in the emulate lung chip, (2) enabling live-cell microscopy imaging, and (3) serving as a medium exchanger in standard tissue culture plates.To demonstrate how TapeTech can enhance biological research through its adaptability. We connected a TapeTech ribbon assembly to the PDMS-based emulate lung chip and successfully applied negative pressure to stretch the porous membrane within the cell chamber (Fig. 6F and G).56 We employed a negative pressure of −70 kPa, as suggested by the device manufacturer. Over the past several decades, there has been growing recognition of the critical role mechanical stretch plays in regulating various cellular processes, including proliferation, signaling, tone, and remodeling.57 Mechanical stretch is known to influence key functions across a range of cell types, such as skeletal, cardiac, vascular, epithelial, and neuronal cells.58 For example, in the lung, low-magnitude stretches of 5–10% are classified as physiological, while high-magnitude stretches of 20% or more are considered pathological, simulating the conditions experienced during diseases like hypertension.59 The TapeTech system enables seamless integration of fluidic connections for medium exchange and negative pressure connections for stretching the porous membrane, making it well-suited for coupling with the emulate lung chip in OoC applications.
One of the key advantages of adapting TapeTech to fluidic chip is its ability to fit into microscope stage-top chambers for extended live-cell imaging. Live-cell imaging of OoC systems is practically challenging with conventional adaptors and fittings because they are often too bulky to fit under the lid of the microscope stage chamber. Additionally, standard tubing cannot be positioned around the chamber lid, making long-term live imaging practically challenging, as the setup must ensure adequate humidity, oxygen and carbon dioxide exchange throughout the process. TapeTech addresses these challenges with its slim footprint, which fits easily around the lid of the microscope stage-top chamber. By eliminating the need for bulky adaptors, the entire chip and reservoir can fit snugly inside the chamber, ensuring proper maintenance of carbon dioxide levels and humidity. We demonstrate this capability by connecting an ibidi 6-channel fluidic chip with our TapeTech assembly, which fits seamlessly under the lid of a microscope stage-top chamber (H301-K-FRAME, Okolabs) without any modifications (Fig. 6H).
TapeTech is not constrained to just OoC applications but can be customized and used as a medium exchanger in standard cultures. To demonstrate this, TapeTech connectors easily fit around the lid of a 6-well plate, enabling its use as a medium exchanger (Fig. 6I, Movie S5†). Since our TapeTech conduits can flex around the well-plate lid, there is no need to drill holes or redesign the lid. By connecting these conduits to pumps, the medium in the well-plate can be efficiently drained (Fig. 6I-i) and replenished (Fig. 6I-ii). By integrating programmable pumps, automated media exchange with TapeTech streamlines tissue culture process by reducing workflow duration, increasing reproducibility, and saving time and labor.
3. Conclusion
In conclusion, the TapeTech microfluidic system presents a robust and adaptable solution to the challenges associated with the world-to-chip interface, particularly in biological research. Traditional methods for connecting fluidic devices rely on bulky adaptors, rigid tubing, and complex fittings, which result in labor-intensive setups prone to connection errors, tangling, and overall inefficiency. These conventional systems are not only cumbersome but also difficult to transport, limiting their practicality in dynamic research environments. TapeTech overcomes these limitations with its streamlined, flexible design, enabling a more efficient, clutter-free setup that reduces user error and enhances portability. The adaptability and ease of integration of TapeTech with existing laboratory setups allow for a more streamlined and efficient experimental workflow, which is critical in the rapidly evolving field of biological research. The utilization of commercially available materials and equipment, such as double-sided tapes and cutting plotters, underscores the versatility, rapidity of design iteration, cost-effectiveness, and accessibility of this approach.Furthermore, the successful demonstration of the TapeTech connectors in maintaining a leak-proof and robust interface under various testing conditions, including high(er) pressures and humid environments, validates its reliability for biological experiments. The ability to customize connector setups to interface seamlessly with standard and specialized fluidic chips, as evidenced by the integration with commercially available fluidic chip systems and well plates, enhances its practicality for diverse research requirements. The application of TapeTech in live-cell imaging and its ability to maintain optimal gas exchange in confined microscopy incubator setups help alleviate practical challenges commonly encountered in the field. This capability, when paired with the potential for automation through programmable pumps, offers added advantages for researchers who require real-time monitoring and observation of in vitro systems. It facilitates continuous data collection, leading to observations that may not be apparent through intermittent monitoring alone.
In summary, the approach presented here simplifies the technical complexity of microfluidic experiments for cell biology and life science applications. The adaptability of TapeTech for multi-channel setups positions it as a powerful tool for advancing into multi-organ models or complex co-culture systems. Additionally, although our demonstrations emphasize the high customizability of the TapeTech system for laboratory settings at small-scale production, the potential for mass manufacturing remains promising. Leveraging existing manufacturing processes, such as stamping double-sided tape used in the electronics industry, could enable broader deployment. However, further studies are needed to assess the repeatability and alignment precision required for high-throughput production. This adaptability, paired with future scalability efforts, could enhance the potential for commercialization, making advanced microfluidic technologies more accessible, cost-effective, and widely available for research and clinical applications.
4. Materials and methods
4.1. Materials
170 μm thick double-sided coated tapes (9495LE) were purchased from 3M Company (Saint Paul, Minnesota, USA). SLICK PAPER silicone release paper, PMMA sheets (thickness = 3 & 5 mm), and Nitto double-coated tape 5302A-50 (Nitto Denko Corporation, Osaka, Japan) were purchased on Amazon (Seattle, WA, USA). PET films of varying thickness (0.001′′, 0.002′′, and 0.004′′) were purchased from CS Hyde (Lake Villa, IL, USA). The cutting plotter employed was a Silhouette Cameo 4 (Silhouette America, Utah, USA). Polymer coverslips (coverslips for sticky-Slides), 6-channel chamber slides (μ-Slide VI 0.4 and μ-Slide VI – Flat), self-adhesive cover film (ibiSeal), were acquired from ibidi GmbH (Gräfelfing, Germany). Silicone tubing (Liveo™ Pharma-50) was acquired from Qosina (Ronkonkoma, NY, USA). GFP expressing human umbilical vein endothelial cells were purchased from Angio Proteomie (Boston, MA, USA). Cell culture medium, EGM2 was purchased from Lonza (Basal, Switzerland). Collagen type I (rat tail), was purchased from Corning. All secondary antibodies (Alexa Fluor), phalloidin stain, and FITC (5/6-fluorescein isothiocyanate) were purchased from ThermoFisher (Waltham, MA, USA). PECAM-1 antibodies were purchased from BD Biosciences (Franklin Lakes, NJ, USA). VE-cadherin antibodies were purchased from Cell Signaling (Danvers, MA, USA). Hoechst nucleic acid stains were purchased from Life Technologies (Carlsbad, CA, USA).4.2. Fabrication of TapeTech fluidics
All tape and polymer thin films were precisely cut to dimension using a Silhouette Cameo cutting plotter. The design files were created in Rhinoceros (Robert McNeel & Associates, WA, USA) and exported in DXF format. These files were then imported into Silhouette Studio® software (Silhouette America, Utah, USA) and sent to the cutting plotter. To facilitate the cutting of the tape, silicone release paper was first attached to the cutting mat with the non-stick side up, ensuring that the double-sided tape could be easily removed post-cutting. For polymer sheets, the sheets were placed directly on the sticky cutting mat before cutting. PMMA sheets of thickness 3 mm and 5 mm were cut using an Epilog Mini 24 laser cutter (Epilog Laser, CO, USA).4.3. Quantitative measurement of flow rate and fluid displacement
Flow rate and fluid displacement were quantified by using a camera to monitor the movement of a blue dye within a silicone tube. Initially, a small drop of blue dye was placed into the tube, followed by a small air column to separate the dye from the subsequent fluid. The tube was then connected to the device, and de-ionized (DI) water at room temperature was used for all flow rate measurements. A camera was positioned directly above the tubing to record a video at 60 frames per second (fps) when the connectors were connected to the device. This video was then converted into a TIFF stack using a Python code, and the motion of the blue dye was tracked with TrackMate in Fiji (ImageJ).60–62 The velocity of the dye drop [mm s−1] was correlated with volumetric displacement [μL min−1], leveraging the known relationship between the volume of fluid in the tube and the distance it occupies within the tube.4.4. Quantitative measurement of fibrin gel expansion due to connectors
Single-channel devices cast with fibrin gel were used for the experiments. The microfluidic devices, fabricated from multiple layers of tape cut using a cutting plotter, consisted of a needle guide and a fluidic chamber for casting the fibrin gel. An acupuncture needle, serving as a sacrificial mold, was inserted into the device. Needles with a 250 μm diameter were pre-soaked in a 0.1% bovine serum albumin (BSA) solution for 30 minutes to facilitate easy removal after gel crosslinking. The fibrin gel precursor, containing 5 mg mL−1 fibrinogen, was mixed with 0.5 μL thrombin (1 U mL−1), and the solution was immediately pipetted into the device through the gel ports. The devices were incubated at 37 °C with 5% CO2 for 15 minutes to allow the fibrin gel to fully crosslink. Once crosslinking was complete, the needle was carefully removed, leaving a 250 μm diameter channel in the fibrin gel.The fibrin gel device was then connected to a separate fluidic chip using plug-based ports and fluidic tubing, which was primed with fluid to eliminate any air bubbles. The device was placed under a microscope, and video was captured during the plug insertion to monitor performance. A similar setup was used for tubing in PDMS and for the tape-based condition by swapping out the fluidic chip accordingly.
4.5. Quantitative measurement of burst pressure duration
For the pressure tolerance test, tapes and PET film of various diameters were cut using a cutting plotter. These tapes and film assemblies were adhered to different substrates, depending on the test parameters. One end of the substrate was connected to an Elveflow (Paris, France) OB1 flow control instrument, with the pressure programmed to follow a five-step pressure ramp profile, consisting of 40 kPa for 60 s, 80 kPa for 60 s, 120 kPa for 60 s, 180 kPa for 60 s, and 200 kPa for 60 s.4.6. Preparation and cultivation of HUVECs for microfluidic device integration
HUVECs (Angio Proteomie, Boston, MA, USA) were cultured in a 37 °C, 5% CO2 incubator and supplemented with EGM-2 endothelial cell growth medium (Lonza, Basel, Switzerland). Cells were passaged at 70–80% confluency, with only cells between passages 3 and 10 being used for experiments. Fluidic devices, such as the ibidi smart slide, μ-Slide VI 0.4, μ-Slide VI – Flat, and self-fabricated chamber slides, were initially coated with type I collagen (5 μg cm−2 in PBS) for 1 hour inside the incubator. Subsequently, the channels were washed three times with PBS, and any remaining PBS was thoroughly aspirated from the fluidic channels. HUVECs were dissociated from the culture flasks, centrifuged to form a cell pellet, and the supernatant was discarded. The cells were then resuspended in EGM-2 at the required concentrations and seeded into the devices: 4 × 106 cells per mL. After allowing 1 hour for cell attachment, the devices were refreshed with fresh EGM-2 medium. The devices were incubated for at least 6 hours to allow the HUVECs to form a confluent layer before attaching the TapeTech connectors to initiate perfusion culture (Fig. 6). The flow rate was determined by first establishing the desired shear stress and then using the microfluidic calculator provided by Elveflow to calculate the required flow rate (https://www.elveflow.com/microfluidic-calculator/).4.7. Quantification of cell alignment
We developed a Python-based image analysis pipeline to quantify cell orientations from a series of microscopy images (converted to JPEG). The script utilizes OpenCV for image processing and contour detection, and matplotlib for data visualization. Images were first converted to grayscale and then smoothed using a Gaussian blur with a tunable kernel size. Adaptive thresholding was applied to the blurred images to generate binary images, with adjustable block size and constant parameters. Contours were detected from the binary images, and cells near the image borders (within 5% of the border width or height) were excluded to avoid artifacts from partial cells. For each detected contour, the minimum area rectangle was fitted, and the orientation angle was calculated, with 0° (and 180°) indicating the longer axis of the cell being perfectly vertical and 90° indicating it being perfectly horizontal.4.8. Immunofluorescence staining and imaging
The HUVECs in the flow chamber were fixed in 4% paraformaldehyde (PFA) for 5 minutes at 37 °C and washed three times with PBS. The cells were then permeabilized with 0.1% Triton-X (Sigma Aldrich, St. Louis, MO, USA) in PBS for 10 minutes. Following permeabilization, the sample was blocked with 2% BSA in PBS for 1 hour. The cells were then stained with fluorescently conjugated antibodies against either VE-cadherin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Alexa Fluor® 647 Mouse Anti-Human CD144, BD, cat. nr. 561567, Franklin Lakes, NJ, USA) or PECAM-1 (1
200, Alexa Fluor® 647 Mouse Anti-Human CD144, BD, cat. nr. 561567, Franklin Lakes, NJ, USA) or PECAM-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Alexa Fluor® 647 Mouse Anti-Human CD31, BD, cat. nr. 558094, Franklin Lakes, NJ, USA), phalloidin (1
200, Alexa Fluor® 647 Mouse Anti-Human CD31, BD, cat. nr. 558094, Franklin Lakes, NJ, USA), phalloidin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Alexa Fluor™ 555 Phalloidin, ThermoFisher, cat. nr. A34055, Waltham, MA, USA), and Hoechst 33342 (1
1000, Alexa Fluor™ 555 Phalloidin, ThermoFisher, cat. nr. A34055, Waltham, MA, USA), and Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Life Technologies, cat. nr. 62249, Carlsbad, CA, USA) in a 2% BSA solution at 4 °C for 48 hours. After this, the devices were washed three times with PBS and stored at 4 °C until imaging. Images were acquired on an LSM980 confocal microscope with Airyscan 2 Module (Zeiss GMBH, Oberkochen, GER) with a 32× objective (C-Achroplan 32×/0.85 NA, Zeiss GMBH, Oberkochen, GER). A z-stack was acquired at 1 AU and 1× Nyquist and the final images were created in ImageJ62 by maximum intensity projection.
000, Life Technologies, cat. nr. 62249, Carlsbad, CA, USA) in a 2% BSA solution at 4 °C for 48 hours. After this, the devices were washed three times with PBS and stored at 4 °C until imaging. Images were acquired on an LSM980 confocal microscope with Airyscan 2 Module (Zeiss GMBH, Oberkochen, GER) with a 32× objective (C-Achroplan 32×/0.85 NA, Zeiss GMBH, Oberkochen, GER). A z-stack was acquired at 1 AU and 1× Nyquist and the final images were created in ImageJ62 by maximum intensity projection.
4.9. Gene expression analysis by quantitative polymerase chain reaction (QPCR)
Total RNA from endothelial cells was isolated using GenElute™ Mammalian Total RNA Miniprep kit (Qiagen, cat# 74106) according to manufacturer's instructions. cDNA was synthesized from RNA using qScript cDNA Supermix (Quanta Sciences, cat# 95048-100) according to manufacturer's instructions and cDNA was then used as a template for QPCR using SYBR™ Green PCR Master Mix (Life Technologies, cat# 4367659). QPCR was performed with the Applied Biosystems ViiA 7 Real-Time PCR system (Life Technologies), with 40 cycles of 95 °C for 3 s, 60 °C for 10 s, and 72 °C for 20 s. Specific gene targets were detected with the primers listed in Table S1.† Relative quantification was performed using the comparative Ct method.4.10. Statistical analysis
All error bars in this paper were calculated using the standard deviation for each condition, reflecting the variability of the data. Statistical differences among conditions were assessed using two-sample t-tests. To mitigate the risk of type I errors from multiple comparisons, a Bonferroni correction was applied, modifying the significance threshold proportionally to the number of tests performed. Levels of significance were indicated by asterisks: **** for p < 0.0001, *** for p < 0.001, ** for p < 0.01, * for p < 0.05, and ‘ns’ for non-significant outcomes (p ≥ 0.05).Data availability
The data supporting this article have been included as part of the ESI.† Data for this article, including metadata, DXF files are available at OSF registry at https://osf.io/42tev.Author contributions
Conceptualization: T. C., A. C. I. V. S., D. G. S., C. S. C. Methodology: T. C., A. C. I. V. S., D. G. S., A. V., C. S. C. Software: T. C. Validation: T. C., A. C. I. V. S., D. G. S., A. V., J. J., J. L. T., J. S., A. P., A. E. S. Formal analysis: T. C., A. C. I. V. S., D. G. S., J. J., J. L. T. Investigation: T. C., A. C. I. V. S., D. G. S., A. V., J. J., J. L. T., J. S., A. P., A. E. S. Resources: C. S. C. Data curation: T. C., A. C. I. V. S., D. G. S., A. V., J. J., J. L. T., J. S., A. P., A. E. S. Writing – original draft: T. C., A. C. I. V. S., D. G. S., J. L. T., C. S. C. Writing – review & editing: T. C., A. C. I. V. S., D. G. S., A. V., J. L. T., J. S., A. P., A. E. S., J. B., J. E., C. S. C. Visualization: T. C., A. C. I. V. S., D. G. S., J. L. T. Supervision: J. E., C. S. C. Project administration: J. E., C. S. C. Funding acquisition: C. S. C.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the NIH (EB00262, EB033821) and the Wellcome Leap HOPE Program to C. S. C.References
- A. Manz, N. Graber and H. á. Widmer, Miniaturized total chemical analysis systems: a novel concept for chemical sensing, Sens. Actuators, B, 1990, 1, 244–248 CrossRef CAS.
- N. Convery and N. Gadegaard, 30 years of microfluidics, Micro Nano Eng., 2019, 2, 76–91 CrossRef.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane), Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- M. A. Unger, H.-P. Chou, T. Thorsen, A. Scherer and S. R. Quake, Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography, Science, 2000, 288, 113–116 CrossRef CAS PubMed.
- A. Lee, The third decade of microfluidics, Lab Chip, 2013, 13, 1660–1661 RSC.
- A. W. Martinez, S. T. Phillips and G. M. Whitesides, Three-dimensional microfluidic devices fabricated in layered paper and tape, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 19606–19611 CrossRef CAS PubMed.
- G. M. Whitesides, Cool, or simple and cheap? Why not both?, Lab Chip, 2013, 13, 11–13 RSC.
- G. Whitesides, The Frugal Way, The Economist - The World in 2012, 2011, p. 154 Search PubMed.
- A. Ainla, M. M. Hamedi, F. Güder and G. M. Whitesides, Electrical textile valves for paper microfluidics, Adv. Mater., 2017, 29, 1702894 CrossRef PubMed.
- C.-C. Wang, J. W. Hennek, A. Ainla, A. A. Kumar, W.-J. Lan, J. Im, B. S. Smith, M. Zhao and G. M. Whitesides, A paper-based “pop-up” electrochemical device for analysis of beta-hydroxybutyrate, Anal. Chem., 2016, 88, 6326–6333 CrossRef CAS PubMed.
- M. M. Thuo, R. V. Martinez, W.-J. Lan, X. Liu, J. Barber, M. B. Atkinson, D. Bandarage, J.-F. Bloch and G. M. Whitesides, Fabrication of low-cost paper-based microfluidic devices by embossing or cut-and-stack methods, Chem. Mater., 2014, 26, 4230–4237 CrossRef CAS.
- J. M. Ramsey, The burgeoning power of the shrinking laboratory, Nat. Biotechnol., 1999, 17, 1061–1062 CrossRef CAS PubMed.
- V. Silverio, S. Guha, A. Keiser, R. Natu, D. R. Reyes, H. van Heeren, N. Verplanck and L. H. Herbertson, Overcoming technological barriers in microfluidics: Leakage testing, Front. Bioeng. Biotechnol., 2022, 10, 958582 CrossRef PubMed.
- Y. Temiz, R. D. Lovchik, G. V. Kaigala and E. Delamarche, Lab-on-a-chip devices: How to close and plug the lab?, Microelectron. Eng., 2015, 132, 156–175 CrossRef CAS.
- H. Becker, One size fits all?, Lab Chip, 2010, 10, 1894–1897 RSC.
- H. Van Heeren, Standards for connecting microfluidic devices?, Lab Chip, 2012, 12, 1022–1025 RSC.
- T. Ching, Y.-C. Toh, M. Hashimoto and Y. S. Zhang, Bridging the academia-to-industry gap: organ-on-a-chip platforms for safety and toxicology assessment, Trends Pharmacol. Sci., 2021, 42, 715–728 CrossRef CAS PubMed.
- L. Etxeberria, U. Aguilera, P. Garcia de Madinabeitia, A. Saez, A. M. Zaldua, J. L. Vilas-Vilela, L. Fernández and A. Llobera, Critical Study on the Tube-to-Chip Luer Slip Connectors, Front. Med. Technol., 2022, 4, 881930 CrossRef PubMed.
- M. I. Mohammed, S. Haswell and I. Gibson, Lab-on-a-chip or Chip-in-a-lab: Challenges of Commercialization Lost in Translation, Procedia Technol., 2015, 20, 54–59 CrossRef.
- J. Ducrée, Efficient development of integrated lab-on-a-chip systems featuring operational robustness and manufacturability, Micromachines, 2019, 10, 886 CrossRef PubMed.
- V. Silverio and S. Cardoso, Lab-on-a-chip: Systems integration at the microscale, Drug delivery devices and therapeutic systems, Elsevier, 2021, pp. 63–87 Search PubMed.
- K. C. Bhargava, B. Thompson and N. Malmstadt, Discrete elements for 3D microfluidics, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 15013–15018 CrossRef CAS PubMed.
- E. W. Young and D. J. Beebe, Fundamentals of microfluidic cell culture in controlled microenvironments, Chem. Soc. Rev., 2010, 39, 1036–1048 RSC.
- D. Mark, S. Haeberle and G. Roth, et al., Microfluidic lab-on-a-chip platforms: Requirements, characteristics and applications, Chem. Soc. Rev., 2010, 39, 1153–1182 RSC.
- P. N. Nge, C. I. Rogers and A. T. Woolley, Advances in microfluidic materials, functions, integration, and applications, Chem. Rev., 2013, 113, 2550–2583 CrossRef CAS PubMed.
- B. Zhang, A. Korolj, B. F. L. Lai and M. Radisic, Advances in organ-on-a-chip engineering, Nat. Rev. Mater., 2018, 3, 257–278 CrossRef.
- K. Ronaldson-Bouchard and G. Vunjak-Novakovic, Organs-on-a-chip: a fast track for engineered human tissues in drug development, Cell Stem Cell, 2018, 22, 310–324 CrossRef CAS PubMed.
- J. Shemesh, I. Jalilian, A. Shi, G. H. Yeoh, M. L. K. Tate and M. E. Warkiani, Flow-induced stress on adherent cells in microfluidic devices, Lab Chip, 2015, 15, 4114–4127 RSC.
- S. Varma and J. Voldman, A cell-based sensor of fluid shear stress for microfluidics, Lab Chip, 2015, 15, 1563–1573 RSC.
- E. VanBavel, Effects of shear stress on endothelial cells: possible relevance for ultrasound applications, Prog. Biophys. Mol. Biol., 2007, 93, 374–383 CrossRef CAS PubMed.
- H. H. G. Song, A. Lammers, S. Sundaram, L. Rubio, A. X. Chen, L. Li, J. Eyckmans, S. N. Bhatia and C. S. Chen, Transient Support from Fibroblasts is Sufficient to Drive Functional Vascularization in Engineered Tissues, Adv. Funct. Mater., 2020, 30(48), 2003777 CrossRef CAS PubMed.
- I. Pereiro, A. F. Khartchenko, L. Petrini and G. V. Kaigala, Nip the bubble in the bud: a guide to avoid gas nucleation in microfluidics, Lab Chip, 2019, 19, 2296–2314 RSC.
- S. N. Bhatia and D. E. Ingber, Microfluidic organs-on-chips, Nat. Biotechnol., 2014, 32, 760–772 CrossRef CAS PubMed.
- T. Satoh, S. Sugiura, K. Shin, R. Onuki-Nagasaki, S. Ishida, K. Kikuchi, M. Kakiki and T. Kanamori, A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform, Lab Chip, 2018, 18, 115–125 RSC.
- S. Shuchat, G. Yossifon and M. Huleihel, Perfusion in Organ-on-Chip Models and Its Applicability to the Replication of Spermatogenesis In Vitro, Int. J. Mol. Sci., 2022, 23, 5402 CrossRef CAS PubMed.
- L. Kim, Y.-C. Toh, J. Voldman and H. Yu, A practical guide to microfluidic perfusion culture of adherent mammalian cells, Lab Chip, 2007, 7, 681–694 RSC.
- D. Huh, G. A. Hamilton and D. E. Ingber, From 3D cell culture to organs-on-chips, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed.
- I. E. Araci and S. R. Quake, Microfluidic very large scale integration (mVLSI) with integrated micromechanical valves, Lab Chip, 2012, 12, 2803–2806 RSC.
- S. Smith, M. Sypabekova and S. Kim, Double-Sided Tape in Microfluidics: A Cost-Effective Method in Device Fabrication, Biosensors, 2024, 14, 249 CrossRef CAS PubMed.
- J.-S. Hwang, S.-Y. Kim, Y.-S. Kim, H.-J. Song, C.-Y. Park and J.-D. Kim, Implementation of PCB-based PCR chip using double-sided tape, Int. J. Control Autom., 2015, 8, 117–124 CrossRef.
- D. A. Chistiakov, A. N. Orekhov and Y. V. Bobryshev, Effects of shear stress on endothelial cells: go with the flow, Acta Physiol., 2017, 219, 382–408 CrossRef CAS PubMed.
- E. Tzima, M. Irani-Tehrani, W. B. Kiosses, E. Dejana, D. A. Schultz, B. Engelhardt, G. Cao, H. DeLisser and M. A. Schwartz, A mechanosensory complex that mediates the endothelial cell response to fluid shear stress, Nature, 2005, 437, 426–431 CrossRef CAS PubMed.
- G. Wang, S. Kostidis, G. L. Tiemeier, W. M. Sol, M. R. de Vries, M. Giera, P. Carmeliet, B. M. van den Berg and T. J. Rabelink, Shear stress regulation of endothelial glycocalyx structure is determined by glucobiosynthesis, Arterioscler., Thromb., Vasc. Biol., 2020, 40, 350–364 CrossRef CAS PubMed.
- W. J. Polacheck, M. L. Kutys, J. Yang, J. Eyckmans, Y. Wu, H. Vasavada, K. K. Hirschi and C. S. Chen, A non-canonical Notch complex regulates adherens junctions and vascular barrier function, Nature, 2017, 552, 258–262 CrossRef CAS PubMed.
- N. Baeyens, C. Bandyopadhyay, B. G. Coon, S. Yun and M. A. Schwartz, Endothelial fluid shear stress sensing in vascular health and disease, J. Clin. Invest., 2016, 126, 821–828 CrossRef PubMed.
- S. R. A. Kratz, C. Eilenberger, P. Schuller, B. Bachmann, S. Spitz, P. Ertl and M. Rothbauer, Characterization of four functional biocompatible pressure-sensitive adhesives for rapid prototyping of cell-based lab-on-a-chip and organ-on-a-chip systems, Sci. Rep., 2019, 9, 9287 CrossRef CAS PubMed.
- L. E. Stallcop, Y. R. Álvarez-García, A. M. Reyes-Ramos, K. P. Ramos-Cruz, M. M. Morgan, Y. Shi, L. Li, D. J. Beebe, M. Domenech and J. W. Warrick, Razor-printed sticker microdevices for cell-based applications, Lab Chip, 2018, 18, 451–462 RSC.
- J. Sun, C. Paddock, J. Shubert, H.-B. Zhang, K. Amin, P. J. Newman and S. M. Albelda, Contributions of the extracellular and cytoplasmic domains of platelet-endothelial cell adhesion molecule-1 (PECAM-1/CD31) in regulating cell-cell localization, J. Cell Sci., 2000, 113, 1459–1469 CrossRef CAS PubMed.
- D. M. McDonald, Endothelial gaps and permeability of venules in rat tracheas exposed to inflammatory stimuli, Am. J. Physiol., 1994, 266, L61–L83 CAS.
- Z. Liu, D. L. Ruter, K. Quigley, N. T. Tanke, Y. Jiang and V. L. Bautch, Single-cell RNA sequencing reveals endothelial cell transcriptome heterogeneity under homeostatic laminar flow, Arterioscler., Thromb., Vasc. Biol., 2021, 41, 2575–2584 CrossRef CAS PubMed.
- R. Tsaryk, N. Yucel, E. V. Leonard, N. Diaz, O. Bondareva, M. Odenthal-Schnittler, Z. Arany, J. M. Vaquerizas, H. Schnittler and A. F. Siekmann, Shear stress switches the association of endothelial enhancers from ETV/ETS to KLF transcription factor binding sites, Sci. Rep., 2022, 12, 4795 CrossRef CAS PubMed.
- M. Yoshizumi, J.-i. Abe, K. Tsuchiya, B. C. Berk and T. Tamaki, Stress and vascular responses: atheroprotective effect of laminar fluid shear stress in endothelial cells: possible role of mitogen-activated protein kinases, J. Pharmacol. Sci., 2003, 91, 172–176 CrossRef CAS PubMed.
- C. Zhang and D. Xing, Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends, Nucleic Acids Res., 2007, 35, 4223–4237 CrossRef CAS PubMed.
- P. Nath, T. S. Maity, F. Pettersson, J. Resnick, Y. Kunde, N. Kraus and N. Castano, Polymerase chain reaction compatibility of adhesive transfer tape based microfluidic platforms, Microsyst. Technol., 2014, 20, 1187–1193 CrossRef CAS.
- D. Patko, Z. Mártonfalvi, B. Kovacs, F. Vonderviszt, M. Kellermayer and R. Horvath, Microfluidic channels laser-cut in thin double-sided tapes: Cost-effective biocompatible fluidics in minutes from design to final integration with optical biochips, Sens. Actuators, B, 2014, 196, 352–356 CrossRef CAS.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Reconstituting organ-level lung functions on a chip, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- C. M. Leung, P. De Haan, K. Ronaldson-Bouchard, G.-A. Kim, J. Ko, H. S. Rho, Z. Chen, P. Habibovic, N. L. Jeon and S. Takayama, A guide to the organ-on-a-chip, Nat. Rev. Methods Primers, 2022, 2, 33 CrossRef CAS.
- C. L. Thompson, S. Fu, M. M. Knight and S. D. Thorpe, Mechanical Stimulation: A Crucial Element of Organ-on-Chip Models, Front. Bioeng. Biotechnol., 2020, 8, 602646 CrossRef PubMed.
- N. F. Jufri, A. Mohamedali, A. Avolio and M. S. Baker, Mechanical stretch: physiological and pathological implications for human vascular endothelial cells, Vasc. Cell, 2015, 7, 8 CrossRef PubMed.
- D. Ershov, M.-S. Phan, J. W. Pylvänäinen, S. U. Rigaud, L. Le Blanc, A. Charles-Orszag, J. R. Conway, R. F. Laine, N. H. Roy and D. Bonazzi, TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines, Nat. Methods, 2022, 19, 829–832 CrossRef CAS PubMed.
- J.-Y. Tinevez, N. Perry, J. Schindelin, G. M. Hoopes, G. D. Reynolds, E. Laplantine, S. Y. Bednarek, S. L. Shorte and K. W. Eliceiri, TrackMate: An open and extensible platform for single-particle tracking, Methods, 2017, 115, 80–90 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld and B. Schmid, Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00970c |
| This journal is © The Royal Society of Chemistry 2025 |